Significance
Snails serve as hosts for the larval development of many medically and agriculturally important parasitic flatworms, including schistosomes, blood flukes that collectively infect more than 260 million people globally. Here, we functionally characterize a granulin-like snail growth factor that drives the development of snail immune cells, thereby making a schistosome-susceptible snail resistant to infection. This study presents the functional characterization of an endogenous gastropod growth factor as well as demonstrated reversal of a susceptible snail phenotype toward resistance using a defined snail factor.
Keywords: Biomphalaria glabrata, Schistosoma mansoni, schistosomiasis, granulin, hematopoiesis
Abstract
Digenean trematodes are a large, complex group of parasitic flatworms that infect an incredible diversity of organisms, including humans. Larval development of most digeneans takes place within a snail (Gastropoda). Compatibility between snails and digeneans is often very specific, such that suitable snail hosts define the geographical ranges of diseases caused by these worms. The immune cells (hemocytes) of a snail are sentinels that act as a crucial barrier to infection by larval digeneans. Hemocytes coordinate a robust and specific immunological response, participating directly in parasite killing by encapsulating and clearing the infection. Hemocyte proliferation and differentiation are influenced by unknown digenean-specific exogenous factors. However, we know nothing about the endogenous control of hemocyte development in any gastropod model. Here, we identify and functionally characterize a progranulin [Biomphalaria glabrata granulin (BgGRN)] from the snail B. glabrata, a natural host for the human blood fluke Schistosoma mansoni. Granulins are growth factors that drive proliferation of immune cells in organisms, spanning the animal kingdom. We demonstrate that BgGRN induces proliferation of B. glabrata hemocytes, and specifically drives the production of an adherent hemocyte subset that participates centrally in the anti-digenean defense response. Additionally, we demonstrate that susceptible B. glabrata snails can be made resistant to infection with S. mansoni by first inducing hemocyte proliferation with BgGRN. This marks the functional characterization of an endogenous growth factor of a gastropod mollusc, and provides direct evidence of gain of resistance in a snail-digenean infection model using a defined factor to induce snail resistance to infection.
Snails of the genus Biomphalaria serve as the intermediate host for Schistosoma mansoni, one of the species of parasitic flatworms that cause schistosomiasis. The extensive amplification of the parasite within the snail host complicates disease control efforts and is one of the reasons that schistosomiasis continues to be one of the most important neglected tropical diseases. Studies investigating compatibility between Biomphalaria snails and S. mansoni suggest that the snail immune response plays a crucial role in determining infection outcome. Although circulating humoral factors are known to play an important role in determining infection outcome via recognition of various parasite targets, or by directly damaging the parasite tegument (1–4), it is ultimately the immune cells of the snail, known as hemocytes, that are primarily responsible for encapsulating and eliminating the parasite (5).
The central role of hemocytes in the anti-schistosome response underscores the importance of maintaining suitable numbers of circulating and tissue-resident immune cells. We currently understand very little about the mechanics of hematopoiesis in gastropods. Our limited understanding of the process suggests that the primary hematopoietic organ in snails is a thin tissue located in the anterior of the pericardial region known as the amoebocyte-producing organ (APO) (6). Following stimulation with excretory/secretory (ES) products of digenean trematodes, such as S. mansoni (7) or Echinostoma paraensei (8), the APO noticeably swells and displays an increase in mitotic events (9). The hemocytes generated during these events tend to be adherent and granulocytic, and exhibit greater capacity to generate humoral factors relevant to parasite killing compared with the preexisting population (2). Following the stimulation of hemocyte development by exogenous factors, the proportion of hemocyte subsets alters to favor the granulocytic morphology (9) that is typically associated with parasite encapsulation (10), phagocytosis (2), and the production of reactive oxygen and nitrogen intermediates (11) important for parasite killing.
Parasite-specific factors that are excreted or secreted upon initial infection of the snail are important drivers of hematopoiesis in B. glabrata. The second, and lesser-understood, drivers are endogenous snail growth factors that, in response to infection, induce hemocyte proliferation/differentiation within the APO. Recent studies screening the transcriptome and proteome of B. glabrata have yielded promising candidates that possess canonical domains associated with hematopoietic factors of other organisms, both invertebrate and vertebrate (12).
Granulins are evolutionarily conserved growth factors. Whereas fungi appear to lack a granulin gene (GRN), other members of the plant and animal kingdoms surveyed so far possess at least one copy of GRN. Granulins are identified by a unique 12-cysteine motif that can be found, often repeatedly, throughout the progranulin protein (PGRN). These are secreted propeptides that can be subsequently cleaved into smaller functional units by elastase (13). Progranulin is involved in numerous biological processes including tissue repair (14), early embryogenesis, inflammation, neurobiology, and cell proliferation (15), via activation of the MAPK pathway (13). The products of PGRN cleavage (usually ∼6 kDa) can also be functional in a variety of roles that includes participation in and activation of the immune response (16).
We report on the identification and functional characterization of a progranulin molecule [B. glabrata granulin (BgGRN)] from the snail B. glabrata. This study represents a number of significant advancements for the B. glabrata–S. mansoni infection model and for the field of gastropod immunobiology. We demonstrate that BgGRN expression is responsive to S. mansoni challenge and induces the proliferation of B. glabrata immune cells, driving the in vivo development of an adherent hemocyte subset distinct from resident hemocytes. This effect is initiated via induction of the MAPK signaling pathway, and can be inhibited by knockdown of BgGRN using RNAi as well as via preincubation with a blocking antibody. Administration of recombinant BgGRN into a B. glabrata snail strain that is highly susceptible to S. mansoni infection before challenge resulted in a significant reversal of the susceptible phenotype. This study demonstrates the induction of resistance phenotype following S. mansoni challenge by using a defined gastropod-specific factor.
Results
In Silico Analysis of BgGRN
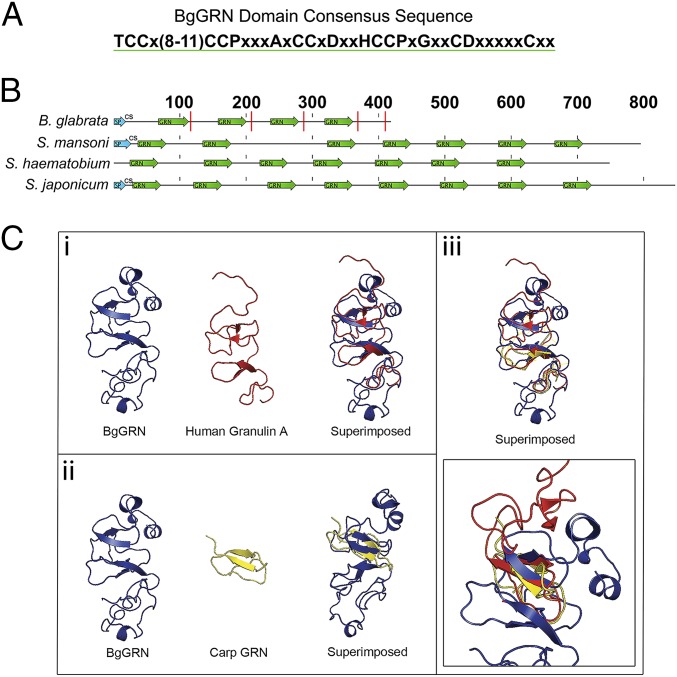
BgGRN domain prediction and comparison with schistosome granulin.
The B. glabrata, Schistosoma mansoni, Schistosoma hematobium, and Schistosoma japonicum progranulin proteins all possess multiple granulin domains (Fig. 1 A and B and Fig. S1). B. glabrata GRN has four predicted granulin domains characterized by amino acid residues 68–112, 158–200, 237–279, and 319–361. Modeling of BgGRN suggests that it shares secondary and tertiary structural properties with known structures of human granulin A and carp granulin. Although the number of granulin domains differ between BgGRN and the human and carp granulins, superimposition reveals predicted overlap of the α-helices and β-sheets associated with the conserved 12-cysteine domains (Fig. 1C).
Fig. 1.
In silico prediction of BgGRN granulin domain architecture compared with some known and predicted granulins. (A) The consensus sequence of the BgGRN granulin domain highlighting the canonical 12-cysteine motif that defines granulin proteins. (B) Reference diagram highlighting the predicted locations of the granulin domain (green arrows) in the predicted progranulin protein sequences of B. glabrata, S. mansoni, S. hematobium, and S. japonicum. Predicted signal peptides (SP) are highlighted in blue arrows and are followed by the predicted cut site (CS) for the signal peptide. Predicted elastase cleavage sites for BgGRN are highlighted with a red line intersecting the protein at the approximate cut site. (C) The predicted 3D structure of BgGRN compared with the two known crystal structures for human (i) and carp (ii) granulins, with a focus on the similar organization of β-sheets and α-helices (iii).
Fig. S1.
Protein alignment of BgGRN with pro-GRNs of S. mansoni, S. hematobium, and S. japonicum. Comparison of the amino acid conservation between the pro-BgGRN and pro-GRN proteins of three schistosomes highlights the conservation of the 12-cysteine motif that defines members of the GRN family. Granulin domains are highlighted in green and highlight the high amino acid conservation of the GRN domains between the three schistosomes. The four GRN domains of BgGRN share a 48.9%, 52.2%, 60.4%, and 50% amino acid identity with the three schistosome pro-GRNs. However, a large stretch of amino acids present in the S. mansoni pro-GRN breaks the second GRN domain, and so does an additional amino acid stretch present in all three schistosome pro-GRNs in domain three. BgGRN and S. japonicum granulin possess predicted secretion signal peptide domain in residues 1–18 and cleavage sites between residues 18 and 19. S. mansoni granulin has a predicted signal peptide from residues 1–26 and cleavage site between 26 and 27. The S. hematobium granulin sequence does not possess a traditional signal peptide that can be identified using predictive software.
Signal peptide and cleavage site annotations.
