Abstract
Immune recognition by T cells relies on the presentation of pathogen-derived peptides by infected cells but the persistence of chronic infections calls for new approaches to modulate immune recognition. Antigen cross-presentation, the process by which pathogen antigens are internalized, degraded and presented by MHC-I is crucial to prime CD8 T cell responses. The original degradation of antigens is performed by pH-dependent endolysosomal cathepsins. Here we show that HIV protease inhibitors (PIs) prescribed to HIV-infected persons variably modulate cathepsin activities in human antigen presenting cells (APCs), dendritic cells and macrophages, and CD4 T cells, three cell subsets infected by HIV. Two HIV PIs acted in two complementary ways on cathepsin hydrolytic activities: directly on cathepsins and indirectly on their regulators by inhibiting Akt kinase activities, reducing NADPH oxidase 2 (NOX2) activation, lowering phagolysosomal ROS production and pH, which altogether led to enhanced cathepsin activities. HIV PIs modified endolysosomal degradation and epitope production of proteins from HIV and other pathogens in a sequence-dependent manner. They altered cross-presentation of antigens by dendritic cells to epitope-specific T cells and T cell-mediated killing. HIV PI-induced modulation of antigen processing partly changed the self MHC-peptidome displayed by primary human cells. This first identification of prescription drugs modifying the regulation of cathepsin activities and the MHC-peptidome may provide an alternate therapeutic approach to modulate immune recognition in immune disease beyond HIV.
Introduction
HIV infection has been considered as a chronic illness since the availability of highly active antiretroviral treatments (HAART), a combination of nucleoside reverse transcriptase inhibitors (NRTIs), non-NRTIs (nNRTIs), protease inhibitors (PIs), and integrase inhibitors given to HIV-infected patients to suppress HIV replication (1). HIV PIs block the HIV aspartyl protease preventing the cleavage of HIV Gag-Pol polyproteins and the conversion of HIV particles into mature infectious virions (2). The ability of proteasomes to cleave similar bonds as HIV-1 protease (3) raised questions about interactions between HIV PIs and proteasomal catalytic sites.
Proteasomes play a central role in the generation of MHC-I peptides (4). They degrade full-length proteins and defective ribosomal products into peptides that can be further shortened by cytosolic aminopeptidases and endopeptidases. Some peptides are translocated by the TAP complex into the endoplasmic reticulum (ER), where they can be further trimmed by ER-resident aminopeptidases (ERAP1 or ERAP2) and, if they contain appropriate anchor residues, loaded onto MHC-I and displayed at the cell surface. HIV PIs ritonavir (RTV), saquinavir (SQV), and nelfinavir (NFV) inhibit the proteolytic activities of the proteasome, a threonine-protease (5, 6), causing intracellular accumulation of polyubiquitinated proteins (7). In mice infected with lymphocytic choriomeningitis virus (LCMV) and treated with RTV, cytotoxic T cell (CTL) responses against LCMV epitopes were changed (6). We previously showed that HIV PIs altered proteasome and aminopeptidase activities in primary human cells, modified HIV protein degradation patterns in a sequence-dependent manner, epitope production and presentation, altering CTL responses (8).
Professional antigen presenting cells (APCs) like dendritic cells (DCs) and macrophages can cross-present exogenous antigens (9, 10). Antigens transiting through endosomes or phagosomes are partially degraded by cathepsins. Degradation fragments can escape into the cytosol for processing and presentation in the direct antigen processing pathway (11), or be processed completely in the phagosomes by cathepsins before loading onto MHC-I (12, 13). The mechanisms of cross-presentation and connection with the direct presentation pathway remain incompletely understood (12-17). For instance hen egg-white lysozyme (HEL) targeted to early endosomes is completely processed and loaded unto MHC-I in the endosomal compartments while HEL targeted to late endosomes is not cross-presented (15). Soluble ovalbumin escapes into the cytosol for proteasomal processing before cross-presentation while HEL is processed and loaded within the endosome (15). The inhibition of cathepsin activities, especially cathepsin S altered epitope cross-presentation, showing the importance of cathepsins in this pathway (13, 15, 16). Cathepsin activities are controlled through pH regulation. Upon phagocytosis, phagosomes fuse with early and late endosomes and with lysosomes, thus acquiring progressively both the acidification machinery (mainly the V-ATPase complex) and lysosomal proteases (18). Acidification results in the activation of cathepsins with optimal proteolytic activities between pH 4-6 (18, 19). The activation and recruitment of NADPH oxidase 2 (NOX2) at the phagosomal membrane generates reactive oxygen species (ROS), increasing pH and limiting cathepsin activities and antigen degradation in DCs (20, 21).
We previously showed that HIV PIs alter the hydrolytic activities of non-aspartyl proteases such as proteasomes and aminopeptidases (8). However the impact of HIV PIs on cathepsins, cysteine and aspartyl proteases involved in cross-presentation, has never been assessed. The aim of this study was to investigate the effect of five HIV PIs (RTV, SQV, NFV, the ubiquitously used lopinavir/ritonavir (LPV/RTV [Kaletra]) and the most recently prescribed darunavir (DRV)) on cathepsin activities in primary APCs and CD4 T cells, the main targets of HIV infection and subsets involved in clearing pathogens or priming immune responses. Our results showed that RTV reduced cathepsin activities while SQV and NFV enhanced them. We identified two complementary mechanisms: a direct effect of PIs on cathepsins and an indirect effect on the regulatory pathway of lysosomal pH through reduction of kinase activities, NOX2 activation, phagolysosomal ROS production and pH, leading to enhanced cathepsin activities. Altered cathepsin activities modified antigen degradation patterns and epitope production in a sequence- and cell type-dependent manner, and was linked to variable cross-presentation of HIV epitopes, altered CTL recognition and partial modifications of the self MHC-peptidome.
Materials and Methods
Study approval
PBMC were isolated from buffy coats collected from anonymous blood donors approved by the Partners Human Research Committee under protocol 2005P001218 (Boston, USA). PBMC from HLA-typed blood donors were obtained after written informed consent and approval under protocol 2010P002121 by the Partners Human Research Committee.
Antiretroviral drugs
HIV PIs were obtained from Selleckchem (RTV, LPV, and DRV), Sigma (SQV), and Santa Cruz (NFV). All drugs were dissolved in 100% DMSO. Kaletra is a combination of LPV and RTV at 5:1 ratio. Kaletra concentrations mentioned in this article correspond to LPV amount. Aliquots of stock solutions of 10 mM were kept at −20C.
Measurement of phagosomal pH and ROS production
Monocytes and CD4 T cells were isolated from PBMC by immunomagnetic enrichment (StemCell). Monocytes were differentiated into DCs and macrophages as in (22). Phagosomal pH and ROS production were measured as described in (23). Briefly, beads were covalently coupled with FITC and FluoProbes 647 DHR 123 (Life Technologies). Cells treated for 30min with different HIV PIs (5uM) or inhibitors (10uM of DPI or 1uM Bafilomycin) were pulsed with beads for 20 min and extensively washed in cold PBS. The cells were then incubated at 37C and analyzed by flow cytometry at different times.
In vitro peptide degradation assay and MS analysis
2nmol of pure peptides were digested with 15μg of whole cell extracts at 37°C in pH4 degradation buffer (24). The degradation was stopped with 2.5μL of 100% formic acid and peptide fragments were purified by 5% trichloroacetic acid precipitation. Degradation peptides were identified by in-house MS as previously described (8, 19).
Proteolytic activities in live cells and cell extracts
Proteolytic activity measurement was performed as we previously described in (8, 19, 25). Briefly, 5x104 DCs, Møs or CD4T cells were pretreated with indicated PIs and omnicathepsin, cathepsin S, cathepsin D or cathepsin E activities were measured using Z-FR-AMC, Z-VVR-AMC, Ac-RGFFP-AMC or MOCAc-GSPAFLAK-Dnp-R specific fluorogenic substrates respectively. Specific inhibitors were used to confirm the specificity of the reactions, E64 for omnicathepsin, Z-FL-COCHO for cathepsin S, pepstatin A for cathepsin D and E. The rate of fluorescence emission, which is proportional to the proteolytic activity, was measured every 5 minutes at 37°C in a Victor-3 Plate Reader (Perkin Elmer).
Protein expression, phosphorylation level and kinase activity measurement
Protein extracts (30 ug/lane) from treated (5uM PIs and/or 0.4ug/mL PMA) or untreated DCs were subjected to SDS-PAGE on 4%–12% gradient gel. Protein bands were visualized by dual infrared fluorophore Odyssey Infrared Imaging System (Li-cor Biosciences) and densitometric quantification was performed as in (22). The “Kinase Selectivity Profiling System: AGC-1” (Promega) was used in addition to “Kinase-Glo® Luminescent kit” (Promega) to measure kinase activities.
Cross-presentation assay
Immature DCs were exposed to recombinant HIV-1 p24-protein (Abcam) for 1hr at 37C. DCs were thoroughly washed and cultured for 4 hours before adding the epitope-specific CTL clones at a 4:1 effector-to-target ratio in 96-well plates. DCs pulsed with indicated peptide concentrations were used as controls for antigen presentation and CTL specificity. Target cell lysis was measured with a fluorescent-based assay developed in the lab (26).
Isolation and identification of MHC-bound peptides
MHC-bound peptides (including MHC-I and MHC-II) from control and drug-treated live PBMCs were isolated directly from live cells and identified by high resolution MS/MS tandem (Rucevic, Kourjian, Le Gall, unpublished data). In brief, 5×107 cells were shortly exposed to mild acid treatment (10% acetic acid, 5 min) and peptide pool obtained was immediately subjected to ultrafiltration (3kDa cut-off) for MHC peptides enrichment. The pools of eluted peptides were further subjected to LC-ESI-MS/MS analysis. They were loaded onto ChromXP C18 5μm cHiPLC capillary column (75μm × 15cm, Eksigent) interfaced with an Orbitrap LTQ XL MS (Thermo Fisher) and resolved with a gradient consisting of an aqueous mobile phase A (0.1% formic acid in water) and organic mobile phase B (0.1% formic acid in 100% acetonitrile). Ten most abundant multiply charged ions were selected for high resolution MS/MS sequencing. Data were analyzed with Proteome Discoverer 1.4 software (Thermo Fisher) and searched against the SwissProt human database using MASCOT search engine. To ensure high accuracy of identified sequences, the mass tolerance on precursor was set to 5ppm and 0.02Da for fragment ions with all assignments made at 1% FDR. The candidate MHC peptides identified were selected based on their presence in triplicate samples. All identified MHC peptides were blasted against non-redundant SwissProt database restricted to human entries to identify their corresponding source proteins.
