Abstract
Humans and animals are unable to synthesize essential amino acids such as branch chain amino acids methionine (Met), lysine (Lys) and tryptophan (Trp). Therefore, these amino acids need to be supplied through the diets. Several essential amino acids are deficient or completely lacking among crops used for human food and animal feed. For example, soybean is deficient in Met; Lys and Trp are lacking in maize. In this mini review, we will first summarize the roles of essential amino acids in animal nutrition. Next, we will address the question: “What are the amino acids deficient in various plants and their biosynthesis pathways?” And: “What approaches are being used to improve the availability of essential amino acids in plants?” The potential targets for metabolic engineering will also be discussed, including what has already been done and what remains to be tested.
Keywords: Protein nutritional quality, Essential amino acids, Lysine, Methionine, Tryptophan, Genetic engineering.
1. INTRODUCTION
One of the primary components of the living cells is protein, whose building blocks are amino acids. Animals, including humans, can only produce about half of the 20 common amino acids needed for life; the rest – known as essential amino acids – must be obtained via diet. Plants are the major source of dietary proteins for both humans and other animals. In plants, amino acids are synthesized from basic elements such as carbon and oxygen from the air, hydrogen from water and nitrogen from the soil. Recent estimates indicate that more than half of the world’s population suffer from diseases caused by inadequate supplies of minerals, vitamins and essential amino acids [1]. In developing countries, these problems are intensified because the food supplies mostly rely on a single crop, which often provides an imbalanced or inadequate nutritional profile [2]. Most plant proteins are incomplete sources of amino acids. Among essential amino acids, methionine (Met), lysine (Lys), and tryptophan (Trp) are present in limited quantity in a variety of plants, particularly in cereals and legumes, the major crops for food and feed. Increasing essential amino acid content in legumes and cereals is a key approach to fighting malnutrition in developing countries.
2. NUTRITIONAL ROLES OF ESSENTIAL AMINO ACIDS
S-ademosylmethionine (SAM), a substrate involved in epigenetics and in fatty acid oxidation, is made from Met. As a direct precursor of SAM, Met is known as an important methyl donor indirectly (via SAM) in human metabolism [3, 4]. In most mammalian cells, homocysteine accumulation causes serious injury to endothelial cells, leading to atherosclerosis [4, 5]. However, the accumulated homocysteine can be reduced by re-methylation into Met [6, 7]. Several diseases in mice caused by methionine deficiency have been reported [4, 8]. Met residues in proteins, which are the most susceptible to oxidation by oxidative stress, are considered natural antioxidants when coupled with methionine sulfoxide reductases [9]. In animals, including humans, lack of Met also leads to methylation-related disorders such as fatty liver, tumorigenesis, neurological disorders and atherosclerosis [10-13]. The limited availability of Met leads to DNA strand breakage and fragmentation, which may be significant to the carcinogenic process [14]. In livestock, Met deficiency limits animal products such as quality of wool in sheep, milk and milk-derived products [15].
Lys plays several important roles in defense mechanism of animals, including humans. Lys deficiency could decrease the defense ability of mammalian cells to viruses [16, 17]. Lys deficiency is also the major cause of the osteoporosis in humans [18]. Stress-induced anxiety in mice was reportedly caused by the lack of Lys in their daily diets [19, 20]. A diet deficient in Met and Lys intake can reduce biological value of plant-based nutrition to 50-70%, compared to a balanced diet with high abundance of essential amino acids [21]. In humans, defects of Lys metabolism may result in familial hyperlysinemia due to genetic disorder. LKR-SDH gene mutation also causes familial hyperlysinemia [17].
Unlike other amino acids, Trp acts as a precursor to several neurochemicals, such as serotonins and melatonine. Dietary requirements for Trp in animals, including humans, were described [22]. Trp deficiency in daily diet leads to several symptoms in animals. Stresses caused by the loss of sleep were reportedly caused by the lack of Trp in daily food [23]. Loss of Met, Lys, and Trp in diet caused several symptoms, include weight loss, decrease in muscle mass and stress caused by losing sleep. These things can happen in both humans and animals. The quality of animal feed is determined by nutritional value of feed proteins, which could enhance the content of essential amino acids. It is possible to contribute a balanced diet for animals and, therefore, offers good growth and reproduction.
3. AMINO ACIDS LACKING IN PLANTS PROTEINS
Grains and grain legumes represent key sources of proteins for human and animal consumption. Grain legumes, such as soybean and soy products, beans, lentils, ground nuts, peas and chickpeas, are staple foods in many parts of the world. The world’s total value of legume crops is estimated at US$200 billion annually. Akibode and Maredia reported that many of the poorest countries in the world derive 10-20% of their total dietary energy supply from legumes [24]. In addition, cereals supply 68% food of nutritional calories worldwide. However, legumes and cereals contain limited levels of several essential amino acids. For example, pulse storage proteins are rich in Lys but lack sulfur-containing amino acids, mainly Met. In contrast, cereal crops are almost devoid of Lys and Trp [21, 25-27].
Lys and Trp levels, according to amino acid analysis and experiment results, are lowest in maize (Zea mays), lower than the minimum requirements for monogastric animals including humans [28]. Alcohol-soluble protein fractions of zein (prolamine), the most abundant protein in maize endosperm (40-60%), contain high levels of glutamine, proline, leucine and alanine but are deficient in Lys and Trp. Maize grains contain between 8.5% to 13.6% in proteins (w:w), of which Lys is 2.5 - 3.6% and Trp is 0.37 - 0.67% [29]. In 2005, Anjum and co-workers suggested that Lys is the most limiting amino acid in whole wheat flour [30].
In legumes, Met is considered the first limiting amino acid. Even soybean protein, considered the best plant protein, is not a complete protein because of the low level of Met content [31]. Scherz et al. [32] reported that stable foods, such as legumes, cereals, and nuts, are significantly deficient in Lys, Trp and Met content relative to animal-derived proteins (Table 1). In addition, according to a report issued in 1985 by WHO expert consultants, cereals supplied only 1.5-4.5 mol% of Lys compared to the requirement of 5.5 mol%, whereas protein from legumes supplied only 1-2 mol% sulfur-amino acid (Met and Cys) compared to the 3.5 mol % recommended by the WHO [33]. Another major crop for protein is potato. Although protein from potato is seen as better than that of cereal and legume crops, it is still suboptimal for Lys and several essential amino acids [34].
Table (1).
Lys, Met and Trp content in the major protein sources worldwide.
| Food | Lys* | Met | Trp |
|---|---|---|---|
| Beans | 1870 | 260 | 230 |
| Peas | 610 | 100 | 100 |
| Soybean | 1900 | 580 | 450 |
| Maize | 290 | 190 | 70 |
| Barley | 380 | 180 | 150 |
| Rice | 290 | 170 | 90 |
| Wheat | 380 | 220 | 150 |
| Potatoes | 130 | 30 | 30 |
| Nuts | 750 | 330 | 450 |
| Bovine meat | 2310 | 650 | 290 |
| Pig meat | 2200 | 720 | 310 |
| Freshwater fish | 2020 | 700 | 260 |
| Marine fish, other | 2050 | 600 | 240 |
According to 32.
milligrams of limiting amino acid per 100 gram of food.
Deficiencies in several essential amino acids are partly due to the nature of their biosynthesis pathways and the long distance transportation from synthesis organs to storage organs. For examples, in plants, asparagine and glutamine were transported from leaves to developing seeds and were then converted to Lys. Another branch in the aspartate pathway can also synthesize Met [35]. In Lys biosynthesis pathway, dihydrodipicolinate synthase (DHDPS) plays an important role for Lys accumulation. The rate-limiting step in the Lys catabolism in vivo is mainly catalyzed by DHDPS, the first unique enzyme in the pathway [36]. Pyruvate and aspartate semi-aldehyde are catalyzed to 4-hydroxy-2,3,4,5-tetrahydrodipicolinate by DHDPS. Recombinant DHDPS from several plant species were characterized and the enzyme was found to be extremely sensitive to feedback inhibition by Lys even at very low levels [36, 37]. Seed-specific overexpression of feedback-insensitive bacterial DHDPS in various plant species was shown to increase accumulation of Lys in seeds [36, 38]. In addition, enhanced Lys levels were also linked to the knockout of Lys catabolic enzymes; lysine-ketoglutarate reductase (LKR) catalyzes the formation of saccharopine and saccharopine dehydrogenase (SDH) hydrolyzes saccharopine to glutamate and α-aminoadipic acid. These two enzymes are present in a single bifunctional polypeptide. The LKR-SDH knockdown Arabidopsis mutant was shown to increase Lys content in the seeds [38] and the combination of overexpressing a bacterial DHDPS and LKR-SDH knockdown resulted in 80-fold increases in free Lys in the seeds. In maize grains, opaque2 transcription factor indirectly regulates the LKR-SDH gene expression as well as some classes of zein-coding genes. In developing grains, the LKR-SDH biofunctional enzyme is localized in the outer endosperm layer and little was detected in the embryo. Hence, there are a low levels of Lys in the outer endosperm layers where Lys may be transported from the embryo to be degraded [39]. Thus, Lys metabolic regulatory in the seeds may be different among plant species [21].
