Abstract
Background
Corynebacterium pseudotuberculosis, a facultative intracellular bacterial pathogen, is the etiological agent of caseous lymphadenitis (CLA), an infectious disease that affects sheep and goats and it is responsible for significant economic losses. The disease is characterized mainly by bacteria-induced caseous necrosis in lymphatic glands. New vaccines are needed for reliable control and management of CLA. Thus, the putative virulence factors SpaC, SodC, NanH, and PknG from C. pseudotuberculosis FRC41 may represent new target proteins for vaccine development and pathogenicity studies.
Results
SpaC, PknG and NanH presented better vaccine potential than SodC after in silico analyses. A total of 136 B and T cell epitopes were predicted from the four putative virulence factors. A cluster analysis was performed to evaluate the redundancy degree among the sequences of the predicted epitopes; 57 clusters were formed, most of them (34) were single clusters. Two clusters from PknG and one from SpaC grouped epitopes for B and T-cell (MHC I and II). These epitopes can thus potentially stimulate a complete immune response (humoral and cellular) against C. pseudotuberculosis. Several other clusters, including two from NanH, grouped B-cell epitopes with either MHC I or II epitopes. The four target proteins were expressed in Escherichia coli. A purification protocol was developed for PknG expression.
Conclusions
In silico analyses show that the putative virulence factors SpaC, PknG and NanH present good potential for CLA vaccine development. Target proteins were successfully expressed in E. coli. A protocol for PknG purification is described.
Electronic supplementary material
The online version of this article (doi:10.1186/s12934-016-0479-6) contains supplementary material, which is available to authorized users.
Keywords: Corynebacterium pseudotuberculosis, Pathogenicity and virulence, Vaccine potential, Epitope prediction, Protein expression, Protein purification
Background
Caseous lymphadenitis (CLA) is a chronic, pyogenic, contagious disease of sheep and goat that imposes considerable economic losses for farmers in many countries [1, 2]. The disease is caused by Corynebacterium pseudotuberculosis (C. pseudotuberculosis): a gram-positive pleomorphic, non-capsulated, non-motile, fimbriated, facultative intracellular bacterium, multiplying within macrophages [1]. Corynebacterium ulcerans and C. pseudotubercu1osis produce phospholipase D (PLD), which is unique among corynebacteria. It promotes the hydrolysis of ester bonds in sphingomyelin in mammalian cell membranes, possibly contributing to the spread of the bacteria from the initial site of infection to the secondary sites within the host. Moreover, it provokes dermonecrotic lesions; and at higher doses it is lethal to a number of different species of laboratory and domestic animals [3–5].
CLA disease is expressed in external and visceral forms, either separately or together [3–5]. External CLA lesions appear initially as abscesses that convert later on to pyogranulomas ranging in size from millimeters to centimeters. These external lesions are mostly located within superficial lymph nodes, but infrequently in subcutaneous tissues. Wool or hair over CLA lesions may be lost due to the weak dermonecrotic action of C. pseudotuberculosis exotoxins and the pressure atrophy of overlying skin by the lesions. Visceral lesions are not detectable clinically but express themselves according to their number, site and effect on the involved organ. Progressive weight loss, respiratory disorders and chronic recurrent ruminal tympany are the most prominent signs that may accompany visceral CLA lesions.
Identification/removal of infected animals is a key factor for success of disease control measures. Vaccination of healthy animals is another strategy broadly recommended for disease control. In fact, control of CLA depends on vaccination in most countries [2, 5–7]. Although bacterin, toxoid, combined, and live vaccines are available, the disease has persisted even after prolonged vaccination, indicating the suppressive nature of CLA vaccination [5, 7]. C. pseudotuberculosis infection of farmer animals can contaminate meat and milk, putting consumers at risk due to its zoonotic potential [7]. The ability of C. pseudotuberculosis to infect both animals and humans makes necessary the development of new vaccines for a reliable control and management of CLA once the currently available commercial vaccines are unable to fully protect susceptible animals against the disease [7, 8]. In this way, the study of other C. pseudotuberculosis virulence factors that might be involved in CLA pathogenesis can provide new vaccine targets.
The complete genome sequence of a C. pseudotuberculois strain (FRC41) isolated from a 12-year-old girl with necrotizing lymphadenitis allowed the identification of spaC and nanH as genes encoding proteins regarded as potential virulence factors [8]. SpaC is a putative adhesive pili tip protein. The pilus structure can probably make the initial contact with host cell receptors to enable additional ligand-receptor interactions and to facilitate the efficient delivery of virulence factors and intracellular invasion [9]. NanH, by its turn, is a putative extracellular neuraminidase [8]. Neuraminidases, or sialidases, belong to a class of glycosyl hydrolases that catalyze the removal of terminal sialic acid residues from a variety of glycoconjugates and can contribute to the recognition of sialic acids exposed on host cell surfaces. Most sialidase-producing microorganisms are pathogenic or commensal when in close contact with mammalian hosts. It has been also suggested that, in some types of pathogenic bacteria, sialidases function as potential virulence factors that contribute to the recognition of sialic acids exposed on the surface of the host cell [10]. A homologous counterpart of C. pseudotuberculois FRC41 NanH was characterized in C. diphtheriae KCTC3075 and shown to be a protein containing neuraminidase and trans-sialidase activities [11].
The C. pseudotuberculosis FRC41 genome also encodes a putative secreted copper,zinc-dependent superoxide dismutase (SodC) that is characterized by a lipobox motif and may be anchored in the cell membrane [8]. The extracellular location of this enzyme suggests that it may protect the surface of C. pseudotuberculosis cells against superoxide generated externally by the mammalian host cells. In Mycobacterium tuberculosis, SodC contributes to the resistance of this microorganism against the oxidative burst products generated by activated macrophages [12, 13]. The protective activity of Cu,Zn-SODs has been associated with virulence in other bacteria, such as Neisseria meningitides and Hemophylus ducreyi [8].
As part of important cell signaling mechanisms, eukaryotic-like serine/threonine protein kinases encountered in bacteria are a class of molecules that also deserves attention since they are part of complex signaling pathways and play a diversity of physiological roles in developmental processes, secondary metabolism, cell division, cell wall synthesis, essential processes, central metabolism, and virulence [14, 15]. Mycobacterium tuberculosis genome encodes 11 eukaryotic-like serine/threonine protein kinases (PknA to PknL, except for PknC). Protein kinase G (PknG) gained particular interest because it affects the intracellular traffic of M. tuberculosis in macrophages. Most microbes and nonpathogenic mycobacteria quickly find themselves in lysosomes, where they are killed. By contrast, M. tuberculosis stays within phagosomes; the bacterium releases PknG to block phagosome-lysosome fusion. Bacteria lacking pknG gene are rapidly transferred to lysosomes and eliminated [16, 17]. The genome of C. pseudotuberculosis FRC41 has a gene encoding for a putative PknG protein [8] but its function in the bacterium still needs to be investigated.
Therefore, C. pseudotuberculosis SpaC, NanH, SodC, and PknG proteins may play important roles in virulence and pathogenicity. In the present work, a characterization and evaluation of the vaccine potential of these proteins were performed in silico. The heterologous expression of these putative virulence factors in Escherichia coli is also described.
Methods
Protein sequences
The amino acid sequences of the target proteins were retrieved from NCBI GenBank: SpaC [gb| ADK29663.1], SodC [gb| ADK28404.1], NanH [gb| ADK28179.1], PknG [gb| ADK29622.1].
Homology searches
NCBI BLASTP [18] searches in UniProtKB database [19] were performed to identify homologues of the target proteins in the CMNR group of microorganisms (from Corynebacterium, Mycobacterium, Nocardi, and Rhodococcus genera): Corynebacteriumn, taxid:1716; Mycobacterium, taxid:1763; Nocardia, taxid:1817; Rhodococcus, taxid:1827. Likewise, BLASTP searches in UniprotKB database were performed to identify homologues of the target proteins in mammalian species of the Ovis (taxid: 9935), Bos (taxid: 9903), Equus (taxid: 9789), Equus (taxid: 35510), Mus (taxid: 10088), Mus (taxid: 862507) genera and in Homo sapiens (taxid: 9606). BLAST Genome [18] searches in C. pseudotuberculosis (taxid: 1719) complete genomes available at NCBI genome database were performed to identify the presence of the target protein genes in other C. pseudotuberculosis strains.
