Abstract
The incidence of methicillin-resistant Staphylococcus aureus (MRSA) pneumonia in otherwise healthy individuals is increasing. To investigate the mechanism underlying the epidemiological success of predominant community-associated (CA)-MRSA strains, we examined their fitness traits during the initial interaction between bacteria and the host occurring in the lower airway. Using a mouse respiratory infection model, we show that clinical isolates often responsible for community-associated infections are highly resistant to clearance from healthy airways, while S. aureus strains not as prevalent or traditionally associated with hospital-associated infections are relatively susceptible. Mechanistically, the competitive fitness of S. aureus is a result of both agr-dependent and -independent resistance to innate bacterial killing. Furthermore, we show that rather than evasion from neutrophil-dependent bactericidal process, the observed S. aureus fitness in the lower airways is due to its intrinsic resistance to resident alveolar macrophage-mediated intracellular killing. Importantly, we demonstrate that the virulence determinants responsible for bacterial persistence in immune-competent mice are dispensable in mice with predisposing conditions such as influenza infection. Together, these novel findings of the improved competence of predominant CA-MRSA strains to survive innate killing in healthy hosts, particularly at the very beginning stage of infection, provide a unique insight into their epidemiological success.
Introduction
Staphylococcus aureus primarily resides in the nasopharynx of healthy hosts, from where it can spread to the lower respiratory tract. Since the host innate immunity is generally competent in ensuring the sterility of lower airways, staphylococcal pneumonia is uncommon. Nonetheless, the incidence of staphylococcal pneumonia in otherwise healthy individuals is increasing, and is currently mediated primarily by community-associated methicillin-resistant S. aureus (CA-MRSA) strain USA300 (1, 2). These observations suggest that USA300, and potentially other S. aureus backgrounds or populations, are capable of evading host innate defenses and thus are more often associated with clinical infections.
Studies in animal models of pneumonia have detected multiple CA-MRSA virulence factors, such as alpha-hemolysin and phenol-soluble modulin (PSM) peptides, associated with disease severity (3, 4). Conversely, virulence determinants that are essential for bacterial survival in healthy hosts, particularly at the earliest stage of infection, have received less attention. Even fewer studies have examined host immune factors responsible for maintaining the sterility of lower airways during subclinical S. aureus infections.
In the present study, we examined the fitness traits of numerous S. aureus strains during the initial interaction between bacteria and the host occurring in the lower airway, and investigated the hypothesis that prevalent CA-MRSA isolates, such as USA300 and USA400, have improved ability to survive host innate immune defenses at the very beginning stage of infection. Indeed, we found that clinical isolates often responsible for community-associated infections are highly resistant to initial bacterial killing mechanisms in the healthy airways, while S. aureus strains not as prevalent or traditionally associated with hospital-associated infections are relatively susceptible. Further, we show that rather than evasion from neutrophil-dependent bactericidal process, the enhanced fitness of S. aureus in the lower airways is a result of its intrinsic resistance to resident alveolar macrophage-mediated intracellular killing. Importantly, we demonstrate that these virulence determinants responsible for bacterial persistence in immune-competent mice are dispensable in hosts with predisposing conditions such as influenza infection.
Materials and Methods
Murine model of bacterial infection
Specific pathogen-free, 6-8 week old C57BL/6 mice were purchased from Charles River Laboratories (Wilmington, MA). Specific pathogen-free, C57BL/6 wild-type, p47phox−/−, Nox2−/− (female) and Nox2−/y (male) mice were initially purchased from the Jackson Laboratory and bred at University of Nebraska medical center following IACUC guidelines.
For bacterial challenge, 8-12 week old, sex and age-matched mice were anesthetized, infected intranasally (i.n.) with 50 μl of PBS containing a sublethal or lethal dose of S. aureus from overnight cultures. Bacterial burdens in the lungs and bronchoalveolar lavage fluids (BALFs) were measured by sacrificing infected mice at the indicated time points, and plating serial 10-fold dilutions of each sample onto blood agar plates. The plates were incubated at 37°C overnight, and CFUs were enumerated 24 h later. To induce post-influenza S. aureus pneumonia, mice were challenged with 50 PFU/mouse A/PR/8 virus seven days prior to the bacterial infection (5). Viral titers were determined by plaque assays on MDCK cell monolayers.
Plasmids expressing DsRed and GFP
The plasmids pCM29 (6) and pVT1 were transduced into BAA-1695 and LAC-JE2 using φ11 phage. For construction of plasmid pVT1, a 5.86kb vector backbone devoid of sGFP gene was initially PCR amplified from pCM29 using primers pCM29-F (5’-GAATTCGTAATCATGTCATAGC-3’) and pCM29-R (5’-AAATAATCATCCTCCTAAGGTAC-3’). Similarly, the optimized DsRed insert (677bp) was PCR amplified from plasmid pDM4 (7) using primers DsREDopt-F (5’-CTTAGGAGGATGATTATTTATGGATAATACAGAAGATGTTATTAAAG-3’) and DsREDopt-R (5’-TGACATGATTACGAATTCTTATAAAAACAAATGATGACGACC-3’). The two fragments were assembled using the NEBuilder high-fidelity DNA assembly cloning kit according to the manufacturer’s instructions and the resulting plasmid (pVT1) was electroporated into E. coli ElectroTen-Blue (Stratagene). Clones were verified by colony PCR and sequencing.
Bronchoalveolar lavage (BAL) cell analysis
BALFs were collected by making a longitudinal incision on the ventral side of the neck exposing the trachea and lavaging the lung twice with 0.8 ml PBS, pH 7.4. Total leukocyte counts were determined using a hemocytometer.
For flow cytometric analysis, BAL cells were incubated with 2.4G2 mAb against FcγRII/III, and stained with APC-conjugated anti-CD11c (Caltag Laboratories), BUV395-, PE-, or FITC-conjugated anti-CD11b (BD Biosciences), FITC-, PE- or PE-Cy7-conjugated anti-Ly6G mAb (Clone 1A8, BD Biosciences), PerCP-Cy5.5-conjugated anti-Ly6C mAb (Biolegend), and BV421-conjugated anti-Siglec-F (BD Biosciences). The stained cells were analyzed on a BD LSRII-green using BD FACSDiva and FlowJo software analysis.
Determination of TNF-α production by ELISA
BALFs and lung homogenates were harvested and assayed for TNF-α by ELISA using commercially available kits from BD Biosciences.
Neutrophil depletion
Neutrophils are depleted using anti-Gr-1 mAb RB6-8C5 (BioXCell). Specifically, at 24 h before bacterial infection, naïve C57BL/6 mice were injected i.p. with 0.1 mg of anti-Gr-1 mAb to deplete neutrophils or with rat IgG as a control. The efficiency of neutrophil depletion in bacterial-infected mice was confirmed by flow cytometry (5).
