Abstract
Current models of the functional architecture of human cortex emphasize areas that capture coarse-scale features of cortical topography but provide no account for population responses that encode information in fine-scale patterns of activity. Here, we present a linear model of shared representational spaces in human cortex that captures fine-scale distinctions among population responses with response-tuning basis functions that are common across brains and models cortical patterns of neural responses with individual-specific topographic basis functions. We derive a common model space for the whole cortex using a new algorithm, searchlight hyperalignment, and complex, dynamic stimuli that provide a broad sampling of visual, auditory, and social percepts. The model aligns representations across brains in occipital, temporal, parietal, and prefrontal cortices, as shown by between-subject multivariate pattern classification and intersubject correlation of representational geometry, indicating that structural principles for shared neural representations apply across widely divergent domains of information. The model provides a rigorous account for individual variability of well-known coarse-scale topographies, such as retinotopy and category selectivity, and goes further to account for fine-scale patterns that are multiplexed with coarse-scale topographies and carry finer distinctions.
Keywords: functional magnetic resonance imaging (fMRI), hyperalignment, multivariate pattern analysis (MVPA), neural decoding, representational similarity analysis (RSA)
Introduction
The information in perceptions, thoughts, and knowledge is represented in patterns of activity in populations of neurons in human cortex. Representation in a cortical field can be modeled as a high-dimensional space in which each dimension is a local measure of neural activity, for example, a neuron's spike rate or a functional magnetic resonance imaging (fMRI) voxel's hemodynamic response, and patterns of activity are vectors in this space (Haxby et al. 2014). Response vectors can be decoded with multivariate pattern analysis (MVPA; Haxby et al. 2001, 2014) of fMRI data in cortical fields that are associated with different domains of information, such as low-level visual features in early visual cortex (Kay et al. 2008), meaningful objects, and faces in ventral temporal (VT) cortex (Haxby et al. 2001; Cox and Savoy 2003; Kriegeskorte, Mur and Bandettini 2008; Kriegeskorte, Mur et al. 2008; Naselaris et al. 2009; Haxby et al. 2011; Connolly et al. 2012; Khaligh-Razavi and Kriegeskorte 2014), biological motion, and facial expression in the superior temporal sulcus (STS; Said et al. 2010; Carlin et al. 2011), speech and music in the superior temporal gyrus (STG; Formisano et al. 2008; Casey et al. 2011), and motor actions in sensorimotor and premotor cortices (Oosterhof et al. 2010, 2013). Representational similarity analysis (RSA; Kriegeskorte, Mur and Bandettini 2008; Kriegeskorte, Mur et al. 2008; Connolly et al. 2012; Kriegeskorte and Kievit 2013) reveals that representational geometry for the same stimuli varies across different cortical fields, reflecting how information is transformed in processing pathways.
Currently dominant models for the functional architecture of human cortex, which are based on tiled, interconnected cortical areas for global functions (Desikan et al. 2006; Power et al. 2011; Yeo et al. 2011; Amunts et al. 2013; Kanwisher 2010; Gordon et al. 2016; Grill-Spector and Weiner 2014; Sporns 2015), do not offer an account for distributed population codes, which are contained within these areas, in a framework that is common across subjects. These population codes are detected with fMRI as variable patterns of activity with a fine spatial scale that carry fine-grained distinctions in the information that they represent. Areal models of the functional architecture of cortex posit a common framework on a spatial scale that captures only a coarser topography of dissociable, more global functions, such as simple motion versus biological motion perception.
Here, we present a common high-dimensional model of representational spaces in the human cortex that is based on a large set of response-tuning basis functions that are common across brains, each of which is associated with individual-specific topographic basis functions. Once individuals’ data are transformed into the model dimensions with common response-tuning basis functions, patterns of response across dimensions afford between-subject multivariate pattern classification (bsMVPC) with high accuracy and markedly enhanced intersubject correlation (ISC) of representational geometry. This model is a radical departure from previous models of cortical functional architecture as it is based on a common representational space rather than a common cortical topography. By modeling functional topographies as weighted sums of overlapping topographic basis functions, our model also accounts for coarse areal topography and goes further to capture fine-scale topographies that coexist with coarse topographies and carry finer distinctions.
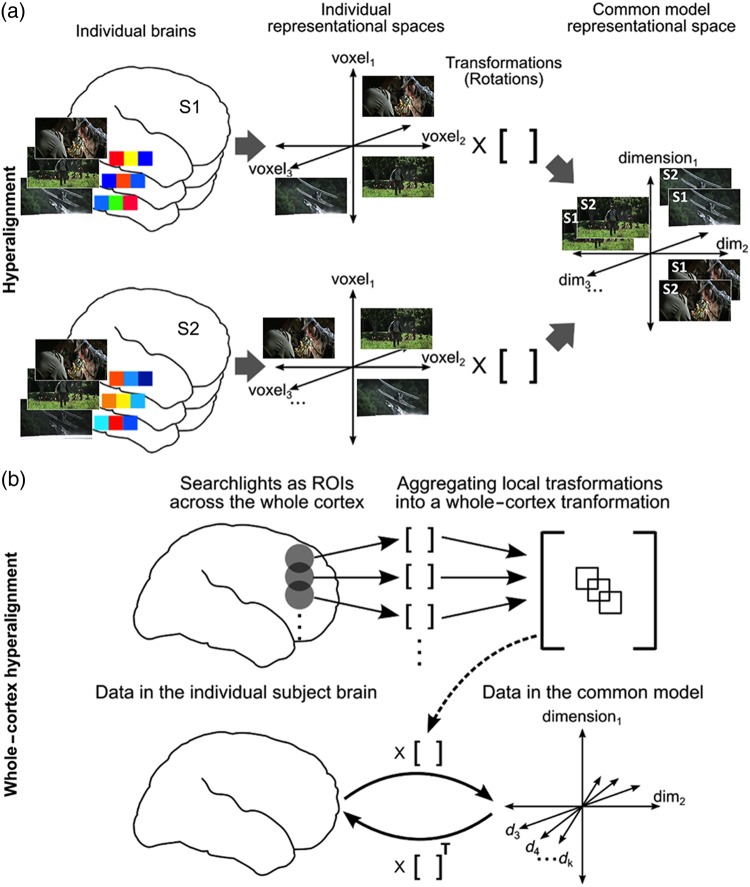
We derive this model using a searchlight hyperalignment algorithm, extending our previous region of interest (ROI) hyperalignment algorithm (Haxby et al. 2011), to produce a model for the whole cortex (Fig. 1). Our new algorithm produces a single transformation matrix for each individual brain, affording conversion of the representational spaces for all cortical fields in that individual into a single common model space (Fig. 1b).
Figure 1.
Schematic of whole-cortex searchlight hyperalignment. (a) Hyperalignment aligns neural representational spaces of ROIs in individual subjects’ into a common model space of that ROI using rotation in a high-dimensional space. The Procrustes transformation finds the optimal orthogonal transformation matrix to minimize the distances between response vectors for the same movie timepoints in different subjects’ representational spaces. An individual subject's transformation matrix rotates that subject's anatomical space into the common space and can be applied to rotate any pattern of activity in that subject's brain into a vector in common space coordinates. (b) Searchlight hyperalignment aligns neural representational spaces in all cortical searchlights. Local searchlight transformation matrices are then aggregated into a single matrix for each subject. Each subject's whole-cortex transformation matrix maps any data from that subject's cortex into the common model space. The transpose of that matrix maps any data from the common model space into individual subject's cortex. Dimensions in the common space have common tuning basis functions, as documented by increased ISC of movie fMRI time series (Supplementary Fig. 1), and individual-specific topographic basis functions (Supplementary Fig. 2).
We derived the transformation matrices based on brain responses measured with fMRI while subjects watch and listen to a full-length movie, affording a rich variety of visual, auditory, and social percepts (Hasson et al. 2010). The algorithm also can be applied to simpler, controlled experimental data, but our previous results showed that the sampling of response vectors from these experiments is impoverished and produces a model representational space that does not generalize well to new stimuli in other experiments (Haxby et al. 2011). Results show that the computational principles underlying this common model have broad general validity for representational spaces in occipital, temporal, parietal, and frontal cortices.
The common model is a high-dimensional representational vector space with the capacity to model a wide variety of response vectors, their representational geometry, and their complex, multiplexed cortical topographies. The model also captures coarse-scale features of human brain functional organization that have been described based on univariate analyses, such as one-dimensional category selectivities (e.g., faces versus objects; Kanwisher et al. 1997; Epstein and Kanwisher 1998; Downing et al. 2001) and 2-dimensional retinotopy (polar angle and eccentricity; Sereno et al. 1995; Nishimoto et al. 2011) and captures their topographic variability across individual brains. Capturing these dimensions in the context of a high-dimensional common model space provides an explicit, computational account for how coarse- and fine-scale topographies that code coarse- and fine-scale distinctions are multiplexed and overlapped.
Materials and Methods
Participants
We scanned 11 healthy young right-handed participants (4 females; mean age: 24.6 ± 3.7 years) with no history of neurological or psychiatric illness. All had normal or corrected-to-normal vision. Informed consent was collected in accordance with the procedures set by the Committee for the Protection of Human Subjects. Subjects were paid for their participation.
MRI Data Acquisition
We collected fMRI data from all subjects, while they watched a full-length feature film “Raiders of the Lost Ark” that we use as the basis for hyperalignment and for the principal validation tests of bsMVPC and intersubject alignment of representational geometry. For additional validation tests of generalization to independent experiments, we collected fMRI data in 10 subjects in a visual category perception experiment (6 animal species; Haxby et al. 2011; Connolly et al. 2012), in 9 subjects in a visual category-selectivity functional localizer study, and in 8 subjects in a retinotopy localizer study. The most relevant aspects of the fMRI data acquisition are presented here, and further details about scanning parameters and preprocessing can be found in Supplementary Material.
