Summary
Early growth response gene-1 (EGR1) widely exists in the cell nucleus of such as, zebrafish, mice, chimpanzees and humans, an it also can be observed in the cytoplasm of some tumors. EGR1 was named just after its brief and rapid expression of different stimuli. Accumulating studies have extensively demonstrated that the widespread dysregulation of EGR1 is involved in hematological malignancies such as human acute myeloid leukemia (AML), chronic myelogenous leukemia, chronic lymphocytic leukemia, multiple myeloma, and B cell lymphoma. With the deep research on EGR1, its expression, function and regulatory mechanism has been gradually elucidated, and provides more possibilities for treatment strategies of patients with leukemia. Herein, we summarize the roles of EGR1 in its biological function and relationship with leukemia.
Keywords: Early growth response gene-1 (EGR1), acute myeloid leukemia, tumor
1. Introduction
Early growth response gene-1 (EGR1), also known as NGFI-A, krox-24, ZIF268 and TIS8, is an immediate early gene which encodes a Cys2-His2-type zinc finger transcription factor widely expressed in eukaryotic cells from yeast to humans (1–3). It is one of the largest studies of tumor-specific proteins, which are located in the 5q31 region (4,5). It has an important role in controlling synaptic plasticity, wound repair, female reproductive capacity, inflammation, growth control, differentiation, apoptosis and tumor progression (6). Experiments have also proved that acute myeloid leukemia and myelodysplastic syndromes are associated with heterozygous loss of EGR1 (7). Here, we focus on the relationship of EGR1 with acute myeloid leukemia.
2. The summarization of EGR1's discovery and function
EGR1 was first discovered in the mid-1980s (8). The EGR family includes EGR1, EGR2, EGR3, EGR4 four related members, that can quickly and briefly be up-regulated through a variety of external stimuli, including activation, proliferation and differentiation signals, tissue damage and apoptosis signals (9). EGR1, EGR2, EGR3 and EGR4 share a highly conserved DNA binding domain, composed of three zinc finger motifs that together bind to a 9-bp G/C-rich consensus sequence (GCGGGGGCG) (10). It has been used extensively as a model system for detecting how TFIIIA-like zinc fingers recognize DNA, and how it has served as a basis for engineering some types of artificial DNA-binding proteins (11). EGRs are involved in regulating the immune response by means of the induction of differentiation of lymphocyte precursors, and activation of B and T cells (12). EGR1 binds to DNA G/C-rich sequences through 3 zinc-finger motifs in its carboxyl terminal and regulates gene transcription through co-operation with other activating or repressing factors (13). It may be divided into three zones. The N-terminal portion (amino acids 1–331) is rich in proline (14.2%) and serine (16%) and has 7.9% alanine and 7.9%, threonine. The C-terminal region (residues 417–533) also contains a very high proportion of proline and serine (15.4% and 26.5%, respectively) as well as 10.3% alanine and 11.1% threonine (14).
3. Biological function and role in tumors
The EGR1 gene encodes a zinc finger protein and its expression is modulated in diverse biological systems with kinetics resembling those of c-fos (14). EGR1 together with c-fos is crucial for normal myeloid cell differentiation through transcriptional regulation (15). Gene expression analysis revealed that EGR1 and c-fos were down-regulated in hematopoietic primitive cells (16). C-fos and EGR1 represent the key transcription factors that are differentially activated by macrophage colony-stimulating factor (M-CSF) and granulocyte colony-stimulating factor (G-CSF) to resolve neutrophil versus monocyte cell fate (17). However, EGR1 has more of an advantage than c-fos because of different structure, which increases its expression and decreases sensitivity to stimulation (18). EGR1 can regulate cell growth, differentiation, growth inhibition, and apoptosis in various kinds of cells (19). Many factors can regulate expression of EGR1, including miR-424, miR-146a, miR-181a, E2h2, wilms tumor suppressor 1 (WT1), and Iron (9,20–25). It's also reported in the literature that EGR1 can be regulated by erythropoietin (EPO) (26,27). MiR675 upregulates long noncoding RNA H19 through activating EGR1 in human liver cancer (28). More importantly, EGR1 can regulate some signaling such as p53, transforming growth factor beta 1 (TGFβ1), phosphatase and tensin homolog deleted on chromosome ten (PTEN), Fibronectin, and enterovirus 71 (EV71) (29–32). The promoter of the human TGFβ1, p53, and the fibronectin gene contains at least two EGR1-binding sites, both of which can bind EGR1 to activate transcription. The proximal promoter of PTEN is GC rich and contains one functional EGR1-binding site (29). Moreover, it plays important roles in decidualization, megakaryocyte differentiation, apoptosis, tendon development, lung injury, liver injury, kidney diseases, chronic obstructive pulmonary disease (COPD), angiogenesis, fibrosis, atherosclerosis, cell cycle and other biological functions (33–52). EGR1 has a critical role in promoting autophagy and apoptosis in response to cigarette smoke exposure in vitro and in vivo (53). EGR1 controls metabolism, especially its suppression of lipolysis and promotes fat accumulation by inhibiting the expression of triglyceride lipase (54). Although the expression of EGR1 is low in most tissues, it is high in islets. EGR1 regulates insulin gene expression by up-regulating Pdx1 (55). EGR1 gene expression may contribute to the decrease of B-cell proliferation and the consequent cell failure observed in the later stages of type 2 diabetes (56). The increase of EGR1 expression in the brain is associated with formation of emotional memory and schizophrenia (57). It has been proved that EGR1 mutant mice had no changes in short-term memory, but long-term memory was severely damaged (58). Ischemia-induced EGR1 expression may exaggerate brain injury by reducing brain-derived neurotrophic factor (BDNF) expression (59). EGR1 exhibited a biphasic expression behavior. It was previously described to be down-regulated in many breast carcinoma tissues while it was upregulated in highly invasive inflammatory breast carcinoma. It started to be upregulated 4 h after SNAI1 induction, and was repressed after 24 h (6). Interestingly, in prostate cancer, kidney cancer and stomach cancer EGR1 stimulates the growth of tumor cells, and is associated with poor prognosis. In contrast, EGR1 is a tumor suppressor in fibrosarcoma, glioblastoma, melanoma, esophageal cancer, lung cancer and breast cancer (60–64).
