Abstract
Chronic Lymphocytic Leukemia (CLL) remains fatal due to the development of resistance to existing therapies. Targeting abnormal glucose metabolism sensitizes various cancer cells to chemotherapy and/or elicits toxicity. Examination of glucose dependency in CLL demonstrated variable sensitivity to glucose deprivation. Further evaluation of metabolic dependencies of CLL cells resistant to glucose deprivation revealed increased engagement of fatty acid oxidation upon glucose withdrawal. Investigation of glucose transporter expression in CLL reveals up-regulation of glucose transporter GLUT4. Treatment of CLL cells with HIV protease inhibitor ritonavir, that inhibits GLUT4, elicits toxicity similar to that elicited upon glucose-deprivation. CLL cells resistant to ritonavir are sensitized by co-treatment with metformin, potentially targeting compensatory mitochondrial complex 1 activity. Ritonavir and metformin have been administered in humans for treatment of diabetes in HIV patients, demonstrating the tolerance of this combination in humans. Our studies strongly substantiate further investigation of FDA approved ritonavir and metformin for CLL.
INTRODUCTION
CLL is the most common leukemia in Western countries and is characterized by the accumulation of a mature monoclonal CD5+ B cells in primary and secondary lymphoid tissues1. Therapeutic modalities employed in the treatment of CLL include single-agent alkylators, purine nucleoside analogs, immunotherapy and combinations of chemotherapy or chemoimmunotherapy (e.g. fludarabine-cyclophopshamide+/−rituximab)2. There has also been a surge of ongoing investigations into BCR-associated kinases – Bruton Tyrosine Kinase (BTK) inhibitors, Spleen tyrosine kinase (Syk) inhibitors, and PI3 – kinase inhibitors, BCL-2 inhibitor as targeted therapies in CLL, which are currently in clinical and preclinical development3–7. Progression free survival (PFS) reported in fludarabine- refractory CLL treated patients has varied; with reports from 5.5 months to 25 months8. Overall response rates reported have been in the range of 14% to 93%, and overall survival (OS) in the range of 11 months to 38 months across different studies8. A phase Ib/II clinical trial of the BTK inhibitor Ibrutinib, which was recently published showed in patient with relapsed/refractory CLL/SLL at 26 months, the estimated PFS, and OS were 75% and 83% respectively. At present allogeneic stem cell transplantation is the only curative treatment for CLL but is fraught with complications resulting in significant morbidity and mortality. In addition, stem cell transplant is not usually an appropriate option for the majority of patients diagnosed with CLL; as the median age at diagnosis is 72 years old. Novel therapeutics with decreased toxicity are thus of great need.
There is an increasing interest in targeting cancer cell metabolism for therapy. Cancer cells exhibit metabolic alterations that facilitate generation of biosynthetic precursors required for the duplication of cellular biomass. In addition, altered metabolism promotes oncogenic signaling and resistance to a wide range of existing treatments9,10 suggesting the utility of targeting cancer metabolism. Elevated glucose uptake and preferential metabolism by aerobic glycolysis was first described by Otto Warburg in the early 1900s11. This property of increased glucose uptake in cancer cells forms the basis of [18F] fluoro-2-deoxyglucose-positron emission tomography (FDG-PET) used in the diagnosis and prognostic monitoring of cancer12, however this phenomenon has not yet been capitalized upon for therapy. Various agents such as genistein, fasentin, STF-31, 2-deoxyglucose and 3 bromopyruvate are being investigated for their ability to target glucose metabolism13–17
Existing FDA approved drugs are increasingly being evaluated for their indirect impact on cellular metabolism. We have previously demonstrated the utility of targeting glucose transport in myeloma cells with the HIV protease inhibitor ritonavir18 that has an off-target inhibitory effect on glucose transporter GLUT419. In this study we explored the glucose dependency of CLL primary patient samples. We detect a range of sensitivity of CLL cells to glucose deprivation. We hypothesize that resistance to glucose deprivation is mediated by compensatory metabolism of alternative mitochondrial substrates. Compensatory mitochondrial metabolism can be targeted with the anti-diabetic metformin due to its inhibitory effect on mitochondrial complex 1. Our pre-clinical data bolsters the rationale for further investigation of ritonavir and metformin for the treatment of CLL.
MATERIALS and METHODS
Patient population
We used primary patient samples for these studies. After obtaining consent on an IRB-approved protocol, peripheral blood was collected from patients with a known diagnosis of CLL who were cared for at the Robert H. Lurie Cancer Center of Northwestern University, Chicago Illinois. Patient’s needed to have an absolute lymphocyte count (ALC) of ≥10,000 K/µL to be eligible.
Cell culture
CLL cells were cultured for 48 hours in a 37°C incubator in glucose-free RPMI 1640 media(Invitrogen)supplemented with 10% dialyzed fetal bovine serum (Life technologies™), 100 units/ml penicillin, 100 mg/ml streptomycin and 4mM glutamine and/or 5mM glucose where needed.
Chemicals and Reagents
Chloroquine, 1,1-dimethylbiguanide hydrochloride (metformin), etomoxir, methylpyruvate, oligomycin, carbonyl cyanide 4-(trifluoromethoxy)phenylhydrazone (FCCP), antimycin and rotenone were obtained from Sigma-Aldrich. Ritonavir was purchased from Euroasia, Euroasian Chemicals Pvt LtD.
Isolation of normal B lymphocytes and CLL cells
CLL cells were isolated by negative selection, using a Human B-cell Enrichment Kit with CD43 depletion (Easy Step® Negative selection). Normal B lymphocytes were purified from peripheral blood mononuclear cells (PBMC) from normal controls using a negative selection kit (Stem Cell Technologies).
RNA extraction and real time RT-PCR
RNA isolation was done with RNeasy Mini Kit purchased from QIAgen. For realtime RT-PCR three loading controls – HPRT1, GUSB, and B2M were selected based on prior work by Valceckiene et al20. The primer probe sets recognizing these three loading controls and Glucose Transporter (GLUT) 1, 3, and 4 were purchased from Applied Biosystems. Loading normalization was achieved using the geNorm method21.
Flow cytometry
All viability studies were assessed using flow cytometry. After specific treatments CLL cells were washed with PBS, and then stained with AnnexinV – fluorescein isothiocyanate and DAPI. The CLL samples were subsequently run on Dako Cyan™ ADP analyzer or the BD LSRFortessa™ cell analyzer. Data analysis was performed with the FCS Express version 3 (De Novo Software, Los Angeles, CA).
Immunoblot Analysis
CLL cell lysates were made with the Complete Lysis-M buffer supplemented with phosphatase inhibitor cocktail tablets (Roche Applied Science). Proteins were initially separated by electrophoresis, and after transfer to nitrocellulose, immunoblots were probed with GLUT1 antibody (Abcam®), GLUT4 antibody (Dr. S Cushman, NIDDK), GAPDH antibody (EMD Millipore), PARP antibody (BD Pharmingen) and MCL-1 antibody (Santa Cruz Biotechnology) followed by secondary horse-radish conjugated antibodies.
Immunofluorescence microscopy
The cells were washed once in PBS, and spun onto coated microscope slides (Shandon Cytoslide™) using a Shandon Cytospin™ 2 cytocentrifuge (Thermo Electron Corp., Pittsburgh, PA). Slides were fixed in 3% paraformaldehyde, permeabilized with 0.03% saponin in PBS, and blocked with 10% normal goat serum containing 0.03% saponin as described previously18. Cells were stained with optimized dilutions of antibodies to human GLUT4 as previously described. Secondary antibodies used for detection were anti-rabbit IgG-Alexa Fluor 594 (Invitrogen). Stained cells were mounted with Ultra Cruz mounting medium (Santa Cruz Biotechnology, Inc., Santa Cruz, CA) containing DAPI for counterstaining. Cells were visualized with a 63× (1.4 numerical apertures) oil objective LSM-510 Meta Carl Zeiss confocal microscope. Image analysis was performed using the Zeiss Axiovision LE image browser.
