Significance
The ability of eukaryotic cells to pass their genomes properly from one cell generation to the next depends on the 1.2-MDa ubiquitin ligase complex APC/C (anaphase-promoting complex/cyclosome) and on the correct timing of its activation by the substrate adaptor CDC20 (cell division cycle 20). Although it has been known for two decades that mitotic APC/C phosphorylation is required for its activation by CDC20, the mechanistic basis of this process remained unknown, in part because the existence of numerous phospho-sites on APC/C made systematic mutagenesis approaches difficult. Here we have used the biGBac technique for the rapid assembly of multigene expression constructs to overcome this limitation and discovered that APC/C contains an autoinhibitory loop region that prevents CDC20 binding until it becomes phosphorylated in mitosis.
Keywords: cell cycle, mitosis, APC/C, CDC20, phosphorylation
Abstract
Chromosome segregation and mitotic exit are initiated by the 1.2-MDa ubiquitin ligase APC/C (anaphase-promoting complex/cyclosome) and its coactivator CDC20 (cell division cycle 20). To avoid chromosome missegregation, APC/CCDC20 activation is tightly controlled. CDC20 only associates with APC/C in mitosis when APC/C has become phosphorylated and is further inhibited by a mitotic checkpoint complex until all chromosomes are bioriented on the spindle. APC/C contains 14 different types of subunits, most of which are phosphorylated in mitosis on multiple sites. However, it is unknown which of these phospho-sites enable APC/CCDC20 activation and by which mechanism. Here we have identified 68 evolutionarily conserved mitotic phospho-sites on human APC/C bound to CDC20 and have used the biGBac technique to generate 47 APC/C mutants in which either all 68 sites or subsets of them were replaced by nonphosphorylatable or phospho-mimicking residues. The characterization of these complexes in substrate ubiquitination and degradation assays indicates that phosphorylation of an N-terminal loop region in APC1 is sufficient for binding and activation of APC/C by CDC20. Deletion of the N-terminal APC1 loop enables APC/CCDC20 activation in the absence of mitotic phosphorylation or phospho-mimicking mutations. These results indicate that binding of CDC20 to APC/C is normally prevented by an autoinhibitory loop in APC1 and that its mitotic phosphorylation relieves this inhibition. The predicted location of the N-terminal APC1 loop implies that this loop controls interactions between the N-terminal domain of CDC20 and APC1 and APC8. These results reveal how APC/C phosphorylation enables CDC20 to bind and activate the APC/C in mitosis.
Eukaryotic cells pass their genomes from one cell generation to the next by first replicating DNA and simultaneously connecting the nascent sister chromatids during S-phase to allow their biorientation on the spindle later in mitosis. Once all chromosomes have become bioriented, the cohesion that holds sister chromatids together is destroyed, enabling chromosome segregation in anaphase and the formation of daughter cells with correct ploidy. A key step in this “chromosome cycle” is initiated by a ubiquitin ligase complex, called the anaphase-promoting complex/cyclosome (APC/C) (1–5). The APC/C initiates sister chromatid separation by assembling chains of the small protein ubiquitin on B-type cyclins, the activating subunits of cyclin-dependent kinase 1 (CDK1), and on securin, an inhibitor of the protease separase (reviewed in ref. 6). The subsequent degradation of these proteins by the 26S proteasome activates separase, which separates sister chromatids by cleaving cohesin, a complex that mediates sister chromatid cohesion.
APC/C is composed of 14 types of subunits (five are present in two copies each) (7–9), which assemble into a 1.2-MDa complex composed of two major structural parts, called the “platform” and the “arc lamp” (9). APC/C recognizes B-type cyclins and securin in metaphase with help of a coactivator protein called CDC20, which associates with APC/C specifically in mitosis, resulting in formation of APC/CCDC20. A related coactivator, CDH1, binds APC/C during mitotic exit to form APC/CCDH1, which remains active throughout the G1-phase and in postmitotic cells (reviewed in ref. 10). Both CDC20 and CDH1 interact with APC/C via a C-terminal “IR tail” (11) and an N-terminal “C-box” (12), which associate with the APC/C subunits APC3 and APC8, respectively (13–15). Once bound to APC/C, the coactivators enable substrate recruitment via their WD40 “propeller” domains, which recognize “degron” sequences in APC/C substrates, such as the destruction (D) box (14, 16). Covalent attachment of ubiquitin to lysine residues in substrates is mediated by the ubiquitin-conjugating (E2) enzymes UBE2D (also known as UBCH5), UBE2C (also known as UBCH10 and UBCx), and UBE2S, with the latter two having specific roles in the initiation and elongation of ubiquitin chain formation on APC/C substrates, respectively (2, 7, 17–20).
The activity of APC/C is tightly controlled to ensure that specific substrates are targeted for degradation only at the appropriate time during the cell cycle. This regulation occurs on at least two levels. The formation of APC/CCDC20 and APC/CCDH1 is regulated by mitotic protein kinases, with phosphorylation of APC/C by CDK1 and to a lesser extent by Polo-like kinase 1 (PLK1) increasing CDC20 binding and APC/CCDC20 activity (2–4, 21–27) and phosphorylation of CDH1 by CDK1 having the opposite effect, preventing binding and activation of APC/C by CDH1 (23, 28–31). The opposing effect of phosphorylation on APC/CCDC20 and APC/CCDH1 ensures that the former can only be active in mitosis when CDK1 activity is high, whereas the latter can only be active after mitotic exit when CDK1 activity is low, as a result of cyclin degradation mediated by APC/CCDC20 (10). CDC20 phosphorylation has also been found to inhibit binding of CDC20 to APC/C and recruitment of UBE2S, and dephosphorylation of CDC20 by protein phosphatase 2A has been proposed to enable APC/CCDC20 activation (32–35).
A second level of APC/C regulation is provided by specific inhibitors of CDC20 and CDH1, called the mitotic checkpoint complex (MCC) (36, 37) and EMI1 (38). Whereas EMI1 contributes to APC/CCDH1 inhibition throughout the S- and G2-phase, MCC is assembled only during prometaphase at kinetochores of chromosomes that have not been properly bioriented on the mitotic spindle yet (reviewed in ref. 39). MCC is composed of CDC20 and three other subunits (MAD2, BUBR1, and BUB3) (36, 37), associates with APC/CCDC20 to form APC/CMCC (40), and prevents recruitment and ubiquitination of B-type cyclins and securin (41). By doing so, MCC delays separase activation until chromosome biorientation has been completed and thereby prevents precocious sister chromatid separation. This surveillance mechanism, called the spindle assembly checkpoint (SAC), is important for avoiding the formation of aneuploid daughter cells.
Although it is well known that APC/C phosphorylation is required for activation of APC/CCDC20 (2, 3, 21–27) and that this process is essential for proper cell division, the precise role of APC/C phosphorylation in APC/CCDC20 activation remains unknown. Mitotic APC/C phosphorylation promotes CDC20 binding (22, 23, 26), but it is unknown how phosphorylation causes this effect, which phospho-sites are responsible for APC/C activation, and if CDC20 recruitment is the only function of APC/C phosphorylation.
To be able to address these and other questions, we have developed a method, called biGBac, to rapidly assemble expression constructs containing all 14 human APC/C cDNAs (42). Here we have used mass spectrometry to identify 68 evolutionarily conserved phosphorylation sites that are present on mitotic APC/C in its CDC20-bound form, and we have used the biGBac technique in combination with site-directed mutagenesis to generate 47 forms of recombinant APC/C in which we systematically changed either all or various subsets of the 68 sites to nonphosphorylatable or phospho-mimicking residues. We characterized the ability of CDC20 to activate these APC/C mutants to mediate the ubiquitination and subsequent proteasomal degradation of cyclin B1 and securin. Our results show that the presence of phospho-mimicking residues in a single loop in APC1 (consisting of residues 296–401) enables CDC20 to bind and activate APC/C in the absence of mitotic APC/C phosphorylation. Importantly, deletion of this loop also allows APC/C activation by CDC20. We therefore propose that this loop normally prevents access of CDC20 to one of its binding sites, presumably formed by APC1 and APC8 with which the C-box containing N-terminal domain (NTD) of CDC20 interacts, and that phosphorylation of the APC1 loop alleviates this inhibitory effect. These results reveal how APC/C phosphorylation enables CDC20 to bind and activate the APC/C in mitosis.
Results
Identification of Phosphorylation Sites on Mitotic APC/C Associated with CDC20.
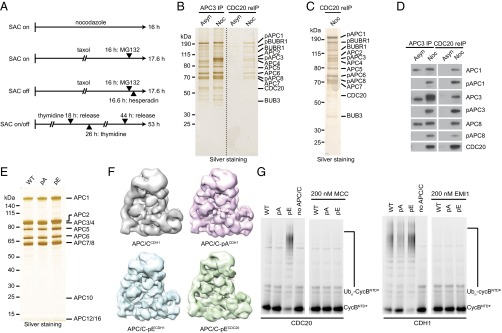
Previous studies identified numerous phosphorylation sites on APC/C isolated from human mitotic cells (22, 43–45), but these contain various forms of APC/C (41), APC/CMCC, and apo-APC/C in prometaphase and APC/CCDC20 and apo-APC/C in metaphase (apo-APC/C is a form of APC/C not bound to MCC, CDC20, or CDH1). To identify phospho-sites that might be required for CDC20 binding, we first analyzed which phospho-sites are present on APC/C in its CDC20-bound forms (APC/CCDC20 and APC/CMCC). For this purpose, we synchronized HeLa cells in mitosis by four different cell-cycle synchronization procedures (Fig. 1A), purified APC/C via immunoprecipitation (IP) with antibodies to APC3 (also known as CDC27), isolated APC/C bound to CDC20 from these samples by re-IP with CDC20 antibodies, and analyzed phospho-sites by in-solution digest–mass spectrometry (44). We enriched cells either in prometaphase, where the SAC is active by treatment with the spindle poisons nocodazole or taxol, or in a metaphase-like state by treatment with taxol and the Aurora B inhibitor hesperadin, which inactivates the SAC (41, 46), or in various stages of mitosis without drug treatments (which have been reported to modulate APC/C phosphorylation) (45) by double-thymidine arrest–release and mitotic “shake-off.” To prevent mitotic exit, which is normally caused by SAC inactivation, cells exposed to taxol with or without hesperadin were also treated with the proteasome inhibitor MG132, as described (41). For comparison, we also analyzed phospho-sites on interphase APC/C isolated by APC3 IP from cells arrested at the onset of S-phase (Fig. S1A). SDS/PAGE followed by silver staining (Fig. 1 B and C) or immunoblotting (Fig. 1D) revealed that re-IP with CDC20 antibodies specifically enriched phosphorylated APC/C from mitotic cells, as could be seen by electrophoretic mobility shifts of APC1, APC3, APC6 (also known as CDC16), and APC8 (CDC23) and by using antibodies specific to phospho-sites on these subunits (22), consistent with the notion that APC/C phosphorylation is required for CDC20 binding (22, 23, 26).
Fig. 1.
Immunopurification of mitotically phosphorylated APC/CCDC20 and purification and characterization of nonphosphorylatable and phospho-mimicking human APC/C. (A) Schematic overview of the synchronization procedures to obtain mitotic HeLa S3 cells. To obtain prometaphase-arrested cells with active SAC (SAC on), cells were synchronized using either the spindle poisons nocodazole or taxol. To obtain metaphase-arrested cells with inactive SAC (SAC off), cells were synchronized using taxol and supplemented with the Aurora B inhibitor hesperadin. Cells exposed to taxol with or without hesperidin were also treated with the proteasome inhibitor MG132 to prevent mitotic exit (41). To enrich for mitotic cells with unperturbed spindle checkpoint, cells were synchronized using a double-thymidine arrest–release procedure (SAC on/off). Mitotic cells were harvested by mitotic shake-off. (B–D) APC/C was immunoprecipitated from extracts of asynchronous (Asyn) or nocodazole-arrested (Noc) HeLa cells using APC3 antibody beads, re-immunoprecipitated using CDC20 antibody beads, and subjected to SDS/PAGE and silver staining (B and C) or Western blotting (D). Positions of subunits that display a mitotic phosphorylation electrophoretic mobility shift are indicated (pAPC1, pAPC3, and pAPC8). Note that in the CDC20 re-IP only the slowly migrating forms of APC1, APC3, and ACP8 were detected and that these three all cross-reacted with phospho-specific antibodies to these subunits. (E) Silver-stained SDS/PAGE gel of purified recombinant WT, nonphosphorylatable (pA), and phospho-mimicking (pE) APC/C. (F) Single-particle reconstruction by negative stain EM of recombinant APC/Ccoactivator–UBCH10–Ub–substrate complexes: APC/CCDH1 (gray, 20 Å resolution) (7), APC/C-pACDH1 (pink, 16 Å resolution), APC/C-pECDH1 (blue, 16 Å resolution), and APC/C-pECDC20 (green, 19 Å resolution). pA and pE maintain APC/C structural integrity. (G) Ubiquitination reactions, carried out in the presence of recombinant APC/C-WT, pA or pE, and fluorescein-labeled substrate N-terminal fragment of cyclin B (CycBNTD*), were analyzed by SDS/PAGE and fluorescence scanning. The reactions with CDC20 with or without MCC are in the Left panel, and the ones with CDH1 with or without EMI1 (EMI1-SKP1) are in the Right panel.
