Abstract
Resting central Tregs (cTregs) and activated effector Tregs (eTregs) are required for self-tolerance, but the heterogeneity and relationships within and between phenotypically distinct subsets of cTregs and eTregs are poorly understood. By extensive immune profiling and deep sequencing of TCRβ V-regions, two subsets of cTregs, based on expression of Ly-6C, and three subsets of eTregs, based on distinctive expression of CD62L, CD69, and CD103, were identified. Ly-6C+ cTregs exhibited lower basal activation, expressed on average lower affinity TCRs, and less efficiently developed into eTregs when compared to Ly-6C− cTregs. The dominant TCR Vβs of Ly-6C+ cTregs were shared by eTregs at a low frequency. A single TCR clonotype was also identified that was largely restricted to Ly-6C+ cTregs, even under conditions that promoted the development of eTregs. Collectively, these findings indicate that some Ly-6C+ cTregs may persist as a lymphoid-specific subset, with minimal potential to develop into highly activated eTregs, while other cTregs readily develop into eTregs. In contrast, subsets of CD62Llo eTregs showed higher clonal expansion and were more highly inter-related than cTreg subsets based on their TCRβ repertoires, but exhibited varied immune profiles. The CD62Llo CD69− CD103− eTreg subset displayed properties of a transitional intermediate between cTregs and more activated eTreg subsets. Thus, eTregs subsets appear to exhibit substantial flexibility, likely in response to environmental cues, to adopt defined immune profiles that are expected to optimize suppression of autoreactive T cells.
Introduction
CD4+ Foxp3+ regulatory T cells (Tregs) are essential for establishing and maintaining self-tolerance and for limiting immune responses (1, 2). To mediate such activity, Foxp3+ Tregs exhibit substantial heterogeneity that is phenotypically distinguishable and appears to be analogous to that described for activated T effector (Teff) cells (3, 4). Recent evidence supports a model whereby effector Tregs (eTregs) with an activated phenotype are derived from a more quiescent pool of “naïve”, central or memory Tregs (5, 6). Central Tregs (cTregs) are characterized by expression of CCR7 and high levels of CD62L and lower expression of CD44 (7, 8). Ly-6C markers at least some cTregs and these cTregs exhibit less suppressive activity and express lower affinity TCRs when compared to Ly-6C− Tregs (9, 10). cTregs are primarily localized in secondary lymphoid tissues and their homeostasis depends in part on signaling through the IL-2R (8). eTregs are characterized by expression of CD69, CD103, and Klrg1, high expression of CD44, GITR, and ICOS, and chemokine receptors that direct tissue localization, e.g. CXCR3, CCR6, and CCR9 (11). Correspondingly, eTregs dominate tissue sites such as the gut mucosa, lung, fat, and acutely injured skeletal muscle (12). The development of eTregs from cTregs depends on TCR signaling and activity associated with the transcriptional regulator IRF4 (6, 7, 13). Genome-wide expression profiling indicates that eTregs are distinguished from cTregs by increased levels of mRNAs for suppressive molecules such as IL-10, tissue-related chemokine receptors, and pro-apoptotic molecules, but lower levels of pro-survival mRNAs (14, 15). Overall, the activation of eTregs leads to their expansion into highly suppressive, but short-lived cells well-suited to regulate inflammatory responses in non-lymphoid tissues.
Much has also been learned concerning the heterogeneity of Tregs based on examining their TCR repertoire, primarily using TCR transgenic models to limit their specificities. This work demonstrated that the Treg TCR repertoire is highly diverse, with only a partial and somewhat limited overlap with the repertoire found on conventional T cells (16–18). These studies indicate that most Tregs in the periphery are thymus derived, with peripherally developed Tregs more obvious in the gut mucosa. Tregs show heterogeneity in their TCR repertoire based on the anatomical location of draining secondary lymphoid tissues (19) or within distinct non-lymphoid tissue sites (20). This finding is consistent with localized responses by Tregs to tissue-specific self-antigens.
The above findings and additional studies indicate that cTregs give rise to eTregs (4, 14) and more robust clonal expansion is associated with eTregs (21). However, the relationship between phenotypically distinct subsets within cTregs and eTregs has not been extensively studied. Indeed, it is not known whether there is additional heterogeneity within cTregs and whether their development into eTregs represents a default pathway for cTregs that undergo clonal expansion. We also do not know whether clonal expansion of eTregs is broadly associated with these cells or rather is a property of an eTreg subset.
In the present study, these issues were addressed in part by examining mRNA expression of immune-related genes using the Nanostring platform, and the TCR repertoire by next generation sequencing for subsets of cTregs and eTregs. Adoptive transfer studies of purified subsets of Tregs were performed to assess their capacity to develop into eTregs and suppress autoimmunity. Our study is consistent with a model where cTregs are not solely precursor cells for eTregs and that extensive clonal expansion in response to self-antigen is restricted to subsets of eTregs.
Material and Methods
Mice
Foxp3/RFP-reporter (22), Foxp3/GFP reporter (23), IL-2Rβ−/− (24), IL-2RβY3 (referred to a Y3) (25), CD45.1-congenic, and TCRβ transgenic Tg(Tcrb)93Vbo/J (26) mice, all on the C57BL/6 genetic background were bred within the specific pathogen-free animal facility at the University of Miami. Purified Tregs (1.5 × 105) or Treg subsets (1 × 105) from Foxp3-reporter mice or CD45.1 spleen cells (10 × 106) were adoptively transferred i.v. All other studies used adult age matched mice typically 8–16 weeks of age. All animal studies were reviewed and approved by the Institutional Animal Care and Use Committee at the University of Miami.
FACS analysis and sorting
The following monoclonal antibodies to the indicated molecules, the fluorescent labels and their sources were used in this study: CD4 (GK1.5, FITC; RM4-5, PE, Biolegend; PE-Cy7, BD Pharmingen); CD62L (MEL-14, APC/Cy-7; Biolegend; MEL-14, eVolve™ 605, eBioscience); CD69 (H1.2F3, PE/Cy7, Biolegend); CD103 (2E7, PE, eBioscience); Klrg1 (2F1/KLRG1, APC, Biolegend, Biotin, eBioscience); Foxp3 (FKJ-16s, PercpCy5.5, eFluor®450, eBioscience); TCR Vβ 8.1, 8.2 (KJ16-133.18, PE, Biolegend); IL-2, (Jes6-5H4, APC, Biolegend); IL-17 (ebio17b7, PE-CY7, eBioscience); IFNγ (XMG1.2, PE, Biolegend); CD25 (PC-61, PE, BD Pharmingen; PE-Cy7, Biolegend); CD103 (2E7, APC, eBioscience); CD39 (24DMS1, PE, eBioscience); Ki67 (B56, Alexa700, BD Pharmingen); Bcl-2 (BCL/10C4, PE, Biolegend); CD44 (PGP-1, FITC, Alexa647, lab made); CD5 (53-7.3, PerCP-Cy5.5, Biolegend); ICOS (15F9, PerCP-eFluor®710, eBioscience); CTLA4 (UC10-4F10-11, PE, BD Pharmingen); CD73 (eBioTY/11.8, PE, PE-Cy7, eBioscience); Thy-1.1 (HIS51, PerCP-Cy5.5, FITC, eBioscience); Ly-6C (AL-21, APC-Cy7, Biotin, BD Pharmingen). Streptavidin conjugates used are: PE-Strep, Biolegend; PE-Cy7 Strep, PerCP-eFluor®710-Strep; APC-Strep, eBioscience. We conjugated anti-CD4 (GK1.5) and anti-CD8 (53.6.7) using Alexa Fluor® 700 NHS Ester from Molecular Probes according to the manufacturer’s instructions. Samples were analyzed on a LSR-Fortessa-HTS. More then 100,000 events were collected per sample. To purify Tregs and Treg subsets, total CD4+ T cells were obtained using anti-CD4 magnetic MicroBeads (Miltenyi Biotec). The cells were then stained and sorted based on the expression of the Foxp3 reporter dye and cell surface markers using FACS Aria-II flow cytometry. Cells were usually >95% pure.
TaqMan PCR for the AY02 clonotype
RNA was isloted from purified T cell populations, typical from 1–6 × 105 cells, using the Trizol® reagent (Life Technologies), followed by ethanol precipitation in the presence of 40 μg of glycogen (Affymetrix USB, ultrapure, MB grade) as a carrier. RNAs were dissolved in 12 μl of RNase free water and 10 μl of this RNA was reverse transcribed in a 20 μl reaction. First the RNA, poly(T)-Oligo(T)16mer (5 μM) (Life Technologies) and dNTPs (500 μM) (Promega) were pre-heated at 65° C for 5 min. After cooling, recombinant RNasin® Ribonuclease inhibitor (20 units) (Promega), DTT (5 mM), SuperScript III Reverse Transcriptase (10 units) (Life Technologies) and its buffer were added and the cDNA was synthesized at 50° C for 1h. The primers and probes for TaqMan PCR are shown in Supplemental Fig. 1A. The AY02 clonotype and Cα probes were synthesized and labeled with 6FAM-TAMAR by Applied Biosystems. TaqMan reactions were perform in a volume of 20 μl containing cDNA (6 μl for the AY02 and 1 μl for the Cα reactions), the appropriate forward and reverse primers (500 nM) and gene-specific probe (250 nM), Mg2+ Cl (2.5 mM), and the 2.5x of 5′ MasterMix containing Taq DNA polymerase, dNTPs and buffer components (Fisher Scientific). TaqMan PCR reaction conditions were a single cycle at 50° C for 2 min and 95° C for 10 min, followed by 40 cycles at 95° C for 15 sec and 60° C for 60 sec using a StepOnePlus Real-Time PCR Systems (Applied Biosystems). A standard ramp speed was used. For each reaction, amplification plots of threshold cycles (CT) were determined. Every experiment contained a positive (plasmid with the AY02 clonotype) and negative control (plasmid with an irrelevant Vα2 clonotype). The AY02 plasmid was used for the standard curves by dilution (102 to 108 copies) of this plasmid.
