SUMMARY
One of the most remarkable innovations of the vertebrate adaptive immune system is the progressive organization of the lymphoid tissues that leads to increased efficiency of immune surveillance and cell interactions. The mucosal immune system of endotherms has evolved organized secondary mucosal lymphoid tissues (O-MALT) such as Peyer’s patches, tonsils, and adenoids. Primitive semi-organized lymphoid nodules or aggregates (LAs) were found in the mucosa of anuran amphibians [1], suggesting that O-MALT evolved from amphibian LAs_250 million years ago [1–4]. This study shows for the first time the presence of O-MALT in the mucosa of the African lungfish, an extant representative of the closest ancestral lineage to all tetrapods. Lungfish LAs are lymphocyte-rich structures associated with a modified covering epithelium and express all IGH genes except for IGHW2L. In response to infection, nasal LAs doubled their size and increased the expression of CD3 and IGH transcripts. Additionally, de novo organogenesis of inducible LAs resembling mammalian tertiary lymphoid structures was observed. Using deep-sequencing transcriptomes, we identified several members of the tumor necrosis factor (TNF) superfamily, and subsequent phylogenetic analyses revealed its extraordinary diversification within sarcopterygian fish. Attempts to find AICDA in lungfish transcriptomes or by RT-PCR failed, indicating the possible absence of somatic hypermutation in lungfish LAs. These findings collectively suggest that the origin of O-MALT predates the emergence of tetrapods and that TNF family members play a conserved role in the organization of vertebrate mucosal lymphoid organs.
RESULTS AND DISCUSSION
Lungfish Have Organized Lymphoid Structures in the Nasopharyngeal Cavity and Gut Primarily Composed of Lymphocytes
Organized mucosa-associated lymphoid tissue (O-MALT) is thought to be an innovation unique to endotherms. Teleost fish lack O-MALT [5], whereas amphibians appear to have primitive lymphoid nodules in their gut and lingual mucosa [1–4, 6]. We investigated the mucosal lymphoid tissues of the African lungfish (Protopterus dolloi and P. annectens; Sarcopterygii: Dipnoi), a taxon that holds a key phylogenetic position as the sister group of modern tetrapods. Previously, Peyer’s patch (PP)-like structures were observed in the gut of the Australian lungfish (Neoceratodus forsteri) using routine histology [7].
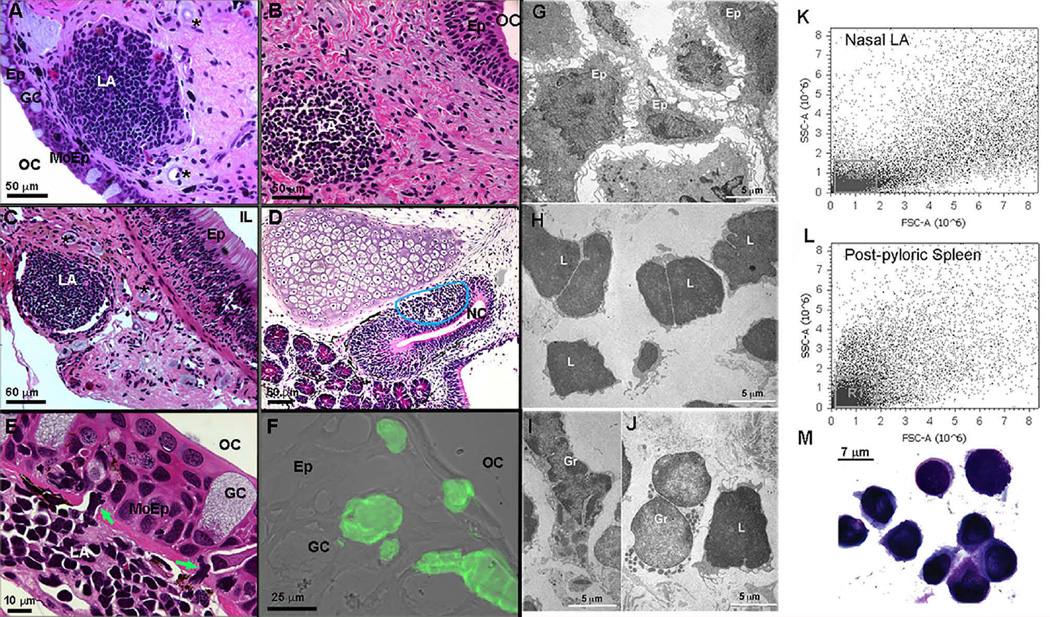
We found both encapsulated and unencapsulated lymphoid aggregates (LAs) in the nasopharyngeal and intestinal mucosa of African lungfish (Figures 1A–1C). The olfactory organs of wild-caught bullfrog tadpoles (Lithobates catesbeianus) also showed unencapsulated LAs at the basal part of the olfactory epithelium (Figure 1D). Lungfish LAs were found either in the submucosa or underneath the epithelium. The covering epithelium had fewer goblet cells present and infiltrating leukocytes in the base of the epithelium (Figure 1E), features typically seen in follicle-associated epithelia (FAEs) such as the epithelium covering anuran tonsillar LAs [6]. Mammalian nasopharynx-associated lymphoid tissue (NALT) M cells are characterized by a specific isolectin-staining pattern, almost exclusively stained with the Griffonia simplicifolia I isolectin-B4 (GSI-B4) directed against α-linked galactoses [8, 9]. Isolectin-B4+ cells were present in lungfish with a goblet cell-like morphology (Figure 1F). Using serial frozen sections, we quantified the number and size of nasal LAs. The mean number of LAs was three (n = 5), with a maximum long diameter, short diameter, and depth of 473.25 ± 11.0 µm, 338.5 ± 19.3 µm, and 185 ± 40.2 µm, respectively. Assuming an ellipsoid shape, the mean total volume of control LAs was 1.55 × 107 µm3.
Figure 1. African Lungfish Posses Organized Mucosal LAs in Mucosal Epithelia.
(A) H&E stain of a P. annectens encapsulated nasal LA. Asterisks denote surrounding blood vessels (micro lymphatic pump).
