Abstract
In chronic lymphocytic leukemia (CLL) the increment in peripheral blood lymphocytes is slower than the expected increment calculated from the cells’ proliferation rate, suggesting that cellular proliferation and apoptosis are concurrent. Exploring this phenomenon, we found overexpression of caspase3, higher cleaved poly (ADP-ribose) polymerase (PARP) levels (P<0.007), and a higher apoptosis rate in cells from patients with high counts compared with cells from patients with low counts. Although we previously found that signal transducer and activator of transcription 3 (STAT3) protects CLL cells from apoptosis, STAT3 levels were significantly higher in cells from patients with high counts than in cells from patients with low counts. Furthermore, overexpression of STAT3 did not protect the cells. Rather, it upregulated caspase3 and induced apoptosis. Remarkably, putative STAT3 binding sites were identified in the caspase3 promoter and a luciferase assay, chromatin immunoprecipitation (ChIP), and an electromobility shift assay (EMSA) revealed that STAT3 activated caspase3. However, caspase3 levels increased only when STAT3 levels were sufficiently high. Using ChIP and EMSA, we found that STAT3 binds with low affinity to the caspase3 promoter, suggesting that at high levels, STAT3 activates proapoptotic mechanisms and induces apoptosis in CLL cells.
Introduction
Chronic lymphocytic leukemia (CLL) is characterized by a gradual accumulation of monoclonal, dysfunctional mature-appearing lymphocytes (1). Traditionally, CLL cells were thought to have a prolonged lifespan because of a deregulated apoptosis pathway (2–4). Studies performed during the past decade revealed that CLL cells proliferate and undergo spontaneous apoptosis. Ki-67, typically expressed in proliferating cells, was found in an appreciable number of CLL bone marrow (BM) cells, and terminal deoxynucleotidyl transferase dUTP nick end labeling also revealed apoptosis in CLL bone marrow cells (5). Deuterated water studies showed that 0.1% to 1.75% of the CLL clone is regenerated daily (6). However, the increment in peripheral blood (PB) CLL cell counts is typically lower than is expected from the cells’ proliferation rate. Therefore, it was assumed that CLL cell proliferation is accompanied by spontaneous apoptosis.
The signal transducer and activator of transcription (STAT)-3 is a latent cytoplasmic transcription factor that upon phosphorylation and dimerization shuttles to the nucleus, binds to DNA (7), and activates STAT3-regulated genes. In various solid tumors and hematologic malignancies, including CLL, STAT3 is constitutively activated and provides neoplastic cells with proliferation signals and a survival advantage (8–17).
Because in CLL cells, STAT3 is constitutively phosphorylated on serine 727 residues and activates anti-apoptotic genes (10, 12–14), we hypothesized that high STAT3 levels inversely correlate with the apoptosis rate in CLL cells. Surprisingly, we found that when STAT3 levels were sufficiently high, STAT3 no longer protected CLL cells from apoptosis. Instead, STAT3 induced the expression of proapoptotic genes, activated the caspase3 gene promoter, and induced apoptosis in CLL cells.
Materials and Methods
Patient characteristics
PB samples were obtained from 64 patients with CLL who were treated at The University of Texas MD Anderson Cancer Center Leukemia Clinic between April 2008 and May 2011, after approval was obtained from the Institutional Review Board approval and written informed consent was obtained from the patients. PB counts of 320 consecutive CLL patients were evaluated to determine the means ± standard deviation (SD) of lymphocyte counts in the 10% of patients with the highest counts (N=32, mean: 140,000 ±49,738 × 109/L) and the 10% of patients with the lowest counts (N=32, mean: 12,800 ±4,564 cells × 109/L). Of the 64 patients, 54 (84%) had not received any prior treatment for CLL. The remaining 10 previously treated patients were evenly distributed between the subgroups of patients with a high or low lymphocyte count. The clinical characteristics of all the patients are depicted in Suppl. Table 1.
Cell fractionation
PB cells were fractionated using Ficoll Hypaque 1077 (Sigma, St. Louis, MO). The low-density cellular fraction was used immediately or frozen for additional studies.
Western Immunoblotting
Western immunoblotting was performed as previously described(18). Briefly, CLL cell extract was prepared. The protein concentration was determined using a Micro BCA protein assay reagent kit (Thermo Scientific, Pierce, Rockford, IL). Cell lysates were denatured and following electrophoresis transferred to a nitrocellulose membranes. The membranes were incubated with either monoclonal mouse anti-human STAT3 (BD Bioscience, Palo Alto, CA), polyclonal rabbit anti-human phosphoserine (serine 727) STAT3 (Cell Signaling Technology, Beverly, MA), caspase3 (Cell Signaling Technology, Beverly, MA), cleaved caspase3 (BD Bioscience), monoclonal STAT1 (BD Bioscience), phosphoserine STAT1 (serine 727) (BD Bioscience) or monoclonal mouse anti-human β-actin (Sigma). Horseradish peroxidase-conjugated secondary antibodies (GE Healthcare, Amersham, Buckinghamshire, UK) and proteins were visualized via an enhanced chemiluminescence detection system (GE Healthcare).
Densitometry analysis was performed using an Epson Expression 1680 scanner (Epson America Inc., Long Beach, CA). Densitometry values were normalized by dividing the numerical value of each sample signal by the numerical value of the signal from the corresponding actin protein levels used as loading control.
Annexin V/propidium iodide assay
The rate of cellular apoptosis was analyzed using double staining with a Cy5-conjugated annexin V kit and propidium iodide (PI; BD Biosciences) according to the manufacturer’s instructions. After incubation for 10 minutes in the dark at room temperature, the samples were analyzed on a FACSCalibur flow cytometer (BD Biosciences). Cell viability was calculated as the percentage of annexin V positive cells.
RNA extraction
After thawing in hot water, cells were washed twice with RPMI (GIBCO) and Trizol (Invitrogen, Carlsbard, CA) was added. The RNA was isolated using an RNeasy purification procedure (Qiagen, Inc., Valencia, CA). RNA quality and concentration were analyzed with a NanoDrop spectrophotometer (ND-1000; NanoDrop Technologies, Wilmington, Delaware).
Quantitative reverse-transcription polymerase chain reaction analysis
We used 500 ng of total RNA in one-step quantitative reverse-transcription polymerase chain reaction (qRT-PCR; Applied Biosystems, Foster City, CA) analysis with an ABI Prism 7700 sequence detection system (Applied Biosystems) using a TaqMan gene expression assay for MAPK8, KRAS, PLCγ2, PERK, calpain9, and caspase3 according to the manufacturer’s instructions. Samples were run in triplicate, and relative quantification was performed by using the comparative CT method.(13)
Microarray analysis
Hybridization was performed with Human Gene 2.0 ST arrays (Affymetrix, Inc, Santa Clara, CA) according to the manufacturer’s instructions. Briefly, 100 ng of total RNA from each sample was reverse transcribed to cDNA, followed by overnight in vitro transcription to generate cRNA, which was also reverse transcribed. The quality of cDNA and fragmented cDNA was assessed using an Agilent Bioanalyzer system. Microarrays were hybridized, washed, stained, and images were scanned using the Affymetrix GeneChip Command Console and analyzed with the Affymetrix Expression Console. The t-test with Welch correction (variance not assumed equal) was used to compare gene expression levels and the analysis was done using Partek Genomics Suite 6.6 (Partek Inc., St. Louis, MO). Network analysis was performed on 1001 genes whose levels were upregulated (P <0.01 in samples with at least a 1.5-fold increase in lymphocyte counts) using the Ingenuity pathway analysis (Ingenuity Systems, www.ingenuity.com). The entire data are provided on (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE68721).
Enzyme-linked immunosorbent assay
Levels of cleaved PARP were determined using an Invitrogen Cleaved PARP enzyme-linked immunosorbent assay (ELISA) kit (Invitrogen) according to the manufacturer’s instructions. Briefly, microtiter wells were coated with mouse anti-rabbit uncleaved (116 Kd) or cleaved (85 Kd) PARP antibodies. Cleaved PARP levels were assessed by comparing the color intensity of the tested samples with that of the standard cleaved PARP protein levels provided in the kit using a microplate autoreader (Model #EL309, Bio-Tek Instruments, Winooski, VT).
Transfection of MM-1 cells with the human STAT3 gene
The human STAT3 sequence was obtained from GenBank NM_139276, and its full-length coding sequence was generated via PCR from the human STAT3 cDNA clone (OriGene, SC124165). The primers used for generation of recombinant STAT3 were: STAT3-Full-5’ GCGGCGGCGGCCGCCCCGCCGCCACCATGGCCCAATGGAATCAGCTACAG; STAT3-3’ GCGGCGCTCGAGTAACATGGGGGAGGTAGCGCAC. The sense primer contained a NotI site, a Kozac codon, and a gene-specific sequence (starting from ATG), and the antisense primers contained an XhoI site and a coding sequence ending at 2313 base pairs (bp) with a mutant stop codon. The PCR products were digested with NotI and XhoI and sub-cloned into the mammalian expression vector pBudCE4.1 (Invitrogen). Green fluorescent protein (GFP) was cloned into another cloning site with HindIII/XbaI. MM-1 cells were transfected via electroporation with a Gene Pulser Xcell electroporation system (Bio-Rad, Hercules, CA) set at 120 mV with 1 pulse. Transfection efficiency was detected using fluorescence microscopy and flow cytometry. After the transfection, cells were incubated for 24 hours or 48 hours with 50 ng/ml recombinant human interleukin (IL)-6 (Life Technologies, Frederick, MD) and harvested for qRT-PCR analysis.
