ABSTRACT
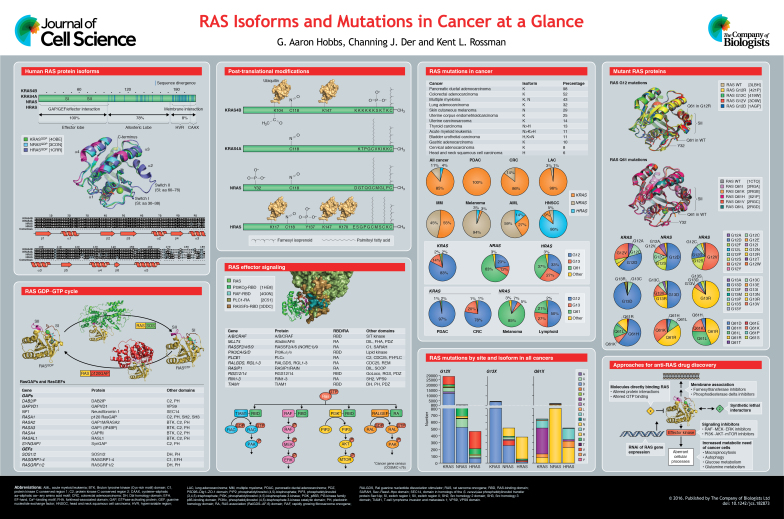
RAS proteins (KRAS4A, KRAS4B, NRAS and HRAS) function as GDP–GTP-regulated binary on-off switches, which regulate cytoplasmic signaling networks that control diverse normal cellular processes. Gain-of-function missense mutations in RAS genes are found in ∼25% of human cancers, prompting interest in identifying anti-RAS therapeutic strategies for cancer treatment. However, despite more than three decades of intense effort, no anti-RAS therapies have reached clinical application. Contributing to this failure has been an underestimation of the complexities of RAS. First, there is now appreciation that the four human RAS proteins are not functionally identical. Second, with >130 different missense mutations found in cancer, there is an emerging view that there are mutation-specific consequences on RAS structure, biochemistry and biology, and mutation-selective therapeutic strategies are needed. In this Cell Science at a Glance article and accompanying poster, we provide a snapshot of the differences between RAS isoforms and mutations, as well as the current status of anti-RAS drug-discovery efforts.
KEY WORDS: GTPase, Oncogene, PI3K, Rac, Raf, Ral
Summary: In this Cell Science at a Glance article, we discuss the distinct roles of and properties of RAS isoforms and mutations in cancer – matters that are current foci of ongoing research studies.
Introduction
Mutations in the RAS gene were first reported in cancer over 30 years ago, and numerous studies have since validated mutant RAS as a driver of tumor initiation and maintenance (Cox and Der, 2010). The three human RAS genes [i.e. Kirsten rat sarcoma viral oncogene homolog (KRAS), neuroblastoma RAS viral (v-ras) oncogene homolog (NRAS) and Harvey rat sarcoma viral oncogene homolog (HRAS)] encode four RAS proteins, with two KRAS isoforms that arise from alternative RNA splicing (KRAS4A and KRAS4B). Although KRAS4B is the predominant splice variant and expressed in many tissues – contributing to its focus in cancer studies – there is significant KRAS4A expression in some tissues (Tsai et al., 2015). RAS GTPases cycle between the GDP-bound inactive and GTP-bound active states with the help of guanine nucleotide exchange factors (RASGEFs) that promote activation, and GTPase-activating proteins (RASGAPs) that inactivate RAS by catalyzing GTP hydrolysis. Once activated, RAS-GTP binds to and activates a spectrum of downstream effectors with distinct catalytic functions (see Box 1).
Box 1. RAS effector signaling.
Activated GTP-bound RAS binds preferentially to its downstream effectors. RAS-GTP preferentially binds to RAS-binding-domain (RBD) or RAS-association (RA)-domain-containing effectors (see poster). Although the RBD and RA domains do not share primary sequence similarity, they are structurally related and share the topology of the ubiquitin superfold (Kiel et al., 2005; Wohlgemuth et al., 2005). There are at least 11 distinct RAS effector families, each of which activates a distinct protein signaling cascade (Vigil et al., 2010). There are substantial cell culture and mouse model analyses that support the driving role of four families (i.e. RAF, PI3K, RalGEF and TIAM1) in RAS-driven oncogenesis (Bryant et al., 2014). Support for effector–driver function is also indicated by the inclusion of components of each effector signaling pathway in the Cancer Gene Census, i.e. those genes for which mutations have been causally implicated in cancers (COSMIC v75). In particular, mutations of BRAF (19%) and PI3KCA (10%) are commonly found in many cancers.
Box 2. Approaches for targeting RAS.
The past and current efforts to develop antagonists of mutant RAS function include direct and indirect approaches. Once considered to be infeasible, recent studies have identified small molecules that directly bind to RAS and disrupt crucial functions of RAS, including (i) GDP–GTP regulation and interaction with its effectors. The development of G12C-selective inhibitors that target the thiol in order to inhibit GTP binding – locking RAS in an inactive state so it cannot interact with its effectors – have been recently described (Hunter et al., 2014; Ostrem et al., 2013). Other molecules that block RAS interaction with the SOS1 RASGEF (Maurer et al., 2012; Sun et al., 2012) or effectors (Shima et al., 2013) have also been identified. Indirect approaches include (ii) inhibition of enzymes that target the RAS CAAX motif in order to prevent membrane association of RAS (e.g. farnesyltransferase) (Cox et al., 2015) or of proteins that facilitate RAS trafficking to the plasma membrane (phosphodiesterase delta) (Zimmermann et al., 2013), (iii) inhibitors of downstream effector signaling (e.g. RAF or PI3K effectors) (Fruman and Rommel, 2014; Samatar and Poulikakos, 2014), (iv) inhibitors of processes that support the increased metabolic needs of cancer cells (e.g. macropinocytosis, autophagy, glucose and glutamine metabolism), (v) unbiased genetic or chemical screens for synthetic lethal interactors (Barbie et al., 2009; Kumar et al., 2012; Luo et al., 2009; Sarthy et al., 2007; Scholl et al., 2009) and (vi) RNA interference (RNAi) of KRAS expression (Pecot et al., 2014; Xue et al., 2014; Yuan et al., 2014).
