Abstract
MOV10 (Moloney leukemia virus 10, homolog) is an interferon-inducible RNA helicase, associated with small RNA-induced silencing. Here, we report that MOV10 exhibits antiviral activity, independent of its helicase function, against a number of positive and negative-strand RNA viruses by enhancing type I interferon (IFN) induction. Using a number of CRISPR/Cas9-mediated knockout human cells, we show that IRF3-mediated IFN induction and downstream IFN signaling through IFN receptor was necessary to inhibit virus replication by MOV10. MOV10 enhanced IRF3-mediated transcription of IFN. However, this IFN induction by MOV10 was unique and independent of the known RIG-I/MAVS-mediated RNA-sensing pathway. Upon virus infection, MOV10 specifically required IKKε not TBK1, for its antiviral activity. The important role of MOV10 in mediating antiviral signaling was further supported by the finding that viral proteases from picornavirus family specifically targeted MOV10 as a possible innate immune evasion mechanism. These results establish MOV10, an evolutionary conserved protein involved in RNA silencing, as an antiviral gene against RNA viruses that uses a RLR-independent pathway to enhance IFN response.
Keywords: MOV10, interferon, ISG, IRF3, VSV, SeV
INTRODUCTION
Cellular innate immunity is the first line of defense mounted by the host upon pathogen invasion. For virus infection this immunity is primarily mediated by type I interferons (IFN) and IFN-stimulated genes (ISG), and provides immediate protection as well as shapes the subsequent adaptive immune response. Upon virus infection, viral nucleic acids are sensed by various innate immune receptors, such as Retinoic acid-inducible gene I (RIG-I)-like receptors (RLR) and Toll-like receptors (TLR) to initiate IFN and ISG induction (1). The cytosolic DExD/H-box family helicases RIG-I and MDA5 (melanoma differentiation-associated gene 5) are crucial components for sensing cytoplasmic viral RNA resulting from RNA virus infection (2). Upon binding to viral RNA, RIG-I and MDA5, through their common adaptor protein MAVS (mitochondrial antiviral-signaling protein) trigger a signaling cascade. This signaling cascade leads to the activation of transcription factors such as IRF3 (IFN regulatory factor 3) and NF-κB resulting in transcriptional induction of IFN (1, 3, 4). Although RLR signaling is primarily responsible for viral RNA sensing and IFN induction, there is still residual IFN induction upon specific RNA virus infection in the absence of the critical adaptor MAVS (5), indicating the existence of additional viral RNA sensor signaling.
DExD/H-box family helicases are involved in various cellular processes, such as nucleic acid metabolism, RNA interference (RNAi) and innate immunity. Besides RIG-I and MDA5, a number of helicases such as DDX3 and DHX9 have been implicated in the regulation of RLR signaling (6, 7). On the other hand, DDX41, DDX60, DHX9 and DHX36 have been proposed to be involved in cytoplasmic DNA and DNA virus sensing (8). Some of these helicases have been shown to be targeted by various viral proteins (7), which supports their role in antiviral innate immunity. Due to their similarities with RIG-I, various DExD/H-box helicases have been examined for their importance in viral RNA sensing. However, none of the helicases with antiviral activity seemed to function independent of RIG-I-MAVS signaling. Further, for most of these helicases, mechanistic details regarding their involvement in antiviral innate immunity and knockout studies have yet to be described.
MOV10 (Moloney leukemia virus 10, homolog (mouse)), a putative member of SF1 family helicase (9), was first identified as a novel Argonaute-associated protein and was shown to be involved in RNA interference (RNAi)(10, 11). MOV10 orthologs in Drosophila, Armitage and in plants, SDE3 are also involved in RNAi (12, 13). MOV10 is known to bind a broad variety of RNA (14); and through its helicase activity participates in mRNA degradation and translation inhibition (15). In addition, MOV10 has been shown to inhibit HIV replication through several different mechanisms (16–19). It has also been shown to inhibit retroviral transpositions via its association with L1 ribonucleoprotein particle in processing (P-) bodies and stress granules (SGs)(20, 21). However, given that MOV10 is induced by IFN, its role in regulating the replication of other RNA viruses besides HIV has been less clear and in some cases conflicting. MOV10 provides antiviral activity against hepatitis C virus (HCV) in an ISG expression library screen (22). In contrast, MOV10 interacts with hepatitis delta virus antigen and enhances viral RNA replication (23).
Herein we describe antiviral activity of MOV10 against RNA viruses, which is mediated through enhanced IFN induction. The physiological significance of the MOV10 antiviral activity is demonstrated by our finding that MOV10 is specifically targeted by viruses to evade the innate immune response. Mechanistically, this antiviral activity of MOV10 is independent of the known intracellular RNA-sensor signaling through RIG-I and MAVS, and required IKKε Taken together, our findings provide a mechanistic basis for the antiviral properties of MOV10.
MATERIALS AND METHODS
Cells, reagents and viruses
Human embryonic kidney (HEK) 293, 293T, HT1080 cells and primary human foreskin fibroblasts (HFF) were cultured in DMEM (Lonza, Rockland, ME) supplemented with 10% FBS (Atlanta Biologicals, Lawrenceville, GA) and Penicillin/Streptomycin. Plasmids pcDNA3-FLAG-MOV10 and G681A/D682A (MOV10-GD) were generously provided by Dr. Vinay K. Pathak (Center for Cancer Research, National Cancer Institute). Plasmids pcDNA3-FLAG-MOV10-Q129A, Q869A, G529A/K530A/T531A (MOV10-GKT) and D645A/E646A (MOV10-DE) were created by site-directed mutagenesis (Genewiz, South Plainfield, NJ). pcDNA3-FLAG-MOV10-Q129A/Q869A (MOV10-DM) was generated using Q129A and Q869A plasmids as backbone. V5-MOV10 plasmid was generously provided by Dr. Yong-Hui Zheng (Michigan State University)(17). FLAG-MAVS, Myc CVB-3Cpro and 3Cpro-C147A have been described previously (24). EMCV 3Cpro plasmid was generously provided by Dr. Takashi Fujita (25). siRNA against MOV10 (23), IKKε and RIG-I were synthesized by Sigma-Aldrich (St. Louis, MO) and have been described previously (26). Control siRNA SMARTpool were obtained from Thermo Scientific (Waltham, MA). Lipofectamine RNAiMAX was obtained from Invitrogen (Grand Island, NY) Low molecular weight p(I):p(C) was from InvivoGen (San Diego, CA). Transfection reagent XtremeGENE 9, protease inhibitors and GFP antibody were purchased from Roche (Indianapolis, IN). FLAG antibody and anti-FLAG beads were from Sigma-Aldrich (St Louis, MO). MOV10 antibody was purchased from Abcam. RIG-I and IKKε antibodies were from Cell Signaling Technology (Danvers, MA). ISG56, ISG60 and actin antibodies; GFP-tagged VSV and Sendai virus C protein antibodies have been described before (27). Sendai virus Cantell strain (SeV) was purchased from Charles River Laboratories (Wilmington, MA). EMCV was from ATCC (Manassas, VA). RSV strain A2 was described before (28). IFNβ in cellular supernatant was detected by Verikine-HS Human Interferon Beta ELISA Kit (PBL Assay Science, (Piscataway, NJ) according to manufacturer’s protocol.
Establishment of different stable and genome-edited cells lines
HEK293, 293T or HT1080 cells were co-transfected with pcDNA3-FLAG-MOV10 or pcDNA3 vector control and selected with G418 (800 μg/ml) and collected as pool stable cells. The MOV10 stable expression was confirmed by immunoblotting using anti-FLAG antibody.