4. Pathogenesis mechanism of AML by EGR1
In the absence of EGR1, a significant increase in cell cycling occurs in hematopoietic stem cells (HSCs), culminating in an increased number of HSCs and an increased frequency of primary reconstitution under limiting dilution conditions. Most interestingly, loss of EGR1 causes efficient mobilization of HSCs out of their niches (65). Abnormalities of chromosome 5 are common aberrations in acute myeloid leukemia (AML), with del(5q) the most frequent (66,67). There is also literature, which shows that EGR-1 was related to recurrent disease following high-dose chemotherapy (68). Nevertheless, EGR1 haploinsufficiency alone in vivo does not lead to expansion of HSCs or abnormalities in adult hematopoiesis. It has been proven that loss of a single allele of more than one gene on 5q contributes to the pathogenesis of AML (69–71). A number of genes and several microRNAs (miRNAs) located on 5q, including miRNA-145, miRNA-146a, the ribosomal protein S14 (RPS14), the cell division cycle 25 (CDC25), the adenomatous polyposis coli gene (APC) have been implicated in the development of myeloid disorders caused by a gene dosage effect (72,73). (Figure 1) EGR1 may play a functional role in the pathogenesis of AML in patients with del(5q) (74,75). The loss of EGR1 or inactivation increases risk of AML (76). Using locus-specific probes, a deletion of the EGR1 locus 5q31, 7q31 and the TP53 gene was observed in 103 (82%), in 57 (46%) and in 66 (53%) patients respectively. Thirty patients (24%) showed a deletion of all three loci, and in only 13 cases (10%), 5q31, 7q31, or 17p13 was not deleted. An EGR1 deletion alone was observed in 19 cases (15%) in only five and four AMLs respectively (77). In an attempt to define the loss of the 5q31.1 region, fluorescence in situ hybridization analysis was performed in HL-60 cells, which spanned the EGR1 and IL9 gene interval, which was previously shown to be a critical region of loss in AML (78). Loss of the EGR1 gene with deletions of 7q31 or TP53 alone played a role in at least two aspects. First, EGR1 directly controls the expression of fibronectin (FN1) through pathways that involve GFB1 and plasminogen activator-1 (PAI1). Thus, FN1 and PAI1 act together to inhibit the growth of cancer cells. Second, EGR1 is required for p53-dependent apoptosis through the mediation of retinoblastoma (79). To examine the role of EGR1 in hematopoiesis, EGR1+/− and EGR1−/− mice was characterized, and found that EGR1+/− and EGR1−/− mice develop T-cell lymphoma or a myeloproliferative disorder (MPD) at an increased rate and a reduced latency over that observed in wild-type littermates. EGR1+/− and EGR1−/− mice develop T-cell lymphoma or MPD at the same rate and latency, suggesting that loss of a single allele of EGR1 is sufficient for disease predisposition. This is consistent with observations in patients with AML characterized by abnormalities of chromosome 5, in that only 1 EGR1 allele is affected (80). Interestingly, EGR1 is regulated by multiple factors in AML. The cyclin-dependent kinases (CDK) CDK6 and Src family kinases (SFKs) inhibit expression of EGR1 (81,82). On the contrary, Llgl1 (lethal giant larvae homolog 1) and PMA (Phorbol 12-myristate 13-acetate) contribute to the differentiation of hematopoietic stem cells (83,84). Andra Schaefer et al. found that the expression of EGR-1 had a regulatory role in Epo signal transduction in leukemia cells (85).
Figure 1.
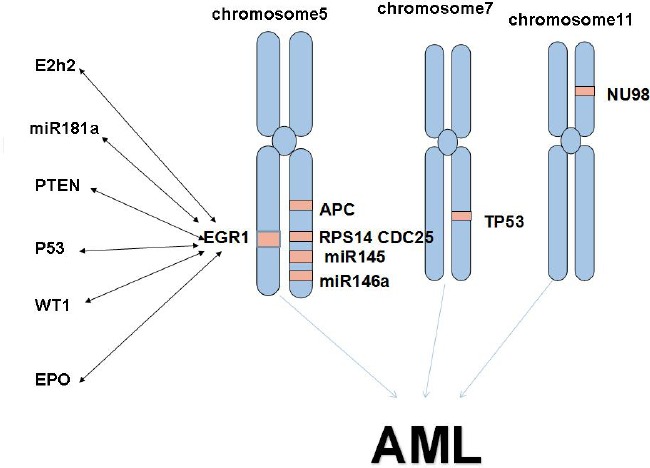
E2h2, miR181a, PTEN, P53, WT1, EPO and EGR1 can regulate each other. The cooperation of EGR1, APC, RPS14, CDC25, miR145, miR146a, TP53 and NU98 may lead to the formation of AML.
