Abstract
Nuclear receptors (NR) act as an integrated conduit for environmental and hormonal signals to govern genomic responses, which relate to cell fate decisions. We review how their integrated actions with each other, shared co-factors and other transcription factors are disrupted in cancer. Steroid hormone nuclear receptors are oncogenic drivers in breast and prostate cancer and blockade of signaling is a major therapeutic goal. By contrast to blockade of receptors, in other cancers enhanced receptor function is attractive, as illustrated initially with targeting of retinoic acid receptors in leukemia. In the post-genomic era large consortia, such as The Cancer Genome Atlas, have developed a remarkable volume of genomic data with which to examine multiple aspects of nuclear receptor status in a pan-cancer manner. Therefore to extend the review of NR function we have also undertaken bioinformatics analyses of NR expression in over 3000 tumors, spread across six different tumor types (bladder, breast, colon, head and neck, liver and prostate). Specifically, to ask how the NR expression was distorted (altered expression, mutation and CNV) we have applied bootstrapping approaches to simulate data for comparison, and also compared these NR findings to 12 other transcription factor families. Nuclear receptors were uniquely and uniformly downregulated across all six tumor types, more than predicted by chance. These approaches also revealed that each tumor type had a specific NR expression profile but these were most similar between breast and prostate cancer. Some NRs were down-regulated in at least five tumor types (e.g. NR3C2/MR and NR5A2/LRH-1)) whereas others were uniquely down-regulated in one tumor (e.g. NR1B3/RARG). The downregulation was not driven by copy number variation or mutation and epigenetic mechanisms maybe responsible for the altered nuclear receptor expression.
Keywords: TCGA, cancer NR3C1/GR NR5A2/LRH-1 NR1B3/RARG, Bootstrap analyses, gene expression, copy number variation, mutation
Introduction
1. Nuclear receptor gene regulatory actions are integrated at multiple levels
The 48 human NRs form a major network to sense lipophilic molecules from diet, metabolism and hormone production, and regulate genes involved in development, metabolism, circadian rhythm, immune function, proliferation and differentiation(1–6). Reflecting this central role, they represent the target for approximately 15% of all pharmacologic drugs (7).
The NR superfamily can be classified in several ways. The superfamily can either be sub-divided by phylogenetics (8), or grouped according to the cellular location and ligand genomic response(9). In this latter classification, Type I receptors are typically cytoplasmic, associated with heatshock proteins, and ligand binding induces nuclear translocation. These receptors include the high affinity steroid receptors. Type II receptors, by contrast, are retained in the nucleus in the absence of ligand and continuously participate in chromatin modification events that are reversed upon ligand binding. These receptors are typified by NRs that bind micronutrient ligands, for example, NR1B1/RARA and NR1I1/VDR. This group can be further sub-divided to include the Type III receptors that bind as homodimers in the absence of ligand at direct repeats and include NR2A1/HNF4A. Finally, Type IV receptors bind to DNA through only a single binding domain (as a receptor monomer or dimer); this group includes the orphan receptors NR1D1/EAR1 and NR1F1/ROR1.
Amongst NRs there are well-established examples of co-operative or antagonistic behavior at the gene regulatory level, for instance resulting in the antagonizing transcriptional effects of ERα and RARs in breast cancer cells(10). Conversely, RXR heterodimerization potentiates the actions of several NRs as illustrated in studies combining 9-cis retinoic acid (RXR ligand) with a range of other ligands which has combinatorial effects on cellular phenotypes (11–14) which are mediated through underlying regulation of the global transcriptome(15–18).
The interactions of NRs with coactivators and corepressors has revealed further levels of integration and suggest that gene regulation is dispersed across NRs by virtue of co-factor sharing. Coactivators, such as NCOA3/AIB1, are vital for transactivation by being a platform for the proteins that govern chromatin remodeling and looping, and the sequestration of the basal transcriptional machinery. Similarly, but in an opposite manner, corepressors act to silence or suppress transcription(19–21).
Outside of NR interactions with one another and with corepressors and coactivators, it is also clear that their signaling actions are guided by the actions of pioneer factors such as Forkhead box (FOX) family members(22–24) and integrated with other transcription factor signaling pathways(25), including WNT(26), p53(27–31), SMADs(32–34) and KLFs(35, 36). One elegant approach to capture such interactions was undertaken by Novershtern et al. (37) who measured the transcriptome profiles of a large number of hematopoietic stem cells, multiple progenitor states and terminally differentiated cell types. They found distinct regulatory circuits in both stem cells and differentiated cells, and identified 80 distinct modules of tightly co-expressed genes in the hematopoietic system. For example, one module was expressed in granulocytes and monocytes and included genes encoding enzymes and cytokine receptors that are essential for inflammatory responses. Major players in this module were VDR together with the factors CEBPα and SPI1/PU.1. This suggests that the VDR works together with this small set of transcription factors, in order to regulate granulocyte and monocyte differentiation. It is reasonable to anticipate that such modules exist in multiple cell types but are guided by the tissue specific expression of NRs and other factors. As genomic-based approaches are increasingly applied to individual receptors, and groups of NRs, it is becoming ever more clear how their combined actions are central to co-ordinate complex gene regulatory programs that govern cell fate decisions (reviewed in(38)).
2. Distorted nuclear receptor function in cancer
The work of Dr. George Beatson(39) in breast cancer and Dr. Charles Huggins(40) in prostate cancer provided very clear evidence of steroid hormone signaling acting as cancer drivers(41, 42). Aside from the well-established therapeutic targeting of AR and ERs there are established roles to target GR as a pro-apoptotic therapeutic approach in lymphoma(43–45). However ligand-activation of the GR has been less effective in other cancers and suggests the biology of the GR is more nuanced with regards to cancer biology. More recently in advanced therapy resistant settings in prostate cancer a role has been revealed for the GR to promote progression by essentially phenocopying the actions of the AR and suggests there are overlapping genomic functions of these receptors(46).
Subsequently, other therapeutic NR roles emerged in cancer and leukemias, not to antagonize but rather to enhance their function. In this case, potentially as pivotal as the discovery of steroid hormone actions as cancer drivers, was the analyses of RAR functions in leukemia that revealed its actions were disrupted, but could be pharmacologically targeted(47).
This was critical for several reasons. Firstly, all-trans retinoic acid (ATRA)-based leukemia therapy represents one of the earliest and most successful examples of targeted therapies and provided a paradigm for other therapies (48–51); although of course clinical trial success with ATRA preceded cloning of the RARs (52, 53). Secondly, pioneering work by the groups of Dr. Pier Pelicci(54) and Dr. Ron Evans(55) revealed mechanisms that corrupted RAR signaling in leukemia, specifically in acute promyelocytic leukemia. This leukemia is characterized by translocations of the RARα, including PML-RAR, which generate chimeric receptors that inappropriately retained association with corepressors. This discovery established the premise that altered NR interactions with coactivators and corepressors might have an epigenetic consequence that in turn could be targeted by co-treatment with epigenetic therapies. More widely, the therapeutic success of ATRA in leukemias, in so-called differentiation therapies, was a major catalyst for the explorations of the anticancer actions of other, principally type II, NRs in a wide range of leukemias and solid tumors. In this manner RARs, VDR, PPARs and more recently LXRs and FXR have been considered as potentially druggable targets across cancers(56–62). With the increasing number of genomic studies it has also emerged that NRs are disrupted in various tumor types; for example DNA CpG methylation of RARB(63–65) and copy number variation of NR1D1(66) and RARA(67)
More recently, many of these earlier findings have been revisited. For example, reflecting the work of Huggins, the role of ERα and ERβ signaling in the prostate have been re-investigated and distortions to expression of these receptors appears important(68–70). Similarly, an appreciation has emerged of the importance of AR signaling in tissues other than the prostate, notably in breast cancer(71, 72).
3. Nuclear receptor network approaches in cancer cells
Given their interactive nature, various workers have examined NR networks in cancer. For example, profiling approaches using high throughput Q-PCR in breast cancer(73) and in silico analyses of prostate cancer data bases(74) both revealed a large complement of NR expressed in tumor and that expression profiles relate to tumor stage. Beyond expression profiling, other investigators have aimed to undertake cistromic analyses of multiple NRs and interacting transcription factors to construct a network level understanding of gene expression programs in breast cancer(10, 75). These approaches identified high complexity enhancer sites that integrated the actions of multiple NRs and other transcription factors in both direct (cis) and indirect (trans) and often absent of canonical motifs but associated with significant levels of clustering(76–78).
Cistromic analyses in breast cancer revealed the interactions and cross-talk of multiple NRs and revealed that RARγ was amongst the most commonly found NR binding site with approximately 12000 RARγ binding sites in MCF-7 breast cancer cells. These sites were significantly enriched with other NRs (e.g. RARα, PPARγ, VDR, HNF4α, ERα), pioneer-type factors (FOXA1, SP1, STAT3) and co-regulators (CTCF). Key aspects of these associations, focused around the RARγ and RARα, were related to clinical outcome in breast cancer patients and supported the role of larger networks of NRs to control cell fates. Specifically, the data support the concept of cross-talk between RARs and VDR, which exert mitotic restraint, and other NRs, such as ERα, that drive proliferation and survival (10, 75). Other workers also identified a comparable number of RARγ binding sites, many contained in a so-called Mega-Trans complex containing ERα and RARγ at important enhancers in breast cancer(79), and specifically identified a significant role for trans RARγ genome binding. The importance of RARγ to regulate ERα, has been supported further by RNAi screens in breast cancer cells aimed at dissecting tamoxifen resistance(80).
There is also evidence that NR interactions with coactivators and corepressors are distorted in cancer, which ultimately disrupts NR function. Elevated levels of NCOA3/AIB1 enhance ERα actions in breast cancer through a variety of actions and are associated with worse disease free survival. This has been primarily examined within the context of ERα signaling but is also associated with the actions of other Type 1 receptors including PR, AR and GR(81–86). Similarly, the genome-wide binding of the transcriptional co-repressors NCOR1 and NCOR2/SMRT maintains distal enhancer regions in an epigenetically repressed, yet poised, state until released(87, 88). These corepressors are distorted in many cancers through altered expression levels(89), splice variants(90, 91), mutation status(92) and genetic variation(93), suggesting a prominent role in driving the onco-epigenome. We, and others, have explored the capacity of NCOR1 and NCOR2/SMRT to drive the onco-epigenome by distorting the transcriptional actions for various NRs including several type II receptors such as VDR, PPARs, RARs (3, 89, 94–105).
It is tempting to speculate that there are perhaps more general rules for these interactions, with specificities of coactivators or corepressors for certain types of receptors. However, there are few ChIP-Seq studies for these coactivators and corepressors and largely they have not been analyzed in an unbiased manner. To address this issue we recently undertook an integrative genomics analyses of the NCOR1 cistrome by exploiting ENCODE data(106). Surprisingly, we found that within the NCOR1 cistrome, NR motifs of any type were not the most commonly enriched, compared to other transcription factors. Of those NR that were enriched, there were both Type 1 (ERα) and Type II (PPARγ) motifs. This suggests that NCOR1 and NCOR2/SMRT involvement with NR function is either not a major aspect of their function or direct DNA interaction by NR in a cis relationship is limited, and that recruitment may be facilitated by pioneer and other integrating transcription factors. Further integrative and unbiased approaches will be critical in resolving the extent and specificities of coactivator and corepressor interactions in an unbiased manner with NRs and other TFs.
4. Integrative analyses of the nuclear receptor network expression in cancer
Given the availability of high quality genomic data for multiple tumors it is possible to investigate NR expression and function individually and in gene networks across different tumor and tissue controls(107–115). Key to such analyses is the work of The Cancer Genome Atlas (TCGA)(116, 117) which provides genome-wide insight in large numbers of multiple tumor types; at the time of writing this 21441 tumors across 91 cancer studies.
Previously, we exploited the Taylor et al. cohort of prostate cancer tumors(118) available through TCGA and mined the NR network (74). These analyses revealed that the NR superfamily expression was significantly lost in primary PCa, more than predicted by chance(74). These findings suggested a global distortion to the NR superfamily expression in PCa, and given the diversity of NRs detected, that prostate tissue maintenance relies on its ability to sense and respond to a range of hormonal and dietary lipophilic compounds. For instance, members of the RARs, RXRs, LXRs and PPARs were deregulated in a substantial proportion of these patients.
In parallel, to identify potential mechanisms driving these disruptions we also mined microRNAs that are predicted to target specific NRs, and revealed a significant and reciprocal gain of expression of NR targeting microRNA that was more than predicted by chance. For example, miR-106b was amongst the most upregulated miRNA in our analysis and targets several down regulated NRs, including PPARA, suggesting that regulation by miRNAs add yet another layer of complexity within the NR network. Together, these observations of reciprocal NR and miRNA co-expression suggest that epigenetic distortion is important to distort the NR superfamily in the prostate(74, 119–121).
