Abstract
Pharmacokinetic analysis of dynamic contrast-enhanced (DCE) MRI data allows estimation of quantitative imaging biomarkers such as Ktrans (rate constant for plasma/interstitium contrast reagent (CR) transfer) and ve (extravascular and extracellular volume fraction). However, the use of quantitative DCE-MRI in clinical practice is limited with uncertainty in arterial input function (AIF) determination being one of the primary reasons. In this multicenter study to assess the effects of AIF variations on pharmacokinetic parameter estimation, DCE-MRI data acquired at one center from 11 prostate cancer patients were shared among nine centers. Individual AIF from each data set was determined by each center and submitted to the managing center. These AIFs, along with a literature population averaged AIF, and their reference-tissue-adjusted variants were used by the managing center to perform pharmacokinetic data analysis using the Tofts model (TM). All other variables, including tumor region of interest (ROI) definition and pre-contrast T1, were kept constant to evaluate parameter variations caused solely by AIF discrepancies. Considerable parameter variations were observed with the within-subject coefficient of variation (wCV) of Ktrans obtained with unadjusted AIFs being as high as 0.74. AIF-caused variations were larger in Ktrans than ve and both were reduced when reference-tissue-adjusted AIFs were used. These variations were largely systematic, resulting in nearly unchanged parametric map patterns. The intravasation rate constant, kep (= Ktrans/ve), was less sensitive to AIF variation than Ktrans (wCV for unadjusted AIFs: 0.45 vs. 0.74), suggesting that it might be a more robust imaging biomarker of prostate microvasculature than Ktrans.
Keywords: DCE-MRI, Arterial Input Function, Prostate Cancer, Variation, Pharmacokinetic Analysis
Introduction
Dynamic contrast-enhanced magnetic resonance imaging (DCE-MRI) is widely used in studies of cancer and other pathologies. Often included as one component of a prostate multiparametric MRI protocol (1), DCE-MRI is routinely used in clinical MRI examination of the prostate. Nevertheless, its use as a quantitative diagnostic imaging modality remains limited. In clinical practice, only qualitative estimations of contrast reagent (CR) wash-in and wash-out are generally used in interpreting prostate DCE-MRI data. Although pharmacokinetic modeling approaches have been under extensive investigation for more than a decade (2–10), pharmacokinetic analysis of prostate DCE time-course data is not currently recommended for routine use under the Prostate Imaging-Reporting and Data System (PI-RADS) version 2 guidelines (11, 12). Improved reproducibility and standardization in pharmacokinetic analysis of prostate DCE-MRI data is needed for the translation of this quantitative data analysis method into clinical settings.
Quantitative DCE-MRI data analysis using pharmacokinetic models allows extraction and mapping of quantitative parameters of tissue biology in vivo. The most commonly estimated parameters are usually variants of Ktrans, a rate constant for passive CR molecule plasma/interstitium transfer, and ve, the volume fraction of interstitial space (extravascular extracellular space, i.e., the putative CR distribution volume). The CR intravasation rate constant, kep, can be calculated as Ktrans/ve. Unlike qualitative or semiquantitative analysis, the parameters derived from pharmacokinetic modeling of DCE-MRI time-course data are, in principle, independent of the MRI scanner platform (vendor and field strength), data acquisition details (pulse sequence and parameters), CR dose and/or injection protocol, personnel skills, etc. This makes them promising imaging biomarkers in multicenter clinical trials as imaging endpoints for standardization and comparison of results. However, the accuracy and precision of these parameters can be affected by a plethora of factors contributing to the process of pharmacokinetic modeling, including errors in quantification of precontrast T1 (13) and determination of arterial input function (AIF) (14–20), inadequate temporal resolution (21), selection of pharmacokinetic models to fit the data (22, 23), and differences in DCE-MRI acquisition time duration (24, 25).
As a requirement in quantitative estimation of DCE-MRI pharmacokinetic parameters, the time dependence of the plasma and tissue CR concentrations, Cp(t) and Ct(t), respectively, needs to be determined from the DCE-MRI images. The former is the AIF, and its direct quantification from an imaged blood vessel is not straightforward because of, for example, the partial volume effect (signal from a selected image voxel, ideally from 100% blood, is contaminated with signal from nonblood tissues) and in-flow effect (the measured blood signal is affected by signal from the “fresh” blood outside the imaging volume that flows into the imaging volume during data acquisition). Nonetheless, as the driving force that causes in vivo DCE-MRI signal changes in the tissue, accurate AIF determination is fundamental for accurate estimation of pharmacokinetic parameters, particularly for in vivo systems like the prostate where CR extravasation is substantial (9). Significant research effort has been devoted to the development of AIF quantification methods. The most commonly used method is to measure AIF directly from a feeding artery if it is clearly detected within the image field of view (FOV). Other methods include blinded AIF estimation (14, 15), reference tissue and double reference tissue approaches (16, 26), direct blood sampling, empirically derived population-averaged AIF (28), and automated vessel region identification (27). However, given these various methods for AIF quantification, there is a lack of studies investigating the impact of variations in AIF determination on pharmacokinetic analysis of DCE-MRI data, which may potentially lead to best-practice guidelines for analysis of DCE-MRI data acquired from different organs.
The National Cancer Institute has recently founded the Quantitative Imaging Network (QIN) for the development and validation of quantitative imaging methods for evaluation of cancer therapy response. The main mission of the QIN Image Analysis and Performance Metrics Working Group is to provide guidance and reach consensus on quantitative image analysis methods through comparison and validation of analysis algorithms. For quantitative DCE-MRI data analysis, it is important to understand the variations of DCE-MRI pharmacokinetic parameters caused by specific error-prone steps encountered during data analysis. A recent QIN multicenter study showed the effects of variations in pharmacokinetic models and software tools on assessment of breast cancer response to neoadjuvant chemotherapy (23). Here, we report the results and experience from a DCE-MRI AIF challenge project, in which 9 QIN centers independently performed AIF quantifications from the same prostate DCE-MRI data sets and submitted the AIFs to one managing center for central pharmacokinetic analysis of the shared data. The goal of this study is to assess variations in estimated prostate Ktrans, ve, and kep parameters that are resulted from various AIF extraction approaches.
Materials and Methods
AIF Challenge Participating QIN Centers
The QIN centers that participated in this DCE-MRI AIF challenge project were Oregon Health and Science University (OHSU) - managing center, Brigham and Women's Hospital (BWH) in collaboration with General Electric Research and Development, Medical College of Wisconsin (MCW), Icahn School of Medicine at Mount Sinai (MS), University of Michigan center #1 (UM1), University of Michigan center #3 (UM3), University of Pittsburgh (UPitt), Vanderbilt University (VU), and University of Washington (UW). Hereafter, except for where it is explicitly indicated, these 9 institutions are denoted as, not necessarily in the order listed above, QIN1 to QIN9.
Although some centers may have used more than one method for AIF determination from the shared data, only one AIF method from each participating center was included in this study. In addition, we also included a population-averaged AIF published by Geoff Parker (GP) et al (28) for comparison. This AIF was selected because it is extensively cited. The analytical expression of the GP AIF was implemented at the managing center and temporally resampled to match the temporal features of the shared prostate DCE-MRI data.