Movie Study
Stimuli and design
Subjects watched the full-length feature film, Raiders of the Lost Ark, divided into 8 parts of ∼14 min 20-s duration. Successive parts repeated the final 20 s of the previous part to account for hemodynamic response. Data collected during overlapping movie segments were discarded from the beginning of each part. Participants viewed the first 4 parts of the movie in one session and were taken out of the scanner for a short break. Participants then viewed the remaining 4 parts after the break. Video was projected onto a rear projection screen with an LCD projector, which the subject viewed through a mirror on the head coil. The video image subtended a visual angle of ∼22.7° horizontally and 17° vertically. Audio was presented through MR-compatible headphones. Subjects were instructed to pay attention to the movie and enjoy.
See Supplementary Material for details about the fMRI scanning protocol and data preprocessing.
Animal Species Perception Study
Stimuli and design
Stimuli consisted of 32 still color images for each of 6 animal species: ladybugs, luna moths, yellow throated warblers, mallards, ring-tailed lemurs, and squirrel monkeys (Connolly et al. 2012). Stimuli were presented in a slow event-related design. Each event started with the presentation of 3 different images of the same species for 500 ms each followed by 4500 ms of fixation cross. Subjects performed a memory task after every 6 events corresponding to 6 species in which they reported whether the task event appeared during the last 6 events.
See Connolly et al. (2012) and Supplementary Material for further details about the task design, fMRI scanning protocol, and data preprocessing.
Retinotopic Mapping
Stimuli and design
Stimuli consisted of high-contrast black and white checkerboard patterns flickering at 8 Hz within symmetrical wedges for polar angle mapping runs and within a ring for eccentricity mapping runs. There were 3 runs each of clockwise and counterclockwise rotating wedges and 2 runs each of expanding and contracting rings. Subjects performed an attention-demanding fixation color-change detection task during all runs.
See Supplementary Material for furthermore details about the fMRI scanning protocol and data preprocessing.
Category-Selectivity Localizer
Stimuli and design
Subjects viewed still images from 6 categories—human faces, human bodies without heads, small objects, houses, outdoor scenes, and scrambled images—to obtain independent measures of category-selective topographies in lateral occipital and VT cortex. We used a block design in which each block consisted of 16 images from a category with 900 ms of image presentation and 100 ms of gray screen. Subjects performed a one-back repetition detection task by reporting when an image repeated in succession. Each subject participated in a total of 4 runs.
See Supplementary Material for furthermore details about the task design, fMRI scanning protocol, and data preprocessing.
T1 Weighted Scans of Anatomy
High-resolution T1-weighted anatomical scans were acquired at the end of each session (MPRAGE, time repetition (TR) = 9.85 s, time echo = 4.53 s, flip angle = 8°, 256 × 256 matrix, field of view = 240 mm, 160 1 mm thick sagittal slices). The voxel resolution was 0.938 mm × 0.938 mm × 1.0 mm.
Between-Session and Anatomical Alignment of MRI Data
fMRI data were preprocessed using AFNI software (Cox 1996; http://afni.nimh.nih.gov) and programs for defining and manipulating cortical surfaces using FreeSurfer (Fischl et al. 1999; http://freesurfer.net) and PyMVPA (Hanke et al. 2009; http://www.pymvpa.org), including definition of cortical surface searchlights (Oosterhof et al. 2011). For all fMRI sessions, functional data were corrected for head motion by aligning to the last volume of the last functional run. We aligned data from all fMRI sessions to the mean functional image from the first half of the movie study. The anatomical scan of each subject was aligned to the MNI 152 brain, and those parameters were used to align fMRI data to the same template. We derived a gray matter mask by segmenting the MNI_avg152T1 brain provided in AFNI and removing any voxel that was outside the cortical surface by more than twice the thickness of the gray matter at each surface node. It included 54 034 3 mm isotropic voxels across both hemispheres. We used this mask for all subsequent analyses of all subjects.
Resampling Data to Cortical Surface
Cortical surface models were derived from anatomical T1-weighted anatomical scans using FreeSurfer (Fischl et al. 1999; http://freesurfer.net). These surfaces were aligned to the standard surface (fsaverage) using cortical curvature and resampled into a regular grid using AFNI's MapIcosahedron (Saad et al. 2004). We used a surface with a total of 20 484 cortical nodes in each hemisphere with 1.9 mm grid spacing to define cortical surface searchlights for the whole-cortex hyperalignment algorithm. We used a higher density grid to map coarse-scale cortical topographies (retinotopy and category selectivity) based on subjects’ own functional localizer data and on other subjects’ data projected into each subject's cortical space based on anatomy, using curvature-based alignment (FreeSurfer; Fischl et al. 1999), and using our common model. Finally, we performed supplementary analyses (see Supplementary Fig. 10) in which we tested bsMVPC in cortical surface node searchlights to compare performance after curvature-based anatomical alignment (Fischl et al. 1999), our cortical functional alignment algorithms using rubber-sheet warping of the cortical surface (Conroy et al. 2009, 2013; Sabuncu et al. 2010), and searchlight whole-cortex hyperalignment. All of these analyses are described in greater detail below and in Supplementary Material.
Functional ROIs
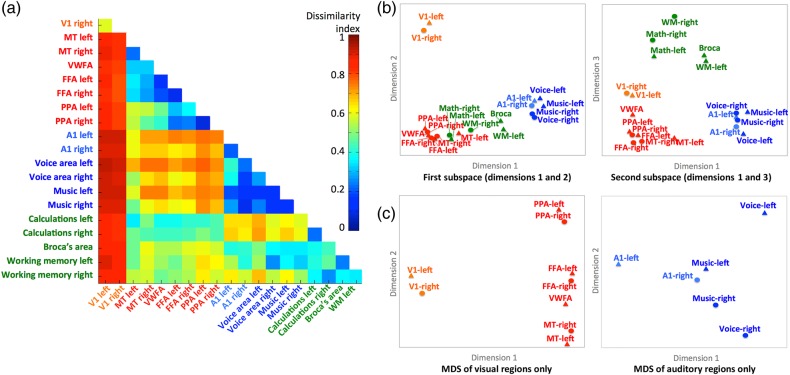
We analyzed the properties of the model in a variety of functionally defined cortical loci to illustrate the general validity of the common model, in particular its applicability in different information domains and the extent to which it captures fine-grained topographies, and to examine the relationships among the representational geometries of distributed cortical regions. We selected 20 loci using Neurosynth, a database derived from meta-analysis of over 10 000 fMRI studies (http://www.neurosynth.org; Yarkoni et al. 2011). We took the coordinates for the peak location associated with selected terms (Supplementary Table 1) and analyzed the properties of the model representational spaces that surrounded these loci in searchlights with a 3-voxel radius (mean volume = 119 voxels; Figs 2–5; Supplementary Figs 8 and 12).
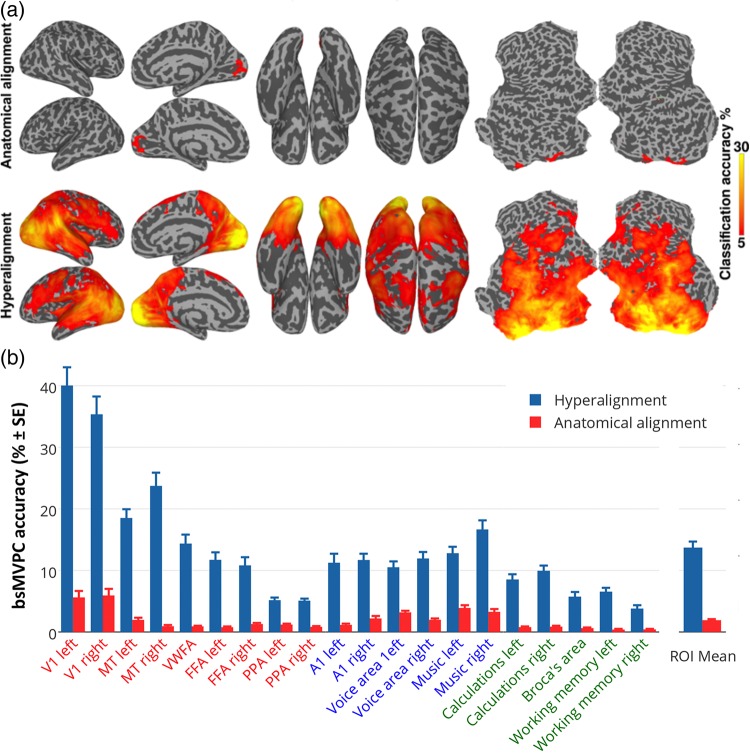
Figure 2.
bsMVPC of movie time segments. (a) bsMVPC accuracies of 15 s movie segments using voxels in 3-voxel radius searchlights before (top) and after whole-cortex hyperalignment (bottom). Model dimensions are assigned to searchlights based on their locations in the reference subject's brain. As in all validation tests on movie data, the common model space and transformation matrices were derived from one half of the movie data and then applied to the other half for cross-validated tests, in this case bsMVPC. (b) bsMVPC accuracies in twenty searchlight ROIs that were selected to cover a range of early and late visual areas in occipital and temporal cortices (V1, MT - middle temporal visual motion area, VWFA—visual word form area, FFA—fusiform face area, PPA—parahippocampal place area), auditory areas in superior temporal cortices (A1, voice areas, music areas), and areas in parietal and prefrontal cortices that are associated with cognitive and language functions (calculations, working memory, expressive speech). We used the coordinates for the peak location associated with selected terms in the Neurosynth database (http://www.neurosynth.org; Supplementary Table 1) as searchlight centers. ROI names are colored to group visual (orange and red), auditory (blue), and cognitive areas (green).