293T cells deficient in RIG-I (293T-RIG-I-KO) were previously described (27). All other knockout cell lines were generated as follows: HEK293T cells were plated at a density of 2×104 cells per well in a 96-well plate. The next day, CRISPR or TALEN plasmids were transfected using GeneJuice transfection reagent (Merk Millipore) according to the manufacturer’s protocol. pRZ-mCherry-Cas9 and pLenti-gRNA constructs were transfected at a ratio of 3:1 (i.e. 150 ng: 50 ng), whereas TALEN pair plasmids were transfected at a ratio of 1:1 (100 ng each). Critical exons of the following genes were targeted using the following gRNA constructs (PAM region in bold letters): IRF3, 5′-GGGGGTCCCGGATCTGGGAGTGG-3′; IFNAR1, 5′-ACAGGAGCGATGA GTCTGTCGGG-3′; MOV10, 5′-GTTCTTCAGACTCGACCGCTGGG-3′; IKBKE, 5′-GCAC AATGCCGTTCTCCCGCAGG-3′. TBK1 deficient cells were created using a TALEN pair targeting the following region 5′-TTCTAATCATCTGTGGCTTTTATCTGATATTTT AGGCCAAGGAGCTACTGCAA-3′ (29). Subsequently, limiting dilution cloning was performed and after 10 days, growing monoclones were selected by bright field microscopy. Thus identified clones were trypsinized and expanded in two separate wells. One well was used to recover gDNA as previously described (30) and subsequently the target region of interest was amplified in a two-step PCR and subjected to deep sequencing. Knockout cell clones were identified as cell clones harboring all-allelic frame shift mutations using OutKnocker (31). Genotypes of the respective knockout cell lines are available upon request.
Quantitative RT–PCR analysis
Total RNA was isolated from transfected and/or stimulated cells by Trizol (Life Technologies) and treated with DNase I at 37°C for 1hr (DNA Free kit, Ambion). cDNA was synthesized using iScript cDNA Synthesis Kit (Bio-Rad, Hercules, CA). One part (1/20th) of the cDNA synthesized from 1 μg RNA was subjected to real-time PCR using EvaGreen Supermix in a CFX96 Real Time System (Bio-Rad) according to the manufacturer’s instructions. All PCR amplification was normalized to ribosomal protein L32 (RPL32) and expressed as fold change with respect to untreated vector control cells, indicated with (◆).
Virus infection and viral growth curve analyses
Cells were seed in 24-well plates and subsequently infected with EMCV or VSV-expressing GFP at different m.o.i. depending on the assay. SeV infection was carried out at different doses ranging from 0.5 to 240 HAU/ml. After infection, the cells were lysed and subjected to immunoblotting using antibody against SeV C protein or GFP protein. For one-step growth curve analysis MOV10 and control cells were infected with different viruses including VSV or EMCV; cell-free medium were collected after infection, and virus titers determined by plaque assay using BHK21, and Vero cells, in 24-well plates. Each virus infection was performed in triplicate. The VSV, EMCV and SeV replication were measured as viral RNA expression using qRT-PCR with the following specific primers: VSV, forward, 5′-GAGGAGTCACCTGGACAATCACT-3′, reverse, 5′-TGCAA GGAAAGCATTGAACAA-3′; EMCV, forward, 5′-TGCAGTGGTTGCTCCCCTGA-3′, reverse, 5′-TGACCGGAATGGGCGACTGT-3′; SeV, forward, 5′-GCTGCCGACAA GGTGAGAGC-3′, reverse, 5′-GCCCGCCATGCCTCTCTCTA-3; RPL32, forward, 5′-GCCAGATCTT GATGCCCAAC-3′, reverse, 5′-CGTGCACATGAGCTGCCTAC-3′; RSV forward, 5′-GCTCTTAGCAAAGTCAAGTTGAATGA-3′, reverse, 5′ TGCTCCGTTGGA TGGTGTATT-3′;. IFIT1, IFT3, ISG15, Cig5 and IFNβ primers were described previously (27). All primers were custom synthesized by Integrated DNA Technologies (Coralville, IA).
Fluorescence microscopy
HEK293-MOV10 or vector cells were plated in 8-well chamber slides (4×104 cells/well). Cells were infected with VSV for up to 16h and fixed with paraformaldehyde (4% v/v), following permeabilization with Triton-X100 0.1%. Nuclei were stained with DAPI Vectashield (Vector Laboratories, Inc. Burlingame, CA). Human primary foreskin fibroblasts were plated in 8-well chamber slides (1×104 cells/well) and transfected with siRNA using Lipofectamine RNAiMAX with either siRNA 10 nM or control siRNA for 72 h. Subsequently, cells were infected with EMCV for 16 h and fixed with paraformaldehyde (4% v/v), following permeabilization with Triton-X100 1%. Infected cells were stained using anti-dsRNA sera (J2) previously described (27) for 16 h at 4 °C. Immunofluorescence detection was carried out with conjugated anti-rabbit Alexa Fluor 488 and conjugated anti-mouse Alexa Fluor 594 (Invitrogen). Representative micrographs were obtained followed by quantitation of the % of infected cells by counting over 500 DAPI positive cells for each condition.
Co-immunoprecipitation and Immunoblotting
Cells were plated in 12-well plates (3×105 cells/well) transfected and/or treated with SeV were lysed in lysis buffer (Triton-X 100 1%, HEPES [pH 7.4] 20mM, NaCl 150mM, MgCl2 1.5mM, EGTA 2 mM, DTT 2 mM, NaF 10 mM, β-Glycerophosphate 12.5 mM, Na3VO4 1 mM, PMSF 1 mM, and Protease Inhibitor). The cleared cell lysates were incubated at 4 °C with antibody and protein A/G agarose beads or anti-FLAG beads overnight, washed five times with lysis buffer, and boiled in 2 × SDS–PAGE loading buffer for elution. Cell lysates boiled in 1 × SDS–PAGE loading buffer and immunoprecipitated samples were subjected to SDS-PAGE electrophoresis and immunoblotting.
Dimerization Assays
Cells were seeded in 12-well plates, and 6 to 24 h post stimulation samples were harvested in 50 μl of native lysis buffer (Tris-HCl 50 mM [pH 7.5], NaCl 75 mM, EDTA 1 mM, NP-40 1%). Native gels (8% without SDS) were pre-run with Tris 25 mM, Glycine 192 mM and Deoxicholate 1% in the cathode chamber for 1 h at 100 V and 4 °C. Prior to loading, equal volume of 2X sample buffer (Tris-HCl 125 mM [pH 6.8], Glycerol 20%, BPB dye 0.1 mg/ml) was added to the samples. Subsequently, samples containing equal amount of total proteins (20 μg) were eletrophoresed for 180 min (100 V at 4 °C) and immunoblotted for IRF3.
Reporter assays
HEK293-MOV10, 293T, 293T-MAVS-KO or 293T-RIG-I-KO (1.5 × 105 cells/well) were seeded in 24-well plates and transfected using X-tremeGENE 9 as indicated. Genome-edited 293T were transfected with MOV10 or vector control plasmids (1 μg each) together with ISRE-luciferase (0.4 μg) and β-actin Renilla luciferase reporter (0.012 μg). Twenty four hours later, the cells from each well were collected by trypsin-EDTA digestion and seeded into 6 wells in 96-well plate. Forty-eight hours post-transfection, the cells were stimulated with SeV (10 HAU/ml for 16 h) or LMW transfection, (1 μg/ml for 16 h), and luciferase activities were measured using the Dual-Glo luciferase assay system from Promega (Madison, WI). The results were expressed as fold induction of firefly luciferase relative to that of non-stimulated control-transfected cells after normalizing to Renilla luciferase, indicated with (◆).
Sub-cellular fractionations
HEK293-MOV10 or vector cells stimulated with different concentrations of SeV (10 to 240 HAU/ml) and 16 h post infection were washed and cell pellets were suspended in hypotonic buffer (HEPES 20 mM [pH8.0], KCl 10 mM, MgCl2 1 mM, Glycerol 20%, Triton-X 100 0.1%) with protease inhibitors (Roche). The cell suspensions (100 μl) were vortexed for 30 s, incubated on ice for 15 min, and centrifuged (16,000g for 10 at 4 °C). The supernatants were collected as soluble cytoplasmic fractions. The remaining nuclear pellets were thoroughly washed in 10 volumes of hypotonic buffer and then resuspended in 100 μl buffer (Tris-HCl 50 mM [pH7.4], NaCl 150 mM, NP-40 1%, Sodium Deoxycholate 0.25%, EDTA 1 mM, PMSF 1 mM, Protease inhibitor) and incubated in ice for 30 min prior to SDS-PAGE analysis.