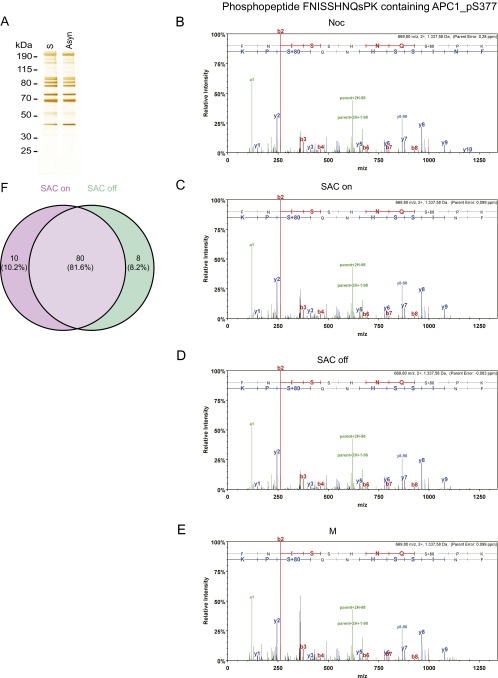
Fig. S1.
Mass spectrometry studies on various forms of APC/C immunopurified from HeLa cells. (A) Silver stain of APC/C immunoprecipitated using APC3 antibody beads from asynchronous cells and double-thymidine arrest–release synchronized cells (S-phase). (B–E) Tandem mass spectra of the phospho-S377 containing phospho-peptide FNISSHNQsPK in APC1 derived from (B) nocodazole-arrested HeLa cells (Noc), (C) Taxol + MG132-arrested cells (SAC on), (D) Taxol + MG132 + hesperadin-arrested cells (SAC off), and (E) cells treated with double-thymidine arrest–release procedure (M). Spectra of the phospho-peptides were acquired by higher energy collisional dissociation of the (M + 2H)2 + precursor, m/z 669.80 (B–E). Fragment ions in the spectra represent single-event preferential cleavage of the peptide bonds resulting in the sequence information recorded simultaneously from both the N and C termini (b- and y-ions, respectively) of the peptide. (F) Venn diagram showing that phospho-sites mapped on SAC on and SAC off samples largely overlap. The SAC on samples contained 10 phospho-peptides that were not found in the SAC off samples, although the corresponding nonphosphorylated peptides were recovered in the SAC off samples. Conversely, the SAC off samples contained seven phospho-peptides that were not found in the SAC on samples, although the corresponding nonphosphorylated peptides were recovered in the SAC on samples. We also identified one phospho-peptide in the SAC off samples for which the corresponding nonphosphorylated peptides were not found in SAC on samples. All phospho-peptides are listed in Dataset S3.
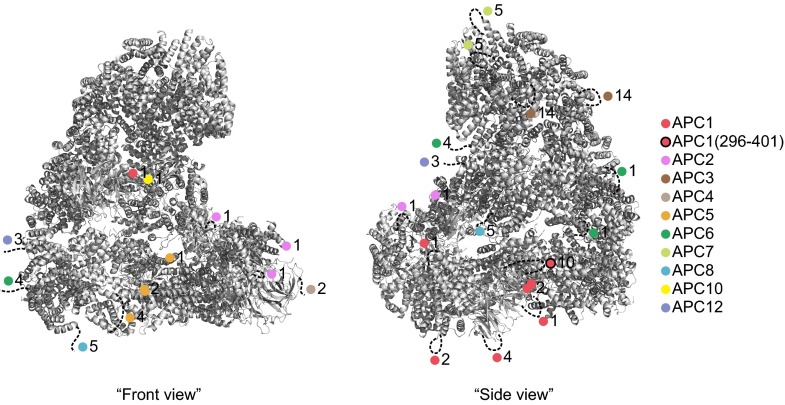
Peptides representing 87% and 72% of the sequences of all APC/C subunits were detected for interphase and mitotic APC/C, respectively, allowing the identification of most, although possibly not all, phospho-sites (Dataset S1). In total, we identified 120 phospho-sites (Dataset S2; see Fig. S1 B–E for tandem mass spectra of a representative phospho-peptide). Of these, we considered as potentially being important for CDC20 binding those that could be found in all four mitotic samples but not on interphase APC/C and that were located on amino acid residues conserved among vertebrate orthologs. These criteria were fulfilled by 68 phospho-sites on serine and threonine residues, 58 of which had previously been reported in the literature (22, 43–45). These 68 sites are located on 10 subunits (APC1, APC2, APC3, APC4, APC5, APC6, APC7, APC8, APC10, and APC12; Table S1). Because APC3, APC6, APC7, APC8, and APC12 are present in two copies each per complex (7–9), the 68 phosphorylated residues identified by mass spectrometry may correspond to up to 100 phospho-sites in APC/C (whether the phospho-peptides measured in our experiments are derived from one or both copies of APC3, APC6, APC7, APC8, and APC12 cannot be concluded from our results). Of these 100 potential sites, 90 are not visible in a known cryo-electron microscopy (cryo-EM) structure of APC/C (Fig. S2) (13). This implies that these sites are present in flexible regions such as loops, consistent with the previous notion that protein kinases preferentially phosphorylate sites that are present in regions that are predicted to be disordered (47). We also noticed some differences in the phosphorylation of APC/C isolated from mitotic cells with an active or an inactive SAC, possibly representing differences between APC/CMCC and APC/CCDC20, but did not analyze these further in this study (Fig. S1F and Dataset S3).
Table S1.
List of mutated phospho-sites of human APC/C subunits
| APC/C subunits | Number of mutations | Mutated phospho-sites |
| APC1 | 21 | S202, S286, T291, S313, T316, S317, S334, S341, S343, S355, S362, S372, S377, T537, S547, S555, S569, S688, S699, S916, S1347 |
| APC2 | 5 | S218, S314, S470, S534, S811 |
| APC3 | 14 | T200, T205, T220, S241, S276, S320, S336, S339, S386, S387, S393, S426, S435, T446 |
| APC4 | 2 | S777, S779 |
| APC5 | 7 | T178, S179, S195, S202, S221, T232, S364 |
| APC6 | 5 | S112, S560, T581, T585, S586 |
| APC7 | 5 | S119, T120, S123, S125, T126 |
| APC8 | 5 | T542, T562, T582, S588, T596 |
| APC10 | 1 | T3 |
| APC12 | 3 | S51, S52, S82 |
Fig. S2.
Location of mutated phospho-sites in APC/C. The location of phospho-sites is visualized in the APC/C–CDH1–EMI1 structure [Protein Data Bank (PDB) ID code 4UI9] (13) as colored spheres and their number in this region indicated. Flexible regions containing phospho-sites are indicated by dotted lines.
Generation of Nonphosphorylatable and Phospho-Mimicking APC/C Mutants.
To analyze the potential role of the conserved mitotic phospho-sites in APC/CCDC20 activation, we mutated all 68 corresponding serine and threonine residues either to alanine (Ala, A) or glutamate (Glu, E) to create mutants in which these sites are nonphosphorylatable or phospho-mimicking, respectively, and used the biGBac technique (42) and expression in baculovirus-infected insect cells to generate recombinant forms of these mutants. We refer to the mutant containing 68 substitutions to alanine and to the one containing 68 substitutions to glutamate as APC/C-pA (poly-Ala) and APC/C-pE (poly-Glu), respectively. Although these complexes contain 100 mutations each (because the subunits APC3, APC6, APC7, APC8, and APC12 are present in two copies each; 7–9), APC/C-pA and APC/C-pE resembled wild-type (WT) APC/C with respect to subunit composition and stoichiometry, as determined by SDS/PAGE and silver staining (Fig. 1E), and with respect to their structure and ability to recruit CDH1 to its normal binding site, as determined by single-particle EM reconstruction (Fig. 1F; compare with WT APC/C in ref. 7). Notably, the electrophoretic mobility of the APC1, APC3, APC6, and APC8 subunits of APC/C-pE was reduced compared with their WT counterparts, which is reminiscent of the reduction in electrophoretic mobility that is caused by mitotic phosphorylation of these subunits (3).
Nonphosphorylatable APC/C-pA Cannot Be Activated by CDC20, Whereas Phospho-Mimicking APC/C-pE Can Be Activated by CDC20 in the Absence of Mitotic Phosphorylation.
To analyze the activity of APC/C-pA and APC/C-pE, we first performed ubiquitination assays. As expected, APC/C-pA behaved similarly as recombinant WT APC/C in that neither complex could efficiently ubiquitinate an N-terminal fragment of cyclin B1 (CycBNTD) in the presence of CDC20 (Fig. 1G, Left). In contrast, both complexes could be activated to some extent by CDH1 in a manner that was sensitive to inhibition by EMI1 (Fig. 1G, Right), indicating that APC/C-pA is not generally deficient in supporting ubiquitination reactions but cannot be activated by CDC20. Importantly, APC/C-pE behaved differently from WT APC/C and APC/C-pA in that APC/C-pE could be activated similarly well by CDC20 and CDH1 in manners that were sensitive to inhibition by MCC and EMI1, respectively (Fig. 1G). The observation that mutation of 68 phospho-sites to glutamate residues enables APC/C activation by CDC20 in the absence of mitotic phosphorylation indicates that phosphorylation of these residues (or of a subset of them) is sufficient for activation of APC/C by CDC20, at least under the assay conditions used.
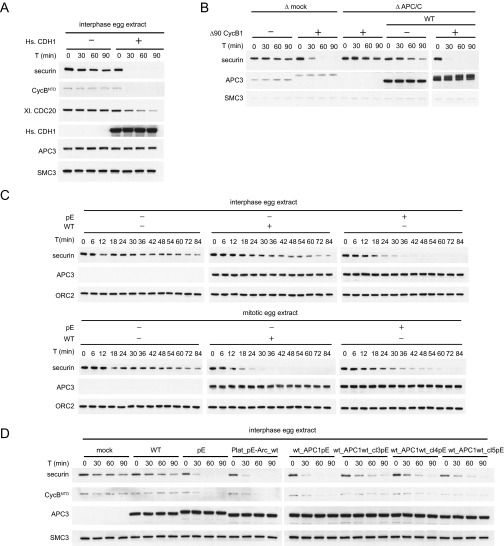
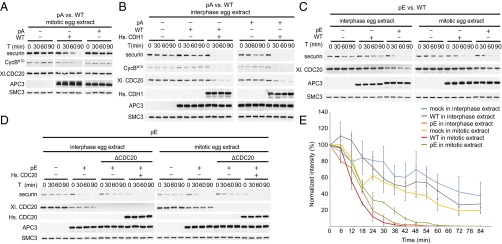
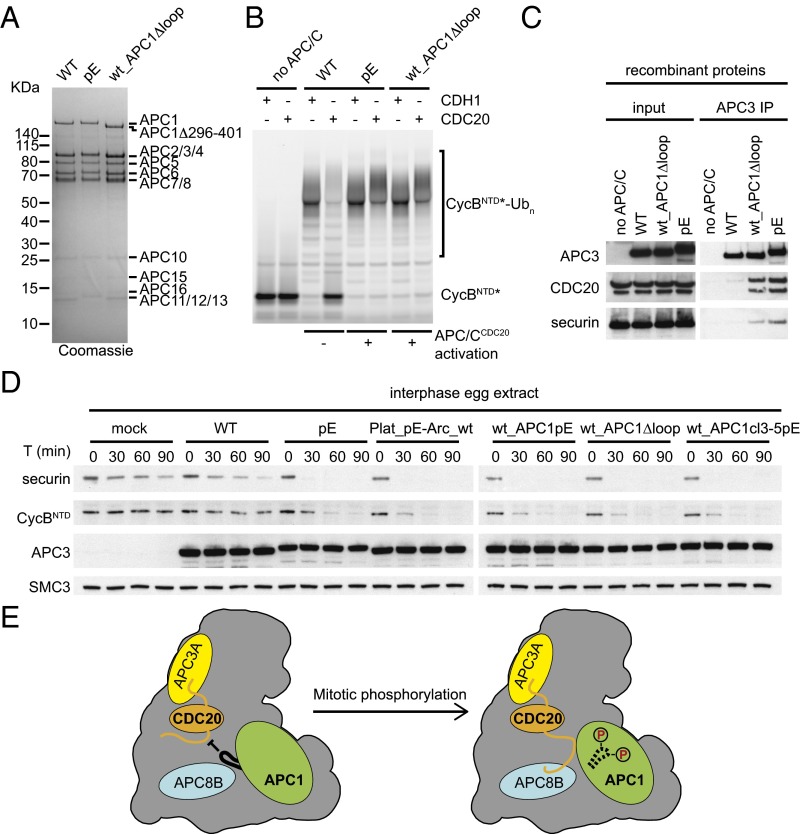
To analyze the effect of these mutations under more physiological conditions, we analyzed the ability of WT APC/C and the pA and pE variants to induce degradation of cyclin B1 and securin in interphase and mitotic Xenopus egg extracts. These extracts recapitulate cell-cycle regulation of APC/CCDC20 activity—that is, mediate ubiquitination and degradation of APC/C substrates in mitosis but not in interphase (16, 48, 49). CDH1 is not present in these extracts (23) but can induce degradation of APC/C substrates if added (14, 50, 51) (Fig. S3A). First, we tested if the 68 mitotic phospho-sites mutated in APC/C-pA are required for mitotic APC/C activation or if phosphorylation of other nonmutated sites might also be sufficient. For this purpose, we immunodepleted endogenous APC/C from mitotic Xenopus extract and replaced it with either recombinant human WT APC/C or APC/C-pA. Whereas WT APC/C could restore degradation of securin and CycBNTD, APC/C-pA could not (Fig. 2A). Similar to the results obtained in ubiquitination assays (Fig. 1G), this was not due to a more general defect of this APC/C mutant, as APC/C-pA was able to induce securin and CycBNTD degradation in interphase extracts if these were also supplemented with CDH1 (Fig. 2B). This indicates that the 68 phospho-sites mutated in APC/C-pA, or a subset of them, are required for mitotic APC/CCDC20 activation.