(B) H&E stain of a P. dolloi lower-jaw unencapsulated LA.
(C) H&E stain of a P. dolloi encapsulated intestinal LA. Asterisks denote surrounding blood vessels (micro lymphatic pump).
(D) H&E stain of a bullfrog tadpole unencapsulated nasal LA.
(E) H&E stain of a modified FAE covering a P. dolloi nasal LA.
(F) Isolectin-B4 stain (green) of a cryosection of the modified epithelium covering a nasal LA in P. dolloi. Note the absence of goblet cells and the presence of infiltrating lymphocytes (green arrows). A fluorescent image was overlaid with a differential interference contrast (DIC) image (n = 3).
(G) Transmission electron microscopy (TEM) shows epithelial-like (Ep) cells in the stroma of a P. dolloi nasal LA.
(H) TEM micrograph of large and small lymphocytes (L) forming the majority of the nasal LA.
(I and J) TEM micrographs of two different types of granulocytes (Gr) found in P. dolloi nasal LA.
(K) Flow cytometry dot plot of a P. dolloi nasal LA single-cell suspension indicates that the majority of the cells have lymphocyte-like morphology (R1), as evidenced by their low side-scatter and forward-scatter values.
(L) Flow-cytometry dot plot of a P. dolloi post-pyloric spleen single-cell suspension known to contain mostly lymphocytes (R1).
(M) Giemsa stain of a P. dolloi nasal LA single-cell suspension demonstrating the presence of lymphocytes in this organ (n = 3).
LA, lymphoid aggregate; OC, oral cavity; GC, goblet cell, Ep, epithelium; IL, intestinal lumen; NC, nasal cavity; MoEp, modified epithelium. See also Figure S1 and Table S2.
By transmission electron microscopy (TEM), we observed stromal cells, such as epithelial cells and fibroblastic reticular cells, lymphocytes (7–10 µm in diameter), and granulocytes in lungfish LAs (Figures 1G–1J). Flow cytometry revealed that the majority of the cells in lungfish LAs belonged to the lymphocyte gate (Figure 1K), a finding confirmed by Giemsa-stained cytospins (Figure 1M). Post-pyloric spleen cell suspensions (Figure 1L) were used as a positive control because they are a B-cell-rich tissue in P. dolloi [10]. High endothelial venules (HEVs), a key feature of functional lymphoid structures [11], were not found in lungfish. Staining with an anti-mouse peripheral lymph node addressin (PNad) antibody (MECA-79) revealed positively stained vessels surrounding nasal LAs (Figures S1A–S1D). No attempt to identify the reactive epitope in lungfish was made.
Lack of Segregation of T and B Cell Areas in Lungfish Nasal LA
Segregation of lymphoid structures into B and T cell areas and germinal center (GC) formation are canonical features of the mammalian immune system. We sought to determine whether lungfish LAs have an organization similar to mammalian O-MALT. We cloned P. dolloi PTPRC (CD45) and CD3E as pan-leukocyte and pan-T cell markers, respectively. African lungfish possess two primordial gnathostome immunoglobulin (Ig) classes (IGHM and IGHW) as well as two unique Ig isotypes (IGHN and IGHQ) [10]. Fluorescence in situ hybridization (FISH) staining revealed that most cells were PTPRC+ (Figure S1E) and that CD3E+ T cells and IGHW1/IGHM1 B cells were abundant in nasal LAs, accounting for 32.7% ± 0.5% and 26.5% ± 0.3% of all cells, respectively (Figures S1F, S1G, S2C, and S2D). We found no evidence for areas with CD3E or IGH restricted expression, indicating that T and B cells are randomly distributed within the LAs.
Presence of GCs in lungfish LAs was studied by peanut agglutinin (PNA) staining using mouse lymph nodes (LNs) as a positive control. Lungfish LAs were negative for PNA stain, suggesting a lack of GCs, whereas mouse LNs were positive (data not shown). These results indicate that sarcopterygian fish MALT does not reach the organization levels found in mammals, as suggested in amphibians [2]. Despite the absence of GCs in the mucosal LAs of ectotherms, these structures must have conferred some evolutionary advantage. We speculate that exposure to both waterborne and airborne microorganisms due to the dual mode of respiration might have been a strong enough evolutionary pressure for lungfishes to acquire more sophisticated mucosal immune structures such as LAs.
No Evidence of AICDA Expression or SHM in Lungfish LAs
Since somatic hypermutation (SHM), a mechanism for generation of antibody diversity can happen in the absence of GC formation [12], we next tested whether SHM was occurring in lungfish nasal LAs. Lungfish were infected with Edwarsiella ictaluri [13], resulting in 2 × 104 E. ictaluri colony forming units (CFU)/200 ng of DNA in infected post-pyloric spleen or nose tissue and 9 × 103 CFU/200 ng of infected pre-pyloric spleen DNA. No E. ictaluri was detected in the respective control tissues.
Nasal LAs from uninfected and infected lungfish were collected by laser capture microdissection (LCM), and RNA was purified. IGHM1 was previously found to be upregulated in infected versus control lungfish and was therefore selected for amplification. The IGHM1 heavy-chain V regions were amplified, and a total of 15 clones from control or infected samples were sequenced. We found that both control and infected fish presented VH1 region (9 and 11, respectively) and VH2 region (6 and 4, respectively) with identical nucleotide sequences (Figure S2A). These sequences had been previously found during an extensive VH family analysis in African lungfish spleen transcriptomes [10].
