Abstract
Monoclonal antibody (mAb) therapeutics targeting cancer, autoimmune diseases, inflammatory diseases, and infectious diseases are growing exponentially. Although numerous panels of mAbs targeting infectious disease agents have been developed, their progression into clinically useful mAbs is often hindered by the lack of sequence information and/or loss of hybridoma cells that produce them. Here we combine the power of crystallography and mass spectrometry to determine the amino acid sequence and glycosylation modification of the Fab fragment of a potent human astrovirus-neutralizing mAb. We used this information to engineer a recombinant antibody single-chain variable fragment that has the same specificity as the parent monoclonal antibody to bind to the astrovirus capsid protein. This antibody can now potentially be developed as a therapeutic and diagnostic agent.
Keywords: astrovirus, antibody, X-ray crystallography, mass spectrometry, protein sequencing, protein engineering
Human astrovirus (HAstV) is a leading cause of diarrhea in children and immunocompromised individuals but not in healthy adults. Currently no vaccines or antiviral therapies exist for HAstV infections. Several lines of evidence suggest that antibodies produced by the adaptive immune response are key to protection against HAstV infection. First, the rarity of HAstV infection in adults suggests that they have developed a protective adaptive immune response and, in fact, >75% of healthy adults have anti-HAstV antibodies.1 Also, clinical studies with healthy volunteers found that those with more severe HAstV diarrheal disease had no detectable anti-HAstV antibodies.2,3 Finally, immunoglobulin therapy was associated with the recovery of an immune-compromised patient with severe and persistent HAstV infection.4 Together, these data suggest that the development of a therapeutic antibody that neutralizes HAstV could provide a solution to treat and/or prevent HAstV infection.
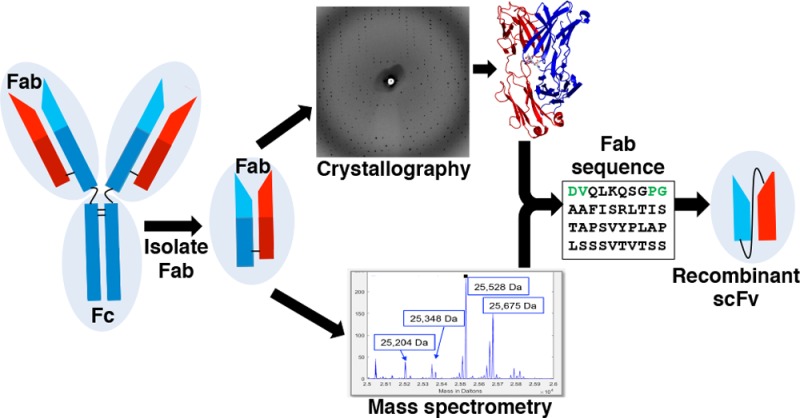
Only two papers published over 20 years ago describe the isolation of monoclonal antibodies (mAbs) that neutralize HAstV in cell culture.5,6 These mouse mAbs bind to the HAstV capsid protein, which forms the T=3 icosahedral shell surrounding the viral RNA genome.5−7 In this study, we investigated one of these mAbs, mAb PL-2, which potently neutralizes HAstV serotype 2 (HAstV-2).5 We obtained limited amounts of mAb PL-2 in ascites fluid; however, the hybridoma cells that produce mAb PL-2 are no longer available. Here, we describe the high-resolution crystal structure and de novo sequencing of the antigen-binding fragment (Fab) of mAb PL-2. Having the Fab PL-2 sequence allowed us to “resurrect” the antibody and engineer a recombinant single-chain variable fragment (scFv) that we find specifically binds to the HAstV-2 capsid protein.
Although de novo sequencing by mass spectrometry and resurrection of antibodies have been reported before,8 the case reported here is remarkable in two ways: (1) prior sequence knowledge—about 95% of the 430 amino acid residues in the Fab PL-2 were already identified or constrained by X-ray crystallographic data; and (2) Fab glycosylation—the Fab PL-2 had an unexpected N-linked glycosylation site. The advanced capabilities of the proteomics search software Byonic (Protein Metrics Inc.) allowed for the rapid amino acid sequence determination of Fab PL-2 and characterization of its glycosylation modifications.
Results
Isolation and Crystallization of Fab PL-2
The HAstV-2-neutralizing mAb PL-2 was purified from mouse ascites fluid using Protein A affinity chromatography. Fab PL-2 fragments were then produced by papain cleavage, followed by Protein A, anion exchange, and size exclusion chromatography purification steps. Fab PL-2 crystallized after 2–4 weeks and formed needle clusters in two different crystallization conditions. Optimization of Fab PL-2 crystals into crystals suitable for X-ray crystallography was initially unsuccessful. We turned to a technique called streak seeding, in which crushed crystal microseeds are used to nucleate new crystallization drops.9 We found that streak-seeding successfully promoted rapid growth of Fab PL-2 crystals within 1–2 days. Surprisingly, we found that single, three-dimensional crystals were obtained only by cross-seeding, whereby microseeds from crystals formed in one crystallization condition are used to nucleate a drop containing a different crystallization condition.10
Crystal Structure Determination of Fab PL-2
Fab PL-2 crystals diffracted to 1.9-Å resolution, and the Fab PL-2 structure was solved by molecular replacement (Figure 1a and Table 1). Although we did not have the sequences of the Fab PL-2 heavy and light chains, the high resolution of our data allowed us to deduce approximately 90% of the amino acids using a combination of electron density maps, neighboring amino acid interactions, and mouse mAb IgG sequence conservation.11 Problematic amino acids were those that had ambiguous electron density and no neighboring interactions with other amino acids. For example, valine and threonine residues had indistinguishable electron density maps (Figure 1b,c). Moreover, amino acid 88 in the heavy chain of Fab PL-2 had perplexing electron density that extended beyond the longest amino acids, and we initially assigned it as lysine (Figure 1d). To obtain the complete sequence of Fab PL-2, we turned to de novo protein sequencing by mass spectrometry (see below). The Fab PL-2 sequence determined by mass spectrometry was used to finalize the structure of Fab PL-2 (Table 1).