Fig. S3.
Western blots of substrate degradation in Xenopus egg extracts. (A) APC/C is not active in interphase egg extracts. Addition of recombinant human CDH1 can activate APC/C and restore securin and CycBNTD (cyclin B1 residues 1–87) degradation. Western blotting as a function of time shows securin and CycBNTD degradation, APC3 controls for levels of the two versions of APC/C in the assays, and SMC3 controls for comparable sample loading. (B) Recombinant human APC/C can restore securin degradation in mitotic extracts depleted of endogenous APC/C. The electrophoretic mobility of the APC3 subunit of recombinant human APC/C is reduced in mitotic Xenopus egg extracts (generated by addition of nondegradable Δ90 CycB1), indicative of mitotic APC/C phosphorylation. (C) APC/C-pE and WT were added back to APC/C-depleted interphase and mitotic egg extracts. Western blotting as a function of time with 6-min intervals provides a high temporal resolution recording of securin degradation. (D) Recombinant APC/C-WT, pE, Plat_pE-Arc_wt, wt_APC1pE, wt_APC1wt_cl3pE, wt_APC1wt_cl4pE, and wt_APC1wt_cl5pE were added back to APC/C-depleted interphase egg extracts. Western blotting as a function of time provides a recording of securin degradation.
Fig. 2.
Substrate degradation in Xenopus egg extracts induced by the addition of WT, pA, or pE. pA, pE, or WT was added back to APC/C-depleted egg extract and incubated with nondegradable Δ90 cyclin B1 for 90 min. Substrates were added, and samples were taken and analyzed at the indicated time points for CycBNTD (cyclin B1 residues 1–87) and securin degradation by Western blotting. Equal amounts of each APC/C variant were used (see APC3 Western blot). SMC3 was used as a loading control. (A) pA vs. WT in mitotic egg extract. (B) pA vs. WT in interphase egg extract supplemented with recombinant human CDH1 protein. (C) pE vs. WT in interphase and mitotic egg extracts. (D) pE in APC/C or APC/C- and CDC20-depleted interphase and mitotic egg extracts supplemented with recombinant human CDC20. (E) Securin degradation time course in mock-depleted interphase (blue) and mitotic (yellow) egg extracts and APC/C-depleted egg extracts following add-back of WT in interphase (gray) or mitosis (red) or add-back of pE in interphase (orange) or mitosis (green). Securin levels were monitored by Western blotting and normalized to ORC2 levels. SEM, n ≥ 2.
To test if mutation of these 68 sites to glutamate is sufficient for APC/CCDC20 activation, we replaced endogenous APC/C with WT APC/C or APC/C-pE in interphase extracts. In these extracts, securin was degraded much more slowly than in mitotic extracts in the presence of endogenous or recombinant WT APC/C (Fig. S3B). However, in the presence of APC/C-pE, interphase extracts degraded securin with kinetics that were similar to the rapid kinetics normally only seen in mitotic extracts (Fig. 2C). The ability of APC/C-pE to induce securin degradation in interphase, where CDC20 is not normally active, was delayed by depletion of endogenous CDC20 and restored by adding back recombinant human CDC20 (Fig. 2D). This implies that APC/C-pE induced securin degradation because this APC/C mutant could be activated by CDC20 in the absence of mitotic phosphorylation, rather than by gaining some other property. Importantly, also when analyzed at higher temporal resolution, the rate of securin degradation in interphase extracts containing APC/C-pE was indistinguishable from the rate of securin degradation in mitotic extracts containing APC/C-WT, indicating that APC/C-pE can be activated to a similar extent by CDC20 as mitotically phosphorylated APC/C (Fig. 2E and Fig. S3C). These results imply that phosphorylation of the 68 sites mutated in APC/C-pE, or phosphorylation of a subset of these, is sufficient for activation of APC/C by CDC20.
Glutamate Substitutions at Mitotic APC/C Phosphorylation Sites Increase CDC20 and Substrate Binding and the Maximal Velocity of Ubiquitination Reactions.
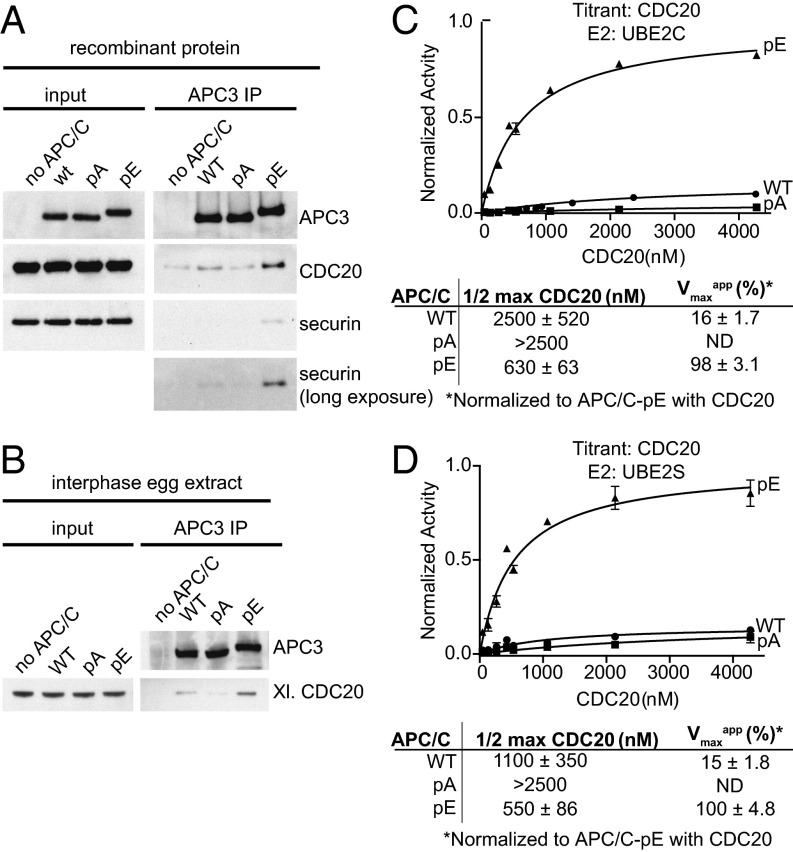
Because previous experiments indicated that mitotic APC/C phosphorylation is required for CDC20 binding (22, 23, 26), we tested if CDC20 can bind to WT APC/C or the pA and pE mutants. Because CDC20 helps to recruit substrates to the APC/C, we also analyzed securin binding to APC/C in these assays. Immunoblotting experiments revealed that more CDC20 and securin bound to APC/C-pE than to WT APC/C and APC/C-pA (Fig. 3A), correlating well with the ability of these complexes to mediate securin ubiquitination and degradation (Fig. 2). The low amount of CDC20 that was retained on WT APC/C and to a lesser degree on APC/C-pA could either represent nonspecific or phosphorylation-independent low-affinity interactions or could have been caused by the presence of some phospho-sites that we detected at low levels on WT APC/C by mass spectrometry (Dataset S4) and that were presumably generated in the insect cells used for APC/C expression and assembly. Similar results were obtained when WT APC/C and the pA and pE mutants were incubated in Xenopus interphase egg extracts, reisolated by IP, and analyzed for the presence of CDC20 by immunoblotting. Also under these conditions, CDC20 bound preferentially to APC/C-pE (Fig. 3B).
Fig. 3.
CDC20 and securin binding assays and kinetics studies of the WT, pA, and pE forms of APC/C. (A) WT, pA, and pE forms of recombinant human APC/C were incubated with recombinant purified CDC20 and securin for 1 h. Bound proteins were isolated using APC3 antibody beads, peptide-eluted, and analyzed by Western blotting. (B) WT, pA, and pE forms of recombinant human APC/C were added to APC/C-depleted interphase egg extract and incubated for 2 h. Bound proteins were analyzed as in A. (C) Curve fits and kinetic parameters from titrating CDC20 to test the role of APC/C mutants pA and pE on catalytic efficiency of Ub–securin ubiquitination with UBE2C. SEM, n ≥ 3. (D) As in C, except UBE2S was used instead of UBE2C. SEM, n ≥ 3.
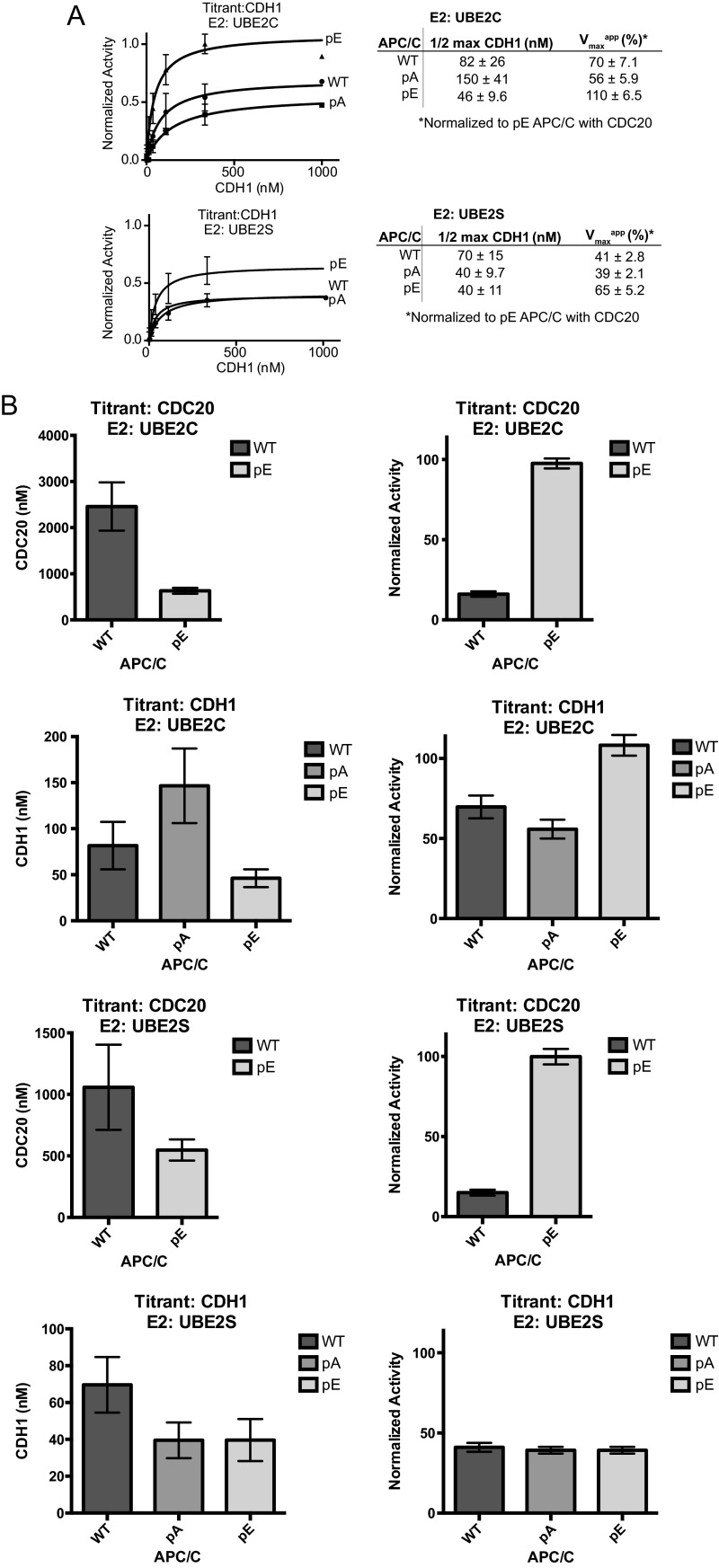
Glutamate substitutions at mitotic phosphorylation sites also made significant contributions to the CDC20-dependent stimulation of APC/C-mediated substrate ubiquitination, as examined by kinetic analyses. WT APC/C and phospho-site variants were analyzed for coactivator-dependent activation over a range of CDC20 or CDH1 concentrations. Although the phospho-site mutants exhibited only small effects on CDH1-dependent activation of APC/C (Fig. S4 A and B), APC/C-pE displayed enhanced catalytic efficiency of substrate (Ub-securin) ubiquitination by 13–24-fold compared with WT levels with either UBE2C or UBE2S, respectively (Fig. 3 C and D and Fig. S4B). Both a decrease in the concentration required for half-maximal activation (2–4-fold) and an increase in the maximum velocity (6–7-fold) were observed in reactions with APC/C-pE and its respective E2s (Fig. 3 C and D and Fig. S4B). In contrast, activation of APC/C-pA by CDC20 was further reduced compared with WT APC/C levels. The simplest interpretation for the decrease in the concentration required for half-maximal activation of APC/C-pE by CDC20 compared with WT APC/C is that CDC20 binding to APC/C-pE is increased, consistent with the results obtained in CDC20 binding experiments (Fig. 3 A and B). This increase in coactivator binding may also enhance E2 catalytic efficiency, as observed in previous studies (7, 8, 19, 20, 52), which could explain the unexpected increase in the maximum velocity of APC/C-pE. These results indicate that phosphorylation of the 68 sites mutated in APC/C-pE, or phosphorylation of a subset of these, increases binding of CDC20 to APC/C, thereby enables substrate recruitment, and in addition increases APC/C’s maximal velocity, possibly by enhancing the catalytic efficiency of the UBE2C and UBE2S.