SHM requires deamination of cytosine to uracyl in DNA by activation-induced (cytidine) deaminase (AICDA). We searched for AICDA sequences in all of our lungfish transcriptome databases (see the Supplemental Experimental Procedures for details), but no sequence matches were found using cartilaginous fish, teleost fish, or tetrapod AICDA sequences, suggesting that African lungfish may have lost AICDA expression during evolution. We ran a number of PCR experiments using different primer sets targeting the conserved motif of all vertebrate AICDA sequences (Table S1) using mammalian spleen cDNA as a positive control. These experiments failed to identify any AICDA transcripts in lungfish despite the positive control producing a PCR band that was confirmed to be AICDA (data not shown). Thus, our results suggest that African lungfish may have lost the expression of AICDA. Next, we searched in the lungfish transcriptomes for other members of the apolipoprotein B mRNA-editing catalytic polypeptide (APOBEC) family to which AICDA belongs. We found that APOBEC1, APOBEC2, APOBEC3C, and APOBEC4 (GenBank: KP792760, KP792761, KP792762, KP792763) are expressed by Protopterus sp. (Figure S2B). However, other members of the APOBEC3 family were not identified, supporting the notion that APOBEC3 diversified in mammals, mostly in primates [14, 15]. AICDA and SHM are believed to be features of all living jawed vertebrates, including cartilaginous fish [15]. AICDA is expressed in teleost fish, but teleosts do not undergo class switching [16]. With respect to SHM, it seems that teleosts utilize SHM, but its function is still not clear [17, 18]. Previous studies have shown that SHM in teleosts does not lead to specific selection of high-affinity B cell mutants, but rather it is used as a mechanism to increase antibody repertoires [17, 19, 20]. Regarding our SHM findings in lungfish, without a genome, it is difficult to ascertain our AICDA and SHM findings. It is possible that E. ictaluri did not prime LAs adequately for SHM to take place, but this is unlikely, given the significant changes in IGH expression recorded here. Alternatively, since the lungfish used in these experiments are not immunologically naive, previous exposures to other pathogens may have masked our results.
The TNFSF Diversified in Sarcopterygian Fish
Tumor necrosis factor superfamily (TNFSF) members are known to govern the development and organization of lymphoid tissues in mammals including PPs and NALT. It has been proposed that TNFSF and TNF receptor superfamily (TNFRSF) paralogues were co-opted by, and are most likely crucial for, the evolution of the adaptive immune system [21–24]. We identified sequence homologs for these molecules in the Protopterus sp. transcriptomes and compared them to human, teleost fish, and coelacanth TNFSF and TNFRSF. We found a total of 13 and 15 genes belonging to TNFSF and TNFRSF, respectively. Three of the TNFSF molecules present in lungfish are also present in human but absent in teleosts (TNFSF1 [lymphotoxin α, LTA], TNFSF3 [lymphotoxon β, LTB], and TNFSF8 [CD30L]). Interestingly, these same three genes were found in the coelacanth genome (Table 1). Out of the 15 members of the TNFRSF found in lungfish, five genes have only been reported to be present in both mammals and coelacanths. Lungfish also possess one gene present in humans but absent in coelacanths (TNFRSF7, CD27) (Table 1), a marker for memory B cells with a central role in T cell and B cell responses in humans [11]. Interestingly, a subset of IgD+ B cells in human tonsils specifically expresses TNFRSF7 cell-surface antigen [25]. Thus, the analysis of TNFRSF revealed the presence of 28 genes in humans; of these, only eight have been reported in mammals and fish (teleost and sarcopterygian fish). This result may indicate that different species have selectively lost paralogues post genome duplication.
Table 1.
Bioinformatic Analysis of TNFSF Genes and TNFRSF Genes Present in Humans, Teleosts, Coelacanths, and Lungfish
| Human | Teleost | Coelacanth | Lungfish | |
|---|---|---|---|---|
| TNFSF (Ligands) | ||||
| EDA | + | + | + | + |
| TNFSF5 | + | + | + | + |
| TNFSF6 | + | + | + | + |
| TNFSF4 | + | − | − | − |
| TNFSF18 | + | + | − | − |
| TNFSF8 | +a | − | +a | +a |
| TNFSF15 | + | + | + | + |
| TNFSF14 | + | + | + | + |
| TNFSF9 | + | − | − | − |
| TNFSF7 | + | − | − | − |
| TNFSF1 | +a | − | +a | +a |
| TNFSF2 | + | + | + | + |
| TNFSF3 | +a | − | +a | +a |
| TNFSF12 | + | + | + | + |
| TNFSF13 | + | + | + | + |
| TNFSF13B | + | + | + | + |
| TNFSF11 | +b | +b | +b | − |
| TNFSF10 | + | + | + | + |
| LTA/TNF-New | − | +c | − | − |
| Total number | 18 | 13 | 14 | 13 |
| TNFRSF (Receptors) | ||||
| TNFRSF16 | + | + | + | + |
| TNFRSF19 | +d | − | +d | − |
| EDAR | +b | +b | +b | − |
| XEDAR | + | − | − | − |
| TNFRSF5 | + | + | + | + |
| TNFRSF6B | +d | − | +d | − |
| TNFRSF6 | + | + | + | + |
| TNFRSF4 | + | − | − | − |
| TNFRSF18 | + | − | − | − |
| TNFRSF8 | + | − | − | − |
| TNFRSF14 | +a | − | +a | +a |
| TNFRSF9 | +a | − | +a | +a |
| TNFRSF1B | + | − | − | − |
| TNFRSF12 | + | − | − | − |
| TNFRSF7 | +e | − | − | +e |
| TNFRSF1A | + | + | + | + |
| TNFRSF3 | + | + | + | + |
| TNFRSF11A | + | − | +a | +a |
| TACI | + | − | − | − |
| TNFRSF17 | +f | +f | − | +f |
| DR6 | + | + | + | + |
| TNFRSF11B | +a | − | +a | +a |
| TNFRSF10A | + | + | + | + |
| TNFRSF10B | + | − | − | − |
| TNFRSF10C | + | − | − | − |
| TNFRSF10D | + | − | − | − |
| IL7RA | + | + | + | + |
| TNFRSF13b | +a | − | +a | +a |
| Total number | 28 | 10 | 16 | 15 |
See also Figure S3.
A gene present in mammals, coelacanth, and lungfish, but not in teleost fish.
A gene present in mammals, teleost, and coelacanth, but not in in lungfish.
A gene only present in teleost, and not sarcopterygian fish or mammals.
A gene present in mammals and coelacanth, but not in in teleost and lungfish.