Figure 1.
Fab PL-2 structure and amino acid sequence interpretation by X-ray crystallography: (a) Fab PL-2 crystal structure light chain (red) and heavy chain (blue) with the glycan highlighted in gray; (b, c) ambiguous electron density where the amino acid could be interpreted as threonine or valine; (d) lysine fitted in ambiguous electron density before mass spectrometry; (e) fitting of N-acetyl-d-glucosamine (GlcNAc) into electron density from (d) after mass spectrometry. The Fab PL-2 structural model and structure factors have been deposited into the online Protein Data Bank (PDB; www.pdb.org) as PDB entry 5I30.
Table 1. Crystallographic Data.
| data collectiona | |
| crystal | Fab PL-2 |
| wavelength (Å) | 0.97741 |
| space group | C2 2 21 |
| a, b, c (Å) | 72.22, 170.47, 78.27 |
| α, β, γ (deg) | 90.0, 90.0, 90.0 |
| resolution (Å) | 38.79–1.90 (2.00–1.90) |
| Rmerge | 0.112 (0.580) |
| I/σI | 13.20 (3.40) |
| completeness (%) | 98.6 (97.4) |
| redundancy | 6.7 (6.4) |
| refinement | |
| resolution (Å) | 85.24–1.90 |
| no. reflections | 35929 |
| Rwork/Rfreeb | 0.191/0.219 |
| Ramachandran (%) | |
| favored | 98.33 |
| allowed | 1.43 |
| outliers | 0.0 |
| rms deviations | |
| bond lengths (Å) | 0.006 |
| bond angles (deg) | 0.929 |
Data were collected from a single crystal. Values for the highest resolution shell are shown in parentheses.
Rfree was calculated using 5% of the reflections.
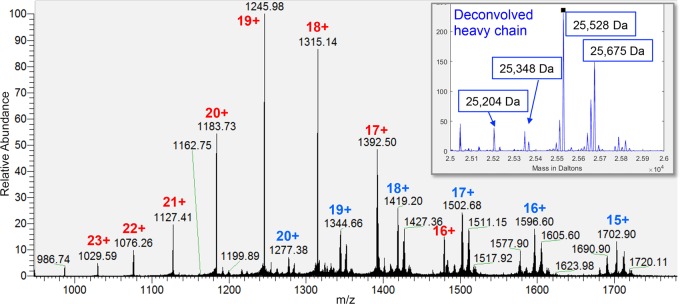
Intact Mass of Fab PL-2
We determined the masses of the full Fab PL-2 and of the Fab PL-2 heavy and light chains from mass spectra of intact and reduced Fab PL-2. We achieved isotope resolution on the Fab PL-2 heavy chain and light chain (Figure 2), but not on the full Fab PL-2, although we identified multiple masses for the intact Fab. The Fab PL-2 heavy chain showed a number of mass variants in both the MS scans and the deconvolved spectra (Figure 2, inset). The mass deltas between variants could be explained by either glycosylation or N- or C-terminal extensions due to inexact cleavage by papain. For example, the delta of ∼147 Da between the most abundant variants could be either fucose or phenylalanine.
Figure 2.
Intact mass analysis of reduced Fab PL-2: MS spectrum of Fab PL-2 light (red) and heavy chain (blue). Deconvolution of the charge states yields intact reduced masses of 23 654 Da for the light chain and 25 528 Da for the heavy chain. The heavy chain gives a series of peaks with mass deltas suggestive of glycosylation; for example, the difference between the two largest peaks (147 Da) is close to the mass of fucose (inset).
De Novo Sequencing of Fab PL-2
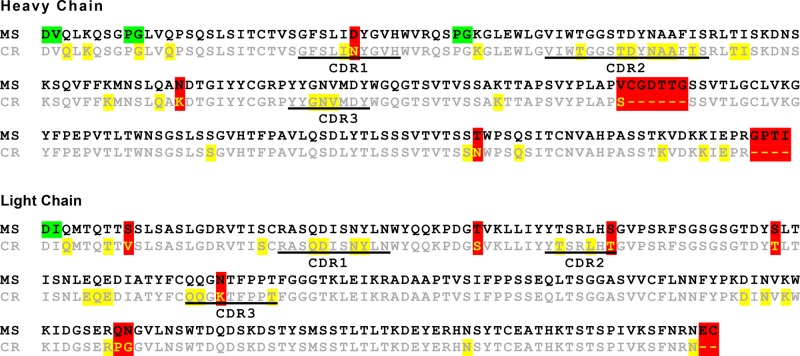
We used four different protease digestions and high-resolution tandem mass spectrometry to obtain the full sequence of Fab PL-2. De novo sequencing was accomplished semiautomatically using Byonic (Protein Metrics Inc.) by iterative error-tolerant “wild card” (mass deviation allowed on any amino acid) searches against the current best protein sequence and manual improvement of the sequence to explain wild card matches. A final gap in the sequence coverage revolved around peptides that contained the Fab PL-2 heavy chain amino acid 88, which was also difficult to identify by electron density maps and was initially assigned as a lysine. In an effort to fill in the missing sequence, the maximum wild card mass was increased from 130 Da, sufficient for single amino acid substitutions, to 500 Da. This search found several wild card matches to the missing stretch with clear peaks at 204.087, the oxonium ion for HexNAc, and wild card masses of 189.026 and 335.083, which are explainable by a single lysine to asparagine substitution (mass delta, −14.052) and a truncated N-glycan, either HexNAc (203.079) or HexNAc-Fuc (349.137). Although the intact mass spectra had given hints that the Fab heavy chain might include glycosylation, the Fab PL-2 glycosylation was initially identified serendipitously, by observation of the characteristic monosaccharide HexNAc oxonium ion in a spectrum with a wild card match to the sequence gap (Figure 3). The discovery of a glycosylated asparagine provided a clear explanation for the electron density map at amino acid 88 (Figure 1e). Merging sequence information from crystallography and mass spectrometry allowed for the determination of the Fab PL-2 sequence with no ambiguities (Figure 4).
