Abstract
The human neocortex is created from diverse intermixed progenitors in the prenatal germinal zones. These progenitors have been difficult to characterize since progenitors—particularly radial glia (RG)—are rare, and are defined by a combination of intracellular markers, position and morphology. To circumvent these problems we developed a method called FRISCR for transcriptome profiling of individual fixed, stained and sorted cells. After validation of FRISCR using human embryonic stem cells, we profiled primary human RG that constitute only 1% of the mid-gestation cortex. These RG could be classified into ventricular zone-enriched RG (vRG) that express ANXA1 and CRYAB, and outer subventricular zone-localized RG (oRG) that express HOPX. Our study identifies the first markers and molecular profiles of vRG and oRG cells, and provides an essential step for understanding molecular networks driving the lineage of human neocortical progenitors. Furthermore, FRISCR allows targeted single-cell transcriptomic profiling of tissues that lack live-cell markers.
Introduction
Several essential progenitor types underpin human brain development. Radial glial cells (RGs) and intermediate progenitor cells (IPCs) are cortical neurogenic and gliogenic progenitors that reside in the ventricular zone (VZ) of the cortex (Fig. 1a,c,d)1–5. RGs are bi-polar epithelial cells with an apical endfoot contacting the ventricular surface, and a basal process that reaches the pial surface. In contrast, IPCs are neurogenic, lack epithelial morphology and have a more limited capacity for proliferation and self-renewal1,3–5. The human brain undergoes a prolonged period of neurogenesis and forms an expanding region of proliferating progenitors called the outer subventricular zone (oSZ)2,5,6. The oSZ contains IPCs as well as outer RGs (oRGs) that express the same canonical transcription factors as RGs in the VZ (vRGs), but are distinguished by their position in the oSZ, lack of an apical endfoot, and the maintenance of a basal process that can extend to the pial surface (Fig. 1a)1,7,8. oRGs are hypothesized to drive the dramatic cortical expansion observed in gyrified brains such as human3,5,9. Understanding the molecular diversity of human RG progenitors is an essential first step to determine 1) if discrete populations of RGs produce specific mature cell types, and 2) what molecular events drive formation of human-specific progenitors and structures (like oRGs and the oSZ). Due to their rarity, human RG analysis has been limited to morphology with a few histological markers to confirm cell identity (Fig. 1b)1,7,8, molecular characterization of microdissected tissue which contains an unknown variety of cell types10,11, or live marker-sorted cells whose purity is unknown12,13. We lack markers of RG progenitor subtypes, which will be critical to understand human corticogenesis.
Figure 1.
Human cortical progenitors are diverse and intermixed during development. (a) Model of the progenitor compartment shows a mixture of ventricular radial glial cells (vRG-light blue), outer RGs (oRGs-purple), intermediate progenitors (IPCs-orange) and other mixed cortical cell types (gray). Known markers for each cell type are shown below. Note RGs identified by antibody staining are called SP (SOX+PAX+EOMES− -dark blue nuclei), and IPCs are called SPE (SOX+PAX+EOMES+ -dark orange nuclei). (b) Immunocytochemistry images of 19 PCW germinal zones. Top left: Low magnification images stitched together (left), or individual micrograph (right) of the VZ, iSZ, and oSZ stained by DAPI, EOMES, PAX6, and SOX2 with the colors indicated. Scale bars are 100 µm. High magnification micrograph of oSZ region (middle) and VZ/iSZ region (bottom) show SP (arrow with filled arrowhead) and SPE (arrowhead) cells. Note many cells the VZ, iSZ, and oSZ lack progenitor markers and are unknown cell types (arrows with open arrowhead). Scale bars are 50 µm. Tables describing human germinal zone regions and symbols (c), and common human cortical progenitors, their germinal zone locations, and their flow cytometry (FACS) phenotypes (d).
Characterizing the full diversity of RG progenitors requires transcriptional profiles of large numbers of single cells, ideally from targeted subpopulations because of low abundance of these progenitors. RGs express SOX2 and PAX6 and lack EOMES (also known as TBR2), while IPCs can express all three of those intracellular markers2,4,5. Sorting cells of these immunophenotypes requires fixation, permeabilization and staining. Many of these steps when done with traditional reagents lead to highly degraded mRNA rendering the cells unusable for transcriptomic profiling. Although new protocols have emerged recently for transcriptional profiling of fixed, permeabilized, stained, and sorted cells, this has only been reported for samples of ≥105 fixed cells, and never for single cells14–18.
Here we present FRISCR (Fixed and Recovered Intact Single Cell RNA), a method for RNA isolation from fixed, permeabilized, stained, and sorted cells suitable for transcriptomic profiling of single cells. We show that the fixation and purification techniques introduce little bias, and yield gene expression data similar to that from living cells. We use this technique to prospectively isolate single RGs from primary human prenatal neocortex and characterize those cells with unbiased transcriptional profiling. Analysis of our single-cell gene expression data identified RG subpopulations that corresponded to human oRGs and vRGs based on position in primary mid-gestation human cortex, and identified the first molecular markers that distinguish oRG cells from vRGs. FRISCR provides a significant advance for single-cell profiling of human primary tissues, will be valuable for profiling rare cell populations in the brain, and will be broadly applicable to tissues that lack live-cell markers.
Results
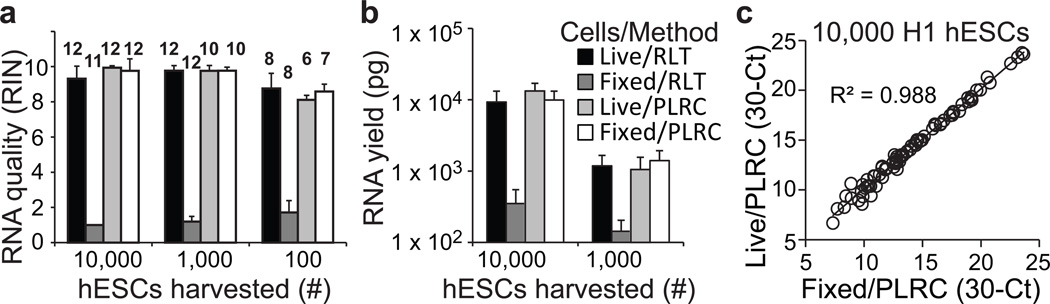
To overcome the challenge of transcriptomic profiling from fixed, stained and sorted cells, we optimized a method to extract RNA from fixed cells. We always handled fixed cells with RNAase-free reagents and included RNase inhibitor. In addition we reverse-crosslinked RNA at 56°C for one hour rather than at 80°C in other reverse crosslinking protocols. Since comparable methods have only been validated with sample of >105 cells, we compared fixed human embryonic stem cell (hESC) RNA to that purified from live hESCs using 10,000-, 1,000-, and 100-cell samples (Supplementary Fig. 1a). Analysis of total RNA showed high RNA integrity numbers (RINs) from batches of fixed cells using this protocol (Fig. 2a, Supplementary Fig. 1b) with no loss of total RNA (Fig. 2b). Fixation did not introduce bias in mRNA detection as assessed by quantitative reverse transcriptase polymerase chain reaction (qRT-PCR); we detected high correlations between live and fixed cells for every gene tested (Fig. 2c).