Met is generally regulated by its synthesis and metabolism as well as a complex network of interactions with the Lys biosynthesis pathway, because they are both derived from the aspartate family pathway. As a result, the Arabidopsis mutant with increased Lys contains lower Met [38]. There is evidence that Met biosynthesis is regulated by the allosteric enzyme threonine synthase (TS) in competition with enzyme cystathionine synthase (CGS) for an important common substrate of the aspartate pathway, O-phosphohomoserine (OPH). OPH can be catalyzed to threonine by TS or cystathionine by CGS [35]. However, OPH is used by TS more than CGS due to higher TS activity in plants [36, 40]. In Arabidopsis, the TS level was found to be seven times greater than that of AtCGS, resulting in a four-fold threonine content compared to Met [40]. TS was higher in level and in activity measurements, indicating that its affinity for OPH is 250 to 500 times higher than that of CGS. This revealed that threonine is produced more than Met in the aspartate pathway [36, 40].
In the Trp biosynthesis pathway, Anthranilate synthase (AS) is an enzyme catalyzing the conversion of chorismate to anthranilate. It is the initial and important rate-limiting step. The enzymatic activity of AS is regulated by feedback inhibition by Trp [41]. Bypassing feedback inhibition was demonstrated to be feasible to increase dietary Trp in the seed of rice [42], in tobacco leaves [43] and in the roots of the forage legume Astragalus sinicus [44]. In addition, it was also found that the sufficiency of AS mRNAs tightly controls Trp accumulation in plants. AS mRNAs was induced under biotic and abiotic stress such as wounding, pathogen or fungal infection. Both AS enzymatic activity and protein level can influence Trp biosynthetic capacity [42].
4. BIOSYNTHESIS PATHWAYS OF ESSENTIAL AMINO ACIDS IN PLANTS AND MICRO-ORGANISMS
4.1. Lysine and Methionine Biosynthesis
Both Lys and Met are two well-known amino acids belonging to the aspartate pathway (Fig. 1) [26]. In the plant kingdom, these are almost always synthesized within the chloroplast and share the three initial steps as a common pathway. The first enzymatic step of the aspartate family is catalyzed by aspartate kinase (AK) which has multiple isoenzymic forms. At least two forms are found in plants. Using ATP and Mg2+, the formation of phosphorylation of aspartate leads to the formation of β-aspartyl phosphate, which is subsequently oxidized to aspartate semialdehyde (ASD) by aspartate semialdehyde dehydrogenase (ASDH). In the last step of the common pathway, ASD forms either dihydrodipicolinate (DHDP), a precursor of diaminopimelic acid and lysine, or O-phosphohomoserine (OPH). OPH may be channeled to threonine or Met [36].
Fig. (1).
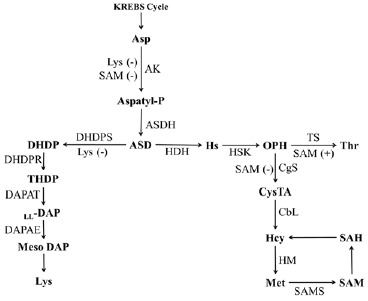
The Aspartate pathway leading to the biosynsthesis of Met and Lys. Asp: Aspatate, AK: Aspartate kinase, ASD:aspartate-semialdehyde, ASDH: aspartic semialdehyde dehydrogenase, DHDP: dihydrodipicolinate, DHDPS: dihydrodipicolinate synthase, HS: homoserine, HDH: homoserine dehydrogenase, OPH: O-phosphohomoserine, HSK: homoserine kinase, Thr: Threonine, TS: Thr synthase, CysTA: cystathionine, CgS: cystathionine γ-synthase, HcY: Homocysteine, CbL: cystathionine β-lyase, SAH: S-adenosylhomocysteine, HM: homocysteine methyltransferase, SAM: S-adenosyl-methionine, SAMS: S-adenosyl-methionine synthetase, THDP: tetrahydro-dipicolinate, DAPAE: DAP epimerase, DAP: diaminopimelate, DAPAT: DAP-aminotransferase.
Among the major regulatory steps controlling Lys synthesis in plants, DHDPS activity is the most prone to feedback inhibition by Lys. Moreover, DHDPS is about 100 times more sensitive to Lys inhibition in plants than in Escherichia coli [45]. Also, E. coli containing lysine-insensitive DHDPS contained increased levels of free Lys [45]. DHDPS cDNAs have been isolated from a range of plant species such as wheat, maize, tobacco, soybean, Arabidopsis, and rice [36, 37, 46]. In plants, diaminopimelate (DAP) has been known as a key intermediate in the Lys biosynthesis pathway. DAP also plays a central role in peptidoglycan biosynthesis, the main component of eubacterial cell walls.
Recent studies indicate differences among pathways to Lys in plants, chlamydia and cyanobacteria [47-49]. After DHDP formation, its reduction to tetrahydrodipicolinate (THDP) is catalyzed by DHDP reductase. At that point, the pathways of plants and bacteria become distinct. In some bacteria including some Bacillus species, THDP directly transfers to meso-DAP by the enzyme DAP dehydrogenase. The majority of bacterial species use a multiple pathway to synthesis Lys: N-succinylation/acetylation followed by transamination and desuccinylation/deacetylation [48]. Plants convert THDP indirectly to L,L-DAP by DAP-aminotransferase. Both plants and bacteria convert L,L-DAP to meso-DAP by DAP epimerase. The final step is decarboxylated to form lysine [47-49].
In the plant kingdom, OPH appears in the common branch point intermediate with the synthesis of Met and threonine. OPH is a common substrate for both enzymes: threonine synthase (TS) and cystathionine gamma-synthase (CgS). Met is converted from OPH in three enzymatic steps: CgS catalyses the formation of the thioether cystathionine from substrates of cysteine, the sulfur atom donor and OPH by trans-sulfuration. This process is only found in plant plastids like Arabidopsis thaliana [35, 50]. The next step converts the intermediate to homocysteine and subsequently to Met. In this mechanism, reactions are catalyzed by the enzymes CgS, cystathionine β-lyase (CbL), and methionine synthase (MS), in that order. Almost 80% of Met is converted into SAM and the remainder takes part as a protein constituent [51]. However, in yeast and bacteria, the nitrogen/carbon precursor for Met synthesis is distinct. In yeast, Met appears to be a direct result from sulphydration of O-acetylhomoserine. In bacteria, succinyl-homoserine as substrate has been investigated in Met synthesis. Another comparative observation between plants and microorganism is that homoserine is the intermediate compound which leads to Met and threonine in microbes. In plants, OPH is the last common intermediates of Met and threonine biosynthesis [52].
4.2. Tryptophan Biosynthesis
The Trp biosynthetic pathway in plants not only produces Trp but also associates with a range of secondary metabolites such as auxin, indole glucosinolates and other secondary compounds. Trp is synthesized from chorismate in chloroplast (Fig. 2). Anthranilate synthase (AS) catalyzes the first reaction of the Trp biosynthesis which converts chorismate and an amine donor (usually glutamine) to form anthranilate; its activity is subject to feedback inhibition by Trp. Microbial and plant anthranilate synthase is a holoenzyme consisting of two α and two β subunits [53]. The β subunits of AS were considered to catalyze glutamine as the donor of ammonia, used by α-subunits of AS to transfer chorismate to anthranilate. In the subsequent step, anthranilate phosphoribosylanthranilate transferase (PAT1) catalyses a conversion of anthranilate and phosphoribosylpyrophosphate to phosphoribosylanthranilate and inorganic pyrophosphate. The third enzyme in the biosynthesis of Trp is phosphoribosylanthranilate isomerase (PAI) activity converting phosphoribosylanthranilate to l-(O-carboxyphenylamino)-l-deoxyribulose-5-phosphate (CDRP). Then, indole-3-glycerol phosphate synthase (IGPS) accepts CDRP as the substrate which is transferred to indole-3-glycerol phosphate [54]. IPGS in plants and micro-organisms exhibit different characteristics. In bacteria and fungi, IPGS is a polyprotein with multiple domains and each domain catalyzes a separate reaction. For exampls, IPGS is fused with PAI to form a bifunctional enzyme in E. coli, Salmonella typhimurium and Vibrio parahaemolyticus. However, in plants IPGS is a monofunctional enzyme [54].