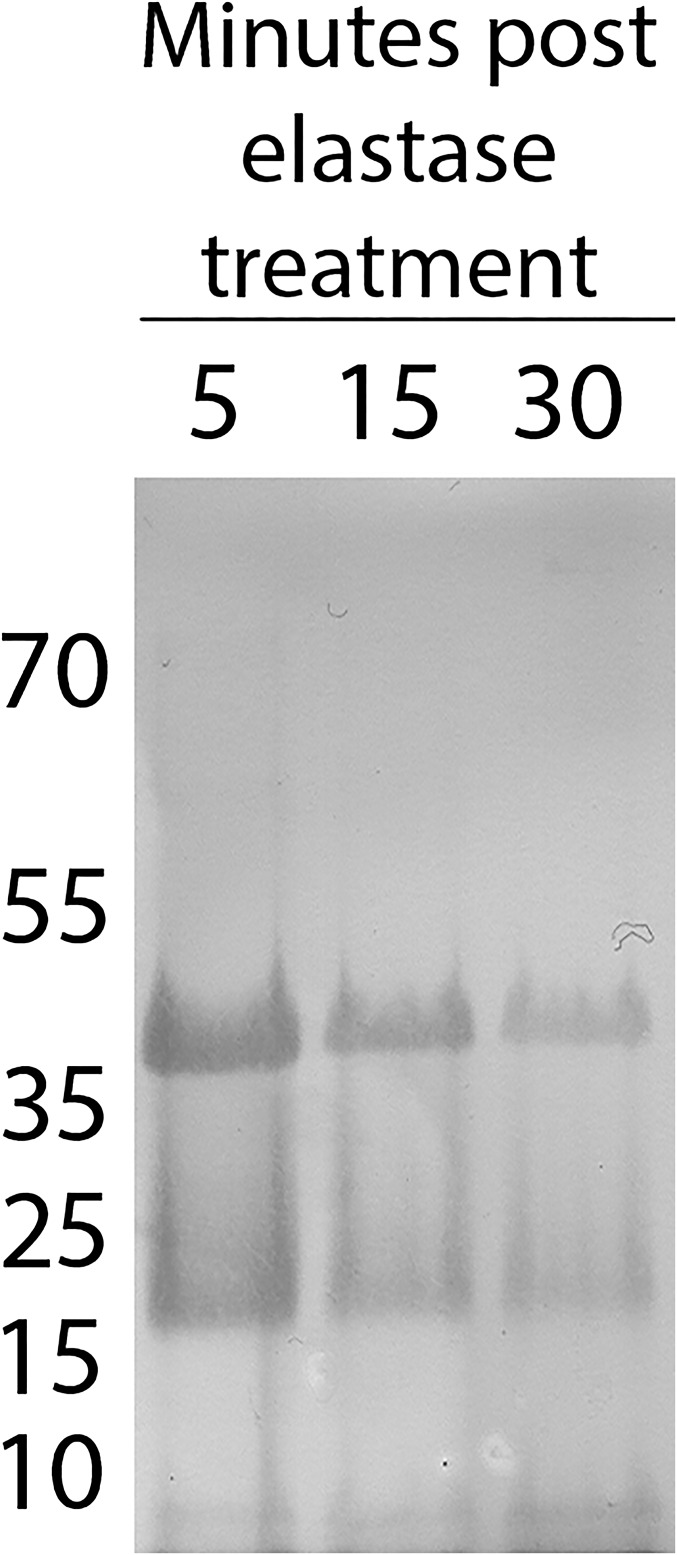
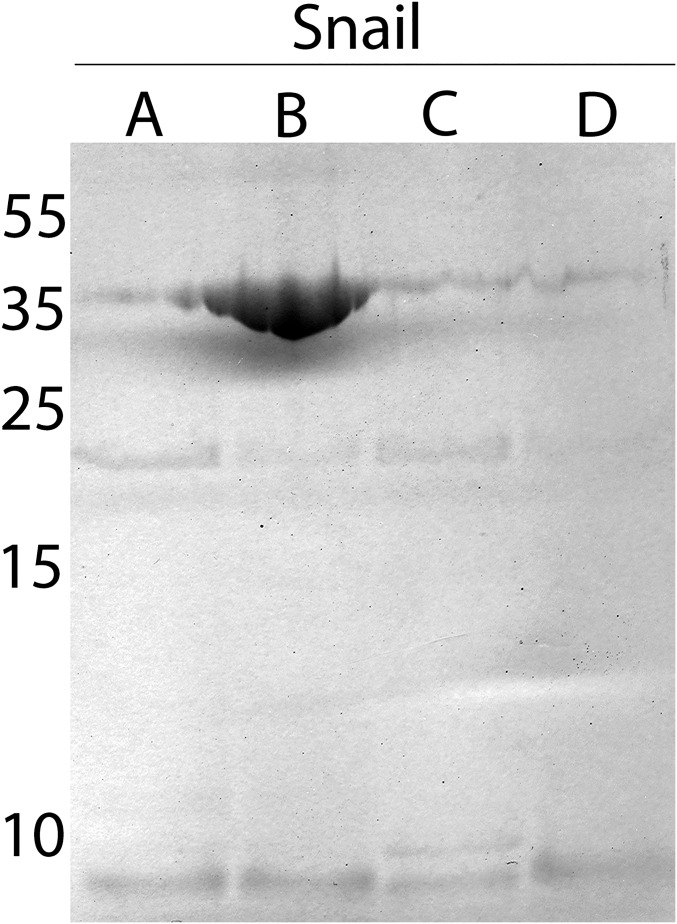
BgGRN has a predicted size of ∼44 kDa, and was consistently observed as a single band in Western blot of nonmanipulated snail plasma, as was recombinant BgGRN (rBgGRN). S. mansoni sporocyst ES products were not positive for GRN when probed with anti-BgGRN antibody. BgGRN possesses a predicted secretion signal domain in residues 1–18 (Fig. 1B), along with five elastase cleavage sites. Four of these sites immediately follow the four granulin domains, and the fifth cleavage site is predicted at the C-terminal end of pro-BgGRN. Incubation of rBgGRN with porcine elastase supports an elastase-mediated mechanism for processing of pro-BgGRN, yielding two distinct ∼18- and ∼8-kDa bands that are consistent with the smaller predicted size of the BgGRN cleavage products (Fig. S2). Longer elastase treatment appeared to produce more of the 8-kDa cleavage product at the expense of the larger pro-BgGRN (Fig. S2). Although cleavage products at these approximate sizes were not detected by Western blot analysis of plasma isolated from nonmanipulated snails, cleavage products were observed in the plasma of S. mansoni-challenged M-line (only ∼18-kDa product; Fig. 2B, Inset), and BS-90 (∼18- and ∼8-kDa products; Fig. 2A, Inset) and hemocyte lysates (∼18- and ∼8-kDa products; Fig. S3).
Fig. S2.
Elastase cleavage of rBgGRN yields two differently sized cleavage products. Anti-BgGRN Western blot of recombinant BgGRN following cleavage with porcine elastase. Cleavage resulted in the appearance of two distinct products at ∼18 kDa and 8 kDa.
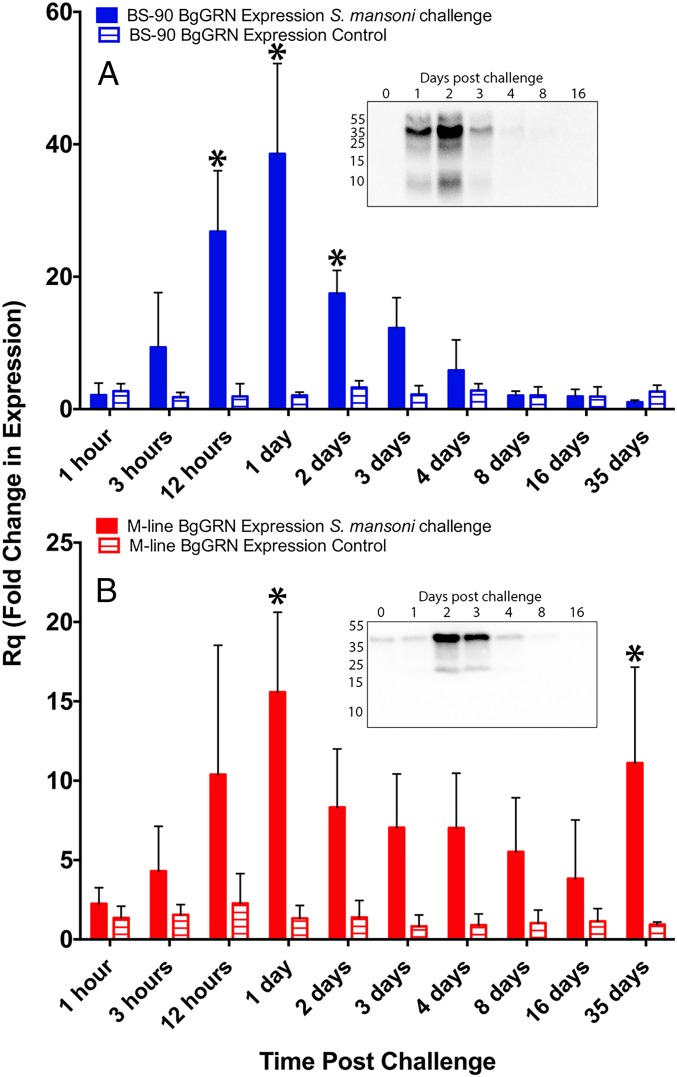
Fig. 2.
Quantitative RT-PCR (qRT-PCR) analysis of BgGRN expression in M-line and BS-90 strains of B. glabrata at critical time points during the entire intramolluscan development of S. mansoni. Fold change of BgGRN expression in BS-90 (A; blue bars; n = 3 for each time point) and M-line (B; red bars; n = 5 for each time point) strains challenged with S. mansoni (full bars) or, not-challenged control (hatched bars) following normalization to a time 0 control group (n = 5 snails) and then to the endogenous control β-actin. Bars represent SE. Asterisks represent within-strain significant difference between S. mansoni-challenged and not-challenged snails at the specific time point (P < 0.05). (Insets) Western blot analyses of S. mansoni-challenged snail plasma at the indicated days post challenge.
Fig. S3.
Cleavage of BgGRN appears to take place intracellularly within hemocytes. Anti-BgGRN Western blot analysis of hemocytes isolated from four different pools (A–D) of five snails suggests that the ∼18-kDa and 8-kDa cleavage products of BgGRN are present within hemocytes.
Analysis of BgGRN Transcript and Protein Expression.
Quantitative RT-PCR analysis of BgGRN transcript at key time points during the intramolluscan development of S. mansoni.
The mean basal transcript abundance of BgGRN as measured by specific primed (Table S1) reactions in BS-90 strain B. glabrata compared with M-line strain was found to be very similar following normalization to β-actin. The mean ΔCτ BgGRN value in nonmanipulated BS-90 snails was 7.78 (SEM = 0.80; n = 5) compared with 7.46 (SEM = 0.11; n = 5) in M-line snails. No change in BgGRN expression was observed at 1 h post challenge (hpc) with S. mansoni in BS-90 or M-line snails (Fig. 2); however, after 1 h, each snail strain displayed different expression profiles. BgGRN transcript abundance increased as early as 3 hpc in BS-90 snails [fold change (Rq) = 9.3, SEM = 4.77], and, by 12 hpc, BgGRN abundance had increased significantly compared with control BS-90 snails (Rq = 26.8, SEM = 5.3). Highest BgGRN transcript levels appeared at 1 d post challenge (dpc; Rq = 38.5, SEM = 7.88), and then decreased consistently from 2 dpc until day 35 (Rq day 2 = 17.5, SEM = 2.01; Rq day 35 = 1.05, SEM = 0.17; Fig. 2A). Compared with M-line snails challenged at the same time, the BS-90 snail BgGRN transcript levels were significantly higher at 12 h and 1 dpc (P < 0.05). None of the BS-90 snails tested positive for S. mansoni GAPDH after 3 dpc, a result consistent with the BS-90 snails being resistant to infection with S. mansoni and clearing the infection within 4 d (17).
Table S1.
List of primers and siRNA oligos
| Gene target | Sequence |
| BgGRN siRNA-1 | 5ʹ-GCAGUAAUCAGAACAAUGUGUUATA-3ʹ |
| BgGRN siRNA-2 | 5ʹ-CUACAGAUGUGAUGUGGAUCAAGGT-3ʹ |
| BgGRN siRNA-3 | 5ʹ-CUGAAAGUGUCGUAUGUCCUGGAGG-3ʹ |
| BgGRN siRNA-4 | 5ʹ-GUAUCAGCAGGAACAUGUAACAAAG-3ʹ |
| GFP siRNA-1 | 5ʹ-CCAUCAUCUUUGAAGAAGGAACAAUCUUCUUCAAAG-3ʹ |
| GFP siRNA-2 | 5ʹ-AGGUAAUAAUACAGGACCCGGUGAUGGUCCUGUAUU-3ʹ |
| GFP siRNA-3 | 5ʹ-AUGUUGUUACUAAUGUAGCCUUGACCUACAUUAGUA-3ʹ |
| BgGRN qRT-PCR | Forward: 5ʹ- TGAAAGTGTCGTATGTCCTGG-3ʹ |
| Reverse: 5ʹ-CATCCATAGTCCCCAGATTGC-3ʹ | |
| BgActin qRT-PCR | Forward: 5ʹ-GCT TCC ACC TCT TCA TCT CTT G-3ʹ |
| Reverse: 5ʹ-GAA CGT AGC TTC TGG ACA TCT G-3ʹ | |
| SmGAPDH | Forward: 5ʹ-TCG TTG AGT CTA CTG GAG TCT TTA CG-3ʹ |
| Reverse: 5ʹ-AAT ATG ATC CTG AGC TTT ATC AAT GG-3ʹ | |
| rBgGRN recombinant expression | Forward: 5ʹ-C ACC AGG AGG ATA TTA CAG ATG TCG TGC CTC AAG ATT GTG-3ʹ |
| Reverse: 5ʹ-GC GTA GAA CTT TGT TTG AGA ATG-3ʹ |
BgGRN also increased in abundance following challenge of M-line B. glabrata with S. mansoni (Fig. 2B). Increases were observed at 3 hpc (Rq = 4.3, SEM = 1.26), peaking at 1 dpc (Rq 1 day = 15.58, SEM = 2.25). Although BgGRN transcript levels declined after 1 dpc, relative expression compared with BS-90 snails remained consistently high, with a second peak in expression at day 35 (Rq day 2 = 8.32, SEM = 1.65; Rq day 35 = 11.11, SEM = 2.70). BgGRN transcript abundance was significantly higher than controls at 1 and 35 dpc (P < 0.05). A total of 96% of the experimental M-line snails were positive for S. mansoni GAPDH, indicating successful infections in these snails. Plasma BgGRN lagged behind transcript expression, with peak expression at 2 dpc in BS-90 and M-line snails but with earlier appearance and higher abundance in the former (Fig. 2, Inset).