Statistical analysis
Data were analyzed using two-tailed paired Student’s t-test. Values were considered significant at p < 0.05, indicated with (*).
RESULTS
Antiviral activity of MOV10 against RNA viruses
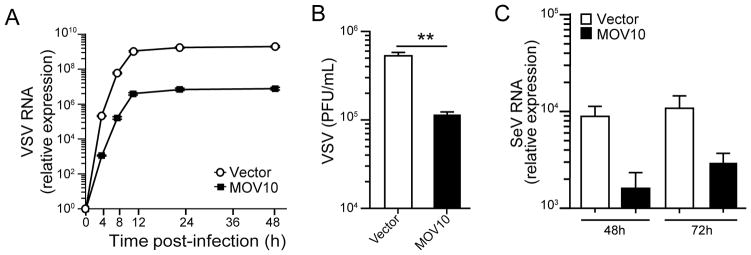
We examined the antiviral activity of MOV10 against RNA viruses from two different families. HEK293 cells stably expressing MOV10 showed substantial suppression of the replication kinetics of vesicular stomatitis virus (VSV), a negative strand RNA virus, compared to vector control cells as measured through viral RNA quantitation (Fig. 1A) and plaque assay (Fig. 1B). MOV10 expression also inhibited the replication of another negative strand RNA virus, Sendai virus (SeV) (Fig. 1C). In addition to negative strand RNA viruses, MOV10 expressing cells markedly restricted replication of positive strand RNA viruses from the picornavirus family such as encephalomyocarditis virus (EMCV, Fig. S1A, S1B) and coxsackievirus B (CVB, Fig. S1C). Taken together these results suggest that MOV10 has antiviral activity against certain RNA viruses.
Fig. 1. Antiviral activity of MOV10 against RNA viruses.
(A–B) MOV10 expression inhibits VSV replication. HEK293-MOV10 and HEK293-vector cells were infected with VSV (0.1 m.o.i.) for indicated times followed by VSV-specific RNA detection by qRT-PCR (A). Cells were infected with VSV (0.1 m.o.i.) for 24 h followed by virus titer determination using plaque assay on BHK21 cells (B). (C) Inhibition of SeV replication in MOV10 expressing cells. HEK293-MOV10 and HEK293-vector stable cells were infected with SeV at 50 HAU/ml for 48 and 72 h followed by detection of SeV RNA by qRT-PCR.
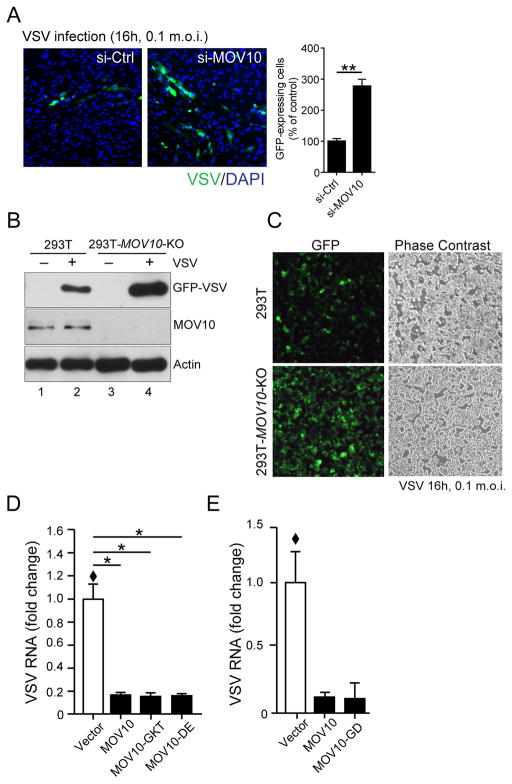
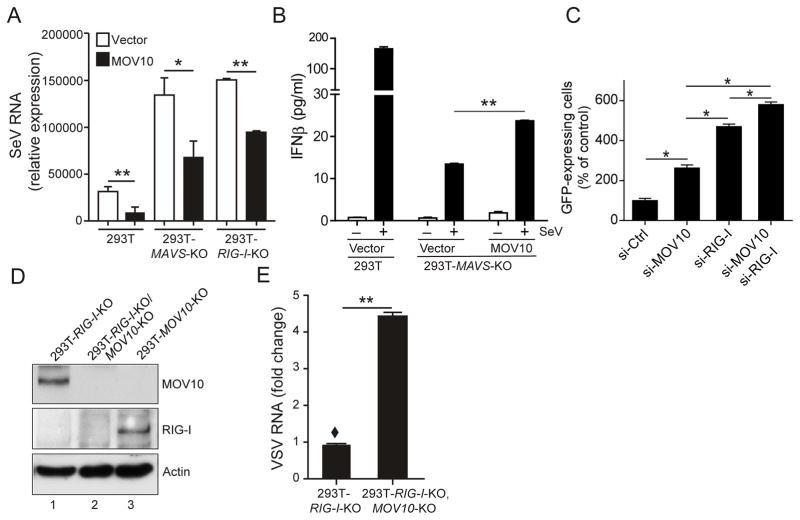
Next, we validated the antiviral activity of MOV10 by silencing its expression in primary human fibroblasts (HFFs), and subsequently infecting these cells with RNA viruses. Transfection of MOV10-specific siRNA enhanced VSV (Fig. 2A) and EMCV (Fig. S1E) replication at least twofold compared to control siRNA transfected cells. The extent of MOV10 silencing in HFFs by siRNA transfection is shown in Fig. S1D. To further prove the antiviral function of human MOV10, we generated MOV10-deficient 293T cells (293T-MOV10-KO) through CRISPR-mediated genome editing. Compared to their wild-type (Wt) counterparts, MOV10-KO cells showed significantly higher levels of virus replication as measured by GFP expression (VSV, Fig. 2B, lane 2 and 4 and Fig. 2C). However, the extent of virus replication enhancement in MOV10-KO cells was not as remarkable for EMCV (at best five-fold, Fig. S1F) as the replication inhibition caused by the ectopic MOV10 expression (well over ten-fold, Fig. S1A). This indicated a possible amplification of the antiviral activity exerted by MOV10 expression, possibly through IFN signaling (see later).
Fig. 2. MOV10 depletion enhances virus replication.
(A) MOV10 silencing enhances VSV replication in primary human fibroblasts (HFF). Primary human foreskin fibroblasts were transfected either with control (si-Ctrl) or MOV10-specific (si-MOV10) siRNA for 72 h. Cells were subsequently infected with VSV as indicated, and analyzed by GFP fluorescence to estimate virus infection. Representative micrographs are shown followed by quantitation of the % of infected cells by approximately counting over 500 DAPI positive cells for each condition.
(B–C) Genomic loss of MOV10 enhances VSV replication. MOV10-deficient 293T cells (293T-MOV10-KO) along with control 293T were infected with VSV at 0.1 m.o.i. and analyzed by immunoblotting (B) and by GFP fluorescence (C).
(D) Inhibition of VSV replication by MOV10 helicase domain mutants. MOV10 Wt and several helicase domain mutants were transiently expressed in 293T-MOV10-KO cells and infected with VSV. Antiviral activities were measured through detection of VSV RNA by qRT-PCR 16 h post-infection.
(E) Inhibition of VSV replication by MOV10 is independent of its anti-HIV activity. MOV10-mutant defective in inhibiting HIV replication (MOV10-GD) was expressed in 293T-MOV10-KO cells and its antiviral activities against VSV were measured as in (D). Plots show mean with standard error bars, where * and ** are P < 0.05 and P < 0.01, respectively, by two-tailed Student’s t test analysis. Sample (◆) was set as 1 for comparison.
