Abstract
The ribosome is a supramolecular ribonucleoprotein complex that functions at the heart of the translation machinery to convert mRNA into protein. Ribosome biogenesis is the primary determinant of translational capacity of the cell and accordingly has an essential role in the control of cell growth in eukaryotes. Cumulative evidence supports the hypothesis that ribosome biogenesis has an important role in the regulation of skeletal muscle mass. The purpose of this review is to, first, summarize the main mechanisms known to regulate ribosome biogenesis and, second, put forth the hypothesis that ribosome biogenesis is a central mechanism used by skeletal muscle to regulate protein synthesis and control skeletal muscle mass in response to anabolic and catabolic stimuli. The mTORC1 and Wnt/β-catenin/c-myc signaling pathways are discussed as the major pathways that work in concert with each of the three RNA polymerases (RNA Pol I, II and III) in regulating ribosome biogenesis. Consistent with our hypothesis, activation of these two pathways has been shown to be associated with ribosome biogenesis during skeletal muscle hypertrophy. Although further study is required, the finding that ribosome biogenesis is altered under catabolic states, in particular during disuse atrophy, suggests that its activation represents a novel therapeutic target to reduce or prevent muscle atrophy. Lastly, the emerging field of ribosome specialization is discussed and its potential role in the regulation of gene expression during periods of skeletal muscle plasticity.
Keywords: muscle hypertrophy, muscle atrophy, mTOR signaling, c-myc, ribosome specialization
Introduction
The ribosome is a supramolecular ribonucleoprotein complex that functions at the heart of the translation machinery to convert the messenger RNA (mRNA) into protein. This ancestral nanomachine consists of two subunits. In eukaryote, the small 40S subunit is composed of the 18S ribosomal RNA (rRNA) and 33 ribosomal proteins (RPs) and processes the decoding of mRNA by aminoacyl–transfer RNA (tRNA). The large 60S subunit is composed of 5S, 5.8S and 28S rRNAs and 47 RPs and promotes the catalysis of peptide bond formation through the peptidyltransferase reaction.
The ribosome was first described in the mid-1950s, with studies on ribosome assembly beginning in the early 1970s in bacteria (Held et al., 1973; Nierhaus and Dohme, 1974). As often is the case, the development of new techniques greatly enhanced our understanding of pre-rRNA processing and ribosome assembly in yeast, a simple eukaryotic organism [for review, see (Woolford and Baserga, 2013)]. In the last couple of decades, studies have revealed that the ribosome has an essential role in the regulation of cell proliferation and growth (Kirn-Safran et al., 2007; Volarevic et al., 2000) and homeostasis in mammalian organisms (Teng et al., 2013). For example, mutations and deletions in genes linked to ribosome biogenesis result in pathologies known as ribosomopathies, which are associated with growth retardation and malformation (Teng et al., 2013). More recently, studies have reported that the activation or alteration in ribosome biogenesis can promote the development of cancer [for review, see (Montanaro et al., 2012)]. Given the necessity of ribosome biogenesis in cell growth, it is not surprising then that the increase in cell size that occurs during muscle hypertrophy requires an increase in ribosome biogenesis. The majority of the studies that have investigated the role of ribosome biogenesis in striated muscle have focused on cardiac muscle; however, novel insights suggest that ribosome biogenesis may also have an important role in the control of skeletal muscle mass in response to changes in contractile activity. The purpose of this review is to, first, summarize the main mechanisms known to regulate ribosome biogenesis and, second, put forth the hypothesis that ribosome biogenesis is a central mechanism used by skeletal muscle to regulate protein synthesis and control skeletal muscle mass in response to anabolic and catabolic stimuli.
Concept of ribosome biogenesis
Ribosome biogenesis and translation capacity
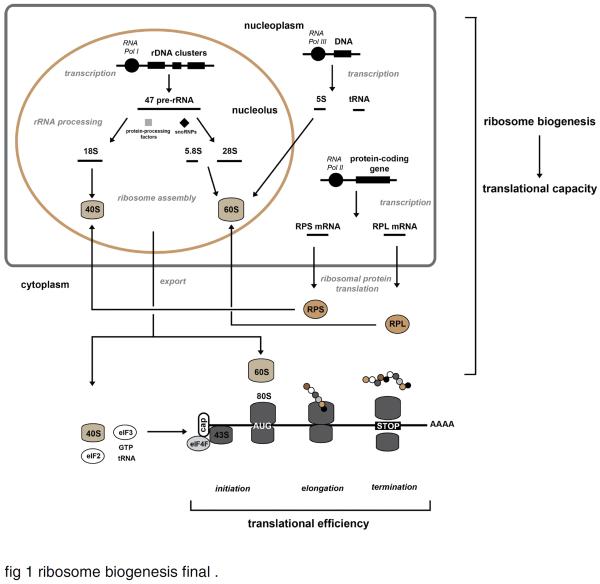
Ribosome biogenesis is a complex, well-orchestrated process that involves precise regulation of transcription of rRNAs, mRNAs and tRNAs, the processing of the polycistronic 47S rRNA precursor (47S pre-rRNA) into several smaller rRNAs (18S, 5.8S and 28S rRNAs), the assembly of the rRNAs and RPs into a ribosome subunit, and the nuclear export of the 40S and 60S subunits into the cytoplasm [for more detailed information, see (Boisvert et al., 2007; Henras et al., 2008; Lindstrom, 2009; Moss et al., 2007; Rodnina and Wintermeyer, 2009; Thomson et al., 2013; Woolford and Baserga, 2013)]. The synthesis of the components that are responsible of ribosome biogenesis required the coordinated actions of the three types of RNA polymerase (RNA Pol I, -II and -III) (Arabi et al., 2005; Grandori et al., 2005; Schlosser et al., 2003). The RNA Pol I-related transcription is thought to be the rate-limiting step in ribosome biogenesis (Moss and Stefanovsky, 1995). RNA Pol I is responsible for the transcription of the polycistronic 47S pre-rRNA in the nucleolus, which is subsequently cleaved into 18S, 5.8S and 28S rRNAs. These three rRNAs are then post-transcriptionally modified through their interaction with small nuclear ribonucleoproteins (snoRNPs) and several protein-processing factors. The nucleoplasmic transcription of 5S rRNA and the tRNAs requires RNA Pol III while the RP encoding-genes are transcribed in the nucleoplasm by RNA Pol II. The proteins of the small (RPS, ribosomal protein small) and large (RPL, ribosomal protein large) ribosomal subunits are subsequently translated in the cytoplasm and imported into the nucleolus to be assembled with their respective ribosomal subunit. The 40S and 60S subunits are then exported to the cytoplasm, where they form the mature 80S ribosome complex (Fig.1).
Fig. 1.
Schematic representation of the cellular translation machinery.
Ribosome biogenesis is a well-orchestrated process that involves the coordinated actions of the three types of RNA polymerase (RNA Pol I, -II and -III). RNA Pol I is responsible for the transcription of the polycistronic 47S pre-rRNA in the nucleolus, which is subsequently spliced and modified by small nuclear ribonucleoproteins (snoRNPs) and several protein-processing factors to form 18S, 5.8S and 28S rRNAs. The nucleoplasmic transcription of 5S rRNA and the tRNAs is achieved by RNA Pol III while the ribosomal protein encoding-genes are transcribed in the nucleoplasm by RNA Pol II. Following the translation in the cytoplasm, the proteins of the small (RPS, ribosomal protein small) and large (RPL, ribosomal protein large) ribosomal subunits are imported into the nucleolus to be assembled with their respective ribosomal subunit. The 5.8S, 28S and 5S rRNAs assemble with RPL proteins to form the 60S ribosome subunit while the 18S rRNA and the RPS proteins form the 40S ribosome subunit. The 40S and 60S subunits are then exported to the cytoplasm, where they form the mature 80S ribosome complex. The process of ribosome biogenesis is primary determinant of the translational capacity of the cell. After nuclear export, the 40S subunit interacts with the eukaryotic initiation factor (eIF) eIF3 and the ternary complex (eIF2, tRNA and GTP) to form the pre-initiation 43S complex. This complex, which is then recruited to the m7GpppN cap structure of the mRNA after its binding to eIF4F complex, scans the 5' UTR (untranslated region) of the mRNA until the initiation codon (AUG) is detected (initiation). The recruitment of 60S subunit at the initiation codon allows the formation of the 80S complex and the start of the elongation of the mRNA. Finally, the finished polypeptide is released from the ribosome when a stop codon is encountered (termination). These three steps of translation (initiation, elongation and termination) determine the translational efficiency. This schematic representation was adapted from (van Riggelen et al., 2010) and (Boisvert et al., 2007).
The rate of protein synthesis within the cell is determined by both the translational efficiency and capacity. Translational capacity is set by the amount of translational machinery per unit tissue, including ribosomes, tRNAs and translational factors, while translational efficiency corresponds to the protein synthesis rate per ribosome. It is generally thought that the increase in the rate of protein synthesis in response to an acute (minutes to hours) anabolic signal primarily results from the activation of the existing translational machinery, thus enhanced translational efficiency. In contrast, chronic (hours to days) anabolic signals are thought to stimulate ribosome biogenesis to generate more ribosomes, leading to an increase in the translational capacity and ultimately in protein synthesis.
The efficiency of translation is influenced by the initiation, elongation and termination steps of translation [for review see (Gebauer and Hentze, 2004; Kapp and Lorsch, 2004)], with the initiation of translation being the primary step at which regulation occurs. The pre-initiation 43S complex, which is composed of the 40S ribosomal subunit, the eukaryotic initiation factor (eIF) eIF3 and the ternary complex (eIF2, tRNA and GTP) is recruited to the m7GpppN cap structure at the 5' end of the mRNA. The pre-initiation complex recognizes the mRNA by the binding of eIF3 to eIF4G subunit of the cap-binding eIF4F complex. This cap-binding complex also contains eIF4E, a subunit that directly binds to the cap, and eIF4A, an RNA helicase that unwinds the secondary structure of the mRNA during transcript scanning. The 43S complex scans the 5' UTR (untranslated region) of the mRNA until the initiation codon is detected, followed by recruitment of the large 60S ribosomal subunit that results in the formation of the 80S complex and the subsequent start of mRNA translation (Fig.1).
The translational capacity of the cell is mainly determined by the quantity of ribosomes present in the cell. The ability of the cell to produce many proteins from one mRNA via polyribosomes (multiple ribosomes on a single transcript) has provided evidence that increasing the translational capacity through the activation of ribosome biogenesis is essential for cell growth. Moreover, the fact that 60% of all transcription is estimated to be devoted to the synthesis of rRNA by RNA Pol I, and 50% of all RNA Pol II transcriptional events involve the production of ribosomal proteins during periods of growth, strongly suggests that ribosome biogenesis is a key process for increasing protein synthesis and cell growth (Granneman and Tollervey, 2007; Warner, 1999).
Pathways involved in the control of ribosome biogenesis
Control of ribosome biogenesis by mTORC1 signaling
mTOR (mechanistic target of rapamycin) is a serine/threonine kinase highly conserved from yeast to mammals that forms two distinct complexes termed mTORC (mTOR complex) 1 and 2. mTORC1 is a rapamycin-sensitive complex known to function as a master regulator of protein synthesis and cell growth. Several studies have demonstrated that mTORC1 signaling has a central role in the control of ribosome biogenesis by promoting the translation of the mRNAs encoding ribosomal proteins and the transcription of rRNAs [for review, see (Iadevaia et al., 2012)]. The relationship between mTORC2 and the ribosome is not clearly understood but it appears that the ribosome may associate with mTORC2 and regulate its activation (Zinzalla et al., 2011). Here, we will focus on the role played by mTORC1 signaling on ribosome biogenesis because this pathway has been shown to be crucial for skeletal muscle hypertrophy (Schiaffino et al., 2013).
