Abstract
Background
Fungal conidia are usually dormant unless the extracellular conditions are right for germination. Despite the importance of dormancy, little is known about the molecular mechanism underlying entry to, maintenance of, and exit from dormancy. To gain comprehensive and inter-species insights, transcriptome analyses were conducted across Aspergillus fumigatus, Aspergillus niger, and Aspergillus oryzae.
Results
We found transcripts of 687, 694, and 812 genes were enriched in the resting conidia compared with hyphae in A. fumigatus, A. niger, and A. oryzae, respectively (conidia-associated genes). Similarly, transcripts of 766, 1,241, and 749 genes were increased in the 1 h-cultured conidia compared with the resting conidia (germination-associated genes). Among the three Aspergillus species, we identified orthologous 6,172 genes, 91 and 391 of which are common conidia- and germination-associated genes, respectively. A variety of stress-related genes, including the catalase genes, were found in the common conidia-associated gene set, and ribosome-related genes were significantly enriched among the germination-associated genes. Among the germination-associated genes, we found that calA-family genes encoding a thaumatin-like protein were extraordinary expressed in early germination stage in all Aspergillus species tested here. In A. fumigatus 63 % of the common conidia-associated genes were expressed in a bZIP-type transcriptional regulator AtfA-dependent manner, indicating that AtfA plays a pivotal role in the maintenance of resting conidial physiology. Unexpectedly, the precocious expression of the germination-associated calA and an abnormal metabolic activity were detected in the resting conidia of the atfA mutant, suggesting that AtfA was involved in the retention of conidial dormancy.
Conclusions
A comparison among transcriptomes of hyphae, resting conidia, and 1 h-grown conidia in the three Aspergillus species revealed likely common factors involved in conidial dormancy. AtfA positively regulates conidial stress-related genes and negatively mediates the gene expressions related to germination, suggesting a major role for AtfA in Aspergillus conidial dormancy.
Electronic supplementary material
The online version of this article (doi:10.1186/s12864-016-2689-z) contains supplementary material, which is available to authorized users.
Keywords: Conidia, Dormancy, Germination, Aspergillus, AtfA, Transcriptome
Background
Conidia are, in general, stress-tolerant reproductive structures, and filamentous fungi vigorously produce conidia under the appropriate conditions [1]. In the presence of water and appropriate nutrients, conidia germinate, whereas conidia enter dormancy when the environment is not appropriate. Dormant conidia are metabolically inactive and are viable for a long time (more than a year) [2]. Therefore, dormant conidia do not consume energy prior to encountering the appropriate conditions for germination. This mechanism allows conidia to find an environment where the fungi can prosper, which consequently contributes to their ubiquity and prosperity in nature. Despite its significance to conidial physiology, however, the molecular mechanisms underlying entering and exiting dormancy remain largely unknown.
To gain insights into dormancy mechanisms, genes and proteins that are specifically highly expressed in conidia, or during germination, have been investigated by transcriptomic or proteomic approaches in this decade. Van Leeuwen et al. [3] found that 4,628 out of 14,253 genes were expressed in Aspergillus niger dormant conidia using Affymetrix microarray chips. Additionally, more than half of the genes changed their expression levels at the beginning of the early germination stage (~2 h). Novodvorska et al. [4] showed that 6,519 genes (42.3 %) were differentially expressed [> twofold fragments per kilobase of transcript per million mapped reads (FPKM)] during the first hour of germination in A. niger, as detected by an RNA-sequencing analysis. Of these, 2,626 genes had increased expression levels, and functional categories related to RNA processing, protein synthesis, and nitrogen metabolism were enriched in the gene set. Proteome analyses regarding Aspergillus conidia were reported by two groups in 2010. Teutschbein et al. [5] presented a proteome of Aspergillus fumigatus’ resting conidia. They detected 449 proteins [4.7 % of predicted open reading frames (ORFs)] in the conidia and 57 were overrepresented compared with in hyphae. Interestingly, pyruvate decarboxylase and alcohol dehydrogenase were found in dormant conidia, suggesting that alcoholic fermentation might occur during dormancy. They also revealed the presence of several A. fumigatus allergens, including Asp f3, Asp f13, Asp f22, Asp f27, and CatA in the conidia. However, Oh et al. [6] investigated the Aspergillus nidulans proteome 1 h after germination and 144 proteins were found to be differentially expressed. These two proteomic data partly overlapped the data from the transcriptomic analysis, even though they were performed using different Aspergillus species under different culture conditions.
Genome sequence data with annotations for four representatives of the Aspergillus species (A. nidulans, A. fumigatus, A. niger, and Aspergillus oryzae) are now available from Aspergillus genome database, AspGD (http://www.aspgd.org/), which is advantageous to the investigation of the universality and diversity of the intra-genus genomes. Indeed, a comparative genome analysis was conducted and provided a vast amount of information on the biology and physiology of filamentous fungi [7–9]. However, studies comparing different species at the transcriptional level have not been undertaken. Because cost-effective RNA-sequencing technology is available, we are now able to determine the transcriptomes of different Aspergillus species and make inter-species comparisons, which could provide new findings.
In the present study, we mainly addressed the issue of dormancy mechanisms in Aspergillus conidia by performing a comparative transcriptome analysis within the genus. Indeed, we focused on the conidia and 1 h-grown conidia of A. fumigatus, A. niger, and A. oryzae for our comparative transcriptome analysis. First, we defined the genes that were dominantly expressed in conidia or germinating conidia as conidia-associated genes (CAGs) or germination-associated genes (GeAGs), respectively. Subsequent comparisons of the CAGs or GeAGs among three species exposed the common conidial features of these filamentous fungi.
Results
Transcriptome determination using RNA-sequencing
To compare the conidial transcriptomes of different Aspergillus species, the conidia of each strain should be harvested from physiologically similar cultivation conditions. We chose potato dextrose agar (PDA) and potato dextrose broth (PDB) for the culture media because all of the strains (A. fumigatus Af293, A. niger IFM 58835, and A. oryzae RIB40) produced a large amount of conidia on PDA. Based on the colony expansion rates, conidia production rates, and germination rates (Additional file 1), we grew them at preferred temperature condition, namely A. fumigatus were grown at 37 °C, A. niger at 30 °C, and A. oryzae at 30 °C on PDA.
We first made comparisons among hyphae, dormant conidia, and 1 h-grown conidia in each species. The conidia were harvested from a 7-day-old culture at temperature specified above, and RNA was extracted from the conidia. Similarly, RNA in hyphae was prepared from mycelia cultivated in PDB at the sub-stationary phase according to dry weight (Additional file 1). To get insight into early response of conidia to the condition preferable to germinate, RNA was extracted from the conidia incubated in PDB at the appropriate temperature for 1 h. At the time-point, no germ tubes were appeared at all and the conidia do not start swelling in all species.
Transcriptomes from three different growth phases (hyphae, conidia, and 1 h-grown conidia) of A. fumigatus, A. niger, and A. oryzae were determined by RNA-sequencing analyses. In A. fumigatus, 97.8, 96.5, and 95.5 % of the ORFs were expressed in hyphae, conidia, and 1 h-grown conidia, respectively (Table 1). In A. niger, 86.7, 83.3, and 72.0 % of ORFs were expressed in hyphae, conidia, and 1 h-grown conidia, respectively, while in A. oryzae, 87.0, 85.8, and 77.1 % of ORFs were expressed in hyphae, conidia, and 1 h-grown conidia, respectively. There were 93, 1,251, and 815 genes whose expression levels were not detected in any of the three phases in A. fumigatus, A. niger, and A. oryzae, respectively (Table 1). The mean FPKM for each transcriptome was calculated and 16.3–19.4 %, 12.0–15.3 %, and 11.0–18.8 % of genes showed FPKM values higher than the mean FPKM in A. fumigatus, A. niger, and A. oryzae, respectively (Table 1).
Table 1.
The numbers of ORFs expressed in each phase of the Aspergillus fungi
| Hyphae | Conidia | 1 h-grown conidia | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| # of ORFs | # of expressed ORFs | % | mean FPKM | # > mean | % | # of expressed ORFs | % | mean FPKM | # > mean | % | # of expressed ORFs | % | mean FPKM | # > mean | % | |
| A. fumigatus | 9783 | 9569 | 97.8 | 69.00 | 1900 | 19.4 | 9445 | 96.5 | 87.67 | 1599 | 16.3 | 9341 | 95.5 | 91.33 | 1906 | 19.5 |
| A. niger | 14056 | 12183 | 86.7 | 40.83 | 2146 | 15.3 | 11711 | 83.3 | 41.82 | 2083 | 14.8 | 10123 | 72.0 | 108.12 | 1682 | 12.0 |
| A. oryzae | 11902 | 10353 | 87.0 | 31.79 | 2234 | 18.8 | 10217 | 85.8 | 49.97 | 1948 | 16.4 | 9173 | 77.1 | 108.5 | 1313 | 11.0 |
Determination of CAGs and GeAGs
To determine the genes whose expression is enriched in conidia or 1 h-grown conidia, we compared transcriptomes between hyphae and conidia, or between conidia and 1 h-grown conidia, respectively. The CAGs and GeAGs were defined as follows: 1) The expression levels in conidia or 1 h-grown conidia increased more than fourfold compared with that in hyphae or conidia, respectively; and 2) The expression levels in conidia or 1 h-grown conidia conidia were higher than the mean FPKM value. Using these criteria, we identified 687, 694, and 812 CAGs in A. fumigatus, A. niger, and A. oryzae, respectively. Similarly, 766, 1,241, and 749 GeAGs were identified in A. fumigatus, A. niger, and A. oryzae, respectively (Fig. 1).