Primary and secondary structure analysis, subcellular localization and prediction of protective antigens
ProtParam [20] and Self-OPtimized prediction method with alignment—SOPMA [21] of expasy server were used to analyze different physiological and physicochemical properties of the target proteins. Molecular weight, theoretical pI, amino acid composition, extinction coefficient, estimated half-life, instability index, aliphatic index and grand average of hydropathicity (GRAVY) were calculated using the ProtParam preset parameters. Solvent accessibility, transmembrane helices, globular regions, bend region, random coil and coiled-coil regions were predicted using SOPMA default parameters. The amino acid sequences were evaluated by PSORTb 3.0.2 [22] to predict subcellular localization of the target proteins. SignalP 4.1 [23] was used to predict the presence and location of signal peptide cleavage sites in the amino acid sequences. The method incorporates a prediction of cleavage sites and a signal peptide/non-signal peptide prediction based on a combination of several artificial neural networks. VaxiJen 2.0 [24] was used for alignment-independent prediction of protective antigens. The tool was developed to allow antigen classification solely based on the physicochemical properties of proteins without the need of sequence alignment.
B-cell epitope prediction
Linear B-cell epitopes were predicted from the target protein sequences using physicochemical properties [25] estimated by in silico methods available in DNASTAR Protean program (Madison, Wisconsin). The Jameson–Wolf method [26] was used to predict the potential antigenic determinants by combining existing methods for protein structural predictions. The results appear as multiple peaks in the antigenic index plot, with each peak signifying a potential antigenic determinant. The emini surface probability method [27] was used to predict the probability that a given region lies on the surface of a protein. The Kyte–Doolittle hydropathy method [28] predicts regional hydropathy of proteins from their amino acid sequences. Hydropathy values are assigned for all amino acids and are then averaged over a user defined window. The average is plotted at the midpoint of the window. The charge density method predicts regions of positive and negative charge by summing charge over a specific range of residues. DNASTAR developed this method using the pK tables of White et al. [29]. Since charged residues tend to lie on the surfaces of proteins, this method aids in predicting surface characteristics. Several wet lab experiments revealed that the antigenic portions were situated in beta turn regions of a protein [30] for these regions the Chou and Fasman beta turn prediction method was used [31, 32]. The Karplus–Schulz flexibility method [33] predicts backbone chain flexibility. The method is useful for resolving antigenic sites, as these regions tend to be among the most flexible in a polypeptide sequence. Conserved domains in the target proteins were identified by searching NCBI’s conserved domain database (CDD) [34]. The results of each method were presented in a graphical frame. The peak of the amino acid residue segment above the threshold value (we used the default) is considered as predicted B-cell epitope. User can select any physicochemical property or a combination of two or more properties for epitope prediction. [35]. We selected amino acid segments in the target protein sequences where peaks above threshold overlapped in four or more methods. B-cell epitopes located in signal peptide or conserved domains were discarded.
T-cell epitope prediction
MHC I binding prediction was performed using the immune epitope database (IEDB) MHC I binding tool [36] and consensus [37] as prediction method which combines predictions from ANN aka NetMHC (3.4), SMM and comblib methods. Mouse MHC alleles (H-2-Db, H-2-Dd, H-2-Kb, H-2-Kd, H-2-Kk, H-2-Ld) and a peptide length of nine mer were selected to make the predictions from target proteins sequences. A median percentile rank of the four predictions methods was the Consensus representative percentile rank used to select the top 1 % of peptides. A small numbered percentile rank indicates high affinity.
MHC II binding predictions for target proteins were performed using NetMHCII 2.2 server [38] to predict binding of 15 mer peptides to two mouse MHC II alleles (H-2-IAb and H-2-IAd) using artificial neuron networks. The prediction values were given in nM IC50 values, and as a %-Rank to a set of 1,000,000 random natural peptides. Strong and weak binding (SB, WB) peptides were indicated in the output. T-cell epitopes located in signal peptide or conserved domains were discarded.
Epitope clustering
Epitope clustering was performed using the IEDB Epitope cluster analysis tool [36]. Clustal omega [39] was used to group predicted B and T-cell epitopes into clusters of similarity based on multiple sequence alignment and visual inspection. Clustal omega alignments were used to double check if single-sequence clusters generated by IEDB epitope cluster analysis tool were in fact composed of unique epitopes (no pairs).
Cloning procedures
Miniprep plasmid purifications, agarose gel electrophoresis, and E. coli media were as described [40]. Amino acids 2–23 and amino acids 2–31 were removed from sodC and nanH ORF sequences, respectively. These regions containing signal peptide were eliminated before cloning in order to improve protein expression since they are relatively rich in hydrophobic amino acids. ORF codons of all four target proteins were replaced by E. coli preferential codons [41]. Optimized ORF sequences were synthesized and individually cloned into pD444-NH expression vector (T5 promoter, IPTG inducible, strong ribosome binding site, His-tag, ampicillin resistance marker, high copy origin of replication, 4027 bp size) by DNA2.0 (Menlo Park, CA). Each ORF-containing plasmid (pD444-NH;pknG, pD444-NH;spaC, pD444-NH;sodC, and pD444-NH;nanH) was transformed into BL21(DE3) E. coli strains according to the OverExpress™ Electrocompetent Cells kit (Lucigen, Middleton) instructions.
Protein expression in E. coli
Protein expression protocol was according to OverExpress™ Electrocompetent Cells kit (Lucigen, Middleton) instructions. Briefly, transformed cell cultures at OD 0.5–0.7 were induced with 1 mM IPTG for 5 h at 37 °C. SDS-PAGE of non-induced and induced cell culture samples and Coomassie blue staining was as described [42].
Purification of PknG
Bacteria transformed with pD444-NH;pknG was induced as described above. Cell pellet was collected by 8000 rpm centrifugation, resuspended in buffer A (10 mM NaH2PO4 pH7.4, 300 mM NaCl, 1 % glycerol, 5 mM imidazole), lysed on ice with ten 15-s sonication pulses using a ultrasonic processor Marconi-MA 103 (Piracicaba, São Paulo) and centrifuged at 15,000×g for 15 min. The supernatant containing recombinant proteins was purified under native conditions using 1 mL of immobilized Ni Sepharose (GE Healthcare). The resin was washed using buffer A with 80 mM imidazole. Recombinant PknG was eluted from the column with buffer A containing 400 mM imidazole. The eluted protein was dialyzed against buffer B (10 mM NaH2PO4 buffer pH 7.4 and 50 mM NaCl) and concentrated by ultrafiltration. The concentrated fraction was injected on a Superdex 75 10/300 GL (GE Healthcare) size exclusion column previously equilibrated with buffer B. The purity of the sample was assessed by SDS–PAGE.
Results and discussion
Traditional vaccination approaches are based on complete pathogen either live attenuated or inactivated. Among the major problems these vaccines brought are crucial safety concerns, because those pathogens being used for immunization may become activated and cause infection. Moreover due to genetic variation of pathogen strains around the world, vaccines are likely to lose their efficacy in different regions or for a specific population. Novel vaccine approaches like DNA vaccines and epitope based vaccines have the potential to overcome these barriers to create more effective, specific, strong, safe and long lasting immune response without all undesired effects [43]. Next-generation sequencing and proteomic techniques have enabled researchers to mine entire microbial genomes, transcriptomes and proteomes to identify novel candidate immunogens [44]. In silico techniques are the best alternative to find out which regions of a protein out of thousands possible candidates are most likely to evoke immune response [35]. This reverse vaccinology approach has enjoyed considerable success in the past decade, beginning with Neisseria meningitides, and continuing with Streptococcus pneumonia, pathogenic E. coli, and antibiotic resistant Staphylococcus aureus [44].