Human Prostate DCE-MRI Data Acquisition and Sharing
As part of the institutional review board-approved BWH quantitative imaging studies, multiparametric magnetic resonance images were collected for the purposes of detection and/or staging of prostate cancer. The images were obtained with a GE Signa HDx 3.0 T system (GE Healthcare, Waukesha, Wisconsin) using a combination of 8-channel abdominal array and endorectal coil (Medrad, Pittsburgh, Pennsylvania). The MRI sequences included T1- and T2-weighted imaging, diffusion-weighted imaging, and DCE-MRI as described by Hegde JV et al (29). The axial DCE-MRI acquisition with full prostate gland coverage used a 3-dimensional SPoiled Gradient Recalled (SPGR) sequence with repetition time (TR)/echo time (TE)/Flip Angle (α) = 3.6 ms/1.3 ms/15°, FOV = (26 cm)2, slice thickness = 6 mm, and reconstructed image voxel size = 1 × 1 × 6 mm. DCE-MRI frames were acquired at approximately 5-second intervals (the number of slices per frame varied between 12 and 16, resulting in time resolution between 4.4 and 5.3 seconds) to achieve a clinically appropriate compromise between spatial and temporal resolutions. Gadopentetate dimeglumine (Magnevist; Berlex Laboratories, Wayne, New Jersey) was injected intravenously (0.15 mmol/kg) using a syringe pump at a rate of 3 mL/s followed by 20 mL of saline flush at the same rate. The DCE protocol included approximately 5 baseline frames before contrast injection.
A subset of the imaging data from the BWH prostate DCE-MRI database was uploaded to The Cancer Imaging Archive (TCIA) server for data sharing (10). From these, 11 data sets from 11 patients with known prostate cancer diagnosis, tumor region of interest (ROI) and relatively consistent acquisition time length (4.5–6 minutes) were downloaded by participating QIN centers for this AIF challenge project.
AIF Determination by QIN Centers
Although the AIF challenge participants were informed of the single-image slice in each data set where the tumor ROI was drawn and for which the ROI time-course data will be subjected to pharmacokinetic analysis by the managing center, there was no restriction on which image slice(s) to be used for the AIF quantification. Participating centers were required to extract an AIF time course using their own methodology for each DCE data set. For example, when measuring AIF directly from a femoral artery within the image FOV, the actual number of voxels used to derive the final AIF time course varied substantially among all centers. In general, the following results were saved and submitted to the managing center: (1) extracted AIF signal intensity time-course saved as a single column text (“.txt” file); (2) converted blood plasma CR concentration time-course (see Equation 1); and (3) screen-captured images showing the ROI/voxel locations for AIF measurement. Item (2) was optional, and both items (2) (if submitted) and (3) served as quality control references for the managing center when performing final pharmacokinetic data analyses with the AIFs from different centers. Details of the AIF determination methods (including references) used by the participating centers are summarized in Table 1.
Table 1.
Arterial Input Function (AIF) Quantification Methods by Participating Quantitative Imaging Network (QIN) Centers
| Center | Method |
|---|---|
| OHSU | A single, fixed-size region of interest (ROI) was manually placed inside the femoral artery within the field of view (FOV). Averaged blood intensity time-course was extracted from the ROI, which is further converted to Cp(t) using the parameter values provided in the Materials and Methods section. |
| BWH | GE's OncoQuant prototype tool was used, which includes the following: (1) AIF Search Region Slice Localization; (2) AIF Search Mask Localization; (3) AIF Detection Using Shape-Based Statistics; and (4) AIF Signal-to-Concentration Conversion. See (27) for more details. |
| MCW | Motion-corrected dynamic contrast-enhanced (DCE) series were processed using probabilistic-independent component analysis implemented in the FSL (FMRIB's Software Library, www.fmrib.ox.ac.uk/fsl). These were further whitened and projected into a 20-dimensional subspace using principal component analysis. The AIFs were manually chosen from the results (4, 36–38). |
| MS | ROIs were manually placed inside the iliac arteries within the field of view (FOV) using Osirix (v5.8; Pixmeo, Switzerland). For each AIF determination, 1 ROI was drawn on 1 DCE frame, and its position was adjusted when necessary to account for interframe subject motion. Blood intensity time-courses were extracted from the ROIs. |
| UM1 | ROIs of 5 × 5 voxels were manually placed in 2 to 4 slices showing the highest artery conspicuity on maximum intensity projection displays of the baseline-subtracted DCE images. Voxel time-courses within the ROI were individually displayed on a 5 × 5 panel. Voxels with time-courses showing an AIF curve shape were then individually selected, and their locations and time-courses were automatically saved. |
| UM3 | ROIs were manually drawn on both left and right femoral arteries on the central 4 slices. To minimize the in-flow effect, the inferior and superior slices were excluded. Further, 20 voxels within the ROIs with the highest signal increases were determined by thresholding the histogram of intensity changes. The average signal intensity time curve of the 20 voxels yielded the final AIF signal intensity time-course. |
| UPitt | Images were loaded into PMOD 3.505 (PMOD Technologies Ltd.), a commercial software package. Images were examined to search for an artery near the lesion. A region including the identified artery was surveyed using the voxel browser of PMOD to identify an area with high signal intensity change, followed by AIF ROI delineation. |
| UW | An adapted version of a positron emission tomography (PET) AIF extraction scheme (39), which does not require user-specified AIF ROI, was used. The approach was implemented in R (open-source). The extracted input function was then scaled so that the apparent extraction of gadolinium contrast reagent (CR) based on the analysis of the entire tissue volume signal is 2.5%. |
| VU | A seed point was placed manually inside an artery and then a region-growing method was applied to automatically select the AIF voxels. The intensity range for the region-growing method was set as 80% to 120% of that of the seed point, and the voxel distance from the seed was <10 voxels. Mean signal intensity time-course of the selected voxels was obtained. |
Before performing pharmacokinetic analysis of the shared DCE-MRI data, the managing center first converted the AIF signal intensity time-course to blood R1 ( 1/T1) time course, R1,b(t), using the SPGR steady-state signal intensity equation (30) and a fixed precontrast blood R1 of 0.61 s−1 (31), and then converted to plasma CR concentration time course, Cp(t), using the following equation:
1/T1) time course, R1,b(t), using the SPGR steady-state signal intensity equation (30) and a fixed precontrast blood R1 of 0.61 s−1 (31), and then converted to plasma CR concentration time course, Cp(t), using the following equation:
| (1) |
where r1 is the CR relaxivity at 3T, set at 3.8 mM−1s−1 ; and h is the hematocrit, set at 0.45.
Pharmacokinetic Analysis of Human Prostate DCE-MRI Data
Using a single in-house Matlab-based software package, the managing center performed pharmacokinetic analysis of the shared 11 sets of prostate DCE-MRI data using the AIFs determined by the 9 QIN centers and the literature-based GP AIF. All AIF arrival times were manually aligned with the uptake phase of the tissue response curves. The most commonly used pharmacokinetic model, the Tofts model (TM) (32), was applied to data analysis, and its basic formulation is shown in Equation (2) as follows:
| (2) |
where Ct(t) represents the tissue CR concentration at time t; Cp(t') is the AIF obtained from Equation (1). The fast exchange limit condition intrinsic to the TM (23) implies a linear relationship between R1(t) and Ct(t):
| (3) |
where R1(t) is tissue R1 measured at time t, and R10 is the precontrast tissue R1 (assumed to be 0.63 s−1) (10).
For each DCE-MRI data set, the voxel intensity time-courses within the predefined prostate tumor ROI on a single image slice, drawn by the center (BWH) that acquired the data, were subjected to the TM pharmacokinetic analysis. The mean values of the tumor ROI pharmacokinetic parameters (Ktrans, ve, and kep) were obtained by averaging the corresponding voxel parameter values.