Bioenergetics Assays
Seahorse bioscience extracellular flux (XF24) analyzer (SABiosciences) was utilized to measure oxygen consumption rates (OCR), and extracellular acidification rates (ECAR). Cells were plated in 24-well plates custom designed for XF24 analysis at a density of 2 × 106 per well. CLL cells treated with ritonavir and or metformin were subject to various perturbations i.e. oligomycin (5 µM), FCCP (5 µM), and antimycin (2 µM) + rotenone (2 µM). Mitochondrial oxygen consumption rate (OCR) was determined by correcting for residual OCR measured upon antimycin + rotenone treatment.
Statistical analysis
The number of CLL samples used in the experiments represented in the figures is specified in the figure legends. Student t-test and chi-square test were used to test for between group differences in means or proportions, correspondingly with 2-sided p values using GraphPad Prism software.
CLL patient sample sensitivity to glucose, ritonavir, metformin and the combination was correlated with baseline patient characteristics, and known prognostic factors for CLL in univariate analysis by logistic regression. CLL patient samples’ sensitivity to glucose, ritonavir, metformin and the combination were further correlated with overexpression of GLUT4 using logistic regression. This was done using SAS statistical software. A p value of ≤ 0.05 was deemed to be statistically significant in all analyses.
RESULTS
CLL sample patient characteristics
Table 1 summarizes characteristics of the 35 patient samples used in the study. The absolute lymphocyte count (ALC), and serum lactate dehydrogenase (LDH) levels were not normally distributed as evidenced by the Kolmogorov-Smirnov (K-S) test for normality, p values of <0.01 and 0.03 respectively. The age was not normally distributed based on the K-S test for normality p value = 0.08. The average age of patients included in the study was 64.5 years, and the average ALC was 107600/mm3.
Table 1.
CLL Patient Characteristics
| No | Sex | Age | RAI | ZAP 70 | FISH | CD 38 | IgVh status | LDH | ALC (K/uL) |
Treatment |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | F | 66 | 2 | − | 13q- | − | Mutated; VH4-4 | 241 | 564.48 | R-Campath |
| 2 | F | 66 | 1 | + | 13q- | +/− | Unmutated; VH1-18 | 211 | 214.13 | none |
| 3 | F | 65 | 1 | + | 13q- | +/− | Unmutated; VH1-18 | 207 | 203.04 | None |
| 4 | F | 64 | 1 | − | 13q- | − | 212 | 94.6 | none | |
| 5 | F | 64 | 1 | − | 13q- | − | 206 | 80.37 | none | |
| 6 | M | 73 | 3 | − | 13q- | − | Unmutated; VH3-48 | 160 | 234.62 | None |
| 7 | M | 73 | 3 | − | 13q- | − | Unmutated; VH3-48 | 182 | 354.47 | None |
| 8 | M | 52 | 2 | − | 13q-13q- | − | Unmutated; VH5-51 | 197 | 89 | None |
| 9 | M | 55 | 2 | − | 17p-13q- | − | Unmutated;VH1-69 | 144 | 163.875 | None |
| 10 | F | 65 | 2 | + | 17p- Tri12 | − | 305 | 121.4 | R-CVP | |
| 11 | F | 64 | 2 | + | 17p- Tri12 | − | 195 | 90.25 | R-CVP | |
| 12 | M | 58 | 1 | − | 13q- | − | Mutated; VH4-4 | 194 | 73.272 | None |
| 13 | M | 59 | 0 | + | Tri 12 | − | Unmutated; VH4-59 | 261 | 45.955 | none |
| 14 | M | 58 | 0 | + | Tri 12 | − | Unmutated; VH4-59 | 264 | 28.85 | None |
| 15 | M | 54 | 0 | + | 13q- | + | Borderline mutated; VH6-61 |
177 | 109.3 | none |
| 16 | F | 63 | 1 | − | 13q-13q- | − | Mutated VH3-23 | 237 | 169.63 | None |
| 17 | M | 59 | 0 | − | 13q- | − | 236 | 119.5 | None | |
| 18 | M | 71 | 2 | − | 13q-13q- | − | 230 | 73.3 | none | |
| 19 | M | 72 | 2 | − | 13q-13q- | − | 150 | 65.87 | none | |
| 20 | F | 66 | 3 | + | 5q-, 7q- del 3 | 225 | 30.4 | FCRX2, ofatumumab,ONO/revlimid, azacitidine |
||
| 21 | F | 65 | 0 | − | none | − | 172 | 61.4 | none | |
| 22 | M | 56 | 1 | − | 13q-13q- | − | Mutated IgVH | 141 | 110 | none |
| 23 | M | 76 | 3 | − | − | 216 | 60.16 | Rituximab | ||
| 24 | M | 58 | 0 | − | − | 153 | 51.8 | None | ||
| 25 | F | 78 | 3 | + | 17p-, mono 13,loss of IgH |
− | 273 | 46.718 | R-bendamustine | |
| 26 | F | 63 | 1 | + | 13q- | − | IgVH unmutated | 210 | 62.035 | none |
| 27 | F | 64 | 1 | + | 13q- | − | IgVH unmutated | 211 | 45.36 | none |
| 28 | M | 59 | 3 | + | Tri 12 | − | IgVH unmutated | 418 | 98.487 | Campath/Rituxan |
| 29 | F | 64 | 0 | 17p- | − | Mutated; VH4-4 | 195 | 35.752 | none | |
| 30 | M | 67 | 1 | + | 11q-,13q- | + | IgVH unmutated | 216 | 107.096 | Camp/Rituxan |
| 31 | M | 73 | 0 | − | 13q- | − | 172 | 23.3 | none | |
| 32 | M | 70 | 1 | + | 13q-, del ATM, IGH loss |
+ | 195 | 15.6 | none | |
| 33 | F | 56 | 0 | + | 13q- | − | 167 | 76.6 | none | |
| 34 | F | 66 | 0 | − | 13q-13q- | − | Mutated; VH2-23 | 190 | 16.3 | none |
| 35 | F | 76 | 0 | − | 13q- | − | Mutated; VH3-53 | 158 | 26.1 | none |
CLL patient samples exhibit variable dependency on glucose for maintenance of viability
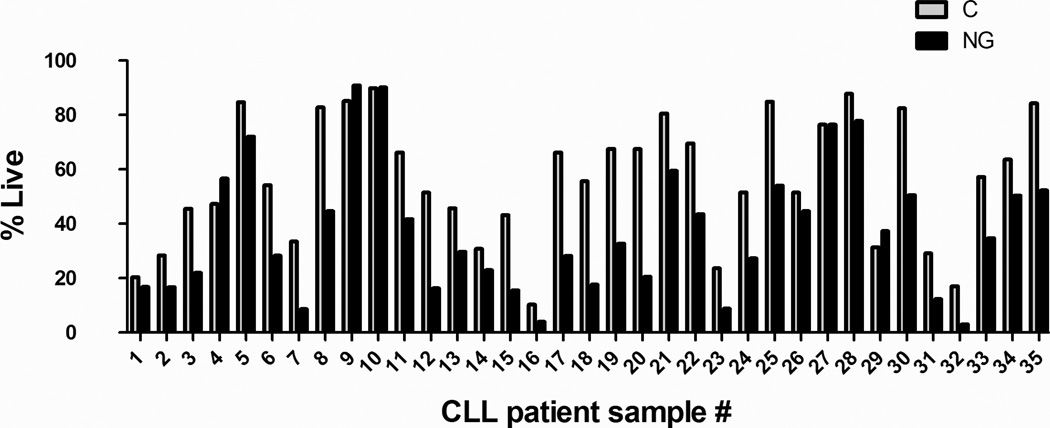
We have previously demonstrated that multiple myeloma cells in contrast to normal B cells exhibit significant cytotoxicity when cultured in glucose-free growth media (GM)18. We similarly sought to determine if CLL patient samples are sensitive to glucose deprivation. CLL patients cell (n=35) were cultured for 48 hours in glucose-free GM. Interestingly, CLL cells exhibited variable sensitivity to glucose deprivation (Figure 1). The Kolmogorov-Smirnov test for normality showed a normally distribution (p value = >0.1500). The mean percent viability upon glucose deprivation was 62.4% after normalization (95%CI= 53.4 – 71.3%) (Supplementary Table 1). If CLL cells had viability of 80% or greater in glucose-free media, they were considered to be resistant to glucose deprivation, and accordingly 23% of CLL patient cells were deemed to be resistant (Figure 1).