Infection of CLL cells with the human STAT3
Full-length human STAT3 was amplified from the human STAT3 cDNA clone (OriGene, SC124165, Rockville, MD) and cloned using SalI/EcoRV into pENTR11(Invitrogen, Grand Island, NY), and sequence-verified, using Gateway LR clonase (Invitrogen). Both GFP and STAT3 were transferred into pInducer20 (plasmid 44012, Addgene, Cambridge, MA) and after overnight infection, protein expression was induced using 1 µg/ml doxycycline (Sigma–Aldrich). The lentiviral vector was produced by transfecting the plasmid into HEK293T cells using the Lipofectamine 2000 transfection reagent (Invitrogen). Then, pINDUCER20 STAT3 and the GFP expression vectors were transfected into HEK293T cells with the packaging vectors pMDLg/pRRE, pRSV-Rev and pMD2.G (Addgene). HEH293T cells were expanded in culture and their supernatant was used to infect the CLL cells as previously described (10).
Transfection of MM-1 cells with caspase3 promoter fragments and luciferase assay
Caspase3 promoter fragments were transfected into MM-1 cells via electroporation as described above. Each construct included a luciferase reporter gene and either 770 bp upstream to the transcription starting site of the caspase3 gene that did not include γ-interferon activation sequence (GAS)-like elements or with 854 bp upstream to the caspase3 gene transcription starting site that included a single GAS-like element. The luciferase activity of unstimulated or IL-6-stimulated MM-1 cells was assessed 24 hours after transfection using a Dual-Luciferase Reporter Assay System (Promega) and a Sirius luminometer V3.1 (Berthold Detection Systems, Pforzheim, Germany). The luciferase activity of each of the human caspase3 promoter constructs was determined by calculating the constructs’ luciferase activity relative to the activity of the Renilla luciferase produced by the pRL-SV40 control vector.
Chromatin immunoprecipitation assay
A chromatin immunoprecipitation (ChIP) assay was performed using a SimpleChIP Enzymatic Chromatin IP Kit (Cell Signaling Technology, Boston, MA) according to the manufacturer’s instructions. Briefly, cells were cross-linked with 1% formaldehyde for 10 minutes at room temperature and then harvested and incubated on ice for 10 minutes in lysis buffer. Nuclei were pelleted and digested with micrococcal nuclease. Following sonication and centrifugation, sheared chromatin was incubated with anti-STAT3 or rabbit serum (negative control) overnight at 4°C. Then, protein G beads were added, and the chromatin was incubated for 2 hours in rotation. Antibody-bound protein-DNA complexes were eluted and subjected to PCR analysis. The primer sets used to amplify the caspase3 promoter putative STAT3 binding sites were as follows: set 1 5’–914TCCCAACAGCCGGCTTAA and 3’ -849AAGAAGCCTGGTTTGGC, set 2: 5’ -1172CACGACCCAAAATAGGAA and 3’ -1047GCTTGTTGGCAAATGCCT, set3: 5’-1299GTCCCTGAATCTGACTTC and 3’-1177CATTTCAGACCCTGAAGC, set 4: 5’-1706GTAGCTGGGATTACAGGT and 3’-1625CAAGATGGTGAAACCCTG.
Electrophoretic mobility shift assay
Non-denatured cellular nuclear extracts were prepared using a NE-PER extraction kit (Thermo Scientific Pierce, Rockford, IL). Nuclear protein extracts were incubated with biotin-labeled caspase3, c-myc, and STAT3 promoters’ DNA probes in binding buffer for 30 minutes on ice. All probes were synthesized by Sigma-Genosys (The Woodlands, TX). Following incubation, the samples were separated on a 5% polyacrylamide gel, transferred onto a nylon membrane, and fixed on the membrane via ultraviolet cross-linking. The biotin-labeled probe was detected with strepavidin-horseradish peroxidase (Gel-Shift Kit; Panomics, Fremont, CA). The control consisted of 7-fold excess unlabeled cold probe. To test the effect of STAT3, anti-STAT3 antibodies (BD Bioscience) mouse IgG1 (BD Bioscience) were added with the nuclear extracts.(10, 19)
Results
High lymphocyte counts are correlated with high levels of STAT3
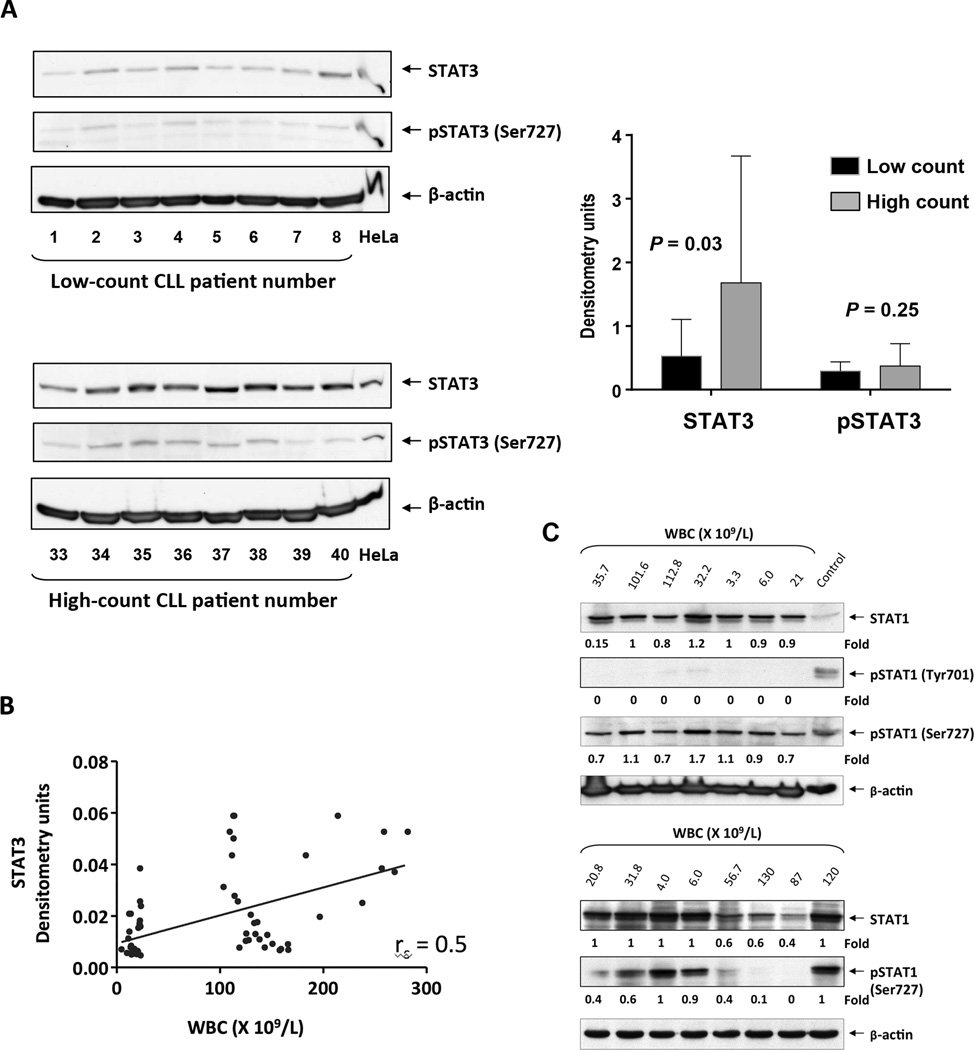
Because in CLL STAT3 is constitutively phosphorylated on serine 727 residues (10), and because STAT3 promotes the proliferation and survival of cancer cells, including CLL cells (20, 21), we assumed that CLL cells from patients with a high tumor burden would harbor high levels of STAT3. To test this hypothesis, we obtained PB cells from patients with a low (N=32) and high (N=32) lymphocyte count. Compared with patients with low lymphocyte counts (mean: 12,800 ±4,654 × 109/L), patients with high counts (mean: 145,000 ±49,738 × 109/L) had advanced disease, high levels of β2M levels, and unmutated Ig heavy chain gene status (Suppl. Table 1). Western blot analysis revealed that CLL cells from patients with a high lymphocyte count harbored higher levels of total STAT3 compared with patients with low counts (P=0.03), whereas overall the levels of serine phosphorylated (p) STAT3 were similar in patients with high and low counts as assessed by densitometry (Figure 1A). High lymphocyte counts correlated with high levels of STAT3 (rs = 0.5, P<0.0001) (Figure 1B). Because STAT1 is constitutively phosphorylated on serine 727 residues (22) and induces apoptosis in CLL cells (23), we assessed the levels of STAT1 and pSTAT1 in patients with high (N = 4) and low (N = 4) lymphocyte counts. Contrary to STAT3, levels of STAT1 and serine pSTAT1 were similar in patients with high and low lymphocyte counts (Figure 1C).
Figure 1. STAT3 levels are higher and STAT1 levels are lower in high-counts compared to low-counts CLLs.