Because of different approaches (Table 1) used by participating centers in direct measurement of the AIF, large variations in the AIF amplitude were observed as a result of differences in measurement locations, number of voxels used, inflow effects, etc. As an alternative approach for pharmacokinetic analysis, an ROI (Figure 1, inset) in the adjacent obturator muscle area on the same image slice as that of the tumor ROI was used as a reference tissue for AIF amplitude adjustment (9, 33). The AIF (including the literature-based GP AIF) amplitude was adjusted until the TM fitting of the muscle ROI DCE-MRI data returned a ve value of 0.1, which is within the range of literature-reported values (34). Therefore, in total, 20 AIFs representing unadjusted and reference tissue-adjusted AIFs from the 9 centers and the GP AIFs were applied for the pharmacokinetic modeling of each prostate DCE-MRI data set using the TM, resulting in 20 sets of Ktrans, ve, and kep parameters that were then separated into two groups of results obtained with adjusted and unadjusted AIFs.
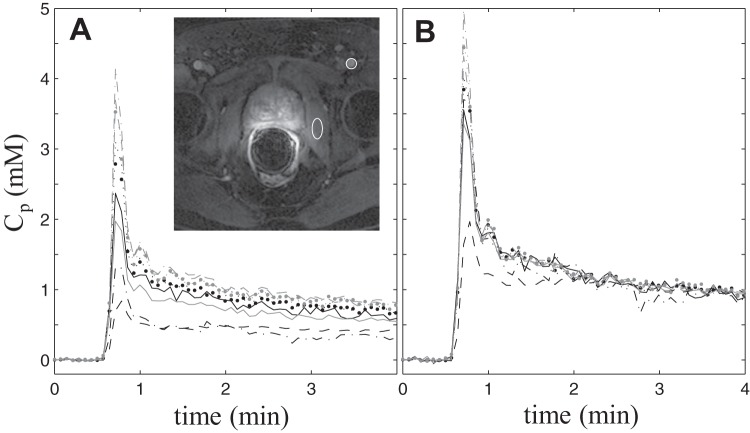
Figure 1.
Individual AIFs extracted from one subject's dynamic contrast-enhanced magnetic resonance imaging (DCE-MRI) data by 9 participating Quantitative Imaging Network centers. The smaller circular region of interest (ROI) in the zoomed image inset (with the prostate in the center of the view) indicates the general location where blood signals were most frequently measured for the final AIF time-courses, and the larger elliptical ROI indicates the general location for the obturator muscle reference tissue ROI. Noticeable variations are evident for both the shape and magnitude of the AIF curves (A). The reference tissue-adjusted AIFs of the same subject (B). The agreement among the individually measured AIFs is clearly improved following the adjustment.
All voxel-fitting results were included in calculating the mean tumor ROI pharmacokinetic parameter values, as it was difficult to set optimal criteria for excluding voxel fittings obtained with AIFs determined by a diverse array of methods. Using the prior knowledge that a physically meaningful ve is between 0.0 and 1.0, the lower and upper boundaries for ve fitting were set accordingly for each voxel. When the reference tissue-adjusted AIFs (from all participating centers) were used, all fitted voxel ve values for all DCE-MRI data sets were within the limits (none returned boundary values). When the unadjusted AIFs were used, on average, there were <3% voxels (ranging from 0%–6.6% when summarized by participating centers), where the returned ve values reached the upper boundary of 1.0. In these limited numbers of voxels, the ve value of 1.0 and the returned Ktrans values were taken as the fitted parameter values.
Statistical Analysis
The original parameter values returned from all fittings were used for statistical analysis. Descriptive statistical analysis was conducted to summarize the pharmacokinetic parameter values returned by different AIFs, with the distribution graphically assessed by boxplots. Intraclass correlation coefficient (ICC), within-subject coefficient of variation (wCV), and concordance correlation coefficient (CCC) were calculated, and these were reported with the corresponding 95% confidence intervals (CIs). Although all the three coefficients were computed to assess the reproducibility of the pharmacokinetic parameter values from different AIFs, each had a different focus. The ICCs measure the proportion of total variation contributed by between-subject differences, with high ICCs indicating good agreement. The wCV is the ratio of within-subject standard deviation to the mean of the corresponding parameter. A smaller wCV suggests good reproducibility. The CCCs are closely related to ICCs. They were estimated to represent the level of pairwise linear agreement to a 45° line of which the intercept is forced to be 0. A larger CCC (close to 1) suggests good reproducibility. Bland–Altman plots were used to graphically demonstrate pairwise agreements of different AIF measures. SAS 9.4 (Cary, New York) was used for all statistical analysis. SAS macro %ICC9 and %mccc were used for the estimations of ICC, wCV, and CCC.
Results
Pharmacokinetic Parameter Variations Due to AIF Differences
Figure 1A plots the AIFs extracted from the DCE-MRI data of one subject by the 9 participating QIN centers. The inset shows a post-CR DCE image slice zoomed to the prostate area. The smaller circular ROI indicates a common location—the femoral artery—where blood signals were measured for AIF determination, whereas the larger elliptical ROI indicates the general location of the reference tissue ROI in the obturator muscle. Noticeable variations are evident in both the shape and the amplitude among the Cp time-courses, converted from the measured signal intensity time-courses using Equation (1). Figure 1B shows the reference tissue-adjusted AIFs of those shown in Figure 1A. The agreement among the individually measured AIFs is clearly improved following the adjustment. The standard deviation of the 9 measured AIFs over the DCE time-course is significantly smaller for the reference tissue-adjusted AIFs than for the unadjusted ones (Wilcoxon signed-rank test, P < .0001). Similar findings are observed for AIFs from the other 10 subjects.
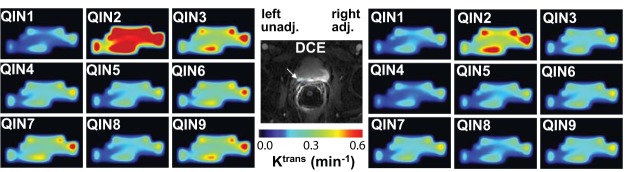
At the center of Figure 2, a zoomed post-CR image slice of the prostate of another subject is shown. The cyan ROI demarks the lesion area used for subsequent TM modeling and parameter comparisons. Ktrans color maps generated by TM analysis of the DCE-MRI data using unadjusted AIFs from the 9 centers are shown on the left panels and those with reference tissue-adjusted AIFs are shown on the right. Under the same color scale, substantial variations, mostly in the magnitude of Ktrans value, can be seen among the Ktrans maps obtained with different unadjusted AIFs (Figure 2, left). These differences are lessened when the Ktrans parameter was derived with reference tissue-adjusted AIFs (Figure 2, right). It is interesting to observe that despite considerable variations in Ktrans value caused by AIF differences, the pattern of voxel Ktrans distribution largely remains the same.
Figure 2.
Grayscale image at the center shows a zoomed DCE-MRI slice of another subject. The cyan-colored ROI (indicated by the white arrow) demarks the lesion area used for subsequent TM modeling and parameter comparisons. Ktrans color maps generated by TM analysis of the DCE-MRI data using unadjusted (unadj.) AIFs from the 9 centers are shown on the left panels and those with reference tissue-adjusted (adj.) AIFs are shown on the right. All 18 panels used the same color scale.