Fig 1. CLL patient cells exhibit varying levels of apoptosis upon glucose deprivation.
CLL cells from patients (n=35), were cultured in complete (C)(5 mM glucose) or glucose-free (NG) media for 48 hours following which cellular viability was assessed by AnnexinV/DAPI staining. All readings were normalized to viability of CLL patient cells cultured in complete media. Average viability of untreated cells at 48 hrs was 56.20% STDEVP 22.86% and for glucose-deprived cells average viability was 37.38 % STDEVP 23.71%. Samples exhibiting greater than 50% death in untreated cells could impact overall responsiveness to therapy.
CLL cells rely on fatty acid oxidation to maintain survival in glucose-deprived conditions
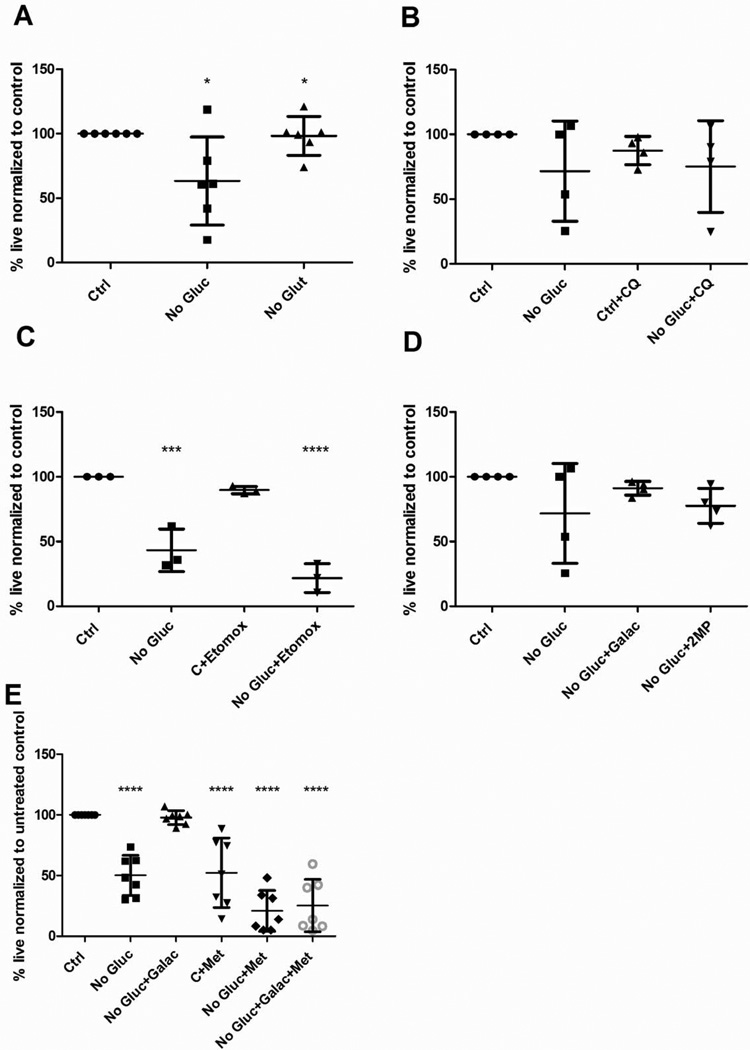
To determine if CLL cells rely on alternative sources of metabolites for maintenance of survival in complete media or in the context of glucose deprivation, we investigated the dependency of CLL cells on glutamine metabolism, fatty oxidation and autophagy. CLL cells (n =6) were cultured in control growth media containing 5mM glucose, or were glucose-free or glutamine-free for 48 hours. Percent viability was normalized to viability of CLL cells cultured in control GM (Fig 2A). We observed that CLL patient cells viability was not impacted by culturing in the absence of glutamine (Figure 2A). There was no significant difference in viability between the CLL cells cultured in control and those cultured in glutamine-free GM, p value = 0.8 (Figure 2A). To determine the role of autophagy in maintaining CLL cell viability upon glucose deprivation, CLL patient samples were cultured in control or glucose-free GM with and without autophagy inhibitor chloroquine (CQ) for 48 hours. Treatment of glucose-deprived CLL cells with CQ did not enhance cytotoxicity (Figure 2B) significantly indicating that glucose-deprived cells were likely not activating autophagy to maintain survival upon glucose deprivation. Next, we wanted to determine if fatty acid oxidation was essential for CLL cell survival. We therefore treated CLL cells with etomoxir (n=3) an inhibitor of fatty acid oxidation, in both the presence and absence of glucose. CLL cell viability was significantly decreased by administration of etomoxir only in the context of glucose-deprivation suggesting that fatty acid oxidation plays a key role maintaining CLL viability when glucose is removed (Figure 2C).
Fig 2. CLL patient cells are sensitized to fatty acid oxidation inhibitor upon glucose deprivation. CLL is not impacted by glutamine deprivation or inhibition of autophagy.
All culture and treatment was for 48 hours. Cell viability assessed by AnnexinV and DAPI staining A - CLL patient cells (n=6), cultured in a) 5mM glucose media, b) glucose-free media and c) glutamine-free media+ 5mM glucose. B - CLL patient cells (n=4) cultured in glucose-containing and glucose-free media in the presence and absence 10µm chloroquine. C - CLL patient cells (n=3), cultured in glucose-containing and glucose-free media in the presence and absence for 10µm etomoxir. D – CLL patient cells (n=4) cultured in glucose containing or glucose-free media in the presence and absence of 10mM galactose or 2 methylpyruvate. All samples were normalized to viability of CLL patient cells cultured in 5mM glucose-containing media. E: CLL patient cells (n=7), cultured in glucose containing or glucose-free media, supplemented with 10mM galactose and treated with 5 mM metformin. *p <0.05 *** p <0.005****p <0.0001.
Having established that CLL cells rely primarily on glucose metabolism for survival we sought to establish whether CLL cells metabolize glucose via glycolysis and/or mitochondrial oxidative phosphorylation (OXPHOS). To determine whether mitochondrial glucose metabolism sustains CLL viability we supplemented glucose-deprived cells with galactose (that is slowly metabolized to glucose to supplement the pentose phosphate pathway and/or mitochondrial OXPHOS) or cell permeant mitochondrial substrate 2-methyl pyruvate (2MP). As demonstrated in Figure 2D galactose and 2MP rescue viability of CLL cells cultured in the absence of glucose, with galactose rescuing viability almost 100%. These results suggest that oxidative mitochondrial metabolism of glucose potentially sustains CLL cell viability.
