Abstract
Early embryonic development features rapid nuclear DNA replication cycles, but lacks mtDNA replication. To meet the high‐energy demands of embryogenesis, mature oocytes are furnished with vast amounts of mitochondria and mtDNA. However, the cellular machinery driving massive mtDNA replication in ovaries remains unknown. Here, we describe a Drosophila AKAP protein, MDI that recruits a translation stimulator, La‐related protein (Larp), to the mitochondrial outer membrane in ovaries. The MDI‐Larp complex promotes the synthesis of a subset of nuclear‐encoded mitochondrial proteins by cytosolic ribosomes on the mitochondrial surface. MDI‐Larp's targets include mtDNA replication factors, mitochondrial ribosomal proteins, and electron‐transport chain subunits. Lack of MDI abolishes mtDNA replication in ovaries, which leads to mtDNA deficiency in mature eggs. Targeting Larp to the mitochondrial outer membrane independently of MDI restores local protein synthesis and rescues the phenotypes of mdi mutant flies. Our work suggests that a selective translational boost by the MDI‐Larp complex on the outer mitochondrial membrane might be essential for mtDNA replication and mitochondrial biogenesis during oogenesis.
Keywords: oogenesis, DNA replication, protein synthesis
Subject Categories: Development & Differentiation; DNA Replication, Repair & Recombination; Protein Biosynthesis & Quality Control
Introduction
Mitochondria contain their own genome, mitochondrial DNA (mtDNA), which encodes 13 key components of the electron‐transport chain (ETC) and is therefore vital for life (reviewed in Wallace, 2007). But mtDNA is prone to accumulating mutations because of its close vicinity to damaging reactive species that arise as by‐products of oxidative reactions (reviewed in Wallace, 2007). To ensure their reproductive success, metazoans must therefore prevent the transmission of deleterious mtDNA mutations from one generation to the next. Since mitochondria are transmitted exclusively through the female germ line in most metazoans, this task falls to the female germ line (reviewed in Stewart & Larsson, 2014). The process appears linked to the regulation of mtDNA replication during oogenesis. In the Drosophila ovary, mtDNA replication commences at the late germarium stage and is dependent on mitochondrial fitness. The selective amplification of wild‐type genomes in healthy mitochondria might help limit the transmission of deleterious mtDNA mutations (Hill et al, 2014). Selective mtDNA replication may also contribute to the genetic bottleneck that facilitates mtDNA segregation and selection in mammals (Wai et al, 2008).
Besides its role in mtDNA quality control, the female germ line is also tasked with providing the massive amounts of mitochondria and mtDNA required to power early embryonic development (Wai et al, 2010; Wolff et al, 2013). In Drosophila, mtDNA replication shuts down completely during early embryogenesis (Rubenstein et al, 1977), perhaps as a consequence of the rapid nuclear divisions without gap phases that mark this stage, as mtDNA is preferentially replicated in late G1 and early G2 phases (Zhang et al, 2015). However, mitochondria undergo massive mtDNA replication in mid‐stage egg chambers (Hill et al, 2014), which furnishes the mature oocyte with millions of copies of mtDNA (Wolff et al, 2013). Mammalian oocytes also display a burst of mtDNA replication prior to fertilization (St John, 2012). A typical mammalian oocyte contains hundreds of thousands of mtDNA molecules that are essential for post‐implantation development (Wai et al, 2010). Despite the essential role of mtDNA replication in mtDNA inheritance across species, the mechanisms of its regulation in the female germ line are largely unknown.
Mitochondria are semi‐autonomous organelles. The majority of mitochondrial proteins, including all components of the mtDNA replication machinery, are encoded in the nuclear genome, synthesized by cytoplasmic ribosomes, and imported into mitochondria (reviewed in Fox, 2012; reviewed in Moraes, 2001). Massive mitochondrial biogenesis therefore demands that nuclear genes produce vast amounts of mtDNA replication factors and mitochondrial proteins in a short developmental window. Even taking into account the fact that each developing oocyte receives the contribution of 16 genomes (from 1 oocyte and 15 accompanying nurse cells), it is perplexing to imagine how single‐copy nuclear genes support the production of the millions of mitochondria found in mature oocytes.
We have been using Drosophila as a model system to investigate mtDNA inheritance, because of the availability of tools to genetically manipulate mtDNA (Xu et al, 2008; Chen et al, 2015). Here, we report on a mitochondrial outer membrane protein, MDI that is required for mtDNA replication and mitochondrial biogenesis in the Drosophila ovary. mdi mutant flies are female semi‐sterile and display impaired mtDNA replication in ovaries. MDI recruits Larp, a translation stimulator to the mitochondrial surface, which then catalyzes the synthesis of a subset of nuclear‐encoded mitochondrial proteins by cytosolic ribosomes. Constitutively targeting Larp to the mitochondrial surface restores local protein synthesis, mtDNA replication, and fertility of mdi mutant female flies. Furthermore, ectopic expression of AKAP1, the human homolog of MDI, in mdi mutant flies rescues their fertility defect. Therefore, the translational regulation of mtDNA replication and mitochondrial biogenesis we observe in Drosophila might in fact be a conserved mechanism guiding mitochondrial inheritance in metazoan.
Results
mdi is essential for mtDNA replication in the ovary
We identified CG3249 from an ongoing RNAi screen for genes required for mtDNA replication in Drosophila ovaries. RNAi against the CG3249 locus significantly reduced mtDNA replication, as indicated by a sharp reduction in the number of mitochondria‐associated EdU puncta in the germarium (Fig EV1A). CG3249 is disrupted in a female‐sterile mutant, spoonbill that was generated by mobilizing a P‐element near CG3249 genomic locus (Hadad et al, 2011). However, the molecular nature of the chromosomal lesion in spoonbill background is uncertain. Another allele of CG3249, yu, was reported to have learning and memory defects, but normal fertility (Lu et al, 2007). Considering the uncertain molecular nature of these alleles and the discrepancies of phenotypes, we generated our own deletion of the CG3249 locus by CRISPR/Cas9 technology as previously described (Gratz et al, 2013). We obtained a 2.4‐kb deletion that removed most of the coding region of CG3249 (Fig 1A). Homozygous flies completely lacked the CG3429 protein product (Fig 1B). Homozygous females were semi‐sterile (Fig 2D), but there was no obvious phenotype in male flies. EdU staining showed that 93% of the ovaries carrying the CG3249 deletion displayed severely reduced mtDNA replication in the germarium, as well as in the egg chambers (Fig 2A and B). The remaining 7% showed normal mtDNA replication in the egg chambers, but reduced replication in the germarium region 2B (Fig 2C). Based on this reduced mtDNA replication phenotype, we named the deletion mutant mdi 1 for mitochondrial DNA insufficient. The mdi 1 mutation also caused mitochondria to clump together in mid‐stage egg chambers and in eggs (Fig EV2A–C), a phenotype that, given its later onset, may be a secondary effect of disrupted mtDNA replication.
Figure EV1. mtDNA replication in mdi RNAi and PKA mutant ovaries.
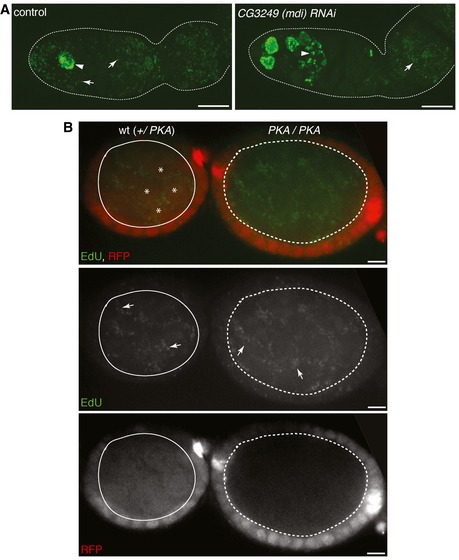
- DNA replication illustrated by EdU incorporation in ovaries of control and mdi RNAi flies. Arrows point to mitochondria and arrowheads point to nuclei. Genotypes: UAS‐dicer; nos‐gal4 (control); UAS‐dicer; nos‐gal4; UAS‐CG3249 IR [CG3249 (mdi) RNAi]. mtDNA replication is inhibited in late germarium stage in the ovariole expressing mdi RNAi. Scale bars, 10 μm.
- mtDNA replication in wt and PKA mutant clones illustrated by EdU incorporation. Arrows point to mtDNA replication. wt clones (solid circle) are labeled with a nuclear‐localized RFP reporter (*). pka null mutant clones that lack RFP signal (dashed circle) display normal mtDNA replication (arrows). Scale bars, 10 μm.
Figure 1. mdi gene, protein, and mdi 1 deletion.
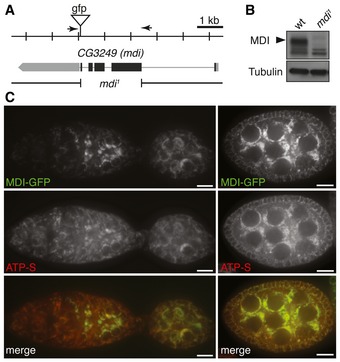
- Schematic drawing of the mdi genomic locus showing the CG3249 (mdi) transcript (gray box) and coding region (black box), and the location of the gfp insertion generated by CRISPR/Cas9‐mediated recombination with two guide RNAs (arrows). The resulting fusion protein is referred to as MDI‐GFP. The mdi 1 mutation is a 2.4‐kb deletion that removes most of the mdi coding region.
- Western blots of MDI protein in wild‐type (wt) and mdi 1 flies, showing that mdi 1 is a protein‐null mutation. Tubulin was used as a loading control.
- Ovarioles expressing MDI‐GFP stained with a mitochondrial marker, ATP synthase (ATP‐S). Overlapping signals (merge) indicate that MDI localizes to mitochondria. Note that MDI is highly expressed in germ cells at late germarium stages (left panels) and in egg chambers (right panels). Scale bars, 10 μm.
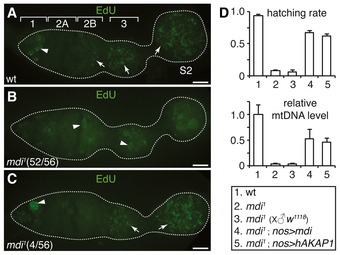
Figure 2. MDI promotes mtDNA replication in the ovary and is essential for female fertility.
