Significance
Late secretory compartments are clearly associated with increased Alzheimer’s disease (AD) risk. We have shown biochemically and in cells that coat protein complex I (COPI)-dependent trafficking, an early Golgi-to-endoplasmic reticulum trafficking step, modifies amyloid precursor protein subcellular localization and cell-surface expression, leading to an altered metabolism. The work presented here demonstrates that a reduced COPI-dependent trafficking in vivo leads to a decrease in the amyloid plaque burden in an AD mouse model, and to an improvement of some memory impairments observed in these mice. Remarkably, human genetic association studies of different AD cohorts led to the identification of genetic markers (SNPs) and mutations in COPI genes linked with an increased AD risk. These results demonstrate in vivo the importance of COPI and early trafficking steps in AD.
Keywords: COPI, Alzheimer, human genetic, EWAS studies, GWAS studies
Abstract
Cellular trafficking and recycling machineries belonging to late secretory compartments have been associated with increased Alzheimer’s disease (AD) risk. We have shown that coat protein complex I (COPI)-dependent trafficking, an early step in Golgi-to-endoplasmic reticulum retrograde transport, affects amyloid precursor protein subcellular localization, cell-surface expression, as well as its metabolism. We present here a set of experiments demonstrating that, by targeting subunit δ-COP function, the moderation of the COPI-dependent trafficking in vivo leads to a significant decrease in amyloid plaques in the cortex and hippocampus of neurological 17 mice crossed with the 2xTg AD mouse model. Remarkably, an improvement of the memory impairments was also observed. Importantly, human genetic association studies of different AD cohorts led to the identification of 12 SNPs and 24 mutations located in COPI genes linked to an increased AD risk. These findings further demonstrate in vivo the importance of early trafficking steps in AD pathogenesis and open new clinical perspectives.
One of the hallmarks of Alzheimer’s disease (AD) is the accumulation of Aβ peptides that aggregate over time to form oligomers and lead ultimately to amyloid plaques. Aβ peptides result from cleavages of the amyloid precursor protein (APP) that occur sequentially (1) and concomitantly with APP trafficking, mainly from the plasma membrane to late endosomes (1, 2). We recently addressed the possible involvement of early trafficking steps and demonstrated that at least one subunit of coat protein complex I (COPI), the main machinery underlying the retrograde transport from the Golgi apparatus to the endoplasmic reticulum (ER), regulates APP trafficking, controlling its maturation and consequently the production of Aβ peptides (3). These biochemical and cellular findings demonstrate the physiological relevance of the COPI complex in AD. All together, these results indicate that the origin of the increased Aβ production in pathological conditions, when trafficking through the endocytic pathway, might be due to APP maturation-state impairments.
Moreover, an in vivo model of impairment in COPI subunit delta (δ-COP), the neurological 17 (Nur17) mouse, was previously developed (4) but not yet studied in the context of AD. This mouse model represents a valuable and unique tool to characterize the relevance of δ-COP in AD etiology. The Nur17 mouse was generated by N-ethyl-N-nitrosourea (ENU) mutagenesis, and positional cloning revealed that it carries a T-to-C missense mutation in δ-COP, leading to partial disruption of intracellular trafficking.
Further highlighting the important role of trafficking in regulating Aβ production, in genome-wide association studies (GWASs) essential components of cellular trafficking and recycling involved in the endocytic or retromer pathways (endosome-to-Golgi retrieval) have been found to be associated with increased AD risk (5, 6).
In our previous study identifying δ-COP as a key regulator of APP biology (3), we demonstrated that δ-COP silencing induced a dramatic decrease of Aβ production in N2A cells. Toxicity of other COPI subunits resulted in inconclusive results regarding their possible influence on Aβ production. Prior studies suggest that the complex as a whole could be important for AD etiology (7–9). We next investigated the selectivity of COPI-dependent trafficking for APP localization and its relevance in vivo, both by looking at amyloid plaque formation in an AD mouse model and, more importantly, by conducting a genetic study on four cohorts of AD patients.
Here we demonstrate that δ-COP regulates APP intracellular trafficking rather selectively. Silencing of δ-COP induced a stronger accumulation of APP in the early secretory pathway (Golgi–ER) compared with three other proteins important for APP metabolism. Moreover, cell-surface APP is significantly decreased after δ-COP silencing, whereas eight other proteins relevant for this study were not significantly affected. In an in vivo AD model crossed with Nur17 mice, reduction of δ-COP function rescues AD pathology by significantly decreasing amyloid plaque load as well as improving memory impairments. Importantly, we report that 12 single-nucleotide polymorphisms (SNPs) located in COPI genes are genetically associated with AD risk. Analysis of whole-genome sequencing (WGS) data identified mutations in each COPI gene initially linked to AD through SNP analysis. Altogether, our findings demonstrate the physiological relevance of the COPI complex in AD pathogenesis, a link further confirmed by our clinical data.
Results
δ-COP Regulates Amyloid-β Production in Various Conditions and APP Trafficking in a Selective Manner.
Our previous study (3) demonstrated that δ-COP regulates the retrograde trafficking of APP, its maturation, and consequently the production of Aβ peptides. We compared the effect of δ-COP silencing in N2a cells transfected with APP wild type (APP-WT) and APP containing the Swedish mutation (APP-SW). δ-COP silencing decreased Aβ40 levels in a similar manner in both systems (Fig. 1 A and B). δ-COP silencing also decreased endogenous Aβ40 production in nontransfected N2a cells (Fig. 1C). We also showed that δ-COP silencing affects the amount of APP associated with the Golgi apparatus (3). Here, using sucrose gradient experiments, we investigated the effect of δ-COP silencing on Golgi-localized proteins (e.g., γ-adaptin) or proteins relevant for APP metabolism (PS1, BACE, and ADAM10). The two main Golgi-containing fractions are shown and were used for quantification purposes. Depletion of δ-COP had a negligible effect on γ-adaptin (increased 1.2-fold in the Golgi apparatus) (Fig. 1D), indicating that the Golgi apparatus had not been broadly compromised. δ-COP silencing moderately increased the amount of PS1, BACE, and ADAM10 associated with the Golgi apparatus (1.57-, 1.56-, and 2.18-fold, respectively) compared with APP (5.74-fold in the Golgi apparatus) (Fig. 1D). Using APP biotinylation assays, we previously showed that δ-COP silencing dramatically decreased the amount of APP at the cell surface (3). Here we demonstrated that this effect was relatively specific for APP as the amount of Notch [gamma-secretase substrate that has a half-life comparable to APP (10, 11)], two other cargo proteins of the COPI complex (NMDAR and Na,K-ATPase) (12, 13), as well as two resident proteins of the cell surface (E-cadherin and syndecan) were unchanged (Fig. 1 E and F). Noticeably, the levels of PS1, BACE, and ADAM10 were also unchanged (Fig. 1 E and F).
Fig. 1.
Effect of δ-COP on Aβ production and protein trafficking. (A–C) Aβ40 measurements of N2a cells pretreated with control or δ-COP siRNAs and transfected with (A) APP-WT or (B) APP-SW or (C) nontransfected. OE, overexpressed. (D) Subcellular localization of various proteins in N2a-695 cells transfected with control siRNA or δ-COP siRNA by sucrose density gradient fractionation. The two main cellular fractions corresponding to the Golgi were analyzed by SDS/PAGE using different antibodies and used for the quantification studies indicated in Results. (E and F) Cell-surface protein measurement after (E) transfection with δ-COP siRNA using cell-surface biotinylation and (F) quantification. (*P < 0.05, **P < 0.01, two-tailed Student’s t test; n = 3. The results are shown as means ± SEM.)