Homology searches
The conservation level between target proteins and proteins of the CMNR group of microorganisms was evaluated by NCBI BLASTP [18] searches in UniprotKB database [19]. This kind of analysis is important for the development of vaccines once they can be used not only for C. pseudotuberculosis FRC41 but for other pathogen strains and pathogens of other species. NCBI BLAST Genome searches show the presence of the target protein genes in all 37 C. pseudotuberculosis strains currently available in NCBI complete genomes database (data not shown). This indicates that SpaC, SodC, NanH and PknG can potentially be expressed not only in a few strains demonstrating the importance of these proteins for this pathogenic bacterium. Well conserved homologous of the target proteins were also found in microorganisms of the CMNR group (Additional files 1, 2 and 3). These findings are a good indication that a vaccine against C. pseudotuberculosis made from the putative virulence factors can be effective not only against numerous strains of the pathogen but also against bacterial pathogens from other species.
The conservation degree among target proteins and mammalian (Ovis, Bos, Equus and Mus genera, Homo sapiens) proteins was also evaluated by BLASTP searches. The analysis was important to reveal the conservation degree among pathogen proteins and host proteins and so the possibility of undesirable immunological cross-reactions which may induce autoimmunity. The results (Additional files 1, 2 and 3) show that C. pseudotuberculosis FRC41 SpaC, SodC, NanH, and PknG sequences share low identity (30 % in average) with mammalian sequences. BLASTP alignments show that most of this weak homology is in conserved domains (data not shown). Thus, regions away from signal peptides and conserved domains are ideal targets for vaccine development.
Primary and secondary structure analysis
The next step was to evaluate the primary and secondary structure features of SpaC, SodC, NanH and PknG as they can predict stability and reveal functional characteristics of the proteins at some extent. Based on ProtParam instability index, SodC was considered the least stable while PknG was the most stable (Table 1). PknG was also the most hydrophilic with the highest GRAVY (−0.211). This same protein also presented the highest aliphatic (92.91) index (Table 1). SOPMA program, used to calculate secondary structure features of the target proteins, reported that SpaC, SodC and NanH were dominated by random coils, consisting in 45.35, 41.26 and 39.05 %, respectively (Table 2). Alpha helix prevailed (44.06 %) in PknG. The differences in secondary structure content and aliphatic character helps to explain the stability indexes estimated for the target proteins. [45].
Table 1.
Physicochemical properties of the target proteins estimated using ProtParam
| Physicochemical property | SpaC | SodC | NanH | PknG |
|---|---|---|---|---|
| Number of amino acids | 796 | 206 | 694 | 749 |
| Molecular weight | 85,964.9 | 21,099.3 | 74,683.3 | 83,349.4 |
| Theoretical pI | 5.13 | 5.96 | 5.05 | 5.13 |
| Total number of negatively charged residues (Asp + Glu) | 96 | 23 | 102 | 101 |
| Total number of positively charged residues (Arg + Lys) | 77 | 17 | 79 | 76 |
| Extinction coefficienta | 93,085 | 4595b | 77,600 | 81,375 |
| Abs 0.1 % (=1 g/l), assuming all pairs of Cys residues form cystines | 1.083 | 0.218b | 1.039 | 0.976 |
| Extinction coefficienta | 92,710 | 4470b | 77,350 | 81,250 |
| Abs 0.1 % (=1 g/l), assuming all Cys residues are reduced | 1.078 | 0.212b | 1.036 | 0.975 |
| The estimated half-life | ||||
| Mammalian reticulocytes, in vitro | 30 h | 30 h | 30 h | 30 h |
| Yeast, in vivo | >20 h | >20 h | >20 h | >20 h |
| Escherichia coli, in vivo | >10 h | >10 h | >10 h | >10 h |
| Instability index (II) | 28.21 (stable) | 19.62 (stable) | 32.92 (stable) | 38.18 (stable) |
| Aliphatic index | 80.16 | 71.65 | 72.58 | 92.91 |
| Grand average of hydropathicity (GRAVY) | −0.442 | −0.245 | −0.485 | −0.211 |
a Extinction coefficients are in units of M−1 cm−1, at 280 nm measured in water
b This protein does not contain any Trp residues. Experience shows that this could result in more than 10 % error in the computed extinction coefficient
Table 2.
Secondary structure content in the target proteins estimated using SOPMA
| Secondary structure | SpaC (%) | SodC (%) | NanH (%) | PknG (%) |
|---|---|---|---|---|
| Alpha helix (Hh) | 111 is 13.94 | 56 is 27.18 | 228 is 32.85 | 330 is 44.06 |
| 310 helix (Gg) | 0.00 | 0.00 | 0.00 | 0.00 |
| Pi helix(Ii) | 0.00 | 0.00 | 0.00 | 0.00 |
| Beta bridge (Bb) | 0.00 | 0.00 | 0.00 | 0.00 |
| Extended strand (Ee) | 244 is 30.65 | 39 is 18.93 | 128 is 18.44 | 114 is 15.22 |
| Beta turn (Tt) | 80 is 10.05 | 26 is 12.62 | 67 is 9.65 | 56 is 7.48 |
| Bend region (Ss) | 0.00 | 0.00 | 0.00 | 0.00 |
| Random coil (Cc) | 361 is 45.35 | 85 is 41.26 | 271 is 39.05 | 249 is 33.24 |
| Ambiguous states (?) | 0.00 | 0.00 | 0.00 | 0.00 |
| Other states | 0.00 | 0.00 | 0.00 | 0.00 |
Subcellular localization and prediction of protective antigens
The candidate molecules from a eukaryotic pathogen expected to induce immunity comprise proteins that are as follows: (i) present on the surface of the pathogen, (ii) excreted/secreted from the pathogen and (iii) homologous to known proteins involved in pathogenesis and virulence [46]. Signal peptide presence and subcellular localization (Table 3) of SpaC (cell wall), SodC (cytoplasmic membrane) and NanH (extracellular) was as predicted before [8]. They were predicted as protective antigens by VaxiJen. Membrane and secreted proteins are considered potential vaccine targets once they are at the host-pathogen interface. These proteins may interact more directly with host molecules for cell adhesion, invasion, multiplication, immune response evasion, damage generation to the host, and survive to host cell defenses [8, 47, 48].
Table 3.
Subcellular localization, signal peptide, and prediction of protective antigen for the target proteins
| Parameter (program) | SpaC | SodC | NanH | PknG |
|---|---|---|---|---|
| Subcellular localization (Psortb) | Cell wall (matched LPXTG; score 9.97) | Cytoplasmic Membrane (matched 61246116: superoxide dismutase Cu–Zn precursor; score 9.68) | Extracellular (matched 585539: sialidase precursor EC 3.2.1.18 NEURAMINIDASE; score 9.70) | Cytoplasmic, (matched 54041713: probable serine/threonine-protein kinase pknG; score 9.89) |
| Signal peptide (signalp 4.1)a | No (D = 0.162 D-cutoff = 0.420) | Yes position: 1–35 (cleavage site between pos. 35 and 36: DSA-DK D = 0.631 D-cutoff = 0.450 networks = signalp-TM) | Yes position: 1–31 (cleavage site between pos. 31 and 32: APA-TL D = 0.562 D-cutoff = 0.450 networks = signalp-TM) | No (D = 0.106 D-cutoff = 0.420) |
| Prediction of protective antigens (VaxiJen) | Probable ANTIGEN (score 0.6912) | Probable ANTIGEN (score 0.7663) | Probable ANTIGEN (score 0.6967) | Probable NON-ANTIGEN score 0.3686) |
a For signal peptide prediction, D-cutoff values were set as sensitive (reproduce SignalP 3.0’s sensitivity)
Like its counterpart in M. tuberculosis, which is predominantly found soluble in the cytoplasm [15], PknG was predicted as a cytoplasmic protein (Table 3). However, VaxiJen predicted this C. pseudotuberculosis putative serine/threonine protein kinase as non-antigenic. In fact, cytoplasmic proteins have not been widely considered as potential immunogens, since they do not have a close contact to many immune systems’ intermediates [49]. Regardless of this, it has been demonstrated that cytoplasmic proteins can be effectively exposed to MHC presentation and may have a key role in the development of a suitable protective immunity. In order to overcome the problem of endogenous antigen access to the MHC II compartment, lysosomal-associated membrane proteins (LAMPs), major lysosomal membrane glycoproteins that contain a cytoplasmic tail targeting sequence that directs the trafficking of the molecule through an endosome/lysossome pathway, including cellular compartments where it is co-localized with MHC II molecules, have been used to induce antigen-trafficking to MHC II compartments and increase the immune response to those antigens [50]. This strategy has shown to elicit enhanced long-term memory response against HIV-1 Gag protein. Besides, a novel mechanism of specific CD8+ T cell-mediated protective immunity can recognize malaria proteins expressed in the cytoplasm of parasites, form clusters around infected hepatocytes, and protect against parasites [51]. This strongly indicates that cellular and molecular mechanisms underlying the protective immune responses against intracellular parasites need further studies.
