SUMMARY
Oncogene activation has been shown to generate replication-born DNA damage, also known as replicative stress (RS). Notably, the ATR kinase –and not ATM- is the primary responder to RS. One limitation for the study of ATR is the lack of potent inhibitors. We here describe a cell-based screening strategy that has allowed us to identify compounds with ATR inhibitory activity in the nanomolar range. Pharmacological inhibition of ATR generates RS, leading to chromosomal breakage in the presence of conditions that stall replication forks. Moreover, ATR inhibition is particularly toxic for p53 deficient cells, this toxicity being exacerbated by RS-generating conditions such as the overexpression of cyclin E. Importantly, one of the compounds is NVP-BEZ235, a dual PI3K/mTOR inhibitor that is currently being tested for cancer chemotherapy, but which we now show is also very potent against ATM, ATR and DNA-PKcs.
INTRODUCTION
The checkpoint response of the so-called DNA damage response (DDR) relies on two members of the PIKK family of protein kinases: Ataxia Telangiectasia Mutated (ATM) and ATM and Rad3-related (ATR)1. Whereas ATM is solely activated by DNA double strand breaks (DSBs), ATR responds to the accumulation of single-stranded DNA (ssDNA) both at resected DSBs as well as at aberrant replicative structures that compromise genome integrity during S phase2. Remarkably, works from the laboratories of Jiri Bartek and Thanos Halazonetis showed evidence of an activated DDR in early stages of tumorigenesis3,4. These findings led the authors to propose that oncogenes could somehow generate DNA damage through replication stress, which would then activate the DDR and limit the expansion of precancerous cells5. Subsequent works confirmed that, indeed, oncogenes generate DNA damage and that the nature of this damage is linked to abnormal replication6–8. In other words, the oncogene-induced DDR would be a cellular response to RS, which is known to be limited by ATR and its downstream kinase Chk19. However, and in contrast to ATM or Chk2, ATR and Chk1 are essential in mammals, which has limited functional studies in this pathway. We here describe our work in the identification of ATR inhibitors and the evaluation of how inhibiting ATR activity affects oncogene-expressing cells.
RESULTS
Screening strategy
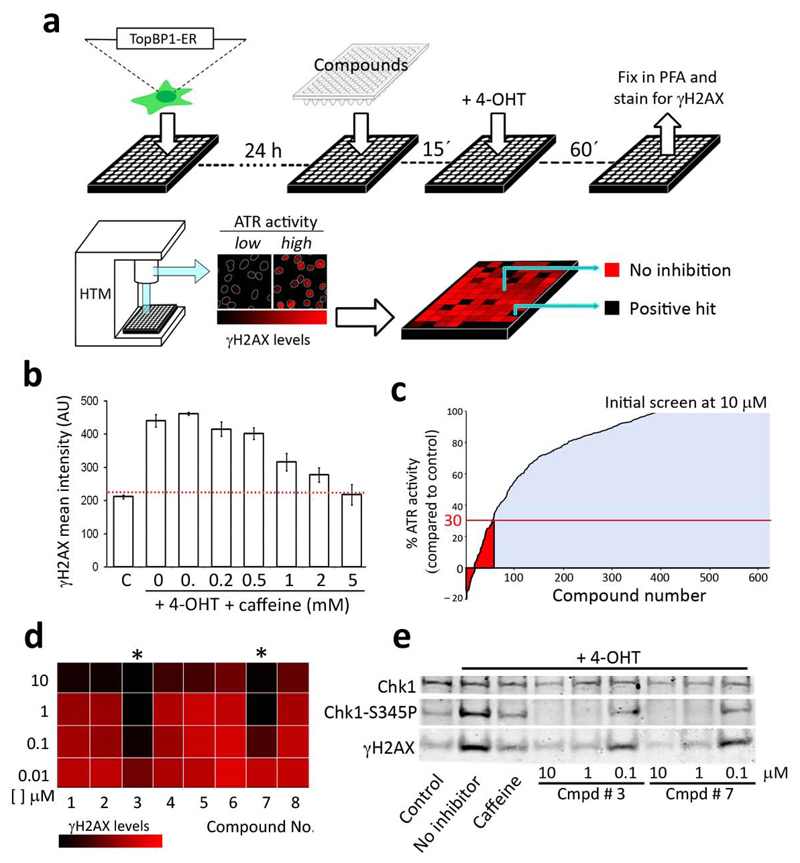
One of the limitations for the discovery of ATR inhibitors is that its kinase activity is restricted to S/G2. This has hindered cell-based screenings due to the large number of false positives that would be identified from an indirect effect of the tested compound on the cell cycle. Overcoming this limitation, we previously developed a cellular system in which ATR activity can be unleashed at will, throughout the cell cycle and in the absence of any actual DNA damage10. The system works due to a fusion of the ATR-activating domain of TopBP111, with a fragment of the estrogen receptor (TopBP1ER). In response to an inert derivative of tamoxifen (4-hydroxy-tamoxifen; 4-OHT), TopBP1ER translocates to the nucleus where it promotes a generalized activation of ATR. Noteworthy, the addition of 4-OHT promotes a pan-nuclear phosphorylation of H2AX (γH2AX), which is strictly dependent on ATR, and independent of ATM or DNA-PKcs10. Hence, looking at 4-OHT induced γH2AX formation in TopBP1ER expressing cells provides a specific and easily measurable readout of ATR activity.