Statistics
Empirical data were analyzed using GraphPad Prism version 6. We compared two variables with t tests. All p values are two-sided, and p values <0.05 were considered significant. Multiple comparisons were done with one-way ANOVA and Dunnett's post-test. p value criteria are assigned as *p<0.05, **p<0.01, and ***p<0.001.
RESULTS
HIV PIs alter cathepsin activities
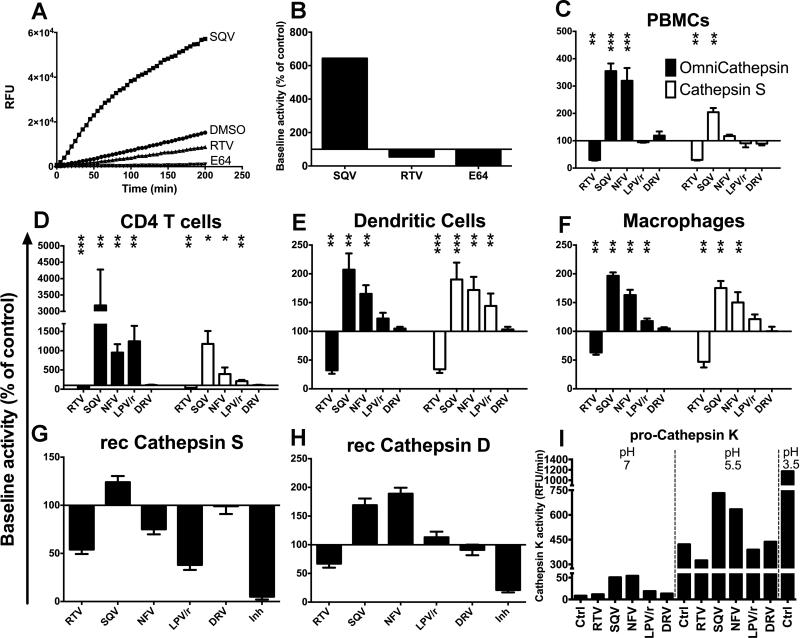
We aimed to assess the effect of five HIV PIs (RTV, SQV, NFV, Kaletra and DRV) on omnicathepsin activities (which measure the combined activities of cathepsin S, L and B) and cathepsin S, D and E, which are expressed in APCs important for antigen processing in the phagosomes (8, 13, 19, 21, 27, 28). The PI concentrations used in this study correspond to plasma levels of ART-treated persons and are not toxic for primary cells (8, 29). The cleavage of a peptidase-specific fluorogenic peptide substrate was measured over time. The specificity of substrate cleavage was checked by preincubation of cells with cognate inhibitors of omnicathepsin (E64) or cathepsin S (Z-FL-COCHO) (1A). The hydrolysis kinetics was calculated as the maximum slope of fluorescence emission after background subtraction (Fig. 1A). 100% represents the maximum slope of fluorescence emission by cells preincubated with DMSO (maximum slope of 67 for control DMSO, 430 for saquinavir, 36 for ritonavir, 5 for E64) (Fig. 1B). The effects of each HIV PI on omnicathepsin and cathepsin S activities were assessed in freshly isolated peripheral blood monocytic cells (PBMCs), CD4 T cells, monocyte-derived DCs and macrophages from at least six different HIV negative donors (Fig. 1 C-F). Baseline cathepsin activities were lower in CD4 T cells and PBMCs than in DCs and highest in macrophages. In PBMCs, RTV reduced both omnicathepsin and cathepsin S activities by 3.3-fold. In contrast, SQV and NFV increased omnicathepsin activity by more than 3-fold but only SQV increased cathepsin S activity. DRV did not change cathepsin activities (Fig. 1C). Similar alterations were seen in CD4 T cells, DCs and macrophages albeit with different fold changes (Fig. 1 D-F).
FIGURE 1. HIV PIs variably alter cathepsin activities in human CD4 T cells, DCs and macrophages.
(A) Omnicathepsin activity was monitored with fluorogenic substrate every 5 min in live PBMCs pretreated with DMSO, 5μM RTV, 5μM SQV or 10μM E64. (B) The maximum slope of fluorescence emission over 1 h in the presence of DMSO was represented as 100% and the effect of each PI was calculated as % of control. (C) PBMCs, (D) CD4 T cells, (E) DCs or (F) macrophages were pretreated for 30min with DMSO (control) or with 5μM of indicated PIs before adding specific cathepsin substrate (Omnicathepsin black bars, Cathepsin S white bars for C, D, E and F). Data represent average +/− SD of cells from 6 healthy donors. (G) Recombinant cathepsin S or (H) recombinant cathepsin D were pretreated with DMSO or 5μM of indicated PIs or inhibitor (10μM ZFL-COCHOO for cathepsin S, 100μM Pepstatin A for cathepsin D) before adding specific substrate for each activity. In each panel, 100% represents the maximum slope of DMSO-treated enzyme. Data represent average +/− SD of 3 independent experiments. (I) Pro-cathepsin K was incubated with 5 μM of DMSO or indicated PIs for 30min at different pH before adding specific substrate. pH 3.5 was used to trigger the maximal procathepsin maturation. The maximum slope of fluorescence emission is represented. *p< 0.05, **p< 0.01, ***p<0.001.
We tested if HIV PIs can directly affect cathepsins. The pH of PBMCs cell extracts was reduced to 4-5.5 in order to specifically activate cathepsins (24). Similar PI-induced alterations of cathepsin activities were observed in PBMC extracts albeit with a lower fold change (unpublished data). The effect of PIs was also tested on recombinant cathepsin S and D. RTV, NFV and Kaletra reduced cathepsin S activity while SQV increased it (Fig. 1G). Cathepsin D activity was reduced by RTV and increased by SQV and NFV (Fig. 1H). Cathepsin D and E activities measured in live DCs showed similar alterations upon PI treatment as purified enzymes (data not shown). We then tested whether HIV PIs affect the maturation of inactive procathepsins into cathepsins. Activation is triggered by a pH drop in endolysosomes, which induces the proteolytic removal of a pro-domain blocking the catalytic site (30). Procathepsin K was treated with 5uM of different PIs at different pH before measuring cathepsin K activity (Fig. 1I). At pH 7 only SQV and NFV activated procathepsin K resulting in a 7.5-fold activity increase. At pH 5.5 SQV and NFV activated procathepsin K by 1.8-fold. Thus HIV PIs have a direct effect on cathepsin maturation and activities. They may bind directly to the catalytic site of cathepsins and inhibit the activity, or interact with noncatalytic effector sites, allosterically modulate the conformation of the enzyme (31), and decrease or increase hydrolytic activities. However differences in fold changes between live cells and recombinant enzymes suggest that other mechanisms might alter cathepsin activities.
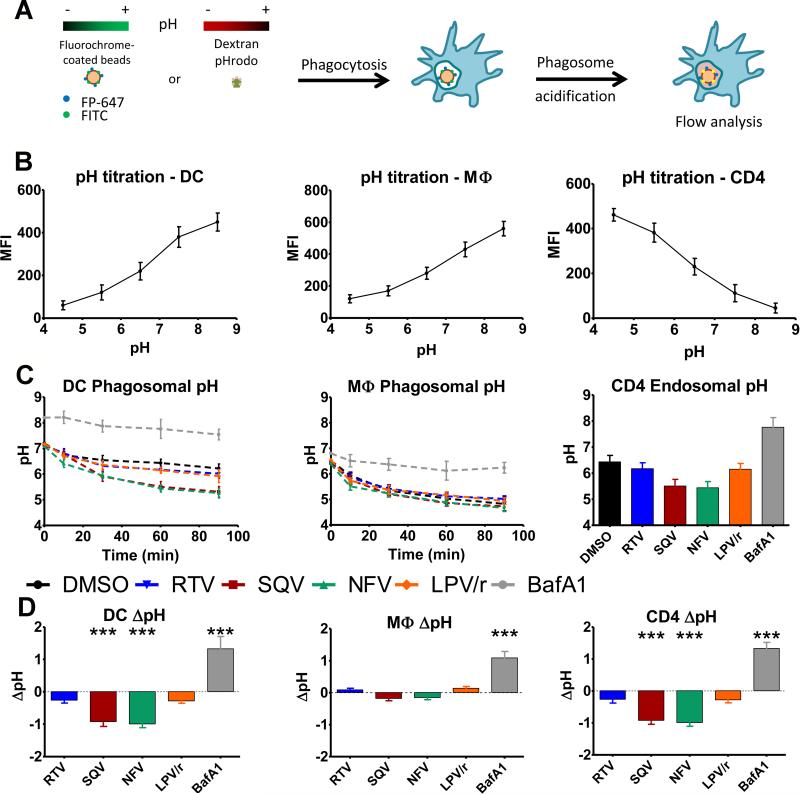
Certain HIV PIs increase phagolysosomal acidification
The activity of endolysosomal cathepsins is partly controlled through pH regulation (18, 19). We assessed whether HIV PI-induced alterations in cathepsin activities are due to changes in phagolysosomal pH in DCs, macrophages and CD4 T cells. We used a flow cytometry-based measurement of phagosomal pH using latex beads coated with pH-sensitive (fluorescein isothiocyanate, FITC) and pH-insensitive (FluoProbe647) fluorescent dyes (23). The fluorescence intensity was quantified by flow cytometry at different times. The pH-insensitive dye allowed the gating on cells that phagocytosed only one bead while the fluorescence intensity of the pH-sensitive dye reflected the phagosomal pH (Fig. 2A). HIV PI treatment did not affect bead uptake (unpublished data). Dextran-pHrodo was used to measure endosomal pH in CD4 T cells (Fig. 2A). Standard curves were obtained by permeabilizing and incubating cells in predetermined pH media. FITC intensity decreased and pHrodo® intensity increased upon acidification (Fig. 2B). The pH values in phagosomes were determined by reporting the mean fluorescence intensity in the cell population to the pH standard curves. As previously shown (20) the pH in DC phagosomes was stable at 6.5 during the 90min monitoring (Fig. 2C left). In contrast the phagosomal pH in macrophages dropped faster and reached pH5 (Fig. 2C middle). Bafilomycin, a specific inhibitor of vacuolar H+ ATPase increased the pH by 1.3 points in all cell types tested. SQV and NFV but not RTV and Kaletra reduced DC phagosomal pH by approximately 1 pH point (Fig. 2C and D left panel). HIV PIs did not significantly affect the already lower phagosomal pH in macrophages (Fig. 2C and D middle panel). CD4 T cell endolysosomal pH was reduced by SQV and NFV treatment by approximately 1 pH point in 60min. RTV and Kaletra did not significantly change the pH (Fig. 2C and D right panel). Thus SQV and NFV but not RTV and Kaletra reduced phagosomal pH, which may contribute the PI-induced increase of cathepsin activities in live cells.