-
A–CmtDNA replication in wt (A) and mdi 1 ovarioles (B, C) as illustrated by EdU incorporation. Arrows point to mitochondrial DNA and arrowheads to nuclei. In wt ovaries (A), mtDNA replication starts at germarium stage 2B and lasts into egg‐chamber stage 2 (S2). In most mdi 1 ovarioles (B), mtDNA replication is undetectable at these stages, whereas in some (C), it appears delayed until germarium stage 3. Scale bars, 10 μm.
-
DHatching rates and mtDNA content of eggs laid by females of different mutant genotypes relative to eggs laid by wt controls. Each data point represents the mean of three independent replicates. Error bars represent SD. Expression of mdi or hAKAP1 in mdi 1 significantly restored the mtDNA level and hatching rate. N = 3 × >100 eggs/genotype for hatching rate. The relative mtDNA level was determined as the average of three biological repeats. P‐values of comparing mdi 1; nos>mdi to mdi 1: hatching rate, P = 1.1995E−05; mtDNA, P = 0.0078. P‐values of comparing mdi 1; nos>hAKAP1 to mdi 1: hatching rate, P = 1.6998E−05; mtDNA level, P = 0.0009.
Figure EV2. Mitochondria are clumped together in the mdi 1 ovary and their eggs.
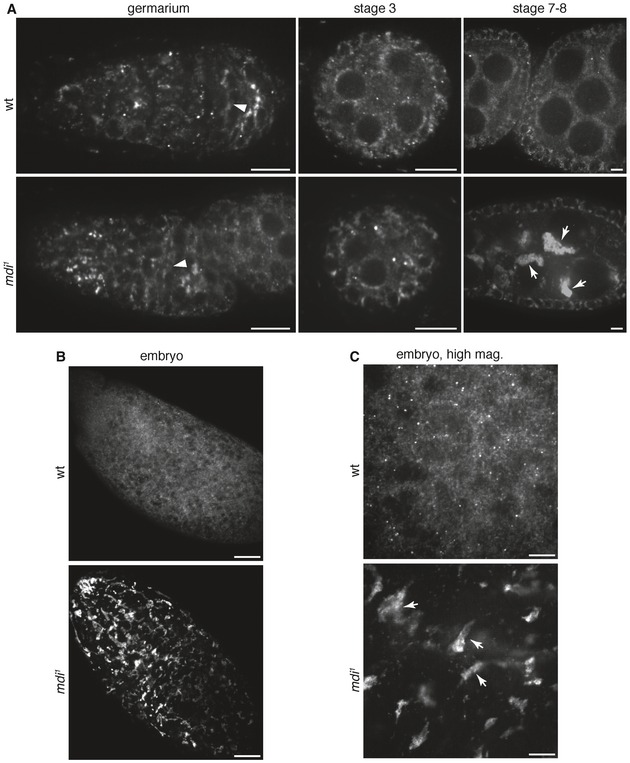
- Representative images of wt and mdi 1 ovarioles stained for ATP‐S, illustrating mitochondrial morphology. Note that mitochondria progressively clump together in mid‐ to late‐stage egg chambers of mdi 1 flies. Scale bars, 10 μm.
- Representative images wt and mdi 1 eggs stained for ATP‐S, illustrating mitochondrial morphology. Scale bars, 100 μm.
- High‐mag images of wt and mdi 1 eggs stained for ATP‐S. Scale bars, 20 μm. Note the clumping mitochondria in eggs produced by mdi 1 flies.
Despite the mitochondrial deficiencies in their ovaries, mdi 1 female flies produced a similar amount of eggs as wild‐type females (251 ± 24 vs. 239 ± 24 per fly). However, only 5% of the eggs produced by mdi 1 females crossed with either mdi 1 or wild‐type males hatched (Fig 2D), demonstrating the maternal‐effect, embryonic lethality of the mdi 1 mutation. Consistent with a reduced level of mtDNA replication during oogenesis, eggs produced by mdi 1 females had only 3% of the mtDNA amount found in the eggs of wild‐type females (Fig 2D). Importantly, the expression of an mdi cDNA transgene in the female germ line driven by nanos‐gal4 restored the mtDNA level and the hatching rate of mdi 1 eggs (Fig 2D). These observations demonstrate that MDI is required for mtDNA replication during oogenesis.
MDI is a multi‐domain protein of the mitochondrial outer membrane
To determine the expression pattern and sub‐cellular localization of MDI in ovaries, we inserted a GFP reporter in‐frame with the mdi ORF at the endogenous locus by CRISPR/Cas9‐mediated recombination (Fig 1A). The resulting fusion protein, MDI‐GFP, was highly expressed in germ cells, and its expression pattern paralleled the pattern of mtDNA replication in the ovary (Fig 1C): commencing at region 2B germarium and remaining active in mid‐stage egg chambers (Hill et al, 2014). The concurrence of MDI expression and mtDNA replication is consistent with the observation that MDI is essential for mtDNA replication in the ovary.
MDI contains a putative mitochondrial targeting sequence (MTS) at its N‐terminus (Fig 3A). Indeed, an MDI‐GFP fusion protein localized exclusively with mitochondria in cultured cells (Fig 3B). In the ovary, MDI‐GFP co‐localized with a mitochondrial marker, ATP synthase (Fig 1C), confirming that MDI is a mitochondrial protein. Moreover, a truncated MDI lacking the putative MTS, MDI∆MTS, had a diffuse cytoplasmic localization (Fig 3B) and failed to rescue the mtDNA levels and the hatching rate of mdi 1 eggs (Fig 3A). Therefore, mitochondrial localization appears essential for MDI's function.
Figure 3. Functional‐genetic analyses of MDI and its interaction with Larp.
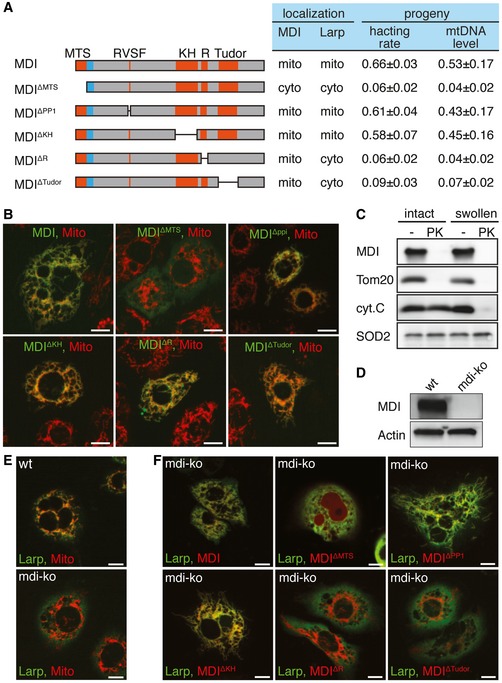
- Schematic drawing of MDI protein and deletion mutants. These proteins were expressed under the nanos‐gal4 driver and tested for their ability to rescue the hatching rate and mtDNA level of eggs from mdi 1 females (progeny column) or for localization and recruitment of partner protein Larp to mitochondria in cultured cells (localization column). N = 3 × >100 eggs/genotype for hatching rate. The relative mtDNA level was the average of three biological repeats.
- Representative images of S2 cells expressing GFP fusions (green) of the MDI proteins diagramed in (A). Staining with MitoTracker Red (red) to label mitochondria shows an overlapping signal (yellow), indicating proper localization to mitochondria for all fusion proteins except MDI∆MTS.
- Submitochondrial localization of MDI. Mitochondria from cultured cells expressing an MDI‐myc fusion protein were kept intact or subjected to swelling to disrupt their outer membranes and digested or not with protease K (PK). Protein extracts were subjected to Western blot analysis. Tom20, cytochrome C (cyt.C), and SOD2 were used as markers of the outer membrane, inter‐membrane space, and matrix, respectively. MDI behaved like an outer membrane protein.
- Western blots of MDI protein in wt and mdi knockout (mdi‐ko) cells confirming the mdi‐ko cells completely lacked MDI protein. Actin was used as a loading control.
- Representative images of wt or mdi‐ko cells expressing a Larp‐GFP fusion protein and stained with MitoTracker Red to label mitochondria. The majority of Larp localized to mitochondria in wt cells (overlapping red and green signal), but diffused into the cytoplasm of mdi‐ko cells.
- Representative images of mdi‐ko cells co‐expressing Larp‐GFP with MDI or the deletion mutants diagramed in (A) fused to the mCherry. Of the 4 MDI deletions that localized to mitochondria, 2 (MDIΔR and MDIΔTudor) did not allow Larp‐GFP to localize to mitochondria. The localization of Larp in mdi 1 flies expressing MDI or MDI deletion mutants, and the hatching rate and mtDNA level in their eggs were determined. The result is summarized in (A).
To determine the sub‐mitochondrial localization of MDI, we purified mitochondria from cultured cells stably expressing an MDI‐Myc fusion protein and digested them with protease K. MDI‐Myc was completely removed by protease K treatment (Fig 3C), as was Tom20, a marker of the outer membrane protein. By contrast, cytochrome C and SOD2, which reside at the inter‐membrane space and the matrix, respectively, were resistant to protease K treatment (Fig 3C), confirming the integrity of the purified mitochondria. We thus conclude that MDI localizes to the mitochondrial outer membrane, with its C‐terminus facing the cytoplasm.
Since mtDNA replication takes place in the matrix, it is puzzling that MDI, an outer membrane protein, should have such profound impact on mtDNA replication. Besides its MTS, MDI is predicted to have a RNA‐binding KH domain, a protein phosphatase 1‐interacting domain (PP1), a R domain that might bind to protein kinase A (PKA) and a Tudor domain (Hadad et al, 2011; Fig 3A). We generated a series of MDI mutants: MDI∆KH, MDI∆PP1, MDI∆R, and MDI∆Tudor with deletion in each of these domains (Fig 3A). All of these mutants localized to mitochondria properly (Fig 3B). The expression of MDI∆KH or MDI∆PP1 in the mdi 1 female germ line rescued the hatching rate and mtDNA level of their eggs, but the expression of MDI∆R and MDI∆Tudor did not (Fig 3A). This observation suggests that the R domain and Tudor domain are essential for MDI's function. MDI's mammalian homolog, AKAP1, recruits PKA through its R domain to the mitochondrial surface and regulates mitochondrial metabolism and dynamics (Wong & Scott, 2004). However, PKA is believed to localize on the plasma membrane, not the mitochondria, in ovaries (Lane & Kalderon, 1995). Additionally, mtDNA replication was normal in PKA mutant ovaries (Fig EV1B), indicating that MDI does not need to interact with PKA to regulate mtDNA replication. Some Tudor domain proteins localize to the mitochondrial outer membrane and regulate piRNA biogenesis (Honda et al, 2013). However, piRNA level was not affected in the CG3249 (mdi) knockdown ovaries (Handler et al, 2011). It is therefore unlikely that MDI regulates mtDNA replication via a piRNA‐related process.
