Abstract
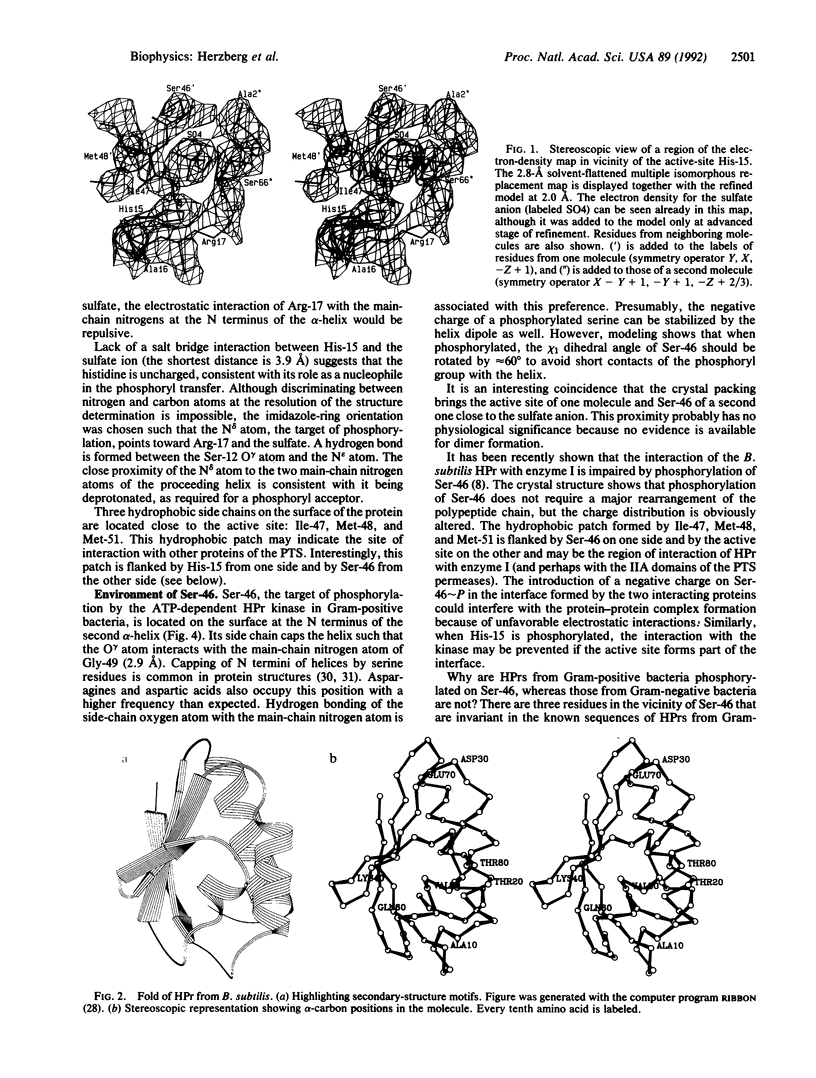
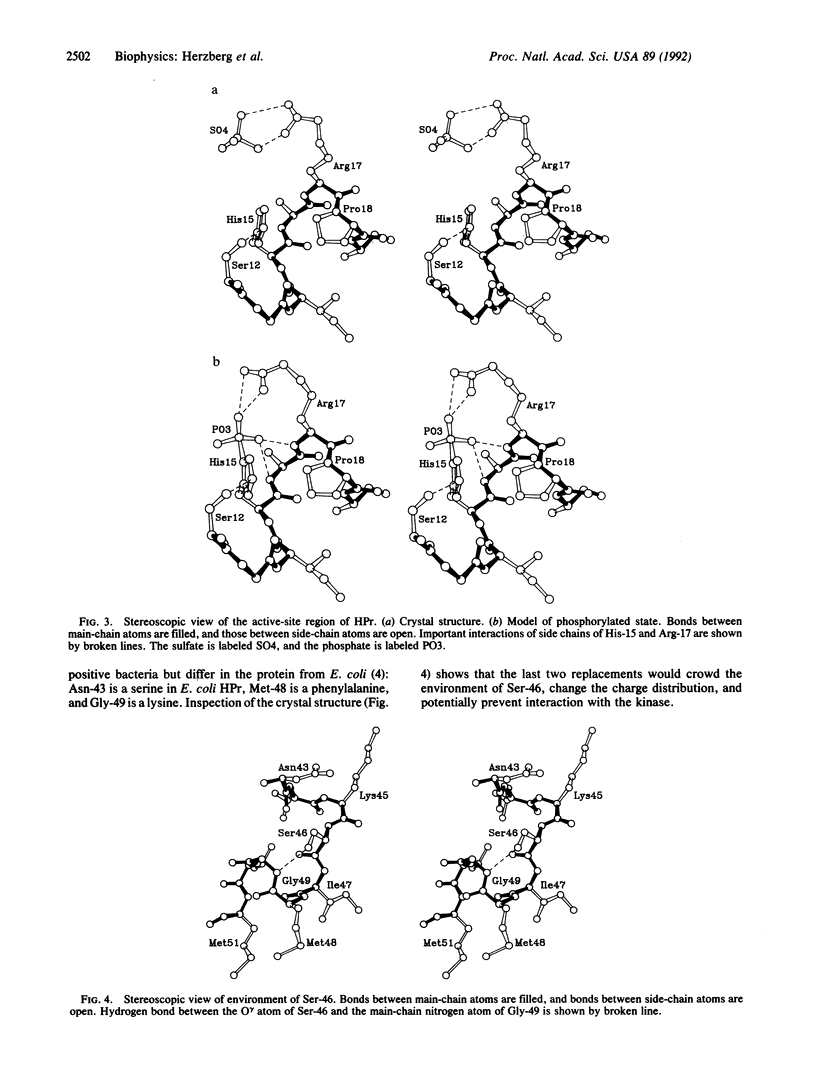
The crystal structure of the histidine-containing phosphocarrier protein (HPr) of the phosphoenolpyruvate:sugar phosphotransferase system (PTS) from Bacillus subtilis has been determined at 2.0-A resolution and refined to a crystallographic residual error R-factor of 0.150. The secondary-structure folding topology of the molecule is that of an open-face beta-sandwich formed by four antiparallel beta-strands packed against three alpha-helices. The active-site histidine, His-15, caps the N terminus of the first helix, suggesting that the helix dipole plays a role in stabilizing the phosphorylated state of the histidine. A sulfate anion located between His-15 and the neighboring Arg-17 has been identified in the electron-density map. Association of this negatively charged species with the two key catalytic residues implies that the crystal structure resembles the phosphorylated state of the protein. A model of the phosphorylated form of the molecule is proposed, in which the negatively charged phosphoryl group interacts with two main-chain nitrogen atoms of the following helix and with the guanidinium group of Arg-17. It is also proposed that the phosphoryl transfer from HPr to the IIA domain of the glucose permease involves Arg-17 switching between two salt bridges: one with the phosphorylated histidyl of HPr and the other with two aspartyl residues associated with the active site of the IIA domain of glucose permease, which are accessible upon complex formation.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Begley G. S., Hansen D. E., Jacobson G. R., Knowles J. R. Stereochemical course of the reactions catalyzed by the bacterial phosphoenolpyruvate:glucose phosphotransferase system. Biochemistry. 1982 Oct 26;21(22):5552–5556. doi: 10.1021/bi00265a026. [DOI] [PubMed] [Google Scholar]
- Brünger A. T., Kuriyan J., Karplus M. Crystallographic R factor refinement by molecular dynamics. Science. 1987 Jan 23;235(4787):458–460. doi: 10.1126/science.235.4787.458. [DOI] [PubMed] [Google Scholar]
- KENDREW J. C., WATSON H. C., STRANDBERG B. E., DICKERSON R. E., PHILLIPS D. C., SHORE V. C. The amino-acid sequence x-ray methods, and its correlation with chemical data. Nature. 1961 May 20;190:666–670. doi: 10.1038/190666a0. [DOI] [PubMed] [Google Scholar]