Plasma BgGRN in B. glabrata before and after siRNA-mediated knockdown.
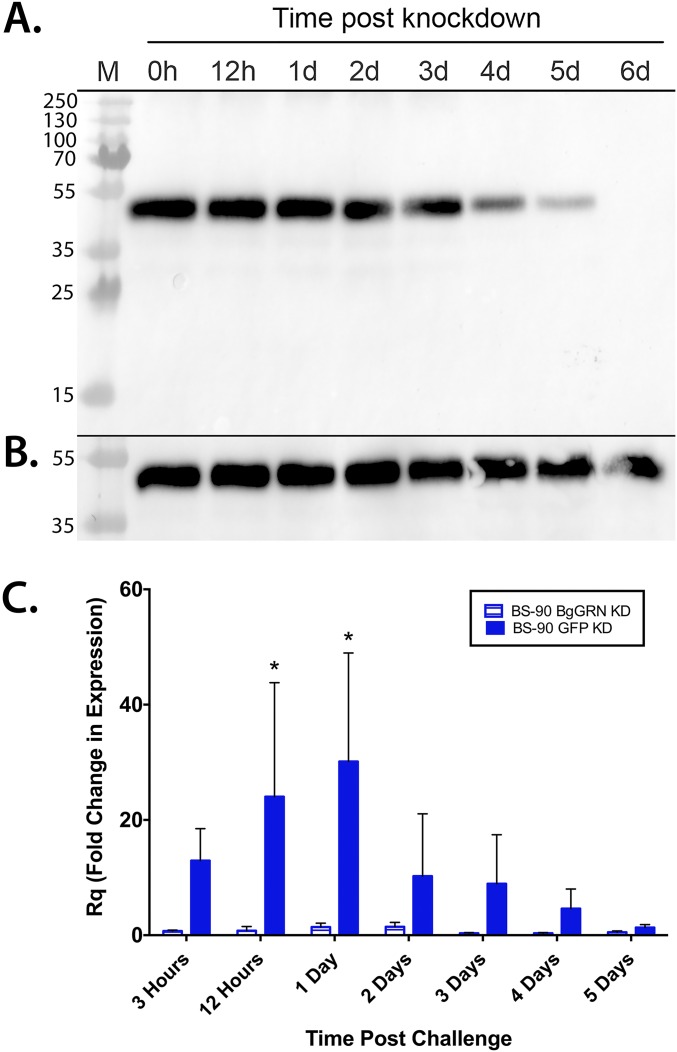
In both snail strains, injection of BgGRN-specific siRNA oligos (Table S1) effectively knocked down BgGRN expression, thereby reducing endogenous BgGRN (Fig. S4). Knockdown impact on plasma BgGRN was noticeable by 2 d post injection, with almost complete loss of detectable BgGRN by 5 d post injection. Knockdown of BgGRN 4 d before challenge with S. mansoni resulted in an almost complete abrogation of any challenge-mediated induction of BgGRN expression in BS-90 snails compared with GFP-knockdown controls (Fig. S4).
Fig. S4.
Kinetics of siRNA-mediated knockdown of BgGRN in vivo. (A) Western blot assessment of B. glabrata progranulin abundance in BS-90 B. glabrata plasma following injection of BgGRN-specific siRNA oligos demonstrates detectable reductions in circulating BgGRN by 3 d post injection and a complete loss of detectable BgGRN by day 6. Knockdown efficacy was compared with control BS-90 snails injected with siRNA oligos specific for GFP (B). The blot shown is representative of five independent trials conducted. (C) qRT-PCR assessment of BgGRN transcript abundance in BS-90 B. glabrata injected with siRNA oligos specific for BgGRN (hatched bars; n = 5 for each time point) or GFP (filled bars; n = 5 for each time point) 4 d before challenge with S. mansoni. Both BgGRN and GFP knockdown samples were normalized to a time 0 control group (n = 5) and to the endogenous control β-actin. Bars represent SE. Asterisk indicates a significant difference between BgGRN knockdown and GFP knockdown group means at the indicated time point.
BgGRN Induces Proliferation of Hemocytes and B. glabrata Embryonic Cells in Vivo and in Vitro via ERK Pathway Signaling.
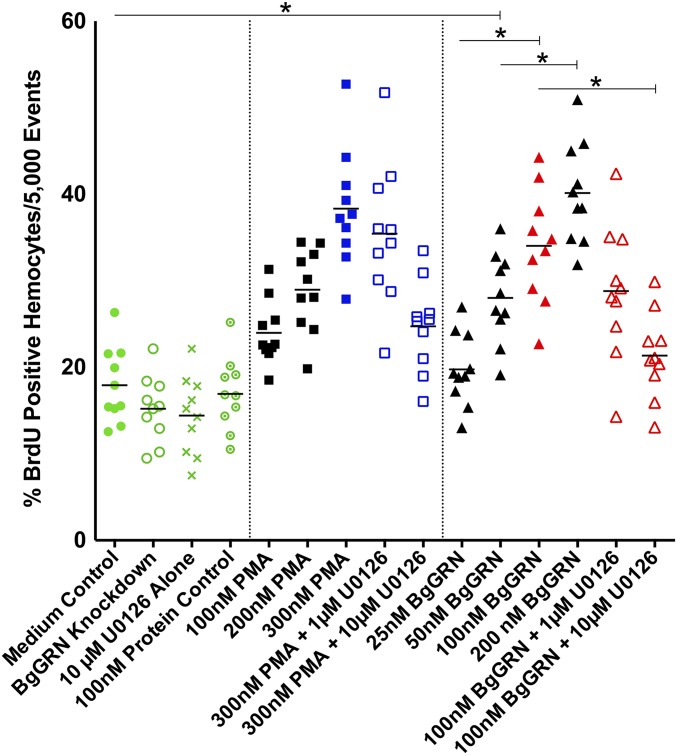
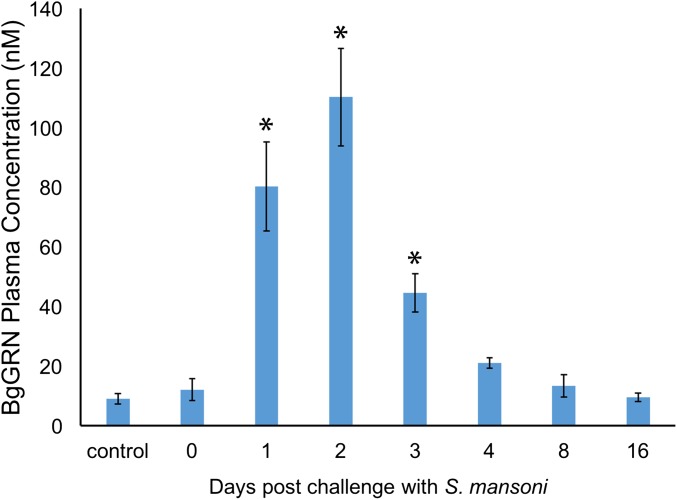
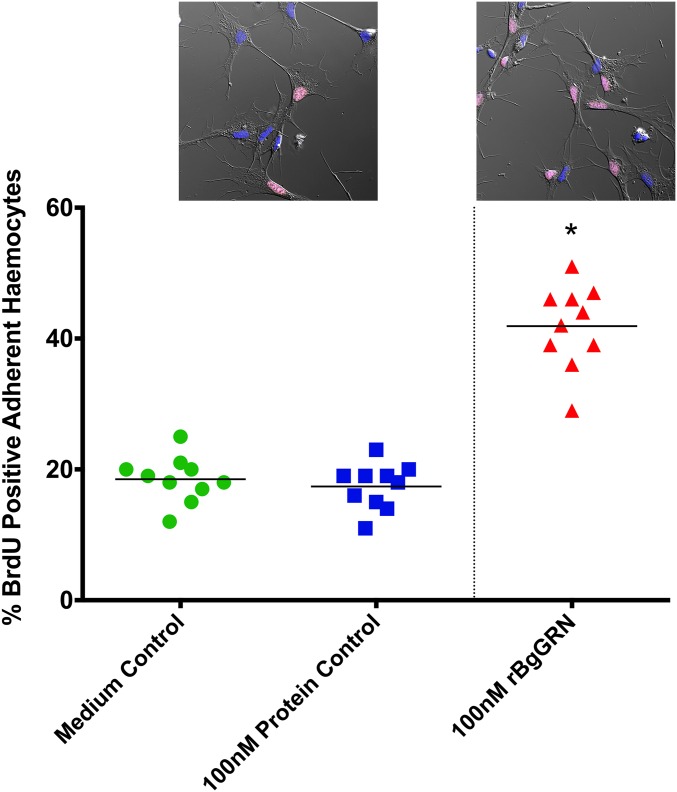
Coinjection of BrdU with rBgGRN into BS-90 snails resulted in an increased percentage of circulating hemocytes positive for BrdU (BrdU+) after 48 h compared with vehicle controls of medium alone (Fig. 3). The mean baseline percentage of BrdU+ circulating hemocytes in BS-90 snails was found to be 17.9% (SEM = 1.4%), compared with 19.8% (SEM = 1.3%), 28% (SEM = 1.6%), 34% (SEM = 2.1%), and 40.1% (SEM = 1.8%) BrdU+ hemocytes following 48 h stimulation with 25, 50, 100, and 200 nM BgGRN, respectively. Natural levels of circulating BgGRN observed in snail plasma (12.1 ± 3.67 nM) increased as a result of S. mansoni challenge in BS-90 snails to a peak of 110.3 ± 16.3 nM at day 2 post challenge. Circulating BgGRN returned to baseline concentrations (13.4 ± 3.76 nM) by day 8 post challenge (Fig. S5). The percentage of cells that had incorporated BrdU following BgGRN injection was comparable to the positive control phorbol 12-myristate 13-acetate (PMA), in which 24% (SEM = 1.2%), 29% (SEM = 1.5%), and 38.3% (SEM = 2.1%) of circulating hemocytes were positive for BrdU after injection of 100, 200, and 300 nM PMA, respectively. Inhibition of the ERK signaling pathway by U0126 at a concentration of 10 µM abrogated BgGRN and PMA-induced proliferation (i.e., BrdU incorporation) in circulating hemocytes (Fig. 3).
Fig. 3.
BgGRN-induced proliferation of circulating BS-90 B. glabrata hemocytes in vivo. Measurement of the percent BrdU-positive BS-90 primary hemocytes (out of 5,000 total events; n = 10 independent trials for each treatment) following 48 h stimulation. Exposures to 25, 50, 100, and 200 nM rBgGRN were compared with 100, 200, and 300 nM PMA (positive controls) and negative controls (medium, knockdown, recombinant protein, and inhibitor-only controls). Increases in the percentage of BrdU-positive cells were rBgGRN dose-dependent, with an apparent plateau at 100 nM. Important comparisons of significance are highlighted and indicated by asterisks.
Fig. S5.
Circulating BgGRN in BS-90 snail plasma. Indirect ELISA was used to assess the concentration of BgGRN in BS-90 snail plasma at 0, 1, 2, 3, 4, 8, and 16 dpc with S. mansoni. Plasma from three different snails was used for independent assessments at each time point. All plasma samples were compared with a standard curve generated using known concentrations of rBgGRN. Bars represent SD. Asterisk represents significance compared with control (not challenged) snails.