The ribosomal proteins (and other components of the translational machinery) are translated from 5'-TOP (terminal tract of pyrimidine) mRNAs. Under basal condition, the translation of these mRNAs is inhibited, while this inhibition is relieved in response to anabolic signals, most probably through the activation of mTORC1 (Fig.2). Rapamycin, a specific inhibitor of mTORC1, was shown to inhibit the translation of 5'-TOP mRNAs (Jefferies et al., 1997) but the mechanism by which mTORC1 controls their translation is still not clearly understood. It was demonstrated that S6K (ribosomal protein S6 kinase), a well-known target of mTORC1, and RPS6 (a target of S6K which is also a 5'-TOP mRNA-encoding protein) promote the translation the 5'-TOP mRNAs (Jefferies et al., 1997; Jefferies et al., 1994) but this concept remains controversial (Pende et al., 2004; Ruvinsky et al., 2005). The AMP-activated protein kinase (AMPK), a metabolic stress sensor known to inhibit mTORC1 activity was also suggested to repress the translation of several 5'-TOP mRNA (RPS6, RPS8 and eEF1α mRNAs) through a mechanism independent of S6K and RPS6 (Reiter et al., 2008).
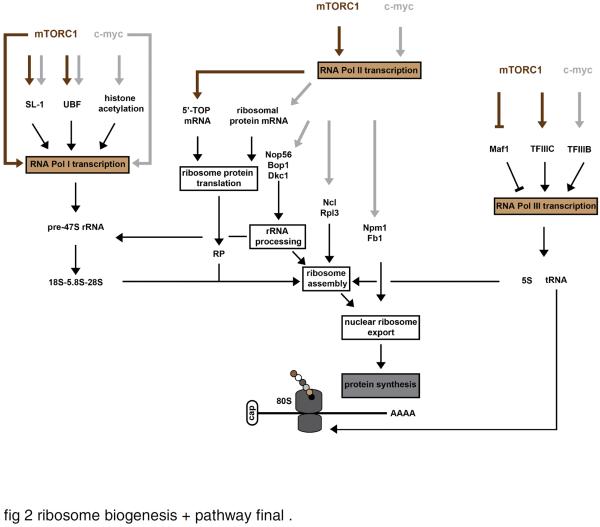
Fig. 2.
Regulation of ribosome biogenesis by mTORC1 and c-myc.
mTORC1 (mechanical target of rapamycin complex 1) and c-myc (c-myelocytomatosis oncogene) work in concert with each of the three RNA polymerases in regulating ribosome biogenesis. mTORC1 and c-myc seem to directly promote the transcription of the pre-rRNA 47S, and can lead to the activation of the RNA Pol I transcriptional factor SL-1 (selectively factor-1), and the rDNA transcription factor UBF (upstream binding factor). Some lines of evidence also suggest that c-myc indirectly increases the RNA Pol I transcription by promoting the opening of the chromatin structure near rDNA loci through histone acetylation. mTORC1 appears to regulate the translation of 5'-TOP (terminal tract of pyrimidine) mRNAs which encode ribosomal proteins and other components of the translational machinery, while c-myc activates the transcription of several ribosomal protein-encoding genes. In addition, c-myc promotes the transcription of several auxiliary factors required for ribosome biogenesis, such as genes involved in rRNA processing [Nop56 (Nop56 ribonucleoprotein), Bop1 (block of proliferation 1) and Dkc1 (dyskeratosis congenital 1, dyskerin)], ribosome assembly [Ncl (nucleolin) and Rpl3 (ribosome protein L3)], and nuclear ribosome export [Npm1 (nucleophosmin 1) and Fbl (fibrillarin)]. Finally, mTORC1 and c-myc activate the RNA Pol III transcription through their interaction with TFIIIC (transcription factor IIIC) and TFIIIB (transcription factor IIIB), respectively. mTORC1 may also inhibit the function of the RNA Pol III repressor Maf1.
mTORC1 is known to promote the transcription of rRNAs by regulating transcription factors that associate with RNA polymerases I and III [for review, see (Iadevaia et al., 2012)] (Fig.2). Inhibition of mTORC1 by rapamycin inactivates TIF-1A, a component of the RNA Pol I transcriptional factor SL-1(selectivity factor-1), and impairs the transcription of the pre-rRNA 47S (Mayer et al., 2004). Moreover, rapamycin induces the inactivation of UBF (upstream binding factor), a rDNA transcription factor which associates with SL-1 (Hannan et al., 2003). Hannan and colleagues (2003) also demonstrated that S6K1 promotes RNA Pol I transcription and activation of UBF. mTORC1 has also been shown to inhibit Rb (retinoblastoma), a tumor suppressor protein involved in the sequestration of UBF, thereby promoting pre-rRNA 47S transcription (Nader et al., 2005). These findings, in addition to the ability of mTORC1 to directly associate with the promoters of RNA Pol I-genes (Tsang et al., 2010), provides strong evidence that mTORC1 signaling regulates Pol I transcription. Furthermore, it was demonstrated that mTORC1 associates with RNA Pol III genes (which synthesize 5S rRNA and tRNA) through the binding with TFIIIC (transcription factor IIIC), a DNA-binding factor that recognizes the promoters of these genes (Kantidakis et al., 2010). Moreover, mTORC1 inactivates Maf1 in yeast, a RNA Pol III repressor that mediates diverse stress signals to inhibit 5S rRNA and tRNA transcription (Upadhya et al., 2002), through its direct phosphorylation and interaction with Sch9, a homologue of the mammalian S6Ks (Wei and Zheng, 2009). mTORC1 appears to repress the binding of Maf1 on the chromatin while Sch9 seems to promote the nuclear export of Maf1 (Wei and Zheng, 2009). In mammalian cells, mTORC1 directly phosphorylates Maf1 and impairs its association with the chromatin. Although the Sch9 phosphorylation sites of Maf1 do not exist in mammalian cells, S6K1 may have a role in the regulation of Maf1 function (Shor et al., 2010).
Control of ribosome biogenesis by Wnt/β-catenin/c-myc signaling
β-catenin is a multifunctional protein that interacts with the cytoskeleton and is capable of translocating to the nucleus to function as a transcription factor (Ben-Ze'ev et al., 2000; Novak and Dedhar, 1999). β-catenin activation is mainly mediated by the canonical Wnt signaling pathway in which Wnt ligands bind Frizzled (Fzd) receptor and other co-receptors, leading to phosphorylation and activation of the Disheveled (Dvl). Dvl can then deactivate GSK-3β (glycogen synthase kinase-3β) through its phosphorylation, thereby resulting in the inability of GSK-3β to phosphorylate and inactivate β-catenin. The accumulation of unphosphorylated β-catenin leads to its stabilization and translocation to the nucleus (Hinoi et al., 2000; Li et al., 1999; Thomas et al., 1999). In the nucleus, β-catenin interacts with TCF/LEF (T cell factor/lymphocyte enhancement factor) DNA-binding proteins to drive expression of the transcription factor c-myc (c-myelocytomatosis oncogene). Although there are a number of different regulatory mechanisms known to maintain tight control of c-myc expression (Meyer and Penn, 2008), the focus of this review will be on the Wnt/β-catenin/c-myc signaling because several components of this pathway are highly expressed during skeletal muscle hypertrophy (Armstrong and Esser, 2005) and β-catenin could have a major role in this process (Armstrong et al., 2006). It is noteworthy that GSK-3β is also inhibited through phosphorylation by Akt (Cross et al., 1995), while Wnt7a is able to activate the Akt/mTOR pathway (von Maltzahn et al., 2012). These finding suggest an interaction between the PI3K (phosphatidylinositol 3-kinase)/Akt/mTOR and the Wnt/β-catenin/c-myc signaling pathways.
c-myc is a transcription factor that is known to regulate the expression of many genes involved in cell growth, cell proliferation and apoptosis (Zeller et al., 2006). It is not surprising then that there is a significant amount of evidence showing that c-myc promotes protein synthesis and cell growth through the regulation of ribosome biogenesis. Over-expression of c-myc induces an increase in cell growth and protein synthesis in tumor cells (Schuhmacher et al., 1999), hepatocytes (Kim et al., 2000) and cardiomyocytes (Olson et al., 2013). Recent studies have identified c-myc as a central player that works in concert with each of the three RNA polymerases in regulating ribosome biogenesis [for review, see (Gomez-Roman et al., 2006; van Riggelen et al., 2010)]. The main role played by c-myc in the regulation of ribosome biogenesis is summarized in Fig.2. Studies have provided evidence that c-myc directly influences Pol I-mediated transcription of rRNA by binding to the promoter of ribosome DNA (rDNA), indirectly by regulating the expression and/or the recruitment of UBF and SL-1 factor and by promoting the opening of the chromatin structure near rDNA loci through the acetylation of nucleosomal histones H3 and H4 (Arabi et al., 2005; Grandori et al., 2005). c-myc also activates the transcription of several ribosomal protein large (Rpl) and small (Rps) genes (Coller et al., 2000; Kim et al., 2000) while c-myc inactivation results in a reduction of ribosomal protein gene expression (Wu et al., 2008). In addition to these ribosomal proteins, a number of Pol II genes which encode auxiliary factors required for ribosome biogenesis are direct transcriptional targets of c-myc (Alawi and Lee, 2007; Zeller et al., 2006). These genes are involved in rRNA processing [Nop56 (Nop56 ribonucleoprotein), Bop1 (block of proliferation 1) and Dkc1 (dyskeratosis congenital 1, dyskerin)], ribosome assembly [Ncl (nucleolin) and Rpl3 (ribosome protein L3)], and nuclear-cytoplasmic shuttling of mature ribosomal subunits [Npm1 (nucleophosmin 1) and Fbl (fibrillarin)]. Finally, c-myc has been reported to activate Pol III transcription through its interaction with TFIIIB factor (Gomez-Roman et al., 2003).
Ribosome biogenesis in skeletal muscle
It has been clearly demonstrated that ribosome biogenesis stimulates protein synthesis, thereby leading to cell growth. The importance of ribosome biogenesis for cell growth has been primarily described in yeast and in tumor cells, while studies reporting such a relationship in striated muscle have been mainly in cardiac muscle. In response to hypertrophic stimuli, 18S transcription by RNA Pol I is increased in cardiomyocytes (Zhang et al., 2013), while the activation of rDNA transcription is associated with an increase in UBF expression (Hannan et al., 1996). The RNA Pol III transcription is also activated in cardiomyocytes during hypertrophic growth (Goodfellow et al., 2006; Zhang et al., 2013), and appears to be mediated by the transcription factor TFIIIB (Goodfellow et al., 2006). These findings provide strong evidence that ribosome biogenesis is stimulated in response to an anabolic stimulus and leads to cardiac cell hypertrophy. In contrast to cardiac muscle, there are a limited number of studies that have investigated ribosome biogenesis in skeletal muscle (Goodman et al., 2011; Machida et al., 2012; Nader et al., 2014; Quy et al., 2013; von Walden et al., 2012). Here, we put forth the idea that ribosome biogenesis has an important role in the control of skeletal muscle mass in response to both anabolic stimuli and under catabolic conditions.
Regulation of ribosome biogenesis in response to high contractile activity
The majority of studies on the regulation of protein synthesis during skeletal muscle hypertrophy have focused on the mechanisms controlling translational efficiency, in particular the step of translation initiation (Csibi et al., 2010; Mayhew et al., 2011). Here, we will provide evidence that ribosome biogenesis (which mainly determines the translational capacity) is activated in response to anabolic stimuli (Table 1) and contributes to increase protein synthesis, thereby leading to skeletal muscle hypertrophy. Given that there is a paucity of information on ribosome biogenesis in skeletal muscle in response to anabolic treatment, i.e., nutritional or pharmaceutical (Nader et al., 2005), we focused our attention on how ribosome biogenesis is affected by the anabolic stimulus provided by high contractile activity.