Fig. 1.

Experimental settings and a summary of the comparative transcriptomic analysis. The conidia-associated genes (CAGs) were determined by comparisons between FPKMs of conidia and hyphae (C/H). The germination-associated genes (GeAGs) were determined by comparisons between FPKMs of 1 h-grown conidia and conidia (Ge/C)
We used freshly harvested conidia to investigate the conidial transcriptome. Thus, whether the transcriptome could change during preservation (7 d) after drying, when the resting conidia should be metabolically inactive, was of interest. To address this, we compared A. fumigatus transcriptomes between the freshly harvested conidia and the dried resting conidia, focusing on 687 CAGs (data not shown). In the resting conidia, only 4 and 6 % of the CAGs showed a more than fourfold increase and a less than ¼ decrease, respectively, compared with the freshly harvested conidia. Whereas 58 % of the CAGs showed an unchanged expression level, less than twofold, during preservation, and 69 % of all of the genes were unchanged in their expression levels. These results suggested that the mRNA profiles of the conidia were largely unaffected during the 7-day incubation, and specific mRNA degradation did not occur during drying.
Functional classifications of CAGs and GeAGs using gene ontology (GO) terms
To gain a comprehensive insight into the specific molecular functions of the CAGs and GeAGs, a functional classification analysis was conducted. The total numbers of GO terms for A. fumigatus, A. niger, and A. oryzae were 233, 244, and 235, respectively. Among them, 8, 2, and 14 terms were significantly overrepresented or underrepresented in the CAGs, respectively (Additional file 2). The three Aspergillus species shared only one CAG-enriched GO term that is associated with glyoxysome (Table 2). In addition, functional classes related to adaptation to the intracellular oxidative state, such as oxidation-reduction process, oxidoreductase activity, and response to oxidative stress, were found in each fungus (Additional file 2). In the same way, 50, 70, and 59 terms were found in the GeAGs, respectively (Additional file 3). The larger number of terms found in GeAGs suggested that more molecular functions were triggered during the beginning of germination in the all three fungi. Indeed, 30 GO terms were commonly overrepresented in the GeAG of the three species (Table 2). Most were associated with ribosome function and primary cellular activities, such as translation, respiration, and metabolism, which supported the previous view that the construction of translational machinery starts at a very early stage in germination [2–4].
Table 2.
Common CAG- and GeAG-enriched GO terms
| Count (changed: not changed) | ||||||
|---|---|---|---|---|---|---|
| GO | terms | A. fumigatus | A. niger | A. oryzae | Over (+)/under (−) | |
| CAG-enriched | ||||||
| # C — Cellular component | ||||||
| GO:0009514 | glyoxysome | 13:24 | 10:28 | 15:21 | + | |
| GeAG-enriched | ||||||
| # C — Cellular component | ||||||
| GO:0005730 | nucleolus | 113:56 | 119:40 | 106:50 | + | |
| GO:0030686 | 90S preribosome | 37:0 | 34:1 | 33:1 | + | |
| GO:0005829 | cytosol | 243:1105 | 333:986 | 225:1050 | + | |
| GO:0032040 | small-subunit processome | 32:2 | 35:0 | 33:2 | + | |
| GO:0005840 | ribosome | 44:22 | 57:16 | 44:21 | + | |
| GO:0005762 | mitochondrial large ribosomal subunit | 26:4 | 30:0 | 21:4 | + | |
| GO:0030687 | preribosome, large subunit precursor | 24:3 | 20:1 | 17:4 | + | |
| GO:0005739 | mitochondrion | 71:304 | 95:297 | 57:315 | + | |
| GO:0009986 | cell surface | 21:51 | 29:46 | 25:52 | + | |
| GO:0031966 | mitochondrial membrane | 12:13 | 14:13 | 11:11 | + | |
| GO:0005886 | plasma membrane | 42:224 | 73:191 | 43:214 | + | |
| GO:0005634 | nucleus | 154:1289 | 238:1331 | 140:1322 | + | |
| GO:0005819 | spindle | 8:13 | 10:12 | 7:13 | + | |
| GO:0016021 | integral component of membrane | 15:555 | 36:695 | 17:780 | - | |
| # F — Molecular function | ||||||
| GO:0003735 | structural constituent of ribosome | 110:18 | 126:3 | 95:9 | + | |
| GO:0003723 | RNA binding | 44:83 | 57:55 | 42:64 | + | |
| GO:0008026 | ATP-dependent helicase activity | 14:25 | 19:22 | 12:27 | + | |
| GO:0003743 | translation initiation factor activity | 12:16 | 23:6 | 15:12 | + | |
| GO:0051082 | unfolded protein binding | 14:30 | 21:25 | 13:28 | + | |
| GO:0005524 | ATP binding | 69:439 | 102:475 | 64:506 | + | |
| GO:0003676 | nucleic acid binding | 51:286 | 76:236 | 49:227 | + | |
| GO:0005525 | GTP binding | 18:75 | 24:68 | 21:81 | + | |
| GO:0000166 | nucleotide binding | 35:208 | 53:208 | 36:244 | + | |
| GO:0016491 | oxidoreductase activity | 8:391 | 20:634 | 12:635 | - | |
| GO:0000981 | sequence-specific DNA binding RNA polymerase II transcription factor activity | 2:238 | 12:326 | 2:248 | - | |
| # P — Biological process | ||||||
| GO:0006412 | translation | 86:14 | 97:3 | 72:11 | + | |
| GO:0006364 | rRNA processing | 26:7 | 24:4 | 23:9 | + | |
| GO:0000447 | endonucleolytic cleavage in ITS1 to separate SSU-rRNA from 5.8S rRNA and LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA) | 20:2 | 21:1 | 19:2 | + | |
| GO:0000027 | ribosomal large subunit assembly | 18:4 | 19:1 | 17:3 | + | |
| GO:0035690 | cellular response to drug | 34:151 | 49:135 | 37:157 | + | |
| GO:0006696 | ergosterol biosynthetic process | 10:16 | 11:15 | 8:19 | + | |
| GO:0006413 | translational initiation | 9:12 | 17:5 | 11:9 | + | |
| GO:0009060 | aerobic respiration | 9:18 | 12:11 | 9:16 | + | |
| GO:0008152 | metabolic process | 9:324 | 16:468 | 6:513 | - | |
| GO:0055085 | transmembrane transport | 16:405 | 21:552 | 16:651 | - | |
Expression profile of genes encoding allergenic proteins
A. fumigatus was reported to possess 22 known allergenic proteins, which is the highest number in fungi found to date [10]. A comprehensive view of the allergenic gene expression would be useful for understanding which cellular forms of the fungus were potential allergens. Our transcriptome data revealed that 14, 9, and 9 allergen-related genes showed FPKM values higher than the corresponding mean FPKM in hyphae, conidia, and 1 h-grown conidia, respectively (Additional file 4). Among the highly expressed genes, 8 genes were commonly found in all cellular forms, 5 transcripts (aspf3, aspf8, cyp4, hsp90, and rpL3) of which were most abundant in 1 h-grown conidia. Notably, previous proteome research revealed that AspF3 and AspF8 were presented in the A. fumigatus germinating conidia [11]. In addition, the other A. fumigatus allergenic genes (Asp f1, Asp f4, sod3, and Asp f7) were highly expressed in hyphae but relatively silent in conidia. These transcriptional data suggest that higher amount of allergenic proteins may be present in hyphae and germinating conidia of A. fumigatus.
Expression profiles of common orthologous genes
In addition to species-specific allergenic genes, the Aspergillus species should share a large number of genes that are involved in common biological functions, such as primary metabolism and biosynthesis for cellular components. To gain more general insights into Aspergillus biology, we focused on the sets of orthologous genes that were found in each genome of A. fumigatus, A. niger, and A. oryzae and that had the highest homology levels. Using the three homology searches, A. fumigatus vs. A. niger, A. fumigatus vs. A. oryzae, and A. niger vs. A. oryzae, we found 6,172 sets of common genes whose counterparts represented 63.1, 43.9, and 51.9 % of all of the genes in A. fumigatus, A. niger, and A. oryzae, respectively (Additional file 5). The RNA-sequencing data showed that 5,972 (96.3 %), 5,758 (93.3 %), and 5,636 (91.3 %) of the common genes were expressed in A. fumigatus, A. niger, and A. oryzae, respectively.
We next sought to identify the common genes in the CAGs and GeAGs. Among the CAGs of the three species, 91 genes were found to be common, which was only 11.2–13.2 % of the CAGs in each species (Fig. 2a). These ratios were lower than those for whole gene sets in each species (43.9–63.1 %), suggesting that the CAGs are more diverse and strain specific compared with the non-CAGs. Of the 91 genes, 43 were greater than 10-fold the FPKM ratio of conidia to hyphae in all of the species (Table 3). While there was no information on 17 of the genes’ encoded protein functions, the rest included several genes that were previously reported to be conidia-specific or expressed during the asexual developmental stage. CatA is a conidia-specific catalase and is responsible for the stress tolerance of conidia, which was well documented in A. fumigatus and A. oryzae, as well as A. nidulans [12–14]. ConJ is a conidia-specific protein that was studied in A. nidulans [15], and was reported to be expressed during asexual development in A. fumigatus [16]. VosA plays an important role in conidia formation and trehalose accumulation in conidia, as shown in A. fumigatus and A. nidulans [17, 18]. PilB is enriched in the conidia of A. fumigatus [5], and fhk1, encoding a hybrid histidine kinase, was reported to be up-regulated during asexual development in A. fumigatus [16]. These results suggested that most of the common CAGs were involved in conidia-specific functions and played an essential role in the conidial biology of Aspergillus species.