Missense gain-of-function mutations in all three RAS genes are found in 27% of all human cancers, with 98% of the mutations at one of three mutational hotspots: G12, G13 and Q61 (COSMIC v75). Conventionally, mutant RAS is considered to be defective in GAP-mediated GTP hydrolysis, which results in an accumulation of constitutively GTP-bound RAS in cells. Additionally, the involvement of RAS in cancer is greater than that indicated by its mutation frequency; perturbations in GDP–GTP regulation, loss of GAPs [e.g. neurofibromin 1 (NF1)] or persistent receptor tyrosine kinase-mediated activation of GEFs [e.g. son of sevenless 1 (SOS1)] are additional mechanisms of RAS activation in cancer. Finally, recent studies support a role for the remaining wild-type (WT) RAS proteins present in RAS-mutant cancers in contributing to cancer growth (Grabocka et al., 2014; Lim et al., 2008; Young et al., 2013), although other studies suggest that WT RAS can act as a tumor suppressor (Bremner and Balmain, 1990; Qiu et al., 2011; To et al., 2013; Weyandt et al., 2015; Zhang et al., 2001).
Historically, the majority of biochemical and structural studies of RAS have focused on HRAS (Vetter, 2014). However, HRAS is the least frequently mutated RAS isoform in human cancers (4%), whereas KRAS is the predominantly mutated isoform (85%), followed by NRAS (11%). Furthermore, the G12V mutation has been traditionally characterized as the ‘poster child’ for oncogenic RAS when defining the biological properties of mutant RAS in cancer. However, there is increasing evidence that mutations at each of the three missense-mutation hotspots (G12, G13 and Q61) have distinct structural and biochemical defects (Buhrman et al., 2007; Burd et al., 2014; Hunter et al., 2015; Smith et al., 2013). Evidence that different amino acid substitutions at any one hotspot can have differential oncogenic potencies as well as distinct functional consequences (Ihle et al., 2012) adds an additional layer of complexity, suggesting that ‘not all RAS mutants are created equal’. Further, there are striking cancer-type-specific and isoform-distinct differences in the observed frequencies of specific RAS missense mutations at the three hotspots (Cox et al., 2014; Prior et al., 2012). With increasing experimental evidence supporting RAS isoform and mutation differences, as well as cell-specific and genetic-context-specific differences, there is growing speculation that there will not be one simple anti-RAS therapeutic approach for all RAS mutant cancers. Instead, cancer-type-specific therapeutic strategies must be determined for different subsets of RAS mutations. Here, we summarize our current understanding of RAS-isoform- and RAS-mutation-specific functional differences.
RAS isoform differences and post-translational modifications
The three RAS genes encode four RAS protein isoforms that are highly similar in primary sequence (82-90% amino acid (aa) sequence identity), structure and biochemical properties (GTP binding, hydrolysis and prenylation) (see poster). The N-terminal 164 residues comprise the G domain, which is involved in GTP binding and hydrolysis. Within the G domain are switch I (SI) and switch II (SII), regions that change in conformation during GDP–GTP cycling and are the main determinants in effector binding. In contrast, the C-terminal hypervariable region (HVR) shares little sequence similarity.
Although the sequence divergence of RAS proteins is typically thought of as residing solely within the HVR, there is a second region of sequence divergence. RAS isoforms share 100% sequence identity in the N-terminus of the G domain termed the effector lobe (aa 1–86), but there is only 82% sequence similarity within residues 87–166, termed the allosteric lobe (Buhrman et al., 2011). Whether these sequence differences contribute to the functional differences of RAS isoforms is still largely understudied. However, the allosteric lobe has been suggested to play a role in SII conformation and membrane orientation (Parker and Mattos, 2015), and RAS membrane orientation has been shown to regulate effector utilization (Abankwa et al., 2008, 2010).
There are striking cancer-type-specific mutational profiles of RAS gene isoforms in cancer, suggesting tissue-distinct roles for RAS in driving oncogenesis. For example, there is near-exclusive mutation of KRAS in pancreatic ductal, lung and colorectal carcinoma, whereas NRAS is the predominant isoform mutated in cutaneous melanoma (Cox et al., 2014). In contrast, HRAS mutations predominate in head and neck squamous cell carcinoma. Recent mouse model studies have begun to address the issue of why specific RAS gene isoforms are preferentially mutated in specific cancers. In one study, the Kras G12D mutation but not the Nras G12D mutation promoted colon cancer development in Apc-deficient mice, supporting the ability of KRAS but not NRAS to initiate the formation of colon cancer (Haigis et al., 2008). In contrast, another mouse carcinogenesis model showed that the preferential basis for Kras mutation in lung cancer was not due to distinct functional differences between RAS isoforms but, rather, the distinct regulation of expression of the RAS gene isoform (Westcott et al., 2015). Thus, whether a specific RAS gene is required for cancer development arising from different tissues remains unresolved.
RAS isoform differences have been identified at the level of protein translation and provide one possible explanation for why KRAS is the predominantly mutated isoform in cancer. Unlike HRAS, the KRAS DNA coding sequence has a high frequency of rare codons, resulting in poor KRAS protein translation and expression (Lampson et al., 2013). As RAS mutations are the initiating genetic events in many cancers, it has been proposed that the high expression of activated HRAS, but not KRAS, induces senescence. Consequently, a cell with mutated KRAS will persist to allow subsequent genetic events in order to promote tumor progression. Supporting this possibility, it was found that mice harboring a codon-optimized KRAS coding sequence – resulting in increased KRAS protein expression – show significantly reduced tumor formation (Pershing et al., 2015).
Currently, differences in RAS isoforms have been ascribed largely to sequence differences within their C-terminal HVRs, a site at which RAS proteins are differentially lipid-modified. KRAS4A and KRAS4B have polybasic sequences that facilitate membrane-association in acidic membrane regions (Gelabert-Baldrich et al., 2014). In addition, KRAS4A and NRAS are covalently modified by a single palmitic acid, whereas HRAS can be palmitoylated at two sites within the HVR. Palmitoylation is reversible and substoichiometric. The lipidation profile of each isoform has been shown to dictate membrane localization (Cox et al., 2015; Jang et al., 2015). Recent evidence indicates that the palmitoylation state of HRAS and NRAS also dictates their distribution within the Golgi membrane, with HRAS distributed throughout and NRAS localized to the cis-Golgi (Lynch et al., 2015). At the plasma membrane, HRAS is in the GTP-bound state when in non-ordered lipid domains and is GDP-bound when in lipid rafts (Rotblat et al., 2004); yet, the opposite seems to be true for NRAS (Eisenberg et al., 2011).
