Abstract
Ovarian cancer is one of the leading female cancers in the United States. Challenges remain in early diagnosis of this deadly disease. Matrix metalloproteinases (MMPs) family genes are paradoxically involved in cancer promotion and suppression. We hypothesize that genetic variants in MMP genes are associated with ovarian cancer development, so they could be potential markers for ovarian cancer diagnosis and prognosis. In this study of 417 ovarian cancer cases and 417 healthy controls, we genotyped a comprehensive panel of 266 single nucleotide polymorphisms (SNPs) in 23 MMP genes and analysed their associations with ovarian cancer risk, overall survival and treatment response in ovarian cancer cases who received platinum-based chemotherapy with surgery. In the analysis on 339 Caucasian cases and 349 Caucasian controls, 4 SNPs were significantly associated with cancer risk. The most significant association was observed for rs2292730 (OR = 2.03, 95% CI = 1.39–2.96, P = 0.0002). Classification and regression tree analysis identified four terminal nodes with differential risk of ovarian cancer. Thirty-four SNPs were significantly associated with overall survival and four of which showed significant association with response to chemotherapy. Unfavourable genotype analysis of top SNPs on overall risk of death showed significant gene-dosage effect, survival tree analysis differentiated patients into distinct risk groups based on their genetic profiles with median survival times (MSTs) ranging from 17.7 to 151.7 months. In conclusion, our results suggest that genetic variants in MMP pathway genes may modulate the risk and clinical outcomes of ovarian cancer, both individually and jointly.
Keywords: MMP, SNP, ovarian cancer
INTRODUCTION
Ovarian cancer is among the 10 leading cancers in women in the United States. In 2013, there are an estimated 22,240 new cases and 14,030 new deaths [1]. Ovarian cancer has the highest mortality rate among all the gynecologic cancers, largely because it is often at an advanced stage by the time of diagnosis. Symptoms reported from ovarian cancer patients include pelvic pain, abnormal vaginal bleeding, and involuntary weight loss, as well as non-specific symptoms such as abdominal pain, back pain, urinary urgency, and fatigue, and etc., which contribute to the difficulties of early diagnosis and the resulting low survival rate [2–5]. Diagnosed patients are usually treated with surgical resection followed by platinum-based chemotherapy [6]. Despite continuous advances in ovarian cancer research, diagnosis, and clinical treatment during the past 30 years [7], no cost-effective screening strategy for early-stage ovarian cancer, which would significantly increase the survival rate, is available. Improved understanding of genetic risk factors and the identification of novel biomarkers will enable a tailored approach for personalized prevention and treatment strategies to be developed for ovarian cancer.
In general, risk factors of ovarian cancer include aging, family history, infertility, hormonal therapy, and etc [8,9]. Germline mutations in BRCA gene (the gene that produces breast cancer susceptibility protein) are significantly associated with hereditary forms of ovarian cancer. Female carriers of germline BRCA1 mutations have an approximately 60% lifetime risk of developing ovarian cancer; carriers of BRCA2 mutations have a moderately increased risk [10]. In addition, the most common histological type of ovarian cancer, serous carcinoma, is reported to be associated with a particularly high frequency of molecular events involving BRCA pathway dysfunction, such as TP53 mutation, chromosomal instability, distinct molecular subtypes, and DNA copy number-driven changes in gene expression [11].
Matrix metalloproteinases (MMPs) are a family of more than 20 zinc-dependent enzymes known to degrade extracellular matrix and basement membrane components [12]. MMPs are important in the development of a variety of inflammatory, neurode-generative, and malignant diseases [13–17]. Previously, they were considered exclusively up-regulated in tumor growth, invasion, and metastasis [18–20]; however, more recent studies have suggested that MMPs, as well as tissue inhibitors of metalloproteinases (TIMPs), are paradoxically involved in both cancer-promoting and cancer-suppressing functions [21]. In a recent report, Delassus et al. [22] presented a detailed map of the known up- and down-regulatory signaling pathways from common cancer progression suppressors to MMPs. They reported that there were increased expressions of MMPs interlinked with known suppressors in the tested ovarian cancer cell line. Specifically, MMP2 was up-regulated by E-cadherin and p53, MMP14 by fibulin ID and Raf kinase inhibitory protein, MMP16 by fibulin ID and phosphatase and tensin homolog, and MMP25 by phosphatase and tensin homolog [22].In addition, studies on ovarian cancer tissue samples demonstrated the positive association of MMP2 and MMP9 high expression with disease aggressiveness, and that MMP 9 mediates EGFR-dependent E-cadherin loss in ovarian carcinoma cells [23,24].
Genetic polymorphisms in the promoter regions from selected MMP have been investigated in three Asian populations—two Chinese populations and one Korean population [25–27]. Study on large scale genetic variants covering a wide scope of MMP family and inhibitor genes has not been reported. In our study, we carefully selected 266 single-nucleotide polymorphisms (SNPs) from the complete gene region and from the flanking regions of 22 MMP genes and the TIMP1 gene. The purpose was to evaluate their association with ovarian cancer risk, disease-specific survival, and treatment response.
MATERIALS AND METHODS
Study Population and Data Collection
In this study, we enrolled 417 patients with newly diagnosed and histologically confirmed ovarian cancer at The University of Texas MD Anderson Cancer Center from August 1991 to January 2009. We define control subjects as individuals in normal health conditions without prior history of cancer, excluding non-melanoma skin cancer. Controls were prospectively recruited in parallel with cases from the Kelsey-Seybold Clinic. Controls were matched to cases by age (±5yr), gender and ethnicity. Written consent forms were obtained from all subjects before interviews on demographic and epidemiologic information, including age, height, weight, ethnicity, alcohol consumption, and smoking history and status. Immediately after each interview, a 40 ml blood sample was drawn into heparinized tubes for lymphocyte isolation and DNA extraction. In this study, a patient’s treatment response was defined by whether there was evidence of residual disease as indicated by various clinical measures, such as positron emission tomography or computed tomography scans, second-look surgery, and post-chemotherapy CA-125 level. Patients who had died during the follow-up period were considered to have poor response. The study was approved by the MD Anderson institutional review board.
SNP Selection and Genotyping
SNP selections were based on previously established criteria [28] through which a customized panel of genes involved in cancer-related cellular pathways, including MMP pathways, was generated using the iSelect platform (Illumina, Inc., San Diego, CA). A priority score was assigned to each gene based on a literature review and a query of the Gene Ontology database (http://www.geneontology.org/). A total of 266 potentially functional and tagging SNPs from 22 genes in the MMP family and the TIMP1 gene were selected via database mining of the International HapMap Project [29] and dbSNP [30]. Tagging SNPs were defined as SNPs with an r2 threshold of 0.80 and a minor allele frequency (MAF) >0.01 in Caucasians. Potentially functional SNPs were chosen from both coding and regulatory regions such as promoter region, splicing site, 5′ untranslated region (UTR), and 3′ UTR. In addition, SNP selection covered flanking regions 10 kb upstream of the transcriptional start site and 10 kb downstream of the transcriptional end site of each gene. A complete set of SNPs was sent to Illumina technical support for iSelect Infinium BeadChip design. Genomic DNA was extracted from peripheral blood lymphocytes using the QIAmp DNA extraction kit (Qiagen, Hilden, Germany) and genotyped according to the manufacturer’s protocol using the Illumina BeadArray platform. Genotypes were automatically called using the BeadStudio software package provided by Illumina.