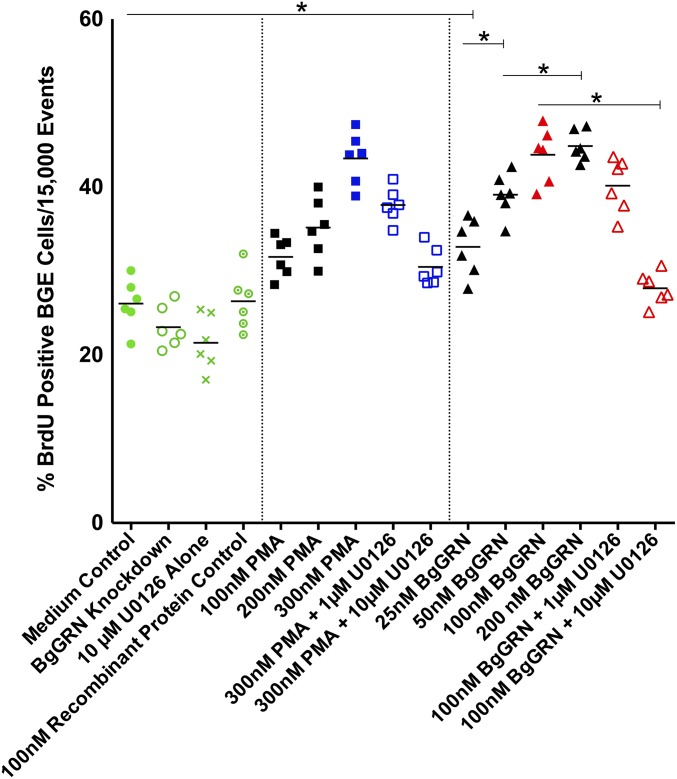
BgGRN elicited very similar results when applied to B. glabrata embryonic (Bge) cells (n = 6 for all treatments) for 48 h under in vitro culture conditions (Fig. S6). The mean percentage of BrdU+ Bge cells cultured in medium alone was 26% (SEM = 1.2%), compared with 32.8% (SEM = 1.4%), 39% (SEM = 1.1%), 43.8% (SEM = 1.3%), and 44.9% (SEM = 0.8%) after treatment with 25, 50, 100, and 200 nM BgGRN. BrdU was incorporated into the DNA of 31.7% (SEM = 1.0%), 35.1% (SEM = 1.5%), and 43.4% (SEM = 1.3%) of Bge cells following treatment with PMA at concentrations of 100, 200, and 300 nM. As with primary hemocytes, the proliferation induced by BgGRN and PMA was abrogated by application of 10 µM U0126.
Fig. S6.
BgGRN-induced proliferation of Bge cells in vitro. Measurement of the percent BrdU-positive Bge cells (out of 15,000 total events; n = 6 independent trials for each treatment) following 24-h stimulation. Exposures of 25, 50, 100, and 200 nM rBgGRN resulted in comparable increases in the percentage of BrdU-positive cells compared with 100, 200, and 300 nM PMA positive controls, and even 25 nM rBgGRN-treated cells displayed a significantly higher incorporation of BrdU compared with medium-only controls. Successive increases in the concentration of rBgGRN resulted in significant increases in the percentage of BrdU-positive cells up to 100 nM, after which BrdU incorporation appeared to plateau. The proliferation-inducing effect of rBgGRN was abrogated by addition of the ERK1/2 blocker U0126 at 10 µM, but not at 1 µM. Important comparisons of significance are highlighted and indicated by asterisks.
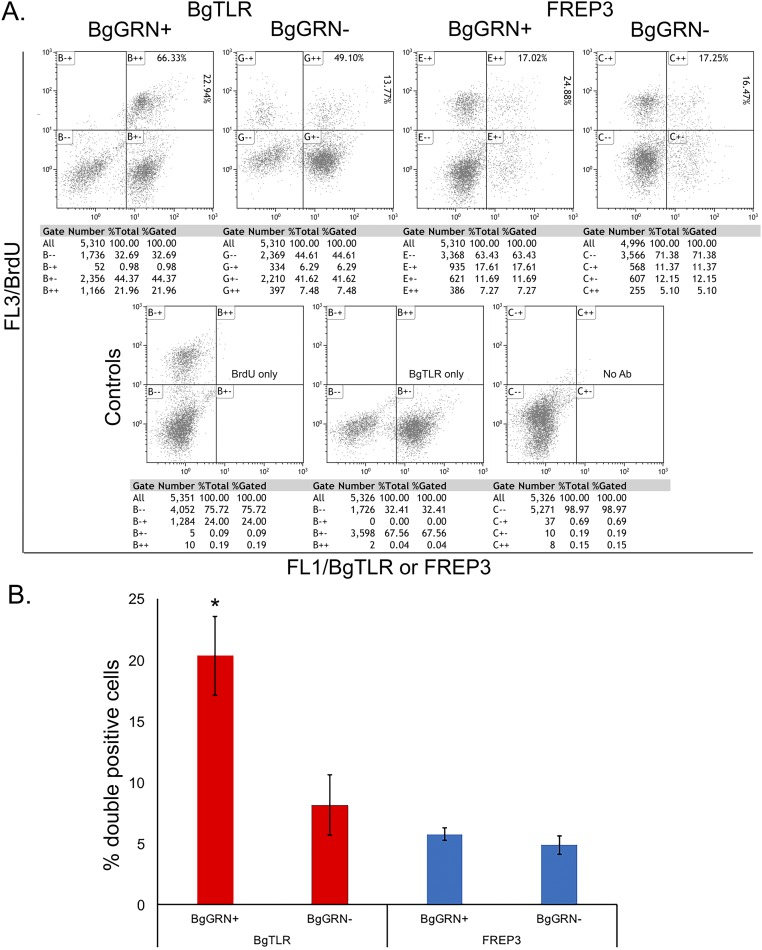
Knockdown of BgGRN in BS-90 and Bge cells 4 d before assessment of BrdU incorporation resulted in a nonsignificant, but observable, reduction of BrdU+ cells in both groups [14.7% (SEM = 0.9%)] and [23.3% (SEM = 1.0%), respectively] compared with controls (as detailed earlier; Fig. 3 and Fig. S6). Assessment of newly proliferated hemocytes using B. glabrata fibrinogen-related protein 3 (BgFREP3) (2) and Bg Toll-like receptor (BgTLR) (18)—proteins known to be associated with resistance to S. mansoni—indicates that BgGRN-induced hemocytes have a higher percentage of BgTLR-positive cells whereas the proportion of BgFREP3-positive cells was unaffected (Fig. S7).
Fig. S7.
BgGRN-induced hemocyte proliferation increases the proportion of BgTLR-positive cells. BS-90 snails were coinjected with BrdU and rBgGRN or BrdU and medium only as control. After 48 h, hemocytes were isolated, processed, and costained for BrdU and immune protein of interest. (A) Colabeling of BrdU and BgTLR or BgFREP3. Controls include single staining for BrdU, BgTLR, or no antibody. For the colabeled profiles, the top right box shows the proportion of double-positive adherent hemocytes that are BgTLR- or BgFREP3-positive, respectively (Top), and BrdU-positive (Right). (B) Plot of percentage of double-positive cells for the different treatments derived from A.
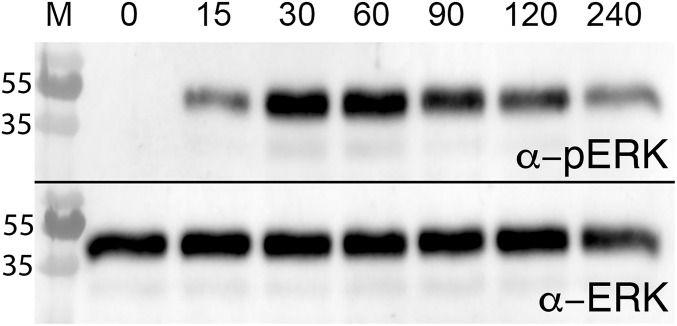
Western blot analysis confirmed that induction of cellular proliferation by BgGRN occurred, in part, via phosphorylation of the p44/42 MAPK in the ERK signaling pathway (Fig. S8). Injection of 100 nM rBgGRN into BS-90 snails resulted in phosphorylation of ERK1/2 at 15 min post injection, with maximal phosphorylated ERK compared with total ERK observed at 30 and 60 min post induction. At 90, 120, and 240 min post treatment, the amount of phosphorylated ERK declined, but remained detectable. Total ERK levels remained constant. As with other assessments of B. glabrata ERK1/2 analyses by Western blot, only a single band was easily visible in our experiments (19).
Fig. S8.
Proliferative effects of rBgGRN rely on signaling via ERK1/2 and the MAPK pathway. Western blot analysis of ERK1/2 was carried out following rBgGRN injection into BS-90 snail hemocoels. Using an anti-phosphorylated ERK1/2 antibody, ERK phosphorylation was assessed over 240 min at the indicated time points in minutes.
BgGRN Stimulates Generation of Adherent Hemocytes in Vivo.
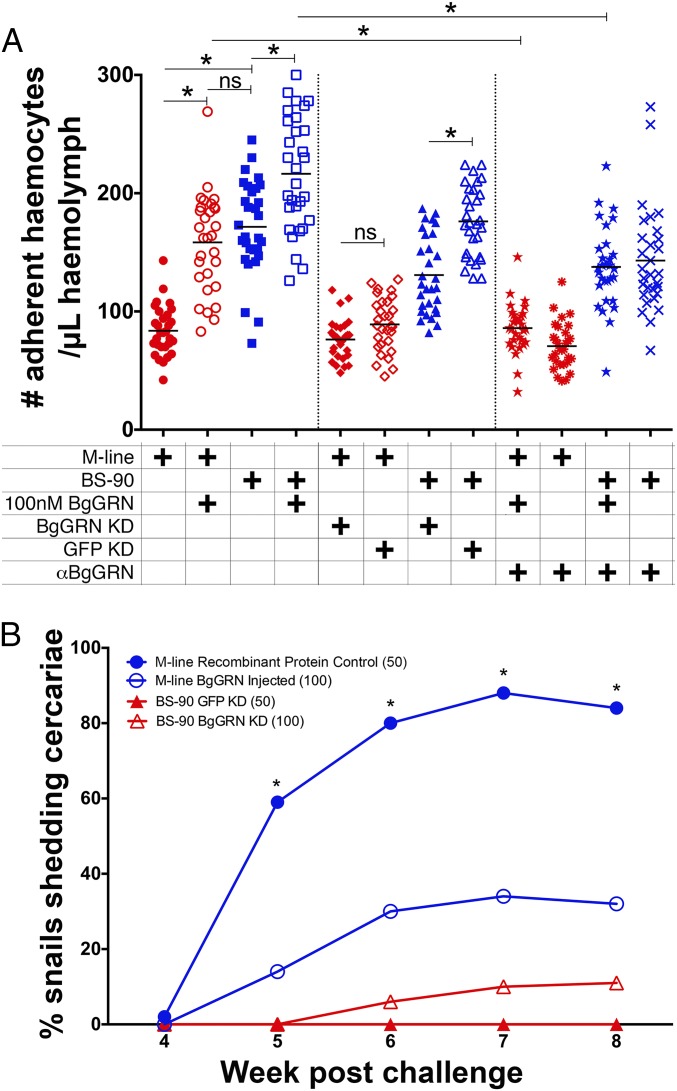
The baseline number of adherent (spread) hemocytes in BS-90 snails was more than double the mean number in M-line snails (171.5; SEM = 7.3; n = 30; and 83.7; SEM = 3.8; n = 30, respectively; Fig. 4A). Injection of rBgGRN to a final concentration of ∼100 nM in the snail hemolymph 48 h before hemocyte isolation significantly increased the number of adherent hemocytes isolated from both snails [BS-90 = 216.5; SEM = 8.9 (n = 30), M-line = 158.4; SEM = 7.5 (n = 30)]. These values reflect 126% and 189% increases in the number of circulating hemocytes in BS-90 and M-line snails, respectively, increasing the number of adherent hemocytes in the M-line snails such that there was no statistically significant difference between the baseline BS-90 and M-line + rBgGRN groups. We confirmed that the increase in adherent hemocytes was the direct result of BgGRN-induced proliferation by enumerating BrdU+ adherent hemocytes following BgGRN injection in snails (Fig. S9). Preincubation of rBgGRN with a polyclonal αBgGRN antibody blocked the increase in adherent cell numbers, confirming that the proliferative effect was the result of application of rBgGRN (Fig. 4A).