IL2-IC treated mice
Mouse IL-2 (eBioscience, Recombinant Protein Carrier-Free) and anti-mIL-2 antibody (JES6-1A12, Bio X cell) were mixed at a 2 to 1 molar ratio and incubated for 30 min at room temperature. Each mouse received 1 μg of IL-2/5 μg of JES6-1A12 complex in 200 μl of PBS i.p. for three consecutive days.
Nanostring gene expression analysis
The indicated Treg subsets were lysed in RTL buffer (Qiagen). The lysates from 1 × 104 cell equivalents were used for RNA profiling using the nCounter Mouse Immunology Gene Expression Code Set (NanoString) at the Oncogenomics Core at the University of Miami. All analysis of expression data, including normalization, log2 transformation, determining fold change ratios between groups, Student’s two-tailed t testing and calculation of False Discovery Rates was carried out using NSolver™ version 2.5, a statistical analysis program provided by Nanostring Technologies®, according to the manufacturer’s recommendations.
Deep sequencing of TCRβ repertoire
DNA was isolated from the indicated Treg subsets using the QIAamp® DNA Micro Kit (QIAGEN) and eluted in 50 μl of AE buffer. Amplification and sequencing of TCRβ CDR3 regions was performed using the immunoSEQ platform (Adaptive Biotechnologies, Seattle, WA). ImmunoSEQ combines multiplex PCR with high throughput sequencing and a sophisticated bioinformatics pipeline for TCRβ CDR3 region analysis (27, 28). Sufficient sequence information was obtained to assign V subgroups and J regions associated with each CDR3.
Intracellular cytokine assay
Cells (1–2 × 106) were cultured in 24 well flat bottom plates with complete RPMI 1640 medium (29) containing PMA (50 ng/ml), ionomycin (1 μM) and brefeldin A (Biolegend) for 4 hr. Cells were harvested, washed, and stained for cell surface markers and then fixed, permeabilized, and stained to detect IL-2, IL-17, and IFNγ by flow cytometry.
Statistical analysis
Data were analyzed by one way ANOVA applying Tukey’s multiple comparison test or unpaired two-tailed Student’s t-test using Graph Pad Prism 6.0 software, as indicated in the Figure legends. All data are reported as the mean ± SD. In all Figures, significant difference are indicated as *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001.
Results
Ly-6C identifies subsets of cTregs
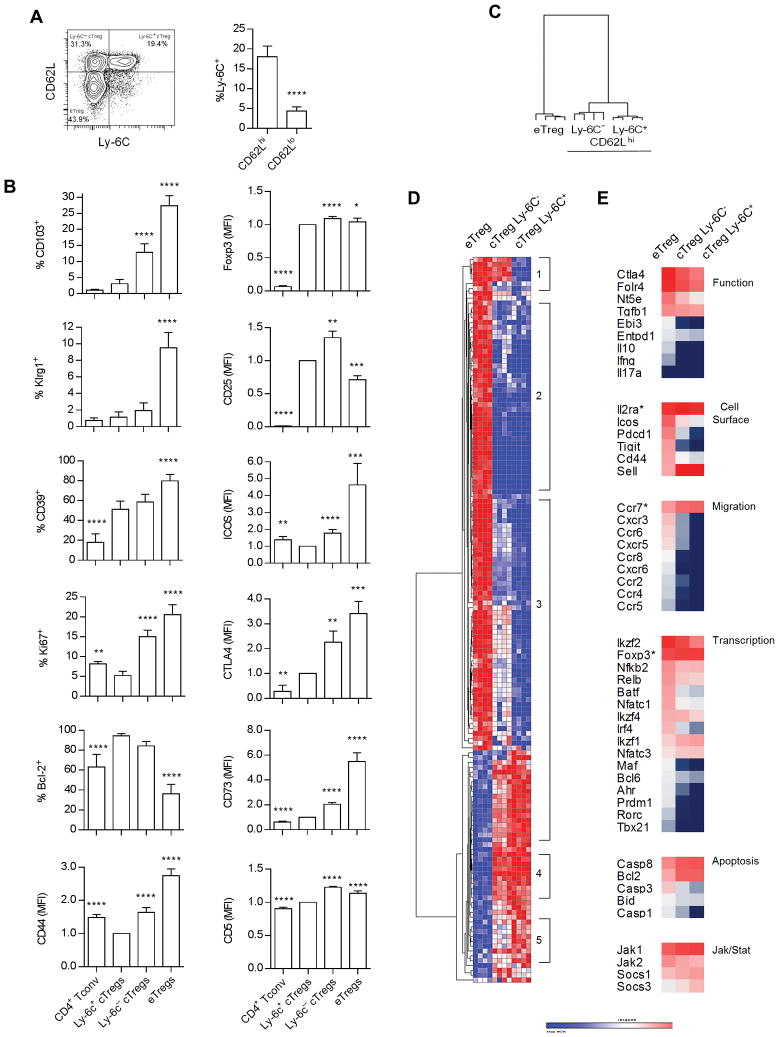
Ly-6C+ Tregs express lower affinity TCRs, are less suppressive, and are cTregs based on expression of high levels of CD62L and low levels of CD44 (9, 10). Nevertheless, it remains unclear whether all cTregs express Ly-6C. FACS analysis of peripheral splenic Tregs showed that only a subset of CD62Lhi cTregs express Ly-6C (Fig. 1A). Representative FACS profiles (Supplemental Fig. 2) and analysis of the resulting data (Fig. 1B) revealed that Ly-6C+ and Ly-6C− cTregs and eTregs expressed similar levels of Foxp3. Ly-6C+ cTregs expressed the lowest levels of the activation molecules CD103, Klrg1, CD44, and ICOS, the proliferative marker Ki67, the Treg functional molecules CD39, CD73, and CTLA4, and CD5, whose levels directly correlate with the affinity of the TCR (30), but expressed the highest levels of Bcl-2. This expression pattern by the Ly-6C+ cTreg subset is consistent with long-lived slowly replicating cells that express TCRs with a relatively low affinity for self-antigen and low suppressive activity. In most cases, Ly-6C− cTregs exhibited an intermediate phenotype when compared to Ly-6C+ cTregs and eTregs. Two exceptions were CD5 and CD25, whose levels were the highest for Ly-6C− Tregs; this phenotype is consistent with a Treg subset that expresses higher affinity TCRs and preferentially responds to IL-2.
Figure 1.
Ly-6C+ Tregs represent a distinct subsets of cTregs. (A) The distribution of Ly-6C on splenic cTregs and eTregs based on expression of CD62L. Data (n=12) were analyzed by a two-tailed unpaired t-test. (B) FACS analysis of the indicated molecules on Ly-6C+ and Ly-6C− CD62Lhi cTregs, Ly-6C− CD62Llo eTregs and CD4+ Foxp3− T cells (Tconv). Data (n=6–12) were analyzed by one way ANOVA using Tukey’s multiple comparison test when represented as % positive or by a one-sample t-test when represented as mean fluorescent intensity (MFI). All comparisons for significant differences were made in reference to Ly-6C+ cTregs. (C–E) Nanostring analysis of mRNA expression by Ly-6C+ and Ly-6C− CD62Lhi cTregs and CD69+ CD62Llo eTregs. Hierarchical clustering (one minus the Pearson correlation) of samples (C) and mRNAs (D) that were expressed at significantly different levels (p≤0.05, corrected for FDR) between any two groups. (E) Expression of selected mRNAs based on log2 intensity values.
To further explore the relationship between these cTreg subsets and eTregs, Nanostring gene expression profiling was performed using an array that measures 561 immunologically relevant genes. Results from all mRNAs are shown in Supplemental Table 1A. Hierarchical clustering of the 150 differentially expressed mRNAs revealed that Ly-6C+ and Ly-6C− cTreg subsets were much more related to each other than to eTregs (Fig. 1C). Many of these genes were similarly expressed by both cTreg subsets (Fig. 1D, regions 2 and 4, and 1E), but for others expression by Ly-6C− cTregs tended to move toward that of eTregs (Fig. 1D, regions 1,3 and 5). Examples of such genes shown in Fig. 1E, which are associated with region 3, include Nt5e, Entpd1, Pdcd1, Cxcr3, Ccr6, Cxcr5, Ccr2, Irf4, Ahr, Casp1 and Casp3. Overall, these data show that cTregs express the lowest levels of molecules associated with Treg suppressive function, activation, tissue-seeking chemokine receptors, transcription factors related to TCR signaling and regulation of suppressive pathways, but have the highest expression of molecules associated with T cell survival and regulation of Jak/Stat signaling. These properties were almost always the most polarized for Ly-6C+ cTregs while the Ly-6C− cTregs showed a tendency for higher activation, progressing toward eTregs.
cTreg subsets distinctly contribute to the TCR repertoire of eTregs
Deep sequencing was performed for TCRβ for splenic Ly-6C+ and Ly-6C− CD62Lhi CD69− CD103− cTregs and CD62Llo CD69+ eTregs to further explore the relationship between these subsets. For these 3 Treg subsets from two replicate experiments, DNA was isolated from approximately 7 × 105 cells (range 4–11 × 105). Productive sequence reads ranged from 3.7–9.5 × 105, yielding ~23,000–54,000 unique sequences with high repertoire diversity, as clonality of the repertoire measured between ~0.02–0.05, where 1 equals monoclonal. Sequence coverage (mean 17.6; range 9.9–30.5) was similar for all samples. No skewing of V genes and J regions were noted between these subsets.