Statistical Analysis
Statistical analyses were performed using Inter-cooled STATA software, version 10. Student’s t-test was used to assess differences in continuous variables between cases and controls. Chi-square analysis was used to evaluate differences in patient characteristics and SNP genotypes. The chi-square test for Hardy–Weinberg equilibrium was applied to each SNP among the control subjects. For each SNP, three genetic models (dominant, recessive, and additive) were tested, and the most significant test among the three was used to report the statistical significance. In all statistical analysis, a P-value ≤ 0.05 was considered significant. To control for the effects of multiple testing, we also calculated the q value, a false discovery rate-adjusted P-value, for each SNP. SNPs in linkage disequilibrium were identified, and the one with the smallest P-value was included for further analysis. Internal validation of the results was performed a bootstrap resampling method. One hundred bootstrap samples were generated for single SNP analysis. Each time, a bootstrap sample was selected from the original dataset and the P-value was obtained for each SNP among the dominant, recessive and additive models. Haplotypes analysis was performed using the HelixTree software v6.4.3 (Golden Helix, Bozeman, MT) and were included in the analysis on top SNPs associated with overall ovarian cancer risk. For unfavourable analysis, 10 000 bootstrap samples were generated, and the bias-corrected bootstrap confidence intervals were reported.
The overall risk of ovarian cancer and likelihood of poor treatment response were estimated as odds ratios with 95% confidence intervals (CIs) for each SNP using unconditional multivariate logistic regression, adjusted for age. The overall risk of death was estimated as hazard ratios (HRs) and 95% CIs for each SNP using the Cox proportional hazards model, adjusted for age, clinical stage, treatment regimen, and histology. The cumulative effects of combined variants were estimated by counting the number of unfavorable genotypes identified from the main effects of a single SNP analysis. Unfavorable genotypes were pooled and categorized into three groups with low, medium, and high risk, with the group having the lowest risk being the reference group. Classification and regression tree (CART) analysis by HelixTree software, and survival tree (STREE) analysis by the STREE program (http://masal.med.yale.edu/stree/) were performed to identify higher order gene-gene interactions involved in determining the overall risk and survival of ovarian cancer, respectively. Kaplan–Meier plots and log-rank tests were applied to estimate differences in overall survival by genotypes from each SNP. Survival time was calculated from the start date of treatment to the date of death or last patient follow-up.
For functional validation, the publicly available web software Genevar [31] was utilized to investigate expression quantitative trait loci (eQTL) associations within the genetic regions of all the MMP pathway SNPs associated ovarian cancer risk, survival and treatment response. cis-eQTL associations were obtained from profiling data in four previous studies incorporated in Genevar [32–35]. The P value threshold for significant eQTL association was set to be 0.05. For SNPs not directly reported by the above mentioned studies, their proxy SNPs were utilized instead, which were determined by SNAP version 2.2, a web-based software that identifies proxy SNPs based on linkage equilibrium, physical distance and commercially available arrays [36].
RESULTS
Population Characteristics
In our study, we collected data for 417 cases and 417 controls with average ages of 60.73 ±10.36 and 60.30 ± 10.71 yr, respectively (Supplementary Table 1). Because of the small number of non-white participants, we limited our statistical analyses for overall risk assessment to the 339 white cases (81.3%) and 349 white controls (83.7%). Among these 339 patients, 317 (94%) had sufficient information about survival time to permit statistical analysis; their median survival time (MST) was 48.3 months, with 146 deaths (46%) and 152 recurrences (48%). Among the 304 patients for whom clinical stage information was available, 202 (66%) were categorized as having FIGO stage III disease, that is, ovarian cancer that has spread outside the pelvis to the abdomen and abdominal lymph nodes [37] (Supplementary Table 1). A total of 295 patients had sufficient follow-up data to allow analysis of treatment outcome; 96 (33%) of them showed no response to chemotherapy.
Association Between MMP SNPs and Overall Risk
Among the 266 SNPs we analyzed, 24 were found to be significantly associated with the risk of ovarian cancer. After adjusting for multiple comparisons using q value at 10% level, four SNPs remained significant, and they are MMP9: rs6094237, MMP20: rs2292730, rs12278250, and rs9787933 (Table 1). The top three SNPs were from MMP20. The most significant association was observed for SNP rs2292730, which resulted in an increased overall risk with an adjusted OR of 2.03 (95% CI, 1.39–2.96, P = 2.23 × 10−4), while MMP9: rs6094237, MMP20: rs12278250, and MMP20: rs9787933 all led to decreased risk of ovarian cancer, with adjusted ORs of 0.53 (95% CI, 0.35–0.79), 0.50 (95% CI, 0.32–0.76), and 0.50 (95% CI, 0.34–0.75), respectively.
Table 1.
Genes and SNPs Associated With Overall Ovarian Cancer Risk
| Gene | SNP | Nucleotide change |
Position | MAF |
Genotypea (WW/WV/VV) |
ORb (95% CI) | P | Statistical model |
Bootstrapc |
||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Case | Control | Case | Control | P <0.05 | P <0.01 | P <0.001 | |||||||
| MMP9 | rs6094237 | T > A | 5’ FGR | 0.366 | 0.441 | 135/160/44 | 118/154/77 | 0.53 (0.35–0.79) | 1.88 × 10−3 | Recessive | 100 | 96 | 25 |
| MMP20 | rs2292730 | G > A | Intron | 0.501 | 0.434 | 91/156/92 | 100/195/54 | 2.03 (1.39–2.96) | 2.23 × 10−4 | Recessive | 100 | 100 | 93 |
| rs12278250 | T > A | 3’ FGR | 0.060 | 0.103 | 302/33/4 | 279/66/3 | 0.50 (0.32–0.76) | 1.41 × 10−3 | Dominant | 100 | 97 | 40 | |
| rs9787933 | G > C | Intron | 0.070 | 0.120 | 294/41/3 | 269/76/4 | 0.50 (0.34–0.75) | 8.49 × 10−4 | Dominant | 100 | 100 | 52 | |
| MP20 Haplotyped | |||||||||||||
| W_W_W | 281 | 301 | 1 (reference) | ||||||||||
| W_W_V | 5 | 13 | 0.41 (0.15–1.17) | 9.72 × 10−2 | |||||||||
| W_V_W | 325 | 294 | 1.18 (0.94–1.48) | 0.147 | |||||||||
| W_V_V | 4 | 0 | N.A.e | ||||||||||
| V_W_W | 14 | 10 | 1.5 (0.65–3.42) | 0.340 | |||||||||
| V_W_V | 27 | 62 | 0.47 (0.29–0.75) | 1.85 × 10−3 | |||||||||
Genotype distribution, WW, wild type homozygous; WV, heterozygous; W, variant homozygous.
Adjusted for age.
Internal validation of the results choosing from the best genetic model using bootstrap 100 times.
Haplotype in the order of rsl2278250, rs2292730, rs9787933, W = wild type allele, V = variant allele.
Not available due to small subgroup population.
Haplotype analysis was performed on the three MMP20 SNPs that presented significant association with ovarian cancer risk. Using the wild-type alleles of all three SNPs as the reference, the haplotype containing the variant alleles of SNP rs12278250 and rs9787933, and the wild-type allele of rs2292730 showed significant association with decreased risk of ovarian cancer (adjusted OR = 0.47, 95% CI = 0.29–0.75, P = 1.85 × 10−3).The haplotype containing only wild-type alleles of rs12278250 and rs2292730, and variant allele of rs9787933 also demonstrated borderline significance in association with decreased ovarian cancer risk (adjusted OR = 0.41, 95% CI = 0.15–1.17, P = 9.72 × 10−2).