KRAS4B is distinguished from other RAS isoforms in having a phosphorylation site (S181) within the HVR that acts as an electrostatic farnesyl switch, causing KRAS4B translocation from the plasma membrane to endomembrane compartments (Barcelo et al., 2014; Quatela et al., 2008) (see poster). This altered subcellular localization differentially influences effector engagement and biological activity of KRAS4B. NRAS has been shown to be phosphorylated by the Src tyrosine kinase at Y32 (Bunda et al., 2014). This modification was shown to decrease NRAS affinity to the RAS-binding domain (RBD) of RAF (see Box 1) and increase the affinity to RasGAP, thereby providing a new level of regulation not previously observed for RAS family GTPases. Additionally, HRAS can be phosphorylated at Y137 by the ABL tyrosine kinase, resulting in increased RAF interaction and decreased intrinsic GTP hydrolysis (Ting et al., 2015).
C-terminal to the HVR, all RAS proteins terminate with a CAAX tetrapeptide motif (C, cysteine; A, aliphatic aa; X, any aa) that signals for three sequential C-terminal post-translational modifications that enhance hydrophobicity and promote plasma membrane association (Ahearn et al., 2012). Whereas all RAS CAAX motifs can be modified by the farnesyltransferase-catalyzed addition of a C15 farnesyl isoprenoid lipid, in the absence of farnesyltransferase activity, KRAS4B and NRAS can be modified by geranylgeranyltransferase-I-catalyzed addition of a C20 geranylgeranyl isoprenoid; this accounts for the failure of farnesyltransferase inhibitors to effectively block the membrane association of the RAS isoforms most commonly mutated in cancer.
Additionally, RAS isoforms have been observed to be differentially ubiquitylated. HRAS has been shown to be mono- and di-ubiquitylated, and ubiquitylation internalizes HRAS from the plasma membrane, limiting HRAS-mediated RAF signaling (Jura et al., 2006). NRAS was also shown to be similarly ubiquitylated, whereas KRAS has later shown to be mono/di- ubiquitylated (Sasaki et al., 2011). Monoubiquitylation of KRAS4B at K147 increased effector binding, whereas ubiquitylation-deficient KRAS G12V showed reduced oncogenic function (Sasaki et al., 2011). However, no consequences regarding subcellular localization were observed. Ubiquitylation at K147 enhanced WT KRAS-GTP formation and a later study identified impaired GAP interaction to account for this altered property (Baker et al., 2013a). HRAS has been shown to be ubiquitylated at K117, and this modification accelerated nucleotide exchange and activation (Baker et al., 2013b). Given the differentially observed ubiquitylation patterns that have been reported between RAS isoforms, RAS ubiquitylation is likely to have distinct roles in different cell types based on the isoform and site of modification.
KRAS4B has also been shown to be acetylated at K104. Acetylation has been proposed to disrupt the conformation of SII, impairing GEF-mediated activation and, consequently, reducing effector activation and transforming potency (Yang et al., 2012). Whether other RAS isoforms become acetylated remains to be determined.
Cancer-specific hotspot frequency variations
The RAS gene isoforms are also distinguished by their striking differences in the mutation frequency at each of the three hotspots (G12, G13 and Q61) (see poster). G12 mutations comprise 83% of all KRAS mutations, followed by G13 mutations (14%), whereas Q61 mutations are rare (2%). In striking contrast, Q61 is the predominantly mutated hotspot in NRAS, followed by G12 and G13. HRAS displays an intermediate pattern, with comparable mutation frequencies of G12, G13 and Q61. Furthermore, the mutation frequency within one RAS isoform can exhibit significant differences between cancer types. NRAS Q61 mutations comprise the most frequently mutated hotspot in melanoma, whereas G12 mutations are rare. In contrast, NRAS G12 mutations are favored in acute myeloid leukemia. KRAS mutations in pancreatic ductal adenocarcinoma (PDAC) are dominated by G12 mutations, whereas G13 and Q61 mutations are rare. However, there is a relatively high frequency of G13 mutations in colorectal adenocarcinoma (CRC). These patterns suggest the intriguing possibility that different RAS mutations have different functional consequences and the properties crucial for their oncogenic functions vary depending on the tissue of origin.
There are probably qualitative and quantitative reasons for hotspot mutation preferences. Mice harboring a codon-optimized KRAS coding sequence showed a shift of chemical carcinogen urethane-induced lung tumor formation, with G12V/D-activating mutations now favored over Q61/R mutations seen in the authentic Kras gene in mice (Pershing et al., 2015). The Kras Q61 mutant tumors showed greater ERK activation, arguing for greater potency within this hotspot. Another study found that, in p16INK4a-deficient mice, the frequency of metastatic melanoma initiation for Nras Q61R was increased more than 20-fold compared with Nras G12D (Burd et al., 2014). A clear mechanistic basis for the enhanced oncogenic activities of NRAS Q61 mutants in melanoma remains to be established.
RAS mutation hotspots – structural, biological and functional differences
Whereas much of the current ‘dogma’ on the consequences of missense mutations on RAS function was established from the study of the HRAS G12V mutation, there is growing evidence and appreciation for the different functional outcomes of RAS mutations at aa positions 12, 13 and 61. In biochemical studies, it has been shown that the G12V mutation leads to a loss of GAP sensitivity, whereas the Q61L mutation leads to reduced intrinsic hydrolysis and GAP sensitivity, as well as increased intrinsic nucleotide exchange (Smith et al., 2013). The RAS G13D mutant shows decreased GAP-mediated hydrolysis and a massively increased rate of intrinsic nucleotide exchange compared to that of WT RAS (Smith et al., 2013).
Structural analysis of several RAS G12 mutant crystal structures revealed that only the G12R mutation alters the structure of RAS relative to the WT structure (see poster). The structural perturbation within SII appears to be a result of the arginine side chain displacing the glutamine residue at position 61 in the nucleotide binding site, a residue that is crucial for GTP hydrolysis. Interestingly, several RAS Q61 mutant structures show alterations in SII (see poster).