Fig. 2.
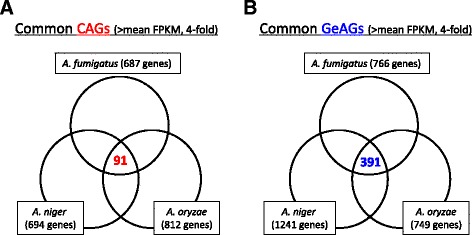
Venn diagrams comparing the CAGs and GeAGs from the three Aspergillus species. The common CAGs (a) and common GeAGs (b) were identified
Table 3.
A list of common CAGs (Ratio > 10)
| FPKM ratio of conidia to hyphae*1 | ||||||||
|---|---|---|---|---|---|---|---|---|
| A. fumigatus | A. niger | A. oryzae | A. fumigatus | A. niger | A. oryzae | atfA-dependency *2 | Gene name in Af | Annotation in A. fumigatus |
| Afu2g00200 | An12g10720 | AO090113000153 | - | 458.5 | - | full | cat3 | Catalase, putative |
| Afu8g01530 | An12g10710 | AO090113000154 | 1751.2 | 1334.9 | 2307.7 | n.d. | - | HHE domain protein |
| Afu1g01490 | An15g07300 | AO090102000259 | 1291.8 | 1040.6 | 467.4 | n.d. | - | Hypothetical protein |
| Afu2g14330 | An17g01885 | AO090009000665 | 1014.6 | 238.2 | 780.3 | n.d. | - | Hypothetical protein |
| Afu6g03210 | An12g10240 | AO090011000656 | 798.1 | 439.3 | 1367.5 | full | conJ | Conidiation-specific protein 10 |
| Afu8g05810 | An15g04670 | AO090005000570 | 584.1 | 1287.1 | 28.3 | n.d. | - | DUF1295 domain protein |
| Afu5g10160 | An14g04530 | AO090010000533 | 560.4 | 133.2 | 22.4 | n.d. | - | ActVA 4 protein |
| Afu8g00600 | An03g00920 | AO090010000696 | 525.8 | - | - | n.d. | - | Conserved hypothetical protein |
| Afu3g01210 | An01g10950 | AO090026000081 | 454.0 | 867.9 | 583.9 | n.d. | - | ThiJ/PfpI family protein |
| Afu5g01160 | An03g02190 | AO090020000259 | 338.5 | 21.7 | 12.7 | full | - | Monosaccharide transporter |
| Afu8g06020 | An15g04770 | AO090005000539 | 275.9 | 74.8 | 297.8 | full | - | Glutamate decarboxylase |
| Afu6g03890 | An09g03130 | AO090701000158 | 249.3 | 350.6 | 194.9 | full | catA | Catalase A |
| Afu1g03580 | An18g04120 | AO090009000418 | 196.6 | 288.5 | 629.2 | n.d. | - | Hypothetical protein |
| Afu3g00640 | An11g08160 | AO090009000116 | 165.1 | 189.5 | 25.2 | n.d. | - | Peptidoglycan binding domain protein |
| Afu2g01590 | An03g04860 | AO090701000790 | 125.5 | 22.0 | 74.4 | full | nce102 | Non-classical export protein (Nce2), putative |
| Afu2g15740 | An15g05990 | AO090012000251 | 103.7 | 261.2 | - | full | - | Oxidoreductase, short chaindehydrogenase/reductase family |
| Afu5g03930 | An09g06270 | AO090102000598 | 100.6 | 220.2 | 70.0 | partial | - | Alcohol dehydrogenase, putative |
| Afu1g13530 | An08g06600 | AO090012000522 | 100.5 | 74.2 | 35.6 | n.d. | - | Hypothetical protein |
| Afu3g03940 | An01g00280 | AO090124000046 | 91.1 | 24.5 | 47.6 | full | - | 2,3-diketo-5-methylthio-1-phosphopentanephosphatase, putative |
| Afu3g11550 | An02g07350 | AO090003000710 | 88.6 | 42.0 | 360.8 | n.d. | - | LEA domain protein |
| Afu5g03269 | An09g05520 | AO090102000529 | 88.6 | 54.5 | 72.9 | n.d. | - | Unknown |
| Afu6g08320 | An11g01750 | AO090003000094 | 87.1 | 81.9 | 134.4 | n.d. | pilB | Putative conserved eisosome component |
| Afu3g12760 | An02g08740 | AO090012000846 | 81.3 | 85.1 | 268.0 | n.d. | - | Hypothetical protein |
| Afu4g03390 | An14g02450 | AO090011000203 | 79.2 | 15.6 | 21.0 | full | - | Aquaporin |
| Afu5g13100 | An14g06050 | AO090120000435 | 77.1 | 16.0 | 78.7 | n.d. | - | Hypothetical protein |
| Afu6g13470 | An08g03030 | AO090020000658 | 60.5 | - | 496.2 | n.d. | - | Conserved hypothetical protein |
| Afu1g13550 | An08g06620 | AO090012000521 | 45.4 | 97.8 | 113.2 | n.d. | - | Hypothetical protein |
| Afu5g14310 | An09g01150 | AO090020000287 | 43.8 | 137.2 | 13.0 | not CAG | - | Short chain dehydrogenase/reductase familyprotein |
| Afu3g10480 | An16g04420 | AO090003000594 | 31.5 | 76.5 | 134.2 | n.d. | - | Conserved hypothetical protein |
| Afu6g08730 | An11g06120 | AO090001000547 | 30.2 | 49.1 | 699.1 | not CAG | - | 6-phosphogluconate dehydrogenase,decarboxylating |
| Afu4g09250 | An04g04280 | AO090023000611 | 29.7 | 11.6 | 18.2 | n.d. | - | Hypothetical protein |
| Afu4g05900 | An04g00100 | AO090023001009 | 25.1 | 35.1 | 24.7 | n.d. | - | Conserved hypothetical protein |
| Afu4g01020 | An18g01860 | AO090003001579 | 24.1 | 28.6 | 99.1 | n.d. | fhk1 | Sensor histidine kinase/response regulator, putative |
| Afu6g13860 | An08g08500 | AO090103000362 | 23.9 | 370.9 | 429.6 | n.d. | - | Conserved hypothetical protein |
| Afu5g09180 | An07g03930 | AO090020000514 | 23.7 | 671.7 | 448.4 | n.d. | - | Hypothetical protein |
| Afu4g10860 | An04g05790 | AO090003001124 | 23.2 | 22.7 | 24.9 | n.d. | vosA | Velvet family protein |
| Afu5g01290 | An14g07380 | AO090010000635 | 22.4 | 680.6 | 42.7 | partial | - | Oxidoreductase, zinc-binding dehydrogenasefamily, putative |
| Afu1g03090 | An01g05320 | AO090005000810 | 21.7 | 17.9 | 16.1 | n.d. | - | Conserved hypothetical protein |
| Afu2g04200 | An11g02200 | AO090003000208 | 20.8 | 16.6 | 37.7 | not CAG | hppD | 4-hydroxyphenylpyruvate dioxygenase |
| Afu7g01430 | An12g01460 | AO090010000221 | 16.5 | 109.6 | 37.5 | full | - | Opsin 1 |
| Afu2g16930 | An04g09030 | AO090102000125 | 14.8 | 33.2 | 103.7 | not CAG | - | Succinate:fumarate antiporter (Acr1), putative |
| Afu2g10020 | An16g05030 | AO090011000634 | 10.1 | 1755.9 | 87.7 | n.d. | - | Hypothetical protein |
| Afu2g02310 | An07g06530 | AO090011000512 | 10.0 | 19.7 | 71.1 | not CAG | sur7 | sur7 protein, putative |
*1: “-”, not expressed in hyphae
*2: AtfA-dependency is clarified by real-time RT PCR. n.d., not determined
Likewise, we found 391 common genes in the GeAGs of the three species (Fig. 2b). This corresponded to 31.5–52.2 % of the GeAGs, and 46 of them showed more than 10-fold the FPKM ratio in 1 h-grown conidia compared with conidia in all of the species (Table 4). There were 13 genes that encoded a putative ribosome protein or a ribosome-related protein, which supported the view that de novo ribosome-complex biosynthesis begins at an early stage of germination. Notably, A. fumigatus calA (Afu3g09690) and the corresponding genes of A. niger (An16g03330) and A. oryzae (AO090005001280) showed quite high expression levels in 1 h-grown conidia (Table 4). The verification of the expression profiles during 2 h of germination using real-time PCR revealed that the A. fumigatus calA expression level was strongly induced at an early stage of germination (Fig. 3), suggesting that the CalA protein functions during the A. fumigatus germination process. Although the detailed function of CalA in germination remains unclear, CalA was detected on the swollen conidial surface by the binding of anti-CalA serum, and recombinant CalA protein was demonstrated to bind with laminin and murine lung cells [19]. CalA is a thaumatin-like protein, and A. fumigatus has the paralogous proteins encoded by Afu8g01710 and Afu3g00510 (calB and calC, respectively) (Additional file 6). Interestingly, the expression levels of calB and calC were induced during germination as well, while the expression level of calB was markedly high in hyphae (Fig. 3). In A. niger and A. oryzae, only a single protein, AoCalA and AnCalA, respectively, with high homology to the thaumatin-like protein was found, and the expression levels were also up-regulated in 1 h-grown conidia (Table 4, Additional file 6). Notably, A. nidulans has two thaumatin-like proteins, CetA and CalA, and the deletion of both genes was reported to result in synthetic lethality [20], suggesting an important role of the thaumatin-like proteins in Aspergillus conidial germination. The genetic distribution among the Aspergillus species (Additional file 6) suggested that the germination-related thaumatin-like protein is duplicated in some species and that their roles might be indispensable for filamentous fungi.
