Significance
African great apes were recently found to host a large diversity of parasites (subgenus Laverania) related to the main agent of human malaria (Plasmodium falciparum). Despite their close genetic relationships, these parasites are highly host-specific, infecting either chimpanzees or gorillas. This host specificity could result from incompatibilities between parasites and hosts or from a strong host tropism of the vectors. To test this second hypothesis, we performed a large entomological survey in the heart of the Gabonese rainforest (central Africa) to identify the vector species involved in ape Plasmodium transmission. Our results demonstrated that all ape parasites are transmitted by the same three vector species, thus rejecting the hypothesis that vectors could be responsible for the Laverania host specificity.
Keywords: Plasmodium, Laverania, Anopheles, ape-to-human infection, African rainforest
Abstract
Recent studies have highlighted the large diversity of malaria parasites infecting African great apes (subgenus Laverania) and their strong host specificity. Although the existence of genetic incompatibilities preventing the cross-species transfer may explain host specificity, the existence of vectors with a high preference for a determined host represents another possibility. To test this hypothesis, we undertook a 15-mo-long longitudinal entomological survey in two forest regions of Gabon, where wild apes live, at different heights under the canopy. More than 2,400 anopheline mosquitoes belonging to 18 species were collected. Among them, only three species of Anopheles were found infected with ape Plasmodium: Anopheles vinckei, Anopheles moucheti, and Anopheles marshallii. Their role in transmission was confirmed by the detection of the parasites in their salivary glands. Among these species, An. vinckei showed significantly the highest prevalence of infection and was shown to be able to transmit parasites of both chimpanzees and gorillas. Transmission was also shown to be conditioned by seasonal factors and by the heights of capture under the canopy. Moreover, human landing catches of sylvan Anopheles demonstrated the propensity of these three vector species to feed on humans when available. Our results suggest therefore that the strong host specificity observed in the Laveranias is not linked to a specific association between the vertebrate host and the vector species and highlight the potential role of these vectors as bridge between apes and humans.
Recent studies on great apes in Africa have revealed the existence of a large diversity of Plasmodium parasites infecting chimpanzees and gorillas, some being related to the most deadly human parasite Plasmodium falciparum (subgenus Laverania), others to the human parasites Plasmodium malariae, Plasmodium ovale, or Plasmodium vivax (subgenus Plasmodium) (1–4).
Within the subgenus Laverania, eight species are currently recognized. Among them, four species (Plasmodium reichenowi, Plasmodium gaboni, Plasmodium billcollinsi, and Plasmodium billbrayi) were observed only in chimpanzees and three (Plasmodium praefalciparum, Plasmodium adleri, and Plasmodium blacklocki) only in gorillas (2, 3, 5). In this subgenus, only P. falciparum infects humans. In natura, although these different host species cooccur in the same habitat where their ranges overlap, no transfer of Laverania parasites was ever documented between humans and apes or between gorillas and chimpanzees despite large sampling efforts (2, 3, 6). Similarly, ancient reciprocal transplant experiments of Laverania parasites between humans and apes (mostly chimpanzees) failed to produce infections (5). On the contrary, for parasites of the subgenus Plasmodium, like P. vivax or P. malariae, transfers were documented in natural populations (2, 4, 7) or during experimental infections (5). All this suggests therefore a strong host specificity of the Laverania parasites.
The origin of this host specificity in the Laverania could result from an incompatibility at the parasite/vertebrate host interface, at the vector/host interface, or at the parasite/vector interface (5). The first hypothesis has already received much attention and some studies have concluded to the potential existence of a genetic barrier precluding the transfer of parasites from one host species to another (especially from great apes to humans) (5, 8). This barrier would be the consequence of specific receptor/ligand interactions at the host red blood cell/parasite interface.
However, this hypothesis is at odds with observations made in conditions of artificial confinement such as in ape sanctuaries, where different host species (humans and great apes) live in close proximity and where human-to-ape transfers were documented. For instance, bonobos (Pan paniscus), from a sanctuary in the Democratic Republic of Congo, were found infected with P. falciparum, a parasite supposed to be human-specific (4). A similar phenomenon was observed in chimpanzees in a Cameroonian sanctuary (9), suggesting that host switches are possible under certain conditions, and hence that the species barrier might be porous. Therefore, other factors could contribute to this strong host/parasite association observed in natura in the Laveranias. One possibility could be that the segregation of the parasites in different hosts (i.e., chimpanzees and gorillas) is the result of specific interactions between parasites and mosquitoes (governed by genetic factors) or mosquitoes and hosts (i.e., feeding behavior).
The previous observations highlight the need to characterize the mode of transmission of the Laveranias among great apes and particularly to identify the mosquito species involved and their biology and dynamics in rainforests of Africa. To date, few studies have addressed this question, and the natural transmission of ape Plasmodium remains poorly understood, as does the transmission of other zoonotic Plasmodium (e.g., rodent Plasmodium). Indeed, most information about Plasmodium mosquito vectors were acquired in human contexts, concerned anthropophagic species (10), and are de facto difficult to transpose to natural rainforest contexts. Nevertheless, several authors have speculated that some of the main human vector species in forested areas of west and central Africa could be also involved in ape Plasmodium transmission (5). This is particularly the case of Anopheles moucheti and species from the Anopheles nili group that have overlapping habitats with great apes (11, 12).
We are aware of only two specific entomological studies aiming to identify vectors of ape Plasmodium undertaken to date. The first was carried out in western Uganda near nests of wild chimpanzees but failed to detect the presence of Plasmodium parasites among collected Anopheles species (13). The second survey, carried out in Gabon in areas where wild or semiwild apes (chimpanzees and gorillas) live, provided valuable preliminary results about the potential role of Anopheles vinckei and An. moucheti as candidate vectors of P. praefalciparum and P. vivax between apes (11). This study was nevertheless insufficient to demonstrate that these mosquitoes were effective vectors involved in transmission in absence of evidence of the presence of infecting stage (sporozoite) in salivary glands, and also because of the low number of infected mosquitoes found (n = 3).
In the present paper, we present the results of a 15-mo-long longitudinal entomological survey we undertook in two natural parks of Gabon (central Africa) to characterize the sylvatic anopheline species, to identify among them those involved in ape Plasmodium transmission and to assess the intensity and dynamics of transmission and their potential as ape-to-human bridge vectors.
Results
Diversity of Trapped Anopheline Mosquitoes.
A total of 2,415 female anopheline mosquitoes were caught all along the survey in both study sites (the park of La Lékédi and the national park of La Lopé; Fig. S1). The majority of the specimens have been assigned to 18 known anopheline species. For the remaining specimens (n = 62), species could not be determined confidently using morphological and Cytochrome Oxydase Subunit II (COII)-based phylogenetic analyses. In the park of La Lékédi, 1,885 and 231 specimens were respectively collected from the three subsites and around the orphan sanctuaries. These specimens belonged to 15 known species, of which the most frequent were Anopheles marshallii (51.18%), An. moucheti (22.59%), and An. vinckei (14.65%). The number of specimens trapped per anopheline species is given in Table S1. In the park of La Lopé (Mikongo), 299 specimens were trapped all along the survey. They belonged to 10 known anopheles species, among which the dominant species were An. vinckei (46.15%), Anopheles carnevalei (18.73%), and Anopheles implexus (9.03%; Table S1).
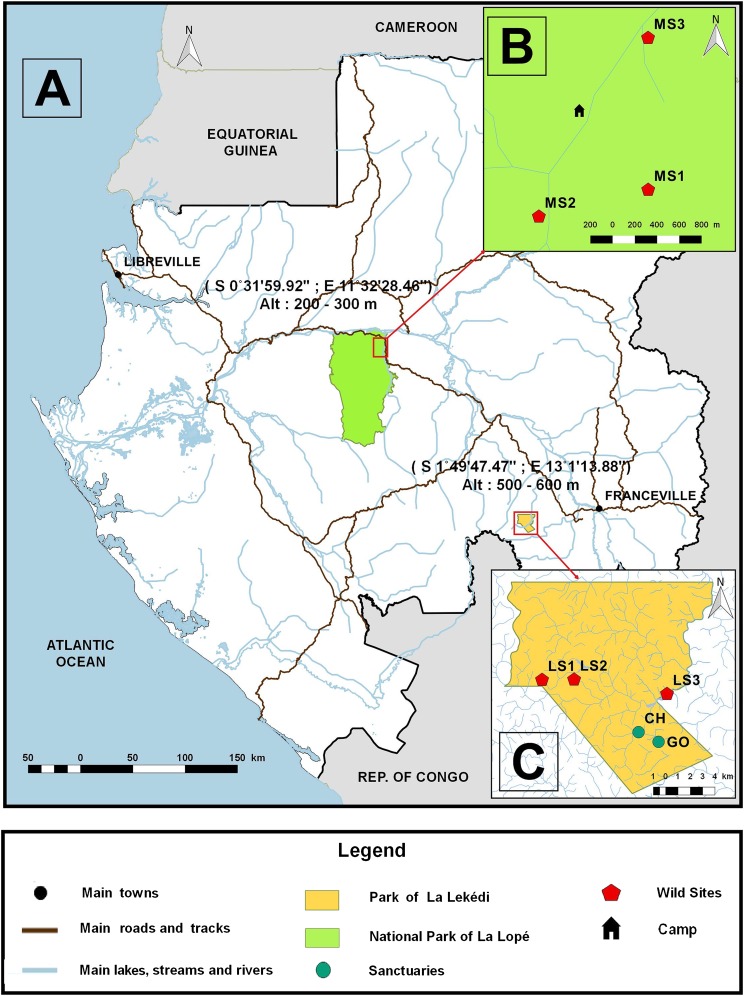
Fig. S1.