5. Pan-cancer post-genomic assessment of the nuclear receptor superfamily
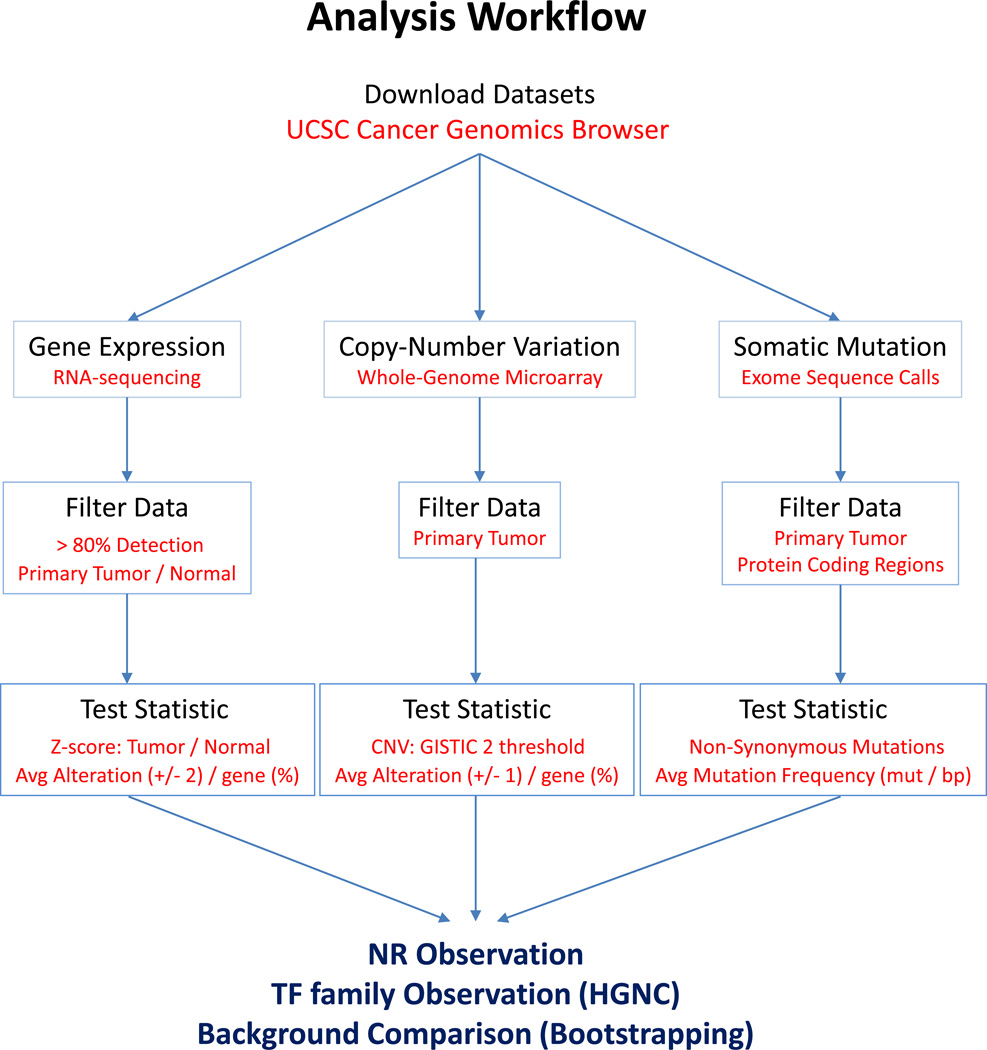
These findings in the literature, and our own work to date, support the concept that the NR superfamily acts in an integrated manner and is disrupted by multiple mechanisms across cancers. To complement this review we have also collated and analyzed NR expression from multiple TCGA cohorts. We have undertaken a comprehensive analysis of the NR superfamily across multiple cancer types in TCGA with the goal of establishing the tumor-specific and pan-cancer extent to which NRs are distorted. Specifically, from over 3000 tumors spread across six different cancer types we have examined NR expression, copy number variation and mutation status (Figure 1).
Figure 1. Workflow diagram summarizing TCGA analyses.
Data were downloaded directly from the UCSC Cancer Genomics Browser (https://genome-cancer.ucsc.edu/), filtered, and global transcriptomic and genomic alterations determined. Observed alterations of NRs and other TF families were directly compared to their genomic background equivalents using a bootstrapping approach.
5.1. Pan-cancer analytical methods
The cancer types selected had matched normal tissue from the tumor-targeted organ and were Bladder (BLCA), Breast (BRCA), Colon (COAD), Head and neck (HNSC), Liver (LIHC), and Prostate (PRAD) cancers. Part of the rationale for choosing these tumor types is that BLCA, COAD, and HNSC are derived from tissues that are all exposed to a wide array of environmental signals, whereas LIHC is derived from a tissue that is the major center of central metabolism and also is exposed to a diverse array of primary metabolites and products of digestion. Finally, PRAD and BRCA are cancers that are driven by steroid hormone signaling.
Data sets
All analyses, unless otherwise indicated, were undertaken using the R platform for statistical computing (version 3.1.0)(67), and a range of library packages were implemented in Bioconductor (122). All transcription factor (TF) family annotations and their inclusive gene identifiers, including NRs were obtained through the HUGO Gene Nomenclature Committee(123). In the first instance normalized RPKM RNA-seq data from the cohorts were downloaded through the UCSC Cancer Genomics Browser(124) (Figure 1, Table 1, Table 2). Only primary, not metastatic, tumor data was considered, and only NRs that were detectible in at least 80% of normal and tumor samples were included in the expression analyses.
Table 1.
Summary of data sets used for integrative analyses of the NR superfamily across the 6 tumor types indicated; Bladder (BLCA), Breast (BRCA), Colon (COAD), Head and neck (HNSC), Liver (LIHC), and Prostate (PRAD). All normalized data were downloaded directly through the UCSC Cancer Genomics Browser (https://genome-cancer.ucsc.edu/).
| Tumor Type | Dataset ID | Data Type | Tumor (n) | Normal (n) |
|---|---|---|---|---|
| PRAD | TCGA_PRAD_exp_HiSeqV2 | Gene Expression | 497 | 52 |
| TCGA_PRAD_gistic2thd | Copy Number Variation | 492 | NA | |
| TCGA_PRAD_mutation_broad_gene | Somatic Mutation | 425 | NA | |
| BRCA | TCGA_BRCA_exp_HiSeqV2 | Gene Expression | 1095 | 113 |
| TCGA_BRCA_gistic2thd | Copy Number Variation | 1079 | NA | |
| TCGA_BRCA_mutation_curated_wustl_gene | Somatic Mutation | 982 | NA | |
| COAD | TCGA_COAD_exp_HiSeqV2 | Gene Expression | 286 | 41 |
| TCGA_COAD_gistic2thd | Copy Number Variation | 450 | NA | |
| TCGA_COAD_mutation_bcm_gene | Somatic Mutation | 217 | NA | |
| LIHC | TCGA_LIHC_exp_HiSeqV2 | Gene Expression | 371 | 42 |
| TCGA_LIHC_gistic2thd | Copy Number Variation | 370 | NA | |
| TCGA_LIHC_mutation_broad_gene | Somatic Mutation | 202 | NA | |
| HNSC | TCGA_HNSC_exp_HiSeqV2 | Gene Expression | 519 | 43 |
| TCGA_HNSC_gistic2thd | Copy Number Variation | 522 | NA | |
| TCGA_HNSC_mutation_broad_gene | Somatic Mutation | 508 | NA | |
| BLCA | TCGA_BLCA_exp_HiSeqV2 | Gene Expression | 407 | 19 |
| TCGA_BLCA_gistic2thd | Copy Number Variation | 408 | NA | |
| TCGA_BLCA_mutation_broad_gene | Somatic Mutation | 238 | NA |
Table 2.
Summary of transcription factor families, including NRs, utilized in TCGA analyses. Gene families and their members were downloaded from the HUGO Gene Nomenclature Committee (www.genenames.org). The average size of TF families examined = 48.15, approximately the size of the NR superfamily.
| Transcription Factor Family | ID | n |
|---|---|---|
| Nuclear hormone receptors | NR | 48 |
| Basic helix-loop-helix proteins | bHLH | 110 |
| Basic leucine zipper proteins | bZIP | 43 |
| E2F transcription factors | E2F | 8 |
| Forkhead boxes | FOX | 43 |
| GATA zinc finger domain containing | GATA | 15 |
| General transcription factors | GeneralTF | 25 |
| Helix-turn-helix ETS type domain containing | ETS | 28 |
| High mobility group | HMG | 15 |
| Homeoboxes | Homeobox | 257 |
| Kruppel-like transcription factors | KLF | 17 |
| SMAD family | SMAD | 8 |
| Sp transcription factors | SP | 9 |
Statistical Analyses
To establish if expression levels of NRs and other transcription factors were different in tumors compared to normal samples, we first established the mean and distribution of expression of all genes in pools of normal samples and then calculated the relative expression of genes in tumor samples using Z scores. In this manner, the tumor expression of all genes, including NRs and other TFs, were converted into normal tissue relative Z-scores and significantly altered tumor expression of detectable genes was determined by considering only values that were elevated (Z ≥ 2) or suppressed (Z ≤ −2). For copy-number analysis, previously determined copy number variation (CNV) estimates (via the GISTIC 2 method) were directly downloaded and utilized.
For mutation analysis, only transcripts containing protein-coding sequences (CDS) were considered. CDS lengths from all exons associated with a given gene were compiled from Ensembl using BioMart(125). To correct for mutation frequencies being proportional to the coding length, all regions exons for a given gene were added (including all alternative exons) to yield the total CDS length for each gene. Mutation frequencies (mutations / protein coding base pair) were then calculated utilizing the number of mutations detected across tumors, the number of tumors, and the CDS lengths for all protein coding genes, including NRs and other indicated TF families.
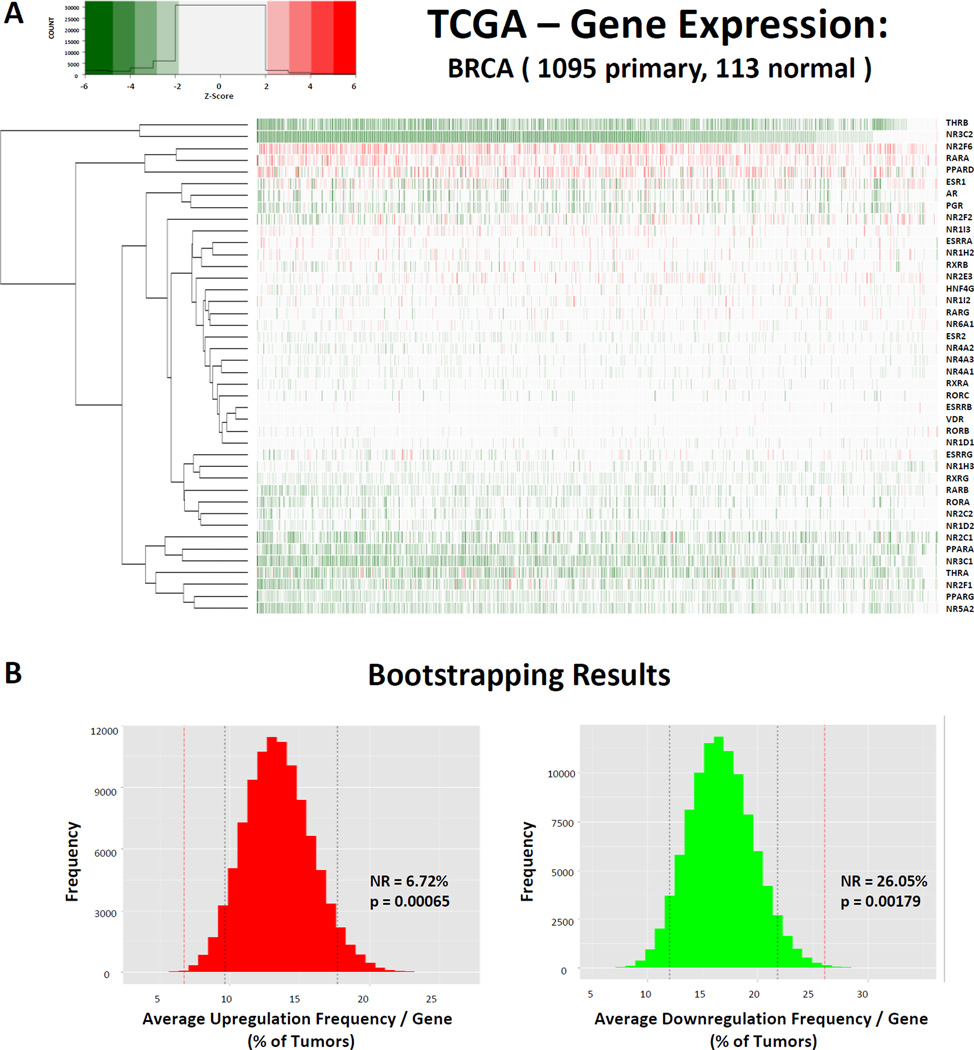
To test if NR superfamily tumor tissue relative expression Z-scores were significantly different than expected by chance we utilized bootstrapping permutation approaches. For instance in BRCA, we determined that the average NR is upregulated (Z ≥ 2) in 6.72% of tumors, while downregulated (Z ≤ −2) in 26.05% of tumors (Figure 2, Table 3). Specifically, to test whether these observations were more or less than would be predicted by chance, we applied bootstrap approaches. This approach applies a random sampling method to simulate the distribution of expression changes across the transcriptome for comparison to observed findings(126). We sampled 100,000 replicates of random gene sets equal to the size of the number of NRs detectable (all NRs detected in > 80% tumors and normal samples) in each respective cancer (e.g. 42 in BRCA), within the detectable transcriptome gene set (e.g. n=16,622 in BRCA), and thus determined the distribution of significant relative expression changes across all genes within the patient cohort allowing us to directly compare the observed alterations of NRs.
Figure 2. Nuclear Receptors are downregulated in cancer.