Table 4.
A list of common GeAGs (Ratio > 10)
| FPKM ratio of 1 h-grown conidia to conidia | |||||||
|---|---|---|---|---|---|---|---|
| A. fumigatus | A. niger | A. oryzae | A. fumigatus | A. niger | A. oryzae | Gene name in Af | Annotation in A. fumigatus |
| Afu3g09690 | An16g03330 | AO090005001280 | 2140.3 | 7365.7 | 1063.5 | - | Extracellular thaumatin domain protein, putative |
| Afu4g08110 | An04g02550 | AO090023000758 | 75.3 | 163.0 | 12.4 | - | Translation elongation factor G1, putative |
| Afu4g07630 | An04g02000 | AO090023000812 | 53.9 | 74.1 | 11.6 | - | Microtubule associated protein (Ytm1), putative |
| Afu6g02690 | An12g08230 | AO090120000155 | 48.5 | 87.3 | 16.2 | mtfA | C2H2 finger domain protein, putative |
| Afu1g10990 | An08g03290 | AO090038000305 | 43.2 | 729.2 | 13.4 | - | Ribosomal RNA processing protein, putative |
| Afu1g04370 | An01g03230 | AO090003000912 | 39.7 | 2627.6 | 18.7 | - | Hypothetical protein |
| Afu1g05310 | An01g04590 | AO090003000828 | 39.1 | 194.0 | 13.0 | - | DUF699 ATPase, putative |
| Afu2g10070 | An16g04970 | AO090011000630 | 35.6 | 11.6 | 26.2 | - | Carbamoyl-phosphate synthase, large subunit |
| Afu4g07540 | An04g01900 | AO090023000821 | 35.1 | 42.8 | 11.6 | - | Small nucleolar ribonucleoprotein complexsubunit, putative |
| Afu2g16040 | An15g06390 | AO090012000197 | 30.2 | 73.6 | 10.6 | - | rRNA biogenesis protein, putative |
| Afu2g09860 | An16g05290 | AO090011000649 | 29.0 | 71.4 | 33.7 | - | Purine-cytosine permease |
| Afu1g10440 | An08g02390 | AO090038000383 | 25.7 | 100.8 | 10.2 | - | Conserved hypothetical protein |
| Afu7g04700 | An13g01230 | AO090005000164 | 25.5 | 11.8 | 28.7 | - | Conserved hypothetical portein |
| Afu5g03870 | An09g06310 | AO090102000594 | 23.7 | 42.9 | 10.7 | - | Conserved hypothetical protein |
| Afu4g06350 | An04g00680 | AO090023000955 | 23.6 | 130.8 | 10.3 | - | rna binding protein |
| Afu3g06320 | An11g10000 | AO090020000118 | 21.3 | 29.8 | 14.0 | - | Conserved hypothetical protein |
| Afu2g02190 | An07g06640 | AO090011000497 | 21.0 | 52.7 | 28.0 | - | Hypothetical nuclear protein |
| Afu6g08720 | An11g06110 | AO090001000546 | 20.3 | 38.3 | 23.7 | - | 5’-methylthioadenosine phosphorylase |
| Afu4g03650 | An14g01560 | AO090011000292 | 20.1 | 68.9 | 12.4 | - | Ribosome associated DnaJ chaperone Zuotin, putative |
| Afu6g13370 | An08g09160 | AO090003000462 | 17.9 | 10.0 | 11.3 | utp10 | SSU processome component (Utp10), putative |
| Afu2g05950 | An02g14340 | AO090001000712 | 17.2 | 28.2 | 14.7 | - | snRNP and snoRNP protein (Snu13), putative |
| Afu3g13320 | An02g09200 | AO090012000801 | 17.0 | 14.3 | 10.7 | rps0 | 40S ribosomal protein S0, putative |
| Afu1g15730 | An01g14080 | AO090005001132 | 16.9 | 31.5 | 10.4 | - | Ribosomal protein S8 |
| Afu2g17200 | An04g09270 | AO090102000099 | 16.8 | 26.2 | 11.1 | - | Hypothetical protein |
| Afu2g11810 | An02g03860 | AO090026000462 | 16.7 | 77.0 | 16.8 | - | Conserved hypothetical protein |
| Afu5g11470 | An18g04470 | AO090026000687 | 16.0 | 83.7 | 24.6 | - | MYB DNA binding protein (Tbf1), putative |
| Afu5g06430 | An17g01270 | AO090009000609 | 15.7 | 76.2 | 10.5 | - | Mitochondrial large ribosomal subunit L7, putative |
| Afu2g02150 | An07g06760 | AO090011000490 | 15.5 | 20.0 | 12.7 | S10a | Ribosomal protein S10 |
| Afu6g04330 | An15g01160 | AO090701000118 | 15.1 | 35.8 | 12.0 | - | DEAH-box RNA helicase (Dhr1), putative |
| Afu1g05390 | An18g04220 | AO090009000405 | 14.8 | 28.8 | 10.6 | - | Mitochondrial ADP,ATP carrier protein (Ant), putative |
| Afu1g11710 | An08g03910 | AO090038000249 | 13.9 | 16.2 | 13.2 | - | 60S ribosomal protein L1 |
| Afu8g05330 | An16g07400 | AO090005000616 | 13.9 | 16.1 | 10.8 | - | Methylenetetrahydrofolate dehydrogenase |
| Afu3g10800 | An02g06530 | AO090003000629 | 13.7 | 10.4 | 10.9 | - | Eukaryotic translation initiation factor 3subunit CLU1/TIF31, putative |
| Afu3g11260 | An02g07010 | AO090003000679 | 13.6 | 37.3 | 16.5 | ubiC | Ubiquitin (UbiC), putative |
| Afu3g12300 | An02g08080 | AO090005000737 | 13.4 | 20.5 | 11.2 | - | Ribosomal L22e protein family |
| Afu5g12360 | An14g06860 | AO090120000354 | 13.3 | 15.2 | 12.8 | - | Mitochondrial oxaloacetate transporter (Oac),putative |
| Afu3g06580 | An11g09740 | AO090020000342 | 13.1 | 38.1 | 14.1 | - | WD repeat protein |
| Afu3g05490 | An11g11150 | AO090020000020 | 13.0 | 72.7 | 13.3 | - | Nrap protein superfamily |
| Afu6g10460 | An11g00990 | AO090023000242 | 12.8 | 10.8 | 15.5 | lag1 | Ceramide synthase membrane component (Lag1),putative |
| Afu6g09990 | An11g04985 | AO090038000466 | 12.1 | 29.5 | 10.7 | - | Importin beta-4 subunit, putative |
| Afu6g08580 | An11g05510 | AO090001000519 | 11.9 | 16.0 | 10.4 | fkbp4 | FKBP-type peptidyl-prolyl isomerase, putative |
| Afu3g10660 | An02g06320 | AO090003000611 | 11.5 | 23.0 | 47.8 | erg13 | Hydroxymethylglutaryl-CoA synthase |
| Afu4g13170 | An01g08850 | AO090009000264 | 11.2 | 23.6 | 16.9 | cpcB | Guanine nucleotide-binding protein subunit, putative |
| Afu3g00880 | An09g03120 | AO090003001496 | 11.1 | 486.0 | 48.1 | - | Conserved hypothetical protein |
| Afu6g09060 | An11g06810 | AO090001000583 | 10.7 | 50.8 | 11.3 | - | Mitochondrial 60S ribosomal protein L6precursor |
| Afu2g03380 | An07g07840 | AO090120000249 | 10.6 | 15.7 | 11.2 | - | Alkaline serine protease |
Fig. 3.
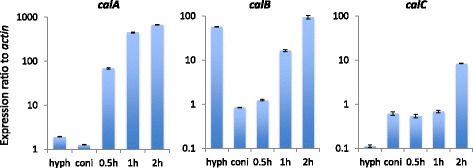
Quantitative real-time RT-PCR analyses of calA-family genes in A. fumgiatus. Expression profiles of calA, calB, and calC during the germination process (~2 h) and hyphae (hyph) were analyzed. Each value represents the expression ratio relative to that of the actin gene. Data presented are the averages of three replicates, and the bar indicates standard deviation
Role of the AtfA transcription factor in regulating CAGs
In our previous study, we demonstrated that A. fumigatus catA, conJ, and fhk1 were up-regulated during the asexual stage in an AtfA-dependent manner [16]. AtfA is a bZip-type transcription factor that is involved in conidial stress tolerance, and the molecular function of AtfA is largely conserved in some Aspergillus species [16, 21, 22]. As described above, the known AtfA-dependent genes were commonly enriched in the conidia of A. oryzae, A. niger, and A. fumigatus (Table 3). Thus, it was of interest to investigate whether conidia-accumulated transcripts of common CAGs were dependent on the AtfA transcription factor. We cultivated the A. fumigatus control strain (AfS35) and atfA deletion mutant, and the expression levels of the selected genes in hyphae and 4- and 8-day-old conidia were analyzed using quantitative real-time RT-PCR. Most of the CAGs tested showed AtfA-dependent expression in resting conidia (Fig. 4). This clearly indicated that AtfA played a major role in conidial biology.