Figure 2.
High quality and yield RNA from fixed and sorted hESCs. (a,b) Quantification of Bioanalyzer RNA integrity numbers (RINs) and total recovered RNA purified from live or fixed cells, and RNA harvested using either the Qiagen microRNeasy Kit (RLT), or using a modified fixed-cell method with protease lysis and reverse crosslinking (PLRC). Sample colors are indicated in b. (a) RINs from 10,000, 1,000, and 100 sorted cells from 2–3 independent experiments. The biological replicates are indicated by the numbers above the bars. (b) Yield from column purified total RNA from 10,000 and 1,000 H1 hESCs. We diluted or concentrated samples such that 66 cell equivalents were loaded per lane of the Bioanalyzer. Data are from 12 biological replicates, except for the 1,000-cell PLRC samples which have 10 replicates, processed over three independent experiments. Barplots in a and b show mean ± SD. (c) Comparison of expression level of 82 genes by qRT-PCR of 10,000 live or fixed hESC samples processed with PLRC. Data are from n = 7–8 biological replicates across two independent experiments, and only genes that were detected in at least 3 replicates from both conditions were included.
We sought to extend this technique to single cells by improving mRNA recovery. Using oligo dT25 beads, we measured better recovery of cell-associated mRNAs and could elute in low volumes after purifying the RNA from the reverse crosslinking buffer (Supplementary Fig. 1c). Eluting in a low volume ensured we could apply the entire sample to the SmartSeq2 reaction without concentration. We called this method FRISCR (Fixed and Recovered Intact Single Cell RNA) (Fig. 3a). To validate FRISCR we sorted either live or fixed single H1 hESCs, and prepared mRNA by either standard Triton X-100 Lysis (TL) or FRISCR. FRISCR followed by SmartSeq219 amplified comparable amounts of cDNA from individual fixed and live cells (Fig. 3b, Supplementary Fig. 2b). We sequenced each sample and then subsampled to 5 million total reads for analysis. Fixed cells prepared with TL gave poor read alignment indicative of much lower mRNA input, whereas FRISCR libraries generated sequencing alignments from fixed cells comparable to live cells (Fig. 3c, Supplementary Fig. 2a). Directly comparing live and fixed cells prepared with FRISCR revealed that the frequency of reads mapping to different transcript classes did not change, and the total number of genes per cell detected was similar (Supplementary Fig. 2c). Reads across all genes showed a similar 3′ to 5′ bias (Fig. 3d), however, fixed cells showed an increased 3′ read bias with longer transcripts (Supplementary Fig. 2g). Spearman correlations of all genes did not discriminate live from fixed cells (Fig. 3e), and only two genes in the genome were differentially detected between sets of single cells (Fig. 3f, Supplementary Fig. 3). Comparing data from TL- versus FRISCR-prepared live cells demonstrated a slight increase in 3′ bias read recovery that was more pronounced with longer transcripts (Fig. 3d, Supplementary Fig. 2g). Analysis of ERCC spike-in mRNAs added at the time of cell collection showed TL and FRISCR samples gave similar linear mRNA amplification (Supplementary Fig. 2d), while TL samples showed moderately higher estimated sensitivity compared to FRISCR, recovering ~25% versus ~16% of transcripts, respectively (Supplementary Fig. 2e). Nevertheless Spearman correlation intermixed FRISCR and TL cells (Fig. 3e), and just over thirty genes were differentially expressed of which many are non-coding genes or irregularly polyadenylated (Fig. 3f, Supplementary Fig. 3)20. No method showed major changes in GC coverage or chromosomal transcript representation bias (Supplementary Fig. 2h–i) or rates of alignment to mRNA (Fig. 3c). We detected a subtle increase in the mutation rate in FRISCR-prepared live cells, but FRISCR substantially reduced the mutation rate of fixed cells compared to standard TL (Supplementary Fig. 2f). Taken together, these data show that FRISCR faithfully recovers RNA from permeabilized, stained and sorted single cells that is similar to mRNA from live cells and useful for transcriptomic profiling.
Figure 3.
FRISCR allows purification and amplification of mRNA from single fixed hESCs that is similar to live cells. (a) Schematic description of steps in the FRISCR method. Asterisks mark steps with stopping points. (b–f) We processed live or fixed single sorted H1 hESCs by standard Triton-X100 lysis (TL) or FRISCR, and sequenced after making SmartSeq2 cDNA libraries. (b) Representative Bioanalyzer traces of SmartSeq2 amplified cDNA are shown from the four experimental conditions indicated. (c) Alignment statistics for HiSeq RNA-Seq data from cells processed by the four experimental conditions. * P < 0.05 and ** P < 0.001 by one-way ANOVA followed by Tukey’s post-hoc test, and plots show mean ± SD. Data in (d–f) are from n = 7–15 biological replicates as shown, processed over two independent days of experiments. Analysis carried out in (d–f) is on libraries subsampled to 5 million reads. (d) Normalized average read counts per experimental preparation are shown across the percentile predicted transcript length (3′ to 5′). Only transcripts not alternatively spliced, and with TPM > 1 in at least 30 samples were selected. Solid line is average and shading illustrates SD. (e) Heatmap with hierarchical clustering for single hES cells showing pairwise Spearman correlation of all expressed genes. Color bars indicate experimental replicate and experimental condition. (f) Table showing the number of differentially expressed genes between hESCs from the different conditions calculated by DESeq with adjusted P < .01.
We leveraged FRISCR to profile rare and unmarked cell types and examined the diversity of RG progenitors from primary fetal human cortex. Antibody staining of human cortical tissues between 15 and 19 post-conception weeks (PCW) reproducibly identified early IPCs (SOX2+PAX6+EOMES+; “SPE”), and RGs (SOX2+PAX6+EOMES−; “SP”) (Fig. 1a,b,d). We used FACS and FRISCR to harvest mRNA from single SP cells from four primary tissues (PCW 14, 16, 18, and 19) and single SPE cells from two primary tissues (PCW 18 and 19) (Fig. 4a, Supplementary Fig. 4a, 5a). We profiled cells in G0-G1 phase to reduce cell cycle-dependent gene expression variation. Additionally, we used DAPI, cell size and light scattering properties to ensure non-nucleated debris and doublets were excluded (Supplementary Fig. 4a). Although RG and IPC progenitors are enriched in the germinal zones, they are rare when the entire cortical thickness is prepared: 3.2 ± 0.9% of cells were SOX2+PAX6+, and this population was further divided into EOMES+ cells (SPE) (2.1 ± 0.6%) and EOMES− cells (SP) (1.0 ± 0.3%) (Fig. 4a, Supplementary Fig. 4a). We amplified cDNA from these sorted cells (207 SP and 48 SPE) using SmartSeq219. Primary cell cDNA showed more low molecular weight products than hESC cDNA (Fig. 3b, Supplementary Fig. 5b), perhaps indicating some RNA degradation due to ≥4-hour post-mortem intervals in these clinical specimens. Single-cell libraries were sequenced at low read depth on the MiSeq (Supplementary Table 1, Supplementary Data 1–2). Single-cell read alignments were typically consistent but some cells displayed poor mapping (Supplementary Fig. 5c), potentially due to primer dimers or contaminating dead cells. Libraries with more than 10,000 mapped reads, >20% reads mapping to mRNA, and detectable GAPDH expression were included in the analysis (157 SP and 29 SPE cells) (Supplementary Fig. 5d, Supplementary Table 1). Approximately 3000–4000 unique genes were detected per progenitor cell (Supplementary Fig. 5e), similar to an independent study of live prenatal human single cells21. As expected, PAX6 and SOX2 were expressed in most SP and SPE cells, and EOMES mRNA was enriched in SPE versus SP cells (86% and 22%, respectively) (Fig. 4b). Similarly, principal component analysis and hierarchical clustering of genes with variance above technical noise demonstrated SP partitioning away from SPE cells independently of read depth, cortical sample, or mapping percentage, despite the low read depth and partial sample degradation (Supplementary Fig. 5f–g). This partition was not observed after correlation with all expressed genes or with only ERCC spike-in RNA reads (Supplementary Fig. 5g). Lastly, our cells clustered with previously identified RGs and IPCs profiled from an independent study on live human cortical cells (Supplementary Fig. 6a)21, and largely lacked expression of common contaminating cell markers (Supplementary Fig. 6b). As a whole, these data demonstrate that FRISCR permits robust directed profiling of rare primary human neuronal progenitors.
