Abstract
Background
Mycobacterium abscessus complex (MABC) is the most drug resistant of the mycobacterial pathogens. M. abscessus subsp. abscessus encodes a functional erythromycin ribosomal methylase gene, erm(41), causing inducible macrolide resistance. However, some clinical isolates of M. abscessus subsp. abscessus harboring nonfunctional erm(41) were susceptible to macrolide, even after extended incubation of 14 days. Loss of function of the erm(41) genes was associated with a T-to-C substitution at position 28 of the gene (T28C), leading to an amino acid change from Trp to Arg at codon 10. Pulmonary disease caused by M. abscessus subsp. abscessus strains with an nonfunctional erm(41) (C28 sequevar) may be responsive to macrolide-containing antibiotic regimens. Therefore, all M. abscessus subsp. abscessus strains with a functional erm(41) (T28 sequevar) were thought to be resistant to macrolide with extended incubation. Here, we report the first case of pulmonary disease caused by a strain of M. abscessus subsp. abscessus which was susceptible to macrolide due to T19 sequevar of erm(41) gene.
Case presentation
A 62-year-old Korean female was referred to our hospital due to chronic cough, sputum, and hemoptysis lasting more than 5 months. The patient’s sputum was positive for acid-fast bacilli staining and nontuberculous mycobacteria (NTM) were isolated twice from sputum specimens. The isolate was identified as M. abscessus subsp. abscessus. The isolate had a point mutation of C → T at position 19 (C19 → T) in the erm(41) gene, instead of expected C28 sequevar of erm(41), and had no rrl mutation. The isolate displayed a clarithromycin susceptible phenotype with an Arg → Stop codon change in erm(41). The patient was successfully treated with a macrolide-containing regimen.
Conclusion
This is the first case of pulmonary disease caused by a strain of M. abscessus subsp. abscessus showing clarithromycin susceptible phenotype due to T19 sequevar of the erm(41) gene. The erm(41) gene is clinically important, and non-functional erm alleles may be an important issue for the management of MABC lung disease. The presence of a non-functional erm(41) allele in M. abscessus subsp. abscessus isolates may be associated with better outcomes.
Keywords: Nontuberculous mycobacteria, Mycobacterium abscessus, Lung diseases, Clarithromycin, Drug resistance
Background
The prevalence of lung diseases caused by nontuberculous mycobacteria (NTM) is increasing worldwide [1, 2]. Mycobacterium abscessus complex (MABC) is a rapidly growing mycobacterium and is the second most common cause of NTM lung disease after M. avium complex in many countries [3–5]. In addition, MABC has emerged as an important pathogen in patients with cystic fibrosis and chronic lung diseases, such as bronchiectasis [6–10].
MABC is the most drug resistant of mycobacterial pathogens, resulting in limited therapeutic options and a high treatment failure rate [11–14]. MABC is comprised of three closely related subspecies: M. abscessus subsp. abscessus, M. abscessus subsp. massiliense and M. abscessus subsp. bolletii [15, 16]. M. abscessus subsp. abscessus encodes a functional erythromycin ribosomal methylase gene, erm(41), which modifies the binding site for macrolide antibiotics, causing inducible macrolide resistance [17–19]. However, some clinical isolates of M. abscessus subsp. abscessus were susceptible to macrolide antibiotics, even after extended incubation of 14 days [20]. Loss of function of erm(41) genes was associated with a T-to-C substitution at position 28 of the gene (T28C), leading to an amino acid change from Trp to Arg at codon 10 [17, 18]. Pulmonary disease caused by these strains with M. abscessus subsp. abscessus with C28 sequevar may be responsive to macrolide-containing antibiotic regimens [20]. Conversely, all M. abscessus subsp. abscessus strains with a T28 sequevar were thought to be resistant to clarithromycin with extended incubation [18].
Although there were multiple polymorphisms associated with amino acid changes, only this T28C substitution resulted in loss of erm(41) gene function and no other nucleotide substitution was known to be associated with macrolide susceptibility [20]. We report the first case of pulmonary disease caused by a strain of M. abscessus subsp. abscessus that was susceptible to clarithromycin due to a T19 sequevar of the erm(41) gene. The patient was treated successfully with macrolide-containing antibiotics. This study was approved by the institutional review board of the Samsung Medical Center.