Oxidative phosphorylation is essential for CLL cell viability that can be targeted with metformin
While we have demonstrated that galactose and 2MP rescue viability of glucose-deprived CLL cells we sought to independently confirm dependency of CLL cells on mitochondrial OXPHOS. We treated CLL cells with 5mM metformin (a mitochondrial complex 1 inhibitor) for 48 hours in the presence or absence of glucose. CLL cells cultured in control media treated with metformin exhibit a reduction in cell viability of 57.6%; (p = <0.0001). Glucose deprivation further sensitizes cells to metformin suggesting that the cells are continuing to rely on OXPHOS upon glucose-deprivation. Addition of galactose to glucose-deprived cells in conjunction with metformin did not rescue viability confirming that galactose rescued CLL cell viability in glucose-free media by maintaining complex 1 activity (Figure 2E).
Role for GLUT4 in glucose transport in CLL
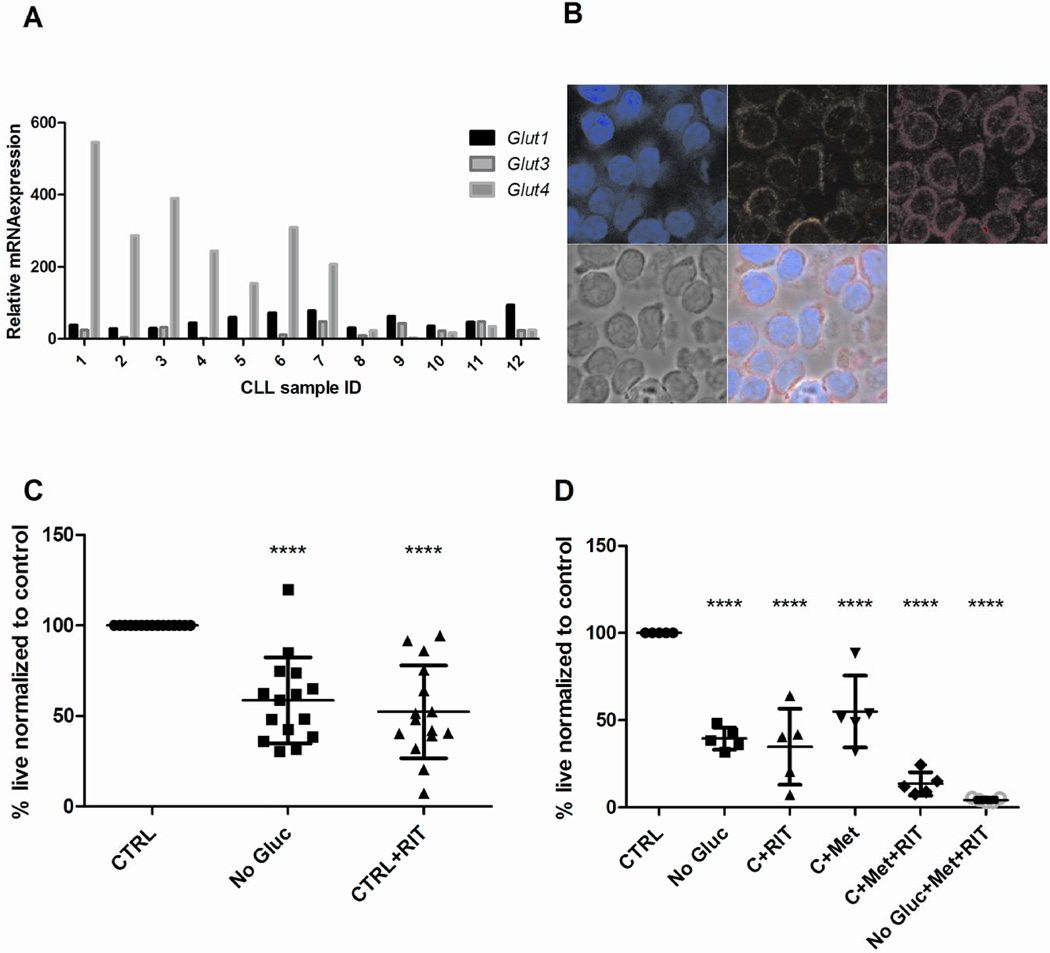
We have previously demonstrated a dependence of multiple myeloma on the insulin responsive glucose transporter (GLUT4)18. GLUT4 was demonstrated to be constitutively localized to the plasma membrane of myeloma cells facilitating glucose transport18. To examine a role for GLUT4 in CLL glucose transport we evaluated mRNA expression of GLUT4, in addition to GLUT1 and GLUT3, normally expressed in B cells. 60% of the CLL cells overexpressed GLUT4, but not GLUT1 and GLUT3 in comparison to normal B cells (Figure 3A). Using immunofluorescence microscopy, we also observed that GLUT4 was highly expressed in CLL cells co-localizing with the plasma membrane (Figure 3B). These results suggest a potential role for GLUT4 in facilitating glucose transport in CLL.
Fig 3. CLL cells express elevated GLUT4 that exhibits plasma membrane co-localization and treatment with GLUT4 targeting agent ritonavir, elicits apoptosis.
A – Expression of GLUT1, GLUT3, GLUT4 in patient samples and normal B lymphocytes by qRT-PCR. Relative quantification is normalized to expression in NBL (not shown). B – GLUT4 plasma membrane localization in CLL cells; clockwise from top left- DAPI, wheat germ agglutinin stain of plasma membrane, GLUT4, phase contrast and Merge. C - CLL patient cells (n=15) cultured glucose-containing media or glucose-free media, in the presence or absence of 20µm ritonavir. Cell viability is determined by AnnexinV and DAPI staining. All samples were normalized to viability of CLL patient cells cultured in 5mM glucose –containing media. D - CLL patient cells (n=5) cultured in glucose-containing and glucose-free media and treated with 20µm ritonavir, 5mM metformin and the combination for 48hours. Cell viability measurement and normalization as in C. ****p <0.0001.
Targeting glucose transport in CLL with HIV protease inhibitor ritonavir that has an off-target inhibitory effect on GLUT4
Next we wanted to determine if GLUT4 could be targeted in CLL cells. We have previously demonstrated that GLUT4 activity in myeloma cells can be targeted with the HIV protease inhibitor ritonavir, which has an off-target inhibitory effect on GLUT418,19. We treated (n= 15) CLL patient samples with ritonavir and observed that ritonavir decreased CLL cell viability in approximately 50% of the samples tested (p = <0.0001) (Figure 3C).
Targeting glucose transport and compensatory mitochondrial metabolism with FDA approved compounds, ritonavir and metformin
Our studies have thus far demonstrated a dependency of CLL cells on mitochondrial metabolism fueled primarily by glucose and oxidation of fatty acids. We therefore tested the impact of co-treatment of ritonavir (potentially targeting GLUT4) and metformin (targeting complex 1 activity) on CLL cell viability. The combination of these two FDA approved agents decreased CLL cell viability more than either compound alone (Figure 3D), and this effect was potentiated when they were cultured in glucose-free media.