Map showing the location of the study sites in Gabon (A). (B and C) Location of different subsites in the national park of La Lopé [three wild sites of Mikongo (MS1–MS3)] and in the park of La Lékédi [three wild sites (LS1–LS3) and two sanctuaries (CH and GO)].
Table S1.
Number of Anopheles specimens (whole body and salivary glands) analyzed per species in each site
| Anopheles species | La Lékédi | La Lopé | ||||||||||
| Wild sites | Sanctuaries | Wild sites | ||||||||||
| WB | SG | WB | SG | WB | SG | |||||||
| n | +PCR | n | +PCR | n | +PCR | n | +PCR | n | +PCR | n | +PCR | |
| An. carnevalei | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 56 | 1 | 56 | 1 |
| An. cinctus | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 2 | 0 |
| An. coustani | 28 | 2 | 10 | 0 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| An. demeillioni | 13 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 5 | 0 | 5 | 0 |
| An. gabonensis | 56 | 5 | 13 | 0 | 1 | 0 | 1 | 0 | 12 | 2 | 12 | 1 |
| An. gambiae | 1 | 0 | 1 | 0 | 2 | 0 | 2 | 0 | 0 | 0 | 0 | 0 |
| An. implexus | 3 | 0 | 3 | 0 | 1 | 0 | 1 | 0 | 27 | 0 | 26 | 0 |
| An. jebudensis | 3 | 0 | 3 | 0 | 2 | 0 | 2 | 0 | 0 | 0 | 0 | 0 |
| An. maculipalpis | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 1 | 0 |
| An. marshallii | 1,033 | 4 | 776 | 6 | 50 | 1 | 50 | 0 | 10 | 0 | 10 | 0 |
| An. moucheti | 325 | 4 | 200 | 1 | 153 | 4 | 135 | 2 | 7 | 0 | 7 | 0 |
| An. nili ss | 6 | 0 | 6 | 0 | 5 | 1 | 5 | 0 | 0 | 0 | 0 | 0 |
| An. obscurus | 9 | 0 | 9 | 0 | 0 | 0 | 0 | 0 | 13 | 3 | 12 | 0 |
| An. paludis | 68 | 0 | 51 | 1 | 8 | 0 | 6 | 0 | 0 | 0 | 0 | 0 |
| An. squamosus | 1 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| An. tenebrosus | 0 | 0 | 0 | 0 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| An. theileri | 1 | 0 | 1 | 0 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| An. vinckei | 309 | 26 | 200 | 4 | 1 | 0 | 1 | 0 | 138 | 8 | 118 | 1 |
| Undetermined | 29 | 0 | 0 | 0 | 5 | 0 | 2 | 0 | 28 | 1 | 27 | 1 |
| Total | 1,885 | 41 | 1,274 | 12 | 231 | 6 | 209 | 2 | 299 | 15 | 276 | 4 |
An., Anopheles; +PCR, number of specimens positive to Plasmodium; SG, salivary gland; WB, whole body.
Vectors of Plasmodium.
In total, 2,415 whole bodies and 1,759 salivary glands of Anopheles specimens were screened for the presence of Plasmodium parasites using a Cytochrome b (Cyt-b)-based nested PCR method. Eighty were positive (62 whole bodies and 18 salivary glands; Table S1). Overall, nine known Anopheles species and one undetermined Anopheles species were found infected (Table S1). The average prevalences of Plasmodium-infected Anopheles whole bodies were 5.02% in La Lopé (Mikongo), 2.06% near the orphan sanctuaries, and 2.17% in the wild sites of La Lékédi (Table S1). The average sporozoite rates in these sites were 1.45%, 0.96%, and 0.94%, respectively.
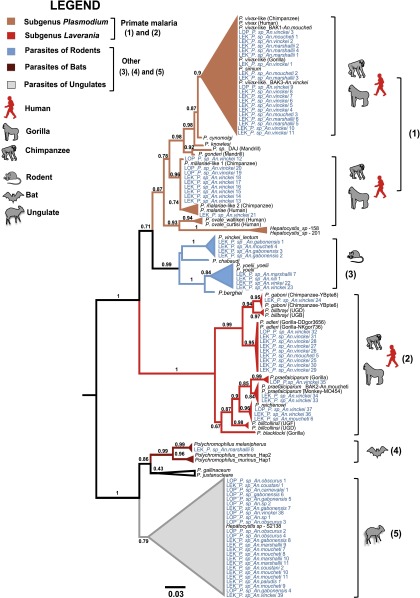
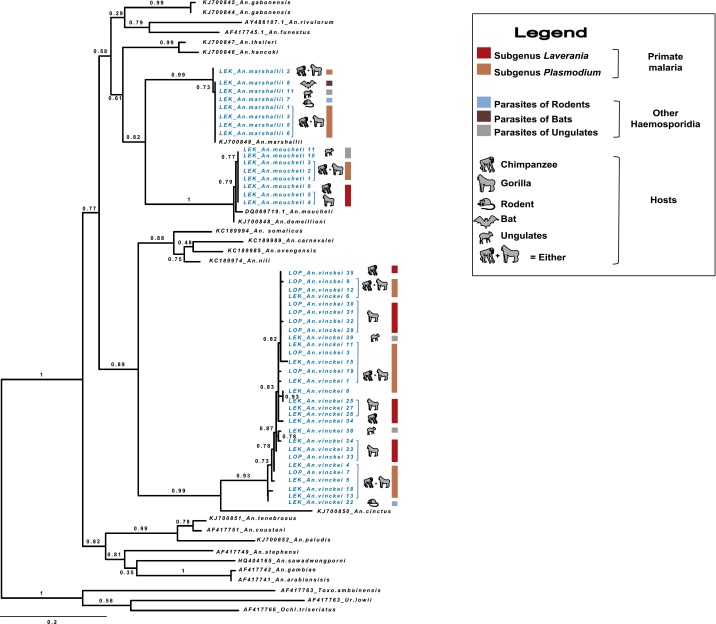
Alignment of the 80 parasite Cyt-b sequences with the references and phylogenetic analyses revealed that they belonged to five major clades (Fig. 1). Four of them match with known parasites infecting several mammal host species (apes, rodents, bats) and one corresponds to parasites recently found in African antelopes (14). Among the sequences belonging to the known parasite clades, some (clades 1 and 2) come from parasites of great apes (57.5%), including parasites from the subgenus Plasmodium (25% of P. vivax-like, 12.5% of P. malariae-like) and from the subgenus Laverania [11.25% of P. adleri, 3.75% of P. reichenowi, 3.75% of P. praefalciparum, and 1.25% of P. gaboni; from (clade 3) parasites of rodents (5.0% of P. vinckei and 5.0% Plasmodium yoelii); and from (clade 4) parasites of bats (1.25% of Polychromophilus spp.)]. The remaining clade (clade 5) encompassed 31.35% of our sequences, and includes only one known reference of a parasite isolated from an African monkey (15). This clade was recently shown to be mostly composed of parasites of African Bovidae (14).
Fig. 1.
Phylogenic position of Haemosporidia infecting Anopheles mosquitoes in this study. The tree was constructed by using partial Cyt-b sequences (773 bp) using maximum-likelihood (ML) methods. Bootstrap values are given at each node. Sequences obtained in this study are in blue.