Fig. (2).
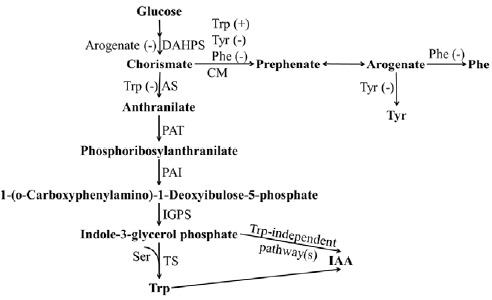
Trp biosynthesis pathway. AS: Anthranilate synthase; PAT: Phosphoribosylanthranilate transferase; PAI: Phosphoribosyl anthranilate isomerase; IGPS: Indole-3-glycerol phosphate synthase; Trp: Tryptophan; TS: Trp Synthase; Tyr: Tyrosine; Ser: Serine; IAA: Indole-3-acetic acid; Phe: Phenylalanine; AH: Arogenate dehydro; DAHPS: DAHP Synthase.
Trp synthase (TS) appears to be a heterodimer of α (TSα) and β (TSβ) subunits. TSα cleaves indole-3-glycerol phosphate to form indole and glyceraldehyde-3-phosphate; this is called the α-reaction. Subsequently, TSβ accepts indole as substrate and catalyzes the conversion of Ser into Trp. This is the β-reaction [55]. Unlike in bacteria and fungi, Trp in plants also serves as a precursor for the biosynthesis of various secondary metabolites, especially the phytohormone IAA [55].
5. CURRENT STATUS ON IMPROVING ESSENTIAL AMINO ACIDS CONTENT IN PLANTS
The success of the genetic approach has been mostly restricted to improving protein quality in model plants with enriched Lys, Trp and Met production (summarized in Tables 2 and 3). So far, this approach has had relatively limited success in crop species. Limited availability of crop genetic resources for plant breeding has been a significant limitation. Knowledge obtained from basic genetics and genetic engineering studies has also been successfully used to enrich the content of essential amino acids in crop plants. Enriching crop plants in essential amino acids has both economical and humanitarian interest. It helps humans and monogastric animals could obtain essentials amino acids through their diets. Here, we discussed several approaches for improving the content of Lys, Met, and Trp in plants.
Table (2).
Proteins rich in Lys and/or Met used to improve essential amino acids in crops.
| Name | Protein Sizes | Amino Acid Content | Crops/Plants | Amino Acid Increases | Ref. | ||
|---|---|---|---|---|---|---|---|
| Lys | Met | Lys | Met | ||||
| Zein | 10 kDa | 20 % | Maize (Zea mays) | 22.5 % | [56] | ||
| Met-rich 2S seed protein | 17 kDa | 18 % | Tobacco (Nicotiana tabacum) | 30 % | [57] | ||
| β-phaseolin | - | 3 residues | Common bean (Phaseolus vulgaris) | 9 residues | [58] | ||
| Prleg | - | Potato (Solanum tuberosum L.) | 3 fold | [59] | |||
| SBgLR | - | 8.7% | Maize (Zea mays) | 30.43% | [60] | ||
| MB-16 | 11 kDa | 11.11 mg/g total protein | Soybean (Glycine max L.) | 12.91 mg/g total protein (16.2%) | [42] | ||
| GhLRP | 24 kDa | Maize (Zea mays) | 16.2-65% | [61] | |||
| lysine-rich sb401 | 240 aa | 40 residues | Maize (Zea mays) | 24% | [62] | ||
Table (3).
Crops modified to bypass feedback regulation of the Lys/Met/Trp biosynthetic pathways.
| Amino Acids | Crops/Plants | Enzymes | Genes | Genebank Accession Number | Increase in Amino Acids Compared with WT (Fold) | Ref. | |
|---|---|---|---|---|---|---|---|
| Free | Total | ||||||
| Lys | Soybean (Glycine max L.) |
Lys insensitive DHDPS | Corynebacterium dapA gene | X53993.1 | 100 | 5 | [76] |
| Lys sensitive E.coli AK | E. coli LysC gene | M11812.1 | |||||
| Canola (Brassica napus L.) |
Lys insensitive DHDPS | Corynebacterium dapA gene | X53993.1 | >100 | 2 | ||
| Corn (Zea mays) | Lys insensitive DHDPS | Corynebacterium dapA gene | >2 | [77] | |||
| Met | Thale cress (Arabidopsis thaliana) |
TS | mto2-1 TS gene | 22 | [78] | ||
| Thale cress (Arabidopsis thaliana) |
Methionine γ-lyase (MGL) | AtMGL gene | At1g64660 | 9.2 | [78] | ||
| Alfalfa (Medicago sativa) |
PEPC kinase (PPCK) | GsPPCK3 gene | 2.29-2.34 | [79] | |||
| Trp | Astragalus (Astragalus sinicus) |
Feedback-insensitive AS | ASA2 gene | 1.3-5.5 | [65] | ||
| Potato (Solanum tuberosum L.) |
Mutant rice α-subunit of AS | OASA1D gene | 2-20 | [75] | |||
| Rice (Oryza sativa L.) | Mutant rice α subunit of AS | OASA1D gene | 80 | [80] | |||
| Tobacco (Nicotiana tabacum) |
Feedback-insensitive AS | ASA2 gene | M92353 | 3 | [44] | ||
| Adzuki bean (Vigna angularis) |
Mutant rice α subunit of AS | OASA1D gene | 6.5-16.5 | 1.3-2.1 | [81] | ||
| Rice (Oryza sativa L.) | AS | OASA2 gene | 230 | [61] | |||
5.1. Improving Met-content in Plants
Like carbohydrates and fats, protein is a “macronutrient”, which is needed in relatively large amounts for human and animals to be healthy. High quality protein sources are found in several crops, including beans, legumes, and nuts. Plants are the primary source of all protein consumed by human and livestock. Thus, improving the quality of plant proteins is an important step to enhance human nutrition. Thus, genetic engineering studies have been performed to increase Met-content in plant proteins by manipulating methionine synthesis.
Most enzymes in biosynthesis pathways leading to amino acids are inhibited by their end-products (allosteric regulation). 2S-sunflower seed protein has been characterized for its IgE-binding capacity, the protein possesses a significant amount of sulfur-containing amino acids [48]. This could be used to improve protein quality of other crops through genetic engineering. A chimeric gene encoding a methionine-rich Brazil nut (Bertholletia excelsa) protein contains over 18 % methionine, whereas most proteins contain only a few percent of methionine. The accumulation of the methionine-rich protein could make a significant enhancement in methionine levels in seed protein of transgenic tobacco [57] and potato [63]. Improving protein quality in the seed is another possibility. The expression of storage genes in seed crops directly affects the nutritional quality of the seed protein. Met-rich proteins were isolated to synthesize cDNA; among the proteins studied are 10kDa zein protein from maize [56], and MRNP34 hydrophobic protein [64]. Using genetic analysis, several genes encoded Met-rich protein in maize were recorded: Gene Zps10/(22) was found to be located on chromosome 9 [65], β-kafirin from sorghum grain [66], polymorphic cDNAs encoded for the Met-rich protein from Manduca sexta [67]. Basically, Met-rich proteins in plants were isolated and characterized to compare with major storage proteins. Another approach is to identify and characterize Met-rich protein from crops. A great deal of effort was focused on edible dry beans (Phaseolus vulgaris L.) [25], maize endosperm [68], Canabis sativa [69], peanuts [70], and soybean (Glycine max) [71].
Increasing biosynthesis of low-abundance, endogenous Met-rich protein was a strategy which could resolve the deficiency of sulfur amino acids in plants [72]. Using this approach, total seed methionine was increased by 6.8 % in transgenic soybean and canola plants. In addition, Gm2S-1 was recorded as a candidate gene for overexpression to improve nutritional quality of protein in soybean and other legumes [21]. Another strategy has been to isolate endogenous genes then modify the nucleotide sequence to accommodate more sulfur-containing amino acids. For instant, a 45-bp sequence encoding a Met-rich region from maize was inserted to a gene coding for β-phaseolin from Phaseolus vulgaris [58]. Using 2-D gel electrophoresis and in vitro labeling, George et al. detected several Met-rich proteins. The identification of a Met-rich protein in soybean seed and its composition were reported as a first important step to cloning its gene [73].