Alveolar macrophage depletion
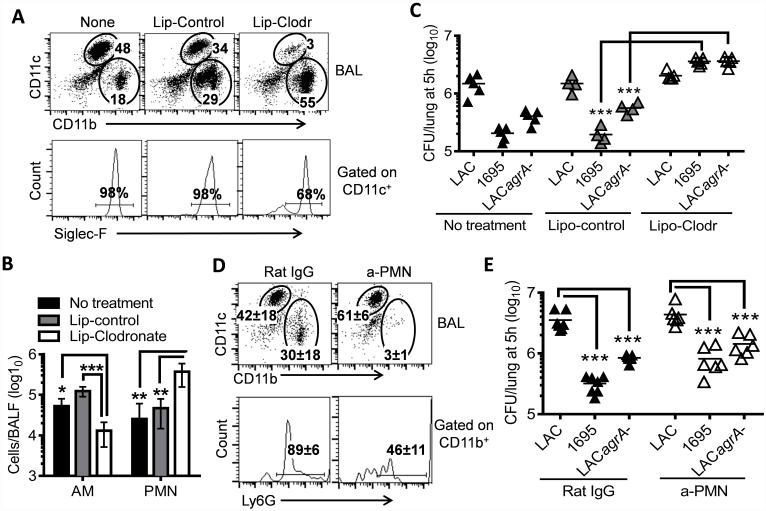
Alveolar macrophages are depleted through liposome-clodronate (Encapsula NanoSciences) treatment (8). Specifically, at 48h before bacterial infection, mice were given i.n. with 50 μl liposome-clodronate or with liposome-PBS as a control. The efficiency of pulmonary macrophage depletion was confirmed by flow cytometry (Fig. 5A-B).
Figure 5. Alveolar macrophages are responsible for initial bacterial killing in the lower airways.
(A) Inflammatory cell profiles, (B) numbers of alveolar macrophages (AM) and neutrophils (PMN) in the airways (4~5 mice/group), and (C) numbers of bacteria in the lungs 5 h after infection of naïve, liposome-control or liposome-clodronate treated C57BL/6 mice with 107 CFU of LAC-JE2 WT, agrA mutant, and 1695 in a 1:1:1 mixture. Numbers (mean of 4~5 mice/group) shown in (A) represent percent of indicated populations in the respective gates. Data shown are representative of two independent experiments. (D) Airway inflammatory cell profiles and (E) numbers of bacteria in the lungs 5 h after infection of α-PMN antibody-treated mice with 107 CFU of LAC-JE2 WT, agrA mutant and 1695 in a 1:1:1 mixture. Control mice were treated with Rat IgG. Data shown were combined from two independent experiments. *P< 0.05, **P< 0.01 and ***P< 0.001, Tukey's multiple comparisons test.
In vitro MH-S cell phagocytosis assay
The MH-S murine alveolar macrophage cell line was grown for 24 h until confluence. GFP-expressing 1695 or LAC-JE2 from overnight cultures were harvested and resuspended in RPMI with 10% FBS. The cells were inoculated with S. aureus at multiplicity of infection (MOI) 10 and co-incubated for 60 min on ice, allowing bacteria binding onto the cells. Thereafter, the cells were washed twice with cold media to remove free bacteria and further incubated at 37°C. At various time points (0, 20, 40, and 60 min), extracellular bacteria were killed by adding 20 μg/ml of lysostaphin and incubating the cell cultures for 10 min at 37 °C. The cells were washed with cold media and detached with scrapers. The CFUs in cell suspensions were determined as described above. The cells were harvested by centrifugation, and re-suspended with 2% BSA for flow cytometry analysis.
Statistical analyses
Results are graphed as means + s.d. Significant differences between experimental groups were determined using a Student’s t-test (to compare two samples), or an ANOVA analysis followed by Tukey's multiple comparisons test (to compare multiple samples) in GraphPad Prism 6 (La Jolla, CA). Survival analyses were performed using the Kaplan-Meier log rank test. For all analyses, a P value <0.05 was considered to be significant.
Results
Prevalent CA-MRSA strains are more resistant to initial clearance from the lower airways
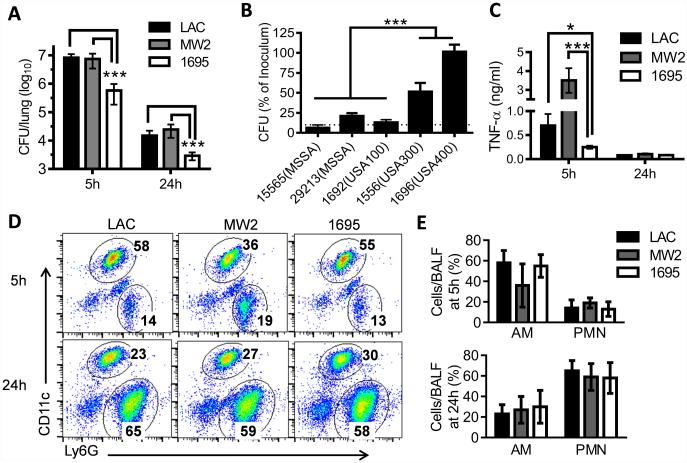
To investigate the mechanism underlying the epidemiological success of predominant US CA-MRSA strains in otherwise healthy hosts, we evaluated the virulence potential of CA-MRSA strains LAC-JE2 (a plasmid-cured derivative of USA300 LAC)(9) and MW2 (USA400)(10), in a comparison with a low-frequency MRSA strain BAA-1695 (non USA100-1100) in a respiratory infection model (Supplemental Table. S1). Considering that the early events in the confrontation between the host and invading bacteria likely decide symptomatic or asymptomatic infection, we assessed host innate immune responses and bacterial survival at early time points after a sublethal dose of S. aureus infection. We found that S. aureus strains exhibited a significant variability in their competency to survive in healthy airways, indicating a variation in their relative resistance to innate bacterial killing (Fig. 1A-B). Significantly, compared with other S. aureus clinical isolates, the known prevalent CA-MRSA strains USA300 and USA400 exhibited improved survival in the lower respiratory tract (Fig. 1A-B). However, lung bacterial burdens and mortality rates were comparable in mice infected with high doses (> 2×108 CFU/mouse) of LAC-JE2 or 1695 (Supplemental Fig. S1). Together, these findings raise the possibility that the resistance to innate bacterial killing contributes to the epidemiological success of prototypical CA-MRSA strains and is not directly associated with their growth or lethality in immune-competent mice.
Figure 1. Prevalent CA-MRSA strains are more resistant to innate clearance from the lower airways.
(A) Lung bacterial burdens at 5 and 24 h after infection of C57BL/6 mice (n=5) with 107 CFU of MRSA LAC-JE2 (LAC), MW2 or BAA-1695. (B) Relative numbers of bacteria remaining in the lungs (n=4) 5 h after S. aureus infection. The CFU mean±s.d. are represented as percentages of the bacterial titer in the inoculum. (C) TNF-α levels, (D) representative flow cytometry plots and (E) frequencies (mean±s.d.) of alveolar macrophages (CD11c+) and neutrophils (CD11c−Ly6G+) in the airways (n=5) at 5 and 24 h after infection of C57BL/6 mice with 107 CFU of MRSA LAC-JE2, MW2 or BAA-1695. *P< 0.05, ***P< 0.001, Tukey's multiple comparisons test. Data shown are representative of two independent experiments.
Furthermore, consistent with their increased bacterial burdens compared with 1695-infected mice, both LAC-JE2 and MW2-infected animals exhibited significantly elevated airway TNF-α production at 5 h after infection (Fig. 1C). Conversely, the relative numbers of alveolar macrophages (CD11c+) were comparable in all MRSA-infected mice (Fig. 1D-E). In addition, there was no significant difference in the kinetics or extent of neutrophil (CD11c−Ly6G+) recruitment among the three MRSA-infected groups (Fig. 1D-E). Together, these results suggest that the defective bacterial killing associated with LAC-JE2 or MW2-infection is not a result of insufficient immune activation.