Figure 3.
De novo sequencing via wild card search. Byonic matched the pictured mass spectrum to MNSLQAK{+189.026}DTGIYYC[+57.021]GR, where { } indicates a wild card (unidentified) mass delta and [ ] indicates a known modification, in this case cysteine alkylation. The peak at 204.086 is the oxonium ion for HexNAc, the nearly universal mark of a glycopeptide. Manual analysis quickly corrected the peptide to MNSLQAN[+203.079]DTGIYYC[+57.021]GR, explaining the peaks at 1218.525, 1289.556, and 1418.607 as ∼y10, ∼y11, and ∼y12 (iso1) with ∼ indicating loss of the glycan.
Figure 4.
Alignment of Fab PL-2 sequences determined by X-ray crystallography (CR) and mass spectrometry (MS). Highlighted in yellow are amino acids whose identities were ambiguous in electron density maps. Highlighted in green are amino acids whose order was ambiguous in mass spectrometry data due to an absence of fragmentation. Highlighted in red are amino acids that were incorrect or absent from the sequence determined by X-ray crystallography but were corrected after mass spectrometry.
Fab PL-2 Glycosylation Modification Determination
Once glycosylation was recognized, the Byonic glycopeptide search found approximately 50 other glycoforms of the same peptide, using a glycopeptide search with a corrected FASTA protein sequence and Byonic’s predefined glycan database of 309 mammalian N-glycan compositions. Some glycans are as large as HexNAc(5)Hex(9)Fuc(1) at 2620 Da, which most likely represents a core-fucosylated glycan with three antennae, each terminating in Gal-α-Gal. Other glycan compositions, along with manual inspection of characteristic spectrum peaks, indicate other glycan motifs not usually found in Fc glycosylation, such as antennal fucosylation and sialylation with either or both NeuAc and NeuGc. Our results are in agreement with previous reports that glycans on the Fab can be larger and more varied than those on the Fc.12 The distance between Asn87 and the closest CDR loop is ∼20 Å, suggesting that the glycan is unlikely to be involved in antigen binding. More likely, the glycan is involved in Fab stability and solubility.17
Engineering of scFv PL-2
Having the Fab PL-2 heavy chain and light chain amino acid sequences allowed us to engineer a recombinant antibody construct comprising a single-chain variable fragment (scFv) of the PL-2 antibody. Our recombinant scFv PL-2 construct is a fusion protein of the variable regions of the heavy and light chains, connected by a flexible 20 amino acid linker. We chose to express the scFv using the Drosophila S2 cell system because these cells can glycosylate protein and secrete the recombinant scFv into the growth media.13 Recombinant scFv PL-2 was purified by Strep-Tactin affinity chromatography followed by size exclusion chromatography to isolate the purified, monomeric form of the scFv (Figure 5a,b). Overall yields are ∼20 mg of scFv PL-2 per liter of S2 cells.
Figure 5.
Detection of HAstV-2 capsid protein by scFv PL-2. (a) Reducing SDS-PAGE analysis of purified proteins. Lanes: M, molecular weight marker; 1, mAb PL-2; 2, scFv PL-2; 3, negative control mAb NegC. (b) Anti-Strep-tag Western blot detection of scFv PL-2, which contains a Strep-tag (lane 4). (c) SDS-PAGE (left) and anti-His-tag Western blot (right) analyses of wheat germ extracts containing recombinant HAstV-2 capsid protein (C) or wheat germ extract alone (−). (d) SDS-PAGE (left) and anti-His-tag Western blot (right) analyses of immunoprecipitation experiments using scFv PL-2 and wheat germ extracts containing recombinant HAstV-2 capsid protein (C) or wheat germ extract alone (−). (e) ELISA detection of antibody binding to HAstV-2 capsid protein. Wells were coated with wheat germ extracts containing recombinant HAstV-2 capsid protein (+ Capsid) or wheat germ extract alone (− Capsid). Binding was detected by a HRP-conjugated goat anti-mouse IgG secondary antibody (for full-length mAbs) or HRP-conjugated Strep-Tactin (for scFv PL-2). Experiments with mAb PL-2 and scFv PL-2 were performed in triplicate. Due to limited amounts of wheat germ extract samples, the negative control experiments with mAb NegC or no primary antibody were performed in duplicate. Error bars represent the standard deviation.