(A) Left panel: Western immunoblot of CLL cells from patients with low and high lymphocyte counts. The figure depicts blots of 8 patients with low lymphocyte counts (upper panel) and 8 patients with high lymphocyte counts (lower panel). Anti-STAT3 and anti-serine pSTAT3 antibodies were used. Actin was used as the loading control, and HeLa cell extract served as the positive control. A similar analysis was performed using all other samples (data not shown). Right panel: Densitometry analysis of total STAT3 and serine pSTAT3 of cells from CLL patients with low (N = 32) and high (N = 32) lymphocyte counts normalized to the β-actin level is depicted. As shown, total STAT3 levels were higher in cells from patients with high lymphocyte counts, whereas serine pSTAT3 levels were not significantly different in patients with high and low lymphocyte counts. (B) STAT3 levels of CLL cells from 64 CLL patients, quantified using densitometry analysis of Western immunoblotting analysis, were plotted against the patients’ WBC counts. Spearman test was used to calculate the correlation between STAT3 and the patients white blood cell counts (WBC). (C) Westren Immunoblot analysis of CLL cells from patients with low (N = 9, median WBC = 20.8; range 3.3 to 35.7 ×109/L) and high (N= 6, median WBC = 107.2; range 56.7 to 130 ×109/L) lymphocyte counts using STAT1 and pSTAT1 antibodies. HeLa and NIH323 cell lines were used as positive controls. The medians of densitometry levels were compared using the Mann-Whitney test. As shown, unphosphorylated STAT1 and serine pSTAT1 levels were similar in cells from patients with high- and low- lymphocyte counts. Tyrosine pSTAT1 was not detected.
High lymphocyte counts are correlated with high rates of spontaneous apoptosis
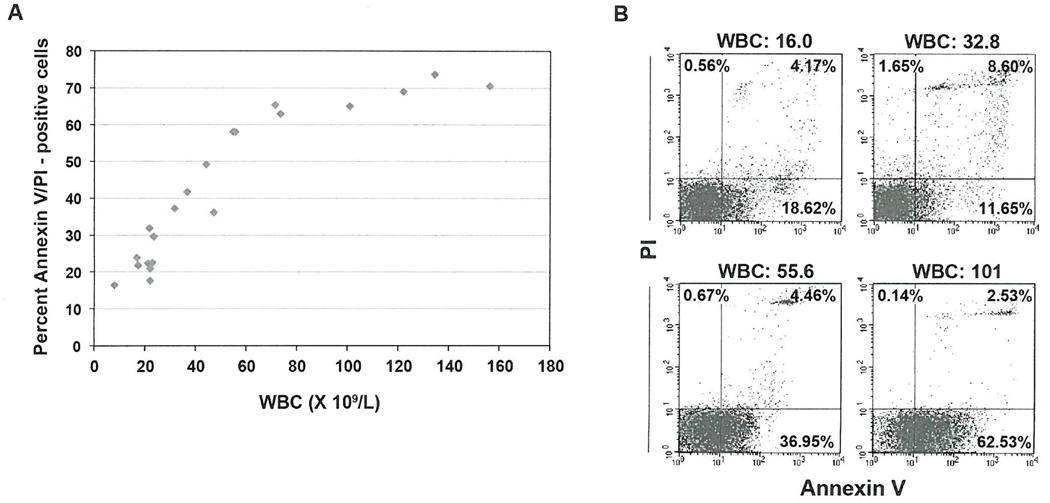
Previous studies showed that approximately 1% of the entire CLL clone divides daily (24). Because the actual increase in the PB lymphocyte count is lower than the expected lymphocyte count estimated using the CLL cell proliferation rate, we hypothesized that CLL cell proliferation is associated with concomitant spontaneous apoptosis. Using Annexin V and PI staining, we assessed the spontaneous apoptosis rate of fresh PB low-density cells obtained from patients with CLL and found that CLL cell apoptosis rates correlated with white blood cell (WBC) counts (rp = 0.88, P = <0.0001) (Figure 2A). For example, the spontaneous apoptosis rate of CLL PB low-density cells (>98% lymphocytes) from a patient with a WBC count of 16 × 109/L was 23%, whereas the spontaneous apoptosis rate of CLL PB cells from a patient with a WBC count of 101 × 109/L was 65% (Figure 2B). Because similar experiments (data not shown) revealed a similar trend, we sought to determine whether high lymphocyte counts correlated with a high apoptosis rate.
Figure 2. Peripheral lymphocytes of patients with CLL undergo spontaneous apoptosis.
(A) Spontaneous apoptosis rates of freshly obtained low-density cells from 19 CLL patients. Cellular apoptosis was assessed using flow cytometry following annexin V/PI staining. Rates of spontaneous apoptosis correlated with WBC counts (rp = 0.88, P = < 0.0001, Figure 2A) (B) Shown are rates of spontaneous apoptosis (early, lower right quadrant; late, upper right quadrant) of 4 patients with low, intermediate and high WBC counts.
To test our hypothesis, we first analyzed the gene signature profile of CLL cells from 9 patients with high (N=4) and low (N=5) lymphocyte counts. The same trend in gene expression was validated by qRT-PCR and pathway analysis revealed that the “apoptosis signaling pathway” was enriched with genes whose levels were high in CLL cells from patients with high lymphocyte counts (P = 0.02). Specifically, caspase3, MAPK 8, KRAS, PLCγ2, PKC, calpain9, and caspase3 were incorporated in this pathway (Supplementary figure 1).
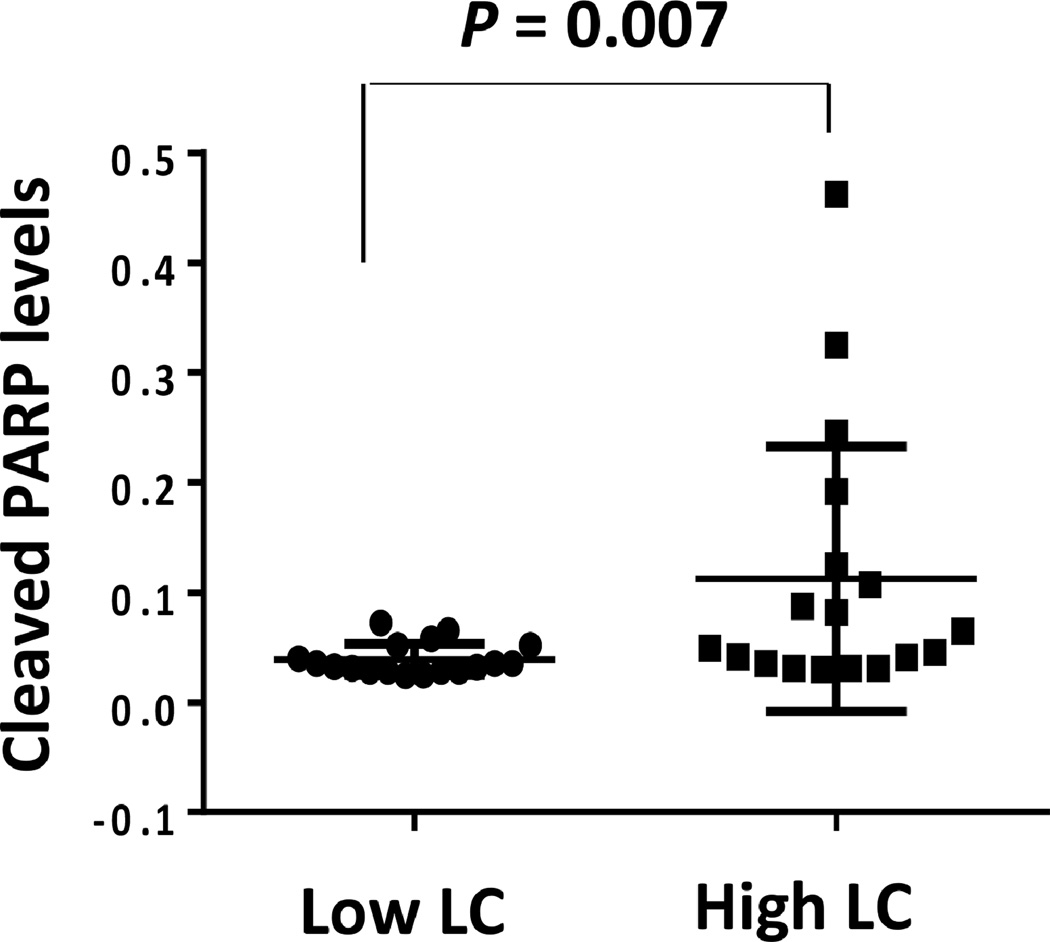
Taken together, these results suggest that the apoptosis signaling pathway is activated in CLL cells from patients with high lymphocyte counts and that those cells undergo spontaneous apoptosis at a high rate. To further delineate these findings, we obtained cells from CLL patients with high (N=16) and low (N=16) lymphocyte counts and, using ELISA, assessed the levels of cleaved PARP. PARP, known to protect cellular integrity, is cleaved and inactivated by cleaved caspase3. We found that the median level of cleaved PARP was significantly higher in cells from patients with high lymphocyte counts (median, 0.03; range, 0.02 – 0.07) than in cells from patients with low lymphocyte counts (median, 0.06; range, 0.03 – 0.46), (P<0.007) (Figure 3).
Figure 3. Cleaved PARP levels are high in CLL cells from patients with high lymphocyte counts.
CLL cell (ng/5 × 105 cells) cleaved PARP levels were measured by ELISA using PB cells from 32 patients. Median cleaved PARP levels in cells of patients with low (N = 16) and high (N = 16) lymphocyte counts were analyzed using the Mann-Whitney test. As shown, the levels of cleaved PARP were significantly higher in cells from patients with high lymphocyte counts (P < 0.007). LC denotes lymphocyte count.