Case presentation
A 62-year-old Korean female was referred to our hospital due to chronic cough, sputum, and hemoptysis lasting more than 5 months. She was a non-smoker and had no history of previous treatment for pulmonary tuberculosis. The patient was 149.7 cm tall and weighed 40.7 kg. The erythrocyte sedimentation rate was 77 mm/h, and C-reactive protein was 2.37 mg/dL. A human immunodeficiency virus antibody test was negative. A chest computed tomography (CT) scan revealed bronchiectasis and bronchiolitis in both lungs, suggesting the nodular bronchiectatic form of NTM lung disease (Fig. 1a).
Fig. 1.
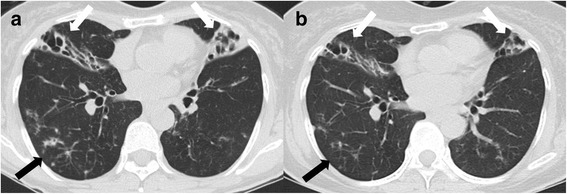
A 62-year-old female with bronchiectasis and nontuberculous mycobacterial lung disease caused by Mycobacterium abscessus subsp. abscessus. a Transverse chest computed tomography scan (2.5-mm-section thickness) at the start treatment revealed bilateral bronchiectasis and consolidations (white arrows) in the right middle lobe and lingular division of the left upper lobe as well as multiple tree-in-bud appearances (black arrow), suggesting bronchiolitis. b Transverse chest computed tomography scan (2.5-mm-section thickness) at 12 months of antibiotic treatment revealed decreased consolidation around the bronchiectasis (white arrows) and decreased bronchiolitis (black arrow)
The patient’s sputum was positive for acid-fast bacilli staining and NTM were isolated twice from sputum specimens in both solid (3 % Ogawa solid media; Shinyang, Seoul, South Korea) and liquid culture system (Bactec MGIT 960 system; BD Diagnostics, Sparks, MD, USA). To identify an etiological agent, bacteria grown in the MGIT 960 culture system were initially propagated in 7H9 broth (Difco Laboratories, Detroit, MI, USA) supplemented with 10 % (vol/vol) oleic acid-albumin-dextrose-catalase (OADC; BD Diagnostics) for 7 days at 37 °C. They were then sub-cultured in egg-based 3 % Ogawa solid media (Shinyang, Seoul, South Korea), and genomic DNA was extracted from cultured bacteria. M. abscessus was the initial species identified using a reverse line blot hybridization assay (REBA Myco-ID; M&D, Inc., Wonju, South Korea) based on the rpoB gene [21].
To confirm the accuracy of this identification, sequencing analyses of rpoB, hsp65, and 16S rRNA were performed using GenBank (http://blast.ncbi.nlm.nih.gov/) with the BLAST algorithm [22–24]. The 16S rRNA sequences were 100 % identical to M. abscessus subsp. abscessus (GenBank accession no. NR074427), M. abscessus subsp. massiliense (GenBank accession no. NR074421), M. chelonae (GenBank accession no. AY457082), and M. abscessus subsp. bolletii (GenBank accession no. NR043236). The rpoB sequences showed 99.7 % similarity to those of the M. abscessus subsp. abscessus type strain, with only a 2-base mismatch (GenBank accession no. CU458896). The hsp65 sequences were 100 % identical to those of the M. abscessus subsp. abscessus type strain (GenBank accession no. CU458896). The isolate was identified as M. abscessus subsp. abscessus by sequencing based method.
Drug susceptibility testing was performed using a broth microdilution method and M. peregrinum ATCC 700686 was used for quality control according to the guidelines [25], revealing that the isolate was susceptible to clarithromycin (minimum inhibitory concentration [MIC], ≤0.5 μg/mL), even after extended incubation for 14 days (Table 1). The isolate was genotyped for erm(41) polymorphism and for rrl mutation, which are known as the main mechanisms of macrolide resistance [26]. The isolate had a point mutation of C → T at position 19 (C19 → T) in the erm(41) gene, instead of the expected C28 sequevar of erm(41). It also had no rrl mutation. To the best of our knowledge, the C19 → T mutation of erm(41) in M. abscessus subsp. abscessus is the first description.
Table 1.