5. The possibility of EGR1 as therapy target of patients with AML
The primary structure of the EGR1 protein suggests that it is a DNA-binding protein with transcriptional regulatory activity, and it may function as a tumor suppressor locus whose absence or loss of function could lead to deregulated cell growth (86). This gives us an inclination that EGR1 or EGR1 target gene is useful for treatment of blood malignant tumors (87). One study mentioned that EGR1 and p21 are key signaling molecules of genipin-induced apoptosis in gastric cancer cells (88). Another article revealed that the down-regulation of EGR1-p21 expression provides a mechanism for improved hematopoiesis (89). Histone deacetylase (HDAC) inhibitors can reactivate EGR1 in various cell types, leading to decreased cell proliferation and increased cell apoptosis (90). HDAC recruitment may participate in the repressive mechanism that EGR1 directly represses myocyte enhancer factor 2 (MEF2) activity for treatment of cardiac disease (91). Experimental evidence has demonstrated that EGR1 diminished the aggressiveness of M1myc leukemia and abrogated the leukemic potential of IL-6-treated M1myc cells. Altered EGR1 expression can work together with deregulated c-Myc in exacerbating the leukemic phenotype (92). It is also reported that EGR-1 plays an indispensable role in the regulation of platycodon D-induced cell death and the 1, 25D3-induced cessation of cell proliferation, which is characteristic of the terminal stage of differentiation of these cells (93,94). EGR1 and WT1 are structurally related transcription factors and bound to quite similar DNA sequences (95). This gives us a revelation that down-regulating the expression of WT1 can up-regulate the expression of EGR1. In this way, inhibition of proliferation and differentiation of leukemia cells is no longer a problem. EGR1 is also important for development of the macrophage lineage (96). It is interesting to note that EGR-1 abrogates the block in M1 terminal differentiation imparted by oncogenic c-Myc or E2F-1, suppressing their leukemia promoting function in nude mice (97). A novel mechanism of thalidomide in the treatment of leukemia is that thalidomide could suppress leukemia cell invasion and migration by upregulation of EGR-1 (98). Also that paeoniflorin (PF) playing a role in human leukemia U937 cells is based on the regulation of EGR1 (99). LY294002 (LY29) is a commonly used pharmacologic inhibitor of phosphatidylinositol 3-kinase (PI3 K) and has shown an antitumorigenic effect. It could suppress leukemia cell invasion and migration at least in part through up-regulation of EGR-1, independent of its PI3 K-Akt inhibitory activity (100). In summary, we believe that EGR is likely to be a target for treatment of AML.
Acknowledgements
This study was supported by the National Natural Science Foundation of China (No. 81101605, 81573467), the ‘Twelfth Five-Year’ National Science and Technology Support Program (2013BAI07B02), the Natural Science Foundation of Shandong Province of China (ZR2011HL045, ZR2015YL028, 2015ZRC03102) and the Project for Laureate of Taishan Scholar (NO. ts201511075). Shandong Scientific and technological project (2013YD18031).
References
- 1. Shelly C, Petruzzelli L, Herrera R. K562 cells resistant to phorbol 12-myristate 13-acetate-induced growth arrest: Dissociation of mitogen-activated protein kinase activation and Egr-1 expression from megakaryocyte differentiation. Cell Growth Differ. 2000; 11:501-506. [PubMed] [Google Scholar]
- 2. Ferguson J, Bird C, Wadhwa M, Burns C. Detection of neutralizing antibodies to erythropoietin by inhibition of rHuEPO-stimulated EGR1 gene expression in the UT-7/EPO cell line. J Immunol Methods. 2013; 387:191-198. [DOI] [PubMed] [Google Scholar]
- 3. Wang D, Guan MP, Zheng ZJ, Li WQ, Lyv FP, Pang RY, Xue YM. Transcription factor Egr1 is involved in high glucose-induced proliferation and fibrosis in rat glomerular mesangial cells. Cell Physiol Biochem. 2015; 36:2093-2107. [DOI] [PubMed] [Google Scholar]
- 4. MacKinnon RN, Kannourakis G, Wall M, Campbell LJ. A cryptic deletion in 5q31.2 provides further evidence for a minimally deleted region in myelodysplastic syndromes. Cancer Genet. 2011; 204:187-194. [DOI] [PubMed] [Google Scholar]
- 5. Brown JM, Nemeth K, Kushnir-Sukhov NM, Metcalfe DD, Mezey E. Bone marrow stromal cells inhibit mast cell function via a COX2-dependent mechanism. Clin Exp Allergy. 2011; 41:526-534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Li J, Zhou J, Zhang D, Song Y, She J, Bai C. Bone marrow-derived mesenchymal stem cells enhance autophagy via PI3K/AKT signalling to reduce the severity of ischaemia/reperfusion-induced lung injury. J Cell Mol Med. 2015; 19:2341-2351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Volkert S, Kohlmann A, Schnittger S, Kern W, Haferlach T, Haferlach C. Association of the type of 5q loss with complex karyotype, clonal evolution, TP53 mutation status, and prognosis in acute myeloid leukemia and myelodysplastic syndrome. Genes Chromosomes Cancer. 2014; 53:402-410. [DOI] [PubMed] [Google Scholar]