Table 1.
Potential regulation of ribosome biogenesis in response to anabolic and catabolic stimuli.
| Stimulus | Type | Characteristic | Model | Marker of ribosome biogenesis | References |
|---|---|---|---|---|---|
| Anabolic | Resistance exercise | 12 sessions in 5 weeks, knee extension | Human | Tendency to increase in [total RNA] | (Haddad et al., 2005) |
| Acute leg exercise (4 sets of 10 reps) | Human | Increase in S6K1 and RPS6 phosphorylation | (Glover et al., 2008a) | ||
| 3 sessions/week, 20 weeks. Microarray | Human | Down-regulation of 14 ribosomal protein genes associated with the mTOR-related expression network in the subjects with the highest muscle hypertrophy | (Phillips et al., 2013) | ||
| Acute resistance exercise (arm) | Human | Increase in RPS6 phosphorylation, 45 pre-rRNA and c-myc mRNA expression | (Nader et al., 2014) | ||
| Neural stimulation: twice a week for 6 weeks | Rat | Increase in S6K phosphorylation | (Baar and Esser, 1999) | ||
| Neural stimulation: 2 bouts of 30min | Rat | Increase in [total RNA] and amount, in S6K1 phosphorylation | (Haddad and Adams, 2006) | ||
|
| |||||
| Resistance exercise + protein ingestion | Leg exercise combined with protein ingestion, 2 sessions/week, 21 weeks | Human | Increase in mTOR, S6K and RPS6 phosphorylation 1h after acute execise combined with protein ingestion | (Hulmi et al., 2009) | |
|
| |||||
| Mechanical overload | Synergist ablation | Mouse | Increase in [total RNA], 47S pre-RNA expression, RNA Pol I regulon expression, c-myc expression, UBF expression | (von Walden et al., 2012) | |
| Mouse | Increase in [total RNA], S6K1 phosphorylation | (Miyazaki et al., 2011) | |||
| Mouse | Increase in [total RNA], 28S + 18S content, S6K1 phosphorylation, UBF protein expression | (Goodman et al., 2011) | |||
| Mouse | Activation of the Wnt/βcatenin/cmyc pathway | (Armstrong et al., 2006) | |||
| Mouse | Increased [total RNA] associated with increased in c-myc expression | (Chaillou et al., 2013) | |||
| Rat | Increase in [total RNA], S6K and RPS6 phosphorylation, total RPS6 protein expression | (Chaillou et al., 2012) | |||
|
| |||||
| Reloading after atrophy | Reloading after hindlimb suspension | Mouse | Increase in [total RNA] and total RNA content | (Dapp et al., 2004) | |
| Rat | Increase in [total RNA] and total RNA content | (Heinemeier et al., 2009) | |||
|
| |||||
| Serum stimulation | Myotubes treated with 20% FBS | L6 myotube | Increase in ribosomal RNA content | (Nader et al., 2005) | |
| Catabolic | Aging | 24 vs 70 years old- Acute resistance exercise | Human | No basal difference in S6K phosphorylation and protein synthesis. Increase in S6K phosphorylation and protein synthesis blunts in elderly after resistance exercise | (Kumar et al., 2009) |
| 27 vs 70 years old- Acute resistance exercise | Human | No increase in S6K1 and protein synthesis in aged patients after resistance exercise | (Fry et al., 2011) | ||
| 18–25 vs 60–75 years old- 12 weeks of resistance training combined with protein ingestion | Human | Increased phosphorylation of S6K after resistance training combined with protein ingestion is less pronounced in elderly | (Farnfield et al., 2012) | ||
| 6 vs 30 month old | Rat | Similar amount of total RNA per muscle | (Haddad and Adams, 2006) | ||
|
| |||||
| Cachexia | COPD | Human | Decrease in S6K phosphorylation in hypoxemic compared with non-hypoxemic COPD patients | (Favier et al., 2010) | |
| Critically ill | Human | Decrease in mTORC1 and S6K phosphorylation in critically ill patients compared to control subjects | (Constantin et al., 2011) | ||
| Colorectal cancer | Mouse | Decrease in S6K phosphorylation in cancer mouse associated with skeletal muscle atrophy and reduced protein synthesis | (White et al., 2011) | ||
| Glucocorticoid administration (dexamethasone) | Rat | Decrease in S6K phosphorylation and β-catenin protein expression | (Schakman et al., 2008) | ||
| Chronic kidney disease | Rat | No changes in [total RNA] and S6K phosphorylation | (Chen et al., 2008) | ||
| Atrophy induced by fasting, diabetes mellitus, renal failure and tumor implantation. Micro-array | Rat | Increase in RPL12 and nucleolin mRNA, two markers of ribosomal assembly | (Lecker et al., 2004) | ||
| Disuse | Unilateral unloading | Human | Decrease in [total RNA] | (Haddad et al., 2005) | |
| Human | Decrease in [total RNA] | (Gamrin et al., 1998) | |||
| Inactivity after abdominal surgery | Human | Decrease in [total RNA] | (Petersson et al., 1990) | ||
| Hindlimb suspension and immobilization | Rat | Decrease in [total RNA] | (Babij and Booth, 1988) | ||
| Hindlimb suspension | Rat | Decrease in [total RNA] | (Heinemeier et al., 2009) | ||
|
| |||||
| Denervation | Long-term spinal cord injury | Human | Decrease in [total RNA] in SCI subjects | (Bickel et al., 2003) | |
| Spinal cord injury | Rat | Decrease in total RNA concentration and content | (Haddad et al., 2003) | ||
| Sciatic nerve transection | Rat | Up-regulation of several ribosomal proteins. Down-regulation of a few ribosomal proteins. | (Sun et al., 2012) | ||
| Sciatic nerve transection | Mouse | Activation of mTORC1 in denervated muscle associated with increased expression of RPS6 and RPL7 | (Quy et al., 2013) | ||
| Sciatic nerve transection | Mouse | Activation of mTORC1 signaling and decrease in rRNA expression | (Machida et al., 2012) | ||
[total RNA], total RNA concentration; COPD, chronic obstructive pulmonary disease; SCI, spinal cord injury.
The mammalian ribosome structure is made up of two-thirds rRNA and one-third protein (Moss et al., 2007). Given that total RNA is primarily composed of the rRNA (>85%), thus an increase in total RNA content indicates an increase in translational capacity through modulation of ribosome biogenesis. We and other groups have reported a significant increase in total RNA concentration during skeletal muscle hypertrophy induced by synergist ablation (Adams et al., 2002; Chaillou et al., 2012; Chaillou et al., 2013; Goodman et al., 2011; Miyazaki et al., 2011; von Walden et al., 2012) (Table 1), and this increase in translational capacity was accordingly associated with a dramatic increase in protein synthesis (Miyazaki et al., 2011) and S6K phosphorylation (Adams et al., 2002; Chaillou et al., 2012; Goodman et al., 2011; Miyazaki et al., 2011). Using the synergist ablation model of hypertrophy, Goodman et al., (2011) showed that inhibition of mTORC1 by rapamycin prevented muscle fiber hypertrophy and the increase in S6K phosphorylation as well as partially blunting the overload-induced increase in total RNA concentration (Goodman et al., 2011). Moreover, the change in S6K phosphorylation in response to resistance exercise in rodents shows a high correlation with the increase in skeletal muscle mass (Baar and Esser, 1999). Therefore, S6K, which has been shown to be a critical component of the signaling cascade that regulates ribosome biogenesis [see “Control of ribosome biogenesis by mTORC1 signaling” section and (Iadevaia et al., 2012)], could be a crucial regulator of ribosome biogenesis during skeletal muscle hypertrophy.
A recent study showed that the expression of the 47S pre-rRNA, which is transcribed by RNA Pol I, was increased during hypertrophy induced by synergist ablation (von Walden et al., 2012). These authors also found that the increase in c-myc protein expression during skeletal muscle hypertrophy correlates with the up-regulation of UBF and several Pol I regulatory factors (POL1RB, TIF-1A, PAF53, TTF1 and TAF1C). We recently published a microarray study confirming that c-myc expression is highly increased using the same model (Chaillou et al., 2013). Based on our microarray data (publicly available at GEO, accession number GSE47098; http://www.ncbi.nlm.nih.gov/geo), we identified other up-regulated genes that potentially regulate ribosome biogenesis (unpublished results presented in Table 2). These genes are involved in the regulation of RNA Pol I transcription (Polr1c, Polr1e, Taf1b and Taf1d), rRNA processing (Nop56 and Bop1), ribosome assembly (Ncl and Rpl3), nuclear ribosome export (Npm1 and Fbl), and c-myc co-regulatory factors (Max, Kat2a and Trrap). In addition, numerous genes encoding ribosomal proteins were moderately up-regulated in this hypertrophic model (data not shown; more detailed information in the legend of Table 2), while the mRNA expression of genes involved in the regulation of RNA Pol III transcription (genes encoding the RNA Pol III enzyme and TFIIIB factor) were only slightly, if at all, increased in hypertrophied muscle (data not shown). Although somewhat speculative, these findings suggest c-myc is a central regulator of ribosome biogenesis during hypertrophy induced by synergist ablation, by orchestrating the transcription of rRNA and ribosomal protein-coding mRNA, and by promoting rRNA processing, ribosome assembly and nuclear ribosome export. The upregulation of c-myc expression in response to synergist ablation was also associated with an increased expression of its upstream regulator, β-catenin (Armstrong and Esser, 2005), while hypertrophy was prevented in myofibers in which β-catenin was inactivated (Armstrong et al., 2006). Although further investigation is required, the Wnt/β-catenin/c-myc pathway represents a viable mechanism required to stimulate ribosome biogenesis in skeletal muscle, thereby promoting protein synthesis and muscle hypertrophy.
Table 2.
Up-regulation of genes involved in ribosome biogenesis during skeletal muscle hypertrophy induced by synergist ablation in mice.
| Function | Symbol | Gene name | Fold change | |||
|---|---|---|---|---|---|---|
| day 1 | day 3 | day 5 | day 7 | |||
| Pol I transcription | Polr1c | Polymerase (RNA) I polypeptide C | 1.50 | 1.33 | 1.41 | 1.48 |
| Polr1e | Polymerase (RNA) I polypeptide E | 1.73 | 1.66 | 1.42 | 1.48 | |
| Taf1b | TATA box binding protein (Tbp)-associated factor, RNA polymerase I, B | 1.30 | 1.39 | 1.50 | 1.46 | |
| Taf1d | TATA box binding protein (Tbp)-associated factor, RNA polymerase I, D | 3.00 | 2.01 | 2.46 | 2.70 | |
|
| ||||||
| rRNA processing | Nop56 | NOP56 ribonucleoprotein | 1.71 | 1.58 | 1.43 | 1.43 |
| Bop1 | Block of proliferation 1 | 1.18 | 1.61 | 1.54 | 1.44 | |
|
| ||||||
| Ribosome assembly | Ncl | Nucleolin | 2.00 | 1.42 | 1.40 | 1.43 |
| Rpl3 | Ribosomal protein L3 | 2.60 | 3.02 | 3.12 | 3.34 | |
|
| ||||||
| Nuclear ribosome export | Npm1 | Nucleophosmin 1 | 2.13 | 1.93 | 2.09 | 2.09 |
| Fbl | Fibrillarin | 1.49 | 1.76 | 1.56 | 1.49 | |
|
| ||||||
| c-myc co-regulatory factors | Max | Max protein | 1.78 | 1.44 | 1.36 | 1.29 |
| Kat2a | K(lysine) acetyltransferase 2A | 1.20 | 1.52 | 1.47 | 1.41 | |
| Trrap | Transformation/transcription domain-associated protein | 1.11 | 1.42 | 1.62 | 1.96 | |
|
| ||||||
| c-myc | c-myelocytomatosis oncogene | 14.54 | 8.43 | 5.93 | 4.89 | |
These data are derived from a microarray analysis previously published by our research group (Chaillou et al., 2013) and are available by using the GEO accession number GSE47098. In this study, we assessed the gene expression in the plantaris muscle in response to 1, 3, 5, 7, 10 and 14 days following synergist ablation. In this table, we present the expression of selected up-regulated genes involved in ribosome biogenesis during the first week of skeletal muscle hypertrophy (days 1, 3, 5 and 7), a period associated with the major changes in total RNA concentration (Chaillou et al., 2013). The selected genes were up-regulated (>50% increase compared to control muscle) at least at one time point between days 1 and 7. These genes were involved in the regulation of RNA Pol I transcription, rRNA processing, ribosome assembly, nuclear ribosome export or were c-myc co-regulatory factors.