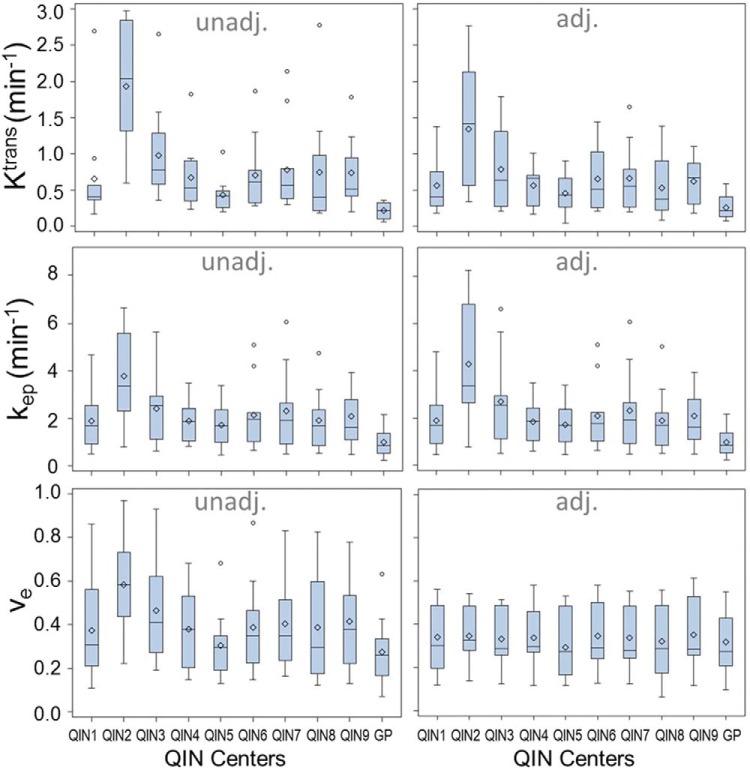
To illustrate the variations seen in Figure 2 for one parameter from a single DCE-MRI data set, Figure 3 shows the boxplots for Ktrans, ve, and kep parameters obtained from the 11 subject data sets with adjusted and unadjusted AIFs (including those from the GP AIF). For most measurements, the mean is greater than the median, which is commonly seen when distributions are skewed toward the right (the larger parameter values). The dispersions of the estimated metrics vary substantially across institutions (or AIFs). Examining the results from the same institution (or from one set of unadjusted and adjusted AIFs), it can be observed that the agreement in parameter dispersion between the unadjusted and adjusted AIFs is better for kep than for Ktrans. In fact, kep dispersion is hardly affected by the difference in AIF scaling.
Figure 3.
Boxplots of the tumor mean Ktrans, ve, and kep parameters for the 11 subjects obtained with unadjusted (unadj.) and adjusted (adj.) AIFs from the 9 centers and the population-averaged GP AIF from the literature (28). The diamond and bar symbols represent the mean and median values, respectively. The body of the box is bounded by the upper 25% and lower 25% quartiles, representing the interquartile range of the middle 50% of the measurements. The upper and lower whiskers define the range of nonoutliers. The outliers are plotted as dots beyond the whiskers.
Figure 4 shows the column graphs of wCV for Ktrans, ve, and kep obtained with the unadjusted (shaded light gray) and adjusted (dark gray) AIFs. The error bars are the 95% CIs. A smaller wCV value indicates less variation in measurements on the same subject by different approaches. In this study, the wCV values range from 0.15 for ve with adjusted AIFs to 0.74 for Ktrans with unadjusted AIFs. The wCV of ve is the smallest, whereas that of Ktrans is the largest among the three pharmacokinetic parameters with either unadjusted or adjusted AIFs. From unadjusted to adjusted AIFs, the parameter variations decrease for Ktrans and ve (wCV values decrease from 0.74 to 0.60 and from 0.33 to 0.15, respectively). On the other hand, it increases slightly for kep (wCV value increases from 0.45 to 0.54).
Figure 4.
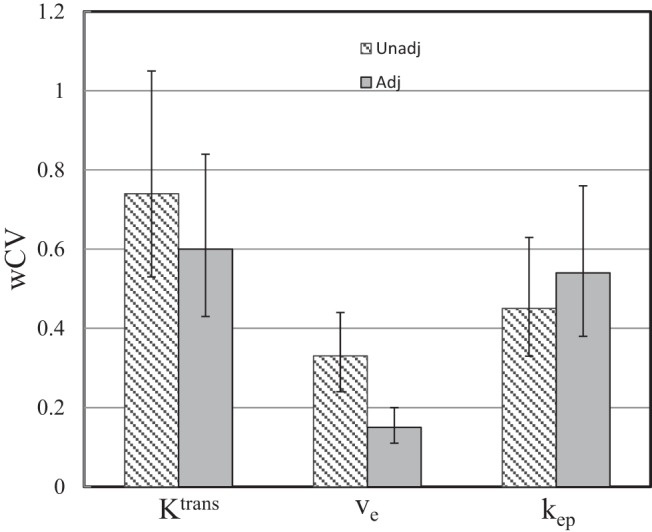
Column graphs of wCV for the Ktrans, ve, and kep parameters obtained with the unadjusted (unadj., shaded light gray) and adjusted (adj., dark gray) AIFs. The respective 95% confidence intervals (CIs) are shown as error bars.
Figure 5 shows the column graphs of ICC values for Ktrans, ve, and kep obtained with unadjusted (shaded light gray) and adjusted (dark gray) AIFs. The respective 95% CIs are shown as error bars. Consistent with the results shown in Figure 4, Ktrans has the smallest ICC value compared with kep and ve with either unadjusted or adjusted AIF, indicating its high dependence on AIF quantification for prostate DCE-MRI. From unadjusted to adjusted AIFs, the ICC value increases from 0.30 to 0.38 and from 0.62 to 0.88 for Ktrans and ve, respectively, whereas it decreases from 0.52 to 0.46 for kep.
Figure 5.

Column graphs of ICC for the Ktrans, ve, and kep parameters obtained with the unadjusted (unadj., shaded light gray) and adjusted (adj., dark gray) AIFs. The respective 95% CIs are shown as error bars.
Concordance Analysis
Concordance correlation analysis was conducted to assess parameter agreement between any two AIFs within the same group (adjusted or unadjusted). The Ktrans CCC values are tabulated in Table 2A. CCC values for the unadjusted AIFs are listed above the dashed diagonal line, and those for the adjusted AIFs are listed below the diagonal line. Tables 2, B and C show the corresponding equivalents for the ve and kep parameters, respectively. The CCC ranges obtained with unadjusted AIFs are 0.031–0.944, 0.334–0.986, and 0.145–0.957 for Ktrans, ve, and kep, respectively; and those obtained with adjusted AIFs are 0.082–0.965, 0.554–0.993, and 0.129–0.965 for Ktrans, ve, and kep, respectively. In general, the CCC values increase from unadjusted to adjusted AIFs for the Ktrans and ve parameters, although few changes are observed for the kep parameter. In addition, the CCC value that resulted from pairing of the GP AIF with any other AIF (unadjusted or adjusted) is usually among the smallest in the CCC ranges described above.
Table 2A.