Cumulative effect of the top four SNPs on ovarian cancer risk was analyzed by counting the number of unfavorable genotypes in each individual and a significant gene-dosage effect (Ptrend < 0.0001) was observed. Compared to patients having ≤ 1 of these unfavorable genotypes, those having 2 or 3 saw their risk of developing ovarian cancer more than doubled (OR, 2.37; 95% CI, 1.46–3.85); for patients having 4 of these unfavorable genotypes, their overall risk increased nearly five-fold (OR, 4.53, 95% CI, 2.53–8.13) (Ptrend = 2.45 × 10−7; Table 2).
Table 2.
SNPs Associated With Overall Ovarian Cancer Risk by Unfavorable Genotype Analysis
| No. of unfavorable genotypes |
Case (%) | Control (%) | ORa (95% CI) |
P-value | Bootstrap 95% CIb |
|---|---|---|---|---|---|
| 0–1 | 27 (8.0) | 66 (19.0) | 1 (reference) | ||
| 2–3 | 233 (68.9) | 240 (69.0) | 2.37 (1.46–3.85) | 4.48 × 10−4 | 1.47–4.04 |
| 4 | 78 (23.1) | 42 (12.1) | 4.53 (2.53–8.13) | 3.94 × 10−7 | 2.53–8.54 |
| Ptrend | 2.45 × 10−7 |
Adjusted for age.
Internal validation of the results using bootstrap 10000 times with bias-corrected model.
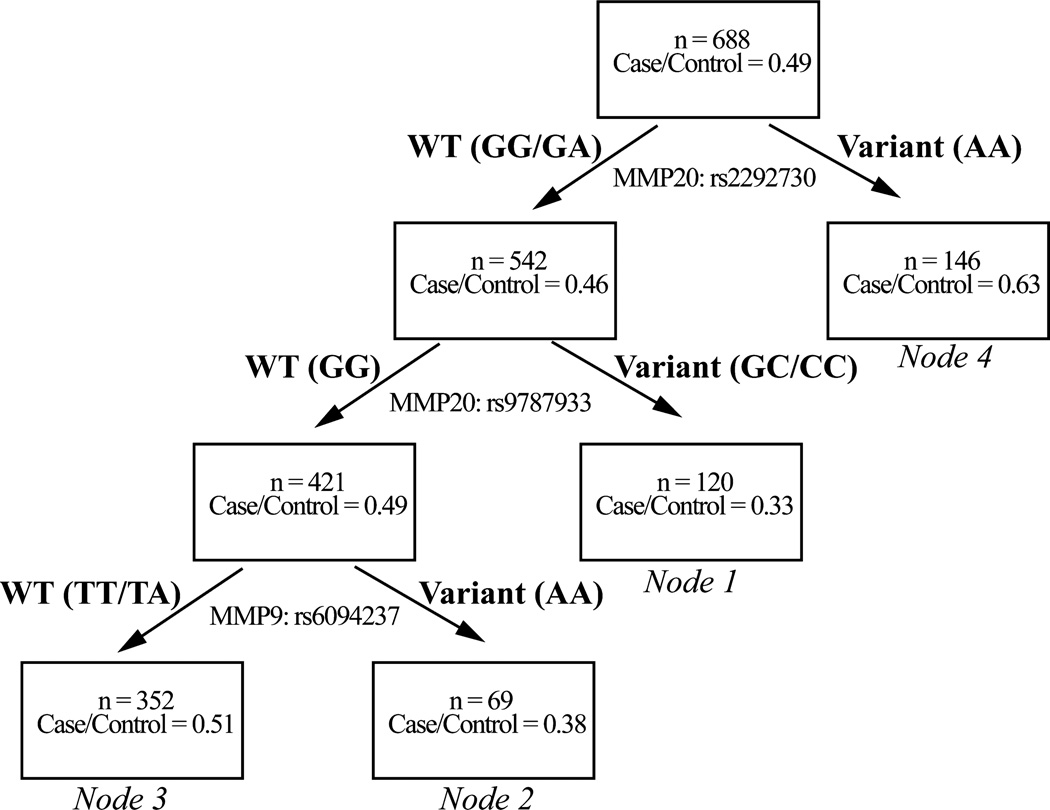
CART analysis was applied, and demonstrated higher order gene interactions among SNPs MMP20: rs2292730, MMP20:rs9787933, and MMP9:rs6094237 (Figure 1). Four terminal nodes with differential risk of ovarian cancer (percentage of cases in terminal nodes ranging from 33% to 63%) were also identified. The reference group (node 1) with the lowest risk was composed of subjects carrying the wild-type allele of MMP20:rs2292730 and the variant allele of MMP20: rs9787933. The initial split determined by MMP20: rs2292730 resulted in the group with the highest risk (node 4), indicating the dominant effect of this risk SNP in determining an individual’s risk for ovarian cancer.
Figure 1.
CART analysis showing interactions of risk-associated MMP SNPs in patients with ovarian cancer. Genotype(s) of each population are presented in parentheses. WT, wild-type.
Association Between MMP SNPs and Survival
In survival analysis, there were 42 SNPs significantly associated with overall survival, 34 of which remained significant after adjusting for multiple comparisons using q value at 10% level (Table 3). The most significant association was observed for SNP rs2239008 from MMP1 (HR = 3.10, 95% CI = 1.54–6.24, P = 1.57 × 10−3).
Table 3.