- KUNDIG W., GHOSH S., ROSEMAN S. PHOSPHATE BOUND TO HISTIDINE IN A PROTEIN AS AN INTERMEDIATE IN A NOVEL PHOSPHO-TRANSFERASE SYSTEM. Proc Natl Acad Sci U S A. 1964 Oct;52:1067–1074. doi: 10.1073/pnas.52.4.1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabsch W., Sander C. Dictionary of protein secondary structure: pattern recognition of hydrogen-bonded and geometrical features. Biopolymers. 1983 Dec;22(12):2577–2637. doi: 10.1002/bip.360221211. [DOI] [PubMed] [Google Scholar]
- Kalbitzer H. R., Hengstenberg W., Rösch P., Muss P., Bernsmann P., Engelmann R., Dörschug M., Deutscher J. HPr proteins of different microorganisms studied by hydrogen-1 high-resolution nuclear magnetic resonance: similarities of structures and mechanisms. Biochemistry. 1982 Jun 8;21(12):2879–2885. doi: 10.1021/bi00541a012. [DOI] [PubMed] [Google Scholar]
- Kapadia G., Reizer J., Sutrina S. L., Saier M. H., Jr, Reddy P., Herzberg O. Crystallization of the Bacillus subtilis histidine-containing phosphocarrier protein HPr and of some of its site-directed mutants. J Mol Biol. 1990 Mar 5;212(1):1–2. doi: 10.1016/0022-2836(90)90296-X. [DOI] [PubMed] [Google Scholar]
- Klevit R. E., Waygood E. B. Two-dimensional 1H NMR studies of histidine-containing protein from Escherichia coli. 3. Secondary and tertiary structure as determined by NMR. Biochemistry. 1986 Nov 18;25(23):7774–7781. doi: 10.1021/bi00371a073. [DOI] [PubMed] [Google Scholar]
- Knowles J. R. Enzyme-catalyzed phosphoryl transfer reactions. Annu Rev Biochem. 1980;49:877–919. doi: 10.1146/annurev.bi.49.070180.004305. [DOI] [PubMed] [Google Scholar]
- Liao D. I., Kapadia G., Reddy P., Saier M. H., Jr, Reizer J., Herzberg O. Structure of the IIA domain of the glucose permease of Bacillus subtilis at 2.2-A resolution. Biochemistry. 1991 Oct 8;30(40):9583–9594. doi: 10.1021/bi00104a004. [DOI] [PubMed] [Google Scholar]
- Meadow N. D., Fox D. K., Roseman S. The bacterial phosphoenolpyruvate: glycose phosphotransferase system. Annu Rev Biochem. 1990;59:497–542. doi: 10.1146/annurev.bi.59.070190.002433. [DOI] [PubMed] [Google Scholar]
- Postma P. W., Lengeler J. W. Phosphoenolpyruvate:carbohydrate phosphotransferase system of bacteria. Microbiol Rev. 1985 Sep;49(3):232–269. doi: 10.1128/mr.49.3.232-269.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reizer J. Regulation of sugar uptake and efflux in gram-positive bacteria. FEMS Microbiol Rev. 1989 Jun;5(1-2):149–156. doi: 10.1016/0168-6445(89)90019-3. [DOI] [PubMed] [Google Scholar]