Aβ Peptide Levels, Amyloid Plaque Density, and Memory Impairment Are Reduced in AD/δ-COP Mutant Mice.
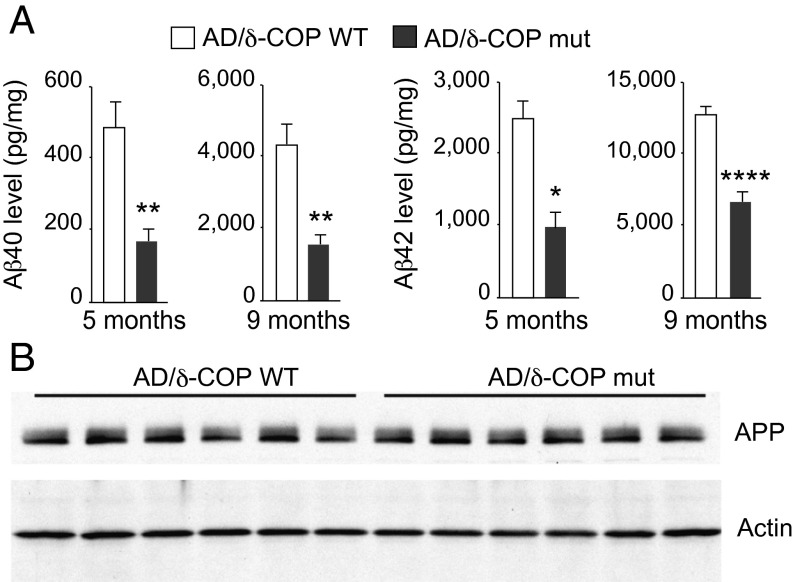
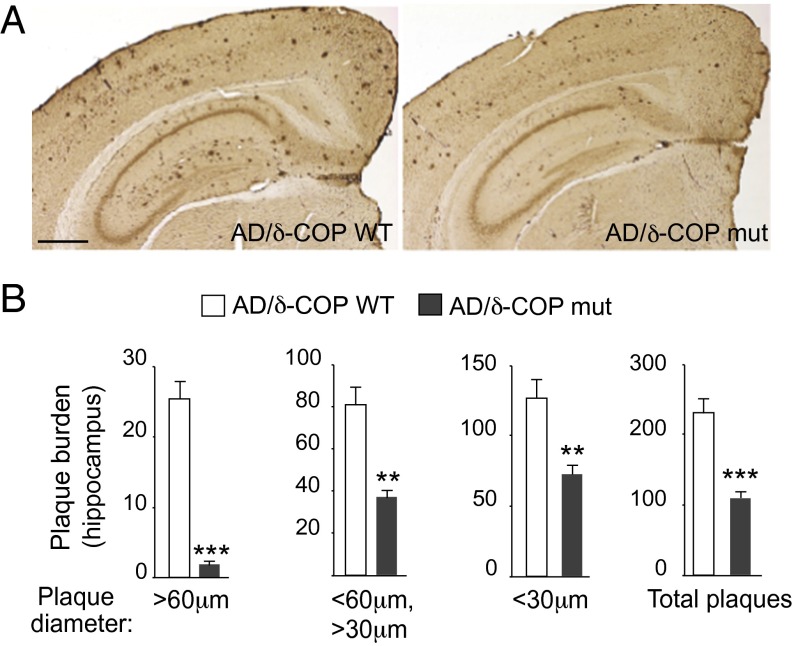
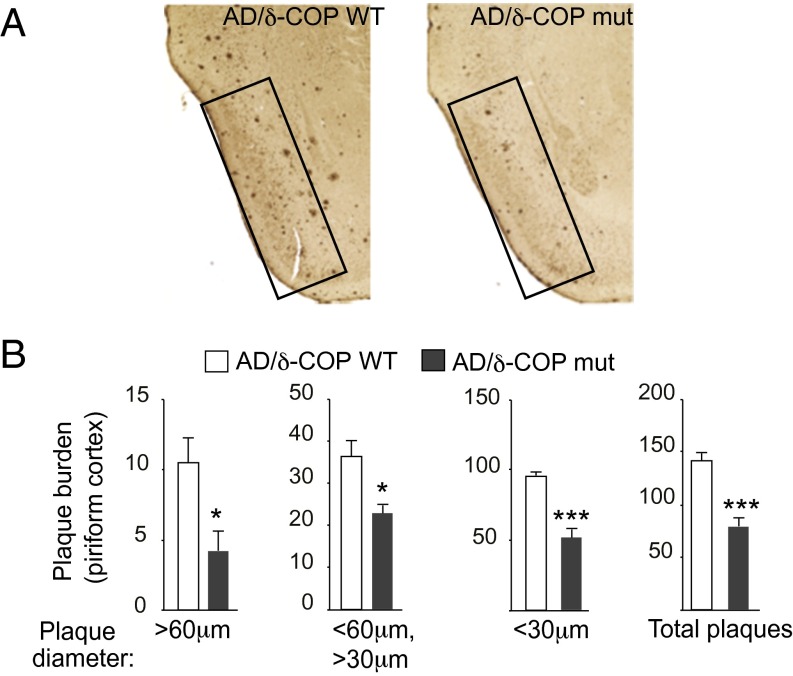
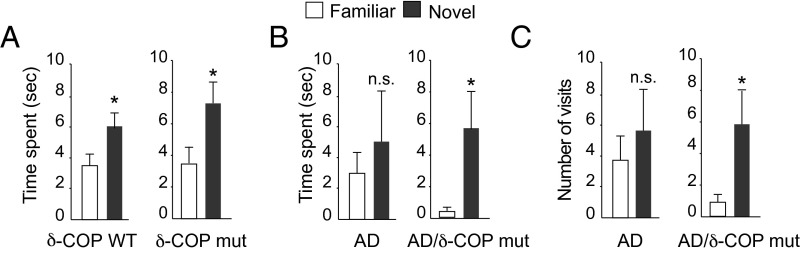
The potential relevance of δ-COP in the pathogenesis of AD was then explored in vivo with mice that harbor a single mutation leading to a partially inactive δ-COP (nur17), referred to in this study as δ-COP mutant (δ-COP mut) mice (δ-COP knockout is lethal) (4). We measured Aβ40 levels in δ-COP mut mice compared with δ-COP WT mice and found that, after quantification, Aβ40 levels were not significantly reduced. However, when crossed with the double-transgenic AD mouse model 2xTg (APP Swedish, PS1Δ9 mutations), δ-COP mutant mice displayed a significant reduction in the levels of Aβ40 and Aβ42 in the hippocampus at 5 and 9 mo of age (Fig. 2A). At 5 mo of age, the total level of APP was unchanged in AD/δ-COP mut mice compared with AD/δ-COP WT mice (Fig. 2B). Amyloid plaque development was significantly reduced in the hippocampus (52.5 ± 4.7%) in 9-mo-old δ-COP mutant mice (Fig. 3 and Fig. S1A). The number of amyloid plaques in the hippocampus was grouped and quantified according to size. The most prominent reduction was observed for the larger plaques (diameter >60 µm; 88.2 ± 3.5%) (Fig. 3B). Amyloid plaque development was also reduced in the piriform cortex in a comparable manner to the hippocampus (44.6 ± 6.4%) in 9-mo-old δ-COP mutant mice (Fig. 4 and Fig. S1B). The most prominent reduction was also observed for the larger plaques in that region (diameter >60 µm; 60.0 ± 13.9%) (Fig. 4B). We next investigated whether partially inactive δ-COP also influences the memory of AD mice using the novel object recognition test. To ensure that the δ-COP mutation does not interfere with the ability to perform this memory test, we compared WT mice with δ-COP mut mice in a non-AD background. Both groups of animals spent significantly more time with the novel object compared with the familiar object, indicating that δ-COP mut mice could perform the task as well as WT mice (Fig. 5A). We also confirmed that novel object recognition was impaired in the 2xTg mice at 9 mo of age, as reported previously (Fig. 5B, Left) (14). Importantly, the partial loss of δ-COP function rescued the capacity to discriminate novel versus familiar objects in AD/δ-COP mut mice compared with AD/δ-COP WT mice (Fig. 5 B and C), which was consistent with plaque load reduction and decrease of Aβ production in these mice.
