Abstract
Computational docking can be used to predict bound conformations and free energies of binding for small molecule ligands to macromolecular targets. Docking is widely used for the study of biomolecular interactions and mechanisms, and is applied to structure-based drug design. The methods are fast enough to allow virtual screening of ligand libraries containing tens of thousands of compounds. This protocol covers the docking and virtual screening methods provided by the AutoDock suite of programs, including a basic docking of a drug molecule with an anticancer target, a virtual screen of this target with a small ligand library, docking with selective receptor flexibility, active site prediction, and docking with explicit hydration. The entire protocol will require approximately 5 hours.
Introduction
Computational docking is widely used for study of protein-ligand interactions and for drug discovery and development. Typically the process starts with a target of known structure, such as a crystallographic structure of an enzyme of medicinal interest. Docking is then used to predict the bound conformation and binding free energy of small molecules to the target. Single docking experiments are useful for exploring the function of the target, and virtual screening, where a large library of compounds are docked and ranked, may be used to identify new inhibitors for drug development.
AutoDock is a suite of free open–source software for the computational docking and virtual screening of small molecules to macromolecular receptors. The suite currently includes several complementary tools:
AutoDock Vina: a turnkey computational docking program based on a simple scoring function and rapid gradient-optimization conformational search. 1
AutoDock: a computational docking program based on an empirical free energy force field and rapid Lamarckian genetic algorithm search method2,3.
Raccoon2: an interactive graphical tool for virtual screening and analysis4.
AutoDockTools: an interactive graphical tool for coordinate preparation, docking and analysis.5
AutoLigand: a program for predicting optimal sites of ligand binding on receptors.6
The AutoDock suite, including source, is freely available, and has been widely used in research and drug discovery.
Comparisons to Other Methods
A variety of academic and commercial methods for computational ligand docking are currently available (see Ref 1 for an extensive review of current methods). Most of these methods simplify the problem in two ways to make the computation tractable. First, the conformational space is reduced by imposing limitations to the system, such as a rigid receptor and fixed bond angles and lengths in the ligand. Second, a simplified scoring function, often based on empirical free energies of binding, is used to score poses quickly at each step of the conformation search.
Both of these are serious limitations, and users must employ tools such as molecular dynamics or free energy perturbation if a more realistic conformational search or energy prediction is necessary. These tools are complementary with computational docking methods, since docking methods generally search a larger conformational space, but more advanced methods can predict conformation and energy more accurately within a local area of the conformational landscape.
Advanced docking methods may be used to improve results in cases where the limitations of requiring a rapid method for energy evaluation are too restrictive. For instance, many docking methods employ a rigid model for the receptor, which often leads to improper results for proteins with appreciable induced fit upon binding. AutoDock includes a method for treating a selection of receptor sidechains explicitly, to account for limited conformational changes in the receptor. In addition, ordered water molecules often mediate interactions between ligands and receptors, and advanced methods for treating selected waters explicitly have been implemented in AutoDock. Both of these advanced methods are demonstrated in this protocol.
Many reports have compared the performance of popular docking methods such as AutoDock (recently reviewed by Sousa et al. 7). Different methods can achieve different success rates depending on specific targets, but in general, they all perform similarly when tested on a series of diverse protein-ligand complexes: they all perform well for the prediction of bound complexes for drug-sized molecules, with estimates of free energies of binding with errors of roughly 2–3 kcal/mol, provided that there is not significant motion required in the receptor. Better results may be obtained by tuning the docking method for a particular system or moving to more sophisticated and computationally-intensive parameterizations of the system.
Applications of the Protocol
The AutoDock suite is used in numerous laboratories: a recent search on PubMed yielded over 1000 citations in the past year. We have applied the AutoDock suite to a number of different problems in collaboration with researchers, showing some of the scope of uses. The first versions of AutoDock were used to explore the binding of substrates to the enzyme aconitase 8, and docked conformations were 8 used to help interpret crystallographic density maps and to explore intermediates in the reaction. Results of several similar studies on proteins and their substrates were reported in the following years 9–11. We have also used the Autodock suite as a tool for drug design and virtual screening against a number of targets, including HIV protease 12–14, tuberculosis targets 15,16, and beta-secretase 17,18. AutoLigand was used to identify druggable sites for stabilizing a protein-protein interface 19 and covalent docking was used to explore covalent inhibitors of protein tyrosine phosphatases 20.
AutoDock and AutoDock Vina
Two docking methods have been developed in parallel, to respond to two different needs. Development began with AutoDock2,3,5,21,22, and it continues to be the platform for experimentation in docking methods. AutoDock Vina was developed more recently to fulfill the need for a turnkey docking method that doesn’t require extensive expert knowledge from users1. It is highly optimized to perform docking experiments using well-tested default methods. Both methods are currently freely available. AutoDock Vina is fast and effective for most systems, while AutoDock is available for systems that require additional methodological enhancements.
Both methods are designed to be generic computational docking tools, accepting coordinate files for receptor and ligand, and predicting optimal docked conformations. Typically, users start with receptor coordinates from crystallography or NMR spectroscopy, and ligand coordinates generated from SMILES strings or other methods.
Because the search methods are stochastic, a set of optimal docked conformations is predicted, then typically clustered spatially to analyze consistency of the results. Highly clustered results are an indication that the conformational search procedure is exhaustive enough to ensure coverage of the accessible conformational space. Due to the stochastic nature of the search, the method cannot ensure that a global minimum has been found. For this reason, it is important to use re-docking experiments with known complexes of similar conformational complexity to evaluate the docking protocol being used.
AutoDock and AutoDockVina currently employ several simplifications that affect the results that are obtained. The most significant simplification is the use of a rigid receptor. This approximation reduces the size of the conformational space, allowing it to be searched reliably, and reduces the computational effort of scoring each trial conformation. When applying these docking methods to a given receptor it is important to consider the possible effects of this limitation, and if the system includes significant receptor motion, a number of methods may be employed, including:
Using receptor structures taken from receptor-ligand complexes, where there is some expectation that the receptor is in the relevant conformation.
Docking to a collection of different receptor structures, which cover the expected range of flexibility in the receptor. These may be obtained from multiple structural determinations or simulation.
Use of explicit receptor side chain flexibility during docking, if information is available on relevant side chains (described in the protocol).
The scoring methods also employ a variety of simplifications that will affect the results. The AutoDock Vina scoring function is highly approximate, with spherically symmetric hydrogen bond potentials, implicit hydrogens, and no electrostatic contribution. It has been demonstrated to perform well with ligands with typical biological size and composition. The AutoDock force field includes physically based contributions, including a directional hydrogen-bonding term with explicit polar hydrogens, and electrostatics. If these contributions are important in a particular system, AutoDock would be the appropriate tool. In addition, the parameterization of the AutoDock scoring function is available to the user, to allow tuning for particular systems if desired. For instance, methods for incorporating explicit solvents and for predicting conformations of covalent complexes were developed by modifying the AutoDock potentials23,24
Raccoon and Virtual Screening
Virtual screening is rapidly becoming the primary application of computational docking methods, with many successes in the discovery of new lead compounds for pharmaceutical development25. The idea is to screen a large library of available ligands to identify a small subset for purchase and experimental testing.
Raccoon is a graphical user interface designed to streamline the steps of performing a virtual screening and analyzing the results. These include:
Automated server connection manager and installation of docking services (such as AutoDock Vina).
Ligand Library for upload and management of large ligand collections.
Receptor management from multiple targets and flexible residues.
Graphical interface for docking parameter setup.
Graphical management of jobs on computational resources.
Automated retrieval and preprocessing of results to extract features of interest.
User-friendly filtering of virtual screening results based on properties and interactions.
Export of filtered results.
Coordinate Preparation with AutoDockTools
Successful docking and virtual screening requires careful attention to the quality of the coordinates used for receptors and ligands. Both AutoDock and AutoDock Vina use a simplified representation of the molecules, which is included in a modified Protein Data Bank (PDB) file format, termed PDBQT:
United atom representation: Both methods require coordinate sets that include the polar hydrogen atoms. AutoDock uses polar hydrogen coordinates during docking; AutoDock Vina uses them to assign the hydrogen bonding state of the heteroatoms, but does not use explicit hydrogens during the docking.