- 8. Yamamoto N, Akamatsu H, Hasegawa S, Yamada T, Nakata S, Ohkuma M, Miyachi E, Marunouchi T, Matsunaga K. Isolation of multipotent stem cells from mouse adipose tissue. J Dermatol Sci. 2007; 48:43-52. [DOI] [PubMed] [Google Scholar]
- 9. Guilak F, Estes BT, Diekman BO, Moutos FT, Gimble JM. 2010 Nicolas Andry Award: Multipotent adult stem cells from adipose tissue for musculoskeletal tissue engineering. Clin Orthop Relat Res. 2010; 468:2530-2540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Chandra A, Lan S, Zhu J, Siclari VA, Qin L. Epidermal growth factor receptor (EGFR) signaling promotes proliferation and survival in osteoprogenitors by increasing early growth response 2 (EGR2) expression. J Biol Chem. 2013; 288:20488-20498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Squires A, Atas E, Meller A. Nanopore sensing of individual transcription factors bound to DNA. Sci Rep. 2015; 5:11643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Chen F, Zhang C, Jia X, Wang S, Wang J, Chen Y, Zhao J, Tian S, Han X, Han L. Transcriptome profiles of human lung epithelial cells A549 interacting with aspergillus fumigatus by RNA-Seq. PLoS One. 2015; 10:e0135720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Charolidi N, Pirianov G, Torsney E, Pearce S, Laing K, Nohturfft A, Cockerill GW. Pioglitazone identifies a new target for aneurysm treatment: Role of Egr1 in an experimental murine model of aortic aneurysm. J Vasc Res. 2015; 52:81-93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Shingyochi Y, Orbay H, Mizuno H. Adipose-derived stem cells for wound repair and regeneration. Expert Opin Biol Ther. 2015; 15:1285-1292. [DOI] [PubMed] [Google Scholar]
- 15. Saeed H, Abdallah BM, Ditzel N, Catala-Lehnen P, Qiu W, Amling M, Kassem M. Telomerase-deficient mice exhibit bone loss owing to defects in osteoblasts and increased osteoclastogenesis by inflammatory microenvironment. J Bone Miner Res. 2011; 26:1494-1505. [DOI] [PubMed] [Google Scholar]
- 16. Jiang C, Hu X, Wang L, Cheng H, Lin Y, Pang Y, Yuan W, Cheng T, Wang J. Excessive proliferation and impaired function of primitive hematopoietic cells in bone marrow due to senescence post chemotherapy in a T cell acute lymphoblastic leukemia model. J Transl Med. 2015; 13:234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hu N, Qiu Y, Dong F. Role of Erk1/2 signaling in the regulation of neutrophil versus monocyte development in response to G-CSF and M-CSF. J Biol Chem. 2015; 290:24561-24573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lee KM, Coehlo M, McGregor HA, Waltermire RS, Szumlinski KK. Binge alcohol drinking elicits persistent negative affect in mice. Behav Brain Res. 2015; 291:385-398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Zhao DY, Jacobs KM, Hallahan DE, Thotala D. Silencing Egr1 attenuates radiation-induced apoptosis in normal tissues while killing cancer cells and delaying tumor growth. Mol Cancer Ther. 2015; 14:2343-2352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Shen X, Tang J, Hu J, Guo L, Xing Y, Xi T. MiR-424 regulates monocytic differentiation of human leukemia U937 cells by directly targeting CDX2. Biotechnol Lett. 2013; 35:1799-1806. [DOI] [PubMed] [Google Scholar]
- 21. Tanaka S, Miyagi S, Sashida G, Chiba T, Yuan J, Mochizuki-Kashio M, Suzuki Y, Sugano S, Nakaseko C, Yokote K, Koseki H, Iwama A. Ezh2 augments leukemogenicity by reinforcing differentiation blockage in acute myeloid leukemia. Blood. 2012; 120:1107-1117. [DOI] [PubMed] [Google Scholar]
- 22. Verduci L, Azzalin G, Gioiosa S, Carissimi C, Laudadio I, Fulci V, Macino G. microRNA-181a enhances cell proliferation in acute lymphoblastic leukemia by targeting EGR1. Leuk Res. 2015; 39:479-485. [DOI] [PubMed] [Google Scholar]
- 23. Cahill EF, Tobin LM, Carty F, Mahon BP, English K. Jagged-1 is required for the expansion of CD4+ CD25+ FoxP3+ regulatory T cells and tolerogenic dendritic cells by murine mesenchymal stromal cells. Stem Cell Res Ther. 2015; 6:19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Signorelli P, Ghidoni R. Resveratrol as an anticancer nutrient: Molecular basis, open questions and promises. J Nutr Biochem. 2005; 16:449-466. [DOI] [PubMed] [Google Scholar]