It is noteworthy that 44% and 26% of the genes encoding RPS and RPL, respectively, were up-regulated (>50% increase compared to control muscle) in hypertrophied muscle (data not shown). The highest increases were about 80% for the RPS-coding genes (80, 81 and 83% for RPS16, RPS19 and RPS4X, respectively) and 3-fold for the RPL-coding genes (2.82 and 3.34-fold for RPL12 and RPL3, respectively).
It is noteworthy that the over-expression of Wnt7a in skeletal muscle results in fiber hypertrophy and hyper-phosphorylation of Akt and RPS6 (von Maltzahn et al., 2012), while rapamycin blunts the hypertrophy of muscle fibers over-expressing Wnt-7a. Moreover, strenuous physical exercise activates Akt in skeletal muscle with the subsequent inactivation of its target GSK-3β through phosphorylation, while these results are associated with a dephosphorylation (i.e. activation) of β-catenin (Aschenbach et al., 2006). These findings would suggest that the Wnt/β-catenin/c-myc and the Akt/mTORC1 signaling pathways interact to regulate, in concert, ribosome biogenesis during skeletal muscle hypertrophy.
As discussed above, ribosome biogenesis is strongly activated in skeletal muscle in response to synergist ablation, and several lines of evidence suggest that its activation is necessary to induce the supra-physiological muscle hypertrophy observed in this model. Unfortunately, there have been limited studies investigating ribosome biogenesis under a more physiological anabolic condition, such as resistance exercise. An increase in total RNA concentration was observed in rats (Haddad and Adams, 2006) while only a tendency toward an increase in total RNA was found in humans (Haddad et al., 2005). Moreover, the phosphorylation of S6K was increased after resistance exercise in rat (Baar and Esser, 1999; Haddad and Adams, 2006), and in humans when resistance exercise was combined (Hulmi et al., 2009) or not (Glover et al., 2008a) with protein ingestion. A recent study also revealed an increase expression of two markers of ribosome biogenesis (47 pre-rRNA and c-myc mRNA) after an acute resistance exercise in humans (Nader et al., 2014). Although it remains to be clearly elucidated, these findings are consistent with the notion that ribosome biogenesis is activated in response to high-resistance exercise.
Somewhat surprising, a microarray study found in humans that after 20 weeks of resistance exercise 14 ribosomal protein genes, associated with mTOR-related network, were down-regulated in those subjects showing the largest skeletal muscle hypertrophy (Phillips et al., 2013). In contrast to these results, the expression of ribosomal proteins was increased during re-growth following atrophy (Heinemeier et al., 2009) and in response to synergist ablation (Chaillou et al., 2012). Despite these disparate findings, the activation of ribosome biogenesis in response to high contractile activity conditions seems to be mainly explained by the stimulation of rRNA transcription while the regulation of ribosomal protein expression requires further study.
Regulation of ribosome biogenesis in catabolic states
Skeletal muscle atrophy results from a negative balance between protein synthesis and protein degradation. Although protein degradation undoubtedly contributes to the development of muscle atrophy under various conditions, there is a growing recognition that the decrease in the rate of protein synthesis in catabolic states has a more important role in the loss of muscle mass than previously thought (Gordon et al., 2013; Phillips et al., 2009; Thomason and Booth, 1990). The objective of this section of the review is to put forth the hypothesis that ribosome biogenesis is negatively affected under muscle wasting conditions, such as sarcopenia, cachexia, disuse atrophy and denervation. A summary of the studies that have reported on some aspects of ribosome biogenesis under catabolic conditions is presented in Table 1.
Sarcopenia
It is generally thought that the loss of muscle mass associated with aging (sarcopenia) is in part the result of a reduction in the rate of protein synthesis. However, the findings are contradictory, with some studies showing a decrease in the basal rates of protein synthesis in skeletal muscle of aged compared to young subjects, while other studies reported no change in protein synthesis rates [for detailed information, see (Gordon et al., 2013)]. Despite this controversy, it has become well-known that skeletal muscle of aged humans is resistant to anabolic stimuli, with the suggestion that the loss of skeletal muscle mass in elderly results from of an inadequate stimulation of protein synthesis in response to anabolic stimuli such as resistance exercise (Fry et al., 2011).
The two markers of ribosome biogenesis described above (S6K phosphorylation and total RNA concentration) do not seem to be affected in basal condition in both humans and animals during aging (Haddad and Adams, 2006; Kumar et al., 2009). In contrast, the increase in S6K phosphorylation in response to resistance exercise was blunted in aged humans, a result consistent with the idea that the elderly become resistant to anabolic stimulation of protein synthesis (Fry et al., 2011; Kumar et al., 2009). In addition, the increase in skeletal muscle mass (Hwee and Bodine, 2009; Thomson and Gordon, 2006) and in protein synthesis rate (Pehme et al., 2004) was impaired in old compared to young animals during hypertrophy induced by synergist ablation, while the increase in S6K phosphorylation observed in this hypertrophic model was reduced in aged animals (Thomson and Gordon, 2006). We also observed a reduced increase in skeletal muscle mass in aged animals using this model of hypertrophy, and this result was accompanied by a diminished increase in total RNA concentration in hypertrophied muscle of aged animals (unpublished observation).
Although further investigation is required to clarify the regulation of ribosome biogenesis in skeletal muscle during aging, we suggest that ribosome biogenesis is unaffected in a basal post-absorptive state during aging, whereas ribosome biogenesis may be impaired in response to anabolic stimuli thereby contributing to the altered stimulation of protein synthesis after resistance exercise.
Cachexia
Skeletal muscle atrophy is observed in several chronic diseases and pathologies such as cancer, chronic obstructive pulmonary disease (COPD), chronic heart failure, chronic kidney disease and stroke (Gordon et al., 2013; Harrington et al., 1997; Schols et al., 2005; Sions et al., 2012; Williams et al., 2012). Several factors (reduced nutrient intake, physical inactivity, sepsis, impaired motor recruitment, cellular hypoxia, acidosis and oxidative stress) are capable of promoting the loss of skeletal muscle mass. Although the underlying mechanisms responsible for muscle atrophy depend on the cachectic state, a reduction in the rate of protein synthesis is generally observed during cachexia. Due to the complexity of studying the regulation of protein synthesis in cachectic patients, the majority of studies investigating the impact of cachexia on skeletal muscle have been carried out using animal models. An alteration of mTORC1 signaling was observed in several models of cachexia (Chen et al., 2011; Schakman et al., 2008; White et al., 2011) while recent evidence indicates that critically ill (Constantin et al., 2011) and hypoxemic COPD (Favier et al., 2010) patients also show altered mTORC1 signaling.
The field's understanding of the regulation of ribosome biogenesis in skeletal muscle during cachexia is very limited. Chen et al., (2008) reported that the total RNA concentration was not modified in skeletal muscle of chronic kidney disease rats (Chen et al., 2008). Unexpectedly, the mRNA expression of two markers of ribosome assembly (RPL12 and nucleolin) was increased during skeletal muscle atrophy induced by fasting, diabetes mellitus, renal failure and tumor implantation (Lecker et al., 2004). Moreover, it was recently proposed that the decrease in protein synthesis observed during sepsis was the result of a decrease in translational efficiency rather than a change in the ribosome content or protein synthetic capacity (Lang et al., 2007; Orellana et al., 2011). Altogether, these findings suggest that ribosome biogenesis may not be altered during cachexia. The mechanisms controlling the loss of skeletal muscle during cachexia are multi-factorial, thus further experiments are required to investigate the regulation of ribosome biogenesis during cachexia under different disease states.
In contrast to aging, the skeletal muscle does not seem to be insensitive to anabolic stimuli during cachexia. Although protein synthesis was not stimulated by feeding in cancer patients, S6K phosphorylation was increased in the post-absorptive state in these subjects (Williams et al., 2012). Moreover, leucine ingestion stimulates the mTORC1 pathway in uremic rats (Chen et al., 2011) and the increase in skeletal muscle mass in response to synergist ablation was similar in uremic and healthy animals (Chen et al., 2008). This latter study also showed that the increase in total RNA concentration in hypertrophied muscle was not impaired in the model of chronic kidney disease used. Recently, it was observed that skeletal atrophy induced by the glucocorticoid dexamethasone was associated with a reduction in S6K phosphorylation and β-catenin expression (Schakman et al., 2008). These authors revealed that over-expression of β-catenin completely blunts dexamethasone-induced fiber atrophy, while the over-expression of a constitutively active form of Akt in dexamethasone-treated mice induced fiber hypertrophy that was associated with an increase in both S6K phosphorylation and β-catenin expression. Although the hypothesis remains to be tested, activating ribosome biogenesis may be a relevant therapeutic target to prevent or attenuate skeletal muscle atrophy during cachexia. The development of therapies that target the Akt/mTORC1 and the β-catenin/c-myc signaling may be effective to stimulate ribosome biogenesis under cachectic conditions.
Disuse atrophy
The loss of skeletal muscle mass following limb immobilization, bed rest and hindlimb suspension, termed disuse atrophy, is mainly attributed to a reduction in the rate of muscle protein synthesis [for a detailed review, see (Phillips et al., 2009)]. During disuse atrophy in both humans and animals, the rate of protein synthesis is reduced in post-absorptive state while its stimulation by nutrients is attenuated (de Boer et al., 2007; Drummond et al., 2012; Glover et al., 2008b; Kelleher et al., 2013). Although the mechanisms responsible for the alteration of protein synthesis and anabolic resistance following disuse atrophy are not completely understood, it appears that deregulation of mTORC1 signaling contributes to this dysfunction. The regulation of ribosome biogenesis during disuse atrophy is currently poorly explored. A reduction of the total RNA concentration was shown in rats during hindlimb suspension (Babij and Booth, 1988; Heinemeier et al., 2009) and immobilization (Babij and Booth, 1988), and in humans during unloading (Gamrin et al., 1998; Haddad et al., 2005) and inactivity after abdominal surgery (Petersson et al., 1990). Although a change in ribosome content was not observed following immobilization in humans (Gibson et al., 1987; Gibson et al., 1988), these results provide evidence that ribosome biogenesis could be impaired during disuse atrophy and contribute to the decrease in protein synthesis.
It was recently shown that the skeletal muscle re-growth following immobilization in the mTOR+/− mice was impaired (Lang et al., 2012). The increase in 4E-BP1 binding to eIF4E suggested that translation initiation was reduced in mTOR+/− muscle, a finding consistent with the observed decrease in protein synthesis. Unfortunately, the regulation of ribosome biogenesis during muscle re-growth was not investigated in this study. Recently, it was shown that total RNA content increased in skeletal muscle during the first few days of reloading following hindlimb suspension to a level higher than in the control muscle, and this change preceded the complete recovery of the skeletal muscle mass (Heinemeier et al., 2009). Although the exact mechanism remains to be described, these findings suggest that the activation of ribosome biogenesis may be beneficial in preventing or attenuating the loss of skeletal muscle mass during disuse atrophy.
