Abstract
Germination of spores into actively growing cells is a process essential for survival and pathogenesis of many microbes. Molecular mechanisms governing germination, however, are poorly understood in part because few tools exist for evaluating and interrogating the process. Here, we introduce an assay that leverages developments in microfluidic technology and image processing to quantitatively measure germination with unprecedented resolution, assessing both individual cells and the population as a whole. Using spores from Cryptococcus neoformans, a leading cause of fatal fungal disease in humans, we developed a platform to evaluate spores as they undergo morphological changes during differentiation into vegetatively growing yeast. The assay uses pipet-accessible microdevices that can be arrayed for efficient testing of diverse microenvironmental variables, including temperature and nutrients. We discovered that temperature influences germination rate, a carbon source alone is sufficient to induce germination, and the addition of a nitrogen source sustains it. Using this information, we optimized the assay for use with fungal growth inhibitors to pinpoint stages of germination inhibition. Unexpectedly, the clinical antifungal drugs amphotericin B and fluconazole did not significantly alter the process or timing of the transition from spore to yeast, indicating that vegetative growth and germination are distinct processes in C. neoformans. Finally, we used the high temporal resolution of the assay to determine the precise defect in a slow-germination mutant. Combining advances in microfluidics with a robust fungal molecular genetic system allowed us to identify and alter key temporal, morphological, and molecular events that occur during fungal germination.
Introduction
Many microorganisms, including bacteria, fungi, and protozoa rely on the ability to produce spores to survive harsh environmental conditions and/or disperse to new environments1. Spores such as those from the bacterium Bacillus subtilis are optimized for long-term survival via a "shelter in place" strategy in which spores are formed to withstand a substandard environment until local conditions improve2. In contrast, other spores, such as those of the fungus Saccharomyces cerevisiae are optimized for dispersal via water or wind in an "evacuation" strategy to escape poor local conditions and attempt to enter a new environment conducive to growth3-5. In both scenarios, spores need to be resistant to environmental stresses, remain relatively metabolically quiescent for long periods of time, and initiate germination in response to nutrient-rich environments or other signals3,6,7. Germination is the process by which spores break dormancy and resume vegetative growth; the ability of spores to differentiate into vegetatively growing cell types only when conditions are favorable is essential for microbial survival.
Fungi are particularly effective at using spore formation, dispersal, and germination as a survival mechanism, resulting in representation of fungi across all nearly ecosystems on earth1. Given the ubiquitous nature of fungal spores in the environment, it is common for humans to be exposed to them via skin contact, ingestion, and inhalation. While most of these interactions are harmless, some can result in the development of fatal human disease. Many different medically relevant fungi, including Aspergillus fumigatus, Blastomyces dermatitidis, Histoplasma capsulatum, Paracoccidioides brasiliensis, Cryptococcus neoformans, and others infect humans via inhalation from environmental sources8,9. The most common cause of fatal fungal disease is C. neoformans, causing hundreds of thousands of deaths worldwide annually9. C. neoformans is associated with soil, trees, and bird droppings10-12. Upon inhalation, C. neoformans establishes a reservoir in the lung from which it can disseminate to the central nervous system and cause fatal meningoencephalitis11,13. Spores from these pathogenic environmental fungi have been shown to cause disease in animal models, and in every case, the ability of spores to cause disease is dependent on the transition from relatively dormant particles (spores) to vegetatively growing cells (yeast or hyphae)14. Despite their importance in disease, very little is known about the basic biology of infectious fungal spores of humans, including how they are formed and how they germinate.
Fungi exhibit diverse mechanisms of germination that generally result in significant morphological changes as compact spores transition into larger, actively growing cell types. The physical changes associated with germination have been documented across diverse fungi6; however, the process by which this differentiation program occurs at a molecular level is poorly understood. Traditionally, fungal germination has been evaluated in model systems such as S. cerevisiae and Aspergillus nidulans15,16. In these fungi, assays for germination have generally relied on either microscopic evaluation of individual cells (low resolution across the population) or the ability of a spore to form a colony on a plate (low resolution at the cellular level). These assays leave major gaps in our understanding of the germination process with respect to the events that occur morphologically and molecularly across a population and the extent to which those events vary in response to different environmental conditions, such as temperature, nutrient availability, and signaling molecule concentrations.
Here we present a quantitative, high-resolution germination assay using spores from C. neoformans, microfluidic devices, and image processing algorithms. We took advantage of the fact that C. neoformans spores undergo a discernable morphological transition as they differentiate from spores into yeast, allowing the development of quantitative visual parameters of germination. We also capitalized on advances in microscale bioengineering that have led to the development of pipet-accessible platforms that are easy to operate without complex fluid handling machinery and that can accommodate non-adherent cell populations17. The germination assay combines a simple microfluidic device that is optimized for imaging fungi and an image-processing algorithm optimized for measuring morphological characteristics of the imaged population on a cell-by-cell basis. Using the assay, we identified the effects of various temperatures, nutrients, and compounds on spore germination and vegetative yeast growth. Yeast and spores responded differently to fungal growth inhibitors indicating that germination and vegetative growth are molecularly distinct biological processes. By using a fungal mutant, we were able to parse the germination process and identify a gene product that impinges on spore differentiation. This assay provides the first high resolution, quantitative assessment of spore germination at both the single cell and population levels.
Materials and Methods
Device fabrication and preparation
Soft lithography was used to fabricate two master molds of arrays of 24 devices, as described previously18. Briefly, silicon wafers were spin coated with SU-8 100 (Microchem, Newton, MA), soft baked at 85°C for 4 hours, and exposed to UV through a transparent mask with the desired pattern before baking again at 85°C for 2 hours. One master formed the culture chamber layer while the other formed the inlet and outlet port layer. Polydimethylsiloxane (PDMS, Sylgard 184 Silicon Elastomer Kit, Dow Corning Corporation, Midland, MI) was applied to the master molds using a 10:1 ratio of base to curing agent and cured at 80°C for 4 hours. Both PDMS layers were soxhlet extracted to remove any unpolymerized PDMS monomer and plasma bonded to a glass microscope slide. Immediately prior to use, the devices were UV sterilized and filled with 1X PBS as a wetting agent.
C. neoformans spore isolation and yeast culture
All strains used were of the serotype D background, JEC20 (a) and JEC21 (α), and were handled using standard techniques and media as previously described19. Spores were isolated from cultures as previously described20. Briefly, yeast of both mating types were grown on YPD for 2 days at 30°C, mixed in a 1:1 ratio in phosphate buffered saline (PBS) and spotted onto V8 pH 7 agar plates. After 5 days at 25°C, the spots were scraped off the plate, resuspended in 65% percoll and 1 X PBS, and centrifuged for 20 minutes at 4°C and 4000 RPM. Spores were recovered from the bottom of the gradient by piercing the tube with a needle. Spores were then wash three times in PBS and counted with a hemoctyometer. Yeast were used after growth for two days on solid YPD media at 30°C.