- 25. Lee SM, Lee SB, Prywes R, Vulpe CD. Iron deficiency upregulates Egr1 expression. Genes Nutr. 2015; 10:468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Fang J, Menon M, Kapelle W, Bogacheva O, Bogachev O, Houde E, Browne S, Sathyanarayana P, Wojchowski DM. EPO modulation of cell-cycle regulatory genes, and cell division, in primary bone marrow erythroblasts. Blood. 2007; 110:2361-2370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Thiel A, Beier M, Ingenhag D, Servan K, Hein M, Moeller V, Betz B, Hildebrandt B, Evers C, Germing U, Royer-Pokora B. Comprehensive array CGH of normal karyotype myelodysplastic syndromes reveals hidden recurrent and individual genomic copy number alterations with prognostic relevance. Leukemia. 2011; 25:387-399. [DOI] [PubMed] [Google Scholar]
- 28. Fricke S. Measurement and illustration of immune interaction after stem cell transplantation. Methods Mol Biol. 2011; 690:315-332. [DOI] [PubMed] [Google Scholar]
- 29. Baron V, Adamson ED, Calogero A, Ragona G, Mercola D. The transcription factor Egr1 is a direct regulator of multiple tumor suppressors including TGFbeta1, PTEN, p53, and fibronectin. Cancer Gene Ther. 2006; 13:115-124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Liu L, Zhang SX, Aeran R, Liao W, Lu M, Polovin G, Pone EJ, Zhao W. Exogenous marker-engineered mesenchymal stem cells detect cancer and metastases in a simple blood assay. Stem Cell Res Ther. 2015; 6:181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Joseph MM, Aravind SR, George SK, Raveendran Pillai K, Mini S, Sreelekha TT. Anticancer activity of galactoxyloglucan polysaccharide-conjugated doxorubicin nanoparticles: Mechanistic insights and interactome analysis. Eur J Pharm Biopharm. 2015; 93:183-195. [DOI] [PubMed] [Google Scholar]
- 32. Song Y, Cheng X, Yang X, Zhao R, Wang P, Han Y, Luo Z, Cao Y, Zhu C, Xiong Y, Liu Y, Wu K, Wu J. Early growth response-1 facilitates enterovirus 71 replication by direct binding to the viral genome RNA. Int J Biochem Cell Biol. 2015; 62:36-46. [DOI] [PubMed] [Google Scholar]
- 33. Liang XH, Deng WB, Li M, Zhao ZA, Wang TS, Feng XH, Cao YJ, Duan EK, Yang ZM. Egr1 protein acts downstream of estrogen-leukemia inhibitory factor (LIF)-STAT3 pathway and plays a role during implantation through targeting Wnt4. J Biol Chem. 2014; 289:23534-23545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Turroni S, Tolomeo M, Mamone G, Picariello G, Giacomini E, Brigidi P, Roberti M, Grimaudo S, Pipitone RM, Di Cristina A, Recanatini M. A natural-like synthetic small molecule impairs bcr-abl signaling cascades and induces megakaryocyte differentiation in erythroleukemia cells. PLoS one. 2013; 8:e57650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Jaluria P, Chu C, Betenbaugh M, Shiloach J. Cells by design: A mini-review of targeting cell engineering using DNA microarrays. Mol Biotechnol. 2008; 39:105-111. [DOI] [PubMed] [Google Scholar]
- 36. Liu H, Zhu S, Zhang C, Lu P, Hu J, Yin Z, Ma Y, Chen X, OuYang H. Crucial transcription factors in tendon development and differentiation: Their potential for tendon regeneration. Cell Tissue Res. 2014; 356:287-298. [DOI] [PubMed] [Google Scholar]
- 37. Ngiam N, Post M, Kavanagh BP. Early growth response factor-1 in acute lung injury. Am J Physiol Lung Cell Mol Physiol. 2007; 293:L1089-L1091. [DOI] [PubMed] [Google Scholar]
- 38. Pritchard MT, Nagy LE. Ethanol-induced liver injury: Potential roles for egr-1. Alcohol Clin Exp Res. 2005; 29:146S-150S. [DOI] [PubMed] [Google Scholar]
- 39. Kushibiki T, Hirasawa T, Okawa S, Ishihara M. Low reactive level laser therapy for mesenchymal stromal cells therapies. Stem Cells Int. 2015; 2015:974864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Tarnawski AS, Jones MK. Inhibition of angiogenesis by NSAIDs: Molecular mechanisms and clinical implications. J Mol Med (Berl). 2003; 81:627-636. [DOI] [PubMed] [Google Scholar]
- 41. Ghosh AK, Quaggin SE, Vaughan DE. Molecular basis of organ fibrosis: Potential therapeutic approaches. Exp Biol Med (Maywood). 2013; 238:461-481. [DOI] [PubMed] [Google Scholar]
- 42. Orbay H, Takami Y, Hyakusoku H, Mizuno H. Acellular dermal matrix seeded with adipose-derived stem cells as a subcutaneous implant. Aesthetic Plast Surg. 2011; 35:756-763. [DOI] [PubMed] [Google Scholar]
- 43. Yi JH, Park SW, Kapadia R, Vemuganti R. Role of transcription factors in mediating post-ischemic cerebral inflammation and brain damage. Neurochem Int. 2007; 50:1014-1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Oh YK, Jang E, Paik DJ, Youn J. Early growth response-1 plays a non-redundant role in the differentiation of B cells into plasma cells. Immune Netw. 2015; 15:161-166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Yamamoto S, Yamane M, Yoshida O, Waki N, Okazaki M, Matsukawa A, Oto T, Miyoshi S. Early growth response-1 plays an important role in ischemia-reperfusion injury in lung transplants by regulating polymorphonuclear neutrophil infiltration. Transplantation. 2015; 99:2285-2293. [DOI] [PubMed] [Google Scholar]