Denervation
A loss of muscle mass is observed in denervated skeletal muscle in both animals (Haddad et al., 2003; Machida et al., 2012; Quy et al., 2013) and humans (Bickel et al., 2003). The rate of protein breakdown is increased during denervation (Goldspink, 1976), while protein synthesis seems to be activated rather than inhibited during the first few days following denervation (Goldspink, 1976; Quy et al., 2013). The regulation of ribosome biogenesis in this catabolic state is poorly elucidated and the results are inconsistent. A reduction of the total RNA concentration was observed after spinal cord injury in humans (Bickel et al., 2003) and rats (Haddad et al., 2003) but this result was not observed after sciatic denervation (Babij and Booth, 1988; Goldspink, 1976). It was recently shown that the expression of rRNA and 47 pre-rRNA was reduced in denervated skeletal muscle, while unexpectedly the mTORC1 signaling was activated (Machida et al., 2012). The activation of this anabolic pathway was confirmed by another group using the same model of sciatic nerve transection and it was proposed to be the result of the accumulation of amino acids released from the proteasome-dependent proteolysis (Quy et al., 2013). However and in contrast to Machida et al. (2012), this latter study suggested that ribosome biogenesis could be activated rather than impaired during denervation, as evidenced by the increased expression of the ribosomal protein RPS6 and RPL7. A recent proteomic analysis also revealed that several ribosomal proteins were up-regulated in skeletal muscle during sciatic denervation, while only a few ribosomal proteins were down-regulated (Sun et al., 2012). The discrepancies between these studies could be explained by the different time points of denervation studied and/or by the markers of ribosome biogenesis investigated (rRNA vs. ribosomal protein).
It was recently shown that the inhibition of mTORC1 signaling by rapamycin results in the prevention of denervation-induced increases in protein synthesis and RPS6 and RPL7 expression, while these findings were associated with an increase in muscle atrophy during denervation (Quy et al., 2013). Moreover, another study revealed that the activation of mTORC1 induced by the deletion of TSC1 (tuberous scleroris complex 1) in skeletal muscle prevents the soleus muscle atrophy induced by denervation (Bentzinger et al., 2013); unfortunately, the authors did not investigate the ribosome biogenesis in this study. Even if the mTOR signaling and protein synthesis are activated during the initial stage following skeletal muscle denervation, most probably as a result of the release of free amino acids from the proteasome-dependent proteolysis (Quy et al., 2013), an additive activation of this pathway may be beneficial to promote ribosome biogenesis and prevent muscle atrophy during denervation.
A few studies have provided information on the regulation of the Wnt/β-catenin/c-myc pathway in denervated skeletal muscle. A transcriptome analysis showed that expression of several genes associated with Wnt signaling was decreased in denervated skeletal muscle while c-myc mRNA expression was increased (Magnusson et al., 2005). Although no change was found at the protein level (Siu and Alway, 2005), the transcriptional activation of c-myc may be explained by the cellular remodeling associated with denervation, such as regeneration, apoptosis, or inflammatory response. Further experiments are required to understand the role played by c-myc during denervation and to investigate whether the activation of this transcription factor could be beneficial to prevent atrophy in denervated skeletal muscle.
Perspectives: Ribosome specialization in skeletal muscle
Mauro and Edelman formalized the concept of ribosome specialization in their ribosome filter hypothesis, proposing the ribosome was not just a “tape head that converts ribonucleotide sequences into amino acid sequences” but rather directly regulates translation of specific mRNAs (Mauro and Edelman, 2007). The direct regulation of translation by the ribosome is influenced by the composition of the ribosome which is determined by post-translational modifications of ribosomal RNAs and/or proteins as well as the proteins associated with the ribosome. The notion of ribosome specialization has generated a great deal of excitement lately because it represents a heretofore unrecognized level of gene regulation – much like microRNAs did 10 years ago (Gilbert, 2011; Xue and Barna, 2012). Recently, the Barna laboratory extended the concept of ribosome specialization to mammals when they reported that the tissue-specific expression of Rpl38 during mouse embryonic development was required for proper patterning via translation of specific Hox transcripts (Kondrashov et al., 2011). Here, we introduce the idea that ribosome specialization exists in skeletal muscle and may be involved in the regulation of skeletal muscle mass.
The best example of ribosome specialization in skeletal muscle is the phosphorylation of RPS6 in response to an anabolic signal (Chaillou et al., 2012; Farnfield et al., 2012; Hulmi et al., 2009). As previously mentioned, it was initially thought that phosphorylation of RPS6 directed the specific translation of mRNAs harboring a 5'-TOP sequence (Jefferies et al., 1997; Jefferies et al., 1994) though later studies have called into questions this idea (Ruvinsky et al., 2005). A recent study found that skeletal muscle mass and strength were decreased in mice in which RPS6 phosphorylation was abolished, and this finding was associated with a decrease in the abundance of contractile proteins (Ruvinsky et al., 2009). Although it remains to be elucidated, a reasonable possibility is that RPS6 phosphorylation modifies ribosome function thereby preferentially translating mRNAs encoding sarcomeric proteins.
A study examining the expression profile of ribosomal proteins made the surprising discovery that some ribosomal proteins are expressed in a tissue-specific manner (Thorrez et al., 2008). Transcriptome analysis of 22 tissues revealed that ribosomal protein large 3 (Rpl3) mRNA was highly enriched in all tissues except striated muscles; expression of the Rpl3 paralogue, Rpl3-like (Rpl3l), was exactly the opposite, being highly expressed only in striated muscle, skeletal muscle in particular (Thorrez et al., 2008). A query of BioGPS (http://biogps.org) also confirmed the skeletal muscle-specific expression patterns of Rpl3 and Rpl3l in both human and rodents. Our microarray analysis during skeletal muscle hypertrophy in response to synergist ablation (GEO accession number GSE47098) revealed that the expression pattern of Rpl3 and Rpl3l was rapidly and highly reversed from control levels such that Rpl3l was down-regulated and Rpl3 was up-regulated. The greatest change in Rpl3 and Rpl3l mRNA expression was observed after 7 days following synergist ablation (+234% and −91%, respectively) while the expression of all other RP genes was only modestly changed during skeletal muscle hypertrophy. The role played by Rpl3 and Rpl3l genes during skeletal muscle hypertrophy is currently unexplored but this example of ribosome specialization suggests that the ribosome function may be modified during skeletal muscle hypertrophy.
Conclusion
Ribosome biogenesis appears to be a key process for increasing protein synthesis and cell growth. The mTORC1 signaling pathway and the transcription factor c-myc have a central role in the control of ribosome biogenesis by promoting the synthesis of ribosomal proteins and the transcription of rRNAs. c-myc also regulates the expression of auxiliary factors involved in rRNA processing, ribosome assembly and nuclear ribosome export. Ribosome biogenesis is strongly activated during skeletal muscle hypertrophy induced by synergist ablation and several lines of evidence suggest that its activation is necessary to induce the supra-physiological hypertrophy observed in this model. Currently, the role played by ribosome biogenesis in response to resistance exercise remains to be clearly elucidated. Ribosome biogenesis does not seem to be affected during sarcopenia and cachexia, while it is most probably altered during disuse atrophy (unloading, physical inactivity, immobilization and hindlimb suspension). Furthermore, the regulation of ribosome biogenesis in response to denervation remains controversial. Activating ribosome biogenesis through the development of therapies that target the mTORC1 and the β-catenin/c-myc signaling pathways may be beneficial in preventing or attenuating the loss of skeletal muscle mass in these catabolic states. Lastly, the concept of ribosome specialization has emerged in the field of skeletal muscle, as evidenced by the specific regulation of Rpl3 and Rpl3l gene expression during skeletal muscle hypertrophy. The potential role of ribosome specialization in the regulation of gene expression during skeletal muscle hypertrophy and atrophy remains to be investigated.
Acknowledgments
Contract grant sponsor : National Institues of Health
Contract grant number : AR061939
Footnotes
This article has been accepted for publication and undergone full peer review but has not been through the copyediting, typesetting, pagination and proofreading process, which may lead to differences between this version and the Version of Record. Please cite this article as doi: [10.1002/jcp.24604]
Conflict of interest Nothing to declare
References
- Adams GR, Caiozzo VJ, Haddad F, Baldwin KM. Cellular and molecular responses to increased skeletal muscle loading after irradiation. Am J Physiol Cell Physiol. 2002;283(4):C1182–1195. doi: 10.1152/ajpcell.00173.2002. [DOI] [PubMed] [Google Scholar]
- Alawi F, Lee MN. DKC1 is a direct and conserved transcriptional target of c-MYC. Biochem Biophys Res Commun. 2007;362(4):893–898. doi: 10.1016/j.bbrc.2007.08.071. [DOI] [PubMed] [Google Scholar]
- Arabi A, Wu S, Ridderstrale K, Bierhoff H, Shiue C, Fatyol K, Fahlen S, Hydbring P, Soderberg O, Grummt I, Larsson LG, Wright AP. c-Myc associates with ribosomal DNA and activates RNA polymerase I transcription. Nat Cell Biol. 2005;7(3):303–310. doi: 10.1038/ncb1225. [DOI] [PubMed] [Google Scholar]
- Armstrong DD, Esser KA. Wnt/beta-catenin signaling activates growth-control genes during overload-induced skeletal muscle hypertrophy. Am J Physiol Cell Physiol. 2005;289(4):C853–859. doi: 10.1152/ajpcell.00093.2005. [DOI] [PubMed] [Google Scholar]
- Armstrong DD, Wong VL, Esser KA. Expression of beta-catenin is necessary for physiological growth of adult skeletal muscle. Am J Physiol Cell Physiol. 2006;291(1):C185–188. doi: 10.1152/ajpcell.00644.2005. [DOI] [PubMed] [Google Scholar]