Fig. 2.
Effect of partial inactivation of δ-COP on Aβ level in 2xTg AD mice. (A) Measurements of Aβ40 and Aβ42 in the hippocampus of age-matched 2xTg AD mice (AD/δ-COP WT vs. AD/δ-COP mut mice) (n = 6–11 per genotype; one outlier has been excluded among the 9-mo-old mice). (B) Analysis of APP and actin levels by Western blotting. (*P < 0.05, **P < 0.01, ****P < 0.0001, two- or one-tailed Student’s t test or Mann–Whitney test. The results are shown as means ± SEM.)
Fig. 3.
Effect of partial inactivation of δ-COP on amyloid plaque formation evaluated by immunohistochemistry in the hippocampus of 2xTg AD mice. (A) Representative images show amyloid plaques in the hippocampus of male mice at 9 mo (n = 6 per genotype). (Scale bar, 500 μm.) (B) Quantification of the number of plaques in the hippocampus (n = 6 per genotype). (**P < 0.01, ***P < 0.001, two- or one-tailed Student’s t test with or without Welch’s correction. The results are shown as means ± SEM.)
Fig. S1.
Replicates of amyloid plaque immunostaining in 9-mo-old AD/δ-COP WT vs. AD/δ-COP mice. Representative images showing amyloid plaques in the brain. Amyloid plaque development was studied in both (A) the hippocampus and (B) the piriform cortex. (Scale bar, 500 μm.)
Fig. 4.
Effect of partial inactivation of δ-COP on amyloid plaque formation evaluated by immunohistochemistry in the piriform cortex of 2xTg AD mice. (A) Representative images show amyloid plaques in the piriform cortex (black boxes) of male mice at 9 mo (n = 6 per genotype). (B) Quantification of the number of plaques in the piriform cortex (n = 6 per genotype). (*P < 0.05, ***P < 0.001, two-tailed Student’s t test. The results are shown as means ± SEM.)
Fig. 5.
Effect of partial inactivation of δ-COP on memory in 2xTg AD mice. (A and B) Effect of δ-COP on novel object recognition in 9-mo-old male mice in (A) a WT or (B) an AD background. Measurement of time spent with a novel versus familiar object. (C) Measurement of the number of visits to the novel versus familiar object (n = 5–14 per genotype). (*P < 0.05, one-tailed Student’s t test or Mann–Whitney test; n.s., not significant. The results are shown as means ± SEM.)
Several SNPs and Variants Are Associated with AD in Multiple COPI Genes.
To determine whether COPI complex components are risk factors for AD, we investigated the possible association of SNPs located in the vicinity of COPI genes with AD (Table S1). The National Institute of Mental Health (NIMH) Genetics Initiative Alzheimer’s Disease Study (15, 16), originally ascertained for the study of genetic risk factors in AD with family-based methods, was used in the WGS analyses in this study. The basis for ascertainment in the NIMH collection was at least two affected individuals within a family, typically siblings. The complete NIMH study cohort contains a total of 1,536 subjects from 457 families. For the purpose of this analysis, only subjects of self-reported European ancestry were included, consisting of 1,345 participants (941 affected and 404 unaffected) from 410 families. Meta-analysis of six independent study cohorts within the candidate gene regions of the COPI complex revealed several SNPs that are associated with AD (Table 1 and Table S2). Seven SNP association signals approached statistical significance (P < 5 × 10−4). The most-significant associations (with meta-P values between 0.002 and 7.E-04) were observed in the COPI subunit delta gene region (COPD; two SNPs), COPI subunit alpha (COPA), COPI subunit zeta 1 (COPZ1), and COPI subunit zeta 2 (COPZ2; three SNPs) (Table 1 and bolded SNPs in Table S2). Five candidate COPI genes (the four genes mentioned above plus COPI subunit beta 1; COPB1) exhibit nominally significant association (P < 0.05) with AD. Interestingly, across the six studies, the SNPs are mainly associated with an increased risk of AD (Table 1 and Tables S2 and S3).
Table S1.
Breakdown of the number of independent tag SNPs from each of the eight candidate coatomer protein complex genes
| Gene symbol | Gene name | Chr | Start | End | Gene length, bp | Total SNPs | Initial filter | Tag SNPs |
| COPA | Coatomer protein complex, subunit alpha | 1 | 160,258,377 | 160,313,354 | 54,977 | 100 | 92 | 16 |
| COPG1 | Coatomer protein complex, subunit gamma 1 | 3 | 128,968,453 | 128,996,616 | 28,163 | 43 | 43 | 4 |
| COPB2 | Coatomer protein complex, subunit beta 2 | 3 | 139,076,433 | 139,108,522 | 32,089 | 73 | 68 | 9 |
| COPB1 | Coatomer protein complex, subunit beta 1 | 11 | 14,479,049 | 14,521,441 | 42,392 | 73 | 68 | 18 |
| COPD | Coatomer protein complex, subunit delta | 11 | 118,443,102 | 118,473,748 | 30,646 | 57 | 53 | 13 |
| COPZ1 | Coatomer protein complex, subunit zeta 1 | 12 | 54,718,911 | 54,745,633 | 26,722 | 30 | 27 | 8 |
| COPZ2 | Coatomer protein complex, subunit zeta 2 | 17 | 46,103,533 | 46,115,152 | 11,619 | 110 | 104 | 18 |
| COPE | Coatomer protein complex, subunit epsilon | 19 | 19,010,323 | 19,030,199 | 19,876 | 44 | 41 | 10 |
| Totals | 530 | 496 | 96 |
Base pair positions from the UCSC February 2009 (GRCh37/hg19) genome build. The total SNPs column contains the total number of SNPs in the gene region imputed plus observed on the Affymetrix 6.0 SNP panel. The initial filter column contains the number of SNPs remaining after removal of SNPs with MAF <0.01 in probands or that deviated from Hardy–Weinberg equilibrium (P < 0.0001) or imputed SNPs with information score <0.40.
Table 1.