To increase Met levels in plants, several transgenic approaches have been used. Cystathionine γ-synthase (CGS), the first committed enzyme in the methionine biosynthesis pathway, was overexpressed in transgenic tobacco plants and showed that high expression level of T-AtCGS could lead to higher total Met contents in the transgenic plants [74]. The transformation of PrLeg gene into potato, which contains low amounts of sulfur-containing amino acids, was found to enhance Met content in the tubers [59]. To improve the quality of protein in soybean seed, research showed that M-16, a 11-kDa protein containing high sulfur amino acids, can be used [42]. On the other hand, the expression of some genes could enhance not only the plant stress tolerance but also its Met content, including genes encoding phosphoenolpyruvate carboxylase (PEPCs) and PEPC kinases (PPCKs) in alfalfa. The overexpression of PPCK could improve plant alkali tolerance as well as the Met content compared to wild type [75]. This approach might be an effective way to improve plant stress tolerance and methionine content.
In summary, several approaches for increasing Met content in plants have been attempted. But the most promising results of Met enhancement in plants came from the approach related to Met-rich proteins. Combining several methods, the improvement of Met content in plants could reach significantly improved levels and opportunity for use as food or feed.
5.2. Improving Lys-content in Plant
Lys plays an important role in nutrition because of the low Lys level in cereals and grains, which are major crops worldwide. Several approaches have been employed for improving Lys content in plants. Research to improve Lys content was conducted via plant breeding, intervention of amino acid metabolism, genetic engineering, and plant transformation techniques. Using a genetic selection for high lysine mutants in maize, barley, and sorghum led to the identification of mutants with high lysine content. Based on biochemical pathways of lysine biosynthesis, different combinations of genetic methods were used to increase synthesis of Lys.
Firstly, the improvement of Lys content could be done by increasing accumulation of Lys in plants. Reducing the activity of Lys-Ketoglutarate Reductase/Saccharopine Dehydrogenase (LKR/SDH) was reported to increase the accumulation of Lys in tobacco seeds [82, 83]. Lys is synthesized through Asp family pathway, which also leads to Met and Thr. Interestingly, dihydrodipicolinate synthase (DHDPS), an enzyme committed to Lys synthesis, is inhibited by Lys through an inhibition loop [84]. Mutant tobacco with high Lys content was developed by modulating the DHDPS gene. In maize, the transgenic lines carrying modified DHDPS exhibited increased Lys content [77]. LY038 transgenic maize line, which has twice as much Lys content as the control, was constructed by using a seed-specific promoter.
To improve Lys level in protein, efforts to synthesize Lys-rich proteins in plants via genetic engineering have been reported. Most of the time, the approaches employed were modifications of endogenous protein sequences or synthetic protein. Recently, the protein translation elongation factor 1α (EF-1α) from cereal embryo was reported to be rich in Lys. Singh et al. investigated the relationship between EF-1α content and several proteins from wheat germ. Using immunochemical to detect Lys content of wheat, the authors could identify the correlation between EF-1α levels and Lys content in plants [85].
Using a different method, genes encoding Lys-rich proteins, such as SBgLR from potato, were tested as a candidate for increasing Lys content in maize. Through gene transformation, a transgenic maize line harboring SBgLR gene had a significantly higher Lys content compared to the control. When treated with NaCl, the transgenic line also displayed enhanced tolerance to salt stress [60]. In maize, the opaque2 mutant was a valuable material for breeding to enhancing the nutritional value of the crops. The accumulation of some Lys-rich protein, such as sorbitol dehydrogenase and glyceraldehyde 3-phosphate dehydrogenase, was found in opaque2 maize line [86]. Sb401, a gene encoding a Lys-rich protein, had been successfully integrated into the genome of maize. Compared to several conventional maize lines, the transgenic lines had a higher Lys content and content of total protein [87]. Therefore, sb401 can be another important gene for the improvement of nutritive value of maize.
In addition, Lys is thought to be the first limiting essential amino acid in rice. Thus, the creation of Lys-rich rice is of interest for the researchers. The first transgenic rice line was constructed from a gene encoding a Lys-rich protein cloned from Psophocarpus tetragonolobus. The transgene was found to express in leaves, stems and roots of the transgenic rice plants [44]. Another achievement was in Brassica napus, where a Lys-rich protein gene from the seeds of Psophocarpus was successfully transformed into the plant [88]. Three genes encoding Lys-rich proteins in Arabidopsis, including AtAGP17, AtAGP18 and AtAGP19 were reported, in which AtAGP18 could be involved in a signal transduction pathway that regulates plant growth and development [89]. Other transgenic rice lines have been developed by over-accumulation of Lys-rich binding proteins [62]. Results from this research offer further approaches to improving Lys content in plants.
5.3. Improving Trp Content in Plants
Trp is the second most limiting essential amino acid in protein metabolism. As described above, Trp biosynthesis in plants is feedback-regulated by its end-products. The first research on Trp in plants was done by Kreps and colleagues, they found a mutant of Arabidopsis, amt-1, that had elevated Trp accumulation in the plants [90]. They suggested that elevated anthranilate synthase (AS) activity found in the mutant caused by an alteration in the α-subunit of the enzyme was responsible for the increased resistance to feedback inhibition by Trp. Following this approach, several studies were conducted to improve Trp content in crop plants. Feedback-insensitive AS and ASA2 isolated from a 5-methyl-tryptophan (5-MTP) resistant cell line were introduced into the forage legume Astragalus sinicus [65]. Results showed that the ASA2 transcript and high Trp were co-detected in the leaves, stems and roots of transgenic plants, indicating that ASA2 could be used to transform plants of different species for increased Trp levels. Maize seeds, deficient in Lys and Trp, become one of the most popular subjects to study. The accumulation of 19- and 22- kDa α-zeins in transgenic plants resulting in higher Lys and Trp content has also been reported [91]. The overexpression of OASA1D, a gene encoding a feedback-insensitive α-subunit of rice AS, was reported to accumulate Trp in calli and leaves of rice. The total Trp content in the seeds of the transgenic rice plants was also increased. Characterization of OASA1D transgenic rice lines demonstrated the feasibility of enhancing the Trp content in the seeds of rice and other crops [81]. The OASA1D transgene was also shown to raise Trp content in transgenic potato [41], adzuki bean (Vigna angularis) [81]. Several OASA1D-overexpressed rice lines were significantly correlated with over-accumulation of Trp [92]. Small changes were also observed in several minor metabolites in the OASA1D-overexpressed lines. AS activity was an important determinant in IAA biosynthesis as well as Trp accumulation [80]. In transgenic potato plants, the expression of OASA1D induced a significant increase in Trp [75]. This increase could be explained by a reduction in the sensitivity of AS to feedback inhibition by Trp.
According to the biosynthesis pathways for essential amino acids, a link between the synthesis of Phe and Trp was found. A mutant of mtr1-D gene encoded a form of prephenate dehydratase (PDT) in rice were created to elucidate the connection between Phe and Trp accumulations [72]. In tobacco, several transgenic lines with ASA2 had shown a significant increase in Trp level [44]. Furthermore, metabolic engineering of crops could be applied by a combination of computational biology and gene targeting. Saika et al. reported the mutation in OASA2, a key enzyme of Trp biosynthesis in rice, led to higher Trp accumulation in plants [61]. Gene targeting could reduce the risk of gene silencing relative to other conventional approaches.
6. FUTURE DIRECTIONS
Research summarized in this review has a significant potential for creating nutritionally enriched plants. The developments of molecular biology techniques will facilitate the improvement of protein quality in plants. Recombinant DNA techniques and plant transformation have become useful tools to determine the factors that could enhance essential amino acids in plants.
The efforts in enhancing the content of essential amino acids in plants had made significant progress. In the future, these results will be important for global agriculture. The approach of overexpressing genes encoding proteins rich in Met, Lys, and Trp, although making slower progress, is a key step for improving nutritional quality of plants. For example, the results in creating transgenic plants with Met-, Lys- or Trp-rich protein by overexpressing several specific genes would make a significant improvement in protein for use as food or feed. Met-, Lys- or Trp-rich proteins were characterized and found to be easily digested, and also safe for consumption [93].
Essential-amino-acids-rich proteins in plants must pass the test for their effects on the human and animal digestion systems before being continued to the next steps. Employing genetics transformation to improve protein quality in plants also encountered a trouble with the global acceptance of genetically modified (GM) crops. Like most GM crops, the development of plants with high levels of essential amino acids in protein depends on the public acceptance of genetically modified organism (GMO) [94]. Recently, there has been acceptance in some countries. For examples, the situation of LY038 maize, which has double the Lys content without changing the grain protein content, is recognized by the USDA to be as safe as other varieties bred through conventional methods. Then, LY038 maize was approved for commercial use in a number of countries [95]. In the future, production of GM crops with enhanced essential amino acids is also expected to combine two or three important amino acids, namely Met, Lys and Trp.