Genes and SNPs Associated With Ovarian Cancer Risk of Death
| Gene | SNP | Nucleotide change |
Position | MAF |
Genotypea (WW/WV/W) |
HRb (95% CI) | P | Statistical model |
Bootstrapc |
||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Dead | Alive | Dead | Alive | P < 0.05 | P < 0.01 | P < 0.001 | |||||||
| MMP1 | rs2239008 | G > A | 3′ UTR | 0.240 | 0.199 | 85/52/9 | 108/61/4 | 3.1 (1.545–6.24) | 1.57 × 10−3 | Recessive | 97 | 42 | 1 |
| rs17293761 | G > A | 3′ FGR | 0.110 | 0.085 | 116/28/2 | 143/27/1 | 1.84 (1.205–2.81) | 4.79 × 10−3 | Dominant | 97 | 27 | 1 | |
| MMP2 | rs2287074 | G > A | T-460 | 0.445 | 0.425 | 43/76/27 | 56/87/30 | 1.54(1.065–2.24) | 2.49 × 10−2 | Dominant | 80 | 3 | 0 |
| rs1005912 | T > A | 5′ FGR | 0.524 | 0.416 | 32/74/39 | 54/94/25 | 1.83 (1.255–2.68) | 1.97 × 10−3 | Recessive | 100 | 66 | 6 | |
| rs1005913 | A > C | 5′ FGR | 0.349 | 0.231 | 62/66/18 | 103/60/10 | 1.38 (1.075–1.78) | 1.38 × 10−2 | Additive | 94 | 8 | 0 | |
| rs1116195 | T > A | 5′ FGR | 0.479 | 0.384 | 35/82/29 | 61/91/21 | 1.33 (1.035–1.72) | 2.79 × 10−2 | Additive | 91 | 13 | 0 | |
| rs11076099 | G > A | 5′ FGR | 0.360 | 0.451 | 58/71/17 | 47/96/30 | 0.76 (0.585–0.98) | 3.55 × 10−2 | Additive | 74 | 3 | 0 | |
| MMP3 | rs3025090 | G > A | 5′ FGR | 0.086 | 0.147 | 121/25/0 | 124/47/2 | 0.62 (0.395–0.96) | 3.39 × 10−2 | Dominant | 37 | 0 | 0 |
| MMP8 | rs17099462 | C > A | 5′ FGR | 0.363 | 0.301 | 61/64/21 | 85/72/16 | 1.33 (1.035–1.70) | 2.57 × 10−2 | Additive | 51 | 0 | 0 |
| MMP9 | rs6073983 | T > A | 5′ FGR | 0.212 | 0.214 | 94/42/10 | 103/66/4 | 2.29 (1.155–4.54) | 1.84 × 10−2 | Recessive | 90 | 19 | 1 |
| MMP11 | rs738791 | G > A | Intron | 0.473 | 0.509 | 45/64/37 | 49/72/52 | 0.72 (0.575–0.90) | 4.05 × 10−3 | Additive | 100 | 45 | 1 |
| MMP12 | rs476185 | T > A | Intron | 0.318 | 0.254 | 64/71/11 | 94/70/9 | 1.68 (1.165–2.43) | 5.58 × 10−3 | Dominant | 100 | 69 | 5 |
| rs11225446 | C > A | 5′ FGR | 0.130 | 0.199 | 111/32/3 | 112/53/8 | 0.65 (0.445–0.96) | 3.13 × 10−2 | Dominant | 41 | 1 | 0 | |
| MMP13 | rs579627 | G > A | 5′ FGR | 0.212 | 0.145 | 93/44/9 | 129/38/6 | 1.49 (1.115–2.00) | 8.35 × 10−3 | Additive | 100 | 75 | 7 |
| rs10791602 | G > A | 5′ FGR | 0.317 | 0.249 | 69/60/16 | 98/64/11 | 1.48 (1.145–1.93) | 3.36 × 10−3 | Additive | 100 | 88 | 16 | |
| rs12577227 | G > A | 3′ FGR | 0.253 | 0.341 | 84/50/12 | 78/72/23 | 0.73 (0.565–0.96) | 2.20 × 10−2 | Additive | 88 | 10 | 0 | |
| MMP14 | rs12893368 | A > G | Intron | 0.209 | 0.130 | 89/53/4 | 130/41/2 | 1.52 (1.075–2.16) | 1.90 × 10−2 | Dominant | 95 | 22 | 0 |
| MMP16 | rs12114224 | C > A | Intron | 0.106 | 0.052 | 118/25/3 | 155/18/0 | 1.97 (1.285–3.04) | 1.98 × 10−3 | Dominant | 100 | 81 | 5 |
| rs17722347 | G > A | Intron | 0.092 | 0.046 | 120/25/1 | 157/16/0 | 1.78 (1.155–2.76) | 9.97 × 10−3 | Dominant | 98 | 27 | 0 | |
| rs7823185 | G > A | Intron | 0.116 | 0.052 | 11 5/28/3 | 155/18/0 | 1.69 (1.115–2.57) | 1.48 × 10−2 | Dominant | 99 | 29 | 1 | |
| rs13257117 | G > A | Intron | 0.178 | 0.176 | 97/46/3 | 116/53/4 | 1.57 (1.105–2.24) | 1.39 × 10−2 | Dominant | 98 | 26 | 0 | |
| rs2616487 | A > G | intron | 0.387 | 0.335 | 58/63/25 | 77/76/20 | 1.37 (1.075–1.75) | 1.13× 10−2 | Additive | 97 | 29 | 0 | |
| rs2664369 | A > C | 3′ FGR | 0.363 | 0.410 | 62/62/22 | 57/90/26 | 0.69 (0.495–0.97) | 3.26 × 10−2 | Dominant | 68 | 1 | 0 | |
| rs10090371 | C > A | Intron | 0.184 | 0.204 | 92/38/6 | 105/51/8 | 0.67 (0.465–0.97) | 3.56 × 10−2 | Dominant | 50 | 0 | 0 | |
| MMP20 | rs17098913 | G > A | Intron | 0.154 | 0.090 | 104/39/3 | 143/29/1 | 1.63 (1.125–2.37) | 1.03 × 10−2 | Dominant | 100 | 78 | 4 |
| rs1612069 | C > A | Intron | 0.462 | 0.390 | 44/69/33 | 63/85/25 | 1.49 (1.035–2.16) | 3.23 × 10−2 | Dominant | 66 | 1 | 0 | |
| rs10895324 | G > A | 5′ FGR | 0.411 | 0.454 | 51/70/25 | 55/79/39 | 0.77 (0.605–0.98) | 3.08 × 10−2 | Additive | 58 | 0 | 0 | |
| rs948138 | A > G | 5′ FGR | 0.472 | 0.425 | 44/65/36 | 58/83/32 | 1.36 (1.075–1.72) | 1.07 × 10−2 | Additive | 99 | 37 | 1 | |
| rs4554864 | G > A | 5′ FGR | 0.318 | 0.379 | 69/61/16 | 69/77/27 | 0.72 (0.565–0.93) | 1.25 × 10−2 | Additive | 95 | 11 | 0 | |
| rs11784408 | G > C | Intron | 0.339 | 0.425 | 64/65/17 | 58/83/32 | 0.65 (0.465–0.91) | 1.34 × 10−2 | Dominant | 100 | 31 | 1 | |
| MMP24 | rs2425019 | A > G | Intron | 0.425 | 0.486 | 46/76/24 | 47/84/42 | 0.75 (0.585–0.97) | 2.60 × 10−2 | Additive | 74 | 3 | 0 |
| MMP27 | rs11225389 | C > A | 5′ UTR | 0.260 | 0.213 | 81/51/12 | 104/61/6 | 2.00 (1.055–3.82) | 3.63 × 10−2 | Recessive | 59 | 0 | 0 |
| rs11607205 | C > A | Intron | 0.209 | 0.165 | 92/47/7 | 122/45/6 | 1.57 (1.105–2.23) | 1.26 × 10−2 | Dominant | 100 | 57 | 0 | |
| TIMP1 | rs5906435 | G > A | Intron | 0.301 | 0.396 | 68/68/10 | 59/91/23 | 0.49 (0.255–0.94) | 3.32 × 10−2 | Recessive | 32 | 0 | 0 |
Genotype distribution, WW = wild type homozygous, WV = heterozygous, W = variant homozygous.
Adjusted for age, clinical stage, chemotherapy regimen, and histology.
Internal validation of the results choosing from the best genetic model using bootstrap 100 times.