Atom typing: Both methods require a simplified typing of atoms, including identification of aromatic and aliphatic carbon atoms and identification of the hydrogen bonding state of heteroatoms.
Atomic charges: AutoDock uses Gasteiger-Marsili atomic charges for calculation of electrostatic interactions and desolvation energies. AutoDock Vina does not use atomic charges.
Ligand flexibility: Both methods require the user to specify the torsional degrees of freedom in ligand molecules and in any receptor side chains that are to be flexible.
Search Space: Both methods require the user to specify a docking box covering the space around the receptor that will be searched.
The graphical user interface AutoDockTools provides methods for creating suitable coordinate files from files in a variety of common formats. Automated scripts are also available for batch processing of libraries of compounds.
Preparation of coordinate files (steps 1–4) is arguably the most important aspect of the process, since the quality of coordinates will affect all results from docking. After preparation, take a critical look at the coordinate files and examine the protonation state and charges (particularly metals, if present) to see if they are consistent with knowledge of the system. For instance, ADT does not provide any charges for metal ions, therefore charges need to be manually added in the PDBQT file using a text editor. Receptor coordinate files deposited in the PDB often have many challenges that need to be addressed. Double-check to make sure that you have the proper biological unit, that essential residues or loops are not missing from the coordinate set, that you have chosen only one set of coordinates where there are alternate conformations in the deposition, and that you have included only essential cofactors and structural waters.
Advanced Methods
The default methods used in AutoDock and AutoDock Vina are highly effective for typical drug-like ligands, and have been widely used for applications such as virtual screening25. Several refinements have been developed, however, to approach problems that present challenges with the default methods.
Receptor Flexibility: Arguably the greatest limitation in these types of docking methods is the rigid model of the receptor. AutoDock and AutoDock Vina both allow limited flexibility of selected receptor sidechains, as described in this protocol. For systems with larger motions of loops or domains, the Relaxed Complex Method26 has shown success by sampling a variety of receptor conformations using molecular dynamics, and then performing docking simulations on these snapshots.
Explicit Hydration: Interactions between biological molecules are often mediated by ordered water molecules. We have developed a method that uses the existing version of AutoDock, but modifies the force field to model explicit water molecules. The ligand is decorated with an ensemble of water molecules, which may then contribute to the interaction or not based on a modified energy evaluation grid. In tests, this hydration has shown improvement in the prediction of bound conformations of small fragment molecules, such as those used in fragment-based drug discovery23.
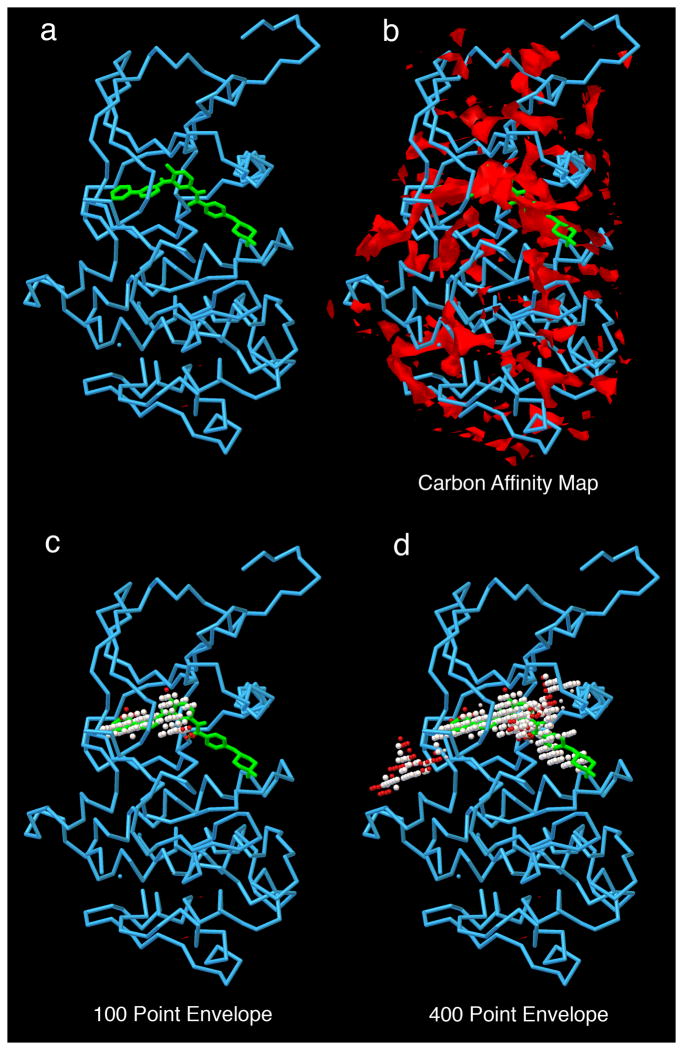
Active Site Prediction: Docking of ligands to the entire surface of a protein is often computationally prohibitive. The AutoDock Suite includes the AutoLigand program for identifying likely binding sites on a receptor surface25. AutoLigand predicts optimal substrate envelopes for binding sites based on the free energy force field of AutoDock. From coordinates for the receptor, AutoLigand calculates and analyzes maps of interaction energy to output coordinate files that show optimal envelopes for ligand binding to the receptor.
Limitations of the Protocol
The AutoDock suite is designed to solve a specific problem: the docking of small, drug-like molecules to biological macromolecules of known structure. It has been implemented, calibrated, and tested with a diverse set of protein-ligand complexes of biological and medicinal interest, and is expected to perform consistently with this type of system, as described in more detail in the “Anticipated Results” below.
Systems that deviate from these design parameters will give variable results and should be approached with caution. Users most often encounter two significant limitations. First, users often want to dock very large ligands, such as a decapeptide. These ligands present too many degrees of freedom and docking methods are not able to search the accessible conformational space. Most often, this problem is solved by breaking the problem into smaller pieces. Second, the protein targets often show significant conformational flexibility, which is not modeled in the AutoDock suite, apart from the selective sidechain motion including in this Protocol. This problem is typically approached by generating conformations of the protein with other methods (such as molecular dynamics) before docking.
About the Protocol
This protocol will describe many of the steps involved in a typical drug discovery project. The target for the protocol is the kinase domain of the proto-oncogene tyrosine protein kinase c-Abl. The protein is an important target for cancer chemotherapy, in particular, the treatment of chronic myelogenous leukemia (CML). This example was chosen because it demonstrates the use of the protocol on an important cancer target with an approved drug Gleevec (Imantinib), and can show the impact of the drug resistant mutations of the target. The protocol will cover:
Redocking experiments using the structure of c-Abl with the anti-cancer drug imatinib (PDB entry 1iep27), using AutoDock Vina and AutoDock.
Virtual screening with Raccoon2 of a library of compounds against c-Abl, using protein coordinates from PDB entry 1iep.
Cross-docking of imatinib with c-Abl coordinates from PDB entry 1fpu28, modeling flexibility in a threonine that interacts with the drug.
Prediction of optimal ligands for c-Abl using AutoLigand.
An example of docking with explicit water molecules, which can improve results for fragment-based drug design.