Fig. S4.
Kinetic analysis showing that phospho-mimetics enhance the activity of APC/CCDC20. (A) Curve fits and kinetic parameters from titrating CDH1 to test the role of APC/C mutants pA and pE on catalytic efficiency of Ub-securin ubiquitination with UBE2S and UBE2C. SEM, n ≥ 3. (B) Bar graphs showing the effect of pA and pE on the concentrations of CDC20 and CDH1 required for half-maximal activation and normalized activity in Ub-securin ubiquitination assays with UBE2S and UBE2C, respectively.
Phosphorylation of an N-Terminal Loop Region in APC1 (Residues 296–401) Is Sufficient for Activation of APC/CCDC20.
To analyze which phospho-sites control APC/CCDC20 activation, we generated 45 additional APC/C variants in which only subsets of the 68 phospho-sites mutated in APC/C-pE were substituted by glutamate residues. Tables S2 and S3 list these complexes and summarize results obtained with them in ubiquitination assays.
Table S2.
Summary of effects of pE mutations in APC/C subunits on APC/CCDC20-dependent GST-CycBNTD ubiquitination activity
| APC/C | APC1 | APC2 | APC3 | APC4 | APC5 | APC6 | APC7 | APC8 | APC10 | APC12 | APC/C-CDC20 activation |
| WT | WT | WT | WT | WT | WT | WT | WT | WT | WT | WT | – |
| pA | pA | pA | pA | pA | pA | pA | pA | pA | pA | pA | – |
| pE | pE | pE | pE | pE | pE | pE | pE | pE | pE | pE | + |
| pE_APC1wt | WT | pE | pE | pE | pE | pE | pE | pE | pE | pE | – |
| pE_APC2wt | pE | WT | pE | pE | pE | pE | pE | pE | pE | pE | + |
| pE_APC3wt | pE | pE | WT | pE | pE | pE | pE | pE | pE | pE | + |
| pE_APC4wt | pE | pE | pE | WT | pE | pE | pE | pE | pE | pE | + |
| pE_APC5wt | pE | pE | pE | pE | WT | pE | pE | pE | pE | pE | + |
| pE_APC6wt | pE | pE | pE | pE | pE | WT | pE | pE | pE | pE | + |
| pE_APC7wt | pE | pE | pE | pE | pE | pE | WT | pE | pE | pE | + |
| pE_APC8wt | pE | pE | pE | pE | pE | pE | pE | WT | pE | pE | + |
| pE_APC10wt | pE | pE | pE | pE | pE | pE | pE | pE | WT | pE | + |
| pE_APC12wt | pE | pE | pE | pE | pE | pE | pE | pE | pE | WT | + |
| Plat_pE-Arc_wt | pE | pE | WT | pE | pE | WT | WT | WT | WT | WT | + |
| Plat_wt-Arc_pE | WT | WT | pE | WT | WT | pE | pE | pE | pE | pE | – |
| APC/C_5E | S1347E | WT | WT | WT | WT | S112E | WT | S542E,T562E | T3E | WT | – |
Bold indicates the subunits in which the effect of phospho-mimicking mutations or wild-type residues was tested.
Table S3.
Summary of effects of pE mutations in APC1 on APC/CCDC20-dependent CycBNTD* ubiquitination activity
| APC1 | |||||||||||
| APC/C | Cl1 | Cl2 | Cl3 | Cl4 | Cl5 | Cl6 | Cl7 | Cl8 | Cl9 | APC2–12 | APC/C–CDC20 activation |
| WT | WT | WT | – | ||||||||
| pE | pE | pE | + | ||||||||
| wt_APC1pE | pE | WT | + | ||||||||
| wt_APC1wt_cl1pE | pE | WT | WT | WT | WT | WT | WT | WT | WT | WT | – |
| wt_APC1wt_cl2pE | WT | pE | WT | WT | WT | WT | WT | WT | WT | WT | n.d. |
| wt_APC1wt_cl3pE | WT | WT | pE | WT | WT | WT | WT | WT | WT | WT | +/− |
| wt_APC1wt_cl4pE | WT | WT | WT | pE | WT | WT | WT | WT | WT | WT | +/− |
| wt_APC1wt_cl5pE | WT | WT | WT | WT | pE | WT | WT | WT | WT | WT | +/− |
| wt_APC1wt_cl6pE | WT | WT | WT | WT | WT | pE | WT | WT | WT | WT | – |
| wt_APC1wt_cl7pE | WT | WT | WT | WT | WT | WT | pE | WT | WT | WT | – |
| wt_APC1wt_cl8pE | WT | WT | WT | WT | WT | WT | WT | pE | WT | WT | – |
| wt_APC1wt_cl9pE | WT | WT | WT | WT | WT | WT | WT | WT | pE | WT | n.d. |
| wt_APC1pE_cl1wt | WT | pE | pE | pE | pE | pE | pE | pE | pE | WT | + |
| wt_APC1pE_cl2wt | pE | WT | pE | pE | pE | pE | pE | pE | pE | WT | + |
| wt_APC1pE_cl3wt | pE | pE | WT | pE | pE | pE | pE | pE | pE | WT | + |
| wt_APC1pE_cl4wt | pE | pE | pE | WT | pE | pE | pE | pE | pE | WT | + |
| wt_APC1pE_cl5wt | pE | pE | pE | pE | WT | pE | pE | pE | pE | WT | + |
| wt_APC1pE_cl6wt | pE | pE | pE | pE | pE | WT | pE | pE | pE | WT | + |
| wt_APC1pE_cl7wt | pE | pE | pE | pE | pE | pE | WT | pE | pE | WT | n.d. |
| wt_APC1pE_cl8wt | pE | pE | pE | pE | pE | pE | pE | WT | pE | WT | + |
| wt_APC1pE_cl9wt | pE | pE | pE | pE | pE | pE | pE | pE | WT | WT | n.d. |
| pE_APC1wt_cl1pE | pE | WT | WT | WT | WT | WT | WT | WT | WT | pE | n.d. |
| pE_APC1wt_cl2pE | WT | pE | WT | WT | WT | WT | WT | WT | WT | pE | n.d. |
| pE_APC1wt_cl3pE | WT | WT | pE | WT | WT | WT | WT | WT | WT | pE | n.d. |
| pE_APC1wt_cl4pE | WT | WT | WT | pE | WT | WT | WT | WT | WT | pE | + |
| pE_APC1wt_cl5pE | WT | WT | WT | WT | pE | WT | WT | WT | WT | pE | + |
| pE_APC1wt_cl6pE | WT | WT | WT | WT | WT | pE | WT | WT | WT | pE | +/− |
| pE_APC1wt_cl7pE | WT | WT | WT | WT | WT | WT | pE | WT | WT | pE | +/− |
| pE_APC1wt_cl8pE | WT | WT | WT | WT | WT | WT | WT | pE | WT | pE | +/− |
| pE_APC1wt_cl9pE | WT | WT | WT | WT | WT | WT | WT | WT | pE | pE | +/− |
| pE_APC1pE_cl1wt | WT | pE | pE | pE | pE | pE | pE | pE | pE | pE | + |
| pE_APC1pE_cl2wt | pE | WT | pE | pE | pE | pE | pE | pE | pE | pE | + |
| pE_APC1pE_cl3wt | pE | pE | WT | pE | pE | pE | pE | pE | pE | pE | + |
| pE_APC1pE_cl4wt | pE | pE | pE | WT | pE | pE | pE | pE | pE | pE | + |
| pE_APC1pE_cl5wt | pE | pE | pE | pE | WT | pE | pE | pE | pE | pE | + |
| pE_APC1pE_cl6wt | pE | pE | pE | pE | pE | WT | pE | pE | pE | pE | + |
| pE_APC1pE_cl7wt | pE | pE | pE | pE | pE | pE | WT | pE | pE | pE | + |
| pE_APC1pE_cl8wt | pE | pE | pE | pE | pE | pE | pE | WT | pE | pE | + |
| pE_APC1pE_cl9wt | pE | pE | pE | pE | pE | pE | pE | pE | WT | pE | + |
| wt_APC1Δloop | WT | WT | Δ | Δ | Δ | WT | WT | WT | WT | WT | + |
| wt_APC1cl3-5pE | WT | WT | pE | pE | pE | WT | WT | WT | WT | WT | + (Xenopus) |
Bold indicates the clusters or subunits in which the effect of phospho-mimicking mutations or wild-type residues was tested. n.d., not determined.
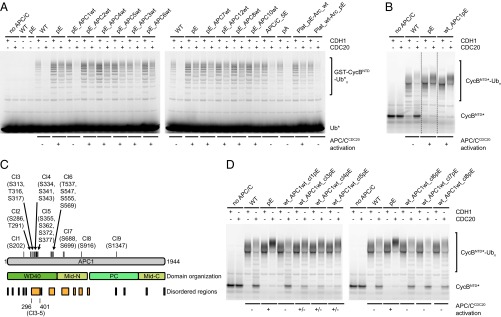
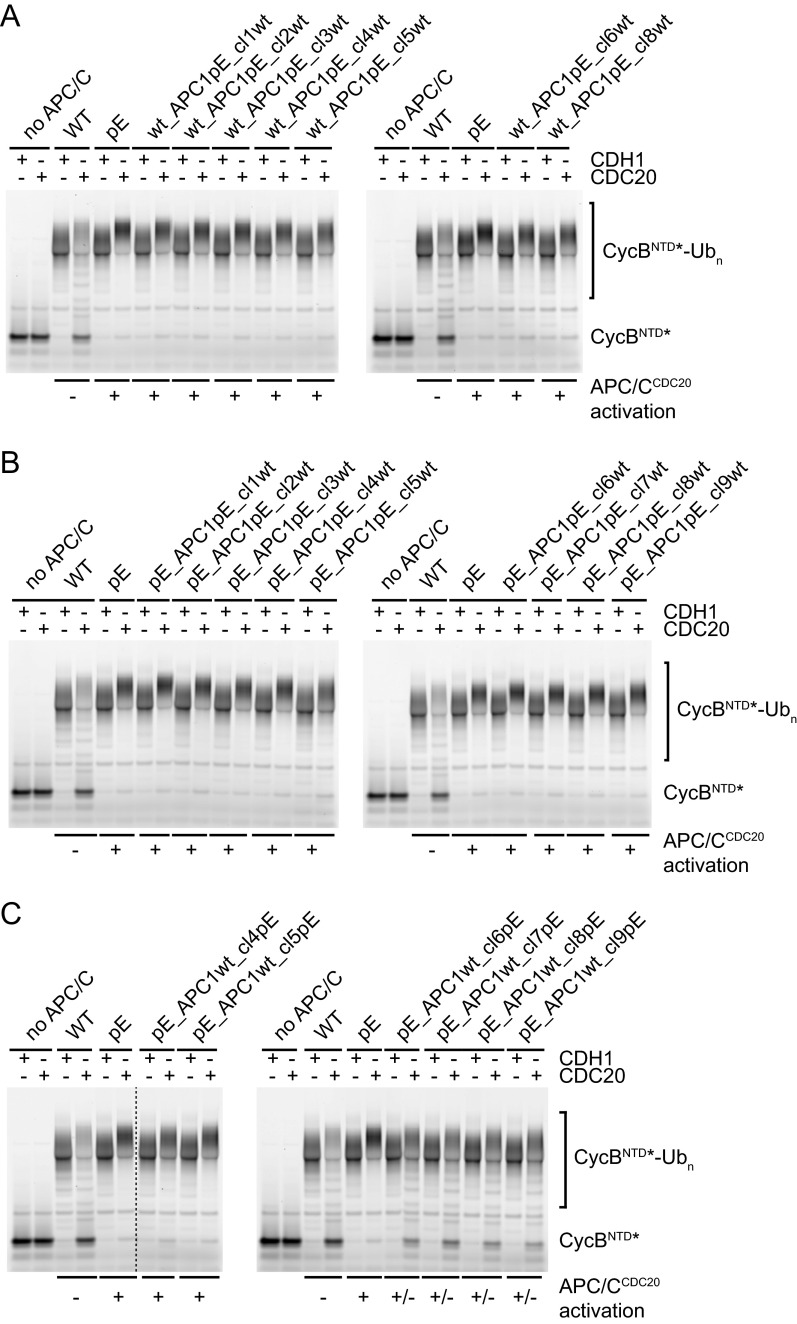
First, we tested if the presence of phospho-mimicking mutations is required in all 10 APC/C subunits mutated in APC/C-pE to enable activation by CDC20. For this purpose, we generated APC/C variants in which mitotic phospho-sites were mutated to glutamate in 9 out of the 10 subunits but the remaining subunit was present in its WT form. Unexpectedly, of the resulting 10 complexes, all could be activated by CDC20 in ubiquitination assays, except APC/C in which WT APC1 was present (Fig. 4A, pE_APC1wt). In parallel, we identified five phospho-sites in the APC/C structure (Fig. S2) (13) that are located in the vicinity of predicted CDC20 interaction sites (APC1S1347, APC6S112, APC8S542,T562, and APC10T3) and mutated these to glutamate residues, but this “best guess” APC/C mutant could not be activated by CDC20 (Fig. 4A, APC/C-5E). Furthermore, we generated and tested complexes that contain phospho-mimicking mutations only in the platform (Plat_pE-Arc_wt) or the arc lamp domain (Plat_wt-Arc_pE) and found that glutamate substitutions in platform subunits (APC1, APC2, APC4, and APC5) were sufficient for APC/CCDC20 activation, whereas phospho-mimicking mutations in arc lamp subunits (APC3, APC6, APC7, APC8, APC10, and APC12) were not (Fig. 4A). These results implied that phosphorylation sites in APC1 are required for APC/CCDC20 activation, that phosphorylation of other APC/C subunits is either not required or that there is redundancy between them, and that substitutions of phospho-sites in APC1, APC2, APC4, and APC5 are sufficient for this process.