Competitive fitness of prevalent CA-MRSA strains
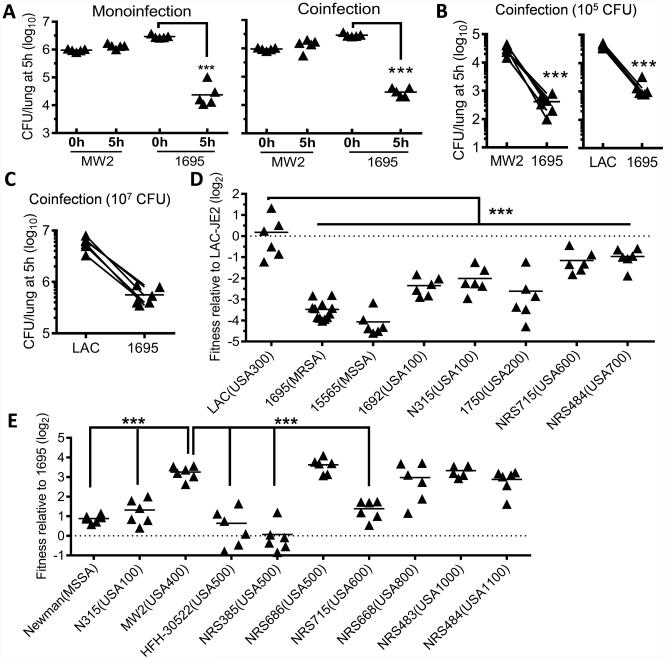
To further investigate the relative competence of S. aureus strains to survive in healthy airways, we assessed lung bacterial burdens at 5 h after infection with MW2 and 1695 in a competition assay (Fig. 2A-B). Similar to monoinfection, the viability of MW2 was significantly higher than 1695 at 5 h after coinfection. Likewise, LAC-JE2 exhibited significantly increased (>10-fold) survival compared with 1695 after various doses of coinfection (Fig. 2B-C). The enhanced competitive fitness of LAC-JE2 and MW2 in healthy airways indicates that rather than compromising host innate defense, the resistance trait of these S. aureus strains is intrinsic to the bacteria.
Figure 2. S. aureus clinical isolates diversify in their competency to survive in the lower airways.
(A) Lung bacterial burdens at 0 and 5 h after infection of C57BL/6 mice with MW2 and 1695 individually (monoinfection), or a 1:1 mixture (coinfection). Data shown are representative of two independent experiments. (B-C) Lung bacterial burdens at 5 h after infection of C57BL/6 mice with 105 (B) or 107 CFU (C) of 1695, MW2 or LAC-JE2 (LAC) in a 1:1 mixture. (D-E) Competitive survival of various S. aureus clinical isolates in the lungs at 5 h after infection of C57BL/6 mice with LAC-JE2 (D) or 1695 (E) in a 1:1 mixture. Each symbol represents the fold change of CFU, i.e., the output CFU ratio divided by the input CFU ratio of co-inoculated strains, in a single mouse. ***P< 0.001, Tukey's multiple comparisons test. Data shown in (B-E) were combined from at least two independent experiments.
To determine whether this resistance trait is generally shared by prevalent S. aureus strains, we assessed the relative fitness of various clinical isolates during coinfection with either LAC-JE2 (Fig. 2D) or the susceptible strain, 1695 (Fig. 2E). A description of these representative clinical isolates can be found in Supplemental Table S1. Notably, the initial survival is comparable between LAC-JE2 and the parental strain LAC (Fig. 2D). As expected, S. aureus clinical isolates exhibited a significant variability in their competitive resistance to innate killing mechanisms in the lower airways. Specifically, compared with representative USA600 and USA700 strains, the parental strain LAC was more resistant to initial clearance from the lower airways (Fig. 2D). Conversely, no significant difference in initial survival was observed among MW2, representative USA800, USA1000 and USA1100 strains (Fig. 2E). Nonetheless, significantly reduced intrapulmonary killing of bacteria was detected in mice infected with LAC (Fig. 2D) or MW2 (Fig. 2E), compared with those infected with classical HA-MRSA strains (i.e., USA100 and USA200). Thus, the competitive fitness of these CA-MRSA strains in healthy airways is consistent with their reported prevalence in causing community-associated S. aureus infections.
Contribution of agr to the competitive fitness of S. aureus
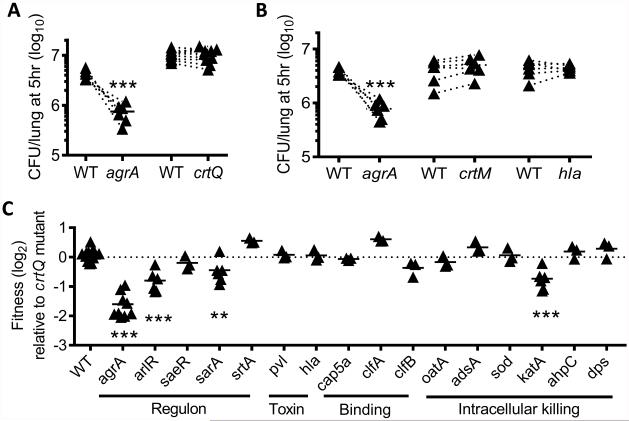
We next investigated the genetic basis that determines the competitive fitness of S. aureus in the lower airways during acute infection. Although this issue has been addressed in other models, including pneumonia, no previous studies have examined host-pathogen interactions at such an acute phase (i.e. 5 h), a critical period that our experiments show dictates the course of infection (Fig. 1). Based on their reported roles during S. aureus pathogenesis (11), we selected 18 well-recognized virulence factors and assessed their respective contribution to S. aureus survival (Supplemental Table. S2). Although S. aureus golden pigment has been shown to be important for resistance to oxidative killing by neutrophils (12, 13), we found that mutations in the staphyloxanthin synthesis pathway, i.e., mutants crtQ and crtM (9), had no effect on the competitive fitness of LAC-JE2 at the very initial stage of infection (Fig. 3A-B). Given that pigment mutants can be easily distinguished from WT and other isogenic mutants based on their colony color and the fact that disruption of crtQ had no fitness defect in our model, crtQ mutant was used as an inoculation control for assessing the competitive survival of other LAC-JE2 mutants from the Nebraska Transposon Mutant Library (9). Interestingly, most mutants did not exhibit impaired fitness compared to crtQ mutant during coinfection (Fig. 3C) or the corresponding isogenic parental strains (Fig. 4). Particularly, no requirement was observed for major cytolytic toxins, i.e., α-hemolysin (Hla), PVL and α-helical phenol-soluble modulin (PSMα) (Figs. 3B-C&Fig. 4A), or surface protein A (Spa) (Fig. 4B), despite their known contributions to the severity of S. aureus pneumonia (14-16). Virulence factors such as catalase (KatA), superoxide dismutase (SOD), and adenosine synthase (AdsA), are known to be important for resistance to oxidative stress (17, 18). However, only katA mutant exhibited significantly reduced survival in the lower airways, suggesting its increased susceptibility to oxidative killing (Fig. 3C).
Figure 3. Contribution of individual virulence factors to LAC-JE2 survival in the lower airways.
(A-B) Lung bacterial burdens at 5 h after infection of C57BL/6 mice with 107 CFU of LAC-JE2 WT in a 1:1 mixture with mutant agrA, crtQ (A), crtM (B) or hla (B). (C) Competitive survival of various LAC-JE2 mutants in the lungs 5 h after infection of C57BL/6 mice in a 1:1 mixture with crtQ mutant. Each symbol represents the fold change of CFU. **P< 0.01, ***P< 0.001, Tukey's multiple comparisons test. Data shown are combined from at least two independent experiments.
Figure 4. AgrA is required for the competitive survival of S. aureus in the lower airways.