- 46. Tain YL, Huang LT, Chan JY, Lee CT. Transcriptome analysis in rat kidneys: Importance of genes involved in programmed hypertension. Int J Mol Sci. 2015; 16:4744-4758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Bhattacharyya S, Ishida W, Wu M, Wilkes M, Mori Y, Hinchcliff M, Leof E, Varga J. A non-Smad mechanism of fibroblast activation by transforming growth factor-beta via c-Abl and Egr-1: Selective modulation by imatinib mesylate. Oncogene. 2009; 28:1285-1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Liao BY, Wang Z, Hu J, Liu WF, Shen ZZ, Zhang X, Yu L, Fan J, Zhou J. PI-88 inhibits postoperative recurrence of hepatocellular carcinoma via disrupting the surge of heparanase after liver resection. Tumour Biol. 2015. 10.1007/s13277-015-4085-8 [DOI] [PubMed] [Google Scholar]
- 49. Petzuch B, Groll N, Schwarz M, Braeuning A. Application of HC-AFW1 hepatocarcinoma cells for mechanistic studies: Regulation of cytochrome P450 2B6 expression by dimethyl sulfoxide and early growth response 1. Drug Metab Dispos. 2015; 43:1727-1733. [DOI] [PubMed] [Google Scholar]
- 50. Liu Q, Du GQ, Zhu ZT, Zhang C, Sun XW, Liu JJ, Li X, Wang YS, Du WJ. Identification of apoptosis-related microRNAs and their target genes in myocardial infarction post-transplantation with skeletal myoblasts. J Transl Med. 2015; 13:270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Leonardi D, Oberdoerfer D, Fernandes MC, Meurer RT, Pereira-Filho GA, Cruz P, Vargas M, Chem RC, Camassola M, Nardi NB. Mesenchymal stem cells combined with an artificial dermal substitute improve repair in full-thickness skin wounds. Burns. 2012; 38:1143-1150. [DOI] [PubMed] [Google Scholar]
- 52. Le Billan F, Khan JA, Lamribet K, Viengchareun S, Bouligand J, Fagart J, Lombes M. Cistrome of the aldosterone-activated mineralocorticoid receptor in human renal cells. FASEB J. 2015; 29:3977-3989. [DOI] [PubMed] [Google Scholar]
- 53. Chen ZH, Kim HP, Sciurba FC, et al. Egr-1 regulates autophagy in cigarette smoke-induced chronic obstructive pulmonary disease. PloS one. 2008; 3:e3316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Singh M, Shin YK, Yang X, Zehr B, Chakrabarti P, Kandror KV. 4E-BPs control fat storage by regulating the expression of Egr1 and ATGL. J Biol Chem. 2015; 290:17331-17338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Cheong MW, Kuo LH, Cheng YN, Tsai PJ, Ho LC, Tai HC, Chiu WT, Chen SH, Lu PJ, Shan YS, Chuang LM, Tsai YS. Loss of Egr-1 sensitizes pancreatic beta-cells to palmitate-induced ER stress and apoptosis. J Mol Med (Berl). 2015; 93:807-818. [DOI] [PubMed] [Google Scholar]
- 56. Rondinone CM. Minireview: Ribonucleic acid interference for the identification of new targets for the treatment of metabolic diseases. Endocrinology. 2006; 147:2650-2656. [DOI] [PubMed] [Google Scholar]
- 57. Cattane N, Minelli A, Milanesi E, Maj C, Bignotti S, Bortolomasi M, Bocchio Chiavetto L, Gennarelli M. Altered gene expression in schizophrenia: Findings from transcriptional signatures in fibroblasts and blood. PLoS one. 2015; 10:e0116686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Bozon B, Kelly A, Josselyn SA, Silva AJ, Davis S, Laroche S. MAPK, CREB and zif268 are all required for the consolidation of recognition memory. Philos Trans R Soc Lond B Biol Sci. 2003; 358:805-814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Yang L, Jiang Y, Wen Z, Xu X, Xu X, Zhu J, Xie X, Xu L, Xie Y, Liu X, Xu G. Over-expressed EGR1 may exaggerate ischemic injury after experimental stroke by decreasing BDNF expression. Neuroscience. 2015; 290:509-517. [DOI] [PubMed] [Google Scholar]
- 60. Yoon TM, Kim SA, Lee DH, Lee JK, Park YL, Lee KH, Chung IJ, Joo YE, Lim SC. EGR1 regulates radiation-induced apoptosis in head and neck squamous cell carcinoma. Oncol Rep. 2015; 33:1717-1722. [DOI] [PubMed] [Google Scholar]
- 61. Sarma SN, Kim YJ, Ryu JC. Differential gene expression profiles of human leukemia cell lines exposed to benzene and its metabolites. Environ Toxicol Pharmacol. 2011; 32:285-295. [DOI] [PubMed] [Google Scholar]
- 62. Abdulkadir SA. Mechanisms of prostate tumorigenesis: Roles for transcription factors Nkx3.1 and Egr1. Ann N Y Acad Sci. 2005; 1059:33-40. [DOI] [PubMed] [Google Scholar]
- 63. Genet F, Kulina I, Vaquette C, et al. Neurological heterotopic ossification following spinal cord injury is triggered by macrophage-mediated inflammation in muscle. J Pathol. 2015; 236:229-240. [DOI] [PubMed] [Google Scholar]