Molecular dynamics simulations have indicated a biophysical rationale for the biochemical differences between G12 and G13 mutations. Using a crystal structure model of HRAS bound to p120RASGAP to model hydrolysis, the G12V mutation resulted in the displacement of the catalytic water and of Q61, leading to a loss of GAP sensitivity. However, an increase in the dynamics of the γ phosphate, SII, catalytic water and arginine finger of p120RASGAP was observed in G13V-mutant RAS. Thus, the increased dynamics of these regions accounted for the decreased GAP sensitivity of this mutant (Khrenova et al., 2014).
HRAS WT and HRAS Q61L also showed altered dynamics when bound to RAF-RBD (Fetics et al., 2015). The RAS Q61L mutation resulted in allosteric changes in the structure of RAS in regions distal from the site of mutation. Using crystal structures of HRAS WT and HRAS Q61L bound to RAF-RBD in conjunction with computational modeling, the Q61L mutation was shown to cause increased flexibility in SII; however, when HRAS Q61L was bound to RAF, SII was significantly more rigid relative to HRAS WT. This is in contrast to the observation made by using HRAS WT, in that its binding to RAF increases the flexibility of SII. Further, the Q61L mutation increased the flexibility of the allosteric lobe (residues R97 and Y137), which increased the rigidity of loop 4 in the RAF-RBD, a region that is of key importance for interaction between RAF and MEK (Fetics et al., 2015). These results support the possibility that the RAS hotspot mutations have unique consequences on RAS structure and function.
There is intriguing evidence that mutations at different hotspots can impact the clinical outcome and treatment of cancer patients. Initial studies using anti-epidermal growth factor receptor (EGFR) therapy to treat CRC prompted the US Food and Drug Administration (FDA) to revise their recommendation to exclude patients with KRAS G12 or G13 mutations from treatment (Allegra et al., 2009). EGFR is a receptor tyrosine kinase that is positioned upstream of RAS. However, subsequent analyses has suggested that CRC patients with KRAS G13 mutations benefit from anti-EGFR therapy (Tejpar et al., 2012). This issue continues to evolve (Van Cutsem et al., 2015) because the National Comprehensive Cancer Network recommendation now indicates that CRC patients with any KRAS or NRAS mutation, including those of Q61 and A146, will not benefit from anti-EGFR therapy (Tran et al., 2015).
A limited number of studies have observed different clinical outcomes for different KRAS mutations in PDAC. One study found that KRAS G12D and G12R mutations are negative prognostic factors for overall survival (Ogura et al., 2013). However, a second study found that the presence of KRAS G12R alone correlates with increased overall survival, whereas the KRAS G12D mutation resulted in the shortest overall survival (Faris et al., 2014). Recent analyses observed that patients with KRAS Q61 mutations showed significantly improved survival (Witkiewicz et al., 2015) and, interestingly, Q61-mutant tumors showed decreased ERK activation in these samples. In summary, these observations indicate that more consideration with regard to the site of mutation in the context of the cancer type of the patient is necessary when determining the prognostic value of the mutation.
Mutation-dependent signaling
Although six possible single-base-change missense mutations can occur at the codons for G12, G13 and Q61, their frequencies are not uniform (see poster). At the codon for G12, G12D is the predominant KRAS (41%) and NRAS (52%) mutation, whereas G12V predominates in HRAS (57%). At the codon for G13, G13D is the most frequent substitution for KRAS (89%) and NRAS (50%); yet it is rare in HRAS (3%), where G13R (85%) is the predominant mutation. Additionally, at the codon for Q61, Q61H is the predominant KRAS mutation (58%), yet it is rare in both NRAS (6%) and HRAS (5%), in which Q61R is the main substitution (47% and 43%, respectably). Finally, there are cancer-type differences regarding the substitutions seen at a given RAS residue. Considering, for example, the KRAS codon for G12, the predominant substitution is G12D in PDAC, followed by G12V. In contrast, in lung adenocarcinoma (LAC), the main substitution is G12C, which is rare in PDAC (3%) (Cox et al., 2014). Although tissue-specific exposure to certain carcinogens is likely to contribute to these distinct frequencies, it is possible that different substitutions at any one position do not have equivalent biological outcomes. These isoform differences also suggest the intriguing possibility that the same mutation does not have equivalent consequences in the different RAS isoforms.
The concept that different aa substitutions have distinct functional consequences at a specific mutation hotspot was first revealed in mutagenesis studies. When the consequences of all 19 possible aa mutations at the codon for G12 were studied in HRAS, a wide range of oncogenic potential was observed (Seeburg et al., 1984). A similar study assessed the consequences of 17 different mutations on HRAS Q61 (Der et al., 1986). Although all 17 mutants shared comparable defects in GTP hydrolysis activity in vitro, the Q61P and Q61E mutations did not show increased transforming ability relative to HRAS WT in mouse fibroblast focus-formation assays.
Emerging evidence suggests that there are mutation-selective consequences on effector signaling. A comparison of KRAS G12C, G12V and G12D mutations in non-small cell lung cancer cell lines revealed mutation-specific alterations regarding effector preference (Ihle et al., 2012). For instance, increased AKT phosphorylation was observed in G12D but not G12C KRAS-expressing cell lines, whereas increased levels of RAL-GTP were detected in G12C KRAS cell lines (Ihle et al., 2012). In another study comparing the effector preference of KRAS G12 and KRAS G13 mutants by using quantitative proteomics to search for non-traditional KRAS-mediated pathways, the colon cancer stem cell marker DCLK1 and the receptor tyrosine kinase MET were both found to be upregulated in G12-mutant-expressing KRAS cells, whereas the tight-junction protein ZO-2 was upregulated in KRAS-G13D-expressing cells when compared to parental lines (Hammond et al., 2015). These studies indicate that individual RAS mutants can signal differently, suggesting that each RAS mutant requires unique pharmacological targeting.
Conclusions
After more than three decades of intense research focus, our understanding of RAS structure, biochemistry and biology is, indeed, very comprehensive. Yet, much remains to be elucidated and many issues are still poorly understood. Without any anti-RAS therapies in the clinic, there is now better recognition of these remaining challenges, prompting a ‘renaissance’ in RAS research and the initiation of the US National Cancer Institute RAS Initiative (Ledford, 2015; Thompson, 2013). Among these issues is the need to delineate the distinct roles that each RAS isoform serves in normal and disease settings. Additionally, until recently, the field has simplistically cataloged cancers as either WT or mutant for RAS; yet, more than one-hundred different missense mutations have been found in cancer. With the arrival of ‘personalized medicine’, a greater attention to and appreciation for RAS-isoform- and RAS-mutation-specific differences will be important in the development of mutation-selective anti-RAS strategies.