In order to establish a screening platform, we optimized the assay in 96 well plates and automatized the quantification of the nuclear γH2AX signal through High-Throughput Microscopy (HTM). The screening strategy was then to expose the cells to the compound to be tested for 15 min, followed by 4-OHT for an additional hr, and then processed for γH2AX immunofluorescence (IF) by HTM. The screening pipeline and its typical readout are illustrated in Figure 1a (see Methods for a full description of the procedure). As proof-of-principle, addition of Caffeine, which is known to inhibit ATR at high concentrations, led to a stepwise decrease in the γH2AX signal (Fig. 1b); and the readouts were highly reproducible from experiment to experiment. Thus, the TopBP1ER based pipeline is sensitive enough to detect ATR inhibitors in a cell-based assay.Next, we decided upon the set of compounds to be tested. The test-library for our screening was a subset of 623 compounds, identified as having some activity towards PI3K in a previous screening that evaluated 33,992 small compounds12. Therefore, due to the similarity of PIKKs with PI3K, we reasoned that such a pool could be enriched in potential inhibitors of ATR. The initial screening was performed at 10 μM, which identified a relatively large set of chemicals with some ATR inhibitory capacity (Fig. 1c). We then set a threshold, and reanalyzed all the compounds that led to a lower than 30% γH2AX signal (or more than 70% inhibition) for further analysis. In this secondary analysis, the compounds were re-analyzed at increasing concentrations that started at 10nM (0.01, 0.1, 1 and 10 μM). Consistent with the previous screen, most compounds showed substantial inhibitory activity at 10 μM. However, most of them failed to display activity below 1 μM. Notably, 2 compounds showed almost 100% inhibition at 100 nM, and even some response at the lowest dose used (Fig. 1d). Moreover, these 2 molecules not only inhibited H2AX but also Chk1 phosphorylation in response to 4-OHT, strongly suggesting their capability to inhibit ATR (Fig. 1e). We thus centred our subsequent analyses on the characterization of these 2 compounds.
Figure 1.
Screening strategy for the identification of ATR inhibitors. (a) Schematic representation of the pipeline followed for the identification of ATR inhibitors (see Methods for a full description). Data were represented in a color-coded table of a 96 well plate, in which potential ATR inhibitors could be identified as black wells that had lost the 4-OHT-induced γH2AX signal. (b) Effect of increasing doses of caffeine on the nuclear γH2AX signal induced by 4-OHT. Error bars indicate s.d. AU: Arbitrary Units.(c) Results of the screening performed at 10 μM for the 623 compounds tested (average of 3 different experiments performed in duplicate). Highlighted in red are compounds that presented more than 70% inhibition and that were taken for further analysis. (d) Image illustrating the relative activity of 8 of the compounds from the secondary screen at decreasing concentrations, 2 of which show almost full activity at 100 nM (asterisks).(e) H2AX and Chk1 phosphorylation in TopBP1-ER cells exposed to 4-OHT, in the presence of absence of the compounds identified in (b). Caffeine (5mM) was used as a positive control of ATR inhibition.