FIGURE 2. HIV PIs alter phagosomal and endosomal pH.
(A) Representation of the experimental method of phagosomal and endosomal pH measurement. (B) pH titration curves obtained using fluorochrome-coated beads in DCs (left panel) and macrophages (middle panel) or using dextran pHrodo in CD4 T cells (right panel). (C) Phagosomal pH measurement over time after 30min pretreatment of cells with 5μM of different PIs or 1μM Bafilomycin A1. Fluorescence measured by flow cytometry was compared to the pH titration curves in (B) to determine pH values. (D) Difference in pH between phagosomes of cells treated with 5μM of different PIs or 1μM Bafilomycin A1 compared to phagosomes of cells treated with DMSO 60min after phagocytosis. Data represent average +/− SD of cells from 5 healthy donors. ***p <0.001.
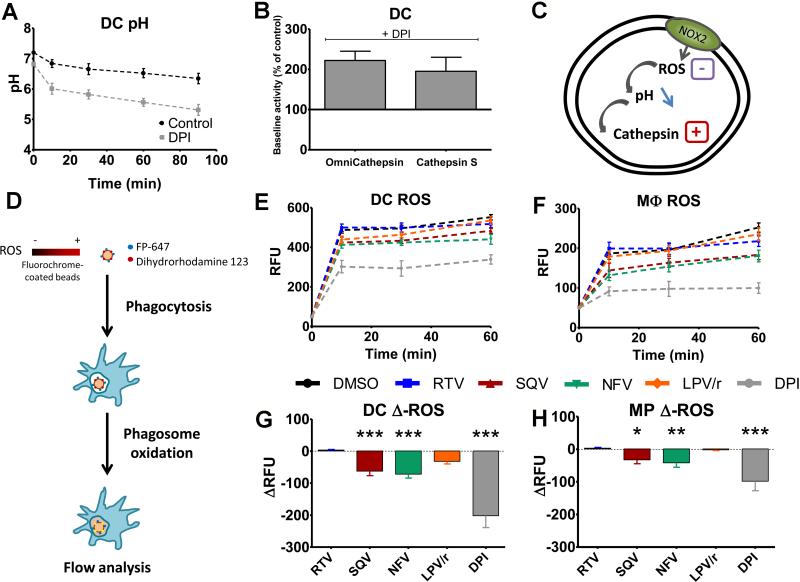
Certain HIV PIs reduce ROS generation in phagolysosomes
The phagosomal pH is in part regulated by the consumption of protons through the generation of ROS by NOX2 (20, 32). To confirm this finding in our experimental model we incubated DCs with diphenylene iodonium (DPI), a specific inhibitor of flavin-containing enzymes such as NOX2. DPI reduced phagosomal pH by approximately 1 pH point (Fig. 3A). DPI treatment increased omnicathepsin and cathepsin S activities in DCs by more than 2-fold, confirming that phagosomal ROS affect cathepsin activities (Fig. 3B). We hypothesized that HIV PIs might modulate ROS generation through NOX2, subsequently affecting phagosomal pH and cathepsin activities (Fig. 3C). Latex beads were coated with dihydrorhodamine 123 (DHR), a dye that only emits fluorescence under oxidative conditions (23), and an oxidation-insensitive (FluoProbe647) fluorescent dye were added to macrophages or DCs. At different times after phagocytosis, we measured ROS generation by quantifying the fluorescent intensity of DHR on cells that phagocytosed only one bead (Fig. 3D). As expected DC phagosomes produced at least twice more ROS than macrophages, and ROS generation was strongly inhibited by DPI treatment in both cell types (20) (Fig. 3E-F). RTV and Kaletra did not significantly alter ROS production in DCs and macrophages. In contrast, SQV and NFV significantly reduced ROS generation in DCs (−63 and −72 RFUs respectively), and to a lesser extent in macrophages (−33 and −42 RFUs respectively) (Fig. 3E-F-G-H), showing that SQV and NFV inhibition of ROS generation in DCs and macrophages led to phagosomal acidification and increased cathepsin activities.
FIGURE 3. HIV PIs alter ROS production in DC phagosomes.
(A) DC phagosome pH measurement over time after 30min treatment with DMSO (black) or 10 μM DPI (gray). Average +/− SD of 3 independent experiments. (B) Omnicathepsin and cathepsin S activities in DCs after 30min treatment with 10μM DPI. 100% represents the maximum slope of DMSO-treated DCs. Average +/− SD of 3 independent experiments. (C) Representation of the hypothesis: PIs inhibit ROS production, leading to phagosome acidification and cathepsin activation. (D) Representation of the method to used measure phagosomal ROS. ROS measurement in DCs (E) and macrophages (F) after a 30min pretreatment with 5μM PI or 10μM DPI. Differences in ROS production in phagosomes of DCs (G) and macrophages (H) after treatment with different PIs or DPI compared to DMSO 60min after phagocytosis. Data represent average of cells from 5 healthy donors. *p< 0.05, **p <0.01, ***p <0.001.
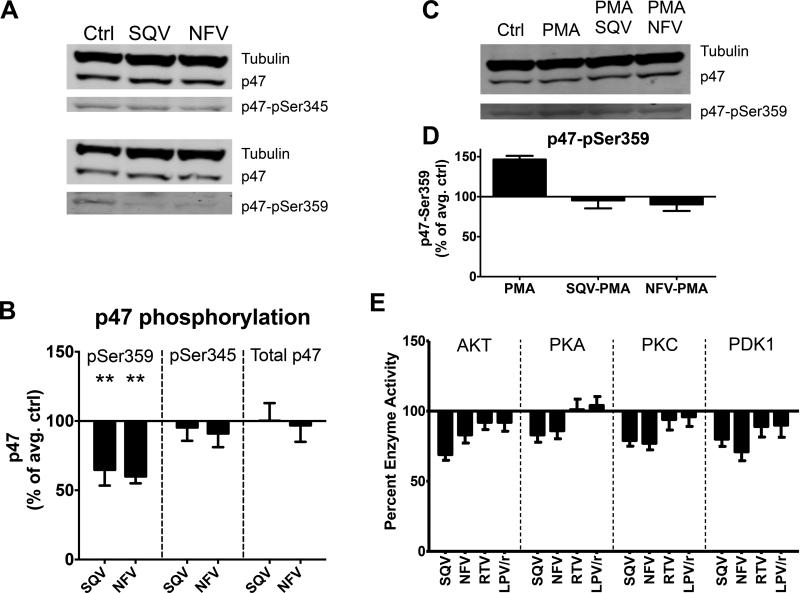
The activation of NOX2 is triggered by the phosphorylation of one of its subunit, phox-p47 at multiple locations and regulated by different kinases, including Akt and PKC (33). To examine whether SQV and NFV-induced reduction of NOX2 activity is linked to changes in phox-p47, we analyzed by western blot the phosphorylation of phox-p47 at serine 345 and serine 359 in DCs after PI treatment (Fig. 4A). Reduction in the phosphorylation through point mutation at either serine was shown to reduce NOX2 activity by at least half (34). SQV and NFV reduced p47-ser359 phosphorylation by 1.7-fold but did not affect p47-ser345 phosphorylation (Fig. 4A-B). To test the significance of this inhibition we assessed whether SQV and NFA could counteract the strong stimulatory effect of Phorbol myristate acetate (PMA) on p47 phosphorylation (Fig. 4C). As previously reported (34, 35), PMA increased p47-ser359 phosphorylation by 1.5-fold. SQV and NFV pretreatment blocked the stimulatory effect of PMA, showing that PMA and PIs exert opposite effects on pathways leading to p47 phosphorylation (Fig. 4D). To assess if this reduction in phosphorylation was caused by direct inhibition of kinases of the PI3K/Akt kinase pathway involved in the upstream regulation of phox-p47, we measured the activities of Akt, PKA, PKC and Phosphoinositide-dependent kinase-1 (PDK1) in presence or absence of different PIs. SQV and NFV but not RTV and Kaletra moderately reduced all four kinase activities by 1.5-fold (Fig. 4E), suggesting that SQV and NFV-induced inhibition of PDK1/Akt caused to reduced phox-p47 phosphorylation, leading to lower NOX2 activity and reduced ROS production in phagosomes. This mechanism is complementary to the direct enhancement of cathepsin activities induced by these two PIs.
FIGURE 4. HIV PIs inhibit kinase activity, reduce p47 phosphorylation and counteract PMA stimulatory effect on p47 phosphorylation.
(A) Western-Blot using extracts of DCs pretreated with DMSO, SQV or NFV and antibodies against tubulin, p47, p47-pSer345 and p47-pSer359. (B) Quantification of tubulin-normalized bands. 100% represents the normalized band value of DMSO control. Data represent average +/− SD of 4 independent experiments using DCs from 4 healthy donors. (C) Western Blot analysis of tubulin, p47, and p47-pSer359 in extracts of DCs pretreated with DMSO, PMA, SQV+PMA or NFV+PMA. (D) Quantification of tubulin-normalized bands. 100% represents the normalized band value of DMSO control. Data represent the average +/− SD of 4 independent experiments using DCs from 4 healthy donors. (E) Kinase activity measurement after 30min pretreatment with 5μM of different PIs. 100% represents the baseline activity of the control. Data represent average +/− SD of 3 independent experiments.