- Aschenbach WG, Ho RC, Sakamoto K, Fujii N, Li Y, Kim YB, Hirshman MF, Goodyear LJ. Regulation of dishevelled and beta-catenin in rat skeletal muscle: an alternative exercise-induced GSK-3beta signaling pathway. Am J Physiol Endocrinol Metab. 2006;291(1):E152–158. doi: 10.1152/ajpendo.00180.2005. [DOI] [PubMed] [Google Scholar]
- Baar K, Esser K. Phosphorylation of p70(S6k) correlates with increased skeletal muscle mass following resistance exercise. Am J Physiol. 1999;276(1 Pt 1):C120–127. doi: 10.1152/ajpcell.1999.276.1.C120. [DOI] [PubMed] [Google Scholar]
- Babij P, Booth FW. Alpha-actin and cytochrome c mRNAs in atrophied adult rat skeletal muscle. Am J Physiol. 1988;254(5 Pt 1):C651–656. doi: 10.1152/ajpcell.1988.254.5.C651. [DOI] [PubMed] [Google Scholar]
- Ben-Ze'ev A, Shtutman M, Zhurinsky J. The integration of cell adhesion with gene expression: the role of beta-catenin. Experimental cell research. 2000;261(1):75–82. doi: 10.1006/excr.2000.5045. [DOI] [PubMed] [Google Scholar]
- Bentzinger CF, Lin S, Romanino K, Castets P, Guridi M, Summermatter S, Handschin C, Tintignac LA, Hall MN, Ruegg MA. Differential response of skeletal muscles to mTORC1 signaling during atrophy and hypertrophy. Skelet Muscle. 2013;3(1):6. doi: 10.1186/2044-5040-3-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bickel CS, Slade JM, Haddad F, Adams GR, Dudley GA. Acute molecular responses of skeletal muscle to resistance exercise in able-bodied and spinal cord-injured subjects. Journal of applied physiology (Bethesda, Md : 1985) 2003;94(6):2255–2262. doi: 10.1152/japplphysiol.00014.2003. [DOI] [PubMed] [Google Scholar]
- Boisvert FM, van Koningsbruggen S, Navascues J, Lamond AI. The multifunctional nucleolus. Nature reviews Molecular cell biology. 2007;8(7):574–585. doi: 10.1038/nrm2184. [DOI] [PubMed] [Google Scholar]
- Chaillou T, Koulmann N, Simler N, Meunier A, Serrurier B, Chapot R, Peinnequin A, Beaudry M, Bigard X. Hypoxia transiently affects skeletal muscle hypertrophy in a functional overload model. Am J Physiol Regul Integr Comp Physiol. 2012;302(5):R643–654. doi: 10.1152/ajpregu.00262.2011. [DOI] [PubMed] [Google Scholar]
- Chaillou T, Lee JD, England JH, Esser KA, McCarthy JJ. Time course of gene expression during mouse skeletal muscle hypertrophy. Journal of applied physiology (Bethesda, Md : 1985) 2013;115(7):1065–1074. doi: 10.1152/japplphysiol.00611.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Sood S, Biada J, Roth R, Rabkin R. Increased workload fully activates the blunted IRS-1/PI3-kinase/Akt signaling pathway in atrophied uremic muscle. Kidney Int. 2008;73(7):848–855. doi: 10.1038/sj.ki.5002801. [DOI] [PubMed] [Google Scholar]
- Chen Y, Sood S, McIntire K, Roth R, Rabkin R. Leucine-stimulated mTOR signaling is partly attenuated in skeletal muscle of chronically uremic rats. Am J Physiol Endocrinol Metab. 2011;301(5):E873–881. doi: 10.1152/ajpendo.00068.2011. [DOI] [PubMed] [Google Scholar]
- Coller HA, Grandori C, Tamayo P, Colbert T, Lander ES, Eisenman RN, Golub TR. Expression analysis with oligonucleotide microarrays reveals that MYC regulates genes involved in growth, cell cycle, signaling, and adhesion. Proc Natl Acad Sci U S A. 2000;97(7):3260–3265. doi: 10.1073/pnas.97.7.3260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Constantin D, McCullough J, Mahajan RP, Greenhaff PL. Novel events in the molecular regulation of muscle mass in critically ill patients. J Physiol. 2011;589(Pt 15):3883–3895. doi: 10.1113/jphysiol.2011.206193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cross DA, Alessi DR, Cohen P, Andjelkovich M, Hemmings BA. Inhibition of glycogen synthase kinase-3 by insulin mediated by protein kinase B. Nature. 1995;378(6559):785–789. doi: 10.1038/378785a0. [DOI] [PubMed] [Google Scholar]
- Csibi A, Cornille K, Leibovitch MP, Poupon A, Tintignac LA, Sanchez AM, Leibovitch SA. The translation regulatory subunit eIF3f controls the kinase-dependent mTOR signaling required for muscle differentiation and hypertrophy in mouse. PLoS One. 2010;5(2):e8994. doi: 10.1371/journal.pone.0008994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dapp C, Schmutz S, Hoppeler H, Fluck M. Transcriptional reprogramming and ultrastructure during atrophy and recovery of mouse soleus muscle. Physiological genomics. 2004;20(1):97–107. doi: 10.1152/physiolgenomics.00100.2004. [DOI] [PubMed] [Google Scholar]
- de Boer MD, Selby A, Atherton P, Smith K, Seynnes OR, Maganaris CN, Maffulli N, Movin T, Narici MV, Rennie MJ. The temporal responses of protein synthesis, gene expression and cell signalling in human quadriceps muscle and patellar tendon to disuse. J Physiol. 2007;585(Pt 1):241–251. doi: 10.1113/jphysiol.2007.142828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond MJ, Dickinson JM, Fry CS, Walker DK, Gundermann DM, Reidy PT, Timmerman KL, Markofski MM, Paddon-Jones D, Rasmussen BB, Volpi E. Bed rest impairs skeletal muscle amino acid transporter expression, mTORC1 signaling, and protein synthesis in response to essential amino acids in older adults. Am J Physiol Endocrinol Metab. 2012;302(9):E1113–1122. doi: 10.1152/ajpendo.00603.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farnfield MM, Breen L, Carey KA, Garnham A, Cameron-Smith D. Activation of mTOR signalling in young and old human skeletal muscle in response to combined resistance exercise and whey protein ingestion. Applied physiology, nutrition, and metabolism = Physiologie appliquee, nutrition et metabolisme. 2012;37(1):21–30. doi: 10.1139/h11-132. [DOI] [PubMed] [Google Scholar]
- Favier FB, Costes F, Defour A, Bonnefoy R, Lefai E, Bauge S, Peinnequin A, Benoit H, Freyssenet D. Downregulation of Akt/mammalian target of rapamycin pathway in skeletal muscle is associated with increased REDD1 expression in response to chronic hypoxia. Am J Physiol Regul Integr Comp Physiol. 2010;298(6):R1659–1666. doi: 10.1152/ajpregu.00550.2009. [DOI] [PubMed] [Google Scholar]
- Fry CS, Drummond MJ, Glynn EL, Dickinson JM, Gundermann DM, Timmerman KL, Walker DK, Dhanani S, Volpi E, Rasmussen BB. Aging impairs contraction-induced human skeletal muscle mTORC1 signaling and protein synthesis. Skelet Muscle. 2011;1(1):11. doi: 10.1186/2044-5040-1-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamrin L, Berg HE, Essen P, Tesch PA, Hultman E, Garlick PJ, McNurlan MA, Wernerman J. The effect of unloading on protein synthesis in human skeletal muscle. Acta Physiol Scand. 1998;163(4):369–377. doi: 10.1046/j.1365-201X.1998.t01-1-00391.x. [DOI] [PubMed] [Google Scholar]
- Gebauer F, Hentze MW. Molecular mechanisms of translational control. Nature reviews Molecular cell biology. 2004;5(10):827–835. doi: 10.1038/nrm1488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson JN, Halliday D, Morrison WL, Stoward PJ, Hornsby GA, Watt PW, Murdoch G, Rennie MJ. Decrease in human quadriceps muscle protein turnover consequent upon leg immobilization. Clin Sci (Lond) 1987;72(4):503–509. doi: 10.1042/cs0720503. [DOI] [PubMed] [Google Scholar]
- Gibson JN, Smith K, Rennie MJ. Prevention of disuse muscle atrophy by means of electrical stimulation: maintenance of protein synthesis. Lancet. 1988;2(8614):767–770. doi: 10.1016/s0140-6736(88)92417-8. [DOI] [PubMed] [Google Scholar]
- Gilbert WV. Functional specialization of ribosomes? Trends Biochem Sci. 2011;36(3):127–132. doi: 10.1016/j.tibs.2010.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glover EI, Oates BR, Tang JE, Moore DR, Tarnopolsky MA, Phillips SM. Resistance exercise decreases eIF2Bepsilon phosphorylation and potentiates the feeding-induced stimulation of p70S6K1 and rpS6 in young men. Am J Physiol Regul Integr Comp Physiol. 2008a;295(2):R604–610. doi: 10.1152/ajpregu.00097.2008. [DOI] [PubMed] [Google Scholar]
- Glover EI, Phillips SM, Oates BR, Tang JE, Tarnopolsky MA, Selby A, Smith K, Rennie MJ. Immobilization induces anabolic resistance in human myofibrillar protein synthesis with low and high dose amino acid infusion. J Physiol. 2008b;586(Pt 24):6049–6061. doi: 10.1113/jphysiol.2008.160333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldspink DF. The effects of denervation on protein turnover of rat skeletal muscle. Biochem J. 1976;156(1):71–80. doi: 10.1042/bj1560071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Roman N, Felton-Edkins ZA, Kenneth NS, Goodfellow SJ, Athineos D, Zhang J, Ramsbottom BA, Innes F, Kantidakis T, Kerr ER, Brodie J, Grandori C, White RJ. Activation by c-Myc of transcription by RNA polymerases I, II and III. Biochemical Society symposium. 2006;(73):141–154. doi: 10.1042/bss0730141. [DOI] [PubMed] [Google Scholar]
- Gomez-Roman N, Grandori C, Eisenman RN, White RJ. Direct activation of RNA polymerase III transcription by c-Myc. Nature. 2003;421(6920):290–294. doi: 10.1038/nature01327. [DOI] [PubMed] [Google Scholar]
- Goodfellow SJ, Innes F, Derblay LE, MacLellan WR, Scott PH, White RJ. Regulation of RNA polymerase III transcription during hypertrophic growth. The EMBO journal. 2006;25(7):1522–1533. doi: 10.1038/sj.emboj.7601040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodman CA, Frey JW, Mabrey DM, Jacobs BL, Lincoln HC, You JS, Hornberger TA. The role of skeletal muscle mTOR in the regulation of mechanical load-induced growth. J Physiol. 2011;589(Pt 22):5485–5501. doi: 10.1113/jphysiol.2011.218255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon BS, Kelleher AR, Kimball SR. Regulation of muscle protein synthesis and the effects of catabolic states. The international journal of biochemistry & cell biology. 2013;45(10):2147–2157. doi: 10.1016/j.biocel.2013.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grandori C, Gomez-Roman N, Felton-Edkins ZA, Ngouenet C, Galloway DA, Eisenman RN, White RJ. c-Myc binds to human ribosomal DNA and stimulates transcription of rRNA genes by RNA polymerase I. Nat Cell Biol. 2005;7(3):311–318. doi: 10.1038/ncb1224. [DOI] [PubMed] [Google Scholar]
- Granneman S, Tollervey D. Building ribosomes: even more expensive than expected? Curr Biol. 2007;17(11):R415–417. doi: 10.1016/j.cub.2007.04.011. [DOI] [PubMed] [Google Scholar]
- Haddad F, Adams GR. Aging-sensitive cellular and molecular mechanisms associated with skeletal muscle hypertrophy. Journal of applied physiology (Bethesda, Md : 1985) 2006;100(4):1188–1203. doi: 10.1152/japplphysiol.01227.2005. [DOI] [PubMed] [Google Scholar]
- Haddad F, Baldwin KM, Tesch PA. Pretranslational markers of contractile protein expression in human skeletal muscle: effect of limb unloading plus resistance exercise. Journal of applied physiology (Bethesda, Md : 1985) 2005;98(1):46–52. doi: 10.1152/japplphysiol.00553.2004. [DOI] [PubMed] [Google Scholar]
- Haddad F, Roy RR, Zhong H, Edgerton VR, Baldwin KM. Atrophy responses to muscle inactivity. I. Cellular markers of protein deficits. Journal of applied physiology (Bethesda, Md : 1985) 2003;95(2):781–790. doi: 10.1152/japplphysiol.00317.2003. [DOI] [PubMed] [Google Scholar]