MATERIALS
EQUIPMENT
Starting Data
Coordinate file for receptor (in a variety of formats, including pdb, mol2, cif & sdf)
Coordinate file for ligand (in a variety of formats, including pdb, mol2, cif & sdf)
Several files are available in Supplementary Data for use as a tutorial for each of the protocols: 1iep_receptorH.pdb (coordinates of c-Abl kinase domain from PDB entry 1iep, with hydrogen atoms added in AutoDockTools), 1iep_ligandH.pdb (coordinates of imatinib from PDB entry 1iep, with hydrogen atoms added in AutoDockTools), 1fpu_receptorH.pdbqt (coordinates of c-Abl kinase domain from PDB entry 1fpu, in PDBQT format), imatinib.pdbqt (coordinates of imatinib in PDBQT format), NCIdivII_subset (a folder that includes 499 compounds from the ZINC library, formatted as PDBQT files)
Hardware and Software
Computer: Linux, Macintosh, or Windows PC; Internet access
For virtual screening with Raccoon, a Linux cluster/HPC with either a PBS or SGE scheduler
Text editor
AutoDock: http://autodock.scripps.edu (information on installation is available at: http://autodock.scripps.edu/downloads/autodock-registration/autodock-4-2-download-page/)
AutoDock Vina: http://vina.scripps.edu (information on installation is available at: http://vina.scripps.edu/manual.html#faq)
AutoDockTools (part of MGLTools): http://mgltools.scripps.edu (information on installation is available at: http://mgltools.scripps.edu/downloads)
AutoLigand is part of AutoDockTools
Raccoon is available at http://autodock.scripps.edu/resources/raccoon
PROCEDURE
Coordinate Preparation with AutoDockTools - timing 10 min
-
1|
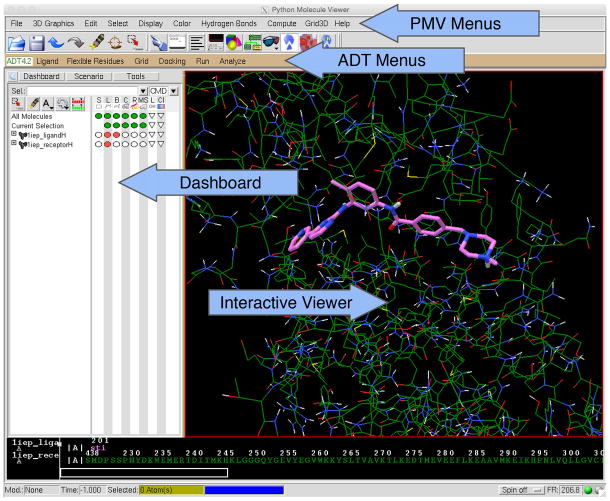
Generate the ligand coordinate file. A coordinate set that includes hydrogen atoms is required. This may be obtained in a variety of ways, including experimental coordinates from the Protein Data Bank (www.pdb.org) or Cambridge Crystallographic Database (ccdc.cam.ac.uk), or structure generation methods such as the CACTUS server (cactus.nci.nih.gov/translate). The file 1iep_ligandH.pdb is provided for use as a tutorial for this protocol (all example files are supplied in Supplementary Data) it includes ligand coordinates taken from PDB entry 1iep, to which all hydrogen atoms have been added and manually adjusted to the known protonation state. Start AutoDockTools (ADT) (Figure 1) and set the working directory by clicking “File->Preferences->Set”. Type your working directory path name into the “Startup Directory” box and click “Set”. Click “Dismiss” at the bottom of the window.
-
2|
Read the atomic coordinates. To do this, first select “Ligand->Input->Open” and use the “Files of type” menu to choose “PDB files”. Click on your coordinate file, in this case, 1iep_ligandH.pdb, and click “Open”. ADT will read the coordinates, add charges if necessary, merge non-polar hydrogens, and assign appropriate atom types. At this point, the ligand will be displayed in the viewer window, with aromatic carbons in green. Click “OK” on the popup to continue.
CRITICAL STEP If your coordinate set does not include hydrogen positions, click “File->Read Molecule” and choose your coordinate file. Then, by clicking “Edit->Hydrogens->Add”, you will add all hydrogens by default. Select “Ligand->Input->Choose” to choose the ligand molecule. -
3|
Prepare a PDBQT file by selecting “Ligand->TorsionTree->DetectRoot”, this will define the center of the torsion tree. Selecting “Ligand->TorsionTree->ChooseTorsions” will launch a window that allows choice of torsional degrees of freedom. Rotatable bonds are in green, rigid portions are in red, and potentially rotatable bonds that are currently set as “not rotatable” (such as the peptide bond at the center of imatinib) are in magenta. Clicking on bonds will switch the rotation flexibility on and off. When finished, click “Done”. Click “Ligand->Output->SaveAsPDBQT”, then select “Save” to write the file 1iep_ligandH.pdbqt.
?TROUBLESHOOTING
-
4|
Generate the receptor coordinate file. A receptor coordinate file with all hydrogen atoms is required. If you are using experimental structures (for instance, from the PDB), use a text editor to remove water, ligands, cofactors, ions, etc. that should not be included in the receptor. The file 1iep_receptorH.pdb is provided for use as a tutorial for this protocol andit includes receptor coordinates taken from PDB entry 1iep. Open the file by selecting “Grid->Macromolecule->Open”, use the “Files of type” menu to choose “all files”. Click on your coordinate file, in this case, 1iep_receptorH.pdb, and click “Open”. ADT will read coordinates, add charges, merge non-polar hydrogens, and assign appropriate atom types. Click “OK” to accept the changes. A window will pop up to write the PDBQT file. Click “Save” to write a file 1iep_receptorH.pdbqt
CRITICAL STEP If your coordinate set does not include hydrogen positions, Click “File->Read Molecule” and choose your coordinate file. Then, selecting “Edit->Hydrogens->Add” will add all hydrogens by default. Click “Grid->Macromolecule->Choose” to choose the receptor molecule.?TROUBLESHOOTING
Figure 1. AutoDockTools.
ADT is built within the Python Molecule Viewer. ADT commands for coordinate preparation, docking, and analysis are available through menus on the lower toolbar. PMV commands for higher level visualization are available on the upper tool bar. The Dashboard controls representation, colors, and labeling of molecular objects, which are displayed in the Interactive Viewer panel. For more information on the capabilities of PMV and ADT, see http://mgltools.scripps.edu/documentation.
Methods for Docking Simulation
-
5|
Once receptor and ligand coordinates are formatted, the AutoDock suite provides a number of methods for docking simulation. This protocol includes six methods, ranging from a simple docking to advanced methods, as described in the following table.
Option Method Description A Single docking experiment with AutoDock Vina Basic docking method for study of a single ligand with a single receptor B Single docking experiment with AutoDock Basic docking method for study of a single ligand with a single receptor, with explicit calculation of affinity maps. C Virtual Screening with Raccoon2 and AutoDock Vina Virtual screen of a library of ligands with a single receptor, often used for drug discovery D AutoDock Vina with Flexible Side Chains Docking method for a single ligand with a single receptor, incorporating limited receptor flexibility E Active Site Prediction with AutoLigand Method for analysis of receptor binding sites, for prediction of drugable sites F Docking with Explicit Waters Advanced docking method for a single ligand with a single receptor incorporating explicit bridging water molecules
(A) Single Docking Experiment with AutoDock Vina - timing 30 min
Generate a configuration file (Box 1) for AutoDock Vina that specifies the PDBQT files for the ligand and receptor, and defines the docking parameters. To do this, restart ADT and set the default working directory (see Step 1). Select “Grid->Macromolecule->Choose/Open”. “Choose” is used when coordinates have already been read into ADT (as in the coordinate setup above), and “Open” is used to read coordinates from a file. For this protocol, we are assuming that we have restarted ADT and need to read from coordinate files. “Open” the receptor PDBQT file, click “Yes” to preserve the existing charges in the file, and “OK” to accept. There may also be a warning window if there are slight irregularities in charges. Click “OK” if it appears.
Select “Grid->SetMapTypes->OpenLigand”, choose the ligand PDBQT file and click “Open”. “Grid->GridBox” opens a window for defining the center and size of the search space. “Center->CenteronLigand” will define the box based on the ligand. Other options are available, or the values may be changed manually with the thumbwheels. Important: when finished, choose “File->CloseSavingCurrent” in the window “Docking->Output->VinaConfig”, write the configuration file (default name: config.txt) by clicking “Save”.
-
Run AutoDock Vina. The imatinib ligand used in this protocol is challenging, and Vina will occasionally not find the correct pose with the default parameters. Vina provides a parameter called “Exhaustiveness” to change the amount of computational effort used during a docking experiment. The default exhaustiveness value is 8; increasing this to about 24 will give a more consistent docking result. There are two ways to run AutoDock Vina, from ADT or from the command line. From ADT select “Run->RunAutoDockVina” and in the popup window, use the “Browse” buttons to specify the path to the AutoDock Vina executable file vina and the path to the configuration file. Edit the “Cmd” line to change the exhaustiveness, such as:
./vina --config config.txt --exhaustiveness=24
Click the “Launch” button to run the program.