Figure 4.
FRISCR allows profiling of primary human cortical progenitors. Color scheme throughout figure is shown in b. (a) FACS plots show gating strategy and frequency of gated cells compared to total cortical cells from three independent experiments. Left plot includes only G0-G1 singlet cortical cells (gated with DAPI) and right plot shows only the SOX2+PAX6+ cells gated on the left plot. (b) Barplots show the frequency of indicated genes detected per cell. Only cells with >10,000 mapped reads, >20% mRNA mapping, and GAPDH expression (n = 157 SP and n = 29 SPE cells) were included in this analysis.
Having profiled an order of magnitude more RGs and IPCs than previous studies by genome-wide transcriptomics21, we could identify progenitor heterogeneity. We identified five modules of co-expressed genes from high variance genes by weighted gene co-expression network analysis (WGCNA)22 (Fig. 5a, Supplementary Fig. 7). We excluded two modules from clustering that contained many cell cycle-related genes; these modules were enriched in SPE cells which were twice as likely found in the S-G2-M phases of the cell cycle (Supplementary Fig. 4b). Genes populating the five WGCNA modules were used to cluster cells into six clusters labeled A through F (Fig. 5a,b, Supplementary Fig. 8), all but one containing cells from each brain. Module 1 reflected the division of the majority of RG from IPC cells, and contained canonical markers of RGs including VIM, and IPCs including EOMES, HES6 and NEUROG1 (Fig. 5a, Supplementary Fig. 7). Modules 2–5 revealed four RG subpopulations as seen by module eigengene values (Fig. 5a). Cluster E cells were enriched for module 3 genes including immediate early genes EGR1 and FOS21, and CXCL12, ANXA1 and CYR61 (Supplementary Fig. 7,8). FACS analysis confirmed that a substantial number of SP, but few SPE cells, stained positively for CYR61 (Supplementary Fig. 9a). A subset of cluster E cells also expressed many genes in module 5 (Supplementary Fig. 7,8). Cells in clusters C and D expressed higher levels of modules 2 and 4 genes but not model 3 and 5 genes. Module 2 is composed of genes such as FAM107A, HOPX and SLCO1C1; and module 4 genes are enriched specifically in cluster C cells. These analyses directly demonstrate an unknown molecular diversity within the human RG compartment.
Figure 5.
Identification of human cortical progenitor cell types with FRISCR. (a) Module eigengene values derived from WGCNA analysis enrichment in cells. (b) Heatmap showing the Spearman correlation from individual cells by genes identified from the 5 WGCNA modules. (c) Heatmap shows correlation of single cell-derived WGCNA module eigengenes with 15 PCW (left) and 21 PCW (right) gene expression data from the BrainSpan Atlas of the Developing Human Brain11. Abbreviations are: subgranular zone (SG), subplate (SP), intermediate zone (IZ), outer subventricular zone (oSZ), inner subventricular zone (iSZ), and VZ (ventricular zone). (d) Heatmaps showing genes both differentially expressed between cell clusters C and D (oRGs) versus cluster E (vRGs) from the FRISCR-prepared single cells (data shown on left), as well as differentially expressed between oSZ and VZ regions of 21 PCW human tissues (data shown on right). Common progenitor markers are included at the top of the heatmap for reference. All heat maps show normalized expression with red indicating high expression, blue indicating low expression; colorbars for a, b and the FRISCR single cell plot in d are indicated in the bottom of a.
To further characterize this diversity we compared RG gene expression signatures to anatomical atlases. The module 2 and 4 eigengenes were enriched in the oSZ relative to the VZ in 21 PCW cortex (P = 6.3×10−11 for module 2 and P = 1.5×10−19 for module 4) but not at earlier time points in the BrainSpan Atlas of the Developing Human Brain11 (Fig. 5c, Supplementary Fig. 10a). Reciprocally, the module 3 eigengene was enriched in the VZ at all timepoints (P = 3.9×10−15, P = 6.2×10−12, and P = 7.8×10−10 for 15, 16, and 21 PCW tissues), while module 5 is only significantly enriched in the VZ in 21 PCW tissue (P = 4.1×10−6) (Fig. 5c, Supplementary Fig. 10a). Strong HOPX expression initiates around embryonic day 70 in macaque according to the NIH Blueprint Non-Human Primate Atlas, coinciding with the emergence of the oSZ (Supplementary Fig. 10c)23. Thus, we hypothesize that vRGs express module 3 and 5 genes, whereas oRGs express module 2 and 4 genes3,4.
To test the distinction between vRG and oRG cells, we identified a set of genes enriched in modules 2 or 3 that were also differentially expressed between the VZ and the oSZ regions in the 21 PCW BrainSpan Atlas data. We identified 10 genes enriched in both the FRISCR-prepared vRG cells and the 21 PCW VZ, and 9 genes enriched both in FRISCR-prepared oRG cells and the 21PCW oSZ (Fig. 5d, Supplementary Fig. 10b). The canonical progenitor markers SOX2, PAX6, EOMES, and HES1 were expressed at similar levels in both germinal zone regions (Fig. 5d). Confirmation of several genes that marked vRG and oRG cells was carried out with antibody staining of human cortical sections (Fig. 6a). CRYAB and ANXA1 showed signal only in the VZ (except for vascular staining), while oRG markers HOPX and F3 (module 2 genes) showed high expression in the oSZ and iSZ, but faint signal in the VZ of a 15 and 16 PCW cortex (Fig. 6a, Supplementary Fig. 9b, 11a). We used SOX2 to mark RGs, since nearly all SOX2+ cells are PAX6+ in the germinal zones (Supplementary Fig. 12a). HOPX+SOX2+ cells in the VZ occasionally exhibited apical process anchored at the ventricular surface at PCW16 (Fig. 6b), and in the oSZ often displayed basally-directed processes as assessed by phospho-VIMENTIN or HOPX staining (Fig. 6c–d, Supplementary Fig. 11b). At later developmental stages such as PCW 19 and 21, HOPX was exclusively detected in the oSZ and almost never in the VZ by both gene expression (Fig. 5d, Supplementary Fig. 10b), and antibody staining (Fig. 6d, Supplementary Fig. 11a). HOPX+ oRGs and ANXA1+ vRGs were proliferative since 17% of HOPX+SOX2+ cells and 28% of ANXA1+SOX2+ cells were also positive for Ki67 (Fig. 6e–f, Supplementary Fig. 11c). The majority of oRG progenitors were marked by HOPX: more than 78% of SOX2+ cells in the oSZ co-stained with HOPX, and less than 1% of HOPX+ cells in the oSZ are SOX2− (Fig. 6g). Reciprocally, 85% of SOX2+ cells in the VZ were ANXA1+, and only 2% of ANXA1+ cells in the VZ lacked SOX2 expression (Fig. 6h). These data confirm that module 2 genes such as HOPX and module 3 genes such as ANXA1 represent first-in-class diagnostic markers for human mid-gestation oRGs and vRGs. Multiplexing these markers reveals distinct novel progenitor populations in the developing human cortex for further analysis (Fig. 6i, Supplementary Fig. 12b–e). Interestingly, mouse HOPX expression is restricted to the medial cortex24, while we detected human HOPX protein and gene expression throughout the human developing cortical germinal zones (Supplementary Fig. 13a–b). This suggests HOPX expression has changed through evolution. Study of these progenitors will help reveal molecular changes that underpin cortical evolution.