- Hannan KM, Brandenburger Y, Jenkins A, Sharkey K, Cavanaugh A, Rothblum L, Moss T, Poortinga G, McArthur GA, Pearson RB, Hannan RD. mTOR-dependent regulation of ribosomal gene transcription requires S6K1 and is mediated by phosphorylation of the carboxy-terminal activation domain of the nucleolar transcription factor UBF. Mol Cell Biol. 2003;23(23):8862–8877. doi: 10.1128/MCB.23.23.8862-8877.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannan RD, Luyken J, Rothblum LI. Regulation of ribosomal DNA transcription during contraction-induced hypertrophy of neonatal cardiomyocytes. J Biol Chem. 1996;271(6):3213–3220. doi: 10.1074/jbc.271.6.3213. [DOI] [PubMed] [Google Scholar]
- Harrington D, Anker SD, Chua TP, Webb-Peploe KM, Ponikowski PP, Poole-Wilson PA, Coats AJ. Skeletal muscle function and its relation to exercise tolerance in chronic heart failure. Journal of the American College of Cardiology. 1997;30(7):1758–1764. doi: 10.1016/s0735-1097(97)00381-1. [DOI] [PubMed] [Google Scholar]
- Heinemeier KM, Olesen JL, Haddad F, Schjerling P, Baldwin KM, Kjaer M. Effect of unloading followed by reloading on expression of collagen and related growth factors in rat tendon and muscle. J Appl Physiol. 2009;106(1):178–186. doi: 10.1152/japplphysiol.91092.2008. [DOI] [PubMed] [Google Scholar]
- Held WA, Mizushima S, Nomura M. Reconstitution of Escherichia coli 30 S ribosomal subunits from purified molecular components. J Biol Chem. 1973;248(16):5720–5730. [PubMed] [Google Scholar]
- Henras AK, Soudet J, Gerus M, Lebaron S, Caizergues-Ferrer M, Mougin A, Henry Y. The post-transcriptional steps of eukaryotic ribosome biogenesis. Cellular and molecular life sciences : CMLS. 2008;65(15):2334–2359. doi: 10.1007/s00018-008-8027-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinoi T, Yamamoto H, Kishida M, Takada S, Kishida S, Kikuchi A. Complex formation of adenomatous polyposis coli gene product and axin facilitates glycogen synthase kinase-3 beta-dependent phosphorylation of beta-catenin and down-regulates beta-catenin. J Biol Chem. 2000;275(44):34399–34406. doi: 10.1074/jbc.M003997200. [DOI] [PubMed] [Google Scholar]
- Hulmi JJ, Tannerstedt J, Selanne H, Kainulainen H, Kovanen V, Mero AA. Resistance exercise with whey protein ingestion affects mTOR signaling pathway and myostatin in men. Journal of applied physiology (Bethesda, Md : 1985) 2009;106(5):1720–1729. doi: 10.1152/japplphysiol.00087.2009. [DOI] [PubMed] [Google Scholar]
- Hwee DT, Bodine SC. Age-related deficit in load-induced skeletal muscle growth. The journals of gerontology Series A, Biological sciences and medical sciences. 2009;64(6):618–628. doi: 10.1093/gerona/glp026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iadevaia V, Huo Y, Zhang Z, Foster LJ, Proud CG. Roles of the mammalian target of rapamycin, mTOR, in controlling ribosome biogenesis and protein synthesis. Biochemical Society transactions. 2012;40(1):168–172. doi: 10.1042/BST20110682. [DOI] [PubMed] [Google Scholar]
- Jefferies HB, Fumagalli S, Dennis PB, Reinhard C, Pearson RB, Thomas G. Rapamycin suppresses 5'TOP mRNA translation through inhibition of p70s6k. The EMBO journal. 1997;16(12):3693–3704. doi: 10.1093/emboj/16.12.3693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefferies HB, Reinhard C, Kozma SC, Thomas G. Rapamycin selectively represses translation of the “polypyrimidine tract” mRNA family. Proc Natl Acad Sci U S A. 1994;91(10):4441–4445. doi: 10.1073/pnas.91.10.4441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kantidakis T, Ramsbottom BA, Birch JL, Dowding SN, White RJ. mTOR associates with TFIIIC, is found at tRNA and 5S rRNA genes, and targets their repressor Maf1. Proc Natl Acad Sci U S A. 2010;107(26):11823–11828. doi: 10.1073/pnas.1005188107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapp LD, Lorsch JR. The molecular mechanics of eukaryotic translation. Annu Rev Biochem. 2004;73:657–704. doi: 10.1146/annurev.biochem.73.030403.080419. [DOI] [PubMed] [Google Scholar]
- Kelleher AR, Kimball SR, Dennis MD, Schilder RJ, Jefferson LS. The mTORC1 signaling repressors REDD1/2 are rapidly induced and activation of p70S6K1 by leucine is defective in skeletal muscle of an immobilized rat hindlimb. Am J Physiol Endocrinol Metab. 2013;304(2):E229–236. doi: 10.1152/ajpendo.00409.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S, Li Q, Dang CV, Lee LA. Induction of ribosomal genes and hepatocyte hypertrophy by adenovirus-mediated expression of c-Myc in vivo. Proc Natl Acad Sci U S A. 2000;97(21):11198–11202. doi: 10.1073/pnas.200372597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirn-Safran CB, Oristian DS, Focht RJ, Parker SG, Vivian JL, Carson DD. Global growth deficiencies in mice lacking the ribosomal protein HIP/RPL29. Dev Dyn. 2007;236(2):447–460. doi: 10.1002/dvdy.21046. [DOI] [PubMed] [Google Scholar]
- Kondrashov N, Pusic A, Stumpf CR, Shimizu K, Hsieh AC, Xue S, Ishijima J, Shiroishi T, Barna M. Ribosome-mediated specificity in Hox mRNA translation and vertebrate tissue patterning. Cell. 2011;145(3):383–397. doi: 10.1016/j.cell.2011.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar V, Selby A, Rankin D, Patel R, Atherton P, Hildebrandt W, Williams J, Smith K, Seynnes O, Hiscock N, Rennie MJ. Age-related differences in the dose-response relationship of muscle protein synthesis to resistance exercise in young and old men. J Physiol. 2009;587(Pt 1):211–217. doi: 10.1113/jphysiol.2008.164483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang CH, Frost RA, Vary TC. Regulation of muscle protein synthesis during sepsis and inflammation. Am J Physiol Endocrinol Metab. 2007;293(2):E453–459. doi: 10.1152/ajpendo.00204.2007. [DOI] [PubMed] [Google Scholar]
- Lang SM, Kazi AA, Hong-Brown L, Lang CH. Delayed recovery of skeletal muscle mass following hindlimb immobilization in mTOR heterozygous mice. PLoS One. 2012;7(6):e38910. doi: 10.1371/journal.pone.0038910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lecker SH, Jagoe RT, Gilbert A, Gomes M, Baracos V, Bailey J, Price SR, Mitch WE, Goldberg AL. Multiple types of skeletal muscle atrophy involve a common program of changes in gene expression. Faseb J. 2004;18(1):39–51. doi: 10.1096/fj.03-0610com. [DOI] [PubMed] [Google Scholar]
- Li L, Yuan H, Weaver CD, Mao J, Farr GH, 3rd, Sussman DJ, Jonkers J, Kimelman D, Wu D. Axin and Frat1 interact with dvl and GSK, bridging Dvl to GSK in Wnt-mediated regulation of LEF-1. The EMBO journal. 1999;18(15):4233–4240. doi: 10.1093/emboj/18.15.4233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindstrom MS. Emerging functions of ribosomal proteins in gene-specific transcription and translation. Biochem Biophys Res Commun. 2009;379(2):167–170. doi: 10.1016/j.bbrc.2008.12.083. [DOI] [PubMed] [Google Scholar]
- Machida M, Takeda K, Yokono H, Ikemune S, Taniguchi Y, Kiyosawa H, Takemasa T. Reduction of ribosome biogenesis with activation of the mTOR pathway in denervated atrophic muscle. J Cell Physiol. 2012;227(4):1569–1576. doi: 10.1002/jcp.22871. [DOI] [PubMed] [Google Scholar]
- Magnusson C, Svensson A, Christerson U, Tagerud S. Denervation-induced alterations in gene expression in mouse skeletal muscle. The European journal of neuroscience. 2005;21(2):577–580. doi: 10.1111/j.1460-9568.2005.03855.x. [DOI] [PubMed] [Google Scholar]
- Mauro VP, Edelman GM. The ribosome filter redux. Cell Cycle. 2007;6(18):2246–2251. doi: 10.4161/cc.6.18.4739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer C, Zhao J, Yuan X, Grummt I. mTOR-dependent activation of the transcription factor TIF-IA links rRNA synthesis to nutrient availability. Genes Dev. 2004;18(4):423–434. doi: 10.1101/gad.285504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayhew DL, Hornberger TA, Lincoln HC, Bamman MM. Eukaryotic initiation factor 2B epsilon induces cap-dependent translation and skeletal muscle hypertrophy. J Physiol. 2011;589(Pt 12):3023–3037. doi: 10.1113/jphysiol.2010.202432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer N, Penn LZ. Reflecting on 25 years with MYC. Nat Rev Cancer. 2008;8(12):976–990. doi: 10.1038/nrc2231. [DOI] [PubMed] [Google Scholar]
- Miyazaki M, McCarthy JJ, Fedele MJ, Esser KA. Early activation of mTORC1 signalling in response to mechanical overload is independent of phosphoinositide 3-kinase/Akt signalling. J Physiol. 2011;589(Pt 7):1831–1846. doi: 10.1113/jphysiol.2011.205658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montanaro L, Trere D, Derenzini M. Changes in ribosome biogenesis may induce cancer by down-regulating the cell tumor suppressor potential. Biochimica et biophysica acta. 2012;1825(1):101–110. doi: 10.1016/j.bbcan.2011.10.006. [DOI] [PubMed] [Google Scholar]
- Moss T, Langlois F, Gagnon-Kugler T, Stefanovsky V. A housekeeper with power of attorney: the rRNA genes in ribosome biogenesis. Cellular and molecular life sciences : CMLS. 2007;64(1):29–49. doi: 10.1007/s00018-006-6278-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moss T, Stefanovsky VY. Promotion and regulation of ribosomal transcription in eukaryotes by RNA polymerase I. Progress in nucleic acid research and molecular biology. 1995;50:25–66. doi: 10.1016/s0079-6603(08)60810-7. [DOI] [PubMed] [Google Scholar]
- Nader GA, McLoughlin TJ, Esser KA. mTOR function in skeletal muscle hypertrophy: increased ribosomal RNA via cell cycle regulators. Am J Physiol Cell Physiol. 2005;289(6):C1457–1465. doi: 10.1152/ajpcell.00165.2005. [DOI] [PubMed] [Google Scholar]
- Nader GA, von Walden F, Liu C, Lindvall J, Gutmann L, Pistilli EE, Gordon PM. Resistance exercise training modulates acute gene expression during human skeletal muscle hypertrophy. Journal of applied physiology (Bethesda, Md : 1985) 2014 doi: 10.1152/japplphysiol.01366.2013. [DOI] [PubMed] [Google Scholar]
- Nierhaus KH, Dohme F. Total reconstitution of functionally active 50S ribosomal subunits from Escherichia coli. Proc Natl Acad Sci U S A. 1974;71(12):4713–4717. doi: 10.1073/pnas.71.12.4713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novak A, Dedhar S. Signaling through beta-catenin and Lef/Tcf. Cellular and molecular life sciences : CMLS. 1999;56(5–6):523–537. doi: 10.1007/s000180050449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson AK, Ledee D, Iwamoto K, Kajimoto M, O'Kelly Priddy C, Isern N, Portman MA. C-Myc induced compensated cardiac hypertrophy increases free fatty acid utilization for the citric acid cycle. Journal of molecular and cellular cardiology. 2013;55:156–164. doi: 10.1016/j.yjmcc.2012.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orellana RA, Wilson FA, Gazzaneo MC, Suryawan A, Davis TA, Nguyen HV. Sepsis and development impede muscle protein synthesis in neonatal pigs by different ribosomal mechanisms. Pediatric research. 2011;69(6):473–478. doi: 10.1203/PDR.0b013e3182176da1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pehme A, Alev K, Kaasik P, Seene T. Age-related changes in skeletal-muscle myosin heavy-chain composition: effect of mechanical loading. Journal of aging and physical activity. 2004;12(1):29–44. doi: 10.1123/japa.12.1.29. [DOI] [PubMed] [Google Scholar]