Programs in the AutoDock suite may also be run at the command line. To do this, instead open a terminal window and change to the directory that contains the coordinate files and configuration file. Then issue the command listed above. This command assumes that the AutoDock Vina executable vina is also located in the same directory.
Running AutoDock Vina will write a docked coordinate file 1iep_ligandH_out.pdbqt and also present docking information to the terminal window. The predicted free energy of binding should be about -13 kcal/mol for poses similar to the crystallographic pose. With exhaustiveness set to 24, Vina will most often give a single docked pose with this energy. With the lower default exhaustiveness, several poses flipped end-to-end, with less favorable energy, may be reported.
?TROUBLESHOOTING
-
To visualize the results from AutoDock Vina (Figure 2), launch ADT and set the working directory (see Step 1). Select “Analyze->Dockings->OpenAutoDockVinaResult” and choose the output file from Step 5(A)iii. Choose the default of “Single molecule with multiple conformations” and click “OK”. This will show coordinates for each docked result. Use the arrow keys on the keyboard to scroll through the poses. Click “Analyze->Macromolecule->Open” to choose the receptor coordinate file to visualize the receptor, 1iep_receptorH.pdbqt. Select “File->ReadMolecule” to choose the ligand coordinate file 1iep_ligandH.pdb, visualize the crystallographic location of the ligand and compare to the docked conformation.
“Analyze->Dockings->ShowInteractions” will launch a visualization that highlights interactions between the ligand and receptor.
Box 1. Vina Configuration File.
receptor = 1iep_receptorH.pdbqt ligand = 1iep_ligandH.pdbqt center_x = 15.19 center_y = 53.903 center_z = 16.917 size_x = 15.0 size_y = 17.25 size_z = 15.0 out = 1iep_ligandH_out.pdbqt
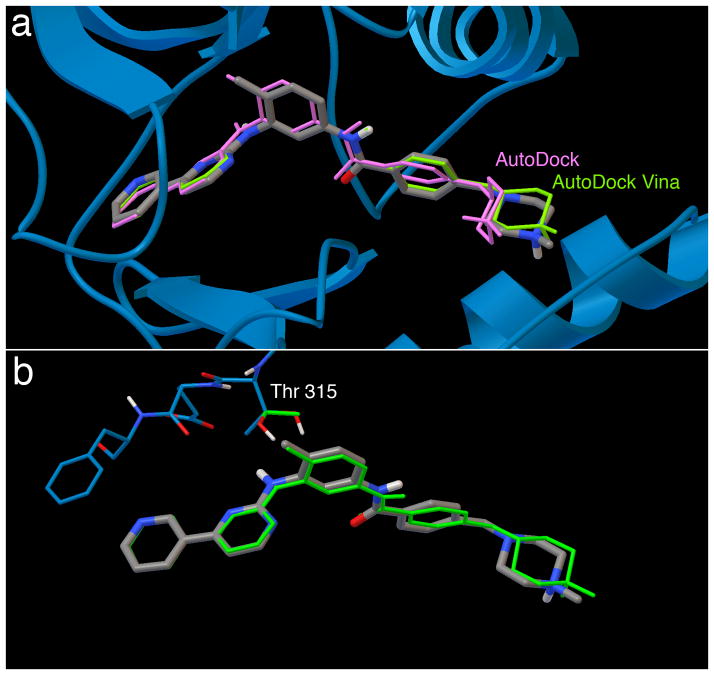
Figure 2. Results of docking imatinib to its receptor in bound and apo conformations.
a. Redocking of flexible imatinib to rigid Abl (PDB entry 1iep) using AutoDock (purple) and AutoDock Vina (green). The X-ray crystallographic ligand position is in silver.
b. Cross docking of flexible imatinib to Abl (PDB entry 1fpu) with a single flexible residue side chain using AutoDock Vina (green). Note that Vina does not retain hydrogen atom positions during docking, so the threonine hydroxyl hydrogen is placed in a random position in the docked coordinate set.
(B) Single Docking Experiment with AutoDock - timing 60 minutes
Generate a grid parameter file for AutoDock that specifies the PDBQT files for the receptor, and parameters for generating the atomic affinity maps. Start ADT and set the default working directory (see Step 1). Select “Ligand->Input->Open” and choose the ligand PDBQT file (1iep_ligandH.pdbqt). Select “Grid->Macromolecule->Open” and choose the receptor PDBQT file (1iep_receptorH.pdbqt). Set the map types by selecting “Grid->SetMapTypes->ChooseLigand” and choosing the ligand PDBQT file (1iep_ligandH.pdbqt). Define the search space by selecting “Grid->GridBox,” which will open a window for specifying the center and size of the search space. Use “Center->CenteronLigand” to define a minimal box. When finished, choose “File->CloseSavingCurrent.” Finally, select “Output->SaveGPF” to save the grid parameter file, which is typically given the extension.gpf (use “1iep.gpf”).
-
Run AutoGrid, either from ADT or from the command line. From ADT, select “Run->RunAutoGrid,” and in the popup window, use the “Browse” buttons to specify the path to the AutoGrid executable file and the path to the grid parameter file. After you specify the grid parameter file, ADT will suggest a name for the log file. Click the “Launch” button to run AutoGrid.
Alternatively, at the command line, open a terminal window and change to the directory that contains the coordinate files and grid parameter file. Then issue the command:
./autogrid4 -p 1iep.gpf -l 1iep.glg
This command assumes that the AutoGrid executable autogrid4 is also located in the same directory.
?TROUBLESHOOTING
Optional: visualize AutoGrid maps in ADT (see section E below). Start ADT and set the default working directory (see Step 1). In the top toolbar, select “Grid3D->Read” and choose a map file (such as 1iep_receptorH.C.map for the carbon affinity map). In the popup control panel, click on the file in the “Grid Name” panel, click on the “Isocontour” button, and ADT will display a histogram of map values. Change the “max” value to 0.0 and press return (or ‘enter’). Shift-click in the histogram to calculate an isocontour; you can then drag the bar in the histogram right and left to change the contour level. Values in the range of −0.5 to −0.1 are effective. The menu option “File->ReadMolecule” can be used to display the receptor.
Generate the docking parameter file that specifies the PDBQT file for the ligand and parameters for the docking simulation. Select “Docking->Macromolecule->SetRigidFilename” and choose the receptor PDBQT file (1iep_receptorH.pdbqt). Select “Docking->Ligand->Choose,” choose the ligand PDBQT file (1iep_ligandH.pdbqt), and accept the default ligand docking parameters, which will randomize the pose of the ligand before docking. Select “Docking->SearchParameters->GeneticAlgorithmParameters” and use the default parameters for most drug-sized ligands, except set “Number of GA Runs” to 50. Additional docking parameters are specified with “Docking->DockingParameters;” use default parameters for most drug-sized ligands. Finally, write the docking parameter file by selecting “Docking->Output->LamarckianGA,” which is typically given the extension.dpf (use “1iep.dpf”).
-
Run AutoDock, either from ADT or at the command line. To run from ADT, select “Run->RunAutoDock,” and in the popup window, use the “Browse” buttons to specify the path to the AutoDock4 executable file and the path to the docking parameter file. After you specify the docking parameter file, ADT will suggest a name for the log file. Click the “Launch” button to run the program.
Alternatively, at the command line, open a terminal window and change to the directory that contains the coordinate files and docking parameter file. Then issue the command:
./autodock4 -p 1iep.dpf -l 1iep.dlg
This command assumes that the AutoDock executable autodock4 is also located in the same directory.