Linear B-cell epitope prediction
The general problem in achieving an effective treatment of C. pseudotuberculosis infections in animals and humans is probably related to the facultative intracellular lifestyle of this bacterium, as it can survive and multiply in macrophages [52]. The knowledge on the immunity induced by C. pseudotuberculosis indicates that the resistance to infection is a complex process involving components of the non-specific and specific host responses, in which humoral and cellular immune responses are both operative [7].
B-cell epitopes can induce both primary and secondary immunity. Although it is believed that the majority of B-cell epitopes are conformational epitopes, experimental determination of epitopes has focused primarily on the identification of linear (non conformational) B-cell epitopes [25]. This is mainly because predictions of conformational epitopes depend on experimentally determined protein structures or homologous protein structures for in silico modeling. So far, there is no protein structure of the target proteins or structures of highly homologous proteins available for modeling.
Most of the existing linear B-cell epitope prediction methods are based on physicochemical properties relating to surface exposure, such as flexibility or hidrophilicity [25, 35], as it is thought that epitopes must lie at the protein surface for antibody binding to occur. Thus, the target proteins were scanned for B-cell epitopes using several methods designed to quantitate protein physicochemical properties. Graphical outputs of the prediction methods are shown in Figs. 1, 2, 3, and 4. High values gave rise to peaks, whereas valleys correspond to negative properties of the protein. Selected B-cell linear epitopes of target proteins are shown in Table 4. The putative adhesive pili tip protein SpaC, seconded by PknG, presented the highest number of B-cell epitopes. We did pick only one B-cell epitope from SodC since the protein is short (206 aa), has a 35 aa long signal peptide (Table 3) and its highly conserved domain occupies most of the amino acid sequence (Fig. 2).
Fig. 1.
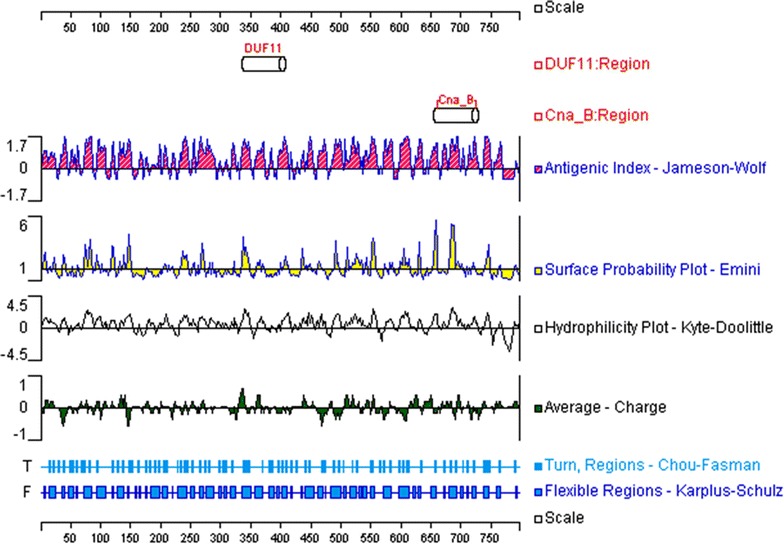
Graphical outputs of the different methods used to quantitate the physicochemical properties used to predict B-cell epitopes from SpaC. On top are the conserved domains of the target protein identified by searching NCBI’s Conserved Domain Database (CDD). The scales indicate the amino acid positions
Fig. 2.
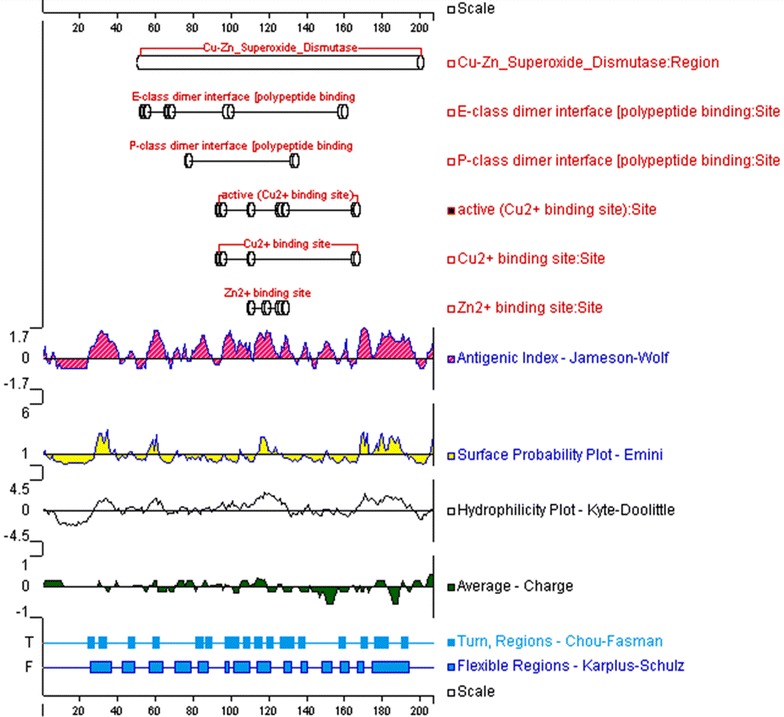
Graphical outputs of the different methods used to quantitate the physicochemical properties used to predict B-cell epitopes from SodC. On top are the conserved domains of the target protein identified by searching NCBI’s Conserved Domain Database (CDD). The scales indicate the amino acid positions
Fig. 3.
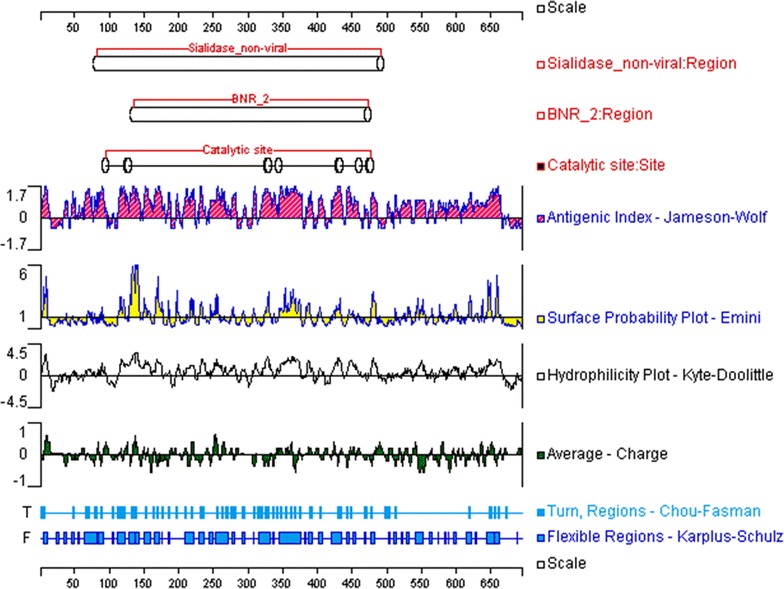
Graphical outputs of the different methods used to quantitate the physicochemical properties used to predict B-cell epitopes from NanH. On top are the conserved domains of the target protein identified by searching NCBI’s Conserved Domain Database (CDD). The scales indicate the amino acid positions
Fig. 4.