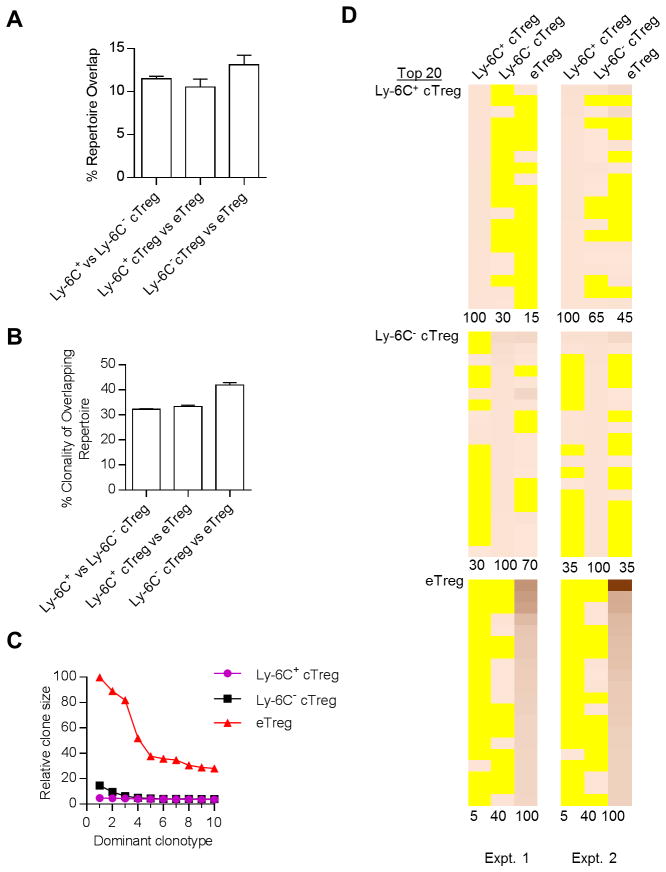
When assessing the entire Vβ repertoire of the deduced productive protein V-region sequences, including the V gene and CDR3s, repertoires for these 3 Treg subsets overlapped by ~10–12% (Fig. 2A). This result is consistent with a modest interrelationship among the specificities expressed by these subsets. The number of individual protein V sequences that overlapped between these subsets were also similar (~3,500–4,000). The abundance of these shared sequences was relatively low, especially between the Ly-6C+ vs. Ly-6C− cTregs. The clonality of the shared sequences, i.e. same nucleotide and amino acid V region sequences, was also low (~32–42%) (Fig. 2B), consistent with convergent recombination characterizing some of the sequences within the overlapping TCRβ repertoires of cTregs and eTregs.
Figure 2.
The relationship of the TCR repertoire between cTreg subsets and eTregs from normal C57BL/6 mice. Spleen, peripheral lymph nodes (PLN) and mesenteric LN (MLN) were pooled from Foxp3/RFP reporter mice. Ly-6C+ and Ly-6C− (CD62Lhi CD69− CD103−) cTregs and eTregs (CD62Llo CD69+) were FACS purified and DNA was isolated for deep sequencing for TCRβ. Data are from 2 independent experiments and represented as the mean ± the range. (A,B) Characterization of repertoire overlap. (A) Repertoire overlap based on the total number of shared productive VDJ protein sequence reads calculated as: (S1s + S2s)/(S1t and S2t) × 100, where S is an individual sample, s and t are the number of shared and total sequences reads, respectively. (B) Clonality was also determined for the overlapping unique productive sequences, where % clonality = (number of shared individual unique VDJ nucleotide sequences/number of shared individual unique VDJ amino acid sequences) × 100. (C, D) Comparison of the dominant clonotypes. (C) The relative clone size of the 10 most dominant TCRβ VDJ sequences within each Treg subset, where the number of sequences reads for the most prevalent clonotype was normalized to a value of 100 and all other sequence reads were proportionally reduced in comparison to this sequence. (D) The frequency of the 20 most dominant clonotypes (protein VDJ) was determined for each subset from both experiments. Heat maps were constructed where yellow indicates the absence of a particular clonotype whereas the presence of a clonotype is quantitatively represented by increased intensity of the orange-like color. The % overlap to the reference sequence, listed as 100%, is shown below each heat map.
The 10 most abundant clonotypes were identified for Ly-6C+ and Ly-6C− cTregs and eTregs. After normalization, eTregs showed much larger clone sizes than cTregs (Fig. 2C). This result is consistent with greater clonal expansion by eTregs. The distribution of the top 20 clonotypes of each subset between the cTreg and eTreg subsets was determined (Fig. 2D). The top sequences for Ly-6C+ cTregs were least frequently detected in eTregs, whereas the most frequent sequences in eTregs were almost entirely excluded from cTregs. Ly-6C− cTregs showed an intermediate pattern where they were more readily found in Ly-6C+ cTregs and eTregs. Overall these data support the notion that some Ly-6C+ and Ly-6C− cTregs develop toward eTregs, which occurs more frequently in the latter, but also raise the possibility that some cTregs, especially Ly-6C+ Tregs, persist mainly as cTregs.
Distinctive properties of eTreg subsets
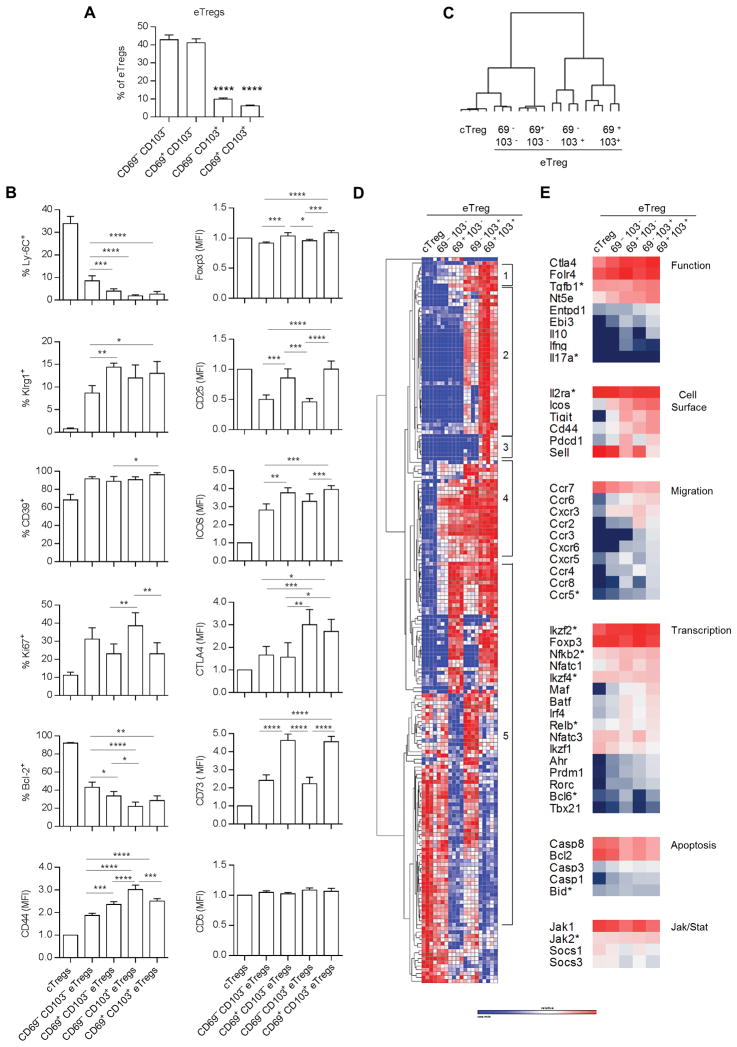
Past genome-wide profiling showed that CD69+, Klrg1+ and CD103+ Tregs appear to be more activated than total Tregs, cTregs, or CD62Llo Tregs (14, 15). We refined this analysis to better quantify immunological properties of eTregs after fractionation into 4 subsets (Fig. 3A); all were CD62Llo but they varied in their expression of CD69 and CD103, with CD103 found on a minority of peripheral eTregs. FACS analysis showed that CD69− CD103− eTregs showed the least activated phenotype based on lower expression of Klrg1, CD44, and ICOS, but higher expression of Ly-6C and Bcl-2 (Fig. 3B). Highest proliferation was associated with CD69− Tregs based on Ki67 expression. These cells showed lower CD25 and Foxp3 levels, consistent with the notion that this response was largely IL-2-independent (21). Treg functional molecules were distinctively expressed such that all subsets expressed equivalent levels of CD39, but the highest levels of CTLA4 and CD73 were associated with CD103+ and CD69+ eTregs, respectively.
Figure 3.
eTregs express heterogeneity consistent with 3 major subsets. (A) The distribution of the indicated CD69 and CD103 subsets within CD62Llo eTregs. Data (n= 5) were analyzed by one way ANOVA using Tukey’s multiple comparison test. All comparisons for significant differences were made in reference to CD62Lhi CD69− CD103− cTregs. (B) FACS analysis of the indicated molecules on eTreg subsets and total cTregs. Data (n=5) were analyzed by one way ANOVA using Tukey’s multiple comparison test when represented as % positive or by a one-sample t-test when represented as mean fluorescent intensity (MFI). Comparisons for significant differences were between all eTreg subsets. (C–E) Nanostring analysis of mRNA expression by CD62Lhi cTregs and eTreg subsets. Hierarchical clustering (one minus the Pearson correlation) of samples (C) and mRNAs (D) that were expressed at significantly different levels (p≤0.05, corrected for FDR) between any two groups. (E) Expression of selected mRNAs based on log2 intensity values.