-
Visualize AutoDock results (Figure 3a). Start ADT and set the default working directory (see Step 1), and to access the results, select “Analyze->Dockings->Open” and choose the 1iep.dlg file. There are several options in ADT for visualizing results: select “Analyze->Conformations->PlayRankedByEnergy” and click on the arrow buttons to scroll through conformations, or select “Analyze->Docking->Load” to open a clickable window with ranked conformations and energies
?TROUBLESHOOTING
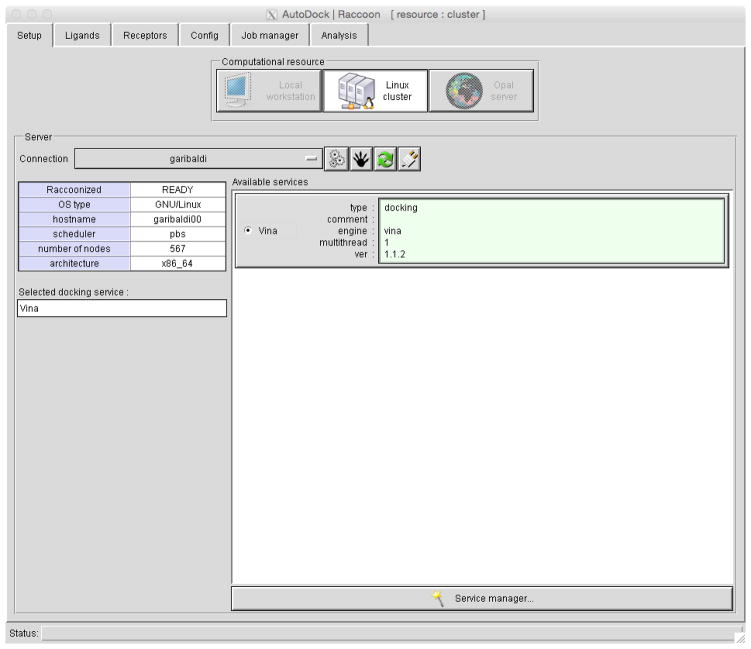
Figure 3. Raccoon2.
Tabs at the top choose each of the steps for setting up, running and analyzing a virtual screen.
(C) Virtual Screening with Raccoon2 and AutoDock Vina - timing 60 min
-
Start Raccoon2 and configure the server. Raccoon is designed to run virtual screening on a large computational resource, such as a Linux cluster. When you add a new server, you must configure the connection and install one or more docking services. Launch Raccoon2 (Figure 3) and create a new server connection by clicking the three-gear icon to open the Connection Manager. Several options need to be set: server name (a name to identify the resource); address (the host name or IP address of the server); and a username and password. Click “Save” and close the Connection Manager.
Install a docking service by selecting the server in the Connection menu. Click on the raccoon-paw icon to prepare (“raccoonize”) the server. Click on the “Service Manager” icon (a wand) and add a new service by clicking on the plus icon, and give it a descriptive name of your choice (“Vina docking service”). Click on “Install AutoDock Vina on the server”, and accept several default selections. Click Save, and close the Service Manager. Finally, from the “Available Services” panel, select the “Vina docking service”
?TROUBLESHOOTING
-
Set up the Ligand Library. This protocol describes virtual screening of C-Abl with a small library of 500 compounds that includes the known inhibitor imatinib. Tools for generating and processing ligand libraries are available at http://autodock.scripps.edu/resources/databases.
Ligand libraries are stored on the server and made available for docking jobs, to reduce redundancy and allow tracing and reproducing experiments.
In the LIGANDS tab, select “AddLigand->ScanDirectory” and browse to the directory containing the ligand library. Close the report window and select “Upload”, type a descriptive library name of your choice and click “Start.” Close after upload is complete, close the Library Manager. Right click to select the desired ligand library.
Set up the receptor coordinates. In the RECEPTORS tab, select “Add->ScanDirectory” browse to the receptor PDBQT file (created in Step 4), and click “OK”. Close the report window.
Configure AutoDock Vina docking parameters. In the CONFIG tab: load a receptor structure in the 3D viewer by selecting files in the Receptor list. Use “Center” and “Size” thumbwheels to define the search space, or place the mouse cursor over boxes and type numeric values. Modify docking parameters if desired or load the config.txt file created in 5(A)i.
Peform the virtual screening calculation. In the JOB MANAGER tab, ensure that all “Cluster submission requirements” are green, then click “Submit.” Several options will need to be specified: set “Project” to “new” and type a descriptive name of your choice; set “Experiment” to “new” and type a descriptive name of your choice; and click “OK” to start the calculation. To follow the status of the docking, right click on the experiment name and select “update status.” When one or more jobs in the experiment are finished, download them individually by right-clicking on the job name, or together by right-clicking on the experiment and select “Download results.”
-
Filter and analyze the results. To assist with filtering and selection, Raccoon calculates docking pose properties such as interaction, score, and ligand efficiency. In the ANALYSIS->DATA SOURCE tab, click on “Add results->Select downloaded results”, and browse to the results directory. Choose the receptor file to be used for processing results and type a descriptive log file name or select the config.txt file generated at Step 5(A)i.
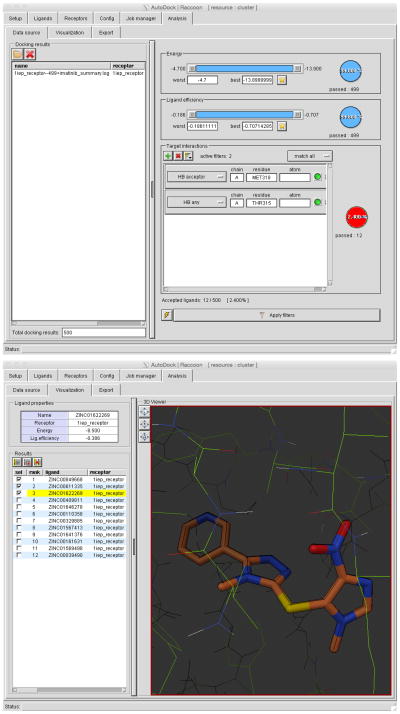
The filter panel allows a variety of filters to be applied to the results (Figure 4): “Energy” will show poses within an energy range; “Ligand Efficiency” will select poses within a range of ligand efficiency; and with “Target Interactions,” use “+” to filter by selected interactions with receptor atoms.
For the c-Abl virtual screen described in this protocol, click on the “+” button and add a “HB acceptor” filter with MET318, and press return (Note: the interface currently requires all capitals for the residue name). Click on the “+” button and add a “HB any” filter with THR315, and press enter. Switch the menu from “match any” to “match all” and update the filters. This will select the small number of molecules that show both of these interactions. The THR319 position of c-Abl is known to mutate to obtain resistance. To find molecules that do not interact with this position, click on the filter button for THR315 added above, and switch the filter from “wanted” (green) to “not wanted” (red)
?TROUBLESHOOTING
Export results. Once a set of ligands is filtered, select interesting ones by clicking on the button in the Results panel. Use the EXPORT tab to export the selected candidates.
Figure 4. Raccoon Results Filtering.
The “Data Source” window (top) has several options for filtering the results of a virtual screen. Two interaction filters are applied here. The “Visualization” window (bottom) allows visualization of the filtered results, and checkboxes can be used to select the desired compounds for export.
(D) AutoDock Vina with Flexible Side Chains - timing 60 min
-
Generate receptor coordinate files. The receptor coordinates are split into two PDBQT files, one for the rigid portion and one for the flexible side chains. As with the rigid docking protocols, the method requires a receptor coordinate file that includes all hydrogen atoms. This protocol describes the cross-docking of imatinib to c-Abl in PDB entry 1fpu, treating Thr315 as flexible.
Start ADT and set the default working directory (see Step 1), select “Grid->Macromolecule->Open,” and choose 1fpu_receptorH.pdbq. Select residue(s) to be treated as flexible by selecting “FlexibleResidues->Input->ChooseMacromolecule,” and browse to the receptor coordinate file. Several selection methods are available, including direct selection and using the Select menu. With “Select->SelectFromString” in the PMV toolbar, choose in “Residue”, type “THR315” and click “Add”. ADT will highlight the selected residue with small yellow crosses. Click “Dismiss”. Choose this selected residue to be treated as flexible with the command “FlexibleResidues->ChooseTorsionsinCurrentlySelectedResidues”. Finally, select “FlexibleResidues->Output->SaveFlexiblePDBQT” to save the flexible coordinate file 1fpu_flex.pdbqt and “FlexibleResidues->Output->SaveRigidPDBQT” to save the rigid receptor coordinate file 1fpu_rigid.pdbqt.