Device design and operation
Device dimensions were chosen to ensure that cells would be retained in the culture well despite multiple fluid replacements as previously described17. Briefly, by having a wide culture well and allowing the cells to settle to the bottom of the well out of the direct path of fluid flow, shear stresses on the cells were minimized. To operate the devices, 5 μL spore suspension at 1 × 107 spores/mL or 5 μL yeast suspension at 1 × 106 yeast/mL was loaded into the input port and allowed to passively pump through the device. Cells were allowed to settle to the bottom of the well for 5 minutes before 5 μL 2X germination medium was added for a final concentration in the device of 1X (as demonstrated in Supplemental Figure 1, a volume replacement displaces half of the liquid in a device). Unless stated otherwise, germination or yeast growth was assessed after 16 hours of incubation at 30°C. All assays were performed in triplicate. At assay points, two images were taken per device. Light microscopy was carried out using a Nikon Diaphot inverted phase contrast microscope fitted with a 20X objective and Nikon D5000 camera.
Germination assay analysis
Images of germinating spores were processed in ImageJ21 using a custom macro. Briefly, each image was converted to grayscale and auto thresholded using the built in IsoData algorithm. A watershed algorithm was applied to visually separate cells clumped together, and all particles in the image were tabulated along with measurements of their area, aspect ratio, circularity, and solidity using the “analyze particles” function. These tables of measurements were exported to Matlab for further processing (MathWorks, Natick, MA). Specifically, particles that were too small (area < 1.35 μm2), too large (area > 25 μm2), too oblong (aspect ratio < 0.4), or that had jagged edges (circularity < 0.7 or solidity < 0.8) were filtered out as being either particulates in the frame or image processing artifacts. All particles remaining were considered to be cells. Cell populations were either graphed as 2D histograms of cell area and aspect ratio or categorized as spores (area < 7.436 μm2 and aspect ratio < 0.8), yeast (area > 7.842 μm2 and aspect ratio > 0.8), or intermediates (all remaining cells) before graphing in Prism (GraphPad, La Jolla, CA) as population fractions.
Generation of gene deletion strains
NHP6A (CNF04730) deletion mutants were created using fusion PCR to generate knockout constructs as described previously22 using the following oligos: NHP6A - 5'-flank with CHO3359 [5'-GCACAGGCGACGCTCAAGGT-3'] and CHO3360 [5'-GTCATAGCTGTTTCCTGGATGGTGAGTGGGCGCCAGGT-3'], 3'-flank with CHO3361 [5'-CATCCTCGCAGCAAGGGCGGCCTCATGTCTTTGCTTACGAGCTT -3'] and CHO3362 [5'-TACGGGAAAAGTTGAGGTGCGAGG-3'], NEOR with CHO3363 [5'-ACCTGGCGCCCACTCACCATCCAGGAAACAGCTATGAC-3'] and CHO3364 [5'-AAGCTCGTAAGCAAAGACATGAGGCCGCCCTTGCTGCGAGGATG-3']. CHO3359 and CHO3362 were used to create the final nhp6a::NEOR deletion cassette. The nhp6a::NEOR deletion cassette was transformed into JEC20 and JEC21 by biolistic transformation, grown on medium containing 1 M sorbitol, and selected on medium containing 200 μg/mL G418. NEOR transformants were screened for correct integration of the deletion construct by PCR and Southern blotting, resulting in a nhp6aΔ strains (CHY2532 and 2533) and independent α nhp6aΔ strains (CHY2534 and 2535). These strains were evaluated for general phenotypes (growth, temperature sensitivity, stress tolerance) and all were found to behave like wild type strains (data not shown and Supplemental Figure 4). CHY2533 was then backcrossed with JEC21 to produce reassorted progeny (CHY2552-CHY2559), and multiple segregants were crossed to produce nhp6aΔ spores for further evaluation. In all cases, the only discernable phenotype of nhp6aΔ spores was a possible delay in germination.
Yeast growth assay analysis
Images of yeast cells growing were processed in ImageJ21 using a custom macro. The macro converted each image to grayscale and auto thresholded the image using the built in IJ_IsoData algorithm. This caused all parts of the frame covered with cells to be colored black and the rest white. The macro calculated what percent of the image frame was covered in black, a measure of the number of cells present in the frame. Percent frame coverage values were exported to Prism (GraphPad, La Jolla, CA) for graphing and statistical comparisons.
Statistical methods
Statistical comparisons of germination were performed on spore, yeast, or intermediate subpopulations separately; if any of the comparisons using unpaired Students t-tests assuming equal variance achieved a p-value of <0.05, the population distributions were considered to be significantly different. Yeast growth comparisons were performed using percent frame coverage measurements in unpaired Student’s t-tests assuming equal variance. For comparing time course datasets, ANOVA was performed using population fractions at each time point (spore and yeast population fractions were analyzed separately). All statistical calculations were done using Prism (GraphPad, La Jolla, CA).
Results
Spore morphology changes consistently and synchronously during germination
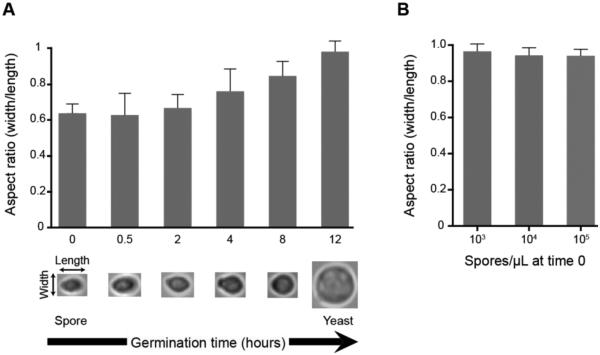
To determine whether observed changes in spore shape and size during germination would be visible and quantifiable using standard light microscopy, we germinated spores and measured them over the course of 12 hours. 104 spores/μL were exposed to a nutrient-rich liquid growth medium (YPD) at 30°C and cells were recovered and fixed in formaldehyde after 0.5, 2, 4, 8 and 12 hours. Each population of fixed cells was observed microscopically using a standard light microscope at 1000X magnification. Ten spores from each sample were selected at random and measured in both width and length. The resulting measurements were used to calculate an aspect ratio of width/length. An ungerminated population of spores showed an average aspect ratio of 0.6 (1.9 μm/3.1 μm), indicating an ovoid shape. There was little variation in aspect ratio across the spores measured. Over time of exposure to nutrients, the aspect ratio of the spores gradually increased as the cells became rounder (Figure 1A). After 12 hours of germination, the population showed an average aspect ratio of 0.95 (4.43 μm/4.65μm) indicating a circular shape consistent with vegetatively growing yeast. This change in aspect ratio was statistically significant over time, and most importantly, largely synchronous, as evidenced by a relatively small standard deviation (at 12 h, S.D. = 0.055) and high reproducibility between experiments. In addition, neither the timeline nor synchronicity of germination was altered by 10-fold increases or decreases in spore concentration (103/μL − 105/μL) (Figure 1B). This consistent and measurable behavior of C. neoformans spores during the germination process enabled quantitative assessment of germination; however, the laborious nature of the measurements necessitated low sample sizes (in this case ten cells per time point) that limited assay resolution and applicability.