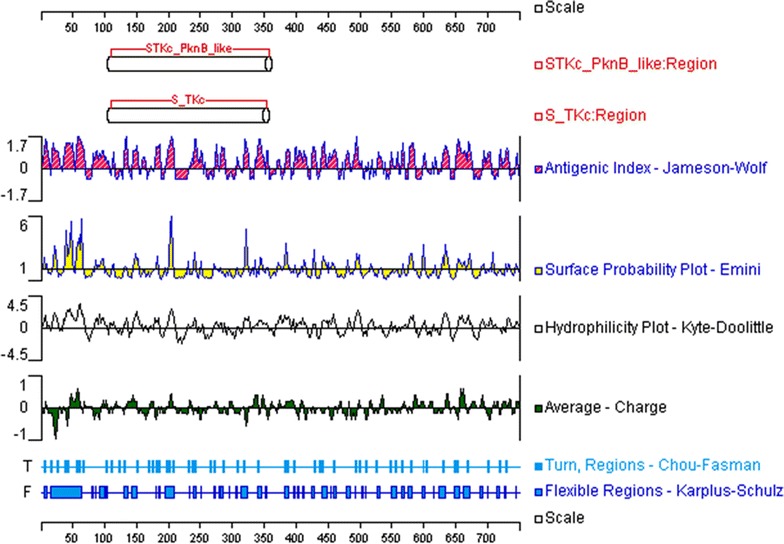
Graphical outputs of the different methods used to quantitate the physicochemical properties used to predict B-cell epitopes from PknG. On top are the conserved domains of the target protein identified by searching NCBI’s Conserved Domain Database (CDD). The scales indicate the amino acid positions
Table 4.
B-cell epitopes predicted from target proteins
| Target Protein | Epitope number | B-cell epitopesa |
|---|---|---|
| SpaC | 1 | 1-MEVPEKTKVEIRFQTGSKISTPSTPSV-27 |
| SpaC | 2 | 70-SQHTNRGETFNDRNSTDLYVQ-90 |
| SpaC | 3 | 116-AYNPKEGYIYAISQGRLKTLQSSKLRIYDEDPNYPAGHLL-155 |
| SpaC | 4 | 234-NDYTSTGKTDSNYVWGI-250 |
| SpaC | 5 | 251-KNSSNPAVLERIDVRDGSRKEFSLDGVKDPLGQNVEKGIYGT-292 |
| SpaC | 6 | 331-IVAKRKGPTSQNNDATSNG-349 |
| SpaC | 7 | 434-KATYKVTANQSISNNEKCLQNTASIYAN-461 |
| SpaC | 8 | 504-GNGLRKVTYKIEVKNPKGFPETKYSLTDTPQFADSV-539 |
| SpaC | 9 | 540-KLERLKVISDYGKKNQEVQAADISV-564 |
| SpaC | 10 | 615-FGLFNSAKLKVGVSEKTSEGCAPIVR-640 |
| SpaC | 11 | 647-QLKKVDAENKETELQATFE-665 |
| SpaC | 12 | 735-PLSKSADQGKDPNLVIL-751 |
| SpaC | 13 | 756-VRVGTLPKTGGHGVAIYLV-774 |
| SodC | 1 | 26-SSSTTTKDSADKAMTS-41 |
| NanH | 1 | 1-MTDSHRRGTRKALVTLTA-18 |
| NanH | 2 | 65-GEGKLPDPVTSEFF-78 |
| NanH | 3 | 520-IEDAKAATAKAEEATAN-536 |
| NanH | 4 | 559-AEAKSAAQDAI-569 |
| NanH | 5 | 595-KAENEAKALAE-605 |
| NanH | 6 | 617-SQDQAKALAEA-627 |
| NanH | 7 | 645-EKEKSGKAGGTDNTENKGFWQE-666 |
| PknG | 1 | 1- MNDPLSRGTEAIPFDPFADDEEDDLSGLLND-31 |
| PknG | 1.1 | 38-DTDTDARSREKSISTFRSRRGTNRDDRTVANG-69 |
| PknG | 1.2 | 79-STAEEMLKDDAYIEQKGLEKPLLHPGD-105 |
| PknG | 2 | 381-SPQRSTFGTKHMVFRTDQLIDGIERNVRITSEEVNA-416 |
| PknG | 3 | 438-YAEPSQTLQTLRDAMAQEEFANSKEIPL-465 |
| PknG | 4 | 479-EARSWLDTLDATLSDDWRHQWYSGVTS-505 |
| PknG | 5 | 576-LTKDPETLRFKALYL-590 |
| PknG | 6 | 627-QVPQNSTHRRMAELTAI-643 |
| PknG | 7 | 651-LSESRIRRAARRLESIPTNEPRFLQIKIA-679 |
| PknG | 8 | 718-DSLRLLARSAPNVHHRYTLV-737 |
a Epitopes in signal peptide and conserved domains were discarded
T-cell epitope prediction
A desirable vaccine preparation should present MHC I and II epitopes for the development of a protective and long lasting immune response to C. pseudotuberculosis. MHC I epitopes are presented to CD8+ T cells by cells infected with C. pseudotuberculosis, leading to the apoptosis of the host cell and interruption of the bacterial multiplication, and it was already described the injection of anti-CD4 or anti-CD8 monoclonal antibody resulted in significantly increased mortality and a marked suppression of IFN-gamma production in mice [53]. MHC II epitopes are involved in the activation of CD4+ T cells, which will drive the host immune response to a Th1 protective response, as well as to a production of IFN-gamma, that will help macrophages in the fusion of phagosomes and lysosomes, resulting in the destruction of bacteria that underwent phagocytic process [54]. Ultimately, specific high affinity binding should be the main concern since the efficiency of an epitope vaccine greatly relies on the precise interaction between epitope and HLA molecule [55]. Table 5 shows nine mer peptides from target proteins with high affinity (Consensus percentile rank <1 %) for mouse MHC I alleles. Most of them were from SpaC and PknG. SodC peptides were discarded since they were located in conserved regions. The few strong binding peptides to MHC II were limited to mouse H-2-IAb allele and most of them were from NanH (Table 6). Only two MHC II strong binding peptides were predicted from SodC but both were discarded because they were located in conserved regions of the protein. Additional file 4 shows the MHC class II epitopes predicted from target proteins.
Table 5.