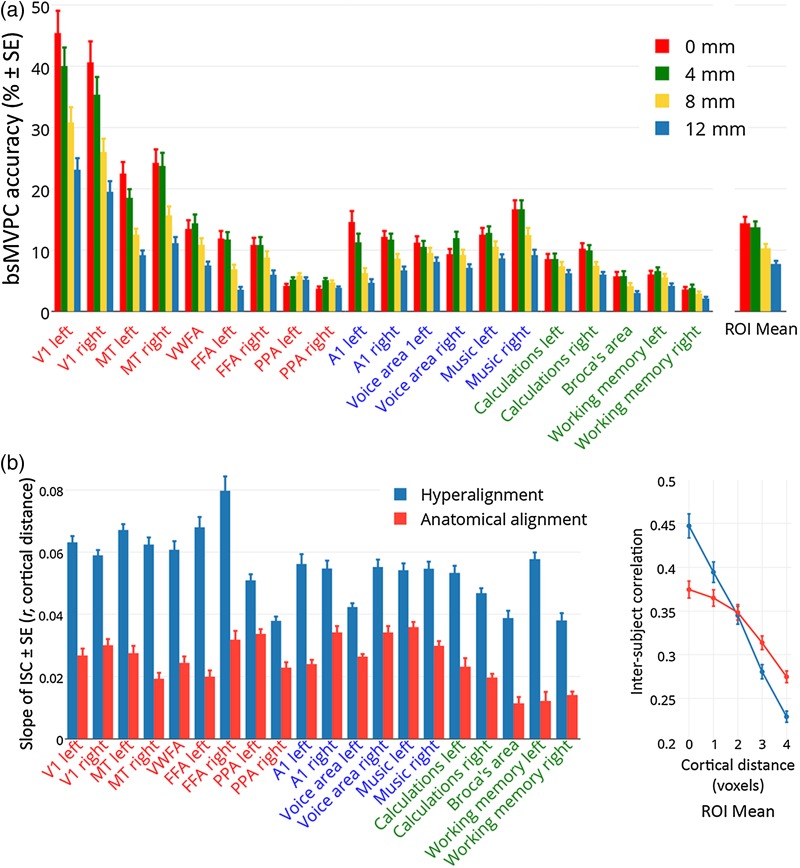
Figure 5.
Estimating the spatial resolution of shared representations in the common model space. (a) Effect of spatial smoothing of fMRI data on accuracies of bsMVPC of 15 s movie time segments in 20 selected searchlight ROIs (same as those used in Figs 2–4, Supplementary Table 1). Smoothing filters had a FWHM of 0 mm, 4 mm, 8 mm, and 12 mm. See Supplementary Figure 7 for brain maps of the effect of smoothing. (b) The slope of ISCs of time series by cortical distance in 20 searchlight ROIs after hyperalignment and anatomical alignment (left). The ROI mean correlations between time series for voxels in the same locations and for voxels at different distances from each other are shown on the right. ISC of time series for anatomically aligned data were calculated on data that were controlled for the effect of filtering (see Supplementary Fig. 1).
We implemented our algorithm and ran our analyses in PyMVPA unless otherwise specified (Hanke et al. 2009; http://www.pymvpa.org). All preprocessing and analyses were carried out on a 64-bit Debian 7.0 (wheezy) system with additional software from NeuroDebian (Halchenko and Hanke 2012; http://neuro.debian.net).
Derivation of A Common Model Representational Space Using Cortical Surface-Searchlight Hyperalignment
The common model of representational spaces in human cortex consists of a single high-dimensional representational space and individual-specific transformation matrices that are used to derive the space and to project data from individual anatomic spaces into the common model space and, conversely, to project data from the common space into individual anatomic spaces. The model uses hyperalignment to derive the transformation matrices and the common model representational space. We base the derivation of the transformation matrices and the common space on responses to the movie—a complex, naturalistic, dynamic stimulus. Although the algorithm also can be applied to fMRI data from more controlled experiments, we found that a common model based on such data has greatly diminished general validity (Haxby et al. 2011), presumably because, relative to a rich and dynamic naturalistic stimulus, such experiments sample an impoverished range of brain states.
Overview of Algorithm
An individual's transformation matrix rotates that subject's voxel space into the common model representational space (with reflections), and its transpose rotates the common model space back into the individual subject's anatomical voxel space (Haxby et al. 2011). Hyperalignment uses the Procrustes transformation (Schönemann 1966) to derive the optimal rotation parameters that minimize intersubject distances between response vectors for the same stimuli, namely the same timepoints in the movie. Consequently, common model dimensions are weighted sums of individual subjects’ voxels. In other words, individual voxels are not simply shuffled or assigned, one-by-one to single common model dimensions.
Derivation of a Common Model Space
To derive a single common representational space, we first hyperalign one subject to a reference subject's representational space. Then, we hyperalign a third subject to the mean response vectors for the first 2 subjects. We then hyperalign each successive subject to the mean vectors for the previously hyperaligned subjects. In a second iteration, we recalculate transformation matrices for each subject to the mean vectors from the first iteration and then recalculate the space of mean vectors. This space is the model space. We then recalculate the transformation matrices for each subject to the mean vectors in this common space. Each dimension is linked to a single location in the reference subject's brain, but the mean response vectors in this space are derived from all subjects and the transformation matrix for the reference subject is not an identity matrix.
Searchlight Hyperalignment
Hyperalignment of a cortical region permits mapping information from a cortical location in one subject into locations anywhere within that region of another subject, making it inappropriate for whole-brain analysis. We developed a surface-searchlight algorithm for hyperalignment of all of human cortex that only permits mapping of information in one subject's cortex into locations in nearby cortex in other subjects (Kriegeskorte et al. 2006; Chen et al. 2011; Oosterhof et al. 2011; Fig. 1b). We calculate hyperalignment transformation matrices for each subject in each cortical disc searchlight then aggregate an individual's searchlight matrices into a single transformation matrix. Thus, data from any cortical location can only be rotated into locations in a reference brain that are within one searchlight diameter.
We defined surface searchlights (as implemented in PyMVPA) in all subjects (Oosterhof et al. 2011) in which each surface node was the center of a searchlight cortical disc of radius 20 mm. We extended the thickness of this disc beyond the gray matter thickness, as computed using the FreeSurfer, by 1.5 times inside the white matter–gray matter boundary and 1.0 times outside the gray matter–pial surface boundary. This dilation was done to account for any misalignment of gray matter as computed on the anatomical scan and the gray matter voxels in the EPI scan due to distortion. To reduce the contribution from noisy or nongray matter voxels that were included due to dilation, we performed voxel selection within these cortical discs of all subjects using a between-subject correlation measure as described in Haxby et al. (2011) and selected the top 70% voxels in each cortical disc searchlight. The mean number of voxels in these cortical surface searchlights was 235. We then performed hyperalignment of searchlights from all subjects at each cortical location using the selected voxels across all subjects. We thus hyperaligned surface searchlights centered over all 20 484 nodes producing a list of matrices for each node (Fig. 1a). For each subject, we aggregated the orthogonal matrices by adding all the weights estimated for a given voxel-to-feature mapping from all surface searchlights that included that voxel-feature pair. This resulted in an N × N transformation matrix for each subject, where N is the number of voxels in the gray matter mask. Each subject's whole-cortex transformation matrix is sparse with each column (a model dimension) populated by nonzero values only for those rows (voxels) that were within one searchlight diameter of the location for that dimension in the reference brain. We used the transpose of each subject's transformation matrix to get a reverse mapping from the common space to each individual subject's cortical voxel space (Fig. 1b). Whole-cortex hyperalignment transformation matrices were applied to new datasets by first normalizing the data in each voxel by z-scoring and multiplying that normalized dataset with the transformation matrix. The same procedure was followed to apply reverse mapping.
Validity Testing
We test the validity of the common model by transforming data from new experiments into the common model space using transformation matrices derived from responses to the movie, then testing whether the representation of information is better aligned across subjects. For validation testing using only movie data, we derived the common model space and transformation matrices using data from one half of the movie and performed validity tests on the other half. We performed these tests twice using each half of the movie for deriving a model, then testing validity on the other half, and averaged the results across these 2 tests for each subject. For validation testing using data from other experiments, we derived the common model space and transformation matrices using all of the movie data then used each individual's transformation matrix to project the data from the independent experiments into the common model space. The other experiments for validation testing were on categorical perception of animal species (Connolly et al. 2012), on functional localizers for category-selective regions (Malach et al. 1995; Kanwisher et al. 1997; Epstein and Kanwisher 1998; Downing et al. 2001; Peelen and Downing 2005), and on the retinotopic organization of early visual cortex (Sereno et al. 1995).
Control for the Effect of Resampling
Mapping any data into the common model space by applying transformation matrices resamples the original data and, thus, is a complex spatial filter. In order to control for the effect of spatial filtering in comparisons between hyperaligned data and anatomically aligned data, we derived a control dataset with equivalent filtering that preserved anatomical variability across subjects. We computed 11 sets of hyperalignment transformations each with a different subject as reference. Each such set maps the data from all subjects to a common model space based on the voxel-space of that set's reference subject. To derive the control dataset, we mapped each subject's data into the common model space derived with the next subject in our order as reference subject, with the data of the last subject in the list mapped to the common model space with first subject as the reference. These datasets are filtered by hyperalignment; but, since their common model spaces have different references, the correspondence across these datasets is based only on anatomical alignment and preserves the anatomical variability in this sample of subjects. In contrast, regular hyperalignment mapped all subjects into a single common model space, with the first subject in the list as the reference in our analyses. We show in supplementary analyses using this spatial filtering control that the effect of spatial filtering is small and does not account for the much larger effect of hyperalignment (Supplementary Figs 1 and 5).
bsMVPC of Movie Time Segments
We tested bsMVPC of movie time segments in each half of the movie separately using searchlights with a radius of 3 voxels (Figs 2 and 5, Supplementary Figs 4, 5, 7, and 10) for both data in the common model space and for anatomically aligned data. Only voxels in the gray matter mask were included for bsMVPC analysis in each searchlight. The mean number of voxels in these volume searchlights was 102. We classified 15-s time segments (6 timepoints or TRs) using a correlation distance one-nearest neighbor classifier (Haxby et al. 2001, 2011). We also tested bsMVPC of shorter time segments in supplemental analyses (Supplementary Fig. 4). The response vectors for each time segment in each subject were compared with the mean response vectors from other subjects for the same time segment and all other time segments of the same length, using a sliding window in single TR increments for a total of over 1300 such time segments in each half of the movie. A subject's time segment response vector was correctly classified if it was most similar to the mean response vector for the same time segment in other subjects (1 out of over 1300 classification, chance <0.1%). Classification accuracies in each searchlight were averaged across both halves of the movie in each subject and mapped to the center voxel for that searchlight for visualization.
