Significance
We show that beak and skull shapes in birds of prey (“raptors”) are strongly coupled and largely controlled by size. This relationship means that, rather than being able to respond independently to natural selection, beak shapes are highly constrained to evolve in a particular way. The main aspects of shape variation seem to correspond with specific genes active during development. Because raptors are not each other’s closest relatives, similar shape constraints may therefore have been present in the ancestors of all modern songbirds, including Darwin’s finches, the classic example of explosive evolution in birds. If this hypothesis is true, then such classic examples may be unusual, needing first to break a genetic lock before their beaks could evolve new shapes.
Keywords: geometric morphometrics, integration, allometry, birds, modularity
Abstract
Bird beaks are textbook examples of ecological adaptation to diet, but their shapes are also controlled by genetic and developmental histories. To test the effects of these factors on the avian craniofacial skeleton, we conducted morphometric analyses on raptors, a polyphyletic group at the base of the landbird radiation. Despite common perception, we find that the beak is not an independently targeted module for selection. Instead, the beak and skull are highly integrated structures strongly regulated by size, with axes of shape change linked to the actions of recently identified regulatory genes. Together, size and integration account for almost 80% of the shape variation seen between different species to the exclusion of morphological dietary adaptation. Instead, birds of prey use size as a mechanism to modify their feeding ecology. The extent to which shape variation is confined to a few major axes may provide an advantage in that it facilitates rapid morphological evolution via changes in body size, but may also make raptors especially vulnerable when selection pressures act against these axes. The phylogenetic position of raptors suggests that this constraint is prevalent in all landbirds and that breaking the developmental correspondence between beak and braincase may be the key novelty in classic passerine adaptive radiations.
The avian beak offers a classic example of adaptation to feeding ecology, with beak morphology frequently considered to represent evolutionary adaptation to specialized trophic niches [e.g., Galápagos finches (1), Hawaiian honeycreepers (2), and Madagascan vangas (3)]. Despite this axiom, we lack quantitative data on the degree to which skull and beak morphology is influenced not only by feeding ecology, but also by other sources of variation or constraint (4). Although the beak is often seen as the target of selection mechanisms closely allied to feeding ecology, such as prey type, feeding style, or beak use, evidence also suggests that beak morphology and variation may be constrained by a number of other factors, including evolutionary history (phylogeny) and development on the component parts of the entire skull. Breakthrough experiments in molecular genetics have shown that the mechanisms driving beak shape variation encompass modifications to the timing of expression of conserved developmental pathways (5–9), resulting in beak diversity described by a few relatively simple geometric transformations (10). However, pleiotropic associations between different skull structures can also contribute to the shape of the avian beak (11), and Sonic hedgehog signaling from the forebrain also relates to the spatial organization of, and changes to, face and beak shape (12–14). Furthermore, assessments of bird skull phenotypic variation suggest that beak morphology may evolve cohesively with cranial morphology (15, 16). Size is also an important consideration when assessing morphological variation. Larger animals generally have access to larger prey due to their increased gape and greater absolute muscular power, and size is further related to morphology via allometry, the tendency of traits to vary with size throughout a morphological structure. Allometry has been demonstrated to be a key contributing factor to craniofacial form across a range of mammalian (17, 18) and avian (15, 19) clades, and evolvability of body size is proposed to be a major evolutionary pathway in the avian stem (20).
In this study, we quantified the role of adaptation versus constraint in avian craniofacial evolution. Using diurnal birds of prey (“raptors”), we quantified the degree to which morphological convergence in feeding ecology can be attributed to variation controlled by evolutionary allometry (size), phylogeny, and integration between the beak and braincase. Raptors are an ideal group to skull avian craniofacial evolution. They possess strong, hook-shaped beaks and powerful talons for holding and tearing flesh, are found in every habitat and continent except Antarctica (21), and vary considerably in size, from 40–12,500 g (22). Although traditionally considered to be monophyletic, recent molecular phylogenies (23–25) recognize that diurnal raptors comprise three nonsister families [Falconidae (falcons and caracaras), Cathartidae (New World vultures), and Accipitridae (the largest clade, including hawks, eagles, kites, harriers, buzzards, and Old World vultures)] and two further monotypic families for the osprey (Pandionidae, Pandion haliaeetus) and secretarybird (Sagittaridae, Sagittarius serpentarius). Despite some differences in the positions of Accipitridae and Falconidae between different topologies (25, 26), raptor families are consistently recovered at the base of both major landbird clades, and a raptorial ancestor for the landbird radiation has been suggested (25). Extensive morphological and dietary convergence is seen between raptor families [for instance, between scavenging Old and New World vultures (27) and the avivore sparrowhawks (e.g., Accipiter nisus) and falcons (e.g., Falco columbarius)] alongside the repeated evolution of recognizable ecomorphotypes (e.g., eagles and kites) within the Accipitridae (28, 29). Additionally, certain species, such as the snail kite (Rostrhamus sociabilis) and hook-billed kite (Chondrohierax uncinatus), show highly specialized, independently derived beak morphologies associated with their diet.
If selection pressures underpinning raptor beak shape are related to feeding ecology, we predict that distantly related birds of the same dietary groups should share similar shaped beaks and skulls, irrespective of phylogeny (i.e., evolutionary convergence). Using 3D shape analysis, we quantified how cranial shape variation is related to size (allometry) and tested the long-standing view that the beak and braincase act as independent modules, enabling birds to adapt their beaks independently to a variety of ecological roles.
Results
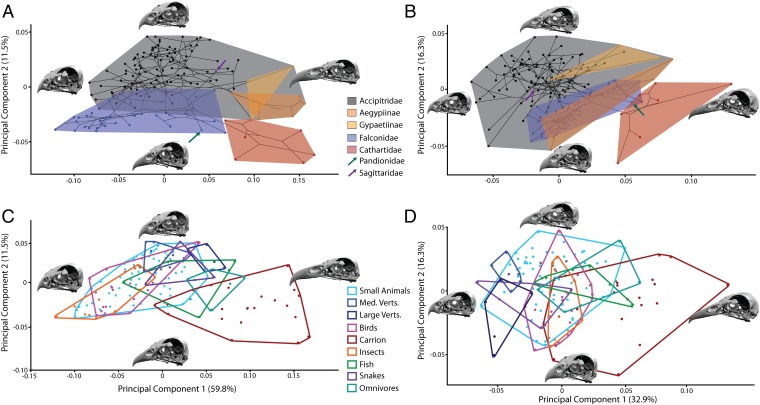
A 3D dataset of 22 landmarks and 40 semilandmarks collected from the skulls of 147 raptor species representing all major radiations (Fig. S1 and Tables S1 and S2) was subjected to Procrustes superimposition and principal components analysis (PCA) to generate a morphospace of skull shape variation (Fig. 1). Accipitrids and falconids occupied similar space on principal component 1 (PC1) (59.8%; positive PC1 scores represent an elongation of the beak, flattening of the skull roof and rotation of the occipital from a ventral to a posterior orientation) (Movies S1 and S2), but separate on PC2 (11.5%; positive PC2 scores represent increased beak curvature with narrowed jugal width) (Movies S3 and S4). Permutation tests rejected the null hypothesis of no phylogenetic signal (P < 0.0001) in skull shape. Pairwise nonparametric multivariate analysis of variance (NPMANOVA) found significant differences in shape between the three main families (Bonferroni-corrected P = 0.0003). The two monotypic families, Pandionidae and Sagittaridae, plotted within the Falconidae and Accipitridae, respectively. Mapping of phylogeny over the morphospace to create a phylomorphospace revealed extensive criss-crossing of branches, yet three distinctive parallel-trending radiations stretched into sparsely populated morphospace at the positive end of PC1 (Fig. 1A): the New World cathartid vultures and the two Old World vulture accipitrid (nonsister) subfamilies, Aegypiinae and Gypaetinae. We therefore uncovered an almost exclusive area of “vulture space” on PC1, with only two nonvulturine taxa falling on the very edge of this region.
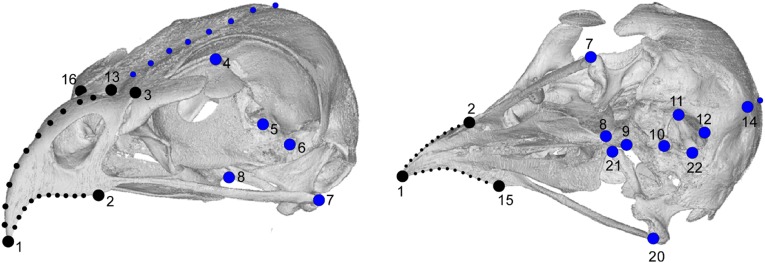
Fig. S1.