Most-significant meta-analysis results from the six datasets for the COPI gene set
| SNP | Gene | Chr | Position | Location | MAF | Overall meta-P | Effect direction | Comments |
| rs7531886 | COPA | 1 | 160,258,242 | Near 3′ UTR | 0.45 | 0.02 | −+++++ | |
| rs12033011 | COPA | 1 | 160,296,055 | Intron | 0.30 | 0.0015 | +++++− | |
| rs72868007 | COPB1 | 11 | 14,479,553 | Intron | 0.03 | 0.03 | ++++++ | |
| rs73022058 | COPD/IFT46 | 11 | 118,423,672 | Intron | 0.09 | 0.02 | ++−+++ | |
| rs3132828 | COPD/IFT46 | 11 | 118,431,003 | Intron | 0.44 | 0.002 | ++++++ | 12 kb from the 5′ UTR, within the intron of IFT46 |
| rs498872 | COPD/PHLDB1 | 11 | 118,477,367 | 3′ UTR | 0.29 | 0.002 | ++++++ | 4 kb from the 3′ UTR, overlapping with the 5′ UTR of PHLDB1 |
| rs34280607 | COPZ1 | 12 | 54,719,968 | Intron | 0.02 | 0.008 | ++++++ | |
| rs61614746 | COPZ1 | 12 | 54,740,214 | Intron | 0.06 | 0.04 | ++++++ | |
| rs757352 | COPZ2 | 17 | 46,097,153 | Intergenic | 0.17 | 7.E-04 | ++++++ | 6 kb from the 3′ UTR |
| rs9898218 | COPZ2 | 17 | 46,106,634 | Intron | 0.38 | 0.001 | ++++++ | |
| rs7216504 | COPZ2 | 17 | 46,117,341 | Intron | 0.20 | 0.03 | −+++++ | |
| rs11650615 | COPZ2/NFE2L1 | 17 | 46,123,698 | Intergenic | 0.27 | 7.E-04 | −+++++ | 9 kb from the 5′ UTR |
Base pair positions were obtained from the UCSC February 2009 (GRCh37/hg19) genome build (https://genome.ucsc.edu/). Chr, chromosome; MAF, minor allele frequency. Overall meta-P includes analysis for all six studies. Effect direction indicates a positive sign for each study that confers a risk effect and a negative sign for each study that confers a protective effect. The positive or negative effect sign corresponds to direction in the following samples (in order): NIMH, NCRAD, GenADA, TGEN2, NIA-LOAD, and ADNI.
Table S2.
Most-significant meta-analysis association results from the six datasets for the eight candidate COPI genes
| SNP | Gene | Overall meta-P | Effect direction | Case-control meta-P | Family meta-P | NIMH Info. Fam. | NCRAD Info. Fam. | NIMH FBAT-P | NCRAD FBAT-P | GenADA P value | TGEN2 P value | NIA-LOAD P value | ADNI P value |
| rs7531886 | COPA | 0.02 | −+++++ | 0.02 | 0.37 | 156 | 128 | −0.87 | 0.13 | 0.16 | 0.51 | 0.08 | 0.46 |
| rs12033011 | COPA | 0.0015 | +++++− | 0.10 | 0.005 | 141 | 136 | 0.015 | 0.12 | 0.76 | 0.40 | 0.03 | 0.80 |
| rs72868007 | COPB1 | 0.03 | ++++++ | 0.26 | 0.40 | 25 | 23 | 0.47 | 0.64 | 0.19 | 0.25 | 0.69 | 0.09 |
| rs73022058 | COPD/IFT46 | 0.02 | ++−+++ | 0.87 | 0.04 | 62 | 75 | 0.04 | 0.40 | 0.29 | 0.47 | 0.05 | 0.06 |
| rs3132828 | COPD/IFT46 | 0.002 | ++++++ | 0.22 | 0.11 | 69 | 76 | 0.37 | 0.17 | 0.43 | 0.20 | 0.11 | 0.012 |
| rs498872 | COPD/PHLDB1 | 0.002 | ++++++ | 0.16 | 0.11 | 153 | 120 | 0.49 | 0.11 | 0.15 | 0.57 | 0.10 | 0.05 |
| rs34280607 | COPZ1 | 0.008 | ++++++ | 0.02 | 0.33 | 20 | 27 | 0.16 | 0.94 | 0.52 | 0.02 | 0.35 | 0.24 |
| rs61614746 | COPZ1 | 0.04 | ++++++ | 0.81 | 0.08 | 50 | 38 | 0.03 | 0.84 | 0.58 | 0.42 | 0.89 | 0.12 |
| rs757352 | COPZ2 | 7.E-04 | ++++++ | 0.04 | 0.09 | 112 | 99 | 0.17 | 0.30 | 0.13 | 0.10 | 0.14 | 0.14 |
| rs9898218 | COPZ2 | 0.0010 | ++++++ | 0.0011 | 0.31 | 208 | 154 | 0.58 | 0.37 | 0.19 | 0.0006 | 0.33 | 0.56 |
| rs7216504 | COPZ2 | 0.03 | −+++++ | 0.02 | 0.75 | 155 | 109 | −0.19 | 0.04 | 0.78 | 0.09 | 0.07 | 0.31 |
| rs11650615 | COPZ2/NFE2L1 | 7.E-04 | −+++++ | 0.0007 | 0.39 | 179 | 129 | −0.50 | 0.03 | 0.16 | 0.003 | 0.32 | 0.11 |
The effect direction column indicates a positive sign for each study that confers a risk effect and a negative sign for the study that confers a protective effect. The positive or negative sign represents the following studies in this order: NIMH, NCRAD, GenADA, TGEN2, NIA-LOAD, and ADNI. The Info. Fam. columns indicate the number of informative families from the FBAT analysis for each family-based study. A negative FBAT P value indicates an undertransmission of the minor allele in affected offspring, whereas a positive FBAT P value indicates an overtransmission of the minor allele in affected offspring.
Table S3.
Study cohort sample size
| Study cohort | Cases | Controls | Subjects | Platform |
| NIMH, 387 families | 857 | 329 | 1,186 | Affymetrix 6.0 |
| NCRAD, 354 families | 884 | 70 | 954 | Affymetrix 6.0 |
| GenADA | 783 | 755 | 1,538 | Affymetrix 500 |
| TGEN2 | 953 | 556 | 1,509 | Affymetrix 1M |
| NIA-LOAD | 371 | 875 | 1,246 | Illumina 370 |
| ADNI | 170 | 192 | 362 | Illumina Human610 |
| Total study sample | 4,018 | 2,777 | 6,795 |
Two family-based study samples and four case-control cohorts were used.
We then searched for rare and highly penetrant variants in the WGS data for the COPI complex in 410 AD families, where no other previously known AD mutations could be established as genetic risk factors. We limited our search to variants that are predicted to have a significant impact on gene functionality based on publicly available algorithms (i.e., KEGG, SIFT, PolyPhen2, HPRD, CADD, fitcon score catalogs) (17). In total, we identified 24 variants across the nine COPI genes. Interestingly, eight COPI genes (the five genes that exhibit SNPs associated with AD as well as COPE, COPG1, and COPG2) display alternate variants that segregate with the disease in the carrier families (Table 2 and Table S4). These variants might be responsible for AD or serve as drivers of pathology in the carrier families.
Table 2.