A gene present in mammals and lungfish, but not in teleost and coelacanth.
A gene present in human, teleost, and lungfish, but not in coelacanth.
Phylogenetic analyses of vertebrate LT molecules (Figure S3) indicate that vertebrate LTA, LTB, and LTBR cluster together, forming two different clades, with coelacanth and lungfish molecules being closely related. Teleost fish lack unambiguous orthologs of LTA and LTB but possess a potentially related ligand (TNF-New) that is physically adjacent to TNF [22]. Altogether, our phylogenetic analyses suggest that TNFSF/TNFRSF (including LT) underwent a significant expansion during the transition of vertebrates from water to land. Alternatively, this diversity may be present earlier on but lost in teleosts, as suggested by others [22].
Adhesion Molecules, Chemokines, TNFSF Members, and ID2 Are Expressed in Lungfish LAs
Organization of lymphocytes in LNs requires signals delivered by lymphoid tissue inducer (LTi) cells such as adhesion molecules, chemokines, cytokines, and several TNFSF members [26]. Using LCM-dissected nasal and intestinal LAs from control lungfish, we were able to detect the expression of immune cell markers, as well as genes responsible for the formation and organization of lymphoid tissue [26]. We found that both nasal and intestinal LAs express the majority of immune markers and MALT-related genes such as IGH genes, interleukin-7 receptor alpha (IL7RA), vascular cell adhesion molecule 1 (VCAM1), intercellular adhesion molecule 1 (ICAM1), and LTA and LTB (TNFSF1 and TNFSF3, respectively) (Table S2). Interestingly, neither express mucosal addressin cell adhesion molecule 1 (MADCAM1), an endothelial cell adhesion marker that directs leukocytes to sites of inflammation [27]. Nasal LAs alone were seen to express the LTBR (TNFRSF3) and the TNF-related activation-induced cytokine receptor (TNFRSF11A), known to play a critical role in antigen-specific T cell responses (Table S2). Overall, intestinal LAs do not express as many “LN-organizing molecules” as nasal LAs, indicating that there are important differences in the development and function of these two O-MALT structures in Dipnoi. This is in agreement with previous studies in mammals that demonstrated that NALT and PP formation and developmental programs are considerably different [28]. Finally, CXCR5, but not CCR7 or CCL19, was expressed within nasal LAs, supporting our observations regarding the absence of a T cell zone in these structures.
Nasal Luminal Antigen Is Transported to Lungfish Nasal LAs via Luminal Sampling and Migrating Leucocytes
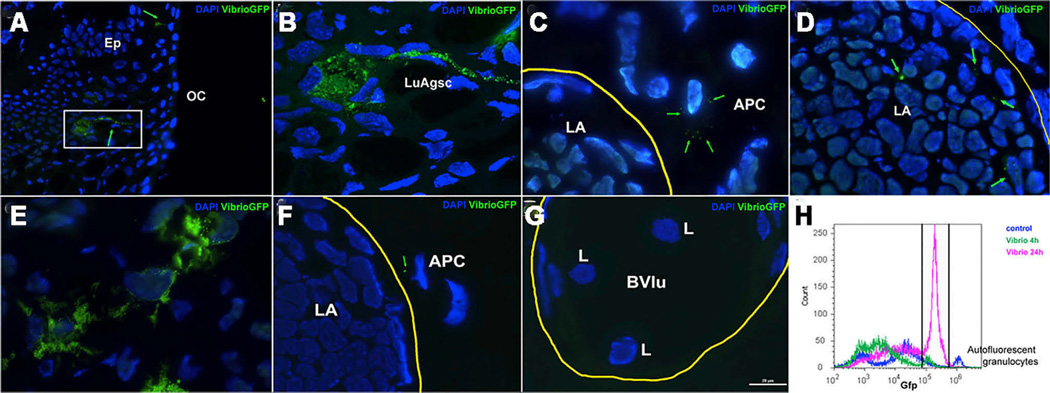
Mucosal lymphoid tissues can collect antigen via a number of mechanisms, including M cells, macrophages, and dendritic cells [29, 30]. Here we used the fish pathogen Vibrio anguillarum labeled with GFP and injected it directly into the nasal passages of P. dolloi. At 4 hr, we identified luminal antigen sampling cells in the nasal epithelium that were loaded with GFP-bacteria (Figures 2A and 2B). Importantly, antigen-loaded leucocytes with migratory morphology and similar to macrophages were observed adjacent to the nasal LA structure (Figure 2C). Additionally, few bacterial cells were observed inside the outer layer of the LA area (Figure 2D). At 24 hr, many bacteria-loaded leucocytes were observed in the epithelium surrounding the nasal LAs (Figure 2E). Migratory leukocytes were also observed immediately adjacent to the LA (Figure 2F), and the surrounding blood vessels contained numerous lymphocyte-like cells (Figure 2G) not present in the controls. By flow cytometry, the number of GFP+ cells was 7.1% at 4 hr and 20% at 24 hr, whereas in control animals it was 4% (due to high-intensity auto-fluorescent granulocytes) (Figure 2H). Thus, lungfish nasal LAs appear to have similar mechanisms to collect antigen as those described in mucosal lymphoid tissues of mammals.
Figure 2. Nasally Delivered Bacterial Pathogens Reach P. dolloi Nasal LAs via Luminal Antigen Sampling Cells and Migratory Macrophages.
Live GFP-V. anguillarum cells were delivered into the nasal cavity of P. dolloi. Nasal LAs and covering epithelium were collected 4 hr and 24 hr later.
(A) Immunofluorescence microscopy image of the covering epithelium of a P. dolloi nasal LA 4 hr after GFP-V. anguillarum delivery. A luminal antigen sampling cell (green arrow) is loaded with GFP-bacteria.
(B) Confocal microscopy image of the boxed area in (A).
(C) Immunofluorescence microscopy image of a P. dolloi nasal LA 4 hr after GFP-V. anguillarum delivery. A migratory macrophage with internalized GFP-bacteria is shown next to the nasal LA.