Antigen Recognition by mAb PL-2 and scFv PL-2
mAb PL-2 binds to the surface of the HAstV-2 virion, which is formed by the virus capsid protein.5 To determine if recombinant scFv PL-2 retains the ability to bind to the HAstV-2 capsid protein, we first used a wheat germ cell-free protein synthesis system to express the full-length HAstV-2 capsid protein.14 Recombinant HAstV-2 capsid protein containing a C-terminal 10-histidine tag (∼90 kDa) was expressed and remained in the soluble fraction of the wheat germ extracts (Figure 5c). Unfortunately, we were unable to purify the recombinant HAstV-2 capsid protein in sufficient amounts for antibody-binding studies. Instead, the binding studies described below were performed with wheat germ extract containing recombinant HAstV-2 capsid protein.
To test for scFv PL-2 binding to the HAstV-2 capsid protein, we first performed an immunoprecipitation experiment using scFv PL-2 immobilized on Strep-Tactin beads (Figure 5d). The scFv fragment was able to associate with recombinant HAstV-2 capsid protein and pull it out of the wheat germ extract. Although the amount of capsid protein was too low to detect by Coomassie-stained SDS-PAGE, an anti-histidine-tag Western blot detected the presence of the His-tagged HAstV-2 capsid protein. As a negative control, we performed the immunoprecipitation experiment with wheat germ extract alone (no HAstV-2 capsid protein), and no His-tagged proteins were immunoprecipitated (Figure 5d).
To further validate the binding of scFv PL-2 to the HAstV-2 capsid protein, we performed an enzyme-linked immunosorbent assay (ELISA) (Figure 5e). ELISA plates were coated with wheat germ extract containing recombinant HAstV-2 capsid protein (+ Capsid) or wheat germ extract alone (− Capsid), and binding by mAb PL-2, a negative control mAb NegC, or scFv PL-2 was determined. Our experiments reveal that both mAb PL-2 and scFv PL-2 bind to wheat germ extract containing recombinant HAstV-2 capsid protein, and no binding was observed to wheat germ extract alone. Furthermore, no binding was observed by negative control mAb NegC. Together, these data suggest that recombinant scFv PL-2 has the same binding specificity as mAb PL-2.
Discussion
In this study, we sought to resurrect the HAstV-neutralizing mAb PL-2, whose amino acid sequence was unknown. Unfortunately, as is often the case for mouse mAbs produced decades ago, the hybridoma cells that produce mAb PL-2 were no longer available for sequencing of antibody heavy and light chain mRNA transcripts. However, several milliliters of mAb PL-2 in ascites fluid still existed for further studies. Here, we determined the Fab PL-2 amino acid sequence and glycosylation modification by combining X-ray crystallography and mass spectrometry. Having the Fab PL-2 amino acid sequence allowed us to produce recombinant scFv PL-2 that retained specificity for binding to its viral antigen, the HAstV-2 capsid protein. Our ability to now produce recombinant forms of mAb PL-2 in endless supplies allows for further characterization of this antibody’s binding-site epitope and mechanism of HAstV neutralization. Having the mAb PL-2 sequence also allows for its humanization and development into a therapeutic antibody for the prevention or treatment of HAstV infection.
Our studies highlight the recent advancements in de novo protein sequencing by mass spectrometry. Advances in mass spectrometer instrumentation enable ultrahigh-resolution studies on challenging, low-abundance, and high-complexity samples. Equally significant are the advances in mass spectrometry search algorithms that facilitate rapid de novo protein sequencing, allowing it to become a routine application. In this study, we needed only a few days to analyze mass spectrometry data and determine the Fab PL-2 sequence; however, we concede that the high degree of prior sequence knowledge by X-ray crystallography facilitated the de novo sequencing by Byonic’s wild card search. Nevertheless, in subsequent studies of other antibodies, we have found that it is quite normal to have >90% identity between an unknown Fab and the closest Fab sequence in GenBank.
An unexpected observation during the course of this study was the identification of a glycosylation site in Fab PL-2 at an asparagine in the sequence QANDT between CDRs H2 and H3. The Byonic glycopeptide search found approximately ∼50 glycoforms at this site. In subsequent studies of other antibodies, we have observed Fab glycosylation at this same site. Retrospective analyses reveal that ∼20% of mouse and human mAbs have Fab glycosylation. The germline sequence is QADDT, and because D → N is one of the most common mutations, this N-glycosylation site is fairly likely to arise during somatic hypermutation. Little is known about the role of Fab glycosylation in antigen binding and autoimmune disease. Recent studies have shown that Fab glycosylation varies in a similar way as Fc glycosylation (more sialylation, less bisecting GlcNAc) during pregnancy, when autoimmune diseases generally go into remission.15 Furthermore, Fab glycosylation has been shown to react to certain leukocytes and affect local immune responses, such as those in the placenta.16 Only a few therapeutic mAbs include Fab glycosylation; one such example is cetuximab, a therapeutic mAb with Fab glycosylation at the homologous sequence QSNDT. Interestingly, engineering of a Fab glycosylation site into the therapeutic antibody bevacizumab has been used to mask hydrophobic amino acids, thereby increasing the antibody stability and decreasing its aggregation.17
Overall, we hope that our studies encourage other infectious disease researchers to characterize mAbs that have been isolated in their laboratories. With advancements in de novo protein sequencing and recombinant antibody production and engineering, these mAbs can now be developed into clinically useful diagnostic antibodies and antimicrobial therapeutic antibodies.
Methods
mAb PL-2 Purification
mAb PL-2 was purified from mouse ascites fluid with Protein A Plus columns (Thermo Scientific). mAb PL-2 was eluted with IgG elution buffer (pH 2.8; amine-based), and elution was neutralized with 1 M Tris, pH 8. mAb PL-2 was dialyzed into phosphate-buffered saline, pH 7.4 (PBS).