CCC Values of the Ktrans Parameter Obtained With Unadjusted and Adjusted AIFs
| QIN1 | QIN2 | QIN 3 | QIN4 | QIN5 | QIN6 | QIN7 | QIN8 | QIN9 | GP | |
|---|---|---|---|---|---|---|---|---|---|---|
| QIN1 | 0.185 | 0.759 | 0.825 | 0.501 | 0.755 | 0.747 | 0.412 | 0.941 | 0.049 | |
| QIN2 | 0.228 | 0.395 | 0.186 | 0.082 | 0.241 | 0.297 | 0.159 | 0.189 | 0.047 | |
| QIN3 | 0.726 | 0.507 | 0.645 | 0.336 | 0.836 | 0.889 | 0.301 | 0.732 | 0.083 | |
| QIN4 | 0.835 | 0.267 | 0.637 | 0.591 | 0.769 | 0.645 | 0.515 | 0.750 | 0.084 | |
| QIN5 | 0.776 | 0.179 | 0.531 | 0.880 | 0.472 | 0.367 | 0.307 | 0.445 | 0.223 | |
| QIN6 | 0.780 | 0.370 | 0.896 | 0.744 | 0.612 | 0.944 | 0.209 | 0.639 | 0.115 | |
| QIN7 | 0.755 | 0.376 | 0.887 | 0.662 | 0.965 | 0.548 | 0.262 | 0.612 | 0.105 | |
| QIN8 | 0.825 | 0.250 | 0.621 | 0.882 | 0.689 | 0.739 | 0.666 | 0.353 | 0.107 | |
| QIN9 | 0.897 | 0.130 | 0.610 | 0.720 | 0.605 | 0.747 | 0.507 | 0.655 | 0.031 | |
| GP | 0.210 | 0.082 | 0.212 | 0.287 | 0.297 | 0.393 | 0.300 | 0.253 | 0.104 |
CCC: concordance correlation coefficient; values from unadjusted AIFs are presented in the top right triangle and those from reference-tissue-adjusted AIFs are presented in bottom left triangle; Ktrans: rate constant for plasma/interstitium contrast reagent (CR) transfer.
Table 2B.
CCC Values of the ve Parameter Obtained With Unadjusted and Adjusted AIFs
| QIN1 | QIN2 | QIN3 | QIN4 | QIN5 | QIN6 | QIN7 | QIN8 | QIN9 | GP | |
|---|---|---|---|---|---|---|---|---|---|---|
| QIN1 | 0.458 | 0.873 | 0.696 | 0.822 | 0.911 | 0.890 | 0.713 | 0.913 | 0.732 | |
| QIN2 | 0.942 | 0.473 | 0.369 | 0.347 | 0.421 | 0.453 | 0.538 | 0.535 | 0.334 | |
| QIN3 | 0.981 | 0.969 | 0.666 | 0.587 | 0.868 | 0.886 | 0.676 | 0.745 | 0.510 | |
| QIN4 | 0.973 | 0.965 | 0.971 | 0.545 | 0.737 | 0.782 | 0.767 | 0.538 | 0.530 | |
| QIN5 | 0.726 | 0.668 | 0.693 | 0.803 | 0.787 | 0.719 | 0.591 | 0.733 | 0.936 | |
| QIN6 | 0.982 | 0.955 | 0.980 | 0.951 | 0.622 | 0.986 | 0.797 | 0.713 | 0.737 | |
| QIN7 | 0.993 | 0.965 | 0.992 | 0.982 | 0.984 | 0.732 | 0.838 | 0.696 | 0.688 | |
| QIN8 | 0.979 | 0.951 | 0.965 | 0.954 | 0.973 | 0.690 | 0.985 | 0.534 | 0.619 | |
| QIN9 | 0.952 | 0.820 | 0.913 | 0.887 | 0.924 | 0.703 | 0.929 | 0.900 | 0.621 | |
| GP | 0.929 | 0.879 | 0.949 | 0.873 | 0.933 | 0.554 | 0.931 | 0.924 | 0.880 |
CCC: concordance correlation coefficient; values from unadjusted AIFs are presented in the top right triangle and those from reference-tissue-adjusted AIFs are presented in bottom left triangle; ve: extravascular and extracellular volume fraction.
Table 2C.
CCC Values of the kep Parameter Obtained With Unadjusted and Adjusted AIFs
| QIN1 | QIN2 | QIN3 | QIN4 | QIN5 | QIN6 | QIN7 | QIN8 | QIN9 | GP | |
|---|---|---|---|---|---|---|---|---|---|---|
| QIN1 | 0.400 | 0.649 | 0.881 | 0.894 | 0.792 | 0.745 | 0.872 | 0.891 | 0.319 | |
| QIN2 | 0.303 | 0.620 | 0.339 | 0.327 | 0.553 | 0.645 | 0.407 | 0.335 | 0.145 | |
| QIN3 | 0.752 | 0.605 | 0.591 | 0.591 | 0.934 | 0.942 | 0.690 | 0.465 | 0.356 | |
| QIN4 | 0.872 | 0.282 | 0.586 | 0.932 | 0.684 | 0.595 | 0.866 | 0.788 | 0.352 | |
| QIN5 | 0.883 | 0.247 | 0.569 | 0.965 | 0.680 | 0.615 | 0.856 | 0.811 | 0.442 | |
| QIN6 | 0.775 | 0.427 | 0.856 | 0.677 | 0.656 | 0.957 | 0.705 | 0.579 | 0.385 | |
| QIN7 | 0.748 | 0.525 | 0.910 | 0.614 | 0.938 | 0.616 | 0.696 | 0.519 | 0.344 | |
| QIN8 | 0.867 | 0.308 | 0.656 | 0.873 | 0.683 | 0.856 | 0.697 | 0.722 | 0.346 | |
| QIN9 | 0.925 | 0.231 | 0.642 | 0.783 | 0.542 | 0.812 | 0.547 | 0.738 | 0.182 | |
| GP | 0.318 | 0.129 | 0.248 | 0.390 | 0.391 | 0.438 | 0.348 | 0.350 | 0.171 |
CCC: concordance correlation coefficient; values from unadjusted AIFs are presented in the top right triangle and those from reference-tissue-adjusted AIFs are presented in bottom left triangle; kep (= Ktrans/ve): CR intravasation rate constant.
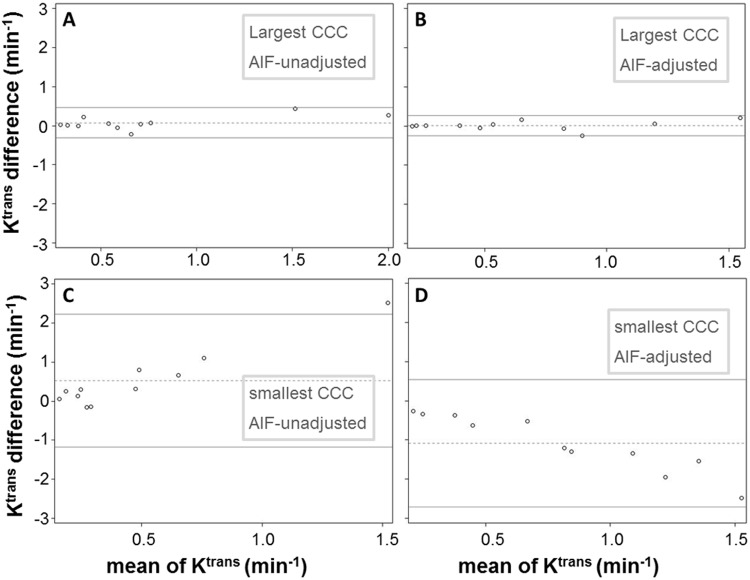
The Bland–Altman plots depicted in Figure 6 show examples of the agreement in Ktrans for AIF pairs with the largest (Figure 6, A and B) and smallest (Figure 6, C and D) CCC values within the unadjusted (Figure 6, A and C) and adjusted (Figure 6, B and D) AIF groups. Although the differences between measurements are mostly within the 95% CIs for all plots, it is rather visually clear (with all 4 plots having the same vertical-axis scale) that the width of confidence bands substantially differs between AIF pairs with greater CCC values (Figure 6, A and B) and those with smaller CCC values (Figure 6, C and D): narrower for the former, wider for the latter. For the AIF pairs with the largest CCCs (Figure 6, A and B), or the best agreements in the estimated Ktrans values, the means of Ktrans differences between the two AIFs represented by the dotted lines are close to 0 at 0.076 and 0.009 min−1, respectively, for unadjusted and adjusted AIFs. For the AIF pairs with the smallest CCCs (Figure 6, C and D), or the worst agreements in the estimated Ktrans values, the means of Ktrans differences are 0.529 and –1.085 min−1, respectively, for unadjusted and adjusted AIFs, considerably different from 0. In addition, in cases of poor Ktrans agreement (Figure 6, C and D), there seems to be a pattern of correlation between the difference in Ktrans and the mean of Ktrans, with larger differences corresponding to larger mean values.