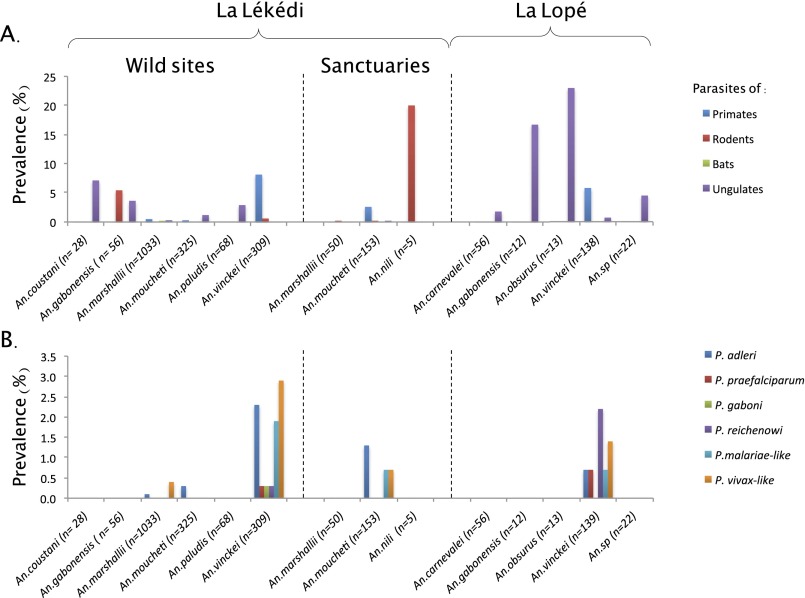
The details of Anopheles species found to be infected by ape Plasmodium (as well as by the other parasite clades) are presented in Fig. 1 and Fig. S2. Over all sites, only three species were detected positive to ape parasites: An. vinckei, An. moucheti, and An. marshallii. Plasmodium infections were the highest for An. vinckei, followed by An. moucheti and An. marshallii (Fig. S2). For each of these mosquito species, at least one ape parasite was detected in their salivary glands, hence confirming their role as effective vectors of these parasites (Table S2). A power test was performed to determine if other anopheline species could also be involved in transmission of ape Plasmodium but remained undetected because of too-small sample sizes. Considering a prevalence of infection similar to the one observed in An. vinckei (the highest prevalence recorded in each site), our test revealed that a potential role in ape Plasmodium transmission of all other Anopheles species could not be excluded (P > 0.05).
Fig. S2.
Relative proportion of mosquitoes infected by (A) the different groups of Haemosporidia from rodents, bats, ungulates, and nonhuman primates and (B) the different species of nonhuman primate Plasmodium.
Table S2.
Anopheles species and number of whole bodies or salivary glands found infected with ape Plasmodium in both sites in Gabon
| Ape Plasmodium | An. vinckei | An. marshallii | An. moucheti | |||
| WB | SG | WB | SG | WB | SG | |
| P. vivax-like | 9 | 2 | 1 | 5 | 2 | 1 |
| P. malariae-like | 8 | 2 | — | — | — | — |
| P. adleri | 8 | — | — | — | — | 1* |
| P. gaboni | 1 | — | — | — | — | — |
| P. praefalciparum | 2 | 1 | — | — | — | — |
| P. reichenowi | 2 | — | — | — | 1* | — |
| Total | 35 | 6 | 5 | |||
P., Plasmodium; SG, salivary gland; WB, whole body.
Infected Anopheles collected in orphan sanctuaries.
Factors Affecting the Transmission of Ape Plasmodium.
A logistic regression was performed to analyze the effects of different factors on the prevalence of ape Plasmodium infections in the mosquitoes. Two models were fitted independently. The first model included as fixed factor only the Anopheles species. The second model included the height of trap and the month of collection. In this second model, the mosquito species was not included to increase the power to detect a link between ecological factors and the infection by ape Plasmodium. Results (Table S3) indicated that all factors had a significant effect (or marginally significant, for height) on transmission. Mosquito infection rates were significantly higher (i) for An. vinckei than for An. moucheti and An. marshallii; (ii) at midheight than on the ground or at canopy level, and (iii) during the rainy season than during the dry season (Fig. S3).
Table S3.
Results of the logistic regressions testing the effect of different ecological factors on the ape Plasmodium infection rate
| Variable | Odds ratio (95% CI) | P value |
| Species | ||
| An. moucheti | 0.60 (0.068–5.23) | 0.642 |
| An. vinckei | 15.16 (4.81–47.82) | 3.61 × 10−6 |
| Rainy season | 3.49 (1.076–11.35) | 0.0374 |
| Midheight | 1.94 (0.98–3.83) | 0.0560 |
| Canopy | 0.50 (0.144–1.73) | 0.2742 |
The variable to be predicted was the presence of an ape Plasmodium infection. For species, the predictive variable (fixed effect) was the Anopheles species. For season and height, the predictive variables were the height of collection (three modalities: ground, midheight, and canopy) and the season (two modalities: dry season or rainy season).
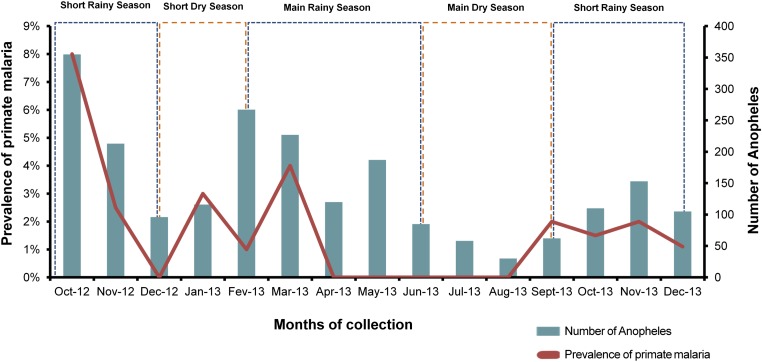
Fig. S3.
Variation of prevalence of infection and Anopheles densities during the survey and relation with the season of collection (rainy or dry season). Values are averaged per month.
Genetic Structure of Ape Plasmodium Vector Populations.
To assess the genetic diversity and to search for a potential structure at an intraspecific level among An. vinckei, An. moucheti, and An. marshallii populations, we constructed a phylogeny based on COII sequences (Fig. S4). The phylogeny showed low levels of genetic diversity for An. moucheti and An. marshallii and a higher level for An. vinckei. For all vector species, we did not found any evidence for the presence of cryptic species or the existence of genetic structuration associated with the parasite host tropism (gorilla or chimpanzee; Fig. S4).
Fig. S4.
Phylogenetic position of the three main ape Plasmodium vectors (Anopheles vinckei, Anopheles moucheti, and Anopheles marshallii). The phylogeny was inferred by using ML methods and 603-bp COII sequences including a subset of infected Anopheles specimens from this study and three mosquito outgroups. An.: Anopheles; Ochl.: Ochlerotatus; Toxo.: Toxorhynchites; Ur.: Uranotaenia. Sequences obtained in this study are in blue.
Tropism of Sylvan Anopheline Mosquitoes Toward Humans.
The propensity of sylvan anopheline mosquitoes to feed on humans was tested by using human landing catches. A total of 395 mosquitoes belonging to two genera (Anopheles and Aedes) were collected. The Anopheles genus was well represented, with 99.24% (n = 392) of the specimens belonging to six known and one undetermined species (referred here as An. sp.): An. marshallii (49%), Anopheles paludis (37.5%), An. moucheti (11.5%), An. vinckei (0.8%), Anopheles cinctus (0.3%), Anopheles gabonensis (0.3%), and undetermined specimens (0.8%). Over all specimens, two were positive for Plasmodium infections and belonged to the species An. vinckei. One was infected with a P. malariae-like parasite, the other by a P. vivax-like parasite.
Discussion
To identify the vectors of ape Plasmodium and determine whether they can be involved in the strong host specificity observed in the Laveranias, we performed a longitudinal survey of the anopheline communities in two wildlife reserves of Gabon (central Africa).
Over the entire survey, only three species of anopheline mosquitoes were detected positive to ape Plasmodium: An. vinckei, An. moucheti, and An. marshallii. Their role in transmission was confirmed by the detection of the parasites in the salivary glands of several specimens (Table S2). All species were shown to be able to transmit both parasites of chimpanzees and gorillas, and, for one of them (An. vinckei), we also have evidence that it can support the transmission of several parasites of different hosts (gorilla and chimpanzee) simultaneously (i.e., same period and same site). Our results therefore suggest that the strong host specificity observed in the Laveranias is not linked to a specific association between the vertebrate host and the vector species but could be caused, as previously suggested, by some more or less permeable genetic barriers involving specific host-parasite ligand/receptor interactions (5, 8).