MOV10 has in vitro directional helicase activity, which has been shown to be necessary for its ability to recruit nonsense-mediated mRNA decay factor UPF1 and promote cellular mRNA degradation (15). However, its RNA-binding property is independent of this helicase activity. To distinguish between different functional properties that were necessary for the MOV10 antiviral activity, we transiently expressed Wt and various helicase domain mutants of MOV10 in MOV10-KO cells and measured their antiviral activities against EMCV and VSV. Two mutants targeting the conserved residues in helicase Motif I, G529A/K530A/T531A (MOV10-GKT), and Motif II, D645A/E646A (MOV10-DE) showed similar antiviral activity compared to the Wt (Fig. 2D). These mutations are known to inactivate the catalytic activity of MOV10 without affecting its RNA binding (15). Therefore, the helicase activity of MOV10 was not necessary for the antiviral activity against VSV. It also indicates that the ability of MOV10 to promote UPF1-mediated mRNA decay is not important to inhibit the replication of these RNA viruses. Another mutant of MOV10, known to be defective in inhibiting HIV (G681A/D682A, MOV10-GD) (16) also showed similar antiviral activity to Wt (Fig. 2E) indicating that the antiviral activity of MOV10 against VSV is independent of its anti-HIV activity. Taken together these results indicate that similar to a number of ISGs, MOV10 provides antiviral activity against a number of RNA viruses and that this antiviral activity is independent of the helicase, and anti-HIV activity of MOV10.
Antiviral activity of MOV10 is mediated through IRF3 signaling
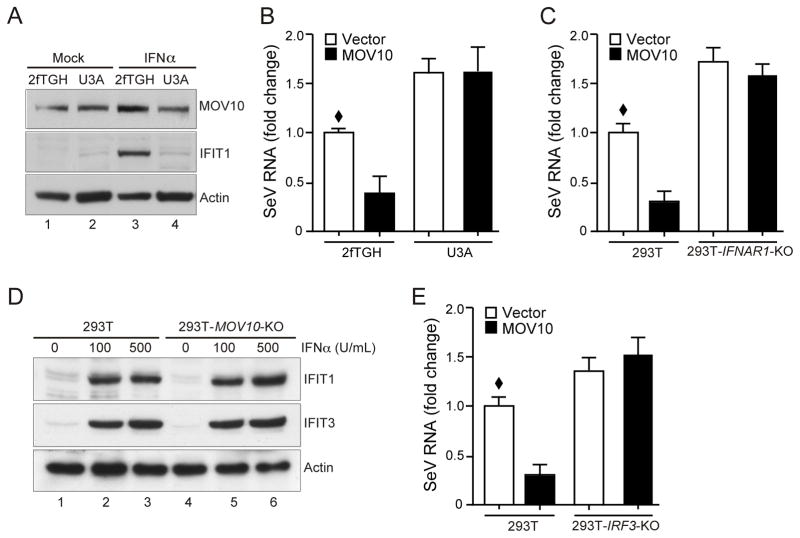
As stated above, the strong antiviral activity observed in MOV10 expressing cells (Fig. 1A) prompted us to investigate the role of IFN signaling, which may work in a feed-forward manner to amplify the antiviral activity. We stably expressed MOV10 in human HT1080 derived U3A cells, which were defective in STAT1 expression and IFN signaling (36). As expected, U3A cells showed loss of IFIT1 (also known as ISG56) induction after IFN treatment compared to control parental 2fTGH cells (Fig. 3A, lanes 3 and 4). MOV10 expression did not affect IFIT1 induction indicating that MOV10 did not influence IFN signaling downstream of STAT1. However, MOV10 expression in U3A cells failed to provide protection against SeV infection (Fig. 3B). To further confirm these results using a different system, we created IFN receptor (IFNAR1)-deficient 293T cells (293T- IFNAR1-KO) that were defective in IFN signaling (Fig. S2A), using CRISPR-mediated genome editing. Transient expression of MOV10 in 293T-IFNAR1-KO cells showed no reduction in subsequent EMCV replication, while the control 293T cells showed reduced virus replication (Fig. 3C). These results, in two different systems, indicated that an intact JAK-STAT-mediated IFN signaling was necessary for the inhibition of virus replication by MOV10.
Fig. 3. Antiviral activity of MOV10 is mediated through IRF3 and requires downstream IFN signaling.
(A) Characterization of 2fTGH and STAT1-deficient U3A cells stably expressing MOV10. Cells were treated with IFN (500 U/ml for 16 h) to induce expression of endogenous IFIT1 and analyzed by immunoblotting with indicated antibodies.
(B) MOV10 expression does not inhibit SeV replication in STAT1-deficient U3A cells. Indicated cells were infected with SeV (50 HAU/ml) for 48 h followed by detection of SeV RNA by qRT-PCR.
(C) IFN signaling is required for antiviral activity of MOV10. IFNAR1-deficient (293T-IFNAR1-KO) along with control 293T cells were transfected with MOV10 followed by SeV infection ((50 HAU/ml for 48 h) and SeV-specific RNA analysis by qRT-PCR.
(D) Loss of MOV10 does not affect IFN signaling. 293T and 293T-MOV10-KO cells were treated with IFNα as indicated for 16 h followed by immunoblotting analysis of endogenous IFIT1 and IFIT3 induction.
(E) IRF3 is needed for MOV10 antiviral activity. IRF3-deficient (293T-IRF3-KO) along with control 293T cells were infected with SeV and analyzed by qRT-PCR similar to (C). Plots show mean with standard error bars, where was set as 1 for comparison.
Having established the necessity of an intact IFN signaling for the MOV10 antiviral activity, we examined whether the absence of MOV10 affected JAK-STAT-mediated IFN signaling by assaying the induction of a number of ISG following IFN treatment in 293T-MOV10-KO cells. However, 293T-MOV10-KO cells did not exhibit any defect in IFN-stimulated gene induction (Fig. 3D and S2B) indicating that MOV10 did not directly affect JAK-STAT-mediated IFN signaling. Thus, we focused on the upstream events of this cascade, IRF3-mediated IFN induction, and examined the role of MOV10 on IRF3 signaling using 293T-IRF3-KO cells (Fig. S2C). As shown in Fig. 3E, transient expression of MOV10 showed the expected inhibition of SeV replication in Wt 293T cells. However, this inhibition was absent in MOV10 expressing 293T-IRF3-KO cells, which suggested that the IRF3-mediated IFN induction was necessary for the inhibition of virus replication by MOV10. Altogether these results indicated that the antiviral activity of MOV10 is mediated through IRF3 signaling by possibly affecting IFN induction.
MOV10 enhances IRF3-mediated IFN induction
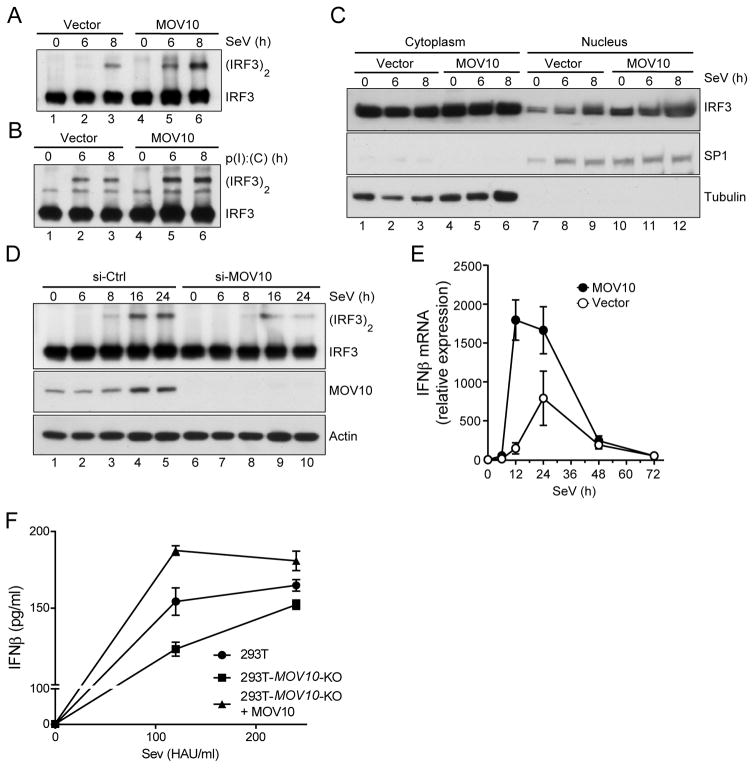
As the antiviral activity of MOV10 was dependent on IRF3, we next examined the effect of MOV10 on IRF3 activation. We used SeV infection and low molecular weight p(I):p(C) transfection, both of which causes RIG-I-dependent IRF3 activation (4). In response to both stimuli MOV10 enhanced IRF3 dimerization kinetics in a dose-dependent manner (Fig. 4A, 4B, lanes 5 and 6 compared to lanes 2 and 3, respectively). The nuclear translocation of IRF3 following its dimerization was also increased in MOV10 expressing cells (Fig. 4C, lanes 11 and 12 compared to lanes 8 and 9). Conversely, silencing of MOV10 with siRNA reduced SeV-mediated IRF3 dimerization (Fig. 4D, lanes 8–10 compared to lanes 3–5). This enhanced IRF3 activation in presence of MOV10 also resulted in enhanced IFNβ induction. MOV10-expressing cells showed significant increase in the kinetics and the amplitudes of endogenous IFNβ mRNA induction (Fig. 4E). Further, SeV-mediated IFNβ protein induction was reduced in 293T-MOV10-KO cells, while that was enhanced upon ectopic expression of MOV10 in these cells (Fig. 4F). These results suggested that MOV10 antiviral activity could be potentially mediated through IRF3 activation, and IRF3-mediated IFN induction.