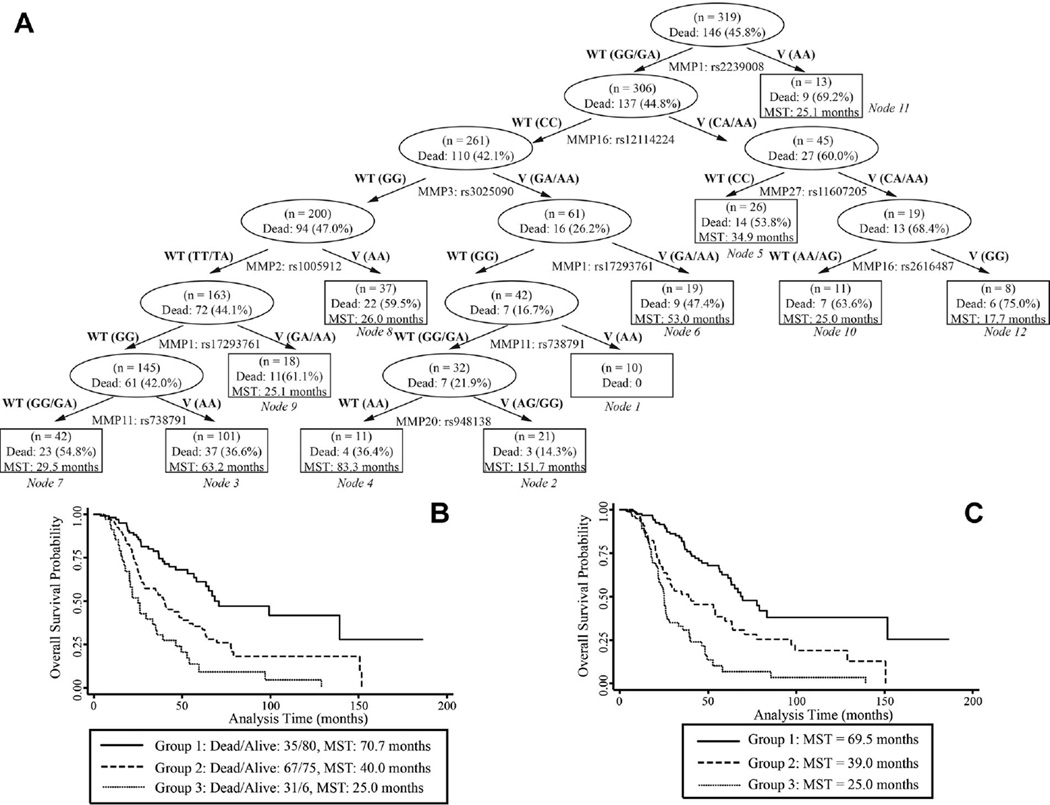
We also studied the cumulative effect of the top SNPs on overall survival. Compared to low risk group of individuals (≤11 unfavorable genotypes), patients in medium (12~18 unfavorable genotypes) high risk groups (≥ 19 unfavorable genotypes) were exposed to significantly increased risk of death (HR = 2.31, 95% CI = 1.51–3.51, and HR = 4.06, 95% CI = 2.45–6.73, respectively). The MST for patients having ≤11, 12–18, and ≥19 unfavorable genotypes were 70.7, 40.0, and 25.0 months, respectively (log rank P < 0.0001) (Figure 2B). In our survival tree analysis, 9 SNPs demonstrated interactions that led to 12 terminal nodes with MSTs ranging from 17.7 to 151.7 months (Figure 2A). These 12 nodes could be further differentiated into three distinct groups (Ptrend < 0.001), with MST of 69.5, 39.0, and 25.0 months for low-, medium-, and high-risk group, respectively (Figure 2C).
Figure 2.
(A) STREE analysis showing gene-gene interactions in the MMP family that regulated overall survival of patients with ovarian cancer between survival-associated SNPs; and Kaplan-Meier curves of survival times in patients with ovarian cancer (B) carrying unfavorable genotypes of survival-associated SNPs in the MMP pathway (median survival times of patients in groups 1, 2, and 3 (carrying 0–11, 12–18, and ≥19 unfavorable genotypes, respectively) are shown with a log-rank P-value of 8.22 × 10−9), and (C) in the 12 terminal nodes categorized into three groups as identified by STREE analysis-groups 1 (nodes 1–4), 2 (nodes 5–8), and 3 (nodes 9–12) with a log-rank P-value of 8.10 × 10−14. WT, wild-type; V, variant.
Association Between MMP SNPs and Response to Chemotherapy
In the evaluation of association between SNPs and chemotherapy response, 9 SNPs were removed due to their failure to pass Hardy–Weinberg equilibrium in controls. 19 SNPs were significantly associated with patients’ response to chemotherapy, and 4 of them also showed significant association with overall survival. After adjusting for multiple comparisons using q value at 10% level, rs7826929, a SNP in the intron region of MMP16 was the only one that remained significant. The variant genotype of rs7826929 increased the risk for poor response compared with the wild-type genotype by 3.3-fold (95% CI, 1.82–6.12), with P = 1 × 10−4 when using the dominant model. Unfavorable genotype analysis was not performed since there was only one SNP that reached statistical significance after adjusting for multiple comparisons.
cis-eQTL Association Between MMP Genes and SNPs
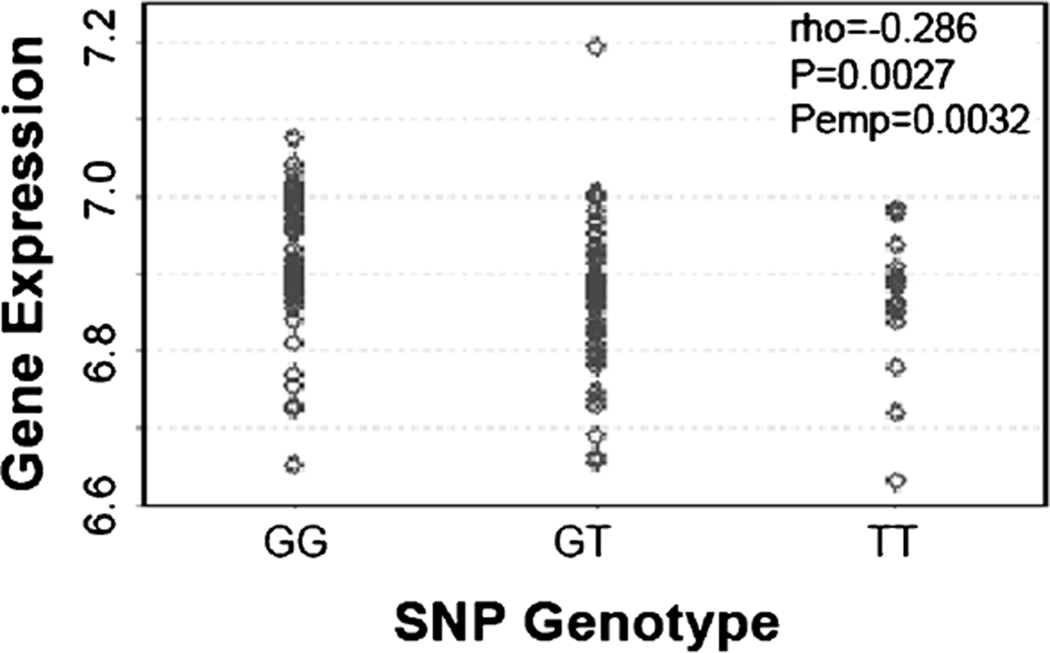
cis-eQTL associations were investigated among the above reported MMP pathway SNPs significantly associated with ovarian cancer risk (4 SNPS), survival (34 SNPs), and treatment response (1 SNP). Among all 39 SNPs investigated, the MMP8 5′ flanking region SNP rs17099462 was identified to have an eQTL association with MMP8 in the HapMap3 population [33], and the variant allele was significantly associated with decreased expression of MMP8 in lymphoblastoid cell lines in the Caucasian subgroup (Figure 3).
Figure 3.
eQTL association between MMP8 expression and SNP MMP8: rs17099462 in the lymphoblastoid cell lines from the HapMap3 Caucasian population.