Figure 6.
Confirmation of vRG and oRG markers. (a) Grayscale (left) or merged (right) micrographs of germinal zone showing co-staining of SOX2 (magenta) and ANXA1, CRYAB or HOPX (green). Tissue is 16 PCW cortex, scales are 100 µm. Abbreviations: marginal zone (MZ), cortical plate (CP), subplate/intermediate zone (SP/IZ), outer subventricular zone (oSZ), inner subventricular zone (iSZ), and ventricular zone (VZ). (b) HOPX expression in the VZ; scale is 50 µm. Co-expression with SOX2 (arrowhead), SOX2-only cells (arrow), and HOPX+ apical endfoot are indicated (asterisk). (c) A mitotic cell co-stained with SOX2, HOPX and phospho-VIM shows basal cell process (asterisk) and cell body (arrowhead); scale is 20 µm. (d) Full cortical thickness (left), and insets (right) from germinal zone showing co-staining of SOX2 (magenta) and HOPX (green). Scales are 100 µm. (e) Quantitation of SOX2+HOPX+ or SOX2+HOPX− cells in oSZ that are Ki67+. (f) Quantification of SOX2+ANXA1+ or SOX2+ANXA1− cells in VZ that are Ki67+. Data is mean ± SD for (e) five or (f) six human tissues at 14–19 PCW; ** P < 0.01 by Wilcoxon rank sum test. Diagrams show the percentage overlap of SOX2 and HOPX in oSZ cells (g), and of SOX2 and ANXA1 in VZ cells (h). Data in g and h from n = 9 and n = 10 human tissues between 14 and 19 PCW, respectively. 200 to 500 cells evaluated per brain. (i) Summary of oRG and vRG cell type markers identified in this study and overlap with existing RG and IPC markers.
Discussion
FRISCR is the first method, to our knowledge, that enables targeted mRNA purification and single-cell transcriptomic profiling of fixed cells that matches data generated from live-sorted cells. FRISCR has three important advantages over current methods for single-cell profiling. First, it allows enrichment of low frequency cell populations without live-cell markers. Without such enrichment, we would have had to profile 20,000 cortical cells in an unbiased way to obtain data for 200 SP cells. Such enrichment is particularly valuable for analyzing primary human tissues that are genetically inaccessible, and where live-cell markers are unknown or incompletely validated. Second, intracellular markers can be multiplexed with genetic reporters or other marking strategies to prospectively isolate and profile precisely defined cell types. Third, the FRISCR procedure allows fixed cells to be stored indefinitely at −80°C (and easily transported), unlike current protocols that require sorting of cells on the same day as tissue harvesting. These advantages make FRISCR an important new tool to investigate cellular heterogeneity at the single-cell level and can be easily implemented by labs already conducting single-cell profiling. Future optimizations of FRISCR will streamline sample processing, could allow both genome and transcriptome sequencing from the same fixed and sorted cell25, and allow single-cell profiling of cells marked by FISH probes15,26.
We demonstrated that FRISCR can be applied to investigate the composition of human neocortical progenitors. Several studies have analyzed gene expression patterns for differential expression or co-expression in microdissected germinal zones including the oSZ10,11,27. Although these studies revealed new RG gene networks, they did not reveal distinct markers of oRGs, likely because those cell types are a minority of the total population of cells sampled. Two recent studies have set out to profile human RG progenitors at the single-cell level. To enrich for live cortical progenitors, the first study sorted cells based on a combination of CD133 expression and DiI labeling to mark apical (including vRGs) and basal progenitors (excluding vRGs and including oRGs)12. ARHGAP11B was identified and shown to regulate human progenitor dynamics. However only their apical progenitors expressed PAX6 and both populations expressed high levels of EOMES. Thus their apical compartment likely contained IPCs and RGs, while their basal compartment likely lacks RGs due to the lack of PAX6. We did not detect ARHGAP11B expression in RG or IPC cells, indicating this gene likely plays a role in different cell types or stages. The second study used differential CD133, GLAST, and CD15 staining to enrich and profiled vRGs or oRGs13. NEUROG2 was identified as a regulator of oRG cells13. High EOMES expression was detected in both vRGs and oRGs similar to the first study. We detect only low expression of NEUROG2 in our RG cells and much higher expression in our IPCs, suggesting that their population of oRGs best corresponds to our IPCs which also express HES6 and EOMES. These studies did not detect our validated and novel vRG- or oRG-specific markers. Together these observations strongly suggest our study is the first to isolate and accurately profile these cells transcriptomically at the single-cell level.
Our results lay a foundation to uncover the developmental potential and molecular control of human RG diversity, maturation, and fate. Using our technique, we discovered validated markers for human vRGs and oRGs, some of which are suggestive of molecular control of RG self-renewal and differentiation. HOPX, the marker for oRGs and a transcriptional regulator, shows expression that is divergent between mouse and human developing cortex24. Thus, understanding the role of HOPX may illuminate differences between mouse and human cortical development. HOPX is expressed in adult neurogenic progenitors in the hippocampus as well as self-renewing multipotent progenitors in the intestine, skin, and certain cancers28–32. In these contexts HOPX functions by suppressing immediate early response genes32,33. Thus, the expansion of the human oSZ compartment could involve the reuse of existing mechanisms that control stem cell homeostasis in different tissues. Indeed, HOPX was recently shown to mark restricted cardiomyocyte progenitors, and to modulate their fate through the integration of niche BMP and WNT signals34, pathways which are also thought to underpin corticogenesis27,35. Building on our findings future work can focus on characterizing the fates of mid-gestation vRG and oRG cells and determining whether they maintain neurogenic potential or have become glial restricted8,36,37. Unraveling the fates of these progenitors and their regulatory mechanisms is central to understanding how the human brain has evolved.