Fig. 4. MOV10 enhances IRF3 activation and IRF3-mediated gene induction.
(A–B) MOV10 enhances RIG-I-mediated IRF3 dimerization. HEK293-MOV10 and HEK293-vector stable cells were either infected with 240 HAU/ml SeV (A), or transfected with 1 μg/ml low molecular weight p(I):p(C) (B) and samples collected at 6 and 8 h post treatment. Cell lysates were analyzed in non-denaturing PAGE followed by immunoblotting with IRF3 antibody.
(C) Increased nuclear translocation of IRF3 in MOV10 expressing cells. HEK293-MOV10 and HEK293-vector stable cells were infected with 240 HAU/ml SeV and samples were collected 6 and 8 h post-infection. Cytoplasmic and nuclear fractions were analyzed by immunobloting as indicated. Tubulin and the nuclear transcription factor SP1 were used as controls for cytoplasmic and nuclear fractions, respectively.
(D) MOV10 silencing reduced SeV-mediated IRF3 dimerization. MOV10 expression was silenced in HEK293 cells by siRNA transfection for 72 h and subsequently infected with SeV (120 HAU/ml) for indicated times and analyzed for IRF3 dimerization as in (A) (top panel). MOV10 silencing was monitored by immunoblotting (bottom panels).
(E) Induction of endogenous IFNβ mRNA is enhanced in MOV10 expressing cells. HEK293-MOV10 and HEK293-vector stable cells were stimulated with SeV (120 HAU/ml) and total RNA were collected at indicated times post-infection. IFNβ mRNAs were detected by qRT-PCR using specific primers.
(F) Modulation of SeV-mediated IFNβ induction by MOV10. Indicated cells were infected with either 120 or 240 HAU/ml SeV for 12 h, followed by IFNβ protein measurement in the culture supernatant.
MOV10 activates innate immune signaling independent of RIG-I-MAVS
RLR are the primary cytosolic receptors that sense viral RNA and initiate the signaling cascade leading to IRF3 activation. Our observation that MOV10 provided antiviral activity against both EMCV and SeV, which are sensed by MDA5 and RIG-I respectively, indicated that it either worked in conjunction with, or independent of RLR-mediated sensing. Thus in the next series of experiments we investigated the role of RIG-I and its adaptor MAVS in mediating MOV10 antiviral activity. First, we determined the effect of MOV10 expression on SeV replication in 293T derived RIG-I-KO (also known as DDX58) cells created by genome targeting (29). As expected, loss of either RIG-I or MAVS markedly increased SeV replication in control vector expressing cells (Fig. 5A). Surprisingly, transient expression of MOV10 inhibited SeV replication in control 293T, as well as in both RIG-I-KO and MAVS-KO cells (Fig. 5A). Further, the presence of MOV10 showed enhancement of IRF3 dimerization in MAVS-KO cells (Fig. S3A, lanes 7–8 compared to lanes 5–6) indicating that MOV10 enables IRF3 activation even in the absence of MAVS. These results suggested that the antiviral activity of MOV10 is independent of RIG-I and MAVS. Indeed, MOV10 expression in MAVS-KO cells showed significant enhancement in IFNβ protein induction (Fig. 5B). However, this finding that antiviral activity of MOV10 is independent of RIG-I-MAVS and may operate in parallel would predict that the antiviral activities of these two pathways should have additive antiviral effects. We examined this prediction in primary human fibroblasts, where VSV replications were further enhanced when MOV10 and RIG-I were silenced together compared to their individual silencing (Fig. 5C and S3B). Finally, we created double deficient cells by genetically targeting MOV10 and RIG-I simultaneously (Fig. 5D). Compared to RIG-I-KO cells, RIG-I-KO/MOV10-KO cells were substantially more permissive to VSV replication (Fig. 5E). Altogether, these results indicated that the MOV10 antiviral activity operated independent, and in parallel to RIG-I and MAVS to provide additional host defense against RNA viruses through IFN induction.
Fig. 5. MOV10 activates innate immune signaling independent of RIG-I-MAVS.
(A) Inhibition of SeV replication by MOV10 in both MAVS-KO and RIG-I-KO 293T cells. MAVS and RIG-I-deficient 293T along with control 293T cells were infected with SeV (50 HAU/ml) for 48 h followed by SeV-specific RNA detection by qRT-PCR.
(B) Enhanced IFNβ induction by MOV10 in 293T-MAVS-KO cells. Cells were transfected either with vector or MOV10 as indicated, followed by SeV (240 HAU/ml) infection for 24 h. IFNβ protein levels in the culture supernatants were assayed as before.
(C) Silencing of MOV10 and RIG-I shows additive enhancement of VSV replication in fibroblasts. Human primary foreskin fibroblasts were transfected with indicated siRNA for 72 h and subsequently infected with VSV (0.1 m.o.i) for 16 h (F). Viral replication was quantitated by GFP-fluorescence as before (Fig. 2A). Representative micrographs are shown in Fig. S3B.
(D–E) Loss of MOV10 further enhances VSV replication in RIG-I-deficient cells. Lysates from 293T-RIG-I-KO, 293T-MOV10-KO/RIG-I-KO and 293T-MOV10-KO cells were analyzed by immunoblotting with indicated antibodies (D). Indicated cells were infected with VSV (0.1 m.o.i.) and VSV RNA analyzed by qRT-PCR 16 h post-infection (G). Plots show mean with standard error bars, where * and ** are P < 0.05 and P < 0.01 respectively by two-tailed Student’s t test analysis, and was set as 1 for comparison.
IKKε is involved in MOV10-mediated activation of IRF3
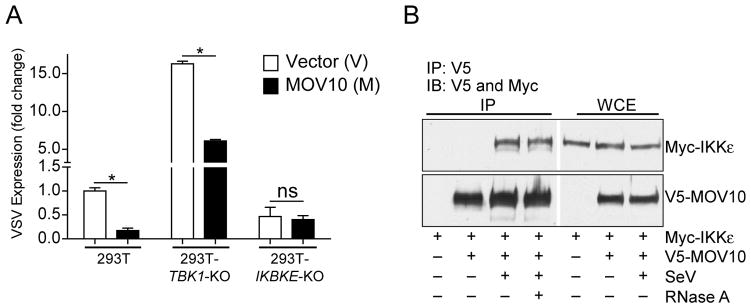
Following engagement of various TLR and RLR signaling, IRF3 is activated by Inhibitor of κB kinase (IKK) family kinases TANK-binding kinase 1 (TBK1) and IKKε through Ser/Thr phosphorylation. As IRF3 was essential for the MOV10 antiviral activity, we next investigated the roles of these two kinases in MOV10-mediated activation of IRF3 and antiviral activity. Once again, we employed genome editing to target IKKε (IKBKE) and TBK1 in 293T cells. Comparison of the VSV replication in the vector transfected samples from these three cell lines showed substantial increase in VSV replication only in TBK1-KO, but not in IKKε–KO cells (Fig. 6A) indicating the critical role of TBK1 in the antiviral activity. As expected, MOV10 expression inhibited VSV (Fig. 6A) replication in control 293T cells. Significant antiviral activities were also seen with MOV10 expression in TBK1-KO cells (Fig. 6A). However, MOV10 expression failed to protect the IKBKE-KO cells against VSV (Fig. 6A). This indicated that MOV10-mediated antiviral activity is most likely mediated through IKKε and not through TBK1. Involvement of IKKε was further established by examining the physical interaction of MOV10 and IKKε. In co-immunoprecipitation assays IKKε co-precipitated with MOV10 in a SeV infection-dependent, but RNA-independent manner (Fig. 6B). Additionally, we were also able to co-precipitate exogenously expressed (Fig. S3C) as well as endogenous IKKε (Fig. S3D) with endogenous MOV10 following SeV infection. Taken together these results suggest involvement of IKKε as the downstream kinase for MOV10-induced IRF3 activation and antiviral activity.