Beyond Laveranias, our study also confirms the high level of transmission among apes of Plasmodium species belonging to the subgenus Plasmodium and closely related to other human parasites like P. vivax or P. malariae (2, 7, 16). The same three vector species (An. vinckei, An. moucheti, and An. marshallii) are involved in their transmission. The host specificity of these parasites seems not as strong as that observed for parasites from the Laverania subgenus, as P. vivax-like and P. malariae were found in chimpanzees and gorillas in addition to humans. Mechanisms explaining the stronger switching capacities of non-Laveranias remain poorly understood, but the lack of host specificity of vectors probably favors the phenomenon.
Factors Affecting Infection Rates in Sylvan Anopheles.
Among the three species that were shown to be involved in ape Plasmodium transmission, the highest prevalences of infection were observed for An. vinckei, followed by An. moucheti and An. marshallii. All other collected sylvan species might also be involved in the transmission of ape Plasmodium parasites, but our sampling effort was not sufficient to reach a conclusion. Variations among vector species of ape Plasmodium infection rates could be explained by several nonexclusive factors, including their trophic behavior, their density, their longevity, their tropism for certain forest microhabitats (e.g., clearing, savannah, or undergrowth), and their susceptibility to infections (17).
Regarding their trophic behavior, very few data are available on these different vectors in strict nonanthropized forest environments. In our study, the three vector species were found infected by a variety of parasite species known to infect different groups of mammals like rodents (P. yoelii and P. vinckei), ungulates (Plasmodium spp.), bats (Polychromophilus spp.), and primates, indicating that these vector species bite a broad range of hosts. Moreover, the COII-based phylogeny of An. vinckei, An. moucheti, and An. marshallii provided no indication for the existence of population substructure that could be related to differences in host tropism at an intraspecific level. Overall, our results suggest that these vectors are opportunistic rather than specialized for their blood meal. Such a propensity to bite a wide range of hosts is probably an adaptive trait in response to temporal fluctuations of diversity and density of hosts available in the forest. For An. vinckei, however, the proportion of ape parasites relative to the other parasites was far higher than for An. moucheti or An. marshallii, suggesting a possible preference to feed on these hosts when available.
Habitat preference could also explain some of the variations of infection rates between species. Very little is known of the ecology of An. vinckei and its habitat preferences. From our data, it seems that this is the dominant species in dense forest environment as in the park of La Lopé (site of Mikongo) or in certain sites of La Lékédi (e.g., site 1). In other places, such as the park of La Lékédi, where forest alternates with savannah (i.e., savannah–forest mosaic), An. marshallii predominates, followed by An. moucheti and finally An. vinckei. Although An. moucheti is also known to be a forest vector, An. marshallii seems to prefer nonforested areas (18), which may therefore explain its lesser implication as an ape Plasmodium vector given that apes generally nest in forest areas.
A higher longevity of An. vinckei compared with the other Anopheles species could also explain the high infection rate observed in this species despite its lower density in certain environments. Indeed, the longer a mosquito lives, the higher its odds to have several blood meals, which, in turn, increases the probability to become infected (19, 20), but also to allow the complete development of the parasite and its subsequent transmission to a novel host.
Finally, Anopheles species may differ in their ability to support the parasite development. This susceptibility, which may vary from complete receptiveness, whereby all individuals support infection, to the opposite, total refractoriness, whereby no individuals support infection, is generally specific of a parasite/vector species combination (21, 22). Thus, a degree of compatibility exists between parasite and mosquito, and the extent of this compatibility determines the success of transmission (23). Past experiments with a chimpanzee infected with P. reichenowi showed that some species of Anopheles were susceptible to the parasite whereas other species could not be infected, suggesting that not all vectors are able to transmit ape Plasmodium (17, 21, 22). Thus, An. vinckei could be the most susceptible vector of ape Plasmodium.
Beyond the nature of the vector, infection prevalence in sylvan mosquitoes was strongly affected by seasons, with peaks of transmission occurring during the rainy seasons (i.e., from October to December and from February to May), as frequently observed in human foci (24). This seasonality can be explained by the fact that, as rain increases, the number of suitable larval breeding sites for Anopheles increases. For An. vinckei, breeding sites are creeks with clear water and very shaded places (18). For An. moucheti, breeding sites are along the borders of slow-moving streams and large rivers, or in pools and ponds (12, 25). Finally, breeding sites for An. marshallii are fresh, clear, shaded water in nonforested environments (18). As the number of breeding sites increases, so does the Anopheles population size, which results into an increased malaria transmission (26). Increased temperatures during rainy seasons are also a factor favoring transmission by reducing the time needed for the parasite to complete its development in the mosquito and by accelerating the development of the mosquito larvae (27–29).
Finally, we showed that height of capture under the canopy also explains variations in the prevalence of infection in anopheline mosquitoes. Transmission was the highest at midlevel (10–12 m) but similar at ground and canopy levels. This could partly be because most nests built by chimpanzees and gorillas fall within this range of heights (between 5 and 15 m) (30), thus favoring malaria transmission. In addition, mosquito flying height was shown to be related to host abundance patterns (31, 32). In La Lopé, An. vinckei mosquitoes are more abundant at midheight than at other heights, a distribution that could also be driven by the abundance of great ape nests in this site.
Risk of Interspecies Transfers.
Several authors have speculated about the possibility that apes could be a source of new Plasmodium infections for humans and vice versa, providing a mosquito species could act as bridge between these different host species (5, 17). Several recent studies have indeed reported transfers of Plasmodium from apes to humans or humans to apes in certain conditions, but the mosquitoes involved in these transfers were never identified (4, 7, 9, 33).
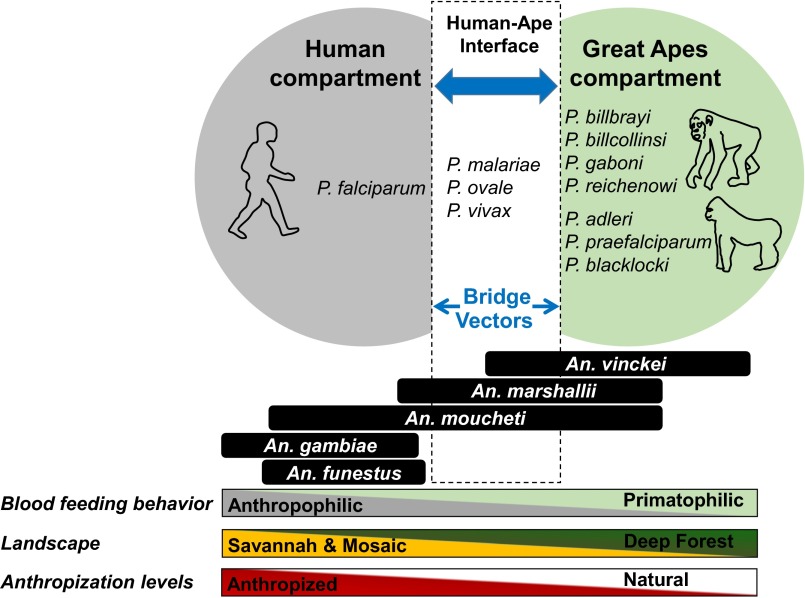
As shown in our study, all three sylvan Anopheles species identified as vectors of ape Plasmodium (An. vinckei, An. moucheti, and An. marshallii) could act as bridge vectors between apes and humans. Indeed, as shown, all species are opportunistic for their blood meals, and, when available, humans may constitute a possible source of blood, as demonstrated by the human landing catches. In light of our results, it seems that the risk of transfers of parasites to humans could be most important in savannah–forest mosaics where the zoo-anthropophilic vectors An. marshallii and An. moucheti are present in high densities in addition to An. vinckei. Moreover, these risks could be amplified by the presence of human-made structures like ponds or lakes, which could favor the development of An. moucheti. Fig. 2 provides a schematic view of the role of the different Anopheles species in the transmission of human and ape malaria parasites and their potential as bridges between the two host compartments according to biological and environmental factors.
Fig. 2.
Schematic view of the role of the Anopheles species in the transmission of human and ape malaria parasites and their potential as bridge vectors between the two host compartments according to biological and environmental factors.
More generally, our study reveals that sylvan mosquitoes may act as bridges between several groups of mammals, including rodents, primates, ungulates, bats, and humans. Beyond the sanitary risk this may represent, this propensity of sylvan anopheline mosquitoes to feed on different hosts may explain the evolutionary history of Plasmodium parasites characterized by numerous host switches that occurred among different mammal groups or species (34–36). Thus, P. falciparum in humans likely evolved following a transfer from gorillas (2, 34). Several host switches occurred in the history of the Laveranias between gorillas and chimpanzees. Finally, several transfers of parasites likely occurred between bats and rodents as well as between bats and monkeys (37). Therefore, our results highlight the key role played by the feeding behavior of vectors in the switches between hosts and therefore in the diversification of malaria parasites during their evolutionary histories.
Absence of Monkey Plasmodium?