- Reizer J., Saier M. H., Jr, Deutscher J., Grenier F., Thompson J., Hengstenberg W. The phosphoenolpyruvate:sugar phosphotransferase system in gram-positive bacteria: properties, mechanism, and regulation. Crit Rev Microbiol. 1988;15(4):297–338. doi: 10.3109/10408418809104461. [DOI] [PubMed] [Google Scholar]
- Reizer J., Sutrina S. L., Saier M. H., Stewart G. C., Peterkofsky A., Reddy P. Mechanistic and physiological consequences of HPr(ser) phosphorylation on the activities of the phosphoenolpyruvate:sugar phosphotransferase system in gram-positive bacteria: studies with site-specific mutants of HPr. EMBO J. 1989 Jul;8(7):2111–2120. doi: 10.1002/j.1460-2075.1989.tb03620.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson J. S., Richardson D. C. Amino acid preferences for specific locations at the ends of alpha helices. Science. 1988 Jun 17;240(4859):1648–1652. doi: 10.1126/science.3381086. [DOI] [PubMed] [Google Scholar]
- Richardson J. S. The anatomy and taxonomy of protein structure. Adv Protein Chem. 1981;34:167–339. doi: 10.1016/s0065-3233(08)60520-3. [DOI] [PubMed] [Google Scholar]
- Sharma S., Georges F., Delbaere L. T., Lee J. S., Klevit R. E., Waygood E. B. Epitope mapping by mutagenesis distinguishes between the two tertiary structures of the histidine-containing protein HPr. Proc Natl Acad Sci U S A. 1991 Jun 1;88(11):4877–4881. doi: 10.1073/pnas.88.11.4877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B. C. Resolution of phase ambiguity in macromolecular crystallography. Methods Enzymol. 1985;115:90–112. doi: 10.1016/0076-6879(85)15009-3. [DOI] [PubMed] [Google Scholar]
- Wittekind M., Reizer J., Deutscher J., Saier M. H., Klevit R. E. Common structural changes accompany the functional inactivation of HPr by seryl phosphorylation or by serine to aspartate substitution. Biochemistry. 1989 Dec 26;28(26):9908–9912. doi: 10.1021/bi00452a005. [DOI] [PubMed] [Google Scholar]
- Wittekind M., Reizer J., Klevit R. E. Sequence-specific 1H NMR resonance assignments of Bacillus subtilis HPr: use of spectra obtained from mutants to resolve spectral overlap. Biochemistry. 1990 Aug 7;29(31):7191–7200. doi: 10.1021/bi00483a006. [DOI] [PubMed] [Google Scholar]
- el-Kabbani O. A., Waygood E. B., Delbaere L. T. Tertiary structure of histidine-containing protein of the phosphoenolpyruvate:sugar phosphotransferase system of Escherichia coli. J Biol Chem. 1987 Sep 25;262(27):12926–12929. [PubMed] [Google Scholar]