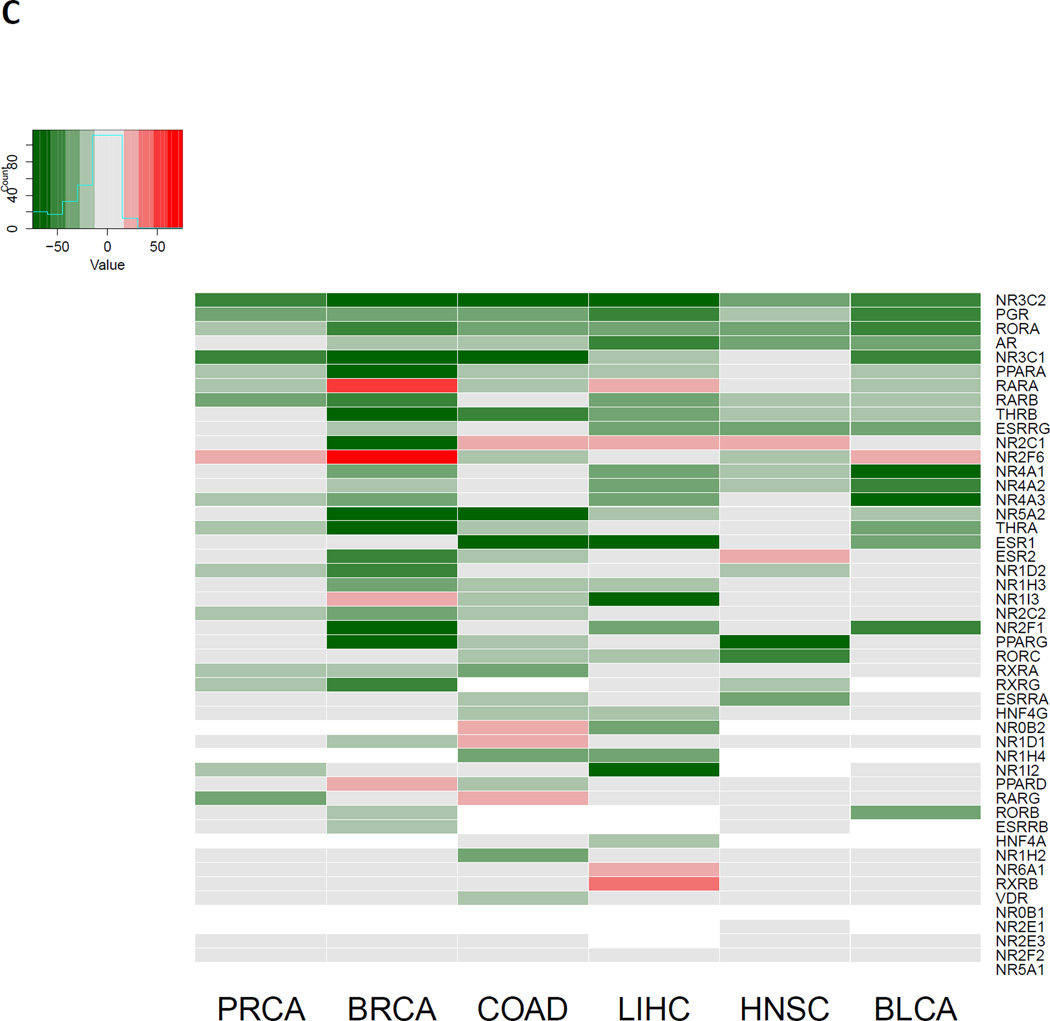
(A) Heatmap depicting the relative expression of all 42 NRs (rows) detectable in 1095 BRCA samples (columns) relative to the expression of NRs observed in a pool of 113 matched normal samples. Tumors with relative expression (Z-score) ≥ +/− 2 are considered significantly upregulated (red) or downregulated (green), respectively. (B) Bootstrapping results comparing the observed mean % of tumors with significantly disrupted expression for NRs relative to the background transcriptome in BRCA. Note that NRs are significantly upregulated less than is predicted by chance (p = 0.00065) and significantly downregulated more than is predicted by chance (p = 0.00179). (C) Pan-Cancer summary of NR expression patterns. Relative expression scores were determined by summing the Z-scores for a given gene in a given cancer and dividing by the square root of the number of tumors available for that tumor type. Shown are the 37 NRs detectable across all cancer types.
Table 3.
Summary of gene expression bootstrapping results comparing the average tumor/normal relative expression (% of tumors displaying overexpression (Z-score ≥ 2) and % of tumors displaying underexpression (Z-score ≤ −2)) of detectable members of 13 TF families, including NRs, across six different tumor types, relative to the background transcriptome. Shown is the observed value for each TF family (TF AVG), including NRs, as well as the mean value of the transcriptome background (TCGA AVG) for each respective cancer, as well as the bootstrapping results comparing the two. Significantly distorted expression patterns are highlighted in yellow (p < 0.05). Note that NRs are less commonly upregulated, and more commonly downregulated, than would be predicted by chance across all six tumor types.
| OVEREXPRESSED |
UNDEREXPRESSED |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF AVG (%) |
TCGA AVG (%) |
p-value: Less |
p-value: More |
TF AVG (%) |
TCGA AVG (%) |
p-value: Less |
p-value: More |
||
| NR |
PRAD |
3.09 | 7.52 | 1.50E-04 | 1.00E+00 | 12.19 | 8.66 | 9.77E-01 | 2.27E-02 |
|
BRCA |
6.72 | 13.30 | 6.60E-04 | 9.99E-01 | 26.06 | 16.86 | 9.98E-01 | 1.79E-03 | |
|
COAD |
4.75 | 18.16 | 1.00E-05 | 1.00E+00 | 30.17 | 19.77 | 9.96E-01 | 3.77E-03 | |
|
LIHC |
8.87 | 18.34 | 5.00E-05 | 1.00E+00 | 26.92 | 16.61 | 9.99E-01 | 5.90E-04 | |
|
HNSC |
3.61 | 9.27 | 1.40E-04 | 1.00E+00 | 13.17 | 7.83 | 9.97E-01 | 3.04E-03 | |
| BLCA | 2.79 | 10.12 | 1.00E-05 | 1.00E+00 | 25.82 | 11.64 | 1.00E+00 | 2.00E-05 | |
| bHLH |
PRAD |
7.12 | 7.52 | 3.68E-01 | 6.31E-01 | 10.93 | 8.66 | 9.62E-01 | 3.76E-02 |
|
BRCA |
10.57 | 13.30 | 5.39E-02 | 9.46E-01 | 20.61 | 16.86 | 9.53E-01 | 4.69E-02 | |
|
COAD |
17.69 | 18.16 | 4.43E-01 | 5.57E-01 | 27.91 | 19.77 | 9.97E-01 | 2.69E-03 | |
|
LIHC |
16.52 | 18.34 | 1.93E-01 | 8.07E-01 | 18.53 | 16.61 | 8.11E-01 | 1.89E-01 | |
|
HNSC |
7.78 | 9.27 | 1.47E-01 | 8.53E-01 | 6.42 | 7.83 | 1.30E-01 | 8.70E-01 | |
| BLCA | 8.27 | 10.12 | 9.35E-02 | 9.06E-01 | 17.14 | 11.64 | 9.97E-01 | 3.38E-03 | |
| bZIP |
PRAD |
4.88 | 7.52 | 2.56E-02 | 9.74E-01 | 10.16 | 8.66 | 8.18E-01 | 1.81E-01 |
|
BRCA |
9.01 | 13.30 | 3.47E-02 | 9.65E-01 | 21.93 | 16.86 | 9.36E-01 | 6.38E-02 | |
|
COAD |
12.15 | 18.16 | 3.72E-02 | 9.63E-01 | 15.93 | 19.77 | 1.42E-01 | 8.58E-01 | |
|
LIHC |
8.80 | 18.34 | 1.00E-05 | 1.00E+00 | 21.90 | 16.61 | 9.62E-01 | 3.84E-02 | |
|
HNSC |
5.22 | 9.27 | 7.85E-03 | 9.92E-01 | 6.51 | 7.83 | 2.25E-01 | 7.75E-01 | |
| BLCA | 4.52 | 10.12 | 3.70E-04 | 1.00E+00 | 19.99 | 11.64 | 9.98E-01 | 1.64E-03 | |
| E2F |
PRAD |
13.30 | 7.52 | 9.43E-01 | 5.72E-02 | 3.60 | 8.66 | 6.55E-02 | 9.34E-01 |
|
BRCA |
41.62 | 13.30 | 1.00E+00 | 6.00E-05 | 4.39 | 16.86 | 1.11E-02 | 9.89E-01 | |
|
COAD |
58.13 | 18.16 | 1.00E+00 | 3.00E-05 | 3.67 | 19.77 | 6.25E-03 | 9.94E-01 | |
|
LIHC |
51.25 | 18.34 | 1.00E+00 | 3.00E-05 | 1.62 | 16.61 | 1.30E-04 | 1.00E+00 | |
|
HNSC |
17.99 | 9.27 | 9.62E-01 | 3.84E-02 | 2.75 | 7.83 | 5.25E-02 | 9.47E-01 | |
| BLCA | 16.40 | 10.12 | 9.11E-01 | 8.84E-02 | 1.44 | 11.64 | 4.02E-03 | 9.96E-01 | |
| FOX |
PRAD |
8.53 | 7.52 | 7.48E-01 | 2.52E-01 | 9.13 | 8.66 | 6.25E-01 | 3.75E-01 |
|
BRCA |
17.00 | 13.30 | 9.30E-01 | 7.02E-02 | 19.97 | 16.86 | 7.85E-01 | 2.15E-01 | |
|
COAD |
20.27 | 18.16 | 7.07E-01 | 2.92E-01 | 21.94 | 19.77 | 7.07E-01 | 2.93E-01 | |
|
LIHC |
22.55 | 18.34 | 8.93E-01 | 1.07E-01 | 13.49 | 16.61 | 1.91E-01 | 8.08E-01 | |
|
HNSC |
12.96 | 9.27 | 9.50E-01 | 5.00E-02 | 6.97 | 7.83 | 3.50E-01 | 6.50E-01 | |
| BLCA | 10.15 | 10.12 | 5.26E-01 | 4.74E-01 | 11.00 | 11.64 | 4.37E-01 | 5.62E-01 | |
| GATA |
PRAD |
6.85 | 7.52 | 4.22E-01 | 5.78E-01 | 6.85 | 8.66 | 2.74E-01 | 7.26E-01 |
|
BRCA |
18.96 | 13.30 | 8.77E-01 | 1.23E-01 | 14.67 | 16.86 | 3.86E-01 | 6.14E-01 | |
|
COAD |
17.10 | 18.16 | 4.66E-01 | 5.34E-01 | 15.06 | 19.77 | 2.57E-01 | 7.42E-01 | |
|
LIHC |
19.68 | 18.34 | 6.27E-01 | 3.73E-01 | 13.17 | 16.61 | 2.66E-01 | 7.34E-01 | |
|
HNSC |
7.97 | 9.27 | 3.85E-01 | 6.15E-01 | 3.88 | 7.83 | 7.29E-02 | 9.27E-01 | |
| BLCA | 8.45 | 10.12 | 3.38E-01 | 6.62E-01 | 15.57 | 11.64 | 8.18E-01 | 1.82E-01 | |
| GeneralTF |
PRAD |
7.70 | 7.52 | 5.65E-01 | 4.35E-01 | 6.80 | 8.66 | 1.96E-01 | 8.04E-01 |
|
BRCA |
15.68 | 13.30 | 7.63E-01 | 2.37E-01 | 8.37 | 16.86 | 6.94E-03 | 9.93E-01 | |
|
COAD |
30.97 | 18.16 | 9.95E-01 | 5.01E-03 | 5.97 | 19.77 | 1.30E-04 | 1.00E+00 | |
|
LIHC |
23.75 | 18.34 | 9.31E-01 | 6.86E-02 | 10.47 | 16.61 | 3.75E-02 | 9.62E-01 | |
|
HNSC |
14.10 | 9.27 | 9.66E-01 | 3.39E-02 | 6.81 | 7.83 | 3.46E-01 | 6.54E-01 | |
| BLCA | 13.79 | 10.12 | 9.24E-01 | 7.58E-02 | 4.66 | 11.64 | 4.35E-03 | 9.96E-01 | |
| ETS |
PRAD |
4.18 | 7.52 | 2.14E-02 | 9.79E-01 | 12.65 | 8.66 | 9.65E-01 | 3.47E-02 |
|
BRCA |
6.31 | 13.30 | 3.90E-03 | 9.96E-01 | 17.77 | 16.86 | 6.28E-01 | 3.71E-01 | |
|
COAD |
18.71 | 18.16 | 5.65E-01 | 4.35E-01 | 23.51 | 19.77 | 7.91E-01 | 2.09E-01 | |
|
LIHC |
8.96 | 18.34 | 1.49E-03 | 9.99E-01 | 18.21 | 16.61 | 6.77E-01 | 3.23E-01 | |
|
HNSC |
6.93 | 9.27 | 1.65E-01 | 8.35E-01 | 8.23 | 7.83 | 6.05E-01 | 3.94E-01 | |
| BLCA | 5.07 | 10.12 | 9.28E-03 | 9.91E-01 | 8.88 | 11.64 | 1.93E-01 | 8.07E-01 | |
| HMG |
PRAD |
9.01 | 7.52 | 7.32E-01 | 2.68E-01 | 10.69 | 8.66 | 7.68E-01 | 2.32E-01 |
|
BRCA |
24.11 | 13.30 | 9.86E-01 | 1.42E-02 | 8.34 | 16.86 | 3.70E-02 | 9.63E-01 | |
|
COAD |
31.29 | 18.16 | 9.76E-01 | 2.35E-02 | 6.19 | 19.77 | 4.71E-03 | 9.95E-01 | |
|
LIHC |
31.43 | 18.34 | 9.92E-01 | 7.97E-03 | 12.96 | 16.61 | 2.53E-01 | 7.46E-01 | |
|
HNSC |
19.89 | 9.27 | 9.97E-01 | 3.34E-03 | 8.02 | 7.83 | 5.67E-01 | 4.33E-01 | |
| BLCA | 18.53 | 10.12 | 9.88E-01 | 1.24E-02 | 6.32 | 11.64 | 9.42E-02 | 9.06E-01 | |
| Homeobox |
PRAD |
7.75 | 7.52 | 6.21E-01 | 3.79E-01 | 7.23 | 8.66 | 5.35E-02 | 9.46E-01 |
|
BRCA |
11.78 | 13.30 | 2.00E-01 | 8.00E-01 | 22.57 | 16.86 | 9.99E-01 | 6.20E-04 | |
|
COAD |
20.88 | 18.16 | 9.10E-01 | 8.99E-02 | 18.99 | 19.77 | 3.56E-01 | 6.43E-01 | |
|
LIHC |
20.29 | 18.34 | 8.50E-01 | 1.50E-01 | 13.73 | 16.61 | 6.81E-02 | 9.32E-01 | |
|
HNSC |
17.46 | 9.27 | 1.00E+00 | 1.00E-05 | 4.51 | 7.83 | 1.00E-05 | 1.00E+00 | |
| BLCA | 11.07 | 10.12 | 8.05E-01 | 1.95E-01 | 13.83 | 11.64 | 9.32E-01 | 6.75E-02 | |
| KLF |
PRAD |
4.01 | 7.52 | 5.79E-02 | 9.42E-01 | 11.00 | 8.66 | 8.07E-01 | 1.93E-01 |
|
BRCA |
0.72 | 13.30 | 2.10E-04 | 1.00E+00 | 39.24 | 16.86 | 1.00E+00 | 5.00E-04 | |
|
COAD |
3.85 | 18.16 | 7.60E-04 | 9.99E-01 | 33.29 | 19.77 | 9.80E-01 | 1.95E-02 | |
|
LIHC |
3.48 | 18.34 | 2.00E-05 | 1.00E+00 | 29.86 | 16.61 | 9.91E-01 | 8.78E-03 | |
|
HNSC |
6.65 | 9.27 | 2.01E-01 | 7.98E-01 | 8.66 | 7.83 | 6.53E-01 | 3.47E-01 | |
| BLCA | 0.97 | 10.12 | 1.00E-05 | 1.00E+00 | 31.85 | 11.64 | 1.00E+00 | 5.00E-05 | |
| SMAD |
PRAD |
1.33 | 7.52 | 5.46E-03 | 9.94E-01 | 24.45 | 8.66 | 9.99E-01 | 6.70E-04 |
|
BRCA |
2.09 | 13.30 | 1.49E-03 | 9.99E-01 | 32.33 | 16.86 | 9.77E-01 | 2.26E-02 | |
|
COAD |
7.78 | 18.16 | 8.74E-02 | 9.12E-01 | 40.08 | 19.77 | 9.86E-01 | 1.39E-02 | |
|
LIHC |
11.46 | 18.34 | 1.29E-01 | 8.71E-01 | 24.29 | 16.61 | 8.74E-01 | 1.26E-01 | |
|
HNSC |
7.03 | 9.27 | 3.40E-01 | 6.60E-01 | 10.91 | 7.83 | 8.02E-01 | 1.98E-01 | |