Bypass of the IR-induced G2/M checkpoint
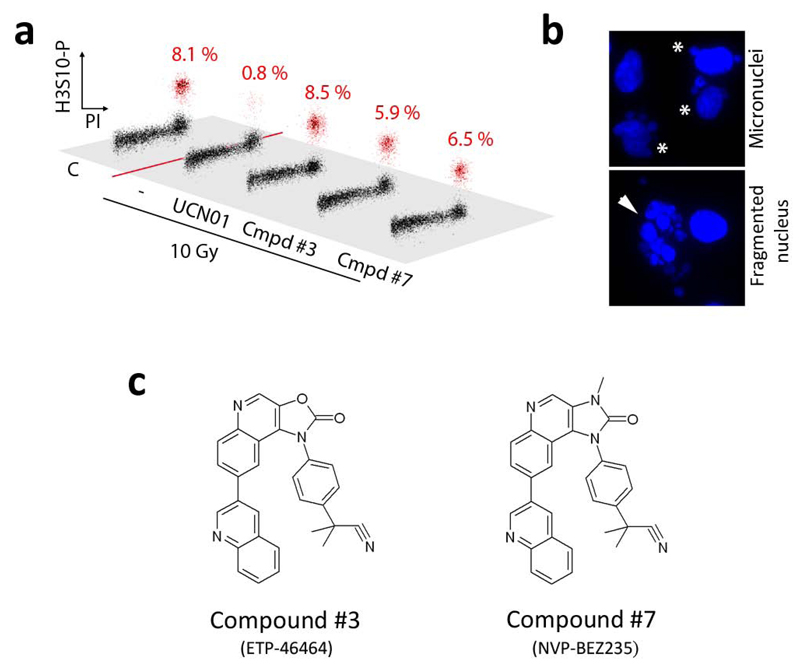
To determine whether the compounds were also capable of inhibiting ATR in response to actual DNA damage, we analysed their activity in U2OS cells exposed to ionizing radiation (IR). In response to IR, ATR activity contributes to the activation of the G2/M checkpoint so that damaged cells do not enter mitosis in the presence of breaks13. Accordingly, one proposed clinical application for Chk1 inhibitors is to combine them with genotoxic agents, so that damaged cells will undergo abnormal mitosis in the absence of a proficient G2/M checkpoint. In agreement with this, and similar to what we observed in the presence of the Chk1 inhibitor UCN-01, the use of the 2 identified compounds abrogated the G2/M checkpoint (Fig. 2a) and led to the presence of micronuclei or completely fragmented nuclei in cells exposed to IR (Fig. 2b). The structures and formal names of these two compounds are provided in Figure 2c. Nevertheless, given that the IR-induced G2/M checkpoint is also controlled by ATM, the effect of these compounds could be not solely dependent on ATR.
Figure 2.
Effect of the compounds in the G2/M checkpoint. (a) Cell cycle distribution of U2OS cells 4 hr after a treatment with 10 Gy of IR, in the presence of the indicated chemicals (UCN-01: 0.3 μM; compounds: 1 μM). H3 Ser10 phosphorylation (y axis) is used to distinguish mitotic cells from G2 cells. Cells were kept on taxol for the 4 hr period to capture all cells undergoing mitosis during that time. The percentage of mitotic cells is indicated in red. (b) Examples of the typical aberrations observed in IR-treated cells in the presence of the drugs. In this case, no taxol was added to prevent the influence of the drug in mitotic abnormalities. (c) Structures and formal names of the 2 compounds that are analyzed in the rest of the manuscript.
Selectivity of the inhibitors for ATR vs ATM and DNA-PKcs
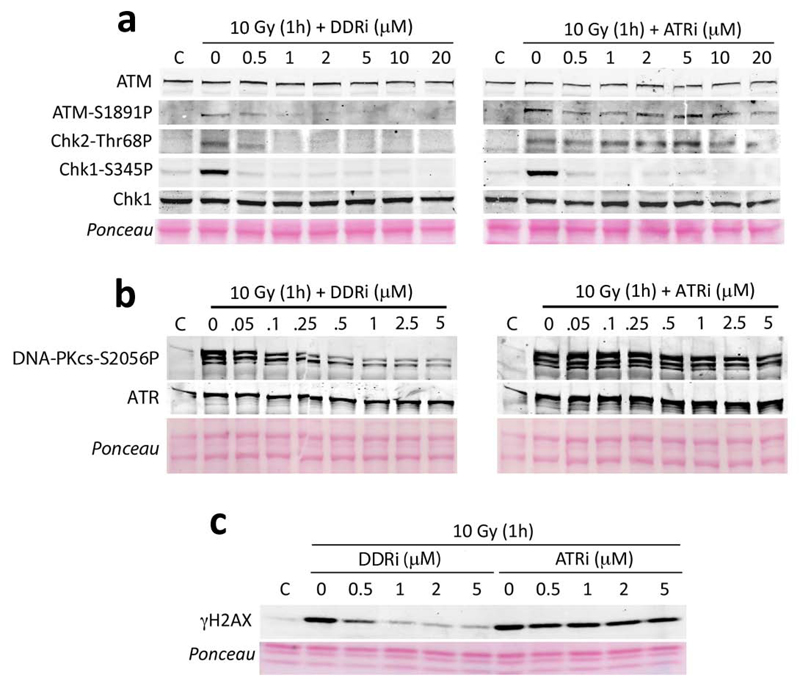
In order to determine the specificity of the compounds and particularly since available inhibitors of ATR are also potent inhibitors of ATM and DNA-PKcs, we analysed the selectivity of the 2 compounds against ATM, ATR and DNA-PKcs. IR-induced ATM and Chk2 vs Chk1 phosphorylation provide a sensitive readout of ATM and ATR activity, respectively14–17. Likewise, IR-induced DNA-PKcs autophosphorylation at Ser 2056 provides an in vivo readout of DNA-PK activity18. Whereas both drugs potently inhibited Chk1 phosphorylation, one of them also inhibited IR-induced ATM, Chk2 and DNA-PKcs phosphorylation, suggesting that it was an efficient inhibitor of all DDR kinases (Fig. 3a,b). In agreement with this, whereas the ATR selective inhibitor did not affect IR-induced γH2AX formation –which is jointly controlled by ATM and DNA-PKcs19-, the other compound also affected this phosphorylation (Fig. 3c). For simplicity, we will name these 2 compounds ATRi (previously, cmpd #3) and DDRi (cmpd #7) from now on.