In addition to the searchlight bsMVPC of movie segments, a whole-cortex bsMVPC analysis of movie segments was performed. Test data in model dimensions or anatomically aligned voxels were projected into singular vector (SV) spaces that were derived from the training data (Supplementary Fig. 3). The SVs were sorted in the descending order of their singular values for both the hyperaligned data in common model space and for the anatomically aligned data. bsMVPC was performed for multiple sets of top SVs in each half of the movie, and the accuracies for the 2 halves were averaged for each subject at each set size. The 95% confidence interval (CI) for the difference between peak bsMVPC of anatomically aligned and peak bsMVPC of hyperaligned data was estimated using a bootstrap procedure by sampling subjects 10 000 times (Kirby and Gerlanc 2013).
ISC of Movie fMRI Time Series Responses
ISCs of time series for each half of the movie were computed between each subject and the average of all others subjects before and after hyperalignment. We present the mean of these values in each voxel for visualization in Supplementary Figure 1. The mean ISC was calculated across all the voxels in the gray matter mask, and the 95% CI for the difference between anatomically aligned and hyperaligned data was estimated using a bootstrap procedure as described above (Kirby and Gerlanc 2013). All operations involving correlations were performed after Fisher transformation and the results were inverse Fisher transformed for presentation. Note that the bootstrapping procedure does not assume any distribution, so we could perform it on correlation values.
ISC of Representational Geometry
ISCs of representational geometries were computed as the correlations between each subject's representational geometry and the mean representational geometry of other subjects for each half of the movie using searchlights with a radius of 3 voxels. In each searchlight, representational geometry was computed as a matrix of correlation coefficients between responses for every pair of timepoints (TRs) in one half of the movie, resulting in representational geometry vectors with over 850 000 timepoint pairs. This representational geometry vector captures the similarity of the responses to timepoints, presumably based on the information that is represented in a cortical searchlight and is shared across timepoints. Different searchlights will have different representational geometries because of differences in the type of information that is represented. ISCs were Fisher-z-transformed before averaging across both halves of the movie in each voxel. These were then averaged across all subjects and inverse Fisher transformed before mapping onto the cortical surface for visualization (Fig. 3 and Supplementary Fig. 6). The same steps were performed to compute ISCs of representational similarity before and after hyperalignment. Fisher transformed correlation values were averaged across all voxels in the gray matter mask in each subject and the 95% CI for the difference between anatomically aligned and hyperaligned data was calculated using bootstrapping as described above (Kirby and Gerlanc 2013).
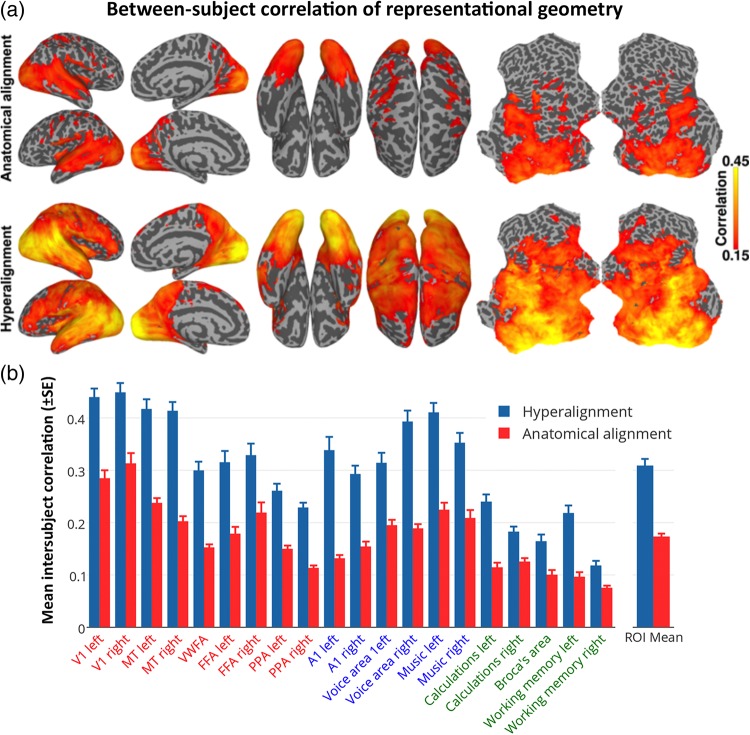
Figure 3.
ISC of neural representational geometry for movie content. (a) ISCs of representational geometries before (top) and after whole-cortex hyperalignment (bottom). Local representational geometries were computed as the similarities among responses to all movie timepoints within a 3-voxel radius searchlight, and the correlations were mapped to the center voxel. (b) The size of these effects is illustrated in the same 20 searchlight ROIs as shown in Figure 2b.
Second Order, Inter-Regional Representational Geometry
For a closer examination of the relationships among selected functional ROIs (Supplementary Table 1) in terms of the dissimilarities of their respective representational geometries, which reflect differences in the information represented by activity in these cortical areas, we analyzed the pairwise distances between ROI representational geometry vectors with multidimensional scaling (MDS) (Fig. 4). Dissimilarities between representational geometries for different ROIs were measured as intersubject dissimilarities, that is, the distance between the representational geometry for one ROI in one subject and the representational geometry for a different ROI in a different subject. Thus, this procedure is not affected by within-subject factors, such as head movements and moments of distraction that can simultaneously alter between timepoint similarities of responses in distant cortical fields but are unrelated to the information that is represented in those fields. Correlation-based inter-regional dissimilarity indices (D) were based on inter-regional ISCs (rROI1×ROI2) adjusted for the maximum possible similarity based on within-region ISCs of representational geometry for each of the 2 regions (rROI1 and rROI2):
Figure 4.
Second-order RSA of functional areas. (a) Matrix of all pairwise dissimilarities between the representational geometries in searchlight ROIs. (b) Multidimensional scaling (MDS) plots of the relationships among the representational geometries in visual (orange and red), auditory (blue), and cognitive (green) ROIs. Searchlight ROIs are the same as those shown in Figures 2 and 3. The plot on the left shows the ROIs plotted on the first and second dimensions. The plot on the right shows the ROIs plotted on the first and third dimensions. (c) Separate MDS analyses restricted to the visual (left) and auditory areas (right).
Note that these within-region and inter-regional distances reflect intersubject similarities, not similarities within-subjects.
Estimating Spatial Granularity of Common Model Maps
Decoding neural representations using MVPC and RSA relies on fine features of patterns that have a finer, within-area spatial scale than do the larger, coarse-scale “functional areas”, such as MT (the motion-sensitive middle temporal area) or the FFA. The goal of the common model is to align these fine-scale features that represent fine information distinctions. To test whether the common model does in fact align information-carrying features of patterns of brain activity that have a fine spatial scale in cortical anatomy, we analyzed the effect of spatially smoothing or blurring data—a common procedure that increases signal-to-noise for coarsely distributed information but blurs fine spatial patterns—and the spatial point-spread function (PSF) of the response-tuning functions that are shared across brains.
Effect of spatial smoothing
We smoothed the movie data prior to estimating the common model space and deriving individual transformation matrices using Gaussian smoothing filters with full-width-at-half-maximums (FWHM) of 0 mm, 4 mm, 8 mm, and 12 mm. The main bsMVPC analysis is identical to the part of this analysis with a spatial smoothing filter of FWHM = 4 mm. Application of the cortical surface-searchlight hyperalignment algorithm and bsMVPC of movie time segments then followed the same procedures as used for the main analysis (Fig. 5a and Supplementary Fig. 7).
Estimating the cortical PSF for intersubject similarity of local response tuning
We calculated ISCs of movie time series for one half of the movie for voxels or model dimensions with the same anatomical location (distance = 0), for voxels or dimensions with adjacent anatomical locations (sharing a face; distance = 1), and for voxels or dimensions separated by 2, 3, or 4 voxels. Note that this analysis examines only ISCs and, thus, does not reflect within-subject factors that can increase spatial autocorrelations and, thereby, flatten the cortical PSF. We calculated the mean ISC for each cortical distance across all voxels/dimensions in each searchlight functional ROI (Supplementary Table 1). The PSF was calculated as the magnitude of ISC of time series as a function of cortical distance. The PSF was calculated for time series from each half of the movie. For the analysis of PSF of data in the common model space, we derived the common space and calculated hyperalignment transformation matrices based on data from the other half of the movie. The PSFs for the 2 halves of the movie were averaged for each subject. We used linear regression to estimate the slope of the relation between cortical distance (in voxels) and ISC of time series (Fig. 5b).
Testing General Validity with an Experiment on Perception of Animal Species
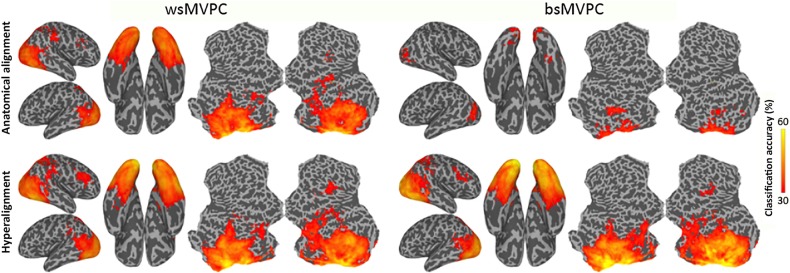
Searchlight within-subject MVPC (wsMVPC), bsMVPC, and representational geometry analyses of the neural representation of 6 animal species categories (Connolly et al. 2012) were performed on data that was rotated into the common model space based on responses to the movie, as well as on anatomically aligned data. MVPC of animal species used a linear SVM classifier (Cortes and Vapnik 1995) and β weights for responses to each species in each run in searchlights of 3 voxel radius (see Supplementary Methods). The SVM classifier used the default soft margin option in PyMVPA that automatically scales the C parameter according to the norm of the data. For within-subject classification, a leave-one-run-out cross validation was used. For bsMVPC, leave-one-subject-and-one-run-out cross validation was used to avoid any effect of run-specific information. Accuracies from all run folds of a subject were averaged, and mean accuracy across all subjects was mapped onto a cortical surface for visualization. The same procedure was used on data before and after hyperalignment. bsMVPC accuracies were averaged across all searchlights with wsMVPC accuracies above 30% (chance = 16.7%) in a ventral visual pathway ROI (see Fig. 6), and the 95% CI for the differences between mean wsMVPC, mean bsMVPC of anatomically aligned data, and mean bsMVPC of hyperaligned data was estimated using a bootstrap procedure (Kirby and Gerlanc 2013).