- Pende M, Um SH, Mieulet V, Sticker M, Goss VL, Mestan J, Mueller M, Fumagalli S, Kozma SC, Thomas G. S6K1(−/−)/S6K2(−/−) mice exhibit perinatal lethality and rapamycin-sensitive 5′-terminal oligopyrimidine mRNA translation and reveal a mitogen-activated protein kinase-dependent S6 kinase pathway. Mol Cell Biol. 2004;24(8):3112–3124. doi: 10.1128/MCB.24.8.3112-3124.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersson B, Wernerman J, Waller SO, von der Decken A, Vinnars E. Elective abdominal surgery depresses muscle protein synthesis and increases subjective fatigue: effects lasting more than 30 days. The British journal of surgery. 1990;77(7):796–800. doi: 10.1002/bjs.1800770725. [DOI] [PubMed] [Google Scholar]
- Phillips BE, Williams JP, Gustafsson T, Bouchard C, Rankinen T, Knudsen S, Smith K, Timmons JA, Atherton PJ. Molecular networks of human muscle adaptation to exercise and age. PLoS genetics. 2013;9(3):e1003389. doi: 10.1371/journal.pgen.1003389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips SM, Glover EI, Rennie MJ. Alterations of protein turnover underlying disuse atrophy in human skeletal muscle. J Appl Physiol. 2009;107(3):645–654. doi: 10.1152/japplphysiol.00452.2009. [DOI] [PubMed] [Google Scholar]
- Quy PN, Kuma A, Pierre P, Mizushima N. Proteasome-dependent activation of mammalian target of rapamycin complex 1 (mTORC1) is essential for autophagy suppression and muscle remodeling following denervation. J Biol Chem. 2013;288(2):1125–1134. doi: 10.1074/jbc.M112.399949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiter AK, Bolster DR, Crozier SJ, Kimball SR, Jefferson LS. AMPK represses TOP mRNA translation but not global protein synthesis in liver. Biochem Biophys Res Commun. 2008;374(2):345–350. doi: 10.1016/j.bbrc.2008.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodnina MV, Wintermeyer W. Recent mechanistic insights into eukaryotic ribosomes. Current opinion in cell biology. 2009;21(3):435–443. doi: 10.1016/j.ceb.2009.01.023. [DOI] [PubMed] [Google Scholar]
- Ruvinsky I, Katz M, Dreazen A, Gielchinsky Y, Saada A, Freedman N, Mishani E, Zimmerman G, Kasir J, Meyuhas O. Mice deficient in ribosomal protein S6 phosphorylation suffer from muscle weakness that reflects a growth defect and energy deficit. PLoS One. 2009;4(5):e5618. doi: 10.1371/journal.pone.0005618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruvinsky I, Sharon N, Lerer T, Cohen H, Stolovich-Rain M, Nir T, Dor Y, Zisman P, Meyuhas O. Ribosomal protein S6 phosphorylation is a determinant of cell size and glucose homeostasis. Genes Dev. 2005;19(18):2199–2211. doi: 10.1101/gad.351605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schakman O, Kalista S, Bertrand L, Lause P, Verniers J, Ketelslegers JM, Thissen JP. Role of Akt/GSK-3beta/beta-catenin transduction pathway in the muscle anti-atrophy action of insulin-like growth factor-I in glucocorticoid-treated rats. Endocrinology. 2008;149(8):3900–3908. doi: 10.1210/en.2008-0439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiaffino S, Dyar KA, Ciciliot S, Blaauw B, Sandri M. Mechanisms regulating skeletal muscle growth and atrophy. The FEBS journal. 2013;280(17):4294–4314. doi: 10.1111/febs.12253. [DOI] [PubMed] [Google Scholar]
- Schlosser I, Holzel M, Murnseer M, Burtscher H, Weidle UH, Eick D. A role for c-Myc in the regulation of ribosomal RNA processing. Nucleic acids research. 2003;31(21):6148–6156. doi: 10.1093/nar/gkg794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schols AM, Broekhuizen R, Weling-Scheepers CA, Wouters EF. Body composition and mortality in chronic obstructive pulmonary disease. The American journal of clinical nutrition. 2005;82(1):53–59. doi: 10.1093/ajcn.82.1.53. [DOI] [PubMed] [Google Scholar]
- Schuhmacher M, Staege MS, Pajic A, Polack A, Weidle UH, Bornkamm GW, Eick D, Kohlhuber F. Control of cell growth by c-Myc in the absence of cell division. Curr Biol. 1999;9(21):1255–1258. doi: 10.1016/s0960-9822(99)80507-7. [DOI] [PubMed] [Google Scholar]
- Shor B, Wu J, Shakey Q, Toral-Barza L, Shi C, Follettie M, Yu K. Requirement of the mTOR kinase for the regulation of Maf1 phosphorylation and control of RNA polymerase III-dependent transcription in cancer cells. J Biol Chem. 2010;285(20):15380–15392. doi: 10.1074/jbc.M109.071639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sions JM, Tyrell CM, Knarr BA, Jancosko A, Binder-Macleod SA. Age- and stroke-related skeletal muscle changes: a review for the geriatric clinician. Journal of geriatric physical therapy (2001) 2012;35(3):155–161. doi: 10.1519/JPT.0b013e318236db92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siu PM, Alway SE. Mitochondria-associated apoptotic signalling in denervated rat skeletal muscle. J Physiol. 2005;565(Pt 1):309–323. doi: 10.1113/jphysiol.2004.081083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun H, Li M, Gong L, Liu M, Ding F, Gu X. iTRAQ-coupled 2D LC-MS/MS analysis on differentially expressed proteins in denervated tibialis anterior muscle of Rattus norvegicus. Mol Cell Biochem. 2012;364(1–2):193–207. doi: 10.1007/s11010-011-1218-2. [DOI] [PubMed] [Google Scholar]
- Teng T, Thomas G, Mercer CA. Growth control and ribosomopathies. Current opinion in genetics & development. 2013;23(1):63–71. doi: 10.1016/j.gde.2013.02.001. [DOI] [PubMed] [Google Scholar]
- Thomas GM, Frame S, Goedert M, Nathke I, Polakis P, Cohen P. A GSK3-binding peptide from FRAT1 selectively inhibits the GSK3-catalysed phosphorylation of axin and beta-catenin. FEBS letters. 1999;458(2):247–251. doi: 10.1016/s0014-5793(99)01161-8. [DOI] [PubMed] [Google Scholar]
- Thomason DB, Booth FW. Atrophy of the soleus muscle by hindlimb unweighting. J Appl Physiol. 1990;68(1):1–12. doi: 10.1152/jappl.1990.68.1.1. [DOI] [PubMed] [Google Scholar]
- Thomson DM, Gordon SE. Impaired overload-induced muscle growth is associated with diminished translational signalling in aged rat fast-twitch skeletal muscle. J Physiol. 2006;574(Pt 1):291–305. doi: 10.1113/jphysiol.2006.107490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson E, Ferreira-Cerca S, Hurt E. Eukaryotic ribosome biogenesis at a glance. J Cell Sci. 2013;126(Pt 21):4815–4821. doi: 10.1242/jcs.111948. [DOI] [PubMed] [Google Scholar]
- Thorrez L, Van Deun K, Tranchevent LC, Van Lommel L, Engelen K, Marchal K, Moreau Y, Van Mechelen I, Schuit F. Using ribosomal protein genes as reference: a tale of caution. PLoS One. 2008;3(3):e1854. doi: 10.1371/journal.pone.0001854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsang CK, Liu H, Zheng XF. mTOR binds to the promoters of RNA polymerase I- and III-transcribed genes. Cell Cycle. 2010;9(5):953–957. doi: 10.4161/cc.9.5.10876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Upadhya R, Lee J, Willis IM. Maf1 is an essential mediator of diverse signals that repress RNA polymerase III transcription. Molecular cell. 2002;10(6):1489–1494. doi: 10.1016/s1097-2765(02)00787-6. [DOI] [PubMed] [Google Scholar]
- van Riggelen J, Yetil A, Felsher DW. MYC as a regulator of ribosome biogenesis and protein synthesis. Nat Rev Cancer. 2010;10(4):301–309. doi: 10.1038/nrc2819. [DOI] [PubMed] [Google Scholar]
- Volarevic S, Stewart MJ, Ledermann B, Zilberman F, Terracciano L, Montini E, Grompe M, Kozma SC, Thomas G. Proliferation, but not growth, blocked by conditional deletion of 40S ribosomal protein S6. Science. 2000;288(5473):2045–2047. doi: 10.1126/science.288.5473.2045. [DOI] [PubMed] [Google Scholar]
- von Maltzahn J, Bentzinger CF, Rudnicki MA. Wnt7a-Fzd7 signalling directly activates the Akt/mTOR anabolic growth pathway in skeletal muscle. Nat Cell Biol. 2012;14(2):186–191. doi: 10.1038/ncb2404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Walden F, Casagrande V, Ostlund Farrants AK, Nader GA. Mechanical loading induces the expression of a Pol I regulon at the onset of skeletal muscle hypertrophy. Am J Physiol Cell Physiol. 2012;302(10):C1523–1530. doi: 10.1152/ajpcell.00460.2011. [DOI] [PubMed] [Google Scholar]
- Warner JR. The economics of ribosome biosynthesis in yeast. Trends Biochem Sci. 1999;24(11):437–440. doi: 10.1016/s0968-0004(99)01460-7. [DOI] [PubMed] [Google Scholar]
- Wei Y, Zheng XF. Sch9 partially mediates TORC1 signaling to control ribosomal RNA synthesis. Cell Cycle. 2009;8(24):4085–4090. doi: 10.4161/cc.8.24.10170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White JP, Baynes JW, Welle SL, Kostek MC, Matesic LE, Sato S, Carson JA. The regulation of skeletal muscle protein turnover during the progression of cancer cachexia in the Apc(Min/+) mouse. PLoS One. 2011;6(9):e24650. doi: 10.1371/journal.pone.0024650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams JP, Phillips BE, Smith K, Atherton PJ, Rankin D, Selby AL, Liptrot S, Lund J, Larvin M, Rennie MJ. Effect of tumor burden and subsequent surgical resection on skeletal muscle mass and protein turnover in colorectal cancer patients. The American journal of clinical nutrition. 2012;96(5):1064–1070. doi: 10.3945/ajcn.112.045708. [DOI] [PubMed] [Google Scholar]
- Woolford JL, Jr., Baserga SJ. Ribosome Biogenesis in the Yeast Saccharomyces cerevisiae. Genetics. 2013;195(3):643–681. doi: 10.1534/genetics.113.153197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu CH, Sahoo D, Arvanitis C, Bradon N, Dill DL, Felsher DW. Combined analysis of murine and human microarrays and ChIP analysis reveals genes associated with the ability of MYC to maintain tumorigenesis. PLoS genetics. 2008;4(6):e1000090. doi: 10.1371/journal.pgen.1000090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue S, Barna M. Specialized ribosomes: a new frontier in gene regulation and organismal biology. Nature reviews Molecular cell biology. 2012;13(6):355–369. doi: 10.1038/nrm3359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeller KI, Zhao X, Lee CW, Chiu KP, Yao F, Yustein JT, Ooi HS, Orlov YL, Shahab A, Yong HC, Fu Y, Weng Z, Kuznetsov VA, Sung WK, Ruan Y, Dang CV, Wei CL. Global mapping of c-Myc binding sites and target gene networks in human B cells. Proc Natl Acad Sci U S A. 2006;103(47):17834–17839. doi: 10.1073/pnas.0604129103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Liu R, Townsend PA, Proud CG. p90(RSK)s mediate the activation of ribosomal RNA synthesis by the hypertrophic agonist phenylephrine in adult cardiomyocytes. Journal of molecular and cellular cardiology. 2013;59:139–147. doi: 10.1016/j.yjmcc.2013.03.006. [DOI] [PubMed] [Google Scholar]
- Zinzalla V, Stracka D, Oppliger W, Hall MN. Activation of mTORC2 by association with the ribosome. Cell. 2011;144(5):757–768. doi: 10.1016/j.cell.2011.02.014. [DOI] [PubMed] [Google Scholar]