6.1. Genome Editing Techniques and the Improvement of Protein Quality in Crops
Targeted genome editing using synthetic nuclease or endonuclease enzymes has recently become a powerful tool in genetic engineering. Zinc finger nucleases (ZFNs) employing a zinc finger domain with FokI endonuclease domain, had been used successfully as genome editing tool in Arabidopsis thaliana [96, 97], Zea mays [98] and legume [99]. More recently, researchers have developed new tools for plant genome editing, like the Transcription Activator-Like Effector Nucleases (TALENs), and the Clustered Regularly Interspaced Short Palindromic Repeat (CRISPR)/Cas9. In 2015, Clasen et al. used TALENs to knock-out vacuolar invertase gene (VInv), which encodes a protein that cleaves sucrose to glucose and fructose. These results improved the characteristics of commercial potato which eliminated reducing sugars, therefore reducing proportion of acrylamide [100]. In nature, CRISPR/Cas9 functions as a bacterial adaptive immunity system. CRISPR/Cas9 applications have been developed for gene modification in a range of plant species including soybean [101], rice, Arabidopsis, tobacco, sorghum [102] and sweet orange [103]. Genome editing techniques, especially TALENs and CRISPR/Cas9 system, are precise approaches to improve traits including enhanced nutritional quality and commercial valuable compounds. Given the high precision nature of genome-editing techniques, genes encoding feedback-sensitive enzymes in the amino acid biosynthesis pathways can be edited to become insensitive to feedback inhibition. Combined with conventional breeding, the transgenes encoding editing machinery can be removed and the “edited plant” with improved protein quality can be considered as “non-GMO” plants.
ACKNOWLEDGEMENTS
We apologize to our colleagues whose original works were not cited due to space limitations. The authors wish to thank Stephanie Dalquist (skd@mit.edu) and Georgina Smith (g.smith@cgiar.org) for correcting English usage in this manuscript. Dung Tien Le received funding support from the National Foundation for Science and Technology Development (NAFOSTED) under the grant number 106-NN.02-2013.46.
CONFLICT OF INTEREST
The authors confirm that this article content has no conflict of interest.
REFERENCES
- 1.Zhu C.F., Naqvi S., Gomez S.G., Pelacho A.M., Capell T., Christou P. Vitamin, Protein and Essential Mineral Enhancement of Cereal Crops for Food Security. GMO Biosafety Res. 2010;1:315–318. [Google Scholar]
- 2.Graham R. D., Gregorio G. Breeding for nutritional characteristics in cereals. Novartis Found. Symp. 2001;236:205–214. doi: 10.1002/9780470515778.ch15. [DOI] [PubMed] [Google Scholar]
- 3.Garcia M.M., Guéant-Rodriguez R.M., Pooya S., Brachet P., Alberto J.M., Jeannesson E., Maskali F., Gueguen N., Marie P.Y., Lacolley P., Herrmann M., Juillière Y., Malthiery Y., Guéant J.L. Methyl donor deficiency induces cardiomyopathy through altered methylation/acetylation of PGC-1α by PRMT1 and SIRT1. J. Pathol. 2011;225(3):324–335. doi: 10.1002/path.2881. [DOI] [PubMed] [Google Scholar]
- 4.Forges T., Monnier-Barbarino P., Alberto J.M., Guéant-Rodriguez R.M., Daval J.L., Guéant J.L. Impact of folate and homocysteine metabolism on human reproductive health. Hum. Reprod. Update. 2007;13(3):225–238. doi: 10.1093/humupd/dml063. [DOI] [PubMed] [Google Scholar]
- 5.Guthikonda S., Haynes W.G. Homocysteine: role and implications in atherosclerosis. Curr. Atheroscler. Rep. 2006;8(2):100–106. doi: 10.1007/s11883-006-0046-4. [DOI] [PubMed] [Google Scholar]
- 6.Scott J.M., Weir D.G. Folic acid, homocysteine and one-carbon metabolism: a review of the essential biochemistry. J. Cardiovasc. Risk. 1998;5(4):223–227. doi: 10.1097/00043798-199808000-00003. [DOI] [PubMed] [Google Scholar]
- 7.Fowler B. Homocysteine: overview of biochemistry, molecular biology, and role in disease processes. Semin. Vasc. Med. 2005;5(2):77–86. doi: 10.1055/s-2005-872394. [DOI] [PubMed] [Google Scholar]
- 8.Swanson D.A., Liu M.L., Baker P.J., Garrett L., Stitzel M., Wu J., Harris M., Banerjee R., Shane B., Brody L.C. Targeted disruption of the methionine synthase gene in mice. Mol. Cell. Biol. 2001;21(4):1058–1065. doi: 10.1128/MCB.21.4.1058-1065.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Luo S., Levine R.L. Methionine in proteins defends against oxidative stress. FASEB J. 2009;23(2):464–472. doi: 10.1096/fj.08-118414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Poirier L.A. The effects of diet, genetics and chemicals on toxicity and aberrant DNA methylation: an introduction. J. Nutr. 2002;132(8) Suppl.:2336S–2339S. doi: 10.1093/jn/132.8.2336S. [DOI] [PubMed] [Google Scholar]
- 11.Fukagawa N.K., Galbraith R.A. Advancing age and other factors influencing the balance between amino acid requirements and toxicity. J. Nutr. 2004;134(6) Suppl.:1569S–1574S. doi: 10.1093/jn/134.6.1569S. [DOI] [PubMed] [Google Scholar]
- 12.Fukagawa N.K. Sparing of methionine requirements: evaluation of human data takes sulfur amino acids beyond protein. J. Nutr. 2006;136(6) Suppl.:1676S–1681S. doi: 10.1093/jn/136.6.1676S. [DOI] [PubMed] [Google Scholar]
- 13.McCully K.S. Vascular pathology of homocysteinemia: implications for the pathogenesis of arteriosclerosis. Am. J. Pathol. 1969;56(1):111–128. [PMC free article] [PubMed] [Google Scholar]
- 14.Lertratanangkoon K., Orkiszewski R.S., Scimeca J.M. Methyl-donor deficiency due to chemically induced glutathione depletion. Cancer Res. 1996;56(5):995–1005. [PubMed] [Google Scholar]
- 15.Xu S., Harrison J.H., Chalupa W., Sniffen C., Julien W., Sato H., Fujieda T., Watanabe K., Ueda T., Suzuki H. The effect of ruminal bypass lysine and methionine on milk yield and composition of lactating cows. J. Dairy Sci. 1998;81(4):1062–1077. doi: 10.3168/jds.S0022-0302(98)75668-1. [DOI] [PubMed] [Google Scholar]
- 16.Gaby A.R. Natural remedies for Herpes simplex. Altern. Med. Rev. 2006;11(2):93–101. [PubMed] [Google Scholar]
- 17.Sacksteder K.A., Biery B.J., Morrell J.C., Goodman B.K., Geisbrecht B.V., Cox R.P., Gould S.J., Geraghty M.T. Identification of the alpha-aminoadipic semialdehyde synthase gene, which is defective in familial hyperlysinemia. Am. J. Hum. Genet. 2000;66(6):1736–1743. doi: 10.1086/302919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Civitelli R., Villareal D.T., Agnusdei D., Nardi P., Avioli L.V., Gennari C. Dietary L-lysine and calcium metabolism in humans. Nutrition. 1992;8(6):400–405. [PubMed] [Google Scholar]
- 19.Smriga M., Kameishi M., Uneyama H., Torii K. Dietary L-lysine deficiency increases stress-induced anxiety and fecal excretion in rats. J. Nutr. 2002;132(12):3744–3746. doi: 10.1093/jn/132.12.3744. [DOI] [PubMed] [Google Scholar]
- 20.Karau A., Grayson I. Amino acids in human and animal nutrition. Adv. Biochem. Eng. Biotechnol. 2014;143:189–228. doi: 10.1007/10_2014_269. [DOI] [PubMed] [Google Scholar]
- 21.Galvez A.F., Revilleza M.J., de Lumen B.O., Krenz D.C. Food for Health in the Pacific Rim. Food & Nutrition Press, Inc.; 2008. Enhancing the Biosynthesis of Endogenous Methionine-Rich Proteins (MRP) to Improve the Protein Quality of Legumes Via Genetic Engineering. pp. 540–552. [Google Scholar]