Acknowledgements
We apologize to colleagues whose work we could not include due to space limitations.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Funding
This work was supported by the National Institutes of Health [F32 CA200313 and NCI 5-T32CA009156 to G.A.H.; CA042978, CA179193, CA175747 and CA199235 to C.J.D.]; the Department of Defense [CA140731 to C.J.D.]; the Pancreatic Cancer Action Network-AACR [to C.J.D.], and the Lustgarten Foundation [to C.J.D.]. Deposited in PMC for release after 12 months.
Cell science at a glance
A high-resolution version of the poster is available for downloading at http://jcs.biologists.org/lookup/suppl/doi:10.1242/jcs.182873/-/DC1. Individual poster panels are available as JPEG files at http://jcs.biologists.org/lookup/suppl/doi:10.1242/jcs.182873/-/DC2.
References
- Abankwa D., Gorfe A. A. and Hancock J. F. (2008). Mechanisms of Ras membrane organization and signaling: Ras on a rocker. Cell Cycle 7, 2667-2673. 10.4161/cc.7.17.6596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abankwa D., Gorfe A. A., Inder K. and Hancock J. F. (2010). Ras membrane orientation and nanodomain localization generate isoform diversity. Proc. Natl. Acad. Sci. USA 107, 1130-1135. 10.1073/pnas.0903907107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahearn I. M., Haigis K., Bar-Sagi D. and Philips M. R. (2012). Regulating the regulator: post-translational modification of RAS. Nat. Rev. Mol. Cell Biol. 13, 39-51. 10.1038/nrm3255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allegra C. J., Jessup J. M., Somerfield M. R., Hamilton S. R., Hammond E. H., Hayes D. F., McAllister P. K., Morton R. F. and Schilsky R. L. (2009). American Society of Clinical Oncology provisional clinical opinion: testing for KRAS gene mutations in patients with metastatic colorectal carcinoma to predict response to anti-epidermal growth factor receptor monoclonal antibody therapy. J. Clin. Oncol. 27, 2091-2096. 10.1200/JCO.2009.21.9170 [DOI] [PubMed] [Google Scholar]
- Baker R., Lewis S. M., Sasaki A. T., Wilkerson E. M., Locasale J. W., Cantley L. C., Kuhlman B., Dohlman H. G. and Campbell S. L. (2013a). Site-specific monoubiquitination activates Ras by impeding GTPase-activating protein function. Nat. Struct. Mol. Biol. 20, 46-52. 10.1038/nsmb.2430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker R., Wilkerson E. M., Sumita K., Isom D. G., Sasaki A. T., Dohlman H. G. and Campbell S. L. (2013b). Differences in the regulation of K-Ras and H-Ras isoforms by monoubiquitination. J. Biol. Chem. 288, 36856-36862. 10.1074/jbc.C113.525691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbie D. A., Tamayo P., Boehm J. S., Kim S. Y., Moody S. E., Dunn I. F., Schinzel A. C., Sandy P., Meylan E., Scholl C. et al. (2009). Systematic RNA interference reveals that oncogenic KRAS-driven cancers require TBK1. Nature 462, 108-112. 10.1038/nature08460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barcelo C., Paco N., Morell M., Alvarez-Moya B., Bota-Rabassedas N., Jaumot M., Vilardell F., Capella G. and Agell N. (2014). Phosphorylation at Ser-181 of oncogenic KRAS is required for tumor growth. Cancer Res. 74, 1190-1199. 10.1158/0008-5472.CAN-13-1750 [DOI] [PubMed] [Google Scholar]
- Bremner R. and Balmain A. (1990). Genetic changes in skin tumor progression: correlation between presence of a mutant ras gene and loss of heterozygosity on mouse chromosome 7. Cell 61, 407-417. 10.1016/0092-8674(90)90523-H [DOI] [PubMed] [Google Scholar]
- Bryant K. L., Mancias J. D., Kimmelman A. C. and Der C. J. (2014). KRAS: feeding pancreatic cancer proliferation. Trends Biochem. Sci. 39, 91-100. 10.1016/j.tibs.2013.12.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buhrman G., Wink G. and Mattos C. (2007). Transformation efficiency of RasQ61 mutants linked to structural features of the switch regions in the presence of Raf. Structure 15, 1618-1629. 10.1016/j.str.2007.10.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buhrman G., O'Connor C., Zerbe B., Kearney B. M., Napoleon R., Kovrigina E. A., Vajda S., Kozakov D., Kovrigin E. L. and Mattos C. (2011). Analysis of binding site hot spots on the surface of Ras GTPase. J. Mol. Biol. 413, 773-789. 10.1016/j.jmb.2011.09.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunda S., Heir P., Srikumar T., Cook J. D., Burrell K., Kano Y., Lee J. E., Zadeh G., Raught B. and Ohh M. (2014). Src promotes GTPase activity of Ras via tyrosine 32 phosphorylation. Proc. Natl. Acad. Sci. USA 111, E3785-E3794. 10.1073/pnas.1406559111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burd C. E., Liu W., Huynh M. V., Waqas M. A., Gillahan J. E., Clark K. S., Fu K., Martin B. L., Jeck W. R., Souroullas G. P. et al. (2014). Mutation-specific RAS oncogenicity explains NRAS codon 61 selection in melanoma. Cancer Discov. 4, 1418-1429. 10.1158/2159-8290.CD-14-0729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox A. D. and Der C. J. (2010). Ras history: the saga continues. Small GTPases 1, 2-27. 10.4161/sgtp.1.1.12178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox A. D., Fesik S. W., Kimmelman A. C., Luo J. and Der C. J. (2014). Drugging the undruggable RAS: mission possible? Nat. Rev. Drug Discov. 13, 828-851. 10.1038/nrd4389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox A. D., Der C. J. and Philips M. R. (2015). Targeting RAS membrane association: back to the future for anti-RAS drug discovery? Clin. Cancer Res. 21, 1819-1827. 10.1158/1078-0432.CCR-14-3214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Der C. J., Finkel T. and Cooper G. M. (1986). Biological and biochemical properties of human rasH genes mutated at codon 61. Cell 44, 167-176. 10.1016/0092-8674(86)90495-2 [DOI] [PubMed] [Google Scholar]