Generate parameter files for AutoDock Vina. To include the mobile residue, a larger grid box must be used: its bounds can be found using the “Grid->GridBox” tool. For this protocol, enter them manually. In ADT, select “Docking->Output->VinaConfig” and use “Browse” to select the rigid (1fpu_rigid.pdbqt) and flexible (1fpu_flex.pdbqt) receptor files. Important: make sure to use the 1fpu_rigid.pdbqt for the rigid file, not the full receptor PDBQT file. Click “Browse” to select the ligand coordinate file 1iep_ligandH.pdbqt (step 3), type 15.19, 53.9, 14.6 into the “Center” boxes, and type 15.0, 17.25, 19.5 into the “Size” boxes. Change the “Output filename” to “config_flex.txt,” and change the “Out” filename (this is the file for results) to 1fpu_ligandH_flex.pdbqt. This will create a configuration file called config_flex.txt (Box 2).
-
Perform flexible side chain docking in AutoDock Vina at the command line:
./vina --config config_flex.txt --exhaustiveness 24
The “exhaustiveness” parameter performs additional docking simulations. The default value is “8”; a value 3 to 4 times larger works well for the current system.
-
Analyze the flexible docking results using ADT (Figure 3b) as described in Step 5A(iv).
?TROUBLESHOOTING
Box 2. AutoDock Vina Configuration File for Flexible Docking.
receptor = 1fpu_rigid.pdbqt ligand = 1iep_ligandH.pdbqt flex = 1fpu_flex.pdbqt center_x = 15.19 center_y = 53.903 center_z = 14.661 size_x = 15.0 size_y = 17.25 size_z = 19.5 out = 1fpu_ligandH_flex.pdbqt
(E) Active Site Prediction with AutoLigand - timing 30 min
-
AutoLigand identifies optimal regions for ligand binding by analyzing AutoGrid interaction energy maps for three atom types (C, OA and HD) and the electrostatics and desolvation maps, all calculated with a 1 Å grid spacing. The first step is to calculate the energy maps. Start ADT and set the default working directory (see Step 1). In “Grid->Macromolecule->Open,” select a coordinate file for receptor (1iep_receptorH.pdbqt). By default, ADT will name the map files based on the name of this file. In “Grid->SetMapTypes->Directly,” edit the atom list to be “C HD OA.”
In “Grid->GridBox,” adjust the size to cover the entire protein, and set the spacing to 1 A. For 1iep, use a size of 60, 60, 70 (put the mouse cursor over the dial, type in values, and press return (or ‘enter’)), and save the parameters using “File->CloseSavingCurrent”. Write the grid parameter file with “Grid->Output->Save GPF” with the filename “1iep.gpf.” Run AutoGrid at the command line (see Step 5(B)ii):
./autogrid4 -p 1iep.gpf -l 1iep.glg &
AutoLigand is not installed by default, and must be loaded into ADT: select “File->BrowseCommands.” Then, in “Select a Package,” select “AutoDockTools;” in “Select a Module”, select “AutoLigand Command;” click “Load Select Module: and then click “OK.” This will add an “AutoLigand” entry to the ADT “Compute” menu.
-
AutoLigand may be run in two modes. This protocol will generate ligand envelopes individually, manually picking a seed point and a volume in ADT. This mode is appropriate if you know the location of the active site of your target. AutoLigand may also be run at the command line to scan the entire surface of the protein, predicting the location of optimal binding sites. This mode is described in more detail on the AutoDock website.
Select “File->ReadMolecule” and read 1iep_receptorH.pdbqt (from step 4), and select “File->ReadMolecule” and read 1iep_ligandH.pdbqt from step 3(the ligand coordinates are not used by AutoLigand, but will be used to locate the active site in this protocol). Start the AutoLigand computation with “Compute->AutoLigand->RunAutoLigand.” This opens a window that allows you to choose the seed point and size of the envelope. Use the sliders to place the yellow seed point marker near the ligand/active site, or shift-click on an atom in the ligand to place the marker there. Use the default of 100 for the volume of the envelope. Start the AutoLigand calculation by clicking “OK.” When AutoLigand finishes, a pop-up window will display the results. The envelope will be displayed in the viewer (Figure 5), and a PDB file will be written with coordinates of the envelope.
Figure 5. AutoLigand Results.
c-Abl is analyzed with AutoLigand. a) c-Abl with imatinib; b) carbon affinity map calculated around the protein--notice the many disconnected areas of strong affinity; c) 100 point envelope identifies the regions of the active site that provide the strongest affinity for the drug; d) 400 point envelope includes the region of high affinity and also extends into adjacent solvent-accessible regions on the protein surface.
(F) Docking with Explicit Waters - timing 60 min
-
This method adds dummy atoms to the ligand that correspond to all possible sites of hydration. A modified AutoGrid map is then used during docking, giving a favorable score when the water is well placed and omitting the water if it overlaps with the receptor. A final script analyzes the docked results, retaining only those waters in appropriate positions.
This protocol assumes that the ligand and receptor have been prepared for a standard AutoDock docking. Two coordinate files, ligand.pdbqt and protein.pdbqt,, created using the same protocol as those described in steps 1–4 in the above example, have been provided for this protocol, along with a modified parameter file for the force field, a modified docking parameter file, and several python scripts for performing each step.
To add water positions to the ligand, type at the command line:
$ python wet.py -i ligand.pdbqt
This script adds the dummy “W” atoms to the ligand PDBQT file, saving it to the file “ligand_HYDRO.pdbqt”
-
Calculate the default atomic grid maps. Start ADT and set the default working directory (see Step 1). Select “Ligand->Input->Open” and choose the ligand PDBQT file (ligand.pdbqt), and select “Grid->Macromolecule->Open” and choose the receptor PDBQT file (receptor.pdbqt). Set the map types in “Grid->SetMapTypes->Directly...” and ensure that the “HD” and “OA” types are in the list. Use “Grid->GridBox” to specify the center and size of the search space. In the popup window, use “Center->CenteronLigand” to define a minimal box. When finished, save the parameters using “File->CloseSavingCurrent.” Write the grid parameter file using “Output->SaveGPF,” with the name “protein.gpf.” Run AutoGrid (see step 5(B)ii for more information) with “Run->RunAutoGrid” or at the command line with:
./autogrid4 -p protein.gpf -l protein.glg.”
-
Generate the “W” map. If standard filenames are used for the maps, only the receptor name must be specified for the script that generates the map for the water energy evaluation:
.$ python mapwater.py -r protein.pdbqt -s protein.W.map
-
Create a modified docking parameter file. Select “Docking->Macromolecule->SetRigidFilename” and choose the receptor PDBQT file (protein.pdbqt). Select “Docking->Ligand->Choose” and choose the ligand PDBQT file (ligand_HYDRO.pdbqt), and accept the default ligand docking parameters, which will randomize the pose of the ligand before docking. Set the search parameters in “Docking->SearchParameters->GeneticAlgorithmParameters,” changing “Number of GA Runs” to 50 and use defaults on the remaining parameters. In “Docking->DockingParameters,” use default parameters for most drug-sized ligands
Write the docking parameter file using “Docking->Output->LamarckianGA,” with the name “ligand_HYDRO_protein.dpf.”
The standard docking parameter file must be modified manually in two ways: “parameter_file AD4_water_forcefield.dat” must be added near the top and “protein.W.map” must be added to the list of maps. Both of these changes are highlighted in Box 3.
?TROUBLESHOOTING
-
Run AutoDock (see step 5(B)v for more information) in ADT with “Run->RunAutoDock” or at the command line with:
./autodock4 -p ligand_HYDRO_protein.dpf -l ligand_HYDRO_protein.dlg.
-
Extract and score the results at the command line with:
$ python dry.py -c -r protein.pdbqt -m protein.W.map -i ligand_HYDRO_protein.dlg
This script will filter the docking results using the receptor to identify displaced water and the W map to rank the conserved ones as STRONG or WEAK. This will write a file called ligand_LELC_DRY_SCORED.pdbqt with the calculated energy.