Drug susceptibility testing results for antimicrobial agents against the isolate
| Drug | MIC (μg/mL) for each category | MIC of isolate (μg/mL) | ||
|---|---|---|---|---|
| Susceptible | Intermediate | Resistant | ||
| Amikacin | ≤16 | 32 | ≥64 | 8 |
| Cefoxitin | ≤16 | 32–64 | ≥128 | 64 |
| Imipenem | ≤4 | 8–16 | ≥32 | 8 |
| Clarithromycin | ≤2 | 4 | ≥8 | ≤0.5 |
| Ciprofloxacin | ≤1 | 2 | ≥4 | >16 |
| Moxifloxacin | ≤1 | 2 | ≥4 | 16 |
| Doxycycline | ≤1 | 2–4 | ≥8 | >32 |
The patient received oral clarithromycin (1,000 mg/d) with an initial 4-week hospitalization for intravenous amikacin and cefoxitin [27, 28]. At day 20 of treatment, clarithromycin was switched to azithromycin (250 mg/d) due to gastrointestinal disturbance. After discharge, the patient received oral azithromycin for a total duration of 15 months. Her sputum cultures converted to and remained negative after 2 months of antibiotic treatment. Chest CT at 12 months of treatment revealed improvement in consolidations and bronchiolitis (Fig. 1b).
Discussion
It is important to distinguish the three subspecies of MABC because of their differences in susceptibility to clarithromycin [29–32]. M. abscessus subsp. abscessus, M. abscessus subsp. massiliense, and M. abscessus subsp. bolletii are commonly inducible resistant, susceptible, and resistant to clarithromycin, respectively [15].
This is the first case of pulmonary disease caused by a strain of M. abscessus subsp. abscessus showing a clarithromycin susceptible phenotype due to the T19 sequevar of erm(41) gene. According to a previously reported paper by Nash et al., M. abscessus subsp. abscessus strains with T28 → C had no inducible resistance to clarithromycin and had low MIC. In the present study, the M. abscessus subsp. abscessus clinical isolate had a C-to-T mutation at position 19 (C19 → T) leading to an Arg → Stop codon change at codon 7 of the erm(41) gene. This strain was named SMC-Mabs-T19. The SMC-Mabs-T19 strain revealed low MIC, similar to the M. abscessus subsp. abscessus strain with a C28 sequevar. Therefore, maintenance of low clarithromycin MIC against the SMC-Mabs-T19 strain might have been a result of the production of non-functional Erm(41). The entire erm(41) sequence of the susceptible isolate SMC-Mabs-C19 was unique and differed from that of M. abscessus subsp. abscessus type strain (GenBank accession no. CU458896) by only two bases: a C-to-T mutation at position 19 (C19 → T), and a T-to-C mutation at position 159 (T159 → C) (Fig. 2a and b). Of these differences, T159 → C was also present in the inducible resistant strains MC719 and UC22 (GenBank accession nos. EU177504 and CP012044, respectively). Therefore, the T19 sequevar was the most likely explanation for the lack of function of erm(41) alleles from the SMC-Mabs-T19 strain. Until now, erm(41) with a T19 sequevar has not been reported in GenBank. However, we found only two M. abscessus subsp. abscessus clinical isolates harboring erm(41) with C19 → G or A point mutations, and more information regarding drug susceptibility was not available (GenBank accession nos. FJ358485 and KP702837, respectively; Fig. 2b).
Fig. 2.