One surprising result of our study was the absence of Anopheles mosquitoes infected by monkey parasites, like Plasmodium gonderi or Plasmodium sp. DAJ. P. gonderi is one of the only two Plasmodium species infecting monkeys in west and central Africa (38, 39). Its natural known hosts are the primates of the genera Cercocebus, Mandrillus, and Cercopithecus, well represented in both study sites (La Lopé and La Lékédi). Plasmodium sp. DAJ is a Plasmodium species that was first discovered in mandrills but that seems to also infect some species of the genus Cercopithecus (34). Although nothing is known about the vectors of P. sp. DAJ (likely Anopheles), previous experimental infections of Anopheles specimens have shown their potential implications as vectors for P. gonderi (40). The absence of these parasites is surprising, as the density of their hosts is higher than that of great apes in the different parks. One possible reason for this absence could be related to the lack of attractiveness of our traps for their mosquito vectors or to the size of the mosquito sample. Moreover, it is possible that other mosquito species (or arthropods) are involved in the transmission of these parasites. Indeed, the Plasmodium of birds are transmitted by mosquitoes belonging to Culex, Aedes, and Culiseta genera, but also by some Anopheles and Mansonia (41). Other Diptera such as sandflies (Psychodidae), bat flies (Streblidae, Nycteribiidae), black flies (Simuliidae), and Culicoides midges (Ceratopogonidae) are involved in the transmission of several malaria parasites infecting lizards, birds, bats, and monkeys (35, 41, 42).
Conclusion
This study shows that the transmission of Plasmodium of great apes would be mainly ensured by An. vinckei, An. moucheti, and An. marshallii in Gabon. Transmission was shown to be highest during the rainy season and at midlevel under the canopy. Because all three species transmit parasites of both chimpanzees and gorillas and do not show phylogenetic structure, our results suggest that the strong host specificity observed in the Laveranias is not linked to a specific association between the vertebrate host and the vector species, but would rather be the consequence of incompatibilities at the parasite/vertebrate interface as previously suggested. In vitro experimental infections of erythrocytes of different host species by genetically modified parasites could help determine the origin of this host specificity.
Importantly, we also showed that these vector species could assume the role of bridge vectors between apes and humans, as well as with species belonging to other mammal groups. This information is of considerable public health importance as it highlights the possibility of transfers of new zoonotic infections from apes to humans. Although such transfers may be rare as a result of the strong host specificity, they have happened in the past and may be at the origin of new epidemics in humans if some variants are able to cross the species barrier. This is all the more true in a context of reduction of malaria transmission in human populations, which opens new niches for zoonotic parasites. Such propensity of these vectors to feed on different host species should also be of concern for the wildlife conservation community, as potential recurrent release of human infectious diseases to great apes, which are already at high risk of extinction, may accelerate their disappearance.
Materials and Methods
The longitudinal survey of Anopheles mosquitoes was performed during 15 mo inside of two wildlife reserves of Gabon. Mosquitoes were identified and screened for the presence of Plasmodium parasites by using molecular methods. Generalized linear models were used to explore the effect of ecological factors on Anopheles infection rates. SI Materials and Methods and Table S4 and S5 provide more details on protocols and methods. Mosquito human landing catches were approved by the national ethic committee of Gabon and informed consent statement was obtained from all participants.
Table S4.
Cyt-b sequences of parasites recovered in this study and those used as references for phylogenetic analyses and their GenBank accession numbers
| Isolate ID | GenBank accession no. |
| Hepatocystis_sp_158 | JQ070951 |
| Hepatocystis_sp_201 | JQ070956 |
| Hepatocystis_sp_S2138 | JQ070884 |
| P. berghei | DQ414646 |
| P. billbrayi (UGB) | GQ355470 |
| P. billbrayi (UGD) | GQ355471 |
| P. billcollinsi (UGF) | GQ355478 |
| P. billcollinsi (UGD) | GQ355477 |
| P. blacklocki (Gorilla) | HM235378 |
| P. chabaudi | DQ414648 |
| P. cynomolgi | AB444126 |
| P. gallinaceum | KP025675 |
| P. gonderi (Mandrill) | JF923752 |
| P. juxtanucleare | NC008279 |
| P. knowlesi | JQ345504 |
| P. malariae (Human) | AB489194 |
| P. malariae-like 1 (Chimpanzee) | GU815517 |
| P. malariae-like 2 (Chimpanzee) | HM234994 |
| P. ovale curtisi (Human) | GU723534 |
| P. ovale wallikeri (Human) | GU723548 |
| P. praefalciparum (Monkey-MO454) | JF923762 |
| P. praefalciparum_BAK2-An. moucheti | KC140103 |
| P. praefalciparum (Gorilla) | HM235308 |
| P. reichenowi | AF069610 |
| P. simium | AF069620 |
| P. gaboni (Chimpanzee-YBpte6) | HM235104 |
| P. gaboni (Chimpanzee-YBpte8) | HM235105 |
| P. sp._DAJ (Mandrill) | JF923753 |
| P. adleri (Gorilla-DDgor3656) | HM234991 |
| P. adleri (Gorilla-NKgor736) | HM235081 |
| P. vinckei lentum | DQ414654 |
| P. vivax (Human) | KF591813 |
| P. vivax-like (Gorilla) | JX444719 |
| P. vivax-like (Chimpanzee) | JX444720 |
| P. vivax-like_BAK3-An. vinckei | KC140105 |
| P. vivax-like_BAK1-An. moucheti | KC140104 |
| P. yoelii | DQ414657 |
| P. yoelii_yoelii | DQ414660 |
| Polychromophilus melanipherus | KF159681 |
| Polychromophilus murinus_Hap2 | KF159700 |
| Polychromophilus murinus_Hap1 | KF159714 |
| LOP_P.sp_An. carnevalei 1 | KU318101 |
| LEK_P.sp_An. coustani 1 | KU318031 |
| LEK_P.sp_An. coustani 2 | KU318032 |
| LEK_P.sp_An. gabonensis 1 | KU318033 |
| LEK_P.sp_ An. gabonensis 2 | KU318034 |
| LEK_P.sp_ An. gabonensis 3 | KU318035 |
| LOP_P.sp_ An. gabonensis 4 | KU318102 |
| LOP_P.sp_ An. gabonensis 5 | KU318103 |
| LOP_P.sp_ An. gabonensis 6 | KU318104 |
| LEK_P.sp_ An. gabonensis 7 | KU318036 |
| LEK_P.sp_ An. gabonensis 8 | KU318037 |
| LEK_P.sp_ An. marshallii 1 | KU318038 |
| LEK_P.sp_ An. marshallii 2 | KU318040 |
| LEK_P.sp_ An. marshallii 3 | KU318041 |
| LEK_P.sp_ An. marshallii 4 | KU318042 |
| LEK_P.sp_ An. marshallii 5 | KU318039 |
| LEK_P.sp_ An. marshallii 6 | KU318043 |
| LEK_P.sp_ An. marshallii 7 | KU318044 |
| LEK_P.sp_ An. marshallii 8 | KU318045 |
| LEK_P.sp_ An. marshallii 9 | KU318046 |
| LEK_P.sp_ An. marshallii 10 | KU318047 |
| LEK_P.sp_ An. marshallii 11 | KU318048 |
| LEK_P.sp_ An. moucheti 1 | KU318049 |
| LEK_P.sp_ An. moucheti 2 | KU318052 |
| LEK_P.sp_ An. moucheti 3 | KU318053 |
| LEK_P.sp_ An. moucheti 4 | KU318054 |
| LEK_P.sp_ An. moucheti 5 | KU318055 |
| LEK_P.sp_ An. moucheti 6 | KU318056 |
| LEK_P.sp_ An. moucheti 7 | KU318057 |