| BLCA | 3.22 | 10.12 | 3.07E-02 | 9.69E-01 | 19.47 | 11.64 | 9.02E-01 | 9.76E-02 | |
| SP |
PRAD |
4.77 | 7.52 | 2.40E-01 | 7.60E-01 | 10.89 | 8.66 | 7.34E-01 | 2.65E-01 |
|
BRCA |
4.69 | 13.30 | 6.00E-02 | 9.40E-01 | 18.84 | 16.86 | 6.39E-01 | 3.60E-01 | |
|
COAD |
24.59 | 18.16 | 7.68E-01 | 2.32E-01 | 12.00 | 19.77 | 2.25E-01 | 7.75E-01 | |
|
LIHC |
23.23 | 18.34 | 7.65E-01 | 2.34E-01 | 7.91 | 16.61 | 1.16E-01 | 8.84E-01 | |
|
HNSC |
3.18 | 9.27 | 7.88E-02 | 9.21E-01 | 6.74 | 7.83 | 4.77E-01 | 5.23E-01 | |
| BLCA | 1.60 | 10.12 | 1.04E-02 | 9.90E-01 | 11.71 | 11.64 | 5.62E-01 | 4.38E-01 | |
Similarly, CNV frequency was determined for all genes across the genome (e.g. percentage of tumors with detectable CNV). For instance in BRCA, we observed that the average NR was amplified to some extent in 24.06% of tumors, while deleted in 20.90% of tumors (Supplemental Table 2). Likewise, random sampling (n = 100,000) was performed to identify the background genomic distribution of copy number alterations in equivalent sized gene sets, to which the NR superfamily observations were directly compared.
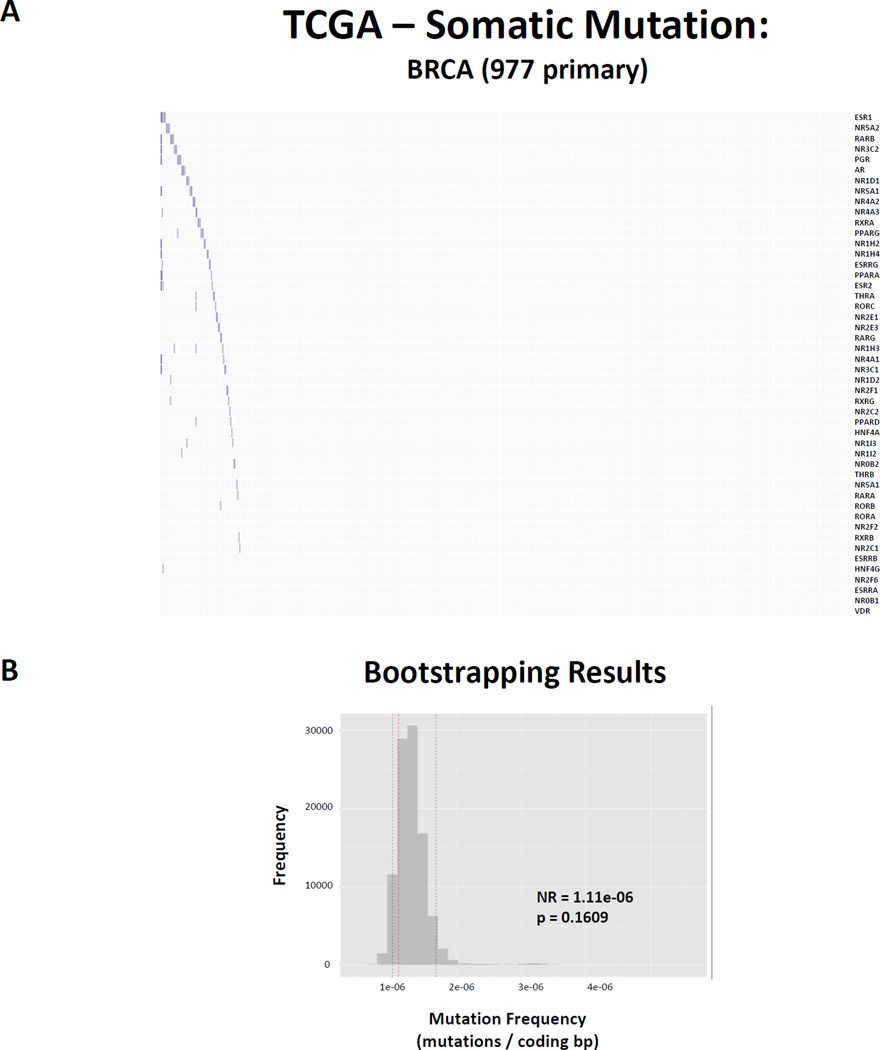
Lastly, mutation frequency was determined for all protein coding genes across the genome as described above (mutations / protein coding base pair (bp)). Again, using BRCA as an example, the average NR has a mutation frequency of 1.11E−06 mutations/bp, with the most commonly mutated member being ESR1 (mutation frequency = 1.72E-06) (Figure 4, Supplemental Table 1). Random sampling approaches were similarly applied to query this observation against the background protein coding genome.
Figure 4. Nuclear Receptors are not common targets of somatic mutation in cancer.
(A) Heatmap depicting non-synonymous mutations found in protein coding regions for all 48 NRs (rows) in 977 primary BRCA samples (columns). Observed mutations are depicted in blue. (B) Bootstrapping results comparing the observed mutation frequency (mutations / protein coding base pair) for NRs relative to the background protein coding genome in BRCA. Note that NRs are not significantly mutated more or less than is predicted by chance.
In all cases, empirical p-values were calculated based on the position of determined NR observations relative to the sampling distribution of the genome, to simply determine how probable the observations would be considered likely to occur by random chance. In the same manner stated above, observations and statistical testing were determined for 12 other transcription factor families (Table 2), to serve as comparison. Results of all comparisons are summarized in Table 3 (Expression), Supplemental Table 1 (CNV), and Supplemental Table 2 (Mutations).
5.2. Nuclear receptor superfamily expression is distorted in both common and unique manners across tumors
The majority of the members of the NR superfamily were expressed across the six tumor types. That is, considering NRs that were expressed at a detectable level (RPKM > 0) in >80% of normal and tumor samples revealed that 37 NRs were detected across all six tumor types, and the smallest number expressed was in BLCA which expressed 40 members, whereas the remainder of tissues expressed 42 members. Few tissues shared a common set of NRs, the exception being prostate and breast. Some NRs were undetectable across all tissues, including NR5A1/SF1 and NR0B1/DAX1, while others had distinct patterns of expression including FXR, which was detectible only in colon and liver.
Staying with BRCA as an example, Figure 2 shows the relative expression of the 42 NRs across 1095 tumors compared to a pool of 113 matched normal samples. The bootstrap analyses reveal that collectively, NRs are significantly less overexpressed than predicted by chance, and more underexpressed than predicted by chance. This was not a unique event to the BRCA cohort, but rather was seen across all tumor types examined (Table 3). This collective, pan-cancer downregulation was unique to NRs and not observed to a significant extent across all cancer types for any of the other 12 TF families examined. The closest were the KLF and SMAD families. For example, KLF members were collectively and significantly underexpressed in breast, colon and liver cancer types. By contrast, the E2Fs displayed the opposite pattern and were collectively overexpressed in at least three of the tumor types. Other TF families, including the FOX and GATA family members, showed little consensus pattern or were not collectively altered in any cancer. These findings would support the concept that NRs, and interacting TFs that control differentiation such as KLFs and SMADs, are significantly downregulated in a wide spectrum of tumors (32–36).
Next, we sought to compare the patterns of NR expression across tumor types (Figure 2C). To achieve this, a relative expression score was calculated by summing the total Z scores across all tumors of a given cancer type and normalizing each by the square root of the number of tumors available for that respective cancer. Reflecting the fact that as a whole the NR superfamily is significantly downregulated, very few individual NRs had a relative expression score that was positive, and none were positive across all tumors. NR2F6/EAR2 was over-expressed in BLCA, BRCA and PRCA which was also reported previously in bladder cancer cell lines(127). Its expression was also elevated in BRCA and there are several reports of the elevated expression and function of this orphan NR in breast cancer(73) (reviewed in(128)). Also in breast cancer RARA and NR1I3 were elevated. RARA is co-amplified with HER2/ERBB2(67) and has been proposed to stratify a novel subtype of BRCA(129). NR1I3/CAR was also overexpressed and has not previously been described in breast cancer. RARA elevation was also identified in LIHC and there is some evidence to support an oncogenic function for this receptor uniquely in the liver(130). Another NR to be elevated in 3/6 tumors was the orphan receptor NR2C1/TR2, but this appears relatively underexplored in the cancer field (reviewed in(128)). Other examples of NRs showing increased expression in cancers (NR6A1/GCN1, RARG, NR1D1/Rev-Erb-alpha, RXRB) are also relatively unexplored.
Conversely, there were many examples of individual NR expression being lost in cancer. In several instances, NR expression was lost in only some tumor types, or focally in single cancers, suggesting tissue specific importance. One example is RARG loss in PRAD, which we identified previously(118) and reflects the prostate metaplasia observed in RARγ null mice(131). Other examples of specific NR loss that have been previously characterized include VDR loss in COAD, Rev-Erb-alpha loss in BRCA, and NR1H2/LXRB loss in COAD(132–134).
However, other NRs were commonly downregulated in all or most cancers examined, suggesting broader tissue importance. These include examples from all types of NRs, from high affinity classical steroid hormone receptors such as PR, MR, GR; Type II NRs including RARB, PPARA; to Type III NRs including NR2F1/COUP-TF1 and NR2C2/TR2; and Type IV receptors such as NR1D2/Rev-erb-beta. In particular there are a group of eight NRs, which are down-regulated in at least five of the six tumor types (GR, MR, PGR, AR, NR1A2/THRB, NR5A2/LRH-1, RARB, NR1F1/ROR-alpha). Some of these are relatively well described and characterized in cancer, such as RARB as well as RORA as a negative regulator of proliferation and an emerging therapeutic target(135, 136). However what is perhaps less obvious is why GR and MR should be so uniformly and strongly reduced in expression. Given the roles for these receptors to contribute to the control of local inflammation, and the importance of inflammation as an early trigger for cancer, it is tempting to speculate that the common loss of these receptors reflects this central role. This surprising finding was also identified in breast cancer by high throughput Q-PCR approaches which revealed that MR, along with THRB and PPARG were altered and significantly predicted metastasis-free survival in patients(73).