Fab PL-2 Production and Purification
mAb PL-2 Fab fragments (Fab PL-2) were generated by incubation of purified mAb PL-2 with immobilized papain overnight at 37 °C. Antibody Fc fragments were removed with Protein A Plus columns, and the flow-through containing the Fab PL-2 fragments was collected. Fab PL-2 was then dialyzed into 20 mM Tris, pH 8.5, and 25 mM NaCl and further purified by anion exchange chromatography on a HiTrap Q FF column (GE Healthcare) with a gradient elution with 20 mM Tris, pH 8.5, and 1000 mM NaCl. Fab PL-2 was further purified by size exclusion chromatography on a Superdex 200 column in PBS.
Fab PL-2 Structure Determination
Purified Fab PL-2 was dialyzed into 10 mM Tris, pH 8.0, and 150 mM NaCl and concentrated to 18.1 mg/mL. A crystal grew by sitting drop vapor diffusion with a well solution of 25% PEG 3350 and 0.1 M citric acid, pH 4.5. This crystal was used for self-seeding into a pre-equilibrated hanging drop with a well solution of 32% PEG 3350 and 0.1 M citric acid, pH 4.5. Needle-like crystals grown in this condition were used for cross-seeding into a pre-equilibrated hanging drop with a well solution of 15% PEG 3350 and 0.2 M ammonium phosphate monobasic, pH 4.6. Crystals were transferred into a cryoprotectant of 30.8% PEG 3350, 0.2 M magnesium acetate, and 30% ethylene glycol and then flash frozen in liquid nitrogen. Diffraction data were collected at cryogenic temperature at the Advanced Light Source Beamline 5.0.1. Diffraction data were processed with iMosflm18 (Table 1). The structure was solved by molecular replacement using the Fab fragment from the influenza virus N9 neuraminidase–NC41 Fab complex structure (PDB ID 1NCA)19 and the program PHASER.20 The Fab PL-2 structure was refined and manually rebuilt using PHENIX21 and Coot,22 respectively. The initial Fab PL-2 sequence was determined by assessment of the electron density maps of each amino acid side chain. Observations of hydrogen bond or van der Waals interactions aided in distinguishing between amino acids with similar electron density maps. If an amino acid could not be determined using the electron density alone, the consensus amino acid from an alignment of 29 different Fab–antigen complex crystal structures was used to predict the initial Fab PL-2 sequence.11 This preliminary Fab PL-2 sequence was used as a starting point for sequence determination by mass spectrometry. The Fab PL-2 structure was corrected and finalized using the sequence determined by mass spectrometry. The Fab PL-2 structural model and structure factors have been deposited into the online Protein Data Bank (PDB; www.pdb.org) as PDB entry 5I30.
Sample Preparation for Mass Spectrometry
Purified Fab PL-2 in PBS at 0.5 mg/mL in a total of 0.5 mL was used for mass spectrometry. Ten micrograms were buffer exchanged into 0.5% acetic acid for intact mass analysis using 7 kDa MWCO Zeba columns (Pierce). Four micrograms were reduced by adding TCEP in 0.5% acetic acid to a final concentration of 20 mM. The reaction was allowed to proceed at room temperature for 1 h, and the sample was desalted using POROS R2 50 beads. For protease digests, 40 μg of Fab PL-2 were buffer exchanged into 100 mM ammonium bicarbonate using 7 kDa MWCO Zeba columns. The sample was reduced in DTT (10 mM final concentration) for 1 h at 57 °C followed by alkylation with iodoacetamide (20 mM final concentration) for 45 min at room temperature in the dark. The sample was divided into four aliquots of 10 μg and digested overnight at room temperature with trypsin, with Glu-C and Lys-C (Promega) overnight, or with chymotrypsin (Promega) for 4 h at room temperature. Samples were acidified with 0.2% TFA/5% formic acid and desalted using POROS R2 50 beads.
Mass Spectrometry
Intact Fab
Fab PL-2 (0.1 μg) was loaded onto a ProSwift RP-4H monolithic column (0.1 × 250 mm) mounted on an EASY nLC-1000 nanoUHPLC coupled to a Thermo Scientific Orbitrap Fusion mass spectrometer. The Fab was eluted off the column using a gradient of 20–100% B (90% acetonitrile, 0.5% acetic acid) over 50 min. MS1 data were acquired by alternating between ion trap scans and Orbitrap scans, with the HCD cell set to intact protein mode (0.003 bar); ion trap scans were performed at m/z 1000–2200 using quadrupole isolation. Maximum injection time was set to 35 ms with an AGC target of 3 × 105 summing 20 microscans. The S-Lens RF was set to 100 with ISD set to 5 eV. Settings for the Orbitrap scan were identical, except the resolution was set to 15 000 and the AGC target was set to 5 × 105.
Reduced Fab
Reduced Fab PL-2 (0.1 μg) was loaded onto a ProSwift RP-4H monolithic column (0.1 × 250 mm), mounted on an EASY nLC-1000 nanoUHPLC coupled to an Orbitrap Fusion mass spectrometer. The Fab was eluted off the column using a gradient of 0–100% B (90% acetonitrile, 0.5% acetic acid) over 50 min. Data were acquired at full MS in the Orbitrap, with an HCD cell set to intact protein mode (0.003 bar). Scans were acquired from m/z 600 to 1800 with 450 000 resolution using quadrupole isolation. Maximum injection time was set to 35 ms with an AGC target of 5 × 105 summing 10 microscans. The S-Lens RF was set to 80 with ISD set to 5 eV.