Figure 6.
Bland–Altman plots are shown to demonstrate agreement in Ktrans for AIF pairs with the largest (A and B) and smallest (C and D) CCC values within the unadjusted (A and C) and adjusted (B and D) AIF groups. The two solid horizontal lines represent the upper and lower limits of the 95% CI, whereas the dotted horizontal line represents the mean value of Ktrans differences between the two measurements.
Discussion
The main goal of this multicenter AIF data analysis challenge was to evaluate variations of estimated pharmacokinetic parameters in prostate cancer due to differences in AIF determination. Individually measured AIFs were obtained for each DCE-MRI data set with different QIN center-specific methods, which include manual AIF–voxel selection, semiautomatic AIF–voxel identification, automated AIF region identification, and AIF determination using commercial software packages. Quality control measures such as fixed tumor ROI definition, fixed tumor T10, and central data analysis with a commonly used pharmacokinetic model were adopted to ensure that DCE-MRI parameter variations are mainly due to AIF differences only. Although the software package used by the managing center for central pharmacokinetic data analysis was an in-house-developed version of the TM (32), its mathematical formulation was validated using digital reference object phantom data in a previous DCE-MRI data analysis challenge (23).
The results from this multicenter study clearly show that variations in AIF quantification result in variations in the estimated pharmacokinetic parameter values for prostate DCE-MRI data. Among the Ktrans, kep, and ve parameters, Ktrans has the largest whereas ve has the smallest AIF uncertainty-caused variations. The wCV values ranged from as low as 0.33 for ve to as high as 0.74 for Ktrans with unadjusted AIFs, whereas the ICC ranged from 0.30 for Ktrans to 0.62 for ve. Similar results of ve being the most “robust” parameter to AIF variation have been reported in a simulation study (35) previously, and its “robustness” may be the direct consequence that ve is the most influential parameter within the Ktrans range (33) seen in prostate cancer. That is, when comparing the effects of the same percentage change in a single parameter on the DCE-MRI time-course, ve change causes the most noticeable DCE-MRI time-course deviation than the other parameters (33), suggesting that the ve parameter is shaped more by the tissue DCE time-course during model fitting. Another probable reason that Ktrans is more susceptible to AIF variation than ve is that quantification of Ktrans strongly depends on the initial AIF spike, whereas ve quantification relies more on the entire AIF time-course. This work complements a recent study comparing AIF determinations with fully automated and semiautomated approaches for prostate DCE-MRI data analysis (10). Both efforts show that Ktrans variation due to AIF uncertainty is the most prominent when compared with variations of other parameters in pharmacokinetic analysis of prostate cancer DCE-MRI data.
It is important to point out that the AIF influence on Ktrans estimation is dependent on CR extravasation (Ktrans magnitude) (33), underscoring the importance of accurate AIF measurement in Ktrans modeling when CR extravasation is extensive. This CR extravasation-dependent characteristic is more clearly illustrated in extreme cases such as normal brain tissue where gadolinium-based CR acts as an intravascular agent during the short period after CR injection. Under this condition of no CR extravasation, the AIF has no effect on Ktrans (which is undetectable). For organs with extensive CR extravasation, like the prostate (10), the initial AIF curve shape strongly influences the estimation of Ktrans. This is possibly the reason that the Ktrans values obtained with a fixed, population-based GP AIF show the least agreement (lowest CCC values) with those obtained with individually measured AIFs. The individually measured AIFs (mostly from the femoral artery voxels) from the actual DCE-MRI data generally captured similar initial AIF curve shapes despite the use of different quantification methods and potential errors from partial volume and inflow effects. The data acquisition-specific details may not be well characterized by the GP AIF, which is modeled on the basis of data from either the aorta or iliac arteries, acquired with different pulse sequence parameters and generated at different field strength. Thus, in cases of substantial CR extravasation, pharmacokinetic parameters should be estimated with individually determined AIFs whenever possible instead of a generic population-averaged AIF unrelated to a specific acquisition protocol.
As shown in this study, there are, however, steps one can take to lessen the effects of AIF variations on estimation of pharmacokinetic parameters. The agreement in the Ktrans and ve parameters obtained with reference tissue-adjusted AIFs is improved when compared with that obtained with unadjusted AIFs. This is a direct result of better agreement in AIF amplitude among the individually measured AIFs following the muscle reference tissue adjustment. However, the reference tissue approach is far from a perfect solution to AIF uncertainty-caused parameter variations. High wCV, low ICC, and low CCC values are still prevalent after the reference tissue method was used, particularly for Ktrans, of which the estimated value is strongly influenced by both the magnitude and the initial curve shape of the AIF.
The results from this multicenter challenge project are supported by findings from a recent simulation study (unpublished results), which aims to identify pharmacokinetic parameters that are relatively insensitive to AIF variations. In fact, the simulations show complete kep insensitivity to AIF magnitude errors (unpublished results). The current study shows that for TM analysis, kep is less sensitive to AIF uncertainty when compared with Ktrans. Defined as the CR intravasation rate constant, kep is predominantly characterized by the washout phase of the DCE time-course. Because kep is often calculated as Ktrans/ve and not as an independent variable in model fitting of the DCE time-course data, it is sometimes underused in clinical DCE-MRI studies. Results from this work, however, suggest that, considering the uncertainties in AIF determination, kep may be a more reproducible DCE-MRI parameter than the Ktrans parameter and thus a more robust imaging biomarker of perfusion and permeability. For prostate DCE-MRI, kep can offer a different perspective of prostate microvasculature, particularly when the Ktrans ranges of benign and cancerous tissue overlap (5).
It is important to note that DCE-MRI parameter variations caused by AIF variations are mostly systematic. As an example shown in Figure 2, the differences among the prostate tumor Ktrans maps obtained with different AIFs are mostly in voxel Ktrans values. The pattern of voxel Ktrans distribution largely remains similar for all the maps. This suggests that assessment of tumor heterogeneity through texture analysis of DCE-MRI parametric maps may not be affected greatly by variations in AIF determination. In addition, for longitudinal DCE-MRI studies to assess cancer therapy response, the systematic errors caused by AIF quantification variations may be largely cancelled in the calculation of percent changes of DCE-MRI parameters before and after therapy. In a multicenter breast DCE-MRI data analysis challenge study (23), we showed that the DCE-MRI parameter percent changes before and after the first cycle of neoadjuvant chemotherapy were substantially less sensitive to variations in the pharmacokinetic model and the software package used for data analysis when compared with the absolute parameter values.
Because of its unique temporal signatures and often superior image contrast, DCE-MRI is widely used in prostate imaging as part of a multiparametric prostate MRI protocol. DCE-MRI data analysis with qualitative and/or semiquantitative assessment is favored in current clinical practice largely because of their relative simplicity. However, the rich information embedded in the DCE-MRI data is likely underutilized. Pharmacokinetic data analysis for estimation of tissue biology-specific parameters has the potential to provide more consistent results for broad cross-vendor and cross-scanner platform applications. Robust and reliable AIF determination remains a real challenge for adoption of pharmacokinetic modeling of prostate DCE-MRI data in clinical settings. The results from this study provide useful information on how to minimize errors in the estimation of prostate DCE-MRI parameters caused by uncertainties in AIF determination and the parameters that are less sensitive to AIF variations.