DISCUSSION
The MMP family of proteases has long been suggested to play a role in the progression of different types of human cancer. Expressions of MMP1, MMP2, MMP7, MMP9, MMP11, MMP13, and MMP14 were previously reported to be up-regulated in tumor cells and to appreciably decrease the rate of cancer survival [21]. In this study, we evaluated the association of MMP genetic variants with ovarian cancer risk, survival, and treatment response. By assessing the effects of 266 SNPs from 23 genes in the MMP family, we found that four SNPs were associated with ovarian cancer risk after adjustment for multiple comparisons. In the evaluation of single SNP associations with overall risk of death, 34 SNPs from 13 MMP and TIMP1 genes were shown to have significant effects after adjustment for multiple comparisons. Only one intronic SNP (rs7826929) showed a significant association with patients’ response to chemotherapy; it was not, however, among the SNPs most strongly associated with overall risk or risk of death.
SNP rs2292730 with the most significant association with ovarian cancer risk resides in the intronic region on gene MMP20, and it was shown to double the overall risk. Interestingly, another MMP20 SNP rs9787933 was suggested to have protective effect on ovarian cancer risk. MMP20 is a relatively newly identified member of the MMP family and was named “human enamelysin” when first studied [38]. MMP20 has been considered a tooth-specific protease, and studies on the association of MMP20 with cancer have been limited to odontogenic, dental, and tongue carcinomas [39–44]. Our results suggest that MMP20 can play both risk-heightening and -mitigating roles, and that MMP20 may be regulatory in ovarian cancer. However, because of the lack of functional studies and the fact that most intronic SNPs are non-functional, these results more likely suggest that other coding SNP(s) tagged by rs2292730 or rs9787933 might be causal to ovarian cancer. Furthermore, the haplotype containing the wild-type allele of the MMP20: rs2292730 and the variant alleles of MMP20: rs12278250 and rs9787933 showed strong association with decreased ovarian cancer risk by almost one half compared to the reference haplotype with all wild-type alleles of these three SNPs. These results are highly consistent with the findings from single SNPs analysis, and demonstrated that the haplotype pattern composed of the wild-type alleles of risk SNPs and the variant alleles of the potentially protective SNPs could be significantly favoured in individuals with relatively lower cancer risk.
In the evaluation of genetic variants on overall survival, SNPs MMP1:rs2239008 and MMP11: rs738791 represented the most significant risk-conferring and risk-protective roles, respectively. MMP1 is expressed in ovarian cancer cells [45], and increased expression of MMP1 has been associated with a lower survival rate in several kinds of cancer, including those of the breast, lung, gastric, and colon [21]. Considering its 3’ UTR location in MMP1, SNP rs2239008 could potentially play a role in regulating MMP1 expression or protein folding and thereby affect ovarian cancer progression and survival. As predicted by PolymiRTS [46], rs2239008 located in the seed region of four human microRNAs (miR-22-5p, miR-4277, miR-4524b-3p, and miR-5584-3p), disrupted the conserved microRNA sites and potentially interfered with microRNA binding. Furthermore, these microRNA binding sites have been shown to be also disrupted by the same SNP in gastric cancer [47]. Although no experimental investigation has been performed to validate these germline and somatic associations, these results from computational prediction have pointed out the potential of this SNP in functional regulation of microRNA binding and MMP1 expression. The variant allele of rs738791 in the MMP11 gene was most strongly associated with decreased risk of death in our ovarian cancer patient population. This result indicates that this intronic SNP is protective and may be functionally correlated with MMP11’s cancer-suppressive effects during metastasis, as suggested by previous studies [48]. In addition, one of our top SNPs MMP8: rs17099462 obtained from the survival analysis was identified to have eQTL association with MMP8, a gene involved in ovarian cancer progression through interleukin and pro-inflammatory cytokine regulated overexpression [49]. Taken into account of the statistical association with survival, as well as the experimentally validated correlation with gene function, a functional mechanism is implied for this MMP8 5′ flanking region SNP in regards to affect ovarian cancer progression and patient survival.
Previous studies on the role of MMP16 in cancer have been relatively limited compared to those for other MMPs. It was reported that MMP16 expression was higher in malignant tumor cells than cells from pre-malignant lesions in melanoma [50], but its expression levels in malignant tumors and in normal tissues of the pancreas were shown to be the same [51]. Our results suggest that, based on the association between the intronic SNP in MMP16 and response to treatment, MMP16 plays a potential role in regulating response to chemotherapy; our results further suggest that future studies that focus on MMP16 function in cancer progression would be beneficial.
MMPs’ cancer-suppressing effects have been previously reported; one study used a mouse model and another applied cell biology techniques to a number of MMPs, including MMP3, MMP8, MMP9, MMP11, MMP12, MMP19, and MMP26 [52,53]. In our study, SNPs from MMP9 and MMP20 associated with overall cancer risk and SNPs from MMP2, MMP3, MMP8, MMP9, MMP11, MMP12, MMP13, MMP16, MMP20, and MMP24 associated with overall risk of death all suggested protective effects in single SNP analysis, as shown by corresponding ORs and HRs <1, respectively. It has been accepted that MMPs are both cancer-promoting and cancer-suppressing in the modulation of cancer progression [21,52,53], and our results indirectly support this notion from the perspective of the impact of sequence variations on cancer risk and survival. In fact, our study is the first one to point out MMPs’ paradoxical effects in ovarian cancer, and our genotypic analysis may provide target loci of interest for further investigations on the specific mechanisms by which the MMP pathway is involved in ovarian cancer.
Higher order gene–gene interactions among members of the MMP family were detected in the association with both overall risk and survival of ovarian cancer. Meanwhile, unfavorable genotype analysis confirmed the cumulative effects of combined variants on overall risk and survival. From our eQTL analysis, we also found that multiple SNPs were functionally associated with distant genes in the MMP family, such as intronic SNP rs9787933 in MMP 20 was in eQTL association with MMP8, another intronic SNP rs476185 in MMP 12 was also in eQTL association with MMP7, and the MMP13 5′ flanking region SNP rs579627 was in eQTL association with MMP1 (unreported data). These findings all pointed out the fact that genes in the MMP family are likely to be inter-correlated and co-function in regulating cancer development. Furthermore, the network of MMP interactions also involves tumor suppressors from other signaling pathways. Considering the strong contribution of MMP2 and MMP16 in our survival analysis, as well as the significant association between the MMP16 SNP and patients’ response to chemotherapy as previously reported [22], it is believed that the MMP family is highly cross-linked with genes from other pathways. To obtain a full map of MMP interactions in terms of ovarian cancer risk, survival, and treatment response, analyses of individual and pooled SNPs from broader gene networks need to be performed. Also needed is a validation study in a larger population on selected SNPs, so as to confirm the significant associations and filter out false positive results.
In conclusion, we evaluated the effect of genetic variants in MMP pathway on ovarian cancer risk, survival and treatment response, utilizing a pathway-based analysis and a respectable population size. For each case or control subject, we obtained detailed demographic, epidemiologic, and clinical information from the same institution according to standard procedures ensuring uniformity in operation. In order to minimize false discoveries, we performed the adjustment of multiple comparisons as well as internal validation by a bootstrap resampling procedure. To our best knowledge, this is the first study to examine the association of MMP polymorphisms with ovarian cancer risk and clinical outcome. Top SNPs resulted from our single SNP analysis presented the potential to serve as markers for ovarian cancer risk and survival prediction. Higher order interactions between the top SNPs were implied and established a novel combination of SNPs for risk assessment and survival prediction. The ultimate goal is to establish a pathway-based network of biomarkers. With further validation, our results could be of potential clinical interest in benefiting the prognosis and treatment of ovarian cancer.