Online Methods
Cell isolation from fetal cortex
We acquired tissue from the Birth Defects Research Laboratory (BDRL) at the University of Washington, who obtained appropriate written informed consent and provided available non-identifying information for each sample. The Human Subjects Division at the University of Washington Tissue approved tissue acquisition, which was performed according to the requirements of the Uniform Anatomical Gift Act and National Organ Transplant Act for the acquisition of human tissue for biomedical research purposes. Fetal age was determined by foot length and date of last menses.
We divided cortical pieces into one half for fixation, sectioning, and ICC staining, and the other half for cell isolation. For dissociation, we minced the tissue into small pieces (approx. 0.25 – 0.5 mL volume) with #5 forceps (Fine Science Tools) in Ca2+- and Mg2+-free HBSS (14175-095, Life Technologies). We treated minced pieces with 2 mL trypsin solution for 20 min at 37°C (Ca2+- and Mg2+-free HBSS, 10 mM HEPES, 2 mM MgCl2, 0.25 mg/ml bovine pancreatic trypsin (EMD Millipore), 10 µg/mL DNase I (Roche), pH 7.6). We quenched digestion with 6 mL of ice-cold Quenching Buffer (440 mL Leibovitz L-15 medium, 50 mL water, 5 mL 1M HEPES pH 7.3–7.4, 5 mL 100× Pen-Strep, 2 mg/mL bovine serum albumin [A7030, Sigma], 100 µg/mL trypsin inhibitor [T6522, Sigma], 10 µg/mL DNase I, 100 nM TTX [Tocris, #1069], 20 µM DNQX [Tocris #0189], and 50 µM DL-AP5 [Tocris #3693]). We then pelleted the samples (220×g, 4 min, 4°C) and resuspended with 1 mL of quenching buffer and triturated on ice with a P1000 pipette set to 1 mL, using 25 gentle cycles up and down without forming bubbles. We then diluted the cell suspension to 30 mL in Staining Medium (440 mL Leibovitz L-15 medium, 50 mL water, 5 mL 1M HEPES pH 7.3–7.4, 5 mL 100× Pen-Strep, 20 mL 77.7 mM EDTA pH 8.0 [prepared from Na2H2EDTA], 1 g bovine serum albumin, 100 nM TTX, 20 µM DNQX, and 50 µM DL-AP5), filtered through a 45 micron cell filter, pelleted (220×g, 10 min, 4°C), resuspended in 5 mL staining medium, and counted on a hemocytometer (typically ~30–50 million live cells isolated per cortical piece at ~50% viability).
Cell isolation from culture
We maintained H1 hESCs (WiCell) on Matrigel (Corning) in mTESR1 media (StemCell Technologies), authenticated them by karyotype analysis (Cell Line Genetics), and periodically tested them for sterility and mycoplasma contamination (IDEXX BioResearch). We dissociated adherent cultures with StemPro Accutase Cell Dissociation Reagent (Life Technologies), and then centrifuged the cells (220×g, 3 min) and the dissociation solution was removed.
FRISCR
Cell wash
We washed then resuspended the cells in RNase-free Staining Buffer (SB) (1×PBS pH 7.4, 1% RNase-free BSA [Gemini Bioproducts], and 0.0025% RNasin Plus [Promega]). We placed cells on ice until fixation or sorting.
Fixation
We fixed the single cell suspension with 4% PFA (Electron Microscopy Sciences) in PBS on ice for 15 minutes, then pelleted (335×g, 3 min, 4°C), washed once with 1 mL SB, then resuspended in SB at 10 million cells/mL, and froze the cells at −80°C in aliquots.
Permeabilization and staining
We thawed and permeabilized the cells by resuspending and incubating for 10 minutes on ice in 1×PBS, 0.1% Triton X-100 (Sigma), 1%BSA, 0.0025% RNasin Plus. We incubated one million cells in staining buffer (SB) (1×PBS, 1%BSA, 0.0025% RNasin Plus) with primary antibodies for 30 min at 4°C. Antibodies used were: Alexa488 or PE-conjugated anti-PAX6 (O18-1330; BD Biosciences), PE-conjugated anti-DCX (30/Doublecortin; BD Biosciences), PerCP-Cy5.5-conjugated anti-SOX2 (O30-678; BD Biosciences), eFluor660-conjugated anti-EOMES (WD1928; eBioscience), rabbit anti-CYR61 (D4H5D; Cell Signaling Technology) followed by PE-conjugated goat anti-rabbit IgG (Life Technologies). We washed cells in SB, resuspended in SB containing 1 µg/mL DAPI (Life Technologies), and filtered prior to sorting.
Single-cell sorting
We carried out cell sorting on a BD FACS ARIA-II SORP (BD Biosciences) using a 130 µm nozzle. We sorted single cells into strip tubes containing 5 µL of PKD Buffer (Qiagen) with 1:16 Proteinase K Solution (Qiagen) and ERCC spike-in synthetic RNAs (Life Technologies).
Cell lysis, reverse-crosslinking, and RNA purification
To lyse cells and purify RNA with dT25 beads, we thawed samples at 25°C – room temperature (RT), mixed, and then incubated at 56°C for 1 h in a thermal cycler with the lid set at 66°C. We vortexed cells for 10 sec, spun down, and placed on ice. We prepared oligo dT25 magnetic beads (Life Technologies) with 3 washes of 1× Hybridization Buffer (2×SSPE, 0.05%Tween20, 0.0025% RNasin Plus) and then resuspended in half of the original volume of 2× Hybridization Buffer. 5 µL of washed dT25 beads (0.05 mg of beads) were used per reaction.
We added beads to reverse crosslinked samples and then heated to 56°C for 1 min, incubated at RT 10 min to allow mRNA hybridization, and then placed on ice. We washed beads two times in 100 µL of ice-cold Hybridization Buffer, followed by a subsequent wash using ice cold 1 × PBS, 0.0025% RNasin Plus. We removed PBS and added 2.8 µL of RNase-free water, then resuspended the beads and incubated the mixture at 80°C for two minutes to elute mRNA, then immediately pelleted on a room temperature magnet. We rapidly removed the supernatant containing mRNA and transferred to a new tube and stored at −80°C.
RNA extraction for populations of cells
Sorting
We sorted cells as described above. We sorted populations up to 1,000 cells into 1.5 mL tubes with 100 µL of PKD solution. We sorted 10,000 cell samples into 1.5 mL tubes containing 500 µL of SB, pelleted, and resuspended in 100 µL PKD solution.
Cell lysis, reverse-crosslinking, and RNA purification
We purified total RNA using either the standard live-cell methods from the RNeasy Micro Kit (RLT method) or a modified protocol for fixed cells using the miRNeasy FFPE Kit (Qiagen) (protease lysis, reverse-crosslinking method (PLRC)). For the PLRC method, we thawed cells in 100µL of PKD/Proteinase K solution at RT, mixed, and incubated at 56°C for 1 h. Samples were then centrifuged at 20,000×g for 20 min. We transferred the supernatant to a new tube and added 10 µL of DNase Booster Buffer and 10 µL of DNase I stock solution and mixed by inversion. Then we incubated samples 15 min at RT then added 320 µL of RBC buffer and 1120 µL of ethanol and mixed. Finally, we applied the samples to MinElute columns, washed twice with 500 µL RPE Buffer, and eluted RNA into 15 µL water. We evaluated and quantified RNA on a Bioanalyzer 2100 (Agilent) using the RNA Pico Kit. To normalize input, we used 1 µL directly from 1,000 cells, 1 µL of 10-fold diluted RNA from 10,000 cells, or 1 µL of 10-fold concentrated RNA from 100 cells (Savant DNA 120 SpeedVac).