Figure 1.
Spore germination is associated with a change in cell shape and size that is concentration independent. (A) Spores (104/μL) were germinated in rich growth medium at 30°C and measured at six time points over 12 hours (x-axis) to generate an average aspect ratio of width/length (y-axis) for each time point. In each sample 10 randomly chosen cells were measured. Representative cells for each time point are shown beneath the x-axis. The results depicted are representative of three independent experiments, all indicating the same trend. (B) Increasing concentrations of spores were germinated for 24 hours, and 10 cells from each concentration were randomly chosen for aspect ratio measurements. Error bars represent standard deviation.
Microscale well device and assay development
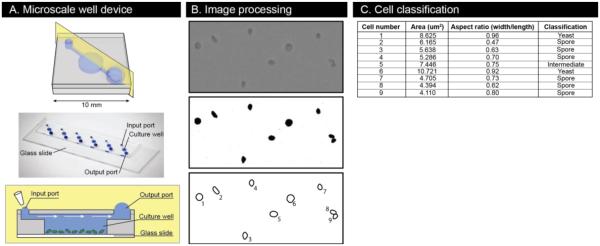
We designed a microscale device that can accommodate non-adherent cells in liquid culture and allow for easy manipulation and imaging. The microscale device is made of PDMS bonded to a glass slide and is comprised of a channel for fluid exchange above a central, depressed culture well (Figure 2A). Surface tension forces are leveraged to passively move fluid added to the input port through the device to the output port23. In a typical germination experiment, a cell suspension is added to the input port, gets pulled through the device and cells that end up above the culture well settle to the bottom and remain there through subsequent fluid exchanges; because of the geometry of the device, the wall shear stress is low enough at the bottom of the well (scales with well depth) for non-adherent cells to stay largely undisturbed while fluid is exchanged in the device17. Each fluid addition replaces approximately half the volume of the culture well (Supplemental Figure 1), so media and other compounds were added at 2X concentrations. The microscale well device facilitates efficient testing of conditions; the low volume of each device, 5 μL, and small footprint (6 devices fit on one microscope slide) make it feasible to efficiently test numerous conditions at the same time.
Figure 2.
Microscale well device operation and quantitative phenotypic information extraction. (A) Schematics of the device demonstrate a platform for culture and imaging of non-adherent cells. The photo shows 6 devices bonded to a glass microscope slide and filled with 5 μL dye. (B) Image processing steps include applying a density threshold to the raw images and then automatically detecting and measuring cells. (C) Measurements are used to bin cells into spore, yeast, or intermediate categories based on size and shape.
The microscale well device enables analysis of hundreds of individual cells throughout the germination process. Loading spores at a concentration of 104/μL resulted in a dispersed monolayer of cells at the bottom of each well that could be imaged over time. Approximately 600 cells were evaluated from each device, ~300 in each image of two separate fields. Performing each condition in technical triplicate then resulted in ~1800 cells to analyze. To facilitate the analysis, we developed an automated algorithm to process the images and record morphological measurements of each individual cell (Figure 2B). The algorithm first applies a filtering threshold based on pixel density, to pick out the cells from the background. It then identifies cells from debris on the basis of size and shape and measures and records attributes of each cell including area and aspect ratio (width/length). Finally, based on these attributes, it can identify cells as spores, yeast, or intermediates (Figure 2C and 3). By combining the microscale well device with the automated image processing algorithm, it is possible, for the first time, to collect large-scale, quantitative phenotypic measurements of the fungal germination process.
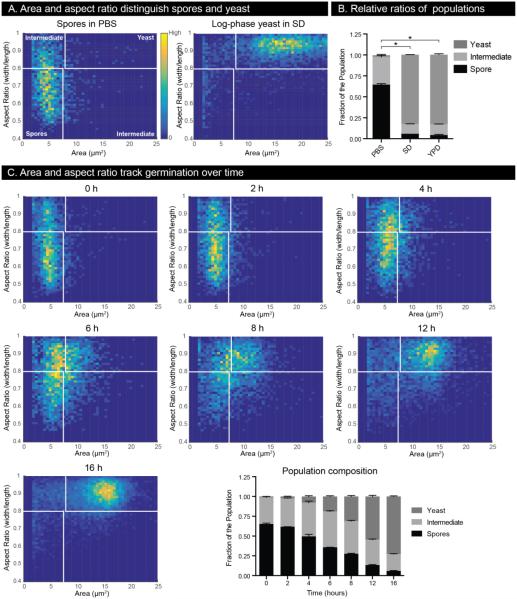
Figure 3.
Germination in microscale devices can be tracked by measuring cell area and aspect ratio. (A) 2D histograms of cell size and aspect ratio for a population of spores in PBS and a population of log-phase yeast in SD. Plots contain cell measurements from three independent wells. Colors are normalized on each plot such that yellow represents the area and aspect ratio combination with the most cells observed and dark blue represents area and aspect ratio combinations that were not observed. Note that cells in the lower left quadrant are defined as spores, those in the upper right quadrant as yeast, and all remaining are classified as intermediates. (B) Stacked bar plots of normalized cell population composition based on the number of cells in each category defined in (A). * indicates p-value <0.05 compared with PBS, and error bars represent standard deviation. (C) Germination dynamics of spores visualized by 2D histograms of cell area and aspect ratio as well as a stacked bar plot of the population composition over time.
Spore germination can be evaluated on a cell-by-cell basis using microdevices
To evaluate the robustness of the microscale assay in detecting differences between distinct cell populations, dormant spores in PBS (saline, non-germinating condition) and actively growing yeast in SD (minimal medium, growth condition) were evaluated in the devices, and the resulting data were represented in two-dimensional (2D) histograms (Figure 3A) and stacked bar plots (Figure 3B). In the 2D histograms, small, ovoid cells reside in the lower left quadrant and large, round cells reside in the upper right quadrant. As anticipated, a population of pure, dormant (un-germinated) spores populates primarily the lower left quadrant of the plot, whereas a population of vegetatively growing yeast populates the upper right quadrant (Figure 3A). Based on the natural separation of the two populations, cutoffs, shown in white, were chosen to define spores, yeast, and intermediate cells (Figure 3A). Small yeast, such as new daughter cells, would be expected to populate the upper left quadrant of the plot (small and round). Because these cells could confound our analysis of spore germination, we made a very stringent cut-off for the aspect ratio of spores of <0.8 to exclude small yeast. While this excludes some cells that are actually spores, we chose this cut-off to limit our analysis to the most clear-cut representatives of the two cell populations. As such, cells falling outside the quadrants defined for spores and yeast were designated as intermediates, representing neither spores nor yeast in our analysis (Figure 3A). These data were also represented in normalized stacked bar plots to reveal the relative ratios of each population (Figure 3B). Populations of pure spores in PBS typically harbored ~60% spores, ~40% intermediates, and <1% yeast. Yeast grown in either minimal medium (SD) or rich medium (YPD) were indistinguishable from one another in the devices and typically harbored <5% cells classified as spores (visualized microscopically as slightly ovoid daughter cells), ~10% intermediates, and ~85% yeast (Figure 3B).