Figure 6.
MVPC of responses to animal species. Searchlight-based classification accuracies of 6 animal species within-subjects (left) and between-subjects (right). Classification is performed on the data after anatomical alignment (top row) and whole-cortex hyperalignment (bottom row). Whole-cortex transformation matrices were derived from the movie data.
Projecting Other Subjects’ Topographic Maps Into A Subject's Cortical Anatomy
Transformation of individual anatomical voxel spaces into the common model space with high-dimensional improper rotations may appear to obscure the coarse-scale functional topography in the individual brain or, on the other hand, may provide a rigorous computational method that preserves that topography and models individual variations of that topography. To investigate the fate of coarse-scale topographies, we examined whether 2 of the most prominent such topographies—retinotopy in early visual cortex and category selectivity in the ventral visual pathway—could be modeled in individual cortical spaces by projecting functional localizer data from other subjects into those spaces via the common model as derived from movie-viewing fMRI data.
Common model maps of retinotopy
Retinotopic tuning maps were projected into common model space dimensions using the transformation matrices derived from movie data. Instead of using preferred polar angle and eccentricity values at each voxel, we computed the tuning function at each voxel for 18 different polar angles, from 0 to 170° in steps of 10, and 18 different eccentricities, from fovea to maximum eccentricity in our experiment. We derived a subject's common model-estimated retinotopic tuning maps by averaging other subjects’ tuning maps in the common model space and projecting them into that subject's brain using reverse mapping—multiplying the tuning maps in common model space by the transpose of that individual's transformation matrix. These common model-estimated maps were then resampled into the curvature-aligned standard cortical surface mesh (Fischl et al. 1999; Saad et al. 2004).
Retinotopic maps for individual subjects also were computed based on other subjects’ data after anatomical alignment. Individual subjects’ tuning maps computed from the retinotopic mapping study were resampled into the same curvature-aligned standard cortical surface mesh (Fischl et al. 1999). We analyzed the similarity of the retinotopic maps as estimated from a subject's own retinotopy and as estimated from other subjects’ data. We fit sinusoids to each node's polar angle and eccentricity tuning functions and computed the cosines of its preferred angles. In order to limit this computation to early visual areas with reliable retinotopy, we selected surface nodes in each subject that were reliable as measured by a correlation between the measurements of polar angle and eccentricity on odd and even runs that was greater than or equal to 0.45 as reported by AFNI's 3dRetinoPhase. Correlation coefficients between tuning maps based on each subject's own data and other subjects’ data were computed and Fisher transformed. We estimated the 95% CI for the difference between these correlations based on anatomically aligned and hyperaligned data using bootstrapping (Kirby and Gerlanc 2013).
Common model maps of category selectivity
To project category-selectivity topographic maps based on other subjects into the cortical anatomy of a new subject, we first transformed all subjects’ data from the category-selective localizer study into the common model space using the transformation matrices derived from the full movie data. For each subject, the functional localizer data from all other subjects were projected into that subject's brain using the transposes of individual transformation matrices derived from the full movie data. These data were then smoothed with a 6 mm FWHM Gaussian filter and mapped to the curvature-aligned cortical surface mesh. Category-selective t-statistic maps were then computed for selectivity for faces, places, objects, and bodies using 3dDeconvolve and 3dREMLfit in AFNI on surface nodes based on each subject's own data and, independently, on the data from other subjects in that subject's anatomical space. For comparison, similar category-selective t-statistic maps also were calculated for each subject based on other subjects’ data in the curvature-aligned cortical surface mesh before hyperalignment, thereby using only anatomical alignment. The similarity of category-selectivity maps calculated from subjects’ own localizer data and from other subjects’ localizer data was computed by calculating correlations (Pearson's r) of the t-statistic maps in a ventral visual pathway surface ROI that included VT and lateral occipital cortices, testing the similarity of each individual to both the hyperaligned and anatomically aligned data from other subjects.
We then contrasted the similarity of maps based on individual and other subjects’ data to the within-subject reliability of category-selective maps. We estimated the within-subject reliability of the category-selectivity maps by computing the correlation between t-statistic maps computed from odd and even runs. To control for the effect of only using half of the localizer data, we also computed the correlations between the t-statistic maps calculated from the odd and the even runs in each subject and the maps calculated from other subjects’ data and averaged these correlations after Fisher transformation. We tested the significance of differences between correlations by calculating 95% CI of these differences using bootstrapping (Kirby and Gerlanc 2013).
Results
Between-Subject Classification of Movie Segments
In the first set of validation tests, we performed searchlight bsMVPC of 15 s movie time segments using a sliding time window with 1 TR increments (1 out of over 1300 classification for each half of the movie, chance accuracy <0.1%). Figure 2a shows the map of searchlight bsMVPC accuracy of movie segments based on anatomically aligned features (top) and common model features (bottom). bsMVPC using common model features yielded accuracies greater than 5% in fields in occipital, temporal, parietal, and prefrontal cortices with an overall peak of 50.3% in VT cortex. In contrast, bsMVPC using anatomically aligned features with accuracies greater than 5% was limited to early visual cortex with a peak of 8.6%.
To illustrate the size of the effect in different cortices, we selected 20 functionally defined loci in occipital, temporal, parietal, and prefrontal cortices for different visual, auditory, and cognitive functions using a meta-analytic database of functional neuroimaging studies (http://www.neurosynth.org; Yarkoni et al. 2011; see Supplementary Table 1). Figure 2b shows that bsMVPC using common model dimensions when compared with anatomically aligned voxels yielded markedly higher accuracies in searchlights surrounding each of these loci. Mean bsMVPC accuracy across these loci is 7-fold higher for response vectors in the common model space (mean = 13.7%) than for vectors in the anatomically aligned voxel space (mean = 1.9%).
Because the information in a movie scene is multimodal, we predicted that aggregating information across all cortical fields would yield substantially better bsMVPC than that for local cortical neighborhoods in searchlights. For whole-cortex bsMVPC, we reduced the dimensionality of the common model with singular value decomposition (SVD) of the movie response vectors, averaged across subjects. Whole-cortex bsMVPC accuracy, using common model SVs as features, peaked at 93.0% (SE = 1.4%) with around 450 SVs, which account for ∼90% of the variance (Supplementary Fig. 3). We performed SVD in a similar way on the anatomically aligned data and found that bsMVPC accuracies were significantly lower with a peak of 74.8% (95% CI for the difference in peak accuracies: [14.8%, 21.8%]) with a much smaller set of dimensions (∼50). These results show that shared information content in the neural representation of the multimodal movie was far greater when information from distributed cortical fields was aggregated in both common model and in anatomical spaces. This whole-cortex analysis combined shared information from coarse-scale topographies, which is captured in both the model and anatomical spaces, and fine-scale patterns, which only the common model captures. Relative to anatomical alignment, hyperalignment reduced whole-cortex bsMVPC errors by 72%, indicating that aggregating the shared, fine-grained local information, which is captured in the common model space, greatly increases the total shared neural information about multimodal experience. The shared information in the common model space is much higher dimensional than is the information that is shared in the anatomically aligned space, providing further evidence that anatomy-based methods align fewer, coarser-grained features of shared functional topographies.
Representational Geometry
ISC of Local Representational Geometries
Whereas MVPC detects distinctions among response vectors, RSA characterizes the representational geometry of the similarities among those vectors, revealing relationships among representations that MVPC does not (Kriegeskorte, Mur and Bandettini 2008; Kriegeskorte, Mur et al. 2008; Connolly et al. 2012; Kriegeskorte and Kievit 2013; Haxby et al. 2014). We analyzed the representational geometry of responses to the movie by calculating representational similarity matrices (RSMs) comprised of all correlations between response vectors for all pairs of timepoints (TRs) in each half of the movie (>850 000 pairs each) in each searchlight. Figure 3a shows cortical maps of ISCs of local representational geometries before and after hyperalignment. Hyperalignment increased mean ISC of searchlight representational geometries from 0.103 to 0.197 (95% CI of difference: [0.081, 0.106]). Figure 3b illustrates the size of the increases of ISCs of representational geometries for the same functional loci used in Figure 2b. Mean ISCs of representational geometries were greater in all of these loci in common model space than in anatomically aligned spaces. Mean ISC across these loci was 1.8 times larger in common model space (r = 0.28) than in anatomical space (r = 0.16).
Second-Order Geometry of Distances Between Regional Representational Geometries
Regional movie RSMs index the similarities and dissimilarities of neural responses to different parts of the movie, reflecting the representational geometry of information that is represented in a cortical field. Cortical fields that encode different types of information, therefore, will show different representational geometries. The similarities and differences among regional representational geometries, therefore, will reflect how the representation of information is structured across these cortical fields. We calculated between-region ISCs of RSMs for all pairs of these 20 functional loci. Mean between-region ISCs were consistently lower than the within-region ISCs (means of 0.11 and 0.28, respectively). We then analyzed the structure of the between-region intersubject dissimilarities—second-order RSA of the inter-regional representational dissimilarity matrix (RDM)—using MDS (Fig. 4). The full inter-regional intersubject RDM is shown in Figure 4a. Note that this second-order geometry for the main analysis is based on intersubject dissimilarities between local representational geometries. The reliability of inter-regional geometry was indexed by ISC of within-subject inter-regional RDMs to assure independence. Inter-regional representational geometry was highly consistent across subjects in anatomically aligned space but significantly more so in common model space (ISC = 0.857 and 0.927, respectively; 95% CI for the difference: [0.064, 0.077]).