Fig. 4.

Quantitative real-time RT-PCR analyses of the common CAGs in A. fumgiatus. Expression levels of the common CAGs in 4- and 8-day-old conidia of Afs35 (WT) and ΔatfA were analyzed. Each value represents a fold increase in the expression ratios compared with those in hyphae. Data presented are averages of three replicates, and the bar indicates standard deviation
In our previous study, the deletion of AtfA resulted in stress-labile conidia and delayed germ-tube formation [16]. To gain further insight into role of AtfA in conidial longevity, conidial transcriptomes were compared between the wildtype and the atfA mutant (Fig. 5). Among the whole genes, 10 and 13 % showed increased (>4 times) and decreased (< ¼ times) expression levels, respectively, in conidia of the atfA deletion mutant. For the CAGs, 54 % showed a decreased level (more than fourfold) of expression in the atfA mutant conidia compared with the wildtype. For the common CAGs, 63 % had lower expression levels in the A. fumigatus atfA mutant conidia (data not shown), which possibly led to deleterious defects in stress-homeostasis. Unexpectedly, a portion of GeAGs showed higher expression levels in the conidia of the atfA mutant. Particularly, calA and calB were highly up-regulated in the atfA conidia (Fig. 6). This derepression of the germination-associated genes in the resting conidia suggested that uncontrolled exit from dormancy might occur in the conidia.
Fig. 5.
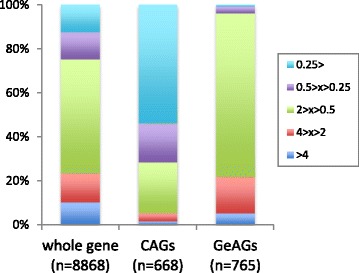
Distribution of genes with differential expression level in the atfA mutant conidia. Comparison between conidia of A. fumgiatus Afs35 (WT) and ΔatfA. From the FPKM values, the relative expression ratios of all of the genes (left), the CAGs (middle), and the GeAGs (right) were calculated as ΔatfA/WT conidia
Fig. 6.
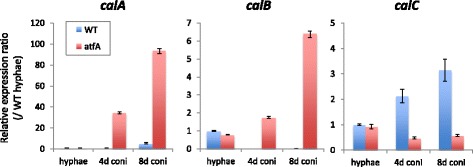
Expression levels of calA, calB, and calC in 4- and 8-day-old conidia of Afs35 (WT) and ΔatfA were analyzed. Each value represents the expression ratios compared with those in WT hyphae. Data presented are averages of three replicates, and the bar indicates standard deviation
Conidia of the null atfA mutant revealed germination-related traits
The first step toward germination, after sensing available carbon sources and water, is isotropic swelling, which is followed by polarized growth with germ tube formation [23]. During the isotropic growth, de novo ergosterol biosynthesis begins and a sterol-rich domain is observed in a polar position of the swollen conidia [24]. To investigate if conidia defective in the atfA gene showed precocious swelling, the freshly harvested conidia of the atfA mutant were microscopically observed by staining with filipin, which binds to ergosterol. Most of the conidia of the atfA mutant, but not of wildtype, were stained with filipin (Fig. 7a). Notably, some atfA mutant conidia showed sterol-rich domain formation and filipin staining at putative organelle membranes (Fig. 7a). These results supported the idea that resting conidia lacking AtfA began germination process without any available nutrients, such as carbon sources.
Fig. 7.

Metabolic activities in ΔatfA resting conidia. a Ergosterols were stained by filipin in the resting conidia of A. fumigatus Afs35 (WT), ΔatfA, and the complemented strain (Co-atfA). The test was repeated twice, and more than three sections were observed. The representative photos were shown. b The resazurin assay indicates metabolic activity in the ΔatfA resting conidia, which are without nutrients. c The XTT assay shows metabolic activity in the ΔatfA resting conidia, which are without nutrients
We then investigated the metabolic activity of the resting conidia using respiration indicators, resazurin, and XTT reagents (Fig. 7b and c). When incubated in water without any carbon sources, the wildtype conidia showed no change in color, indicating a metabolically inactive state. On the contrary, the water containing atfA mutant conidia changed color based on the respiration markers, suggesting that the conidia defective in AtfA were somehow metabolically active even in the absence of nutrients. Collectively, these germination-related traits suggested that AtfA has a role in maintaining conidial dormancy, and the deletion of AtfA led to the derepression of dormancy.
Discussion
Dormant conidia of filamentous fungi are easy-to-disperse, stress-tolerant structures [1]. These properties enable the fungi to survive in a variety of harsh environments. Thus, there is a keen interest in the molecular mechanisms of conidial dormancy and germination. We used Aspergillus conidia as a model to gain a transcriptomic view of dormancy and the exit from dormancy. We used three Aspergillus species for transcriptome comparisons, which enabled us to achieve more universal findings.
The comparative analysis identified the common CAGs and GeAGs, which revealed that transcripts associated with stress tolerance and ribosome biogenesis are abundant in the conidia and 1 h-grown conidia, respectively. In fact, CatA, Cu/Zn superoxide dismutase (sod1), and trehalose synthase (tpsA) genes were found to be highly expressed in the conidia, which was consistent with the previous studies [13, 25, 26]. The conidia devoid of CatA showed an increased sensitivity to hydrogen peroxide in A. fumigatus and A. nidulans, indicating that oxidative stress tolerance in the Aspergillus conidia requires CatA [12, 13]. Our transcriptome data also showed that unstudied putative catalase genes (Afu2g00200, An12g10720, and AO090113000153) were commonly up-regulated in the conidia (Table 3). The amino acid sequences of the proteins are well conserved among the Aspergillus species, and they have a catalase core domain (IPRO: 11614), suggesting that it may play an important role in oxidative stress adaptation in the conidia. Notably, this putative catalase (here designated as Cat3), as well as CatA, was expressed in an AtfA-dependent manner (Fig. 4). Another catalase gene, cat1/catB, showed mycelia-specific expression in A. niger and A. oryzae, but not in A. fumigatus in which the expression level of cat1 (Afu3g02270) was quite low in all of the cell forms tested (Additional file 5). The bifunctional catalase-peroxidase cat2/cpeA/catD was highly expressed in the conidia of A. fumigatus (cat2), whereas the expression levels were quite low in A. oryzae (Additional file 5). Collectively, A. fumigatus conidia contained high transcript levels of the three catalase genes, catA, cat2, and cat3, which might contribute to the protection of resting conidia from oxidative stresses in the environments.
In addition to catalases, the CAGs included an array of genes associated with conidial biology. VosA, a velvet-family protein, is involved in trehalose accumulation in the conidia and UV-tolerance [18]. The opsin 1 gene encodes a protein homologous to Neurospora crassa rhodopsin NOP-1 that is expressed in a conidiation-related manner [27]. The most recent report demonstrated that Fusarium fujikuroi rhodopsin was highly accumulated in the conidia produced under light conditions and the F. fujikuroi rhodopsin functioned as a light-dependent proton pump [28]. One could hypothesize that light-dependent biological roles are conserved across most fungal spores/conidia. We also found that aquaporin genes were specifically expressed in the conidia. Although the function of aquaporin remains to be investigated in filamentous fungi, it was demonstrated that S. cerevisiae aquaporin was required for normal sporulation and freeze tolerance [29, 30], from which one could hypothesize that aquaporin might play an important role in conidial maturation by allowing water outflow. Taking these results into account, our comparative transcriptome analysis provided a novel insight into the molecular mechanisms underlying conidial dormancy and maturation. However, the GeAGs mainly included genes involved in fundamental cellular processes, such as ribosome biogenesis, nucleotide biogenesis, ubiquitin, and translation factors (Table 4). This corroborated previous transcriptome and proteome studies for Aspergillus species [2, 4, 6, 11].
In general, comparisons between transcriptome and proteome data provide results that may aid in understanding cellular functions and mechanisms. Teutschbein et al. [5] previously presented 449 proteins from the A. fumigatus resting conidia. We compared the conidial proteins against the transcriptome data from the present study, revealing that 448 had transcripts in the conidia and 284 (63.4 %) exhibited no less than the mean FPKM value. The genes corresponding to the proteins from the 40 most abundant spots in the conidial proteome analysis all showed greater than average FPKM values, except rodA, and 21 (52.5 %) were more highly expressed in the conidia than in the hyphae (> fourfold). This suggested a good accordance between the proteins expressed and the genes present in the resting conidia. However, when we compared the 100 most highly expressed CAGs against the 448 conidial proteins, only 26 corresponding proteins were found. When we compared the 687 CAGs against the proteins, 104 of the CAGs (15.1 %) were found in the conidial proteome. This comparison suggested that the transcripts with high FPKM levels were not necessarily translated in the resting conidia and that they might exist in an mRNA form. If this hypothesis is true, the conidial dormancy (metabolic inactive state) could be resulted from the absence of proteins necessary for metabolic and cellular activities in the resting conidia. Furthermore, the pre-packed mRNAs in resting conidia could contribute to an immediate response to exit from dormancy when the conidia sense appropriate nutrients in the extracellular environment. However, there is an open question regarding the mechanism underlying the preservation of mRNA in the resting conidia during dormancy.