HIV PIs alter degradation patterns of antigenic peptides
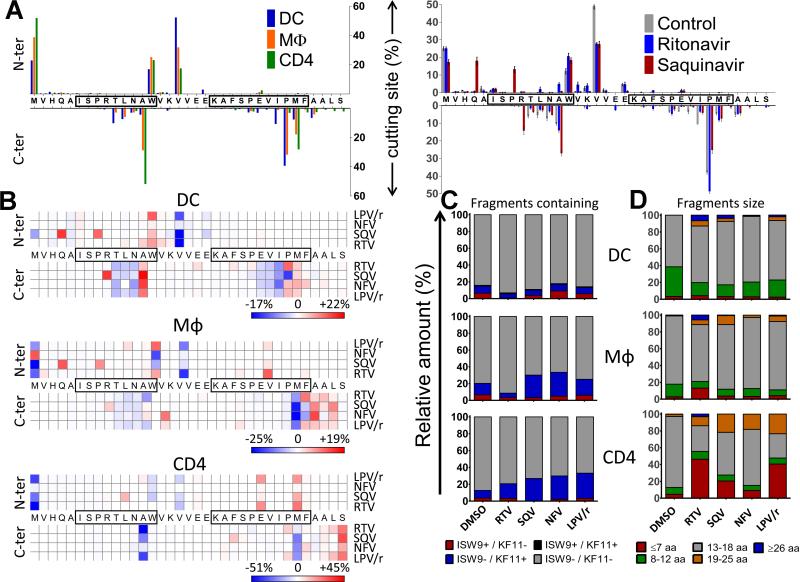
In order to analyze how PI-induced changes in cathepsin activities alter antigen processing in compartments involved in cross-presentation we used our in vitro antigen degradation assay (19, 24). We previously showed that the reduction of cellular extracts pH to 4 activates cathepsins, inhibits proteasome and aminopeptidase activities and mimics degradation conditions in purified endolysosomes (19, 24). DC, macrophage and CD4 T cell whole cell extracts placed at acidic pH were used to degrade a synthetic 35-mer peptide in HIV-1 Gag p24 (MVHQAISPRTLNAWVKVVEEKAFSPEVIPMFAALS, aa10-44 in Gag p24) containing the HLA-B57-restricted epitopes ISW9 and KF11 (36). Supplemental Fig.1 shows all degradation peptides from the p24 35-mer identified by LC-MS/MS after a 60-min degradation in DC, macrophage or CD4 T cell extracts in the absence of PIs, where each bar corresponds to a peptide with a defined peak intensity. We quantified the relative amount of each fragment by measuring their contribution to the total intensity of all degradation fragments as previously done (19). We displayed the relative amount of peptides starting at any N-terminal residue or ending at any C-terminal residue (top and bottom bars respectively), thus showing the relative frequency of cleavage sites in the fragment (Fig. 5A). For instance a frequent N-terminal cleavage site is observed at a Valine in between boxed epitopes ISW9 and KF11. We observed differences in the frequency of cutting sites between DC, macrophage and CD4 T cell extracts, confirming the differential antigen processing activities by different cell types previously reported by our group (19, 22, 25) (Fig. 5A left panel). A similar analysis was performed for degradations done in the presence of different HIV PIs. In DCs SQV introduced new cutting sites within the ISW9 epitope (for instance, N-terminal of proline position 8 and C-terminal of arginine position 9). RTV and SQV also changed the frequency of certain cutting sites (for example N-terminal of valine position 17 and C-terminal of proline position 29) (Fig. 5A right panel). We generated heat maps showing the % changes in the frequency of cleavages at each aa induced by each PI over DMSO. Figure 5B shows that HIV PIs increased cleavages at some locations and decreased it at others. For instance SQV increased the cleavage within the ISW9 epitope in DCs (Fig. 5B top panel), potentially leading to the destruction of ISW9. Similar changes were observed when the full HIV p24 protein was used for degradation (Supplemental Fig.2). To further analyze the effect of PIs on epitope production, we measured the amount of epitope-containing fragments. In DCs, SQV reduced the production of B57-ISW9-containing fragments by 1.7-fold which was expected because of the increased cleavage within the epitope seen in Figure 5B top panel. RTV reduced the production of B57-KF11 containing fragments by 1.8-fold and B57-ISW9 containing fragments by 6-fold. In macrophages, both RTV and SQV reduced the amount of B57-ISW9 containing fragment by 2-fold. The amount of B57-KF11-containing fragments was reduced 2.6-fold by RTV and increased 2-fold by SQV, 2-fold by NFV and 1.4-fold by Kaletra. In CD4 T cells, the amount of B57-ISW9 containing fragments was reduced 6-fold by SQV. The amount of B57-KF11 containing fragments was increased 2-fold by RTV and 3-fold by SQV, NFV and Kaletra (Fig. 5C). Beside the PI-induced changes in the amount of ISW9 and KF11-containing fragments, HIV PI treatments changed the size distribution of the degradation products. The amount of 8-12aa long peptides was reduced by all PIs by an average of 2-fold in DCs and macrophages. In CD4 T cells the amount of 13-18aa long peptides was reduced 2.7-fold by RTV, 1.7-fold by SQV, 1.4-fold by NFV and 3-fold by Kaletra (Fig. 5D). Since cathepsins are involved in MHC-II epitope processing (37), we assessed the effect of PIs on MHC-II epitope production. DC whole cell extracts placed at acidic pH were used to degrade a synthetic 24-mer peptide in HIV-1 Gag p24 (FRDYVDRFYKTLRAEQASQEVKNW, aa 161-185 in Gag p24) rich in MHC-II epitopes (36) in presence or absence of PIs. Supplemental Figure 3 shows that SQV and NFV increased cleavages at some locations and decreased it at others. These changes in the cutting sites patterns altered the amount of MHC-II epitope production. For instance the amount of MHC-II DQA1*01/DQB1*05-restricted FT11 (FRDYVDRFYKT) was increased upon SQV treatment and MHC-II DRB1*01/DRB1*04-restricted FE13 (FYKTLRAEQASQE) was decreased by NFV (Supplemental Fig.3). Together these results suggest that by altering cathepsin activities PIs changed the patterns and frequency of cutting sites, affecting the amount of MHC-I and MHC-II epitopes produced.
FIGURE 5. HIV PIs change protein degradation patterns and epitope production in lysosomal compartments.
(A) Right panel: Cleavage patterns of p24–35mer incubated with DC (blue), macrophage (orange) or CD4 (green) cell extracts for 60 minutes at pH4.0. Left panel: Relative amount of fragments starting or ending at each aa (top: N-terminal, bottom: C-terminal) after a 60min degradation in DC cell extracts preincubated with 5uM DMSO (gray), RTV (blue) or SQV (red) at pH4.0. (B) Cleavage patterns of p24–35mer incubated with DC (upper panel), Macrophages (middle panel) or CD4 T (lower panel) are shown as heat maps of cleavage site intensity compared to DMSO control. Red represents increased cleavage and blue reduced cleavage compared to control. (C) Degradation products from B identified by LC-MS/MS were grouped into fragments containing B57-ISW9 and B57-KF11 epitopes (black), only B57-KF11 epitope (blue), only B57-ISW9 epitope (red), or neither epitope (gray), respectively. The contribution of each category of peptides to the total intensity of all degradation products is shown for each condition. (D) All peptides from B were grouped according to their lengths of fragments: ≥ 26 aa (blue), 19–25 aa (orange), 13–18 aa (gray), 8–12 aa (green), and fragments ≤ 7 aa (red). The contribution of each category of peptides to the total intensity of all degradation products is shown for each condition. For (A-D) data are representative of three independent experiments with three different donors’ DCs.
HIV PIs alter degradation patterns of antigens from other pathogens co-infecting HIV positive individuals
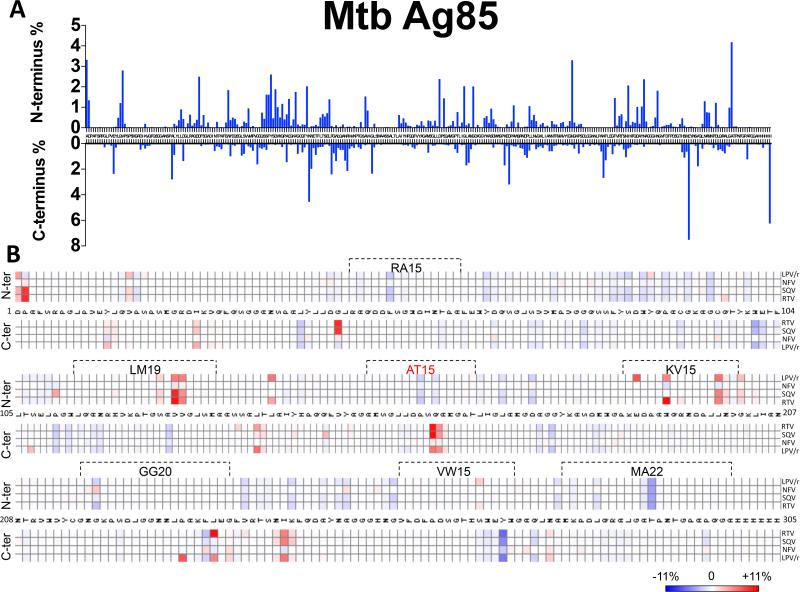
One third of HIV infected individuals on ART are co-infected with other pathogens such as Mycobacterium Tuberculosis (Mtb) or Hepatitis C virus (HCV) (38, 39). We hypothesized that PIs by altering cathepsin activities might alter the processing of epitopes from co-infecting pathogens in APCs. We assessed the effect of the PIs on the processing of two antigens: Mtb Ag85 and HCV NS3 that both contain multiple MHC-I and MHC-II epitopes (40-45). DC whole cell extracts placed at acidic pH were used to degrade a Mtb Antigen 85 and HCV NS3 in presence of different PIs. The frequency of cleavage sites (fig. 6A) and the percentage changes in the frequency of each cutting site upon PI treatment (fig. 6B) were analyzed as previsouly described. HIV PIs increased the cleavage at some locations, decreased it at others and created new cleavages sites. These alterations in the degradation patterns changed the amount of epitopes produced during Mtb Antigen 85 and HCV NS3 degradation in presence of different PIs. Out of the seven epitopes exactly generated during Mtb Antigen 85 degradation, PIs changed the production amount of all of them. For instance, all the PIs except NFV reduced the cleavage within the RA15 (RAQDDFSGWDINTPA 47-61aa) epitope which as expected led to a 2-5 fold increase of RA15 epitope production (fig6. B). In contrast, all the PIs tested increased the cleavage within the LM19 (LQANRHVKPTGSAVVGLSM 113-131aa) epitope, leading to 2-4 fold decrease in epitope production. In addition to aterations in epitope production, RTV and SQV by inctroducing a new cleavage site within the AT15 (AMSGLLDPSQAMGPT 152-166aa) epitope completely abolished its production (fig6. B). PI treatment also altered the degradation of HCV NS3 protein causing increased or decreased cutting frequencies at various locations (data not shown). Together these results suggest that by altering cathepsin activities PIs changed the patterns and frequency of cutting sites in Mtb and HCV antigens leading to changes in epitope production. How these PI-induced variations in epitope production will modify the hierarchy or specificity immune reponses against co-infecting pathogens in HIV+ individuals remains to be assessed.