The first dimension of the MDS solution for all 20 ROIs (Fig. 4b) separated visual from auditory areas with cognitive areas in between but with calculation areas overlapping visual areas. The second dimension separates V1 from all other areas. The third dimension separates the cognitive areas from the visual and auditory sensory/perceptual areas. Because this MDS analysis attempts to account for pairwise similarities among all 20 ROIs, the relationships among areas within the visual and auditory systems are not shown clearly. Separate MDS analyses of the 9 visual ROIs and the 6 auditory ROIs revealed a structure that reflects their functional relationships. The first dimension of the MDS plot of visual ROIs (Fig. 4c left panel) separates V1 from the higher-level visual ROIs, whereas the second dimension captures distinctions among these higher-level visual ROIs with MT at one end, PPA at the other, and the FFAs and VWFA in the middle. Right and left visual areas clustered most closely with each other. The first dimension of the MDS plot of auditory ROIs (Fig. 4c right panel) captures distinctions among A1, voice areas, and music areas. The second dimension separates left hemisphere areas from right hemisphere areas with a minimal difference between left and right A1 and maximal difference between left and right voice areas. Both right and left music areas are closer to the right voice area than to the left voice area.
The results of this second-order RSA show that these regional geometries reflect representations of different types of information, show that the inter-regional relationships reflect the large-scale organization of the human cortex into sensory, perceptual, and cognitive systems, and reveal more detailed organization in visual and auditory pathways. These results demonstrate that the local representational spaces that are modeled with common basis functions in the common model do indeed represent different domains of information, not global information that is spread across these cortical fields.
Spatial Resolution of Shared Information Content
bsMVPC of movie time segments showed that the benefit of using features in the common model space, rather than anatomically aligned voxels, was greater for searchlights, which are local, than for the whole cortex. This difference suggests that the effect of hyperaligning data into the common model space has a much larger effect on fine-scale local patterns than on inter-regional, coarse-scale global patterns. We performed 2 analyses to examine more directly the spatial resolution of patterns of activity that are shared across subjects in common model space.
The Effect of Smoothing on bsMVPC
In the first analysis, we tested the effect of smoothing the data before hyperalignment, and, thereby, blurring the fine-scale patterns of response. The highest bsMVPC accuracies using model space features were achieved with no spatial smoothing (Fig. 5a and Supplementary Fig. 7). A 4 mm FWHM filter produced a small decrement that was inconsistent across areas. An 8 mm FWHM filter, in contrast, produced a large decrement in bsMVPC accuracies in all regions except right and left PPA. On average, 8 mm smoothing decreased bsMVPC accuracy by 28.3%, and 12 mm smoothing decreased bsMVPC accuracy by 46.2% in these searchlight ROIs. These results indicate that the information that is aligned in the common model space carries information in patterns that are shared across subjects with a spatial resolution as fine as 2 voxels. Analysis of the effect of spatial smoothing, however, is confounded with the effect of noise suppression that smoothing affords. Because of this effect, we found that the very low bsMVPC accuracies for anatomically aligned data actually increased with spatial smoothing. Mean bsMVPC accuracy across ROIs for anatomically aligned data increased from 1.14% for unsmoothed data to 3.54% for data smoothed with a 12 mm FWHM filter, a 2.3-fold increase.
The Spatial PSF for Time Series ISCs
In the second analysis, we made a more direct and unconfounded test of the spatial resolution of distinctive response-tuning functions that are aligned across subjects in the common model space by analyzing the spatial PSF of ISCs of tuning functions. Note that this analysis examines only ISCs and, thus, does not reflect within-subject factors, such as spatial autocorrelation, that can increase spatial smoothness and, thereby, flatten the cortical PSF. The slopes relating ISCs of voxel time series by distances between voxels were markedly steeper for data in common model space than for data in a common anatomical space in all ROIs (Fig. 5b). Examination of the PSF for hyperaligned and anatomically aligned data, averaged across ROIs, shows that ISCs of time series are higher for hyperaligned data than for anatomically aligned data in voxels/dimensions with the same location in the reference subject's anatomical space and in voxels/dimensions that are in adjacent locations. Moreover, whereas ISCs in anatomical space do not differ for voxels with the same locations when compared with adjacent voxels, ISCs for the same common model dimension are substantially higher than for adjacent common model dimensions. ISCs of time series for voxels separated by more than 2 voxels were higher for anatomically aligned data than for data in the common model space. These results indicate that the alignment of response-tuning profiles in the common model space is highly spatially specific capturing distinctions between the response-tuning functions in adjacent dimensions.
Overall, the results of these analyses of the spatial resolution or granularity of the information content that is shared across subjects in the common model space indicates that hyperalignment aligns response-tuning functions across subjects at a fine spatial scale. The analysis of the effect of smoothing the data revealed that reducing the differences between voxels that are adjacent or separated by a single voxel—the effect of smoothing with an 8 mm FWHM filter—significantly degrades bsMVPC of hyperaligned responses to movie time segments, indicating that bsMVPC relies on inter-voxel differences of tuning functions at this fine spatial scale. The analysis of the intersubject PSF revealed that hyperalignment captures distinctions in response-tuning functions for adjacent voxels that are common across subjects. Thus, the common model is capturing variations in shared response-tuning functions with a fine-grained spatial distribution—carrying information in patterns of activity with granularity as fine as a single voxel.
Analysis of Data From Other Experiments in the Common Model Space
Perception of Animal Species
We asked if the whole-cortex hyperalignment derived from the movie data generalizes to an independent experiment and how the shared information captured by bsMVPC compares with the information captured by wsMVPC, in which a new classifier model is derived for each subject. Since subjects only watched the movie once, wsMVPC of movie time segments was not possible. Figure 6 shows average wsMVPC and bsMVPC accuracy maps using anatomically aligned and hyperaligned data. Whole-cortex hyperalignment, when compared with anatomical alignment, dramatically increased bsMVPC accuracies in bilateral early visual, lateral occipital, VT, and posterior lateral temporal cortices and in right parietal and prefrontal cortices. The cortical areas in which the 6 animal species can be classified with accuracies >30% (chance = 16.67%) are strikingly similar for wsMVPC and bsMVPC of hyperaligned data. Mean bsMVPC of hyperaligned data in the occipitotemporal searchlights (44.6%) is slightly, but significantly, higher than mean wsMVPC of hyperaligned data in the same searchlights (41.2%) (95% CI for this difference: [1.5%, 6.3%]). Thus, whole-cortex hyperalignment affords bsMVPC in local cortical fields that is equivalent or better than that of wsMVPC, suggesting that a classifier model trained on a set of subjects in the common model space can predict categorical information in a new subject better than a classifier that was trained on that test subject's own data, presumably because bsMVPC analysis affords larger datasets for training classifiers.
Retinotopic Maps in Common Model Space
We tested the capacity of the hyperalignment-derived common model to capture functional topography and individual topographic variation by delineating retinotopy in an individual brain by projecting other subjects’ data into that brain. In our previous report (Haxby et al. 2011), we showed that we could predict individual-specific topographies for category-selective regions in VT cortex using region-of-interest hyperalignment, and we found similar results in the current data using whole-cortex hyperalignment (see Supplementary Fig. 8).
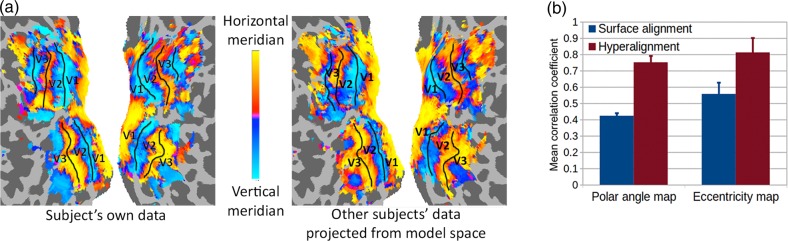
We computed retinotopic maps of receptive field polar angle and eccentricity in early visual cortex for 8 subjects using a standard fMRI retinotopy localizer, then compared the maps computed from each subject's own retinotopy scan with maps computed based on other subjects’ retinotopy data. We hyperaligned each subject's retinotopy data into the common model space using whole-cortex hyperalignment transformation matrices derived from the movie data. We then averaged the data from all but one subject in the common model space and projected the average data back into the left-out subject's brain, using the inverse mapping computed from the movie data. These projected data were used to estimate polar angle and eccentricity maps in that subject based on other subjects’ retinotopy. Figure 7a shows a subject's polar angle maps with estimated boundaries between early visual areas based on the locations of the horizontal and vertical meridia (Sereno et al. 1995), derived from that subject's own retinotopy data and as estimated from other subjects’ data projected into that subject's cortical anatomy (see Supplementary Fig. 9 for all subjects). The common model-estimated polar angle map is highly similar to the map based on the subject's own retinotopy scan data and preserves the topography enough to draw the boundaries between multiple early visual areas. Figure 7b shows the average spatial correlations of the measured maps with the maps estimated based on other subjects’ data after hyperalignment (mean correlation for polar angle = 0.75 and eccentricity = 0.81) and after surface-curvature-based alignment (mean correlation for polar angle = 0.42 and eccentricity = 0.56). Hyperalignment significantly increases the correlation between measured and estimated maps for both polar angle and eccentricity (95% CIs for differences: [0.295, 0.345] and [0.185, 0.326], respectively).
Figure 7.
Modeling individual retinotopy maps using the common model. Polar angle and eccentricity maps were estimated in each subject based on corresponding maps of other subjects aligned into the common model derived from the movie data. (a) Polar angle maps in a subject based on that subject's own retinotopy scan and based on others’ retinotopy that was rotated into common model dimensions then projected in this subject's cortical anatomy. See Supplementary Figure 9 for all subjects. (b) Mean correlations between the polar angle and eccentricity maps of each subject and maps estimated from other subjects after anatomical alignment (surface alignment using sulcal curvature, blue) and whole-cortex hyperalignment (red). Retinotopic maps based on the common model are significantly better predictors of individual retinotopy than are maps based on anatomical features.