| LEK_P.sp_ An. moucheti 8 | KU318058 |
| LEK_P.sp_ An. moucheti 9 | KU318059 |
| LEK_P.sp_ An. moucheti 10 | KU318050 |
| LEK_P.sp_ An. moucheti 11 | KU318051 |
| LEK_P.sp_ An. nili 1 | KU318060 |
| LOP_P.sp_ An. obscurus 1 | KU318105 |
| LOP_P.sp_ An. obscurus 2 | KU318106 |
| LOP_P.sp_ An. obscurus 3 | KU318107 |
| LOP_P.sp_ An. obscurus 4 | KU318108 |
| LEK_P.sp_ An. paludis 1 | KU318061 |
| LOP_P.sp_ An. sp 1 | KU318109 |
| LOP_P.sp_ An. sp 2 | KU318110 |
| LEK_P.sp_ An. vinckei 1 | KU318062 |
| LEK_P.sp_ An. vinckei 2 | KU318063 |
| LOP_P.sp_ An. vinckei 3 | KU318069 |
| LEK_P.sp_ An. vinckei 4 | KU318064 |
| LEK_P.sp_ An. vinckei 5 | KU318065 |
| LEK_P.sp_ An. vinckei 6 | KU318066 |
| LEK_P.sp_ An. vinckei 7 | KU318067 |
| LEK_P.sp_ An. vinckei 8 | KU318068 |
| LOP_P.sp_ An. vinckei 9 | KU318070 |
| LEK_P.sp_ An. vinckei 10 | KU318071 |
| LEK_P.sp_ An. vinckei 11 | KU318072 |
| LOP_P.sp_ An. vinckei 12 | KU318075 |
| LEK_P.sp_ An. vinckei 13 | KU318074 |
| LEK_P.sp_ An. vinckei 14 | KU318076 |
| LEK_P.sp_ An. vinckei 15 | KU318077 |
| LEK_P.sp_ An. vinckei 16 | KU318078 |
| LEK_P.sp_ An. vinckei 17 | KU318080 |
| LEK_P.sp_ An. vinckei 18 | KU318073 |
| LOP_P.sp_ An. vinckei 19 | KU318081 |
| LOP_P.sp_ An. vinckei 20 | KU318079 |
| LEK_P.sp_ An. vinckei 21 | KU318082 |
| LEK_P.sp_ An. vinckei 22 | KU318083 |
| LEK_P.sp_ An. vinckei 23 | KU318084 |
| LEK_P.sp_ An. vinckei 24 | KU318085 |
| LEK_P.sp_ An. vinckei 25 | KU318087 |
| LEK_P.sp_ An. vinckei 26 | KU318088 |
| LEK_P.sp_ An. vinckei 27 | KU318090 |
| LEK_P.sp_ An. vinckei 28 | KU318089 |
| LEK_P.sp_ An. vinckei 29 | KU318091 |
| LEK_P.sp_ An. vinckei 30 | KU318093 |
| LEK_P.sp_ An. vinckei 31 | KU318086 |
| LOP_P.sp_ An. vinckei 32 | KU318092 |
| LEK_P.sp_ An. vinckei 33 | KU318094 |
| LEK_P.sp_ An. vinckei 34 | KU318095 |
| LOP_P.sp_ An. vinckei 35 | KU318096 |
| LEK_P.sp_ An. vinckei 36 | KU318097 |
| LOP_P.sp_ An. vinckei 37 | KU318098 |
| LOP_P.sp_ An. vinckei 38 | KU318099 |
| LEK_P.sp_ An. vinckei 39 | KU318100 |
An., Anopheles; P., Plasmodium.
Table S5.
COII sequences of mosquitoes recovered in this study and those used as references for phylogenetic analyses and their GenBank accession numbers
| Culicidae mosquitoes | GenBank accession no. |
| An. arabiensis | AF417741.1 |
| An. carnevalei | KC189989.1 |
| An. cinctus | KJ700841.1 |
| An. coustani | AF417751.1 |
| An. demeilloni | KJ700848 0.1 |
| An. funestus | AF417745.1 |
| An. gabonensis | KJ700844 0.1 |
| An. gabonensis | KJ700845.1 |
| An. gambiae | AF417742.1 |
| An. hancocki | KJ700846 0.1 |
| An. marshallii | KJ700849.1 |
| LEK_An. marshallii 2 | KU317988 |
| LEK_An. marshallii 3 | KU317990 |
| LEK_An. marshallii 5 | KU317984 |
| LEK_An. marshallii 6 | KU317985 |
| LEK_An. marshallii 7 | KU317989 |
| LEK_An. marshallii 8 | KU317980 |
| LEK_An. marshallii 11 | KU317983 |
| An. moucheti | DQ069719.1 |
| LEK_An. moucheti 1 | KU318001 |
| LEK_An. moucheti 2 | KU317995 |
| LEK_An. moucheti 3 | KU317990 |
| LEK_An. moucheti 5 | KU317993 |
| LEK_An. moucheti 6 | KU317992 |
| LEK_An. moucheti 10 | KU317994 |
| LEK_An. moucheti 11 | KU318000 |
| An. nili | KC189974.1 |
| An. ovengensis | KC189985.1 |
| An. paludis | KJ700852.1 |
| An. sawadwongporni | HQ404165.1 |
| An. somalicus | KC189994.1 |
| An. stephensi | AF417749.1 |
| An. tenebrosus | KJ700851.1 |
| An. theileri | KJ700847 0.1 |
| LEK_An. vinckei 1 | KU318018 |
| LOP_An. vinckei 3 | KU318030 |
| LEK_An. vinckei 4 | KU318013 |
| LEK_An. vinckei 5 | KU318014 |
| LEK_An. vinckei 6 | KU318015 |
| LOP_An. vinckei 7 | KU318016 |
| LEK_An. vinckei 8 | KU318017 |
| LOP_An. vinckei 9 | KU318023 |
| LEK_An. vinckei 11 | KU318004 |
| LOP_An. vinckei 12 | KU318026 |
| LEK_An. vinckei 13 | KU318002 |
| LEK_An. vinckei 15 | KU318021 |
| LEK_An. vinckei 18 | KU318025 |
| LOP_An. vinckei 19 | KU318029 |
| LEK_An. vinckei 22 | KU318003 |
| LEK_An. vinckei 23 | KU318008 |
| LEK_An. vinckei 24 | KU318010 |
| LEK_An. vinckei 25 | KU318005 |
| LEK_An. vinckei 26 | KU318009 |
| LEK_An. vinckei 27 | KU318006 |
| LOP_An. vinckei 29 | KU318007 |
| LOP_An. vinckei 30 | KU318022 |
| LOP_An. vinckei 31 | KU318020 |
| LOP_An. vinckei 32 | KU318019 |
| LOP_An. vinckei 33 | KU318024 |
| LEK_An. vinckei 34 | KU318011 |
| LOP_An. vinckei 35 | KU318027 |
| LEK_An. vinckei 38 | KU318012 |
| LEK_An. vinckei 39 | KU318028 |
| An. rivulorum | AY486107.1 |
| Ochlerotatus triseriatus | AF417766.1 |
| Toxorhynchites amboinensis | AF417763.1 |
| Uranotaenia lowii | AF417764.1 |
An., Anopheles. Phylogenetic position of the three main ape Plasmodium vectors is provided in Fig. S4.
SI Materials and Methods
Study Sites.
From October 2012 to December 2013 (15 mo), we carried out a longitudinal survey of Anopheles mosquitoes inside of two wildlife reserves of Gabon (Fig. S1): the private park of La Lékédi (Bakoumba, Haut-Ogooué province) and the National park of La Lopé (Mikongo, Ogooué Ivindo Province; Fig. S1). Both reserves are natural home of wild populations of gorillas (Gorilla gorilla gorilla) and chimpanzees (Pan troglodytes troglodytes), and a previous study (6) has shown the circulation of ape Plasmodium in both host species inside these reserves.
In each reserve, following a prospection for great ape activity (direct visual observation or presence of nests, feeding traces or feces), three subsites, spaced several kilometers apart, were selected for mosquito collections (Fig. S1). In the park of La Lékédi, all subsites (LS1–LS3; Fig. S1) were located in savannah-forest-mosaic landscapes. In the park of La Lopé, all subsites (MS1–MS3; Fig. S1) were located south of Mikongo village in a dense equatorial forest (Fig. S1).
In the park of La Lékédi, mosquito collections were also performed near two semifree ranging enclosures supporting groups of orphan chimpanzees (n = 10) and gorillas (n = 2; hereafter referred to as orphan sanctuaries CH and GO, respectively; Fig. S1) known to be infected by ape Plasmodium (43).
Mosquito Sampling.
In each reserve and each of the three subsites, mosquitoes were sampled monthly for two consecutive nights from 5:00 PM to 7:00 AM. Because great apes usually build sleeping nests at different heights (chimpanzees at 5–35 m, gorillas at 0.5–15 m) (30, 44), and assuming that sylvan mosquitoes can seek animal blood sources from the ground to the canopy level, traps were placed at different heights of the tree layer: ground (1.5 m), midheight (8–10 m in La Lékédi and 10–12 m at La Lopé), and canopy levels (15–22 m in La Lékédi and 19–25 m at La Lopé). In each subsite, we defined a grid of nine trap positions. For each grid, the number of traps per height was constant (n = 3), but the positions of traps were randomly selected and maintained along the survey. All traps were separated by 50 m along the grid. Each of the 27 traps was run monthly during two successive nights during 15 months, representing an overall sampling effort (in both sites) of 1,620 night-traps.