FIGURE 6. HIV PIs change M. Tuberculosis Ag85 degradation patterns and epitope production in lysosomal compartments.
Mtb Ag85 was degraded for 60 minutes at pH4.0 in DC extracts preincubated or not with different PIs (5 μM). Degradation products were analyzed by LC-MS/MS and the contribution of the cleavage of each amino acid position (N-ter and C-ter) to the total intensity of all degradation products was quantified. (A) Cleavage pattern of Mtb Ag85 by DC without PI treatment. (B) Heat map representing the increase (red) or decrease (blue) in cutting intensity at each amino acid of Mtb Ag85 by DCs induced by each PI compared to control. The boxed areas represent MHC-I and MHC-II epitopes produced following the degradation. AT15 marked in red represents the epitope whose production was abolished by RTV and SQV. Data are representative of two independent experiments with two different donors’ DCs each analyzed in duplicate by LC-MS/MS.
HIV PIs alter the cross-presentation of Gag p24 epitopes
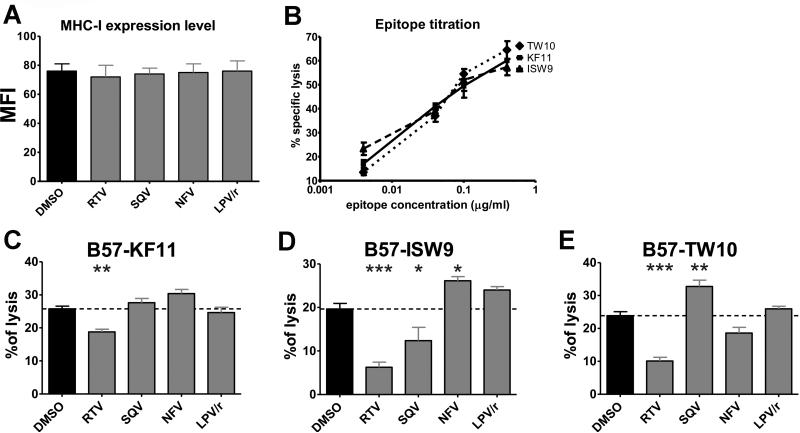
In order to understand if these PI-induced alterations of epitope production measured in vitro also affect endogenous processing and presentation by DCs we analyzed the cross-presentation of the three optimally defined HLA-B57 restricted HIV-epitopes originating from HIV-1 p24 protein: HLA-B57-restricted ISW9 (ISPRTLNAW, aa15-23 in Gag p24), HLA-B57-restricted KF11 (KAFSPEVIPMF, aa30-40 in Gag p24), and HLA-B57-restricted TW10 (TSTLQEQIGW, aa108-117 in Gag p24) (36). Monocyte-derived DCs from HLA-B57+ patients loaded with HIV-1 p24 protein in presence of different HIV PIs were challenged with epitope-specific CTLs (19). Target cell killing was monitored using a fluorescent-based assay developed in the lab (26). As previously seen in B cell lines (8), HIV PIs had no effect on the level of MHC-I surface expression in DCs (Fig. 7A). The three epitopes tested were restricted by HLA-B57 and shown to have similar affinities (36, 46). DCs pulsed with increasing amount of peptides were increasingly lysed after addition of cognate CTLs, showing the similar sensitivity of these CTL clones for their cognate peptides (Fig. 7B). RTV reduced by 1.4-fold the B57-KF11 cross-presentation by DCs while the other PIs had no effect (Fig. 7C). B57-ISW9 cross-presentation was reduced by RTV and SQV (3.2-fold and 1.6-fold respectively). In contrast, NFV increased killing by 1.35-fold (Fig. 7D). RTV reduced B57-TW10 cross-presentation by 2.4-fold while SQV increased it by 1.4-fold (Fig. 7E). During endogenous processing and presentation of p24, the effect of PIs, the asynchronous timing of epitope production (19, 47), the relative amount of each epitope and number of B57 molecules available for loading will also define the relative amount of the three epitope presented by HLA-B57. Although we cannot quantify the relative amount of each of the three B57 epitopes endogenously processed and presented by infected cells, the changes in CTL recognition induced by HIV PIs are in accordance with changes in epitope production measured with our degradation assays. For instance, SQV enhanced cleavages within the B57-ISW9 epitope (Fig. 5B), leading to reduced production of B57-ISW9 containing fragments (Fig. 5C), which resulted in lower endogenous cross-presentation of B57-ISW9 by DCs to CTL (Fig. 7D). These results demonstrate the link between drug-induced alteration of epitope production and subsequent changes in epitope-specific CTL responses to cross-presented epitopes.
FIGURE 7. HIV PIs variably alter the processing and cross-presentation of HIV p24.
(A) Flow cytometry MFI representing surface MHC-I expression levels of DCs pretreated with 5μM of the indicated PIs. (B) Lysis percentage of DCs pulsed with different concentrations of peptides B57-TW10, B57-KF11 or B57-ISW9 in a fluorescence-based killing assay with epitope-specific CTLs. Data represents the average +/− SD of 3 experiments. (C-E) Monocyte-derived DCs from HLA-B57+ donors were pretreated with 5μM of the indicated PIs and loaded with HIV p24 for 1h. After extensive wash and 4h incubation B57-KF11 (C), B57-ISW9 (D) or B57-TW10 (E) CTLs were added and the cell lysis was measured using a fluorescence-based assay. Data represents the average +/− SD of 5 independent experiments with 5 different donors. *p<0.05, **p<0.01, ***p<0.001.
HIV PI SQV alters PBMC MHC self peptidome
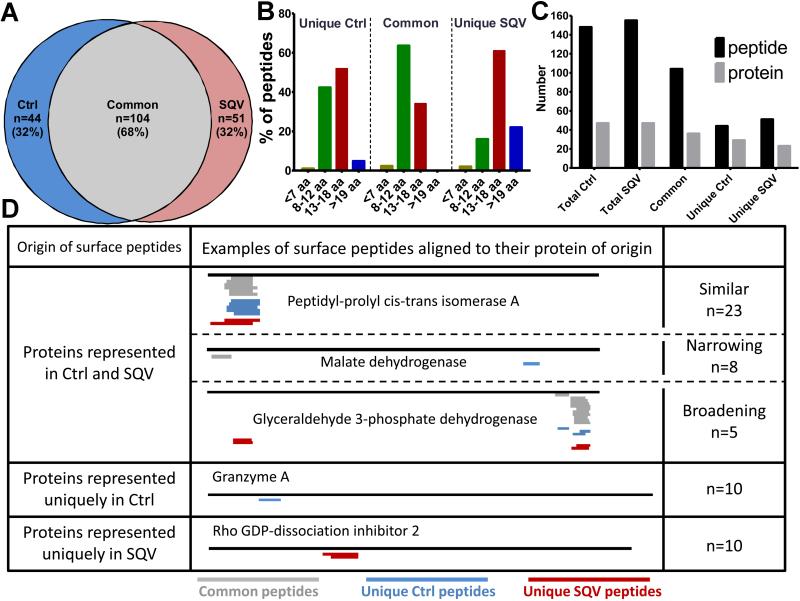
This and our previous studies (8) showed that HIV PIs affect both the direct endogenous antigen processing and cross-presentation pathways. In addition these alterations were not limited to HIV proteins but affected the processing of Mtb and HCV antigens. Therefore, we wondered if HIV PIs also affect self-derived MHC-bound peptidome (MHC-I and MHC-II) of uninfected cells. We analyzed the self-derived total MHC-bound peptidome (MHC-I and MHC-II) of healthy PBMCs pretreated with DMSO or SQV for 2 days. MHC-bound peptides were acid-eluted from live PBMCs and 686 and 680 human peptides were identified in triplicate runs with high mass accuracy by in-house LC-MS/MS on Ctrl-PBMC and SQV-PBMC respectively (Rucevic, Kourjian, Le Gall, unpublished data). We performed an in-depth comparative analysis of the peptides commonly identified with the highest confidence scores among repeated MS runs (148 Ctrl-PBMC and 155 SQV-PBMC peptides listed in Supplemental Table 1). The comparative mapping revealed that 68% of peptides were commonly presented by Ctrl- and SQV-treated PBMCs. 32% of peptides were uniquely presented by Ctrl- or SQV-treated cells (Fig. 8A). As expected the majority of common MHC-bound peptides were 8-12aa long (compatible with MHC-I Ioading) and 40% were 13-18aa compatible with MHC-II loading (Fig. 8B). Intriguingly >80% of peptides uniquely presented on SQV-PBMCs were longer than 13aa compared to only 60% of peptides presented uniquely on Ctrl-PBMCs (Fig. 8B). Since SQV treatment did not change the surface expression of MHC-I or MHC-II (unpublished data), these results suggest that SQV may increase the diversity of MHC-II peptidome and/or lead to the presentation of longer peptides by MHC-I. The total peptidome of Ctrl- and SQV-PBMCs were sampled from 56 host proteins. Many of the commonly identified MHC peptides were derived from the same proteins and corresponded to nested peptides with an average of 3 peptides per protein (Fig. 8C-D). However peptides uniquely identified from SQV- or Ctrl-PBMCs were sampled from diverse proteins with one peptide per one protein (Fig. 8C).
FIGURE 8. HIV PIs alter the MHC self peptidome.