COPI variants in AD families
| Gene | Identifier | Variant | No. of families | No. of affected carriers | No. of unaffected carriers | MAF (WGS) |
| COPA | rs57425682 | T146A | 14 | 20/29 | 2/6 | 0.007 |
| 1:160278885 | 5′ splice | 1 | 2/2 | 0/0 | 0.0007 | |
| rs75190422 | 5′ splice | 6 | 8/10 | 0/1 | 0.003 | |
| 1:160303386 | 5′ splice | 1 | 3/4 | 0/0 | 0.001 | |
| 1:160283843 | R260C | 1 | 2/3 | 0/1 | 0.0007 | |
| COPB1 | 11:14521151 | 3′ splice | 6 | 9/16 | 1/12 | 0.003 |
| rs201467424 | V789I | 1 | 3/3 | 0/0 | 0.001 | |
| rs375670951 | 5′ splice | 1 | 3/3 | 0/0 | 0.001 | |
| 11:14520389 | D29Y | 1 | 2/2 | 0/0 | 0.0007 | |
| 11:14502548 | R351K | 1 | 2/3 | 0/2 | 0.0007 | |
| rs144780995 | P625S | 1 | 3/3 | 0/0 | 0.0007 | |
| COPG1 | 3:128968592 | 5′ splice | 1 | 2/2 | 0/0 | 0.0007 |
| COPG2 | rs143820112 | 3′ splice | 1 | 2/2 | 0/0 | 0.0007 |
| COPD | rs78730658 | 5′ splice | 4 | 7/11 | 0/0 | 0.002 |
| rs188303468 | 3′ splice | 2 | 3/5 | 0/2 | 0.001 | |
| rs138863361 | A6S | 2 | 3/5 | 0/2 | 0.001 | |
| COPE | rs2231987 | S13C | 18 | 27/40 | 4/13 | 0.01 |
| rs199731661 | 5′ splice | 6 | 6/11 | 2/7 | 0.002 | |
| rs141039416 | R96Q | 1 | 2/2 | 0/0 | 0.0007 | |
| rs34510432 | R85H | 1 | 2/3 | 0/0 | 0.0007 | |
| COPZ1 | 12:54741793 | E136Q | 1 | 2/2 | 0/0 | 0.0007 |
| COPZ2 | rs115870363 | 3′ splice | 9 | 13/17 | 2/3 | 0.005 |
| 17:46115120 | frameshift | 1 | 2/2 | 0/0 | 0.001 | |
| 17:46115121 | W6R | 1 | 2/2 | 0/0 | 0.001 |
The variants resulting from the analyses of the nine COPI genes are listed. Identifier: dbSNP (The Single Nucleotide Polymorphism Database; www.ncbi.nlm.nih.gov/snp) ID when available or the physical location of the variant in the genome (hg19 reference panel). Variant: impact of the mutation. No. of families: number of families where at least one subject is a carrier. No. of affected carriers: ratio of the number of subjects showing the presence of the alternate allele per the total number of affected patients in the carrier families. No. of unaffected carriers: ratio of the number of unaffected family members carrying the alternate allele per the total number of those unaffected in the carrier families. MAF (WGS): frequency of the minor alternate allele in the whole-genome sequencing reported study population.
Table S4.
Variants from WGS analyses of the COPI complex genes
| Gene | Identifier | Alt. Ex | Variant | No. of families | No. of affected carriers | Med. AAO | No. of unaffected carriers | MAF (dbSNP) | MAF (WGS) |
| COPA | ENST00000368069 | nEx 33 | nAA 1,233 | ||||||
| rs57425682 | 6 | T146A | 14 | 20/29 | 73 | 2/6 | 0.003 | 0.007 | |
| 1:160278885 | 5 | 5′ splice | 1 | 2/2 | 72.5 | 0/0 | N/A | 0.0007 | |
| rs75190422 | 8 | 5′ splice | 6 | 8/10 | 65.5 | 0/1 | 0.001 | 0.003 | |
| 1:160303386 | 28 | 5′ splice | 1 | 3/4 | 75 | 0/0 | N/A | 0.001 | |
| 1:160283843 | 9 | R260C | 1 | 2/3 | 68 | 0/1 | N/A | 0.0007 | |
| COPB1 | ENST00000249923 | nEx 22 | nAA 953 | ||||||
| 11:14521151 | 1 | 3′ splice | 6 | 9/16 | 77 | 1/12 | N/A | 0.003 | |
| rs201467424 | 18 | V789I | 1 | 3/3 | 76 | 0/0 | N/A | 0.001 | |
| rs375670951 | 8 | 5′ splice | 1 | 3/3 | 76 | 0/0 | N/A | 0.001 | |
| 11:14520389 | 2 | D29Y | 1 | 2/2 | 76.5 | 0/0 | N/A | 0.0007 | |
| 11:14502548 | 9 | R351K | 1 | 2/3 | 78.5 | 0/2 | N/A | 0.0007 | |
| rs144780995 | 15 | P625S | 1 | 3/3 | 70.5 | 0/0 | N/A | 0.0007 | |
| COPG1 | ENST00000314797 | nEx 24 | nAA 874 | ||||||
| 3:128968592 | 1 | 5′ splice | 1 | 2/2 | 70.5 | 0/0 | N/A | 0.0007 | |
| COPG2 | ENST00000445977 | nEx 9 | nAA 246 | ||||||
| rs143820112 | 4 | 3′ splice | 1 | 2/2 | 64 | 0/0 | N/A | 0.0007 | |
| COPD | ENST00000264028 | nEx 10 | nAA 511 | ||||||
| rs78730658 | 3 | 5′ splice | 4 | 7/11 | 78 | 0/0 | 0.001 | 0.002 | |
| rs188303468 | 1 | 3′ splice | 2 | 3/5 | 70 | 0/2 | 0.005 | 0.001 | |
| rs138863361 | 2 | A6S | 2 | 3/5 | 70 | 0/2 | 0.005 | 0.001 | |
| COPE | ENST00000262812 | nEx 10 | nAA 308 | ||||||
| rs2231987 | 1 | S13C | 18 | 27/40 | 74 | 4/13 | 0.07 | 0.01 | |
| rs199731661 | 10 | 5′ splice | 6 | 6/11 | 72 | 2/7 | 0.003 | 0.02 | |
| rs141039416 | 3 | R96Q | 1 | 2/2 | 79 | 0/0 | N/A | 0.0007 | |
| rs34510432 | 3 | R85H | 1 | 2/3 | 66 | 0/0 | 0.005 | 0.0007 | |
| COPZ1 | ENST00000262061 | nEx 9 | nAA 177 | ||||||
| 12:54741793 | 7 | E136Q | 1 | 2/2 | 69 | 0/0 | N/A | 0.0007 | |
| COPZ2 | ENST00000006101 | nEx 10 | nAA 208 | ||||||
| rs115870363 | 6 | 3′ splice | 9 | 13/17 | 75 | 2/3 | 0.004 | 0.005 | |
| 17:46115120 | 2 | 6, frameshift | 1 | 2/2 | 87.5 | 0/0 | N/A | 0.001 | |
| 17:46115121 | 2 | W6R | 1 | 2/2 | 87.5 | 0/0 | N/A | 0.001 |
The variants resulting from the analyses of the nine COPI genes are listed above. Identifier: dbSNP (The Single Nucleotide Polymorphism Database) ID when available or the physical location of the variant in the genome (hg19 reference panel). Alt. Ex: exon affected by the variation. Variant: impact of the mutation. No. of families: number of families where at least one subject is a carrier. No. of affected carriers: ratio of the number of subjects showing the presence of the alternate allele/total number of affected patients in the carrier families. Med. AAO: median onset age of the AD patients. No. of unaffected carriers: ratio of the number of unaffected family members carrying the alternate allele/total number of unaffected in the carrier families. MAF (dbSNP) and MAF (WGS): frequency of the minor alternate allele in the reported study population. ENST, Ensembl Canonical Transcript ID for the gene of interest; N/A, not available; nAA, number of amino acids in the transcribed protein; nEx, number of exons in the gene transcript.