- 64. Lu J, Li XP, Dong Q, Kung HF, He ML. TBX2 and TBX3: The special value for anticancer drug targets. Biochim Biophys Acta. 2010; 1806:268-274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Wilson A, Laurenti E, Trumpp A. Balancing dormant and self-renewing hematopoietic stem cells. Curr Opin Genet Dev. 2009; 19:461-468. [DOI] [PubMed] [Google Scholar]
- 66. Galvan AB, Mallo M, Arenillas L, Salido M, Espinet B, Pedro C, Florensa L, Serrano S, Sole F. Does monosomy 5 really exist in myelodysplastic syndromes and acute myeloid leukemia? Leuk Res. 2010; 34:1242-1245. [DOI] [PubMed] [Google Scholar]
- 67. Zou C, Song G, Luo Q, Yuan L, Yang L. Mesenchymal stem cells require integrin beta1 for directed migration induced by osteopontin in vitro. In Vitro Cell Dev Biol Anim. 2011; 47:241-250. [DOI] [PubMed] [Google Scholar]
- 68. Staber PB, Linkesch W, Zauner D, Beham-Schmid C, Guelly C, Schauer S, Sill H, Hoefler G. Common alterations in gene expression and increased proliferation in recurrent acute myeloid leukemia. Oncogene. 2004; 23:894-904. [DOI] [PubMed] [Google Scholar]
- 69. Qian Z, Joslin JM, Tennant TR, Reshmi SC, Young DJ, Stoddart A, Larson RA, Le Beau MM. Cytogenetic and genetic pathways in therapy-related acute myeloid leukemia. Chem Biol Interact. 2010; 184:50-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Lee HR, Oh B, Hong DS, et al. Cytogenetic features of 5q deletion and 5q-syndrome in myelodysplastic syndrome in Korea; marker chromosomes proved to be chromosome 5 with interstitial deletion by fluorescence in situ hybridization. Cancer Genet Cytogenet. 2010; 203:193-202. [DOI] [PubMed] [Google Scholar]
- 71. Kwon WK, Lee JY, Mun YC, Seong CM, Chung WS, Huh J. Clinical utility of FISH analysis in addition to G-banded karyotype in hematologic malignancies and proposal of a practical approach. Korean J Hematol. 2010; 45:171-176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Stoddart A, Fernald AA, Wang J, Davis EM, Karrison T, Anastasi J, Le Beau MM. Haploinsufficiency of del(5q) genes, Egr1 and Apc, cooperate with Tp53 loss to induce acute myeloid leukemia in mice. Blood. 2014; 123:1069-1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Kavanagh H, Mahon BP. Allogeneic mesenchymal stem cells prevent allergic airway inflammation by inducing murine regulatory T cells. Allergy. 2011; 66:523-531. [DOI] [PubMed] [Google Scholar]
- 74. Ebert BL. Molecular dissection of the 5q deletion in myelodysplastic syndrome. Semin Oncol. 2011; 38:621-626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Wang H, Wen J, Chang CC, Zhou X. Discovering transcription and splicing networks in myelodysplastic syndromes. PLoS one. 2013; 8:e79118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Tang G, Goswami RS, Liang CS, Bueso-Ramos CE, Hu S, DiNardo C, Medeiros LJ. Isolated del(5q) in Patients Following Therapies for Various Malignancies May Not All Be Clinically Significant. Am J Clin Pathol. 2015; 144:78-86. [DOI] [PubMed] [Google Scholar]
- 77. Schoch C, Haferlach T, Bursch S, Gerstner D, Schnittger S, Dugas M, Kern W, Loffler H, Hiddemann W. Loss of genetic material is more common than gain in acute myeloid leukemia with complex aberrant karyotype: A detailed analysis of 125 cases using conventional chromosome analysis and fluorescence in situ hybridization including 24-color FISH. Genes Chromosomes Cancer. 2002; 35:20-29. [DOI] [PubMed] [Google Scholar]
- 78. Fainardi E, Castellazzi M, Stignani M, Morandi F, Sana G, Gonzalez R, Pistoia V, Baricordi OR, Sokal E, Pena J. Emerging topics and new perspectives on HLA-G. Cell Mol Life Sci. 2011; 68:433-451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Wang Y, Xue Y, Chen S, Wu Y, Pan J, Zhang J, Shen J. A novel t(5; 11)(q31; p15) involving the NUP98 gene on 11p15 is associated with a loss of the EGR1 gene on 5q31 in a patient with acute myeloid leukemia. Cancer Genet Cytogenet. 2010; 199:9-14. [DOI] [PubMed] [Google Scholar]