B. buteo showing landmarks and semilandmark curves used in analysis. Black, “beak” block; blue, “braincase” block. Left shows the skull and landmarks in oblique left lateral view; Right in oblique ventral view.
Table S1.
Landmark list and semilandmark curves
| Landmark | Location | Block |
| LM1 | Tip of the beak | Beak |
| LM2 | Anteriormost position of antorbital fenestra, projected perpendicular to the tomial edge (L) | Beak |
| LM3 | Center of the craniofacial hinge, projected perpendicular to the lacrimal articulation (L) | Beak |
| LM4 | Anteriormost point of the olfactory nerve (CN 1) opening (L) | Braincase |
| LM5 | Lateralmost point of the trigeminal nerve (CN V) opening (L) | Braincase |
| LM6 | Lateralmost point of the facial nerve (CN VII) opening (L) | Braincase |
| LM7 | Articulation between jugal and quadrate (L) | Braincase |
| LM8 | Articulation between palatine and pterygoid (L) | Braincase |
| LM9 | Center of nuchal crest | Braincase |
| LM10 | Center of occipital condyle | Braincase |
| LM11 | Lateralmost point of foramen magnum (L) | Braincase |
| LM12 | Posteriormost point of foramen magnum | Braincase |
| LM13 | Center of craniofacial hinge | Beak |
| LM14 | Center of nuchal crest | Braincase |
| LM15 | Anteriormost position of antorbital fenestra, projected perpendicular to the tomial edge (R) | Beak |
| LM16 | Center of the craniofacial hinge, projected perpendicular to the lacrimal articulation (R) | Beak |
| LM17 | Anteriormost point of the olfactory nerve (CN 1) opening (R) | Braincase |
| LM18 | Lateralmost point of the trigeminal nerve (CN V) opening (R) | Braincase |
| LM19 | Lateralmost point of the facial nerve (CN VII) opening (R) | Braincase |
| LM20 | Articulation between jugal and quadrate (R) | Braincase |
| LM21 | Articulation between palatine and pterygoid (R) | Braincase |
| LM22 | Lateralmost point of foramen magnum (R) | Braincase |
| Curve 1 | Dorsal profile of beak, between landmarks 1–13 | Beak |
| Curve 2 | Dorsal profile of braincase, between landmarks 13–14 | Braincase |
| Curve 3 | Left tomial edge, between landmarks 1–2 | Beak |
| Curve 4 | Right tomial edge, between landmarks 1–15 | Beak |
CN, cranial nerve; L, left hand side; R, right hand side.
Table S2.
Specimens used in analysis
| Scientific name | Family | Diet | Sex | Mass, g* | NMNH specimen no. | Surface |
| Accipiter badius polyzonoides | Accipitridae | Small animals | F | 196.0 | 430530 | NE |
| Accipiter bicolor | Accipitridae | Birds | M | 245.0 | 622236 | NE |
| Accipiter cooperii | Accipitridae | Birds | M | 349.0 | 636924 | NE |
| Accipiter fasciatus | Accipitridae | Small animals | M | 510.0 | 620189 | NE |
| Accipiter gentilis | Accipitridae | Birds | M | 912.0 | 610353 | NE |
| Accipiter haplochorus | Accipitridae | Small animals | F | 254.0 | 561511 | NE |
| Accipiter henicogrammus | Accipitridae | Small animals | F | — | 556987 | NE |
| Accipiter melanochlamys | Accipitridae | Birds | M | 294.0 | 561484 | NE |
| Accipiter melanoleucus | Accipitridae | Birds | M | 695.0 | 291786 | NE |
| Accipiter minullus | Accipitridae | Birds | M | 75.7 | 490283 | NE |
| Accipiter nisus | Accipitridae | Birds | F | 325.0 | 344423 | NE |
| Accipiter novaehollandiae griseogularis | Accipitridae | Small animals | U | 258.5 | 558270 | NE |
| Accipiter poliogaster | Accipitridae | No information | F | — | 622941 | NE |
| Accipiter striatus velox | Accipitridae | Birds | M | 103.0 | 553261 | NE |
| Accipiter tachiro | Accipitridae | Birds | M | 202.0 | 622998 | NE |
| Accipiter virgatus confusus | Accipitridae | Birds | F | 143.0 | 488909 | NE |
| Aegypius monachus | Accipitridae | Carrion | U | 9,625.0 | 614152 | PG |
| Aquila audax | Accipitridae | Large vertebrates | M | 3,500.0 | 620192 | PG |
| Aquila rapax | Accipitridae | Medium vertebrates | F | 2,250.0 | 430406 | PG |
| Aviceda subcristata | Accipitridae | Small animals | F | 294.0 | 558306 | NE |
| Busarellus nigricollis | Accipitridae | Fish | M | 614.0 | 345773 | NE |
| Butastur indicus | Accipitridae | Small animals | U | 397.0 | 223986 | NE |
| Buteo albicaudatus | Accipitridae | Small animals | F | 884.0 | 632372 | NE |
| Buteo albonotatus | Accipitridae | Small animals | M | 628.0 | 621080 | NE |
| Buteo buteo | Accipitridae | Small animals | F | 969.0 | 554270 | NE |
| Buteo jamaicensis | Accipitridae | Small animals | U | 1,126.0 | 290346 | NE |
| Buteo lagopus s.-johannis | Accipitridae | Small animals | M | 847.0 | 291309 | NE |
| Buteo lineatus | Accipitridae | Small animals | M | 475.0 | 614338 | NE |
| Buteo magnirostris | Accipitridae | Insects | M | 269.0 | 288766 | NE |
| Buteo nitidus | Accipitridae | Small animals | M | — | 623049 | NE |
| Buteo platypterus | Accipitridae | Small animals | F | 490.0 | 613957 | NE |
| Buteo polyosoma (poecilochorus) | Accipitridae | Small animals | M | — | 346398 | NE |
| Buteo regalis | Accipitridae | Medium vertebrates | M | 1,059.0 | 289973 | NE |
| Buteo ridgwayi† | Accipitridae | Small animals | F | — | 226132 | NE |
| Buteo rufinus | Accipitridae | Small animals | U | 1,174.5 | 019535 | NE |
| Buteo rufofuscus | Accipitridae | Small animals | U | 1,164.3 | 431785 | NE |
| Buteo solitarius | Accipitridae | Small animals | F | 606.0 | 622623 | NE |
| Buteo swainsoni | Accipitridae | Small animals | M | 908.0 | 321986 | NE |
| Buteogallus aequinoctialis | Accipitridae | Small animals | F | 715.0 | 621054 | NE |
| Buteogallus anthracinus | Accipitridae | Small animals | M | 793.0 | 344053 | NE |
| Buteogallus meridionalis | Accipitridae | Small animals | U | 808.0 | 560138 | NE |
| Buteogallus urubitinga | Accipitridae | Small animals | M | 925.0 | 621696 | NE |
| Caracara cheriway | Falconidae | Carrion | F | — | 321805 | NE |
| Caracara plancus | Falconidae | Carrion | U | 893.5 | 630187 | NE |
| Cathartes aura | Cathartidae | Carrion | U | 1,467.0 | 354339 | NE |
| Cathartes burrovianus | Cathartidae | Carrion | M | 953.0 | 622341 | NE |
| Cathartes melambrotus | Cathartidae | Carrion | F | 1,200.0 | 621939 | NE |
| Chondrohierax u. uncinatus | Accipitridae | Small animals‡ | U | 278.0 | 289784 | NE |
| Circaetus cinereus | Accipitridae | Snakes | M | 2,048.0 | 430776 | PG |
| Circaetus gallicus | Accipitridae | Snakes | F | 1,703.0 | 430827 | NE |
| Circus aeruginosus | Accipitridae | Small animals | M | 492.0 | 344419 | NE |
| Circus approximans | Accipitridae | Small animals | U | 705.0 | 492471 | NE |
| Circus buffoni | Accipitridae | Small animals | M | 410.0 | 623127 | NE |
| Circus cinereus | Accipitridae | Birds | U | 420.0 | 321772 | NE |
| Circus cyaneus hudsonius | Accipitridae | Small animals | M | 358.0 | 291684 | NE |
| Circus maurus | Accipitridae | Birds | M | — | 558448 | NE |
| Coragyps atratus | Cathartidae | Carrion | M | 2,172.0 | 559659 | PG |
| Daptrius ater | Falconidae | Carrion | F | 342.0 | 226167 | NE |
| Elanoides forficatus | Accipitridae | Insects | M | 442.0 | 289686 | NE |
| Elanus caeruleus | Accipitridae | Small animals | F | 350.0 | 558447 | NE |