Fig. 4.
BgGRN induces the proliferation and development of adherent hemocytes to the point that susceptible M-line B. glabrata are able to defend against S. mansoni infection. (A) Effects of rBgGRN, knockdown of BgGRN, and anti-BgGRN polyclonal antibody on the number of adherent hemocytes. Snails were injected with 100 nM rBgGRN (induction) or rBgGRN preincubated with anti-BgGRN antibody (abrogation treatment), and, 48 h later, hemocytes were isolated and used for adherent hemocyte counts. Knockdown snails received siRNA oligos against BgGRN or GFP (control) 48 h before the treatments. Important comparisons of significance are highlighted and indicated by asterisks. (B) Injection of rBgGRN into M-line B. glabrata 4 d before challenge with S. mansoni significantly impacted S. mansoni infection success. Complementary knockdown of BgGRN in BS-90 B. glabrata also altered S. mansoni infection success rate, albeit not significantly.
Fig. S9.
Increase in adherent hemocytes are the direct result of BgGRN-stimulated hemocyte proliferation. BS-90 snails were coinjected with BrdU and rBgGRN or BrdU and medium only as control. After 48 h, hemocytes were isolated and placed directly onto microscope slides to adhere for 2 h in a humidified chamber. Following incubation, hemocytes were fixed, permeabilized, and stained with anti-BrdU mouse monoclonal antibody. BrdU-positive hemocytes were counted from among 100 adherent hemocytes from random fields of view (Insets), and percentages are expressed in graphical form.
Knockdown of BgGRN significantly reduced the number of adherent hemocytes in BS-90 [130.8 (SEM = 6.0)], but not in M-line [76.3 (SEM = 3.1)], snails compared with GFP-specific knockdown controls (BS-90 = 176.2; SEM = 5.5; M-line = 89.1; SEM = 4.1; Fig. 4A). No significant changes in adherent hemocyte numbers isolated from BS-90 or M-line snails were measured following treatment with 100 nM recombinant protein control (BgTemptin produced in same expression system) or a polyclonal mouse-derived isotype control (αHSP70).
Prior Stimulation of Hemocyte Proliferation by BgGRN in Naturally Susceptible B. glabrata Snails Significantly Protects Against S. mansoni Infection.
Significantly fewer M-line snails injected with rBgGRN, before exposure to S. mansoni, developed patent infections compared with snails injected with a recombinant protein control (Fig. 4B). This trend was observed throughout the course of the intramolluscan infection. The percentages of the experimental snail group (n = 100) that shed S. mansoni cercariae were 0%, 14%, 30%, 34%, and 32% at weeks 4, 5, 6, 7, and 8, respectively. In contrast, 2%, 59%, 80%, 88%, and 84% of the control snails (n = 50) shed cercariae at the same time post challenge. The percentage of infected snails was significantly different between rBgGRN-injected M-line snails and those injected with the recombinant protein control from week 5 post challenge onward (Fig. 4B). Sporocysts were not found in any of the M-line snails that did not shed cercariae. The knockdown of BgGRN in BS-90 snails had a small impact on S. mansoni challenge outcome: 6%, 10%, and 11% shed S. mansoni cercariae on weeks 6, 7, and 8, respectively (n = 100), whereas none of the GFP-knockdown controls shed cercariae at any point (n = 50; Fig. 4B).
Discussion
Encapsulation of schistosome sporocysts by snail hemocytes is critical for the elimination of the parasite (5, 10). This highlights the importance of maintaining a sufficient population of readily mobilized hemocytes within a snail at all times, and also the ability to drive production of new cells rapidly in response to challenge. The results of this study demonstrate that BgGRN participates in the hematopoietic events that control the production of the circulating hemocyte population. Introduction of BgGRN into B. glabrata snails that are naturally susceptible to S. mansoni infection resulted in a 54% reduction in the number of snails successfully infected by week 7 post challenge. That sporocysts were not observed upon dissection of these refractory M-line snails implies that the snails had successfully cleared any parasites. Underpinning this impressive change in infection outcome is a significant increase in the generation of circulating adherent hemocytes, which is the hemocyte subset that participates in the encapsulation of S. mansoni sporocysts (10). Adherent hemocytes are also highly phagocytic (2) and are responsible for the production of reactive and nitrogen intermediates (11) and soluble immune factors that are important for successful clearance of digenetic trematodes (1, 2). This cell population nearly doubled following administration of rBgGRN.
Whether the snails in this study defended against infection by S. mansoni because they had higher circulating cell numbers, or because those additional cells were immunologically distinct and conveyed new immune functionality, is a central question. To address it, we used two known immunological markers of snail resistance to S. mansoni infection: BgFREP3 (3) and BgTLR (18). That BgTLR was found to be proportionally higher on BrdU-positive cells induced by BgGRN, and BgFREP3 was not, suggests that the hemocyte population induced by BgGRN may be more geared toward the cellular immune response than the humoral. Increase in the BgGRN-induced adherent hemocyte numbers in the susceptible M-line strain of B. glabrata paralleled the mean number of hemocytes present in the naturally refractory BS-90 strain before stimulation. These findings support the hypothesis that B. glabrata resistance to S. mansoni infection is influenced by the hemocyte numbers and repertoire of the snail (5).
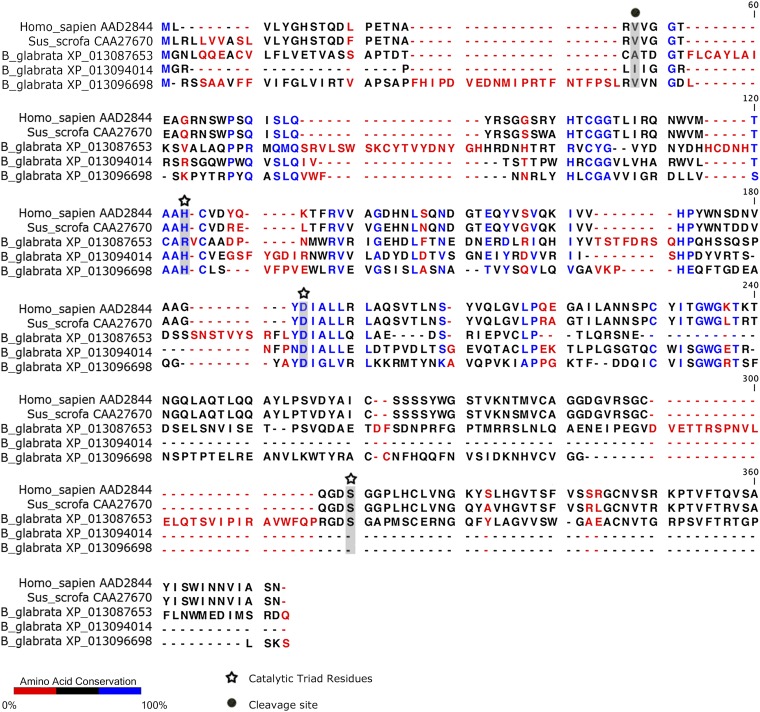
BgGRN possesses five predicted elastase cleavage sites that are each located immediately following one of the four 12-cysteine granulin domains. Similar to observations that have been made in studies focusing on human PGRN (16), incubation of rBgGRN with elastase results in the production of ∼8-kDa granulin fragments that would be predicted should all cleavage sites be processed; at least one intermediate product is consistently present in BgGRN elastase digests that resides at ∼18 kDa, similar in size to the ∼15-kDa intermediate fragment observed when human PGRN is digested with elastase (16). As porcine elastase was used to cleave rBgGRN, it is possible that differences between porcine elastase and the elastase of B. glabrata underpin the cleavage patterns observed. B. glabrata elastases were found to possess catalytic sites known to be active in human and porcine elastases (Fig. S10), indicating that they do possess the canonical motifs relevant for traditional elastase function (20). However, two of the three B. glabrata elastase variants found had only incomplete sequences available, and, consequently, the final catalytic residue could not be compared.
Fig. S10.
Amino acid alignment of human, porcine, and B. glabrata elastases. Analysis of the predicted amino acid sequence of B. glabrata elastase suggests that it possesses all relevant catalytic sites known to be active in human and porcine elastases. Only incomplete sequences were available for two of the three B. glabrata elastase variants, so the final catalytic residue could not be compared. Homo sapiens elastase (AAD2844.1), Sus scrofa elastase (CAA27670), and the three B. glabrata predicted elastase proteins (XP_013096698, XP_13094014, and XP_013087653) were aligned in CLC Genomics workbench using the alignment tool with a gap open cost of 10, a gap extension cost of 1.0, and an end gap cost of “as any other” using the very accurate (slow) option. The active sites were identified using the NCBI conserved domains database.
Native BgGRN was detectable in the plasma of unexposed B. glabrata only in its progranulin form; the ∼18-kDa cleavage product was observed only in plasma (of both M-line and BS-90 snails) following S. mansoni challenge, and the ∼8-kDa product was seen only in BS-90 plasma following challenge. In addition, the intermediate ∼18-kDa and smaller ∼8-kDa fragments were observed in the cytoplasmic fractions of B. glabrata hemocyte lysates. These observations suggest that activation or stimulation of the hemocytes (or other BgGRN producing cells) leads to the cleavage of BgGRN. These observations are supported by studies demonstrating that, in some cases, cleavage of PGRN by enzymes such as matrix metalloproteinase-12 occurs within the cytoplasm (21). Whether the cytoplasmic BgGRN and observed fragments differ functionally from extracellular BgGRN remains unknown. The source of the cytoplasmic BgGRN, which could derive from extracellular BgGRN that enters hemocytes or posttranslational processing of BgGRN before secretion, remains to be determined.
Although the hemocyte receptor mediating the proliferative effects of BgGRN observed in this study is unknown, there are a number of candidate receptors that warrant investigation. Even within the mammalian PGRN literature, there remains uncertainty regarding a definitive GRN receptor. Sortilin (22) and TNF receptors (23) have been implicated in recognition of PGRN, although recent evidence contradicts these initial reports (24). TLR9 activation has also been found to be reliant on PGRN as a cofactor (25). At present, we know very little about whether homologs of these molecules exist in B. glabrata. Analysis of the B. glabrata genome highlights the presence of genes that possess high predicted amino acid identity to the canonical domains that characterize TNF receptors and sortilin. However, whether these genes are expressed, or encode for proteins that function in a similar manner to known mammalian molecules, has not yet been investigated to our knowledge. Of the possible candidate molecules that may interact with BgGRN, TLRs are the best characterized in B. glabrata. A number of leucine-rich repeat domain-containing transcripts have been identified in the B. glabrata genome, and at least 13 of these appear to be bona fide TLRs possessing a toll-interleukin-1 domain. Although little is known about the functions of these TLRs, the fact that B. glabrata possesses many of the downstream components associated with TLR signaling in other organisms (26), and that at least one TLR in B. glabrata is highly abundant on BgGRN-induced adherent hemocytes, is indicative that they likely retain their role as pattern-recognition receptors in B. glabrata and are important participants in the immune response.