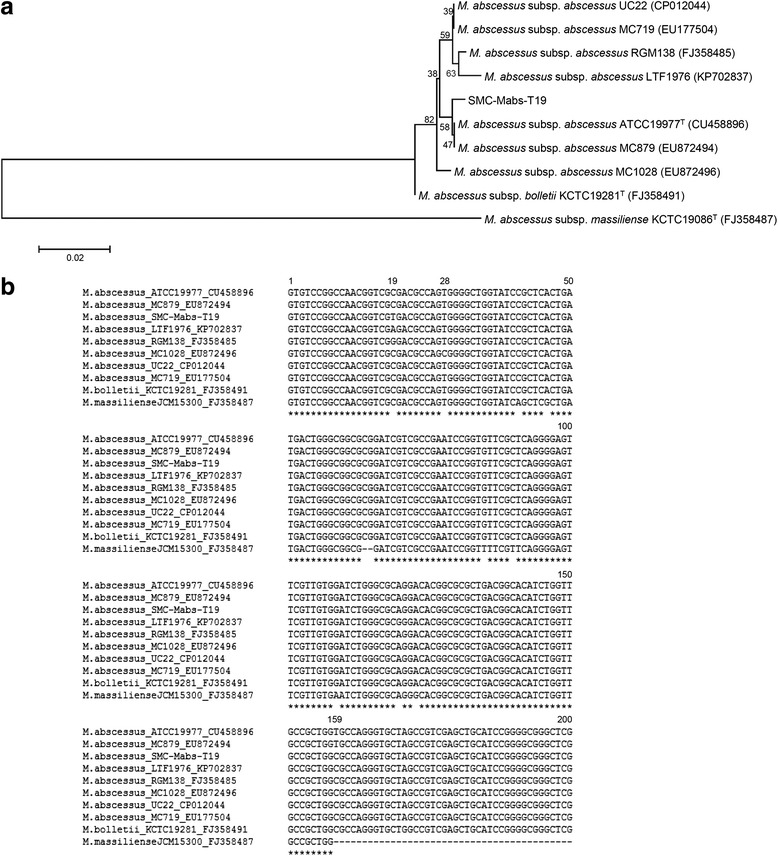
Analysis of DNA sequences in the erm(41) of Mycobacterium abscessus subsp. abscessus clinical isolate SMC-Mabs-T19. a Phylogenetic position of isolate SMC-Mabs-T19 and other strains belonging to M. abscessus complex based on entire erm(41) gene sequences. This tree was constructed using the neighbor-joining method. Percentages indicated at nodes represent the bootstrap levels supported by 1,000 re-sampled datasets. Scale bars indicate evolutionary distance in base substitutions per site. b Sequence alignment of erm(41) from SMC-Mabs-T19 and other strains belonging to the M. abscessus complex. Base numbering is from the first base of erm(41). Identical nucleotides are indicated by an asterisk below sequences. Two deletion sites of M. abscessus subsp. massiliense are indicated by dashes
Conclusions
The erm(41) gene is clinically important, and non-functional erm alleles may be an important issue for management of MABC lung disease. The presence of a non-functional erm(41) allele in M. abscessus subsp. abscessus isolates may be associated with better outcomes.
Ethics and consent to participate
This study protocol was approved by the institutional review board of the Samsung Medical Center (IRB approval 2008-09-016).
Consent to publish
Written informed consent was obtained from the patient for publication of this Case report and any accompanying images. A copy of the written consent is available for review by the Editor of this journal.
Availability of data and materials
All the data supporting the findings is contained within the manuscript.
Acknowledgments
Funding
This research was supported by the Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Science, ICT and future Planning (NRF-2015R1A2A1A01003959) and by a grant of the Korea Health technology R&D Project through the Korea Health Industry Development Institute (KHIDI), funded by the Ministry of Health & Welfare, Republic of Korea (HI15C2778). The funders had no role in the design of the study, the collection and analysis of the data, or the preparation of the manuscript.
Abbreviations
- CT
computed tomography
- erm(41)
erythromycin ribosomal methylase gene
- MABC
Mycobacterium abscessus complex
- NTM
nontuberculous mycobacteria
Footnotes
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
WJK designed the study. SYK and BHJ drafted the manuscript. WJK contributed to the diagnosis and treatment. SYK carried out microbiological analysis. SJS reviewed and edited the manuscript. WJK reviewed and supervised the manuscript. All the authors approved the final version of the manuscript.