Discussion
The discovery of a genetic link between several subunits of the COPI complex and AD in humans and the confirmation of the importance of the COPI complex in amyloid plaque burden and memory impairments in an AD mouse model strongly support the importance of the COPI complex in APP processing and amyloidogenesis.
The important reduction in beta amyloid plaques observed in AD/δ-COP mutant mice, although not complete, induces a significant behavioral improvement compared with AD mice. These results are in agreement with other studies showing that a strong reduction of amyloid plaques is sufficient to translate into behavioral consequences without necessarily requiring a total clearance of the plaques (18–20).
Numerous AD mouse models have been developed over the years with different degrees of success, especially when considering cognitive impairments. We chose to work with one of the broadly used AD mouse models, namely the 2xTg model. The ataxic condition observed in Nur17 mice, a condition slightly worsening with age, did not allow us to fully explore all cognitive functions typically measured in mice. We described here the use of the novel object recognition test, as we showed by comparing WT mice with δ-COP mut mice that both cohorts spent significantly more time with the novel object compared with the familiar object, indicating that the general condition of δ-COP mut mice was compatible with this test. More importantly, we showed that the partial loss of δ-COP activity rescued the capacity to discriminate novel versus familiar objects in AD/δ-COP mut mice, consistent with plaque load reduction and decrease of Aβ production in these mice. The AD 2xTg mice develop cognitive impairments at a relatively advanced age (12 mo). It would be interesting to evaluate the impact of δ-COP function on other cognitive aspects, and this could perhaps be achieved in AD mouse models developing cognitive impairments earlier, such as 3xTg or 5xFAD.
In terms of therapeutic application, due to the biological importance of COPI in general and δ-COP specifically (mice partially lacking δ-COP activity display some phenotypes, such as ataxia), any therapeutic intervention targeting the COPI complex will have to target this biological function rather moderately and/or indirectly, perhaps just occasionally, to circumvent issues similar to the ones raised using γ-secretase inhibitors (21). To achieve selectivity, it may be possible to identify and target a regulator of APP trafficking through COPI.
The fact that several members of the COPI complex are associated with AD risk in our genetic studies reinforces the importance of the COPI pathway for AD progression, as found for several subunits of the γ-secretase complex. It is important to note that, in small late-onset families, unaffected family members are often not available and in almost all cases the rare functional variants are either novel or have never been observed in control subjects. As a consequence of the scarcity, in the databases, of individuals carrying the mutation, it is exceedingly rare to reach significance and the mutations identified require full characterization. However, we note that even with APP and PSEN1/2, we have observed families with mixed genetic etiologies in which all affected patients do not carry the pathogenic mutation. For future work, it will be interesting to test the relevance for AD of each of the variants identified genetically in several COPI subunits to ascertain their biological role in the progression of the disease.
In summary, we are providing evidence confirming the relevance of COPI for AD pathology in vivo, in a mouse model of AD, where we showed that both the burden of plaques and plaque-related memory impairments can be improved after reducing δ-COP activity. This notion has been further confirmed using a genetic approach, studying four different cohorts of human patients and identifying several associations between different COPI subunits and AD. Altogether, the in vivo data presented here in combination with our previous findings (3) unveil new avenues for therapeutic interventions.
Materials and Methods
Cell Culture, Knockdown, and Overexpression.
N2a and N2a-695 (overexpressing APP695) cells were grown in 1:1 DMEM/Opti-MEM (Life Technologies) in the presence of 5% (vol/vol) FBS (Sigma). N2a and N2a-695 cells were transfected using DharmaFECT 2 (Thermo Fisher Scientific). Small interfering RNA (siRNA) of the δ-COP subunit or nontargeting control siRNA was purchased from Thermo Fisher Scientific.
Sequences for δ-COP siRNA are as follows: sense sequence: 5′-GGAGAAUGUUAACCUGGCAUU-3′; and antisense sequence: 5′-P-UGCCAGGUUAACAUUCUCCUU-3′.
Lipofectamine 2000 (Life Technologies) was used for all transient transfections. Mammalian expression vectors used for protein overexpression were purchased from GeneCopoeia. Constructs were based on a pReceiver-M07 plasmid, and cDNAs were tagged with a C-terminal HA sequence. pcDNA4-β-CTF expression vector was a kind gift from Y.-M. Li, Memorial Sloan-Kettering Cancer Center, New York, and pcDNA3-APP was provided by H. Rebholz, The City College of New York, New York.
Aβ Quantification.
For cell cultures, the medium was replaced 6 h before collecting supernatants for Aβ40 and Aβ42 peptide measurements. Aβ levels were normalized to total protein levels. For in vivo experiments, brains were dissected on ice, homogenized in 70% (vol/vol) formic acid solution, and sonicated, and homogenates were centrifuged for 1 h at 180,000 × g. The supernatants were collected and neutralized with 1 M Tris base. The concentrations of the Aβ40 and Aβ42 peptides were determined using the sandwich ELISA technique (Life Technologies).
Western Blotting Analysis and Antibodies.
Protein samples were analyzed by SDS/PAGE and Western blot after BCA quantification. Antibodies used were APP-CTF (in house and Abcam), γ-adaptin (BD Biosciences), β-actin, Na,K-ATPase (Cell Signaling Technology), PS1 (in house) ADAM10 (Abcam), and BACE (Abcam). HA antibody for immunoprecipitation (GenScript) and HA antibody for Western blotting (Roche) were used.
Subcellular Fractionation.
Subcellular fractionation was performed as previously described (3). Briefly, cells were homogenized in a sucrose buffer (0.25 M sucrose) using a ball-bearing cell cracker. Homogenates were separated on a sucrose gradient (0.25–2 M sucrose) by centrifugation (2.5 h at 250,000 × g). Fractions corresponding to the Golgi were analyzed by SDS/PAGE and Western blotting using the corresponding antibodies for APP-CTF, actin, γ-adaptin, PS1, ADAM10, and BACE (see above).
Cell-Surface Biotinylation.
Cells were washed with ice-cold PBS and incubated with Sulfo-NHS-SS-Biotin (Thermo Fisher Scientific) in PBS for 30 min at 4 °C. Biotin solution was then removed and cells were washed once with 50 mM Tris and twice with ice-cold PBS.
Precipitation of Biotinylated Proteins.
Cells were lysed in RIPA buffer followed by centrifugation (10,000 × g) and lysates were mixed with streptavidin-Dynabeads (Life Technologies). After an hour of incubation at 4 °C, the beads were washed five times with ice-cold PBS/0.01% Tween. Biotinylated proteins were resolved by SDS/PAGE and analyzed by Western blot with different antibodies as indicated in the text.
Mouse Strains.
δ-COP wild-type and δ-COP mutant mice (mixed background of C57BL/6J and 129S6/SvEvTac) were kindly provided by A. Ikeda from the Baylor College of Medicine, Houston. The mutation was identified previously as a single-nucleotide substitution in δ-COP (4). These mice were crossed with the 2xTg AD mouse model (APPswe; PS1ΔE9) (Jackson Laboratory) for Aβ analyses and behavioral experiments.