Thus, whole-cortex hyperalignment based on neural activity measured during free viewing of a movie affords precise delineation of retinotopy in the early visual cortices of individual subjects, even though the transformation matrices are computed using large overlapping cortical searchlights and subjects do not maintain fixation to this complex, cluttered and dynamic visual stimulus.
Discussion
Common Model Structure, Components, and Principles
We present here a linear model of the functional organization of human cortex. The model is a high-dimensional representational vector space. Model dimensions have distinctive tuning functions—profiles of responses to a wide variety of stimuli—that are common across brains and serve as response-tuning basis functions for modeling any pattern of activity in an individual brain as a response vector in the common model representational space. Model dimensions are associated with local, individual-specific patterns of activity that serve as topographic basis functions for modeling individual variability of functional topographies (Fig. 7 and Supplementary Figs 2, 8 and 9). The model space is not a 2- or 3-dimensional anatomical space. We use a new searchlight hyperalignment algorithm to derive transformation matrices for the whole cortex that rotate individual brain spaces into the common representational space and, conversely, to project data from the common representational space into individual cortical topographies.
The common representational space affords a valid model for widely divergent domains of information. In order to derive a common model space with general validity, we sampled a broad range of functional brain states by measuring patterns of cortical activity evoked by viewing and listening to a complex, dynamic stimulus, namely a full-length adventure movie, Raiders of the Lost Ark. After transformation into common space coordinates, neural response vectors in occipital, temporal, parietal, premotor, and prefrontal cortices showed markedly increased bsMVPC of brain states and ISCs of representational geometries. The representational geometries for different cortical loci are distinctive and have a second-order inter-regional geometry that reflects relationships among areas in visual and auditory pathways and in other cognitive areas. Importantly, the lawful variation in local representational geometries revealed by this second-order analysis shows that the common model aligns representations of diverse domains of information and is not simply aligning global factors that are common across these areas. Analyses of the effect of spatial smoothing and the spatial PSF of shared local tuning functions revealed that the common model captures shared information-bearing patterns of neural activity with fine-grained spatial resolution. Projection of data from the common model space back into individual subjects’ anatomy captures individual variability in retinotopy in early visual cortex and category selectivity in occipitotemporal cortex.
This common model of representational spaces can afford better classification of an individual's brain states based on other subjects’ data (bsMVPC) than on that subject's own data (wsMVPC), because it makes it possible to have an arbitrarily large, multisubject training dataset. In this report, we show this effect with only 10 subjects’ data from an animal category perception experiment. Increasing the number of subjects should improve further the precision of the model and the power of bsMVPC (see Supplementary Fig. 2c in Haxby et al. 2011).
Overall, these results suggest that the common model captures common, underlying principles of representation that are valid across widely divergent domains of information. The broad-based validity of the model rests on 3 factors: its general conceptual framework—a linear, high-dimensional representational space; the computational algorithm for deriving transformation matrices—whole-cortex hyperalignment; and the use of rich, naturalistic stimuli for broad sampling of response vectors in representational spaces.
A Straightforward, High-Dimensional Linear Model
The general structure of the model is a linear model that rests on common bases for modeling tuning functions, population codes, and functional topographies. The common model space is a high-dimensional representational vector space, and any pattern of activity in an individual brain can be transformed into a vector in this common model space. Conversely, any response vector in model space coordinates can be mapped into the anatomy of any subject's cortex as a weighted sum of the local topographies for model dimensions. Thus, the tuning functions for model dimensions are a basis set that can model the functional response of any voxel, and the local topographies for model dimensions are a basis set that can model any pattern of cortical activity.
The Hyperalignment Algorithm
Hyperalignment is based on the Procrustes transformation and finds the optimal rotation that aligns a pattern of high-dimensional response vectors in a searchlight from one subject to the pattern of response vectors for the same brain states in other subjects. A critical factor underlying the success of this algorithm is that it preserves the similarity structure of local neural representations—the pairwise distances between response vectors (see Supplementary Fig. 6). Similarity structures for neural response vectors are highly consistent across subjects and suggest commonality of representation (Kriegeskorte, Mur and Bandettini 2008; Kriegeskorte, Mur et al. 2008; Connolly et al. 2012; Kriegeskorte and Kievit 2013), and preserving that similarity structure prevents losing that commonality.
A Broad Sampling of Brain States
We used a large sample of response vectors for a broad range of brain states to derive the model space and transformation matrices. Neural representational spaces in human cortex are high-dimensional. Discovery of a rich variety of common tuning functions and finely tuned estimation of parameters in transformation matrices rest on extensive sampling of vectors in these very large spaces. We measure a broad sample of response vectors while subjects view and listen to a complex, dynamic stimulus that contains a rich variety of visual objects, scenes, and motion; auditory speech, music, and other sounds; human actions, and social interactions.
Dimensionality of the Common Model Space
A lower-dimensional subspace is adequate to capture the distinctions among response vectors. SVD of the movie data indicated that the information contained in those data could be captured in a common model space with ∼450 orthogonal dimensions. In our initial report on a common model of VT cortex, whose volume is ∼10% of all cortex, we found that ∼35 dimensions were sufficient to capture the movie information content contained in the fMRI data, as well as the information in 2 category perception experiments. These dimensionality estimates are a function of the spatial and temporal resolution of fMRI and the number and variety of response vectors used to derive the common space. The true dimensionality of representation in human cortex surely involves vastly more distinct tuning functions. Estimates of the dimensionality of cortical representation, therefore, will almost certainly be much higher as data with higher spatial and temporal resolution for larger and more varied samples of response vectors are used to build new common models.
Other Common Bases for Neural Representation
The model that we present here is based on the discovery of neural tuning basis functions that are common across brains. Others have developed models that rest on other common bases. RSA (Kriegeskorte, Mur and Bandettini 2008; Kriegeskorte, Mur et al. 2008; Kriegeskorte and Kievit 2013) is based on finding a common pattern of pairwise similarities between stimulus vectors based on neural response patterns, models, or behavioral measures. RSA affords comparison of representational geometry across brain areas, measurement modalities, and species (Kriegeskorte, Mur and Bandettini 2008; Kriegeskorte, Mur et al. 2008; Connolly et al. 2012; Kriegeskorte and Kievit 2013; Cichy et al. 2014; Khaligh-Razavi and Kriegeskorte 2014), but models of this type are limited to the stimuli that are included in the experiment for discovering similarity structure, whereas our approach provides a basis for recasting any new response vector in the dimensions of the common model space. Stimulus model based encoding (Kay et al. 2008; Mitchell et al. 2008; Naselaris et al. 2009, 2010; Nishimoto et al. 2011; Huth et al. 2012) finds the common basis in the features of the stimuli themselves, then derives mappings to predict the responses in each voxel in each subject's brain based on these stimulus feature bases. This approach affords predictions of response patterns for new stimuli but provides no systematic procedure to discover shared neural population codes for the stimulus feature bases. In fact, our method complements stimulus encoding models by providing a common model of brain responses to map stimulus feature models, thus affording a single common encoding model across subjects.
Individual Deviations From the Common Model Space
The common model of representational spaces that we present here is designed to discover the commonalities of neural representation that are shared across brains, and the results show that such commonality is stronger and more detailed than was previously known. Neural representations, however, clearly vary across brains. Similarly, an individual's cortical topography can have idiosyncratic features, such as those described by Laumann et al. (2015), which presumably would not be modeled well by projection of other subjects’ data from the common model space into that subject's cortical space. Because the common model accounts for individual topographies based on other subjects’ data better than other methods can, it may provide a better basis for describing topographic variations that are true deviations from group norms.
Why A Common Model?
The transformation matrix for an individual is the key that unlocks that person's neural code. Once a subject's neural responses are in common model space coordinates, they can be decoded based on other subjects’ data. Thus, neural decoding can be based on very large datasets from unlimited numbers of other subjects. Our results show that the common model captures fine-grained distinctions among neural response vectors that encode a broad range of stimuli in diverse domains of information. This common model representational space offers a framework for multisubject neural decoding that captures the shared fine-grained structure of representations encoded in population responses. Moreover, by accounting for between-subject variability of function–anatomy relationship at fine-scale, our model can provide a basis for more sensitive investigations of individual differences in neural representations and cortical topographies.
Supplementary Material
Supplementary material can be found at: http://www.cercor.oxfordjournals.org/.
Funding
This work was supported by grants from the National Institute of Mental Health (5R01MH075706) and the National Science Foundation (NSF1129764). Funding to pay the Open Access publication charges for this article was provided by Dartmouth College.
Notes
Conflict of Interest. None declared.