- 80. Joslin JM, Fernald AA, Tennant TR, Davis EM, Kogan SC, Anastasi J, Crispino JD, Le Beau MM. Haploinsufficiency of EGR1, a candidate gene in the del(5q), leads to the development of myeloid disorders. Blood. 2007; 110:719-726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Scheicher R, Hoelbl-Kovacic A, Bellutti F, Tigan AS, Prchal-Murphy M, Heller G, Schneckenleithner C, Salazar-Roa M, Zochbauer-Muller S, Zuber J, Malumbres M, Kollmann K, Sexl V. CDK6 as a key regulator of hematopoietic and leukemic stem cell activation. Blood. 2015; 125:90-101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Jones JE, Wang L, Kropf PL, Duan R, Johnson DE. Src family kinase gene targets during myeloid differentiation: Identification of the EGR-1 gene as a direct target. Leukemia. 2009; 23:1933-1935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Kumbrink J, Kirsch KH. p130Cas acts as survival factor during PMA-induced apoptosis in HL-60 promyelocytic leukemia cells. Int J Biochem Cell Biol. 2013; 45:531-535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Heidel FH, Bullinger L, Arreba-Tutusaus P, Wang Z, Gaebel J, Hirt C, Niederwieser D, Lane SW, Dohner K, Vasioukhin V, Fischer T, Armstrong SA. The cell fate determinant Llgl1 influences HSC fitness and prognosis in AML. J Exp Med. 2013; 210:15-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Schaefer A, Kosa F, Bittorf T, Magocsi M, Rosche A, Ramirez-Chavez Y, Marotzki S, Marquardt H. Opposite effects of inhibitors of mitogen-activated protein kinase pathways on the egr-1 and beta-globin expression in erythropoietin-responsive murine erythroleukemia cells. Cell Signal. 2004; 16:223-234. [DOI] [PubMed] [Google Scholar]
- 86. Uccelli A, de Rosbo NK. The immunomodulatory function of mesenchymal stem cells: Mode of action and pathways. Ann N Y Acad Sci. 2015; 1351:114-126. [DOI] [PubMed] [Google Scholar]
- 87. Gibbs JD, Liebermann DA, Hoffman B. Egr-1 abrogates the E2F-1 block in terminal myeloid differentiation and suppresses leukemia. Oncogene. 2008; 27:98-106. [DOI] [PubMed] [Google Scholar]
- 88. Ko H, Kim JM, Kim SJ, Shim SH, Ha CH, Chang HI. Induction of apoptosis by genipin inhibits cell proliferation in AGS human gastric cancer cells via Egr1/p21 signaling pathway. Bioorg Med Chem Lett. 2015; 25:4191-4196. [DOI] [PubMed] [Google Scholar]
- 89. Zhang QS, Deater M, Schubert K, Marquez-Loza L, Pelz C, Sinclair DA, Grompe M. The Sirt1 activator SRT3025 expands hematopoietic stem and progenitor cells and improves hematopoiesis in Fanconi anemia mice. Stem Cell Res. 2015; 15:130-140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Sobolewski C, Sanduja S, Blanco FF, Hu L, Dixon DA. Histone deacetylase inhibitors activate tristetraprolin expression through induction of early growth response protein 1 (EGR1) in colorectal cancer cells. Biomolecules. 2015; 5:2035-2055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Feng Y, Desjardins CA, Cooper O, Kontor A, Nocco SE, Naya FJ. EGR1 functions as a potent repressor of MEF2 transcriptional activity. PLoS one. 2015; 10:e0127641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Shafarenko M, Liebermann DA, Hoffman B. Egr-1 abrogates the block imparted by c-Myc on terminal M1 myeloid differentiation. Blood. 2005; 106:871-878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Shin DY, Kim GY, Li W, Choi BT, Kim ND, Kang HS, Choi YH. Implication of intracellular ROS formation, caspase-3 activation and Egr-1 induction in platycodon D-induced apoptosis of U937 human leukemia cells. Biomed Pharmacother. 2009; 63:86-94. [DOI] [PubMed] [Google Scholar]
- 94. Chen F, Wang Q, Wang X, Studzinski GP. Up-regulation of Egr1 by 1,25-dihydroxyvitamin D3 contributes to increased expression of p35 activator of cyclin-dependent kinase 5 and consequent onset of the terminal phase of HL60 cell differentiation. Cancer Res. 2004; 64:5425-5433. [DOI] [PubMed] [Google Scholar]
- 95. Manieri NA, Stappenbeck TS. Mesenchymal stem cell therapy of intestinal disease: are their effects systemic or localized? Curr Opin Gastroenterol. 2011; 27:119-124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Heo SC, Jeon ES, Lee IH, Kim HS, Kim MB, Kim JH. Tumor necrosis factor-α-activated human adipose tissue-derived mesenchymal stem cells accelerate cutaneous wound healing through paracrine mechanisms. J Invest Dermatol. 2011; 131:1559-1567. [DOI] [PubMed] [Google Scholar]
- 97. Gibbs JD, Liebermann DA, Hoffman B. Leukemia suppressor function of Egr-1 is dependent on transforming oncogene. Leukemia. 2008; 22:1909-1916. [DOI] [PubMed] [Google Scholar]
- 98. Liu P, Li J, Lu H, Xu B. Thalidomide inhibits leukemia cell invasion and migration by upregulation of early growth response gene 1. Leuk Lymphoma. 2009; 50:109-113. [DOI] [PubMed] [Google Scholar]
- 99. Salunga TL, Tabuchi Y, Takasaki I, Feril LB, Jr., Zhao QL, Ohtsuka K, Tsuneyama K, Kondo T. Identification of genes responsive to paeoniflorin, a heat shock protein-inducing compound, in human leukemia U937 cells. Int J Hyperthermia. 2007; 23:529-537. [DOI] [PubMed] [Google Scholar]
- 100. Liu P, Xu B, Li J, Lu H. LY294002 inhibits leukemia cell invasion and migration through early growth response gene 1 induction independent of phosphatidylinositol 3-kinase-Akt pathway. Biochem Biophys Res Commun. 2008; 377:187-190. [DOI] [PubMed] [Google Scholar]