Nanostring profiling was also performed for total cTregs and eTreg subsets. The expression of all mRNAs is shown in Supplemental Table 1B. Hierarchical clustering of the 193 differentially expressed mRNAs revealed that CD103− eTreg subsets are more related to cTregs, with CD62Llo CD69− CD103− Tregs the closest subset to cTregs (Fig. 3C). CD62Llo CD69+ CD103+ Tregs were more distantly related to cTregs. Inspection of the clustering of individual mRNAs resulted in 3 major patterns of mRNA expression. One pattern is generally increasing expression of mRNAs from CD62Llo CD69− CD103− → CD62Llo CD69+ CD103+ eTregs (Fig. 3D, regions 1 and 4). Representative mRNAs include Ctla4, Nt5e, Icos, Tigit, Ccr2, Ccr6, Cxcr3, Ahr, Prdm1, Maf, Rorc (Fig. 3E), A second pattern of mRNAs is relatively high expression by CD103+ eTreg subsets (Fig. 3D, region 2), which include Entpd1 and Ccr3 (Fig. 3E). The third pattern is a somewhat favored expression of mRNAs related to the CD69+ eTreg subset, unrelated to whether they express CD103 (Fig. 3D, region 5). Some genes in this group include Irf4, Ebi3, Pdcd1, Tbx21, Ccr8, Il10, Cxcr5, and Sell (Fig. 3E). Very few mRNAs were found distinctly expressed only by the most highly active eTregs, CD62Llo CD69+ CD103+ (Fig. 3D, region 3). A notable mRNA in this group is Pparg, which is important for tissue-residing Tregs (31). Collectively, these data support the existence of 3 main eTreg subsets: a transitional less activated CD62Llo CD69− CD103− subset, and more highly activated CD62Llo CD69+ and CD62Llo CD103+ subsets.
TCR repertoire relationship between phenotypically distinct eTreg subsets
Deep sequencing was performed for TCRβ for Treg subsets isolated based on distinctive expression of CD62L, CD69 and CD103 from WT mice. DNA was isolated from approximately 4 × 105 cells (range 2–6 × 105). The total number of sequence reads (~275,000–350,000), the number of unique sequences (~16,000–32,000), and the repertoire diversity (~0.04–0.09), where 1 equals monoclonal, and sequence coverage (mean 10.8; range 7.0–15.4) were similar for all samples. No skewing of V genes and J regions were noted between these subsets.
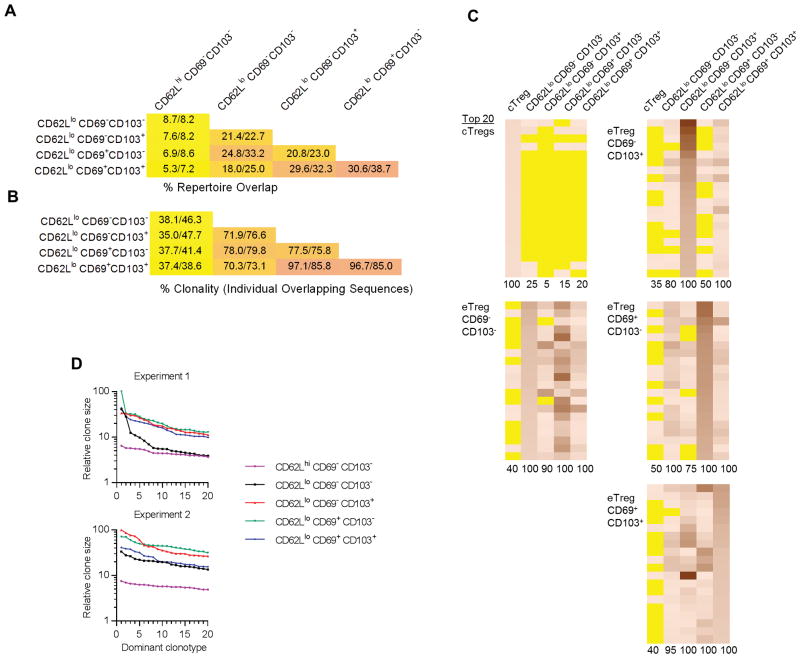
Comparing the deduced TCRβ protein V-region sequences, including the V gene and CDR3s, in the entire repertoire between all samples showed that the 4 eTreg subsets were more related to each other than cTregs (Fig. 4A). In accordance with the mRNA expression analysis (Fig. 3D), the highest relationship of TCRβ repertoires was noted between CD62Llo CD69+ CD103− and CD62Llo CD69+ CD103+ subsets and then between CD62Llo CD69− CD103+ and CD62Llo CD69+ CD103+ subsets (Fig. 4A). The clonality of these shared sequences, i.e. clonotypes encoded by a single DNA sequence, followed a similar hierarchy with the lowest clonality for cTregs and the highest for CD62Llo CD69+ CD103−, CD62Llo CD69− CD103+, and CD62Llo CD69+ CD103+ eTregs (Fig. 4B). Thus, these data show that these eTreg subsets are related to each other, with relatedness and clonality increasing for Tregs that express a more activated phenotype.
Figure 4.
The relationship of the TCR repertoire between eTreg subsets from normal C57BL/6 mice. The TCR repertoire was determined for the indicated Treg subsets from 2 independent experiments and analyzed as described in the legend to Fig. 2. (A–B) Characterization of repertoire overlap. (A) Repertoire overlap based on the total number of shared productive VDJ protein sequences. (B) Clonality (nucleotide/amino acid VDJ sequences) of individual shared unique productive sequences. The numbers within each box represent the % overlap from each experiment. (C–D) Comparison of the dominant clonotypes. (C) The frequency of the 20 most dominant clonotypes (protein VDJ) was determined for each subset from 1 of 2 experiments with similar results. Heat maps were constructed where yellow indicates the absence of a particular clonotype whereas the presence of a clonotype is quantitatively represented by increased intensity of the orange-like color. (D) The relative clone size of the 20 most dominant TCRβ VDJ sequences within each Treg subset, where the number of sequences reads for the most prevalent clonotype was normalized to a value of 100 and all other sequence reads were proportionally reduced in comparison to this sequence.
To further examine the relationship among Treg subsets, the 20 most abundant clonotypes within each subset were identified and evaluated for the extent to which they appeared in other Treg subsets. Heat maps, where yellow represents the absence of a specificity, revealed that the dominant clonotypes associated with cTregs were largely absent in the other 4 Treg subsets whereas the dominant specificities for the more activated Tregs were largely absent from the CD62Lhi Tregs (Fig. 4C). Substantial overlap in individual clonotypes was found between the 4 eTreg subsets. As the most dominant sequences likely represent TCR specificities associated with clonal expansion, this finding provides further evidence supporting a unique composition of some of the Treg specificities within the CD62Lhi cTregs. The relatively high relatedness and clonality of TCRβ clonotypes for the CD62Llo CD69+ CD103−, CD62Llo CD69− CD103+, and CD62Llo CD69+ CD103+ and their high presence within all CD62Llo subsets (Fig. 4A–C) is also consistent with highly inter-related cell populations that have undergone clonal expansion.
To estimate which Treg subsets underwent the greatest clonal expansion we plotted the numbers of sequence reads for the 20 top clonotypes for each Treg subset (Fig. 4D). The relative clone sizes were the largest for the CD62Llo CD69+ CD103−, CD62Llo CD69− CD103+, and CD62Llo CD69+ CD103+ Treg subsets whereas lower clonal expansion is noted for CD62Llo CD69− CD103− Tregs. Thus, the highly activated phenotype of these cells correlates with a potential for high clonal expansion in response to self-antigens.
Ly-6C− cTregs more readily develop into eTregs
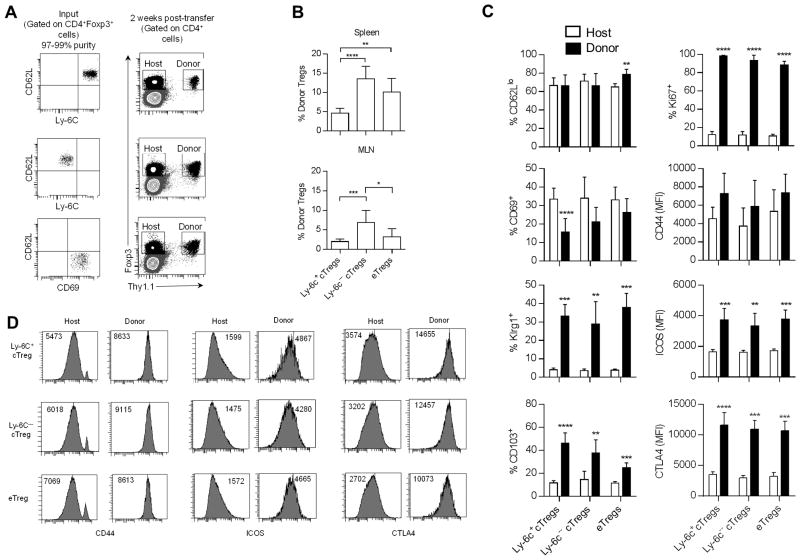
The distinctive repertoire of cTregs vs. eTreg subsets raised the question of the overall capacity of cTregs to generate eTregs. To test this point, highly purified Ly-6C+ and Ly-6C− cTregs and CD62Llo CD69+ eTregs were transferred to IL-2RβY3 (Y3) mice (Fig. 5A) and their development into eTregs was assessed 2 weeks post-transfer. Y3 mice express a mutant IL-2Rβ in T lineage cells, where 3 critical tyrosine residues in the cytoplasmic tail were mutated to phenylalanine. The low IL-2R signaling by the mutant IL-2Rβ is sufficient to support largely normal Treg development and homeostasis (25). Engraftment of donor WT Tregs in lympho-replete Y3 mice is much more extensive when compared to WT recipients, probably due to favorable competition for IL-2 and permits following the development of small numbers (1 × 105) of donor Treg subsets.
Figure 5.
Ly-6C− cTregs more efficiently develop into eTregs. CD62Lhi Ly-6C+ (n=8) and Ly-6C− cTregs (n=5) and CD62Llo CD69+ eTregs (n=4) were FACS purified from spleen cells obtained from Thy-1.1 Foxp3/RFP-reporter mice. 1 × 105 Tregs were adoptively transferred to Y3 mice. Two weeks later the engraftment of Thy-1.1+ Tregs was assessed for the spleen and MLN. (A) Representative dot plots of the purified donor Treg subsets (left) and of the overall level of donor Tregs in the spleens of Y3 mice (right). (B) Evaluation of the proportion of donor Tregs in total Tregs. The x-axis refers to the input donor Treg subset. Data were analyzed by one way ANOVA using Tukey’s multiple comparison test. (C) Phenotype of the host and donor Tregs and (D) representative histograms. The x-axis in (C) refers to the input donor Treg subset. Data were analyzed by an unpaired two-way t-test.