Competitive survival of (A) LACΔpsmα, (B) LACΔspa, (C) 502AΔagrA and (D) 1695ΔagrA in the lungs at 5 h after infection of C57BL/6 mice in a 1:1 mixture with the corresponding parent strains (A-C), 1695 or LAC-JE2 (D). Each symbol represents the fold change of CFU. **P< 0.01, ***P< 0.001, t test. Data shown were combined from two independent experiments.
The most dramatic effect on fitness was elicited by a mutation in the agrA virulence regulator gene (Fig. 3C). AgrA is a response regulator encoded by the agr operon that plays a critical role in the coordinated expression of many staphylococcal virulence factors (19). Considering that individual S. aureus strains may use different virulence strategies during this process, we assessed the requirement for agrA in a methicillin-susceptible S. aureus (MSSA) strain 502A (20-22). Indeed, disruption of agrA significantly compromised the competitive fitness of 502A in the lower airways (Fig. 4C). In sharp contrast, an agrA mutant did not further reduce 1695 survival (Fig. 4D). Together, these results indicate that virulence proteins whose expression is under the control of AgrA promote the competitive fitness of S. aureus strains in the lower airways.
Alveolar macrophages are essential and sufficient for initial killing of susceptible strains
We next sought to identify host factors responsible for effective killing of susceptible S. aureus isolates. We have previously shown that both alveolar macrophages and neutrophils are required for efficient S. aureus control in vivo (5). In the current study, we investigated the extent to which alveolar macrophages versus neutrophils contribute to early (5 h) bacterial killing in the lower airways. Alveolar macrophages (CD11c+Siglec-F+) were depleted though i.n. liposome-clodronate treatment (Fig. 5A-B). Interestingly, we found that temporary macrophage-depletion completely abolished early clearance of both the susceptible strain 1695 and LAC-JE2 agrA mutant (Fig. 5C), highlighting an essential role of alveolar macrophages in this process, and in addition, supporting a role for agrA in resistance to killing by macrophages. In contrast, neutrophil depletion did not appear to have a significant effect on the competitive fitness of LAC-JE2 compared with 1695 and LAC-JE2 agrA mutant during coinfection (Fig. 5D-E). Together, these results indicate that the competitive fitness of LAC-JE2 in the lower airways is a result of its resistance to alveolar macrophage-dependent bactericidal activity.
Resistance to intracellular killing by alveolar macrophages
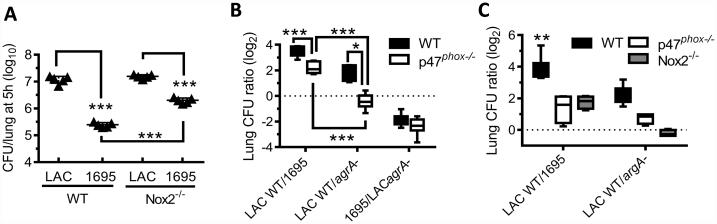
It is known that phagocytes are capable of intracellular bacterial killing through oxidative burst (5, 23). NADPH oxidase is responsible for production of reactive oxygen species in this process, and both Nox2 (i.e. gp91phox) and p47phox are critical subunits for its enzyme activity. In agreement with our previous findings (5), early clearance of 1695 was significantly reduced in mice defective in oxidative burst (Fig. 6A). However, compared with strain 1695, the fitness advantage of LAC-JE2 was still evident in Nox2−/− females (Fig. 6A). These results suggest that the susceptible strain 1695 is vulnerable to both Nox2-dependent and -independent killing by alveolar macrophages. In contrast, compared to the LAC-JE2 agrA mutant, the LAC-JE2 wild-type strain failed to display a fitness advantage in Nox2−/− or p47phox−/− mice (Fig. 6B-C), indicating that agrA expression is essential for LAC-JE2 resistance to oxidative killing. As a result, strain 1695 exhibited decreased fitness in both WT and oxidative burst-deficient mice even compared with the LAC-JE2 agrA mutant (Fig. 6B-C). Taken together, these findings indicate that alveolar macrophages are capable of both oxidative burst-dependent and -independent killing of the 1695 susceptible strain.
Figure 6. Oxidative burst-dependent bacterial killing in the lower airways.
(A) Numbers of bacteria in the lungs 5 h after infection of C57BL/6 WT and Nox2−/− females with 107 CFU of LAC-JE2 and 1695 in a 1:1 mixture. Data shown are representative of two independent experiments. (B-C) Competitive survival of bacteria in the lungs 5 h after infection of C57BL/6 WT, p47phox−/− and Nox2−/− mice (6~9 mice/group) with 107 CFU of LAC-JE2 WT, agrA mutant and/or 1695 in a 1:1 (B) or 1:1:1 mixture (C). Data shown were combined from two independent experiments. *P< 0.05, ***P< 0.001, Tukey's multiple comparisons test.
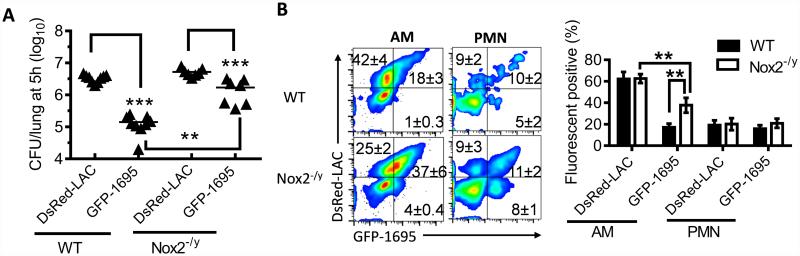
To further explore the mechanism for this resident macrophage-mediated, initial control of S. aureus, we utilized GFP or DsRed-expressing 1695 and LAC-JE2 to quantitate bacterial internalization and intracellular killing by phagocytes (Fig. 7A). In vitro lysostaphin protection assays established that alveolar macrophages exhibited fluorescence signals only after bacterial internalization (Supplemental Fig. S2). If the improved survival of LAC-JE2 was due to its evasion from recognition and/or internalization by alveolar macrophages, we would expect reduced uptake of DsRed-LAC-JE2. On the contrary, the percentages of DsRed+ alveolar macrophages (CD11c+) were significantly higher than those of GFP-1695+ macrophages in both WT and Nox2-deficient (Nox2−/y) males (Fig. 7B). Furthermore, consistent with LAC-JE2 resistance to oxidative killing, the relative numbers of DsRed+ alveolar macrophages in Nox2−/y males were similar to those in WT controls (Fig. 7B). Conversely, due to defective oxidative killing, Nox2−/y mice exhibited significant increases in the relative numbers of GFP-1695+ alveolar macrophages compared with WT animals (Fig. 7B). The defective killing of GFP-1695 in Nox2−/y mice led to an increase in the proportion of DsRed+GFP+ and therefore a corresponding decrease in DsRed+GFP− alveolar macrophages (Fig. 7B). In contrast, the percentages of both DsRed+ and GFP+ neutrophils were comparable in WT and Nox2−/y airways (Fig. 7B). Taken together, these data establish that rather than evading phagocytic uptake, the competitive fitness of strain LAC-JE2 is a result of its ability to survive within alveolar macrophages.
Figure 7. LAC-JE2 persists within alveolar macrophages after internalization.
(A) Lung bacterial burdens, and (B) flow cytometry analysis and quantification of S. aureus phagocytosis (mean±s.d.) by alveolar macrophages (gated on CD11c+) and neutrophils (gated on CD11c−CD11b+) at 5 h after infection of C57BL/6 WT and Nox2−/y males (4~5 mice/group) with 107 CFU of DsRed-LAC-JE2 (DsRed-LAC) and GFP-1695 in a 1:1 mixture. **P< 0.01, ***P< 0.001, Tukey's multiple comparisons test. Data shown are representative of two independent experiments.