(D) Immunofluorescence microscopy image of a P. dolloi nasal LA 4 hr after GFP-V. anguillarum delivery. Bacterial cells are present inside the LA.
(E) Immunofluorescence microscopy image of the covering epithelium of a P. dolloi nasal LA 24 hr after GFP-V. anguillarum delivery. The amount of GFP fluorescence inside phagocytic cells is greater than at 4 hr.
(F) A GFP+ leucocyte (putative antigen-presenting cell, APC) can be observed adjacently to the nasal LA at 24 hr.
(G) Blood vessels surrounding nasal LAs carry abundant lymphocyte like cells 24 hr after bacteria delivery.
(H) Flow-cytometry histogram of nasal cell suspensions from control (dark-blue line), 4 hr GFP-V. anguillarum (green line), and 24 hr GFP-V. anguillarum (pink line). The gate indicates the percentage of GFP+ cells. The small but high-intensity peak found in the control sample may be due to auto-fluorescent granulocytes (n = 1).
OC, oral cavity; Ep, epithelium; LuAgsc, luminal antigen sampling cell; LA, lymphoid aggregate; APC, putative antigen-presenting cell; L, lymphocyte; Bvlu, blood vessel lumen. Yellow lines circle LAs in (C), (D), and (F) and a blood vessel in (G). Green arrows point GFP-bacteria. Cell nuclei were stained with DAPI (blue).
Microbial Exposure Leads to Architectural Changes in Nasal LAs and Formation of Tertiary Lymphoid Structures in Lungfish
Unprogrammed lymphoid structures known as tertiary lymphoid structures such as isolated lymphoid follicles (ILFs) can also develop in a number of body sites in response to chronic inflammation, cancer, or microbial exposure [31]. Moreover, it has been proposed that ILFs may be the evolutionary forerunners of specialized O-MALT [21].
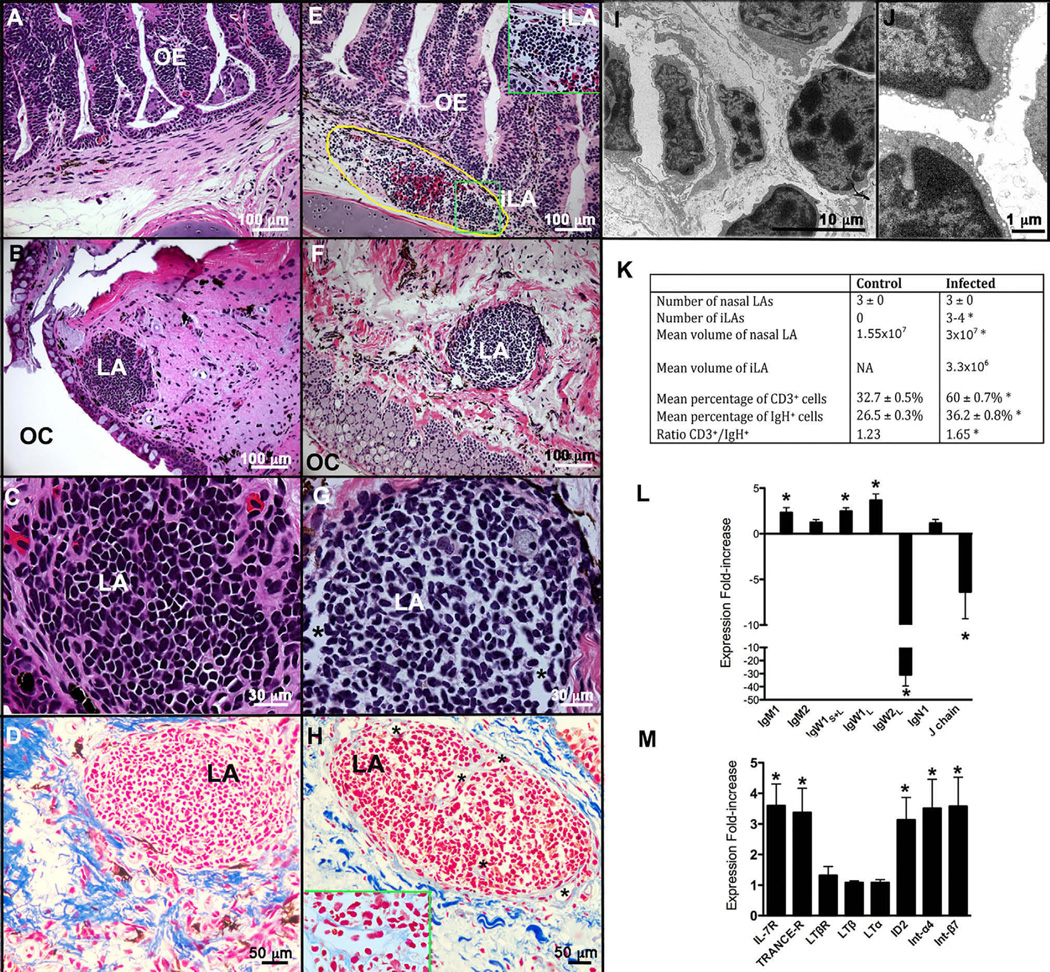
Changes in the architecture of nasal LAs were examined after nasal infection with E. ictaluri (Figures 3A–3F). The packing density of the cells was reduced in infected compared to control LAs, with the presence of numerous gaps among lymphocytes (Figures 3B–3D and 3F–3H). H&E stains showed that lymphocyte morphology was less homogenous in infected than in uninfected LAs, with elongated and invaginated cell shapes dominating the sample. Trichrome stains further confirmed the changes in architecture, with abundant capillaries within the infected LA not seen in controls (Figures 3A–3H). These capillaries were also observed by TEM (Figure 3I). The lymphocytes inside the infected LAs also had a different ultrastructure compared to controls. Specifically, their chromatin was less densely packed, and they had abundant cytoplasmic vesicles at the rim of the cell (Figures 3I and 3J).
Figure 3. P. dolloi NALT Responds to Nasal Infection with Enterobacteria.