- Eisenberg S., Beckett A. J., Prior I. A., Dekker F. J., Hedberg C., Waldmann H., Ehrlich M. and Henis Y. I. (2011). Raft protein clustering alters N-Ras membrane interactions and activation pattern. Mol. Cell. Biol. 31, 3938-3952. 10.1128/MCB.05570-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faris J. E., Borger D. R., Fernandez-del Castillo C., Clark J. W., Blaszkowsky L. S., Zhu A. X., Allen J. N., Murphy J. E., Ferrone C., Goyal L. et al. (2014). Effect of molecular genotyping to predict outcomes in patients with metastatic pancreatic cancer. J. Clin. Oncol. 32, 5s; abstract 4128. [Google Scholar]
- Fetics S. K., Guterres H., Kearney B. M., Buhrman G., Ma B., Nussinov R. and Mattos C. (2015). Allosteric effects of the oncogenic RasQ61L mutant on Raf-RBD. Structure 23, 505-516. 10.1016/j.str.2014.12.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fruman D. A. and Rommel C. (2014). PI3K and cancer: lessons, challenges and opportunities. Nat. Rev. Drug Discov. 13, 140-156. 10.1038/nrd4204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelabert-Baldrich M., Soriano-Castell D., Calvo M., Lu A., Vina-Vilaseca A., Rentero C., Pol A., Grinstein S., Enrich C. and Tebar F. (2014). Dynamics of KRas on endosomes: involvement of acidic phospholipids in its association. FASEB J. 28, 3023-3037. 10.1096/fj.13-241158 [DOI] [PubMed] [Google Scholar]
- Grabocka E., Pylayeva-Gupta Y., Jones M. J. K., Lubkov V., Yemanaberhan E., Taylor L., Jeng H. H. and Bar-Sagi D. (2014). Wild-type H- and N-Ras promote mutant K-Ras-driven tumorigenesis by modulating the DNA damage response. Cancer Cell 25, 243-256. 10.1016/j.ccr.2014.01.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haigis K. M., Kendall K. R., Wang Y., Cheung A., Haigis M. C., Glickman J. N., Niwa-Kawakita M., Sweet-Cordero A., Sebolt-Leopold J., Shannon K. M. et al. (2008). Differential effects of oncogenic K-Ras and N-Ras on proliferation, differentiation and tumor progression in the colon. Nat. Genet. 40, 600-608. 10.1038/ng.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond D. E., Mageean C. J., Rusilowicz E. V., Wickenden J. A., Clague M. J. and Prior I. A. (2015). Differential reprogramming of isogenic colorectal cancer cells by distinct activating KRAS mutations. J. Proteome Res. 14, 1535-1546. 10.1021/pr501191a [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter J. C., Gurbani D., Ficarro S. B., Carrasco M. A., Lim S. M., Choi H. G., Xie T., Marto J. A., Chen Z., Gray N. S. et al. (2014). In situ selectivity profiling and crystal structure of SML-8-73-1, an active site inhibitor of oncogenic K-Ras G12C. Proc. Natl. Acad. Sci. USA 111, 8895-8900. 10.1073/pnas.1404639111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter J. C., Manandhar A., Carrasco M. A., Gurbani D., Gondi S. and Westover K. D. (2015). Biochemical and structural analysis of common cancer-associated KRAS mutations. Mol. Cancer Res. 13, 1325-1335. 10.1158/1541-7786.MCR-15-0203 [DOI] [PubMed] [Google Scholar]
- Ihle N. T., Byers L. A., Kim E. S., Saintigny P., Lee J. J., Blumenschein G. R., Tsao A., Liu S., Larsen J. E., Wang J. et al. (2012). Effect of KRAS oncogene substitutions on protein behavior: implications for signaling and clinical outcome. J. Natl. Cancer Inst. 104, 228-239. 10.1093/jnci/djr523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang H., Abraham S. J., Chavan T. S., Hitchinson B., Khavrutskii L., Tarasova N. I., Nussinov R. and Gaponenko V. (2015). Mechanisms of membrane binding of small GTPase K-Ras4B farnesylated hypervariable region. J. Biol. Chem. 290, 9465-9477. 10.1074/jbc.M114.620724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jura N., Scotto-Lavino E., Sobczyk A. and Bar-Sagi D. (2006). Differential modification of Ras proteins by ubiquitination. Mol. Cell 21, 679-687. 10.1016/j.molcel.2006.02.011 [DOI] [PubMed] [Google Scholar]
- Khrenova M. G., Mironov V. A., Grigorenko B. L. and Nemukhin A. V. (2014). Modeling the role of G12V and G13V Ras mutations in the Ras-GAP-catalyzed hydrolysis reaction of guanosine triphosphate. Biochemistry 53, 7093-7099. 10.1021/bi5011333 [DOI] [PubMed] [Google Scholar]
- Kiel C., Wohlgemuth S., Rousseau F., Schymkowitz J., Ferkinghoff-Borg J., Wittinghofer F. and Serrano L. (2005). Recognizing and defining true Ras binding domains II: in silico prediction based on homology modelling and energy calculations. J. Mol. Biol. 348, 759-775. 10.1016/j.jmb.2005.02.046 [DOI] [PubMed] [Google Scholar]
- Kumar M. S., Hancock D. C., Molina-Arcas M., Steckel M., East P., Diefenbacher M., Armenteros-Monterroso E., Lassailly F., Matthews N., Nye E. et al. (2012). The GATA2 transcriptional network is requisite for RAS oncogene-driven non-small cell lung cancer. Cell 149, 642-655. 10.1016/j.cell.2012.02.059 [DOI] [PubMed] [Google Scholar]
- Lampson B. L., Pershing N. L. K., Prinz J. A., Lacsina J. R., Marzluff W. F., Nicchitta C. V., MacAlpine D. M. and Counter C. M. (2013). Rare codons regulate KRas oncogenesis. Curr. Biol. 23, 70-75. 10.1016/j.cub.2012.11.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ledford H. (2015). Cancer: the Ras renaissance. Nature 520, 278-280. 10.1038/520278a [DOI] [PubMed] [Google Scholar]