There are limitations in this multicenter effort. The study cohort size was small (11 patients), and the AIF determination methods were mostly constrained to the approach of direct measurement from an artery. In addition, no longitudinal data were available, and, therefore, the effects of AIF variation on the DCE-MRI assessment of prostate cancer progression and/or response to treatment were not investigated. The current study only summarizes the results of AIF variations for a single pharmacokinetic model (TM); thus, parameter reproducibility from different models and relevant model comparisons with AIF variations are beyond the scope of this work. Finally, voxel DCE-MRI parameter distribution patterns were assessed visually without the use of a quantitative texture analysis method.
Conclusion
In conclusion, this multicenter data analysis study highlights one (not all) significant challenge in the quantitative pharmacokinetic analysis of DCE-MRI data, i.e., considerable variations in DCE-MRI parameter values were observed because of variations in AIF determination. The AIF-caused parameter variations are higher in Ktrans than in ve. As a solution to reduce parameter variation, AIF amplitude can be adjusted after its measurement by using the reference tissue method. Compared with Ktrans, kep is less sensitive to AIF uncertainty, suggesting that kep may be a more robust pharmacokinetic parameter for characterization of prostate microvasculature. The variations in parameter estimates caused by differences in AIF are systematic, and thus, the patterns of voxel-based DCE-MRI parametric maps were largely unaffected. In multicenter clinical trials involving quantitative DCE-MRI, central data analysis with a fixed AIF determination method should be adopted for a single time-point study to minimize the undesirable effects due to uncertainty in AIF determination. This approach may introduce systematic errors in estimated pharmacokinetic parameters, but it avoids random errors resulted from data analysis by individual centers with different AIF determination methods, which could be detrimental in addressing biological questions. In a longitudinal multicenter study, percent changes of pharmacokinetic parameters, instead of their absolute values, should be used as imaging endpoints to more accurately evaluate biological changes.
Acknowledgments
This study was supported by National Institutes of Health grants U01-CA154602, U01-CA151261, U01-CA183848, U01-CA154601, U01-CA148131, U01-CA176110, U01-CA172320, U01-CA142565, U01-CA166104, and U01-CA140230. Other than OHSU (which was the managing center) and BWH (which provided the shared prostate DCE-MRI data), all other participating QIN centers contributed equally to this study.
Conflict of Interest: None reported.
Disclosure: No disclosures to report.
Footnotes
- AIF
- Arterial input function
- CCC
- concordance correlation coefficient
- CR
- contrast reagent
- DCE-MRI
- dynamic contrast-enhanced magnetic resonance imaging
- ICC
- intraclass correlation coefficient
- Ktrans
- rate constant for plasma/interstitium contrast reagent transfer
- kep
- rate constant for contrast reagent intravasation
- R10 = 1/T10
- pre-contrast tissue longitudinal relaxation rate constant
- ROI
- region of interest
- TM
- Tofts model
- ve
- extravascular, extracellular volume fraction
- wCV
- within-subject coefficient of variation
References
- 1. Kayat Bittencourt L, Litjens G, Hulsbergen-van de Kaa CA, Turkbey B, Gasparetto EL, Barentsz JO. Prostate Cancer: The European Society of Urogenital Radiology prostate imaging reporting and data system criteria for predicting extraprostatic extension by using 3-T multiparametric MR imaging. Radiology. 2015. August;276(2):479–489. [DOI] [PubMed] [Google Scholar]
- 2. Buckley DL, Roberts C, Parker GJ, Logue JP, Hutchinson CE. Prostate cancer: evaluation of vascular characteristics with dynamic contrast-enhanced T1-weighted MR imaging–initial experience. Radiology. 2004. December;233(3):709–715. [DOI] [PubMed] [Google Scholar]
- 3. Alonzi R, Taylor NJ, Stirling JJ, d'Arcy JA, Collins DJ, Saunders MI, Hoskin PJ, Padhani AR. Reproducibility and correlation between quantitative and semiquantitative dynamic and intrinsic susceptibility-weighted MRI parameters in the benign and malignant human prostate. J Magn Reson Imaging. 2010. July;32(1):155–164. [DOI] [PubMed] [Google Scholar]
- 4. Koh TS, Thng CH, Ho JT, Tan PH, Rumpel H, Khoo JB. Independent component analysis of dynamic contrast-enhanced magnetic resonance images of breast carcinoma: a feasibility study. J Magn Reson Imaging. 2008. July;28(1):271–277. [DOI] [PubMed] [Google Scholar]
- 5. Oto A, Kayhan A, Jiang Y, Tretiakova M, Yang C, Antic T, Dahi F, Shalhav AL, Karczmar G, Stadler WM. Prostate cancer: differentiation of central gland cancer from benign prostatic hyperplasia by using diffusion-weighted and dynamic contrast-enhanced MR imaging. Radiology. 2010. December;257(3):715–723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Türkbey B, Thomasson D, Pang Y, Bernardo M, Choyke PL. The role of dynamic contrast-enhanced MRI in cancer diagnosis and treatment. Diagn Interv Radiol. 2010. September;16(3):186–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Franiel T, Hamm B, Hricak H. Dynamic contrast-enhanced magnetic resonance imaging and pharmacokinetic models in prostate cancer. Eur Radiol. 2011. March;21(3):616–626. [DOI] [PubMed] [Google Scholar]
- 8. Bonekamp D, Macura KJ. Dynamic contrast-enhanced magnetic resonance imaging in the evaluation of the prostate. Top Magn Reson Imaging. 2008. December;19(6):273–284. [DOI] [PubMed] [Google Scholar]
- 9. Li X, Priest RA, Woodward WJ, Tagge IJ, Siddiqui F, Huang W, Rooney WD, Beer TM, Garzotto MG, Springer CS Jr. Feasibility of shutter-speed DCE-MRI for improved prostate cancer detection. Magn Reson Med. 2013. March;27;69:171–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Fedorov A, Fluckiger J, Ayers GD, Li X, Gupta SN, Tempany C, Mulkern R, Yankeelov TE, Fennessy FM. A comparison of two methods for estimating DCE-MRI parameters via individual and cohort based AIFs in prostate cancer: a step towards practical implementation. Magn Reson Imaging. 2014. May;32(4):321–329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Eberhardt SC, Carter S, Casalino DD, Merrick G, Frank SJ, Gottschalk AR, Leyendecker JR, Nguyen PL, Oto A, Porter C, Remer EM, Rosenthal SA. ACR Appropriateness Criteria prostate cancer–pretreatment detection, staging, and surveillance. J Am Coll Radiol. 2013. February;10(2):83–92. [DOI] [PubMed] [Google Scholar]
- 12. The American College of Radiology. MR Prostate Imaging Reporting and Data System version 2.0. Accessed February 2016, from http://www.acr.org/Quality-Safety/eNews/Issue-09-March-2015/PIRADS-Version-2.