Figure 3.
ATM-, ATR- and DNA-PKcs-dependent phosphorylations in vivo. Levels of ATM, Chk2 and Chk1 (a); DNA-PKcs (b) and H2AX (c) phosphorylation analysed in U2OS cells upon exposure to 10 Gy at increasing concentrations of the inhibitors. Note that ATR levels are not affected by the use of these compounds.
To determine whether the compounds were able to directly inhibit the kinase activities of ATM and ATR, we performed in vitro kinase assays with immonuprecipitated wild type and kinase-dead versions of flag-ATM and flag-ATR using GST-p53 as a substrate (Supplementary Fig. 1a and Supplementary Methods). Both compounds were similarly potent against ATR, but the ATRi was more efficient in inhibiting ATR than ATM also in vitro. In addition to ATM and ATR, we also analysed the in vitro selectivity of these compounds by running kinase assays against a panel of 26 kinases (Supplementary Fig. 1b). As is the case for most kinase inhibitors, in vitro the compounds presented inhibitory activity towards several other kinases. Noteworthy, the DDRi identified in this screen was originally described in 2008 as a dual PI3K and mTOR inhibitor with potent anti-tumoral activity (NVP-BEZ235)20. Thus, we specifically calculated the in vitro IC50 of the 2 compounds against recombinant PI3Kα, mTOR and DNA-PKcs, as well as the in vitro IC50 for ATM and ATR through the IP assay described above (Supplementary Fig. 1c). In vitro, both compounds presented substantial activity against ATM, ATR, PI3Kα, mTOR and DNA-PKcs, although the ATRi was less effective on ATM and DNA-PKcs. It should be noted that, as shown in Fig. 3, the ATRi is even more selective for ATR vs ATM and DNA-PKcs in cellulo than it is in vitro. This is not unusual for kinase inhibitors. For instance, whereas the Tyr kinase inhibitor NVP-AEW541 is equally potent on IGF-IR and InsR kinases in vitro, it is 26 fold more selective against IGF-IR in cellulo, a property that is been explored for cancer chemotherapy21. Nevertheless, and having these data in mind, we went on to characterize the cellular effects of these compounds, but specifically focusing on those effects that are known to be associated to ATR deficiency and not to ATM, DNA-PKcs, PI3K or mTOR.