Overexpression of STAT3 upregulates caspase3 levels and induces apoptosis
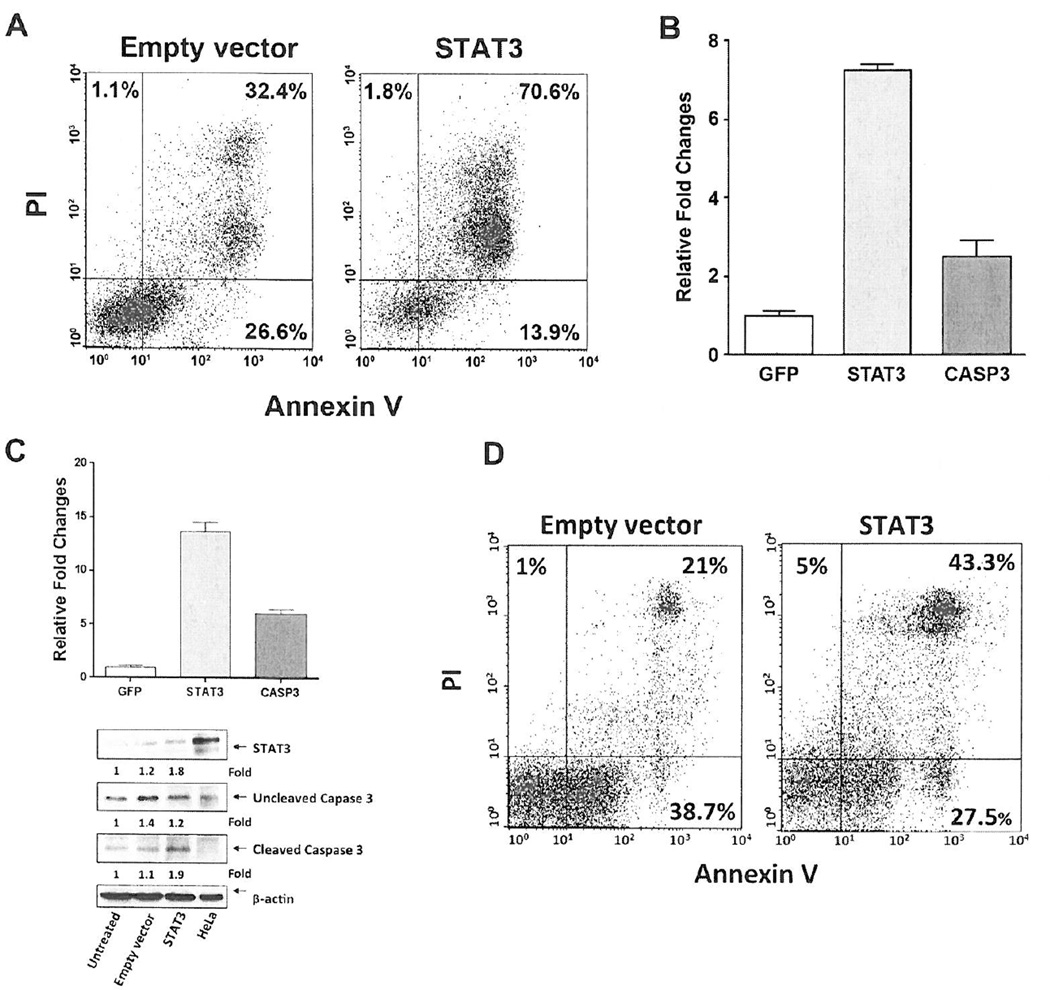
Although STAT3 is a transcription factor that provides CLL cells with a survival advantage, and its levels are high in lymphocytes from CLL patients with high lymphocyte counts (Figure 1), CLL cells from patients with high lymphocyte counts undergo spontaneous apoptosis at a higher rate than cells obtained from patients with low lymphocyte counts who harbor lower STAT3 levels (Figures 2 and 3). To determine whether high levels of STAT3 induce apoptosis, we transfected MM-1 cells with a vector containing the recombinant STAT3 DNA construct and used Annexin/PI staining to assess the cellular apoptosis rate. As shown in Figure 4A, 85% of cells transfected with STAT3 underwent apoptosis, as opposed to 59% of cells that were transfected with the empty vector (44% difference).
Figure 4. Overexpression of STAT3 upregulates caspase3 and induces apoptosis in MM-1 and CLL cells.
(A) MM-1 cells were transfected via electroporation with a mammalian vector that coexpressed the full-length STAT3 sequence and GFP or with the same vector expressing only GFP (empty vector) and stimulated with IL-6. Using double staining for annexinV/PI, we detected the cellular apoptosis rate via flow cytometry 24 hours after the transfection. As shown, at a transfection rate of 42%, 85% of CLL cells transfected with STAT3 underwent apoptosis compared to a 59% apoptosis rate of cells transfected with the empty vector. A representative Figure from 3 different experiments is depicted. In all experiments apoptosis rate was higher in MM-1 cell transfected with recombinant STAT3 DNA (P< 0.001; paired t-test). (B) Relative expression of STAT3 and caspase3 mRNA assessed via qRT-PCR in MM-1 cells transfected with STAT3 and stimulated with IL-6. Compared to control untransfected cells, STAT3 and caspase3 RNA levels were markedly upregulated. RNA levels of STAT3 and caspase3 in STAT3-transfected MM-1 cells. As shown, transfection of MM-1 cells with STAT3 induced a 7.5-fold increase in STAT3 and a 2.5-fold increase in caspase3 RNA levels. The means ± SD of gene expression fold change from 3 different experiments are depicted. (C) Upper panel: Relative expression of STAT3 and caspase3 mRNA assessed using qRT-PCR in CLL cells infected with a lentivirus harboring the human STAT3 gene. CLL cells were infected with the full-length STAT3 gene sequence. As shown, overexpression of STAT3 (≥ 30% infection efficiency) induced a 13.5-fold increase in STAT3 and a 6-fold increase in caspase3 RNA levels. The means ± SD of gene expression fold change from 3 different experiments using the same patient’s cells are depicted. Lower panel: Protein levels of STAT3 and uncleaved and cleaved Caspase3 of the same patient’s cells assessed by western immunoblotting and quantitated by densitometry. As shown, STAT3 levels increased by 0.6-fold and cleaved Caspase3 levels by 0.8-fold compared with their corresponding levels in cells infected with the empty vector. Similar results were obtained using CLL cells from two other patients with low lymphocyte counts. (D) Apoptosis rate of the CLL cells infected with the full-length STAT3 gene or the empty vector assessed by flow cytometry 24 hours after infection. As shown, at an infection rate of 30%, the apoptosis rate of CLL cells transfected with STAT3 was 70.8% whereas that of cells infected with the empty vector was 59.7%. Similar results were obtained using cells from two other patients with low lymphocyte counts.
Because the protein levels of caspase3, an essential pro-apoptotic protein activated downstream of both the intrinsic and extrinsic apoptotic pathways, were significantly increased in CLL cells from patients with a high lymphocyte count, and overexpression of STAT3 in MM-1 cells induced apoptosis (Figure 4A), we sought to determine whether there is a cause-and-effect association between STAT3 and caspase3. Therefore, we transfected MM-1 cells with recombinant STAT3 DNA and using qRT-PCR assessed STAT3 and caspase3 RNA levels 24 hours after the transfection. Transfection of MM-1 cells with STAT3 induced a 7.5-fold increase in STAT3 expression levels and a 2.5-fold increase in caspase3 expression levels (Figure 4B). Similarly, infection of CLL cells with a lentivirus harboring the full-length human STAT3 induced an increase in STAT3 and caspase3 expression, an increase in STAT3 and cleaved caspase3 protein levels (Figure 4C), and an increase in apoptosis rate of CLL cells (Figure 4D).
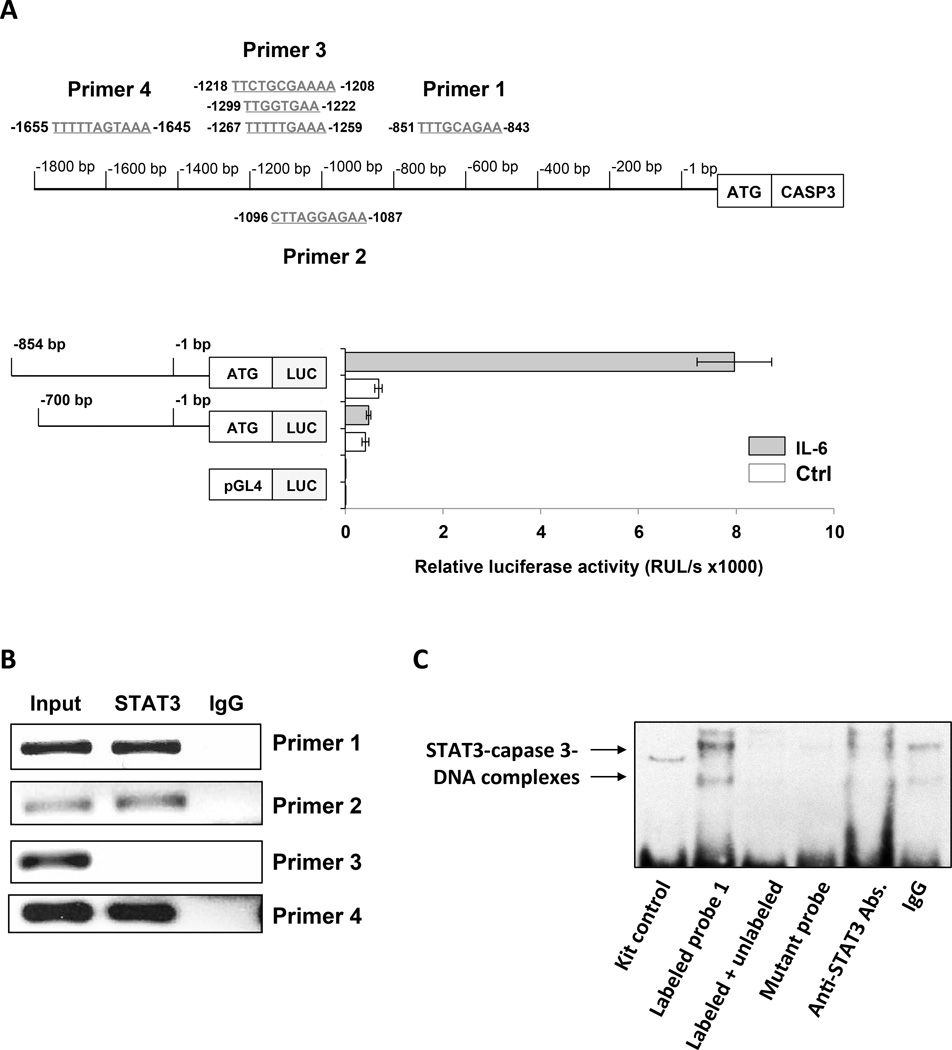
STAT3 binds and activates the promoter of the caspase3 gene
Because overexpression of STAT3 resulted in increased caspase3 RNA levels, we sought to determine whether STAT3 directly activates the caspase3 gene. We conducted a sequence analysis of the caspase3 gene promoter and identified within the promoter six GAS-like elements thought to be STAT3 binding sites (Figure 5A, upper panel). Then, to determine whether STAT3 activates the caspase3 promoter, we transfected MM-1 cells with the luciferase reporter gene driven by truncated fragments of the caspase3 promoter. Because in unstimulated MM-1 cells STAT3 is not activated (25), we incubated the cells with IL-6 that induces phosphorylation of STAT3 on tyrosine 705 residues (10, 26, 27). IL-6-induced luciferase activity was detected in the 854 bp fragment that harbors putative STAT3 binding sites but not in shorter fragments lacking those sites (Figure 5A, lower panel). Then, using ChIP, we found that DNA fragments detected by primers 1, 2, and 4, but not primer 3, each corresponding to a single GAS-like element (Figure 5A), co-immunoprecipitated with STAT3 (Figure 5B), suggesting that STAT3 binds to sites 1, 2, and 4 (but not site 3) of the caspase3 promoter. To confirm that STAT3 binds to the caspase3 gene promoter, we performed EMSA using a biotinylated DNA probe of binding site 1 of the caspase3 promoter. MM-1 nuclear protein extract bound to and formed a complex with the caspase3 DNA promoter, and the binding was completely reversed by the addition of excess unlabeled probe or attenuated by anti-STAT3 antibodies, whereas no binding was detected when a biotinylated mutated site 1 DNA probe was used (Figure 5C).