Influenza infection suppresses bacterial killing by alveolar macrophages
We show above that prevalent CA-MRSA strains are more resistant to early macrophage-mediated clearance from healthy airways. However, given that S. aureus is the most common cause of nosocomial infections (24), we hypothesized that strains predominating in patients, often with predisposing conditions, may be subject to different selective pressures that could account for their decreased ability to resist innate immune clearance in our animal model. For example, secondary S. aureus infection is a major complication during influenza pneumonia (25), due to the suppressive effect of viral infection on antibacterial immune responses (5, 26-28). We thus investigated whether influenza infection undermines the requirement for S. aureus competitive fitness in the healthy airways, which, in turn, increases host susceptibility to secondary bacterial infection, particularly by S. aureus strains that can be readily cleared by alveolar macrophages in healthy hosts.
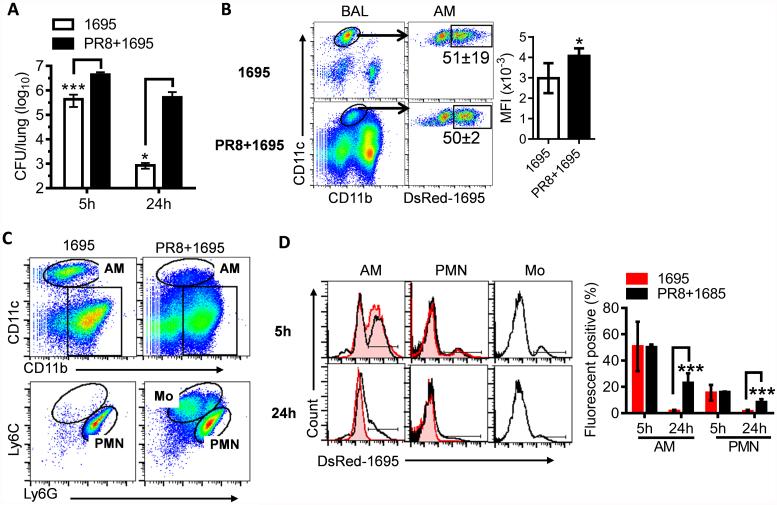
We have previously demonstrated that influenza infection inhibits oxidative bacterial killing during subsequent S. aureus infection (5). Indeed, innate clearance of 1695 was significantly inhibited in influenza-infected mice (Fig. 8). In agreement, even though the percentage of DsRed-1695+ alveolar macrophages at 5 h was not considerably affected by preceding influenza infection, their mean fluorescence intensity (MFI) was significantly increased in influenza-infected mice, indicating a defect in intracellular killing by alveolar macrophages (Fig. 8B). The numbers of bacteria recovered from lungs after MRSA infection alone or post-influenza MRSA infection further confirmed that the survival of 1695 and LAC-JE2 agrA mutant was reduced only in healthy but not influenza-infected mice (Fig. 9A). As a result, compared with nonvirally infected controls, influenza-infected mice exhibited increased susceptibility to secondary infection with either MRSA LAC-JE2 or 1695 (Fig. 9B). Together, these data establish that influenza infection alleviates requirements for the fitness trait of S. aureus in healthy airways, including agr-dependent resistance to oxidative killing by alveolar macrophages. Consequently, in contrast to the differences observed during primary bacterial infection, dysregulation of macrophage function following influenza infection results in comparable host susceptibility to secondary infection with MRSA LAC-JE2 and 1695.
Figure 8. Influenza infection inhibits intracellular bacterial killing by alveolar macrophages.
(A) Numbers of bacteria in the lungs, (B) flow cytometry analysis of S. aureus phagocytosis (mean±s.d.) by alveolar macrophages at 5 h, (C) inflammatory cell profiles at 24 h, and (D) representative histograms and quantification of S. aureus phagocytosis by alveolar macrophages and neutrophils at 5 and 24 h after infection of C57BL/6 mice (n=5) with 107 CFU of DsRed-1695 alone (1695) or on day 7 after PR8 infection (PR8 + 1695). *P< 0.05, ***P< 0.001, t test. Data shown are representative of two independent experiments.
Figure 9. Influenza infection induces susceptibility to secondary S. aureus infection.
(A) Numbers of bacteria in the lungs 5 h after infection of naïve (-Flu) or day 7 postinfluenza infected (+Flu) C57BL/6 mice with 107 CFU of LAC-JE2 WT, agrA mutant and 1695 in a 1:1:1 mixture. ***P< 0.001, t test. Data shown are combined from two independent experiments. (B) Survival of C57BL/6 mice after infection with 2×108 CFU of 1695 or LAC-JE2 (LAC) alone or on day 7 after PR8 infection. Data shown are representative of two independent experiments.
Discussion
We show in this report that low-frequency, hospital- and community-associated S. aureus strains differ in their intrinsic resistance to macrophage-mediated clearance from healthy airways at the earliest stage of infection. Importantly, this resistance trait is in concordance with the prevalence of US MRSA isolates in community-associated infections. Mechanistically, the competitive fitness of S. aureus is a result of both agr-dependent and independent resistance to intracellular killing by alveolar macrophages. These novel findings of the improved competency of predominant CA-MRSA strains to survive innate killing, particularly at the very beginning stage of infection, provide a unique insight into their epidemiological success.
The objective of our present study was to gain new insights into why certain MRSA strains predominate in community settings. To this end, we revealed that the prevalence of MRSA isolates in community-associated infections was strongly correlated with their capacity to resist innate immune killing. However, we found that the resistance potential of USA500 strain NRS686 (HA-MRSA), but not HFH-30522 (CA-MRSA) or NRS385 (HA-MRSA), was not significantly different from that of MW2. Although considered to be the progenitor of the predominant CA-MRSA clone USA300, USA500 only causes sporadic diseases in both community and hospital settings. In addition, USA500 strains are known to vary in their virulence properties due to insertion of IS256 into the rot promoter and subsequent alternation of cytotoxin expression (29). Nonetheless, the prototypical US CA-MRSA strains, i.e., USA300 and USA400, are significantly more resistant to innate killing mechanisms than traditional HA-MRSA isolates such as USA100 and USA200.
From a bacterial perspective, the function of S. aureus virulence factors during pathogenic progress has been extensively studied in various infection models (Supplemental Table. S2). The agr quorum-sensing system is known to be essential for regulating virulence factor expression and therefore, pathogenesis (30). Several S. aureus toxins, including α-hemolysin, PSM peptides, and PVL, which have been shown to impact on experimental pneumonia caused by CA-MRSA, are controlled by the Agr regulatory system (31). Moreover, CA-MRSA strains commonly have higher Agr activity than HA-MRSA strains (32). In agreement, our findings suggest that AgrA-regulated virulence genes contribute to the competitive fitness of S. aureus in the lower airways during acute infection; however, we found that at the very initial stage of lung infection, no single toxin, including α-hemolysin, PSMα peptide, or PVL, accounts for the fitness trait of S. aureus. Furthermore, we show that the fitness trait is not only determined by agrA-dependent resistance to oxidative killing but also influenced by other agrA-independent mechanisms and therefore, it is likely a result of the combined contribution of many virulence genes. Nonetheless, even though additional virulence genes have yet to be definitively demonstrated, our findings indicate that these bacterial factors are capable of intracellularly inhibiting macrophage-mediated killing.