Fig. 6. IKKε is required for MOV10 antiviral activity.
(A) MOV10-mediated protection from VSV infection is lost in IKKε-deficient cells. MOV10 was expressed in control 293T, 293T-TBK1-KO and 293T-IKBKE-KO cells, and subsequently infected with VSV (0.1 m.o.i.) for 16 h followed by virus-specific RNA detection by qRT-PCR. * P < 0.05 and ns = not significant by two-tailed Student t test analysis.
(B) MOV10 co-precipitates with IKKε upon SeV infection. 293T cells were transfected with MOV10-V5 and Myc-IKKε plasmids as indicated followed by infection with SeV (240 HAU/ml). Cell lysates from infected and un-infected cells were immunoprecipitated with FLAG antibody and immunoblotted with FLAG and Myc antibodies. SeV infected sample was split in two parts and one part was subjected to RNase A treatment before immunoprecipitation. Whole cell extracts (WCE) were from each sample were also analyzed for expression.
MOV10 is targeted for degradation by picornavirus proteases
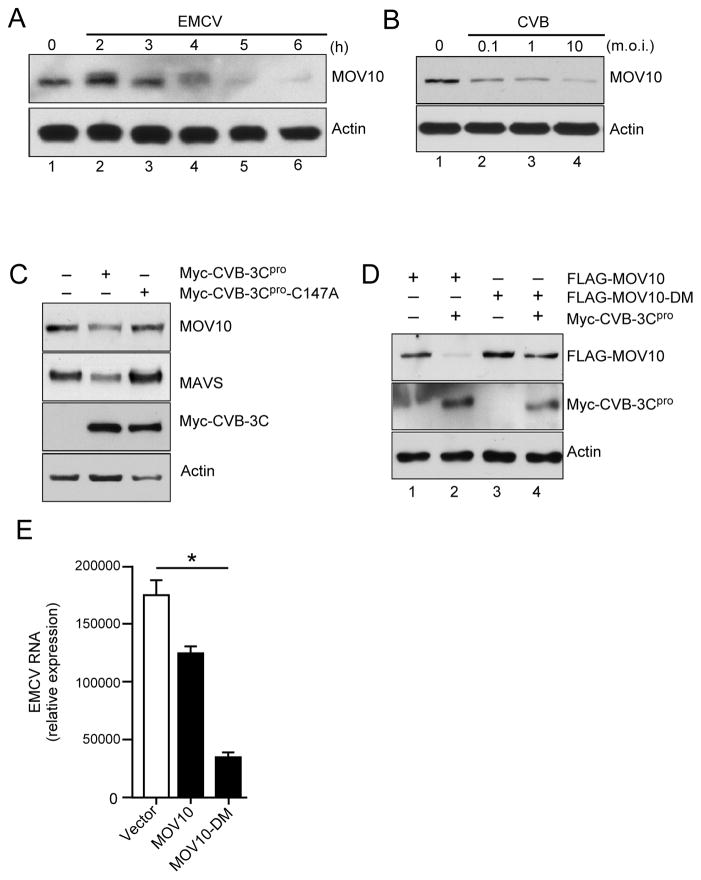
Targeting of the host proteins involved in innate immune response is an evolutionarily conserved mechanism among many RNA viruses, which helps evade the host response (32, 33). Examination of this feature has allowed to identify physiological importance of a number of host proteins involved in the host protection (34). Picornaviruses accomplish this by utilizing the virally-encoded proteases to cleave crucial components of the RLR and IFN signaling pathways (35). To find out whether these viruses also target MOV10, we examined the amounts of endogenous MOV10 protein during two different types of picornavirus infection, EMCV and CVB. Endogenous MOV10 protein gradually decreased during EMCV infection (Fig. 7A). Similar reductions in protein amounts were observed with CVB infection in a viral dose dependent manner (Fig. 7B). To further investigate this viral antagonism of MOV10 we examined MOV10 degradation by viral proteases. Because CVB 3Cpro is known to target and degrade a variety of innate immune signaling proteins (24), we tested whether CVB 3Cpro expression also affected the steady state levels of MOV10. Transfection of CVB 3Cpro in 293T cells induced a reduction in the total levels of both endogenous (Fig. 7C) and ectopically expressed (Fig. S4A) MOV10 and MAVS – a known target of 3Cpro degradation (24). In contrast, expression of a catalytically inactive 3Cpro mutant (C147A) did not induce degradation (Fig. 7C). Expression of the 3Cpro from EMCV also showed a similar targeting of MOV10 signifying its importance in promoting innate immunity against these viruses (Fig. S4B). Analysis of MOV10 primary sequence identified two glutamine residues (Q129 and Q169) that could serve as possible cleavage sites for the picornavirus 3C proteases (37, 38). To identify the 3Cpro target site(s) in MOV10, we created point mutants of MOV10 using site-directed mutagenesis. As shown in Fig. S4C, both single-mutants of MOV10 showed partial protection from 3Cpro-mediated loss of expression. However, co-expression of the double-mutant Q129A/Q869A (MOV10-DM) with both CVB (Fig. 7C) and EMCV (Fig. S4B) 3Cpro, showed almost complete protection from the 3Cpro-mediated degradation. Indeed, when we tested the functional activity of this mutant in MOV10-KO cells by ectopically expressing MOV10, the Q129A/Q869A mutant showed substantially higher antiviral activity against EMCV compared to the Wt MOV10 (Fig. 7E). Together, these results further support the notion that MOV10 is a physiologically important host protein that enhances in innate defense against certain RNA virus infections.
Fig. 7. MOV10 is targeted for degradation by picornavirus proteases.
(A–B) Loss of MOV10 expression upon EMCV (A) and CVB (B) infection. HEK293 cells were infected with EMCV (1 m.o.i) or CVB as indicated and analyzed by immunoblotting for MOV10 expression.
(C) MOV10 is targeted by CVB protease 3Cpro. 293T cells were transfected with either Myc-tagged CVB 3Cpro or the catalytically inactive mutant 3Cpro-C147A and analyzed for MOV10 and MAVS expression by immunoblotting.
(D) Protection of MOV10 Q129A/Q869A double-mutant (MOV10-DM) from CVB 3Cpro-mediated degradation. 293T cells were co-transfected with Wt or MOV10-DM along with CVB3 3Cpro as indicated followed immunoblotting.
(E) MOV10-DM expression leads to enhanced protection against EMCV replication. 293T-MOV10-KO cells were transfected either with Wt or MOV10-DM for 24 h later and subsequently challenged with EMCV (0.1 m.o.i.) for 16 h. EMCV RNA was analyzed by qRT-PCR.
DISCUSSION
Various helicases have been known to participate in innate antiviral immunity, with RIG-I and MDA5 being the most important ones for RNA virus mediated IFN induction. However, a number of these helicases with antiviral activity, either positively or negatively regulate RLR signaling. Here, using a series of genome-edited human cell lines we provide evidence that MOV10 enhances IFN induction to inhibit viral replication in a unique RLR-independent pathway. The observation that VSV replication is significantly increased in RIG-I-KO, MOV10-KO double deficient cells compared to only RIG-I deficient, RIG-I-KO cells, provides further evidence for this independence and existence of a parallel MOV10-mediated antiviral signaling pathway against specific viruses. Similar results in primary fibroblasts using MOV10 and RIG-I silencing provided support for this notion in a physiological context. However, the role of MOV10 in modulating TLR-mediated IFN induction is not yet clear.