Recruitment of Larp to mitochondria by MDI is essential for mtDNA replication
To identify potential interacting partners of MDI, we expressed MDI‐myc fusion protein in S2 cells, purified MDI complexes using anti‐myc antibody, and subjected them to mass‐spectrum (MS) analysis. We arbitrarily set the threshold of twofold enrichment in MDI‐myc immunoprecipitates compared to the control immunoprecipitates from non‐transfected S2 cells. To increase the confidence of the MS analysis, we also filtered out hits with less than 25% sequence coverage. Among the 15 proteins that met these criteria were SesB, the mitochondrial ATP:ADP antiporter, and Bor, a mitochondrial AAA protein (Table EV1). The remaining 13 are either ribosomal proteins or proteins involved in translational regulation (Table EV1). The top three candidates (those with the most peptide counts) are eukaryotic translation initiation factor 4G (eIF4G), Larp, and poly‐A binding protein (PABP). Mammalian homologs of these three proteins are all involved in initiating or boosting protein translation (Kahvejian et al, 2005; Sonenberg & Hinnebusch, 2009; Tcherkezian et al, 2014). A previous high‐throughput proteomic study also found that MDI (CG3249) interacts with CG7414 (Guruharsha et al, 2011), which encodes the translation initiation factor 2A. All these observations suggest that MDI might regulate protein translation.
We decided to focus on Larp for two reasons: (i) most bona fide ribosomal proteins are essential for cell viability, which would complicate the genetic analyses; (ii) mutations in larp, like the loss of function of mdi, cause maternal‐effect lethality (Blagden et al, 2009). Importantly, mtDNA replication was severely impaired in larp mutant ovaries (Fig 4B), implying that MDI and Larp might function in the same pathway. We confirmed the MDI‐Larp interaction by co‐immunoprecipitation from ovary extracts (Fig 4A). Although Larp is not annotated as a mitochondrial protein, it associates with mitochondria in spermatocytes (Ichihara et al, 2007). We also found that the majority of Larp coalesced around mitochondria in wild‐type ovaries (Figs 4C and EV3A), but had a diffuse cytoplasmic localization in the mdi 1 ovaries (Figs 4C and EV3A). This observation suggests that MDI is necessary for recruiting Larp to mitochondria.
Figure 4. MDI recruits Larp to mitochondria.
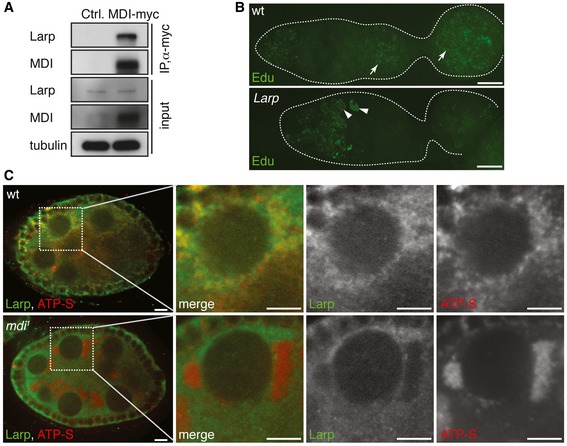
- Co‐immunoprecipitation of Larp with MDI‐myc in transfected S2 cells. Tubulin was used as a loading control.
- mtDNA replication, as illustrated by EdU incorporation, in wt and larp mutant (larp) ovarioles. mtDNA replication is dramatically reduced in the larp ovariole. Arrows: mtDNA; arrowheads: nuclei.
- wt and mdi 1 egg chambers stained for Larp (green) and ATP‐S (red) to reveal mitochondria. Larp closely associates with mitochondria in wt egg chambers. Mitochondria in mdi 1 flies are clumped together and completely lack Larp staining.
Figure EV3. Larp localization in ovaries and expression of mitochondrial proteins in ovary and somatic tissues.
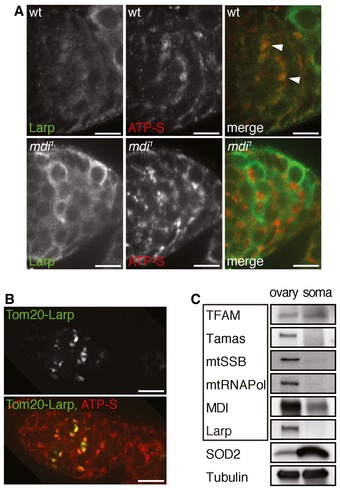
- Wild‐type and mdi 1 germaria were stained for Larp (green) and ATP‐S (red) to reveal mitochondria. Note that Larp closely associates with mitochondria in wt germarium (arrowheads). Mitochondria in mdi 1 flies completely lack Larp staining. Scale bars, 10 μm.
- Tom20‐LarpGFP (Tom20‐Larp) fusion protein was expressed under the control of nanos‐gal4 in an mdi 1 egg chamber that was stained with ATP‐S (red) to mark mitochondria. Note that Tom20‐Larp is concentrated around mitochondria (ATP‐S) in mdi 1 background. Scale bars, 10 μm.
- Western blots of several mitochondrial proteins in ovary and somatic tissues of wild‐type flies. Boxed are mtDNA replication factors, including TFAM, mtDNA polymerase (Tamas), mitochondrial single‐strand DNA binding protein (mtSSB), mitochondrial RNA polymerase (mtRNAPol), MDI, and Larp. Except for TFAM, most proteins required for mtDNA replication are upregulated in ovary mitochondria. Tubulin was used as a loading control.
We next examined whether Larp's association with mitochondria was required for MDI's function. Larp localized to the mitochondria in wild‐type S2 cells, but diffused into the cytoplasm of mdi‐ko S2 cells (Fig 3E), in which the endogenous mdi locus was disrupted by CRISPR/Cas9 technology (Fig 3D). We introduced a series of MDI deletion mutants into mdi‐ko cells and tested their ability to recruit Larp to mitochondria (Fig 3A and F). The same set of MDI mutants were also expressed in the mdi 1 background to test whether they could rescue the fertility of female mdi 1 flies. MDI mutants that failed to recruit Larp to mitochondria also failed to restore the fertility of female mdi 1 flies (Fig 3A and F). By contrast, the MDI mutants that were able to recruit Larp to mitochondria significantly restored the fertility of female mdi 1 flies and the amount of mtDNA in their eggs (Fig 3A and F). Taken together, these results demonstrate that MDI promotes mtDNA replication in ovaries by recruiting Larp to mitochondria.
Cytosolic ribosomes synthesize proteins on the mitochondrial surface
Many mRNAs encoded by the nuclear genome are associated with the mitochondrial outer membrane in yeast and animal cells (Sylvestre et al, 2003; Fox, 2012). This localization is thought to facilitate the import of their protein products into mitochondria. Interestingly, Larp interacts with PABP and eIF4G (Tcherkezian et al, 2014), through which it may stabilize mRNAs and stimulate protein synthesis. Given the interaction between Larp and MDI and their mitochondrial localization, we hypothesized that MDI‐Larp might promote protein synthesis on the mitochondrial surface in the ovary. To our knowledge, protein synthesis on the mitochondrial surface has not been demonstrated previously.
We thus visualized nascent protein synthesis by incorporation of a methionine analog homopropargylglycine (HpG) and subsequent fluorescence‐click chemistry (Dieterich et al, 2010). After incubating dissected ovaries with HpG for 30 min, we observed strong HpG labeling in germaria and egg chambers of wild‐type ovaries (Fig EV4A). Both ER and mitochondria are tightly associated with the fusome in the late germarium stages (Cox & Spradling, 2003; Snapp et al, 2004), which makes it difficult to clearly distinguish ER labeling from mitochondrial labeling. By contrast, mitochondria and ER are distinct from each other except for a few contacting sites, in the mid‐stage egg chambers (Fig 5B). We thus focused on the mid‐stage egg chambers to assess the potential association of protein synthesis with mitochondria.
Figure EV4. Nascent protein synthesis in ovary visualized by HpG incorporation.
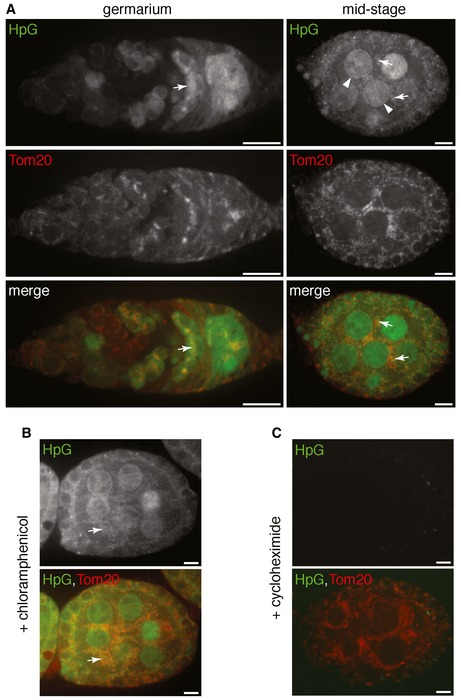
- Representative images of ovarioles expressing Tom20‐mCherry that were incubated with a methionine analog, L‐homopropargylglycine (HpG) for a pulse of 30 min to label the nascent protein synthesis. HpG was visualized with Alexa Fluor 488 through click‐it chemistry. Alexa Fluor 488 signal of the nascent protein synthesis was sensitive to detergent wash used in immunostaining assay. Thus, Tom20‐mCherry was used to mark mitochondria.
- Representative image of HpG incorporation in ovarioles in the presence of chloramphenicol that inhibits mitochondrial ribosomes specifically. Chloramphenicol has no impact on the HpG incorporation in mid‐stage egg chamber.
- Representative image of HpG incorporation in ovarioles in the presence of cycloheximide that inhibits cytosolic ribosomes. Note that cycloheximide greatly reduces HpG signal.