Although the receptor for BgGRN may remain unknown, it appears that the effects of BgGRN are mediated by signaling via the MAPK pathway and ERK1/2. The signaling pathway used by PGRN appears to be a highly conserved aspect of its biology (15). This is noteworthy, as it has been demonstrated that S. mansoni is able to interfere with the phosphorylation of ERK1/2 in susceptible snails, but not those that are phenotypically resistant (19). The kinetics of ERK1/2 phosphorylation following rBgGRN stimulation of hemocytes has also been observed by using human recombinant PGRN (27); however, phosphorylated ERK1/2 is detectable for a longer time (4 h vs. ∼2 h).
The 12-cysteine motif that characterizes granulins defines the protein and confers its conserved secondary and tertiary structure. It may also be responsible for certain functions (28). Investigations in carp identified three isoforms of carp GRN (29) that were later associated with mononuclear phagocytes (30). Functional studies of the carp granulin homolog in the goldfish demonstrated that this teleost granulin shares hematopoietic functions with its mammalian counterparts, inducing proliferation of myeloid cells in vitro (31). Recent studies focusing on a PGRN produced by the flatworm parasite Opisthorchis viverrini demonstrate OvGRN can induce proliferation in fibroblasts of the parasite’s human host and fibroblast cell lines, thereby providing a plausible mechanism for the high rates of liver cancer in individuals infected by O. viverrini (32, 33). The OvGRN studies provide plausibility for a functional role of parasite GRNs on certain host tissues or, alternatively, host GRNs on certain parasite tissues, and highlight the functional and structural conservation of granulins.
The variety of biological functions that PGRN is capable of is impressive, and it is likely that BgGRN may also participate in wound healing, immunological defense, and neuronal development in B. glabrata. Despite its pleiotropic function, we have demonstrated that BgGRN is an important growth factor in B. glabrata, driving hemocyte proliferation and development of an adherent hemocyte subset that is central to the defense of the snail against S. mansoni. Although the list of factors and genes that influence compatibility between B. glabrata and S. mansoni continues to grow, we demonstrate in this study that hematopoietic events regulated by endogenous factors such as granulin can significantly impact the outcome of S. mansoni infection, and that not all hemocytes are equal when considering their role in the immune response.
Materials and Methods
Additional description of materials and methods is provided in SI Materials and Methods.
Snail and Bge Cell Culture and Maintenance.
The M-line and BS-90 strains of the snail B. glabrata are susceptible and refractory to infection with S. mansoni, respectively. Both snail populations are maintained in colonies at the University of Alberta as previously described (34). The Bge cell line (35) (American Type Culture Collection NR-21959) was maintained in Bge cell medium by using established protocols (36).
Anti-BgGRN Polyclonal Antibody Generation and Validation.
A mouse anti-BgGRN polyclonal antibody was generated against the recombinant BgGRN (GenScript). The antibody was affinity purified using a Protein G affinity column (no. 17–0404-01; GE Healthcare) and then further purified against recombinant BgGRN. It was effective for Western blot detection of native BgGRN at concentrations of 1:5,000, and was found to block the proliferation-inducing effects of rBgGRN at concentrations of 1:250. All protocols involving animals were carried out in accordance with the Canadian Council on Animal Care Guidelines and Policies with approval from the Animal Care and Use Committee (Biosciences) for the University of Alberta AUP00000057.
SI Materials and Methods
Schistosoma mansoni Lifecycle and Infection Studies.
The lifecycle of PR-1 strain Schistosoma mansoni is maintained at the University of Alberta as described previously (37). Studies involving S. mansoni challenge consisted of age matched Biomphalaria glabrata snails (∼8-mm shell diameter) that were challenged with five S. mansoni miracidia over a 24-h period in individual wells of 12-well plates containing artificial spring water (ASW) (38). Following a 24-h challenge, snails were transferred into tanks of ASW for the remainder of the study.
Annotation of Predicted Sites and Domains.
Using the GRN-like amino acid sequences for B. glabrata (BgGRN; ADX33287.1), S. mansoni (CCD75905.1), Schistosoma hematobium (KGB36375.1), and Schistosoma japonicum (CAX73857.1), as acquired from GenBank [National Center for Biotechnology Information (NCBI)], we used three different programs/methods [SignalP 4.1 (39), TargetP 1.1 (40, 41), and Phobius (42)] to predict and annotate the signal peptide and cleavage sites, as recommended by UniProt (www.uniprot.org/help/signal). At least two of the methods had to agree to convince us that the prediction was correct. In our case, all three methods used agreed for every sequence. To annotate the granulin domains for each of the sequences, we used the Pfam (43) sequence search tool.
Sequence Alignments.
Amino acid sequences for B. glabrata, S. mansoni, S. hematobium, and S. japonicum, as previously listed, were imported into the CLC Genomics Workbench 7.5.1 (www.clcbio.com). The sequences were aligned by using the CLC proprietary alignment algorithm with a gap open cost of 10, gap extension cost of 1, end gap cost “as any other,” and using the very accurate (slow) setting. From there, the alignment was visually inspected for obvious inaccuracies, modified if need be, and then annotated using the same software. To determine the amino acid conservation among granulin domains within the B. glabrata sequence, each of the four domains were extracted as separate sequences and aligned using the same methods as described in the previous paragraph.
Secondary Structure Predictions and Visualization.
The BgGRN amino acid sequence was submitted to the Robetta Server (robetta.bakerlab.org) (44) for homology modeling. The Ginzu domain prediction resulted in five domains, all using the reference parent Human Granulin A [Protein Data Bank (PDB) ID code 2JYE]. We modeled the first predicted domain (confidence = 0.45), which resulted in five predicted structures. The first structure was used for comparison purposes to human granulin A (PDB ID code 2JYE) and a partial carp granulin (PDB ID code 1QGM), with PDB files extracted from the Research Collaboratory for Structural Bioinformatics Protein Databank (www.rcsb.org) (45). The PDB structural information for all three proteins was loaded into PyMOL (PyMOL Molecular Graphics System, version 1.5.0.4; Schrödinger) for visualization and superimposition.
Identification of BgGRN.
B. glabrata pro-GRN (GenBank accession no. HQ661843) was identified as part of a B. glabrata protein screen in which hemolymph protein abundance was compared between M-line and BS-90 strain snails. Peptide fragments possessing the canonical cysteine-repeat pattern of GRN proteins were identified in high abundance in BS-90 snail hemolymph following S. mansoni challenge. The peptide fragments were used to identify the full pro-BgGRN transcript sequence from the unannotated B. glabrata genome. Expression of BgGRN was assessed using qRT-PCR, and sequence analysis of a full-length RT-PCR amplicon of BgGRN confirmed that the sequence was identical to the transcript predicted from the genome.
qRT-PCR Assessment of BgGRN Transcript Expression.
Assessment of the transcriptional expression of BgGRN over the complete course of S. mansoni intramolluscan development in B. glabrata (M-line and BS-90 strains) was assessed using semiquantitative PCR. The expression of BgGRN was assessed at 0, 1, 3, and 12 h and 1, 2, 3, 4, 8, 16, and 35 d post S. mansoni challenge. Snails were exposed to five S. mansoni miracidia at time 0, and, at each experimental time point, the mRNA of five (M-line) and three (BS-90) snails from the experimental and control groups was collected. Total RNA was extracted from whole snails using the TRIzol reagent with PureLink RNA mini kit according to the manufacturer’s instructions (Life Technologies). RNA concentration was determined using the NanoVue spectrophotometer, and 1 µg was used for first-strand cDNA synthesis using the qScript cDNA synthesis kit (Quanta Biosciences). The cDNA was diluted fivefold, and 5 µL was used as template in qRT-PCR using primers specific for BgGRN and B. glabrata β-actin (Table S1), and the SYBR Green detection system (PerfeCTa SYBR Green FastMix; Quanta Biosciences). Snails were confirmed to be S. mansoni-positive or -negative using a qRT-PCR assay targeting the parasite GAPDH gene (46). All qRT-PCRs were performed on the ABI 7500 Fast Real-Time PCR system (Applied Biosystems) using the following thermocycling conditions: initial hold at 95 °C for 10 min, followed by 40 cycles of 95 °C for 15 s and 60 °C for 1 min, with data collection every cycle. Specificity for the qRT-PCR amplicons was confirmed by continuous melt curve analysis. A ΔΔCτ analysis was performed comparing BgGRN Cτ values to those of the endogenous control β-actin, the transcript expression of which we confirmed to be stable and unaffected by parasite exposure. The ΔCτ values were then normalized to the time 0 samples representing each snail strain, and then converted into Rq (i.e., fold change) values.
Recombinant BgGRN Synthesis and Purification.
Recombinant BgGRN was generated by using the Gateway cloning system according to the manufacturer’s instructions (Life Technologies). Briefly, the coding region was amplified with Phusion high-fidelity DNA polymerase and cloned into the pENTR/d-TOPO vector. Plasmid DNA from this entry clone was isolated and cloned into pIB/V5-His-DEST vector in a Clonase recombination reaction to produce the expression clone. The identity of BgGRN was sequence-verified after cloning in the entry and expression vectors. Plasmid DNA was extracted from the expression vector and transfected into the Sf9 insect cell line using Cellfectin reagent (Life Technologies). Expression of BgGRN was confirmed by Western blot using antibodies against the V5 and histidine tags on the recombinant protein.
Sf9 cells stably expressing BgGRN were selected and maintained in a medium containing blasticidin. Cultures were scaled up in 1,000 mL Corning roller bottles, and the medium containing secreted BgGRN was filtered through 0.45-µm filters. BgGRN protein was purified from the culture medium by using HisPur Ni-NTA magnetic beads (Thermo Scientific) according to the manufacturer’s instructions. Purified BgGRN was dialyzed against PBS buffer twice for 2 h each and then once overnight using Slide-A-Lyzer dialysis kit (Thermo Scientific).
Anti-BgGRN Polyclonal Antibody Generation and Validation.
A mouse anti-BgGRN polyclonal antibody was generated against the recombinant BgGRN (GenScript). The antibody was affinity-purified using a Protein G affinity column (no. 17–0404-01; GE Healthcare), and then further purified against recombinant BgGRN. This antibody was effective for Western blot detection of native BgGRN at concentrations of 1:5,000, and was found to block the proliferation-inducing effects of rBgGRN at 1:250.
Western Blot Analysis of BgGRN.
Western blot detection of BgGRN was done using 250 ng protein suspended in Laemmli protein loading buffer. Samples were heated at 95 °C for 10 min, then loaded on 10% (vol/vol) SDS/PAGE gels and run on the Mini PROTEAN Tetra system (Bio-Rad) at 200 V and 180 mA. Samples were then blotted for 2.5 h onto 0.45 µm supported nitrocellulose membranes (Bio-Rad). Blocking was done for 1 h at room temperature in 5% (wt/vol) skimmed milk prepared in Tris-buffered saline (TBS) solution plus 0.1% Tween-20 (TBS-T buffer) before staining for 1 h in anti-V5 mouse IgG (recombinant BgGRN) or mouse anti-BgGRN IgG (native BgGRN) primary antibody at a concentration of 1:5,000 in blocking buffer. Membranes were washed in TBS-T buffer for 10 min, then twice for 5 min each and once in TBS solution for 5 min. Membranes were then stained for 1 h in HRP-conjugated rabbit anti-mouse IgG antibody diluted 1:5,000 in blocking buffer followed by a wash step as described earlier. Detection was accomplished by incubating the membranes in SuperSignal West Dura Extended Duration substrate (Thermo Scientific). Chemiluminescent signals were acquired on the ImageQuant LAS 4000 machine (GE Healthcare).
Indirect ELISA Measurement of Plasma BgGRN.