Two weeks post-transfer, donor Tregs were noted in the spleen (Fig. 5B) and MLN of IL-Y3 mice. This analysis revealed that the large majority (>95%) of the donor cells expressed Foxp3 (not shown), indicating that donor Tregs were stable. Quantitative assessment indicated greater engraftment by Ly-6C− cTregs when compared to Ly-6C+ cTregs. Phenotypic analysis (Fig. 5C) is consistent with essentially all donor cTregs developing into eTregs. Notably, donor Tregs showed uniformly high expression of CD44, ICOS, and CTLA4, a characteristic phenotype of eTregs (Fig. 5D).
Most donor Tregs were actively proliferating based on Ki67 expression, and this likely accounts for the increase in Klrg1, which marks Tregs that have undergone extensive proliferation and terminal differentiation. The heterogeneous expression of CD62L, CD69, and CD103 by the donor cells, regardless whether the input Tregs were derived from a cTreg or an eTreg, suggests that the expression of these molecules is under the control of environmental cues. Somewhat surprisingly, Ly-6C+ and Ly-6C− donor Tregs yielded increased development into CD103+ Tregs when compared to recipients that received CD62Llo CD69+ eTregs. This finding suggests that the programming of cTregs and eTregs is distinctive with respect to the subtypes of eTregs that are supported. Overall, these data indicate that within the environment of IL-2RβY3 recipients, at least some cTregs develop into eTregs and that eTreg appear not to revert into cTregs.
Some Ly-6C+ cTregs cannot develop into eTregs
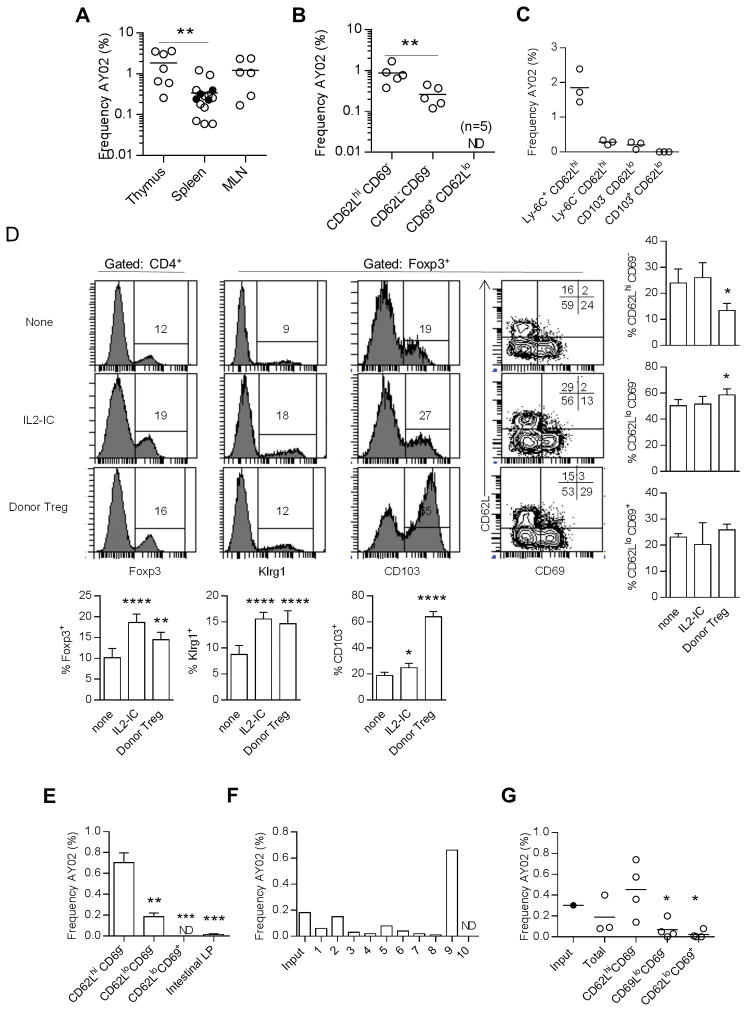
A public TCRα clonotypes was identified for peripheral Tregs from a TCRβ transgenic mouse line and after their transfer into IL-2Rβ−/− mice (32). A TaqMan RT-PCR assay was developed to quantify this clonotype, designated AY02, in Tregs (33) using the scheme and primers and probes as shown in (Supplemental Fig. 1A and 1B). This TaqMan assay is specific for AY02 as a PCR product was not detected when 6 other clonotypes were assayed, some with CDR3s closely related to the AY02 clonotype (Supplemental Fig. 1C). This TaqMan assay accurately measured the frequency of the AY02 clonotype in the context of irrelevant Vα2 TCRs except in a large excess of an irrelevant V region that also contained Jα23, which is also utilized by AY02 (Supplemental Fig. 1D), which is likely due to primer competition. This condition is not expected when assessing the AY02 frequency in a polyclonal Vα2 TCR repertoire because the Jα23 containing TCRs will not be dominant.
The frequency of the AY02 clonotype was determined for unfractionated Tregs and cTreg and eTreg subsets, using standard curves established with plasmid DNA containing the AY02 clonotype (Supplemental Fig. 1E). The AY02 clonotype was detected in Tregs from the thymus, spleen, and mesenteric lymph node (MLN) (Fig. 6A) at a frequency usually between ~0.2–2% of the total Tregs. Although we typically used Tg+ TCRα+/+ mice, the potential for bi-allelic expression of AY02 did not obviously affect our determination of AY02 because this clonotype was detected at an equivalent frequency for splenic Tregs from Tg+ TCRα+/− mice (Fig. 6A). The detection of AY02+ Tregs in the thymus indicates that this clonotype marks Tregs of thymic origin. AY02 was not detected in thymic (n=5) or peripheral (n=8) CD4+ Foxp3− T cells (not shown), indicating that this specificity is unique to Tregs. After fractionation into Treg subsets, the AY02 clonotype was most evident in CD62Lhi CD69− cTregs, but was also detected at a significantly lower amount in CD62Llo CD69− eTregs; AY02 was not detected in CD62Llo CD69+ eTregs (Fig. 6B). When cTregs were fractionated into Ly-6C+ and Ly-6C− cTregs and CD103+ and CD103− eTregs, AY02 was highly associated only with Ly-6C+ cTregs (Fig. 6C). Thus, in the steady state, Tregs expressing the AY02 clonotype are largely restricted to Ly-6C+ cTregs while being largely absent from CD69+ and/or CD103+ eTregs.
Figure 6.
A common TCRβ clonotype is absent from highly activated eTregs. Tregs were FACS purified from the indicated populations of total Tregs or Treg subsets from TCRβ transgenic Foxp3-reporter mice and quantitative RT-PCR was performed using the TaqMan assay as shown in Fig. S1. (A–C)The frequency of the AY02 clonotype was determined for (A) the indicated tissue or (B,C) within the indicated Treg subset using Foxp3/RFP-reporter mice. Horizontal lines in each graph represent the mean. Data were analyzed by one-way ANOVA, using Tukey’s multiple comparison test. ND refers to not detected. (D–G) Effect of IL-2 or homeostatic expansion in IL-2Rβ−/− mice on the AY02 clonotype. Normal TCRβ TCR transgenic Foxp3/RFP mice received 3 daily injections of IL-IC and the mice were analyzed 3 days later. In other experiments purified TCRβ transgenic Tregs were adoptively transferred into neonatal IL-2Rβ−/− recipients and the donor Tregs were assessed 10 to 19 weeks post-transfer. (D) Enumeration of Foxp3+ Tregs and their phenotype. Representative FACS plots and quantitative data for all mice are shown in the bar graphs. Data were analyzed by one-way ANOVA, using Tukey’s multiple comparison test. Significant differences are in relationship to normal untreated (none) B6 mice. (E) The frequency of the AY02 clonotype was determined for splenic Treg subsets or total Tregs from the LP of the SI and LI from 2 independent pools (n=3 to 4/pool) of IL2-IC treated Foxp3-RFP reporter mice. Data are the mean ± range and were analyzed by one-way ANOVA, using Tukey’s multiple comparison test. Significant differences are in relationship to CD62Lhi CD69− cTregs. ND= not detected. (F,G) For purified Foxp3/GFP donor Tregs from IL-2Rβ−/− recipients, the frequency of the AY02 clonotype was determined from individual recipients (F) or from the indicated donor Treg subsets (G), typically from pools of two mice/determination. The horizontal line in the graphs represents the mean. Data in (G) were analyzed by one-way ANOVA, using Tukey’s multiple comparison test. Significant differences were compared in reference to the CD62Lhi CD69− subset. ND=not detected.
The fortuitous detection of the AY02 specificity in Ly-6C+ cTregs provided an opportunity to test whether conditions that support extensive Treg proliferation and activation drove the development of AY02-expressing cTregs into eTregs. One approach was to treat mice with an agonist complex of IL-2 bound to anti-IL-2 (IL2-IC) to enhance the proliferation of Tregs and increase eTregs as assessed by expression of CD103 and Klrg1 (Fig. 6D). When the distribution of the AY02 clonotype in IL2-IC treated mice was assessed in Treg subsets based on expression of CD62L and CD69, the AY02 clonotype remained highly preferentially associated with the CD62Lhi CD69− subpopulation (Fig. 6E).
The detection of AY02+ Tregs in the MLN (Fig. 6A) raised the possibility that eTregs with this clonotype might be readily found in the intestinal lamina propria (LP). The expansion of Tregs by IL2-IC facilitated the isolation of sufficient numbers of Tregs to test this point. However, the intestinal lamina propria contained very few AY02+ Tregs (Fig. 6E), providing additional data that this Treg specificity is not substantially associated with tissue-associated activated eTregs.