The combination of ritonavir with metformin effectively reduces OCR and uncouples OXPHOS
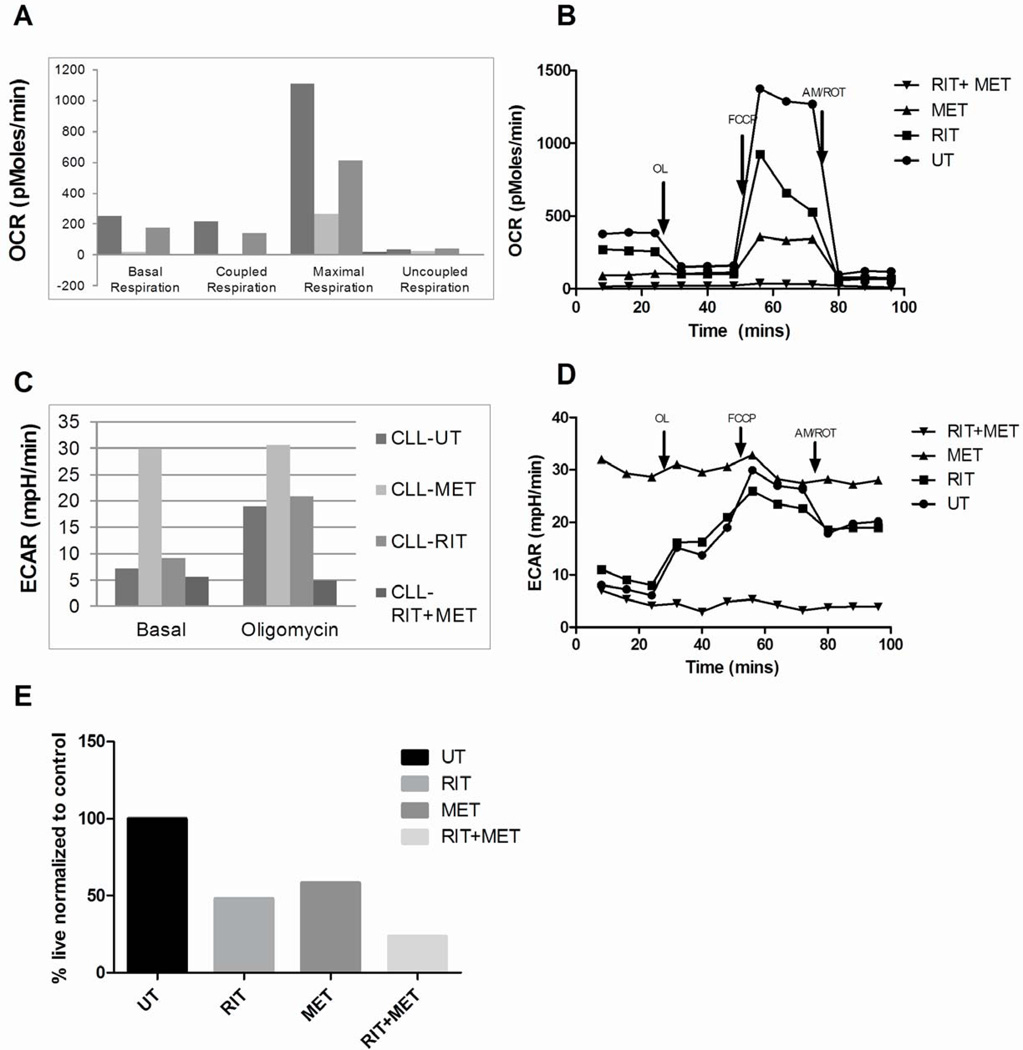
To evaluate the impact of ritonavir and metformin on OCR, CLL cells were left untreated or treated with ritonavir, metformin or the combination for 17 hours and then OCR evaluated in equal numbers of viable cells using a Seahorse XF24 bioenergetics analyzer. OCR was also evaluated subsequent to specific perturbations i.e. oligomycin (ATP synthase inhibitor) to determine coupled respiration (amount of oxygen linked to ATP synthesis), FCCP to determine maximal respiratory capacity and antimycin/rotenone treatments to determine residual oxygen consumption. Figure 4A is a representative data set demonstrating net OCR after correcting for residual oxygen consumption. Ritonavir as anticipated reduces OCR and coupled respiration bolstering our previous results that glucose is metabolized by the tricyclic acid cycle (TCA) cycle coupled to OXPHOS (Figure 4A–B). Ritonavir reduces ECAR (an indirect measure of glycolysis) (Figure 4C and D) suggestive of an impact of ritonavir on glucose transport and glycolysis. Metformin on the other hand significantly reduces OCR and coupled respiration however increases ECAR. Metformin is known to increase glucose transport22 and likely maintains viability due to a switch to glycolysis that can account for the increase in ECAR (Figure 4C and D). It is the combination of both ritonavir with metformin that effectively reduces OCR, uncouples OCR from ATP synthesis, reverses the increase in ECAR induced by metformin to elicit maximal cytotoxicity (Figure 4A and B). Flow cytometric evaluation of apoptosis of the same CLL sample confirmed decreased viability when treated with ritonavir, metformin and the combination by 48 hrs (Figure 4E).
Fig 4. Ritonavir and metformin co-treatment of CLL cells uncouples and suppresses oxygen consumption.
A&B - OCR and C&D - ECAR in CLL patient cells cultured in glucose-containing media pre-treated with 20µm ritonavir, 5mM metformin or the combination for 17 hrs. Equal numbers of viable cells were run on the Seahorse Analyzer and measurements were recorded after treatment with 5um Oligomycin (OL), 5µm FCCP and 2µm Antimycin /2µm Rotenone (AM/ROT). (UT – Untreated, RIT- ritonavir, MET- metformin). Data presented is from one of three representative patient samples. E - CLL patient cells including patient sample used in (A) were cultured in 5 mM glucose-containing media treated with 20µm ritonavir, 5mM metformin and the combination for 48hours. Cell viability is determined by AnnexinV and DAPI staining. All samples were normalized to viability of untreated CLL patient cells.
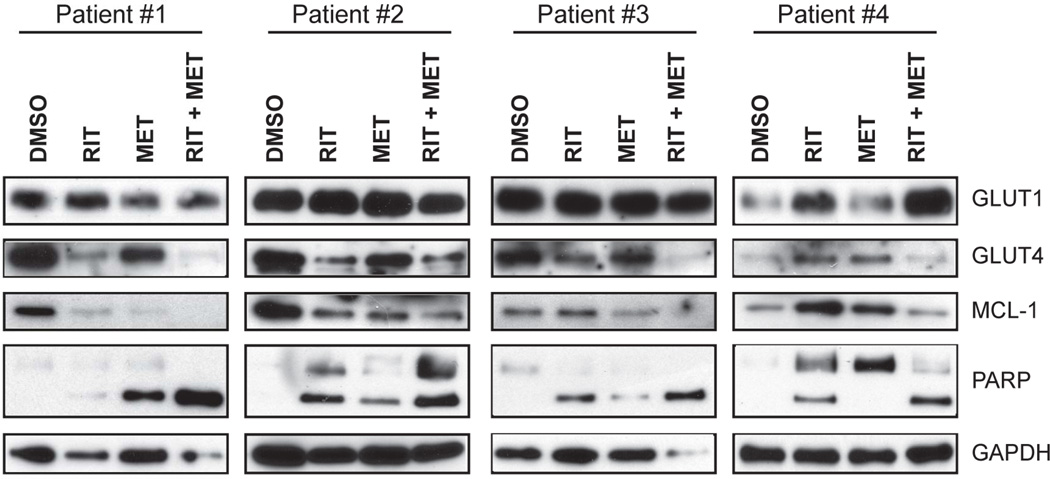
Ritonavir and metformin effectively reduce GLUT4 expression, MCL-1 expression and induce PARP cleavage
Previous studies have demonstrated that HIV protease inhibitors selectively inhibit glucose transport through GLUT4 as well as reduce GLUT4 protein expression19. We evaluated GLUT1 and GLUT4 expression and note that GLUT4 is selectively down-regulated upon treatment with ritonavir. The pro-survival BCL-2 family member MCL-1 is a key resistance promoting factor in a myriad of cancers including CLL23. We and others have demonstrated that increased glucose metabolism attenuates degradation of Mcl-118,24. We then evaluated GLUT4 expression at protein level following treatment of cells with ritonavir, metformin and the combination for 24 hours (Figure 5A). As hypothesized, we observed a decrease in expression of GLUT4 in CLL samples treated with ritonavir, and the combination, whereas GLUT1 is maintained or increased. Our results demonstrate that ritonavir and metformin effectively reduce MCL-1 expression and induce PARP cleavage corroborating our AnnexinV/DAPI apoptosis assessment with these treatments.