Supplementary Material
Acknowledgments
Grant sponsor: Department of Defense Ovarian Cancer Research Program; Grant number: W81XWH-07-0449; Grant sponsor: NIH/ NIMHD; Grant number: 5G12RR003045-21
Abbreviations
- MMP
matrix metalloproteinase
- TIMP
tissue inhibitors of metalloproteinase
- SNP
single-nucleotide polymorphism
- MAF
minor allele frequency
- UTR
untranslated region
- CI
confidence interval
- HR
hazard ratio
- CART
classification and regression tree
- STREE
survival tree
- eQTL
expression quantitative trait loci
- MST
median survival time
Footnotes
Conflict of interest: None.
Additional supporting information may be found in the online version of this article.
REFERENCES
- 1.Siegel R, Naishadham D, Jemal A. Cancer statistics, 2013. Cancer J Clin. 2013;63:11–30. doi: 10.3322/caac.21166. [DOI] [PubMed] [Google Scholar]
- 2.Bankhead CR, Kehoe ST, Austoker J. Symptoms associated with diagnosis of ovarian cancer: A systematic review. BJOG-Int J Obstet Gy. 2005;112:857–865. doi: 10.1111/j.1471-0528.2005.00572.x. [DOI] [PubMed] [Google Scholar]
- 3.Ryerson AB, Eheman C, Burton J, et al. Symptoms, diagnoses, and time to key diagnostic procedures among older US women with ovarian cancer. Obstet Gynecol. 2007;109:1053–1061. doi: 10.1097/01.AOG.0000260392.70365.5e. [DOI] [PubMed] [Google Scholar]
- 4.Goff BA, Mandel LS, Melancon CH, Muntz HG. Frequency of symptoms of ovarian cancer in women presenting to primary care clinics. J Am Med Assoc. 2004;291:2705–2712. doi: 10.1001/jama.291.22.2705. [DOI] [PubMed] [Google Scholar]
- 5.Goff BA, Mandel L, Muntz HG, Melancon CH. Ovarian carcinoma diagnosis—Results of a National Ovarian Cancer Survey. Cancer. 2000;89:2068–2075. doi: 10.1002/1097-0142(20001115)89:10<2068::aid-cncr6>3.0.co;2-z. [DOI] [PubMed] [Google Scholar]
- 6.Cannistra SA. Cancer of the ovary. New Engl J Med. 2004;351:2519–2529. doi: 10.1056/NEJMra041842. [DOI] [PubMed] [Google Scholar]
- 7.Ozols RF. Treatment goals in ovarian cancer. Int J Gynecol Cancer. 2005;15:3–11. doi: 10.1111/j.1525-1438.2005.15351.x. [DOI] [PubMed] [Google Scholar]
- 8.Bandera CA. Advances in the understanding of risk factors for ovarian cancer. J Reprod Med. 2005;50:399–406. [PubMed] [Google Scholar]
- 9.Vo C, Carney ME. Ovarian cancer hormonal and environmental risk effect. Obstet Gyn Clin N Am. 2007;34:687. doi: 10.1016/j.ogc.2007.09.008. [DOI] [PubMed] [Google Scholar]
- 10.Lancaster JM, Carney ME, Futreal PA. BRCA 1 and 2-A Genetic Link to Familial Breast and Ovarian Cancer. Medscape Womens Health. 1997;2:7. [PubMed] [Google Scholar]
- 11.Bowtell D. The genesis and evolution of high-grade serous ovarian cancer. Nat Rev Cancer. 2010;10:803–808. doi: 10.1038/nrc2946. [DOI] [PubMed] [Google Scholar]
- 12.Overall CM, Kleifeld O. Tumour microenvironment—Opinion—Validating matrix metalloproteinases as drug targets and anti-targets for cancer therapy. Nat Rev Cancer. 2006;6:227–239. doi: 10.1038/nrc1821. [DOI] [PubMed] [Google Scholar]
- 13.Close DR. Matrix metalloproteinase inhibitors in rheumatic diseases. Ann Rheum Dis. 2001;60:Iii62–Iii67. doi: 10.1136/ard.60.90003.iii62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hojilla CV, Mohammed FF, Khokha R. Matrix metalloproteinases and their tissue inhibitors direct cell fate during cancer development. Br J Cancer. 2003;89:1817–1821. doi: 10.1038/sj.bjc.6601327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kelly EA, Jarjour NN. Role of matrix metalloproteinases in asthma. Curr Opin Pulm Med. 2003;9:28–33. doi: 10.1097/00063198-200301000-00005. [DOI] [PubMed] [Google Scholar]
- 16.Sung JY, Park SM, Lee CH, et al. Proteolytic cleavage of extracellular secreted alpha-synuclein via matrix metalloproteinases. J Biol Chem. 2005;280:25216–25224. doi: 10.1074/jbc.M503341200. [DOI] [PubMed] [Google Scholar]
- 17.Thompson RW, Parks WC. Role of matrix metalloproteinases in abdominal aortic aneurysms. Ann NY Acad Sci. 1996;800:157–174. doi: 10.1111/j.1749-6632.1996.tb33307.x. [DOI] [PubMed] [Google Scholar]
- 18.Coussens LM, Werb Z. Matrix metalloproteinases and the development of cancer. Chem Biol. 1996;3:895–904. doi: 10.1016/s1074-5521(96)90178-7. [DOI] [PubMed] [Google Scholar]
- 19.Nelson AR, Fingleton B, Rothenberg ML, Matrisian LM. Matrix metalloproteinases: Biologic activity and clinical implications. J Clin Oncol. 2000;18:1135–1149. doi: 10.1200/JCO.2000.18.5.1135. [DOI] [PubMed] [Google Scholar]
- 20.Vihinen P, Kahari VM. Matrix metalloproteinases in cancer: Prognostic markers and therapeutic targets. Int J Cancer. 2002;99:157–166. doi: 10.1002/ijc.10329. [DOI] [PubMed] [Google Scholar]
- 21.Egeblad M, Werb Z. New functions for the matrix metalloproteinases in cancer progression. Nat Rev Cancer. 2002;2:161–174. doi: 10.1038/nrc745. [DOI] [PubMed] [Google Scholar]
- 22.Delassus GS, Cho H, Hoang S, Eliceiri GL. Many new down-and up-regulatory signaling pathways, from known cancer progression suppressors to matrix metalloproteinases, differ widely in cells of various cancers. J Cell Physiol. 2010;224:549–558. doi: 10.1002/jcp.22157. [DOI] [PubMed] [Google Scholar]
- 23.Kamat AA, Fletcher M, Gruman LM, et al. The clinical relevance of stromal matrix metalloproteinase expression in ovarian cancer. Clin Cancer Res. 2006;12:1707–1714. doi: 10.1158/1078-0432.CCR-05-2338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cowden Dahl KD, Symowicz J, Ning Y, et al. Matrix metalloproteinase 9 is a mediator of epidermal growth factor-dependent e-cadherin loss in ovarian carcinoma cells. Cancer Res. 2008;68:4606–4613. doi: 10.1158/0008-5472.CAN-07-5046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li XL, Kang S, Zhao XW, et al. [Association of SNPs in the promoter of MMP-2 and TIMP-2 genes with epithelial ovarian cancer] Yi chuan=Hereditas/Zhongguo yi chuan xue huibian ji. 2008;30:455–462. doi: 10.3724/sp.j.1005.2008.00455. [DOI] [PubMed] [Google Scholar]
- 26.Ju W, Kim JW, Park NH, et al. Matrix metalloproteinase-1 promoter polymorphism and epithelial ovarian cancer: Does ethnicity matter? J Obstet Gynaecol Res. 2007;33:155–160. doi: 10.1111/j.1447-0756.2007.00511.x. [DOI] [PubMed] [Google Scholar]
- 27.Li Y, Jin X, Kang S, et al. Polymorphisms in the promoter regions of the matrix metalloproteinases-1, −3, −7, and −9 and the risk of epithelial ovarian cancer in China. Gynecol Oncol. 2006;101:92–96. doi: 10.1016/j.ygyno.2005.09.058. [DOI] [PubMed] [Google Scholar]