To assess the changes in a spore population over the course of germination, we incubated spores in the devices in SD medium at 30°C and evaluated the cells every 2 hours for 16 hours. The population composition underwent a clear and steady transformation from comprising primarily spores to comprising primarily yeast (Figure 3C). To visualize the characteristics of the cell population during germination, 2D histograms were generated for the population of evaluated cells at each time point. We observed that in response to germination conditions, spores as a population initiate the germination process by first shifting in aspect ratio to become slighter rounder (Figure 3C, 2h and 4h) and then increasing in size (Figure 3C, 6h and 8h). The population continues to increase in both size and aspect ratio until 12 hours of germination at which time they have nearly all transitioned to round cells, which then enlarge to fully complete the germination process. After 16 hours of germination, the population exhibits the properties of a population of yeast (aspect ratio approaching ~0.9 and area approaching ~15 μm2). This process is also captured in a stacked bar plot of the relative proportions of cell types present over the course of the assay (Figure 3C, last panel). The population transition is highly reproducible across different preparations of both spores and devices and is facilitated by the largely synchronous response of the spore population to germination conditions.
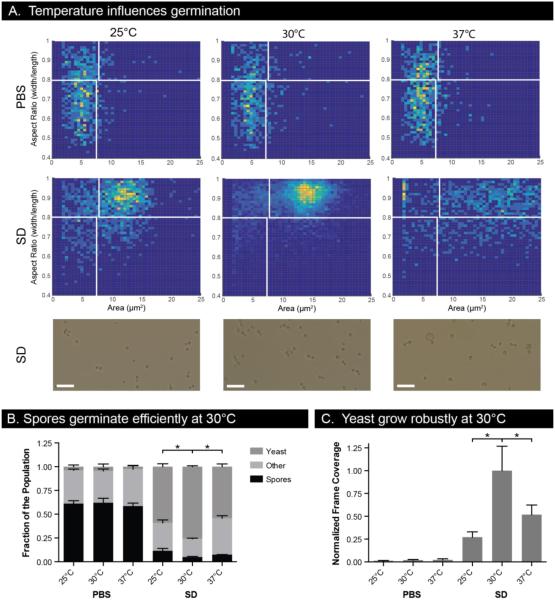
Figure 4.
Temperature dependence of spore germination and yeast growth. (A) Cell size and aspect ratio histograms of spores grown in PBS or SD for 16 hours at 25°C, 30°C, and 37°C. 2D histograms include cell populations from three independent wells. Images show representative cells in SD after 16 hours. Scale bars are 25 μm. (B) Cell population compositions for conditions in (A). * indicates p-value <0.05 in an unpaired t-test comparing subpopulations to those at 30°C. (C) Yeast growth after 16 hours as measured by frame coverage normalized to SD medium at 30°C. Error bars represent standard deviation.
Figure 6.
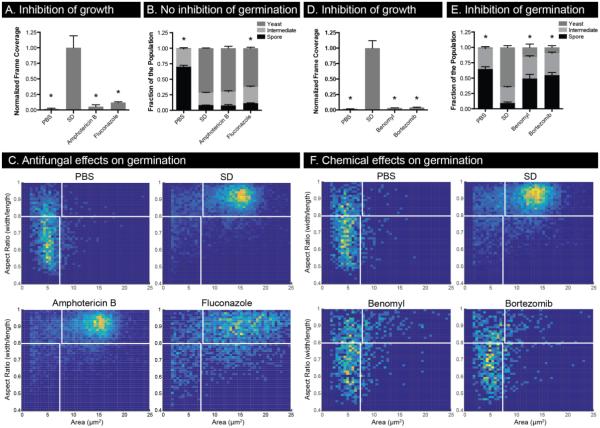
Spore germination and yeast growth are affected differently by antifungals and other chemical compounds. SD containing amphotericin B (1 μg/mL), fluconazole (16 μg/mL), benomyl (62.5 μg/mL), or bortezomib (960 μg/mL) was applied to yeast (A, D) and spores (B, E). Yeast growth was measured by normalized frame coverage (A, D) while germination was measured both with population composition (B, E) and 2D histograms of cell area and aspect ratio (C, F). 2D histograms include cell populations from three independent wells. * indicates p-value <0.05 compared to SD, and error bars represent standard deviation.
To ensure that the changes in spore populations were dependent on the introduction of growth medium as opposed to temperature or PDMS contact, we incubated spores in PBS at 30°C for 15 hours and assessed the population every three hours. At each time point and after 15 hours total, we recorded a population distribution identical to that of the original population, indicating that no germination occurred in response to incubation of spores in the microscale well devices (Supplemental Figure 2A).
To determine the ability of the assay to discriminate among similar germination conditions, C. neoformans spores were exposed to increasing concentrations of SD medium. After incubation for 16 hours at 30°C, the cell population profiles showed a steady decrease in fraction of spores and an increase in fraction of yeast with increasing concentrations of SD (Supplemental Figure 2B). The assay clearly distinguishes the germination effects of 0.05X SD from PBS and 0.1X SD indicating that the resolution of the assay is appropriate for quantitatively comparing similar conditions. Additionally, the spore population germinated in devices in response to 1X SD was compared to a population of yeast grown in macroscale liquid culture in 1X SD at 30°C revealing highly similar population distributions (Supplemental Figure 2B), indicating complete germination of the spores.
Finally, the microscale well device was applied to assay yeast in order to enable comparisons between germination of spores and vegetative growth of yeast. Yeast were added to the devices at a concentration of 103/μL to give the cells ample room to grow and growth was quantitatively assessed in the microscale devices by simply measuring the percent of each image covered with cells following 16 hours of incubation. As anticipated, yeast growth was greater in rich YPD medium than in minimal SD medium, and no growth occurred in PBS (Supplemental Figure 2C).
Thus, we established the baseline conditions under which both spore germination and yeast growth could occur in the microscale well devices and verified the parameters for the assay necessary for high resolution measurements. The algorithms developed for analysis can now provide a much higher resolution view of germination on a large scale, enabling quantitative, population-based assessments of spores and yeast in response to a variety of conditions.