5.3. Neither copy number variation nor mutation fully explain the changes in NR expression
Subsequently, we examined how genomic alterations including CNV and mutations associated with the expression changes observed within the NR superfamily. Collectively, CNV changes in the NR superfamily were not significantly different from the genome background in each of the six tumor types considered (Supplemental Table 1). This was generally true of the other TF families considered with the exception of the SMADs, which were significantly less amplified and more deleted than expected by chance across all the tumor types. This suggests a common mechanism driving downregulation of some transcription factors including SMADs may be result of genomic instability, while others including NRs may be results of additional mechanisms.
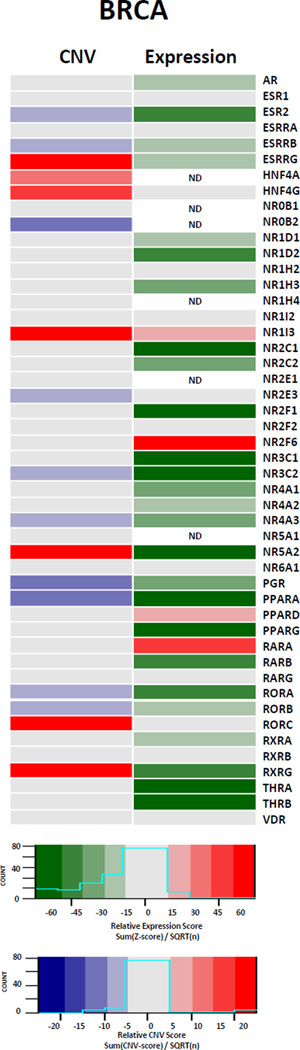
However, comparing the results of CNV alterations with gene expression changes within an individual tumor reveals some NR specific associations for individual receptors, as can be seen comparing relative expression scores to relative CNV scores (calculated in a similar fashion to relative expression scores, as described above) in BRCA (Figure 3). From this comparison it appears that whilst there is a reasonable agreement between copy number loss and reduced or absent gene expression, the reverse is not true. For instance, in the BRCA cohort, nine out of ten NRs that are significantly reduced at the genomic level for the receptor are also reduced or absent in expression, including NR3A2/ESR2 and PGR. By contrast of the seven NRs that are amplified, only one results in elevated gene expression, namely NR1I3. Meanwhile for the other six NRs associated with amplified regions there is no significant gain, and in fact there is still significant loss of expression in several cases, suggesting other regulatory events control expression from the amplicon.
Figure 3. Comparing Nuclear Receptor CNV alterations to expression changes in BRCA.
Relative scores were determined for CNV alterations (Sum of GISTIC 2 threshold alteration values / square root(number of tumors)) and expression changes (Sum of Z-scores / square root(number of tumors)) for each member of the nuclear receptor superfamily. NRs which were not detectable in >80% of samples are considered not detectable (ND).
Finally, examination of the mutational status of the NR superfamily did not reveal a significant relationship when considering the number of mutations, after normalizing for the length of the total coding sequence, as compared to the background genome (Figure 4, Supplemental Table 2). There were not many consistent patterns across cancers concerning TF family mutation frequencies, with the exception of Homeobox TFs which are more commonly mutated than predicted by chance in 3 cancers. Of the focal examples of commonly mutated families in specific cancers such as FOXs in PRAD and GATAs in BRCA, they are largely driven by one or two commonly mutated members, of which there is some precedent in the literature; FOXA1 is commonly mutated in prostate cancer(137), GATA3 is commonly mutated in breast cancer(138), although for others such as ELF3 (ETS family) in BLCA there doesn’t appear to be any literature to date.
6. Conclusions and future perspectives
The present review has described how the NR superfamily is integrated through shared genomic binding, shared pathways of genomic signaling, shared cofactor interactions and cross-talk with other TF pathways. Signaling by NRs is central to many cell fate decisions and as a consequence these actions are corrupted in many cancer cell types. Indeed the history of cancer research is intimately interwoven with elucidation of NR function from the earliest studies of steroidal signaling in breast and prostate cancer, to the identification of targeted therapies, to discovery of epigenetic mechanisms that distort gene regulation function and now, in the post-genomic era, to development of integrative genomic workflows that combine genomic, epigenomic and transcriptomic data to develop dynamic maps of NR signaling.
We sought to build on the review of NR function by developing a pan-cancer view of NR expression by exploiting the remarkable volume of genomic data developed by TCGA. Transcriptomic and genomic alterations of the NR superfamily across six tumor types were examined and by exploiting bootstrapping approaches we were able to generate robust statistical statements concerning the expression, CNV and mutation status of the NR superfamily, alongside 12 other TF families as comparison.
A clear finding from these approaches was that the detected members of the NR superfamily are more downregulated than predicted by chance, an observation which was uniform across the cancers examined. No other TF family displayed this phenomenon, although KLFs and SMADs mirrored it to a more restricted and limited statistical extent and reflects studies that identified cross-talk between NRs/SMADs/KLFs. Within each tumor however the precise up and down regulated NRs varied, and comparing across cancers revealed the common downregulation of GR, MR, PGR and THRB, whereas other changes in expression appeared be unique to a specific tumor type; RARG loss in prostate and gain in colon cancer, gain of NR6A1 and RXRB in liver cancer, loss of VDR in colon cancer; the gain in colon and loss in breast cancer of Rev-erb-alpha and the loss in colon of LXRB.
Interestingly, whilst NRs were strongly downregulated, this was only to a small extent, if at all, explained by genomic causes such as CNV or mutation, as opposed to other TF families including SMADs whose expression changes reflected CNV alterations. Therefore, an interesting implication of this observation is the idea that epigenomic, rather than genomic, mechanisms may be the drivers for this phenomenon, and possibly that while NRs are down-regulated in cancer, they may remain functional. There are well-established roles for DNA methylation to down-regulate NRs, most notably RARB(63–65), and the current findings suggest that targeted DNA methylation may be responsible for suppression of other NRs. Previously, we have considered roles for microRNA to explain NR expression levels and established that certain cohorts of NR targeting miRNA were more up-regulated than predicted by chance(74). Whilst undertaking these studies is statistically more challenging, given the many to many relationships between miRNA and mRNA, it seems reasonable to suggest that networks of miRNA may play a significant role to distort NR network expression and therefore function across cancers.
Interestingly, all tissues examined expressed a broad array of NRs, with BLCA expressing the fewest at 40. This observation coincides with recent undertakings profiling NRs in tissues. For instance, the 42 NRs detected in BRCA correlate well with findings from a recent study examining NR expression in an independent cohort of normal breast tissue and breast tumors of varying stage(73). In this study, 41 NRs were detectable via TaqMan low-density array across breast tissues, with 6 of the 7 undetected NRs also not found to be expressed by our criteria in TCGA samples (NR0B1/DAX1, NR0B2/SHP, HNF4A, NR2E1/TLX, NR1H4/FXR and NR5A1/SF1). The only discrepancy between detectible NRs in breast tissue between the former study and our TCGA analysis was the detectible expression of NR1I3/CAR in TCGA samples, which had amongst the lowest expression of NRs in breast tumors in our analysis. Also in validation of our analysis, this study found a general pan-repression of NRs in breast tumors relative to normal breast tissues, with almost half of detected NRs having significantly lower expression.
The RoadMap Epigenome(113) genomics consortia have underscored the significance of NR-enhancer interactions. Specifically, the Roadmap Epigenome investigators developed an algorithm entitled ChromHMM based on Hidden Markov Models. This was applied to the ~ 3000 genomic and epigenomic data sets generated from over 100 cell types to identify 15 different chromatin states(113). Within these states, enhancer regions(139) represented ~ 3% of the genome, and were typified by genomic location, DNAse sensitivity, and histone modifications (e.g. H3K4me1, H3K27me3 and H3K36me3) (140–143). Within the enhancer modules over 1500 transcription factor motifs were examined for enrichment and revealed 84 significant transcription factor-enhancer modules. Ten of these modules were centered on NRs. Therefore NR are over-represented, and the current study has revealed roles for five of these NRs (GR, GCN1, LRH-1, THRB, RARG) as being more altered than predicted by chance across thousands of tumor samples. These different NR-motif relationships were not identified in the same chromatin states and therefore they perhaps represent high priority receptors for ChIP-Seq based studies to define how loss of expression (but not deletion or mutation) alters the distribution within chromatin states and modulates enhancer associations and/or responses.
Furthermore, in parallel, large scale genome-wide association studies (GWAS) of genetic variation has revealed that the vast majority of SNPs are contained in areas of the genome that are outside of gene exons, and therefore do not have the potential to make a direct contribution to protein structure and function (144). It is emerging that many phenotype- and disease-associated SNPs at distal regions impact transcription factor activity that in turn is associated with disease (144, 145), and therefore integration of frequently altered NRs, in the most parsimonious cancer phenotypes, may prove to be a powerful approach to reveal how genetic variation impacts NR function.
Supplementary Material
Acknowledgments
Funding – This work was supported by the Prostate program of the Department of Defense Congressionally Directed Medical Research Programs [W81XWH-14-1-0608, W81XWH-11-2-0033 to MJC], the European Union-United States Atlantis Program [P116J090011 to MJC] and in part by National Institute of Health [R01 CA095367-06 and 2R01-CA-095045-06 to MJC]. It was also supported, in part, of the National Cancer Institute Cancer Center Support Grant to the Roswell Park Cancer Institute [CA016056]. It was also supported by the Molecular Pharmacology and Experimental Therapeutics NRSA T32 program [T32CA009072 to MDL].