Another approach was to adoptively transfer TCRβ transgenic Tregs into neonatal IL-2Rβ−/− mice, which promotes narrowing and reshaping of the donor Treg TCR repertoire (32) and more effectively promoted the development into eTregs (Fig. 6D). The AY02 clonotype was detected in donor TCRβ transgenic Tregs from 9 of 10 individual IL-2Rβ−/− recipients (Fig. 6F), in spite of repertoire reshaping. The typically lower frequency of AY02 when compared to input Tregs might reflect some tendency to select against this specificity or might reflect that Treg activation and proliferation favors CD62Llo CD103+ eTregs and that AY02+ Tregs were excluded from this Treg subset. To assess this possibility, we examined the distribution of the AY02 clonotype among subsets of engrafted donor Tregs based on expression of CD62L and CD69 (Fig. 6G). Again, the AY02 clonotype was largely restricted to the CD62Lhi CD69− subsets with a frequency similar to the input Tregs. Collectively, these data indicate that CD62Lhi Tregs do not simply represent a pool of cells that ultimately give rise to eTregs but may also contain a population of Tregs whose specificities remain largely restricted to cTregs, perhaps with optimal activity in lymphoid tissues.
eTregs derived from cTregs do not completely suppress autoimmunity in IL-2Rβ−/− mice
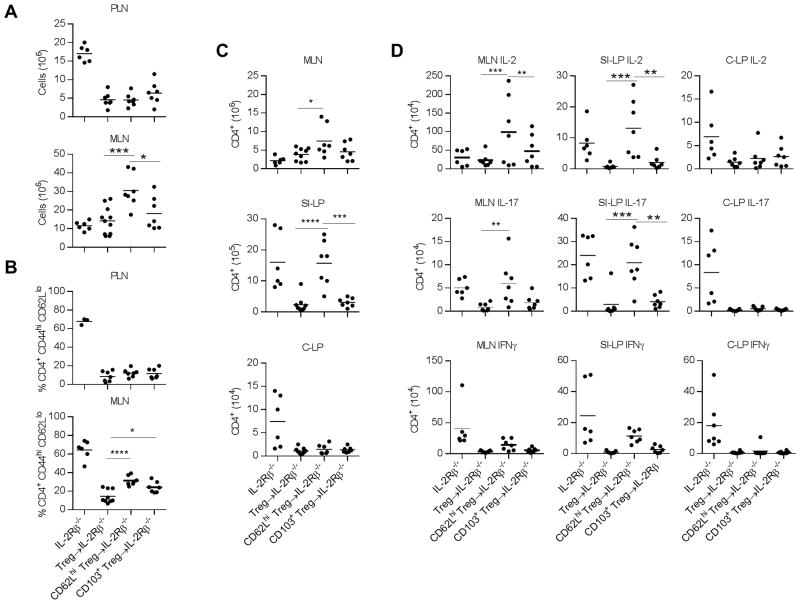
To test the suppressive activity of cTregs and eTregs, we compared the capacity of purified CD62Lhi CD69− CD103− Klrg1− cTregs to CD62Llo CD103+ eTregs to prevent autoimmunity after transfer into neonatal IL-2Rβ−/− mice. Controls included untreated IL-2Rβ−/− mice and IL-2Rβ−/− mice that received either total purified Tregs or unfractionated spleen cells as a source of total Tregs. Donor Tregs readily engrafted and persisted within the PLN, MLN, as well as lamina propria of the small intestine (SI-LP) and colon (C-LP) of IL-2Rβ−/− recipients at largely equivalent levels (Supplemental Fig. 3A). For the PLN and MLN, an increased frequency of Tregs was noted for recipients that received cTregs, consistent with some preference for lymphoid tissues. However, the transferred cTregs readily developed into CD62Llo CD103+ eTregs that were present at only slightly lower levels than occurred for recipients of CD103+ eTregs (Supplemental Fig. 3B). Thus, these data further confirm that some cTregs are precursors to eTregs and that the resulting eTregs adopted a similar phenotype in comparison to mice that received purified eTregs.
Unlike untreated IL-2Rβ−/− mice develop rapid systemic autoimmunity that leads to a wasting syndrome and inflammation in multiple tissues, particularly the lung, liver, and intestines, that typically leads to death by 8–12 weeks of age. Neonatal IL-2Rβ−/− mice that were adoptively transferred with CD62Lhi cTregs or CD103+ eTregs were outwardly normal without evidence of wasting when analyzed 10–12 week post-transfer. Therefore, we focused on early changed associated with autoimmunity in IL-2Rβ−/− mice, i.e. increased lymph node cellularity and increased proportion of activated T cells. With respect to suppression of these autoimmune symptoms, lymphoproliferation in the PLN and a high frequency of CD4+ CD44hi CD62Llo T effector cells (Teff) in the PLN and MLN was not seen for recipients that received cTregs or CD103+ eTregs (Fig. 7A, B). However, increased cellularity in the MLN was noted for IL-2Rβ−/− recipients that received cTregs (Fig. 7A). This increase was much greater than what was seen for untreated IL-2Rβ−/− mice. This result likely reflects the age difference at the time of assay, 4–6 weeks for the untreated IL-2Rβ−/− mice and 10–12 weeks for the Tregs transferred mice, as the former, due to severe autoimmunity, must be tested by this age.
Figure 7.
Control of autoimmunity associated with IL-2Rβ−/− mice after adoptive transfer of Treg subsets. CD62Lhi CD69− CD103− Klrg1− (designed as CD62Lhi) and CD62Llo CD103+ (designated as CD103+) Tregs were purified (generally >95% pure) using Foxp3/RFP reporter mice and adoptively transferred into neonatal IL-2Rβ−/− recipients. These recipients were evaluated 10–12 weeks post-transfer. As a control, some recipients were adoptively transferred with either purified total Tregs or unfractionated spleen cells as a source of total Tregs and the results from these recipients were combined and shown as Treg→IL-2Rβ−/−. Untreated IL-2Rβ−/− mice were also evaluated but at 4–6 weeks of age due to the severity of the autoimmune disease. (A–D) The (A) cellularity, (B) proportion of conventional CD4+ T cells with an activated CD44hi CD62Llo phenotype, (C) number of CD4+ T cells, and (D) cytokine producing CD4+ T cells were determined for the indicated tissues of untreated or Treg-transferred IL-2Rβ−/− mice. Shown are the results from individual mice where a horizontal line within each graph represents the mean. Data were analyzed by one-way ANOVA, using Tukey’s multiple comparison test. Significant differences are only shown for the comparisons between mice receiving donor Tregs.
The increased MLN cellularity within cTreg-treated IL-2Rβ−/− mice raised the possibility that that this Treg subset is less effective in controlling tolerance within the gut mucosa. Indeed, the MLN from these mice showed a slight increased numbers of CD4+ T cells (Fig. 7C), including IL-2- and IL-17-secreting Teff cells (Fig. 7D). Furthermore, when comparing the cTreg vs. CD103+ eTreg transferred IL-2Rβ−/− recipients, the SI-LP, but not C-LP, of the cTreg-treated mice showed a significant increase in the total number of CD4+ T cells (Fig. 7C) and IL-2 and IL-17 producing Teff cells (Fig. 7D). Thus, eTregs derived from cTregs are less effective in maintaining immune homeostasis in the SI-LP.
Discussion
Tregs from secondary lymphoid tissues, which are dominated by thymic-derived Tregs, are highly heterogeneous cells that are classified into two major subsets, cTregs and eTregs, based on their distinctive phenotype, homeostasis, anatomical preference, and transcriptional regulation (5–8, 12, 13). This study refines this view by showing that there are distinctive subpopulations of cTregs and eTregs and by directly considering the inter-relationships between these subsets. These data support the view that there are two main populations of cTregs that are distinguished by expression of Ly-6C and three subpopulations of eTregs that are distinguished by their distinctive expression of CD62L, CD103, and CD69. These three eTreg subsets are distinct from a fourth terminal short-lived eTreg subpopulation, marked by expression of Klrg1 (14).
Extensive immune profiling and TCR repertoire analysis demonstrates that Ly-6C+ and Ly-6C− cTregs are related to each other but represent distinctive cTreg subsets. Many immunological properties are similarly exhibited by these two subsets. However, a set of immune-related mRNAs were identified that show expression by Ly-6C− cTregs moving toward eTregs. On average Ly-6C− Tregs express TCRs with a higher affinity for self-antigens, based on higher expression of CD5, which reflects the intrinsic affinity of the TCR for positively selected self-antigens (30). This greater responsiveness to self-antigens provides one condition to promote a somewhat more activated immune profile by Ly-6C− cTregs. Another difference between Ly-6C+ and Ly-6C− cTreg subsets is that at the steady state the Ly-6C− Tregs showed greater proliferation as measured by Ki67 expression. This increase correlated with heightened expression of CD25 that would promote responsiveness to IL-2. Indeed, the homeostasis of cTregs has been reported to be largely dependent on IL-2 (21). Our findings suggest that IL-2-dependent homeostasis of cTregs primarily reflects the action of IL-2 on Ly-6C− cTregs.
One function of Ly-6C+ and Ly-6C− cTregs is to provide a pool of “naïve” Tregs that become activated into CD62Llo eTregs. This conclusion is based on following the fate of cTregs after transfer into Y3 or IL-2Rβ−/− mice and shared TCR repertoires between cTreg subsets and eTregs. This latter result is in line with other studies that have found the same TCR clonotype in resting and activated Tregs (34–37). However, our study shows that Ly-6C+ cTregs are less efficient in developing into eTregs when directly transferred in vivo. Furthermore, the dominant TCR Vβ clonotypes of Ly-6C+ cTregs were detected at a low frequency in eTregs and reciprocally the prevalent TCR Vβ clonotypes of eTregs were also at a low frequency in Ly-6C+ cTregs. In comparison to eTregs, the dominant TCR Vβs of Ly-6C+ cTregs were more often found in Ly-6C− cTregs. Thus, expression of Ly-6C marks a quiescent and relatively unactivated population of Tregs with lower affinity TCRs that have a greater tendency to develop into Ly-6C− cTregs rather than into eTregs. Thus, one simple interpretation of our data is that cTregs serve primarily as precursors of eTregs and the latter mediate suppressive function required for self-tolerance.