In the two orphan sanctuary sites, two light traps were placed at 1.5 m of height during two successive nights each month. For these collection sites, the sampling effort accounted for 120 night-traps over the entire period.
Over the entire survey, mosquito collection was performed by using CDC light traps with incandescent light (Bioquip Products) supplemented with carbon dioxide. The carbon dioxide was generated by a mixture of yeast (13.5 g), sugar (27 g), and water (166 mL) contained in a 0.5-L plastic bottle placed under the lid of the trap. The mixture was renewed each day before turning on the traps.
Mosquito Identification, Dissection, and Storage.
Sampled arthropods were killed by a passage at −20 °C during 1 h and observed under a Leica M80 binocular tube. Anopheles specimens were retrieved and identified using taxonomic key (45) before proceeding to dissection to isolate salivary glands. For this, female specimens were placed on a glass slide to separate the head from the body in a drop (60 μL) of 1× PBS solution and isolate the salivary glands. Bodies and PBS solution containing salivary glands were then transferred separately into cryovials and stored in field-portable liquid nitrogen containers. All cryovials were transferred to a −80 °C freezer at Centre International de Recherches Médicales de Franceville until molecular analysis. To prevent cross-contaminations, forceps and needles were decontaminated between dissections by successive baths of sodium hypochlorite solution and water.
DNA Extraction and PCR Amplification.
DNA was isolated and purified from the whole mosquito body and salivary glands by using the DNeasy Blood and Tissue kit (Qiagen) according to the manufacturer’s instructions. To search for the presence of Plasmodium DNA, total DNA was used as a template for amplifying a 950-bp fragment of the parasite Cyt-b gene according to a previously described nested PCR protocol (46). For all mosquitoes found infected with Plasmodium parasites, total DNA was also used to amplify a portion of the COII (47) to confirm the mosquito species, especially for specimens for which the morphological diagnose was doubtful or impossible because of poor preservation of individuals’ morphological characters. This mtDNA marker was also used to assess the genetic variability and the population structure of mosquitoes species found infected with ape Plasmodium.
All PCR amplifications were done by using a GeneAmp 9700 thermal cycler (Applied Biosystems), and all PCR-amplified products (10 μL) were run on 1.5% (15 g/L) agarose gels in Tris-Borate-EDTA buffer. PCR products (Cyt-b of Plasmodium and COII of mosquitoes) were sent to Beckman Coulter Genomics for sequencing in forward and reverse directions after purification.
Phylogenetic Analyses.
We performed phylogenetic analyses by using (i) the Cyt-b sequences and (ii) mosquito COII sequences. Each analysis was performed by using a set of published sequences from different Plasmodium and mosquito species (Tables S4 and S5). Multiple alignments of all sequences were carried out by using ClustalW (v 1.8.1 in BioEdit v.7.0.9.0. software) (48). ML tree construction was performed based on the Cyt-b and mosquito COII sequences of 773 and 603 nt, respectively. The best-fitting ML models under the Akaike information criterion was general time reversible + γ-distribution for nucleotides as identified by ModelTest (49). The highest-likelihood DNA trees and corresponding bootstrap support values were obtained by PhyML (www.atgc-montpellier.fr/) using nearest-neighbor interchange plus subtree pruning regrafting branch swapping and 100 bootstrap replicates (50).
Statistical Analyses.
We used R to perform the different statistical analyses (51). A power analysis was performed to determine the probability (binomial probability) of observing no infected mosquitoes given the sample size, considering similar prevalence as the ones observed in the most infected mosquito species.
A logistic binomial generalized linear model (logit link function) was used to analyze the effect of different factors on the level of ape Plasmodium infections in Anopheles. In this regression, the variable to be predicted was the presence of an ape Plasmodium infection. The predictive variables were (i) the site of collection (random effect), (ii) the subsite of collection (random effect), (iii) the height of collection (ground, midlevel, or canopy; fixed effect), (iv) the Anopheles species (fixed effect), and finally, (v) the season of capture (dry vs. rainy season; fixed effect). An ANOVA was done to test the effect of each variable in the model.
Human Landing Catches.
To assess the propensity of the sylvan anopheline mosquitoes to feed on humans when present in their environment, we performed one session of captures on humans in the park of La Lékédi. These captures were done in December 2013 during four consecutive nights (from 6:00 PM to 12:00 PM) in two subsites (LS2 and LS3). Captures were made by five volunteers of our team. Mosquitoes were collected by using hemolysis tubes on the denuded legs of the volunteers (all protected with malaria prophylaxis).
Acknowledgments
We thank the Agence Nationale des Parcs Nationaux and the Centre National de la Recherche Scientifique et Technologique of Gabon who authorized this study and facilitated the access to the national park of Lopé, Société d'Exploitation du Parc de la Lékédi for its support all along this survey in the park of La Lékédi, and Mr. Mesam Ngady Parfait for map conception. This work was funded by Institut de Recherhe pour le Développement (Laboratoire Mixte International ZOFAC), Centre National de la Recherche Scientifique, Centre International de Recherches Médicales de Franceville, as well as the Agence Nationale de la Recherche (ANR) programme Jeunes Chercheuses Jeunes Chercheurs (JCJC) Sciences de la Vie, de la Santé et des Ecosystèmes 7-2012 project ORIGIN (ANR JCJC SVSE 7-2012 ORIGIN).
Footnotes
The authors declare no conflict of interest.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database [accession nos. KU317980–KU318030 (Cytochrome Oxydase Subunit II) and KU318031–KU318110 (Cytochrome b)].
See Commentary on page 5153.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1603008113/-/DCSupplemental.
References
- 1.Duval L, et al. African apes as reservoirs of Plasmodium falciparum and the origin and diversification of the Laverania subgenus. Proc Natl Acad Sci USA. 2010;107(23):10561–10566. doi: 10.1073/pnas.1005435107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Liu W, et al. Origin of the human malaria parasite Plasmodium falciparum in gorillas. Nature. 2010;467(7314):420–425. doi: 10.1038/nature09442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Prugnolle F, et al. African great apes are natural hosts of multiple related malaria species, including Plasmodium falciparum. Proc Natl Acad Sci USA. 2010;107(4):1458–1463. doi: 10.1073/pnas.0914440107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Krief S, et al. On the diversity of malaria parasites in African apes and the origin of Plasmodium falciparum from Bonobos. PLoS Pathog. 2010;6(2):e1000765. doi: 10.1371/journal.ppat.1000765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rayner JC, Liu W, Peeters M, Sharp PM, Hahn BH. A plethora of Plasmodium species in wild apes: A source of human infection? Trends Parasitol. 2011;27(5):222–229. doi: 10.1016/j.pt.2011.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boundenga L, et al. Diversity of malaria parasites in great apes in Gabon. Malar J. 2015;14(1):111. doi: 10.1186/s12936-015-0622-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Prugnolle F, et al. Diversity, host switching and evolution of Plasmodium vivax infecting African great apes. Proc Natl Acad Sci USA. 2013;110(20):8123–8128. doi: 10.1073/pnas.1306004110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wanaguru M, Liu W, Hahn BH, Rayner JC, Wright GJ. RH5-Basigin interaction plays a major role in the host tropism of Plasmodium falciparum. Proc Natl Acad Sci USA. 2013;110(51):20735–20740. doi: 10.1073/pnas.1320771110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pacheco MA, Cranfield M, Cameron K, Escalante AA. Malarial parasite diversity in chimpanzees: The value of comparative approaches to ascertain the evolution of Plasmodium falciparum antigens. Malar J. 2013;12:328. doi: 10.1186/1475-2875-12-328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sinka ME, et al. The dominant Anopheles vectors of human malaria in Africa, Europe and the Middle East: Occurrence data, distribution maps and bionomic précis. Parasit Vectors. 