Temperature affects efficiency and synchronicity of spore germination and yeast growth
C. neoformans strains can grow over a wide range of temperatures, but the effect of temperature on germination is unknown. To test the effect of temperature on germination, we incubated spores at 25°C, 30°C, and 37°C in SD medium for 16 hours in the microscale well devices. Overall, we observed clear effects of temperature on the rate, synchronicity, and completion of germination. 2D histograms demonstrate that germination at 30°C leads to the most homogenous and completely germinated population compared with germination at either 25°C or 37°C (Figure 4A). All temperatures led to a significant reduction in the fraction of the population classified as spores, but the 30°C condition led to significantly more cells classified as yeast instead of intermediate cells (Figure 4B). It appears that germination simply occurs more slowly at 25°C than 30°C, resulting in a less fully-germinated population after 16 hours. In contrast, germination at 37°C led to a population of round cells that exhibited significant variations in size (Figure 4A), consistent with our previous observations of cells of the JEC21 strain background at 37°C (data not shown).
We assessed the effect of temperature alone on germination in the absence of nutrients (PBS) and detected no significant differences (Figure 4A, B). These data show that although temperature may play a role in the kinetics of germination of C. neoformans spores, it is not a trigger or requirement for germination as it is in other fungal systems. We also assayed the effect of temperature on vegetative growth of yeast in the devices, and observed that 30°C allowed for the most robust growth (Figure 4C). Optimal germination and growth temperatures will likely be different among different strain backgrounds and isolates, but for the laboratory strains used in these studies, 30°C was the optimal temperature for both robust yeast growth and efficient and synchronous germination.
Germination relies on synergy of multiple nutrient sources
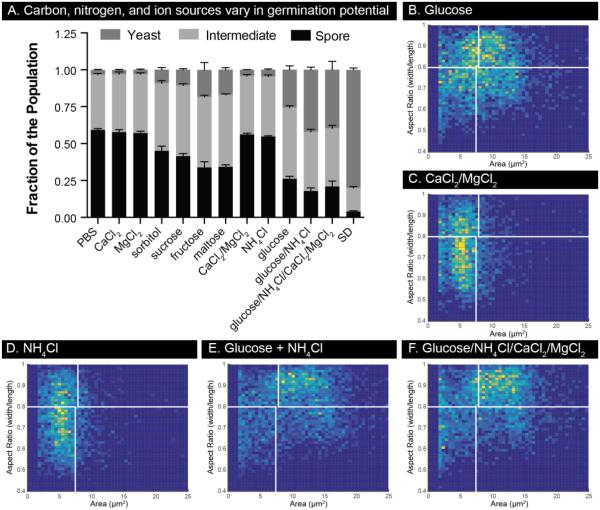
To determine the media components necessary to initiate and sustain the germination process, we measured germination under different nutrient conditions (Figure 5A). Spores were seeded into the devices and given PBS supplemented with different compounds. After 16 hours at 30°C, we observed that all of the carbon sources tested initiated changes consistent with germination; however, even the most effective sugar, glucose, was not able to facilitate complete germination on its own (Figure 5B). Calcium and magnesium, alone or in combination, were not initiators of germination, nor was ammonium chloride (Figures 5C, D, Supplemental Figure 3). A combination of glucose and ammonium chloride led to the most complete germination profile of the panel, with and without ions; however, this condition fell well short of the germination induced by SD medium (Figures 5A, E, F, Supplemental Figure 3). The ability of spores to germinate to completion in SD indicates that germination requires conditions beyond simply having sources of carbon and nitrogen. SD is made with yeast nitrogen base (lacking amino acids), which contains a host of vitamins, trace elements and salts that appear to be important for germination.
Figure 5.
Germination of spores requires both carbon and nitrogen sources. (A) The germination potential of a panel of compounds was tested on spores. All compounds were diluted in PBS to a working concentration of 100 mM except CaCl2 and MgCl2 which were used at 1mM. Germination was allowed to proceed for 16 hours at 30°C after which population composition was assessed. Error bars represent standard deviation. (B)-(F) Histograms of cell size and aspect ratio for a subset of conditions from (A). 2D histograms include cell populations from three independent wells.
Chemical compounds differentially affect germination and vegetative growth
Inhibitors of C. neoformans have been identified solely based on the inhibition of yeast growth, but they have not been tested for their effects on germination. Using the microscale well device and algorithm, we tested the ability of the antifungal drugs amphotericin B and fluconazole as well as the laboratory compounds benomyl and bortezomib to prevent spore germination. As expected, both amphotericin B and fluconazole inhibited yeast growth in the devices at previously identified inhibitory concentrations (Figure 6A)24,25. In sharp contrast, however, neither drug had a substantial effect on the process of germination (Figure 6B). 2D histograms reveal that germination in the presence and absence of amphotericin B are indistinguishable; germination in the presence of fluconazole results in nearly complete germination, but a much larger distribution of cell size in the resulting yeast population (Figure 6C). Both amphotericin B and fluconazole are known to act via effects on fungal cell membrane components, suggesting that perhaps there is little membrane synthesis and/or exposure during germination. A key aspect of this finding is that the processes of spore germination and vegetative growth appear to progress via different mechanisms, leading to the possibility that germination could provide specific, unique targets for inhibition.
In addition to evaluation of the clinical drugs amphotericin B and fluconazole, benomyl and bortezomib, cell toxins selected for their orthogonal mechanisms of action, both inhibited yeast growth in the devices (Figure 6D). However, both benomyl and bortezomib also inhibited germination (Figure 6E). In both cases it appears that the compounds inhibited germination almost entirely as shown by the 2D histograms (Figure 6F). Benomyl and bortezomib are microtubule and proteasome inhibitors, respectively, suggesting that microtubule reorganization and protein degradation are required for germination in addition to yeast growth. In all cases, use of the high-resolution, quantitative assay in conjunction with chemical probes revealed effects on germination and growth that can be used to determine functions and pathways important during germination.