Figure 5. STAT3 binds to and activates the caspase3 gene promoter in MM-1 cells.
(A) Upper panel: Schematic diagram of the 1.8-kb DNA sequence located at the 5’ transcription starting site of the caspase3 gene promoter. In this region, we identified 6 GAS-like elements. Primers 1 to 4 were designed to capture these putative STAT3 binding sites. Lower panel: MM-1 cells were transfected with a reporter construct that included the luciferase gene and either the 770 bp that did not include a GAS-like element or the 854 bp sequence that included a single GAS-like element flanking the 5’ region of the caspase3 gene. After incubation with IL-6, which induces the phosphorylation of STAT3, luciferase activity was detected only in cells transfected with the longer caspase3 promoter construct that included STAT3 binding sites. The means ± SD of 4 different experiments are depicted. (B) MM-1 cell were incubated for 30 minutes with 30 ng/ml IL-6 and DNA was extracted prior to (input) or after chromatin immunoprecipitation with anti-STAT3 antibodies. As shown, DNA from the caspase3 promoter was detected via PCR with primers 1, 2, and 4, but not primer 3, corresponding to the STAT3 putative binding sites 1 to 4. (C) MM-1 cell nuclear extract was incubated with a biotinylated DNA probe that included a putative STAT3 binding site detected via ChIP with primer set 1 in the caspase3 promoter. EMSA showed that the CLL cell nuclear extract bound the biotinylated DNA probe, excess unlabeled probe reversed the binding, anti-STAT3 antibodies (but not IgG) attenuated the binding, and there was no binding to the biotinylated mutated DNA probe, suggesting that CLL cell nuclear STAT3 bound the caspase3 promoter. Representative data from 3 duplicate experiments that yielded identical results are depicted.
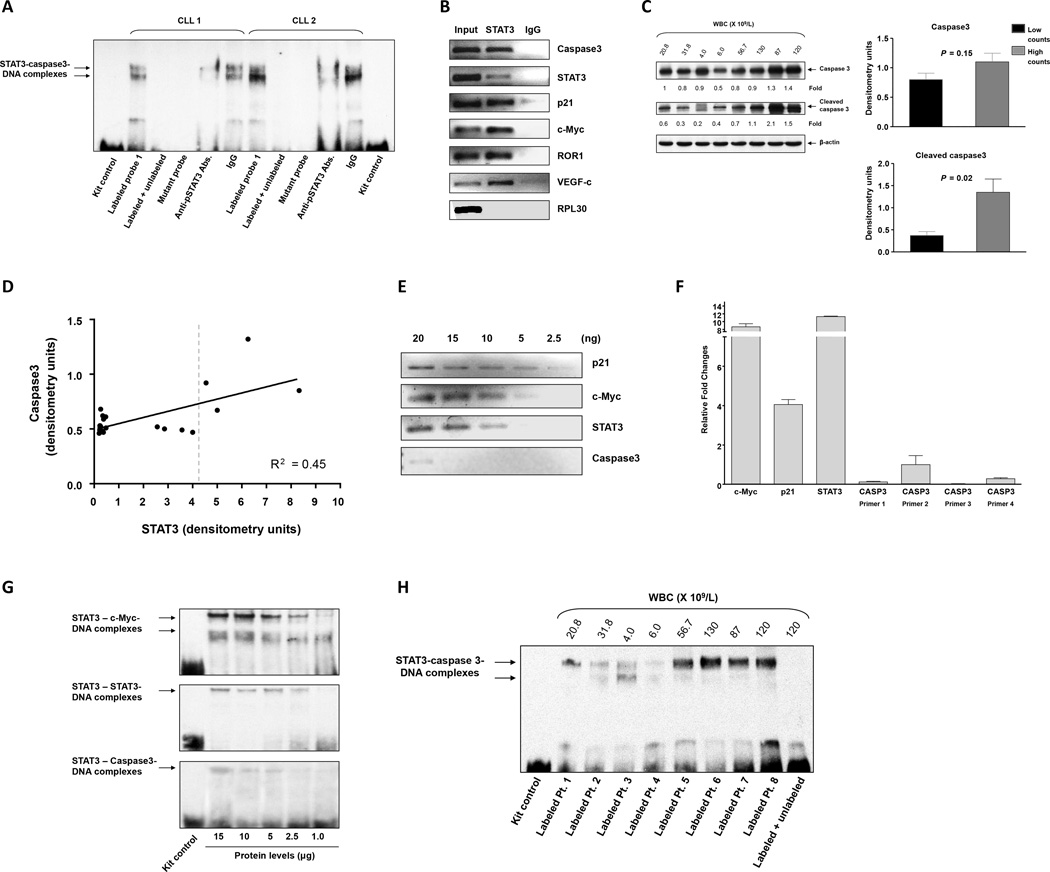
STAT3 binds with low affinity to the caspase3 gene promoter of CLL cells
To validate that STAT3 binds to the caspase3 gene promoter in CLL cells as well, we incubated nuclear extracts of PB CLL cells from two patients with high lymphocyte counts with the biotinylated DNA probe of binding site 1 as described in the previous section. CLL cell nuclear extracts bound to and formed complexes with the caspase3 DNA promoter fragment, and the binding was completely reversed by the addition of excess unlabeled probe (Figure 6A). Unlike the isotype control IgG, anti-serine pSTAT3 antibodies, attenuated the binding, and no binding was detected when a biotinylated mutated site 1 DNA probe was used instead of the site 1 probe. In addition, using ChIP, we found that CLL cell chromatin fragments that were immunoprecipitated with anti-STAT3 antibodies coimmunoprecipitated with DNA of caspase3 and the STAT3-regulated genes STAT3, p21, c-Myc, ROR1, and VEGF-C (Figure 6B). Taken together, these data suggest that STAT3 binds to the caspase3 promoter.
Figure 6. STAT3 binds with low affinity to the caspase3 gene promoter in CLL cells.