| Elanus leucurus | Accipitridae | Small animals | U | 300.0 | 19603 | NE |
| Falco berigora | Falconidae | Small animals | F | 625.0 | 347646 | NE |
| Falco biarmicus | Falconidae | Birds | U | 593.0 | 620138 | NE |
| Falco cherrug | Falconidae | Small animals | F | 1,050.0 | 500262 | NE |
| Falco columbarius | Falconidae | Birds | F | 218.0 | 554550 | NE |
| Falco eleonorae | Falconidae | Insects | M | 390.0 | 488786 | NE |
| Falco femoralis | Falconidae | Birds | F | 407.0 | 622320 | NE |
| Falco longipennis | Falconidae | Birds | M | 213.0 | 347645 | NE |
| Falco mexicanus | Falconidae | Small animals | M | 554.0 | 610758 | NE |
| Falco moluccensis | Falconidae | Small animals | F | — | 558272 | NE |
| Falco naumanni | Falconidae | Insects | F | 164.0 | 603409 | NE |
| Falco perigrinus anatum | Falconidae | Birds | M | 611.0 | 291186 | NE |
| Falco rufigularis | Falconidae | Birds | M | 129.0 | 644063 | NE |
| Falco rupicoloides | Falconidae | Small animals | M | 260.0 | 430626 | NE |
| Falco rusticolis | Falconidae | Small animals | F | 1,752.0 | 567722 | NE |
| Falco sparverius dominicensis | Falconidae | Small animals | M | 111.0 | 555741 | NE |
| Falco subbuteo | Falconidae | Insects | M | 204.0 | 603410 | NE |
| Falco tinnunculus | Falconidae | Small animals | F | 217.0 | 610374 | NE |
| Falco verspertinus amurensis | Falconidae | Insects | U | 165.5 | 289434 | NE |
| Gampsonyx swainsonii | Accipitridae | Small animals | F | 92.5 | 623084 | NE |
| Geranoaetus melanoleucus | Accipitridae | Medium vertebrates | U | 2,252.0 | 318388 | NE |
| Geranospiza caerulescens gracilis | Accipitridae | Small animals | M | 338.0 | 345774 | NE |
| Gymnogyps californianus† | Cathartidae | Carrion | U | 10,104.0 | 492447 | PG |
| Gypaetus barbatus | Accipitridae | Carrion§ | F | 5,680.0 | 345684 | PG |
| Gypohierax angolensis | Accipitridae | Generalist/omnivore | F | 1,600.0 | 291078 | NE |
| Gyps africanus | Accipitridae | Carrion | U | 5,300.0 | 19991 | PG |
| Gyps coprotheres | Accipitridae | Carrion | U | 8,177.0 | 561314 | PG |
| Gyps ruppelli | Accipitridae | Carrion | U | 7,400.0 | 430178 | PG |
| Haliaeetus albicilla | Accipitridae | Fish | M | 4,014.0 | 292774 | PG |
| Haliaeetus leucocephalus | Accipitridae | Generalist/omnivore | U | 7,415.0 | 4882 | PG |
| Haliaeetus vocifer | Accipitridae | Fish | M | 2,212.5 | 488146 | NE |
| Haliastur indus | Accipitridae | Small animals | F | 450.0 | 556984 | NE |
| Haliastur sphenurus | Accipitridae | Small animals | M | 800.0 | 610563 | NE |
| Harpagus bidentatus | Accipitridae | Small animals | F | 239.0 | 612259 | NE |
| Harpia harpjya | Accipitridae | Large vertebrates | U | 6,200.0 | 432244 | PG |
| Herpetotheres cachinnans | Falconidae | Snakes | F | 715.0 | 289775 | NE |
| Hieraeetus spilogaster | Accipitridae | Medium vertebrates | M | 1,225.0 | 430796 | NE |
| Ibycter americanus | Falconidae | Generalist/omnivore | F | 586.0 | 632410 | NE |
| Icthyophaga humilis | Accipitridae | Fish | M | 782.5 | 224807 | NE |
| Icthyophaga icthyaetus | Accipitridae | Fish | U | 2,037.5 | 468555 | NE |
| Ictinia misisippiensis | Accipitridae | Insects | M | 245.0 | 610729 | NE |
| Ictinia plumbea | Accipitridae | Insects | M | 247.0 | 613355 | NE |
| Kaupifalco monogrammicus | Accipitridae | Insects | M | 311.5 | 322456 | NE |
| Leptodon cayanensis | Accipitridae | Small animals | M | 484.0 | 613953 | NE |
| Leucopternis albicollis | Accipitridae | Small animals | M | 600.0 | 613956 | NE |
| Leucopternis melanops | Accipitridae | No information | F | 307.0 | 432181 | NE |
| Leucopternis princeps | Accipitridae | No information | M | 1,000.0 | 613281 | NE |
| Leucopternis semiplumbea | Accipitridae | No information | F | 325.0 | 613955 | NE |
| Lophaetos occipitalis | Accipitridae | Small animals | M | 1,140.0 | 291451 | NE |
| Macheiramphus alcinus | Accipitridae | Birds¶ | U | 650.0 | 559816 | NE |
| Melierax canorus | Accipitridae | Small animals | M | 684.0 | 620139 | NE |
| Melierax metabates mechawi | Accipitridae | Small animals | M | 598.0 | 430326 | NE |
| Micrastur gilvicollis | Falconidae | Small animals | M | 204.0 | 637213 | NE |
| Micrastur ruficollis | Falconidae | Small animals | M | 161.0 | 621387 | NE |
| Micrastur semitorquatus | Falconidae | Small animals | M | 562.0 | 289773 | NE |
| Microhierax caerculescens | Falconidae | Insects | F | 40.0 | 499825 | NE |
| Microhierax erythrogenys | Falconidae | Insects | F | 43.5 | 613010 | NE |
| Milvago chimachima cordatus | Falconidae | Carrion | U | 332.5 | 343844 | NE |
| Milvago chimango | Falconidae | Carrion | M | 296.0 | 635870 | NE |
| Milvus migrans | Accipitridae | Generalist/omnivore | F | 827.0 | 557810 | NE |
| Necrosyrtes monachus | Accipitridae | Carrion | F | 1,813.0 | 291441 | NE |
| Neophron percnopterus | Accipitridae | Carrion | U | 2,120.0 | 17835 | PG |
| Pandion haliaetus | Pandionidae | Fish | F | 1,568.0 | 492597 | NE |
| Parabuteo unicinctus | Accipitridae | Medium vertebrates | M | 690.0 | 630259 | NE |
| Pernis ptilorhynchus gurneyi | Accipitridae | Insects | M | 1,066.0 | 343983 | NE |
| Phalcoboenus australis | Falconidae | Carrion | F | 1,187.0 | 490890 | NE |
| Phalcoboenus carunculatus | Falconidae | Generalist/omnivore | F | — | 614838 | NE |
| Phalcoboenus megalopterus | Falconidae | Small animals | U | 795.0 | 500273 | NE |
| Pithecophaga jefferyi† | Accipitridae | Large vertebrates | M | 4,041.0 | 499879 | PG |
| Polemaetus bellicosus | Accipitridae | Large vertebrates | M | 4,230.0 | 430533 | NE |
| Polihierax insignis | Falconidae | Small animals | M | 98.0 | 490664 | NE |
| Polihierax semitorquatus | Falconidae | Small animals | F | 57.0 | 322394 | NE |
| Polyboroides typus | Accipitridae | Small animals | F | 570.0 | 291787 | NE |
| Rostrhamus sociabilis | Accipitridae | Small animals‡ | M | 378.0 | 631216 | NE |
| Sagittarius serpentarius | Sagittaridae | Insects | F | 3,405.0 | 490786 | PG |
| Sarcorhamphus papa | Cathartidae | Carrion | F | 3,400.0 | 320860 | NE |
| Spilornis cheela | Accipitridae | Snakes | U | 1,072.0 | 19474 | NE |
| Spizaetus ornatus | Accipitridae | Medium vertebrates | M | 1,069.0 | 430495 | NE |
| Spizaetus tyrannus | Accipitridae | Medium vertebrates | M | 1,025.0 | 623090 | NE |
| Spizastur melanoleucus | Accipitridae | Small animals | U | 850.0 | 321507 | NE |
| Spiziapteryx circumcinctus | Falconidae | Small animals | M | 152.0 | 319445 | NE |
| Stephanoaetus coronatus | Accipitridae | Large vertebrates | F | 3,640.0 | 346655 | NE |
| Terathopius ecuadatus | Accipitridae | Small animals | F | 2,438.5 | 319919 | PG |
| Torgos tracheliotus | Accipitridae | Carrion | M | 7,500.0 | 347597 | PG |
| Trigonoceps occipitalis | Accipitridae | Carrion | U | 5,900.0 | 347358 | PG |
| Urotuorchis macrourus batesi | Accipitridae | Small animals | M | 492.0 | 292398 | NE |
| Vultur gryphus | Cathartidae | Carrion | M | 12,500.0 | 346633 | PG |
F, female; M, male; NE, NextEngine laser scanner; NMNH, Smithsonian Institution National Museum of Natural History; PG, photogrammetry; U, unknown.