Cell-free plasma was isolated from BS-90 B. glabrata at 0, 1, 2, 3, 4, 8, and 16 dpc with S. mansoni. Hemolymph from three snails for each time point was collected by head-foot retraction and immediately placed in 1.5-mL tubes on ice. Samples were centrifuged at 1,000 × g for 10 min and the cell-free fraction was collected. Plasma was diluted 1:10, 1:50, or 1:250 with 0.2 M NaHCO3 (pH 9.4), and then 100 µL of sample was pipetted into a 96-well polystyrene plate. Plates were incubated at 4 °C for 24 h and then washed three times for 5 min with wash buffer (25 mM Tris, 0.15 M NaCl, 0.05% Tween-20, pH 7.2). Then, plates were blocked with 2% (wt/vol) BSA in wash buffer for 3 h at room temperature. Following blocking, the buffer was replaced with 100 µL of blocking buffer containing anti-BgGRN antibody (1:500), and incubated at room temperature overnight. Plates were then washed three times for 5 min with wash buffer and then incubated with blocking buffer containing a biotinylated secondary antibody (1:250). Plates were then covered with aluminum foil and incubated at room temperature for 1 h, washed six times for 5 min, and then incubated in the substrate solution containing streptavidin conjugated to DyLight 649 (Thermo Scientific). The reaction was allowed to proceed for 15 min and was then read using a 96-well plate reader (Molecular Devices). All plasma samples were compared with a standard curve generated using a serial dilution series of rBgGRN ranging from 0.001 ng/mL to 10 µg/mL.
Hemocyte/Bge Cell Lysis and Detection of ERK1/2 and Intracellular BgGRN.
Cultured Bge cells and primary hemocytes isolated from pools of five B. glabrata snails were centrifuged once at 1,000 × g for 10 min. Cells were resuspended in 1× sterile snail saline buffer (47) and centrifuged at 1,000 × g for 5 min. Following one wash, cells were lysed using a cell lysis buffer containing 20 mM Tris⋅HCl (pH 7.5), 150 mM NaCl, 1 mM EDTA, 1 mM EGTA, 1% Triton X-100, 2.5 mM sodium pyrophosphate, 1 mM β-glycerophosphate, 30 mM NaF, and phosphatase inhibitor mixtures 2 and 3 (Sigma). Lysates were run on 12% (vol/vol) SDS gels at a concentration of 30 ng per lane. Samples were then blotted for 2.5 h onto 0.45-µm supported nitrocellulose membranes (Bio-Rad). Blocking was done for 4 h at room temperature in 5% (wt/vol) BSA (Sigma) prepared in TBS-T buffer before probing with the primary antibody [αphospho ERK1/2, αtotal ERK1/2 at 1:2,500 dilution (Cell Signaling Technology), or αBgGRN at 1:1,000 dilution]. Visualization of blots was accomplished as described earlier.
Elastase Cleavage of BgGRN.
rBgGRN (25 µg) was treated with 0.3 U/mL porcine elastase (Alfa Aeser) and used to assess whether the predicted elastase cleavage sites on BgGRN were accurate. Elastase incubations took place over 5, 15, and 30 min, after which samples were run on SDS/PAGE and then blotted for 2.5 h onto 0.45-µm supported nitrocellulose membranes (Bio-Rad). Blocking was done for 4 h at room temperature in 5% (wt/vol) BSA (Sigma) prepared in TBS-T buffer before probing with the αBgGRN at 1:1,000 dilution. Visualization of blots was accomplished as described earlier.
Flow-Cytometric BrdU Assay of Hemocyte and Bge Cell Proliferation.
Labeling of B. glabrata hemocytes in vivo with BrdU was accomplished by first making a BrdU stock solution. BrdU (Sigma) was diluted in 1× PBS to a final working concentration of 10 mg/mL This solution was then combined with experimental and control treatments and then injected through the shell into the snail hemocoel using a 27-gauge needle in a 10–20-µL volume that was adjusted depending on the snail shell size (8-mm shell diameter was injected with 10 µL, 16-mm shell diameter was injected with 20 µL). The resulting hole in the snail shell was covered with wax, and the snail was returned to a clean tank of ASW for 48 h before the next manipulation. Primary B. glabrata hemocytes were isolated for use in proliferation bioassays using the head-foot retraction method (48).
Labeling of Bge cells with BrdU in vitro was accomplished by diluting a 1-mM BrdU working solution to a final working concentration of 10 µM in Bge cell culture medium. Bge cells were seeded in six-well culture plates at a density of 1 × 106 cells per milliliter in 2-mL volumes and allowed to adhere for 4 h before treatment and addition of BrdU. The treatments (identified later) and BrdU were administered in the same medium, which replaced normal Bge cell medium following the 4-h pretreatment period. Labeling and treatment occurred over a 24-h period, after which the medium was replaced with a 0.05% Trypsin-EDTA solution (Life Technologies) for 10 min to suspend the adherent Bge cells.
Treatments for Bge cells and B. glabrata hemocytes in vivo consisted of four experimental concentrations of rBgGRN (25, 50, 100, and 200 nM). These treatments were compared with 100, 200, and 300 nM phorbol 12-myristate 13-acetate (PMA) (Sigma) as a proliferation control treatment and with 100 nM rBgGRN or 300 nM PMA combined with 1 µM or 10 µM of the MEK [MAPK (ERK 1/2)] inhibitor U0126 (Promega). These concentrations were exactly calculated for Bge cell assays and were estimated based on predicted hemolymph volume as indicated earlier for studies involving B. glabrata hemocytes.
For Bge cells and isolated primary hemocytes of B. glabrata, the isolated cells were centrifuged at 1,000 × g and resuspended in 1 mL of 1× PBS + 3.7% (vol/vol) formaldehyde. Following a 15-min incubation, cells were washed three times (2 min for each wash) in 1× PBS solution. Cells were then permeabilized using a solution of 0.1% Triton X-100 (Sigma) in 1× PBS solution for 20 min, after which cells were centrifuged and resuspended in 1 mL of 1 N HCl for 10 min, followed by resuspension and incubation for 10 min in phosphate/citric acid buffer, pH 7.4 (made by combining 182 mL of 0.2 M Na2HPO4 with 18 mL of 0.1 M citric acid). Cells were washed three times with Triton X-100 permeabilization buffer (2 min per wash). Cells were prepared for incubation with the anti-BrdU antibody by replacing the final wash of Triton X-100 buffer with a solution of 0.1% Triton X-100, 5% (vol/vol) FCS (Corning) in 1× PBS. Cells were incubated in this solution for 1 h before replacing it with the antibody solution containing 5 µL of the provided anti-BrdU mouse monoclonal antibody conjugated to Alexa Fluor 488 (Life Technologies). Cells were incubated overnight with the antibody solution, after which they were washed three times using 1× PBS (2 min per wash), and then analyzed by flow cytometry. In experiments involving colabeling, hemocytes were incubated with anti-BrdU at the same time with rabbit anti-BgTLR or anti-BgFREP3 antibodies and further stained after washes with a goat anti-rabbit IgG secondary antibody conjugated to Alexa Fluor 680 (Life Technologies). Flow-cytometric analysis of B. glabrata hemocytes and Bge cells was undertaken using a Beckman-Coulter Gallios flow cytometry platform. Size, granularity, and fluorescence were analyzed using the Kaluza software package (Beckman-Coulter). All of these wash and incubation steps took place at room temperature.
siRNA-Mediated Knockdown of BgGRN.
Juvenile snails (∼8-mm shell diameter) were injected with a mixture of 27-mer siRNA oligonucleotides (Integrated DNA Technologies) designed to specifically target four different regions of the BgGRN transcript. The oligonucleotide sequences were confirmed to be unique to BgGRN by comparison with the B. glabrata genome. The oligonucleotide mix was suspended in Xfect transfection reagent (Clontech) to enhance delivery, and 10–20 µL was injected directly into the snail hemocoel at an approximate final concentration of 6 nM, which was determined by estimating the volume of hemolymph within the snail. Control snails received siRNA oligo targeting GFP; these oligos have no known homologies to expressed transcripts in B. glabrata. Sequences for siRNA oligonucleotides are shown in Table S1.
siRNA-mediated knockdown of BgGRN was examined at the transcriptional and protein levels. For the knockdown of BgGRN transcript, RNA was extracted from five snails, converted into cDNA, and used for qRT-PCR as described earlier. Protein knockdown was determined by Western blot on cell-free plasma used directly and probed with a mouse anti-BgGRN polyclonal antibody as described earlier. All proteins from hemocytes and cell-free plasma were quantified with Qubit protein assay kit (Thermo Fisher Scientific) according to the manufacturer’s protocol.
Adherent Hemocyte Counts.
Primary B. glabrata hemocytes were isolated for use in proliferation bioassays using the head-foot retraction method (48). Exactly 10 µL of hemolymph was mixed with 10 µL of sterile Chernin balanced salt solution (CBSS) (49), and 10 µL of the 1:1 mixture was then placed directly onto microscope slides and incubated within a humidified chamber for 2 h at 25 °C. Following incubation, the slides were gently washed twice using sterile CBSS that was warmed to 25 °C. The adherent cells were enumerated, and the values adjusted to reflect the number of adherent (spread) hemocytes per microliter of hemolymph.
BgGRN Modulation of Infection Success.
Two studies were designed to assess the importance of BgGRN, and thus hemocyte number, on S. mansoni–B. glabrata compatibility. The first experiment used M-line B. glabrata snails that were injected through the shell into the hemocoel with 100 nM BgGRN (n = 100 snails) or 100 nM of a recombinant protein control (B. glabrata Temptin produced identically to rBgGRN; n = 50 snails). Holes in the shell resulting from the injection technique were filled over with wax. Injections took place 48 h before challenging each snail with five S. mansoni miracidia for 24 h. Following challenge, snails from each experimental group were placed within independent tanks filled with ASW at a snail density of 25 per tank. Infection success was measured by placing snails into 12-well plates and monitoring for the shedding of cercariae over a 6-h period. Cercarial shedding was assessed once per week at 4, 5, 6, 7, and 8 wk post challenge.
The second experiment used BS-90 strain of B. glabrata that were injected with siRNA oligos specific for the knockdown of BgGRN (n = 100 snails) or GFP as a control (n = 50 snails). For this study, injections took place 24 h before challenge with five S. mansoni miracidia. Challenge was allowed to continue for 24 h before snails were placed in tanks, as outlined earlier, at a density of 25 snails per tank. Cercarial shedding was assessed at 4, 5, 6, 7, and 8 wk post challenge, for a 6-h period in 12-well plates.
Statistical Analysis.
Statistical significance was determined using one-way ANOVA with Tukey’s post hoc tests performed using GraphPad Prism version 6.0f for Mac OS X (GraphPad; www.graphpad.com) unless otherwise indicated. Statistical significance threshold was set at P ≤ 0.05. For the qRT-PCR assay, all statistical comparisons were performed on mean ΔCτ values for each time point (i.e., Cτ values of BgGRN normalized to actin).
Acknowledgments
Schistosome-infected mice were provided by the NIAID Schistosomiasis Resource Center at the Biomedical Research Institute Rockville, MD, through NIH-NIAID Contract HHSN272201000005I for distribution through BEI Resources. This work was supported by the Natural Sciences and Engineering Council of Canada Grant NSERC 418540 (to P.C.H.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. T.Y. is a guest editor invited by the Editorial Board.
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. HQ661843).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1521239113/-/DCSupplemental.