Digested Fab
Each digest (0.2 μg) was loaded onto an EASY-Spray C18 Pepmap RSLC column (500 × 0.075 mm) mounted on an EASY nLC-1000 nanoUHPLC coupled to a Q-Exactive mass spectrometer. The sample was eluted off the column using a gradient of 0–30% B (90% acetonitrile, 0.5% acetic acid) over 90 min. MS1 data were acquired from m/z 400 to 1500, 70 000 resolution, and one microscan. Data-dependent MS2 scans were performed on the top 20 precursors with dynamic exclusion activated for 30 s. Precursors were isolated using a m/z 2 window and HCD fragmented at 27% of normalized collision energy (NCE). Underfill ratio was set to 5%, with a minimum intensity of 2 × 104. Precursors with charges of 1 and 5–8 were excluded from fragmentation, as well as charge unassigned precursors. First mass at MS2 was fixed at m/z 150.
Fab PL-2 Amino Acid Sequence Determination
Unknown amino acids were determined by iteratively searching the spectra against the current best protein sequence (FASTA format) using Byonic v.2.6 (Protein Metrics Inc.). Byonic searches used mass tolerances of 10 ppm for precursors and 20 ppm for fragments and digestion specificities appropriate for the protease, for example, semispecific cutting after Arg and Lys for the trypsin digest and semispecific cutting after Trp, Tyr, Phe, Met, and Leu for chymotrypsin. Initial searches allowed a wild card modification (any mass delta from −130 to +130 Da on any one amino acid residue), along with a few of the most common known modifications, namely, oxidized methionine, deamidated asparagine, pyro-glu N-terminal Glu and Gln, and overalkylated His, Lys, and N-terminus. Recognizable wild cards were manually converted to known variants, usually single amino acid substitutions, by editing the FASTA, and spectra were then searched against the improved FASTA database. After about five iterations, the complete Fab PL-2 light chain sequence was determined, as well as the complete heavy chain sequence, except for a stretch of ∼10 residues between the heavy chain CDR2 and CDR3, around the N-linked glycosylation site, as described above.
Production of Recombinant scFv PL-2
The pMT-BiP-scFv-PL-2 expression plasmid was constructed on the basis of previous studies.13 Briefly, a synthetic gene codon-optimized for Drosophila melanogaster (Integrated DNA Technologies) containing the Fab PL-2 heavy chain residues Asp1– Ser117, a Kpnl restriction site, a GGS(GGGGS)2GGG linker, a Nhel restriction site, and the Fab PL-2 light chain residues Asp1–Arg108, was cloned into a pMT-BiP vector between the BglII and Sacll restriction sites in frame with an N-terminal BiP signal sequence and a C-terminal thrombin protease cleavage site followed by a Twin Strep-tag. The plasmid contains a metallothionein promoter for induction of gene expression. The resulting pMT-BiP-scFv-PL-2 expression plasmid along with a hydrogmycin resistance plasmid (pCoHygro) was used to obtain stably transfected D. melanogaster Schneider 2 (S2) cells. The S2 cells were grown in shaker flasks to 4.2 × 106 cells/mL in Insect-XPRESS medium containing 1× Pen/Strep and 400 μg/mL hygromycin, and expression of scFv PL-2 was induced with 500 μM cupric chloride. After 5 days, cells were pelleted and media containing secreted scFv PL-2 was 0.22 μM filtered and 100-fold concentrated by tangential flow filtration. The medium sample was supplemented with 80 mM Tris pH 8.0 and BioLock (IBA) to mask biotin in the media and 0.22uM filtered again. scFv PL-2 was affinity purified on a StrepTrap column followed by size exclusion chromatography using a Superdex200 column in PBS. Final yields of purified protein were ∼20 mg/L.
Production of Recombinant HAstV-2 Capsid Protein
A synthetic gene codon-optimized for Spodoptera frugiperda (Genewiz) encoding the HAstV-2 capsid protein (UniProt accession no.Q82446) in-frame with a C-terminal thrombin protease cleavage site and a 10× His-tag was cloned into the pEU vector between the KpnI and Notl restriction sites. The resulting pEU-HAstV-2-capsid expression plasmid was isolated following instructions from a DNA purification kit (Clontech Laboratories, Inc.), yet excluding the RNase from the resuspension buffer A1. To separate the DNA plasmid from bacterial RNA contaminants, an RNA and DNA precipitation protocol was adapted from an online protocol.23 Briefly, the sample was diluted with RNase-free water to 90 μLn and 30 μL of 8 M LiCl was added. Sample was incubated for 30 min at −20 °C and spun at 4 °C at 15000g for 15 min. The supernatant was isolated, and 80 μL of isopropanol was added. The sample was incubated for 30 min at −20 °C and spun at 4 °C at 15000g for 15 min. The supernatant was discarded, and the pellet was carefully washed with 100 μL of 70% ethanol. The sample was spun at 4 °C at 15000g for 5 min. The supernatant was discarded, and the pellet was dried at 50 °C for 15 min. DNA plasmid in the pellet was resuspended with 8 μL of 5 mM Tris, pH 8.5. In vitro transcription and translation of recombinant HAstV-2 capsid protein were performed with a wheat germ cell-free protein synthesis system (WEPRO7240 Expression kit, CellFree Sciences). Transcription was carried out at 37 °C for 6 h with a 20 μL reaction containing 2 μg of pEU-HAstV-2-capsid expression plasmid, 1× transcription buffer, 25 mM NTP, 20 U of RNase inhibitor, and 20 U of SP6 RNA polymerase. The translation mixture containing 10 μL of transcribed mRNA, 0.8 μL of creatine kinase at a concentration of 1 mg/mL, and 10 μL of the WEPRO wheat germ extract was incubated under 206 μL of 1× SUB_AMIX. Translation was allowed to proceed for 20 h at 15 °C. Protein expression was evaluated by using RFP as a positive control. The wheat germ extract reaction was mixed by pipetting and then spun down at 14000g for 10 min. The insoluble (pellet) and soluble (supernatant) fractions were analyzed by reducing SDS-PAGE and Western blot with a HRP-conjugated anti-His-tag antibody.