Our finding of MOV10 targeting by viruses further substantiates the physiological significance of this phenomenon. Both EMCV and CVB effectively targeted MOV10 for degradation presumably using the respective 3Cpro viral proteases. Similar to some of the previous studies, we did not detect specific cleavage fragments of MOV10 (39), but the protection of the MOV10 Q129A/Q869A mutants from cleavage and the resulting inhibition of virus replication suggest specific targeting of MOV10 by these viruses. Although our results indicate that for SeV and VSV, the RLR pathway may be dominant over the MOV10 pathway, it is possible that for other viruses that target RLR pathway, MOV10 might be important.
Similar to other helicase family members, MOV10 is a multifunctional protein, and has been implicated in a diverse range of cellular functions including RNA silencing, mRNA translation, and tumor suppression (10, 11, 15, 23, 40, 41). It is also known to restrict HIV replication and retrotransposon mobility (17, 18, 20). Some of these functions are dependent on different functional properties of MOV10, such as the helicase activity and P-body localization (15, 16, 20). Here we describe another unique function of MOV10, antiviral activity against a number of RNA viruses that is independent of its helicase and anti-HIV activity. Although MOV10 has been shown to promote mRNA degradation by associating with UPF1, this activity requires the helicase activity of MOV10. Our demonstration that the helicase mutants of MOV10 provide antiviral activity indicates that this is independent of the UPF1-mediated RNA degradation activity of MOV10. The loss of this antiviral activity of MOV10 in either IRF3 or IFNAR1 deficient cells further supports its role in IFN-ISG-mediated inhibition of virus replication. Mechanistically, MOV10-signaling pathway specifically uses IKKε as the possible mediator kinase for IRF3 activation (Fig. S4D).
Although we define how MOV10 can promote IFN induction, its activation mechanism remains unknown. Similar to MDA5 and RIG-I, MOV10 expression by itself could promote IFN induction, but the biochemical activation mechanism of MOV10 remains to be determined. The helicase mutants we used are known to destroy MOV10 RNA unwinding activity, but retain its RNA binding activity (15). Thus, the nature of the RNA that activates MOV10, either uniquely or in addition to RLR, is not yet clear. The presence of MOV10 in complexes with IFIT proteins, known for their role in the detection of 5′-triphosphate containing uncapped, as well as capped viral RNA lacking 2′O methylation (42, 43), suggest that MOV10 can either directly or along with IFIT proteins bind to viral RNA to promote IFN induction. Although, we did not detect any contribution of IFIT1 and IFIT3 in MOV10-mediated ISG induction by coexpression (not shown), the biochemical basis of self non-self discrimination of RNA by MOV10 requires further clarifications. In this regard, the unique localization of MOV10 in cytoplasmic P-bodies may play an important role. In summary, this study establishes MOV10-mediated IFN induction as an antiviral signaling mechanism.
Supplementary Material
Acknowledgments
We thank Dr. Vinay K. Pathak (National Cancer Institute) and Dr. Yong-Hui Zheng (Michigan State University) for sharing crucial MOV10 Wt and mutant expression plasmids; Dr. Takashi Fujita (Kyoto University) for the EMCV 3Cpro plasmid; and Dr. Jennifer Bomberger for the RSV stock and primers.
This work was supported in part by NIH 1U24AI082673, 1R01AI118896 (SNS). This project used the UPCI core facilities and was supported in part by award P30CA047904.
ABBREVIATIONS
- MOV10
Moloney leukemia virus 10, homolog
- IFN
Interferon
- IRF
Interferon Regulatory Factor
- ISG
Interferon Stimulated Gene
- VSV
vesicular stomatitis virus
- SeV
Sendai virus
- EMCV
encephalomyocarditis virus
Footnotes
CONFLICT OF INTEREST
The authors declare that they have no conflict of interest.
AUTHOR CONTRIBUTION
RAC and AG performed experiments; CW, VH helped create the genome engineered KO cells, SNS, CBC and VH designed experiments and wrote the manuscript.
References
- 1.Goubau D, Deddouche S, Reis e Sousa C. Cytosolic sensing of viruses. Immunity. 2013;38:855–869. doi: 10.1016/j.immuni.2013.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yoneyama M, Kikuchi M, Matsumoto K, Imaizumi T, Miyagishi M, Taira K, Foy E, Loo YM, Gale M, Jr, Akira S, Yonehara S, Kato A, Fujita T. Shared and unique functions of the DExD/H-box helicases RIG-I, MDA5, and LGP2 in antiviral innate immunity. J Immunol. 2005;175:2851–2858. doi: 10.4049/jimmunol.175.5.2851. [DOI] [PubMed] [Google Scholar]
- 3.Takeuchi O, Akira S. Innate immunity to virus infection. Immunol Rev. 2009;227:75–86. doi: 10.1111/j.1600-065X.2008.00737.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Loo YM, Gale M., Jr Immune signaling by RIG-I-like receptors. Immunity. 2011;34:680–692. doi: 10.1016/j.immuni.2011.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bhoj VG, Sun Q, Bhoj EJ, Somers C, Chen X, Torres JP, Mejias A, Gomez AM, Jafri H, Ramilo O, Chen ZJ. MAVS and MyD88 are essential for innate immunity but not cytotoxic T lymphocyte response against respiratory syncytial virus. Proc Natl Acad Sci U S A. 2008;105:14046–14051. doi: 10.1073/pnas.0804717105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bruns AM, Horvath CM. Activation of RIG-I-like receptor signal transduction. Crit Rev Biochem Mol Biol. 2012;47:194–206. doi: 10.3109/10409238.2011.630974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fullam A, Schroder M. DExD/H-box RNA helicases as mediators of anti-viral innate immunity and essential host factors for viral replication. Biochim Biophys Acta. 2013;1829:854–865. doi: 10.1016/j.bbagrm.2013.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Paludan SR, Bowie AG. Immune sensing of DNA. Immunity. 2013;38:870–880. doi: 10.1016/j.immuni.2013.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fairman-Williams ME, Guenther UP, Jankowsky E. SF1 and SF2 helicases: family matters. Curr Opin Struct Biol. 2010;20:313–324. doi: 10.1016/j.sbi.2010.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chendrimada TP, Finn KJ, Ji X, Baillat D, Gregory RI, Liebhaber SA, Pasquinelli AE, Shiekhattar R. MicroRNA silencing through RISC recruitment of eIF6. Nature. 2007;447:823–828. doi: 10.1038/nature05841. [DOI] [PubMed] [Google Scholar]
- 11.Meister G, Landthaler M, Peters L, Chen PY, Urlaub H, Luhrmann R, Tuschl T. Identification of novel argonaute-associated proteins. Curr Biol. 2005;15:2149–2155. doi: 10.1016/j.cub.2005.10.048. [DOI] [PubMed] [Google Scholar]
- 12.Cook HA, Koppetsch BS, Wu J, Theurkauf WE. The Drosophila SDE3 homolog armitage is required for oskar mRNA silencing and embryonic axis specification. Cell. 2004;116:817–829. doi: 10.1016/s0092-8674(04)00250-8. [DOI] [PubMed] [Google Scholar]
- 13.Tomari Y, Du T, Haley B, Schwarz DS, Bennett R, Cook HA, Koppetsch BS, Theurkauf WE, Zamore PD. RISC assembly defects in the Drosophila RNAi mutant armitage. Cell. 2004;116:831–841. doi: 10.1016/s0092-8674(04)00218-1. [DOI] [PubMed] [Google Scholar]
- 14.Castello A, Fischer B, Eichelbaum K, Horos R, Beckmann BM, Strein C, Davey NE, Humphreys DT, Preiss T, Steinmetz LM, Krijgsveld J, Hentze MW. Insights into RNA biology from an atlas of mammalian mRNA-binding proteins. Cell. 2012;149:1393–1406. doi: 10.1016/j.cell.2012.04.031. [DOI] [PubMed] [Google Scholar]
- 15.Gregersen LH, Schueler M, Munschauer M, Mastrobuoni G, Chen W, Kempa S, Dieterich C, Landthaler M. MOV10 Is a 5′ to 3′ RNA helicase contributing to UPF1 mRNA target degradation by translocation along 3′ UTRs. Molecular cell. 2014;54:573–585. doi: 10.1016/j.molcel.2014.03.017. [DOI] [PubMed] [Google Scholar]