Fig. 4.
Phosphorylation sites in the APC1(296–401) loop are a major regulatory element of APC/CCDC20 activity. (A) Ubiquitination reactions using fluorescein-labeled ubiquitin (Ub*) and substrate GST-CycBNTD carried out in the presence of WT or APC/C containing glutamate mutations in 9 of the 10 subunits or only in the platform (APC1/2/4/5) or arc lamp domain (APC3/6/7/8/10/12), using either CDH1 or CDC20, were analyzed by SDS/PAGE and fluorescence scanning. (B) Ubiquitination reactions using fluorescein-labeled substrate cyclin B NTD (CycBNTD*) in the presence of WT, pE, or APC/C containing glutamate mutations only in APC1 (wt_APC1pE). Cropped lanes are indicated by dashed lines. (C) Domain organization of APC1. The 21 phospho-mimicking mutations in APC1 are grouped into nine clusters (Cl, cluster). Cl3–Cl5 are located in the APC1 (296–401) loop. (D) Ubiquitination reactions as in B in the presence of WT or APC/C containing glutamate mutations in individual APC1 clusters.
Together, these observations raised the possibility that phosphorylation of APC1 might be sufficient for APC/CCDC20 activation. To test this, we analyzed a complex in which all 21 phospho-sites in APC1 were mutated to glutamate residues but all other subunits were present in their WT form. Remarkably, this complex (wt_APC1pE) could be activated by CDC20 similarly well as APC/C-pE (Fig. 4B), indicating that APC1 phosphorylation may indeed be sufficient for APC/CCDC20 activation. To determine which APC1 sites control APC/CCDC20 activation, we grouped the 21 phospho-sites into nine clusters (Fig. 4C) and analyzed most of these clusters individually (some of these cluster mutants could not be generated for technical reasons; Table S3). Phospho-mimicking mutations were not required in any of the tested individual clusters, either in the absence or presence of phospho-mimicking mutations in other subunits, provided that phospho-mimicking mutations were present in other APC1 phospho-site clusters (Fig. S5 A and B). This indicates that there is redundancy between different phospho-sites within APC1. In contrast, glutamate substitutions only in clusters 3, 4, or 5 of APC1 were sufficient for activation of APC/C by CDC20 to an intermediate level, despite the fact that only three (clusters 3 and 4) or four residues (cluster 5) were mutated. Remarkably, this was the case not only in the presence of phospho-mimicking mutations in other APC/C subunits (Fig. S5C) but also if all other subunits were present in their WT form (Fig. 4D). Also, when tested in Xenopus interphase extracts, APC/C-containing glutamate substitutions only in clusters 3, 4, or 5 mediated securin degradation more rapidly than WT APC/C, although it occurred less rapidly than APC/C in which all phospho-sites in APC1 had been mutated to glutamate (wt_APC1pE; Fig. S3D). An APC/C variant in which the phospho-site mutations in clusters 3, 4, and 5 were combined (wt_APC1cl3-5pE) supported degradation of securin and CycBNTD similarly well as wt_APC1pE and APC/C-pE (see Fig. 5D). Interestingly, the phospho-site clusters 3, 4, and 5 are all located next to each other in an N-terminal region of APC1 comprising residues 296–401. This region is not visible in existing APC/C structures (13) and is thus predicted to form a flexible loop.
Fig. S5.
Redundancy between clusters of phospho-sites within APC1. Fluorescence scans of SDS/PAGE gels loaded with ubiquitination reaction products using fluorescent substrate cyclin B NTD (CycBNTD*). (A) Phospho-mimicking mutations in none of the tested individual APC1 clusters are required for APC/C–CDC20 activation in the absence of phospho-mimicking mutations in other subunits, suggesting redundancy between at least two APC1 clusters. (B) As in A, but in the presence of phospho-mimicking mutations in other subunits. (C) Phospho-mimicking mutations in APC1 clusters 4 or 5, located in the APC1(296–401) loop, are sufficient for APC/C–CDC20 activation in the presence of phospho-mimicking mutations in other subunits. The other pE_APC1wt_clXpE complexes show only slightly higher APC/CCDC20 activity than WT. Cropped lanes are indicated by a dashed line.
Fig. 5.
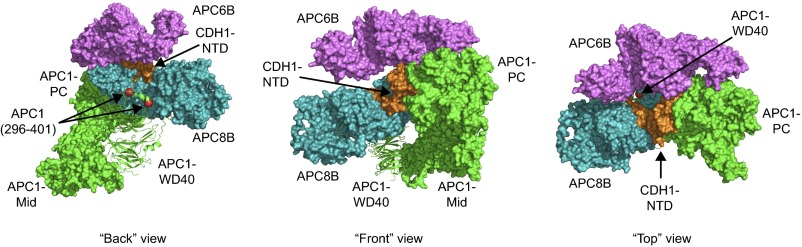
Deletion of the APC1(296–401) loop enables APC/CCDC20 activation. (A) Coomassie-stained SDS/PAGE gel showing WT, pE, and APC/C in which the APC1(296–401) loop is deleted (wt_APC1Δloop). (B) Ubiquitination reactions using CycBNTD* in the presence of WT, pE, or wt_APC1Δloop analyzed by SDS/PAGE and fluorescence scanning. (C) WT, wt_APC1Δloop, and pE were incubated with recombinant purified CDC20 and securin for 1 h, and bound proteins were isolated using APC3 antibody beads, peptide-eluted, and analyzed by Western blotting. (D) Substrate degradation in interphase Xenopus egg extracts induced by the addition of APC/C mutants. WT, pE, Plat_pE-Arc_wt, wt_APC1pE, wt_APC1Δloop, or wt_APC1cl3–5pE was added back to APC/C-depleted interphase egg extract. Substrates were added, and samples were taken and analyzed at the indicated time points for CycBNTD (cyclin B1 residues 1–87) and securin degradation by Western blotting. Equal amounts of each APC/C variant were used (see APC3 Western blot). SMC3 was used as a loading control. (E) Model for APC/CCDC20 activation via APC1(296–401) loop phosphorylation. The IR tail of CDC20 (orange) associates with APC3 (yellow), and the NTD of CDC20 binds to both APC1 (green) and APC8 (blue). The nonphosphorylated APC1(296–401) loop (black) inhibits the association of CDC20 NTD with APC1 and APC8, possibly via limiting the accessibility of APC1 and APC8 binding sites. Following mitotic phosphorylation of the loop, this inhibition is relieved via a conformation change (dashed), enabling APC/C–CDC20 association and activation.
These results suggest that phosphorylation of a flexible N-terminal loop in APC1 is sufficient for activation of APC/C by CDC20. However, our results do not rule out the possibility that phosphorylation of other sites on APC/C can also contribute to the rate or efficiency of APC/CCDC20 activation. Consistent with this possibility, we observed that glutamate substitutions in APC1 clusters 6, 7, or 8 slightly increased APC/CCDC20 activity in the presence of phospho-mimicking mutations in other subunits (Fig. S5C) but not in their absence (Fig. 4D), implying that phosphorylation of other APC/C subunits might be able to contribute to APC/CCDC20 activation.
Deletion of an N-Terminal Loop Region of APC1 Enables Activation of APC/CCDC20 in the Absence of Mitotic Phosphorylation.
The observation that only three to four glutamate substitutions in an N-terminal loop region of APC1 are sufficient for APC/CCDC20 partial activation could be explained by several different possibilities. The simplest of these would be that phosphorylation of this loop would create a binding site that is required for functional interactions between APC/C and CDC20, although in this case it would be unusual that glutamate substitutions of various subsets of phospho-sites would be able to create such a binding site. Alternatively, phosphorylation of this loop could cause structural changes that would enable access of CDC20 to a preexisting binding site. To distinguish between these possibilities, we generated an APC/C variant in which we deleted the N-terminal loop from APC1, whereas all other subunits were present in their WT form (Fig. 5A). Remarkably, this APC/C variant (wt_APC1Δloop), which does not contain any glutamate substitutions, was activated by CDC20 in ubiquitination assays as well as APC/C-pE (Fig. 5B) and also bound CDC20 and securin similarly well as APC/C-pE (Fig. 5C). Also, in interphase Xenopus egg extracts, the wt_APC1Δloop complex supported degradation of securin and CycBNTD similarly well as APC/C-pE and APC/C variants in which either all phospho-sites in the platform domain or in APC1 or the phospho-sites in clusters 3, 4, and 5 had been substituted by glutamate (Fig. 5D). These results imply that the N-terminal APC1 loop has an autoinhibitory function and that its phosphorylation enables binding of CDC20 to a site on APC/C that otherwise is not accessible.
Discussion
Although ubiquitin-dependent proteolysis mediated by APC/CCDC20 is essential for sister chromatid separation, mitotic exit, and proper cell division in presumably all eukaryotes, it is not well understood how APC/C is activated by CDC20 in mitosis. Inactivation of the SAC following biorientation of all chromosomes on the mitotic spindle is required for APC/CCDC20 activation but is not sufficient, as previous observations indicated that phosphorylation of the APC/C is also needed for binding of CDC20 to APC/C and APC/C activation (2–4, 21–27). However, the role of mitotic phosphorylation in formation and activation of APC/CCDC20 has remained unknown so far, in part because methods for the generation of recombinant human APC/C complexes have been developed only recently (53, 54) and because more than 100 different phospho-sites have been identified on the 14 different types of APC/C subunits (refs. 22, 43–45 and this study). The latter situation implied that mutagenic approaches to study APC/C phospho-regulation would be labor intensive, that numerous different combinations of phospho-sites mutations might have to be tested to identify the functionally relevant ones, and that simultaneous mutation of multiple sites could inactivate APC/C by means other than making it nonphosphorylatable.
To overcome the first two limitations, we developed a method (biGBac) for the rapid assembly of multiple cDNAs or genes into baculoviral genomes (42). Here, we used this method to generate and functionally characterize 47 different recombinant versions of the APC/C in which we simultaneously changed up to 68 phospho-site amino acid residues at once. Importantly, the nonphosphorylatable APC/C-pA mutant that we have generated as part of this series lost ubiquitination activity specifically in the presence of CDC20 but not in the presence of CDH1, whereas several phospho-mimicking mutants that we generated, such as APC/C-pE, gained CDC20-dependent activity in the absence of mitotic phosphorylation. These findings eliminate the possibility that the simultaneous mutation of multiple residues (possibly up to 100 sites in APC/C-pA and APC/C-pE given that some APC/C subunits are present in two copies per complex) might have altered APC/C through more general structural defects, a possibility that could not be excluded in a previous study in which mutagenesis of 12 CDK1 consensus recognition sites in budding yeast Cdc27 (orthologous to APC3), Cdc16 (APC6), and Cdc23 (APC8) caused a delay in APC/CCDC20 activation (55).
Our functional analyses of these APC/C variants revealed several important results. Two of these were very unexpected—namely, that mutation of only three to four phospho-sites in an N-terminal loop of APC1 is sufficient for partial activation of APC/C by CDC20, whereas mutation of 10 sites in this loop created a form of APC/C that supported substrate degradation in interphase Xenopus egg extracts as well as APC/C-pE did. Even more strikingly, we found that deletion of this loop enables APC/CCDC20 activation also in the absence of mitotic APC/C phosphorylation. If so, why is APC/C phosphorylated during mitosis on so many other sites, which are present on at least another nine subunits?