References
- 1.Kendall BA, Winthrop KL. Update on the epidemiology of pulmonary nontuberculous mycobacterial infections. Semin Respir Crit Care Med. 2013;34:87–94. doi: 10.1055/s-0033-1333567. [DOI] [PubMed] [Google Scholar]
- 2.Prevots DR, Marras TK. Epidemiology of human pulmonary infection with nontuberculous mycobacteria: a review. Clin Chest Med. 2015;36:13–34. doi: 10.1016/j.ccm.2014.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Prevots DR, Shaw PA, Strickland D, Jackson LA, Raebel MA, Blosky MA, et al. Nontuberculous mycobacterial lung disease prevalence at four integrated health care delivery systems. Am J Respir Crit Care Med. 2010;182:970–6. doi: 10.1164/rccm.201002-0310OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hoefsloot W, van Ingen J, Andrejak C, Angeby K, Bauriaud R, Bemer P, et al. The geographic diversity of nontuberculous mycobacteria isolated from pulmonary samples: an NTM-NET collaborative study. Eur Respir J. 2013;42:1604–13. doi: 10.1183/09031936.00149212. [DOI] [PubMed] [Google Scholar]
- 5.Koh WJ, Chang B, Jeong BH, Jeon K, Kim SY, Lee NY, et al. Increasing recovery of nontuberculous mycobacteria from respiratory specimens over a 10-year period in a tertiary referral hospital in South Korea. Tuberc Respir Dis (Seoul) 2013;75:199–204. doi: 10.4046/trd.2013.75.5.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Leung JM, Olivier KN. Nontuberculous mycobacteria in patients with cystic fibrosis. Semin Respir Crit Care Med. 2013;34:124–34. doi: 10.1055/s-0033-1333574. [DOI] [PubMed] [Google Scholar]
- 7.Leung JM, Olivier KN. Nontuberculous mycobacteria: the changing epidemiology and treatment challenges in cystic fibrosis. Curr Opin Pulm Med. 2013;19:662–9. doi: 10.1097/MCP.0b013e328365ab33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Martiniano SL, Sontag MK, Daley CL, Nick JA, Sagel SD. Clinical significance of a first positive nontuberculous mycobacteria culture in cystic fibrosis. Ann Am Thorac Soc. 2014;11:36–44. doi: 10.1513/AnnalsATS.201309-310OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Adjemian J, Olivier KN, Prevots DR. Nontuberculous mycobacteria among patients with cystic fibrosis in the United States: screening practices and environmental risk. Am J Respir Crit Care Med. 2014;190:581–6. doi: 10.1164/rccm.201405-0884OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Koh WJ, Stout JE, Yew WW. Advances in the management of pulmonary disease due to Mycobacterium abscessus complex. Int J Tuberc Lung Dis. 2014;18:1141–8. doi: 10.5588/ijtld.14.0134. [DOI] [PubMed] [Google Scholar]
- 11.Kasperbauer SH, De Groote MA. The treatment of rapidly growing mycobacterial infections. Clin Chest Med. 2015;36:67–78. doi: 10.1016/j.ccm.2014.10.004. [DOI] [PubMed] [Google Scholar]
- 12.Kang YA, Koh WJ. Antibiotic treatment for nontuberculous mycobacterial lung disease. Expert Rev Respir Med. 2016;10:557–68. doi: 10.1586/17476348.2016.1165611. [DOI] [PubMed] [Google Scholar]
- 13.Stout JE, Koh WJ, Yew WW. Update on pulmonary disease due to non-tuberculous mycobacteria. Int J Infect Dis. 2016;45:123–34. doi: 10.1016/j.ijid.2016.03.006. [DOI] [PubMed] [Google Scholar]
- 14.Ryu YJ, Koh WJ, Daley CL. Diagnosis and treatment of nontuberculous mycobacterial lung disease: clinicians’ perspectives. Tuberc Respir Dis (Seoul) 2016;79:74–84. doi: 10.4046/trd.2016.79.2.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Griffith DE, Brown-Elliott BA, Benwill JL, Wallace RJ., Jr Mycobacterium abscessus. “pleased to meet you, hope you guess my name…”. Ann Am Thorac Soc. 2015;12:436–9. doi: 10.1513/AnnalsATS.201501-015OI. [DOI] [PubMed] [Google Scholar]
- 16.Lee MR, Sheng WH, Hung CC, Yu CJ, Lee LN, Hsueh PR. Mycobacterium abscessus complex infections in humans. Emerg Infect Dis. 2015;21:1638–46. doi: 10.3201/2109.141634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nash KA, Brown-Elliott BA, Wallace RJ., Jr A novel gene, erm(41), confers inducible macrolide resistance to clinical isolates of Mycobacterium abscessus but is absent from Mycobacterium chelonae. Antimicrob Agents Chemother. 2009;53:1367–76. doi: 10.1128/AAC.01275-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bastian S, Veziris N, Roux AL, Brossier F, Gaillard JL, Jarlier V, et al. Assessment of clarithromycin susceptibility in strains belonging to the Mycobacterium abscessus group by erm(41) and rrl sequencing. Antimicrob Agents Chemother. 2011;55:775–81. doi: 10.1128/AAC.00861-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Choi GE, Shin SJ, Won CJ, Min KN, Oh T, Hahn MY, et al. Macrolide treatment for Mycobacterium abscessus and Mycobacterium massiliense infection and inducible resistance. Am J Respir Crit Care Med. 2012;186:917–25. doi: 10.1164/rccm.201111-2005OC. [DOI] [PubMed] [Google Scholar]