(A) Nuclear extracts of CLL cells from two patients with high lymphocyte counts were incubated with biotinylated probe that included the putative STAT3 binding site detected by primer 1. EMSA showed that that the protein extracts from both patients bound the caspase3 DNA probe. The addition of excess unlabeled probe reversed the binding, anti-serine pSTAT3 antibodies (but not IgG) attenuated the binding, and there was no binding to the biotinylated mutated DNA probe. (B) ChIP of CLL cells. CLL cell chromatin fragments were immunoprecipitated with anti-STAT3 antibodies, the co-immunoprecipitated DNA was isolated, and STAT3 target genes were detected via PCR. As shown, the DNA of caspase3, STAT3, p21, c-Myc, ROR1, and VEGF-c co-immunoprecipitated with STAT3. RPL30 promoter was used as a negative control. This experiment was repeated 4 time using samples from 4 different patients. Similar results were obtained in all experiments. The Figure depicts results of two experiments using CLL cells from two different patients. (C) Left panel: Western immunoblot analysis of CLL cells from patients with low (N = 4) and high (N = 4) lymphocyte counts. The membrane depicted in Figure 1B was re-probed using caspase3 and cleaved caspase3 antibodies. Right Panel: Densitometry analysis of the Western bolt data shown in the left panel. (D) STAT3 and caspase3 levels were assessed simultaneously in CLL cells from 24 patients via Western blot analysis and quantitated using densitometry, whereby the level obtained in every lane was normalized to the levels of actin. The figure depicts a correlation between the levels of STAT3 and caspase3. As shown, the levels of caspase3 remained constant across a wide range of STAT3 levels and linearly increased with increasing STAT3 levels behind a threshold (vertical dotted line). A linear regression model (black line) that fits 45% of the variance (R2 = 0.45) is depicted. (E) CLL cell chromatin fragments were immunoprecipitated with anti-STAT3 antibodies. Serial dilutions of the DNA (2.5–20 ng) that were co-immunoprecipitated with STAT3 were prepared, and STAT3-regulated genes were detected via PCR. As shown, caspase3 was detected only in the least diluted (20 ng) DNA preparation. In contrast, p21, c-Myc, and STAT3 were also detected in the 5 ng (p21 and c-Myc) and 10 ng (STAT3) dilutions. This experiment was repeated 4 times. The Figure depicts results of two experiments using CLL cells from two different patients. (F) The binding affinity of 4 putative STAT3 binding sites in the caspase3 promoter of CLL cells was compared to that of c-Myc p21 and STAT3 using qRT-PCR. As in MM-1 cells, caspase3 was detected by primers 1, 2 and 4 but not by primer 3, and the relative fold change in c-Myc, p21 and STAT3 gene expression was significantly higher than that of caspase3, indicating that the binding affinity of STAT3 to c-Myc p21 and STAT3 promoters is stronger than the binding affinity of STAT3 to the caspase3 promoter. The data depicted are from 3 separate experiments. (G) EMSA comparing the binding of serial dilutions (1–15 µg) of CLL cell nuclear protein to biotinylated promoter probes of the STAT3-regulated genes c-Myc, STAT3, and caspase3. As shown, only a high concentration (15 µg) of nuclear protein bound to the caspase3 promoter probe, whereas lower concentrations of nuclear protein bound c-Myc or STAT3 biotinylated promoter probes. Similar data were generated using a sample from another patient. (H) EMSA of CLL cell nuclear extracts from patients with high and low lymphocyte counts. A labeled caspase3 promoter probe was used. As shown, DNA binding of STAT3 to the Caspase3 promoter probe was significantly lower in nuclear extracts from patients with low (median: 13.3 ×109/L, range: 4 – 31.8) vs. high (median: 103 ×109/L, range: 56.7 – 130) WBC counts. The addition of an unlabeled probe (right lane) attenuated the binding of STAT3 to the caspase3 labeled probe.
Like in other cancers, in CLL STAT3 provides the neoplastic cells with a survival advantage (10). However, overexpression of STAT3 induced a significant increase in caspase3 expression and induced apoptotic cell death. Furthermore, CLL cells from patients with a high, but not low, lymphocyte count expressed high levels of STAT3 and underwent spontaneous apoptosis at a high rate. Therefore, using Western immunoblotting, we assessed the levels of caspase3 in CLL cells from patients with high and low lymphocyte counts and found that caspase3 and cleaved caspase3 levels were significantly higher in cells from patient with high counts (Figure 6C). We then plotted the levels of caspase3 as a function of the levels of total STAT3 and found that caspase3 remained constant across a wide range of STAT3 levels and increased only when a threshold of STAT3 protein level was reached (Figure 6D), suggesting that the binding affinity of pSTAT3 to the caspase3 promoter is relatively weak and that pSTAT3 activates the transcription of caspase3 only when its levels are sufficiently high.
To test this hypothesis, we first used ChIP. Serial dilutions of CLL cell DNA of chromatin fragments coimmunoprecipitated with anti-STAT3 antibodies were prepared, and STAT3 target genes were detected via PCR. As opposed to p21, c-Myc, or STAT3, caspase3 was detected only in the least diluted fraction, suggesting that STAT3 binds to the caspase3 promoter with a low affinity (Figure 6E). We also analyzed the DNA that coimmunoprecipitated with STAT3 by using qRT-PCR. High c-Myc, p21 and STAT3 but low caspase3 levels were detected. As in MM-1 cells, DNA of the Caspase3 promoter was detected using primer sets 1, 2 and 4 but not with primer set 3 (Figure 6F). These results confirmed that STAT3 binds to the caspase3 promoter with low affinity.
To further delineate these findings we performed EMSA. Using serial dilutions of CLL cell nuclear extracts we found that the binding of those extracts to the biotinylated caspase3 promoter DNA probe was low compared to the binding of c-Myc or STAT3 promoter probes across all tested dilutions (Figure 6G). Furthermore, the binding of nuclear extracts of CLL cells from patients with low lymphocyte counts (N = 4) was significantly lower than that of patients with high counts (N = 4) (Figure 6H), suggesting that nuclear extracts from CLL patients with high lymphocyte counts bind to the caspase3 promoter with a higher affinity.
Discussion
Herein, we show that when present at high levels, pSTAT3 activates caspase3 and induces apoptosis. For decades the relatively slow accumulation of CLL cells in most patients was attributed to a prolonged lifespan of the leukemia cells (2–4, 28). However, evidence from recent years suggests that CLL cells proliferate and die, that cell birth and death rates are similar and that the actual CLL cell count represents a net effect of clonal turnover (24).
Like in several cancers, in CLL STAT3 is constitutively activated (8, 11, 17, 19, 29, 30). Activated STAT3 functions as an oncogene that induces proliferation and protects the neoplastic cells from apoptosis (10, 14, 31, 32). STAT3 activates the STAT3 gene, and as a result, CLL cells harbor high levels of STAT3 (10). As anticipated, STAT3 levels correlated with the patients’ lymphocyte count. However in patients with high counts STAT3 induced apoptosis rather than providing the cells with a survival advantage. This functional switch occurred only in cells with significantly high STAT3 levels. Likewise, overexpression of STAT3 induced apoptosis in MM-1 cells and in CLL cells from patients with relatively low lymphocyte counts.
In various neoplasms STAT1 and STAT3 undergo similar post-translation modifications (33). Consistent with our data, in CLL both STAT1 and STAT3 are constitutively phosphorylated on serine 727 residues (10, 22) but not on tyrosine residues (34–36). However, whereas activated STAT3 under most conditions protects the cells from apoptosis (10, 12, 13, 22), activated STAT1 induces apoptosis (37). Remarkably, unlike STAT3, STAT1 or serine pSTAT1 levels did not correlated with CLL patients’ lymphocyte counts, suggesting that the proapoptotic effect of STAT3 is independent of STAT1.
Theoretically, at high levels, STAT3 might promote apoptosis either directly or indirectly by no longer activating anti-apoptotic genes. In a previous study, we found that in acute myeloid leukemia granulocyte-macrophage colony-stimulating factor, known to induce myeloid cell proliferation, also upregulated and activated pro-apoptotic caspases by activating the Janus kinase-STAT signaling pathway (38). The results of that study suggested that STAT activation induces apoptosis by directly activating the apoptotic cell death machinery. In the current study, we found that the transcript levels of the STAT3-regulated anti-apoptotic genes Bcl-2 and MCL-1 were similar in CLL cells from patients with high and low lymphocyte counts, suggesting that these anti-apoptotic genes are not downregulated in cells from patients with high lymphocyte counts. However, in cells from patients with high lymphocyte counts, caspase3 levels were significantly high, suggesting that when present at high levels, STAT3 activates caspase3.
In mammalian cells STAT3 is ubiquitously expressed as a latent isoform and is activated upon phosphorylation on tyrosine 705 residues (39). In circulating CLL cells STAT3 is constitutively phosphorylated on serine but not tyrosine residues (10, 22) and both serine- and tyrosine-pSTAT3 activate a similar repertoire of genes (12, 13, 19, 36). By using a luciferase assay of IL-6-stimulated MM-1 cells, in which IL-6 induces tyrosine-pSTAT3 (25), we found that pSTAT3 activates the caspase3 promoter. These data were confirmed by ChIP and an EMSA with antibodies that detect phosphorylated and unphosphorylated STAT3 isoforms. In CLL cells ChIP confirmed that STAT3 binds with low affinity to the caspase3 promoter, and an EMSA revealed that specific anti-serine pSTAT3 antibodies significantly attenuated the binding of pSTAT3 to the caspase3 promoter, suggesting that serine-pSTAT3 rather than its unphosphorylated form activates the caspase3 promoter in CLL cells.
Caspase3 levels increased only when STAT3 levels reached a threshold. High levels of STAT3 were required to induce this effect because the binding affinity of pSTAT3 to the caspase3 gene promoter is relatively low. Hence, the functional switch of STAT3 depends on its DNA-binding affinity. However DNA-binding affinity cannot be predicted by a consensus sequence per-se. STAT3-DNA binding may vary depending on background DNA sequences in the vicinity to the GAS element, the chromatin status and concomitant binding of STAT or DNA structures to other proteins, adaptor molecules, and other factors (40). For example, we identified several GAS-like elements in the caspase3 promoter and used primers to detect them. However one primer (primer 3), constructed to detect a STAT3-specific (TTCN3GAA) binding element, did not detect STAT3-caspase3-DNA binding in MM-1 or CLL cells. While analyzing STAT3 binding sites in the genome of different species, we did not detect conserved sequence blocks within the Caspase3 promoter. However we identifid several putative STAT3 binding sites in the caspase3 promoter of all examined organisms.
STAT3 binds with high affinity to the anti-apoptotic Bcl-2 and MCL-1 gene promoters, and with low affinity to the caspase3 gene promoter, suggesting that in CLL cells from patients with high lymphocyte counts, the activation of the caspase3 gene overcomes in part the protective effect of anti-apoptotic genes, and apoptotic cell death ensues.