Fig 5. Ritonavir and metformin combination suppresses GLUT4 and MCL-1 expression.
A - CLL patient cells treated with 20µm ritonavir, 5mM metformin or the combination for 24 hrs were evaluated for GLUT4, GLUT1, MCL-1, PARP and GAPDH expression by immunoblot analysis.
GLUT4 overexpression does not correlate with sensitivity to glucose deprivation, treatment with ritonavir, metformin, the combination or known prognostic indicators in CLL patients
We collected prognostic data of the patients with regards to RAI stage, ZAP 70 status, CD 38 status, FISH for common CLL genetic abnormalities, IgVH mutational status, age and ALC. Using linear regression analysis we correlated the prognostic data with CLL patient cells sensitivity to glucose deprivation, treatment with ritonavir, metformin, and the combination. The only statistical significance between these prognostic indices and treatment was seen between cytogenetic abnormalities in CLL cells and response to metformin (p = 0.04) (Supplementary Table 2).
Using logistic regression we assessed if there was correlation between the overexpression of GLUT4 in CLL samples (n=12) and sensitivity to glucose deprivation, ritonavir, metformin or the combination treatment. We evaluated the RAI stage disease, ZAP 70 status, CD38 status, cytogenetic abnormalities, IgVH mutational status, age, sex, serum LDH and ALC. Overall we did not determine a statistical significant correlation between these prognostic factors and GLUT4 expression in CLL patient cells (Supplementary table 2).
DISCUSSION
There has been a recent surge of interest investigating tumor cell metabolism for the identification of alternative cancer therapies. Abnormalities in tumor cell metabolism, thought to be subsequent to genomic and epigenomic modifications, are critical to the maintenance and progression of cancer10,25,26. The caveat to targeting tumor cell metabolism remains identification of tumor selective non-redundant rate-limiting metabolic check points. Various studies including those from our laboratory, demonstrate the utility of directly targeting glucose transport and metabolism for chemo-sensitization and therapy27,28. We have previously demonstrated that inhibition of glucose transport (a key rate-limiting check point) inhibits glucose metabolism and glucose regulated signaling to elicit apoptosis in the plasma cell malignancy, multiple myeloma. CLL, also a B-cell malignancy of (mature) CD5+ B cells, has a reduced proliferative index unlike myeloma and is not typically identified by PET imaging29,30. Since glucose regulates a myriad of functions, including ATP synthesis, production of precursors for duplication of cellular biomass and maintenance of redox homeostasis we sought to determine CLL dependency on glucose metabolism.
Our studies suggest that CLL cells are largely dependent upon glucose that is metabolized via OXPHOS. This observation is supported by the fact that CLL cells cultured in the absence of glucose supplemented with galactose maintain viability. Galactose is metabolized to glucose-1-phosphate by a UTP-dependent process, followed by glycolysis that is too inefficient to maintain glycolytic ATP synthesis and lactate production31. Galactose maintenance of cell survival is thus by maintenance of the pentose phosphate pathway and/or mitochondrial OXPHOS31. This rescue effect was also observed (although not to the same extent as with galactose) when CLL cells in the absence of glucose are treated with cell permeant 2-methylpyruvate. Methylpyruvate can bypass the glycolytic pathway supplementing mitochondrial metabolism. In addition, with ritonavir treatment we observe a reduction in OCR strongly suggesting that glucose metabolism sustaining CLL viability is primarily mediated via oxidative phosphorylation. Importantly, ritonavir does reduce ECAR, an indirect correlate of glycolysis suggestive of an inhibition of glucose transport and basal glycolysis. In sum, our results suggest that glycolysis however may not be a key pathway of glucose metabolism sustaining CLL cell survival.
We outline a potential role for the insulin-responsive GLUT4 in facilitating glucose transport in CLL that is likely targeted with the HIV protease inhibitor ritonavir. Ritonavir has an off-target inhibitory effect on GLUT4 preventing glucose uptake by non-competitive reversible inhibition of glucose transport through GLUT419. GLUT4 is normally retained in intra-cellular compartments and recruited to the surface in response to activating stimuli such as upon insulin treatment. While we detect increased GLUT4 content and plasma membrane localization based on immune-staining in CLL cells we have not determined absolute plasma membrane content. This in turn can explain a lack of correlation between GLUT4 expression and sensitivity to ritonavir among the samples tested. Additional mechanisms for ritonavir mediated cytotoxicity that have been investigated in other cancer cell contexts include inhibition of NFκB, inhibition of Akt, enhancement of protein ubiquitination, HDAC suppression and histone deacetylation32–34. These additional targets of ritonavir could explain the observed cytotoxicity in the samples tested and lack of correlation with GLUT4 expression. While our studies do not conclusively prove that ritonavir is targeting GLUT4, the elevated expression of GLUT4 in our CLL samples combined with selective reduction in GLUT4 protein, not GLUT1, the reduction of ECAR upon ritonavir treatment indirectly suggests that glucose transport through GLUT4 may indeed be targeted.
The inability to decrease expression of the pro-survival BCL-2 family member, Mcl-1 correlates with resistance to bortezomib35, rapamycin36, cyclin-dependent kinase inhibitors37, the Bcl-2/Bcl-XL/Bcl-w selective antagonist ABT 73738 and death receptor (Fas/TRAIL)39-induced apoptosis in various cell types including CLL. Our results demonstrate the ability of ritonavir and metformin to effectively reduce MCL-1 protein expression that could potentially be a result of regulation at the mRNA level.
Our results demonstrate that CLL cells cultured in glucose-free media have variation in sensitivity to glucose deprivation. The variation in response to glucose deprivation is similar to that observed in other solid and hematological malignancies40,41 suggesting reliance/engagement of compensatory metabolic pathways for sustaining CLL cell viability. Our studies have demonstrated a role for fatty acid oxidation as potentially maintaining OXPHOS. Targeting glucose metabolism sensitizes CLL to a fatty acid oxidation inhibitor warranting further investigation into the engagement of fatty acid metabolism in CLL. While various metabolites can feed into the TCA cycle to sustain the electron transport chain and OXPHOS we have tested the complex 1 inhibitor metformin with the rationale of potentially targeting all compensatory metabolism that would sustain OXPHOS. Our results demonstrate the uncoupling activity of metformin and an increase in ECAR that may result from increased glucose transport subsequent to AMPK activation by metformin22. However co-treatment with ritonavir, that blocks glucose transport reverses the increase in ECAR and completely uncouples OCR correlating with significant apoptosis in all the CLL patient samples tested.