We show in this study that compared with the predominant CA-MRSA strains such as LAC-JE2 and MW2, MRSA strain 1695 is more readily cleared from the lower airways, and presumably less effective at initiating symptomatic infection in healthy hosts. Of note, S. aureus virulence is not merely determined by resistance to initial killing by hosts. As shown in this report, 1695-infected mice did not show a mortality benefit after high doses of infection. Therefore, virulence determinants that are responsible for S. aureus infectivity are likely different from those that dictate disease severity. Indeed, epidemiological studies indicate that there is no difference in the capacity of CA-MRSA or HA-MRSA strains to cause severe diseases (24).
From a host perspective, it is known that S. aureus outgrowth in vivo is mainly prevented through phagocyte-dependent bactericidal processes. Moreover, intracellular killing has been proposed to be the limiting step of S. aureus control in vivo (33). However, due to relatively low numbers of resident macrophages and the fact that their killing capacity is often overwhelmed after exposure to high bacterial inoculums, the role of macrophages in immune defense against S. aureus infection is often overlooked (22, 34). Furthermore, here we show that alveolar macrophages are ineffective as the first line of defense against S. aureus strains such as LAC-JE2. In line with this observation, investigations of the virulence mechanisms of predominant CA-MRSA strains often focus on neutrophils as the most important phagocyte type responsible for bacterial killing (16, 33, 35). Of note, although it has been shown that lung macrophages from female mice are better in killing of ingested pneumococci (36), we did not observe any significant differences between male and female mice in our model.
We show in this study that the competitive fitness of LAC-JE2 is due to extended survival inside alveolar macrophages in the lower airways. Presumably, this fitness trait permits adaptation of surviving bacteria to in vivo environment, and thereby aids in the initiation of infection in immune-competent hosts (37). Interestingly, in contrast to this strain-dependent differences detected in healthy lungs, biofilm formation by 1695, LAC-JE2 and MW2 was identical in an orthopedic implant biofilm infection model (data not shown) (38-40). Furthermore, we show that influenza infection negates the requirement for bacterial virulence factors conferring this immune evasion property, as revealed by comparable host susceptibility to secondary infection with either MRSA LAC-JE2 or 1695. Together, these findings provide new insights into the distinct epidemiological prevalence of MRSA strains in community settings versus in hosts with predisposing risk factors.
In summary, we show that LAC-JE2, a representative strain of CA-MRSA USA300, persists within alveolar macrophages whereas hospital-acquired (e.g., USA100 and USA200) and low frequency (non USA100-1100) MRSA strains are relatively susceptible to the innate killing mechanism in the lower airways of healthy hosts. Collectively, this discovery reveals a previously uncharacterized virulence trait of S. aureus that may account for the differential infectivity of MRSA strains in otherwise healthy individuals.
Supplementary Material
Acknowledgements
1. The authors thank Dr. Alexander Horswill (University of Iowa) for providing pCM29, Dr. Michael Otto (NIAID) for providing LAC WT and Δpsmα, Dr. Paul J. Planet (Columbia University) for providing strain 502A, and BEI Resources for providing S. aureus strains as listed in Supplemental Table S1. The authors also thank Dr. Philip Hexley, Victoria Smith, and Samantha Wall in the University of Nebraska Medical Center Flow Cytometry Research Facility for assistance with FACS analysis and cell sorting.
This work was supported by National Institutes of Health/National Heart, Lung, and Blood Institute R01 HL118408 to K.S.
Reference
- 1.Defres S, Marwick C, Nathwani D. MRSA as a cause of lung infection including airway infection, community-acquired pneumonia and hospital-acquired pneumonia. The European respiratory journal. 2009;34:1470–1476. doi: 10.1183/09031936.00122309. [DOI] [PubMed] [Google Scholar]
- 2.David MZ, Daum RS. Community-associated methicillin-resistant Staphylococcus aureus: epidemiology and clinical consequences of an emerging epidemic. Clinical microbiology reviews. 2010;23:616–687. doi: 10.1128/CMR.00081-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Otto M. Basis of virulence in community-associated methicillin-resistant Staphylococcus aureus. Annual review of microbiology. 2010;64:143–162. doi: 10.1146/annurev.micro.112408.134309. [DOI] [PubMed] [Google Scholar]
- 4.DeLeo FR, Otto M, Kreiswirth BN, Chambers HF. Community-associated meticillin-resistant Staphylococcus aureus. Lancet. 2010;375:1557–1568. doi: 10.1016/S0140-6736(09)61999-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sun K, Metzger DW. Influenza infection suppresses NADPH oxidase-dependent phagocytic bacterial clearance and enhances susceptibility to secondary methicillin-resistant Staphylococcus aureus infection. Journal of immunology. 2014;192:3301–3307. doi: 10.4049/jimmunol.1303049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pang YY, Schwartz J, Thoendel M, Ackermann LW, Horswill AR, Nauseef WM. agr-Dependent interactions of Staphylococcus aureus USA300 with human polymorphonuclear neutrophils. Journal of innate immunity. 2010;2:546–559. doi: 10.1159/000319855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Moormeier DE, Endres JL, Mann EE, Sadykov MR, Horswill AR, Rice KC, Fey PD, Bayles KW. Use of microfluidic technology to analyze gene expression during Staphylococcus aureus biofilm formation reveals distinct physiological niches. Applied and environmental microbiology. 2013;79:3413–3424. doi: 10.1128/AEM.00395-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sun K, Metzger DW. Inhibition of pulmonary antibacterial defense by interferon-gamma during recovery from influenza infection. Nature medicine. 2008;14:558–564. doi: 10.1038/nm1765. [DOI] [PubMed] [Google Scholar]
- 9.Fey PD, Endres JL, Yajjala VK, Widhelm TJ, Boissy RJ, Bose JL, Bayles KW. A genetic resource for rapid and comprehensive phenotype screening of nonessential Staphylococcus aureus genes. MBio. 2013;4:e00537–00512. doi: 10.1128/mBio.00537-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Baba T, Takeuchi F, Kuroda M, Yuzawa H, Aoki K, Oguchi A, Nagai Y, Iwama N, Asano K, Naimi T, Kuroda H, Cui L, Yamamoto K, Hiramatsu K. Genome and virulence determinants of high virulence community-acquired MRSA. Lancet. 2002;359:1819–1827. doi: 10.1016/s0140-6736(02)08713-5. [DOI] [PubMed] [Google Scholar]
- 11.Foster TJ. Immune evasion by staphylococci. Nature reviews. Microbiology. 2005;3:948–958. doi: 10.1038/nrmicro1289. [DOI] [PubMed] [Google Scholar]