Mass estimates taken from ref. 22. Mass was taken for same-sex birds wherever possible. Otherwise, species averages, opposite sex, or birds of unknown sex were used to estimate mass. Dashes indicate data unavailable.
Critically endangered [International Union for Conservation of Nature and Natural Resources (IUCN) Red List, 2015].
Chondrohierax uncinatus and Rostrhamus sociabilis are both specialist predators of snails but are classified here as small animal predators due to their small sample size.
Gypaetus barbatus is classified as a carrion eater although its diet almost exclusively comprises bone.
Macheirhamphus alcinus is a specialist predator of bats but is here classified as a bird eater.
Fig. 1.
Principal components analyses of raptor skulls. Phylomorphospaces of the original (A) and nonallometric (B) shape data, colored to indicate family (two Accipitrid subfamilies of Old World vulture are also highlighted). Morphospaces of the original (C) and nonallometric (D) shape data, colored to indicate dietary preference.
Despite clustering of vultures, we found limited evidence for wholesale separation of groups on the basis of feeding ecology (Fig. 1C). Carrion feeders were statistically distinct from all other ecological groups, except large vertebrate feeders, fish eaters, and generalists/omnivores (Table 1), but avivores and insectivores (birds specializing in aerial prey capture) were the most distinct in the pairwise comparisons, being significantly different in seven of the eight possible pairings, but not distinct from each other. Contrary to our predictions, no dietary groups were significantly different from all of the others. Piscivores were the least distinctive, with only two significantly different pairings.
Table 1.
Euclidean NPMANOVA of PC scores, with Bonferroni-corrected P values showing differences between dietary groups
| Small animals | Medium vertebrates | Large vertebrates | Birds | Carrion | Insects | Fish | Snakes | Generalist/omnivore | |
| Small animals | 1 | 0.0036 | 0.0144 | 0.0036 | 0.018 | 0.0504 | 0.72 | 0.0036 | |
| Medium vertebrates | 1 | 0.0792 | 0.0036 | 0.0036 | 0.0144 | 1 | 1 | 0.0216 | |
| Large vertebrates | 0.0036 | 0.0792 | 0.0036 | 0.1152 | 0.0108 | 0.8172 | 1 | 0.1728 | |
| Birds | 0.0144 | 0.0036 | 0.0036 | 0.0036 | 1 | 0.0252 | 0.0216 | 0.0036 | |
| Carrion | 0.0036 | 0.0036 | 0.1152 | 0.0036 | 0.0036 | 0.342 | 0.0252 | 1 | |
| Insects | 0.018 | 0.0144 | 0.0108 | 1 | 0.0036 | 0.0252 | 0.0252 | 0.0072 | |
| Fish | 0.0504 | 1 | 0.8172 | 0.0252 | 0.342 | 0.0252 | 1 | 1 | |
| Snakes | 0.72 | 1 | 1 | 0.0216 | 0.0252 | 0.0252 | 1 | 0.6444 | |
| Generalist/omnivore | 0.0036 | 0.0216 | 0.1728 | 0.0036 | 1 | 0.0072 | 1 | 0.6444 |
Bold values indicate significantly different pairings (P < 0.05).
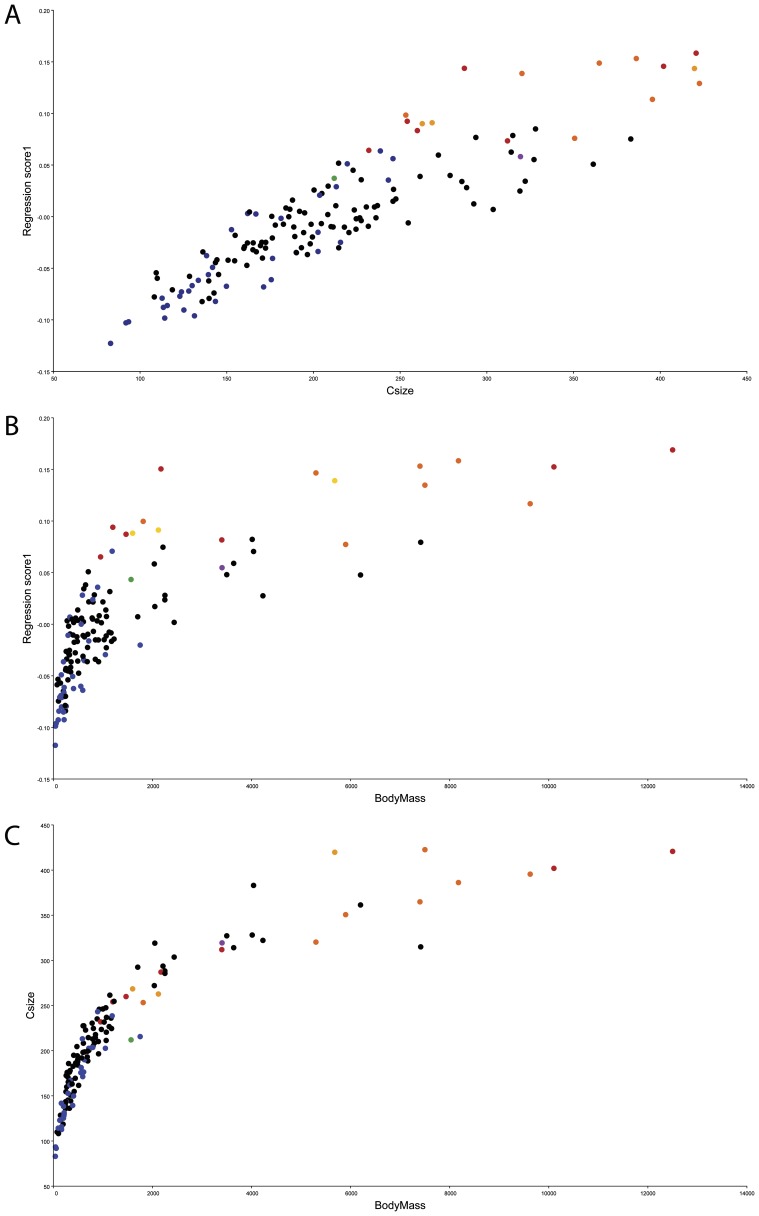
The observation that birds with long beaks and flat, narrow skulls (birds with positive PC1 values) are larger than birds with negative PC1 values was confirmed by a regression of shape data against centroid size; 47.5% of the variation in shape can be predicted from size (P < 0.0001), indicating a very strong allometric relationship between skull shape and size (Fig. S2 A and B). The separation of vultures from other clades on PC1 therefore suggests that part of the dietary signal recovered from the morphospace is size-related (i.e., allometric), and that vultures’ dietary adaptation is achieved by virtue of their increased size.
Fig. S2.
(A) Regression of the symmetric component of shape change to centroid size. (B) Regression of the symmetric component of shape change to estimated body mass. (C) Regression of centroid size to estimated body mass. Black, Accipitridae [orange, Accipitridae: Gypaetiinae (Old World vultures); yellow, Accipitridae: Aegypiinae (Old World vultures)]; blue, Falconidae; red, Cathartidae (New World vultures); green, Pandionidae (Osprey); purple, Sagittaridae (Secretarybird).