SmartSeq2
We prepared sequencing libraries as previously reported19. After reverse transcription and template switching, we amplified cDNA with KAPA HotStart HIFI 2× ReadyMix (Kapa Biosystems) for 19 or 22 cycles for RNA from single hESC or cortical progenitor cells, respectively. We purified PCR products using Ampure XP beads (Beckman Coulter). We quantified cDNA using a High Sensitivity DNA Chip (Agilent) on a Bioanalyzer 2100, or with the Quant-iT PicoGreen dsDNA Assay Kit (Life Technologies) on an Enspire plate reader (PerkinElmer). We used 1 ng of cDNA to generate RNA-Seq libraries using the Nextera XT library prep system (Illumina). Single cells from primary tissue contained a lower amount of mRNA compared to H1 hESCs, requiring a reduction in ERCC spike-in RNAs by 10-fold and addition of three extra PCR cycles. We carried out sequencing of human cortical progenitors on Illumina MiSeq using 31 base paired-end reads. We carried out sequencing of hESCs on the HiSeq using 50 base paired-end reads. We saw few global differences in read statistics between MiSeq runs and HiSeq runs when samples were assessed on both instruments.
RNA-Seq data analysis
We aligned raw read data to GRCh37 (hg19) using the RefSeq annotation gff file downloaded on 4/23/2013. We performed transcriptome alignment first using RSEM38, then we aligned unmapped reads to hg19 using Bowtie39, and then we aligned remaining unmapped reads to the ERCC sequences. Using a custom script we calculated the read mapping % of each cell to mRNA (RefSeq), mitochondrial RNA (mtRNA), noncoding RNA (mRNA NC), ribosomal RNA (rRNA), genome, and ERCC RNA. We performed principal component and clustering analysis using transcripts per million (TPM) values (log2 transformed). We used only high variance genes with adjusted p value smaller than 0.05 and present (mapped reads > 0) in more than 10 cells for analysis. H1 hESC data are deposited at GEO with accession number GSE71858. RSEM-generated gene count and TPM data from primary fetal human tissue is supplied as supplemental data 1 and 2, and raw data will be provided upon request.
Computational analysis
Normalization
All gene expression heatmaps show data normalized across all samples for each gene or eigengene. Single-cell gene expression data and eigengene values are TPM, while microarray data from BrainSpan Atlas of the Developing Human Brain11 is Expression Value. Heatmap values show log2(TPM or Expression Value+1) − average TPM or Expression Value.
WGCNA analysis
We included only high variance genes based on DESeq2 with an adjusted P-value of <0.01 and expressed in at least five cells. We performed WGCNA clustering using soft power of four, cut height of 0.995, and minimum module size of ten. To filter gene modules, we used genes of each module to cluster cells into two clusters, and then evaluated to determine which of the module genes are differentially expressed between the two clusters. We removed a gene module if one of the two clusters contained less than five cells, or if the total differential score (−log10 adjusted P-value of differentially expressed genes) was less than 40. This filtering criterion eliminates gene modules driven by very small numbers of cells, and gene modules with little discriminating power. We also eliminated two gene modules corresponding to cell cycle states. We clustered cells using hclust based on filtered module genes and Ward’s distance measure. After the initial clusters were determined, we detected differentially expressed genes between every pair of cell clusters using the Bioconductor limma package. The union of all such genes was defined as cluster-specific markers. We then re-clustered the cells based on the marker genes using hclust to determine the final clusters.
Mutational analysis
To detect mutations in each sample, we filtered alignment bam files to exclude reads with more than two errors or ambiguous mapping (RSEM ZW score < 0.95). Variance calling was performed by comparing reads to the reference genome using samtools mpileup followed by bcftools call40. We filtered detected mutations to exclude mutations within three base pairs of insertion or deletion mutations, with a quality score of < 10, depth of < 5 or an alignment mapping quality (MQ) score of < 50, using bcftools. We defined common variants as nucleotide changes where the variant is shared by more than 10 samples, and by more than two thirds of all samples that pass the quality filter threshold. The mutation rate for each sample is defined as non-common variant mutations divided by the number of genomic positions that pass the above-mentioned filter.
GC and Chromosomal bias analysis
To assess the GC bias between TL and FRISCR prepared live and fixed single hESCs, we pooled genes into ten equally sized bins based on %GC content. For each bin, we calculated the fraction of the genes detected in each sample out of all annotated genes. To test for any chromosomal bias, we binned genes by their chromosomal locations. We calculated the fraction of the genes detected in each sample out of all annotated genes for each chromosome.
Human Atlas Comparisons
For the BrainSpan Atlas of the Developing Human Brain11 laser capture microdissection (LCM) microarray datasets eigengene expression was determined using the same gene modules described above. In both datasets we compared side-by-side a subset of genes differentially expressed between VZ, iSZ and oSZ and differentially expressed between cell clusters C and D (oRG cells) versus cell cluster E (vRG cells) (Fig. 5d). We determined the statistical significance of eigengene enrichment in human LCM data by unpaired ttest comparing the VZ and the oSZ.
Tissue immunocytochemistry
We fixed cortical tissue sections by immersion in 4% paraformaldehyde in PBS for 24 h at 4°C. We washed tissue in PBS, then transferred to and stored in 30% sucrose in PBS (Sigma) for 2 days (or until tissue sank). We embedded tissue in Tissue-Tek O.C.T (Sakura Finetek) and stored at –80°C. We cut 25 µm coronal cryosections and stored at –80°C. For staining we thawed slides and dried for 15 min at RT, washed briefly with PBS to remove OCT, and blocked in 5% goat or donkey serum (Jackson Immunoresearch), 0.1% Triton X-100 in PBS for 1 h at RT. We diluted primary antibodies in blocking solution and applied to slides 4–12 hrs rocking at RT. We next washed slides in PBS and incubated in secondary antibodies diluted 1:1000 plus 0.5 µg/mL DAPI (Life Technologies) in blocking solution for 1–3 hrs rocking at RT, then applied coverslips with Prolong Gold mounting media (Life Technologies). We used primary antibodies: goat anti-SOX2 (Santa Cruz Biotechnology; SC17320), mouse anti-SOX2 (BD Biosciences, 245610), rabbit anti-PAX6 (BioLegend; PRB-278P); mouse anti-EOMES (eBioscience; WD1928); rabbit anti-HOPX (Santa Cruz Biotechnology; FL-73), mouse anti-CRYAB (Abcam; 1B6.1-3G4); Rabbit anti-c-JUN (Cell Signaling Technology; 60A8), Goat anti-human Coagulation Factor III/Tissue Factor (F3) (R&D Systems; AF2339), mouse anti-Ki67 (BD Biosciences, B56), rabbit anti-Ki67 (Abcam, ab15580), mouse anti-phospho-vimentin (Enzo; 4A4), and mouse anti-Lipocortin-1 (ANXA1) (BioLegend; 74/3). Alexa 488-, 555-, and 647-conjugated secondary antibodies were from Life Technologies.