Supplementary Material
References
- Amunts K, Lepage C, Borgeat L, Mohlberg H, Dickscheid T, Rousseau M-É, Bludau S, Bazin P-L, Lewis LB, Oros-Peusquens A-M et al. 2013. BigBrain: an ultra-high resolution 3D human brain model. Science. 340:1472–1475. [DOI] [PubMed] [Google Scholar]
- Carlin JD, Calder AJ, Kriegeskorte N, Nili NH, Rowe JB. 2011. A head view-invariant representation of gaze direction in anterior superior temporal sulcus. Curr Biol. 21:1817–1821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casey M, Thompson J, Kang O, Raizada R, Wheatley T. 2011. Population codes representing musical timbre for high-level fMRI categorization of music genres. In: Langs G, Rish I, Grosse-Wentrup M, Murphy B, editors. Machine Learning and Interpretation in Neuroimaging. Berlin (Germany): Springer; p. 34–41. [Google Scholar]
- Chen Y, Namburi P, Elliott LT, Heinzle J, Soon CS, Chee MWL. 2011. Cortical surface-based searchlight decoding. Neuroimage. 56:582–592. [DOI] [PubMed] [Google Scholar]
- Cichy RM, Pantazis D, Oliva A. 2014. Resolving human object recognition in space and time. Nat Neurosci. 17:455–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connolly AC, Guntupalli JS, Gors J, Hanke M, Halchenko YO, Wu YC, Abdi H, Haxby JV. 2012. The representation of biological classes in the human brain. J Neurosci. 32:2608–2618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conroy BR, Singer BD, Haxby JV, Ramadge PR. 2009. MRI-Based inter-subject cortical alignment using functional connectivity. In: Bengio Y, Schuurmans D, Lafferty J, Williams CKI, Culotta A, editors. Advances in Neural Information Processing Systems. Vol. 22, p. 378–386. [PMC free article] [PubMed] [Google Scholar]
- Conroy BR, Singer BD, Guntupalli JS, Ramadge PR, Haxby JV. 2013. Inter-subject alignment of human cortical anatomy using functional connectivity. Neuroimage. 81:400–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortes C, Vapnik V. 1995. Support-vector networks. Mach Learn. 20:273–297. [Google Scholar]
- Cox DD, Savoy RL. 2003. Functional magnetic resonance imaging (fMRI) “brain reading”: detecting and classifying distributed patterns of fMRI activity in human visual cortex. Neuroimage. 19:261–270. [DOI] [PubMed] [Google Scholar]
- Cox RW. 1996. AFNI: software for analysis and visualization of functional magnetic resonance neuroimages. Comput Biomed Res. 29:162–173. [DOI] [PubMed] [Google Scholar]
- Desikan RS, Ségonne F, Fischl B, Quinn BT, Dickerson BC, Blacker D, Buckner RL, Dale AM, Maquire RP, Hyman BT et al. 2006. An automated labeling system for subdividing the human cerebral cortex on MRI scans into gyral based regions of interest. Neuroimage. 31:968–980. [DOI] [PubMed] [Google Scholar]
- Downing PE, Jiang Y, Shuman M, Kanwisher N. 2001. A cortical area selective for visual processing of the human body. Science. 293:2470–2473. [DOI] [PubMed] [Google Scholar]
- Epstein R, Kanwisher N. 1998. A cortical representation of the local visual environment. Nature. 392:598–601. [DOI] [PubMed] [Google Scholar]
- Fischl B, Sereno MI, Tootell RBH, Dale AM. 1999. High-resolution intersubject averaging and a coordinate system for the cortical surface. Hum Brain Mapp. 8:272–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Formisano E, De Martino F, Bonte M, Goebel R. 2008. “Who” is saying “What”? Brain-based decoding of human voice and speech. Science. 322:970–973. [DOI] [PubMed] [Google Scholar]
- Gordon EM, Laumann TO, Adeyemo B, Huckins JF, Kelley WM, Petersen SE. 2016. Generation and evaluation of a cortical area parcellation from resting-state correlations. Cereb Cortex. 26:288–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grill-Spector K, Weiner KS. 2014. The functional architecture of the ventral temporal cortex and its role in categorization. Nat Rev Neurosci. 15:536–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halchenko YO, Hanke M. 2012. Open is not enough. Let's take the next step: an integrated, community-driven computing platform for neuroscience. Front Neuroinformatics. 6:22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanke M, Halchenko YO, Sederberg PB, Hanson SJ, Haxby JV, Pollman S. 2009. PyMVPA: a Python toolbox for multivariate pattern analysis of fMRI data. Neuroinformatics. 7:37–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasson U, Malach R, Heeger DJ. 2010. Reliability of cortical activity during natural stimulation. Trends Cogn Sci. 14:40–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haxby JV, Connolly AC, Guntupalli JS. 2014. Decoding neural representational spaces using multivariate pattern analysis. Annu Rev Neurosci. 37:435–456. [DOI] [PubMed] [Google Scholar]
- Haxby JV, Gobbini MI, Furey ML, Ishai A, Schouten JL, Pietrini P. 2001. Distributed and overlapping representations of faces and objects in ventral temporal cortex. Science. 293:2425–2430. [DOI] [PubMed] [Google Scholar]
- Haxby JV, Guntupalli JS, Connolly AC, Halchenko YO, Conroy BR, Gobbini MI, Hanke M, Ramadge PJ. 2011. A common, high-dimensional model of the representational space in human ventral temporal cortex. Neuron. 72:404–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huth AG, Nishimoto S, Vu AT, Gallant JL. 2012. A continuous semantic space describes the representation of thousands of object and action categories across the human brain. Neuron. 76:1210–1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanwisher N. 2010. Functional specificity in the human brain: a window into the functional architecture of the mind. Proc Natl Acad Sci U S A. 107:11163–11170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanwisher N, McDermott J, Chun MM. 1997. The fusiform face area: a module in human extrastriate cortex specialized for face perception. J Neurosci. 17:4302–4311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kay KN, Naselaris T, Prenger RJ, Gallant JL. 2008. Identifying natural images from human brain activity. Nature. 452:352–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khaligh-Razavi S-M, Kriegeskorte N. 2014. Deep supervised, but not unsupervised, models explain IT representation. PLoS Comput Biol. 10:1–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirby KN, Gerlanc D. 2013. BootES: an R package for bootstrap confidence intervals on effect sizes. Behav Res. 45:905–927. [DOI] [PubMed] [Google Scholar]
- Kriegeskorte N, Goebel R, Bandettini P. 2006. Information-based functional brain mapping. Proc Natl Acad Sci U S A. 103:3863–3868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kriegeskorte N, Kievit RA. 2013. Representational geometry: integrating cognition, computation, and the brain. Trends Cogn Sci. 17:401–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kriegeskorte N, Mur M, Bandettini P. 2008. Representational similarity analysis—connecting the branches of systems neuroscience. Front Syst Neurosci. 2:4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kriegeskorte N, Mur M, Ruff DA, Kiani R, Bodurka J, Esteky H, Tanaka K, Bandettini PA. 2008. Matching categorical object representations in inferior temporal cortex of man and monkey. Neuron. 60:1126–1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laumann TO, Gordon EM, Adeyemo B, Snyder AZ, Joo SJ, Chen M-Y, Gilmoe AW, McDermott KB, Nelson SM, Dosenbach NUF et al. 2015. Functional system and areal organization of a highly sampled individual human brain. Neuron. 87:1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malach R, Reppas JB, Benson RR, Kwong KK, Jiang H, Kennedy WA, Ledden PJ, Brady TJ, Rosen BR, Tootell RBH. 1995. Object-related activity revealed by functional magnetic resonance imaging in human occipital cortex. Proc Natl Acad Sci U S A. 92:8135–8139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell TM, Shinkareva SV, Carlson A, Chang KM, Malave VL, Mason RA, Just MA. 2008. Predicting human brain activity associated with the meanings of nouns. Science. 320:1191–1195. [DOI] [PubMed] [Google Scholar]
- Naselaris T, Kay KN, Nishimoto S, Gallant JL. 2010. Encoding and decoding in fMRI. Neuroimage. 56:400–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naselaris T, Prenger RJ, Kay KN, Oliver M, Gallant JL. 2009. Bayesian reconstruction of natural images from human brain Activity. Neuron. 63:902–915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimoto S, Vu AT, Naselaris T, Benjamini Y, Yu B, Gallant JL. 2011. Reconstructing visual experiences from brain activity evoked by natural movies. Curr Biol. 21:1641–1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oosterhof NN, Tipper SP, Downing PE. 2013. Cross-modal and action specific: neuroimaging the human mirror neuron system. Trends Cogn Sci. 17:311–318. [DOI] [PubMed] [Google Scholar]
- Oosterhof NN, Wiestler T, Downing PE, Diedrichsen J. 2011. A comparison of volume-based and surface-based multi-voxel pattern analysis. Neuroimage. 56:593–600. [DOI] [PubMed] [Google Scholar]
- Oosterhof NN, Wiggett AJ, Diedrichsen J, Tipper SP, Downing PE. 2010. Surface-based information mapping reveals cross-modal vision – action representations in human parietal and occipitotemporal cortex. J Neurophysiol. 104:1077–1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peelen MV, Downing PE. 2005. Selectivity for the human body in the fusiform gyrus. J Neurophysiol. 93:603–608. [DOI] [PubMed] [Google Scholar]
- Power JD, Cohen AL, Nelson SM, Wig GS, Barnes KA, Church JA, Vogel AC, Laumann TO, Miezin FM, Schlaggar BL et al. 2011. Functional network organization of the human brain. Neuron. 72:665–678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saad ZS, Reynolds RC, Argall B, Japee S, Cox RW. 2004. in IEEE International Symposium on Biomedical Imaging: Nano to Macro 2:1510–1513. [Google Scholar]
- Sabuncu M, Singer BD, Conroy B, Bryan RE, Ramadge PJ, Haxby JV. 2010. Function-based intersubject alignment of human cortical anatomy. Cereb Cortex. 20:130–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Said CS, Moore CD, Engell AD, Todorov AD, Haxby JV. 2010. Distributed representations of dynamic facial expressions in the superior temporal sulcus. J Vis. 10:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schönemann PH. 1966. A generalized solution of the orthogonal Procrustes problem. Psychometrika. 31:1–10. [Google Scholar]
- Sereno MI, Dale AM, Reppas JB, Kwong KK, Belliveau JW, Brady TJ, Rosen BR, Tootell RB. 1995. Borders of multiple visual areas in humans revealed by functional magnetic resonance imaging. Science. 268:889–893. [DOI] [PubMed] [Google Scholar]
- Sporns O. 2015. Cerebral cartography and connectomics. Philos Trans R Soc Lond B. 370:20140173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yarkoni T, Poldrack RA, Nichols TE, Van Essen DC, Wager TD. 2011. Large-scale automated synthesis of human functional neuroimaging data. Nat Methods. 8:665–670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeo BT, Krienen FM, Sepulcre J, Sabuncu MR, Lashkari D, Hollinshead M, Roffman JL, Smoller JW, Zöllei L, Polimeni JR et al. 2011. The organization of the human cerebral cortex estimated by intrinsic functional connectivity. J Neurophysiol. 106:1125–1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.