- 22.Moehn S., Pencharz P.B., Ball R.O. Lessons learned regarding symptoms of tryptophan deficiency and excess from animal requirement studies. J. Nutr. 2012;142(12):2231S–2235S. doi: 10.3945/jn.112.159061. [DOI] [PubMed] [Google Scholar]
- 23.Badawy A.A. Tryptophan: the key to boosting brain serotonin synthesis in depressive illness. J. Psychopharmacol. (Oxford) 2013;27(10):878–893. doi: 10.1177/0269881113499209. [DOI] [PubMed] [Google Scholar]
- 24.Akibode S., Maredia M. Global and Regional Trends in Production, Trade and Consumption of Food Legume Crops. Report submitted to SPIA. 2011:83p. [Google Scholar]
- 25.Apostolatos G. Isolation and characterization of a methionine-rich protein fraction from edible dry bean (Phaseolus vulgaris L.). Plant Sci. Lett. 1984;33:39–46. doi: 10.1016/0304-4211(84)90066-X. [DOI] [Google Scholar]
- 26.Galili G., Amir R., Hoefgen R., Hesse H. Improving the levels of essential amino acids and sulfur metabolites in plants. Biol. Chem. 2005;386(9):817–831. doi: 10.1515/BC.2005.097. [DOI] [PubMed] [Google Scholar]
- 27.Wenefrida I., Utomo H.S., Blanche S.B., Linscombe S.D. Enhancing essential amino acids and health benefit components in grain crops for improved nutritional values. Recent Pat. DNA Gene Seq. 2009;3(3):219–225. doi: 10.2174/187221509789318405. [DOI] [PubMed] [Google Scholar]
- 28.Energy and protein requirements. Report of a joint FAO/WHO/UNU Expert Consultation. World Health Organ. Tech. Rep. Ser. 1985;724:1–206. [PubMed] [Google Scholar]
- 29.Coleman C., Larkins B. The Prolamins of Maize. In: Shewry P.R., Casey R., editors. Seed Proteins. Springer Netherlands; 1999. pp. 109–139. [DOI] [Google Scholar]
- 30.Anjum F.M., Ahmad I., Butt M.S., Sheikh M.A., Pasha I. Amino acid composition of spring wheats and losses of lysine during chapati baking. J. Food Compos. Anal. 2005;18:523–532. doi: 10.1016/j.jfca.2004.04.009. [DOI] [Google Scholar]
- 31.Erickson P.S., Schauff D.J., Murphy M.R. Diet digestibility and growth of holstein calves fed acidified milk replacers containing soy protein concentrate. J. Dairy Sci. 1989;72(6):1528–1533. doi: 10.3168/jds.S0022-0302(89)79263-8. [DOI] [PubMed] [Google Scholar]
- 32.Scherz H., Senser F., Souci S.W. Food composition and nutrition tables. Boca Raton, FL: CRC Press/Medpharm; 2000. Bundesministerium für Ernährung, L.u.F., Deutsche Forschungsanstalt für, L. [Google Scholar]
- 33.W. H. O. Protein and amino acid requirements in human nutrition. World Health Organ. Tech. Rep. Ser. 2007:1–265. [PubMed] [Google Scholar]
- 34.Jaynes J.M., Yang M.S., Espinoza N., Dodds J.H. Plant protein improvement by genetic engineering: use of synthetic genes. Trends Biotechnol. 1986;4:314–320. doi: 10.1016/0167-7799(86)90183-6. [DOI] [Google Scholar]
- 35.Hesse H., Kreft O., Maimann S., Zeh M., Hoefgen R. Current understanding of the regulation of methionine biosynthesis in plants. J. Exp. Bot. 2004;55(404):1799–1808. doi: 10.1093/jxb/erh139. [DOI] [PubMed] [Google Scholar]
- 36.Azevedo R.A., Lancien M., Lea P.J. The aspartic acid metabolic pathway, an exciting and essential pathway in plants. Amino Acids. 2006;30(2):143–162. doi: 10.1007/s00726-005-0245-2. [DOI] [PubMed] [Google Scholar]
- 37.Kong F., Jiang S., Meng X., Song C., Shi J., Jin D., Jiang S., Wang B. Cloning and Characterization of the DHDPS Gene Encoding the Lysine Biosynthetic Enzyme Dihydrodipocolinate Synthase from Zizania latifolia (Griseb). Plant Mol. Biol. Rep. 2009;27:199–208. doi: 10.1007/s11105-008-0073-0. [DOI] [Google Scholar]
- 38.Zhu X., Galili G. Increased lysine synthesis coupled with a knockout of its catabolism synergistically boosts lysine content and also transregulates the metabolism of other amino acids in Arabidopsis seeds. Plant Cell. 2003;15(4):845–853. doi: 10.1105/tpc.009647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Galili G., Tang G., Zhu X., Gakiere B. Lysine catabolism: a stress and development super-regulated metabolic pathway. Curr. Opin. Plant Biol. 2001;4(3):261–266. doi: 10.1016/S1369-5266(00)00170-9. [DOI] [PubMed] [Google Scholar]
- 40.Curien G., Ravanel S., Dumas R. A kinetic model of the branch-point between the methionine and threonine biosynthesis pathways in Arabidopsis thaliana. Eur. J. Biochem. 2003;270(23):4615–4627. doi: 10.1046/j.1432-1033.2003.03851.x. [DOI] [PubMed] [Google Scholar]
- 41.Yamada T., Tozawa Y., Hasegawa H., Terakawa T., Ohkawa Y., Wakasa K. Use of a feedback-insensitive α subunit of anthranilate synthase as a selectable marker for transformation of rice and potato. Mol. Breed. 2004;14:363–373. doi: 10.1007/s11032-004-0184-8. [DOI] [Google Scholar]
- 42.Zhang Y., Schernthaner J., Labbé N., Hefford M.A., Zhao J., Simmonds D.H. Improved protein quality in transgenic soybean expressing a de novo synthetic protein, MB-16. Transgenic Res. 2014;23(3):455–467. doi: 10.1007/s11248-013-9777-5. [DOI] [PubMed] [Google Scholar]
- 43.Zhang X.H., Brotherton J.E., Widholm J.M., Portis A.R., Jr Targeting a nuclear anthranilate synthase alpha-subunit gene to the tobacco plastid genome results in enhanced tryptophan biosynthesis. Return of a gene to its pre-endosymbiotic origin. Plant Physiol. 2001;127(1):131–141. doi: 10.1104/pp.127.1.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gao Y., Jing Y., Shen S., Tian S., Kuang T., Samuel S.M. Transfer of lysine-rich protein gene into rice and production of fertile transgenic plants. Acta Bot. Sin. 2001;43:506–511. [Google Scholar]
- 45.Vauterin M., Frankard V., Jacobs M. Functional rescue of a bacterial dapA auxotroph with a plant cDNA library selects for mutant clones encoding a feedback-insensitive dihydrodipicolinate synthase. Plant J. 2000;21(3):239–248. doi: 10.1046/j.1365-313x.2000.00668.x. [DOI] [PubMed] [Google Scholar]
- 46.Sarrobert C., Thibaud M.C., Contard-David P., Gineste S., Bechtold N., Robaglia C., Nussaume L. Identification of an Arabidopsis thaliana mutant accumulating threonine resulting from mutation in a new dihydrodipicolinate synthase gene. Plant J. 2000;24(3):357–367. doi: 10.1046/j.1365-313x.2000.00884.x. [DOI] [PubMed] [Google Scholar]
- 47.Hudson A.O., Singh B.K., Leustek T., Gilvarg C. An LL-diaminopimelate aminotransferase defines a novel variant of the lysine biosynthesis pathway in plants. Plant Physiol. 2006;140(1):292–301. doi: 10.1104/pp.105.072629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hudson A. O., Bless C., Macedo P., Chatterjee S. P., Singh B. K., Gilvarg C., Leustek T. Biosynthesis of lysine in plants: evidence for a variant of the known bacterial pathways. Biochim. Biophys. Acta. 2005;1721:27–36. doi: 10.1016/j.bbagen.2004.09.008. [DOI] [PubMed] [Google Scholar]
- 49.McCoy A.J., Adams N.E., Hudson A.O., Gilvarg C., Leustek T., Maurelli A.T. L,L-diaminopimelate aminotransferase, a trans-kingdom enzyme shared by Chlamydia and plants for synthesis of diaminopimelate/lysine. Proc. Natl. Acad. Sci. USA. 2006;103(47):17909–17914. doi: 10.1073/pnas.0608643103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Droux M. Sulfur assimilation and the role of sulfur in plant metabolism: a survey. Photosynth. Res. 2004;79(3):331–348. doi: 10.1023/B:PRES.0000017196.95499.11. [DOI] [PubMed] [Google Scholar]