Destabilization of stalled replication forks
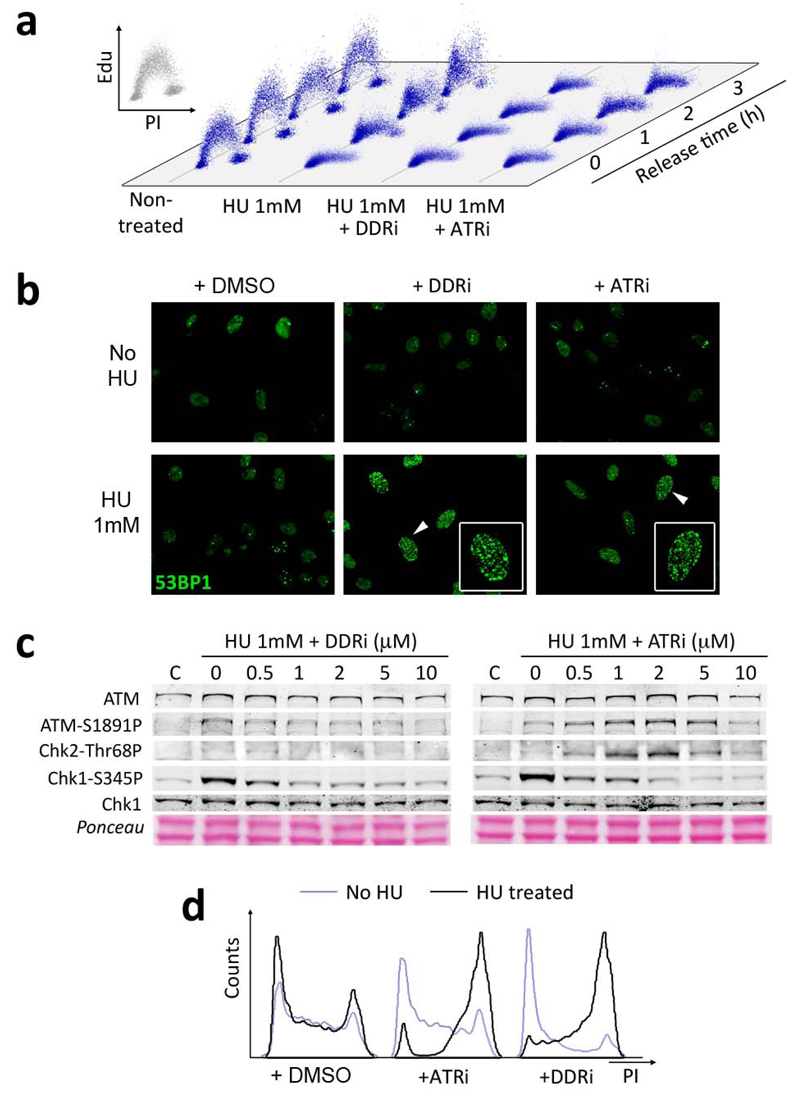
One of the best-established roles of ATR, and which is not shared by ATM or DNA-PKcs, is that it is necessary to prevent the collapse of stalled replication forks9. In order to test this, we exposed U2OS cells to HU for 3 hrs (which promotes fork stalling due to dNTP depletion) in the presence or absence of the inhibitors, and then released them into fresh media. In contrast to the untreated cells, which readily resumed DNA synthesis, cells treated with the inhibitors were unable to do so (Fig. 4a). This observation would be consistent with fragmentation of the stalled replication forks, which has been suggested to make them replication incompetent22. Accordingly, there was a dramatic increase in DSB markers such as 53BP1 foci when the inhibitors were added together with HU (Fig. 4b and Supplementary Fig. 2). Moreover, and concomitant to the inhibition of Chk1 phosphorylation, cells treated simultaneously with HU and ATRi presented elevated levels of ATM and Chk2 phosphorylation. These phosphorylations are indicative of chromosomal breakage, and were not detectable with the DDRi due to the simultaneous inhibition of ATM and DNA-PKcs (Fig. 4c). As a consequence of this damage, there was a very robust accumulation of inhibitor treated cells in G2 16 hrs after the release from HU (Fig. 4d). Noteworthy, G2 accumulation is a hallmark of ATR23 but not ATM24–26 or DNA-PKcs27 deficiency. This reflects that, in contrast to ATM and DNA-PKcs, the DNA damage linked to ATR inhibition occurs during S phase, so that the cells would arrest in the next checkpoint at G2. Importantly, none of these effects could be observed with PI3K, mTOR, ATM or DNA-PKcs inhibitors (Supplementary Fig. 2). Altogether, these data support that the ATR inhibitors identified in our screen promote the breakage of stalled replication forks.
Figure 4.
The identified compounds prevent the breakage of stalled replication forks. (a) The capability to restart DNA synthesis from stalled replication forks was quantified by a pulse of EdU (1 hr), performed at different times after the release from a 3hr exposure to HU. Note that whereas replication can readily restart in control U2OS cells, the presence of either inhibitor stops fork progression. (b) Cells taken from (a) were stained for 53BP1 to visualize the presence of DNA breaks 16 hr after the release from HU. The presence of the ATR inhibitors together with HU leads to a very high accumulation of DNA breaks (HTM quantification of these data is available in Supplementary Fig. 2).(c) Western blot analysis of HU treated U2OS cells (3hrs) in the presence of the inhibitors. (d) Cell cycle profiles of HU treated U2OS cells (1mM, 3hrs) 16 hrs after being released in fresh media free of HU and inhibitors. A dramatic G2 arrest is observed in the inhibitor treated cells.