PBMC were pretreated for 2 days with DMSO or SQV. Total surface peptides (MHC-I and MHC-II peptides) were isolated by acid elution and identified by LC-MS/MS. (A) Venn diagram representation of the number and percentage of peptides commonly or uniquely present on PBMCs treated with DMSO or SQV (peptides listed in table S1). (B) Uniquely or commonly presented peptides were grouped according to their lengths: equal or longer than 19 aa (blue), 13–18 aa (red), 8–12 aa (green), and fragments equal or shorter than 7 aa (gray). (C) Number of total, common or uniquely presented peptides on PBMCs treated with DMSO or SQV and the number of proteins they originate from. (D) The proteins of origin of the surface peptides were grouped into: proteins represented in DMSO and SQV treatment, proteins represented uniquely in DMSO treatment and proteins represented uniquely in SQV treatment. The group of proteins represented in each treatment was further divided into 3 subgroups: similar (unique and common peptides coming from similar location on the protein), narrowing (SQV narrowing the number of peptides coming from this group of proteins), broadening (SQV broadening the number of the peptides coming from this group of proteins). This experiment is representative of two independent peptide elution performed on PBMC from two different donors.
64% of all proteins identified were represented in both untreated and SQV-treated peptidome, whereas 18% were uniquely represented on Ctrl-PBMCs and another 18% only on SQV-PBMCs, suggesting that SQV partly changed the origin of proteins represented on the surface (Fig. 8D). This might be partly explained by a SQV-induced alteration of transcription and expression. However we also identified changes in the number and location of peptides originating from a common protein in line with PI-induced changes in protein degradation patterns (Fig. 8D). The group of proteins represented by both treated and untreated PBMCs was divided into subgroups according to the diversity of peptide location. Peptides presented commonly or uniquely were derived from the same location in 64% of proteins represented by both treated and untreated PBMCs (Fig. 8D - similar). In 22% of the proteins from this group SQV treatment narrowed the locations from which the peptides were derived (Fig. 8D - narrowing), whereas in the remaining 14% of the proteins SQV broadened the locations of the peptides (Fig. 8D –broadening). We concluded that SQV-induced changes in peptidase activities and degradation patterns contributed to partly change the self MHC-bound peptidome.
DISCUSSION
Any perturbation of protease activities could modify protein degradation patterns and consequently epitope presentation and immune recognition. In this study, we showed that several HIV PIs modified cathepsin activities through two complementary mechanisms and altered the MHC-peptidome and recognition of infected cells by epitope-specific CTL.
Four of the five PIs tested variably affected cathepsin activities. RTV mostly reduced all cathepsin activities tested while NFV reduced cathepsin S activity and enhanced cathepsin D activity, suggesting different interactions between each drug and cathepsins. It is intriguing that HIV PIs designed to inhibit the HIV aspartyl protease alter the activities of not only aspartyl protease like cathepsin D and E, but also cysteine proteases such as cathepsin S. HIV PIs might bind directly to the catalytic site of cathepsins or interact with non-catalytic effector or allosteric sites known to regulate the hydrolytic activities (31) or may induce the maturation of procathepsin into cathepsin. These off-target effects of HIV PIs might explain either inhibition or enhancement of cathepsin activities, although molecular modeling and structural studies are required to test this hypothesis.
An increase in cathepsin activity could also be triggered by a drug-induced maturation of inactive procathepsins into cathepsins. At neutral pH Glycoaminoglycans (GAG) can loosen the binding of the propeptide on the catalytic site and accelerate its autocatalytic removal, subsequently activating cathepsins (48, 49). We discovered that SQV, NFV but not RTV facilitated the maturation of procathepsin K even at neutral pH. SQV and NFV might be using the same mechanism as GAG to trigger cathepsin maturation at neutral pH. Cathepsin deregulation and extracellular presence of active cathepsins are linked to several disease, including damage to the musocal barrier in inflammatory bowel disease (50), osteoporosis and osteopenia, cancer, rheumatoid arthritis, atherogenesis and muscular dystrophy (30). Additional research is needed to investigate if PI-induced cathepsin alterations contribute to chronic disease development in patients using long term ART.
First-generation PIs like RTV, SQV, and NFV showed stronger effect on cathepsin activities and its upstream regulators than newer PIs like DRV. First-generation PIs also induced more rapid and profound adverse effects on lipid metabolism (51). The half-life of perilipin, a protein involved in regulating adipocyte lipolysis was reduced NFV by 6-fold in NVF-treated adipocytes, an effect reversed by NH4Cl but not by proteasome inhibition (52). NFV-induced increase of lysosomal cathepsin activity identified in this study might provide the missing link between NFV and perilipin lysosomal proteolysis in lipid metabolism disorders.
SQV and NFV have pleiotropic anti-cancer activities and are being repurposed for cancer treatment (53). The anticancer effects of some PIs observed in vitro has been linked to the suppression of the Akt signaling pathway, but the actual molecular targets remain unknown (54, 55). The direct inhibition of multiple members of the protein kinase-like superfamily (PDK1, Akt and PKA/C) by SQV and NFV provide support to a drug-induced inhibition of cellular processes vital for carcinogenesis and metastasis, and provides a molecular basis to explain the broad-spectrum anti-cancer effect of SQV and NFV (53, 55). Akt and PKC are also involved in regulation the activation of the NOX2 complex (33). NOX2 produces ROS in the phagosomes of neutrophils, eosinophils, monocytes and macrophages. It contributes to the destruction of engulfed pathogens (32), and to the regulation of phagosomal pH in DCs (20). Whether PI-induced NOX2 inhibition affects the capacity of macrophages to kill phagocytosed pathogens requires further investigation.
SQV and NFV present the unique property of activating cathepsin activities in APCs through two independent mechanisms: inhibiting NOX2 activity and increasing phagosome acidification, and directly enhancing cathepsin activities. In CD4 T cells the mechanism of PI-induced enhanced cathepsin activities is less clear but involves direct activation of cathepsins and pH acidification. Enhanced cathepsin activities modified the degradation patterns of HIV, HCV and Mtb proteins, altering frequency of cleavage sites, size distribution of the fragments and epitope production. It altered both MHC-I and MHC-II epitope production and the self-derived peptidome. The increase in 13-18 aa long peptides presented by PBMCs upon SQV treatment suggest that SQV might broaden the MHC-II surface peptidome. Whether this HIV PI-induced modulation of the self peptidome would be sufficient to induce autoimmune symptoms is not known. No increase of autoimmune disease in HIV patients receiving ART including Kaletra has been reported in the literature so far. Thymic selection of T cells involves negative and positive selection in medular and cortical epithelial cells (mTEC/cTEC). The unique expression of thymoproteasome in cTEC but not mTEC (56) contributes to shaping the immunocompetent repertoire of CD8 T cells (57) and determines the antigen responsiveness of CD8 T cells (58). One can speculate that the variable and broad peptide displayed in the thymus during selection (59) might cover some variations in the self. A direct comparison of the thymus peptidome and PI-treated or untreated PBMC are needed to address this question.
The alterations in degradation patterns were drug- and sequence-dependent and variable according to cell subsets, in line with our previous findings on cell subset-specific variations of peptidase activities (22, 25). Given the sequence-specificity of cathepsins (30), each PI may variably increase the cleavage of certain motifs and decrease others. Interestingly HIV PIs modified the degradation of HIV, HCV and Mtb proteins in accordance with sequence-specific rather than virus-specific modifications in degradation patterns. A computational analysis of the degradation patterns as done in (8, 60) may allow us to predict the effect of each PI on epitope production.
PI-induced changes in cathepsin, proteasome and aminopeptidase activities (here and (8)) modified protein degradation, the MHC-peptidome and altered CTL recognition of HIV cross-presenting or infected cells. In ART-treated persons with ongoing replication of drug-resistant viruses or co-infected with Mtb or HCV, PIs might alter the presentation of pathogen-derived epitopes by APCs or target cells. Drug-induced reduction or impairment of epitope presentation could render some pathogen-specific T cell responses useless or less efficient at recognizing target cells or might affect the T cell memory compartment if an epitope is never presented. Conversely increased presentation of others or presentation of new peptides. The effects would be partly determined by the amount of MHC-peptides displayed and the binding affinity of peptides for their MHC and corresponding TCRs. Although the lack of appropriate clinical samples precludes us to test this hypothesis, PI-induced modifications of Mtb and HCV antigen degradation patterns and epitope production showed in this study suggest that HIV PIs may modify the spectrum of pathogen-specific T cell compartment. Mouse models of viral of bacterial infections will allow us to test this hypothesis. Shock-and-kill approaches to clear HIV reservoirs propose to pharmacologically reactivate provirus in the presence of ART to induce immune clearance (61). If ART includes PIs such as Kaletra it will be important to assess the impact of HIV PIs on epitope presentation by CD4 T cells and DC when designing therapeutic to eliminate reactivated HIV-infected cells (62).
By directly acting on cathepsins and altering the physiology of phagocytic compartments HIV PIs modify antigen processing, the MHC peptidome and immune recognition. If HIV PIs allow the diversification of epitope presentation, a temporary PI treatment that would not induce adverse effects observed in long-term HAART may provide complementary approaches to modify or improve immune recognition in the context of various immune disease.
Supplementary Material
Acknowledgments
We thank Drs. F. Pereyra, D. Kavanagh, B. Walker and S. Pillai for stimulating discussions and input on the manuscript.
The project was funded by R01 grants AI084753, AI084106, AI112493 from NIAID to SLG.