- 51.Hesse H., Hoefgen R. Molecular aspects of methionine biosynthesis. Trends Plant Sci. 2003;8(6):259–262. doi: 10.1016/S1360-1385(03)00107-9. [DOI] [PubMed] [Google Scholar]
- 52.Datko A.H., Giovanelli J., Mudd S.H. Homocysteine biosynthesis in green plants. O-Phosphorylhomoserine as the physiological substrate for cystathionine gamma-synthase. J. Biol. Chem. 1974;249(4):1139–1155. [PubMed] [Google Scholar]
- 53.Poulsen C., Bongaerts R.J., Verpoorte R. Purification and characterization of anthranilate synthase from Catharanthus roseus. Eur. J. Biochem. 1993;212(2):431–440. doi: 10.1111/j.1432-1033.1993.tb17679.x. [DOI] [PubMed] [Google Scholar]
- 54.Tzin V., Galili G. The Biosynthetic Pathways for Shikimate and Aromatic Amino Acids in Arabidopsis thaliana. Arabidopsis Book. 2010;8:e0132. doi: 10.1199/tab.0132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Radwanski E.R., Last R.L. Tryptophan biosynthesis and metabolism: biochemical and molecular genetics. Plant Cell. 1995;7(7):921–934. doi: 10.1105/tpc.7.7.921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kirihara J.A., Hunsperger J.P., Mahoney W.C., Messing J.W. Differential expression of a gene for a methionine-rich storage protein in maize. Mol. Gen. Genet. 1988;211(3):477–484. doi: 10.1007/BF00425704. [DOI] [PubMed] [Google Scholar]
- 57.Altenbach S.B., Pearson K.W., Meeker G., Staraci L.C., Sun S.M. Enhancement of the methionine content of seed proteins by the expression of a chimeric gene encoding a methionine-rich protein in transgenic plants. Plant Mol. Biol. 1989;13(5):513–522. doi: 10.1007/BF00027311. [DOI] [PubMed] [Google Scholar]
- 58.Hoffman L.M., Donaldson D.D., Herman E.M. A modified storage protein is synthesized, processed, and degraded in the seeds of transgenic plants. Plant Mol. Biol. 1988;11(6):717–729. doi: 10.1007/BF00019513. [DOI] [PubMed] [Google Scholar]
- 59.Goo Y.M., Kim T.W., Lee M.K., Lee S.W. Accumulation of PrLeg, a Perilla legumin protein in potato tuber results in enhanced level of sulphur-containing amino acids. C. R. Biol. 2013;336(9):433–439. doi: 10.1016/j.crvi.2013.09.002. [DOI] [PubMed] [Google Scholar]
- 60.Wang M., Liu C., Li S., Zhu D., Zhao Q., Yu J. Improved nutritive quality and salt resistance in transgenic maize by simultaneously overexpression of a natural lysine-rich protein gene, SBgLR, and an ERF transcription factor gene, TSRF1. Int. J. Mol. Sci. 2013;14(5):9459–9474. doi: 10.3390/ijms14059459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Saika H., Oikawa A., Matsuda F., Onodera H., Saito K., Toki S. Application of gene targeting to designed mutation breeding of high-tryptophan rice. Plant Physiol. 2011;156(3):1269–1277. doi: 10.1104/pp.111.175778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kawakatsu T., Wang S., Wakasa Y., Takaiwa F. Increased lysine content in rice grains by over-accumulation of BiP in the endosperm. Biosci. Biotechnol. Biochem. 2010;74(12):2529–2531. doi: 10.1271/bbb.100619. [DOI] [PubMed] [Google Scholar]
- 63.Tu H.M., Godfrey L.W., Sun S.S. Expression of the Brazil nut methionine-rich protein and mutants with increased methionine in transgenic potato. Plant Mol. Biol. 1998;37(5):829–838. doi: 10.1023/A:1006098524887. [DOI] [PubMed] [Google Scholar]
- 64.Marie B., Joubert C., Belliard C., Tayale A., Zanella-Cléon I., Marin F., Gueguen Y., Montagnani C. Characterization of MRNP34, a novel methionine-rich nacre protein from the pearl oysters. Amino Acids. 2012;42(5):2009–2017. doi: 10.1007/s00726-011-0932-0. [DOI] [PubMed] [Google Scholar]
- 65.Cho H.J., Brotherton J.E., Song H.S., Widholm J.M. Increasing tryptophan synthesis in a forage legume Astragalus sinicus by expressing the tobacco feedback-insensitive anthranilate synthase (ASA2) gene. Plant Physiol. 2000;123(3):1069–1076. doi: 10.1104/pp.123.3.1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Chamba E.B., Halford N.G., Forsyth J., Wilkinson M., Shewry P.R. Molecular cloning of β-kafirin, a methionine-rich protein of sorghum grain. J. Cereal Sci. 2005;41:381–383. doi: 10.1016/j.jcs.2004.09.004. [DOI] [Google Scholar]
- 67.Wang X.Y., Frohlich D.R., Wells M.A. Polymorphic cDNAs encode for the methionine-rich storage protein from Manduca sexta. Insect Mol. Biol. 1993;2(1):13–20. doi: 10.1111/j.1365-2583.1993.tb00120.x. [DOI] [PubMed] [Google Scholar]
- 68.Goyer A., Collakova E., Shachar-Hill Y., Hanson A.D. Functional characterization of a methionine gamma-lyase in Arabidopsis and its implication in an alternative to the reverse trans-sulfuration pathway. Plant Cell Physiol. 2007;48(2):232–242. doi: 10.1093/pcp/pcl055. [DOI] [PubMed] [Google Scholar]
- 69.Odani S., Odani S. Isolation and primary structure of a methionine- and cystine-rich seed protein of Cannabis sativa. Biosci. Biotechnol. Biochem. 1998;62(4):650–654. doi: 10.1271/bbb.62.650. [DOI] [PubMed] [Google Scholar]
- 70.Basha S.M. Deposition pattern of methionine-rich protein in peanuts. J. Agric. Food Chem. 1991;39:88–91. doi: 10.1021/jf00001a016. [DOI] [Google Scholar]
- 71.Revilleza M.J., Galvez A.F., Krenz D.C., de Lumen B.O. An 8 kDa Methionine-Rich Protein from Soybean (Glycine max) Cotyledon: Identification, Purification, and N-Terminal Sequence. J. Agric. Food Chem. 1996;44:2930–2935. doi: 10.1021/jf960063u. [DOI] [Google Scholar]
- 72.Yamada T., Matsuda F., Kasai K., Fukuoka S., Kitamura K., Tozawa Y., Miyagawa H., Wakasa K. Mutation of a rice gene encoding a phenylalanine biosynthetic enzyme results in accumulation of phenylalanine and tryptophan. Plant Cell. 2008;20(5):1316–1329. doi: 10.1105/tpc.107.057455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.George A.A., De Lumen B.O. A novel methionine-rich protein in soybean seed: identification, amino acid composition, and N-terminal sequence. J. Agric. Food Chem. 1991;39:224–227. doi: 10.1021/jf00001a046. [DOI] [Google Scholar]
- 74.Matityahu I., Godo I., Hacham Y., Amir R. Tobacco seeds expressing feedback-insensitive cystathionine gamma-synthase exhibit elevated content of methionine and altered primary metabolic profile. BMC Plant Biol. 2013;13:206. doi: 10.1186/1471-2229-13-206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Matsuda F., Yamada T., Miyazawa H., Miyagawa H., Wakasa K. Characterization of tryptophan-overproducing potato transgenic for a mutant rice anthranilate synthase alpha-subunit gene (OASA1D). Planta. 2005;222(3):535–545. doi: 10.1007/s00425-005-1565-x. [DOI] [PubMed] [Google Scholar]
- 76.Falco S.C., Guida T., Locke M., Mauvais J., Sanders C., Ward R.T., Webber P. Transgenic canola and soybean seeds with increased lysine. Biotechnology (N. Y.) 1995;13(6):577–582. doi: 10.1038/nbt0695-577. [DOI] [PubMed] [Google Scholar]
- 77.Huang S., Kruger D.E., Frizzi A., D’Ordine R.L., Florida C.A., Adams W.R., Brown W.E., Luethy M.H. High-lysine corn produced by the combination of enhanced lysine biosynthesis and reduced zein accumulation. Plant Biotechnol. J. 2005;3(6):555–569. doi: 10.1111/j.1467-7652.2005.00146.x. [DOI] [PubMed] [Google Scholar]