ATR-mediated suppression of RS
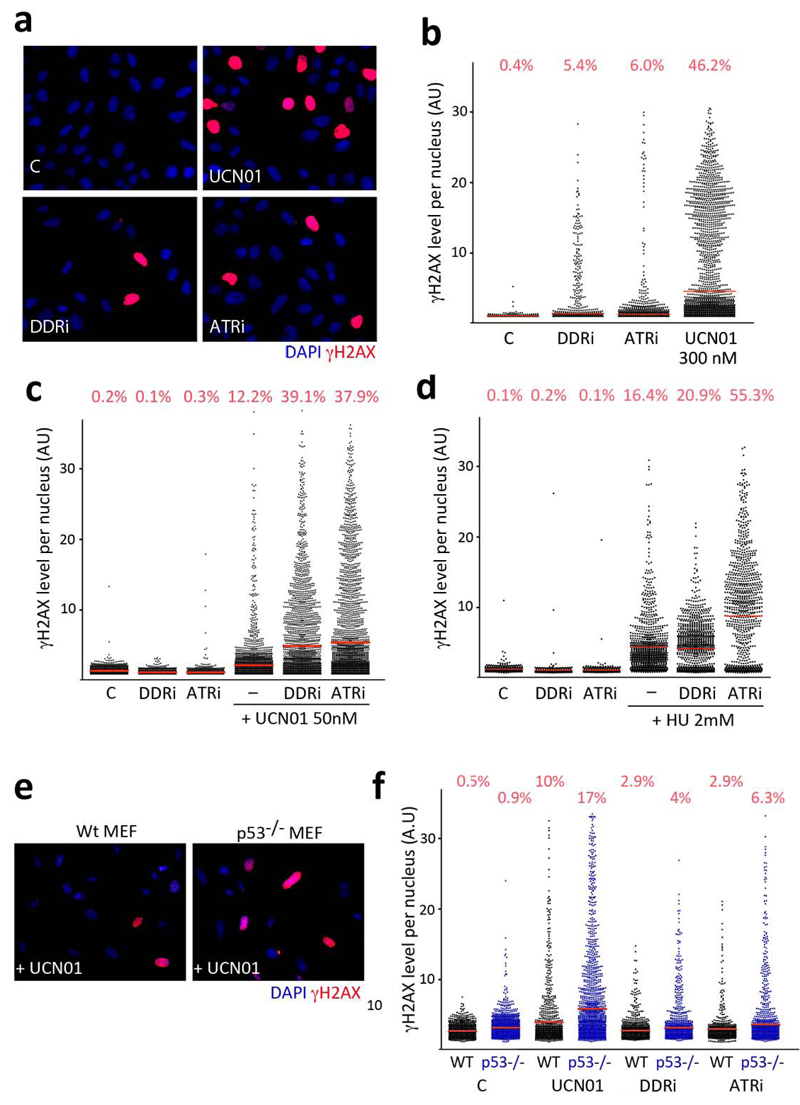
ATR activity is also necessary for replicating cells in the absence of exogenous DNA damage. Accordingly, ATR28,29 or Chk130,31 deletions lead to early embryonic lethality in the mouse. The reason for this essential nature is that ATR and Chk1 prevent the spontaneous generation of replicative stress (RS) during S phase. In mammalian cells, the presence of RS can be detected by a pan-nuclear phosphorylation of H2AX, which contrasts with the foci like pattern observed at DSB. This pan-nuclear staining can be readily detected upon treatment with Chk1 inhibitors32 and is also widespread on embryos and MEF from a hypomorphic ATR mouse strain (ATR-Seckel)23. We thus evaluated whether these inhibitors were capable of inducing RS. Indeed, both compounds led to the appearance of cells with pan-nuclear γH2AX, whereas not as many as with UCN-01 (Fig. 5a,b). As further proof that the pan-nuclear γH2AX staining is indicative of RS, this staining pattern was restricted to S-phase cells (Supplementary Fig. 3a). Moreover, the use of the inhibitors also led to the accumulation of RPA foci (Supplementary Fig. 3b,c). In what regards as to how ATR inhibition could lead to the generation of RS, a recent report revealed that the S-phase-restricted γH2AX observed in the presence of Chk1 inhibitors was dependent on CDC25A33. Similarly, RNAi mediated depletion of CDC25A substantially limited the amount of RS induced by our ATR inhibitors (Supplementary Fig. 3d,e). In summary, these data reveal that the exposure to the inhibitors identified in this work leads the generation of substantial amounts of DNA damage in replicating cells, which are dependent on the Chk1 target CDC25A.
Figure 5.
Inhibition of ATR generates RS. (a) Illustration of the type of pan-nuclear γH2AX staining that is observed upon exposure of U2OS cells to ATR inhibitors (5 μM) or UCN-01 (0.3 μM) for 8 hrs. (b) Representation of the levels of pan-nuclear γH2AX signal obtained in (a) as automatically identified through HTM. (c,d) Representation of the levels of pan-nuclear γH2AX observed when combining lower doses of the ATR inhibitors (1 μM) with low doses of UCN-01 (c), or HU (d). (e) Illustration of the type of pan-nuclear γH2AX staining that is observed upon exposure of wt and p53-/- MEF to UCN-01 (0.3 μM). (f) Pan-nuclear γH2AX in wt and p53 deficient MEF treated with ATR inhibitors (5 μM) or UCN-01 (0.3 μM), as represented in (a). AU: Arbitrary Units.