Architectural changes in nasal LAs, as well as formation of new LAs (named inducible LAs, iLAs), was observed in infected individuals compared to control lungfish.
(A) H&E stain of a control nasal epithelium.
(B) H&E stain of a nasal LA of a control P. dolloi.
(C) Magnified view of the architecture of a control nasal LA (H&E stain).
(D) Trichrome Heidenhain’s azan stain of a control nasal LA. Cell nuclei are in red, and collagen fibers are in dark blue.
(E) H&E stain of the nasal epithelium of an infected lungfish showing the presence of iLA at the base of the epithelium. Circled yellow line shows an area of bleeding containing an iLA. The green box inset shows a magnified view of an iLA.
(F) H&E stain of a nasal LA of an infected P. dolloi.
(G) Magnified view of the architecture of an infected nasal LA (H&E stain).
(H) Trichrome Heidenhain’s azan stain of a control nasal LA. Cell nuclei are in red, and collagen fibers are in dark blue. The inset shows a capillary formed in the center of an infected LA.
(I) Transmission electron micrograph of an infected LA showing a capillary (black asterisk) and lymphocytes (L).
(J) Transmission electron micrograph of the lymphocytes (L) of an infected LA showing small vesicles in their cytoplasm.
(K) Summary table of the main differences between control and infected LAs and their architecture. Asterisks denote statistically significant differences after unpaired Student’s t test (p < 0.05).
(L) Expression of IGH and J chain genes in the upper jaw of P. dolloi 10 days after secondary immunization with E. ictaluri by qPCR.
(M) Expression of IL-7R, TNFRSF11A, LTBR, and ID2 in the upper jaw of P. dolloi 10 days after secondary immunization with E. ictaluri by qPCR.
OE, olfactory epithelium; iLA, inducible lymphoid aggregate; LA, lymphoid aggregate; OC, oral cavity. Black asterisks denote capillaries within the LA of an infected individual. Data were normalized to elongation factor 1α (EF-1α) expression and are shown as the fold increase in mRNA with respect to mock-infected lungfish. Data are represented as mean ± SEM, and the significance of differences was determined by unpaired Student’s t tests (p < 0.05). Asterisks denote statistically significant differences (n = 5). See also Figure S2 and Table S2.
The total number of LAs per fish was not different from control specimens (mean = 3; n = 5), however the size of the LAs increased. LAs from infected lungfish had a mean long diameter of 447.8 ± 35.76 µm, a mean short diameter of 362.6 ± 37.4 µm, and a maximum depth of 353.3 ± 46.45 µm, for a total mean volume of 3 × 107 µm3 (Figure 3K). FISH staining revealed that 60% ± 0.7% of all cells were CD3+ and 36.2% ± 0.8% were IGH+, both significantly higher (p < 0.0001 and p = 0.002, respectively) than in the control LAs (Figured 3K and S2C–S2F). Collectively, these results suggest that microbial exposure induced lymphocyte migration into the lungfish LAs or the proliferation and/or differentiation of lymphocytes within the LA.
Importantly, exposure to the enteropathogen triggered formation of new lymphoid structures at the base of the olfactory epithelium (Figures 3A and 3D). We named these structures inducible LAs (iLAs), and we did not observe them in any control samples. We observed three to four iLAs in infected specimens (Figure 3K). These were unencapsulated and mostly adjacent to areas of bleeding and inflammation. iLAs had a diameter of ~100 µm and a depth of ~80 µm, for a total volume of ~3.3 × 106 µm3 (Figure 3K). FISH staining with IGH and CD3 probes showed that both B and T cells are present in iLAs (mean percentage of CD3+ cells = 31%, mean percentage of IGH+ cells = 16%; data not shown). These results indicate that lungfish iLAs may be equivalent to mammalian tertiary lymphoid structures since they form in response to microbial exposure [31].
Local Nasal Immune Responses Take Place in Response to Enterobacterial Infection in Lungfish
After nasal bacterial infection, qRT-PCR was used to measure the expression of IGH, integrins, TNF-related genes, and the transcription factor ID2 in the nasal mucosa of lungfish. We detected significant changes in the expression patterns of IGH genes. In particular, IGHM1 and IGHW1 were both upregulated, whereas the expression of IGHW2L was significantly downregulated (Figure 3L). However, the VH usage by IGHM1 heavy chain did not differ between infected and control nasal LA samples (Figure S2A). In addition, the expression of two integrins (ITGA44 and ITGB7) was significantly higher in the nose of infected fish compared to controls (Figure 3M). We investigated the expression patterns of IL-7R, TNFRSF11A, LTA, LTB, LTBR, and ID2 due to their importance in NALT organogenesis and lymphoid tissue development and organization. The expression levels of IL7-R, TNFRSF11A, and ID2 were all significantly higher in infected than in control specimens (Figure 3M). However, the expression levels of LTA, LTB, and LTBR remained unchanged at this time point. Since our sampling scheme was limited to one time point, we cannot rule out that changes in LTA, LTB, and LTBR occurred at different time points. It is also worth noting that infection led to formation of iLAs and that infected tissue samples therefore contained both LAs and iLAs. As a consequence, up- or downregulation of gene expression in one structure or the other may be masked in our samples. Our results are nevertheless in agreement with previous studies using mouse models that demonstrated the dependence on ID2, but not LT, for NALT formation [32].
Conclusions
The present study revisits the evolutionary origins of organized lymphoid structures in vertebrate mucosal immune systems. Our findings break the current paradigm that these structures originated in early tetrapods and reveal that the sister group of modern tetrapods, the lungfish, already acquired this immunological innovation. Furthermore, our findings suggest that the diversification of the TNFSF molecules in sarcopterygian fish may have been a critical factor that led to the emergence of mucosal lymphoid tissue genesis and organization in vertebrates.
EXPERIMENTAL PROCEDURES
Juvenile Nigerian spotted lungfish (P. dolloi) and P. annectens (total length: 9–12 in) were obtained from Segrest farms and maintained in the laboratory as described elsewhere [13]. All animal studies were reviewed and approved by the Institutional Animal Care and Use Committee at the University of New Mexico (protocol number 11-100744-MCC). For detailed protocols and statistical analyses, please refer to the Supplemental Experimental Procedures.