nhp6aΔ strains show delays during germination
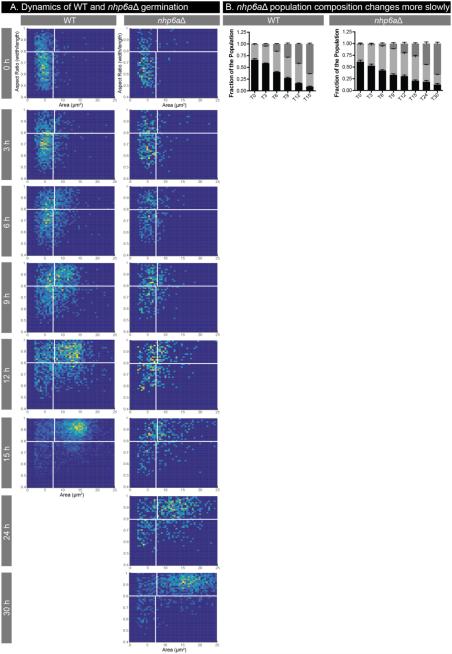
Another approach to probing the molecular mechanisms of germination is via the use of genetic mutants, which allow us to compare germination in the presence or absence of a given protein. Based on gene expression data from spores during germination26, we hypothesized that a mutant harboring a deletion in the gene CNF04730 would show phenotypes in spore processes, such as germination. We deleted CNF04730, which is similar to the chromatin remodeler NHP6A in S. cerevisiae, and assessed the resulting knockout strains and their spores for phenotypes. Deletion strains of the C. neoformans NHP6A showed no discernable phenotypes, including vegetative growth, high temperature growth, growth in the presence of cell wall stressors, sexual development, or spore formation (data not shown and Supplemental Figure 4). To assess germination of nhp6aΔ spores, we carried out comparative germination assays of wild type and nhp6aΔ spores in the devices in SD at 30°C. We determined that the nhp6aΔ spores were capable of germinating to completion, but they did so more slowly than wild type spores, requiring nearly twice as long to complete the process (15 hours vs. 30 hours). 2D histograms show the wild type populations of cells transitioning from spores to yeast as expected. However, in the nhp6aΔ cells, there appears to be slower germination overall with a pause in germination beginning between 6 and 9 hours in which the cells do not progress (Figure 7A); while the 6 hour and 9 hour profiles for the nhp6aΔ cells are nearly identical, for wild type cells, there is a clear progression to rounder, larger cells. By 12 hours, the wild type cells are nearing the end of germination, but the nhp6aΔ cells are still transitioning. This timing indicates an approximately 6 hour delay in germination resulting in profiles in which the 15 hour mutant population is roughly equivalent to the 9 hour wild type population (Figure 7A). The germination delay is evident in stacked bar plots (Figure 7B) in which the population of nhp6aΔ mutant cells remains relatively constant between 9 and 12 hours, and although germination continues, proceeds more slowly than wild type until germination is complete after 30 hours. These data indicate that NHP6A plays a role in germination (after initiation) that is important for sustaining and completing the process. As NHP6A is predicted to be involved in chromatin remodeling in S. cerevisiae, this result suggests that chromatin remodeling is part of the molecular process of germination. Prior to developing our assay in the microscale well devices, it would have been extremely difficult to determine the extent or timing of the nhp6aΔ germination defect. Using the devices, we now have the capacity to evaluate mutant spores during germination in detail across a population and determine the contributions of specific genes to the germination process.
Figure 7.
Germination dynamics differ between WT and nhp6aΔ spores. WT spores and nhp6aΔ spores were incubated at 30°C in SD and imaged periodically for germination. (A) 2D histograms of WT and mutant spores germinating in SD. All plots include cell populations from three independent wells. (B) Population composition of each condition over time. Error bars represent standard deviation. At 9 hours, the WT and nhp6aΔ spore fractions are not significantly different though the yeast fractions are different (p-value <0.001). At 12 hours, both spore and yeast population fractions are significantly different for WT compared to nhp6aΔ (spore p-value <0.01, yeast p-value <0.001). At the end of germination, 15 hours for WT and 30 hours for nhp6aΔ, both spore and yeast population fractions are not statistically different between WT and nhp6aΔ.
Discussion
Germination is essential for spore-forming organisms, but surprisingly little is known about this process, particularly in pathogenic fungi. Here we describe the development of a microscale device-based assay for interrogating the process of fungal germination using spores of the human pathogen C. neoformans. By bringing together the biology of C. neoformans spores with advances in microfluidic technologies, we created a platform with which we can now evaluate a eukaryotic differentiation process at a level of resolution not possible in other systems. The assay reproducibly quantifies the morphological transition at both the cell- and population-level on a large scale, opening up myriad possibilities for probing the molecular events that drive the germination process.
There have been several previous efforts to apply microscale technologies to spore germination assays in both bacteria and fungi27-31. Some rely on changes in electrical conductivity of media27 or polyaniline mesh28 to track germination of Bacillus spp. spores. While these assays are very sensitive on a population basis, they cannot take single-cell measurements into account. In contrast, live cell imaging of germinating Bacillus spores provided single-cell information but could not integrate well across a population29. This was also the case for microscopy-based approaches to evaluate Aspergillus ochraceous in microbioreactors30 and microdevice channels to assess filamentous fungal responses to plant defense proteins31. All of these approaches provide important microscale tools to probe germination, but each has limitations. The microscale germination assay presented here provides many advantages, including the ability to make quantitative measurements of thousands of spores at both the single-cell and population levels, a small scale that enables testing the effects of valuable or rare chemicals or metabolites for their effects on cells, and a device design that can be accessed with a simple micropipette. Avoiding the sophisticated fluid handling equipment common in most microfluidics approaches allows microscale assays to move out of specialized engineering labs and into mainstream biology32,33.
The biology of spore germination is integral to understanding survival of spore-forming fungi, and the spores of many fungal pathogens must germinate in the host to cause disease. However, a facile system with the tools to interrogate germination in detail has not been available. C. neoformans serves as an optimal model organism to probe spore biology because of the general molecular, genetic, animal, and biochemical tools available in the system as well as the specific properties of C. neoformans spores. Spores can be purified in sufficient quantities for large-scale studies, they are stable as a dormant population (in the absence of nutrients) for prolonged periods and across a range of temperatures, and the morphological transition from spore to yeast is measurable, consistent, and most importantly, synchronous. By combining these features with the microscale devices, we have created a system in which to discover, parse, and manipulate a fundamental differentiation process in a medically relevant eukaryotic organism.
Experiments presented here to develop and optimize the microscale assay assessed the effects of temperature and nutrient conditions on germination, leading to several basic discoveries about C. neoformans germination. First, unlike some fungi, extreme conditions such as exposure to very high temperature or pressures are not required to initiate or sustain germination of C. neoformans. Second, germination proceeded across a range of temperatures (25°C to 37°C) representative of conditions C. neoformans encounters both in the environment and in the human host, suggesting that germination could occur in these environments, given the proper nutrients. Third, a small screen of potential nutrients necessary for germination confirmed the need for a carbon source to initiate germination, but it also revealed that germination could not continue without a source of nitrogen. Thus, the metabolic requirements for different stages of germination vary over time. Future studies with the microscale germination assay will be able to test germination across a much wider range of temperatures and conditions to determine the plasticity and robustness of the process, pinpoint precise metabolic requirements, and identify molecular events and pathways required for robust germination.