Interestingly, in Xenopus egg extracts, phosphorylation and activation of APC/CCDC20 occur with ultrasensitive kinetics; that is, APC/CCDC20 does not respond with Michaelis–Menten kinetics but instead responds in an all-or-none “switch-like” fashion to an increasing level of CDK1 activity (56). This unusual behavior results in a delay between CDK1 and APC/CCDC20 activation (56, 57), which might be required to allow sufficient time for spindle assembly in early Xenopus embryos where APC/CCDC20 activity is not tightly controlled by the SAC (58). It is possible that mitotic multisite phosphorylation of the APC/C on subunits other than APC1 is required for the ultrasensitivity or other kinetic properties of APC/CCDC20 activation, for example if phospho-sites on other subunits increased the affinity for mitotic kinases, which then subsequently could phosphorylate APC1. Consistent with this possibility, CDK1 contains a phospho-site binding subunit, called CKS1 (also known as p9 in Xenopus and Suc1 in fission yeast), which is required for efficient binding and phosphorylation of APC/C by CDK1, activation of APC/CCDC20, and cyclin B degradation (25, 59–61). Likewise, PLK1 contains a phospho-site binding domain, the Polo-box, which enables the kinase to bind and phosphorylate preferentially substrates (including APC/C) (62) that have been “primed” for recognition by prior phosphorylation (63). Consistent with the possibility that phosphorylation of APC/C subunits other than APC1 affects the interaction of APC/C with mitotic kinases and alters the kinetics of APC/CCDC20 activation, we observed previously that deletion of a loop region in APC3 containing most of its phospho-sites delayed but did not abolish APC/CCDC20 activation in Xenopus egg extracts (64). In addition, mitotic phosphorylation could control APC/CCDC20 by other mechanisms. Support for this possibility comes from the observation that CDC20 phosphorylation inhibits APC/CCDC20 activation and UBE2S recruitment (32–35, 65, 66). Furthermore, it has been reported that the ability of APC/C to bind the D-box becomes activated in mitosis in a CDC20-independent manner, raising the possibility that APC/C phosphorylation could increase APC/C–substrate interactions by means other than promoting binding of CDC20 to the APC/C (67).
Our findings also raise the important question of how phosphorylation of the N-terminal loop region of APC1 allows APC/CCDC20 activation. Our finding that deletion of this loop enables binding and activation of APC/C by CDC20 in the absence of mitotic phosphorylation suggests that these modifications expose a CDC20 binding site on the APC/C that is not accessible if APC1 is not phosphorylated. What could this binding site be? The autoinhibitory loop of APC1 is located close to the C-box binding site and also close to contacts between APC1 and the NTD of CDC20 (Fig. S6). It is therefore possible that APC1 phosphorylation controls the access of CDC20’s NTD to these binding sites, possibly by steric hindrance, which is only relieved if APC1’s N-terminal loop region becomes phosphorylated (Fig. 5E). If so, it will be interesting to test why phosphorylation of APC1’s autoinhibitory loop is required for binding of CDC20, but not binding of CDH1, and if this differential behavior is determined by structural differences in their NTDs.
Fig. S6.
Location of the APC1(296–401) loop relative to CDH1 NTD. Parts of the APC/C–CDH1–EMI1 structure (PDB ID code 4UI9) (13) are shown with APC1 (green), APC8B (blue), APC6B (purple), and CDH1 NTD (orange). Subunits are shown as surface representations, except for the APC1 WD40 domain, which is shown as a cartoon. The APC1 WD40 domain containing the flanking residues of the APC1(296–401) loop (red spheres) is close to CDH1 NTD interaction sites with APC1 PC domain, APC6B, and APC8B.
Materials and Methods
Antibodies against pApc1 (phosphorylated S355), APC1, APC3, pAPC3, pAPC8, CDH1, and CDC20 have been previously described (22, 68). Polyclonal SMC3 antibodies were generated by Eurogenetec against a synthetic peptide CEMAKDFVEDDTTHG. Polyclonal securin antibodies were generated by Gramsch Laboratories against full-length recombinant protein. In addition, the following commercial antibodies were used: APC8 (1:200, A301-181A; Bethyl Laboratories), Xenopus laevis CDC20 (1:1,000, ab18217; Abcam), and cyclin B1 (1:1,000, MS-868-PABX; Thermo Scientific). Recombinant APC/C, CDH1, CDC20, and E1 were expressed in insect cells. Baculoviral APC/C expression vectors were generated using biGBac (42). All other recombinant proteins were produced in Escherichia coli as previously described (7, 19, 64). Details of ubiquitination reactions, cryo-EM, and substrate degradation assays in Xenopus egg extracts can be found in SI Materials and Methods.
SI Materials and Methods
Cell Culture.
HeLa S3 cells were grown in DMEM including 10% (vol/vol) FBS (Invitrogen), 2 mM l-glutamine, and 100 μg/mL penicillin/streptomycin (both from Sigma) and plated on 245 mm × 245 mm tissue culture dishes. Cells were arrested in S-phase by addition of 2.5 mM thymidine for 24 h. To obtain a cell population arrested in mitosis, we supplemented the culture media with nocodazole (Sigma) (100 nM, 16 h) or taxol (Sigma) (10 μM, 17.6 h). HeLa cells were arrested in prometaphase when SAC is active by a 17.6-h taxol block and by adding the proteasome inhibitor MG132 (10 μM) for the last 1.6 h (SAC on). The SAC arrest was overcome by addition of hesperadin (46) (100 nM) for the last hour, and cell-cycle progression was halted in metaphase due to the presence of MG132 (SAC off). We also used a double-thymidine arrest–release procedure followed by mitotic shake-off to collect mitotic cells.
Sample Preparation for Mass Spectrometry by IP.
Antibody cross-linking to beads (Affi-prep protein A beads; BioRad) was performed as previously described (69–71). APC/C was affinity-purified from synchronized mitotic HeLa cells using APC3 antibody–cross-linked beads. The native complex was eluted with two-bead volumes of 1 mg/mL antigenic peptide dissolved in IP buffer [20 mM Tris·HCl pH 7.5, 150 mM NaCl, 10% (vol/vol) glycerol, 0.1% Tween-20]. Two volumes of APC/C peptide eluate were incubated with one volume of CDC20 antibody-coupled beads (CDC20–re-IP). Beads were washed twice with 20-bead volumes of IP buffer and twice with 20-bead volumes of wash buffer [20 mM Tris·HCl pH 7.5, 100 mM NaCl, 5% (vol/vol) glycerol] for 5 min at 4 °C, and APC/C was recovered by elution with 100 mM glycine–HCl pH 2.2.
Nano-Liquid Chromatography–MS Analysis.
Immunopurified APC/C was digested in solution with different proteases (trypsin, chymotrypsinogen, Lys-C, and Glu-C) according to ref. 72. Proteolytic peptides were separated by nano-HPLC, which was an UltiMate 3000 HPLC RSLC nano system (Thermo Fisher Scientific) coupled to a Q Exactive mass spectrometer (Thermo Fisher Scientific), equipped with a Proxeon nanospray source (Thermo Fisher Scientific). Peptides were loaded onto a trap column [Thermo Fisher Scientific; PepMap C18, 5 mm × 300 μm inner diameter (ID), 5 μm particles, 100 Å pore size] at a flow rate of 25 μL·min−1 using 0.1% TFA as mobile phase. After 10 min, the trap column was switched in line with the analytical column (Thermo Fisher Scientific; PepMap C18, 500 mm × 75 μm ID, 2 μm, 100 Å). Peptides were eluted using a flow rate of 230 nL·min−1 and a binary 2-h gradient, respectively, for 165 min. The gradient starts with the mobile phases: 98% A (water/formic acid, 99.9/0.1, vol/vol) and 2% B (water/acetonitrile/formic acid, 19.92/80/0.08, vol/vol/v) increases to 35% B over the next 120 min, followed by a gradient in 5 min to 90% B; stays there for 5 min; and decreases in 5 min back to the gradient 98% A and 2% B for equilibration at 30 °C. The Q Exactive mass spectrometer was operated in data-dependent mode, using a full scan (m/z range 380–1650, nominal resolution of 70, 000, target value 3E6) followed by MS/MS scans of the 12 most abundant ions. MS/MS spectra were acquired using a normalized collision energy of 27%, an isolation width of 2, and the target value set to 1E5. Precursor ions selected for fragmentation (charge state 2 and higher) were put on a dynamic exclusion list for 30 s. Additionally, the underfill ratio was set to 20%, resulting in an intensity threshold of 4E4. The peptide match feature and the exclude isotopes feature were enabled.
For peptide identification, the .RAW files were loaded into Proteome Discoverer (version 1.4.0.288; Thermo Scientific). All created MS/MS spectra were searched using Mascot 2.2.07 (Matrix Science) and, for additional confidence, the search node MSAmanda v1.4.14.5290 (73) against the Human Swissprot Protein Sequence Database. The following search parameters were used: Beta-methylthiolation on cysteine was set as a fixed modification, and oxidation on methionine, acetylation on lysine and protein N terminus, and phosphorylation on serine, threonine, and tyrosine were set as variable modifications. Monoisotopic masses were searched within unrestricted protein masses for tryptic, chymotryptic, Lys-C, and Glu-C enzymatic specificity. The peptide mass tolerance was set to ±5 ppm and the fragment mass tolerance to ±30 mmu. The maximal number of missed cleavages was set to 2. The result was filtered to 1% FDR (false discovery rates) using Percolator algorithm integrated in Thermo Proteome Discoverer. The localization of the phosphorylation sites within the peptides was performed with the tool phosphoRS (74). All of the identified phosphorylation sites were validated by manual inspection of the corresponding MS spectra.
Recombinant Proteins.
Baculoviral multigene expression transfer vectors were generated using biGBac (42). All recombinant APC/C complexes were expressed in High Five (Thermo Fisher Scientific) or Sf9 insect cells (Thermo Fisher Scientific) either by coinfection with APC/C (14 subunits) and Platform (APC1/2/4-Strep/5/11/15) baculoviruses or by coinfection with Arclamp (APC3/6/7/8/10/12/13/16) and Platform baculoviruses. In wt_APC1Δloop, residues 296–401 of APC1 were replaced by the linker AT(GGS)4TE.
For experiments other than EM, recombinant APC/C was purified by a three-step scheme as previously described: (i) affinity purification with Strep-Tactin Sepharose (IBA Life Sciences) and elution with desthiobiotin (Sigma-Aldrich), (ii) anion exchange with gradient NaCl elution, and (iii) size exclusion chromatography (SEC) into a final buffer of 20 mM Hepes pH 8.0, 200 mM NaCl, and 1 mM DTT (19, 75). UBA1, UBE2C, UBE2S, and donor Ub were purified as described (7, 19, 64). Ub-securin*, CycBNTD*, and Ub* were purified and fluorescently labeled, as denoted by an asterisk, with fluorescein-5 maleimide, as described previously (7, 19, 64). The 3xFLAG-CDC20 was expressed in High Five insect cells and purified by FLAG affinity chromatography (GenScript). The 3xMyc-His6-CDC20 was expressed in Sf9 insect cells and purified by nickel affinity chromatography. The 3xFLAG tag or 3xMyc-His6 tag was cleaved by TEV (tobacco etch virus) protease, and CDC20 was further purified by cation exchange chromatography and SEC. The 3xMyc-His6-CDH1 was expressed in High Five or Sf9 insect cells and purified by nickel affinity chromatography, affinity tag cleavage by HRV13 3C protease, and SEC. For CDC20 and CDH1, the final sizing buffer was 20 mM Hepes pH 7.0, 300 mM (NH4)2SO4, 2 mM DTT, and 2.5% (vol/vol) glycerol. Δ90 cyclin B1, cyclin B1(1–87), and securin were purified as described (7, 16, 19, 64).
Sample Preparation for EM.
Preparation of recombinant APC/C for use in structural studies by negative stain EM was described previously (7). APC/C-WT, APC/C-pA, or APC/C-pE was expressed in High Five insect cells and colysed with recombinant CDC20 or CDH1, expressed in the same cell line. The complexes were then purified by affinity chromatography using a HRV13 3C protease cleavable Twin-Strep tag at the C terminus of APC4. The eluate was incubated with a three-way cross-linked complex of UBE2C, FLAG-Ub, and HSL1 peptide (7), anti-FLAG affinity gel, and HRV13 3C protease for 1 h. The three-way cross-linked complex was formed as previously described, except the Strep affinity tag was removed from UBE2C by HRV13 3C protease. The complexes were thoroughly washed and then eluted by the addition of FLAG peptide. We loaded 110 μg of purified APC/C onto a GraFix gradient (76), consisting of 10–40% (vol/vol) glycerol, 0.025–0.1% glutaraldehyde, 50 mM Hepes pH 8.0, 200 mM NaCl, and 2 mM MgCl2, and centrifugation was performed at 34,000 rpm in a SW55TI rotor (Beckman) for 15 h at 4 °C. The peak protein fraction of the gradient, as determined by BioRad protein assay, was used for EM studies.
Enzyme Assays.
Qualitative APC/C-mediated ubiquitination assays were performed as previously described (7, 19, 64). For kinetic analysis, ubiquitination product bands were quantitated based on the fluorescently labeled substrate using a Typhoon FLA PhosphorImager. Coactivator-independent ubiquitination products were subtracted as background to determine the coactivator-dependent activity.