In the present study, more than half of the A. fumigatus CAGs were regulated for their expression in an AtfA-dependent manner (Fig. 5). Compared with the A. fumigatus CAGs, the common CAGs contained a higher percentage (64 %) of AtfA-dependent genes, indicating that AtfA plays a more crucial role in the biology of Aspergillus resting conidia than was previously thought. The function of AtfA has been intensely studied in A. nidulans, A. oryzae, and A. fumigatus, in which the deletion of the atfA gene commonly resulted in extremely stress-labile conidia and the down-regulation of catA expression in the conidia [16, 21, 22]. As stated above, our data showed that not only catA, but also other catalase genes were highly expressed in the conidia of A. fumigatus in an AtfA-dependent manner. The dehydrin-like proteins DprA, DprB, and DprC, which play roles in the stress adaptation of conidia, were also enriched in the A. fumigatus conidia, and the transcriptional regulation of DprA and DprC was largely dependent on AtfA (Additional file 7). Notably, the atfA deletion mutants of A. fumigatus and A. nidulans are able to produce as many conidia as the wildtype [16, 21]. This clearly indicated that AtfA has no role in conidiation, and suggested that AtfA is likely to be essential for the conidial maturation process, in which the conidia enter dormancy and become more tolerant to environmental stresses.
In the germination process, calA was highly up-regulated in all of the species tested. This suggested that CalA protein commonly play an important role in initiating the germination of Aspergillus conidia. In fact, it was previously reported in A. nidulans that CalA and the paralog protein CetA were required for normal germination and that the GFP-fused proteins were localized to the hyphal periphery [20]. Intriguingly, plant thaumatin-like proteins are able to bind to polysaccharides like beta-glucan and showed beta-1,3-glucanase activity [31–33]. Some were identified as antifungal proteins induced by pathogen exposure. From these characteristics, one could hypothesize that fungal thaumatin-like proteins are involved particularly in conidial isotropic growth (swelling) by loosening cell walls with hydrolyzing activity. This hypothesis was partly supported by the findings that the abnormal expression of calA and calB (Fig. 6) and an abnormal progression of swelling (Fig. 7a) were found in the atfA mutant’s resting conidia. The fungal thaumatin-like proteins were widely conserved in ascomycetes to basidiomycetes, including plant pathogens, which suggested an important role in the biology of filamentous fungi [34]. Therefore, a further functional analysis of the CalA-family protein may increase our understanding of the germination mechanism, as well as the interactions, between plants and pathogens.
One of the notable findings is the derepression of dormancy in the conidia of the A. fumigatus atfA deletion mutant, in which filipin-positive staining and metabolic activity, that were absent in the wildtype conidia, were found. These findings clearly indicated an abnormal exit from dormancy and the partial progression of the germination process. In general, the germination process of Aspergillus conidia starts when the cells perceive carbohydrates, like glucose. The nutrient cues are sensed and signaled via G-proteins and the cAMP-dependent protein kinase A signaling pathway [35, 36]. In the atfA mutant conidia, the signaling pathway might be constitutively activated by an unknown mechanism and subsequently the germination process starts without any extracellular nutrients. The source of the energy for boosting the metabolism in the resting conidia is unknown. One possibility is the glucose derived from the degradation of conidia-accumulated trehalose, which is thought to be used during germination in wildtype conidia. In fact, a lowered level of trehalose was found in the atfA deleted conidia compared with the wildtype conidia [16]. It may be possible that trehalose is abnormally degraded in the resting conidia of the atfA mutant, which in turn results in the exit from dormancy. Besides extracellular glucose, precocious consumption of the immobilized intracellular glucose may account for delayed germination phenotype of the atfA mutant. Although the detailed mechanisms are still obscure, AtfA is essential for the maintenance of metabolically inactive dormancy in resting conidia.
Conclusions
Comparisons among transcriptomes of hyphae, conidia, and 1 h-grown conidia of Aspergillus species provided a wide array of genes differentially expressed in each form. These findings further highlighted the essential role of the AtfA transcription factor in conidial dormancy. In addition, the thaumatin-like proteins are exclusively expressed at the germination stage, suggesting an important role in the germination of Aspergillus species. These results will facilitate the further investigation of molecular mechanisms underlying conidial dormancy and germination.
Methods
Strains and growth conditions
A. fumigatus strain Af293, A. niger strain IFM 58835, and A. oryzae strain RIB40 were used for the transcriptome analysis. To investigate AtfA roles, ΔatfA and the corresponding parental strain Afs35 of A. fumigatus were used [16]. All strains were cultivated on PDA plates. To collect hyphae, the strains were cultured in PDB for 18 h (A. fumigatus) or 26 h (A. niger and A. oryzae).
Conidia preparation
Conidia of each strain were stored in 20 % glycerol in a −80 °C freezer. To prepare fresh conidia, the stored conidia were inoculated on a PDA slant and incubated at 37 °C (A. fumigatus) or 30 °C (A. niger and A. oryzae) for 1 week. The conidia were harvested with phosphate-buffered saline (PBS) containing 0.1 % Tween 20, and the concentration was calculated by counting the conidia with a hemocytometer (Watoson, Kobe, Japan). To collect conidia for RNA purification, conidia were mixed with 15 mL of PDA (final concentration, approximately 104 conidia/mL) before the medium solidified in a 100-mL flask, and they were incubated at 30 or 37 °C for 7 d in the dark. After cultivation, conidia were harvested with PBS-Tween 20, filtered through a Miracloth, counted with a hemocytometer, and washed once with PBS-Tween 20. Microscopic observations revealed that there were no hyphal fragment contaminants in the conidial suspension, which contained exclusively conidia.
When 1 h-grown conidia were collected, approximately 109 conidia were shaken in 10 mL PDB for 1 h and then centrifuged. To prepare the dried conidia, freshly harvested conidia were centrifuged and the supernatant was discarded. The conidia were dried in a vacuum for 2 h, and then incubated at room temperature for 7 d.
RNA and cDNA preparation
Mycelia, conidia, and 1 h-grown conidia from duplicate independent culture were pooled and frozen in liquid nitrogen, and total RNA was isolated using the FastRNA Pro Red Kit (MP Biomedicals, Santa Ana, CA, USA). To obtain cDNA pools from the total RNA, possible contaminating genomic DNA was removed, and reverse transcription was performed using the ReverTra Ace qPCR RT Master Mix with gDNA remover (Toyobo).
RNA-sequencing analysis
An indexed cDNA library was prepared using the SureSelect Strand-Specific RNA Seq (Agilent Technologies) according to standard protocols. Briefly, each total RNA sample (1 μg) was enriched for mRNA using oligo(dT)-tagged beads. RNA samples were fragmented into smaller pieces and used to synthesize cDNAs. The library construction involved end repair, A-tailing, adapter ligation, and amplification. The mean length of each library was approximately 290 bp. Sequencing was performed in a single-end 60-base pair mode on a Hiseq system (Illumina). The RNA-sequencing read data was deposited to the DDBJ Sequence Read Archive under accession No. PRJDB4747.
Expression analysis
Illumina data sets were trimmed using fastq-mcf in ea-utils (v1.1.2-484) [37], where sequencing adapters and sequences with low-quality scores (Phred score Q < 20) were removed. Cleaned reads were mapped to the genome sequence of A. fumigatus Af293 (29,420,142 bp; genome version: s03-m04-r31), A. niger CBS (33,975,768 bp; genome version: s01-m06-r19), or A. oryzae RIB40 (37,912,014 bp; genome version: s01-m08-r26) from AspGD (http://www.aspgd.org/) using TopHat (v2.0.4) with the default parameters [38]. FPKMs were calculated using cuffdiff in Cufflinks (v2.1.1) with default parameters [39]. Data analyses were conducted using the R programming language (https://www.r-project.org/), and cummeRbund software [40].
GO analysis
Genes were functionally categorized using their GO information [41] obtained from AspGD, and overrepresented GO terms were identified using Fisher’s exact test. The one-tailed Fisher’s exact p-value corresponding to the overrepresentation of GO categories with equal to, or greater than, 20 genes was calculated based on counts in 2 × 2 contingency tables [42]. p-values were corrected by the false discovery rate method [43], and the threshold was set as 0.01.
Identification of orthologous genes
BLASTP (v2.2.28+) analyses [44] querying all of the protein sequences from one Aspergillus species against those of the other Aspergillus species were conducted using BLOSUM 80. The reciprocal best-hit pairs between two species—7,302 genes between A. fumigatus and A. niger; 7,216 between A. fumigatus and A. oryzae, and 7,636 between A. niger and A. oryzae—were extracted. Finally, 6,172 orthologous gene sets were identified among the three species and used in this study.
Quantitative real-time RT-PCR
Real-time RT-PCR was performed using SYBR Green detection as described previously [45]. The primer sets used in this study are listed in Additional file 8. The relative expression ratios were calculated by the comparative cycle threshold (Ct) (ΔΔCt) method. The actin gene was used as a normalization reference (internal control). Each sample was tested in triplicate.
Filipin staining
The resting conidia of interest were harvested as described above. 108 conidia were centrifuged and then washed once with PBS + Tween 20. They were dissolved in 200 μL of PBS + Tween20 (final concentration, 5 × 108/mL). A stock solution (1 mM) of filipin (Sigma) was prepared in DMSO, and 5 μL of the solution was added to 50 μL of the conidial suspensions. After incubation at room temperature for 3 min, they were centrifuged, immediately dissolved by an equivalent volume of PBS + Tween20, and observed under a fluorescent microscope.