- 78.Bartlem D., Lambein I., Okamoto T., Itaya A., Uda Y., Kijima F., Tamaki Y., Nambara E., Naito S. Mutation in the threonine synthase gene results in an over-accumulation of soluble methionine in Arabidopsis. Plant Physiol. 2000;123(1):101–110. doi: 10.1104/pp.123.1.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Sun M., Sun X., Zhao Y., Zhao C., Duanmu H., Yu Y., Ji W., Zhu Y. Ectopic expression of GsPPCK3 and SCMRP in Medicago sativa enhances plant alkaline stress tolerance and methionine content. PLoS One. 2014;9(2):e89578. doi: 10.1371/journal.pone.0089578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Morino K., Matsuda F., Miyazawa H., Sukegawa A., Miyagawa H., Wakasa K. Metabolic profiling of tryptophan-overproducing rice calli that express a feedback-insensitive alpha subunit of anthranilate synthase. Plant Cell Physiol. 2005;46(3):514–521. doi: 10.1093/pcp/pci051. [DOI] [PubMed] [Google Scholar]
- 81.Hanafy M.S., Rahman S.M., Khalafalla M.M., El-Shemy H.A., Nakamoto Y., Ishimoto M., Wakasa K. Accumulation of free tryptophan in azuki bean (Vigna angularis) induced by expression of a gene (OASA1D) for a modified α-subunit of rice anthranilate synthase. Plant Sci. 2006;171:670–676. doi: 10.1016/j.plantsci.2006.06.016. [DOI] [Google Scholar]
- 82.Karchi H., Shaul O., Galili G. Lysine synthesis and catabolism are coordinately regulated during tobacco seed development. Proc. Natl. Acad. Sci. USA. 1994;91(7):2577–2581. doi: 10.1073/pnas.91.7.2577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Karchi H., Miron D., Ben-Yaacov S., Galili G. The lysine-dependent stimulation of lysine catabolism in tobacco seed requires calcium and protein phosphorylation. Plant Cell. 1995;7(11):1963–1970. doi: 10.1105/tpc.7.11.1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Galili G., Höfgen R. Metabolic engineering of amino acids and storage proteins in plants. Metab. Eng. 2002;4(1):3–11. doi: 10.1006/mben.2001.0203. [DOI] [PubMed] [Google Scholar]
- 85.Singh J., Sharp P.J., Skerritt J.H. A new candidate protein for high lysine content in wheat grain. J. Sci. Food Agric. 2001;81:216–226. doi: 10.1002/1097-0010(20010115)81:2<216::AID-JSFA794>3.0.CO;2-X. [DOI] [Google Scholar]
- 86.Jia M., Wu H., Clay K.L., Jung R., Larkins B.A., Gibbon B.C. Identification and characterization of lysine-rich proteins and starch biosynthesis genes in the opaque2 mutant by transcriptional and proteomic analysis. BMC Plant Biol. 2013;13:60. doi: 10.1186/1471-2229-13-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Tang M., He X., Luo Y., Ma L., Tang X., Huang K. Nutritional assessment of transgenic lysine-rich maize compared with conventional quality protein maize. J. Sci. Food Agric. 2013;93(5):1049–1054. doi: 10.1002/jsfa.5845. [DOI] [PubMed] [Google Scholar]
- 88.Wang J., Chen L., Liu Q.Q., Sun S.S., Sokolov V., Wang Y.P. Transformation of LRP gene into Brassica napus mediated by Agrobacterium tumefaciens to enhance lysine content in seeds. Genetika. 2011;47(12):1616–1621. [PubMed] [Google Scholar]
- 89.Zhang Y., Yang J., Showalter A.M. AtAGP18, a lysine-rich arabinogalactan protein in Arabidopsis thaliana, functions in plant growth and development as a putative co-receptor for signal transduction. Plant Signal. Behav. 2011;6(6):855–857. doi: 10.4161/psb.6.6.15204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kreps J.A., Ponappa T., Dong W., Town C.D. Molecular basis of alpha-methyltryptophan resistance in amt-1, a mutant of Arabidopsis thaliana with altered tryptophan metabolism. Plant Physiol. 1996;110(4):1159–1165. doi: 10.1104/pp.110.4.1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Huang S., Frizzi A., Florida C.A., Kruger D.E., Luethy M.H. High lysine and high tryptophan transgenic maize resulting from the reduction of both 19- and 22-kD alpha-zeins. Plant Mol. Biol. 2006;61(3):525–535. doi: 10.1007/s11103-006-0027-6. [DOI] [PubMed] [Google Scholar]
- 92.Dubouzet J.G., Ishihara A., Matsuda F., Miyagawa H., Iwata H., Wakasa K. Integrated metabolomic and transcriptomic analyses of high-tryptophan rice expressing a mutant anthranilate synthase alpha subunit. J. Exp. Bot. 2007;58(12):3309–3321. doi: 10.1093/jxb/erm179. [DOI] [PubMed] [Google Scholar]
- 93.Sun S.M., Liu Q. Transgenic approaches to improve the nutritional quality of plant proteins. In Vitro Cell. Dev. Biol. Plant. 2004;40:155–162. doi: 10.1079/IVP2003517. [DOI] [Google Scholar]
- 94.Ufaz S., Galili G. Improving the content of essential amino acids in crop plants: goals and opportunities. Plant Physiol. 2008;147(3):954–961. doi: 10.1104/pp.108.118091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.United S. Animal, Plant Health Inspection, S.; Biotechnology, Regulatory, S.; Monsanto, C. USDA/APHIS environmental assessment in response to Monsanto petition 04-229-01P seeking a determination of nonregulated status for lysine maize line LY038, OECD unique identified REN-00038-3. [Animal and Plant Health Inspection Service, U.S. Dept. of Agriculture]: [Riverdale, MD]; 2004. [Google Scholar]
- 96.Zhang F., Voytas D.F. Targeted mutagenesis in Arabidopsis using zinc-finger nucleases. Methods Mol. Biol. 2011;701:167–177. doi: 10.1007/978-1-61737-957-4_9. [DOI] [PubMed] [Google Scholar]
- 97.de Pater S., Pinas J.E., Hooykaas P.J., van der Zaal B.J. ZFN-mediated gene targeting of the Arabidopsis protoporphyrinogen oxidase gene through Agrobacterium-mediated floral dip transformation. Plant Biotechnol. J. 2013;11(4):510–515. doi: 10.1111/pbi.12040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Shukla V.K., Doyon Y., Miller J.C., DeKelver R.C., Moehle E.A., Worden S.E., Mitchell J.C., Arnold N.L., Gopalan S., Meng X., Choi V.M., Rock J.M., Wu Y.Y., Katibah G.E., Zhifang G., McCaskill D., Simpson M.A., Blakeslee B., Greenwalt S.A., Butler H.J., Hinkley S.J., Zhang L., Rebar E.J., Gregory P.D., Urnov F.D. Precise genome modification in the crop species Zea mays using zinc-finger nucleases. Nature. 2009;459(7245):437–441. doi: 10.1038/nature07992. [DOI] [PubMed] [Google Scholar]
- 99.Curtin S.J., Anderson J.E., Starker C.G., Baltes N.J., Mani D., Voytas D.F., Stupar R.M. Targeted mutagenesis for functional analysis of gene duplication in legumes. Methods Mol. Biol. 2013;1069:25–42. doi: 10.1007/978-1-62703-613-9_3. [DOI] [PubMed] [Google Scholar]
- 100.Clasen B.M., Stoddard T.J., Luo S., Demorest Z.L., Li J., Cedrone F., Tibebu R., Davison S., Ray E.E., Daulhac A., Coffman A., Yabandith A., Retterath A., Haun W., Baltes N.J., Mathis L., Voytas D.F., Zhang F. Improving cold storage and processing traits in potato through targeted gene knockout. Plant Biotechnol. J. 2016;14(1):169–176. doi: 10.1111/pbi.12370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Jacobs T.B., LaFayette P.R., Schmitz R.J., Parrott W.A. Targeted genome modifications in soybean with CRISPR/Cas9. BMC Biotechnol. 2015;15:16. doi: 10.1186/s12896-015-0131-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Jiang W., Zhou H., Bi H., Fromm M., Yang B., Weeks D.P. Demonstration of CRISPR/Cas9/sgRNA-mediated targeted gene modification in Arabidopsis, tobacco, sorghum and rice. Nucleic Acids Res. 2013;41(20):e188. doi: 10.1093/nar/gkt780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Jia H., Wang N. Targeted genome editing of sweet orange using Cas9/sgRNA. PLoS One. 2014;9(4):e93806. doi: 10.1371/journal.pone.0093806. [DOI] [PMC free article] [PubMed] [Google Scholar]