Amyloid Plaque Analysis from Mouse Brain Immune-Labeled Sections.
Mouse brains were fixed in 4% (vol/vol) paraformaldehyde (Electron Microscopy Sciences), washed with PBS, and incubated with different solutions of sucrose. Brains were then frozen on dry ice with freezing medium and sliced coronally in sections of 40 μm. Sections were labeled with Aβ antibody (6E10). Plaques from nine slices per animal were counted.
Behavioral Characterization.
Procedures were adapted from a previously described protocol (4). All procedures were approved by The Rockefeller University Institutional Animal Care and Use Committee animal institutional review board and performed accordingly. Briefly, the task was conducted in a clear Plexiglas open arena (58 × 58 × 46 cm3) using two objects (a 50-mL Falcon tube and a Lego object constructed of two Lego pieces: one square piece centered and attached to the top of one rectangular piece). The task consisted of three phases: habituation, sample, and choice, conducted successively. During habituation, the mouse was placed in the empty arena and allowed to freely explore for 5 min. The sample phase was conducted 24 h later. Two identical objects were fixed to the floor in the northeast and southeast corners of the box (10 cm from the wall) and the mice were allowed to explore for 10 min. Half of the mice in each group were randomly presented with either the Falcon tube or the Lego object during the sample phase. The choice phase was conducted 1 h after the sample phase. One familiar object (identical to that used in the sample phase) and one novel object were placed in the northeast and southeast corners of the box and the mice were allowed to explore for 5 min. The location of the novel object was counterbalanced across groups. The arena and objects were cleaned with 30% (vol/vol) alcohol between each phase. EthoVision tracking software (Noldus Information Technology) was used during the task. Object exploration was manually scored in real time using a computer keyboard. Object exploration was scored only when the mouse’s nose touched the object.
Human Genetic SNP Association Study.
Family-based study samples.
Candidate gene analysis was performed on the National Institute of Mental Health Genetics Initiative Alzheimer’s Disease Study family sample (6, 15) and the National Cell Repository for Alzheimer’s Disease (NCRAD) family sample (study sample characteristics are available at https://www.ncrad.org). Only families of self-reported Caucasian ethnicity from each study sample were included in these analyses. Recruitment for each study was based on families containing at least two affected siblings; most families are typically composed of multiple siblings. The NIMH cohort contains 1,186 subjects from 387 families and the NCRAD cohort contains 954 subjects from 354 families (Table S2). The genotypes from both familial cohorts used to derive imputation-based genotypes were generated with the Affymetrix 6.0 SNP array.
Case-control study samples.
The four case-control cohorts included in these analyses are the Genetics Alzheimer’s Disease Association Study sample (GenADA; GlaxoSmithKline) (22), the Translational Genomics Research Institute Study sample (TGEN2; https://www.tgen.org), the National Institute on Aging Genetics Initiative for Late Onset Alzheimer’s Disease Family Study sample (23) (NIA-LOAD), and the Alzheimer’s Disease Neuroimaging Initiative Study sample (ADNI; https://aibl.csiro.au/adni). In total, the case-control cohorts contain 2,277 cases and 2,378 controls of self-reported Caucasian ethnicity (Table S2).
Data Analysis.
Genotype imputation was carried out with the IMPUTE2 algorithm (24); we used the most recent release of the integrated 1,000 Genomes Project data as the reference population (mathgen.stats.ox.ac.uk/impute/data_download_1000G_phase1_integrated.html).
Selection of Independent Markers.
To determine the number of independent (i.e., linkage equilibrium) SNPs available for association testing from each of the COPI gene regions (plus 20 kb from the 3′ UTR and 5′ UTR), we used a pairwise linkage disequilibrium (LD) test approach implemented in Haploview (version 4.2) software (https://www.broadinstitute.org/scientific-community/science/programs/medical-and-population-genetics/haploview/haploview), whereby all alleles captured are highly correlated (r2 > 0.8) to a minimal set of tag SNPs. After imputation, a total of 530 SNPs were available among the eight COP genes (Table S2). The LD structure for selecting the tag SNPs was based on genotypes derived by imputation from 387 unrelated affected NIMH subjects. SNPs that deviated from Hardy–Weinberg equilibrium (P < 0.0001) or imputed SNPs with information scores less than 40% or that had a minor allele frequency less than 0.01 (imputation accuracy is generally poor for rare alleles) in our NIMH AD patient subset were excluded from the LD analysis (34 SNPs removed, 6%; Table S3). These analyses led to a total of 96 independent SNPs among the eight COP gene regions (Table S2).
Statistical Analysis.
To assess for allelic association within the two family-based samples among our eight candidate gene regions, we used an extension (25) of the family-based association analysis test (FBAT) approach implemented in PBAT (version 3.6). The FBAT approach evaluates whether the minor allele is over- or undertransmitted (i.e., risk or protection) in affected offspring compared with the expected distribution under Mendel’s law of segregation. FBAT identifies association as well as linkage and minimizes false positive associations due to population stratification. FBAT analysis was performed only on SNPs with at least 10 informative families in each of the two family studies. An additive genetic model was assumed for both family-based and case-control association analysis. We report P values from the Liptak test statistic, which uses all available information. The Liptak method attains higher power levels than the traditional FBAT approach by combining the Z statistics that correspond to the P values of the family-based test (the within-family information) with the rank-based P values for population-based analysis (the between-family information).
To assess for allelic association in the case-control datasets, we performed logistic regression analysis, adjusting for known confounds such as sex, age, and population structure, as implemented in the SNPTEST (version 2) software package (https://mathgen.stats.ox.ac.uk/genetics_software/snptest/old/snptest.html). Principal component analysis was performed with EIGENSTRAT (genepath.med.harvard.edu/∼reich/EIGENSTRAT.htm) to assess population structure, and the first three principal components were included as covariates in the logistic model. As an overall summary measure of association, we performed a meta-analysis of the test results from the two family-based studies (NIMH and NCRAD) plus the four case-control studies (GenADA, TGEN2, NIA-LOAD, and ADNI) using the statistical package METAL (https://www.sph.umich.edu/csg/abecasis/metal). Agnostic analyses of total GWAS datasets generally require a Bonferroni correction of 5 × 10−8. Because a limited set of gene candidates and their SNPs (96 SNPs) were chosen to test for association with AD, an experimental-wide Bonferroni correction for 96 independent tests was required (P < 5 × 10−4).
Human Genetic Variant Association Study.
Standard protocol approvals, registrations, and patient consents.
Diagnosis of AD dementia was established according to NINCDS-ADRDA criteria (National Institute of Neurological and Communicative Disorders and Stroke and the Alzheimer's Disease and Related Disorders Association). Informed consent was provided by all participants, and research approval was established by the relevant institutional review boards in the study cohort. No human samples were generated or used for this study; instead only databases generated previously were used, and therefore institutional review board approval was not necessary.
WGS data generation.
Three micrograms of total genomic DNA (150 ng/μL) obtained from the Cell and DNA Repository, Rutgers University was sequenced at Illumina using their latest HiSeq 2500 paired-end sequencing platform. An average of 48-fold coverage of 98% of the genome was observed in the resulting 120-Gb data from each sample. Genomic variants were called in-house using FreeBayes (version 0.9.9.2-18) and the Genome Analysis Toolkit (GATK) best practices method (26), resulting in close to 400 Tb of high-quality sequencing data. A fully annotated GEMINI database (17) with additional information (e.g., functional effect predictions) was created for querying specific genes and loci for analysis purposes (https://bcbio.wordpress.com/2013/10/21/updated-comparison-of-variant-detection-methods-ensemble-freebayes-and-minimal-bam-preparation-pipelines/).