We acquired high-resolution confocal images on a Leica TCS SP8 confocal microscope. Representative maximum intensity projections were created from two to three adjacent optical sections from the middle of a z-stack. We acquired epifluorescence images of the full cortical thickness on a Nikon Eclipse Ti with a motorized stage using a 10× objective, and we stitched together images using MetaMorph software. Brightness and contrast adjustments were made using Adobe Photoshop CS6 and we assembled figures using Adobe Illustrator CS6.
Statistics
Sample inclusion/exclusion
For analysis of RNA from populations of hESCs (Fig. 2, Supplementary Fig 1a), we sorted four cell samples per condition/number on three independent days (12 total samples per condition/number) for the 10,000 and 1,000 cell experiments, and we conducted two independent experiments for the 100 cell experiment (eight total samples per condition/number). Two outliers from both 1,000 hESC Live/PLRC and Fixed/PLRC samples were excluded from analysis due to process failure and no observed RNA. The Bioanalyzer was unable to call a RIN for one 10,000 cell Fixed/RLT sample, and one 100 cell Live/PLRC sample; and one 100 cell Live/PLRC, and Fixed/PLRC samples were lost due to mechanical failure.
For sequencing single hESCs (Fig. 3, Supplementary Figs. 2 and 3), we sorted eight individual hESCs on two days of experiments for each condition (16 cells total per condition). Library generation failed for some cells, as judged by insufficient cDNA for tagmentation (0.1 ng/µL) yielding n = 12–15 cells per condition (Supplementary Fig. 2b). We processed cells with sufficient cDNA into libraries for sequencing by tagmentation, however, samples from only one experiment of Fixed/TL samples were processed for sequencing, and we used the entire sample for library generation due to low cDNA yield. We included all sequenced single hESCs for analysis after subsampling to five million reads per cell. Importantly, the failure rate of live cells verses fixed cells is comparable.
For sequencing single human cortex progenitors (Figs. 4 and 5, Supplementary Figs. 5–8), we sorted 207 total SP cells from across four brains and 48 single SPE cells from across two brains for library preparation and all were sequenced. Of these, cells with more than 10,000 mapped reads, >20% reads mapping to mRNA, and detectable GAPDH expression were included in further analysis (157 SP and 29 SPE cells analyzed), all reads are included in analysis.
For analysis of human cortex by ICC, in Fig. 6e we stained six brains and excluded one due to poor staining (n = 5 total analyzed), in Fig. 6f we stained six brains and analyzed all, in Fig. 6g we stained ten brains and excluded one due to poor staining (n = 9 total analyzed), in Fig. 6h we stained nine brains and analyzed all, in Supplementary Fig. 12a we stained four brains and excluded one due to poor staining (n = 3 total analyzed), and in Supplementary Fig. 12b–c we stained four brains and analyzed all.
Power analysis and statistical tests
In Fig. 2b–c prospective power for the comparisons was 1.0 given effect size = 5*SD (~2-fold change), n = 28 total samples, and α = .05.
In Fig. 3d we used one-way ANOVA followed by Tukey’s post-hoc test for the indicated comparisons. Data within each group followed normal distributions as indicated by Shapiro-Wilk test. Prospective power for the comparisons were 0.99–1.0 given effect size = 1–9*SD (~2-fold changes), n = 44 total samples, and α = .05.
In Fig. 6e, we used Wilcoxon rank sum test because SOX2+HOPX− data did not follow a normal distribution by the Shapiro-Wilk test. Prospective power for this comparison was 0.98 given effect size = 3*SD (~2-fold change), n = 5 brains per group, and α = .05.
For Fig. 6f prospective power for the comparisons was 1.0 given effect size = 5*SD (~2-fold change), n = 5 brains per group, and α = .05. We conducted all power analyses using G*Power41.
Supplementary Material
Acknowledgments
We wish to thank the Allen Institute founders, P. G. Allen and J. Allen, for their vision, encouragement and support. We wish to thank A. Bernard and E. Lein for assistance with specimen procurement, J. Phillips for technical core support and leadership, A-R. Krostag for assistance in SmartSeq2 methods, Y. Wang and WiCell, the sources of the H1 hESCs, C. Thompson for program management, B. Tasic, T. Nguyen, and V. Menon for technical and scientific assistance, J. Miller for assistance in analysis of BrainSpan Atlas of the Developing Human Brain, and T. Bakken for assistance with the National Institutes of Health (NIH) Blueprint Non-Human Primate Atlas. Human primary samples were received from the “Laboratory of Developmental Biology”, supported by NIH Award Number 5R24HD000836 from the Eunice Kennedy Shriver National Institute of Child Health and Human Development. SJ, AMD and SR were supported in part by the NIH Directors Pioneer Award 5DP1MH099906-03 and National Science Foundation grant PHY-0952766.
Footnotes
Accession codes
Gene Expression Omnibus: GSE71858 (H1 hESC data). dbGaP: phs001016.v1.p1 (human single-cell data). Data from donor H14.4010 is available upon request.
Author contributions
ERT established the FRISCR based on prior methods from AMD and SJ, and conducted all FRISCR RNA-Seq experiments. Primary tissues was harvested and analyzed by JKM, RDH and SIS. H1 hESCs were cultured and provided by AMN and all sorting was done by NVS. ZY conducted all RNA-Seq primary analysis and computational analysis of gene expression. BPL, ERT, JKM and SR conceived all experiments and BPL, JKM and SR wrote the manuscript.
Statement of Competing Financial Interests
The authors have filed a provisional patent for the FRISCR method US Serial No. 62/140,438.