Abbreviations
- PI
protease inhibitor
- NOX2
NADPH oxidase 2
- HAART
highly active antiretroviral treatments
- NRTI
nucleoside reverse transcriptase inhibitor
- nNRTI
non- nucleoside reverse transcriptase inhibitor
- ER
endoplasmic reticulum
- ERAP
ER-resident aminopeptidases
- RTV
ritonavir
- SQV
saquinavir
- NFV
nelfinavir
- LCMV
lymphocytic choriomeningitis virus
- DC
dendritic cells
- ROS
reactive oxygen species
- LPV
Lopinavir
- DRV
darunavir
- DPI
diphenylene iodonium
- DHR
dihydrorhodamine 123
- PMA
Phorbol myristate acetate
- Mtb
Mycobacterium Tuberculosis
- HCV
Hepatitis C virus
- GAG
Glycoaminoglycans
REFERENCES
- 1.Autran B, Carcelain G, Li TS, Blanc C, Mathez D, Tubiana R, Katlama C, Debre P, Leibowitch J. Positive effects of combined antiretroviral therapy on CD4+ T cell homeostasis and function in advanced HIV disease. Science. 1997;277:112–116. doi: 10.1126/science.277.5322.112. [DOI] [PubMed] [Google Scholar]
- 2.Flexner C. HIV-protease inhibitors. N Engl J Med. 1998;338:1281–1292. doi: 10.1056/NEJM199804303381808. [DOI] [PubMed] [Google Scholar]
- 3.Diez-Rivero CM, Lafuente EM, Reche PA. Computational analysis and modeling of cleavage by the immunoproteasome and the constitutive proteasome. BMC Bioinformatics. 2010;11:479. doi: 10.1186/1471-2105-11-479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Neefjes J, Jongsma ML, Paul P, Bakke O. Towards a systems understanding of MHC class I and MHC class II antigen presentation. Nat Rev Immunol. 2011;11:823–836. doi: 10.1038/nri3084. [DOI] [PubMed] [Google Scholar]
- 5.Gaedicke S, Firat-Geier E, Constantiniu O, Lucchiari-Hartz M, Freudenberg M, Galanos C, Niedermann G. Antitumor effect of the human immunodeficiency virus protease inhibitor ritonavir: induction of tumor-cell apoptosis associated with perturbation of proteasomal proteolysis. Cancer research. 2002;62:6901–6908. [PubMed] [Google Scholar]
- 6.Andre P, Groettrup M, Klenerman P, de Giuli R, Booth BL, Jr., Cerundolo V, Bonneville M, Jotereau F, Zinkernagel RM, Lotteau V. An inhibitor of HIV-1 protease modulates proteasome activity, antigen presentation, and T cell responses. Proceedings of the National Academy of Sciences of the United States of America. 1998;95:13120–13124. doi: 10.1073/pnas.95.22.13120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pajonk F, Himmelsbach J, Riess K, Sommer A, McBride WH. The human immunodeficiency virus (HIV)-1 protease inhibitor saquinavir inhibits proteasome function and causes apoptosis and radiosensitization in non-HIV-associated human cancer cells. Cancer research. 2002;62:5230–5235. [PubMed] [Google Scholar]
- 8.Kourjian G, Xu Y, Mondesire-Crump I, Shimada M, Gourdain P, Le Gall S. Sequence-specific alterations of epitope production by HIV protease inhibitors. Journal of immunology. 2014;192:3496–3506. doi: 10.4049/jimmunol.1302805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guermonprez P, Valladeau J, Zitvogel L, Thery C, Amigorena S. Antigen presentation and T cell stimulation by dendritic cells. Annual review of immunology. 2002;20:621–667. doi: 10.1146/annurev.immunol.20.100301.064828. [DOI] [PubMed] [Google Scholar]
- 10.Bernhard CA, Ried C, Kochanek S, Brocker T. CD169+ macrophages are sufficient for priming of CTLs with specificities left out by cross-priming dendritic cells. Proceedings of the National Academy of Sciences of the United States of America. 2015;112:5461–5466. doi: 10.1073/pnas.1423356112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kovacsovics-Bankowski M, Rock KL. A phagosome-to-cytosol pathway for exogenous antigens presented on MHC class I molecules. Science. 1995;267:243–246. doi: 10.1126/science.7809629. [DOI] [PubMed] [Google Scholar]
- 12.Bertholet S, Goldszmid R, Morrot A, Debrabant A, Afrin F, Collazo-Custodio C, Houde M, Desjardins M, Sher A, Sacks D. Leishmania antigens are presented to CD8+ T cells by a transporter associated with antigen processing-independent pathway in vitro and in vivo. Journal of immunology. 2006;177:3525–3533. doi: 10.4049/jimmunol.177.6.3525. [DOI] [PubMed] [Google Scholar]
- 13.Shen L, Sigal LJ, Boes M, Rock KL. Important role of cathepsin S in generating peptides for TAP-independent MHC class I crosspresentation in vivo. Immunity. 2004;21:155–165. doi: 10.1016/j.immuni.2004.07.004. [DOI] [PubMed] [Google Scholar]
- 14.Giodini A, Rahner C, Cresswell P. Receptor-mediated phagocytosis elicits cross-presentation in nonprofessional antigen-presenting cells. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:3324–3329. doi: 10.1073/pnas.0813305106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Belizaire R, Unanue ER. Targeting proteins to distinct subcellular compartments reveals unique requirements for MHC class I and II presentation. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:17463–17468. doi: 10.1073/pnas.0908583106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rock KL, Shen L. Cross-presentation: underlying mechanisms and role in immune surveillance. Immunological reviews. 2005;207:166–183. doi: 10.1111/j.0105-2896.2005.00301.x. [DOI] [PubMed] [Google Scholar]
- 17.Chatterjee B, Smed-Sorensen A, Cohn L, Chalouni C, Vandlen R, Lee BC, Widger J, Keler T, Delamarre L, Mellman I. Internalization and endosomal degradation of receptor-bound antigens regulate the efficiency of cross presentation by human dendritic cells. Blood. 2012;120:2011–2020. doi: 10.1182/blood-2012-01-402370. [DOI] [PubMed] [Google Scholar]
- 18.Claus V, Jahraus A, Tjelle T, Berg T, Kirschke H, Faulstich H, Griffiths G. Lysosomal enzyme trafficking between phagosomes, endosomes, and lysosomes in J774 macrophages. Enrichment of cathepsin H in early endosomes. J Biol Chem. 1998;273:9842–9851. doi: 10.1074/jbc.273.16.9842. [DOI] [PubMed] [Google Scholar]
- 19.Dinter J, Duong E, Lai NY, Berberich MJ, Kourjian G, Bracho-Sanchez E, Chu D, Su H, Zhang SC, Le Gall S. Variable processing and cross-presentation of HIV by dendritic cells and macrophages shapes CTL immunodominance and immune escape. PLoS Pathog. 2015;11:e1004725. doi: 10.1371/journal.ppat.1004725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Savina A, Jancic C, Hugues S, Guermonprez P, Vargas P, Moura IC, Lennon-Dumenil AM, Seabra MC, Raposo G, Amigorena S. NOX2 controls phagosomal pH to regulate antigen processing during crosspresentation by dendritic cells. Cell. 2006;126:205–218. doi: 10.1016/j.cell.2006.05.035. [DOI] [PubMed] [Google Scholar]
- 21.Delamarre L, Pack M, Chang H, Mellman I, Trombetta ES. Differential lysosomal proteolysis in antigen-presenting cells determines antigen fate. Science. 2005;307:1630–1634. doi: 10.1126/science.1108003. [DOI] [PubMed] [Google Scholar]
- 22.Dinter J, Gourdain P, Lai NY, Duong E, Bracho-Sanchez E, Rucevic M, Liebesny PH, Xu Y, Shimada M, Ghebremichael M, Kavanagh DG, Le Gall S. Different antigen-processing activities in dendritic cells, macrophages, and monocytes lead to uneven production of HIV epitopes and affect CTL recognition. Journal of immunology. 2014;193:4322–4334. doi: 10.4049/jimmunol.1400491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Savina A, Vargas P, Guermonprez P, Lennon AM, Amigorena S. Measuring pH, ROS production, maturation, and degradation in dendritic cell phagosomes using cytofluorometry-based assays. Methods Mol Biol. 2010;595:383–402. doi: 10.1007/978-1-60761-421-0_25. [DOI] [PubMed] [Google Scholar]
- 24.Vaithilingam A, Lai NY, Duong E, Boucau J, Xu Y, Shimada M, Gandhi M, Le Gall S. A simple methodology to assess endolysosomal protease activity involved in antigen processing in human primary cells. BMC Cell Biol. 2013;14:35. doi: 10.1186/1471-2121-14-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lazaro E, Godfrey SB, Stamegna P, Ogbechie T, Kerrigan C, Zhang M, Walker BD, Le Gall S. Differential HIV epitope processing in monocytes and CD4 T cells affects cytotoxic T lymphocyte recognition. J Infect Dis. 2009;200:236–243. doi: 10.1086/599837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gourdain P, Boucau J, Kourjian G, Lai NY, Duong E, Le Gall S. A real-time killing assay to follow viral epitope presentation to CD8 T cells. J Immunol Methods. 2013;398-399:60–67. doi: 10.1016/j.jim.2013.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chain BM, Free P, Medd P, Swetman C, Tabor AB, Terrazzini N. The expression and function of cathepsin E in dendritic cells. Journal of immunology. 2005;174:1791–1800. doi: 10.4049/jimmunol.174.4.1791. [DOI] [PubMed] [Google Scholar]
- 28.Zaidi N, Herrmann T, Baechle D, Schleicher S, Gogel J, Driessen C, Voelter W, Kalbacher H. A new approach for distinguishing cathepsin E and D activity in antigen-processing organelles. The FEBS journal. 2007;274:3138–3149. doi: 10.1111/j.1742-4658.2007.05846.x. [DOI] [PubMed] [Google Scholar]
- 29.Acosta EP, Kakuda TN, Brundage RC, Anderson PL, Fletcher CV. Pharmacodynamics of human immunodeficiency virus type 1 protease inhibitors. Clinical infectious diseases : an official publication of the Infectious Diseases Society of America. 2000;30(Suppl 2):S151–159. doi: 10.1086/313852. [DOI] [PubMed] [Google Scholar]