The fitting of the initial velocities to the hyperbolic, v = Vmaxapp[X]/(1/2 max coactivator + [X]), equation using GraphPad Prism 6 software, where X is either CDC20 or CDH1, allowed for the concentration resulting in half-maximal stimulation (1/2 max) and apparent Vmax (Vmaxapp) values for coactivator-mediated ubiquitination activity with the APC/C to be determined. In summary, the 1/2 max and Vmaxapp of the coactivators were determined by titrating either CDC20 or CDH1 against either 20 or 10 nM APC/C, respectively, supplemented with 1 μM Ub-securin*, 5 mM MgCl2, 5 mM ATP, 0.25 mg/mL BSA, and 1 μM UBA1, respectively. UBE2C and UBE2S were used at 2 μM and 1 μM, respectively. The reactions were initiated by the addition of 0.3 mM Ub and quenched at single time points under conditions that satisfy initial velocity regimes.
For screening of APC/C versions containing combinations of WT and glutamate substitutions, qualitative ubiquitination assays were performed using either fluorescein-labeled ubiquitin (Ub*) or fluorescein-labeled substrate cyclin B NTD (CycBNTD*), as previously described (19, 75). For fluorescein-labeled ubiquitin assays, 1 µM coactivator, 0.25 µM UBE2C, 0.25 µM UBE2S, 2 µM GST-CycBNTD, 0.25 mg/mL BSA, 2.5 mM MgATP, and ∼10 nM APC/C were mixed, and reactions were initiated by addition of 0.1 mM Ub* and 0.1 µM UBA1 and quenched after 20 min. For fluorescein-labeled substrate assays, 20 nM APC/C, 1 µM coactivator, 0.65 µM UBE2C, 0.125 µM UBE2S, 0.5 µM CycBNTD*, 0.25 mg/mL BSA, and 2.5 mM MgATP were mixed, and reactions were initiated by addition of 0.1 mM Ub and 0.1 µM UBA1 and quenched after 30 min.
CDC20 and Securin Binding Assay.
We mixed 7.5 nM recombinant APC/C with 100 nM CDC20 and 250 nM or 2.5 μM securin in binding buffer (20 mM Tris pH 7.5, 250 mM KCl, 0.05% Tween-20, and 4 mg/mL BSA) and immunoprecipitated it with APC3 antibody beads at room temperature for 1 h. The beads were washed four times with wash buffer (20 mM Tris pH 7.5, 250 mM KCl, 0.05% Tween-20) at 4 °C for 5 min, and then the bound proteins were eluted from the antibody beads using three-bead volumes of 2 mg/mL immunogenic peptide in wash buffer for 1 h rotating at 4 °C.
For binding assays in Xenopus egg extracts, 25 μL APC/C-depleted interphase egg extracts were mixed with 1.66 μg recombinant APC/C and 20 μL APC3 antibody beads and then rotated at room temperature for 2 h. The bound proteins were eluted by two-bead volumes of 2 mg/mL immunogenic peptide in TBST (Tris-buffered saline and Tween 20, 20 mM Tris pH 7.5, 150 mM NaCl, 0.1% Tween-20) and rotating for 1 h at 4 °C.
Substrate Degradation Assays in Xenopus Egg Extracts.
Recombinant human APC/C-dependent substrate degradation experiments in Xenopus egg extracts depleted of endogenous APC/C were performed as previously described (64). In brief, interphase egg extracts were prepared as previously described (77), except that eggs were activated with Ca2+ ionophore A23187 (Calbiochem) at 0.6 μg/mL in Marc’s modified Ringer’s for 4 min. Cycloheximide was added to 50 μg/mL to arrest the extract in interphase. To deplete APC/C, we mixed 70 μL of egg extracts with 2.5 μg of anti-APC3 antibody coupled to 10.5 μL of Affiprep Protein A beads and incubated it at 4 °C for 40 min, twice. To deplete CDC20, we mixed 50 μL of egg extracts with 50 μg of anti-Xenopus CDC20 antibody (78) coupled to 12 μL of Affiprep Protein A beads and incubated it at 4 °C for 40 min, twice. Approximately 1.05 μg of recombinant APC/C was added to 15 μL of APC/C-depleted extract. Interphase egg extracts were induced to enter mitosis by addition of nondegradable cyclin B(Δ90) at 300 nM for 90–120 min before assay. Reactions were incubated at 22 °C for the indicated times after recombinant human securin and CycBNTD addition, and the reactions were quenched with SDS/PAGE sample buffer and boiled for 3 min.
Supplementary Material
Acknowledgments
We thank H. Yamano for CDC20 antibodies, J.J. Blow for ORC2 antisera, G. Petzold for recombinant securin, O. Hudecz for mass spectrometry data analysis, and M. Madalinski for peptide synthesis. For funding, we thank Boehringer Ingelheim, the Austrian Research Promotion Agency (Laura Bassi Centre for Optimized Structural Studies), the European Union (Seventh Framework Programme Grant 227764 MitoSys), the Austrian Science Fund (SFB-F34 and Wittgenstein award, to J.-M.P.), Hertha Firnberg Program of the Austrian Science Fund (R.Q.), Jane Coffin Childs Foundation, Leukemia & Lymphoma Society (N.G.B.), Japan Society for the Promotion of Science (M.Y.), Deutsche Forschungsgemeinschaft (DFG) Sonderforschungsbereich 860 (to H.S.), American Lebanese Syrian Associated Charities, NIH Grants R37GM065930 and P30CA021765, and the Howard Hughes Medical Institute (B.A.S.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1604929113/-/DCSupplemental.
References
- 1.Irniger S, Piatti S, Michaelis C, Nasmyth K. Genes involved in sister chromatid separation are needed for B-type cyclin proteolysis in budding yeast. Cell. 1995;81(2):269–278. doi: 10.1016/0092-8674(95)90337-2. [DOI] [PubMed] [Google Scholar]
- 2.King RW, et al. A 20S complex containing CDC27 and CDC16 catalyzes the mitosis-specific conjugation of ubiquitin to cyclin B. Cell. 1995;81(2):279–288. doi: 10.1016/0092-8674(95)90338-0. [DOI] [PubMed] [Google Scholar]
- 3.Peters JM, King RW, Höög C, Kirschner MW. Identification of BIME as a subunit of the anaphase-promoting complex. Science. 1996;274(5290):1199–1201. doi: 10.1126/science.274.5290.1199. [DOI] [PubMed] [Google Scholar]
- 4.Sudakin V, et al. The cyclosome, a large complex containing cyclin-selective ubiquitin ligase activity, targets cyclins for destruction at the end of mitosis. Mol Biol Cell. 1995;6(2):185–197. doi: 10.1091/mbc.6.2.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zachariae W, Shin TH, Galova M, Obermaier B, Nasmyth K. Identification of subunits of the anaphase-promoting complex of Saccharomyces cerevisiae. Science. 1996;274(5290):1201–1204. doi: 10.1126/science.274.5290.1201. [DOI] [PubMed] [Google Scholar]
- 6.Primorac I, Musacchio A. Panta rhei: The APC/C at steady state. J Cell Biol. 2013;201(2):177–189. doi: 10.1083/jcb.201301130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brown NG, et al. RING E3 mechanism for ubiquitin ligation to a disordered substrate visualized for human anaphase-promoting complex. Proc Natl Acad Sci USA. 2015;112(17):5272–5279. doi: 10.1073/pnas.1504161112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chang L, Zhang Z, Yang J, McLaughlin SH, Barford D. Molecular architecture and mechanism of the anaphase-promoting complex. Nature. 2014;513(7518):388–393. doi: 10.1038/nature13543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dube P, et al. Localization of the coactivator Cdh1 and the cullin subunit Apc2 in a cryo-electron microscopy model of vertebrate APC/C. Mol Cell. 2005;20(6):867–879. doi: 10.1016/j.molcel.2005.11.008. [DOI] [PubMed] [Google Scholar]
- 10.Peters JM. SCF and APC: The yin and yang of cell cycle regulated proteolysis. Curr Opin Cell Biol. 1998;10(6):759–768. doi: 10.1016/s0955-0674(98)80119-1. [DOI] [PubMed] [Google Scholar]
- 11.Vodermaier HC, Gieffers C, Maurer-Stroh S, Eisenhaber F, Peters JM. TPR subunits of the anaphase-promoting complex mediate binding to the activator protein CDH1. Curr Biol. 2003;13(17):1459–1468. doi: 10.1016/s0960-9822(03)00581-5. [DOI] [PubMed] [Google Scholar]
- 12.Schwab M, Neutzner M, Möcker D, Seufert W. Yeast Hct1 recognizes the mitotic cyclin Clb2 and other substrates of the ubiquitin ligase APC. EMBO J. 2001;20(18):5165–5175. doi: 10.1093/emboj/20.18.5165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chang L, Zhang Z, Yang J, McLaughlin SH, Barford D. Atomic structure of the APC/C and its mechanism of protein ubiquitination. Nature. 2015;522(7557):450–454. doi: 10.1038/nature14471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kraft C, Vodermaier HC, Maurer-Stroh S, Eisenhaber F, Peters JM. The WD40 propeller domain of Cdh1 functions as a destruction box receptor for APC/C substrates. Mol Cell. 2005;18(5):543–553. doi: 10.1016/j.molcel.2005.04.023. [DOI] [PubMed] [Google Scholar]
- 15.Matyskiela ME, Morgan DO. Analysis of activator-binding sites on the APC/C supports a cooperative substrate-binding mechanism. Mol Cell. 2009;34(1):68–80. doi: 10.1016/j.molcel.2009.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Glotzer M, Murray AW, Kirschner MW. Cyclin is degraded by the ubiquitin pathway. Nature. 1991;349(6305):132–138. doi: 10.1038/349132a0. [DOI] [PubMed] [Google Scholar]
- 17.Williamson A, et al. Identification of a physiological E2 module for the human anaphase-promoting complex. Proc Natl Acad Sci USA. 2009;106(43):18213–18218. doi: 10.1073/pnas.0907887106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yu H, King RW, Peters JM, Kirschner MW. Identification of a novel ubiquitin-conjugating enzyme involved in mitotic cyclin degradation. Curr Biol. 1996;6(4):455–466. doi: 10.1016/s0960-9822(02)00513-4. [DOI] [PubMed] [Google Scholar]
- 19.Brown NG, et al. Mechanism of polyubiquitination by human anaphase-promoting complex: RING repurposing for ubiquitin chain assembly. Mol Cell. 2014;56(2):246–260. doi: 10.1016/j.molcel.2014.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kelly A, Wickliffe KE, Song L, Fedrigo I, Rape M. Ubiquitin chain elongation requires E3-dependent tracking of the emerging conjugate. Mol Cell. 2014;56(2):232–245. doi: 10.1016/j.molcel.2014.09.010. [DOI] [PubMed] [Google Scholar]
- 21.Golan A, Yudkovsky Y, Hershko A. The cyclin-ubiquitin ligase activity of cyclosome/APC is jointly activated by protein kinases Cdk1-cyclin B and Plk. J Biol Chem. 2002;277(18):15552–15557. doi: 10.1074/jbc.M111476200. [DOI] [PubMed] [Google Scholar]
- 22.Kraft C, et al. Mitotic regulation of the human anaphase-promoting complex by phosphorylation. EMBO J. 2003;22(24):6598–6609. doi: 10.1093/emboj/cdg627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kramer ER, Scheuringer N, Podtelejnikov AV, Mann M, Peters JM. Mitotic regulation of the APC activator proteins CDC20 and CDH1. Mol Biol Cell. 2000;11(5):1555–1569. doi: 10.1091/mbc.11.5.1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lahav-Baratz S, Sudakin V, Ruderman JV, Hershko A. Reversible phosphorylation controls the activity of cyclosome-associated cyclin-ubiquitin ligase. Proc Natl Acad Sci USA. 1995;92(20):9303–9307. doi: 10.1073/pnas.92.20.9303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Patra D, Dunphy WG. Xe-p9, a Xenopus Suc1/Cks protein, is essential for the Cdc2-dependent phosphorylation of the anaphase-promoting complex at mitosis. Genes Dev. 1998;12(16):2549–2559. doi: 10.1101/gad.12.16.2549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rudner AD, Hardwick KG, Murray AW. Cdc28 activates exit from mitosis in budding yeast. J Cell Biol. 2000;149(7):1361–1376. doi: 10.1083/jcb.149.7.1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shteinberg M, Protopopov Y, Listovsky T, Brandeis M, Hershko A. Phosphorylation of the cyclosome is required for its stimulation by Fizzy/cdc20. Biochem Biophys Res Commun. 1999;260(1):193–198. doi: 10.1006/bbrc.1999.0884. [DOI] [PubMed] [Google Scholar]
- 28.Blanco MA, Sánchez-Díaz A, de Prada JM, Moreno S. APC(ste9/srw1) promotes degradation of mitotic cyclins in G(1) and is inhibited by cdc2 phosphorylation. EMBO J. 2000;19(15):3945–3955. doi: 10.1093/emboj/19.15.3945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jaspersen SL, Charles JF, Morgan DO. Inhibitory phosphorylation of the APC regulator Hct1 is controlled by the kinase Cdc28 and the phosphatase Cdc14. Curr Biol. 1999;9(5):227–236. doi: 10.1016/s0960-9822(99)80111-0. [DOI] [PubMed] [Google Scholar]