References
- 1.Battaglia S, Maguire O, Thorne JL, Hornung LB, Doig CL, Liu S, Sucheston LE, Bianchi A, Khanim FL, Gommersall LM, et al. Elevated NCOR1 disrupts PPARalpha/gamma signaling in prostate cancer and forms a targetable epigenetic lesion. Carcinogenesis. 2010;31:1650–1660. doi: 10.1093/carcin/bgq086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baranowski M. Biological role of liver X receptors. J Physiol Pharmacol. 2008;59(Suppl 7):31–55. [PubMed] [Google Scholar]
- 3.Abedin SA, Thorne JL, Battaglia S, Maguire O, Hornung LB, Doherty AP, Mills IG, Campbell MJ. Elevated NCOR1 disrupts a network of dietary-sensing nuclear receptors in bladder cancer cells. Carcinogenesis. 2009;30:449–456. doi: 10.1093/carcin/bgp005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kacevska M, Downes MR, Sharma R, Evans RM, Clarke SJ, Liddle C, Robertson GR. Extrahepatic Cancer Suppresses Nuclear Receptor-Regulated Drug Metabolism. Clinical Cancer Research. 2011;17:3170–3180. doi: 10.1158/1078-0432.CCR-10-3289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kaeding J, Bouchaert E, Bélanger J, Caron P, Chouinard S, Verreault M, Larouche O, Pelletier G, Staels B, Bélanger A, et al. Activators of the farnesoid X receptor negatively regulate androgen glucuronidation in human prostate cancer LNCAP cells. Biochemical Journal. 2008;410:245. doi: 10.1042/BJ20071136. [DOI] [PubMed] [Google Scholar]
- 6.Okada S, Markle JG, Deenick EK, Mele F, Averbuch D, Lagos M, Alzahrani M, Al-Muhsen S, Halwani R, Ma CS, et al. IMMUNODEFICIENCIES. Impairment of immunity to Candida and Mycobacterium in humans with bi-allelic RORC mutations. Science. 2015;349:606–613. doi: 10.1126/science.aaa4282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Overington JP, Al-Lazikani B, Hopkins AL. How many drug targets are there? Nature reviews. Drug discovery. 2006;5:993–996. doi: 10.1038/nrd2199. [DOI] [PubMed] [Google Scholar]
- 8.Nuclear Receptors Nomenclature, C. A unified nomenclature system for the nuclear receptor superfamily. Cell. 1999;97:161–163. doi: 10.1016/s0092-8674(00)80726-6. [DOI] [PubMed] [Google Scholar]
- 9.Mangelsdorf DJ, Thummel C, Beato M, Herrlich P, Schutz G, Umesono K, Blumberg B, Kastner P, Mark M, Chambon P, et al. The nuclear receptor superfamily: the second decade. Cell. 1995;83:835–839. doi: 10.1016/0092-8674(95)90199-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hua S, Kittler R, White KP. Genomic antagonism between retinoic acid and estrogen signaling in breast cancer. Cell. 2009;137:1259–1271. doi: 10.1016/j.cell.2009.04.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hanson AM, Gambill J, Phomakay V, Staten CT, Kelley MD. 9-cis-retinoic acid and troglitazone impacts cellular adhesion, proliferation, and integrin expression in K562 cells. PloS one. 2014;9:e93005. doi: 10.1371/journal.pone.0093005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Elstner E, Linker-Israeli M, Umiel T, Le J, Grillier I, Said J, Shintaku IP, Krajewski S, Reed JC, Binderup L, et al. Combination of a potent 20-epi-vitamin D3 analogue (KH 1060) with 9-cis-retinoic acid irreversibly inhibits clonal growth, decreases bcl-2 expression, and induces apoptosis in HL-60 leukemic cells. Cancer research. 1996;56:3570–3576. [PubMed] [Google Scholar]
- 13.James SY, Mackay AG, Colston KW. Vitamin D derivatives in combination with 9-cis retinoic acid promote active cell death in breast cancer cells. Journal of molecular endocrinology. 1995;14:391–394. doi: 10.1677/jme.0.0140391. [DOI] [PubMed] [Google Scholar]
- 14.Hallenbeck PL, Phyillaier M, Nikodem VM. Divergent effects of 9-cis-retinoic acid receptor on positive and negative thyroid hormone receptor-dependent gene expression. The Journal of biological chemistry. 1993;268:3825–3828. [PubMed] [Google Scholar]
- 15.Chatagnon A, Veber P, Morin V, Bedo J, Triqueneaux G, Semon M, Laudet V, d'Alche-Buc F, Benoit G. RAR/RXR binding dynamics distinguish pluripotency from differentiation associated cis-regulatory elements. Nucleic acids research. 2015;43:4833–4854. doi: 10.1093/nar/gkv370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mendoza-Parra MA, Walia M, Sankar M, Gronemeyer H. Dissecting the retinoid-induced differentiation of F9 embryonal stem cells by integrative genomics. Molecular systems biology. 2011;7:538. doi: 10.1038/msb.2011.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brazda P, Krieger J, Daniel B, Jonas D, Szekeres T, Langowski J, Toth K, Nagy L, Vamosi G. Ligand binding shifts highly mobile retinoid X receptor to the chromatin-bound state in a coactivator-dependent manner, as revealed by single-cell imaging. Molecular and cellular biology. 2014;34:1234–1245. doi: 10.1128/MCB.01097-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Daniel B, Nagy G, Hah N, Horvath A, Czimmerer Z, Poliska S, Gyuris T, Keirsse J, Gysemans C, Van Ginderachter JA, et al. The active enhancer network operated by liganded RXR supports angiogenic activity in macrophages. Genes & development. 2014;28:1562–1577. doi: 10.1101/gad.242685.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jepsen K, Hermanson O, Onami TM, Gleiberman AS, Lunyak V, McEvilly RJ, Kurokawa R, Kumar V, Liu F, Seto E, et al. Combinatorial roles of the nuclear receptor corepressor in transcription and development. Cell. 2000;102:753–763. doi: 10.1016/s0092-8674(00)00064-7. [DOI] [PubMed] [Google Scholar]
- 20.Chen JD. Steroid/nuclear receptor coactivators. Vitamins and hormones. 2000;58:391–448. doi: 10.1016/s0083-6729(00)58032-7. [DOI] [PubMed] [Google Scholar]
- 21.Lonard DM, O'Malley BW. Nuclear receptor coregulators: modulators of pathology and therapeutic targets. Nature reviews. Endocrinology. 2012;8:598–604. doi: 10.1038/nrendo.2012.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lupien M, Eeckhoute J, Meyer CA, Wang Q, Zhang Y, Li W, Carroll JS, Liu XS, Brown M. FoxA1 translates epigenetic signatures into enhancer-driven lineage-specific transcription. Cell. 2008;132:958–970. doi: 10.1016/j.cell.2008.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Serandour AA, Avner S, Percevault F, Demay F, Bizot M, Lucchetti-Miganeh C, Barloy-Hubler F, Brown M, Lupien M, Metivier R, et al. Epigenetic switch involved in activation of pioneer factor FOXA1-dependent enhancers. Genome research. 2011;21:555–565. doi: 10.1101/gr.111534.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sahu B, Laakso M, Pihlajamaa P, Ovaska K, Sinielnikov I, Hautaniemi S, Janne OA. FoxA1 specifies unique androgen and glucocorticoid receptor binding events in prostate cancer cells. Cancer research. 2013;73:1570–1580. doi: 10.1158/0008-5472.CAN-12-2350. [DOI] [PubMed] [Google Scholar]
- 25.Wang L, Zhang X, Farrar WL, Yang X. Transcriptional crosstalk between nuclear receptors and cytokine signal transduction pathways in immunity. Cell Mol Immunol. 2004;1:416–424. [PubMed] [Google Scholar]
- 26.Katoh M, Katoh M. WNT signaling pathway and stem cell signaling network. Clinical cancer research : an official journal of the American Association for Cancer Research. 2007;13:4042–4045. doi: 10.1158/1078-0432.CCR-06-2316. [DOI] [PubMed] [Google Scholar]
- 27.Ellison TI, Smith MK, Gilliam AC, MacDonald PN. Inactivation of the vitamin D receptor enhances susceptibility of murine skin to UV-induced tumorigenesis. J Invest Dermatol. 2008;128:2508–2517. doi: 10.1038/jid.2008.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Saramaki A, Banwell CM, Campbell MJ, Carlberg C. Regulation of the human p21(waf1/cip1) gene promoter via multiple binding sites for p53 and the vitamin D3 receptor. Nucleic acids research. 2006;34:543–554. doi: 10.1093/nar/gkj460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Maruyama R, Aoki F, Toyota M, Sasaki Y, Akashi H, Mita H, Suzuki H, Akino K, Ohe-Toyota M, Maruyama Y, et al. Comparative Genome Analysis Identifies the Vitamin D Receptor Gene as a Direct Target of p53-Mediated Transcriptional Activation. Cancer Res. 2006;66:4574–4583. doi: 10.1158/0008-5472.CAN-05-2562. [DOI] [PubMed] [Google Scholar]
- 30.Lambert JR, Kelly JA, Shim M, Huffer WE, Nordeen SK, Baek SJ, Eling TE, Lucia MS. Prostate derived factor in human prostate cancer cells: gene induction by vitamin D via a p53-dependent mechanism and inhibition of prostate cancer cell growth. J Cell Physiol. 2006;208:566–574. doi: 10.1002/jcp.20692. [DOI] [PubMed] [Google Scholar]
- 31.Kommagani R, Caserta TM, Kadakia MP. Identification of vitamin D receptor as a target of p63. Oncogene. 2006;25:3745–3751. doi: 10.1038/sj.onc.1209412. [DOI] [PubMed] [Google Scholar]
- 32.Zerr P, Vollath S, Palumbo-Zerr K, Tomcik M, Huang J, Distler A, Beyer C, Dees C, Gela K, Distler O, et al. Vitamin D receptor regulates TGF-beta signalling in systemic sclerosis. Annals of the rheumatic diseases. 2014 doi: 10.1136/annrheumdis-2013-204378. in press. [DOI] [PubMed] [Google Scholar]
- 33.Ito I, Waku T, Aoki M, Abe R, Nagai Y, Watanabe T, Nakajima Y, Ohkido I, Yokoyama K, Miyachi H, et al. A nonclassical vitamin D receptor pathway suppresses renal fibrosis. The Journal of clinical investigation. 2013;123:4579–4594. doi: 10.1172/JCI67804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ding N, Yu RT, Subramaniam N, Sherman MH, Wilson C, Rao R, Leblanc M, Coulter S, He M, Scott C, et al. A vitamin D receptor/SMAD genomic circuit gates hepatic fibrotic response. Cell. 2013;153:601–613. doi: 10.1016/j.cell.2013.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Karp JE, Rassool FV. KLFs and ATRA-induced differentiation: new pathways for exploitation. Leukemia research. 2011;35:846–847. doi: 10.1016/j.leukres.2011.04.002. [DOI] [PubMed] [Google Scholar]
- 36.Chen Z, Brand NJ, Chen A, Chen SJ, Tong JH, Wang ZY, Waxman S, Zelent A. Fusion between a novel Kruppel-like zinc finger gene and the retinoic acid receptor-alpha locus due to a variant t(11;17) translocation associated with acute promyelocytic leukaemia. The EMBO journal. 1993;12:1161–1167. doi: 10.1002/j.1460-2075.1993.tb05757.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Novershtern N, Subramanian A, Lawton LN, Mak RH, Haining WN, McConkey ME, Habib N, Yosef N, Chang CY, Shay T, et al. Densely interconnected transcriptional circuits control cell states in human hematopoiesis. Cell. 2011;144:296–309. doi: 10.1016/j.cell.2011.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mendoza-Parra MA, Gronemeyer H. Genome-wide studies of nuclear receptors in cell fate decisions. Seminars in cell & developmental biology. 2013;24:706–715. doi: 10.1016/j.semcdb.2013.07.001. [DOI] [PubMed] [Google Scholar]
- 39.Paul J. Sir George Beatson and the Royal Beatson Memorial Hospital. Med Hist. 1981;25:200–201. [PMC free article] [PubMed] [Google Scholar]
- 40.Machtens S, Schultheiss D, Kuczyk M, Truss MC, Jonas U. The history of endocrine therapy of benign and malignant diseases of the prostate. World journal of urology. 2000;18:222–226. doi: 10.1007/s003450000120. [DOI] [PubMed] [Google Scholar]
- 41.van 't Veer LJ, Dai H, van de Vijver MJ, He YD, Hart AA, Mao M, Peterse HL, van der Kooy K, Marton MJ, Witteveen AT, et al. Gene expression profiling predicts clinical outcome of breast cancer. Nature. 2002;415:530–536. doi: 10.1038/415530a. [DOI] [PubMed] [Google Scholar]
- 42.Heinlein CA, Chang C. Androgen receptor in prostate cancer. Endocrine reviews. 2004;25:276–308. doi: 10.1210/er.2002-0032. [DOI] [PubMed] [Google Scholar]
- 43.Bloomfield CD, Munck AU, Smith KA. Glucocorticoid receptor levels predict response to treatment in human lymphoma. Progress in clinical and biological research. 1984;142:223–233. [PubMed] [Google Scholar]
- 44.Kim J, Jeong D, Nam J, Aung TN, Gim JA, Park KU, Kim SW. MicroRNA-124 regulates glucocorticoid sensitivity by targeting phosphodiesterase 4B in diffuse large B cell lymphoma. Gene. 2015;558:173–180. doi: 10.1016/j.gene.2015.01.001. [DOI] [PubMed] [Google Scholar]
- 45.Bloomfield CD, Smith KA, Peterson BA, Hildebrandt L, Zaleskas J, Gajl-Peczalska KJ, Frizzera G, Munck A. In-vitro glucocorticoid studies for predicting response to glucocorticoid therapy in adults with malignant lymphoma. Lancet. 1980;1:952–956. doi: 10.1016/s0140-6736(80)91405-1. [DOI] [PubMed] [Google Scholar]
- 46.Arora VK, Schenkein E, Murali R, Subudhi SK, Wongvipat J, Balbas MD, Shah N, Cai L, Efstathiou E, Logothetis C, et al. Glucocorticoid receptor confers resistance to antiandrogens by bypassing androgen receptor blockade. Cell. 2013;155:1309–1322. doi: 10.1016/j.cell.2013.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Huang ME, Ye YC, Chen SR, Chai JR, Lu JX, Zhoa L, Gu LJ, Wang ZY. Use of all-trans retinoic acid in the treatment of acute promyelocytic leukemia. Blood. 1988;72:567–572. [PubMed] [Google Scholar]
- 48.Douer D, Koeffler HP. Retinoic acid. Inhibition of the clonal growth of human myeloid leukemia cells. The Journal of clinical investigation. 1982;69:277–283. doi: 10.1172/JCI110450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Breitman TR, Selonick SE, Collins SJ. Induction of differentiation of the human promyelocytic leukemia cell line (HL-60) by retinoic acid. Proceedings of the National Academy of Sciences of the United States of America. 1980;77:2936–2940. doi: 10.1073/pnas.77.5.2936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Castaigne S, Chomienne C, Daniel MT, Berger R, Miclea JM, Ballerini P, Degos L. Retinoic acids in the treatment of acute promyelocytic leukemia. Nouv Rev Fr Hematol. 1990;32:36–38. [PubMed] [Google Scholar]