- 13. Schabel MC, Morrell GR. Uncertainty in T(1) mapping using the variable flip angle method with two flip angles. Phys Med Biol. 2009. January;54(1):N1–N8. [DOI] [PubMed] [Google Scholar]
- 14. Schabel MC, Fluckiger JU, DiBella EV. A model-constrained Monte Carlo method for blind arterial input function estimation in dynamic contrast-enhanced MRI: I. Simulations. Phys Med Biol. 2010. August;55(16):4783–4806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Fluckiger JU, Schabel MC, DiBella EV. Toward local arterial input functions in dynamic contrast-enhanced MRI. J Magn Reson Imaging. 2010. October;32(4):924–934. [DOI] [PubMed] [Google Scholar]
- 16. Yang C, Karczmar GS, Medved M, Stadler WM. Estimating the arterial input function using two reference tissues in dynamic contrast-enhanced MRI studies: fundamental concepts and simulations. Magn Reson Med. 2004. November;52(5):1110–1117. [DOI] [PubMed] [Google Scholar]
- 17. Yang C, Karczmar GS, Medved M, Stadler WM. Multiple reference tissue method for contrast agent arterial input function estimation. Magn Reson Med. 2007. December;58(6):1266–1275. [DOI] [PubMed] [Google Scholar]
- 18. Shi L, Wang D, Liu W, Fang K, Wang YX, Huang W, King AD, Heng PA, Ahuja AT. Automatic detection of arterial input function in dynamic contrast enhanced MRI based on affinity propagation clustering. J Magn Reson Imaging. 2014. May;39(5):1327–1337. [DOI] [PubMed] [Google Scholar]
- 19. Li X, Welch EB, Arlinghaus LR, Chakravarthy AB, Xu L, Farley J, Loveless ME, Mayer IA, Kelley MC, Meszoely IM, Means-Powell JA, Abramson VG, Grau AM, Gore JC, Yankeelov TE. A novel AIF tracking method and comparison of DCE-MRI parameters using individual and population-based AIFs in human breast cancer. Phys Med Biol. 2011. September;56(17):5753–5769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Chen J, Yao J, Thomasson D. Automatic determination of arterial input function for dynamic contrast enhanced MRI in tumor assessment. Med Image Comput Comput Assist Interv. 2008;11(Pt 1):594–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Leach MO, Morgan B, Tofts PS, Buckley DL, Huang W, Horsfield MA, Chenevert TL, Collins DJ, Jackson A, Lomas D, Whitcher B, Clarke L, Plummer R, Judson I, Jones R, Alonzi R, Brunner T, Koh DM, Murphy P, Waterton JC, Parker G, Graves MJ, Scheenen TW, Redpath TW, Orton M, Karczmar G, Huisman H, Barentsz J, Padhani A; Experimental Cancer Medicine Centres Imaging Network Steering Committee. Imaging vascular function for early stage clinical trials using dynamic contrast-enhanced magnetic resonance imaging. Eur Radiol. 2012. July;22(7):1451–1464. [DOI] [PubMed] [Google Scholar]
- 22. Khalifa F, Soliman A, El-Baz A, Abou El-Ghar M, El-Diasty T, Gimel'farb G, Ouseph R, Dwyer AC. Models and methods for analyzing DCE-MRI: a review. Med Phys. 2014. December;41(12):124301. [DOI] [PubMed] [Google Scholar]
- 23. Huang W, Li X, Chen Y, Li X, Chang MC, Oborski MJ, Malyarenko DI, Muzi M, Jajamovich GH, Fedorov A, Tudorica A, Gupta SN, Laymon CM, Marro KI, Dyvorne HA, Miller JV, Barbodiak DP, Chenevert TL, Yankeelov TE, Mountz JM, Kinahan PE, Kikinis R, Taouli B, Fennessy F, Kalpathy-Cramer J. Variations of dynamic contrast-enhanced magnetic resonance imaging in evaluation of breast cancer therapy response: a multicenter data analysis challenge. Transl Oncol. 2014. February;7(1):153–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Cao Y, Li D, Shen Z, Normolle D. Sensitivity of quantitative metrics derived from DCE MRI and a pharmacokinetic model to image quality and acquisition parameters. Acad Radiol. 2010. April;17(4):468–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Jena A, Mehta SB, Taneja S. Optimizing MRI scan time in the computation of pharmacokinetic parameters (K(trans)) in breast cancer diagnosis. J Magn Reson Imaging. 2013. September;38(3):573–579. [DOI] [PubMed] [Google Scholar]
- 26. Kovar DA, Lewis M, Karczmar GS. A new method for imaging perfusion and contrast extraction fraction: input functions derived from reference tissues. J Magn Reson Imaging. 1998. Sep-Oct;8(5):1126–1134. [DOI] [PubMed] [Google Scholar]
- 27. Shanbhag DD, Gupta SN, Rajamani KT, Zhu Y, Mullick R. A generalized methodology for detection of vascular input function with dynamic contrast enhanced perfusion data. Proc Intl Soc Magn Reson Med. 2012. 20:3524. [Google Scholar]
- 28. Parker GJ, Roberts C, Macdonald A, Buonaccorsi GA, Cheung S, Buckley DL, Jackson A, Watson Y, Davies K, Jayson GC. Experimentally-derived functional form for a population-averaged high-temporal-resolution arterial input function for dynamic contrast-enhanced MRI. Magn Reson Med. 2006. November;56(5):993–1000. [DOI] [PubMed] [Google Scholar]
- 29. Hegde JV, Mulkern RV, Panych LP, Fennessy FM, Fedorov A, Maier SE, Tempany CM. Multiparametric MRI of prostate cancer: an update on state-of-the-art techniques and their performance in detecting and localizing prostate cancer. J Magn Reson Imaging. 2013. May;37(5):1035–1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Gupta R. A new look at the method of variable nutation angle for the measurement of spin-lattice relaxation time using Fourier transform NMR. J Magn Reson. 1977;25:231–235. [Google Scholar]
- 31. Lu H, Clingman C, Golay X, van Zijl PC. Determining the longitudinal relaxation time (T1) of blood at 3.0 Tesla. Magn Reson Med. 2004. September;52(3):679–682. [DOI] [PubMed] [Google Scholar]
- 32. Tofts PS, Brix G, Buckley DL, Evelhoch JL, Henderson E, Knopp MV, Larsson HB, Lee TY, Mayr NA, Parker GJ, Port RE, Taylor J, Weisskoff RM. Estimating kinetic parameters from dynamic contrast-enhanced T(1)-weighted MRI of a diffusable tracer: standardized quantities and symbols. J Magn Reson Imaging. 1999. September;10(3):223–232. [DOI] [PubMed] [Google Scholar]
- 33. Li X, Priest RA, Woodward WJ, Siddiqui F, Beer TM, Garzotto MG, Rooney WD, Springer CS Jr. Cell membrane water exchange effects in prostate DCE-MRI. J Magn Reson. 2012. May;218:77–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Padhani AR, Hayes C, Landau S, Leach MO. Reproducibility of quantitative dynamic MRI of normal human tissues. NMR Biomed. 2002. April;15(2):143–153. [DOI] [PubMed] [Google Scholar]
- 35. Cheng HL. Investigation and optimization of parameter accuracy in dynamic contrast-enhanced MRI. J Magn Reson Imaging. 2008. September;28(3):736–743. [DOI] [PubMed] [Google Scholar]
- 36. Hyvärinen A. Fast and robust fixed-point algorithms for independent component analysis. IEEE Trans Neural Netw. 1999;10(3):626–634. [DOI] [PubMed] [Google Scholar]
- 37. Beckmann CF, Smith SM. Probabilistic independent component analysis for functional magnetic resonance imaging. IEEE Trans Med Imaging. 2004. February;23(2):137–152. [DOI] [PubMed] [Google Scholar]
- 38. Mehrabian H, Chandrana C, Pang I, Chopra R, Martel AL. Arterial input function calculation in dynamic contrast-enhanced MRI: an in vivo validation study using co-registered contrast-enhanced ultrasound imaging. Eur Radiol. 2012. August;22(8):1735–1747. [DOI] [PubMed] [Google Scholar]
- 39. Huang J, O'Sullivan F. An analysis of whole body tracer kinetics in dynamic PET studies with application to image-based blood input function extraction. IEEE Trans Med Imaging. 2014. May;33(5):1093–1108. [DOI] [PMC free article] [PubMed] [Google Scholar]