Metabolic activity assay
To compare the respiration activity in conidia, a resazurin assay was conducted. The conidia (2 × 107–2 × 106) of each strain were incubated in 1 mL of distilled water with resazurin (final concentration, 0.1 mM) at 37 °C for 20 h and then photographed. The tetrazolium salt 2,3-bis(2-methoxy-4-nitro-5-sulfophenyl)-2H-tetrazolium-5-carbox-anilide (XTT) assay was conducted. The conidia (2 × 107–2 × 106) of each strain were incubated in 0.4 mL of distilled water with 0.1 mL of XTT solution, containing 1 mg/mL XTT and 50 μM menadione, at 37 °C for 4 h and then photographed.
Ethics approval
Not applicable.
Consent for publication
Not applicable.
Availability of data and material
The datasets, supporting the conclusions of this article, is available in a BioProject PRJDB4747 deposited to DDBJ. All fungal strains are available from National BioResource Project of Mycology Research Center of Chiba University (https://daphne.pf.chiba-u.jp/distribution/catalog).
Acknowledgments
We would like to thank Dr. Atsushi Iwama, Dr. Motohiko Ohshima, and Dr. Atsunori Saraya (Chiba University) for technical support with the Illumina HiSeq 1500.
Funding
This work was supported by a grant from the Noda Institute for Science Research to D.H. This work was also supported in part by the Japanese Ministry of Education, Culture, Sports, Science and Technology (MEXT) Special Budget for Research Project: The Project on Controlling Aspergillosis and the Related Emerging Mycoses, a Cooperative Research Grant of NEKKEN (2014–2015), and the National BioResource Project for Pathogenic Microbes funded by the MEXT, Japan (http://www.nbrp.jp/). MEXT provided funding to H.T. and Y.K. under grant number 221S0002.
Abbreviations
- PDA
potato dextrose agar
- BLOSUM
Blocks substitution matrix
- bZIP
basic leucine zipper
- CAG
conidia-associated gene
- DMSO
dimethyl sulfoxide
- FPKM
fragments per kilobase of transcript per million mapped reads
- GeAG
germination-associated gene
- GFP
green fluorescent protein
- GO
gene ontology
- PBS
phosphate-buffered saline
- PDB
potato dextrose broth
- RT-PCR
reverse transcription polymerase chain reaction
Additional files
Growth test for the strains used in this study. (A) The conidia (approximately 105) of each strain were point-inoculated on PDA, and the palates were incubated for 5 d at the indicated temperatures before photographing. (B) The numbers of conidia were calculated. Conidia of each strain were mixed with 3 mL of PDA (final concentration, approximately 104 conidia/mL) before the medium solidified in a well of 6-well plate, and they were incubated at indicated temperatures for 7 d in the dark. After cultivation, conidia were harvested with PBS-Tween 20, filtered through a Miracloth, and counted with a hemocytometer. The data were obtained in triplicate and the mean value was shown. An error bar indicates standard deviation. (C) Germination rate was calculated in each strain. The conidia (final concentration, 106 conidia/mL) were incubated in 0.5 mL of PDB in an eppendorf tube and were shaken at 160 rpm. At each indicated time-point, the conidia were checked if the germ tube appeared. The data were obtained in at least 5 replicates and the mean value was shown. An error bar indicates standard deviation. (D) Growth in liquid culture was investigated. The conidia (final concentration, 2 × 105 conidia/mL) were incubated in 30 mL of PDB in a 100 mL flask and were agitated at 160 rpm. At each indicated time-point, mycelia were harvested, frozen, and preserved in a −80 °C freezer before being analyzed. The mycelia were lyophilized and weighed. The data were obtained in triplicate and the mean value was shown. An error bar indicates standard deviation. (PPTX 2850 kb)
GO terms that were enriched in CAGs. (XLSX 11 kb)
GO terms that were enriched in GeAGs. (XLSX 56 kb)
List of A. fumigatus allergen-related genes and their FPKM values. (XLSX 12 kb)
FPKM values of the 6172 common genes. (XLSX 986 kb)
Aspergillus CalA-family proteins. (A) The protein lengths and gene IDs are shown. (B) Alignment of the CalA-family protein sequences. This alignment was constructed by ClustalW. (PPTX 79 kb)
Quantitative real-time RT-PCR analyses of dehydrin-like genes in A. fumgiatus. Expression profiles of the dprA, dprB, and dprC in the resting conidia of Afs35 (WT) and ΔatfA were analyzed. Each value represents the expression ratio relative to that of actin. Data presented are averages of three replicates, and the bar indicates standard deviation. (PPTX 55 kb)
List of primers used in this study. (XLSX 51 kb)
Footnotes
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
DH conceived the study, participated in designing and coordinating the study, carried out the RNA-seq and the real-time qRT-PCR experiments, evaluated the transcriptome data, and wrote the manuscript. HT and YK carried out the RNA-seq data analyses and GO analysis. SK, KK, and TG coordinated the study. All of the authors read and approved the final manuscript.
References
- 1.Wyatt TT, Wösten HA, Dijksterhuis J. Fungal spores for dispersion in space and time. Adv Appl Microbiol. 2013;85:43–91. doi: 10.1016/B978-0-12-407672-3.00002-2. [DOI] [PubMed] [Google Scholar]
- 2.Lamarre C, Sokol S, Debeaupuis JP, Henry C, Lacroix C, Glaser P, Coppée JY, François JM, Latgé JP. Transcriptomic analysis of the exit from dormancy of Aspergillus fumigatus conidia. BMC Genomics. 2008;9:417. doi: 10.1186/1471-2164-9-417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.van Leeuwen MR, Krijgsheld P, Bleichrodt R, Menke H, Stam H, Stark J, Wösten HA, Dijksterhuis J. Germination of conidia of Aspergillus niger is accompanied by major changes in RNA profiles. Stud Mycol. 2013;74:59–70. doi: 10.3114/sim0009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Novodvorska M, Hayer K, Pullan ST, Wilson R, Blythe MJ, Stam H, Stratford M, Archer DB. Trancriptional landscape of Aspergillus niger at breaking of conidial dormancy revealed by RNA-sequencing. BMC Genomics. 2013;14:246. doi: 10.1186/1471-2164-14-246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Teutschbein J, Albrecht D, Pötsch M, Guthke R, Aimanianda V, Clavaud C, Latgé JP, Brakhage AA, Kniemeyer O. Proteome profiling and functional classification of intracellular proteins from conidia of the human-pathogenic mold Aspergillus fumigatus. J Proteome Res. 2010;9:3427–42. doi: 10.1021/pr9010684. [DOI] [PubMed] [Google Scholar]
- 6.Oh YT, Ahn CS, Kim JG, Ro HS, Lee CW, Kim JW. Proteomic analysis of early phase of conidia germination in Aspergillus nidulans. Fungal Genet Biol. 2010;47:246–53. doi: 10.1016/j.fgb.2009.11.002. [DOI] [PubMed] [Google Scholar]
- 7.Rispail N, Soanes DM, Ant C, Czajkowski R, Grünler A, Huguet R, Perez-Nadales E, Poli A, Sartorel E, Valiante V, Yang M, Beffa R, Brakhage AA, Gow NA, Kahmann R, Lebrun MH, Lenasi H, Perez-Martin J, Talbot NJ, Wendland J, Di Pietro A. Comparative genomics of MAP kinase and calcium-calcineurin signalling components in plant and human pathogenic fungi. Fungal Genet Biol. 2009;46:287–98. doi: 10.1016/j.fgb.2009.01.002. [DOI] [PubMed] [Google Scholar]
- 8.Gibbons JG, Rokas A. The function and evolution of the Aspergillus genome. Trends Microbiol. 2013;21:14–22. doi: 10.1016/j.tim.2012.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Perez-Nadales E, Nogueira MF, Baldin C, Castanheira S, El Ghalid M, Grund E, Lengeler K, Marchegiani E, Mehrotra PV, Moretti M, Naik V, Oses-Ruiz M, Oskarsson T, Sch├ñfer K, Wasserstrom L, Brakhage AA, Gow NA, Kahmann R, Lebrun MH, Perez-Martin J, Di Pietro A, Talbot NJ, et al. Fungal model systems and the elucidation of pathogenicity determinants. Fungal Genet Biol. 2014;70:42–67. [DOI] [PMC free article] [PubMed]
- 10.Low SY, Dannemiller K, Yao M, Yamamoto N, Peccia J. The allergenicity of Aspergillus fumigatus conidia is influenced by growth temperature. Fungal Biol. 2011;115:625–32. doi: 10.1016/j.funbio.2011.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Suh MJ, Fedorova ND, Cagas SE, Hastings S, Fleischmann RD, Peterson SN, Perlin DS, Nierman WC, Pieper R, Momany M. Development stage-specific proteomic profiling uncovers small, lineage specific proteins most abundant in the Aspergillus fumigatus conidial proteome. Proteome Sci. 2012;10:30. doi: 10.1186/1477-5956-10-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Navarro RE, Stringer MA, Hansberg W, Timberlake WE, Aguirre J. catA, a new Aspergillus nidulans gene encoding a developmentally regulated catalase. Curr Genet. 1996;29:352–9. [PubMed] [Google Scholar]