- 20.Brown-Elliott BA, Vasireddy S, Vasireddy R, Iakhiaeva E, Howard ST, Nash K, et al. Utility of sequencing the erm(41) gene in isolates of Mycobacterium abscessus subsp. abscessus with low and intermediate clarithromycin MICs. J Clin Microbiol. 2015;53:1211–5. doi: 10.1128/JCM.02950-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang HY, Bang H, Kim S, Koh WJ, Lee H. Identification of Mycobacterium species in direct respiratory specimens using reverse blot hybridisation assay. Int J Tuberc Lung Dis. 2014;18:1114–20. doi: 10.5588/ijtld.14.0140. [DOI] [PubMed] [Google Scholar]
- 22.Adekambi T, Colson P, Drancourt M. rpoB-based identification of nonpigmented and late-pigmenting rapidly growing mycobacteria. J Clin Microbiol. 2003;41:5699–708. doi: 10.1128/JCM.41.12.5699-5708.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim H, Kim SH, Shim TS, Kim MN, Bai GH, Park YG, et al. Differentiation of Mycobacterium species by analysis of the heat-shock protein 65 gene (hsp65) Int J Syst Evol Microbiol. 2005;55:1649–56. doi: 10.1099/ijs.0.63553-0. [DOI] [PubMed] [Google Scholar]
- 24.Turenne CY, Tschetter L, Wolfe J, Kabani A. Necessity of quality-controlled 16S rRNA gene sequence databases: identifying nontuberculous Mycobacterium species. J Clin Microbiol. 2001;39:3637–48. doi: 10.1128/JCM.39.10.3638-3648.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.CLSI . Susceptibility testing of mycobacteria, nocardiae, and other aerobic actinomycetes; approved standard-second edition, Document no. M24-A2. Wayne: CLSI; 2011. [PubMed] [Google Scholar]
- 26.van Ingen J, Boeree MJ, van Soolingen D, Mouton JW. Resistance mechanisms and drug susceptibility testing of nontuberculous mycobacteria. Drug Resist Updat. 2012;15:149–61. doi: 10.1016/j.drup.2012.04.001. [DOI] [PubMed] [Google Scholar]
- 27.Jeon K, Kwon OJ, Lee NY, Kim BJ, Kook YH, Lee SH, et al. Antibiotic treatment of Mycobacterium abscessus lung disease: a retrospective analysis of 65 patients. Am J Respir Crit Care Med. 2009;180:896–902. doi: 10.1164/rccm.200905-0704OC. [DOI] [PubMed] [Google Scholar]
- 28.Koh WJ, Jeon K, Lee NY, Kim BJ, Kook YH, Lee SH, et al. Clinical significance of differentiation of Mycobacterium massiliense from Mycobacterium abscessus. Am J Respir Crit Care Med. 2011;183:405–10. doi: 10.1164/rccm.201003-0395OC. [DOI] [PubMed] [Google Scholar]
- 29.Kim HY, Kook Y, Yun YJ, Park CG, Lee NY, Shim TS, et al. Proportions of Mycobacterium massiliense and Mycobacterium bolletii strains among Korean Mycobacterium chelonae-Mycobacterium abscessus group isolates. J Clin Microbiol. 2008;46:3384–90. doi: 10.1128/JCM.00319-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee SH, Yoo HK, Kim SH, Koh WJ, Kim CK, Park YK, et al. The drug resistance profile of Mycobacterium abscessus group strains from Korea. Ann Lab Med. 2014;34:31–7. doi: 10.3343/alm.2014.34.1.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nie W, Duan H, Huang H, Lu Y, Bi D, Chu N. Species identification of Mycobacterium abscessus subsp. abscessus and Mycobacterium abscessus subsp. bolletii using rpoB and hsp65, and susceptibility testing to eight antibiotics. Int J Infect Dis. 2014;25:170–4. doi: 10.1016/j.ijid.2014.02.014. [DOI] [PubMed] [Google Scholar]
- 32.Kim SY, Kim CK, Bae IK, Jeong SH, Yim JJ, Jung JY, et al. The drug susceptibility profile and inducible resistance to macrolides of Mycobacterium abscessus and Mycobacterium massiliense in Korea. Diagn Microbiol Infect Dis. 2015;81:107–11. doi: 10.1016/j.diagmicrobio.2014.10.007. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All the data supporting the findings is contained within the manuscript.