Box 3. Modified Docking Parameter File for docking with explicit waters.
autodock_parameter_version 4.2 # version control parameter_file AD4_water_forcefield.dat #***CUSTOM PARAMETER FILE outlev 1 # diagnostic output level intelec # calculate internal estat seed pid time # seeds for random generator ligand_types A C HD N NA W #***atom types in ligand, add W fld protein.maps.fld # grid_data_file map protein.A.map # atom-specific affinity map map protein.C.map # atom-specific affinity map map protein.HD.map # atom-specific affinity map map protein.N.map # atom-specific affinity map map protein.NA.map # atom-specific affinity map map protein.W.map #***W-specific affinity map elecmap protein.e.map # electrostatics map desolvmap protein.d.map # desolvation map move ligand_HYDRO.pdbqt #***small molecule, with W atoms about 65.7163 39.7789 -2.5985 # small molecule center tran0 random # initial coordinates/A or random axisangle0 random # initial orientation dihe0 random # initial dihedrals rmstol 2.0 # cluster_tolerance/A rmsref ligand_xray_HYDRO.pdbqt # reference ligand conformation ga_pop_size 150 # individuals in population ga_num_evals 2500000 # maximum energy evaluations ga_num_generations 27000 # maximum number of generations ga_elitism 1 # set elitism level ga_mutation_rate 0.02 # rate of gene mutation ga_crossover_rate 0.8 # rate of crossover ga_window_size 10 # ga_cauchy_alpha 0.0 # Cauchy alpha parameter ga_cauchy_beta 1.0 # Cauchy beta parameter set_ga # set the above parameters sw_max_its 300 # Solis & Wets iterations sw_max_succ 4 # rho parameter sw_max_fail 4 # rho parameter sw_rho 1.0 # size of local search space sw_lb_rho 0.01 # lower bound on rho ls_search_freq 0.06 # local search probability set_psw1 # set pseudo-Solis & Wets unbound_model bound # state of unbound ligand ga_run 10 # do this many hybrid GA-LS runs analysis # ranked cluster analysis
TROUBLESHOOTING
| Step | Problem | Possible Reason | Solution |
|---|---|---|---|
| 3 | Incorrect protonation of ligand | ADT currently uses Babel to add hydrogen positions, which makes occasional errors | Double-check the PDBQT file and manually curate errors in protonation |
| 4 | Missing coordinates in receptor | Experimental coordinate sets are heterogeneous. Common problems include missing side chains and loops, asymmetric units that include only a portion of the biological unit, alternate conformations, essential cofactors and structural waters. | Double-check the PDBQT file and manually curate to include the proper biological unit. In general, use the biological unit and omit ligands and waters except for strongly-bound prosthetic groups. |
| 5(A)iii | Vina does not run | ADT “Start Vina” GUI or command line is not using the proper path to the Vina executable, or file is incorrect CPU or operating system type. | Locate the Vina executable on your computer, and use the appropriate path name. Refer to online installation notes to locate the executable. |
| 5(A)iii | No low energy conformations are found | Grid box does not enclose the active site | Choose an appropriate location for the grid box in ADT |
| 5(B)ii | AutoGrid does not run | ADT “Run AutoGrid” GUI or command line is not using the proper path name to the AutoGrid4 executable, or file is incorrect CPU or operating system type. | Locate the AutoGrid executable on your computer, and use the appropriate path name. Refer to online installation notes to locate the executable. |
| 5(B)v | AutoDock does not run | ADT “Run AutoDock” GUI or command line is not using the proper path name to the AutoDock4 executable, or file is incorrect CPU or operating system type. | Locate the AutoDock executable on your computer, and use the appropriate path name. Refer to online installation notes to locate the executable. |
| 5(B)vi | Re-docking experiments do not reproduce the known pose | System has too many degrees of freedom for AutoDock conformational search | Use longer search protocols, or simplify the system by reducing degrees of freedom (docking in parts or rigidifying part of the ligand) |
| 5(C)i | Problems with selection | Right-click is used in several places in Raccoon for selection – use control-click on Macintosh computers | Use “System Preferences” on Macintosh computers to enable right clicking for the mouse or trackpad, |
| 5(C)vi | No results selected during filtering by interaction | Residue names are case sensitive | Use all capitals, such as THR315 |
| 5(D)iv | No low energy conformations are found | Grid box does not enclose all of the side chains | Choose a larger grid box that encloses the binding side and all of the flexible side chains |
| 5(F)iv | Error message about missing W parameters | Water dummy atom parameters not defined in force field parameter file and/or docking parameter files | Ensure that the modified files are used |
TIMING
The indicated timing of each step is a rough estimate--the actual times will depend on the complexity of the system being docked, and the equipment being used for the computation.
Steps 1–4, coordinate preparation: 10 min
Steps 5A, docking with AutoDock Vina: 30 min
Steps 5B, docking with AutoDock: 60 min
Steps 5C, virtual screening with Raccoon2: 60 min
Steps 5D, flexible side chain docking with AutoDock Vina: 60 min
Steps 5E, active site prediction with AutoLigand: 30 min
Steps 5F, docking with explicit hydration: 60 min
ANTICIPATED RESULTS
AutoDock Vina (Step 5A) will provide coordinates for one or more optimized poses for the ligand (Figure 3a). In our tests of the docking of imatinib with c-Abl, the default docking parameters are sufficient to give a consistent solution in most cases. The conformational flexibility of this system is at the limit of the default docking protocol, which may be indicated by a docking result with multiple less favorable poses. For challenging systems with high degrees of conformational flexibility, the exhaustiveness parameter can be used to perform additional docking simulations, often giving more consistent results. This is described in Step 5(A)iii. When analyzing results from AutoDock Vina, note that it uses the input hydrogen positions to assign hydrogen-bonding types to heteroatoms, but does not optimize them during docking simulation, so the hydrogen positions in the output pose are in random conformations.
The number of evaluations used in AutoDock will determine the exhaustiveness of the conformational search (Step 5B). For ligands with 1–4 torsional degrees of freedom, short (250,000) or medium (2,500,000) lengths should provide adequate searching, but for larger ligands, such as imatinib used in this protocol, a longer search is needed, with 10,000,000–25,000,000 evaluations. The clustering analysis is the best way to determine if the simulation has adequately searched the available conformation space: perform multiple docking simulations and confirm that the best conformation is found multiple times.
Virtual screening with Raccoon2 allows the docking and ranking of tens of thousands of compounds to a macromolecular target (Step 5C). Analysis and filtering is one of the most challenging aspects of virtual screening, since the goal is to identify a few promising leads from a large body of docked results. We have obtained effective results by filtering based on predicted docking energy combined with presence of key interactions in the system. Addition of this type of expert knowledge greatly improves the success rate when compounds are tested.
In our hands, roughly half of systems we have used for docking simulations will give useful results using the default rigid model for the receptor. In other cases, protein motion will cause problems for prediction of reasonable poses. AutoDock and AutoDock Vina may be configured to dock ligands with selected receptor residues treated explicitly as flexible (Step 5D). Docking with flexible side chains is difficult and often requires additional computational effort. The test system included with this protocol is an example. PDB entry 1fpu includes a structure of Abl with an inhibitor similar to imatinib. Threonine 315 in the structure is flipped relative to that in 1iep and unable to form a hydrogen bond with the drug. The protocol performs a cross-docking experiment of imatinib docking to Abl from 1fpu, treating Thr315 as flexible. For best results, AutoDock Vina needs to be run with a more exhaustive search to find the proper pose. For systems with larger motion of loops or domains, separate docking simulations may be run for different conformations of the protein, obtained from multiple experimental structures or dynamics simulations of individual structures.
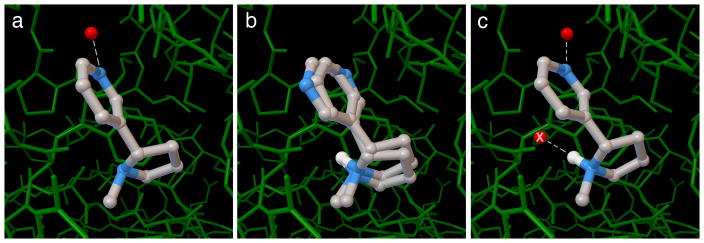
We have found explicit hydration (Step 5F) to be particularly useful for the docking of small ligands and fragment molecules, since water-mediated interactions may form a larger portion of their interaction that with larger ligands. In the example included here (Figure 6), the pyridine ring on nicotine forms a water-mediated interaction with the acetylcholine-binding protein. When the default methods in AutoDock are used, the ligand is placed in the proper position, but two conformations are found with very similar predicted association energies, with the pyridine in the observed position in one and rotated by 180 degrees in the other. In the hydrated docking experiment, two water dummy atoms are placed on the ligand, interacting with the pyridine and interacting with a tertiary amino group. The best pose places the pyridine and associated water in the experimentally observed position, and deletes the other water, since it clashes with the protein.