Data information: Arrows point to the HpG signal associated with mitochondria. Arrowheads point to the HpG signal at perinuclear region. Scale bars in (A–C), 10 μm.
Figure 5. Protein synthesis on the mitochondrial surface.
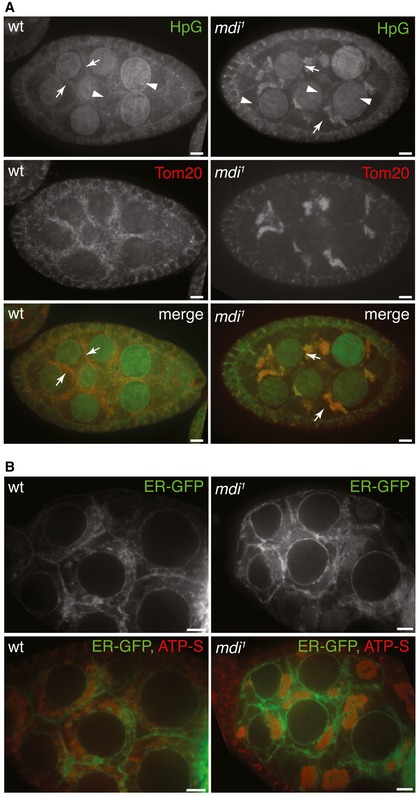
- Nascent protein synthesis revealed by HpG incorporation (green) in wt and mdi 1 egg chambers. Arrowheads point to the HpG signal on the ER in the perinuclear region and cell periphery. Arrows point to the HpG signal associated with mitochondria. Mitochondria are marked by a Tom20‐mCherry (red) created by inserting mCherry at the endogenous Tom20 locus.
- ER location is unaltered in mdi 1 egg chambers. An ER marker (ER‐GFP) was expressed in wt and mdi 1 egg chambers that were co‐stained with ATP‐S to mark mitochondria. ER localizes to the perinuclear region, cytoplasm, and cell periphery in both wt and mdi 1 egg chambers.
In mid‐stage egg chambers, strong HpG signal was detected in the perinuclear region where rough ER, the main site of cytoplasmic protein synthesis, is located (Figs 5A and EV4A). There were also many HpG puncta closely associated with the mitochondrial marker Tom20‐mCherry in the cytoplasm (Figs 5A and EV4A). HpG labeling associated with mitochondria could have resulted from the translation of mtDNA‐encoded mRNAs inside the matrix or the translation of cytoplasmic RNAs at the mitochondrial surface. To distinguish these two possibilities, we treated the ovaries with two ribosomal inhibitors, chloramphenicol, or cycloheximide. Chloramphenicol inhibits mitochondrial ribosomes, but not cytosolic ribosomes (Millis & Suyama, 1972). Chloramphenicol treatment had little effect on HpG labeling in mid‐stage egg chambers (Fig EV4B). In contrast, cycloheximide, which inhibits cytoplasmic ribosomes (Millis & Suyama, 1972), greatly abolished HpG labeling (Fig EV4C). These results demonstrate that the HpG puncta associated with mitochondria are mainly derived from proteins synthesized by cytosolic ribosomes on the mitochondrial surface.
MDI promotes the synthesis of specific proteins on the mitochondrial surface
mdi 1 egg chambers also displayed strong HpG labeling in the perinuclear region and on many cytoplasmic clumps that perfectly overlapped with Tom20‐mCherry (Fig 5A). Meanwhile, the ER structure appeared normal in mdi 1 ovary (Fig 5B). These results further prove that protein synthesis by cytosolic ribosomes occurs on the mitochondrial surface. They also demonstrate that protein synthesis at the mitochondrial surface is still present in mdi mutants.
Since mitochondria clump together in mdi 1 ovaries (Figs 5A and EV2), it is difficult to normalize and quantify the level of HpG incorporation from fluorescence microscopy images. We thus used Western blotting to achieve some quantification of de novo protein synthesis. We incubated the isolated ovaries for 4 h with another methionine analog, L‐azidohomoalanine (AHA), which was subsequently labeled with biotin by click chemistry. The newly synthesized proteins can then be probed with an anti‐biotin antibody on the blot. We first confirmed that most biotin‐reactive bands on the blot were indeed proteins newly synthesized by cytosolic ribosomes, as co‐incubation with cycloheximide greatly blocked the AHA incorporation (Fig EV5A). We next separated the mitochondrial fraction from cytosolic fraction by differential centrifugation. The AHA signal in the cytosolic fractions was comparable between wild‐type and mdi mutant extracts (Fig EV5B), suggesting that the overall protein synthesis by cytoplasmic ribosomes was not affected in mdi 1 ovary. However, the AHA signal associated with the mitochondria fraction was reduced in mdi 1 ovary (Fig 6A), even though not all bands were affected. This observation suggests that MDI promotes the de novo synthesis of a subset of proteins.
Figure EV5. Western blot analyses of nascent protein synthesis and steady‐state protein levels in ovaries.
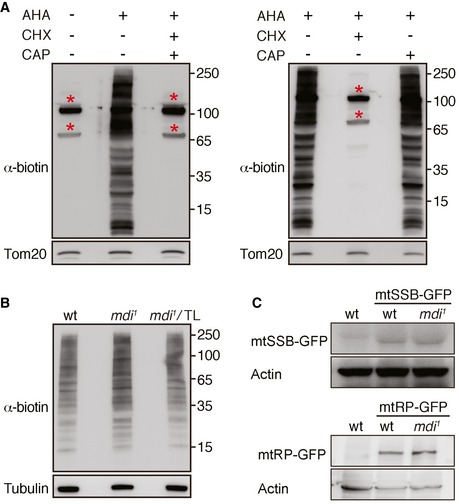
- Western blot analyses of nascent protein synthesis in the mitochondrial fraction of the ovary. The nascent protein synthesis was labeled by AHA incorporation and detected by anti‐biotin antibody. Tom20 was used as a loading control. There were two strong bands (*) in the mitochondria fraction without the AHA incubation, indicating two endogenously biotinylated mitochondrial proteins. The AHA signal was mostly blocked in the presence of a cytosolic translation inhibitor, cycloheximide (CHX). Whereas chloramphenicol (CAP) has no impact on the HpG incorporation, indicating the nascent protein synthesis is mainly derived from cytosolic ribosomes associated with mitochondria.
- Western blot analyses of nascent protein synthesis in the cytosolic fraction of wt, mdi 1, and mdi 1 expressing Tom20‐Larp (mdi 1/TL) ovary. Tubulin was used as a loading control. The overall AHA signals indicating the nascent protein synthesis in the three genotypes are comparable.
- Western blot analyses of mtSSB‐GFP and mtRNApol‐GFP (mtRP‐GFP) in wt and mdi 1 ovary. Actin was used as a loading control.
Figure 6. MDI‐Larp complex promotes the de novo synthesis or import of a subset of nuclear‐encoded mitochondrial proteins.
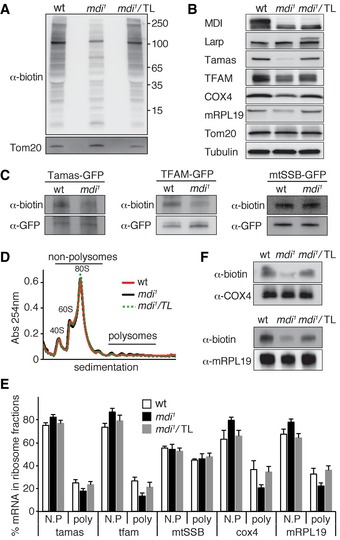
- Detection of nascent protein synthesis in the mitochondrial fraction of the ovary. Nascent protein synthesis was monitored by AHA incorporation and detected by anti‐biotin antibody. Tom20 served as a loading control. Note that the synthesis of nuclear‐encoded mitochondrial proteins was decreased in the mdi 1 background and was restored by overexpressing Tom20‐Larp (mdi 1/TL).
- Western blots of mitochondrial proteins in ovarioles of wt, mdi 1, and mdi 1 flies expressing Tom20‐Larp (mdi 1/TL) fusion protein. Note that levels of Tamas, TFAM, mRpL19, and COX4 were reduced in mdi 1 flies, but restored in mdi 1/TL flies.
- Nascent protein synthesis of Tamas and TFAM was decreased in mdi 1 ovary, whereas mtSSB was not affected. Tfamgfp, Tamasgfp, and mtSSBgfp were expressed in wt or mdi 1 background. Nascent proteins were labeled by AHA incorporation, and then, the GFP‐tagged protein was immunopurified with a GFP antibody and the nascent protein synthesis was detected by anti‐biotin antibody.
- Representative profile of 254 nm absorbance of wt, mdi 1, and mdi 1/TL ovary extracts.
- Polysome mRNA profiling for tamas, tfam, mtSSB, cox4, and mRPL19 in wt, mdi 1, and mdi 1/TL ovary. The percentage of mRNA for each gene in non‐polysomal fractions (N.P, including ribosomal subunits and monosome‐associated) and polysomal fractions (poly) was calculated and plotted. The fractions of tamas, tfam, cox4, and mRPL19 mRNAs in the polysomal fractions were significantly decreased in mdi 1 compared to wt, but were restored in mdi 1/TL flies. N = 4 for all samples. P‐values of comparing wt to mdi 1: tamas, P = 0.0055; tfam, P = 0.001; cox4, P = 0.0066; mRPL19, P = 0.0097. P‐values of comparing mdi 1/TL to mdi 1: tamas, P = 0.0206; tfam, P = 0.0346; cox4, P = 0.0036; mRPL19, P = 0.0016.
- Nascent protein synthesis of COX4 and mRPL19 was decreased in mdi 1 ovary, but restored by overexpressing Tom20‐Larp (mdi 1/TL). The proteins were immunopurified with antibodies against the endogenous proteins.
Given MDI's impact on mtDNA replication, we expected that the proteins affected in mdi mutant would be involved in mtDNA replication. We checked the steady‐state protein levels of mitochondrial DNA polymerase (Tamas) and TFAM on Western blots probed with antibodies against endogenous proteins (Matsuda et al, 2013; Zhang et al, 2015). For two other replication factors, mtSSB and mitochondrial RNA polymerase (mtRNAP), against which there are no effective antibodies, we generated GFP fusion proteins by inserting GFP reporter into the genomic loci. We then crossed these transgenes into mdi mutant background and probed mtSSB‐GFP and mtRNAP‐GFP using a GFP antibody. We found that the amounts of both TFAM and Tamas were markedly reduced in mdi 1 compared to wild‐type ovaries (Fig 6B), while those of mtSSB‐GFP and mtRNAP‐GFP remained unchanged (Fig EV5B).