Next, we explored whether the use of low doses of the inhibitors could boost the development of RS in the presence of other conditions that generate it, such as HU or UCN-01. In fact, the addition of either compound to HU or UCN-01 treated cells lead to a further increase in RS (Fig. 5c,d), suggesting that the combination of these drugs could potentiate their cytotoxic effects. Moreover, the use of low doses of ATR inhibitors, at concentrations in which they do not generate detectable RS, enhanced the effects of low doses of UCN-01 (Supplementary Fig. 4). This information is relevant on the context of in vivo uses of Chk1 or ATR inhibitors, where reaching pharmacologically effective doses of a given compound is a common problem. Importantly, the generation of RS is not observed in the presence of PI3K, mTOR, ATM or DNA-PK inhibitors, not even in the presence of HU (Supplementary Fig. 5a). On the contrary, the RS-associated pan-nuclear γH2AX signal is abrogated in the presence of ATM and DNA-PKcs inhibitors (Supplementary Fig. 5b), as previously shown for ATR-Seckel MEF23. This might explain why the detectable levels of HU-induced γH2AX are lower in the DDRi than in the ATRi, while other effects associated to RS (lethality, RPA foci, G2 arrest, 53BP1 foci in response to HU or the recovery of stalled replication forks) are at least as potent with this inhibitor. Altogether, these results support that the compounds identified in this screen are bona fide ATR inhibitors.
ATR inhibition in the presence of oncogene-induced RS
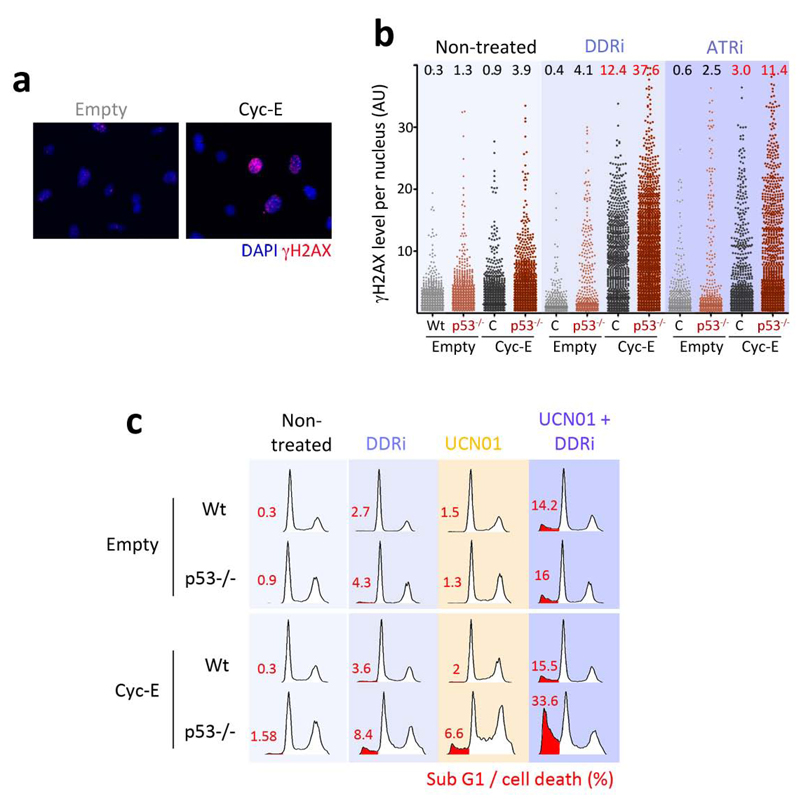
Previous evidence showed that limiting ATR23,34 or Chk135 function is particularly toxic for p53 deficient cells. We recently showed that the underlying reason behind this synthetic lethal interaction is that in conditions of limited ATR function, the increased replication rates linked to p53 absence lead to even further amounts of RS23. In agreement with this, the levels of RS generated by the inhibitors were further increased in p53 deficient MEF (Fig. 5e,f). If our interpretation is correct, then targeting ATR or Chk1 should not only be more toxic for p53 deficient cells, but also for other mutations that promote RS. To test this idea we analyzed the effect of the inhibitors on cyclin E overexpressing cells, alone or in combination with the absence of p53. The reasons for selecting cyclin E overexpression were, first, that this is a common event in human carcinomas and, second, that previous data had already shown that cyclin E overexpression could generate DNA damage3,36,37. We thus evaluated whether targeting ATR and/or Chk1 could be particularly toxic for cyclin E overexpressing cells.