Several recent studies have shown that eTregs are more suppressive than cTregs, including Ly-6C+ Tregs (7, 9, 14). Consistent with this view, the transfer of CD103+ eTregs more fully protected IL-2Rβ−/− mice from autoimmunity than recipients of cTregs. This result was somewhat surprising since in this model cTregs readily gave rise to eTregs, and full protection was expected. IL-2Rβ−/− mice that received cTregs were effectively protected from systemic autoimmunity and colitis, but they showed symptoms consistent with inflammation in the lamina propria of the small intestine. Thus, greater suppressive activity of the CD103+ eTregs may not fully explain their high efficacy. The effective control of autoimmunity associated with IL-2Rβ−/− mice that received donor eTregs may also be the result of distinctive TCR repertoires of cTregs and eTregs, whereby cTregs lack TCR specificities for some tissue-associated antigens while the TCR repertoires of eTregs were already selected to be more responsive to such antigens. Nevertheless, we cannot rule out that the initial distinctive immune properties of the donor cTregs may also contribute to inflammation in the small intestine of IL-2Rβ−/− recipients, even though they developed into eTregs. In particular, the inefficient development of eTregs from Ly-6C+ Tregs within the donor cTregs may have effectively lowered the number of Tregs available to suppress disease relative to those mice that received the CD103+ eTregs.
Beside their role of precursors to eTregs, some of our findings also raise the possibility of a more active role of cTregs, including Ly-6C+ cTregs, in maintaining self-tolerance. In particular, the public AY02 clonotype, which was identified as largely restricted to Ly-6C+ cTregs, did not substantially shift toward more highly activated eTregs, even when Tregs expressing this clonotype were placed under conditions that support substantial proliferation and activation, i.e. stimulation with IL2-IC or transfer into IL-2Rβ-deficient mice. The detection of the AY02 in the latter situation suggests that AY02 may be a useful Treg clonotype, as this specificity persisted under conditions where substantial reshaping of the Treg TCR repertoire occurs to suppress autoreactive T cells (32), i.e. adoptive transfer of Tregs into IL-2Rβ−/− recipients. Given the lower affinity of Ly-6C+ Tregs and the inability to drive AY02 into highly activated Tregs, AY02+ Tregs were not expected to be maintained at a relatively high frequency by cTregs, if these Tregs did not promote immune tolerance. In addition, the public AY02 clonotype was readily found in Tregs within the spleen and especially in the MLN, but was poorly detected in the lamina propria of the small intestine and colon. This finding markedly differs from past work where Treg specificities associated with a tissue site were predominately found in the lymph nodes that drain that tissue (19) and suggests that some Tregs prefer to persist within lymphoid tissue.
We propose that CD62Lhi cTregs may also function to directly suppress autoreactive T cells within secondary lymphoid tissues. The distinctive immune profiles and TCR repertoire of cTregs raise the possibility that these Tregs utilize a distinctive mechanism to maintain tolerance, perhaps based on the action of TGFβ and CTLA4, whose mRNAs are abundant in these cells. The action of these cells may be particularly important under homeostatic non-autoimmune conditions when the clonal frequency of many autoreactive T cells is expected to be low and may be effectively regulated by individual cTregs at low frequencies. Once the immune system is perturbed in a manner that causes particular autoreactive T cells to become more abundant, some cTregs develop into eTregs to try to contain their activity in lymphoid tissues as well as the target tissues that express the relevant autoantigens.
Our immune profiling and TCR repertoire analysis identified 3 main inter-related subsets of eTregs, i.e. CD62Llo CD69− eTregs, with an immune profile more closely related to cTregs, and more activated CD62Llo CD69+ and CD62Llo CD103+ eTregs. Each subset showed a preference for expression of particular mRNAs. However, these expression patterns do not support the existence of mutually exclusive subsets, but a preference for somewhat distinctive gene expression most likely based on environmental or niche signaling received by the eTregs. For example, CD62Llo CD69− eTregs generally showed low expression of many mRNAs related to T cell activation, suppressive function, and tissue homing. Nevertheless, CD62Llo CD69− CD103+ eTregs expressed some features of a more activated Treg. Levels of other mRNAs showed a gradient of expression from low to high in comparison to cTregs across all the eTreg subsets. Collectively, these data imply two concurrent process are ongoing to shape the eTreg compartment. One is moving from a lower (cTreg) to a progressively higher (eTreg) state of activation that may be dependent on a common set of external stimuli. TCR signaling and transcriptional regulation by IRF4 represent required signals for production of eTregs (6, 7, 13). These and other niche-related processes then may act in concert to shape the distinctive immune profiles of eTregs subsets. For example, recent TCR signaling and type-1 interferons favor expression of CD69+ (38) and TGFβ upregulates CD103 (39). A clear interrelationship among these eTreg subsets is illustrated by the high degree of overlap of their TCR Vβ repertoires and the dominant clonotypes. This finding is analogous to that reported for Teff subsets (40).
Our data support a model whereby at least some eTregs are derived from cTregs. One key step is the development of CD62Llo CD69− CD103− eTregs from either Ly-6C+ or Ly-6C− cTregs. CD62Llo CD69− CD103− eTregs might be considered a key transitional step where depending upon the strength of TCR signaling and other niche signals, they develop into CD69+ and/or CD103+ eTregs. Highly activated eTregs are more likely to be derived from Ly-6C− cTregs due to their higher affinity TCRs. Due to the lower affinity of the TCRs expressed by Ly-6C+ Tregs, they are less apt to develop into highly activated Tregs and instead their development may sometimes aborted as Ly-6C− cTregs or transitional CD62Llo CD69− CD103− eTregs, as seen for Tregs expressing the AY02 clonotype. Furthermore, when Ly-6C+ or Ly-6C− wild-type cTregs were transferred into Y3 mice, these Tregs uniformly developed into eTregs, but some still expressed high levels of CD62L. Thus, some eTregs, perhaps at the transitional eTregs stage, may support the re-expression of CD62L to promote traffic of some eTregs into secondary lymphoid tissue.
Supplementary Material
Acknowledgments
This work was supported by National Institutes of Health Grants R01 AI055815 and R01 DK093866.
We thank the flow cytometry cores of the Sylvester Comprehensive Cancer Center and the Diabetes Research Institute at the University of Miami for help with FACS analysis and sorting.
Abbreviations used in this article
- C-LP
colon lamina propria
- cTreg
central regulatory T cell
- eTreg
effector regulatory T cell
- IL2-IC
IL-2/anti-IL2 agonist complexes
- MLN
mesenteric lymph node
- PLN
peripheral lymph mode
- SI-LP
small intestine lamina propria
- Teff cell
T effector
- Treg
regulatory T cell
Footnotes
Disclosures
The authors declare no competing financial interests
References
- 1.Ohkura N, Kitagawa Y, Sakaguchi S. Development and maintenance of regulatory T cells. Immunity. 2013;38:414–423. doi: 10.1016/j.immuni.2013.03.002. [DOI] [PubMed] [Google Scholar]
- 2.Josefowicz SZ, Lu LF, Rudensky AY. Regulatory T cells: mechanisms of differentiation and function. Annu Rev Immunol. 2012;30:531–564. doi: 10.1146/annurev.immunol.25.022106.141623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shevach EM. From vanilla to 28 flavors: multiple varieties of T regulatory cells. Immunity. 2006;25:195–201. doi: 10.1016/j.immuni.2006.08.003. [DOI] [PubMed] [Google Scholar]
- 4.Yuan X, Cheng G, Malek TR. The importance of regulatory T-cell heterogeneity in maintaining self-tolerance. Immunol Rev. 2014;259:103–114. doi: 10.1111/imr.12163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rosenblum MD, I, Gratz K, Paw JS, Lee K, Marshak-Rothstein A, Abbas AK. Response to self antigen imprints regulatory memory in tissues. Nature. 2011;480:538–542. doi: 10.1038/nature10664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cretney E, Xin A, Shi W, Minnich M, Masson F, Miasari M, Belz GT, Smyth GK, Busslinger M, Nutt SL, Kallies A. The transcription factors Blimp-1 and IRF4 jointly control the differentiation and function of effector regulatory T cells. Nat Immunol. 2011;12:304–311. doi: 10.1038/ni.2006. [DOI] [PubMed] [Google Scholar]
- 7.Levine AG, Arvey A, Jin W, Rudensky AY. Continuous requirement for the TCR in regulatory T cell function. Nat Immunol. 2014;15:1070–1078. doi: 10.1038/ni.3004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Smigiel KS, Srivastava S, Stolley JM, Campbell DJ. Regulatory T-cell homeostasis: steady-state maintenance and modulation during inflammation. Immunol Rev. 2014;259:40–59. doi: 10.1111/imr.12170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Delpoux A, Yakonowsky P, Durand A, Charvet C, Valente M, Pommier A, Bonilla N, Martin B, Auffray C, Lucas B. TCR signaling events are required for maintaining CD4 regulatory T cell numbers and suppressive capacities in the periphery. J Immunol. 2014;193:5914–5923. doi: 10.4049/jimmunol.1400477. [DOI] [PubMed] [Google Scholar]