References
- 1.Galinier R, et al. Biomphalysin, a new β pore-forming toxin involved in Biomphalaria glabrata immune defense against Schistosoma mansoni. PLoS Pathog. 2013;9(3):e1003216. doi: 10.1371/journal.ppat.1003216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hanington PC, et al. Role for a somatically diversified lectin in resistance of an invertebrate to parasite infection. Proc Natl Acad Sci USA. 2010;107(49):21087–21092. doi: 10.1073/pnas.1011242107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Moné Y, et al. A large repertoire of parasite epitopes matched by a large repertoire of host immune receptors in an invertebrate host/parasite model. PLoS Negl Trop Dis. 2010;4(9):e813. doi: 10.1371/journal.pntd.0000813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baeza Garcia A, et al. Involvement of the cytokine MIF in the snail host immune response to the parasite Schistosoma mansoni. PLoS Pathog. 2010;6(9):e1001115. doi: 10.1371/journal.ppat.1001115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Larson MK, Bender RC, Bayne CJ. Resistance of Biomphalaria glabrata 13-16-R1 snails to Schistosoma mansoni PR1 is a function of haemocyte abundance and constitutive levels of specific transcripts in haemocytes. Int J Parasitol. 2014;44(6):343–353. doi: 10.1016/j.ijpara.2013.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jeong KH, Lie KJ, Heyneman D. The ultrastructure of the amebocyte-producing organ in Biomphalaria glabrata. Dev Comp Immunol. 1983;7(2):217–228. doi: 10.1016/0145-305x(83)90003-4. [DOI] [PubMed] [Google Scholar]
- 7.Sullivan JT, Pikios SS, Alonzo AQ. Mitotic responses to extracts of miracidia and cercariae of Schistosoma mansoni in the amebocyte-producing organ of the snail intermediate host Biomphalaria glabrata. J Parasitol. 2004;90(1):92–96. doi: 10.1645/GE-3266. [DOI] [PubMed] [Google Scholar]
- 8.Noda S. Effects of excretory-secretory products of Echinostoma paraensei larvae on the hematopoietic organ of M-line Biomphalaria glabrata snails. J Parasitol. 1992;78(3):512–517. [PubMed] [Google Scholar]
- 9.Lie JK, Heyneman D, Jeong KH. Studies on resistance in snails. 4. Induction of ventricular capsules and changes in the amebocyte-producing organ during sensitization of Biomphalaria glabrata snails. J Parasitol. 1976;62(2):286–291. [PubMed] [Google Scholar]
- 10.LoVerde PT, Shoulberg N, Gherson J. Role of cellular and humoral components in the encapsulation response of Biomphalaria glabrata to Schistosoma mansoni sporocysts in vitro. Prog Clin Biol Res. 1984;157:17–29. [PubMed] [Google Scholar]
- 11.Humphries JE, Yoshino TP. Regulation of hydrogen peroxide release in circulating hemocytes of the planorbid snail Biomphalaria glabrata. Dev Comp Immunol. 2008;32(5):554–562. doi: 10.1016/j.dci.2007.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hanington PC, Lun CM, Adema CM, Loker ES. Time series analysis of the transcriptional responses of Biomphalaria glabrata throughout the course of intramolluscan development of Schistosoma mansoni and Echinostoma paraensei. Int J Parasitol. 2010;40(7):819–831. doi: 10.1016/j.ijpara.2009.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zanocco-Marani T, et al. Biological activities and signaling pathways of the granulin/epithelin precursor. Cancer Res. 1999;59(20):5331–5340. [PubMed] [Google Scholar]
- 14.He Z, Bateman A. Progranulin (granulin-epithelin precursor, PC-cell-derived growth factor, acrogranin) mediates tissue repair and tumorigenesis. J Mol Med (Berl) 2003;81(10):600–612. doi: 10.1007/s00109-003-0474-3. [DOI] [PubMed] [Google Scholar]
- 15.Ong CH, Bateman A. Progranulin (granulin-epithelin precursor, PC-cell derived growth factor, acrogranin) in proliferation and tumorigenesis. Histol Histopathol. 2003;18(4):1275–1288. doi: 10.14670/HH-18.1275. [DOI] [PubMed] [Google Scholar]
- 16.Zhu J, et al. Conversion of proepithelin to epithelins: Roles of SLPI and elastase in host defense and wound repair. Cell. 2002;111(6):867–878. doi: 10.1016/s0092-8674(02)01141-8. [DOI] [PubMed] [Google Scholar]
- 17.Sullivan JT, Spence JV, Nuñez JK. Killing of Schistosoma mansoni sporocysts in Biomphalaria glabrata implanted with amoebocyte-producing organ allografts from resistant snails. J Parasitol. 1995;81(5):829–833. [PubMed] [Google Scholar]
- 18.Pila EA, Tarrabain M, Kabore AL, Hanington PC. A novel Toll-like receptor (TLR) influences compatibility between the gastropod Biomphalaria glabrata, and the digenean trematode Schistosoma mansoni. PLoS Pathog. 2016;12(3):e1005513. doi: 10.1371/journal.ppat.1005513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zahoor Z, Davies AJ, Kirk RS, Rollinson D, Walker AJ. Disruption of ERK signalling in Biomphalaria glabrata defence cells by Schistosoma mansoni: Implications for parasite survival in the snail host. Dev Comp Immunol. 2008;32(12):1561–1571. doi: 10.1016/j.dci.2008.05.014. [DOI] [PubMed] [Google Scholar]
- 20.Korkmaz B, Horwitz MS, Jenne DE, Gauthier F. Neutrophil elastase, proteinase 3, and cathepsin G as therapeutic targets in human diseases. Pharmacol Rev. 2010;62(4):726–759. doi: 10.1124/pr.110.002733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Suh HS, Choi N, Tarassishin L, Lee SC. Regulation of progranulin expression in human microglia and proteolysis of progranulin by matrix metalloproteinase-12 (MMP-12) PLoS One. 2012;7(4):e35115. doi: 10.1371/journal.pone.0035115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zheng Y, Brady OA, Meng PS, Mao Y, Hu F. C-terminus of progranulin interacts with the beta-propeller region of sortilin to regulate progranulin trafficking. PLoS One. 2011;6(6):e21023. doi: 10.1371/journal.pone.0021023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jian J, et al. Progranulin directly binds to the CRD2 and CRD3 of TNFR extracellular domains. FEBS Lett. 2013;587(21):3428–3436. doi: 10.1016/j.febslet.2013.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen X, et al. Progranulin does not bind tumor necrosis factor (TNF) receptors and is not a direct regulator of TNF-dependent signaling or bioactivity in immune or neuronal cells. J Neurosci. 2013;33(21):9202–9213. doi: 10.1523/JNEUROSCI.5336-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Moresco EM, Beutler B. Special delivery: Granulin brings CpG DNA to Toll-like receptor 9. Immunity. 2011;34(4):453–455. doi: 10.1016/j.immuni.2011.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang SM, Coultas KA. Identification and characterization of five transcription factors that are associated with evolutionarily conserved immune signaling pathways in the schistosome-transmitting snail Biomphalaria glabrata. Mol Immunol. 2011;48(15-16):1868–1881. doi: 10.1016/j.molimm.2011.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Monami G, et al. Proepithelin promotes migration and invasion of 5637 bladder cancer cells through the activation of ERK1/2 and the formation of a paxillin/FAK/ERK complex. Cancer Res. 2006;66(14):7103–7110. doi: 10.1158/0008-5472.CAN-06-0633. [DOI] [PubMed] [Google Scholar]
- 28.Bateman A, Bennett HP. The granulin gene family: From cancer to dementia. BioEssays. 2009;31(11):1245–1254. doi: 10.1002/bies.200900086. [DOI] [PubMed] [Google Scholar]
- 29.Belcourt DR, Lazure C, Bennett HP. Isolation and primary structure of the three major forms of granulin-like peptides from hematopoietic tissues of a teleost fish (Cyprinus carpio) J Biol Chem. 1993;268(13):9230–9237. [PubMed] [Google Scholar]
- 30.Belcourt DR, Okawara Y, Fryer JN, Bennett HP. Immunocytochemical localization of granulin-1 to mononuclear phagocytic cells of the teleost fish Cyprinus carpio and Carassius auratus. J Leukoc Biol. 1995;57(1):94–100. doi: 10.1002/jlb.57.1.94. [DOI] [PubMed] [Google Scholar]
- 31.Hanington PC, Barreda DR, Belosevic M. A novel hematopoietic granulin induces proliferation of goldfish (Carassius auratus L.) macrophages. J Biol Chem. 2006;281(15):9963–9970. doi: 10.1074/jbc.M600631200. [DOI] [PubMed] [Google Scholar]
- 32.Smout MJ, et al. A granulin-like growth factor secreted by the carcinogenic liver fluke, Opisthorchis viverrini, promotes proliferation of host cells. PLoS Pathog. 2009;5(10):e1000611. doi: 10.1371/journal.ppat.1000611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Papatpremsiri A, et al. Suppression of Ov-grn-1 encoding granulin of Opisthorchis viverrini inhibits proliferation of biliary epithelial cells. Exp Parasitol. 2015;148:17–23. doi: 10.1016/j.exppara.2014.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stibbs HH, Owczarzak A, Bayne CJ, DeWan P. Schistosome sporocyst-killing Amoebae isolated from Biomphalaria glabrata. J Invertebr Pathol. 1979;33(2):159–170. doi: 10.1016/0022-2011(79)90149-6. [DOI] [PubMed] [Google Scholar]
- 35.Hansen EL. 1976. A cell line from embryos of Biomphalaria glabrata (Pulmonata): Establishment and characteristics. Invertebrate Tissue Culture: Research Applications, 1-393 (Academic, New York), pp 75–99.
- 36.Yoshino TP, Bickham U, Bayne CJ. Molluscan cells in culture: Primary cell cultures and cell lines. Can J Zool. 2013;91(6):391–404. doi: 10.1139/cjz-2012-0258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Adema CM, et al. Differential transcriptomic responses of Biomphalaria glabrata (Gastropoda, Mollusca) to bacteria and metazoan parasites, Schistosoma mansoni and Echinostoma paraensei (Digenea, Platyhelminthes) Mol Immunol. 2010;47(4):849–860. doi: 10.1016/j.molimm.2009.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ulmer MJ. Experiments and Techniques in Parasitology. W. H. Freeman.; San Francisco: 1970. Notes on rearing of snails in the laboratory; pp. 143–144. [Google Scholar]
- 39.Petersen TN, Brunak S, von Heijne G, Nielsen H. SignalP 4.0: Discriminating signal peptides from transmembrane regions. Nat Methods. 2011;8(10):785–786. doi: 10.1038/nmeth.1701. [DOI] [PubMed] [Google Scholar]
- 40.Emanuelsson O, Nielsen H, Brunak S, von Heijne G. Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J Mol Biol. 2000;300(4):1005–1016. doi: 10.1006/jmbi.2000.3903. [DOI] [PubMed] [Google Scholar]
- 41.Nielsen H, Engelbrecht J, Brunak S, von Heijne G. Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng. 1997;10(1):1–6. doi: 10.1093/protein/10.1.1. [DOI] [PubMed] [Google Scholar]
- 42.Kall L, Krogh A, Sonnhammer EL. Advantages of combined transmembrane topology and signal peptide prediction–the Phobius web server. Nucleic Acids Res. 2007;35(Web server issue):W429–432. doi: 10.1093/nar/gkm256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Finn RD, et al. Pfam: The protein families database. Nucleic Acids Res. 2014;42(database issue):D222–D230. doi: 10.1093/nar/gkt1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kim DE, Chivian D, Baker D. Protein structure prediction and analysis using the Robetta server. Nucleic Acids Res. 2004;32(Web server issue):W526–531. doi: 10.1093/nar/gkh468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Berman HM, et al. The Protein Data Bank. Nucleic Acids Res. 2000;28(1):235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Boyle JP, Wu XJ, Shoemaker CB, Yoshino TP. Using RNA interference to manipulate endogenous gene expression in Schistosoma mansoni sporocysts. Mol Biochem Parasitol. 2003;128(2):205–215. doi: 10.1016/s0166-6851(03)00078-1. [DOI] [PubMed] [Google Scholar]
- 47.Adema CM, van Deutekom-Mulder EC, van der Knaap WP, Sminia T. NADPH-oxidase activity: The probable source of reactive oxygen intermediate generation in hemocytes of the gastropod Lymnaea stagnalis. J Leukoc Biol. 1993;54(5):379–383. doi: 10.1002/jlb.54.5.379. [DOI] [PubMed] [Google Scholar]
- 48.Sminia T, Barendsen L. A comparative morphological and enzyme histochemical-study on blood-cells of the fresh-water snails Lymnaea-stagnalis, Biomphalaria-glabrata, and Bulinus-truncatus. J Morphol. 1980;165(1):31–39. doi: 10.1002/jmor.1051650104. [DOI] [PubMed] [Google Scholar]
- 49.Chernin E. Observations on hearts explanted in vitro from the snail Australorbis glabratus. J Parasitol. 1963;49(3):353–364. [PubMed] [Google Scholar]