To assess the effect of allometric (size) signal in our dataset, we conducted a PCA on the residuals of the regression of shape data against centroid size (henceforth R_PC). R_PC1 almost halved the variation of PC1 to represent 32.9% of the variation, but R_PC2 increased to 16.3%. When the nonallometric shape was analyzed (Fig. 1 B and D), falconids and accipitrids were less distinctive but a strong phylogenetic signal was still present [P < 0.0001; significant pairwise NPMANOVAs between three main families (Bonferroni-corrected P = 0.0003)]. Little separation of ecologies was apparent in the regression residuals, and there was considerable resemblance between different dietary groups. Scavengers were the most statistically distinct from the other dietary groups (Table 2), even though the two Old World vulture clades no longer radiated out to join the New World vultures in an exclusively vulturine area of morphospace.
Table 2.
Euclidean NPMANOVA of regression residuals’ PC scores, with Bonferroni-corrected P values showing differences between dietary groups
| Small animals | Medium vertebrates | Large vertebrates | Birds | Carrion | Insects | Fish | Snakes | Generalist/omnivore | |
| Small animals | 0.2988 | 0.0108 | 0.0108 | 0.0036 | 0.054 | 1 | 1 | 0.0648 | |
| Medium vertebrates | 0.2988 | 0.3384 | 0.0036 | 0.0036 | 0.0036 | 0.018 | 1 | 0.036 | |
| Large vertebrates | 0.0108 | 0.3384 | 0.0144 | 0.0036 | 0.0108 | 0.1584 | 1 | 0.1188 | |
| Birds | 0.0108 | 0.0036 | 0.0144 | 0.0036 | 0.1944 | 0.9036 | 0.1008 | 0.0108 | |
| Carrion | 0.0036 | 0.0036 | 0.0036 | 0.0036 | 0.0072 | 0.7668 | 0.0036 | 1 | |
| Insects | 0.054 | 0.0036 | 0.0108 | 0.1944 | 0.0072 | 0.2556 | 0.252 | 0.0612 | |
| Fish | 1 | 0.018 | 0.1584 | 0.9036 | 0.7668 | 0.2556 | 0.5904 | 1 | |
| Snakes | 1 | 1 | 1 | 0.1008 | 0.0036 | 0.252 | 0.5904 | 1 | |
| Generalist/omnivore | 0.0648 | 0.036 | 0.1188 | 0.0108 | 1 | 0.0612 | 1 | 1 |
Bold values indicate significantly different pairings (P < 0.05).
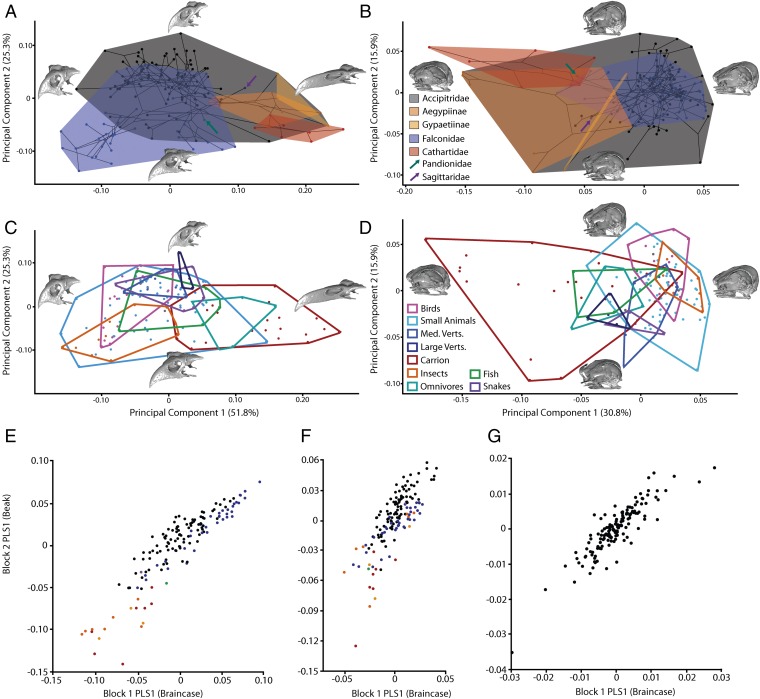
When the landmark configurations were divided in subsets that separately outline the beak and the braincase, we found that braincase morphology was more conservative (less variable) than the beak (Fig. 2, Fig. S1, and Table S1); however, both morphospaces again showed significant allometric and phylogenetic signal and weak ecological clustering (Tables S3–S6). Rather than acting as separate modules, we found that the beak and braincase were highly integrated structures, meaning that almost any change in beak morphology was associated with a correlated and predictable change in braincase morphology. Partial least squares (PLS) analysis of the beak and braincase subsets demonstrated this high degree of correlation (Fig. 2E), with PLS1 representing 97.2% of the covariation (P < 0.001; correlation = 0.91; RV = 0.78). Strikingly, beak–braincase covariation remained even after removal of the allometric (via regression to centroid size) or phylogenetic signal (via phylogenetic independent contrasts) (30), highlighting a conserved developmental constraint on avian craniofacial morphology [PLS1 represents 63.7% of the nonallometric covariation, P < 0.001, correlation = 0.79; RV = 0.48 (Fig. 2F); PLS1 represents 68.0% of the nonphylogenetic covariation, P < 0.001, correlation = 0.88 (Fig. 2G)].
Fig. 2.
Relationship between the beak and braincase. Phylomorphospaces of the beak (A) and braincase (B) individually, colored to indicate family (two Accipitrid subfamilies of Old World vulture are also highlighted). Morphospaces of the beak (C) and braincase (D) individually, colored to indicate dietary preference. Partial least squares analyses showing covariation between the beak and braincase blocks in the original shape data (E), the nonallometric shape data (F), and the phylogenetic independent contrasts (G).
Table S3.
Euclidean NPMANOVA of PC scores from the beak block only
| Accipitridae | Cathartidae | Falconidae | |
| Accipitridae | 0.0003 | 0.0003 | |
| Cathartidae | 0.0003 | 0.0003 | |
| Falconidae | 0.0003 | 0.0003 |
Bonferroni-corrected P values showing differences between families.
Table S6.
Euclidean NPMANOVA of PC scores from the braincase block only, with Bonferroni-corrected P values showing differences between dietary groups
| Small animals | Medium vertebrates | Large vertebrates | Birds | Carrion | Insects | Fish | Snakes | Generalist/omnivore | |
| Small animals | 1 | 0.0036 | 0.0072 | 0.0036 | 0.8856 | 0.0684 | 0.018 | 0.0036 | |
| Medium vertebrates | 1 | 1 | 0.0036 | 0.0756 | 0.018 | 1 | 1 | 1 | |
| Large vertebrates | 0.0036 | 1 | 0.0036 | 0.4824 | 0.0108 | 0.1152 | 1 | 0.1836 | |
| Birds | 0.0072 | 0.0036 | 0.0036 | 0.0036 | 0.3888 | 0.0324 | 0.0072 | 0.0072 | |
| Carrion | 0.0036 | 0.0756 | 0.4824 | 0.0036 | 0.0036 | 1 | 0.1692 | 1 | |
| Insects | 0.8856 | 0.018 | 0.0108 | 0.3888 | 0.0036 | 0.0108 | 0.0144 | 0.0036 | |
| Fish | 0.0684 | 1 | 0.1152 | 0.0324 | 1 | 0.0108 | 0.108 | 1 | |
| Snakes | 0.018 | 1 | 1 | 0.0072 | 0.1692 | 0.0144 | 0.108 | 0.8676 | |
| Generalist/omnivore | 0.0036 | 1 | 0.1836 | 0.0072 | 1 | 0.0036 | 1 | 0.8676 |
Bold values indicate significantly different pairings (P < 0.05).
Table S4.
Euclidean NPMANOVA of PC scores from the braincase block only
| Accipitridae | Cathartidae | Falconidae | |
| Accipitridae | 0.0003 | 0.0003 | |
| Cathartidae | 0.0003 | 0.0003 | |
| Falconidae | 0.0003 | 0.0003 |
Bonferroni-corrected P values showing differences between families.
Table S5.