- 10.Martin B, Auffray C, Delpoux A, Pommier A, Durand A, Charvet C, Yakonowsky P, de Boysson H, Bonilla N, Audemard A, Sparwasser T, Salomon BL, Malissen B, Lucas B. Highly self-reactive naive CD4 T cells are prone to differentiate into regulatory T cells. Nat Commun. 2013;4:2209. doi: 10.1038/ncomms3209. [DOI] [PubMed] [Google Scholar]
- 11.Cretney E, Kallies A, Nutt SL. Differentiation and function of Foxp3+ effector regulatory T cells. Trends Immunol. 2013;34:74–80. doi: 10.1016/j.it.2012.11.002. [DOI] [PubMed] [Google Scholar]
- 12.Burzyn D, Benoist C, Mathis D. Regulatory T cells in nonlymphoid tissues. Nat Immunol. 2013;14:1007–1013. doi: 10.1038/ni.2683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vahl JC, Drees C, Heger K, Heink S, Fischer JC, Nedjic J, Ohkura N, Morikawa H, Poeck H, Schallenberg S, Riess D, Hein MY, Buch T, Polic B, Schonle A, Zeiser R, Schmitt-Graff A, Kretschmer K, Klein L, Korn T, Sakaguchi S, Schmidt-Supprian M. Continuous T cell receptor signals maintain a functional regulatory T cell pool. Immunity. 2014;41:722–736. doi: 10.1016/j.immuni.2014.10.012. [DOI] [PubMed] [Google Scholar]
- 14.Cheng G, Yuan X, Tsai MS, Podack ER, Yu A, Malek TR. IL-2 receptor signaling is essential for the development of Klrg1+ terminally differentiated T regulatory cells. J Immunol. 2012;189:1780–1791. doi: 10.4049/jimmunol.1103768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Feuerer M, Hill JA, Kretschmer K, von Boehmer H, Mathis D, Benoist C. Genomic definition of multiple ex vivo regulatory T cell subphenotypes. Proc Natl Acad Sci U S A. 2010;107:5919–5924. doi: 10.1073/pnas.1002006107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hsieh CS, Zheng Y, Liang Y, Fontenot JD, Rudensky AY. An intersection between the self-reactive regulatory and nonregulatory T cell receptor repertoires. Nat Immunol. 2006;7:401–410. doi: 10.1038/ni1318. [DOI] [PubMed] [Google Scholar]
- 17.Pacholczyk R, Ignatowicz H, Kraj P, Ignatowicz L. Origin and T cell receptor diversity of Foxp3+CD4+CD25+ T cells. Immunity. 2006;25:249–259. doi: 10.1016/j.immuni.2006.05.016. [DOI] [PubMed] [Google Scholar]
- 18.Wong J, Mathis D, Benoist C. TCR-based lineage tracing: no evidence for conversion of conventional into regulatory T cells in response to a natural self-antigen in pancreatic islets. J Exp Med. 2007;204:2039–2045. doi: 10.1084/jem.20070822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lathrop SK, Santacruz NA, Pham D, Luo J, Hsieh CS. Antigen-specific peripheral shaping of the natural regulatory T cell population. J Exp Med. 2008;205:3105–3117. doi: 10.1084/jem.20081359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lathrop SK, Bloom SM, Rao SM, Nutsch K, Lio CW, Santacruz N, Peterson DA, Stappenbeck TS, Hsieh CS. Peripheral education of the immune system by colonic commensal microbiota. Nature. 2011;478:250–254. doi: 10.1038/nature10434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Smigiel KS, Richards E, Srivastava S, Thomas KR, Dudda JC, Klonowski KD, Campbell DJ. CCR7 provides localized access to IL-2 and defines homeostatically distinct regulatory T cell subsets. J Exp Med. 2014;211:121–136. doi: 10.1084/jem.20131142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wan YY, Flavell RA. Identifying Foxp3-expressing suppressor T cells with a bicistronic reporter. Proc Natl Acad Sci U S A. 2005;102:5126–5131. doi: 10.1073/pnas.0501701102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fontenot JD, Rasmussen JP, Williams LM, Dooley JL, Farr AG, Rudensky AY. Regulatory T cell lineage specification by the forkhead transcription factor foxp3. Immunity. 2005;22:329–341. doi: 10.1016/j.immuni.2005.01.016. [DOI] [PubMed] [Google Scholar]
- 24.Suzuki H, Kundig TM, Furlonger C, Wakeham A, Timms E, Matsuyama T, Schmits R, Simard JJL, Ohashi PS, Griesser H, Taniguchi T, Paige CJ, Mak TW. Deregulated T cell activation and autoimmunity in mice lacking interleukin-2 receptor β. Science. 1995;268:1472–1476. doi: 10.1126/science.7770771. [DOI] [PubMed] [Google Scholar]
- 25.Yu A, Zhu L, Altman NH, Malek TR. A low interleukin-2 receptor signaling threshold supports the development and homeostasis of T regulatory cells. Immunity. 2009;30:204–217. doi: 10.1016/j.immuni.2008.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Uematsu Y, Ryser S, Dembic Z, Borgulya P, Krimpenfort P, Berns A, von Boehmer H, Steinmetz M. In transgenic mice the introduced functional T cell receptor beta gene prevents expression of endogenous β genes. Cell. 1988;52:831–841. doi: 10.1016/0092-8674(88)90425-4. [DOI] [PubMed] [Google Scholar]
- 27.Carlson CS, Emerson RO, Sherwood AM, Desmarais C, Chung MW, Parsons JM, Steen MS, LaMadrid-Herrmannsfeldt MA, Williamson DW, Livingston RJ, Wu D, Wood BL, Rieder MJ, Robins H. Using synthetic templates to design an unbiased multiplex PCR assay. Nat Commun. 2013;4:2680. doi: 10.1038/ncomms3680. [DOI] [PubMed] [Google Scholar]
- 28.Robins HS, Campregher PV, Srivastava SK, Wacher A, Turtle CJ, Kahsai O, Riddell SR, Warren EH, Carlson CS. Comprehensive assessment of T-cell receptor beta-chain diversity in alphabeta T cells. Blood. 2009;114:4099–4107. doi: 10.1182/blood-2009-04-217604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dalyot-Herman N, Bathe OF, Malek TR. Reversal of CD8+ T cell ignorance and induction of anti-tumor immunity by peptide-pulsed antigen presenting cells. J Immunol. 2000;165:6731–6737. doi: 10.4049/jimmunol.165.12.6731. [DOI] [PubMed] [Google Scholar]
- 30.Hogquist KA, Jameson SC. The self-obsession of T cells: how TCR signaling thresholds affect fate ‘decisions’ and effector function. Nat Immunol. 2014;15:815–823. doi: 10.1038/ni.2938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cipolletta D, Feuerer M, Li A, Kamei N, Lee J, Shoelson SE, Benoist C, Mathis D. PPAR-γ is a major driver of the accumulation and phenotype of adipose tissue Treg cells. Nature. 2012;486:549–553. doi: 10.1038/nature11132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Adeegbe D, Matsutani T, Yang J, Altman NH, Malek TR. CD4+ CD25+ Foxp3+ T regulatory cells with limited TCR diversity in control of autoimmunity. J Immunol. 2010;184:56–66. doi: 10.4049/jimmunol.0902379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gallard A, Foucras G, Coureau C, Guery JC. Tracking T cell clonotypes in complex T lymphocyte populations by real-time quantitative PCR using fluorogenic complementarity-determining region-3-specific probes. J Immunol Methods. 2002;270:269–280. doi: 10.1016/s0022-1759(02)00336-8. [DOI] [PubMed] [Google Scholar]
- 34.Miyara M, Yoshioka Y, Kitoh A, Shima T, Wing K, Niwa A, Parizot C, Taflin C, Heike T, Valeyre D, Mathian A, Nakahata T, Yamaguchi T, Nomura T, Ono M, Amoura Z, Gorochov G, Sakaguchi S. Functional delineation and differentiation dynamics of human CD4+ T cells expressing the FoxP3 transcription factor. Immunity. 2009;30:899–911. doi: 10.1016/j.immuni.2009.03.019. [DOI] [PubMed] [Google Scholar]
- 35.Bergot AS, Chaara W, Ruggiero E, Mariotti-Ferrandiz E, Dulauroy S, Schmidt M, von Kalle C, Six A, Klatzmann D. TCR sequences and tissue distribution discriminate the subsets of naive and activated/memory Treg cells in mice. Eur J Immunol. 2015;45:1524–1534. doi: 10.1002/eji.201445269. [DOI] [PubMed] [Google Scholar]
- 36.Lei H, Kuchenbecker L, Streitz M, Sawitzki B, Vogt K, Landwehr-Kenzel S, Millward J, Juelke K, Babel N, Neumann A, Reinke P, Volk HD. Human CD45RA− FoxP3hi memory-type regulatory T cells show distinct TCR repertoires with conventional T cells and play an important role in controlling early immune activation. Am J Transplant. 2015;15:2625–2635. doi: 10.1111/ajt.13315. [DOI] [PubMed] [Google Scholar]
- 37.Scheinberg P, Melenhorst JJ, Hill BJ, Keyvanfar K, Barrett AJ, Price DA, Douek DC. The clonal composition of human CD4+CD25+Foxp3+ cells determined by a comprehensive DNA-based multiplex PCR for TCRβ gene rearrangements. J Immunol Methods. 2007;321:107–120. doi: 10.1016/j.jim.2007.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shiow LR, Rosen DB, Brdickova N, Xu Y, An J, Lanier LL, Cyster JG, Matloubian M. CD69 acts downstream of interferon-α/β to inhibit S1P1 and lymphocyte egress from lymphoid organs. Nature. 2006;440:540–544. doi: 10.1038/nature04606. [DOI] [PubMed] [Google Scholar]
- 39.El-Asady R, Yuan R, Liu K, Wang D, Gress RE, Lucas PJ, Drachenberg CB, Hadley GA. TGFβ-dependent CD103 expression by CD8+ T cells promotes selective destruction of the host intestinal epithelium during graft-versus-host disease. J Exp Med. 2005;201:1647–1657. doi: 10.1084/jem.20041044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang C, Sanders CM, Yang Q, Schroeder HW, Jr, Wang E, Babrzadeh F, Gharizadeh B, Myers RM, Hudson JR, Jr, Davis RW, Han J. High throughput sequencing reveals a complex pattern of dynamic interrelationships among human T cell subsets. Proc Natl Acad Sci U S A. 2010;107:1518–1523. doi: 10.1073/pnas.0913939107. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.