- 51.Huang ME, Ye YC, Chen SR, Chai JR, Lu JX, Zhao L, Gu LJ, Wang ZY. Use of all-trans retinoic acid in the treatment of acute promyelocytic leukemia. Haematol Blood Transfus. 1989;32:88–96. [PubMed] [Google Scholar]
- 52.Arveiler B, Petkovich M, Mandel JL, Chambon P. A PstI RFLP for the human retinoic acid receptor in 17q21. Nucleic acids research. 1988;16:6252. doi: 10.1093/nar/16.13.6252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mattei MG, Petkovich M, Mattei JF, Brand N, Chambon P. Mapping of the human retinoic acid receptor to the q21 band of chromosome 17. Human genetics. 1988;80:186–188. doi: 10.1007/BF00702866. [DOI] [PubMed] [Google Scholar]
- 54.Grignani F, De Matteis S, Nervi C, Tomassoni L, Gelmetti V, Cioce M, Fanelli M, Ruthardt M, Ferrara FF, Zamir I, et al. Fusion proteins of the retinoic acid receptor-alpha recruit histone deacetylase in promyelocytic leukaemia. Nature. 1998;391:815–818. doi: 10.1038/35901. [DOI] [PubMed] [Google Scholar]
- 55.Lin RJ, Nagy L, Inoue S, Shao W, Miller WH, Jr, Evans RM. Role of the histone deacetylase complex in acute promyelocytic leukaemia. Nature. 1998;391:811–814. doi: 10.1038/35895. [DOI] [PubMed] [Google Scholar]
- 56.Viennois E, Mouzat K, Dufour J, Morel L, Lobaccaro JM, Baron S. Selective liver X receptor modulators (SLiMs): what use in human health? Molecular and cellular endocrinology. 2012;351:129–141. doi: 10.1016/j.mce.2011.08.036. [DOI] [PubMed] [Google Scholar]
- 57.Chuu CP, Lin HP. Antiproliferative effect of LXR agonists T0901317 and 22(R)-hydroxycholesterol on multiple human cancer cell lines. Anticancer research. 2010;30:3643–3648. [PubMed] [Google Scholar]
- 58.Chuu CP, Kokontis JM, Hiipakka RA, Liao S. Modulation of liver X receptor signaling as novel therapy for prostate cancer. J Biomed Sci. 2007;14:543–553. doi: 10.1007/s11373-007-9160-8. [DOI] [PubMed] [Google Scholar]
- 59.Thorne J, Campbell MJ. The vitamin D receptor in cancer. The Proceedings of the Nutrition Society. 2008;67:115–127. doi: 10.1017/S0029665108006964. [DOI] [PubMed] [Google Scholar]
- 60.Campbell MJ, Carlberg C, Koeffler HP. A Role for the PPARgamma in Cancer Therapy. PPAR research. 2008;2008:314974. doi: 10.1155/2008/314974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Huang H, Xu Y, Zhu J, Li J. Recent advances in non-steroidal FXR antagonists development for therapeutic applications. Current topics in medicinal chemistry. 2014;14:2175–2187. doi: 10.2174/1568026614666141112101840. [DOI] [PubMed] [Google Scholar]
- 62.Swales KE, Korbonits M, Carpenter R, Walsh DT, Warner TD, Bishop-Bailey D. The farnesoid X receptor is expressed in breast cancer and regulates apoptosis and aromatase expression. Cancer research. 2006;66:10120–10126. doi: 10.1158/0008-5472.CAN-06-2399. [DOI] [PubMed] [Google Scholar]
- 63.Campbell MJ, Park S, Uskokovic MR, Dawson MI, Koeffler HP. Expression of retinoic acid receptor-beta sensitizes prostate cancer cells to growth inhibition mediated by combinations of retinoids and a 19-nor hexafluoride vitamin D3 analog. Endocrinology. 1998;139:1972–1980. doi: 10.1210/endo.139.4.5943. [DOI] [PubMed] [Google Scholar]
- 64.Liu Y, Lee MO, Wang HG, Li Y, Hashimoto Y, Klaus M, Reed JC, Zhang X. Retinoic acid receptor beta mediates the growth-inhibitory effect of retinoic acid by promoting apoptosis in human breast cancer cells. Molecular and cellular biology. 1996;16:1138–1149. doi: 10.1128/mcb.16.3.1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhang XK, Liu Y, Lee MO. Retinoid receptors in human lung cancer and breast cancer. Mutation research. 1996;350:267–277. doi: 10.1016/0027-5107(95)00102-6. [DOI] [PubMed] [Google Scholar]
- 66.Dressman MA, Baras A, Malinowski R, Alvis LB, Kwon I, Walz TM, Polymeropoulos MH. Gene expression profiling detects gene amplification and differentiates tumor types in breast cancer. Cancer research. 2003;63:2194–2199. [PubMed] [Google Scholar]
- 67.Doi A, Ishikawa K, Shibata N, Ito E, Fujimoto J, Yamamoto M, Shiga H, Mochizuki H, Kawamura Y, Goshima N, et al. Enhanced expression of retinoic acid receptor alpha (RARA) induces epithelial-to-mesenchymal transition and disruption of mammary acinar structures. Molecular oncology. 2015;9:355–364. doi: 10.1016/j.molonc.2014.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Serenaite I, Daniunaite K, Jankevicius F, Laurinavicius A, Petroska D, Lazutka JR, Jarmalaite S. Heterogeneity of DNA methylation in multifocal prostate cancer. Virchows Archiv : an international journal of pathology. 2015;466:53–59. doi: 10.1007/s00428-014-1678-3. [DOI] [PubMed] [Google Scholar]
- 69.Lau KM, LaSpina M, Long J, Ho SM. Expression of estrogen receptor (ER)-alpha and ER-beta in normal and malignant prostatic epithelial cells: regulation by methylation and involvement in growth regulation. Cancer research. 2000;60:3175–3182. [PubMed] [Google Scholar]
- 70.Li LC, Chui R, Nakajima K, Oh BR, Au HC, Dahiya R. Frequent methylation of estrogen receptor in prostate cancer: correlation with tumor progression. Cancer research. 2000;60:702–706. [PubMed] [Google Scholar]
- 71.Vera-Badillo FE, Templeton AJ, de Gouveia P, Diaz-Padilla I, Bedard PL, Al-Mubarak M, Seruga B, Tannock IF, Ocana A, Amir E. Androgen receptor expression and outcomes in early breast cancer: a systematic review and meta-analysis. Journal of the National Cancer Institute. 2014;106:djt319. doi: 10.1093/jnci/djt319. [DOI] [PubMed] [Google Scholar]
- 72.Robinson JL, Macarthur S, Ross-Innes CS, Tilley WD, Neal DE, Mills IG, Carroll JS. Androgen receptor driven transcription in molecular apocrine breast cancer is mediated by FoxA1. The EMBO journal. 2011;30:3019–3027. doi: 10.1038/emboj.2011.216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Muscat GE, Eriksson NA, Byth K, Loi S, Graham D, Jindal S, Davis MJ, Clyne C, Funder JW, Simpson ER, et al. Research resource: nuclear receptors as transcriptome: discriminant and prognostic value in breast cancer. Molecular endocrinology. 2013;27:350–365. doi: 10.1210/me.2012-1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Long MD, Thorne JL, Russell J, Battaglia S, Singh PK, Sucheston-Campbell LE, Campbell MJ. Cooperative behavior of the nuclear receptor superfamily and its deregulation in prostate cancer. Carcinogenesis. 2014;35:262–271. doi: 10.1093/carcin/bgt334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kittler R, Zhou J, Hua S, Ma L, Liu Y, Pendleton E, Cheng C, Gerstein M, White KP. A comprehensive nuclear receptor network for breast cancer cells. Cell reports. 2013;3:538–551. doi: 10.1016/j.celrep.2013.01.004. [DOI] [PubMed] [Google Scholar]
- 76.Siersbaek R, Baek S, Rabiee A, Nielsen R, Traynor S, Clark N, Sandelin A, Jensen ON, Sung MH, Hager GL, et al. Molecular architecture of transcription factor hotspots in early adipogenesis. Cell Rep. 2014;7:1434–1442. doi: 10.1016/j.celrep.2014.04.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Siersbaek R, Rabiee A, Nielsen R, Sidoli S, Traynor S, Loft A, La Cour Poulsen L, Rogowska-Wrzesinska A, Jensen ON, Mandrup S. Transcription factor cooperativity in early adipogenic hotspots and super-enhancers. Cell reports. 2014;7:1443–1455. doi: 10.1016/j.celrep.2014.04.042. [DOI] [PubMed] [Google Scholar]
- 78.Rada-Iglesias A, Bajpai R, Prescott S, Brugmann SA, Swigut T, Wysocka J. Epigenomic annotation of enhancers predicts transcriptional regulators of human neural crest. Cell Stem Cell. 2012;11:633–648. doi: 10.1016/j.stem.2012.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Liu Z, Merkurjev D, Yang F, Li W, Oh S, Friedman MJ, Song X, Zhang F, Ma Q, Ohgi KA, et al. Enhancer activation requires trans-recruitment of a mega transcription factor complex. Cell. 2014;159:358–373. doi: 10.1016/j.cell.2014.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Mendes-Pereira AM, Sims D, Dexter T, Fenwick K, Assiotis I, Kozarewa I, Mitsopoulos C, Hakas J, Zvelebil M, Lord CJ, et al. Genome-wide functional screen identifies a compendium of genes affecting sensitivity to tamoxifen. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:2730–2735. doi: 10.1073/pnas.1018872108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wargon V, Riggio M, Giulianelli S, Sequeira GR, Rojas P, May M, Polo ML, Gorostiaga MA, Jacobsen B, Molinolo A, et al. Progestin and antiprogestin responsiveness in breast cancer is driven by the PRA/PRB ratio via AIB1 or SMRT recruitment to the CCND1 and MYC promoters. International journal of cancer. Journal international du cancer. 2015;136:2680–2692. doi: 10.1002/ijc.29304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Lanz RB, Bulynko Y, Malovannaya A, Labhart P, Wang L, Li W, Qin J, Harper M, O'Malley BW. Global characterization of transcriptional impact of the SRC-3 coregulator. Molecular endocrinology. 2010;24:859–872. doi: 10.1210/me.2009-0499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Fereshteh MP, Tilli MT, Kim SE, Xu J, O'Malley BW, Wellstein A, Furth PA, Riegel AT. The nuclear receptor coactivator amplified in breast cancer-1 is required for Neu (ErbB2/HER2) activation, signaling, and mammary tumorigenesis in mice. Cancer research. 2008;68:3697–3706. doi: 10.1158/0008-5472.CAN-07-6702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Osborne CK, Bardou V, Hopp TA, Chamness GC, Hilsenbeck SG, Fuqua SA, Wong J, Allred DC, Clark GM, Schiff R. Role of the estrogen receptor coactivator AIB1 (SRC-3) and HER-2/neu in tamoxifen resistance in breast cancer. Journal of the National Cancer Institute. 2003;95:353–361. doi: 10.1093/jnci/95.5.353. [DOI] [PubMed] [Google Scholar]
- 85.Chung AC, Zhou S, Liao L, Tien JC, Greenberg NM, Xu J. Genetic ablation of the amplified-in-breast cancer 1 inhibits spontaneous prostate cancer progression in mice. Cancer research. 2007;67:5965–5975. doi: 10.1158/0008-5472.CAN-06-3168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Grenier J, Trousson A, Chauchereau A, Cartaud J, Schumacher M, Massaad C. Differential recruitment of p160 coactivators by glucocorticoid receptor between Schwann cells and astrocytes. Molecular endocrinology. 2006;20:254–267. doi: 10.1210/me.2005-0061. [DOI] [PubMed] [Google Scholar]
- 87.Raghav SK, Waszak SM, Krier I, Gubelmann C, Isakova A, Mikkelsen TS, Deplancke B. Integrative Genomics Identifies the Corepressor SMRT as a Gatekeeper of Adipogenesis through the Transcription Factors C/EBPbeta and KAISO. Molecular cell. 2012;46:335–350. doi: 10.1016/j.molcel.2012.03.017. [DOI] [PubMed] [Google Scholar]
- 88.Barish GD, Yu RT, Karunasiri MS, Becerra D, Kim J, Tseng TW, Tai LJ, Leblanc M, Diehl C, Cerchietti L, et al. The Bcl6-SMRT/NCoR cistrome represses inflammation to attenuate atherosclerosis. Cell metabolism. 2012;15:554–562. doi: 10.1016/j.cmet.2012.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Khanim FL, Gommersall LM, Wood VH, Smith KL, Montalvo L, O'Neill LP, Xu Y, Peehl DM, Stewart PM, Turner BM, et al. Altered SMRT levels disrupt vitamin D3 receptor signalling in prostate cancer cells. Oncogene. 2004;23:6712–6725. doi: 10.1038/sj.onc.1207772. [DOI] [PubMed] [Google Scholar]
- 90.Mengeling BJ, Goodson ML, Bourguet W, Privalsky ML. SMRTepsilon, a corepressor variant, interacts with a restricted subset of nuclear receptors, including the retinoic acid receptors alpha and beta. Molecular and cellular endocrinology. 2012;351:306–316. doi: 10.1016/j.mce.2012.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Goodson ML, Mengeling BJ, Jonas BA, Privalsky ML. Alternative mRNA splicing of corepressors generates variants that play opposing roles in adipocyte differentiation. The Journal of biological chemistry. 2011;286:44988–44999. doi: 10.1074/jbc.M111.291625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Stephens PJ, Tarpey PS, Davies H, Van Loo P, Greenman C, Wedge DC, Nik-Zainal S, Martin S, Varela I, Bignell GR, et al. The landscape of cancer genes and mutational processes in breast cancer. Nature. 2012;486:400–404. doi: 10.1038/nature11017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Haiman CA, Garcia RR, Hsu C, Xia L, Ha H, Sheng X, Le Marchand L, Kolonel LN, Henderson BE, Stallcup MR, et al. Screening and association testing of common coding variation in steroid hormone receptor co-activator and co-repressor genes in relation to breast cancer risk: the Multiethnic Cohort. BMC Cancer. 2009;9:43. doi: 10.1186/1471-2407-9-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Chang TH, Szabo E. Enhanced growth inhibition by combination differentiation therapy with ligands of peroxisome proliferator-activated receptor-gamma and inhibitors of histone deacetylase in adenocarcinoma of the lung. Clin.Cancer Res. 2002;8:1206–1212. [PubMed] [Google Scholar]
- 95.Battaglia S, Thorne J, Maguire O, Hornung L, Sucheston LE, McCabe C, Bunce CM, Campbell MJ. Elevated NCOR1 disrupts PPAR signaling in prostate cancer and forms a targetable epigenetic lesion. Carcinogenesis. 2010 doi: 10.1093/carcin/bgq086. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Girault I, Lerebours F, Amarir S, Tozlu S, Tubiana-Hulin M, Lidereau R, Bieche I. Expression analysis of estrogen receptor alpha coregulators in breast carcinoma: evidence that NCOR1 expression is predictive of the response to tamoxifen. Clin.Cancer Res. 2003;9:1259–1266. [PubMed] [Google Scholar]