Using the assay to test the effects of well-characterized antifungal drugs, amphotericin B and fluconazole, revealed that although these drugs inhibited growth, neither prevented germination. The differential effects of these drugs on germination and vegetative growth suggest that at least some of the molecular components of the two processes are distinct. This contrasts with studies in S. cerevisiae suggesting that germination is not a specific differentiation process but is instead similar to an exit from stationary phase growth34,35. If germination is indeed molecularly distinct in C. neoformans, it could present a new target for the development of antifungal drugs, an exciting possibility given the extremely limited arsenal of drugs currently available to prevent and treat fungal disease in humans. The microscale devices provide a tested and ready platform for the identification and assessment of new potential antifungal drugs targeting germination.
The use of antifungal compounds with known mechanisms in the microscale devices can also help identify the specific molecular events and pathways that are important during germination. For example, amphotericin B is a polyene drug that binds and permeabilizes membranes that contain ergosterol (the fungal equivalent of cholesterol)36. Resistance of germination to amphotericin B suggests that germinating spores either do not harbor ergosterol in their cell membranes or that existing ergosterol in germinating spores is not accessible to amphotericin B during germination. It is not until the spores become actively growing yeast that amphotericin B can exert its fungicidal effects. A parallel result occurred with fluconazole, which inhibits the biosynthesis of ergosterol. The azole class of drugs inhibits 14α-demethylase (fungal cytochrome P450), leading to fungistatic decreases in ergosterol synthesis37. Again, based on the resistance of germination to fluconazole, it appears that either new membrane synthesis is not required for germination or that existing stores of ergosterol are sufficient for new membrane production. Benomyl and bortezomib both inhibit fundamental cell processes (microtubule polymerization and protein degradation, respectively)36,38,39, so we anticipated that they would inhibit both germination and growth. However, because of the extremely high resolution of the assay, we can also place the timing of inhibition in the time course of germination. In the cases of benomyl and bortezomib, inhibition occurs very early during germination as evidenced by very little change in the cell population over time. These data suggest that dynamic microtubule changes and protein turnover are required very early in the germination process. This approach allows for the first time the development of specific, testable hypotheses of the molecular mechanisms of spore differentiation. Screens with other specific cell inhibitors could pinpoint later times in germination when inhibition occurs, adding resolution to molecular events and identifying tools for additional testing.
In concert with a chemical screening approach, we also have the tools to evaluate mutants of C. neoformans to determine the mechanisms of germination. The nhp6aΔ strain was generated based on the finding that spores harbor relatively high levels of NHP6A transcript compared to yeast. We observed that in C. neoformans the nhp6aΔ spores form slightly smaller colonies than wild type spores on solid agar. The nature of this apparently minor defect became clear only after quantifying germination with the microscale assay, using its high resolution capabilities to detect differences in cell populations. In the case of nhp6aΔ, we determined that germination progresses significantly slower than wild type but ultimately goes to completion. The homologs of NHP6A in S. cerevisiae (NHP6A and B) appear to be involved in chromatin remodeling40,41. Based on the role of NHP6A and B in S. cerevisiae, we hypothesize that the time at which germination slows in the C. neoformans nhp6aΔ is a time at which chromatin decondensation and/or remodeling is occurring. Using this information and overall approach, we will be able to carry out epistasis analysis with different germination mutants, screen for mutants in the process using sensitized strains, and use mutants in concert with inhibitory drugs to identify the nature and order of molecular events that occur during germination.
These studies showed how a developed microscale assay is useful in quantifying and assessing the progression of germination in spores from the fungal pathogen, C. neoformans. In addition, with simple adaptations of the image processing algorithms, the assay could be adapted to study morphological changes in other organisms, including filamentous fungi, while providing the benefit of high cell- and population-level resolution that comes with using the microscale platform. The assay could be adapted to monitor germling development in Aspergiulls spp. or early hyphal development in C. albicans. This platform could even be used to monitor and measure developmental transitions in the life cycle of parasites such as Trypanosoma brucei. The germination assay is a tool with which to discover, parse, and manipulate a fundamental eukaryotic differentiation process and one that will be extended to further studies in C. neoformans and other systems with key biology dependent on morphological transitions.
Summary
We have developed a germination assay, composed of a microfluidic platform and an image processing pipeline, that enables quantitative, statistically rigorous single cell and population-based assessments of germination in high resolution for the first time. We used the assay to test conditions needed for C. neoformans spore germination, a representative model of pathogenic fungal spore germination and overall eukaryotic differentiation. The results demonstrate the power of the platform to be used with chemical and genetic approaches to parse the germination process at the molecular level and offer insights into eukaryotic survival in the environment and in the mammalian host.
Supplementary Material
Innovation statement.
Spore formation is a common strategy used by microbes to survive harsh conditions, but this is effective only if spores can return to normal growth (germinate) when conditions improve. Germination is therefore critical for survival of spore-forming fungi, including human pathogens, but it is poorly understood. Here we integrate advances in microfluidics and image analysis with a well-developed fungal molecular/genetic system in Cryptococcus neoformans to develop a high-resolution assay of germination. This technologic innovation allowed us to systematically explore factors that influence germination and gain biological insights into the key conditions and events required for germination. Using this assay to discover fundamental germination biology opens up the possibility of designing better antifungal drugs to prevent and/or treat deadly fungal diseases.
Acknowledgements
The authors wish to thank Mingwei Huang for experimental expertise and both Mingwei Huang and Robert Brazas for critical comments on the manuscript. This work was supported in part by the National Institute of Health: R01 AI064287 (C.M.H.), Molecular Biosciences Training Grant T32 GM007215 (N.M.W.), P30 CA014520 (D.J.B.); and in part by the National Library of Medicine: Computation and Informatics in Medicine and Biology Training Grant 5T15LM007359 (L.B.). L.B. is a student in the UW-Madison MSTP (T32 GM008692).
Footnotes
ESI is available
Competing financial interests
David J. Beebe holds equity in Bellbrook Labs, LLC, Tasso, Inc., Stacks to the Future, LLC and Salus Discovery, LLC. Christina M. Hull has a financial interest in Lucigen Corporation. The remaining authors declare no competing financial interests.