- Lim K.-H., Ancrile B. B., Kashatus D. F. and Counter C. M. (2008). Tumour maintenance is mediated by eNOS. Nature 452, 646-649. 10.1038/nature06778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo J., Emanuele M. J., Li D., Creighton C. J., Schlabach M. R., Westbrook T. F., Wong K.-K. and Elledge S. J. (2009). A genome-wide RNAi screen identifies multiple synthetic lethal interactions with the Ras oncogene. Cell 137, 835-848. 10.1016/j.cell.2009.05.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch S. J., Snitkin H., Gumper I., Philips M. R., Sabatini D. and Pellicer A. (2015). The differential palmitoylation states of N-Ras and H-Ras determine their distinct golgi subcompartment localizations. J. Cell. Physiol. 230, 610-619. 10.1002/jcp.24779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurer T., Garrenton L. S., Oh A., Pitts K., Anderson D. J., Skelton N. J., Fauber B. P., Pan B., Malek S., Stokoe D. et al. (2012). Small-molecule ligands bind to a distinct pocket in Ras and inhibit SOS-mediated nucleotide exchange activity. Proc. Natl. Acad. Sci. USA 109, 5299-5304. 10.1073/pnas.1116510109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogura T., Yamao K., Hara K., Mizuno N., Hijioka S., Imaoka H., Sawaki A., Niwa Y., Tajika M., Kondo S. et al. (2013). Prognostic value of K-ras mutation status and subtypes in endoscopic ultrasound-guided fine-needle aspiration specimens from patients with unresectable pancreatic cancer. J. Gastroenterol. 48, 640-646. 10.1007/s00535-012-0664-2 [DOI] [PubMed] [Google Scholar]
- Ostrem J. M., Peters U., Sos M. L., Wells J. A. and Shokat K. M. (2013). K-Ras(G12C) inhibitors allosterically control GTP affinity and effector interactions. Nature 503, 548-551. 10.1038/nature12796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker J. A. and Mattos C. (2015). The Ras-membrane interface: isoform-specific differences in the catalytic domain. Mol. Cancer Res. 13, 595 10.1158/1541-7786.mcr-14-0535 [DOI] [PubMed] [Google Scholar]
- Pecot C. V., Wu S. Y., Bellister S., Filant J., Rupaimoole R., Hisamatsu T., Bhattacharya R., Maharaj A., Azam S., Rodriguez-Aguayo C. et al. (2014). Therapeutic silencing of KRAS using systemically delivered siRNAs. Mol. Cancer Ther. 13, 2876-2885. 10.1158/1535-7163.MCT-14-0074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pershing N. L. K., Lampson B. L., Belsky J. A., Kaltenbrun E., MacAlpine D. M. and Counter C. M. (2015). Rare codons capacitate Kras-driven de novo tumorigenesis. J. Clin. Invest. 125, 222-233. 10.1172/JCI77627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prior I. A., Lewis P. D. and Mattos C. (2012). A comprehensive survey of Ras mutations in cancer. Cancer Res. 72, 2457-2467. 10.1158/0008-5472.CAN-11-2612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu W., Sahin F., Iacobuzio-Donahue C. A., Garcia-Carracedo D., Wang W. M., Kuo C.-Y., Chen D., Arking D. E., Lowy A. M., Hruban R. H. et al. (2011). Disruption of p16 and activation of Kras in pancreas increase ductal adenocarcinoma formation and metastasis in vivo. Oncotarget 2, 862-873. 10.18632/oncotarget.357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quatela S. E., Sung P. J., Ahearn I. M., Bivona T. G. and Philips M. R. (2008). Analysis of K-Ras phosphorylation, translocation, and induction of apoptosis. Small Gtpases Dis. Pt. B 439, 87-102. 10.1016/S0076-6879(07)00407-7 [DOI] [PubMed] [Google Scholar]
- Rotblat B., Prior I. A., Muncke C., Parton R. G., Kloog Y., Henis Y. I. and Hancock J. F. (2004). Three separable domains regulate GTP-dependent association of H-ras with the plasma membrane. Mol. Cell. Biol. 24, 6799-6810. 10.1128/MCB.24.15.6799-6810.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samatar A. A. and Poulikakos P. I. (2014). Targeting RAS-ERK signalling in cancer: promises and challenges. Nat. Rev. Drug Discov. 13, 928-942. 10.1038/nrd4281 [DOI] [PubMed] [Google Scholar]
- Sarthy A. V., Morgan-Lappe S. E., Zakula D., Vernetti L., Schurdak M., Packer J. C. L., Anderson M. G., Shirasawa S., Sasazuki T. and Fesik S. W. (2007). Survivin depletion preferentially reduces the survival of activated K-Ras-transformed cells. Mol. Cancer Ther. 6, 269-276. 10.1158/1535-7163.MCT-06-0560 [DOI] [PubMed] [Google Scholar]
- Sasaki A. T., Carracedo A., Locasale J. W., Anastasiou D., Takeuchi K., Kahoud E. R., Haviv S., Asara J. M., Pandolfi P. P. and Cantley L. C. (2011). Ubiquitination of K-Ras enhances activation and facilitates binding to select downstream effectors. Sci. Signal. 4, ra13 10.1126/scisignal.2001518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholl C., Fröhling S., Dunn I. F., Schinzel A. C., Barbie D. A., Kim S. Y., Silver S. J., Tamayo P., Wadlow R. C., Ramaswamy S. et al. (2009). Synthetic lethal interaction between oncogenic KRAS dependency and STK33 suppression in human cancer cells. Cell 137, 821-834. 10.1016/j.cell.2009.03.017 [DOI] [PubMed] [Google Scholar]
- Seeburg P. H., Colby W. W., Capon D. J., Goeddel D. V. and Levinson A. D. (1984). Biological properties of human c-Ha-ras1 genes mutated at codon 12. Nature 312, 71-75. 10.1038/312071a0 [DOI] [PubMed] [Google Scholar]
- Shima F., Yoshikawa Y., Ye M., Araki M., Matsumoto S., Liao J., Hu L., Sugimoto T., Ijiri Y., Takeda A. et al. (2013). In silico discovery of small-molecule Ras inhibitors that display antitumor activity by blocking the Ras-effector interaction. Proc. Natl. Acad. Sci. USA 110, 8182-8187. 10.1073/pnas.1217730110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith M. J., Neel B. G. and Ikura M. (2013). NMR-based functional profiling of RASopathies and oncogenic RAS mutations. Proc. Natl. Acad. Sci. USA 110, 4574-4579. 10.1073/pnas.1218173110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Q., Burke J. P., Phan J., Burns M. C., Olejniczak E. T., Waterson A. G., Lee T., Rossanese O. W. and Fesik S. W. (2012). Discovery of small molecules that bind to K-Ras and inhibit Sos-mediated activation. Angew. Chem. Int. Ed. Engl. 51, 6140-6143. 10.1002/anie.201201358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tejpar S., Celik I., Schlichting M., Sartorius U., Bokemeyer C. and Van Cutsem E. (2012). Association of KRAS G13D tumor mutations with outcome in patients with metastatic colorectal cancer treated with first-line chemotherapy with or without cetuximab. J. Clin. Oncol. 30, 3570-3577. 10.1200/JCO.2012.42.2592 [DOI] [PubMed] [Google Scholar]