- 97.Zhang Z, Yamashita H, Toyama T, Sugiura H, Ando Y, Mita K, Hamaguchi M, Hara Y, Kobayashi S, Iwase H. NCOR1 mRNA is an independent prognostic factor for breast cancer. Cancer Lett. 2005 doi: 10.1016/j.canlet.2005.05.046. in press. [DOI] [PubMed] [Google Scholar]
- 98.Banwell CM, MacCartney DP, Guy M, Miles AE, Uskokovic MR, Mansi J, Stewart PM, O'Neill LP, Turner BM, Colston KW, et al. Altered nuclear receptor corepressor expression attenuates vitamin D receptor signaling in breast cancer cells. Clin Cancer Res. 2006;12:2004–2013. doi: 10.1158/1078-0432.CCR-05-1218. [DOI] [PubMed] [Google Scholar]
- 99.Kim JY, Son YL, Lee YC. Involvement of SMRT Corepressor in Transcriptional Repression by the Vitamin D Receptor. Mol Endocrinol. 2009;23:251–264. doi: 10.1210/me.2008-0426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Li Y, Zhang DY, Ren Q, Ye F, Zhao X, Daniels G, Wu X, Dynlacht B, Lee P. Regulation of a Novel Androgen Receptor Target Gene, the Cyclin B1 Gene, through Androgen-Dependent E2F Family Member Switching. Molecular and cellular biology. 2012;32:2454–2466. doi: 10.1128/MCB.06663-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Trtkova K, Paskova L, Matijescukova N, Strnad M, Kolar Z. Binding of AR to SMRT/N-CoR complex and its co-operation with PSA promoter in prostate cancer cells treated with natural histone deacetylase inhibitor NaB. Neoplasma. 2010;57:406–414. doi: 10.4149/neo_2010_05_406. [DOI] [PubMed] [Google Scholar]
- 102.Hodgson MC, Astapova I, Hollenberg AN, Balk SP. Activity of androgen receptor antagonist bicalutamide in prostate cancer cells is independent of NCoR and SMRT corepressors. Cancer research. 2007;67:8388–8395. doi: 10.1158/0008-5472.CAN-07-0617. [DOI] [PubMed] [Google Scholar]
- 103.Hodgson MC, Astapova I, Cheng S, Lee LJ, Verhoeven MC, Choi E, Balk SP, Hollenberg AN. The androgen receptor recruits nuclear receptor CoRepressor (N-CoR) in the presence of mifepristone via its N and C termini revealing a novel molecular mechanism for androgen receptor antagonists. The Journal of biological chemistry. 2005;280:6511–6519. doi: 10.1074/jbc.M408972200. [DOI] [PubMed] [Google Scholar]
- 104.Linja MJ, Porkka KP, Kang Z, Savinainen KJ, Janne OA, Tammela TL, Vessella RL, Palvimo JJ, Visakorpi T. Expression of androgen receptor coregulators in prostate cancer. Clinical cancer research : an official journal of the American Association for Cancer Research. 2004;10:1032–1040. doi: 10.1158/1078-0432.ccr-0990-3. [DOI] [PubMed] [Google Scholar]
- 105.Banwell CM, MacCartney DP, Guy M, Miles AE, Uskokovic MR, Mansi J, Stewart PM, O'Neill LP, Turner BM, Colston KW, et al. Altered nuclear receptor corepressor expression attenuates vitamin D receptor signaling in breast cancer cells. Clinical cancer research : an official journal of the American Association for Cancer Research. 2006;12:2004–2013. doi: 10.1158/1078-0432.CCR-05-1218. [DOI] [PubMed] [Google Scholar]
- 106.Long MD, van den Berg PR, Russell JL, Singh PK, Battaglia S, Campbell MJ. Integrative genomic analysis in K562 chronic myelogenous leukemia cells reveals that proximal NCOR1 binding positively regulates genes that govern erythroid differentiation and Imatinib sensitivity. Nucleic acids research. 2015 doi: 10.1093/nar/gkv642. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Consortium GT. Human genomics. The Genotype-Tissue Expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science. 2015;348:648–660. doi: 10.1126/science.1262110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Ponten F, Jirstrom K, Uhlen M. The Human Protein Atlas--a tool for pathology. The Journal of pathology. 2008;216:387–393. doi: 10.1002/path.2440. [DOI] [PubMed] [Google Scholar]
- 109.Rosenbloom KR, Sloan CA, Malladi VS, Dreszer TR, Learned K, Kirkup VM, Wong MC, Maddren M, Fang R, Heitner SG, et al. ENCODE data in the UCSC Genome Browser: year 5 update. Nucleic acids research. 2013;41:D56–D63. doi: 10.1093/nar/gks1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Stamatoyannopoulos JA. What does our genome encode? Genome research. 2012;22:1602–1611. doi: 10.1101/gr.146506.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Maher B. ENCODE: The human encyclopaedia. Nature. 2012;489:46–48. doi: 10.1038/489046a. [DOI] [PubMed] [Google Scholar]
- 112.Birney E. The making of ENCODE: Lessons for big-data projects. Nature. 2012;489:49–51. doi: 10.1038/489049a. [DOI] [PubMed] [Google Scholar]
- 113.Roadmap Epigenomics C, Kundaje A, Meuleman W, Ernst J, Bilenky M, Yen A, Heravi-Moussavi A, Kheradpour P, Zhang Z, Wang J, et al. Integrative analysis of 111 reference human epigenomes. Nature. 2015;518:317–330. doi: 10.1038/nature14248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Katayama S, Kanamori M, Hayashizaki Y. Integrated analysis of the genome and the transcriptome by FANTOM. Briefings in bioinformatics. 2004;5:249–258. doi: 10.1093/bib/5.3.249. [DOI] [PubMed] [Google Scholar]
- 115.Sanli K, Karlsson FH, Nookaew I, Nielsen J. FANTOM: Functional and taxonomic analysis of metagenomes. BMC bioinformatics. 2013;14:38. doi: 10.1186/1471-2105-14-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, Jacobsen A, Byrne CJ, Heuer ML, Larsson E, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer discovery. 2012;2:401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, Sun Y, Jacobsen A, Sinha R, Larsson E, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Science signaling. 2013;6:11. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Taylor BS, Schultz N, Hieronymus H, Gopalan A, Xiao Y, Carver BS, Arora VK, Kaushik P, Cerami E, Reva B, et al. Integrative genomic profiling of human prostate cancer. Cancer cell. 2010;18:11–22. doi: 10.1016/j.ccr.2010.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Singh PK, Long MD, Battaglia S, Hu Q, Liu S, Sucheston-Campbell LE, Campbell MJ. VDR regulation of microRNA differs across prostate cell models suggesting extremely flexible control of transcription. Epigenetics : official journal of the DNA Methylation Society. 2015;10:40–49. doi: 10.4161/15592294.2014.989088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Singh PK, Campbell MJ. The Interactions of microRNA and Epigenetic Modifications in Prostate Cancer. Cancers. 2013;5:998–1019. doi: 10.3390/cancers5030998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Thorne JL, Maguire O, Doig CL, Battaglia S, Fehr L, Sucheston LE, Heinaniemi M, O'Neill LP, McCabe CJ, Turner BM, et al. Epigenetic control of a VDR-governed feed-forward loop that regulates p21(waf1/cip1) expression and function in non-malignant prostate cells. Nucleic acids research. 2011;39:2045–2056. doi: 10.1093/nar/gkq875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J, et al. Bioconductor: open software development for computational biology and bioinformatics. Genome biology. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Gray KA, Yates B, Seal RL, Wright MW, Bruford EA. Genenames.org: the HGNC resources in 2015. Nucleic acids research. 2015;43:D1079–D1085. doi: 10.1093/nar/gku1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Goldman M, Craft B, Swatloski T, Cline M, Morozova O, Diekhans M, Haussler D, Zhu J. The UCSC Cancer Genomics Browser: update 2015. Nucleic acids research. 2015;43:D812–D817. doi: 10.1093/nar/gku1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Smedley D, Haider S, Durinck S, Pandini L, Provero P, Allen J, Arnaiz O, Awedh MH, Baldock R, Barbiera G, et al. The BioMart community portal: an innovative alternative to large, centralized data repositories. Nucleic acids research. 2015;43:W589–W598. doi: 10.1093/nar/gkv350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Franke J, Neumann MH. Bootstrapping neural networks. Neural computation. 2000;12:1929–1949. doi: 10.1162/089976600300015204. [DOI] [PubMed] [Google Scholar]
- 127.Okegawa T, Ushio K, Imai M, Morimoto M, Hara T. Orphan nuclear receptor HNF4G promotes bladder cancer growth and invasion through the regulation of the hyaluronan synthase 2 gene. Oncogenesis. 2013;2:e58. doi: 10.1038/oncsis.2013.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Safe S, Jin UH, Hedrick E, Reeder A, Lee SO. Minireview: role of orphan nuclear receptors in cancer and potential as drug targets. Molecular endocrinology. 2014;28:157–172. doi: 10.1210/me.2013-1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Paroni G, Fratelli M, Gardini G, Bassano C, Flora M, Zanetti A, Guarnaccia V, Ubezio P, Centritto F, Terao M, et al. Synergistic antitumor activity of lapatinib and retinoids on a novel subtype of breast cancer with coamplification of ERBB2 and RARA. Oncogene. 2012;31:3431–3443. doi: 10.1038/onc.2011.506. [DOI] [PubMed] [Google Scholar]
- 130.Khetchoumian K, Teletin M, Tisserand J, Herquel B, Ouararhni K, Losson R. Trim24 (Tif1 alpha): an essential 'brake' for retinoic acid-induced transcription to prevent liver cancer. Cell cycle. 2008;7:3647–3652. doi: 10.4161/cc.7.23.7123. [DOI] [PubMed] [Google Scholar]
- 131.Lohnes D, Mark M, Mendelsohn C, Dolle P, Decimo D, LeMeur M, Dierich A, Gorry P, Chambon P. Developmental roles of the retinoic acid receptors. The Journal of steroid biochemistry and molecular biology. 1995;53:475–486. doi: 10.1016/0960-0760(95)00094-g. [DOI] [PubMed] [Google Scholar]
- 132.Derangere V, Chevriaux A, Courtaut F, Bruchard M, Berger H, Chalmin F, Causse SZ, Limagne E, Vegran F, Ladoire S, et al. Liver X receptor beta activation induces pyroptosis of human and murine colon cancer cells. Cell death and differentiation. 2014;21:1914–1924. doi: 10.1038/cdd.2014.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Vedin LL, Gustafsson JA, Steffensen KR. The oxysterol receptors LXRalpha and LXRbeta suppress proliferation in the colon. Molecular carcinogenesis. 2013;52:835–844. doi: 10.1002/mc.21924. [DOI] [PubMed] [Google Scholar]
- 134.Palmer HG, Larriba MJ, Garcia JM, Ordonez-Moran P, Pena C, Peiro S, Puig I, Rodriguez R, de la Fuente R, Bernad A, et al. The transcription factor SNAIL represses vitamin D receptor expression and responsiveness in human colon cancer. Nature medicine. 2004;10:917–919. doi: 10.1038/nm1095. [DOI] [PubMed] [Google Scholar]
- 135.Kojetin DJ, Burris TP. REV-ERB and ROR nuclear receptors as drug targets. Nature reviews. Drug discovery. 2014;13:197–216. doi: 10.1038/nrd4100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Jetten AM. Retinoid-related orphan receptors (RORs): critical roles in development, immunity, circadian rhythm, and cellular metabolism. Nuclear receptor signaling. 2009;7:e003. doi: 10.1621/nrs.07003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Grasso CS, Wu YM, Robinson DR, Cao X, Dhanasekaran SM, Khan AP, Quist MJ, Jing X, Lonigro RJ, Brenner JC, et al. The mutational landscape of lethal castration-resistant prostate cancer. Nature. 2012;487:239–243. doi: 10.1038/nature11125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Cancer Genome Atlas, N. Comprehensive molecular portraits of human breast tumours. Nature. 2012;490:61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Hnisz D, Abraham BJ, Lee TI, Lau A, Saint-Andre V, Sigova AA, Hoke HA, Young RA. Super-enhancers in the control of cell identity and disease. Cell. 2013;155:934–947. doi: 10.1016/j.cell.2013.09.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Reilly SK, Yin J, Ayoub AE, Emera D, Leng J, Cotney J, Sarro R, Rakic P, Noonan JP. Evolutionary genomics. Evolutionary changes in promoter and enhancer activity during human corticogenesis. Science. 2015;347:1155–1159. doi: 10.1126/science.1260943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Markenscoff-Papadimitriou E, Allen WE, Colquitt BM, Goh T, Murphy KK, Monahan K, Mosley CP, Ahituv N, Lomvardas S. Enhancer interaction networks as a means for singular olfactory receptor expression. Cell. 2014;159:543–557. doi: 10.1016/j.cell.2014.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Shlyueva D, Stelzer C, Gerlach D, Yanez-Cuna JO, Rath M, Boryn LM, Arnold CD, Stark A. Hormone-responsive enhancer-activity maps reveal predictive motifs, indirect repression, and targeting of closed chromatin. Molecular cell. 2014;54:180–192. doi: 10.1016/j.molcel.2014.02.026. [DOI] [PubMed] [Google Scholar]
- 143.Xie M, Hong C, Zhang B, Lowdon RF, Xing X, Li D, Zhou X, Lee HJ, Maire CL, Ligon KL, et al. DNA hypomethylation within specific transposable element families associates with tissue-specific enhancer landscape. Nature genetics. 2013;45:836–841. doi: 10.1038/ng.2649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Schaub MA, Boyle AP, Kundaje A, Batzoglou S, Snyder M. Linking disease associations with regulatory information in the human genome. Genome research. 2012;22:1748–1759. doi: 10.1101/gr.136127.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Al Olama AA, Kote-Jarai Z, Berndt SI, Conti DV, Schumacher F, Han Y, Benlloch S, Hazelett DJ, Wang Z, Saunders E, et al. A meta-analysis of 87,040 individuals identifies 23 new susceptibility loci for prostate cancer. Nature genetics. 2014 doi: 10.1038/ng.3094. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.