Caspase3 is an inactive zymogen that is usually activated by the intrinsic and the extrinsic apoptotic pathways (41). By activating the caspase3 gene, STAT3 increases the level of caspase3 and predisposes it to proteolytic cleavage by upstream caspases such as caspase 8 or caspase 9. In CLL cells STAT3 levels correlated with the levels of cleaved- but not uncleaved-caspase3 possibly because as its levels increased, caspase3 underwent rapid cleavage. Unlike other caspases that possess autocatalytic activity, an intrinsic “safety catch” prevents the autocatalysis of caspase3. Upon removal of the safety catch by acidification or other means, caspase3 undergoes autocatalytic cleavage (42). Whether autocatalysis or upstream caspases activate caspase3 in CLL cells remains to be determined.
Whereas in normal cells extracellular stimuli activate STAT3, in CLL STAT3 is constitutively phosphorylated and activated. pSTAT3 forms dimers, shuttles to the nucleus using the karypherin nuclear transport system, binds to DNA, and, following nuclear phosphatase-induced dephosphorylation, STAT3 translocates to the cytosol using nuclear export chromosome region maintenance 1 (10). In the nucleus, pSTAT3 binds primarily to high-affinity DNA-binding sites, and when present at excess, pSTAT3 dimers bind to low-affinity DNA-binding sites. In the cytosol, a cytoplasmic serine kinase phosphorylates the USTAT3 on serine 727 residues (Estrov et al., unpublished data); in addition, USTAT3 binds to the nuclear factor (NF)-κB in competition with IκB (19). The levels of serine pSTAT detected via western immunoblotting were similar in CLL cells of patients with high or low lymphocyte counts, most likely because this assay is not suitable to detect miniscule, albeit biologically significant, changes in pSTAT3 levels. Given that pSTAT3 activates the STAT3 gene and induces the production of STAT3 protein (10), cellular levels of total STAT3 protein represent a better parameter of the activity of STAT3. Therefore, it is very likely that in CLL cells from patients with high lymphocyte counts, both USTAT3 and pSTAT3 levels are high.
USTAT3 activates NF-κB (19) and, like the nuclear factor of activated T cells (NFAT), NF-κB is constitutively activated in unstimulated CLL cells. Together, these transcription factors orchestrate a transcription program that further protects CLL cells from apoptosis (19, 43–45). Like STAT3, NF-κB levels correlate with disease burden (46), and although generally viewed as an anti-apoptotic transcription factor, NF-κB also harbors pro-apoptotic properties (47).
Excessive activation of STAT3 induces apoptosis of CLL cells. Whether this versatile effect represents a physiological negative feedback mechanism used in normal cells to counteract extreme proliferation or unwarranted prolonged cellular survival remains to be determined.
Supplementary Material
Acknowledgments
We thank Markeda Wade of the Department of Scientific Publications at The University of Texas MD Anderson Cancer Center for editing the manuscript.
This study was supported by a grant from the CLL Global Research Foundation.
The University of Texas MD Anderson Cancer Center is supported in part by the National Institutes of Health through a Cancer Center Support Grant (P30CA16672).
Footnotes
Authorship
Conception and design: M.K., Z.E., U.R.
Statistical and bioinformatics analysis: U.R.
Provision of study materials or patients: Z.E., M.K, S.V, W.W., A.F., S.F.
Collection and assembly of data: J.Y.W. performed the western blot and IP experiments;
D.H. performed the PI/Annexin assay; P.L. qRTPCR EMSA and ChiP, experiments; Z.L.,S.G., D.H. and M.M. performed the ELISA experiments
Manuscript writing: U.R., Z.E.
Final approval of manuscript: All authors.
Conflict-of-interest disclosure
The authors declare no competing financial interests.
References
- 1.Chiorazzi N, Rai KR, Ferrarini M. Chronic lymphocytic leukemia. N Engl J Med. 2005;352:804–815. doi: 10.1056/NEJMra041720. [DOI] [PubMed] [Google Scholar]
- 2.Bentley DP, Pepper CJ. The apoptotic pathway: a target for therapy in chronic lymphocytic leukemia. Hematol Oncol. 2000;18:87–98. doi: 10.1002/1099-1069(200009)18:3<87::aid-hon661>3.0.co;2-8. [DOI] [PubMed] [Google Scholar]
- 3.Dameshek W. Chronic lymphocytic leukemia--an accumulative disease of immunolgically incompetent lymphocytes. Blood. 1967;29(Suppl):566–584. [PubMed] [Google Scholar]
- 4.Kitada S, Pedersen IM, Schimmer AD, Reed JC. Dysregulation of apoptosis genes in hematopoietic malignancies. Oncogene. 2002;21:3459–3474. doi: 10.1038/sj.onc.1205327. [DOI] [PubMed] [Google Scholar]
- 5.Bueso-Ramos CE, Ferrajoli A, Medeiros LJ, Keating MJ, Estrov Z. Aberrant morphology, proliferation, and apoptosis of B-cell chronic lymphocytic leukemia cells. Hematology. 2004;9:279–286. doi: 10.1080/10245330410001727046. [DOI] [PubMed] [Google Scholar]
- 6.Chiorazzi N. Cell proliferation and death: forgotten features of chronic lymphocytic leukemia B cells. Best practice & research. Clinical haematology. 2007;20:399–413. doi: 10.1016/j.beha.2007.03.007. [DOI] [PubMed] [Google Scholar]
- 7.Wang X, Crowe PJ, Goldstein D, Yang JL. STAT3 inhibition, a novel approach to enhancing targeted therapy in human cancers (review) International journal of oncology. 2012;41:1181–1191. doi: 10.3892/ijo.2012.1568. [DOI] [PubMed] [Google Scholar]
- 8.Alvarez JV, Greulich H, Sellers WR, Meyerson M, Frank DA. Signal transducer and activator of transcription 3 is required for the oncogenic effects of non-small-cell lung cancer-associated mutations of the epidermal growth factor receptor. Cancer Res. 2006;66:3162–3168. doi: 10.1158/0008-5472.CAN-05-3757. [DOI] [PubMed] [Google Scholar]
- 9.Garcia R, Bowman TL, Niu G, Yu H, Minton S, Muro-Cacho CA, Cox CE, Falcone R, Fairclough R, Parsons S, Laudano A, Gazit A, Levitzki A, Kraker A, Jove R. Constitutive activation of Stat3 by the Src and JAK tyrosine kinases participates in growth regulation of human breast carcinoma cells. Oncogene. 2001;20:2499–2513. doi: 10.1038/sj.onc.1204349. [DOI] [PubMed] [Google Scholar]
- 10.Hazan-Halevy I, Harris D, Liu Z, Liu J, Li P, Chen X, Shanker S, Ferrajoli A, Keating MJ, Estrov Z. STAT3 is constitutively phosphorylated on serine 727 residues, binds DNA, and activates transcription in CLL cells. Blood. 2010;115:2852–2863. doi: 10.1182/blood-2009-10-230060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kusaba T, Nakayama T, Yamazumi K, Yakata Y, Yoshizaki A, Inoue K, Nagayasu T, Sekine I. Activation of STAT3 is a marker of poor prognosis in human colorectal cancer. Oncol Rep. 2006;15:1445–1451. [PubMed] [Google Scholar]
- 12.Li P, Grgurevic S, Liu Z, Harris D, Rozovski U, Calin GA, Keating MJ, Estrov Z. Signal Transducer and Activator of Transcription-3 Induces MicroRNA-155 Expression in Chronic Lymphocytic Leukemia. PLoS One. 2013;8:e64678. doi: 10.1371/journal.pone.0064678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li P, Harris D, Liu Z, Liu J, Keating M, Estrov Z. Stat3 activates the receptor tyrosine kinase like orphan receptor-1 gene in chronic lymphocytic leukemia cells. PLoS One. 2010;5:e11859. doi: 10.1371/journal.pone.0011859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu Y, Li PK, Li C, Lin J. Inhibition of STAT3 signaling blocks the anti-apoptotic activity of IL-6 in human liver cancer cells. J Biol Chem. 2010;285:27429–27439. doi: 10.1074/jbc.M110.142752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Malyukova A, Dohda T, von der Lehr N, Akhoondi S, Corcoran M, Heyman M, Spruck C, Grander D, Lendahl U, Sangfelt O. The tumor suppressor gene hCDC4 is frequently mutated in human T-cell acute lymphoblastic leukemia with functional consequences for Notch signaling. Cancer Res. 2007;67:5611–5616. doi: 10.1158/0008-5472.CAN-06-4381. [DOI] [PubMed] [Google Scholar]
- 16.Morikawa T, Baba Y, Yamauchi M, Kuchiba A, Nosho K, Shima K, Tanaka N, Huttenhower C, Frank DA, Fuchs CS, Ogino S. STAT3 expression, molecular features, inflammation patterns, and prognosis in a database of 724 colorectal cancers. Clin Cancer Res. 2011;17:1452–1462. doi: 10.1158/1078-0432.CCR-10-2694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yin W, Cheepala S, Roberts JN, Syson-Chan K, DiGiovanni J, Clifford JL. Active Stat3 is required for survival of human squamous cell carcinoma cells in serum-free conditions. Molecular cancer. 2006;5:15. doi: 10.1186/1476-4598-5-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ferrajoli A, Faderl S, Van Q, Koch P, Harris D, Liu Z, Hazan-Halevy I, Wang Y, Kantarjian HM, Priebe W, Estrov Z. WP1066 disrupts Janus kinase-2 and induces caspase-dependent apoptosis in acute myelogenous leukemia cells. Cancer Res. 2007;67:11291–11299. doi: 10.1158/0008-5472.CAN-07-0593. [DOI] [PubMed] [Google Scholar]