HAstV-2 Capsid Immunoprecipitation Using scFv PL-2-Coated Beads
One hundred microliters of Strep-Tactin Sepharose resin (IBA) was incubated at 4 °C for 15 min with 0.6 mg of purified scFv PL-2 in PBS. Beads were washed twice with 500 μL of buffer W (100 mM Tris, 150 mM NaCl, 1 mM EDTA, pH8). Beads were then incubated at 4 °C for 15 min with 100 μL of soluble fraction of wheat germ extract containing recombinant HAstV-2 capsid protein (+ Capsid) or 100 μL wheat germ extract alone (− Capsid) containing all of the components for translation but no transcribed mRNA. Beads were washed twice with 500 μL of buffer W, and bound protein was eluted with 40 μL of buffer E (buffer W containing 2.5 mM d-desthibiotin).
Enzyme-Linked Immunosorbant Assay (ELISA)
Fifty microliters of soluble fraction of wheat germ extract containing recombinant HAstV-2 capsid protein (+ Capsid) or 50 μL of wheat germ extract alone (− Capsid) was diluted with 100 μL of PBS and incubated overnight at room temperature in a 96-well microtiter plate. The plate was then washed four times with 1× PBS containing 0.05% Tween 20 (PBST). Wells were blocked by adding 150 μL of 5% BSA in PBS and incubated for 1 h, followed by four PBST washes. Wells were then incubated with 150 μL of primary antibody at room temperature for 1 h. Full-length antibodies mAb PL-2 and the negative control IgG1 mAb (mAb NegC) that recognizes the V1 V2 domain in the HIV envelope glycoprotein were incubated at 5 μg/mL in 1% BSA in PBS (blocking buffer). The scFv PL-2 was incubated at 20 μg/mL in blocking buffer. Plates were washed four times with PBST and then incubated for 1 h at room temperature with HRP-conjugated goat anti-mouse IgG secondary antibody (for full-length mAbs) or HRP-conjugated Strep-Tactin (for scFv), each diluted 1:5000 in blocking buffer. Plates were washed four times with PBST and then developed by adding peroxidase substrate o-phenylenediamine dihydrochloride (OPD) in phosphate–citrate buffer and 1.5% hydrogen peroxide for 5 min at room temperature. The reactions were stopped with 2 N sulfuric acid, and the absorbance was measured at 490 nm.
Acknowledgments
This work was supported by NIH Grant K22 AI095369 to R.M.D. This work was also supported by NIH Grant R42 GM103362 to Dr. David Fenyő. The mass spectrometer used in this study was purchased with funds from NIH/ORIP Grant S10OD010582 to B.M.U. Diffraction data on Fab PL-2 crystals were collected at beamline 5.0.1 of the Advanced Light Source, which is supported by the Director, Office of Science, Office of Basic Energy Sciences, U.S. Department of Energy, under Contract DE-AC02-05CH11231. We thank Dr. Roger Glass for his contributions in the original isolation of mAb PL-2. We also thank Dr. Phillip Berman for sharing his ELISA equipment.
Glossary
Abbreviations
- HAstV
human astrovirus
- mAb
monoclonal antibody
- Fab
fragment antigen binding
- scFv
single-chain fragment variable
- PBS
phosphate-buffered saline
Author Contributions
W.A.B. performed crystallographic and protein-binding experiments and co-wrote the manuscript. D.M. analyzed mass spectrometry data. M.B. analyzed mass spectrometry data and co-wrote the manuscript. B.M.U. performed mass spectrometry experiments. A.S. contributed the unique reagent mAb PL-2. R.M.D. analyzed crystallographic data and co-wrote the manuscript.
The authors declare no competing financial interest.