- 16.Izumi T, Burdick R, Shigemi M, Plisov S, Hu WS, Pathak VK. Mov10 and APOBEC3G localization to processing bodies is not required for virion incorporation and antiviral activity. J Virol. 2013;87:11047–11062. doi: 10.1128/JVI.02070-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Abudu A, Wang X, Dang Y, Zhou T, Xiang SH, Zheng YH. Identification of molecular determinants from Moloney leukemia virus 10 homolog (MOV10) protein for virion packaging and anti-HIV-1 activity. J Biol Chem. 2012;287:1220–1228. doi: 10.1074/jbc.M111.309831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Burdick R, Smith JL, Chaipan C, Friew Y, Chen J, Venkatachari NJ, Delviks-Frankenberry KA, Hu WS, Pathak VK. P body-associated protein Mov10 inhibits HIV-1 replication at multiple stages. J Virol. 2010;84:10241–10253. doi: 10.1128/JVI.00585-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang X, Han Y, Dang Y, Fu W, Zhou T, Ptak RG, Zheng YH. Moloney leukemia virus 10 (MOV10) protein inhibits retrovirus replication. J Biol Chem. 2010;285:14346–14355. doi: 10.1074/jbc.M110.109314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li X, Zhang J, Jia R, Cheng V, Xu X, Qiao W, Guo F, Liang C, Cen S. The MOV10 helicase inhibits LINE-1 mobility. J Biol Chem. 2013;288:21148–21160. doi: 10.1074/jbc.M113.465856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Goodier JL, Cheung LE, Kazazian HH., Jr MOV10 RNA helicase is a potent inhibitor of retrotransposition in cells. PLoS Genet. 2012;8:e1002941. doi: 10.1371/journal.pgen.1002941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schoggins JW, Wilson SJ, Panis M, Murphy MY, Jones CT, Bieniasz P, Rice CM. A diverse range of gene products are effectors of the type I interferon antiviral response. Nature. 2011;472:481–485. doi: 10.1038/nature09907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Haussecker D, Cao D, Huang Y, Parameswaran P, Fire AZ, Kay MA. Capped small RNAs and MOV10 in human hepatitis delta virus replication. Nat Struct Mol Biol. 2008;15:714–721. doi: 10.1038/nsmb.1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mukherjee A, Morosky SA, Delorme-Axford E, Dybdahl-Sissoko N, Oberste MS, Wang T, Coyne CB. The coxsackievirus B 3C protease cleaves MAVS and TRIF to attenuate host type I interferon and apoptotic signaling. PLoS Pathog. 2011;7:e1001311. doi: 10.1371/journal.ppat.1001311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ng CS, Jogi M, Yoo JS, Onomoto K, Koike S, Iwasaki T, Yoneyama M, Kato H, Fujita T. Encephalomyocarditis virus disrupts stress granules, the critical platform for triggering antiviral innate immune responses. J Virol. 2013;87:9511–9522. doi: 10.1128/JVI.03248-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Morosky SA, Zhu J, Mukherjee A, Sarkar SN, Coyne CB. Retinoic acid-induced gene-I (RIG-I) associates with nucleotide-binding oligomerization domain-2 (NOD2) to negatively regulate inflammatory signaling. The Journal of biological chemistry. 2011;286:28574–28583. doi: 10.1074/jbc.M111.227942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhu J, Zhang Y, Ghosh A, Cuevas RA, Forero A, Dhar J, Ibsen MS, Schmid-Burgk JL, Schmidt T, Ganapathiraju MK, Fujita T, Hartmann R, Barik S, Hornung V, Coyne CB, Sarkar SN. Antiviral activity of human OASL protein is mediated by enhancing signaling of the RIG-I RNA sensor. Immunity. 2014;40:936–948. doi: 10.1016/j.immuni.2014.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Durbin JE, Hackenmiller R, Simon MC, Levy DE. Targeted disruption of the mouse Stat1 gene results in compromised innate immunity to viral disease. Cell. 1996;84:443–450. doi: 10.1016/s0092-8674(00)81289-1. [DOI] [PubMed] [Google Scholar]
- 29.Schmid-Burgk JL, Schmidt T, Kaiser V, Honing K, Hornung V. A ligation-independent cloning technique for high-throughput assembly of transcription activator-like effector genes. Nat Biotechnol. 2013;31:76–81. doi: 10.1038/nbt.2460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ablasser A, Schmid-Burgk JL, Hemmerling I, Horvath GL, Schmidt T, Latz E, Hornung V. Cell intrinsic immunity spreads to bystander cells via the intercellular transfer of cGAMP. Nature. 2013;503:530–534. doi: 10.1038/nature12640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schmid-Burgk JL, Schmidt T, Gaidt MM, Pelka K, Latz E, Ebert TS, Hornung V. OutKnocker: a web tool for rapid and simple genotyping of designer nuclease edited cell lines. Genome Res. 2014;24:1719–1723. doi: 10.1101/gr.176701.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gack MU. Mechanisms of RIG-I-like receptor activation and manipulation by viral pathogens. Journal of virology. 2014;88:5213–5216. doi: 10.1128/JVI.03370-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bowie AG, Unterholzner L. Viral evasion and subversion of pattern-recognition receptor signalling. Nat Rev Immunol. 2008;8:911–922. doi: 10.1038/nri2436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hoffmann HH, Schneider WM, Rice CM. Interferons and viruses: an evolutionary arms race of molecular interactions. Trends Immunol. 2015;36:124–138. doi: 10.1016/j.it.2015.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Harris KG, Coyne CB. Enter at your own risk: how enteroviruses navigate the dangerous world of pattern recognition receptor signaling. Cytokine. 2013;63:230–236. doi: 10.1016/j.cyto.2013.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McKendry R, John J, Flavell D, Muller M, Kerr IM, Stark GR. High-frequency mutagenesis of human cells and characterization of a mutant unresponsive to both alpha and gamma interferons. Proc Natl Acad Sci U S A. 1991;88:11455–11459. doi: 10.1073/pnas.88.24.11455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Blom N, Hansen J, Blaas D, Brunak S. Cleavage site analysis in picornaviral polyproteins: discovering cellular targets by neural networks. Protein Sci. 1996;5:2203–2216. doi: 10.1002/pro.5560051107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rawlings ND, Barrett AJ, Bateman A. MEROPS: the database of proteolytic enzymes, their substrates and inhibitors. Nucleic Acids Res. 2012;40:D343–350. doi: 10.1093/nar/gkr987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li K, Foy E, Ferreon JC, Nakamura M, Ferreon AC, Ikeda M, Ray SC, Gale M, Jr, Lemon SM. Immune evasion by hepatitis C virus NS3/4A protease-mediated cleavage of the Toll-like receptor 3 adaptor protein TRIF. Proc Natl Acad Sci U S A. 2005;102:2992–2997. doi: 10.1073/pnas.0408824102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Banerjee S, Neveu P, Kosik KS. A coordinated local translational control point at the synapse involving relief from silencing and MOV10 degradation. Neuron. 2009;64:871–884. doi: 10.1016/j.neuron.2009.11.023. [DOI] [PubMed] [Google Scholar]
- 41.El Messaoudi-Aubert S, Nicholls J, Maertens GN, Brookes S, Bernstein E, Peters G. Role for the MOV10 RNA helicase in polycomb-mediated repression of the INK4a tumor suppressor. Nat Struct Mol Biol. 2010;17:862–868. doi: 10.1038/nsmb.1824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Diamond MS, Farzan M. The broad-spectrum antiviral functions of IFIT and IFITM proteins. Nat Rev Immunol. 2013;13:46–57. doi: 10.1038/nri3344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pichlmair A, Lassnig C, Eberle CA, Gorna MW, Baumann CL, Burkard TR, Burckstummer T, Stefanovic A, Krieger S, Bennett KL, Rulicke T, Weber F, Colinge J, Muller M, Superti-Furga G. IFIT1 is an antiviral protein that recognizes 5′-triphosphate RNA. Nat Immunol. 2011;12:624–630. doi: 10.1038/ni.2048. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.