References
- 1.Brown JKM, Hovmøller MS. Science. 2002;297:537–541. doi: 10.1126/science.1072678. [DOI] [PubMed] [Google Scholar]
- 2.Setlow P. J. Appl. Microbiol. 2006;101:514–525. doi: 10.1111/j.1365-2672.2005.02736.x. [DOI] [PubMed] [Google Scholar]
- 3.Freese EB, Chu MI, Freese E. Journal of Bacteriology. 1982;149:840. doi: 10.1128/jb.149.3.840-851.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Neiman AM. Microbiology and Molecular Biology Reviews. 2005;69:565–584. doi: 10.1128/MMBR.69.4.565-584.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Neiman AM. Genetics. 2011;189:737–765. doi: 10.1534/genetics.111.127126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wyatt TT, Wösten HAB, Dijksterhuis J. In: Advances in Applied Microbiology. Gadd G, Sariaslani S, editors. Vol. 1. Elsevier; Amsterdam: 2013. pp. 43–91. 2. [DOI] [PubMed] [Google Scholar]
- 7.Krijgsheld P, Bleichrodt R, van Veluw GJ, Wang F, Muller WH, Dijksterhuis J, Wösten HAB. Studies in Mycology. 2013;74:1–29. doi: 10.3114/sim0006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nemecek JC, Wüthrich M, Klein BS. Science. 2006;312:583–588. doi: 10.1126/science.1124105. [DOI] [PubMed] [Google Scholar]
- 9.Brown GD, Denning DW, Gow NAR, Levitz SM, Netea MG, White TC. Sci Transl Med. 2012;4:165rv13, 165rv13. doi: 10.1126/scitranslmed.3004404. [DOI] [PubMed] [Google Scholar]
- 10.Levitz SM. Rev. Infect. Dis. 1991;13:1163–1169. doi: 10.1093/clinids/13.6.1163. [DOI] [PubMed] [Google Scholar]
- 11.Casadevall A, Perfect JR. Cryptococcus neoformans. ASM Press; Washington, DC: 1998. [Google Scholar]
- 12.Perfect JR. FEMS Immunology & Medical Microbiology. 2005;45:395–404. doi: 10.1016/j.femsim.2005.06.005. [DOI] [PubMed] [Google Scholar]
- 13.Driver JA, Saunders CA, Heinze-Lacey B, Sugar AM. J Acquir Immune Defic Syndr Hum Retrovirol. 1995;9:168–171. [PubMed] [Google Scholar]
- 14.Kwon-Chung KJ. Mycologia. 1976;68:821–833. [PubMed] [Google Scholar]
- 15.Geijer C, Joseph-Strauss D, Simchen G, Barkai N, Hohmann S. In: Dormancy and Resistance in Harsh Environments. Lubzens E, Cerda J, Clark M, editors. Vol. 1. Springer; Heidelberg: 2010. pp. 29–41. 3. [Google Scholar]
- 16.Osherov N, May GS. FEMS Microbiol Lett. 2001;199:153–60. doi: 10.1111/j.1574-6968.2001.tb10667.x. [DOI] [PubMed] [Google Scholar]
- 17.Young EWK, Pak C, Kahl BS, Yang DT, Callander NS, Miyamoto S, Beebe DJ. Blood. 2012;119:e76–e85. doi: 10.1182/blood-2011-10-384347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Duffy DC, McDonald JC, Schueller OJA, Whitesides GM. Anal. Chem. 1998;70:4974–4984. doi: 10.1021/ac980656z. [DOI] [PubMed] [Google Scholar]
- 19.Sherman F, Fink GR, Hicks JB. Laboratory course manual for methods in yeast genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor; 1987. [Google Scholar]
- 20.Botts MR, Giles SS, Gates MA, Kozel TR, Hull CM. Eukaryotic Cell. 2009;8:595–605. doi: 10.1128/EC.00352-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schneider CA, Rasband WS, Eliceiri KW. Nat Meth. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Davidson RC, Blankenship JR, Kraus PR, de Jesus Berrios M, Hull CM, D'Souza C, Wang P, Heitman J. Microbiology. 2002;148:2607–15. doi: 10.1099/00221287-148-8-2607. [DOI] [PubMed] [Google Scholar]
- 23.Walker GM, Beebe DJ. Lab Chip. 2002;2:131–134. doi: 10.1039/b204381e. [DOI] [PubMed] [Google Scholar]
- 24.Archibald LK, Tuohy MJ, Wilson DA, Nwanyanwu O, Kazembe PN, Tansuphasawadikul S, Eampokalap B, Chaovavanich A, Reller LB, Jarvis WR, Hall GS, Procop GW. Emerging Infect. Dis. 2004;10:143–145. doi: 10.3201/eid1001.020779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Govender NP, Patel J, van Wyk M, Chiller TM, Lockhart SR, Group for Enteric, Respiratory and Meningeal Disease Surveillance in South Africa (GERMS-SA) Antimicrob. Agents Chemother. 2011;55:2606–2611. doi: 10.1128/AAC.00048-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Botts MR, Huang M, Borchardt RK, Hull CM. Eukaryot Cell. 2014;13:1158–1168. doi: 10.1128/EC.00152-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu YS, Walter TM, Chang WJ, Lim KS, Yang L, Lee SW, Aronson A, Bashir R. Lab Chip. 2007;7:603–610. doi: 10.1039/b702408h. [DOI] [PubMed] [Google Scholar]
- 28.Zabrocka L, Langer K, Michalski A, Kocik J, Langer JJ. Lab Chip. 2015;15:274–282. doi: 10.1039/c4lc01009d. [DOI] [PubMed] [Google Scholar]
- 29.Pandey R, Ter Beek A, Vischer NOE, Smelt JPPM, Brul S, Manders EMM. PLoS ONE. 2013;8:e58972. doi: 10.1371/journal.pone.0058972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Demming S, Sommer B, Llobera A, Rasch D, Krull R, Büttgenbach S. Biomicrofluidics. 2011;5 doi: 10.1063/1.3553004. 014104-1-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mattupalli C, Spraker JE, Bertheir E, Charkowski AO, Keller NP, Shepherd RW. Plant Health Research. 2014;15:130–134. [Google Scholar]
- 32.Meyvantsson I, Warrick JW, Hayes S, Skoien A, Beebe DJ. Lab Chip. 2008;8:717–724. doi: 10.1039/b715375a. [DOI] [PubMed] [Google Scholar]
- 33.Sackmann EK, Fulton AL, Beebe DJ. Nature. 2014;507:181–189. doi: 10.1038/nature13118. [DOI] [PubMed] [Google Scholar]
- 34.Joseph-Strauss D, Zenvirth D, Simchen G, Barkai N. Genome Biol. 2007;8:R241. doi: 10.1186/gb-2007-8-11-r241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kloimwieder A, Winston F. G3 (Bethesda) 2011;1:143–149. doi: 10.1534/g3.111.000323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cvek B, Dvorak Z. CDM. 2009;10:1483–1499. [Google Scholar]
- 37.Mahmoud A Ghannoum LBR. Clin. Microbiol. Rev. 1999;12:501. doi: 10.1128/cmr.12.4.501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Singh P, Rathinasamy K, Mohan R, Panda D. IUBMB Life. 2008;60:368–375. doi: 10.1002/iub.42. [DOI] [PubMed] [Google Scholar]
- 39.Yang C, Hamel C, Vujanovic V, Gan Y. ISRN Ecology. 2011;2011:1–8. [Google Scholar]
- 40.Paull TT, Carey M, Johnson RC. Genes and Development. 1996;10:2769–2781. doi: 10.1101/gad.10.21.2769. [DOI] [PubMed] [Google Scholar]
- 41.Stillman DJ. Biochimica et biophysica acta. 2010;1799:175. doi: 10.1016/j.bbagrm.2009.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.