References
- Kurtz J.; Lee T. (1978) Astrovirus gastroenteritis age distribution of antibody. Med. Microbiol. Immunol. 166, 227–230. 10.1007/BF02121154. [DOI] [PubMed] [Google Scholar]
- Kurtz J. B.; Lee T. W.; Craig J. W.; Reed S. E. (1979) Astrovirus infection in volunteers. J. Med. Virol. 3, 221–230. 10.1002/jmv.1890030308. [DOI] [PubMed] [Google Scholar]
- Mitchell D. K. (2002) Astrovirus gastroenteritis. Pediatr. Infectious Dis. J. 21, 1067–1069. 10.1097/00006454-200211000-00018. [DOI] [PubMed] [Google Scholar]
- Bjorkholm M.; Celsing F.; Runarsson G.; Waldenstrom J. (1995) Successful intravenous immunoglobulin therapy for severe and persistent astrovirus gastroenteritis after fludarabine treatment in a patient with Waldenstrom’s macroglobulinemia. Int. J. Hematol. 62, 117–120. 10.1016/0925-5710(95)00396-A. [DOI] [PubMed] [Google Scholar]
- Sanchez-Fauquier A.; Carrascosa A. L.; Carrascosa J. L.; Otero A.; Glass R. I.; Lopez J. A.; San Martin C.; Melero J. A. (1994) Characterization of a human astrovirus serotype 2 structural protein (VP26) that contains an epitope involved in virus neutralization. Virology 201, 312–320. 10.1006/viro.1994.1296. [DOI] [PubMed] [Google Scholar]
- Bass D. M.; Upadhyayula U. (1997) Characterization of human serotype 1 astrovirus-neutralizing epitopes. J. Virol. 71, 8666–8671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dryden K. A.; Tihova M.; Nowotny N.; Matsui S. M.; Mendez E.; Yeager M. (2012) Immature and mature human astrovirus: structure, conformational changes, and similarities to hepatitis E virus. J. Mol. Biol. 422, 650–658. 10.1016/j.jmb.2012.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castellana N. E.; McCutcheon K.; Pham V. C.; Harden K.; Nguyen A.; Young J.; Adams C.; Schroeder K.; Arnott D.; Bafna V.; Grogan J. L.; Lill J. R. (2011) Resurrection of a clinical antibody: template proteogenomic de novo proteomic sequencing and reverse engineering of an anti-lymphotoxin-alpha antibody. Proteomics 11, 395–405. 10.1002/pmic.201000487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira-Rodriguez A.; Fernandez-Leiro R.; Gonzalez Siso M. I.; Cerdan M. E.; Becerra M.; Sanz-Aparicio J. (2010) Crystallization and preliminary X-ray crystallographic analysis of beta-galactosidase from Kluyveromyces lactis. Acta Crystallogr., Sect. F: Struct. Biol. Cryst. Commun. 66, 297–300. 10.1107/S1744309109054931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obmolova G.; Malia T. J.; Teplyakov A.; Sweet R. W.; Gilliland G. L. (2014) Protein crystallization with microseed matrix screening: application to human germline antibody Fabs. Acta Crystallogr., Sect. F: Struct. Biol. Commun. 70, 1107–1115. 10.1107/S2053230X14012552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X.; Singh S. K.; Kumar S. (2010) Potential aggregation-prone regions in complementarity-determining regions of antibodies and their contribution towards antigen recognition: a computational analysis. Pharm. Res. 27, 1512–1529. 10.1007/s11095-010-0143-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abès R.; Teillaud J.-L. (2010) Impact of glycosylation on effector functions of therapeutic IgG. Pharmaceuticals 3, 146. 10.3390/ph3010146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilmartin A. A.; Lamp B.; Rumenapf T.; Persson M. A.; Rey F. A.; Krey T. (2012) High-level secretion of recombinant monomeric murine and human single-chain Fv antibodies from Drosophila S2 cells. Protein Eng., Des. Sel. 25, 59–66. 10.1093/protein/gzr058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takai K.; Sawasaki T.; Endo Y. (2010) Practical cell-free protein synthesis system using purified wheat embryos. Nat. Protoc. 5, 227–238. 10.1038/nprot.2009.207. [DOI] [PubMed] [Google Scholar]
- Bondt A.; Rombouts Y.; Selman M. H.; Hensbergen P. J.; Reiding K. R.; Hazes J. M.; Dolhain R. J.; Wuhrer M. (2014) Immunoglobulin G (IgG) Fab glycosylation analysis using a new mass spectrometric high-throughput profiling method reveals pregnancy-associated changes. Mol. Cell. Proteomics 13, 3029–3039. 10.1074/mcp.M114.039537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu J.; Lei Y.; Huang Y.; Zhao Y.; Li J.; Huang T.; Zhang J.; Wang J.; Deng X.; Chen Z.; Korteweg C.; Deng R.; Yan M.; Xu Q.; Dong S.; Cai M.; Luo L.; Huang G.; Wang Y.; Li Q.; Lin C.; Su M.; Yang C.; Zhuang Z. (2015) Fab fragment glycosylated IgG may play a central role in placental immune evasion. Hum. Reprod. 30, 380–391. 10.1093/humrep/deu323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courtois F.; Agrawal N. J.; Lauer T. M.; Trout B. L. (2016) Rational design of therapeutic mAbs against aggregation through protein engineering and incorporation of glycosylation motifs applied to bevacizumab. mAbs 8, 99–112. 10.1080/19420862.2015.1112477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Battye T. G.; Kontogiannis L.; Johnson O.; Powell H. R.; Leslie A. G. (2011) iMOSFLM: a new graphical interface for diffraction-image processing with MOSFLM. Acta Crystallogr., Sect. D: Biol. Crystallogr. 67, 271–281. 10.1107/S0907444910048675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tulip W. R.; Varghese J. N.; Laver W. G.; Webster R. G.; Colman P. M. (1992) Refined crystal structure of the influenza virus N9 neuraminidase-NC41 Fab complex. J. Mol. Biol. 227, 122–148. 10.1016/0022-2836(92)90687-F. [DOI] [PubMed] [Google Scholar]
- McCoy A. J.; Grosse-Kunstleve R. W.; Adams P. D.; Winn M. D.; Storoni L. C.; Read R. J. (2007) Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674. 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams P. D.; Afonine P. V.; Bunkoczi G.; Chen V. B.; Davis I. W.; Echols N.; Headd J. J.; Hung L. W.; Kapral G. J.; Grosse-Kunstleve R. W.; McCoy A. J.; Moriarty N. W.; Oeffner R.; Read R. J.; Richardson D. C.; Richardson J. S.; Terwilliger T. C.; Zwart P. H. (2010) PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr., Sect. D: Biol. Crystallogr. 66, 213–221. 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P.; Cowtan K. (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr., Sect. D: Biol. Crystallogr. 60, 2126–2132. 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- See http://www.molbi.de/protocols/separate_rna_and_dna_v1_0.htm.