Highlights.
African lungfish possess O-MALT in their nasal and intestinal mucosa
Lungfish O-MALT is rich in lymphocytes but lacks segregation of B and T cells
O-MALT changes its architecture in response to bacterial infection
TNF superfamily members such as lymphotoxins diversified in lungfish
In Brief.
Organized MALT is thought to be an innovation of endotherms, whereas ectothermic amphibians have primitive O-MALT in the form of lymphoid aggregates. Tacchi et al. show that primitive O-MALT first appeared in sarcopterygian fish like the African lungfish, and therefore its origins predate the transition of vertebrates from water to land.
Acknowledgments
Authors wish to thank Dr. YaoFeng Zhao for kindly sharing lungfish Illumina transcriptome databases and for help with AICDA PCRs. We thank Dr. Javier Santander and Dr. Debra Milton for the E. ictaluri and V. anguillarum strains, respectively. Dr. Nancy Ruddle and Dr. Judy Cannon kindly provided mouse lymph nodes, and Dr. Helen Hathaway helped with the Azan stains. We thank Dr. Greg Wiens for insightful discussions of TNF evolution and Kyle Crossey for help with histology samples. This work was funded by National Science Foundation award NSF IOS 1456940 and the NIH (CETI COBRE grant P20GM103452).
Footnotes
ACCESSION NUMBERS
The accession numbers for the sequences reported in this paper are GenBank: KP297823 (P. dolloi PTPRC), KP297824 (P. dolloi CD4), KP297825 (P. dolloi TCRA), KP297826 (P. dolloi TCRB), KP297827 (P. dolloi TCRD), KP297828 (P. dolloi CD74), KP297829 (P. dolloi VCAM1), KP297830 (P. dolloi ICAM1), KP297831 (P. dolloi MADCAM1), KP297832 (P. dolloi CCL19), KP297833 (P. dolloi CCR7), KP297834 (P. dolloi CXCR5), KP297835 (P. dolloi IL7RA), KP297836 (P. dolloi LTBR), KP297837 (P. dolloi TNFRSF11A), KP297838 (P. dolloi ITGA4), KP297839 (P. dolloi ITGB7), KP297840 (P. dolloi LTB), KC812389 (P. dolloi IGHM1), JX999963 (P. dolloi IGHM2), KC152447 (P. dolloi IGHW1L), JX999964 (P. dolloi IGHW1s), KC812390 (P. dolloi IGHW2L), KC174714 (P. dolloi IGHN1), JX999962 (P. dolloi J chain), KP792760 (P. dolloi APOBEC-1), KP792761 (P. dolloi APOBEC-2), KP792762 (P. dolloi APOBEC-3C), and KP792763 (P. dolloi APOBEC-4).
SUPPLEMENTAL INFORMATION
Supplemental Information includes Supplemental Experimental Procedures, three figures, and two tables and can be found with this article online at http://dx.doi.org/10.1016/j.cub.2015.07.066.
AUTHOR CONTRIBUTIONS
L.T. performed in vivo infections, RT-PCRs, bioinformatics, and FISH experiments; P.M. performed TEM; E.T.L. performed LCM; C.T.A. performed transcriptome sequencing; and I.S. designed the study and performed confocal microscopy, histology, cell isolation, and flow cytometry.
REFERENCES
- 1.Goldstine SN, Manickavel V, Cohen N. Phylogeny of gut-associated lymphoid tissue. Am. Zool. 1975;15:107–118. [Google Scholar]
- 2.Manning M. Symposium : Antimicrobial defence mechanisms. J. R. Soc. Med. 1979;72:683–705. [PMC free article] [PubMed] [Google Scholar]
- 3.Ardavín CF, Zapata A, Garrido E, Villena A. Ultrastructure of gut-associated lymphoid tissue (GALT) in the amphibian urodele, Pleurodeles waltlii. Cell Tissue Res. 1982;224:663–671. doi: 10.1007/BF00213761. [DOI] [PubMed] [Google Scholar]
- 4.Ardavin CF, Zapata A, Villena A, Solas MT. Gut-associated lymphoid tissue (GALT) in the amphibian urodele Pleurodeles waltl. J. Morphol. 1982;173:35–41. doi: 10.1002/jmor.1051730105. [DOI] [PubMed] [Google Scholar]
- 5.Tacchi L, Musharrafieh R, Larragoite ET, Crossey K, Erhardt EB, Martin SA, LaPatra SE, Salinas I. Nasal immunity is an ancient arm of the mucosal immune system of vertebrates. Nat. Commun. 2014;5:5205. doi: 10.1038/ncomms6205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Myers MA. A study of the tonsillar developments in the lingual region of anurans. J. Morphol. 1928;45:399–439. [Google Scholar]
- 7.Hassanpour M, Joss J. Anatomy and histology of the spiral valve intestine in juvenile Australian lungfish, Neoceratodus forsteri. Open Zool. J. 2009;2:62–85. [Google Scholar]
- 8.Giannasca PJ, Boden JA, Monath TP. Targeted delivery of antigen to hamster nasal lymphoid tissue with M-cell-directed lectins. Infect. Immun. 1997;65:4288–4298. doi: 10.1128/iai.65.10.4288-4298.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Takata S, Ohtani O, Watanabe Y. Lectin binding patterns in rat nasal-associated lymphoid tissue (NALT) and the influence of various types of lectin on particle uptake in NALT. Arch. Histol. Cytol. 2000;63:305–312. doi: 10.1679/aohc.63.305. [DOI] [PubMed] [Google Scholar]