To test whether the reduced steady‐state protein levels are truly caused by reduced protein synthesis, we performed polysome profiling on several candidates. We prepared ribosomal fractions by gradient sedimentation of ovary extracts and quantified the relative abundance of mRNA in each ribosomal fraction by real‐time PCR analyses. Overall, only a small fraction of ribosomes were assembled into polysome complexes (Fig 6D). This is consistent with a previous work showing a lack of protein translation in mature oocytes (Kronja et al, 2014). Additionally, the ribosome peaks in wt and mdi 1 ovaries overlapped almost perfectly (Fig 6D), suggesting that overall translational activity is not impaired in mdi 1. Of primary significance, the levels of tfam and tamas mRNAs in polysome fractions were significantly reduced in mdi 1 compared to wild‐type extracts (Fig 6E), suggesting a reduced translation of these specific mRNAs in mdi 1 ovary.
To further confirm that de novo synthesis of these proteins was reduced, we directly assessed the AHA incorporation after metabolic labeling. Because the endogenous antibodies of TFAM and Tamas did not work effectively for immunopurification, we generated BAC clone transgenes expressing TFAM‐GFP and Tamas‐GFP in both wild type and mdi mutant. We purified these GFP fusion proteins using a GFP antibody from AHA‐labeled ovaries and probed with an anti‐biotin antibody to visualize the newly synthesized proteins. We found that AHA labeling on both Tamas and TFAM was markedly reduced in mdi mutant (Fig 6C), which reflects the reduced synthesis and/or import of these two proteins.
The MDI‐Larp complex promotes mitochondrial biogenesis
To systematically identify the targets of the MDI‐Larp complex, we compared the proteomes of mature eggs produced by wild‐type and mdi 1 mothers using quantitative mass‐spectrum analysis. Of 2,182 proteins detected in both extracts, 406 were nuclear‐encoded mitochondrial proteins (Tables EV2, EV3 and EV4). There were a total of 65 proteins reduced more than twofold in mdi 1 compared to wild‐type eggs. Among these, 64 proteins were nuclear‐encoded mitochondrial proteins including 21 mitochondrial ribosomal proteins and 23 electron‐transport chain (ETC) subunits (Table EV3). To validate the proteomics results, we probed two candidates, a mitochondrial ribosomal protein, mRPL19, and cytochrome C oxidase subunit 4 (COX4), by Western blot. Indeed, the abundance of both proteins was decreased in mdi 1 compared to wild‐type eggs (Fig 6B).
mtDNA encodes the 16S and 12S rRNAs of the mitochondrial ribosomes and 13 core components of the ETC complexes. Given the reduced amount of mtDNA in mdi 1 ovary and embryo, the reduced steady‐state amounts of nuclear‐encoded mitochondrial ribosomal proteins and ETC subunits could result from a lack of mtDNA‐encoded partners to assemble full complexes, or reduced de novo synthesis. To distinguish between these possibilities, we performed the same ribosome profiling on these two genes as described above. We found that cox4 and mRPL19 mRNAs were less abundant in the polysome fractions of mdi 1 ovaries than of wild‐type ovaries (Fig 6E). Furthermore, AHA labeling confirmed that newly synthesized COX4 and mRPL19 proteins were both markedly reduced in mutant ovaries (Fig 6F).
We next examined whether Larp mediated MDI's role in mitochondrial protein synthesis. A Tom20‐Larp fusion protein that constitutively targeted Larp to the mitochondria outer membrane independently of MDI (Figs 7A and EV3B) restored the steady‐state levels and the de novo protein synthesis of Tamas, TFAM, mRPL19, and COX4 (Figs 6B, C and F) in mdi 1 ovary. Tom20‐Larp also rescued the mtDNA level and the hatching rate of mdi 1 eggs (Fig 7B). These results suggest that Larp mediates most, and perhaps all, of MDI's roles in mtDNA replication and mitochondrial biogenesis in the ovary.
Figure 7. Targeting Larp to the mitochondrial surface partially rescues the mdi 1 phenotype.
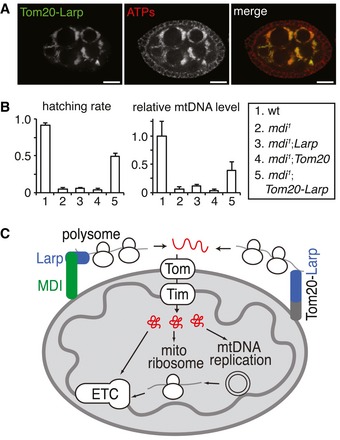
- Mitochondrial localization of a Tom20‐LarpGFP (Tom20‐Larp) fusion protein in an mdi 1 egg chamber stained with ATP‐S to reveal mitochondria. Scale bars, 20 μm.
- Hatching rates and mtDNA contents of eggs produced by flies with different genotypes relative to wt control; each data point represents the mean of three independent replicates. Error bars represent SD. Expression of Tom20‐Larp in mdi 1 (mdi 1 ; Tom20‐Larp) significantly restored the mtDNA level and hatching rate. N = 3 × >100 eggs/genotype for hatching rate. The relative mtDNA level was the average of three biological repeats. P‐values of comparing mdi 1 ; Tom20‐Larp to mdi 1: for mtDNA level, P = 0.0209; for hatching rate, P = 3.4758E−05.
- Model of the role of MDI and Larp in mitochondrial biogenesis. MDI (green rectangle) localizes at the mitochondrial outer membrane and recruits Larp (blue rectangle) to the mitochondrial surface. Larp interacts with polysome (blobs) and translation stimulators and stimulates the translation of a subset of nuclear‐encoded mRNAs. Proteins synthesized on the mitochondrial surface are in close vicinity of the Tom–Tim mitochondrial transporter complexes, which would facilitate their rapid transport into the matrix. The targets of MDI‐Larp complexes include most nuclear‐encoded ETC subunits, mitochondrial ribosomal proteins, TFAM, and mtDNA polymerase (Tamas) (collectively represented in red). Mitochondrial ribosomes are required for the biogenesis of proteins encoded by mtDNA, all of which are ETC subunits. Thus, MDI‐Larp complex seem to coordinate the expression of both nuclear and mitochondrial genome to promote the biogenesis of ETC complexes.
Discussion
Here, we demonstrate that MDI‐Larp complex on mitochondrial surface promotes the translation of a subset of nuclear‐encoded mitochondrial proteins and is required for mtDNA replication and mitochondrial biogenesis in Drosophila ovaries. Our evidence that MDI and Larp work as a complex is multifold: They have similar loss‐of‐function phenotypes, they interact physically, and mitochondrially targeted Larp overexpression can rescue most of the mdi mutant phenotypes that we tested. However, the mechanisms by which they promote the translation of a subset of mitochondrial proteins remain puzzling. Although MDI contains an RNA‐binding KH domain, it is dispensable for MDI's function and thus unlikely to contribute to the specificity of MDI‐Larp complex (Fig 3A). The Larp homolog in yeast binds to a subset of cytosolic mRNAs including many nuclear‐encoded mitochondrial proteins (Schenk et al, 2012; Kershaw et al, 2015). This suggests that Larp might dictate the specificity of the MDI‐Larp complex by binding to a subset of mRNAs and promoting their translation. We also note that the list of MDI‐Larp's targets derived from our proteomic analyses appears incomplete. Many mitochondrial proteins, including most mtDNA replication factors, were not recovered, presumably because their low abundance makes them difficult to detect in our proteomics analyses. Nonetheless, our work demonstrates that the MDI‐Larp complex promotes the biogenesis of a subset of nuclear‐encoded mitochondria proteins including ETC subunits, mitochondrial ribosomal proteins, and mtDNA replication factors.
The mitochondrial ribosomes are responsible for synthesizing the ETC subunits encoded on the mtDNA. Thus, MDI‐Larp participates in ETC biogenesis by both promoting mtDNA replication and coordinating the expression of the nuclear‐ and mitochondria‐encoded components of the ETC complexes. In mdi 1 ovary, reduced biogenesis of ETC should render mitochondria less active, which, together with the reduced supply of mitochondrial DNA replication factors, would impair mtDNA replication (Fig 7C). We recognize that the full spectrum of MDI‐Larp's impact on mitochondrial biogenesis remains to be explored. Nonetheless, the fact that a mitochondrially targeted Larp fusion protein (Tom20‐Larp) can restore local protein synthesis and mtDNA levels in mdi 1 ovaries suggests that local protein synthesis on the mitochondrial surface is likely essential to drive mitochondrial DNA replication and biogenesis during oogenesis.
Our data suggest that post‐transcriptional regulation of gene expression is the major driver of massive mitochondrial biogenesis in the late stages of oogenesis.
Massive mtDNA replication demands vast amounts of replication factors (Moraes, 2001). In fact, several of these factors were found to be more abundant in the female germ line than in somatic tissues (Fig EV3C). However, the mRNA levels of mtDNA replication factor are either unchanged, or only slightly increased in the Drosophila ovary relative to somatic tissues (Chintapalli et al, 2007). Thus, the boost in mitochondrial biogenesis in the ovary must rely primarily on post‐transcriptional mechanisms. Moreover, this post‐transcriptional regulation appears localized to the mitochondrial surface. Localized protein synthesis within a cell has been demonstrated in various biological systems (reviewed in Lesnik et al, 2015). The local translation of specific mRNAs may help to compartmentalize proteins at their active sites (Besse & Ephrussi, 2008), provide fast responses to local needs, and enable protein synthesis under repressive conditions (reviewed in Lesnik et al, 2015). In Drosophila oogenesis, translational control is prevalent during oogenesis and confines the temporal and spatial pattern of various developmental factors (reviewed in Lasko, 2012). Several key factors of germ cell development, including Nanos, are translational inhibitors. Interestingly, there is massive mitochondrial proliferation in a nanos mutant ovary (Bhat, 1999), implying an inhibitory role of Nanos on mitochondrial biogenesis. It is possible that local protein synthesis on mitochondrial surface mediated by MDI‐Larp might relieve the translational inhibition by Nanos.