- 30.Yamaguchi S, Okayama H, Nurse P. Fission yeast Fizzy-related protein srw1p is a G(1)-specific promoter of mitotic cyclin B degradation. EMBO J. 2000;19(15):3968–3977. doi: 10.1093/emboj/19.15.3968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zachariae W, Schwab M, Nasmyth K, Seufert W. Control of cyclin ubiquitination by CDK-regulated binding of Hct1 to the anaphase promoting complex. Science. 1998;282(5394):1721–1724. doi: 10.1126/science.282.5394.1721. [DOI] [PubMed] [Google Scholar]
- 32.Yudkovsky Y, Shteinberg M, Listovsky T, Brandeis M, Hershko A. Phosphorylation of Cdc20/fizzy negatively regulates the mammalian cyclosome/APC in the mitotic checkpoint. Biochem Biophys Res Commun. 2000;271(2):299–304. doi: 10.1006/bbrc.2000.2622. [DOI] [PubMed] [Google Scholar]
- 33.Labit H, et al. Dephosphorylation of Cdc20 is required for its C-box-dependent activation of the APC/C. EMBO J. 2012;31(15):3351–3362. doi: 10.1038/emboj.2012.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Craney A, et al. Control of APC/C-dependent ubiquitin chain elongation by reversible phosphorylation. Proc Natl Acad Sci USA. 2016;113(6):1540–1545. doi: 10.1073/pnas.1522423113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hein JB, Nilsson J. Interphase APC/C-Cdc20 inhibition by cyclin A2-Cdk2 ensures efficient mitotic entry. Nat Commun. 2016;7:10975. doi: 10.1038/ncomms10975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hardwick KG, Johnston RC, Smith DL, Murray AW. MAD3 encodes a novel component of the spindle checkpoint which interacts with Bub3p, Cdc20p, and Mad2p. J Cell Biol. 2000;148(5):871–882. doi: 10.1083/jcb.148.5.871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sudakin V, Chan GK, Yen TJ. Checkpoint inhibition of the APC/C in HeLa cells is mediated by a complex of BUBR1, BUB3, CDC20, and MAD2. J Cell Biol. 2001;154(5):925–936. doi: 10.1083/jcb.200102093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Reimann JD, et al. Emi1 is a mitotic regulator that interacts with Cdc20 and inhibits the anaphase promoting complex. Cell. 2001;105(5):645–655. doi: 10.1016/s0092-8674(01)00361-0. [DOI] [PubMed] [Google Scholar]
- 39.Jia L, Kim S, Yu H. Tracking spindle checkpoint signals from kinetochores to APC/C. Trends Biochem Sci. 2013;38(6):302–311. doi: 10.1016/j.tibs.2013.03.004. [DOI] [PubMed] [Google Scholar]
- 40.Izawa D, Pines J. The mitotic checkpoint complex binds a second CDC20 to inhibit active APC/C. Nature. 2015;517(7536):631–634. doi: 10.1038/nature13911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Herzog F, et al. Structure of the anaphase-promoting complex/cyclosome interacting with a mitotic checkpoint complex. Science. 2009;323(5920):1477–1481. doi: 10.1126/science.1163300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Weissmann F, et al. biGBac enables rapid gene assembly for the expression of large multisubunit protein complexes. Proc Natl Acad Sci USA. 2016;113:E2564–E2569. doi: 10.1073/pnas.1604935113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hegemann B, et al. Systematic phosphorylation analysis of human mitotic protein complexes. Sci Signal. 2011;4(198):rs12. doi: 10.1126/scisignal.2001993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Herzog F, Mechtler K, Peters JM. Identification of cell cycle-dependent phosphorylation sites on the anaphase-promoting complex/cyclosome by mass spectrometry. Methods Enzymol. 2005;398:231–245. doi: 10.1016/S0076-6879(05)98019-1. [DOI] [PubMed] [Google Scholar]
- 45.Steen JA, et al. Different phosphorylation states of the anaphase promoting complex in response to antimitotic drugs: A quantitative proteomic analysis. Proc Natl Acad Sci USA. 2008;105(16):6069–6074. doi: 10.1073/pnas.0709807104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hauf S, et al. The small molecule Hesperadin reveals a role for Aurora B in correcting kinetochore-microtubule attachment and in maintaining the spindle assembly checkpoint. J Cell Biol. 2003;161(2):281–294. doi: 10.1083/jcb.200208092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jiménez JL, Hegemann B, Hutchins JR, Peters JM, Durbin R. A systematic comparative and structural analysis of protein phosphorylation sites based on the mtcPTM database. Genome Biol. 2007;8(5):R90. doi: 10.1186/gb-2007-8-5-r90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Félix MA, Cohen P, Karsenti E. Cdc2 H1 kinase is negatively regulated by a type 2A phosphatase in the Xenopus early embryonic cell cycle: Evidence from the effects of okadaic acid. EMBO J. 1990;9(3):675–683. doi: 10.1002/j.1460-2075.1990.tb08159.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Murray AW, Solomon MJ, Kirschner MW. The role of cyclin synthesis and degradation in the control of maturation promoting factor activity. Nature. 1989;339(6222):280–286. doi: 10.1038/339280a0. [DOI] [PubMed] [Google Scholar]
- 50.Fang G, Yu H, Kirschner MW. Direct binding of CDC20 protein family members activates the anaphase-promoting complex in mitosis and G1. Mol Cell. 1998;2(2):163–171. doi: 10.1016/s1097-2765(00)80126-4. [DOI] [PubMed] [Google Scholar]
- 51.Rankin S, Ayad NG, Kirschner MW. Sororin, a substrate of the anaphase-promoting complex, is required for sister chromatid cohesion in vertebrates. Mol Cell. 2005;18(2):185–200. doi: 10.1016/j.molcel.2005.03.017. [DOI] [PubMed] [Google Scholar]
- 52.Van Voorhis VA, Morgan DO. Activation of the APC/C ubiquitin ligase by enhanced E2 efficiency. Curr Biol. 2014;24(13):1556–1562. doi: 10.1016/j.cub.2014.05.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Uzunova K, et al. APC15 mediates CDC20 autoubiquitylation by APC/C(MCC) and disassembly of the mitotic checkpoint complex. Nat Struct Mol Biol. 2012;19(11):1116–1123. doi: 10.1038/nsmb.2412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhang Z, et al. Recombinant expression, reconstitution and structure of human anaphase-promoting complex (APC/C) Biochem J. 2013;449(2):365–371. doi: 10.1042/BJ20121374. [DOI] [PubMed] [Google Scholar]
- 55.Rudner AD, Murray AW. Phosphorylation by Cdc28 activates the Cdc20-dependent activity of the anaphase-promoting complex. J Cell Biol. 2000;149(7):1377–1390. doi: 10.1083/jcb.149.7.1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yang Q, Ferrell JE., Jr The Cdk1-APC/C cell cycle oscillator circuit functions as a time-delayed, ultrasensitive switch. Nat Cell Biol. 2013;15(5):519–525. doi: 10.1038/ncb2737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Georgi AB, Stukenberg PT, Kirschner MW. Timing of events in mitosis. Curr Biol. 2002;12(2):105–114. doi: 10.1016/s0960-9822(01)00662-5. [DOI] [PubMed] [Google Scholar]
- 58.Minshull J, Sun H, Tonks NK, Murray AW. A MAP kinase-dependent spindle assembly checkpoint in Xenopus egg extracts. Cell. 1994;79(3):475–486. doi: 10.1016/0092-8674(94)90256-9. [DOI] [PubMed] [Google Scholar]
- 59.Patra D, Dunphy WG. Xe-p9, a Xenopus Suc1/Cks homolog, has multiple essential roles in cell cycle control. Genes Dev. 1996;10(12):1503–1515. doi: 10.1101/gad.10.12.1503. [DOI] [PubMed] [Google Scholar]
- 60.Shteinberg M, Hershko A. Role of Suc1 in the activation of the cyclosome by protein kinase Cdk1/cyclin B. Biochem Biophys Res Commun. 1999;257(1):12–18. doi: 10.1006/bbrc.1999.0409. [DOI] [PubMed] [Google Scholar]
- 61.Sudakin V, Shteinberg M, Ganoth D, Hershko J, Hershko A. Binding of activated cyclosome to p13(suc1). Use for affinity purification. J Biol Chem. 1997;272(29):18051–18059. doi: 10.1074/jbc.272.29.18051. [DOI] [PubMed] [Google Scholar]
- 62.May KM, Reynolds N, Cullen CF, Yanagida M, Ohkura H. Polo boxes and Cut23 (Apc8) mediate an interaction between polo kinase and the anaphase-promoting complex for fission yeast mitosis. J Cell Biol. 2002;156(1):23–28. doi: 10.1083/jcb.200106150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Elia AE, Cantley LC, Yaffe MB. Proteomic screen finds pSer/pThr-binding domain localizing Plk1 to mitotic substrates. Science. 2003;299(5610):1228–1231. doi: 10.1126/science.1079079. [DOI] [PubMed] [Google Scholar]
- 64.Yamaguchi M, et al. Structure of an APC3-APC16 complex: Insights into assembly of the anaphase-promoting complex/cyclosome. J Mol Biol. 2015;427(8):1748–1764. doi: 10.1016/j.jmb.2014.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chung E, Chen RH. Phosphorylation of Cdc20 is required for its inhibition by the spindle checkpoint. Nat Cell Biol. 2003;5(8):748–753. doi: 10.1038/ncb1022. [DOI] [PubMed] [Google Scholar]
- 66.Tang Z, Shu H, Oncel D, Chen S, Yu H. Phosphorylation of Cdc20 by Bub1 provides a catalytic mechanism for APC/C inhibition by the spindle checkpoint. Mol Cell. 2004;16(3):387–397. doi: 10.1016/j.molcel.2004.09.031. [DOI] [PubMed] [Google Scholar]
- 67.Yamano H, Gannon J, Mahbubani H, Hunt T. Cell cycle-regulated recognition of the destruction box of cyclin B by the APC/C in Xenopus egg extracts. Mol Cell. 2004;13(1):137–147. doi: 10.1016/s1097-2765(03)00480-5. [DOI] [PubMed] [Google Scholar]
- 68.Kramer ER, Gieffers C, Hölzl G, Hengstschläger M, Peters JM. Activation of the human anaphase-promoting complex by proteins of the CDC20/Fizzy family. Curr Biol. 1998;8(22):1207–1210. doi: 10.1016/s0960-9822(07)00510-6. [DOI] [PubMed] [Google Scholar]
- 69.Buschhorn BA, et al. Substrate binding on the APC/C occurs between the coactivator Cdh1 and the processivity factor Doc1. Nat Struct Mol Biol. 2011;18(1):6–13. doi: 10.1038/nsmb.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Gieffers C, Dube P, Harris JR, Stark H, Peters JM. Three-dimensional structure of the anaphase-promoting complex. Mol Cell. 2001;7(4):907–913. doi: 10.1016/s1097-2765(01)00234-9. [DOI] [PubMed] [Google Scholar]
- 71.Herzog F, Peters JM. Large-scale purification of the vertebrate anaphase-promoting complex/cyclosome. Methods Enzymol. 2005;398:175–195. doi: 10.1016/S0076-6879(05)98016-6. [DOI] [PubMed] [Google Scholar]
- 72.Yates JR, 3rd, Link AJ, Schieltz D. Direct analysis of proteins in mixtures. Application to protein complexes. Methods Mol Biol. 2000;146:17–26. doi: 10.1385/1-59259-045-4:17. [DOI] [PubMed] [Google Scholar]
- 73.Dorfer V, et al. MS Amanda, a universal identification algorithm optimized for high accuracy tandem mass spectra. J Proteome Res. 2014;13(8):3679–3684. doi: 10.1021/pr500202e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Taus T, et al. Universal and confident phosphorylation site localization using phosphoRS. J Proteome Res. 2011;10(12):5354–5362. doi: 10.1021/pr200611n. [DOI] [PubMed] [Google Scholar]
- 75.Jarvis MA, et al. Measuring APC/C-dependent ubiquitylation in vitro. Methods Mol Biol. 2016;1342:287–303. doi: 10.1007/978-1-4939-2957-3_18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kastner B, et al. GraFix: Sample preparation for single-particle electron cryomicroscopy. Nat Methods. 2008;5(1):53–55. doi: 10.1038/nmeth1139. [DOI] [PubMed] [Google Scholar]
- 77.Gillespie PJ, Gambus A, Blow JJ. Preparation and use of Xenopus egg extracts to study DNA replication and chromatin associated proteins. Methods. 2012;57(2):203–213. doi: 10.1016/j.ymeth.2012.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Yamano H, Trickey M, Grimaldi M, Kimata Y. In vitro assays for the anaphase-promoting complex/cyclosome (APC/C) in Xenopus egg extracts. Methods Mol Biol. 2009;545:287–300. doi: 10.1007/978-1-60327-993-2_18. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.