- 28.Wu XF, Spitz MR, Lee JJ, et al. Novel susceptibility loci for second primary tumors/recurrence in head and neck cancer patients: Large-scale evaluation of genetic variants. Cancer Prev Res. 2009;2:617–624. doi: 10.1158/1940-6207.CAPR-09-0025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Altshuler D, Brooks LD, Chakravarti A, et al. A haplotype map of the human genome. Nature. 2005;437:1299–1320. doi: 10.1038/nature04226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sherry ST, Ward MH, Sirotkin K. dbSNP—Database for single nucleotide polymorphisms and other classes of minor genetic variation. Genome Res. 1999;9:677–679. [PubMed] [Google Scholar]
- 31.Yang TP, Beazley C, Montgomery SB, et al. Genevar: A database and Java application for the analysis and visualization of SNP-gene associations in eQTL studies. Bioinformatics. 2010;26:2474–2476. doi: 10.1093/bioinformatics/btq452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Grundberg E, Small KS, Hedman Å, et al. Mapping cis- and trans-regulatory effects across multiple tissues in twins. Nat Genet. 2012;44:1084–1089. doi: 10.1038/ng.2394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stranger BE, Montgomery SB, Dimas AS, et al. Patterns of cis regulatory variation in diverse human populations. PLoS Genet. 2012;8:e1002639. doi: 10.1371/journal.pgen.1002639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nica AC, Parts L, Glass D, et al. The architecture of gene regulatory variation across multiple human tissues: The MuTHER study. PLoS Genet. 2011;7:e1002003. doi: 10.1371/journal.pgen.1002003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dimas AS, Deutsch S, Stranger BE, et al. Common regulatory variation impacts gene expression in a cell type-dependent manner. Science (New York, NY) 2009;325:1246–1250. doi: 10.1126/science.1174148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Johnson AD, Handsaker RE, Pulit SL, Nizzari MM, O’Donnell CJ, de Bakker PIW. SNAP: A web-based tool for identification and annotation of proxy SNPs using HapMap. Bioinformatics. 2008;24:2938–2939. doi: 10.1093/bioinformatics/btn564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kosary CL. Figo Stage, histology, histologic grade, age and race as prognostic factors in determining survival for cancers of the female gynecological system - an analysis of 1973–87 seer cases of cancers of the endometrium, cervix, ovary, vulva, and vagina. Semin Surg Oncol. 1994;10:31–46. doi: 10.1002/ssu.2980100107. [DOI] [PubMed] [Google Scholar]
- 38.Llano E, Pendas AM, Knauper V, et al. Identification and structural and functional characterization of human enamelysin (MMP-20) Biochemistry-US. 1997;36:15101–15108. doi: 10.1021/bi972120y. [DOI] [PubMed] [Google Scholar]
- 39.Hu JCC, Simmer JP. Developmental biology and genetics of dental malformations. Orthod Craniofac Res. 2007;10:45–52. doi: 10.1111/j.1601-6343.2007.00384.x. [DOI] [PubMed] [Google Scholar]
- 40.Lee SK, Seymen F, Kang HY, et al. MMP20 hemopexin domain mutation in amelogenesis imperfecta. J Dent Res. 2010;89:46–50. doi: 10.1177/0022034509352844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sekine S, Takata T, Shibata T, et al. Expression of enamel proteins and LEF1 in adamantinomatous craniopharyngioma: Evidence for its odontogenic epithelial differentiation. Histopathology. 2004;45:573–579. doi: 10.1111/j.1365-2559.2004.02029.x. [DOI] [PubMed] [Google Scholar]
- 42.Vaananen A, Srinivas R, Parikka M, et al. Expression and regulation of MMP-20 in human tongue carcinoma cells. J Dent Res. 2001;80:1884–1889. doi: 10.1177/00220345010800100501. [DOI] [PubMed] [Google Scholar]
- 43.Vaananen A, Tjaderhane L, Eklund L, et al. Expression of collagen XVIII and MMP-20 in developing teeth and odontogenic tumors. Matrix Biol. 2004;23:153–161. doi: 10.1016/j.matbio.2004.04.003. [DOI] [PubMed] [Google Scholar]
- 44.Takata T, Zhao M, Uchida T, et al. Immunohistochemical detection and distribution of enamelysin (MMP-20) in human odontogenic tumors. J Dent Res. 2000;79:1608–1613. doi: 10.1177/00220345000790081401. [DOI] [PubMed] [Google Scholar]
- 45.Davidson B, Reich R, Berner A, et al. Ovarian carcinoma cells in serous effusions show altered MMP-2 and TIMP-2 mRNA levels. Eur J Cancer. 2001;37:2040–2049. doi: 10.1016/s0959-8049(01)00235-0. [DOI] [PubMed] [Google Scholar]
- 46.Ziebarth JD, Bhattacharya A, Chen A, Cui Y. PolymiRTS Database 2.0: Linking polymorphisms in microRNA target sites with human diseases and complex traits. Nucleic Acids Res. 2012;40:D216–D221. doi: 10.1093/nar/gkr1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bhattacharya A, Ziebarth JD, Cui Y. SomamiR: A database for somatic mutations impacting microRNA function in cancer. Nucleic Acids Res. 2013;41:D977–D982. doi: 10.1093/nar/gks1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Andarawewa KL, Boulay A, Masson W, et al. Dual stromelysin-3 function during natural mouse mammary tumor virus-ras tumor progression. Cancer Res. 2003;63:5844–5849. [PubMed] [Google Scholar]
- 49.Stadlmann S, Pollheimer J, Moser PL, et al. Cytokine-regulated expression of collagenase-2 (MMP-8) is involved in the progression of ovarian cancer. Eur J Cancer. 2003;39:2499–2505. doi: 10.1016/j.ejca.2003.08.011. [DOI] [PubMed] [Google Scholar]
- 50.Ohnishi Y, Tajima S, Ishibashi A. Coordinate expression of membrane type-matrix metalloproteinases-2 and 3 (MT2-MMP and MT3-MMP) and matrix metalloproteinase-2 (MMP-2) in primary and metastatic melanoma cells. Eur J Dermatol. 2001;11:420–423. [PubMed] [Google Scholar]
- 51.Ellenrieder V, Alber B, Lacher U, et al. Role of MT-MMPs and MMP-2 in pancreatic cancer progression. Int J Cancer. 2000;85:14–20. doi: 10.1002/(sici)1097-0215(20000101)85:1<14::aid-ijc3>3.0.co;2-o. [DOI] [PubMed] [Google Scholar]
- 52.Lopez-Otin C, Matrisian LM. Tumour micro environment— Opinion—Emerging roles of proteases in tumour suppression. Nat Rev Cancer. 2007;7:800–808. doi: 10.1038/nrc2228. [DOI] [PubMed] [Google Scholar]
- 53.Martin MD, Matrisian LM. The other side of MMPs: Protective roles in tumor progression. Cancer Metast Rev. 2007;26:717–724. doi: 10.1007/s10555-007-9089-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.