- 10.Zhang T, Tacchi L, Wei Z, Zhao Y, Salinas I. Intraclass diversification of immunoglobulin heavy chain genes in the African lungfish. Immunogenetics. 2014;66:335–351. doi: 10.1007/s00251-014-0769-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hayasaka H, Taniguchi K, Fukai S, Miyasaka M. Neogenesis and development of the high endothelial venules that mediate lymphocyte trafficking. Cancer Sci. 2010;101:2302–2308. doi: 10.1111/j.1349-7006.2010.01687.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wilson M, Hsu E, Marcuz A, Courtet M, Du Pasquier L, Steinberg C. What limits affinity maturation of antibodies in Xenopus–the rate of somatic mutation or the ability to select mutants? EMBO J. 1992;11:4337–4347. doi: 10.1002/j.1460-2075.1992.tb05533.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tacchi L, Larragoite E, Salinas I. Discovery of J chain in African lungfish (Protopterus dolloi, Sarcopterygii) using high throughput transcriptome sequencing: implications in mucosal immunity. PLoS ONE. 2013;8:e70650. doi: 10.1371/journal.pone.0070650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bieniasz PD. Intrinsic immunity: a front-line defense against viral attack. Nat. Immunol. 2004;5:1109–1115. doi: 10.1038/ni1125. [DOI] [PubMed] [Google Scholar]
- 15.Litman GW, Rast JP, Fugmann SD. The origins of vertebrate adaptive immunity. Nat. Rev. Immunol. 2010;10:543–553. doi: 10.1038/nri2807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stavnezer J, Amemiya CT. Seminars in Immunology. Vol. 16. Elsevier; 2004. Evolution of isotype switching; pp. 257–275. [DOI] [PubMed] [Google Scholar]
- 17.Yang F, Waldbieser GC, Lobb CJ. The nucleotide targets of somatic mutation and the role of selection in immunoglobulin heavy chains of a teleost fish. J. Immunol. 2006;176:1655–1667. doi: 10.4049/jimmunol.176.3.1655. [DOI] [PubMed] [Google Scholar]
- 18.Marianes AE, Zimmerman AM. Targets of somatic hypermutation within immunoglobulin light chain genes in zebrafish. Immunology. 2011;132:240–255. doi: 10.1111/j.1365-2567.2010.03358.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jiang N, Weinstein JA, Penland L, White RA, 3rd, Fisher DS, Quake SR. Determinism and stochasticity during maturation of the zebrafish antibody repertoire. Proc. Natl. Acad. Sci. USA. 2011;108:5348–5353. doi: 10.1073/pnas.1014277108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fillatreau S, Six A, Magadan S, Castro R, Sunyer JO, Boudinot P. The astonishing diversity of Ig classes and B cell repertoires in teleost fish. Front. Immunol. 2013;4:28. doi: 10.3389/fimmu.2013.00028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lane PJ, McConnell FM, Withers D, Gaspal F, Saini M, Anderson G. Lymphoid tissue inducer cells: bridges between the ancient innate and the modern adaptive immune systems. Mucosal Immunol. 2009;2:472–477. doi: 10.1038/mi.2009.111. [DOI] [PubMed] [Google Scholar]
- 22.Wiens GD, Glenney GW. Origin and evolution of TNF and TNF receptor superfamilies. Dev. Comp. Immunol. 2011;35:1324–1335. doi: 10.1016/j.dci.2011.03.031. [DOI] [PubMed] [Google Scholar]
- 23.Collette Y, Gilles A, Pontarotti P, Olive D. A co-evolution perspective of the TNFSF and TNFRSF families in the immune system. Trends Immunol. 2003;24:387–394. doi: 10.1016/s1471-4906(03)00166-2. [DOI] [PubMed] [Google Scholar]
- 24.Glenney GW, Wiens GD. Early diversification of the TNF superfamily in teleosts: genomic characterization and expression analysis. J. Immunol. 2007;178:7955–7973. doi: 10.4049/jimmunol.178.12.7955. [DOI] [PubMed] [Google Scholar]
- 25.Klein U, Rajewsky K, Küppers R. Human immunoglobulin (Ig)M+IgD+ peripheral blood B cells expressing the CD27 cell surface antigen carry somatically mutated variable region genes: CD27 as a general marker for somatically mutated (memory) B cells. J. Exp. Med. 1998;188:1679–1689. doi: 10.1084/jem.188.9.1679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cupedo T, Mebius RE. Cellular interactions in lymph node development. J. Immunol. 2005;174:21–25. doi: 10.4049/jimmunol.174.1.21. [DOI] [PubMed] [Google Scholar]
- 27.Shyjan AM, Bertagnolli M, Kenney CJ, Briskin MJ. Human mucosal addressin cell adhesion molecule-1 (MAdCAM-1) demonstrates structural and functional similarities to the alpha 4 beta 7-integrin binding domains of murine MAdCAM-1, but extreme divergence of mucin-like sequences. J. Immunol. 1996;156:2851–2857. [PubMed] [Google Scholar]
- 28.Kiyono H, Fukuyama S. NALT-versus Peyer’s-patch-mediated mucosal immunity. Nat. Rev. Immunol. 2004;4:699–710. doi: 10.1038/nri1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Eberl G, Lochner M. The development of intestinal lymphoid tissues at the interface of self and microbiota. Mucosal Immunol. 2009;2:478–485. doi: 10.1038/mi.2009.114. [DOI] [PubMed] [Google Scholar]
- 30.Chang S-Y, Ko H-J, Kweon M-N. Mucosal dendritic cells shape mucosal immunity. Exp. Mol. Med. 2014;46:e84. doi: 10.1038/emm.2014.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Randall TD, Mebius RE. The development and function of mucosal lymphoid tissues: a balancing act with micro-organisms. Mucosal Immunol. 2014;7:455–466. doi: 10.1038/mi.2014.11. [DOI] [PubMed] [Google Scholar]
- 32.Fukuyama S, Hiroi T, Yokota Y, Rennert PD, Yanagita M, Kinoshita N, Terawaki S, Shikina T, Yamamoto M, Kurono Y, Kiyono H. Initiation of NALT organogenesis is independent of the IL-7R, LTbetaR, and NIK signaling pathways but requires the Id2 gene and CD3(−)CD4(+)CD45(+) cells. Immunity. 2002;17:31–40. doi: 10.1016/s1074-7613(02)00339-4. [DOI] [PubMed] [Google Scholar]