References for main text
- 1.Betizeau M, et al. Precursor diversity and complexity of lineage relationships in the outer subventricular zone of the primate. Neuron. 2013;80:442–457. doi: 10.1016/j.neuron.2013.09.032. [DOI] [PubMed] [Google Scholar]
- 2.Borrell V, Reillo I. Emerging roles of neural stem cells in cerebral cortex development and evolution. Developmental neurobiology. 2012;72:955–971. doi: 10.1002/dneu.22013. [DOI] [PubMed] [Google Scholar]
- 3.Dehay C, Kennedy H, Kosik, Kenneth S. The Outer Subventricular Zone and Primate-Specific Cortical Complexification. Neuron. 2015;85:683–694. doi: 10.1016/j.neuron.2014.12.060. [DOI] [PubMed] [Google Scholar]
- 4.Florio M, Huttner WB. Neural progenitors, neurogenesis and the evolution of the neocortex. Development. 2014;141:2182–2194. doi: 10.1242/dev.090571. [DOI] [PubMed] [Google Scholar]
- 5.Lui JH, Hansen DV, Kriegstein AR. Development and evolution of the human neocortex. Cell. 2011;146:18–36. doi: 10.1016/j.cell.2011.06.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rakic P. Evolution of the neocortex: a perspective from developmental biology. Nature reviews. Neuroscience. 2009;10:724–735. doi: 10.1038/nrn2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fietz SA, et al. OSVZ progenitors of human and ferret neocortex are epithelial-like and expand by integrin signaling. Nature neuroscience. 2010;13:690–699. doi: 10.1038/nn.2553. [DOI] [PubMed] [Google Scholar]
- 8.Hansen DV, Lui JH, Parker PR, Kriegstein AR. Neurogenic radial glia in the outer subventricular zone of human neocortex. Nature. 2010;464:554–561. doi: 10.1038/nature08845. [DOI] [PubMed] [Google Scholar]
- 9.Geschwind DH, Rakic P. Cortical evolution: judge the brain by its cover. Neuron. 2013;80:633–647. doi: 10.1016/j.neuron.2013.10.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fietz SA, et al. Transcriptomes of germinal zones of human and mouse fetal neocortex suggest a role of extracellular matrix in progenitor self-renewal. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:11836–11841. doi: 10.1073/pnas.1209647109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Miller JA, et al. Transcriptional landscape of the prenatal human brain. Nature. 2014;508:199–206. doi: 10.1038/nature13185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Florio M, et al. Human-specific gene ARHGAP11B promotes basal progenitor amplification and neocortex expansion. Science. 2015;347:1465–1470. doi: 10.1126/science.aaa1975. [DOI] [PubMed] [Google Scholar]
- 13.Johnson MB, et al. Single-cell analysis reveals transcriptional heterogeneity of neural progenitors in human cortex. Nature neuroscience. 2015;18:637–646. doi: 10.1038/nn.3980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hrvatin S, Deng F, O'Donnell CW, Gifford DK, Melton DA. MARIS: method for analyzing RNA following intracellular sorting. PloS one. 2014;9:e89459. doi: 10.1371/journal.pone.0089459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Klemm S, et al. Transcriptional profiling of cells sorted by RNA abundance. Nature methods. 2014;11:549–551. doi: 10.1038/nmeth.2910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Molyneaux BJ, et al. DeCoN: genome-wide analysis of in vivo transcriptional dynamics during pyramidal neuron fate selection in neocortex. Neuron. 2015;85:275–288. doi: 10.1016/j.neuron.2014.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pan Y, Ouyang Z, Wong WH, Baker JC. A new FACS approach isolates hESC derived endoderm using transcription factors. PloS one. 2011;6:e17536. doi: 10.1371/journal.pone.0017536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pechhold S, et al. Transcriptional analysis of intracytoplasmically stained, FACS-purified cells by high-throughput, quantitative nuclease protection. Nature biotechnology. 2009;27:1038–1042. doi: 10.1038/nbt.1579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Picelli S, et al. Smart-seq2 for sensitive full-length transcriptome profiling in single cells. Nature methods. 2013;10:1096–1098. doi: 10.1038/nmeth.2639. [DOI] [PubMed] [Google Scholar]
- 20.Yang L, Duff MO, Graveley BR, Carmichael GG, Chen LL. Genomewide characterization of non-polyadenylated RNAs. Genome Biol. 2011;12:R16. doi: 10.1186/gb-2011-12-2-r16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pollen AA, et al. Low-coverage single-cell mRNA sequencing reveals cellular heterogeneity and activated signaling pathways in developing cerebral cortex. Nature biotechnology. 2014;32:1053–1058. doi: 10.1038/nbt.2967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang B, Horvath S. A general framework for weighted gene co-expression network analysis. Stat Appl Genet Mol Biol. 2005;4 doi: 10.2202/1544-6115.1128. Article17. [DOI] [PubMed] [Google Scholar]
- 23.Smart IH, Dehay C, Giroud P, Berland M, Kennedy H. Unique morphological features of the proliferative zones and postmitotic compartments of the neural epithelium giving rise to striate and extrastriate cortex in the monkey. Cerebral cortex. 2002;12:37–53. doi: 10.1093/cercor/12.1.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Muhlfriedel S, Kirsch F, Gruss P, Stoykova A, Chowdhury K. A roof plate-dependent enhancer controls the expression of Homeodomain only protein in the developing cerebral cortex. Developmental biology. 2005;283:522–534. doi: 10.1016/j.ydbio.2005.04.033. [DOI] [PubMed] [Google Scholar]
- 25.Macaulay IC, et al. G&T-seq: parallel sequencing of single-cell genomes and transcriptomes. Nature methods. 2015;12:519–522. doi: 10.1038/nmeth.3370. [DOI] [PubMed] [Google Scholar]
- 26.Bushkin Y, et al. Profiling T cell activation using single-molecule fluorescence in situ hybridization and flow cytometry. Journal of immunology. 2015;194:836–841. doi: 10.4049/jimmunol.1401515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lui JH, et al. Radial glia require PDGFD-PDGFRbeta signalling in human but not mouse neocortex. Nature. 2014;515:264–268. doi: 10.1038/nature13973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.De Toni A, et al. Regulation of survival in adult hippocampal and glioblastoma stem cell lineages by the homeodomain-only protein HOP. Neural development. 2008;3:13. doi: 10.1186/1749-8104-3-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shin J, et al. Single-Cell RNA-Seq with Waterfall Reveals Molecular Cascades underlying Adult Neurogenesis. Cell stem cell. 2015;17:360–372. doi: 10.1016/j.stem.2015.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Takeda N, et al. Hopx expression defines a subset of multipotent hair follicle stem cells and a progenitor population primed to give rise to K6+ niche cells. Development. 2013;140:1655–1664. doi: 10.1242/dev.093005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Takeda N, et al. Interconversion between intestinal stem cell populations in distinct niches. Science. 2011;334:1420–1424. doi: 10.1126/science.1213214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yamashita K, Katoh H, Watanabe M. The homeobox only protein homeobox (HOPX) and colorectal cancer. International journal of molecular sciences. 2013;14:23231–23243. doi: 10.3390/ijms141223231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Katoh H, et al. Epigenetic silencing of HOPX promotes cancer progression in colorectal cancer. Neoplasia. 2012;14:559–571. doi: 10.1593/neo.12330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jain R, et al. HEART DEVELOPMENT. Integration of Bmp and Wnt signaling by Hopx specifies commitment of cardiomyoblasts. Science. 2015;348 doi: 10.1126/science.aaa6071. aaa6071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chenn A, Walsh CA. Regulation of cerebral cortical size by control of cell cycle exit in neural precursors. Science. 2002;297:365–369. doi: 10.1126/science.1074192. [DOI] [PubMed] [Google Scholar]
- 36.Gertz CC, Lui JH, LaMonica BE, Wang X, Kriegstein AR. Diverse behaviors of outer radial glia in developing ferret and human cortex. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2014;34:2559–2570. doi: 10.1523/JNEUROSCI.2645-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Noctor SC, Martinez-Cerdeno V, Ivic L, Kriegstein AR. Cortical neurons arise in symmetric and asymmetric division zones and migrate through specific phases. Nature neuroscience. 2004;7:136–144. doi: 10.1038/nn1172. [DOI] [PubMed] [Google Scholar]
Online Methods References
- 38.Li B, Dewey CN. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics. 2011;12:323. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li H. A statistical framework for SNP calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data. Bioinformatics. 2011;27:2987–2993. doi: 10.1093/bioinformatics/btr509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Faul F, Erdfelder E, Lang AG, Buchner A. G*Power 3: a flexible statistical power analysis program for the social, behavioral, and biomedical sciences. Behavior research methods. 2007;39:175–191. doi: 10.3758/bf03193146. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.