Protein synthesis on the mitochondrial outer membrane has been proposed as an effective way to couple translation and import of nuclear‐encoded mitochondrial proteins (reviewed in Fox, 2012). Such coupling might be particularly crucial for the rapid biogenesis of mitochondria during oogenesis and might be another important function of the MDI‐Larp complex. Many mRNAs encoding mitochondrial proteins are localized to the mitochondrial surface (Marc et al, 2002; Sylvestre et al, 2003), and cytoplasmic ribosomes have been known to associate with mitochondria for a long time (Kellems et al, 1974). Recent work on proximity‐specific ribosome profiling identified over 100 mitochondrial proteins that are translated at the vicinity of the outer mitochondrial membrane (Williams et al, 2014). In animal cells, mRNAs encoding subunits of the ETC are recruited to the mitochondrial surface through a PINK1/Parkin‐regulated process (Gehrke et al, 2015). PINK1 is also suggested to promote the translation of these mRNAs. Despite increased evidences demonstrating the association of mRNAs with mitochondria and its proposed implication in co‐translational import, direct evidence for protein synthesis at the mitochondrial surface has so far been lacking. Moreover, the physiological significance of coupled translation–import has yet to be explored. Our experiments applying metabolic labeling to visualize protein synthesis demonstrate that protein synthesis does in fact take place on the mitochondrial surface. The locally synthesized proteins including mtDNA replication factors would be perfectly poised for efficient translocation into the mitochondria to drive massive mitochondrial biogenesis (Fig 7C).
Aside from its association with Larp, MDI appears to interact with other proteins that may mediate other functions. For instance, we found that MDI associates with many proteins involved in ribosomal biogenesis or translational control (Table EV1). MDI appears to be a scaffold protein that recruits ribosomes and translation regulators to the mitochondrial surface. We found that Bor, the Drosophila ATAD3 protein, also co‐purified with MDI (Table EV1). ATAD3, a nucleoid protein, is thought to tether mtDNA to cholesterol‐rich membrane structures at the ER–mitochondria contacting sites in mammalian cells (Gerhold et al, 2015). Interestingly, two outer membrane proteins, Mmm1‐p and Mmm2‐p, are required for mtDNA maintenance in yeast (Hobbs et al, 2001; Youngman et al, 2004). Mmm1‐p associates with the nucleoid protein Mgm101P and forms two‐membrane spanning structure where actively replicating nucleoids localize (Meeusen & Nunnari, 2003). It is an intriguing idea that MDI may also complex with ATAD3 or other unidentified nucleoid proteins and interact with mtDNA directly. However, loss of MDI impairs mtDNA replication in ovary specifically, and this impairment can be rescued by mitochondrially targeted Larp (Tom20‐Larp). Thus, the physiological significance of the potential association between MDI and nucleoid proteins remains to be explored, even if the potential association was confirmed.
MDI belongs to a family of AKAP proteins that are highly conserved among metazoans (reviewed in Wong & Scott, 2004). Some AKAPs function as scaffold to tether PKA and its downstream effectors on the mitochondrial outer membrane, regulating diverse mitochondrial processes including mitochondrial protein import (Schmidt et al, 2011), mitochondrial fission, and apoptosis (Cribbs & Strack, 2007). However, PKA is not required for mtDNA replication in the Drosophila ovary, arguing against the idea that MDI regulates mtDNA replication via a cAMP‐PKA pathway. AKAP1, the mammalian homolog of MDI, also localizes to the mitochondria. Interestingly, AKAP1 knockout mice are female semi‐sterile (Newhall et al, 2006), which is similar to the phenotype of mdi mutant flies. Interaction between PKA and AKAP1 is essential for maintaining meiotic arrest of developing oocytes. However, whether AKAP1 is required for mtDNA replication and mitochondrial biogenesis in mammals has not been examined. In the final stages of mammalian oogenesis prior to fertilization, mtDNA undergoes a burst of replication that significantly increases mtDNA copy number (reviewed in St John, 2012). This massive replication would demand a large amount of mtDNA replication factors, which, in mammals like in flies, are encoded in the nucleus. Interestingly, ectopic expression of human AKAP1 rescues the fertility of mdi 1 female flies (Fig 2F). Thus, the post‐transcriptional regulation on mitochondrial biogenesis by MDI/AKAP1 likely represents an evolutionarily conserved mechanism that may be crucial for mitochondria inheritance across species, even though the details of oogenesis differ between mammals and flies.
Materials and Methods
Fly genetics and husbandry
All flies were maintained on cornmeal medium at 25°C. w 1118 was used as the wild‐type control. MDI‐GFP and Tom20‐mCherry were constructed by inserting GFP or mCherry cDNA into endogenous loci by CRISPR/Cas9‐mediated recombination. Tfam‐gfp, tamas‐gfp, mtSSB‐gfp, and mtRNApol‐gfp reporter lines were generated by integrating BAC clones carrying gfp cDNA to landing sites of VK37(2L)22A3 (tfam and mtSSB) or VK31(3L)62E1(tamas and mtRNApol) using phi‐C31‐mediated transgenesis (Venken et al, 2009). The larp mtr‐null and PKA E95 were described previously (Blagden et al, 2009; Xia et al, 2012). Fecundity test and the embryo‐hatching test were carried out as previously described (Von Stetina et al, 2011; Chen & Wagner, 2012).
CRISPR/Cas9 in flies and cells
To generate mdi deletion, two chiRNA plasmids containing targeting sequences GAGGTAGAGTAGAGGACGAC and GCTAGTTGAGTTGTTCACTA were injected into PBac{y[+mDint2]=vas‐Cas9}VK00027 embryos. The genomic DNA of G1 adults flies was prepared and screened for deletion by PCR using oligos: AACGCATAACCCAGCTGATCCCTA; GCGAAGTTGTTGTGCCCTTATCTTAC.
To insert GFP or mCherry into the endogenous loci of mdi and Tom20, respectively, chiRNA plasmids containing targeting sequences GCTAGTTGAGTTGTTCACTA (mdi) or GTCCGGCTAGAACATGGCAT (Tom20) and a donor plasmid were injected into the embryos of PBac{y[+mDint2]=vas‐Cas9}VK00027 (mdi), or M{vas‐Cas9}ZH2A (Tom20). Donor plasmids contain a 1‐kb upstream and a 1‐kb downstream fragment flanking stop codons of target genes in POT2 vectors. The GFP or mCherry was inserted in front of stop codons. G1 adults were screened for insertion events by PCR using primers for mdi: GATTTCATGAATGTGCCCTTCCA, TTACTTGTACAGCTCGTCCATG; and Tom20: CTTATTGCCTCAGGCATCTA, TTACTTGTACAGCTCGTCCATG.
To generate mdi knockout cell line, S2 cells were transfected with an mdi chiRNA plasmid containing targeting sequence GAGGTAGAGTAGAGGACGAC, a Cas9 expression plasmid (Addgene #42230) and pCoBlast (Invitrogen). The cells were seeded to 96‐well plate 36 h after the transfection and selected with blasticidin (100 μg/ml) for 7 days. The knockout clones were screened using Western blot for the loss of MDI protein.
Molecular biology
BAC clones carrying gfp reporters in the genomic loci were constructed by recombineering (Venken et al, 2009). cDNAs or gene fragments of mdi, mdi truncations, hAKAP1, Tom20, and Tom20‐Larp were cloned into pA‐myc, pA‐GFP, pA‐mCherry, and pUASp‐GFP expression vectors using Drosophila Gateway Cloning system (T. Murphy laboratory, Carnegie Institute of Washington). Quantitative real‐time PCR analysis of mtDNA level was performed as previously described (Zhang et al, 2015).
Immunohistochemistry
EdU incorporation was preformed as previously described (Hill et al, 2014). For detecting nascent protein synthesis, ovaries (5–10 pairs) from 4‐ to 5‐day‐old female flies were dissected in methionine‐free media (MFM) and washed three times with MFM. The ovaries were equilibrated in MFM for 45 min and then incubated with or without 20 μM of chloramphenicol or cycloheximide in MFM for 30 min. The ovaries were then incubated in MFM containing 50 μM HPG with or without 20 μM chloramphenicol or cycloheximide for 30 min. Click‐iT HpG labeling was performed according to the manufacturer's instructions (Life Technology). All images were collected on a Perkin Elmer Ultraview system and processed with Volocity software. Antibodies used in this study were as follows: mouse α‐ATP synthase subunit α (Abcam, 15H4C4, 1:1,000), rabbit α‐Larp (provided by David Glover, 1:500), Alexa Fluor 488 goat α‐rabbit IgG (Invitrogen, 1:200), Alexa Fluor 568 goat α‐mouse IgG (Invitrogen, 1:200).
Biochemistry
Primary antibodies used for Western blot in this study were as follows: α‐Cox4 (ab16056, Abcam), α‐β tubulin (E7, DHSB), α‐actin (C4, Millipore), α‐SOD2 (NB100‐1992, Novus Biologicals), α‐ATP synthase 5α (15H4C4, Abcam), α‐Tom20 (#13929, CST), α‐biotin (#7075, CST), α‐mRPL19 (PA5‐31240, Life Technologies), α‐cytochrome c (7H8.2C12, Novus Biologicals), α‐GFP (11814460001, Roche), α‐Tamas (Zhang et al, 2015), and α‐TFAM (Matsuda et al, 2013). A rabbit polyclonal α‐MDI antibody was raised against a GST‐tagged MDI truncation containing residues 100–585.
Mitochondrial isolation was performed as previously described (Zhang et al, 2015). For protease protection assay, intact mitochondria (200 μg of protein) were resuspended in 20 mM HEPES‐KOH, pH 7.4, 0.6 M sorbitol. Swollen mitochondria were prepared by incubating mitochondria (200 μg of protein) in 20 mM HEPES‐KOH, pH 7.4 for 10 min. Protease K (100 μg/ml) was added to mitochondria preparation and kept on ice for 20 min in protease K treatment experiments. The reaction was stopped by the addition of 2 mM phenylmethylsulfonyl fluoride. Mitochondria were collected by centrifugation at 16,000 g for 5 min at 4°C and followed by SDS–PAGE and Western blot analyses.