2010;3:117. doi: 10.1186/1756-3305-3-117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Paupy C, et al. Anopheles moucheti and Anopheles vinckei are candidate vectors of ape Plasmodium parasites, including Plasmodium praefalciparum in Gabon. PLoS One. 2013;8(2):e57294. doi: 10.1371/journal.pone.0057294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ayala D, et al. Habitat suitability and ecological niche profile of major malaria vectors in Cameroon. Malar J. 2009;8(1):307. doi: 10.1186/1475-2875-8-307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Krief S, et al. Investigations on anopheline mosquitoes close to the nest sites of chimpanzees subject to malaria infection in Ugandan highlands. Malar J. 2012;11:116. doi: 10.1186/1475-2875-11-116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Boundenga L, et al. Haemosporidian parasites of antelopes and other vertebrates from Gabon, Central Africa. PLoS One. 2016;11(2):e0148958. doi: 10.1371/journal.pone.0148958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ayouba A, et al. Ubiquitous Hepatocystis infections, but no evidence of Plasmodium falciparum-like malaria parasites in wild greater spot-nosed monkeys (Cercopithecus nictitans) Int J Parasitol. 2012;42(8):709–713. doi: 10.1016/j.ijpara.2012.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu W, et al. African origin of the malaria parasite Plasmodium vivax. Nat Commun. 2014;5:3346. doi: 10.1038/ncomms4346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Verhulst NO, Smallegange RC, Takken W. Mosquitoes as potential bridge vectors of malaria parasites from non-human primates to humans. Front Physiol. 2012;3:197. doi: 10.3389/fphys.2012.00197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gillies M, De Meillon B. The Anophelinae of Africa south of the Sahara (Ethiopian Zoogeographical Region) Publ S Afr Inst Med Res. 1968;54:265–314. [Google Scholar]
- 19.Hurd H. Manipulation of medically important insect vectors by their parasites. Annu Rev Entomol. 2003;48(1):141–161. doi: 10.1146/annurev.ento.48.091801.112722. [DOI] [PubMed] [Google Scholar]
- 20.Lehane MJ. The Biology of Blood-Sucking in Insects. Cambridge Univ Press; Cambridge, UK: 2005. [Google Scholar]
- 21.Blacklock B, Adler S. A parasite resembling Plasmodium falciparum in a chimpanzee. Ann Trop Med Parasitol. 1922;160:99–106. [Google Scholar]
- 22.Collins WE, Skinner JC, Pappaioanou M, Broderson JR, Mehaffey P. Thesporogonic cycle of Plasmodium reichenowi. J Parasitol. 1986;72(2):292–298. [PubMed] [Google Scholar]
- 23.Jaramillo-Gutierrez G, et al. Mosquito immune responses and compatibility between Plasmodium parasites and anopheline mosquitoes. BMC Microbiol. 2009;9(1):154. doi: 10.1186/1471-2180-9-154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mourou JR, et al. Malaria transmission in Libreville: Results of a one year survey. Malar J. 2012;11:40. doi: 10.1186/1475-2875-11-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Antonio-Nkondjio C, et al. Distribution and larval habitat characterization of Anopheles moucheti, Anopheles nili, and other malaria vectors in river networks of southern Cameroon. Acta Trop. 2009;112(3):270–276. doi: 10.1016/j.actatropica.2009.08.009. [DOI] [PubMed] [Google Scholar]
- 26.Githeko AK, Ndegwa W. Predicting malaria epidemics in the Kenyan highlands using climate data: A tool for decision makers. Glob Change Hum Health. 2001;2(1):54–63. [Google Scholar]
- 27.Lindblade KA, Walker ED, Onapa AW, Katungu J, Wilson ML. Highland malaria in Uganda: Prospective analysis of an epidemic associated with El Niño. Trans R Soc Trop Med Hyg. 1999;93(5):480–487. doi: 10.1016/s0035-9203(99)90344-9. [DOI] [PubMed] [Google Scholar]
- 28.Reiter P. Climate change and mosquito-borne disease. Environ Health Perspect. 2001;109(suppl 1):141–161. doi: 10.1289/ehp.01109s1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Alemu A, Abebe G, Tsegaye W, Golassa L. Climatic variables and malaria transmission dynamics in Jimma town, South West Ethiopia. Parasit Vectors. 2011;4:30. doi: 10.1186/1756-3305-4-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Furuichi T, Inagaki H, Angoue-Ovono S. Population density of chimpanzees and gorillas in the Petit Loango Reserve, Gabon: Employing a new method to distinguish between nests of the two species. Int J Primatol. 1997;18(6):1030–1048. [Google Scholar]
- 31.Lee HI, Seo BY, Burkett DA, Lee WJ, Shin YH. Study of flying height of culicid species in the northern part of the Republic of Korea. J Am Mosq Control Assoc. 2006;22(2):239–245. doi: 10.2987/8756-971X(2006)22[239:SOFHOC]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- 32.Swanson DA, Adler PH. Vertical distribution of haematophagous Diptera in temperate forests of the southeastern U.S.A. Med Vet Entomol. 2010;24(2):182–188. doi: 10.1111/j.1365-2915.2010.00862.x. [DOI] [PubMed] [Google Scholar]
- 33.Duval L, Ariey F. Ape Plasmodium parasites as a source of human outbreaks. Clin Microbiol Infec. 2012;18(6):528–532. doi: 10.1111/j.1469-0691.2012.03825.x. [DOI] [PubMed] [Google Scholar]
- 34.Prugnolle F, et al. A fresh look at the origin of Plasmodium falciparum, the most malignant malaria agent. PLoS Pathog. 2011;7(2):e1001283. doi: 10.1371/journal.ppat.1001283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Martinsen ES, Perkins SL, Schall JJ. A three-genome phylogeny of malaria parasites (Plasmodium and closely related genera): Evolution of life-history traits and host switches. Mol Phylogenet Evol. 2008;47(1):261–273. doi: 10.1016/j.ympev.2007.11.012. [DOI] [PubMed] [Google Scholar]
- 36.Gilbert C, Schaack S, Pace JK, 2nd, Brindley PJ, Feschotte C. A role for host-parasite interactions in the horizontal transfer of transposons across phyla. Nature. 2010;464(7293):1347–1350. doi: 10.1038/nature08939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schaer J, et al. High diversity of West African bat malaria parasites and a tight link with rodent Plasmodium taxa. Proc Natl Acad Sci USA. 2013;110(43):17415–17419. doi: 10.1073/pnas.1311016110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Warren M, Wharton RH. The vectors of simian malaria: Identity, biology, and geographical distribution. J Parasitol. 1963;49:892–904. [PubMed] [Google Scholar]
- 39.Prugnolle F, et al. African monkeys are infected by Plasmodium falciparum nonhuman primate-specific strains. Proc Natl Acad Sci USA. 2011;108(29):11948–11953. doi: 10.1073/pnas.1109368108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sullivan JS, et al. Infection of Aotus and Saimiri monkeys with Plasmodium gonderi. J Parasitol. 2002;88(2):422–425. doi: 10.1645/0022-3395(2002)088[0422:IOAASM]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- 41.Valkiunas G. Avian Malaria Parasites and other Haemosporidia. CRC Press; Boca Raton, FL: 2004. [Google Scholar]
- 42.Bertola PB, et al. Bat flies (Diptera: Streblidae, Nycteribiidae) parasitic on bats (Mammalia: Chiroptera) at Parque Estadual da Cantareira, São Paulo, Brazil: Parasitism rates and host-parasite associations. Mem Inst Oswaldo Cruz. 2005;100(1):25–32. doi: 10.1590/s0074-02762005000100005. [DOI] [PubMed] [Google Scholar]
- 43.Herbert A, et al. Malaria-like symptoms associated with a natural Plasmodium reichenowi infection in a chimpanzee. Malar J. 2015;14(1):220. doi: 10.1186/s12936-015-0743-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Blom A, Almasi A, Heitkonig IMA, Kpanou JB, Prins HHT. A survey of the apes in the Dzanga-Ndoki National between the census and survey methods of estimating the gorilla (Gorilla gorilla gorilla) and Chimpanzee (Pan troglodytes) nest group density. East African Wild Life Society. Afr J Ecol. 2001;39:98–105. [Google Scholar]
- 45.Gillies MT, Coetzee M. A supplement to the Anophelinae of Africa south of the Sahara (Afrotropical region) South Africa Institute of Medical Reseach; Johannesburg: 1987. [Google Scholar]
- 46.Ollomo B, et al. A new malaria agent in African hominids. PLoS Pathog. 2009;5(5):e1000446. doi: 10.1371/journal.ppat.1000446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rahola N, et al. Description of Anopheles gabonensis, a new species potentially involved in rodent malaria transmission in Gabon, Central Africa. Infect Genet Evol. 2014;28:628–634. doi: 10.1016/j.meegid.2014.05.012. [DOI] [PubMed] [Google Scholar]
- 48.Hall TA. BioEdit: A user-friendly biological sequence alignment editor and analysis program for windows 95/98/NT. Nucleic Acids Symp. 1999;41:95–98. [Google Scholar]
- 49.Posada D, Crandall KA. MODELTEST: Testing the model of DNA substitution. Bioinformatics. 1998;14(9):817–818. doi: 10.1093/bioinformatics/14.9.817. [DOI] [PubMed] [Google Scholar]
- 50.Guindon S, et al. New algorithms and methods to estimate maximum-likelihood phylogenies: Assessing the performance of PhyML 3.0. Syst Biol. 2010;59(3):307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- 51.R Development Core Team . R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing; Vienna: 2013. [Google Scholar]