- Thompson H. (2013). US National Cancer Institute's new Ras project targets an old foe. Nat. Med. 19, 949-950. 10.1038/nm0813-949 [DOI] [PubMed] [Google Scholar]
- Ting P. Y., Johnson C. W., Fang C., Cao X., Graeber T. G., Mattos C. and Colicelli J. (2015). Tyrosine phosphorylation of RAS by ABL allosterically enhances effector binding. FASEB J. 29, 3750-3761. 10.1096/fj.15-271510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- To M. D., Rosario R. D., Westcott P. M. K., Banta K. L. and Balmain A. (2013). Interactions between wild-type and mutant Ras genes in lung and skin carcinogenesis. Oncogene 32, 4028-4033. 10.1038/onc.2012.404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran N. H., Cavalcante L. L., Lubner S. J., Mulkerin D. L., LoConte N. K., Clipson L., Matkowskyj K. A. and Deming D. A. (2015). Precision medicine in colorectal cancer: the molecular profile alters treatment strategies. Ther. Adv. Med. Oncol. 7, 252-262. 10.1177/1758834015591952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai F. D., Lopes M. S., Zhou M., Court H., Ponce O., Fiordalisi J. J., Gierut J. J., Cox A. D., Haigis K. M. and Philips M. R. (2015). K-Ras4A splice variant is widely expressed in cancer and uses a hybrid membrane-targeting motif. Proc. Natl. Acad. Sci. USA 112, 779-784. 10.1073/pnas.1412811112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Cutsem E., Lenz H.-J., Kohne C.-H., Heinemann V., Tejpar S., Melezinek I., Beier F., Stroh C., Rougier P., van Krieken J. H. et al. (2015). Fluorouracil, leucovorin, and irinotecan plus cetuximab treatment and RAS mutations in colorectal cancer. J. Clin. Oncol. 33, 692-700. 10.1200/JCO.2014.59.4812 [DOI] [PubMed] [Google Scholar]
- Vetter I. R. (2014). The Structure of the G Domain of the Ras Superfamily. Vienna, Austria: Spinger-Verlag Wien. [Google Scholar]
- Vigil D., Cherfils J., Rossman K. L. and Der C. J. (2010). Ras superfamily GEFs and GAPs: validated and tractable targets for cancer therapy? Nat. Rev. Cancer 10, 842-857. 10.1038/nrc2960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westcott P. M. K., Halliwill K. D., To M. D., Rashid M., Rust A. G., Keane T. M., Delrosario R., Jen K.-Y., Gurley K. E., Kemp C. J. et al. (2015). The mutational landscapes of genetic and chemical models of Kras-driven lung cancer. Nature 517, 489-492. 10.1038/nature13898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weyandt J. D., Lampson B. L., Tang S., Mastrodomenico M., Cardona D. M. and Counter C. M. (2015). Wild-type Hras suppresses the earliest stages of tumorigenesis in a genetically engineered mouse model of pancreatic cancer. PLoS ONE 10, e0140253 10.1371/journal.pone.0140253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witkiewicz A. K., McMillan E. A., Balaji U., Baek G., Lin W.-C., Mansour J., Mollaee M., Wagner K.-U., Koduru P., Yopp A. et al. (2015). Whole-exome sequencing of pancreatic cancer defines genetic diversity and therapeutic targets. Nat. Commun. 6, 6744 10.1038/ncomms7744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wohlgemuth S., Kiel C., Krämer A., Serrano L., Wittinghofer F. and Herrmann C. (2005). Recognizing and defining true Ras binding domains I: biochemical analysis. J. Mol. Biol. 348, 741-758. 10.1016/j.jmb.2005.02.048 [DOI] [PubMed] [Google Scholar]
- Xue W., Dahlman J. E., Tammela T., Khan O. F., Sood S., Dave A., Cai W., Chirino L. M., Yang G. R., Bronson R. et al. (2014). Small RNA combination therapy for lung cancer. Proc. Natl. Acad. Sci. USA 111, E3553-E3561. 10.1073/pnas.1412686111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang M. H., Nickerson S., Kim E. T., Liot C., Laurent G., Spang R., Philips M. R., Shan Y., Shaw D. E., Bar-Sagi D. et al. (2012). Regulation of RAS oncogenicity by acetylation. Proc. Natl. Acad. Sci. USA 109, 10843-10848. 10.1073/pnas.1201487109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young A., Lou D. and McCormick F. (2013). Oncogenic and wild-type Ras play divergent roles in the regulation of mitogen-activated protein kinase signaling. Cancer Discov. 3, 112-123. 10.1158/2159-8290.CD-12-0231 [DOI] [PubMed] [Google Scholar]
- Yuan T. L., Fellmann C., Lee C.-S., Ritchie C. D., Thapar V., Lee L. C., Hsu D. J., Grace D., Carver J. O., Zuber J. et al. (2014). Development of siRNA payloads to target KRAS-mutant cancer. Cancer Discov. 4, 1182-1197. 10.1158/2159-8290.CD-13-0900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z., Wang Y., Vikis H. G., Johnson L., Liu G., Li J., Anderson M. W., Sills R. C., Hong H. L., Devereux T. R. et al. (2001). Wildtype Kras2 can inhibit lung carcinogenesis in mice. Nat. Genet. 29, 25-33. 10.1038/ng721 [DOI] [PubMed] [Google Scholar]
- Zimmermann G., Papke B., Ismail S., Vartak N., Chandra A., Hoffmann M., Hahn S. A., Triola G., Wittinghofer A., Bastiaens P. I. et al. (2013). Small molecule inhibition of the KRAS-PDEdelta interaction impairs oncogenic KRAS signalling. Nature 497, 638-642. 10.1038/nature12205 [DOI] [PubMed] [Google Scholar]