Metformin has recently become an agent which has shown interesting potential as an anti-cancer agent. The first epidemiological report of metformin and its association with cancer was in 2005 when Evan et. Al. reported a reduced incidence of cancer in patients with diabetes who were being treated with metformin compared to those who were on other therapies42. Biguanides were first reported in 2006 to have antineoplastic activity in vitro, and several mechanisms of action have been postulated for this activity43. The inhibition of mitochondrial oxidative phosphorylation and complex 1 activity, by metformin results in a decrease in ATP levels44. This reduction in ATP levels leads to activation of AMP-activated protein kinase (AMPK), leading to decreased mTOR signaling and protein synthesis, also contributing to the anti-proliferative effect43. In vivo studies in mice confirm anti-tumor effects of metformin in breast cancer, lung cancer and the growth of intestinal polyps42,45. Our study also demonstrates the anti-tumor effect of metformin. This effect was further enhanced by the treatment of CLL cells with the combination of ritonavir and metformin. While our in vitro assessment of metformin involved administration of metformin doses higher than that can be achieved in vivo we believe given the anti-cancer properties of metformin that are currently being detected upon in vivo administration in humans and in pre-clinical models we have significant rationale to test this combination in vivo42,43,45.
Recent studies from the Rathmell group have reported that high-risk CLL exhibit elevated glucose metabolism46. Another group observed that patients’ leukemic cells that were dependent on aerobic glycolysis had an inferior progression free survival at 2 years compared to those whose cells utilized oxidative phosphorylation; independent of the presence of good or poor risk cytogenetic features47. In our analysis we did not observe any relationship between IgVh mutational status, ZAP-70 expression or other prognostic indices with CLL cell sensitivity to glucose deprivation, although it has been reported that the IgVH high risk feature in CLL correlates with increased glucose metabolism46. This group also reports increased expression of GLUT1 in CLL cells, which we did not observe in our patient population.
In sum, our studies describe a fundamental role for glucose metabolism in sustaining CLL. We have provided strong rationale for re-purposing ritonavir and metformin to target glucose metabolism and the ensuing metabolic plasticity in CLL. The combination of ritonavir and metformin has been administered to HIV patients (for the treatment of diabetes) and well tolerated in humans bolstering further evaluation of this promising combinatorial regimen for primary or adjunctive CLL therapy.
Supplementary Material
Acknowledgments
The authors thank Vivian Liu for her assistance with purification of CLL cells from patient samples. We thank Dr. Elena A. Monclus and the Chandel Laboratory at Northwestern University for assistance with the Seahorse analyzer and helpful discussions.
This work was made possible by the Northwestern University Cell Imaging Facility and a Cancer Center Support Grant (NCI CA060553), the Robert H. Lurie Comprehensive Cancer Center Flow Cytometry Facility and a Cancer Center Support Grant (NCI CA060553), National Institutes of Health/National Cancer Institute grant T32CA079447-13 to K.U. Adekola., American Cancer Society Illinois Division, grant 188679 to M. Shanmugam, American Cancer Society Research Scholar grant RSG-11-254-01-CSM to M. Shanmugam and the R.H.L.C Cancer Center Gift fund to S. Rosen.
Footnotes
Authors’ contributions
KUAA and MS conceived and performed the research; SDA assisted with experimentation and figure generation and manuscript editing; STR, SM and KUAA identified CLL patients, assisted with sample harvest and provided conceptual advice; ZZ assisted with regression analysis; KUAA and MS wrote the manuscript and MS supervised the project. All authors read and approved the final manuscript.
Disclosure of Conflicts of Interests
The authors declare no competing financial interests.
References
- 1.Riches JC, Ramsay AG, Gribben JG. Chronic lymphocytic leukemia: an update on biology and treatment. Current oncology reports. 2011;13:379–385. doi: 10.1007/s11912-011-0188-6. [DOI] [PubMed] [Google Scholar]
- 2.Jaglowski S, Jones JA. Choosing first-line therapy for chronic lymphocytic leukemia. Expert review of anticancer therapy. 2011;11:1379–1390. doi: 10.1586/era.11.132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brown JR. Final results of a phase I study of idelalisib (GS-1101) a selective inhibitor of PI3Kδ, in patients with relapsed or refractory CLL. J Clin Oncol. 2013;31 (suppl; abstr 7003) [Google Scholar]
- 4.O'Brien SM. A phase II study of the selective phosphatidylinositol 3-kinase delta (PI3Kδ) inhibitor idelalisib (GS-1101) in combination with rituximab (R) in treatment-naive patients (pts) ≥65 years with chronic lymphocytic leukemia (CLL) or small lymphocytic lymphoma (SLL) J Clin Oncol. 2013;31 (suppl; abstr 7005) [Google Scholar]
- 5.Quiroga MP, et al. B-cell antigen receptor signaling enhances chronic lymphocytic leukemia cell migration and survival: specific targeting with a novel spleen tyrosine kinase inhibitor, R406. Blood. 2009;114:1029–1037. doi: 10.1182/blood-2009-03-212837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Niedermeier M, et al. Isoform-selective phosphoinositide 3'-kinase inhibitors inhibit CXCR4 signaling and overcome stromal cell-mediated drug resistance in chronic lymphocytic leukemia: a novel therapeutic approach. Blood. 2009;113:5549–5557. doi: 10.1182/blood-2008-06-165068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Byrd JC, et al. Targeting BTK with ibrutinib in relapsed chronic lymphocytic leukemia. The New England journal of medicine. 2013;369:32–42. doi: 10.1056/NEJMoa1215637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brown JR. The treatment of relapsed refractory chronic lymphocytic leukemia. Hematology / the Education Program of the American Society of Hematology. American Society of Hematology. Education Program. 2011;2011:110–118. doi: 10.1182/asheducation-2011.1.110. [DOI] [PubMed] [Google Scholar]
- 9.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 10.Vander Heiden MG. Targeting cancer metabolism: a therapeutic window opens. Nature reviews. Drug discovery. 2011;10:671–684. doi: 10.1038/nrd3504. [DOI] [PubMed] [Google Scholar]
- 11.Warburg O. On the origin of cancer cells. Science. 1956;123:309–314. doi: 10.1126/science.123.3191.309. [DOI] [PubMed] [Google Scholar]
- 12.Kaida H, et al. Glucose transporter expression of an esophageal gastrointestinal tumor detected by F-18 FDG PET/CT. Clin. Nucl. Med. 2010;35:505–509. doi: 10.1097/RLU.0b013e3181e05d79. [DOI] [PubMed] [Google Scholar]
- 13.Chan DA, et al. Targeting GLUT1 and the Warburg effect in renal cell carcinoma by chemical synthetic lethality. Sci Transl Med. 2011;3:94ra70. doi: 10.1126/scitranslmed.3002394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vera JC, et al. Genistein is a natural inhibitor of hexose and dehydroascorbic acid transport through the glucose transporter, GLUT1. J. Biol. Chem. 1996;271:8719–8724. doi: 10.1074/jbc.271.15.8719. [DOI] [PubMed] [Google Scholar]