- 12.Liu GY, Essex A, Buchanan JT, Datta V, Hoffman HM, Bastian JF, Fierer J, Nizet V. Staphylococcus aureus golden pigment impairs neutrophil killing and promotes virulence through its antioxidant activity. J Exp Med. 2005;202:209–215. doi: 10.1084/jem.20050846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Clauditz A, Resch A, Wieland KP, Peschel A, Gotz F. Staphyloxanthin plays a role in the fitness of Staphylococcus aureus and its ability to cope with oxidative stress. Infect Immun. 2006;74:4950–4953. doi: 10.1128/IAI.00204-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bubeck Wardenburg J, Patel RJ, Schneewind O. Surface proteins and exotoxins are required for the pathogenesis of Staphylococcus aureus pneumonia. Infection and immunity. 2007;75:1040–1044. doi: 10.1128/IAI.01313-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Inoshima I, Inoshima N, Wilke GA, Powers ME, Frank KM, Wang Y, Bubeck Wardenburg J. A Staphylococcus aureus pore-forming toxin subverts the activity of ADAM10 to cause lethal infection in mice. Nature medicine. 2011;17:1310–1314. doi: 10.1038/nm.2451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang R, Braughton KR, Kretschmer D, Bach TH, Queck SY, Li M, Kennedy AD, Dorward DW, Klebanoff SJ, Peschel A, DeLeo FR, Otto M. Identification of novel cytolytic peptides as key virulence determinants for community-associated MRSA. Nature medicine. 2007;13:1510–1514. doi: 10.1038/nm1656. [DOI] [PubMed] [Google Scholar]
- 17.Thammavongsa V, Kern JW, Missiakas DM, Schneewind O. Staphylococcus aureus synthesizes adenosine to escape host immune responses. The Journal of experimental medicine. 2009;206:2417–2427. doi: 10.1084/jem.20090097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Das D, Saha SS, Bishayi B. Intracellular survival of Staphylococcus aureus: correlating production of catalase and superoxide dismutase with levels of inflammatory cytokines. Inflammation research : official journal of the European Histamine Research Society … [et al.] 2008;57:340–349. doi: 10.1007/s00011-007-7206-z. [DOI] [PubMed] [Google Scholar]
- 19.Said-Salim B, Dunman PM, McAleese FM, Macapagal D, Murphy E, McNamara PJ, Arvidson S, Foster TJ, Projan SJ, Kreiswirth BN. Global regulation of Staphylococcus aureus genes by Rot. Journal of bacteriology. 2003;185:610–619. doi: 10.1128/JB.185.2.610-619.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Parker D, Planet PJ, Soong G, Narechania A, Prince A. Induction of type I interferon signaling determines the relative pathogenicity of Staphylococcus aureus strains. PLoS pathogens. 2014;10:e1003951. doi: 10.1371/journal.ppat.1003951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Parker D, Narechania A, Sebra R, Deikus G, Larussa S, Ryan C, Smith H, Prince A, Mathema B, Ratner AJ, Kreiswirth B, Planet PJ. Genome Sequence of Bacterial Interference Strain Staphylococcus aureus 502A. Genome announcements. 2014:2. doi: 10.1128/genomeA.00284-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nugent KM, Pesanti EL. Staphylococcal clearance and pulmonary macrophage function during influenza infection. Infection and immunity. 1982;38:1256–1262. doi: 10.1128/iai.38.3.1256-1262.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kohler J, Breitbach K, Renner C, Heitsch AK, Bast A, van Rooijen N, Vogelgesang S, Steinmetz I. NADPH-oxidase but not inducible nitric oxide synthase contributes to resistance in a murine Staphylococcus aureus Newman pneumonia model. Microbes and infection / Institut Pasteur. 2011;13:914–922. doi: 10.1016/j.micinf.2011.05.004. [DOI] [PubMed] [Google Scholar]
- 24.Diep BA, Carleton HA, Chang RF, Sensabaugh GF, Perdreau-Remington F. Roles of 34 virulence genes in the evolution of hospital- and community-associated strains of methicillin-resistant Staphylococcus aureus. The Journal of infectious diseases. 2006;193:1495–1503. doi: 10.1086/503777. [DOI] [PubMed] [Google Scholar]
- 25.Centers for Disease C, Prevention Severe methicillin-resistant Staphylococcus aureus community-acquired pneumonia associated with influenza--Louisiana and Georgia, December 2006-January 2007. MMWR. Morbidity and mortality weekly report. 2007;56:325–329. [PubMed] [Google Scholar]
- 26.Robinson KM, Choi SM, McHugh KJ, Mandalapu S, Enelow RI, Kolls JK, Alcorn JF. Influenza A exacerbates Staphylococcus aureus pneumonia by attenuating IL-1beta production in mice. Journal of immunology. 2013;191:5153–5159. doi: 10.4049/jimmunol.1301237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Iverson AR, Boyd KL, McAuley JL, Plano LR, Hart ME, McCullers JA. Influenza virus primes mice for pneumonia from Staphylococcus aureus. The Journal of infectious diseases. 2011;203:880–888. doi: 10.1093/infdis/jiq113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kudva A, Scheller EV, Robinson KM, Crowe CR, Choi SM, Slight SR, Khader SA, Dubin PJ, Enelow RI, Kolls JK, Alcorn JF. Influenza A inhibits Th17-mediated host defense against bacterial pneumonia in mice. Journal of immunology. 2011;186:1666–1674. doi: 10.4049/jimmunol.1002194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Benson MA, Ohneck EA, Ryan C, Alonzo F, 3rd, Smith H, Narechania A, Kolokotronis SO, Satola SW, Uhlemann AC, Sebra R, Deikus G, Shopsin B, Planet PJ, Torres VJ. Evolution of hypervirulence by a MRSA clone through acquisition of a transposable element. Molecular microbiology. 2014;93:664–681. doi: 10.1111/mmi.12682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shompole S, Henon KT, Liou LE, Dziewanowska K, Bohach GA, Bayles KW. Biphasic intracellular expression of Staphylococcus aureus virulence factors and evidence for Agr-mediated diffusion sensing. Molecular microbiology. 2003;49:919–927. doi: 10.1046/j.1365-2958.2003.03618.x. [DOI] [PubMed] [Google Scholar]
- 31.Otto M. Community-associated MRSA: what makes them special? International journal of medical microbiology : IJMM. 2013;303:324–330. doi: 10.1016/j.ijmm.2013.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li M, Cheung GY, Hu J, Wang D, Joo HS, Deleo FR, Otto M. Comparative analysis of virulence and toxin expression of global community-associated methicillin-resistant Staphylococcus aureus strains. The Journal of infectious diseases. 2010;202:1866–1876. doi: 10.1086/657419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rigby KM, DeLeo FR. Neutrophils in innate host defense against Staphylococcus aureus infections. Seminars in immunopathology. 2012;34:237–259. doi: 10.1007/s00281-011-0295-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Martin FJ, Parker D, Harfenist BS, Soong G, Prince A. Participation of CD11c(+) leukocytes in methicillin-resistant Staphylococcus aureus clearance from the lung. Infection and immunity. 2011;79:1898–1904. doi: 10.1128/IAI.01299-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu W, Yan M, Sugui JA, Li H, Xu C, Joo J, Kwon-Chung KJ, Coleman WG, Rodgers GP. Olfm4 deletion enhances defense against Staphylococcus aureus in chronic granulomatous disease. The Journal of clinical investigation. 2013;123:3751–3755. doi: 10.1172/JCI68453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yang Z, Huang YC, Koziel H, de Crom R, Ruetten H, Wohlfart P, Thomsen RW, Kahlert JA, Sorensen HT, Jozefowski S, Colby A, Kobzik L. Female resistance to pneumonia identifies lung macrophage nitric oxide synthase-3 as a therapeutic target. eLife. 2014:3. doi: 10.7554/eLife.03711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hamza T, Li B. Differential responses of osteoblasts and macrophages upon Staphylococcus aureus infection. BMC microbiology. 