Euclidean NPMANOVA of PC scores from the beak block only, with Bonferroni-corrected P values showing differences between dietary groups
| Small animals | Medium vertebrates | Large vertebrates | Birds | Carrion | Insects | Fish | Snakes | Generalist/omnivore | |
| Small animals | 1 | 0.0504 | 0.2088 | 0.0036 | 0.0072 | 1 | 1 | 0.0036 | |
| Medium vertebrates | 1 | 0.1188 | 0.234 | 0.0036 | 0.0108 | 1 | 1 | 0.0252 | |
| Large vertebrates | 0.0504 | 0.1188 | 0.0072 | 0.0036 | 0.0252 | 0.9504 | 1 | 0.1908 | |
| Birds | 0.2088 | 0.234 | 0.0072 | 0.0036 | 0.1512 | 0.0936 | 0.1764 | 0.0036 | |
| Carrion | 0.0036 | 0.0036 | 0.0036 | 0.0036 | 0.0036 | 0.1116 | 0.0324 | 1 | |
| Insects | 0.0072 | 0.0108 | 0.0252 | 0.1512 | 0.0036 | 0.1404 | 0.1008 | 0.0072 | |
| Fish | 1 | 1 | 0.9504 | 0.0936 | 0.1116 | 0.1404 | 1 | 1 | |
| Snakes | 1 | 1 | 1 | 0.1764 | 0.0324 | 0.1008 | 1 | 0.8964 | |
| Generalist/omnivore | 0.0036 | 0.0252 | 0.1908 | 0.0036 | 1 | 0.0072 | 1 | 0.8964 |
Bold values indicate significantly different pairings (P < 0.05).
Shape variation associated with the original PLS1 matched the allometric trend of posterior rotation of the occipital and dorsoventral compression of the braincase with increased beak length. Using PLS and regression, we calculated the amount of integrated variation that was independent of allometry as 32.4%. Together, therefore, allometry (47.5%) and integration (32.4%) predicted 79.9% of the total shape variation. The remaining 20.1% still had a significant phylogenetic signal (P < 0.0001), but neither phylogeny nor diet formed clear groups in morphospace (Fig. 3). Applying this same logic to the phylogenetic independent contrasts suggested that this integration was phylogenetically conserved: A large portion of the allometric variation was phylogenetically controlled (allometry predicted only 18.9% of the nonphylogenetic variation, instead of the 47.5% obtained earlier), but similar amounts of integration remained (27.6%).
Fig. 3.
Variation remaining when allometry and integration are removed. (A) Phylomorphospace colored to indicate family (two Accipitrid subfamilies of Old World vulture are also highlighted). (B) Morphospace colored to indicate dietary preference.
Discussion
Beak shape is often viewed as the target for natural selection, independent of the rest of the skull (4, 31, 32). Contrary to this belief, we find that, in raptors (a polyphyletic group at the base of the landbird radiation), beak and braincase morphology are tightly integrated. The beak cannot evolve as a morphologically independent module; changes to beak shape result in predictable changes to braincase morphology, and vice versa. Our findings challenge the long-standing notion of the avian beak as a discrete, adaptable structure. In fact, integration of the beak and braincase, coupled to a strong allometric signal, can explain nearly 80% of skull shape variation. Moreover, this pattern of predictable skull shape changes is shared by all of the families studied, pointing toward an underlying developmental control (33) and a deep, pervasive evolutionary origin for regulatory controls on bird beak shape. Our major axes of beak shape variation (long and narrow vs. short and wide) parallel changes to beak shape in finches linked to signaling molecules such as calmodulin (7) and bone morphogenetic protein 4 (BMP4) (6, 8).
We find a strong relationship between skull shape and size, showing that size is an effective mechanism by which raptors may modify their feeding ecology. For example, niche partitioning and adaptation to certain diets, such as carrion or aerial prey capture, is achieved by changes to body size and subsequently skull size, with the resulting shape being constrained and defined by the nature of beak–braincase integration. However, at body masses above ∼3 kg, skull size and shape plateau (Fig. S2 B and C), indicating a constraint on maximum head size. The analyses show that all vultures look alike despite their different ancestry. Although the vulture clades do not completely converge in shape, this clustering of nonsister taxa based on diet (after refs. 34 and 35) is strong evidence for “incomplete convergence” (36, 37), as has been recognized in other animal groups (e.g., lizards) (38). Taxa that capture aerial prey (insects and birds) are distinct from many other ecological groups, but no ecological grouping is significantly different from all others. A number of raptors are generalist opportunist predators and will vary their diets to reflect prey availability (21), thus perhaps limiting the extent to which the skull can afford to be morphologically specialized toward particular prey. Other behavioral factors, such as hunting strategy (e.g., sit-and-wait vs. aerial pursuit), may also exert an influence on skull morphology. Birds have highly mobile skulls comprising multiple parts that are able to move during feeding, including a flexible region, or “hinge,” separating the beak and braincase into two kinetic modules (32, 39, 40). Despite generating beak movement, the adductor muscles never exert force directly on to the upper beak. Further research is therefore warranted to investigate whether biomechanical function is similarly integrated (41) and how the shape of the upper beak is affected by the skull musculature as it develops. Finally, the phenomena of “many-to-one” and “one-to-many” mapping between form and function mean that similarities in shape do not necessarily imply similarity in function (42–44), further justifying the need for biomechanical analyses of avian skulls.
Evolutionary history plays a significant role in dictating skull shape. Statistically the accipitrids, falconids, and cathartids are morphologically distinct, despite some overlap in morphospace. Further, a strong phylogenetic signal is observed in the beak, as well as in the braincase, despite the fact that the beak should intuitively be the target of intense selection pressure toward convergence due to its role in feeding. This result undoubtedly reflects the strong integration observed between the beak and braincase. The considerable crossing of clades over morphospace indicates low disparity of forms relative to the number of species (45), indicating that raptors are thoroughly exploring a tightly constrained morphological space, through either extensive convergence or, alternatively, limited shape change from a basal morphological state.
The shape change associated with beak–cranium integration mirrors that of allometry although size alone does not explain this trend and phylogeny plays a key role. The trend for the face to elongate with allometry has also been noted in mammals (17) and is postulated to be related to heterochrony, an important factor in the evolution of birds from dinosaurs (46) and a demonstrated mode of generating diversity of beak forms in Darwin’s finches (6, 9). In mammals, it has been shown that integration constrains evolution along paths of least evolutionary resistance, meaning that heterochronic or allometric changes offer a simple mechanism by which evolution can act to produce high disparity (47, 48). The fact that two nonsister clades of accipitrid vultures achieve a vulturine morphology solely by increasing skull size provides a new, nonmammalian example of this phenomenon. A consequence of this mechanism is that skull morphology is highly constrained. Interestingly, animals that demonstrate high levels of integration are less able to respond to shifting selective pressures because they are locked in to a particular dimension of variation (47, 49): in this case, size. Consequently, raptors may be particularly vulnerable if changing environmental conditions result in an adaptive peak that they cannot reach by simply sliding along their allometric trajectory.
Our study was conducted across a polyphyletic group bracketing the base of the landbird radiation. Regardless of whether raptors occur at the base of two major radiations of monophyletic landbirds (25) or whether Accipitridae are found at the base of all landbirds, with Falconidae sister to the parrots and Passeriformes (26), it raises the question of whether integration and allometric control on form is basal to landbirds or has been independently acquired in all raptorial groups from a modular plesiomorphic condition. Given that integration accounts for the same proportion of the variation in the original shape data as in the phylogenetically controlled dataset, we believe that beak–braincase integration as basal to the landbird radiation is the most parsimonious explanation. However, to confirm this hypothesis, more data are needed from other landbirds. Widespread beak–braincase integration has significant ramifications for the notion that bird beaks are independent agents of selection and adaptation and raises the possibility that release from this constraint is a necessary precursor to facilitate classic “textbook” avian adaptive radiations, such as finches, vangas, and Hawaiian honeycreepers.