To identify MDI interacting proteins, cells expressing MDI‐myc were lysed in 150 mM NaCl, 1% Triton X‐100, 50 mM Tris–HCl (pH 8.0) and incubated on ice for 30 min with occasional mixing. Cell lysates were centrifuged for 10 min at 10,000 g at 4°C. Supernatants were collected and incubated with 50 μl α‐Tag MicroBeads (Miltenyi Biotec) for 2 h at 4°C. The beads were washed four times with 150 mM NaCl, 1% Igepal CA‐630, 0.5% sodium deoxycholate, 0.1% SDS, 50 mM Tris–HCl (pH 8.0), and one time with 20 mM Tris–HCl (pH 7.5). The proteins were eluted with 50 mM Tris–HCl (pH 6.8), 50 mM DTT, 1% SDS, 1 mM EDTA, 0.005% bromphenol blue, 10% glycerol and subjected to SDS–PAGE. Coomassie‐stained bands were excised from the gel, destained with acetonitrile, reduced with dithiothreitol, alkylated with iodoacetamide, and digested with trypsin overnight. For embryonic mass spectrometry, triplicates of wt and mutant embryo lysates were sequentially reduced, alkylated, digested overnight with trypsin, and labeled with 6‐plex Tandem Mass Tag (TMT) reagents (Thermo Fisher Scientific; Dayon et al, 2008). Six labeled protein digests were pooled and then separated into 12 fractions using high‐pH reverse‐phase liquid chromatography (Wang et al, 2011). All fractions and protein digests were analyzed using a nanoLCMS system equipped with an LTQ Orbitrap Elite mass spectrometer (Thermo Fisher Scientific). Peptide and protein IDs were assigned by searching LCMS raw data against Uniprot Drome database (http://www.uniprot.org) using Sequest HT algorithm on Proteome Discoverer 1.4 platform (Thermo Fisher Scientific). The results were compiled and quantitatively compared using Scaffold 4.0 software (Proteome Software, Inc, Portland OR). The relative protein abundance in corresponding bands from IP pull‐down samples was quantified using spectral counting method. TMT‐labeled samples were normalized using the total reported ion intensities of their corresponding channels, and then, individual proteins were compared using the normalized report ion intensities.
Polysomal profiling
The polysome profiling was performed as previously described (Baird et al, 2014). Briefly, sucrose gradients ranging from 10 to 50% in 10 mM Tris–HCl (pH 7.5), 75 mM KCl, 1.5 mM MgCl2, and 50 μg/ml cycloheximide were prepared with a tilted tube rotation method on a gradient station (BioComp). Fifty pairs of ovaries were dissected in PBS containing 100 μg/ml cycloheximide and then incubated with 100 μg/ml cycloheximide in PBS for 10 min on ice. The ovaries were homogenized in 10 mM Tris–HCl, pH 7.5, 75 mM KCl, 1.5 mM MgCl2, 1% Triton X‐100, 1% deoxycholate, 2% Tween‐20, 100 μg/ml cycloheximide, 1 mg/ml heparin, and 50 units/ml RNasin and incubated on ice for 10 min. The debris were removed by centrifugation at 12,000 g for 10 min at 4°C, and supernatants were loaded onto 10–50% sucrose gradients and subjected to centrifugation in a Beckman SW41Ti rotor at 200,000 g for 2 h at 4°C. Sucrose fractions and the resulting polysome profiles for each sample were then collected using a Piston Gradient Fractionator and a 254‐nm ultraviolet monitor with Data Quest software. Samples were then immediately mixed with 750 μl of TRIzol Reagent LS. About 5 ng/ml firefly luciferase control RNA (Promega) was added to each pooled sample before RNA isolation, allowing for normalizing the transcript of interest to an exogenous RNA control. RNA isolation and quantitative RT–PCR were performed as described previously (Zhang et al, 2015). Oligonucleotides used for qPCR: Cox4: GGGCGTTTCACTCCTCTTC, GTGCTCCTCATCGAAGGTAAC; mRPL19: TTGTGACCTTCTCCACCAAA, GGAATGATTGTCTTCCGGTT; Tfam: CTCCGAGAAGGAGGTCTACAT, GGATCATCTTCTCCTCCCAAAC; Tamas: CCCTGCTCCGTCAGTTTAAT, CTCCTCTCGCAATCGATACAC; mtSSB: TGCTACACACACCAACTACAA, CGCTGTCCCTTCTTCAAGTAT; firefly luciferase: ATCCGGAAGCGACCAACGCC, GTCGGGAAGACCTGCCACGC.
Detection of nascent protein synthesis by Western blot
AHA labeling of nascent protein synthesis in ovaries was carried out using the same protocol as HpG labeling described above, except that HpG was replaced with AHA in the media and incubated for 4 h. After labeling, ovaries were homogenized in 20 mM HEPES‐KOH, pH 7.4, 0.25 M sucrose, and then centrifuged at 150 g for 10 min at 4°C to remove tissue debris. Supernatants were centrifuged at 9,000 g for 15 min at 4°C to separate mitochondrial pellets from soluble cytosolic fractions. The mitochondrial fraction was resuspended in 1% SDS in 50 mM Tris–HCl (pH 8.0). The cytosolic fraction was first precipitated by methanol and then solubilized in 1% SDS, 50 mM Tris–HCl (pH 8.0). To pull down a specific protein, the ovaries were homogenized in 150 mM NaCl, 1% Triton X‐100, 50 mM Tris–HCl (pH 8.0), incubated with a specific antibody conjugated with magnetic beads for 2 h, and then washed with 150 mM NaCl, 1% Igepal CA‐630, 0.5% sodium deoxycholate, 0.1% SDS, 50 mM Tris–HCl (pH 8.0). The proteins were eluted with 1% SDS in 50 mM Tris–HCl (pH 8.0). AHA was labeled with biotin according to the manufacturer's instructions (Life Technology) and probed with α–biotin (#7075, CST).
Statistical analysis
Error bars represent standard deviations in all the charts. Data were analyzed using two‐tailed Student's t‐test. The difference was considered statistically significant when P < 0.05.
Author contributions
HX and YZ conceived the idea and designed the research; YZ and YC performed the experiments; YZ, YC, MG, and HX analyzed data; HX and YZ wrote the manuscript.
Conflict of interest
The authors declare that they have no conflict of interest.
Supporting information
Expanded View Figures PDF
Table EV1
Table EV2
Table EV3
Table EV4
Review Process File
Acknowledgements
We thank F Chanut and T Finkel for their comments and editing on the manuscript, R Balaban for the discussion on the work, Bloomington Drosophila Stock Center for various fly lines, B Tom and R Hogg for technical assistance on the polysomal analyses, E Matsuura for Drosophila TFAM antibody, D Glover for larp mutant and antibody, J Jia for PKA mutant fly, Developmental Hybridoma Bank for various antibodies, and Bestgene Inc. and Genetivision Inc. for the Drosophila embryo injection service. This work is supported by NHLBI Intramural Research Program.
The EMBO Journal (2016) 35: 1045–1057
References
- Baird TD, Palam LR, Fusakio ME, Willy JA, Davis CM, McClintick JN, Anthony TG, Wek RC (2014) Selective mRNA translation during eIF2 phosphorylation induces expression of IBTKα. Mol Biol Cell 25: 1686–1697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besse F, Ephrussi A (2008) Translational control of localized mRNAs: restricting protein synthesis in space and time. Nat Rev Mol Cell Biol 9: 971–980 [DOI] [PubMed] [Google Scholar]
- Bhat KM (1999) The posterior determinant gene nanos is required for the maintenance of the adult germline stem cells during Drosophila oogenesis. Genetics 151: 1479–1492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blagden SP, Gatt MK, Archambault V, Lada K, Ichihara K, Lilley KS, Inoue YH, Glover DM (2009) Drosophila Larp associates with poly(A)‐binding protein and is required for male fertility and syncytial embryo development. Dev Biol 334: 186–197 [DOI] [PubMed] [Google Scholar]
- Chen B, Wagner A (2012) Hsp90 is important for fecundity, longevity, and buffering of cryptic deleterious variation in wild fly populations. BMC Evol Biol 12: 25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z, Qi Y, French S, Zhang G, Covian Garcia R, Balaban R, Xu H (2015) Genetic mosaic analysis of a deleterious mitochondrial DNA mutation in Drosophila reveals novel aspects of mitochondrial regulation and function. Mol Biol Cell 26: 674–684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chintapalli VR, Wang J, Dow JAT (2007) Using FlyAtlas to identify better Drosophila melanogaster models of human disease. Nat Genet 39: 715–720 [DOI] [PubMed] [Google Scholar]
- Cox RT, Spradling AC (2003) A Balbiani body and the fusome mediate mitochondrial inheritance during Drosophila oogenesis. Development 130: 1579–1590 [DOI] [PubMed] [Google Scholar]
- Cribbs JT, Strack S (2007) Reversible phosphorylation of Drp1 by cyclic AMP‐dependent protein kinase and calcineurin regulates mitochondrial fission and cell death. EMBO Rep 8: 939–944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dayon L, Hainard A, Licker V, Turck N, Kuhn K, Hochstrasser DF, Burkhard PR, Sanchez J‐C (2008) Relative quantification of proteins in human cerebrospinal fluids by MS/MS using 6‐plex isobaric tags. Anal Chem 80: 2921–2931 [DOI] [PubMed] [Google Scholar]
- Dieterich DC, Hodas JJL, Gouzer G, Shadrin IY, Ngo JT, Triller A, Tirrell DA, Schuman EM (2010) In situ visualization and dynamics of newly synthesized proteins in rat hippocampal neurons. Nat Neurosci 13: 897–905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox TD (2012) Mitochondrial protein synthesis, import, and assembly. Genetics 192: 1203–1234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehrke S, Wu Z, Klinkenberg M, Sun Y, Auburger G, Guo S, Lu B (2015) PINK1 and Parkin control localized translation of respiratory chain component mrnas on mitochondria outer membrane. Cell Metab 21: 95–108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerhold JM, Cansiz‐Arda Ş, Lõhmus M, Engberg O, Reyes A, van Rennes H, Sanz A, Holt IJ, Cooper HM, Spelbrink JN (2015) Human mitochondrial DNA‐protein complexes attach to a cholesterol‐rich membrane structure. Sci Rep 5: 15292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gratz SJ, Cummings AM, Nguyen JN, Hamm DC, Donohue LK, Harrison MM, Wildonger J, O'Connor‐Giles KM (2013) Genome engineering of Drosophila with the CRISPR RNA‐guided Cas9 nuclease. Genetics 194: 1029–1035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guruharsha KG, Rual J‐F, Zhai B, Mintseris J, Vaidya P, Vaidya N, Beekman C, Wong C, Rhee DY, Cenaj O, McKillip E, Shah S, Stapleton M, Wan KH, Yu C, Parsa B, Carlson JW, Chen X, Kapadia B, VijayRaghavan K et al (2011) A protein complex network of Drosophila melanogaster. Cell 147: 690–703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadad M, Bresler‐Musikant T, Neuman‐Silberberg FS (2011) Drosophila spoonbill encodes a dual‐specificity A‐kinase anchor protein essential for oogenesis. Mech Dev 128: 471–482 [DOI] [PubMed] [Google Scholar]