2014;14:207. doi: 10.1186/s12866-014-0207-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Heim CE, Vidlak D, Kielian T. Interleukin-10 production by myeloid-derived suppressor cells contributes to bacterial persistence during Staphylococcus aureus orthopedic biofilm infection. Journal of leukocyte biology. 2015;98:1003–1013. doi: 10.1189/jlb.4VMA0315-125RR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Heim CE, Vidlak D, Scherr TD, Hartman CW, Garvin KL, Kielian T. IL-12 promotes myeloid-derived suppressor cell recruitment and bacterial persistence during Staphylococcus aureus orthopedic implant infection. Journal of immunology. 2015;194:3861–3872. doi: 10.4049/jimmunol.1402689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Scherr TD, Hanke ML, Huang O, James DB, Horswill AR, Bayles KW, Fey PD, Torres VJ, Kielian T. Staphylococcus aureus Biofilms Induce Macrophage Dysfunction Through Leukocidin AB and Alpha-Toxin. mBio. 2015:6. doi: 10.1128/mBio.01021-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xiong YQ, Willard J, Yeaman MR, Cheung AL, Bayer AS. Regulation of Staphylococcus aureus alpha-toxin gene (hla) expression by agr, sarA, and sae in vitro and in experimental infective endocarditis. The Journal of infectious diseases. 2006;194:1267–1275. doi: 10.1086/508210. [DOI] [PubMed] [Google Scholar]
- 42.Sharma-Kuinkel BK, Wu Y, Tabor DE, Mok H, Sellman BR, Jenkins A, Yu L, Jafri HS, Rude TH, Ruffin F, Schell WA, Park LP, Yan Q, Thaden JT, Messina JA, Fowler VG, Jr., Esser MT. Characterization of alpha-toxin hla gene variants, alpha-toxin expression levels, and levels of antibody to alpha-toxin in hemodialysis and postsurgical patients with Staphylococcus aureus bacteremia. J Clin Microbiol. 2015;53:227–236. doi: 10.1128/JCM.02023-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Reyes D, Andrey DO, Monod A, Kelley WL, Zhang G, Cheung AL. Coordinated regulation by AgrA, SarA, and SarR to control agr expression in Staphylococcus aureus. Journal of bacteriology. 2011;193:6020–6031. doi: 10.1128/JB.05436-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gong J, Li D, Yan J, Liu Y, Li D, Dong J, Gao Y, Sun T, Yang G. The accessory gene regulator (agr) controls Staphylococcus aureus virulence in a murine intracranial abscesses model. Braz J Infect Dis. 2014;18:501–506. doi: 10.1016/j.bjid.2014.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mazmanian SK, Liu G, Jensen ER, Lenoy E, Schneewind O. Staphylococcus aureus sortase mutants defective in the display of surface proteins and in the pathogenesis of animal infections. Proceedings of the National Academy of Sciences of the United States of America. 2000;97:5510–5515. doi: 10.1073/pnas.080520697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mazmanian SK, Liu G, Ton-That H, Schneewind O. Staphylococcus aureus sortase, an enzyme that anchors surface proteins to the cell wall. Science. 1999;285:760–763. doi: 10.1126/science.285.5428.760. [DOI] [PubMed] [Google Scholar]
- 47.Cosgrove K, Coutts G, Jonsson IM, Tarkowski A, Kokai-Kun JF, Mond JJ, Foster SJ. Catalase (KatA) and alkyl hydroperoxide reductase (AhpC) have compensatory roles in peroxide stress resistance and are required for survival, persistence, and nasal colonization in Staphylococcus aureus. J Bacteriol. 2007;189:1025–1035. doi: 10.1128/JB.01524-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Martinez A, Kolter R. Protection of DNA during oxidative stress by the nonspecific DNA-binding protein Dps. Journal of bacteriology. 1997;179:5188–5194. doi: 10.1128/jb.179.16.5188-5194.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Loffler B, Niemann S, Ehrhardt C, Horn D, Lanckohr C, Lina G, Ludwig S, >Peters G. Pathogenesis of Staphylococcus aureus necrotizing pneumonia: the role of PVL and an influenza coinfection. Expert Rev Anti Infect Ther. 2013;11:1041–1051. doi: 10.1586/14787210.2013.827891. [DOI] [PubMed] [Google Scholar]
- 50.Labandeira-Rey M, Couzon F, Boisset S, Brown EL, Bes M, Benito Y, Barbu EM, Vazquez V, Hook M, Etienne J, Vandenesch F, Bowden MG. Staphylococcus aureus Panton-Valentine leukocidin causes necrotizing pneumonia. Science. 2007;315:1130–1133. doi: 10.1126/science.1137165. [DOI] [PubMed] [Google Scholar]
- 51.Ballal A, Manna AC. Regulation of superoxide dismutase (sod) genes by SarA in Staphylococcus aureus. Journal of bacteriology. 2009;191:3301–3310. doi: 10.1128/JB.01496-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mainiero M, Goerke C, Geiger T, Gonser C, Herbert S, Wolz C. Differential target gene activation by the Staphylococcus aureus two-component system saeRS. Journal of bacteriology. 2010;192:613–623. doi: 10.1128/JB.01242-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nygaard TK, Pallister KB, Ruzevich P, Griffith S, Vuong C, Voyich JM. SaeR binds a consensus sequence within virulence gene promoters to advance USA300 pathogenesis. The Journal of infectious diseases. 2010;201:241–254. doi: 10.1086/649570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fournier B, Klier A, Rapoport G. The two-component system ArlS-ArlR is a regulator of virulence gene expression in Staphylococcus aureus. Molecular microbiology. 2001;41:247–261. doi: 10.1046/j.1365-2958.2001.02515.x. [DOI] [PubMed] [Google Scholar]
- 55.Zielinska AK, Beenken KE, Joo HS, Mrak LN, Griffin LM, Luong TT, Lee CY, Otto M, Shaw LN, Smeltzer MS. Defining the strain-dependent impact of the Staphylococcal accessory regulator (sarA) on the alpha-toxin phenotype of Staphylococcus aureus. Journal of bacteriology. 2011;193:2948–2958. doi: 10.1128/JB.01517-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Liu CZ, Shih MH, Tsai PJ. ClfA(221-550), a fibrinogen-binding segment of Staphylococcus aureus clumping factor A, disrupts fibrinogen function. Thromb Haemost. 2005;94:286–294. doi: 10.1160/TH05-03-0205. [DOI] [PubMed] [Google Scholar]
- 57.McDevitt D, Nanavaty T, House-Pompeo K, Bell E, Turner N, McIntire L, Foster T, Hook M. Characterization of the interaction between the Staphylococcus aureus clumping factor (ClfA) and fibrinogen. Eur J Biochem. 1997;247:416–424. doi: 10.1111/j.1432-1033.1997.00416.x. [DOI] [PubMed] [Google Scholar]
- 58.Nanra JS, Buitrago SM, Crawford S, Ng J, Fink PS, Hawkins J, Scully IL, McNeil LK, Aste-Amezaga JM, Cooper D, Jansen KU, Anderson AS. Capsular polysaccharides are an important immune evasion mechanism for Staphylococcus aureus. Hum Vaccin Immunother. 2013;9:480–487. doi: 10.4161/hv.23223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.O'Riordan K, Lee JC. Staphylococcus aureus capsular polysaccharides. Clinical microbiology reviews. 2004;17:218–234. doi: 10.1128/CMR.17.1.218-234.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Thammavongsa V, Schneewind O, Missiakas DM. Enzymatic properties of Staphylococcus aureus adenosine synthase (AdsA) BMC Biochem. 2011;12:56. doi: 10.1186/1471-2091-12-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bera A, Herbert S, Jakob A, Vollmer W, Gotz F. Why are pathogenic staphylococci so lysozyme resistant? The peptidoglycan O-acetyltransferase OatA is the major determinant for lysozyme resistance of Staphylococcus aureus. Molecular microbiology. 2005;55:778–787. doi: 10.1111/j.1365-2958.2004.04446.x. [DOI] [PubMed] [Google Scholar]
- 62.Thoendel M, Kavanaugh JS, Flack CE, Horswill AR. Peptide signaling in the staphylococci. Chem Rev. 2011;111:117–151. doi: 10.1021/cr100370n. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.