Materials and Methods
Fourteen landmarks were collected from the midline and left-hand side of the beaks and braincases of 147 raptor species, representing all of the major radiations (Fig. S1 and Tables S1 and S2), using a MicroScribe G2LX digitizer (Revware Systems, Inc.). These landmarks were then reflected along the midline landmarks and realigned using FileConverter (www.flywings.org.uk/fileConverter_page.htm) to give 22 landmarks in total. Surfaces of the same specimens were obtained using a NextEngine laser scanner and MultiDrive running ScanStudio HD Pro 1.3.2 (NextEngine, Inc.) or with digital photogrammetry (Photoscan 0.9.0; AgiSoft) and were used to place landmarks along the dorsal margins of the beak and braincase, and bilaterally on the tomial edges of the beak in HyperMesh 11.0 (Altair Engineering Inc.). Landmarks were then resampled (resample.exe; life.bio.sunysb.edu/morph/soft-utility.html) to give 10 equally spaced semilandmarks along each curve. Specimens without a keratinous rhamphotheca were selected because this preparation is most commonly found in museum collections. All data were collected during a single visit to the Smithsonian Institution National Museum of Natural History.
The 62 landmarks and semilandmarks were collated for each specimen, and the semilandmarks were slid to minimize bending energy in the Geomorph package for R (43). The slid configurations for all birds were then imported to MorphoJ (44) and subjected to a Procrustes superimposition. Principle components analysis (PCA) was used to explore shape variation within the sample. The skull of a common buzzard (Buteo buteo) was CT scanned (0.0581 mm resolution, 95 kV, 60 μA, X-Tek HMX 160 μCT system; University of Hull), and the bones were segmented in Avizo (version 7.0; Visualization Science Group). The resulting surface was landmarked in Avizo and used to create warps of the maximum and minimum PC scores in all morphospaces using the plotRefToTarget function in Geomorph based on the PC scores from MorphoJ. Significant morphological differences were assessed by Euclidean NPMANOVA to the principal component (PC) scores across all PCs (PAST 2.17) (45) between the three largest families. Pandionidae and Sagittaridae were excluded from these analyses because each had only one representative, invalidating the sample size criteria of the statistical tests (Table S1). Each species was also assigned to 1 of 10 dietary categories based on their preferred prey as determined from ref. 21 (Table S2). NPMANOVA was performed using these groupings to determine significant morphological differences between birds with different dietary preferences (Table 1). Birds of unknown dietary preference were excluded from these analyses.
A maximum clade credibility tree of the species in the analysis was constructed from a set of 1,000 molecular trees (ref. 24 and birdtree.org/) using the TreeAnnotator package in BEAST 2.1.2 (ref. 46 and Fig. S3). This phylogeny was mapped on to the PC scores in MorphoJ using unweighted square-change parsimony (47), and a permutation test for phylogenetic signal was performed over 10,000 iterations. The Phytools package in R (48) was used to generate a phylomorphospace based on the PC scores from MorphoJ.
Fig. S3.
Tree used in phylomorphospace analyses. Black, Accipitridae [orange, Accipitridae: Gypaetiinae (Old World vultures); yellow, Accipitridae: Aegypiinae (Old World vultures)]; blue, Falconidae; red, Cathartidae (New World vultures); green, Pandionidae (Osprey); purple, Sagittaridae (Secretarybird).
After noticing that position on PC1 seemed to be correlated with size, the symmetric component of shape variation was regressed in MorphoJ on to the centroid sizes of the specimens (Fig. S2A) and on to an estimate of body mass (Fig. S2B) taken from ref. 22. Body mass estimates were not available for some species (Table S2), so these species were excluded from the regression to body mass. Significance was assessed over 10,000 permutations (P < 0.0001) in both regressions.
To assess the effects of size-related variation in shape (allometry) on our results, all analyses were repeated on the residuals of the regression to centroid size. NPMANOVA results for dietary differences are shown in Table 2.
The landmark configuration was separated into two subsets (blocks) representing the beak and braincase (Fig. S1), and morphospaces were generated for each block independently (Fig. 2 A–D). The degree of covariation between the two blocks was assessed over 250 permutations using two-block within-configuration partial least squares (PLS) analysis in MorphoJ (Fig. 2E). This analysis was also repeated on the regression residuals to see how the two blocks covaried in the absence of allometry (Fig. 2F). NPMANOVA of the PC scores of both the beak and the braincase individually gave similar results to the whole skull (Tables S3–S6).
Phylogenetic independent contrasts (PICs) (29) were calculated in MorphoJ to remove the aspects of shape associated with relatedness. The PLS analysis was repeated on the PICs to assess whether covariation was associated with phylogenetic structure (Fig. 2G).
Partial least squares evaluates only the amount of covariation, but it does not make any assessment of the amount of overall variation explained by the covariation. To determine how much of the nonallometric shape (the residuals from the regression of the original shape data to centroid size) was explained by the covariation between the braincase and the beak, we first regressed the PLS1 scores of the nonallometric data (which explained 63.7% of the covariation) of block 1 against the nonallometric PLS1 block 2 to obtain an eigenvector for the nonallometric PLS1. The predication of this regression was then itself regressed against the nonallometric shape to give the nonallometric, nonintegrated shape data (representing 32.4% of the overall variation) presented in Fig. 3. This same method was applied to the PICs to assess the degree to which these relationships were affected by relatedness.
Supplementary Material
Acknowledgments
Chris Milensky (Smithsonian Institution National Museum of Natural History) and Jo Cooper and Judith White (Natural History Museum at Tring) are thanked for providing access to specimens. CT scanning for morphometric warps was provided by Michael Fagan and Sue Taft (University of Hull). Funding was provided by BBSRC Grants BB/I011668/1 and BB/I011714/1.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1602683113/-/DCSupplemental.
References
- 1.Grant BR, Grant PR. Evolution of Darwin’s finches caused by a rare climatic event. Proc Biol Sci. 1993;251(1331):111–117. [Google Scholar]
- 2.Lovette IJ, Bermingham E, Ricklefs RE. Clade-specific morphological diversification and adaptive radiation in Hawaiian songbirds. Proc Biol Sci. 2002;269(1486):37–42. doi: 10.1098/rspb.2001.1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jønsson KA, et al. Ecological and evolutionary determinants for the adaptive radiation of the Madagascan vangas. Proc Natl Acad Sci USA. 2012;109(17):6620–6625. doi: 10.1073/pnas.1115835109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zusi RL. Patterns of diversity in the avian skull. In: Hanken J, Hall BK, editors. The Skull: Patterns of Structural and Systematic Diversity. Vol 2. Univ of Chicago Press; Chicago: 1993. pp. 391–437. [Google Scholar]
- 5.Schneider RA, Helms JA. The cellular and molecular origins of beak morphology. Science. 2003;299(5606):565–568. doi: 10.1126/science.1077827. [DOI] [PubMed] [Google Scholar]
- 6.Abzhanov A, Protas M, Grant BR, Grant PR, Tabin CJ. Bmp4 and morphological variation of beaks in Darwin’s finches. Science. 2004;305(5689):1462–1465. doi: 10.1126/science.1098095. [DOI] [PubMed] [Google Scholar]
- 7.Abzhanov A, et al. The calmodulin pathway and evolution of elongated beak morphology in Darwin’s finches. Nature. 2006;442(7102):563–567. doi: 10.1038/nature04843. [DOI] [PubMed] [Google Scholar]
- 8.Wu P, Jiang TX, Shen JY, Widelitz RB, Chuong CM. Morphoregulation of avian beaks: Comparative mapping of growth zone activities and morphological evolution. Dev Dyn. 2006;235(5):1400–1412. doi: 10.1002/dvdy.20825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mallarino R, et al. Two developmental modules establish 3D beak-shape variation in Darwin’s finches. Proc Natl Acad Sci USA. 2011;108(10):4057–4062. doi: 10.1073/pnas.1011480108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fritz JA, et al. Shared developmental programme strongly constrains beak shape diversity in songbirds. Nat Commun. 2014;5:3700. doi: 10.1038/ncomms4700. [DOI] [PubMed] [Google Scholar]
- 11.Bhullar BA, et al. A molecular mechanism for the origin of a key evolutionary innovation, the bird beak and palate, revealed by an integrative approach to major transitions in vertebrate history. Evolution. 2015;69(7):1665–1677. doi: 10.1111/evo.12684. [DOI] [PubMed] [Google Scholar]
- 12.Marcucio RS, Cordero DR, Hu D, Helms JA. Molecular interactions coordinating the development of the forebrain and face. Dev Biol. 2005;284(1):48–61. doi: 10.1016/j.ydbio.2005.04.030. [DOI] [PubMed] [Google Scholar]
- 13.Young NM, Chong HJ, Hu D, Hallgrímsson B, Marcucio RS. Quantitative analyses link modulation of sonic hedgehog signaling to continuous variation in facial growth and shape. Development. 2010;137(20):3405–3409. doi: 10.1242/dev.052340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hu D, et al. Signals from the brain induce variation in avian facial shape. Dev Dyn. 2015;244(9):1133–1143. doi: 10.1002/dvdy.24284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kulemeyer C, Asbahr K, Gunz P, Frahnert S, Bairlein F. Functional morphology and integration of corvid skulls: A 3D geometric morphometric approach. Front Zool. 2009;6:2. doi: 10.1186/1742-9994-6-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Klingenberg CP, Marugán-Lobón J. Evolutionary covariation in geometric morphometric data: Analyzing integration, modularity, and allometry in a phylogenetic context. Syst Biol. 2013;62(4):591–610. doi: 10.1093/sysbio/syt025. [DOI] [PubMed] [Google Scholar]
- 17.Cardini A, Polly PD. Larger mammals have longer faces because of size-related constraints on skull form. Nat Commun. 2013;4:2458. doi: 10.1038/ncomms3458. [DOI] [PubMed] [Google Scholar]
- 18.Singh N, Harvati K, Hublin JJ, Klingenberg CP. Morphological evolution through integration: A quantitative study of cranial integration in Homo, Pan, Gorilla and Pongo. J Hum Evol. 2012;62(1):155–164. doi: 10.1016/j.jhevol.2011.11.006. [DOI] [PubMed] [Google Scholar]
- 19.Marugán-Lobón J, Buscalioni ÁD. Geometric morphometrics in macroevolution: Morphological diversity of the skull in modern avian forms in contrast to some theropod dinosaurs. In: Elewa AMT, editor. Morphometrics: Applications in Biology and Palaeontology. Springer; New York: 2004. pp. 157–173. [Google Scholar]
- 20.Benson RB, et al. Rates of dinosaur body mass evolution indicate 170 million years of sustained ecological innovation on the avian stem lineage. PLoS Biol. 2014;12(5):e1001853. doi: 10.1371/journal.pbio.1001853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.del Hoyo J, Elliott A, Sargatal J. 1994. New World Vultures to Guineafowl, Handbook of the Birds of the World (Lynx Edicions, Barcelona), Vol 2.