- 15.Wood TE, et al. A novel inhibitor of glucose uptake sensitizes cells to FAS-induced cell death. Mol Cancer Ther. 2008;7:3546–3555. doi: 10.1158/1535-7163.MCT-08-0569. [DOI] [PubMed] [Google Scholar]
- 16.Pelicano H, Martin DS, Xu RH, Huang P. Glycolysis inhibition for anticancer treatment. Oncogene. 2006;25:4633–4646. doi: 10.1038/sj.onc.1209597. [DOI] [PubMed] [Google Scholar]
- 17.Ko YH, et al. Advanced cancers: eradication in all cases using 3-bromopyruvate therapy to deplete ATP. Biochem. Biophys. Res. Commun. 2004;324:269–275. doi: 10.1016/j.bbrc.2004.09.047. [DOI] [PubMed] [Google Scholar]
- 18.McBrayer SK, et al. Multiple myeloma exhibits novel dependence on GLUT4, GLUT8, and GLUT11: implications for glucose transporter-directed therapy. Blood. 2012;119:4686–4697. doi: 10.1182/blood-2011-09-377846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Murata H, Hruz PW, Mueckler M. The mechanism of insulin resistance caused by HIV protease inhibitor therapy. J. Biol. Chem. 2000;275:20251–20254. doi: 10.1074/jbc.C000228200. [DOI] [PubMed] [Google Scholar]
- 20.Valceckiene V, Kontenyte R, Jakubauskas A, Griskevicius L. Selection of reference genes for quantitative polymerase chain reaction studies in purified B cells from B cell chronic lymphocytic leukaemia patients. Br. J. Haematol. 2010;151:232–238. doi: 10.1111/j.1365-2141.2010.08363.x. [DOI] [PubMed] [Google Scholar]
- 21.Vandesompele J, et al. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome biology. 2002;3 doi: 10.1186/gb-2002-3-7-research0034. RESEARCH0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee JO, et al. Metformin induces Rab4 through AMPK and modulates GLUT4 translocation in skeletal muscle cells. J. Cell. Physiol. 2011;226:974–981. doi: 10.1002/jcp.22410. [DOI] [PubMed] [Google Scholar]
- 23.Liu J, et al. Loss of p53 and altered miR15-a/16-1short right arrowMCL-1 pathway in CLL: insights from TCL1-Tg:p53 mouse model and primary human leukemia cells. Leukemia. 2013 doi: 10.1038/leu.2013.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhao Y, et al. Glycogen synthase kinase 3alpha and 3beta mediate a glucose-sensitive antiapoptotic signaling pathway to stabilize Mcl-1. Mol Cell Biol. 2007;27:4328–4339. doi: 10.1128/MCB.00153-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jones RG, Thompson CB. Tumor suppressors and cell metabolism: a recipe for cancer growth. Genes Dev. 2009;23:537–548. doi: 10.1101/gad.1756509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324:1029–1033. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shanmugam M, et al. Targeting glucose consumption and autophagy in myeloma with the novel nucleoside analogue 8-aminoadenosine. J. Biol. Chem. 2009;284:26816–26830. doi: 10.1074/jbc.M109.000646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xu RH, et al. Inhibition of glycolysis in cancer cells: a novel strategy to overcome drug resistance associated with mitochondrial respiratory defect and hypoxia. Cancer Res. 2005;65:613–621. [PubMed] [Google Scholar]
- 29.Bartel TB, et al. F18-fluorodeoxyglucose positron emission tomography in the context of other imaging techniques and prognostic factors in multiple myeloma. Blood. 2009;114:2068–2076. doi: 10.1182/blood-2009-03-213280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zamagni E, et al. Prognostic relevance of 18-F FDG PET/CT in newly diagnosed multiple myeloma patients treated with up-front autologous transplantation. Blood. 2011;118:5989–5995. doi: 10.1182/blood-2011-06-361386. [DOI] [PubMed] [Google Scholar]
- 31.Reitzer LJ, Wice BM, Kennell D. The pentose cycle. Control and essential function in HeLa cell nucleic acid synthesis. J. Biol. Chem. 1980;255:5616–5626. [PubMed] [Google Scholar]
- 32.Ikezoe T, et al. HIV-1 protease inhibitor, ritonavir: a potent inhibitor of CYP3A4, enhanced the anticancer effects of docetaxel in androgen-independent prostate cancer cells in vitro and in vivo. Cancer Res. 2004;64:7426–7431. doi: 10.1158/0008-5472.CAN-03-2677. [DOI] [PubMed] [Google Scholar]
- 33.Srirangam A, et al. Effects of HIV protease inhibitor ritonavir on Akt-regulated cell proliferation in breast cancer. Clin. Cancer Res. 2006;12:1883–1896. doi: 10.1158/1078-0432.CCR-05-1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sato A, Asano T, Ito K. Ritonavir interacts with bortezomib to enhance protein ubiquitination and histone acetylation synergistically in renal cancer cells. Urology. 2012;79:966 e913–966 e921. doi: 10.1016/j.urology.2011.11.033. [DOI] [PubMed] [Google Scholar]
- 35.Podar K, et al. A pivotal role for Mcl-1 in Bortezomib-induced apoptosis. Oncogene. 2008;27:721–731. doi: 10.1038/sj.onc.1210679. [DOI] [PubMed] [Google Scholar]
- 36.Mills JR, et al. mTORC1 promotes survival through translational control of Mcl-1. Proc Natl Acad Sci U S A. 2008;105:10853–10858. doi: 10.1073/pnas.0804821105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Eguchi T, et al. Expression levels of p18INK4C modify the cellular efficacy of cyclin-dependent kinase inhibitors via regulation of Mcl-1 expression in tumor cell lines. Mol Cancer Ther. 2009;8:1460–1472. doi: 10.1158/1535-7163.MCT-08-1159. [DOI] [PubMed] [Google Scholar]
- 38.Nguyen M, et al. Small molecule obatoclax (GX15-070) antagonizes MCL-1 and overcomes MCL-1-mediated resistance to apoptosis. Proc Natl Acad Sci U S A. 2007;104:19512–19517. doi: 10.1073/pnas.0709443104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pradelli LA, et al. Glycolysis inhibition sensitizes tumor cells to death receptors-induced apoptosis by AMP kinase activation leading to Mcl-1 block in translation. Oncogene. 2009 doi: 10.1038/onc.2009.448. [DOI] [PubMed] [Google Scholar]
- 40.Shanmugam M, McBrayer SK, Rosen ST. Targeting the Warburg effect in hematological malignancies: from PET to therapy. Curr. Opin. Oncol. 2009;21:531–536. doi: 10.1097/CCO.0b013e32832f57ec. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Altenberg B, Greulich KO. Genes of glycolysis are ubiquitously overexpressed in 24 cancer classes. Genomics. 2004;84:1014–1020. doi: 10.1016/j.ygeno.2004.08.010. [DOI] [PubMed] [Google Scholar]
- 42.Dowling RJ, Niraula S, Stambolic V, Goodwin PJ. Metformin in cancer: translational challenges. J. Mol. Endocrinol. 2012;48:R31–R43. doi: 10.1530/JME-12-0007. [DOI] [PubMed] [Google Scholar]
- 43.Pollak MN. Investigating metformin for cancer prevention and treatment: the end of the beginning. Cancer discovery. 2012;2:778–790. doi: 10.1158/2159-8290.CD-12-0263. [DOI] [PubMed] [Google Scholar]
- 44.Guigas B, et al. Metformin inhibits mitochondrial permeability transition and cell death: a pharmacological in vitro study. Biochem. J. 2004;382:877–884. doi: 10.1042/BJ20040885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Phoenix KN, Vumbaca F, Claffey KP. Therapeutic metformin/AMPK activation promotes the angiogenic phenotype in the ERalpha negative MDA-MB-435 breast cancer model. Breast Cancer Res. Treat. 2009;113:101–111. doi: 10.1007/s10549-008-9916-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liu T, Weinberg JB, Davis ED, Volkheimer AD, Rathmell J. The Metabolic Signature of CLL: Enhanced Glucose Metabolism in A Subset of High-Risk CLL Patients. ASH Annual Meeting Abstracts. 2012;120:1785. [Google Scholar]
- 47.Gardner JR, et al. Aerobic Glycolysis Predicts Outcome in Early Chronic Lymphocytic Leukemia. ASH Annual Meeting Abstracts. 2012;120:2482. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.