Fig. S1 provides additional images related to amyloid plaque analysis in vivo. Tables S1–S3 provide further information about the human genetic SNP association study and Table S4 provides further information about the variant association study.
Acknowledgments
We are grateful to Dr. Jean-Pierre Roussarie for his critical reading of the manuscript. We thank Dr. Akihiro for providing the δ-COP mutant mice. This work was supported, in part, by grants from the National Institutes of Health (AG/NIA: AG-09464 to P.G.; NIMH-MH060009 to R.E.T.), Fisher Center for Alzheimer’s Research Foundation (P.G. and M.F.), and Cure Alzheimer’s Fund (R.E.T.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1604176113/-/DCSupplemental.
References
- 1.Haass C, Kaether C, Thinakaran G, Sisodia S. Trafficking and proteolytic processing of APP. Cold Spring Harb Perspect Med. 2012;2(5):a006270. doi: 10.1101/cshperspect.a006270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Choy RW, Cheng Z, Schekman R. Amyloid precursor protein (APP) traffics from the cell surface via endosomes for amyloid β (Aβ) production in the trans-Golgi network. Proc Natl Acad Sci USA. 2012;109(30):E2077–E2082. doi: 10.1073/pnas.1208635109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bettayeb K, et al. δ-COP modulates Aβ peptide formation via retrograde trafficking of APP. Proc Natl Acad Sci USA. 2016;113:5412–5417. doi: 10.1073/pnas.1604156113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Xu X, et al. Mutation in archain 1, a subunit of COPI coatomer complex, causes diluted coat color and Purkinje cell degeneration. PLoS Genet. 2010;6(5):e1000956. doi: 10.1371/journal.pgen.1000956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rogaeva E, et al. The neuronal sortilin-related receptor SORL1 is genetically associated with Alzheimer disease. Nat Genet. 2007;39(2):168–177. doi: 10.1038/ng1943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tanzi RE. The genetics of Alzheimer disease. Cold Spring Harb Perspect Med. 2012;2(10):a006296. doi: 10.1101/cshperspect.a006296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen F, et al. TMP21 is a presenilin complex component that modulates gamma-secretase but not epsilon-secretase activity. Nature. 2006;440(7088):1208–1212. doi: 10.1038/nature04667. [DOI] [PubMed] [Google Scholar]
- 8.Réchards M, et al. Presenilin-1-mediated retention of APP derivatives in early biosynthetic compartments. Traffic. 2006;7(3):354–364. doi: 10.1111/j.1600-0854.2006.00388.x. [DOI] [PubMed] [Google Scholar]
- 9.Selivanova A, Winblad B, Farmery MR, Dantuma NP, Ankarcrona M. COPI-mediated retrograde transport is required for efficient gamma-secretase cleavage of the amyloid precursor protein. Biochem Biophys Res Commun. 2006;350(1):220–226. doi: 10.1016/j.bbrc.2006.09.043. [DOI] [PubMed] [Google Scholar]
- 10.Morales-Corraliza J, et al. In vivo turnover of tau and APP metabolites in the brains of wild-type and Tg2576 mice: Greater stability of sAPP in the beta-amyloid depositing mice. PLoS One. 2009;4(9):e7134. doi: 10.1371/journal.pone.0007134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Savage MJ, et al. Turnover of amyloid beta-protein in mouse brain and acute reduction of its level by phorbol ester. J Neurosci. 1998;18(5):1743–1752. doi: 10.1523/JNEUROSCI.18-05-01743.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Morton MJ, et al. Association with beta-COP regulates the trafficking of the newly synthesized Na,K-ATPase. J Biol Chem. 2010;285(44):33737–33746. doi: 10.1074/jbc.M110.141119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jeyifous O, et al. SAP97 and CASK mediate sorting of NMDA receptors through a previously unknown secretory pathway. Nat Neurosci. 2009;12(8):1011–1019. doi: 10.1038/nn.2362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McClean PL, Parthsarathy V, Faivre E, Hölscher C. The diabetes drug liraglutide prevents degenerative processes in a mouse model of Alzheimer’s disease. J Neurosci. 2011;31(17):6587–6594. doi: 10.1523/JNEUROSCI.0529-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Blacker D, et al. NIMH Genetics Initiative Alzheimer’s Disease Study Group Results of a high-resolution genome screen of 437 Alzheimer’s disease families. Hum Mol Genet. 2003;12(1):23–32. doi: 10.1093/hmg/ddg007. [DOI] [PubMed] [Google Scholar]
- 16.Hooli BV, et al. Role of common and rare APP DNA sequence variants in Alzheimer disease. Neurology. 2012;78(16):1250–1257. doi: 10.1212/WNL.0b013e3182515972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Paila U, Chapman BA, Kirchner R, Quinlan AR. GEMINI: Integrative exploration of genetic variation and genome annotations. PLOS Comput Biol. 2013;9(7):e1003153. doi: 10.1371/journal.pcbi.1003153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Janus C, et al. A beta peptide immunization reduces behavioural impairment and plaques in a model of Alzheimer’s disease. Nature. 2000;408(6815):979–982. doi: 10.1038/35050110. [DOI] [PubMed] [Google Scholar]
- 19.Scholtzova H, et al. Memantine leads to behavioral improvement and amyloid reduction in Alzheimer’s-disease-model transgenic mice shown as by micromagnetic resonance imaging. J Neurosci Res. 2008;86(12):2784–2791. doi: 10.1002/jnr.21713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hartman RE, et al. Treatment with an amyloid-beta antibody ameliorates plaque load, learning deficits, and hippocampal long-term potentiation in a mouse model of Alzheimer’s disease. J Neurosci. 2005;25(26):6213–6220. doi: 10.1523/JNEUROSCI.0664-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Selkoe DJ. Resolving controversies on the path to Alzheimer’s therapeutics. Nat Med. 2011;17(9):1060–1065. doi: 10.1038/nm.2460. [DOI] [PubMed] [Google Scholar]
- 22.Li H, et al. Candidate single-nucleotide polymorphisms from a genomewide association study of Alzheimer disease. Arch Neurol. 2008;65(1):45–53. doi: 10.1001/archneurol.2007.3. [DOI] [PubMed] [Google Scholar]
- 23.Wijsman EM, et al. NIA-LOAD/NCRAD Family Study Group Genome-wide association of familial late-onset Alzheimer’s disease replicates BIN1 and CLU and nominates CUGBP2 in interaction with APOE. PLoS Genet. 2011;7(2):e1001308. doi: 10.1371/journal.pgen.1001308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Marchini J, Howie B, Myers S, McVean G, Donnelly P. A new multipoint method for genome-wide association studies by imputation of genotypes. Nat Genet. 2007;39(7):906–913. doi: 10.1038/ng2088. [DOI] [PubMed] [Google Scholar]
- 25.Won S, et al. On the analysis of genome-wide association studies in family-based designs: A universal, robust analysis approach and an application to four genome-wide association studies. PLoS Genet. 2009;5(11):e1000741. doi: 10.1371/journal.pgen.1000741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McKenna A, et al. The Genome Analysis Toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20(9):1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]