- 22.Dunning JB., Jr . CRC Handbook of Avian Body Masses. CRC; Boca Raton, FL: 1993. [Google Scholar]
- 23.Hackett SJ, et al. A phylogenomic study of birds reveals their evolutionary history. Science. 2008;320(5884):1763–1768. doi: 10.1126/science.1157704. [DOI] [PubMed] [Google Scholar]
- 24.Jetz W, Thomas GH, Joy JB, Hartmann K, Mooers AO. The global diversity of birds in space and time. Nature. 2012;491(7424):444–448. doi: 10.1038/nature11631. [DOI] [PubMed] [Google Scholar]
- 25.Jarvis ED, et al. Whole-genome analyses resolve early branches in the tree of life of modern birds. Science. 2014;346(6215):1320–1331. doi: 10.1126/science.1253451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Prum RO, et al. A comprehensive phylogeny of birds (Aves) using targeted next-generation DNA sequencing. Nature. 2015;526(7574):569–573. doi: 10.1038/nature15697. [DOI] [PubMed] [Google Scholar]
- 27.Hertel F. Diversity in body size and feeding morphology within past and present vulture assemblages. Ecology. 1994;75(4):1074–1084. [Google Scholar]
- 28.Lerner HR, Mindell DP. Phylogeny of eagles, Old World vultures, and other Accipitridae based on nuclear and mitochondrial DNA. Mol Phylogenet Evol. 2005;37(2):327–346. doi: 10.1016/j.ympev.2005.04.010. [DOI] [PubMed] [Google Scholar]
- 29.Griffiths CS, Barrowclough GF, Groth JG, Mertz LA. Phylogeny, diversity, and classification of the Accipitridae based on DNA sequences of the RAG-1 exon. J Avian Biol. 2007;38(5):587–602. [Google Scholar]
- 30.Felsenstein J. Phylogenies and the comparative method. Am Nat. 1985;125(1):1–15. [Google Scholar]
- 31.Zusi RL. The role of the depressor mandibulae muscle in kinesis of the avian skull. Proc U S Natl Mus. 1967;123(3607):1–28. [Google Scholar]
- 32.Beecher WJ. The bio-mechanics of the bird skull. Bull Chicago Acad Sci. 1962;11(2):10–33. [Google Scholar]
- 33.Hallgrímsson B, et al. Deciphering the palimpsest: Studying the relationship between morphological integration and phenotypic covariation. Evol Biol. 2009;36(4):355–376. doi: 10.1007/s11692-009-9076-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hertel F. Ecomorphological indicators of feeding behavior in recent and fossil raptors. Auk. 1995;112(4):890–903. [Google Scholar]
- 35.Guangdi SI, Dong Y, Ma Y, Zhang Z. Shape similarities and differences in the skulls of scavenging raptors. Zoolog Sci. 2015;32(2):171–177. doi: 10.2108/zs130253. [DOI] [PubMed] [Google Scholar]
- 36.Herrel A, Vanhooydonck B, Van Damme R. Omnivory in lacertid lizards: Adaptive evolution or constraint? J Evol Biol. 2004;17(5):974–984. doi: 10.1111/j.1420-9101.2004.00758.x. [DOI] [PubMed] [Google Scholar]
- 37.Losos JB. Convergence, adaptation, and constraint. Evolution. 2011;65(7):1827–1840. doi: 10.1111/j.1558-5646.2011.01289.x. [DOI] [PubMed] [Google Scholar]
- 38.Stayton CT. Testing hypotheses of convergence with multivariate data: Morphological and functional convergence among herbivorous lizards. Evolution. 2006;60(4):824–841. [PubMed] [Google Scholar]
- 39.Gussekloo SW, Bout RG. Cranial kinesis in palaeognathous birds. J Exp Biol. 2005;208(Pt 17):3409–3419. doi: 10.1242/jeb.01768. [DOI] [PubMed] [Google Scholar]
- 40.Dawson MM, Metzger KA, Baier DB, Brainerd EL. Kinematics of the quadrate bone during feeding in mallard ducks. J Exp Biol. 2011;214(Pt 12):2036–2046. doi: 10.1242/jeb.047159. [DOI] [PubMed] [Google Scholar]
- 41.Klingenberg CP. Studying morphological integration and modularity at multiple levels: Concepts and analysis. Philos Trans R Soc Lond B Biol Sci. 2014;369(1649):20130249. doi: 10.1098/rstb.2013.0249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lauder GV. On the inference of function from structure. In: Thomason JJ, editor. Functional Morphology in Vertebrate Paleontology. Cambridge Univ Press; Cambridge, UK: 1995. pp. 1–18. [Google Scholar]
- 43.Anderson PS, Friedman M, Brazeau MD, Rayfield EJ. Initial radiation of jaws demonstrated stability despite faunal and environmental change. Nature. 2011;476(7359):206–209. doi: 10.1038/nature10207. [DOI] [PubMed] [Google Scholar]
- 44.Stubbs TL, Pierce SE, Rayfield EJ, Anderson PS. Morphological and biomechanical disparity of crocodile-line archosaurs following the end-Triassic extinction. Proc Biol Sci. 2013;280(1770):20131940. doi: 10.1098/rspb.2013.1940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sidlauskas B. Continuous and arrested morphological diversification in sister clades of characiform fishes: A phylomorphospace approach. Evolution. 2008;62(12):3135–3156. doi: 10.1111/j.1558-5646.2008.00519.x. [DOI] [PubMed] [Google Scholar]
- 46.Bhullar BA, et al. Birds have paedomorphic dinosaur skulls. Nature. 2012;487(7406):223–226. doi: 10.1038/nature11146. [DOI] [PubMed] [Google Scholar]
- 47.Marroig G, Shirai LT, Porto A, de Oliveira FB, De Conto V. The evolution of modularity in the mammalian skull. II. Evolutionary consequences. Evol Biol. 2009;36(1):136–148. [Google Scholar]
- 48.Goswami A, Smaers JB, Soligo C, Polly PD. The macroevolutionary consequences of phenotypic integration: From development to deep time. Philos Trans R Soc Lond B Biol Sci. 2014;369(1649):20130254. doi: 10.1098/rstb.2013.0254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Villmoare B. Morphological Integration, Evolutionary Constraints, and Extinction: A Computer Simulation-Based Study. Evol Biol. 2012;40(1):76–83. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.