Figure 6. Docking with Explicit Hydration.
a) crystallographic structure of acetylcholine binding protein with nicotine. An ordered water (red sphere) mediates interaction with the protein. b) The default protocol in AutoDock finds two conformation of the pyridine ring with equal predicted energy. c) hydrated docking predicts the observed conformation. The second water molecule marked with an X is included during the docking, but is removed because it clashes with the protein.
AutoLigand analyzes the atomic affinity maps to predict the optimal locations for substrate binding (Step 5E). The computation time is dependent on the size of the envelope, and typical drug-sized molecules fall in the range of 50–500 Å3 grid points. The method combines information from three maps--carbon, oxygen and hydrogen--to predict the optimal atom type at each point. The oxygen map also represents likely locations for nitrogen atoms that accept hydrogen bonds, since the force field parameters for nitrogen atom types are similar to oxygen. We have used AutoLigand as a tool to identify the regions of a given ligand that are providing the most affinity, and to identify locations on lead molecules that would provide the most additional affinity during functionalization.
Supplementary Material
Acknowledgments
This work was supported by grant R01 GM069832 from the National Institutes of Health. This is manuscript number 29118 from the Scripps Research Institute.
Footnotes
Author Contributions
All authors contributed equally to this work. D.S.G and S.F. authored the protocol manuscript with extensive input from the other authors, based on tutorials developed by all authors. All authors have been instrumental in development of the AutoDock suite and training of users.
Competing Financial Interests
The authors declare no competing financial interests
References
- 1.Trott O, Olson AJ. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem. 2010;31:455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Goodsell DS, Olson AJ. Automated docking of substrates to proteins by simulated annealing. Proteins: Struct Funct Genet. 1990;8:195–202. doi: 10.1002/prot.340080302. [DOI] [PubMed] [Google Scholar]
- 3.Morris GM, et al. Automated docking using a Lamarckian genetic algorithm and an empirical binding free energy function. J Comp Chem. 1998;19:1639–1662. [Google Scholar]
- 4.Forli Raccoon for Processing Virtual Screening. 2013 < http://autodock.scripps.edu/resources/raccoon>.
- 5.Morris GM, et al. AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. J Comput Chem. 2009;30:2785–2791. doi: 10.1002/jcc.21256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Harris R, Olson AJ, Goodsell DS. Automated prediction of ligand-binding sites in proteins. Proteins. 2008;70:1506–1517. doi: 10.1002/prot.21645. [DOI] [PubMed] [Google Scholar]
- 7.Sousa SF, et al. Protein-ligand docking in the new millennium--a retrospective of 10 years in the field. Curr Med Chem. 2013;20:2296–2314. doi: 10.2174/0929867311320180002. [DOI] [PubMed] [Google Scholar]
- 8.Goodsell DS, Lauble H, Stout CD, Olson AJ. Automated docking in crystallography: analysis of the substrates of aconitase. Proteins: Struct Funct Genet. 1993;17:1–10. doi: 10.1002/prot.340170104. [DOI] [PubMed] [Google Scholar]
- 9.Goodsell DS, Morris GM, Olson AJ. Automated docking of flexible ligands: applications of AutoDock. J Molec Recognition. 1996;9:1–5. doi: 10.1002/(sici)1099-1352(199601)9:1<1::aid-jmr241>3.0.co;2-6. [DOI] [PubMed] [Google Scholar]
- 10.Soares TA, Goodsell DS, Briggs JM, Ferreira R, Olson AJ. Docking of 4-oxalocrotonate tautomerase substrates: implications for the catalytic mechanism. Biolpolymers. 1999;50:319–328. doi: 10.1002/(SICI)1097-0282(199909)50:3<319::AID-BIP7>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- 11.Rosenfeld RJ, et al. Automated docking of ligands to an artificial active site: augmenting crystallographic analysis with computer modeling. J Comput Aided Mol Des. 2003;17:525–536. doi: 10.1023/b:jcam.0000004604.87558.02. [DOI] [PubMed] [Google Scholar]
- 12.Brik A, et al. Rapid diversity-oriented synthesis in microtiter plates for in situ screening of HIV protease inhibitors. ChemBioChem. 2003;4:1246–1248. doi: 10.1002/cbic.200300724. [DOI] [PubMed] [Google Scholar]
- 13.Brik A, et al. 1,2,3-Triazole as a peptide surrogate in the rapid synthesis of HIV-1 protease inhibitors. ChemBioChem. 2005;6:1167–1169. doi: 10.1002/cbic.200500101. [DOI] [PubMed] [Google Scholar]
- 14.Perryman AL, et al. Fragment-based screen against HIV protease. Chem Biol Drug Des. 2010;75:257–268. doi: 10.1111/j.1747-0285.2009.00943.x. JPP943 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cosconati S, et al. Structure-based virtual screening and biological evaluation of Mycobacterium tuberculosis adenosine 5′-phosphosulfate reductase inhibitors. J Med Chem. 2008;51:6627–6630. doi: 10.1021/jm800571m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Perryman AL, et al. A virtual screen discovers novel, fragment-sized inhibitors of Mycobacterium tuberculosis InhA. J Chem Inf Model. 2015;55:645–659. doi: 10.1021/ci500672v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cosconati S, et al. Identification of novel beta-secretase inhibitors through inclusion of protein flexibility in virtual screening calculations. FASEB Journal. 2008;22:791–798. [Google Scholar]
- 18.Cosconati S, et al. Protein flexibility in virtual screening: the BACE-1 case study. J Chem Inf Model. 2012;52:2697–2704. doi: 10.1021/ci300390h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jiang H, et al. Stabilizers of the Max homodimer identified in virtual ligand screening inhibit Myc function. Mol Pharmacol. 2009;76:491–502. doi: 10.1124/mol.109.054858. mol.109.054858 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Leonard SE, Garcia FJ, Goodsell DS, Carroll KS. Redox-Based Probes for Protein Tyrosine Phosphatases. Angewandte Chemie International Edition. 2011;50:4423–4427. doi: 10.1002/anie.201007871. [DOI] [PubMed] [Google Scholar]
- 21.Huey R, Goodsell DS, Morris GM, Olson AJ. Grid-based hydrogen bond potentials in improved directionality. Lett Drug Design Discov. 2004;1:178–183. [Google Scholar]
- 22.Huey R, Morris GM, Olson AJ, Goodsell DS. A semiempirical free energy force field with charge-based desolvation. J Comput Chem. 2006;28:1145–1152. doi: 10.1002/jcc.20634. [DOI] [PubMed] [Google Scholar]
- 23.Forli S, Olson AJ. A force field with discrete displaceable waters and desolvation entropy for hydrated ligand docking. J Med Chem. 2012;55:623–638. doi: 10.1021/jm2005145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bianco G, Forli S, Goodsell DS, Olson AJ. Covalent docking using autodock: Two-point attractor and flexible side chain methods. Protein Science. 2016;25:295–301. doi: 10.1002/pro.2733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cosconati S, et al. Virtual Screening with AutoDock: Theory and Practice. Expert Opin Drug Discov. 2010;5:597–607. doi: 10.1517/17460441.2010.484460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lin JH, Perryman AL, Schames JR, McCammon JA. The relaxed complex method: Accommodating receptor flexibility for drug design with an improved scoring scheme. Biopolymers. 2003;68:47–62. doi: 10.1002/bip.10218. [DOI] [PubMed] [Google Scholar]
- 27.Nagar B, et al. Crystal structures of the kinase domain of c-Abl in complex with the small molecule inhibitors PD173955 and imatinib (STI-571) Cancer Res. 2002;62:4236–4243. [PubMed] [Google Scholar]
- 28.Schindler T, et al. Structural mechanism for STI-571 inhibition of abelson tyrosine kinase. Science. 2000;289:1938–1942. doi: 10.1126/science.289.5486.1938. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.