- Handler D, Olivieri D, Novatchkova M, Gruber FS, Meixner K, Mechtler K, Stark A, Sachidanandam R, Brennecke J (2011) A systematic analysis of Drosophila TUDOR domain‐containing proteins identifies Vreteno and the Tdrd12 family as essential primary piRNA pathway factors. EMBO J 30: 3977–3993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill JH, Chen Z, Xu H (2014) Selective propagation of functional mitochondrial DNA during oogenesis restricts the transmission of a deleterious mitochondrial variant. Nat Genet 46: 389–392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobbs AEA, Srinivasan M, McCaffery JM, Jensen RE (2001) Mmm1p, a mitochondrial outer membrane protein, is connected to mitochondrial DNA (mtDNA) nucleoids and required for mtDNA stability. J Cell Biol 152: 401–410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honda S, Kirino Y, Maragkakis M, Alexiou P, Ohtaki A, Murali R, Mourelatos Z, Kirino Y (2013) Mitochondrial protein BmPAPI modulates the length of mature piRNAs. RNA 19: 1405–1418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichihara K, Shimizu H, Taguchi O, Yamaguchi M, Inoue YH (2007) A Drosophila orthologue of larp protein family is required for multiple processes in male meiosis. Cell Struct Funct 32: 89–100 [DOI] [PubMed] [Google Scholar]
- Kahvejian A, Svitkin YV, Sukarieh R, M'Boutchou M‐N, Sonenberg N (2005) Mammalian poly(A)‐binding protein is a eukaryotic translation initiation factor, which acts via multiple mechanisms. Genes Dev 19: 104–113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellems RE, Allison VF, Butow RA (1974) Cytoplasmic type 80 S ribosomes associated with yeast mitochondria. II. Evidence for the association of cytoplasmic ribosomes with the outer mitochondrial membrane in situ. J Biol Chem 249: 3297–3303 [PubMed] [Google Scholar]
- Kershaw CJ, Costello JL, Castelli LM, Talavera D, Rowe W, Sims PFG, Ashe MP, Hubbard SJ, Pavitt GD, Grant CM (2015) The yeast La related protein Slf1p is a key activator of translation during the oxidative stress response. PLoS Genet 11: e1004903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kronja I, Yuan B, Eichhorn SW, Dzeyk K, Krijgsveld J, Bartel DP, Orr‐Weaver TL (2014) Widespread changes in the posttranscriptional landscape at the Drosophila oocyte‐to‐embryo transition. Cell Rep 7: 1495–1508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane ME, Kalderon D (1995) Localization and functions of protein kinase A during Drosophila oogenesis. Mech Dev 49: 191–200 [DOI] [PubMed] [Google Scholar]
- Lasko P (2012) mRNA localization and translational control in Drosophila oogenesis. Cold Spring Harb Perspect Biol 4: a012294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lesnik C, Golani‐Armon A, Arava Y (2015) Localized translation near the mitochondrial outer membrane: an update. RNA Biol 12: 801–809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y, Lu Y‐S, Shuai Y, Feng C, Tully T, Xie Z, Zhong Y, Zhou H‐M (2007) The AKAP Yu is required for olfactory long‐term memory formation in Drosophila . Proc Natl Acad Sci USA 104: 13792–13797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marc P, Margeot A, Devaux F, Blugeon C, Corral‐Debrinski M, Jacq C (2002) Genome‐wide analysis of mRNAs targeted to yeast mitochondria. EMBO Rep 3: 159–164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda T, Kanki T, Tanimura T, Kang D, Matsuura ET (2013) Effects of overexpression of mitochondrial transcription factor A on lifespan and oxidative stress response in Drosophila melanogaster. Biochem Biophys Res Commun 430: 717–721 [DOI] [PubMed] [Google Scholar]
- Meeusen S, Nunnari J (2003) Evidence for a two membrane‐spanning autonomous mitochondrial DNA replisome. J Cell Biol 163: 503–510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millis AJT, Suyama Y (1972) Effects of chloramphenicol and cycloheximide on the biosynthesis of mitochondrial ribosomes in Tetrahymena. J Biol Chem 247: 4063–4073 [PubMed] [Google Scholar]
- Moraes CT (2001) What regulates mitochondrial DNA copy number in animal cells? Trends Genet 17: 199–205 [DOI] [PubMed] [Google Scholar]
- Newhall KJ, Criniti AR, Cheah CS, Smith KC, Kafer KE, Burkart AD, McKnight GS (2006) Dynamic anchoring of PKA is essential during oocyte maturation. Curr Biol 16: 321–327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubenstein JL, Brutlag D, Clayton DA (1977) The mitochondrial DNA of Drosophila melanogaster exists in two distinct and stable superhelical forms. Cell 12: 471–482 [DOI] [PubMed] [Google Scholar]
- Schenk L, Meinel DM, Strässer K, Gerber AP (2012) La‐motif‐dependent mRNA association with Slf1 promotes copper detoxification in yeast. RNA 18: 449–461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt O, Harbauer AB, Rao S, Eyrich B, Zahedi RP, Stojanovski D, Schönfisch B, Guiard B, Sickmann A, Pfanner N, Meisinger C (2011) Regulation of mitochondrial protein import by cytosolic kinases. Cell 144: 227–239 [DOI] [PubMed] [Google Scholar]
- Snapp EL, Iida T, Frescas D, Lippincott‐Schwartz J, Lilly MA (2004) The fusome mediates intercellular endoplasmic reticulum connectivity in Drosophila ovarian cysts. Mol Biol Cell 15: 4512–4521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonenberg N, Hinnebusch AG (2009) Regulation of translation initiation in eukaryotes: mechanisms and biological targets. Cell 136: 731–745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- St John JC (2012) Transmission, inheritance and replication of mitochondrial DNA in mammals: implications for reproductive processes and infertility. Cell Tissue Res 349: 795–808 [DOI] [PubMed] [Google Scholar]
- Stewart JB, Larsson N‐G (2014) Keeping mtDNA in shape between generations. PLoS Genet 10: e1004670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sylvestre J, Vialette S, Corral Debrinski M, Jacq C (2003) Long mRNAs coding for yeast mitochondrial proteins of prokaryotic origin preferentially localize to the vicinity of mitochondria. Genome Biol 4: R44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tcherkezian J, Cargnello M, Romeo Y, Huttlin EL, Lavoie G, Gygi SP, Roux PP (2014) Proteomic analysis of cap‐dependent translation identifies LARP1 as a key regulator of 5′TOP mRNA translation. Genes Dev 28: 357–371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venken KJT, Carlson JW, Schulze KL, Pan H, He Y, Spokony R, Wan KH, Koriabine M, de Jong PJ, White KP, Bellen HJ, Hoskins RA (2009) Versatile P(acman) BAC Libraries for Transgenesis Studies in Drosophila melanogaster. Nat Methods 6: 431–434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Von Stetina JR, Lafever KS, Rubin M, Drummond‐Barbosa D (2011) A Genetic Screen for Dominant Enhancers of the Cell‐Cycle Regulator α‐Endosulfine Identifies Matrimony as a Strong Functional Interactor in Drosophila . G3 (Bethesda) 1: 607–613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wai T, Teoli D, Shoubridge EA (2008) The mitochondrial DNA genetic bottleneck results from replication of a subpopulation of genomes. Nat Genet 40: 1484–1488 [DOI] [PubMed] [Google Scholar]
- Wai T, Ao A, Zhang X, Cyr D, Dufort D, Shoubridge EA (2010) The role of mitochondrial DNA copy number in mammalian fertility. Biol Reprod 83: 52–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace DC (2007) Why do we still have a maternally inherited mitochondrial DNA? Insights from evolutionary medicine. Annu Rev Biochem 76: 781–821 [DOI] [PubMed] [Google Scholar]
- Wang Y, Yang F, Gritsenko MA, Wang Y, Clauss T, Liu T, Shen Y, Monroe ME, Lopez‐Ferrer D, Reno T, Moore RJ, Klemke RL, Camp DG, Smith RD (2011) Reversed‐phase chromatography with multiple fraction concatenation strategy for proteome profiling of human MCF10A cells. Proteomics 11: 2019–2026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams CC, Jan CH, Weissman JS (2014) Targeting and plasticity of mitochondrial proteins revealed by proximity‐specific ribosome profiling. Science 346: 748–751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolff JN, Sutovsky P, Ballard JWO (2013) Mitochondrial DNA content of mature spermatozoa and oocytes in the genetic model Drosophila . Cell Tissue Res 353: 195–200 [DOI] [PubMed] [Google Scholar]
- Wong W, Scott JD (2004) AKAP signalling complexes: focal points in space and time. Nat Rev Mol Cell Biol 5: 959–970 [DOI] [PubMed] [Google Scholar]
- Xia R, Jia H, Fan J, Liu Y, Jia J (2012) USP8 promotes smoothened signaling by preventing its ubiquitination and changing its subcellular localization. PLoS Biol 10: e1001238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H, DeLuca SZ, O'Farrell PH (2008) Manipulating the metazoan mitochondrial genome with targeted restriction enzymes. Science 321: 575–577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youngman MJ, Hobbs AEA, Burgess SM, Srinivasan M, Jensen RE (2004) Mmm2p, a mitochondrial outer membrane protein required for yeast mitochondrial shape and maintenance of mtDNA nucleoids. J Cell Biol 164: 677–688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F, Qi Y, Zhou K, Zhang G, Linask K, Xu H (2015) The cAMP phosphodiesterase Prune localizes to the mitochondrial matrix and promotes mtDNA replication by stabilizing TFAM. EMBO Rep 16: 520–527 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Expanded View Figures PDF
Table EV1
Table EV2
Table EV3
Table EV4
Review Process File