- 19.Liu Z, Hazan-Halevy I, Harris DM, Li P, Ferrajoli A, Faderl S, Keating MJ, Estrov Z. STAT-3 activates NF-kappaB in chronic lymphocytic leukemia cells. Mol Cancer Res. 2011;9:507–515. doi: 10.1158/1541-7786.MCR-10-0559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bromberg JF, Wrzeszczynska MH, Devgan G, Zhao Y, Pestell RG, Albanese C, Darnell JE., Jr Stat3 as an oncogene. Cell. 1999;98:295–303. doi: 10.1016/s0092-8674(00)81959-5. [DOI] [PubMed] [Google Scholar]
- 21.Sansone P, Bromberg J. Targeting the Interleukin-6/Jak/Stat Pathway in Human Malignancies. Journal of Clinical Oncology. 2012;30:1005–1014. doi: 10.1200/JCO.2010.31.8907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Frank DA, Mahajan S, Ritz J. B lymphocytes from patients with chronic lymphocytic leukemia contain signal transducer and activator of transcription (STAT) 1 and STAT3 constitutively phosphorylated on serine residues. J Clin Invest. 1997;100:3140–3148. doi: 10.1172/JCI119869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Avalle L, Pensa S, Regis G, Novelli F, Poli V. STAT1 and STAT3 in tumorigenesis: A matter of balance. Jak-Stat. 2012;1:65–72. doi: 10.4161/jkst.20045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chiorazzi N, Ferrarini M. Evolving view of the in-vivo kinetics of chronic lymphocytic leukemia B cells. Hematology Am Soc Hematol Educ Program. 2006:273–278. 512. doi: 10.1182/asheducation-2006.1.273. [DOI] [PubMed] [Google Scholar]
- 25.Amit-Vazina M, Shishodia S, Harris D, Van Q, Wang M, Weber D, Alexanian R, Talpaz M, Aggarwal BB, Estrov Z. Atiprimod blocks STAT3 phosphorylation and induces apoptosis in multiple myeloma cells. Br J Cancer. 2005;93:70–80. doi: 10.1038/sj.bjc.6602637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Berishaj M, Gao SP, Ahmed S, Leslie K, Al-Ahmadie H, Gerald WL, Bornmann W, Bromberg JF. Stat3 is tyrosine-phosphorylated through the interleukin-6/glycoprotein 130/Janus kinase pathway in breast cancer. Breast cancer research : BCR. 2007;9:R32. doi: 10.1186/bcr1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Leu CM, Wong FH, Chang C, Huang SF, Hu CP. Interleukin-6 acts as an antiapoptotic factor in human esophageal carcinoma cells through the activation of both STAT3 and mitogen-activated protein kinase pathways. Oncogene. 2003;22:7809–7818. doi: 10.1038/sj.onc.1207084. [DOI] [PubMed] [Google Scholar]
- 28.Robertson LE, Plunkett W, McConnell K, Keating MJ, McDonnell TJ. Bcl-2 expression in chronic lymphocytic leukemia and its correlation with the induction of apoptosis and clinical outcome. Leukemia. 1996;10:456–459. [PubMed] [Google Scholar]
- 29.Chung SS, Aroh C, Vadgama JV. Constitutive Activation of STAT3 Signaling Regulates hTERT and Promotes Stem Cell-Like Traits in Human Breast Cancer Cells. PLoS One. 2013;8:e83971. doi: 10.1371/journal.pone.0083971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Garner JM, Fan M, Yang CH, Du Z, Sims M, Davidoff AM, Pfeffer LM. Constitutive activation of signal transducer and activator of transcription 3 (STAT3) and nuclear factor kappaB signaling in glioblastoma cancer stem cells regulates the Notch pathway. J Biol Chem. 2013;288:26167–26176. doi: 10.1074/jbc.M113.477950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Glienke W, Hausmann E, Bergmann L. Downregulation of STAT3 signaling induces apoptosis but also promotes anti-apoptotic gene expression in human pancreatic cancer cell lines. Tumour Biol. 2011;32:493–500. doi: 10.1007/s13277-010-0143-4. [DOI] [PubMed] [Google Scholar]
- 32.Uckun F, Ozer Z, Vassilev A. Bruton’s tyrosine kinase prevents activation of the anti-apoptotic transcription factor STAT3 and promotes apoptosis in neoplastic B-cells and B-cell precursors exposed to oxidative stress. Br J Haematol. 2007;136:574–589. doi: 10.1111/j.1365-2141.2006.06468.x. [DOI] [PubMed] [Google Scholar]
- 33.Regis G, Pensa S, Boselli D, Novelli F, Poli V. Ups and downs: the STAT1:STAT3 seesaw of Interferon and gp130 receptor signalling. Seminars in cell & developmental biology. 2008;19:351–359. doi: 10.1016/j.semcdb.2008.06.004. [DOI] [PubMed] [Google Scholar]
- 34.Battle TE, Frank DA. STAT1 mediates differentiation of chronic lymphocytic leukemia cells in response to Bryostatin 1. Blood. 2003;102:3016–3024. doi: 10.1182/blood-2002-09-2972. [DOI] [PubMed] [Google Scholar]
- 35.Battle TE, Wierda WG, Rassenti LZ, Zahrieh D, Neuberg D, Kipps TJ, Frank DA. In vivo activation of signal transducer and activator of transcription 1 after CD154 gene therapy for chronic lymphocytic leukemia is associated with clinical and immunologic response. Clin Cancer Res. 2003;9:2166–2172. [PubMed] [Google Scholar]
- 36.Rozovski U, Wu JY, Harris DM, Liu Z, Li P, Hazan-Halevy I, Ferrajoli A, Burger JA, O’Brien S, Jain N, Verstovsek S, Wierda WG, Keating MJ, Estrov Z. Stimulation of the B-cell receptor activates the JAK2/STAT3 signaling pathway in chronic lymphocytic leukemia cells. Blood. 2014;123:3797–3802. doi: 10.1182/blood-2013-10-534073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liang X, Moseman EA, Farrar MA, Bachanova V, Weisdorf DJ, Blazar BR, Chen W. Toll-like receptor 9 signaling by CpG-B oligodeoxynucleotides induces an apoptotic pathway in human chronic lymphocytic leukemia B cells. Blood. 2010;115:5041–5052. doi: 10.1182/blood-2009-03-213363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Faderl S, Harris D, Van Q, Kantarjian HM, Talpaz M, Estrov Z. Granulocyte-macrophage colony-stimulating factor (GM-CSF) induces antiapoptotic and proapoptotic signals in acute myeloid leukemia. Blood. 2003;102:630–637. doi: 10.1182/blood-2002-06-1890. [DOI] [PubMed] [Google Scholar]
- 39.Darnell JE., Jr STATs and gene regulation. Science. 1997;277:1630–1635. doi: 10.1126/science.277.5332.1630. [DOI] [PubMed] [Google Scholar]
- 40.Ehret GB, Reichenbach P, Schindler U, Horvath CM, Fritz S, Nabholz M, Bucher P. DNA binding specificity of different STAT proteins. Comparison of in vitro specificity with natural target sites. J Biol Chem. 2001;276:6675–6688. doi: 10.1074/jbc.M001748200. [DOI] [PubMed] [Google Scholar]
- 41.Liu H, Chang DW, Yang X. Interdimer processing and linearity of procaspase-3 activation. A unifying mechanism for the activation of initiator and effector caspases. J Biol Chem. 2005;280:11578–11582. doi: 10.1074/jbc.M414385200. [DOI] [PubMed] [Google Scholar]
- 42.Roy S, Bayly CI, Gareau Y, Houtzager VM, Kargman S, Keen SL, Rowland K, Seiden IM, Thornberry NA, Nicholson DW. Maintenance of caspase-3 proenzyme dormancy by an intrinsic "safety catch" regulatory tripeptide. Proc Natl Acad Sci U S A. 2001;98:6132–6137. doi: 10.1073/pnas.111085198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chandra-Kuntal K, Singh SV. Diallyl trisulfide inhibits activation of signal transducer and activator of transcription 3 in prostate cancer cells in culture and in vivo. Cancer prevention research. 2010;3:1473–1483. doi: 10.1158/1940-6207.CAPR-10-0123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lee ES, Ko KK, Joe YA, Kang SG, Hong YK. Inhibition of STAT3 reverses drug resistance acquired in temozolomide-resistant human glioma cells. Oncology letters. 2011;2:115–121. doi: 10.3892/ol.2010.210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Real PJ, Sierra A, De Juan A, Segovia JC, Lopez-Vega JM, Fernandez-Luna JL. Resistance to chemotherapy via Stat3-dependent overexpression of Bcl-2 in metastatic breast cancer cells. Oncogene. 2002;21:7611–7618. doi: 10.1038/sj.onc.1206004. [DOI] [PubMed] [Google Scholar]
- 46.Hewamana S, Alghazal S, Lin TT, Clement M, Jenkins C, Guzman ML, Jordan CT, Neelakantan S, Crooks PA, Burnett AK, Pratt G, Fegan C, Rowntree C, Brennan P, Pepper C. The NF-kappaB subunit Rel A is associated with in vitro survival and clinical disease progression in chronic lymphocytic leukemia and represents a promising therapeutic target. Blood. 2008;111:4681–4689. doi: 10.1182/blood-2007-11-125278. [DOI] [PubMed] [Google Scholar]
- 47.Radhakrishnan SK, Kamalakaran S. Pro-apoptotic role of NF-kappaB: implications for cancer therapy. Biochimica et biophysica acta. 2006;1766:53–62. doi: 10.1016/j.bbcan.2006.02.001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.