MHC class I epitopes predicted from target proteins
| Target Protein | Mouse HLA Allele | Epitope number | Start | End | Peptide (9 mer) | Consensus rank (%) |
|---|---|---|---|---|---|---|
| SpaC | H-2-Db | 1 | 615 | 623 | FGLFNSAKL | 0.3 |
| SpaC | H-2-Kk | 2 | 34 | 42 | EEFENTEPI | 0.3 |
| SpaC | H-2-Kb | 3 | 90 | 98 | QSFNRNTGL | 0.35 |
| SpaC | H-2-Kd | 4 | 124 | 132 | IYAISQGRL | 0.4 |
| SpaC | H-2-Kd | 5 | 116 | 124 | AYNPKEGYI | 0.5 |
| SpaC | H-2-Kd | 6 | 199 | 207 | RYLVSNSSQ | 0.5 |
| SpaC | H-2-Kd | 7 | 771 | 779 | IYLVMGVLL | 0.5 |
| SpaC | H-2-Db | 8 | 450 | 458 | KCLQNTASI | 0.6 |
| SpaC | H-2-Db | 9 | 208 | 216 | SGTHNLYTL | 0.7 |
| SpaC | H-2-Dd | 10 | 48 | 56 | VGPSVDPTV | 0.7 |
| SpaC | H-2-Kd | 11 | 458 | 466 | IYANEKDLI | 0.8 |
| SpaC | H-2-Kb | 12 | 785 | 793 | SWSLYRNQL | 0.85 |
| SpaC | H-2-Kb | 13 | 774 | 782 | VMGVLLVLV | 0.95 |
| NanH | H-2-Kk | 1 | 44 | 52 | SEFFDSKVI | 0.3 |
| NanH | H-2-Dd | 2 | 39 | 47 | PDPVTSEFF | 0.4 |
| NanH | H-2-Dd | 3 | 55 | 63 | VDPAGQRCF | 0.4 |
| NanH | H-2-Kk | 4 | 634 | 642 | QELLRIFPG | 0.5 |
| NanH | H-2-Dd | 5 | 655 | 663 | GGMQKLLAF | 0.6 |
| NanH | H-2-Kb | 6 | 645 | 653 | PIFSFLASI | 0.8 |
| PknG | H-2-Kd | 1 | 437 | 445 | SYAEPSQTL | 0.2 |
| PknG | H-2-Kk | 2 | 455 | 463 | EEFANSKEI | 0.2 |
| PknG | H-2-Db | 3 | 678 | 686 | IAIMNAALT | 0.5 |
| PknG | H-2-Ld | 4 | 525 | 533 | LPGEAAPKL | 0.5 |
| PknG | H-2-Kb | 5 | 586 | 594 | KALYLYALV | 0.55 |
| PknG | H-2-Dd | 6 | 665 | 673 | SIPTNEPRF | 0.6 |
| PknG | H-2-Kb | 7 | 685 | 693 | LTWLRQSRL | 0.6 |
| PknG | H-2-Db | 8 | 504 | 512 | TSLFLDDYV | 0.7 |
| PknG | H-2-Kd | 9 | 379 | 387 | LYSPQRSTF | 0.8 |
| PknG | H-2-Kb | 10 | 632 | 640 | STHRRMAEL | 0.85 |
| PknG | H-2-Db | 11 | 457 | 465 | FANSKEIPL | 0.9 |
| PknG | H-2-Kk | 12 | 21 | 29 | EEDDLSGLL | 0.9 |
| PknG | H-2-Kk | 13 | 353 | 361 | LETQLFGIL | 0.9 |
Epitopes in signal peptide and conserved domains were discarded
Table 6.
Total numbers of MHC class II epitope prediction from target proteins
| Target protein | Mouse MHC HLA allele | Number of strong bindersa | Number of weak bindersa | Number of peptidesb |
|---|---|---|---|---|
| PknG | H-2-IAb | 9 | 35 | 735 |
| SpaC | H-2-IAb | 4 | 48 | 782 |
| SodC | H-2-IAb | 0 | 12 | 192 |
| NanH | H-2-IAb | 22 | 64 | 680 |
| PknG | H-2-IAd | 0 | 29 | 735 |
| SpaC | H-2-IAd | 0 | 13 | 782 |
| SodC | H-2-IAd | 2 | 6 | 192 |
| NanH | H-2-IAd | 0 | 32 | 680 |
See epitope sequences in Additional file 4
a Strong binder threshold 50.00. Weak binder threshold 500.00
b Peptide length 15 mer
Epitope clustering
All B and T-cell epitopes (MHC I and II) predicted from the target proteins were grouped in clusters of sequence similarity in order to evaluate the redundancy degree among them. A total of 57 clusters were formed from a set of 136 epitopes predicted (Additional file 5). Most of them (34) were single-sequence clusters. Clusters 4 and 5 (PknG) and cluster 12 (SpaC) grouped epitopes for both B and T-cell (MHC I and II). These groups of epitopes can thus potentially stimulate a complete immune response against C. pseudotuberculosis. The main goal of vaccination is to induce humoral and cellular immunity by selectively stimulating antigen specific CTLs or B cells together with TH cells [56]. Several clusters containing B-cell and either MHC I or II epitopes were also formed. Among them are clusters 9 and 19 formed by epitopes from NanH (Additional file 5). Cluster 14 grouped all SodC weak binding epitopes to H-2-IAb allele.
Protein expression
Large amounts of SpaC, SodC, NanH, and PknG are necessary for future studies on the role of these proteins in C. pseudotuberculosis pathogenicity and virulence. Escherichia coli remains as one of the most attractive hosts among many systems available for heterologous protein production [57]. Thus, pknG, spaC, sodC, and nanH codon-optimized ORFs were cloned into the same expression vector system and individually transformed into BL21(DE3) E. coli strains. SDS-PAGE analyses show the successful expression of the target proteins (Fig. 5a). Purification of PknG using affinity and gel chromatography is shown in Fig. 5b.
Fig. 5.
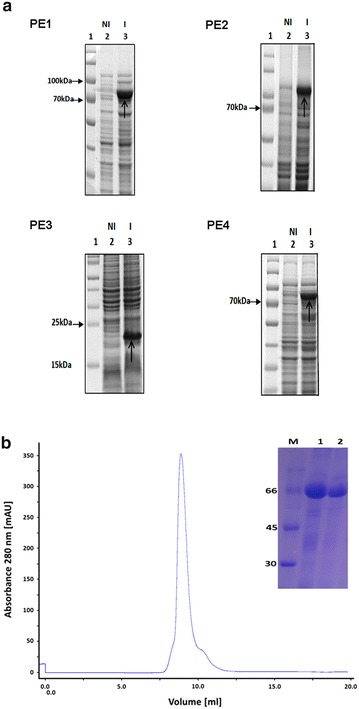
Heterologous expression of the C. pseudotuberculosis FRC41 putative virulence factors in E. coli and rPknG purification. a Coomassie blue-stained SDS-PAGE analyses of the protein expression experiments: PE1, rPknG expression (83 kDa, 10 % gel) in E. coli strain BL21 Star (DE3); PE2, rSpaC expression (86 kDa, 10 % gel) in E. coli strain C43 (DE3); PE3, rSodC expression (18 kDa, 15 % gel) in E. coli strain BL21 Star (DE3); PE4, rNanH expression (71.5 kDa, 10 % gel) in E. coli strain C43 (DE3). 1, pre-stained protein ladder; 2 (NI), non-induced time 0; 3 (I), induced with 1 mM IPTG for 5 h at 37 °C. Arrows indicate the recombinant protein position in the gels. b Chromatogram of the rPknG purification by gel filtration. SDS–PAGE shows an analysis of the purification steps. M molecular-weight markers (kDa); 1, rPknG after affinity chromatography by Ni Sepharose; 2, rPknG purified by gel filtration
From the current study we have suggested that several B and T-cell epitopes predicted from SpaC, SodC, NanH and PknG can be used for the development of a multi peptide vaccine to induce a complete immune response against C. pseudotuberculosis. The next step will be to evaluate experimentally these epitopes in vitro and in vivo to assess their real protective potential.
Conclusions
The in silico analyses performed show that SpaC, PknG and NanH present good potential as targets for vaccine development. Several epitopes from these proteins can potentially induce both humoral and cellular immune responses against C. pseudotuberculosis. The four target proteins were successfully expressed in E. coli. The production of these proteins in large amounts represents an important step for future studies on 3-D structure, pathogenicity, virulence, and vaccine development.
Authors’ contributions
TMS, KTOSJ and ELA performed the bioinformatic analyses. TMS and RWP made the immunological approaches for the epitopes prediction study. KTOSJ, NRT, RFSS and RBM carried out the protein expression and purification experiments. TMS and KTOSJ drafted the manuscript. VA, TMS, RM and RKA participated in the design and coordination of the study. All authors have read and approved the manuscript.
Acknowledgements
This study was supported by CAPES, CNPq, FAPEMIG, and FAPESB.
Competing interests
The authors declare that they have no competing interests.
Abbreviations
- CLA
caseous lymphadenitis
- CMNR
microorganisms from Corynebacterium, Mycobacterium, Nocardi, and Rhodococcus genera
- PLD
phospholipase D
- SOD
superoxide dismutase
- ORF
open reading frame
- SDS-PAGE
sodium dodecyl sulfate polyacrylamide gel electrophoresis
Additional file
10.1186/s12934-016-0479-6 Homologous proteins of C. pseudotuberculosis FRC41 putative virulence factor PknG in CMNR microorganism and mammalians.