- 30.Turk V, Stoka V, Vasiljeva O, Renko M, Sun T, Turk B, Turk D. Cysteine cathepsins: from structure, function and regulation to new frontiers. Biochim Biophys Acta. 2012;1824:68–88. doi: 10.1016/j.bbapap.2011.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Novinec M, Korenc M, Caflisch A, Ranganathan R, Lenarcic B, Baici A. A novel allosteric mechanism in the cysteine peptidase cathepsin K discovered by computational methods. Nat Commun. 2014;5:3287. doi: 10.1038/ncomms4287. [DOI] [PubMed] [Google Scholar]
- 32.Segal AW. How neutrophils kill microbes. Annual review of immunology. 2005;23:197–223. doi: 10.1146/annurev.immunol.23.021704.115653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.El-Benna J, Dang PM, Gougerot-Pocidalo MA, Marie JC, Braut-Boucher F. p47phox, the phagocyte NADPH oxidase/NOX2 organizer: structure, phosphorylation and implication in diseases. Exp Mol Med. 2009;41:217–225. doi: 10.3858/emm.2009.41.4.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Faust LR, el Benna J, Babior BM, Chanock SJ. The phosphorylation targets of p47phox, a subunit of the respiratory burst oxidase. Functions of the individual target serines as evaluated by site-directed mutagenesis. The Journal of clinical investigation. 1995;96:1499–1505. doi: 10.1172/JCI118187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Johnson JL, Park JW, Benna JE, Faust LP, Inanami O, Babior BM. Activation of p47(PHOX), a cytosolic subunit of the leukocyte NADPH oxidase. Phosphorylation of ser-359 or ser-370 precedes phosphorylation at other sites and is required for activity. J Biol Chem. 1998;273:35147–35152. doi: 10.1074/jbc.273.52.35147. [DOI] [PubMed] [Google Scholar]
- 36.Yusim K. HIV Molecular Immunology 2013. Theoretical Biology and Biophysics. Los Alamos National Laboratory; Los Alamos, New Mexico: 2013. [Google Scholar]
- 37.Blum JS, Wearsch PA, Cresswell P. Pathways of antigen processing. Annual review of immunology. 2013;31:443–473. doi: 10.1146/annurev-immunol-032712-095910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Soriano V, Vispo E, Labarga P, Medrano J, Barreiro P. Viral hepatitis and HIV co-infection. Antiviral research. 2010;85:303–315. doi: 10.1016/j.antiviral.2009.10.021. [DOI] [PubMed] [Google Scholar]
- 39.Getahun H, Gunneberg C, Granich R, Nunn P. HIV infection-associated tuberculosis: the epidemiology and the response. Clinical infectious diseases : an official publication of the Infectious Diseases Society of America. 2010;50(Suppl 3):S201–207. doi: 10.1086/651492. [DOI] [PubMed] [Google Scholar]
- 40.Huygen K. The Immunodominant T-Cell Epitopes of the Mycolyl-Transferases of the Antigen 85 Complex of M. tuberculosis. Frontiers in immunology. 2014;5:321. doi: 10.3389/fimmu.2014.00321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Smith SM, Brookes R, Klein MR, Malin AS, Lukey PT, King AS, Ogg GS, Hill AV, Dockrell HM. Human CD8+ CTL specific for the mycobacterial major secreted antigen 85A. Journal of immunology. 2000;165:7088–7095. doi: 10.4049/jimmunol.165.12.7088. [DOI] [PubMed] [Google Scholar]
- 42.Tabatabai NM, Bian TH, Rice CM, Yoshizawa, Gill J, Eckels DD. Functionally distinct T-cell epitopes within the hepatitis C virus non-structural 3 protein. Human immunology. 1999;60:105–115. doi: 10.1016/s0198-8859(98)00102-5. [DOI] [PubMed] [Google Scholar]
- 43.Ciuffreda D, Comte D, Cavassini M, Giostra E, Buhler L, Perruchoud M, Heim MH, Battegay M, Genne D, Mulhaupt B, Malinverni R, Oneta C, Bernasconi E, Monnat M, Cerny A, Chuard C, Borovicka J, Mentha G, Pascual M, Gonvers JJ, Pantaleo G, Dutoit V. Polyfunctional HCV-specific T-cell responses are associated with effective control of HCV replication. European journal of immunology. 2008;38:2665–2677. doi: 10.1002/eji.200838336. [DOI] [PubMed] [Google Scholar]
- 44.Wertheimer AM, Miner C, Lewinsohn DM, Sasaki AW, Kaufman E, Rosen HR. Novel CD4+ and CD8+ T-cell determinants within the NS3 protein in subjects with spontaneously resolved HCV infection. Hepatology. 2003;37:577–589. doi: 10.1053/jhep.2003.50115. [DOI] [PubMed] [Google Scholar]
- 45.Vita R, Overton JA, Greenbaum JA, Ponomarenko J, Clark JD, Cantrell JR, Wheeler DK, Gabbard JL, Hix D, Sette A, Peters B. The immune epitope database (IEDB) 3.0. Nucleic acids research. 2015;43:D405–412. doi: 10.1093/nar/gku938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kloverpris HN, Stryhn A, Harndahl M, van der Stok M, Payne RP, Matthews PC, Chen F, Riddell L, Walker BD, Ndung'u T, Buus S, Goulder P. HLA-B*57 Micropolymorphism shapes HLA allele-specific epitope immunogenicity, selection pressure, and HIV immune control. Journal of virology. 2012;86:919–929. doi: 10.1128/JVI.06150-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Le Gall S, Stamegna P, Walker BD. Portable flanking sequences modulate CTL epitope processing. The Journal of clinical investigation. 2007;117:3563–3575. doi: 10.1172/JCI32047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Caglic D, Pungercar JR, Pejler G, Turk V, Turk B. Glycosaminoglycans facilitate procathepsin B activation through disruption of propeptide-mature enzyme interactions. J Biol Chem. 2007;282:33076–33085. doi: 10.1074/jbc.M705761200. [DOI] [PubMed] [Google Scholar]
- 49.Vasiljeva O, Dolinar M, Pungercar JR, Turk V, Turk B. Recombinant human procathepsin S is capable of autocatalytic processing at neutral pH in the presence of glycosaminoglycans. FEBS Lett. 2005;579:1285–1290. doi: 10.1016/j.febslet.2004.12.093. [DOI] [PubMed] [Google Scholar]
- 50.Menzel K, Hausmann M, Obermeier F, Schreiter K, Dunger N, Bataille F, Falk W, Scholmerich J, Herfarth H, Rogler G. Cathepsins B, L and D in inflammatory bowel disease macrophages and potential therapeutic effects of cathepsin inhibition in vivo. Clin Exp Immunol. 2006;146:169–180. doi: 10.1111/j.1365-2249.2006.03188.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Carr A, Samaras K, Burton S, Law M, Freund J, Chisholm DJ, Cooper DA. A syndrome of peripheral lipodystrophy, hyperlipidaemia and insulin resistance in patients receiving HIV protease inhibitors. AIDS. 1998;12:F51–58. doi: 10.1097/00002030-199807000-00003. [DOI] [PubMed] [Google Scholar]
- 52.Kovsan J, Ben-Romano R, Souza SC, Greenberg AS, Rudich A. Regulation of adipocyte lipolysis by degradation of the perilipin protein: nelfinavir enhances lysosome-mediated perilipin proteolysis. J Biol Chem. 2007;282:21704–21711. doi: 10.1074/jbc.M702223200. [DOI] [PubMed] [Google Scholar]
- 53.Gantt S, Casper C, Ambinder RF. Insights into the broad cellular effects of nelfinavir and the HIV protease inhibitors supporting their role in cancer treatment and prevention. Current. 2013;25:495–502. doi: 10.1097/CCO.0b013e328363dfee. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Plastaras JP, Vapiwala N, Ahmed MS, Gudonis D, Cerniglia GJ, Feldman MD, Frank I, Gupta AK. Validation and toxicity of PI3K/Akt pathway inhibition by HIV protease inhibitors in humans. Cancer Biol Ther. 2008;7:628–635. doi: 10.4161/cbt.7.5.5728. [DOI] [PubMed] [Google Scholar]
- 55.Gills JJ, Lopiccolo J, Dennis PA. Nelfinavir, a new anti-cancer drug with pleiotropic effects and many paths to autophagy. Autophagy. 2008;4:107–109. doi: 10.4161/auto.5224. [DOI] [PubMed] [Google Scholar]
- 56.Tomaru U, Ishizu A, Murata S, Miyatake Y, Suzuki S, Takahashi S, Kazamaki T, Ohara J, Baba T, Iwasaki S, Fugo K, Otsuka N, Tanaka K, Kasahara M. Exclusive expression of proteasome subunit {beta}5t in the human thymic cortex. Blood. 2009;113:5186–5191. doi: 10.1182/blood-2008-11-187633. [DOI] [PubMed] [Google Scholar]
- 57.Nitta T, Murata S, Sasaki K, Fujii H, Ripen AM, Ishimaru N, Koyasu S, Tanaka K, Takahama Y. Thymoproteasome shapes immunocompetent repertoire of CD8+ T cells. Immunity. 2010;32:29–40. doi: 10.1016/j.immuni.2009.10.009. [DOI] [PubMed] [Google Scholar]
- 58.Takada K, Van Laethem F, Xing Y, Akane K, Suzuki H, Murata S, Tanaka K, Jameson SC, Singer A, Takahama Y. TCR affinity for thymoproteasome-dependent positively selecting peptides conditions antigen responsiveness in CD8(+) T cells. Nat Immunol. 2015;16:1069–1076. doi: 10.1038/ni.3237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Adamopoulou E, Tenzer S, Hillen N, Klug P, Rota IA, Tietz S, Gebhardt M, Stevanovic S, Schild H, Tolosa E, Melms A, Stoeckle C. Exploring the MHC-peptide matrix of central tolerance in the human thymus. Nat Commun. 2013;4:2039. doi: 10.1038/ncomms3039. [DOI] [PubMed] [Google Scholar]
- 60.Lazaro E, Kadie C, Stamegna P, Zhang SC, Gourdain P, Lai NY, Zhang M, Martinez SA, Heckerman D, Le Gall S. Variable HIV peptide stability in human cytosol is critical to epitope presentation and immune escape. The Journal of clinical investigation. 2011;121:2480–2492. doi: 10.1172/JCI44932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Siliciano JD, Siliciano RF. HIV-1 eradication strategies: design and assessment. Curr Opin HIV AIDS. 2013;8:318–325. doi: 10.1097/COH.0b013e328361eaca. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Deng K, Pertea M, Rongvaux A, Wang L, Durand CM, Ghiaur G, Lai J, McHugh HL, Hao H, Zhang H, Margolick JB, Gurer C, Murphy AJ, Valenzuela DM, Yancopoulos GD, Deeks SG, Strowig T, Kumar P, Siliciano JD, Salzberg SL, Flavell RA, Shan L, Siliciano RF. Broad CTL response is required to clear latent HIV-1 due to dominance of escape mutations. Nature. 2015;517:381–385. doi: 10.1038/nature14053. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.