- 13.Paris S, Wysong D, Debeaupuis JP, Shibuya K, Philippe B, Diamond RD, Latgé JP. Catalases of Aspergillus fumigatus. Infect Immun. 2003;71:3551–62. doi: 10.1128/IAI.71.6.3551-3562.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hisada H, Hata Y, Kawato A, Abe Y, Akita O. Cloning and expression analysis of two catalase genes from Aspergillus oryzae. J Biosci Bioeng. 2005;99:562–8. doi: 10.1263/jbb.99.562. [DOI] [PubMed] [Google Scholar]
- 15.Suzuki S, Sarikaya Bayram Ö, Bayram Ö, Braus GH. conF and conJ contribute to conidia germination and stress response in the filamentous fungus Aspergillus nidulans. Fungal Genet Biol. 2013;56:42–53. doi: 10.1016/j.fgb.2013.04.008. [DOI] [PubMed] [Google Scholar]
- 16.Hagiwara D, Suzuki S, Kamei K, Gonoi T, Kawamoto S. The role of AtfA and HOG MAPK pathway in stress tolerance in conidia of Aspergillus fumigatus. Fungal Genet Biol. 2014;73:138–49. doi: 10.1016/j.fgb.2014.10.011. [DOI] [PubMed] [Google Scholar]
- 17.Ni M, Yu JH. A novel regulator couples sporogenesis and trehalose biogenesis in Aspergillus nidulans. PLoS One. 2007;2 doi: 10.1371/journal.pone.0000970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Park HS, Ni M, Jeong KC, Kim YH, Yu JH. The role, interaction and regulation of the velvet regulator VelB in Aspergillus nidulans. PLoS One. 2012;7 doi: 10.1371/journal.pone.0045935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Upadhyay SK, Mahajan L, Ramjee S, Singh Y, Basir SF, Madan T. Identification and characterization of a laminin-binding protein of Aspergillus fumigatus: extracellular thaumatin domain protein (AfCalAp) J Med Microbiol. 2009;58:714–22. doi: 10.1099/jmm.0.005991-0. [DOI] [PubMed] [Google Scholar]
- 20.Belaish R, Sharon H, Levdansky E, Greenstein S, Shadkchan Y, Osherov N. The Aspergillus nidulans cetA and calA genes are involved in conidial germination and cell wall morphogenesis. Fungal Genet Biol. 2008;45:232–42. doi: 10.1016/j.fgb.2007.07.005. [DOI] [PubMed] [Google Scholar]
- 21.Hagiwara D, Asano Y, Yamashino T, Mizuno T. Characterization of bZip-type transcription factor AtfA with reference to stress responses of conidia of Aspergillus nidulans. Biosci Biotechnol Biochem. 2008;72:2756–60. doi: 10.1271/bbb.80001. [DOI] [PubMed] [Google Scholar]
- 22.Sakamoto K, Iwashita K, Yamada O, Kobayashi K, Mizuno A, Akita O, Mikami S, Shimoi H, Gomi K. Aspergillus oryzae atfA controls conidial germination and stress tolerance. Fungal Genet Biol. 2009;46:887–97. doi: 10.1016/j.fgb.2009.09.004. [DOI] [PubMed] [Google Scholar]
- 23.Osherov N, May GS. The molecular mechanisms of conidial germination. FEMS Microbiol Lett. 2001;199:153–60. doi: 10.1111/j.1574-6968.2001.tb10667.x. [DOI] [PubMed] [Google Scholar]
- 24.Van Leeuwen MR, Smant W, de Boer W, Dijksterhuis J. Filipin is a reliable in situ marker of ergosterol in the plasma membrane of germinating conidia (spores) of Penicillium discolor and stains intensively at the site of germ tube formation. J Microbiol Methods. 2008;74:64–73. doi: 10.1016/j.mimet.2008.04.001. [DOI] [PubMed] [Google Scholar]
- 25.Lambou K, Lamarre C, Beau R, Dufour N, Latge JP. Functional analysis of the superoxide dismutase family in Aspergillus fumigatus. Mol Microbiol. 2010;75:910–23. doi: 10.1111/j.1365-2958.2009.07024.x. [DOI] [PubMed] [Google Scholar]
- 26.Al-Bader N, Vanier G, Liu H, Gravelat FN, Urb M, Hoareau CM, Campoli P, Chabot J, Filler SG, Sheppard DC. Role of trehalose biosynthesis in Aspergillus fumigatus development, stress response, and virulence. Infect Immun. 2010;78:3007–18. doi: 10.1128/IAI.00813-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bieszke JA, Li L, Borkovich KA. The fungal opsin gene nop-1 is negatively-regulated by a component of the blue light sensing pathway and influences conidiation-specific gene expression in Neurospora crassa. Curr Genet. 2007;52:149–57. doi: 10.1007/s00294-007-0148-8. [DOI] [PubMed] [Google Scholar]
- 28.Garcia-Martinez J, Brunk M, Avalos J, Terpitz U. The CarO rhodopsin of the fungus Fusarium fujikuroi is a light-driven proton pump that retards spore germination. Sci Rep. 2015;5:7798. doi: 10.1038/srep07798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tanghe A, Van Dijck P, Dumortier F, Teunissen A, Hohmann S, Thevelein JM. Aquaporin expression correlates with freeze tolerance in baker’s yeast, and overexpression improves freeze tolerance in industrial strains. Appl Environ Microbiol. 2002;68:5981–9. doi: 10.1128/AEM.68.12.5981-5989.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sidoux-Walter F, Pettersson N, Hohmann S. The Saccharomyces cerevisiae aquaporin Aqy1 is involved in sporulation. Proc Natl Acad Sci U S A. 2004;101:17422–7. doi: 10.1073/pnas.0404337101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Trudel J, Grenier J, Potvin C, Asselin A. Several thaumatin-like proteins bind to beta-1,3-glucans. Plant Physiol. 1998;118:1431–8. doi: 10.1104/pp.118.4.1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Grenier J, Potvin C, Trudel J, Asselin A. Some thaumatin-like proteins hydrolyse polymeric beta-1,3-glucans. Plant J. 1999;19:473–80. doi: 10.1046/j.1365-313X.1999.00551.x. [DOI] [PubMed] [Google Scholar]
- 33.Osmond RI, Hrmova M, Fontaine F, Imberty A, Fincher GB. Binding interactions between barley thaumatin-like proteins and (1,3)-beta-D-glucans. Kinetics, specificity, structural analysis and biological implications. Eur J Biochem. 2001;268:4190–9. doi: 10.1046/j.1432-1327.2001.02331.x. [DOI] [PubMed] [Google Scholar]
- 34.Franco Sde F, Baroni RM, Carazzolle MF, Teixeira PJ, Reis O, Pereira GA, Mondego JM. Genomic analyses and expression evaluation of thaumatin-like gene family in the cacao fungal pathogen Moniliophthora perniciosa. Biochem Biophys Res Commun. 2015;466:629–36. doi: 10.1016/j.bbrc.2015.09.054. [DOI] [PubMed] [Google Scholar]
- 35.Lafon A, Seo JA, Han KH, Yu JH, d’Enfert C. The heterotrimeric G-protein GanB(alpha)-SfaD(beta)-GpgA(gamma) is a carbon source sensor involved in early cAMP-dependent germination in Aspergillus nidulans. Genetics. 2005;171:71–80. doi: 10.1534/genetics.105.040584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fuller KK, Richie DL, Feng X, Krishnan K, Stephens TJ, Wikenheiser-Brokamp KA, Askew DS, Rhodes JC. Divergent Protein Kinase A isoforms co-ordinately regulate conidial germination, carbohydrate metabolism and virulence in Aspergillus fumigatus. Mol Microbiol. 2011;79:1045–62. doi: 10.1111/j.1365-2958.2010.07509.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Aronesty E. Comparison of sequencing utility programs. Open Bioinf J. 2013;9:1–8. doi: 10.2174/1875036201307010001. [DOI] [Google Scholar]
- 38.Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013;14:R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, Pimentel H, Salzberg SL, Rinn JL, Pachter L. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc. 2012;7:562–78. doi: 10.1038/nprot.2012.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Goff L, Trapnell C, Kelley D. cummeRbund: Analysis, exploration, manipulation, and visualization of Cufflinks high-throughput sequencing data. 2013, R package version 2.12.0
- 41.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, Harris MA, Hill DP, Issel-Tarver L, Kasarskis A, Lewis S, Matese JC, Richardson JE, Ringwald M, Rubin GM, Sherlock G. Gene ontology: tool for the unification of biology. The gene ontology consortium. Nat Genet. 2000;25:25–9. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Takahashi H, Morioka R, Ito R, Oshima T, Altaf-Ul-Amin M, Ogasawara N, Kanaya S. Dynamics of time-lagged gene-to-metabolite networks of Escherichia coli elucidated by integrative omics approach. OMICS. 2011;15:15–23. doi: 10.1089/omi.2010.0074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Statist Soc B. 1995;57:289–300. [Google Scholar]
- 44.Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hagiwara D, Takahashi-Nakaguchi A, Toyotome T, Yoshimi A, Abe K, Kamei K, Gonoi T, Kawamoto S. NikA/TcsC histidine kinase is involved in conidiation, hyphal morphology, and responses to osmotic stress and antifungal chemicals in Aspergillus fumigatus. PLoS One. 2013;8 doi: 10.1371/journal.pone.0080881. [DOI] [PMC free article] [PubMed] [Google Scholar]