10.1186/s12934-016-0479-6 Homologous proteins of C. pseudotuberculosis FRC41 putative virulence factors SpaC and NanH in CMNR microorganism and mammalians.
10.1186/s12934-016-0479-6 Homologous proteins of C. pseudotuberculosis FRC41 putative virulence factor SodC in CMNR microorganism and mammalians.
10.1186/s12934-016-0479-6 MHC class II epitopes§ predicted from target proteins.
10.1186/s12934-016-0479-6 Clusters of B and T-cell epitopes predicted from target proteins.
Footnotes
Karina T. O. Santana-Jorge and Túlio M. Santos share first-author credit
Contributor Information
Karina T. O. Santana-Jorge, Email: karinasantana.bqi@gmail.com
Túlio M. Santos, Email: santos_tm@hotmail.com
Natayme R. Tartaglia, Email: nrtbiomed@gmail.com
Edgar L. Aguiar, Email: edgarlaguiar@gmail.com
Renata F. S. Souza, Email: refasi@hotmail.com
Ricardo B. Mariutti, Email: ricardomariutti@yahoo.com.br
Raphael J. Eberle, Email: eberleraphael@gmail.com
Raghuvir K. Arni, Email: arni@sjrp.unesp.br
Ricardo W. Portela, Email: rwportela@oi.com.br
Roberto Meyer, Email: rmeyer@ufba.br.
Vasco Azevedo, Phone: 55 31 3409-2610, Email: vasco@icb.ufmg.br.
References
- 1.Dorella FA, Pacheco LGC, Oliveira SC, Miyoshi A, Azevedo V. Corynebacterium pseudotuberculosis: microbiology, biochemical properties, pathogenesis and molecular studies of virulence. Vet Res. 2006;37:201–218. doi: 10.1051/vetres:2005056. [DOI] [PubMed] [Google Scholar]
- 2.de Sá Guimarães A, do Carmo FB, Pauletti RB, Seyffert N, Ribeiro D, Lage AP, et al. Caseous lymphadenitis: epidemiology, diagnosis, and control. IIOAB J. 2011;2:33–43. [Google Scholar]
- 3.Baird GJ, Fontaine MC. Corynebacterium pseudotuberculosis and its role in Ovine Caseous Lymphadenitis. J Comp Pathol. 2007;137:179–210. doi: 10.1016/j.jcpa.2007.07.002. [DOI] [PubMed] [Google Scholar]
- 4.Fontaine MC, Baird GJ. Caseous lymphadenitis. Small Rumin Res. 2008;76:42–48. doi: 10.1016/j.smallrumres.2007.12.025. [DOI] [Google Scholar]
- 5.Windsor PA. Control of caseous lymphadenitis. Vet Clin North Am-Food Anim Pract. 2011;27:193–202. doi: 10.1016/j.cvfa.2010.10.019. [DOI] [PubMed] [Google Scholar]
- 6.Oreiby AF. Diagnosis of caseous lymphadenitis in sheep and goat. Small Rumin Res. 2015;123:160–6.
- 7.Bastos BL, Dias Portela RW, Dorella FA, Ribeiro D, Seyffert N, et al. Corynebacterium pseudotuberculosis: immunological responses in animal models and zoonotic potential. J Clin Cell Immunol. 2012;S4:005. doi:10.4172/2155-9899.S4-005
- 8.Trost E, Ott L, Schneider J, Schröder J, Jaenicke S, Goesmann A, et al. The complete genome sequence of Corynebacterium pseudotuberculosis FRC41 isolated from a 12-year-old girl with necrotizing lymphadenitis reveals insights into gene-regulatory networks contributing to virulence. BMC Genom. 2010;11:728. doi: 10.1186/1471-2164-11-728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rogers EA, Das A, Ton-That H. Adhesion by pathogenic corynebacteria. Adv Exp Med Biol. 2011;715:91–103. [DOI] [PubMed]
- 10.Kim S, Oh DB, Kang HA, Kwon O. Features and applications of bacterial sialidases. Appl Microbiol Biotechnol. 2011;91:1–15. doi: 10.1007/s00253-011-3307-2. [DOI] [PubMed] [Google Scholar]
- 11.Kim S, Oh DB, Kwon O, Kang HA. Identification and functional characterization of the NanH extracellular sialidase from Corynebacterium diphtheriae. J Biochem. 2010;147:523–533. doi: 10.1093/jb/mvp198. [DOI] [PubMed] [Google Scholar]
- 12.Dussurget O, Stewart G, Neyrolles O, Pescher P, Young D, Marchal G. Role of Mycobacterium tuberculosis copper-zinc superoxide dismutase. Infect Immun. 2001;69:529–33. [DOI] [PMC free article] [PubMed]
- 13.Piddington DL, Fang FC, Laessig T, Cooper M, Orme IM, Buchmeier NA, et al. Cu, Zn Superoxide Dismutase of Mycobacterium tuberculosis contributes to survival in activated macrophages that are generating an oxidative burst cu, zn superoxide dismutase of Mycobacterium tuberculosis contributes to survival in activated macrophages. Infect Immun. 2001;69:4980–4987. doi: 10.1128/IAI.69.8.4980-4987.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pereira SFF, Goss L, Dworkin J. Eukaryote-like serine/threonine kinases and phosphatases in bacteria. Microbiol Mol Biol Rev. 2011;75:192–212. doi: 10.1128/MMBR.00042-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Forrellad MA, Klepp LI, Gioffré A, Sabio y García J, Morbidoni HR, de la Paz Santangelo M, et al. Virulence factors of the Mycobacterium tuberculosis complex. Virulence. 2013;4:3–66. [DOI] [PMC free article] [PubMed]
- 16.Walburger A, Koul A, Ferrari G, Nguyen L, Prescianotto-Baschong C, Huygen K, et al. Protein kinase G from pathogenic mycobacteria promotes survival within macrophages. Science. 2004;304:1800–1804. doi: 10.1126/science.1099384. [DOI] [PubMed] [Google Scholar]
- 17.Warner DF, Mizrahi V. The survival kit of Mycobacterium tuberculosis. Nat Med. 2007;13:282–284. doi: 10.1038/nm0307-282. [DOI] [PubMed] [Google Scholar]
- 18.Altschul SF, Gish W, Pennsylvania T, Park U. Basic local alignment search tool 2Department of computer science. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 19.Consortium TU. UniProt: a hub for protein information. Nucleic Acids Res. 2014;43:D204–12. [DOI] [PMC free article] [PubMed]
- 20.Gasteiger E, Hoogland C, Gattiker A, Duvaud S, Wilkins MR, Appel RD, et al. Protein identification and analysis tools on the ExPASy server. Proteomics protocols handbook. New York City: Humana Press; 2005. p. 571–607.
- 21.Geourjon C, Deléage G. SOPMA: significant improvements in protein secondary structure prediction by consensus prediction from multiple alignments. Comput Appl Biosci. 1995;11:681–684. doi: 10.1093/bioinformatics/11.6.681. [DOI] [PubMed] [Google Scholar]
- 22.Yu NY, Wagner JR, Laird MR, Melli G, Rey S, Lo R, et al. PSORTb 3.0: improved protein subcellular localization prediction with refined localization subcategories and predictive capabilities for all prokaryotes. Bioinformatics. 2010;26:1608–1615. doi: 10.1093/bioinformatics/btq249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Petersen TN, Brunak S, von Heijne G, Nielsen H. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat Methods. 2011;8:785–6. [DOI] [PubMed]
- 24.Doytchinova IA, Flower DR. VaxiJen: a server for prediction of protective antigens, tumour antigens and subunit vaccines. BMC Bioinformatics. 2007;8:4. doi: 10.1186/1471-2105-8-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.El-Manzalawy Y, Honavar V. Recent advances in B-cell epitope prediction methods. Immunome Res. London: BioMed Central Ltd; 2010;6:S2. [DOI] [PMC free article] [PubMed]
- 26.Jameson BA, Wolf H. The antigenic index: a novel algorithm for predicting antigenic determinants. Comput Appl Biosci. 1988;4:181–186. doi: 10.1093/bioinformatics/4.1.181. [DOI] [PubMed] [Google Scholar]
- 27.Emini EA, Hughes JV, Perlow DS, Boger J. Induction of hepatitis A virus-neutralizing antibody by a virus-specific synthetic peptide. J Virol. 1985;55:836–9. [DOI] [PMC free article] [PubMed]
- 28.Kyte J, Doolittle RF. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 1982;157:105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- 29.White A, Handler P, Smith EL, editors. Principles of Biochemistry. 3rd ed. 1964.
- 30.Rini JM, Schulze-Gahmen U, Wilson IA. Structural evidence for induced fit as a mechanism for antibody-antigen recognition. Science. 1992;255:959–965. doi: 10.1126/science.1546293. [DOI] [PubMed] [Google Scholar]
- 31.Chou BPY, Fasman GD. Prediction of the secondary structure of proteins from their amino acid sequence. Adv Enzymol Relat Areas Mol Biol. 1978;47:45–148. doi: 10.1002/9780470122921.ch2. [DOI] [PubMed] [Google Scholar]
- 32.Chou PY. Prediction of protein structural classes from amino acid composition. Predict Protein Struct Princ Protein Conform. 1989;549–86.
- 33.Karplus PA, Schulz GE. Prediction of chain flexibility in proteins synthesis of acetylcholine receptors in xenopus oocytes induced by poly (A)+ -mRNA from locust nervous tissue. Naturwissenschaften. 1985;72:2–3. doi: 10.1007/BF01195768. [DOI] [Google Scholar]
- 34.Marchler-Bauer A, Derbyshire MK, Gonzales NR, Lu S, Chitsaz F, Geer LY, et al. CDD: NCBI’s conserved domain database. Nucleic Acids Res. 2015;43:D222–D226. doi: 10.1093/nar/gku1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Saha S, Saha S, Raghava GPS, Raghava GPS. Prediction methods for B-cell epitopes. Methods Mol Biol. 2007;409:387–94. [DOI] [PubMed]
- 36.Vita R, Overton JA, Greenbaum JA, Ponomarenko J, Clark JD, Cantrell JR, et al. The immune epitope database (IEDB) 3.0. Nucleic Acids Res. 2015;43:D405–D412. doi: 10.1093/nar/gku938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Moutaftsi M, Peters B, Pasquetto V, Tscharke DC, Sidney J, Bui H-H, et al. A consensus epitope prediction approach identifies the breadth of murine TCD8+ -cell responses to vaccinia virus. Nat Biotechnol. 2006;24:817–9. [DOI] [PubMed]
- 38.Nielsen M, Lund O. NN-align. An artificial neural network-based alignment algorithm for MHC class II peptide binding prediction. BMC Bioinform. 2009;10:296. doi: 10.1186/1471-2105-10-296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, et al. Fast, scalable generation of high-quality protein multiple sequence alignments using clustal omega. Mol Syst Biol. 2011;7:539. [DOI] [PMC free article] [PubMed]
- 40.Sambrook J, Russell DW, editors. Molecular cloning: a laboratory manual. 3rd ed. Cold Spring Harbor: Cold Spring Harbor Laboratory Press; 2001.
- 41.Welch M, Govindarajan S, Ness JE, Villalobos A, Gurney A, Minshull J, et al. Design parameters to control synthetic gene expression in Escherichia coli. PLoS One. 2009;4:e7002. [DOI] [PMC free article] [PubMed]
- 42.Saïda F, Uzan M, Odaert B, Bontems F. Expression of highly toxic genes in E. coli: special strategies and genetic tools. Curr Protein Pept Sci. 2006;7:47–56. doi: 10.2174/138920306775474095. [DOI] [PubMed] [Google Scholar]
- 43.Arnon R. A novel approach to vaccine design-pitope-based vaccines. FEBS J. 2006;273:33–34. [Google Scholar]
- 44.Grimm SK, Ackerman ME. Vaccine design: emerging concepts and renewed optimism. Curr Opin Biotechnol. 2013;24:1078–1088. doi: 10.1016/j.copbio.2013.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Trivedi S, Gehlot HS, Rao SR. Protein thermostability in archaea and eubacteria. Genet Mol Res. 2006;5:816–827. [PubMed] [Google Scholar]
- 46.Goodswen SJ, Kennedy PJ, Ellis JT. A guide to in silico vaccine discovery for eukaryotic pathogens. Brief Bioinform. 2013;14:753–774. doi: 10.1093/bib/bbs066. [DOI] [PubMed] [Google Scholar]
- 47.Bhavsar AP, Guttman JA, Finlay BB. Manipulation of host-cell pathways by bacterial pathogens. Nature. 2007;449:827–834. doi: 10.1038/nature06247. [DOI] [PubMed] [Google Scholar]
- 48.Krachler AM, Orth K. Targeting the bacteria–host interface. Virulence. 2013;4:284–94. [DOI] [PMC free article] [PubMed]
- 49.Kaufmann SHE, Hess J. Impact of intracellular location of and antigen display by intracellular bacteria: implications for vaccine development. Immunol Lett. 1999;65:81–84. doi: 10.1016/S0165-2478(98)00128-X. [DOI] [PubMed] [Google Scholar]
- 50.De Arruda LB, Chikhlikar PR, August JT, Marques ETA. DNA vaccine encoding human immunodeficiency virus-1 Gag, targeted to the major histocompatibility complex II compartment by lysosomal-associated membrane protein, elicits enhanced long-term memory response. Immunology. 2004;112:126–135. doi: 10.1111/j.1365-2567.2004.01823.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kimura K, Kimura D, Matsushima Y, Miyakoda M, Honma K, Yuda M, et al. CD8+ T cells specific for a malaria cytoplasmic antigen form clusters around infected hepatocytes and are protective at the liver stage of infection. Infect Immun. 2013;81:3825–3834. doi: 10.1128/IAI.00570-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.McKean S, Davies J, Moore R. Identification of macrophage induced genes of Corynebacterium pseudotuberculosis by differential fluorescence induction. Microbes Infect. 2005;7:1352–1363. doi: 10.1016/j.micinf.2005.05.002. [DOI] [PubMed] [Google Scholar]
- 53.Lan DT, Taniguchi S, Makino S, Shirahata T, Nakane A. Role of endogenous tumor necrosis factor alpha and gamma interferon in resistance to Corynebacterium pseudotuberculosis infection in mice. Microbiol Immunol. 1998;42:863–870. doi: 10.1111/j.1348-0421.1998.tb02362.x. [DOI] [PubMed] [Google Scholar]
- 54.Lan DT, Makino S, Shirahata T, Yamada M, Nakane A. Tumor necrosis factor alpha and gamma interferon are required for the development of protective immunity to secondary Corynebacterium pseudotuberculosis infection in mice. J Vet Med Sci. 1999;61:1203–1208. doi: 10.1292/jvms.61.1203. [DOI] [PubMed] [Google Scholar]
- 55.Chakraborty S, Chakravorty R, Ahmed M, Rahman A, Waise TZ, Hassan F, et al. A computational approach for identification of epitopes in dengue virus envelope protein: a step towards designing a universal dengue vaccine targeting endemic regions. In Silico Biol. 2010;10:235–246. doi: 10.3233/ISB-2010-0435. [DOI] [PubMed] [Google Scholar]
- 56.Dudek NL, Perlmutter P, Aguilar M-I, Croft NP, Purcell AW. Epitope discovery and their use in peptide based vaccines. Curr Pharm Des. 2010;16:3149–57. [DOI] [PubMed]
- 57.Sugiki T, Fujiwara T, Kojima C. Latest approaches for efficient protein production in drug discovery. Expert Opin Drug Discov. 2014;1–16. [DOI] [PubMed]