To evaluate the interaction between ATR and Chk1 with cyclin E overexpression we decided to conduct our experiments in primary MEF. The main reason was that human cell lines contain several other mutations that could confound the interpretation of the data. First, we tested whether cyclin E overexpression led to RS in MEF. Indeed, infection with a cyclin E expressing retrovirus led to the appearance of cells with a pan-nuclear staining of γH2AX (Fig. 6a). The amount of cyclin E-induced RS was further increased by the ATR inhibitors and even more so when the experiment was done in p53 deficient MEF (Fig. 6b). The clinical relevance of this approach is that, as previously shown, RS-induced apoptosis is independent of p5323,34,35,38,39. More than that, whereas cyclin E overexpression in MEF is not pro-apoptotic per se, when combined with ATR inhibitors and p53 absence led to a marked increase in the apoptotic elimination of the cells (Fig. 6c). In agreement with the apoptosis being due to RS, this response was further boosted by UCN-01. In contrast, this selective and p53-independent toxicity of ATR inhibitors towards cells overexpressing cyclin E was not observed upon PI3K or mTOR inhibition, not even when both signalling pathways were simultaneously targeted with a dual PI3K and mTOR inhibitor (Supplementary Fig. 6a,b). The same observations were also made in human primary BJ cells, although to a lesser extent which is likely due to the slower proliferation rates of these cells (Supplementary Fig. 6c,d). Altogether, these results support that targeting the cellular response against RS sensitizes cells harbouring cancer-associated mutations that promote RS.
Figure 6.
Synthetic lethality of ATR inhibition with cyclin E overexpression and/or p53 loss. (a) Example of the type of pan-nuclear γH2AX staining that is observed upon overexpression of cyclin E in wt primary MEF. (b) Quantification of the type of staining shown in (a) through HTM, in wt and p53-/- MEF in the presence or absence of the ATR inhibitors (5 μM) for 8 hrs. AU: Arbitrary Units. (c) Cell cycle profiles of cyclin E overexpressing wt and p53-/- MEF in the presence or absence of the DDRi (5μM) and/or UCN-01 (0.3 μM) for 48 hrs.
DISCUSSION
We here describe a pipeline to successfully identify inhibitors of ATR, and describe two of the most potent compounds identified in this screen. The in vivo IC50 for ATR inhibition calculated with the ATR-specific assay explained in Figure 1 was 100 nM for the DDRi and 25 nM for the ATRi (Supplementary Fig. 7). It should be noted that this IC50 was determined in cellulo, where IC50-s are usually higher than in vitro due to compound permeability and other availability issues. For our cellular assays, the inhibitors were used at 1-5 μM. As a measure of their activity, widely accepted potent inhibitors of ATM (KU5593)40 or DNA-PKcs (NU7026)41 are normally used at 5-10 and 20-30 μM, respectively; and the recently described ATR inhibitor schisandrin B is used at 30 μM for cellular assays42.
In addition to their potency, the two compounds were selected on the following reasons. The ATRi was selected on the basis of the academic interest of providing a potent ATR inhibitor, which is selective for ATR vs ATM and DNA-PKcs even at very high doses. The chemical synthesis route of this compound is also provided here, to allow the generation of this reagent in other laboratories (Supplementary Fig. 8). Unfortunately, this compound has poor pharmacological properties in mice, which limit its use for therapy. In contrast, the reason to work with the DDRi has an important clinical perspective. This compound is already been studied and tested in clinical trials as a dual PI3K and mTOR inhibitor under the name of NVP-BEZ23520. In fact, and as is the case in most PIKK inhibitors available, the compound does show potent activity towards PI3K and mTOR. However, neither PI3K and/or mTOR inhibitors showed any of the effects described in this work including RS, G2 arrest or destabilization of stalled forks. On the contrary, the antiproliferative activity of the compounds due to their effect on mTOR is counterproductive for the generation of RS, and future efforts in improving these compounds should try to limit this effect. Most importantly, not even a dual inhibition of PI3K and mTOR shows a synthetic lethal effect with cyclin-E overexpression and/or p53 deficiency. Recent data have shown that NVP-BEZ235 is a potent radiosensitizing compound for Ras overexpressing tumors43, an effect which seems more likely to be mediated by inhibition of the DDR kinases rather than PI3K or mTOR. In agreement with this, a recent report revealed that RNAi-mediated depletion of ATR is particularly toxic for cells overexpressing oncogenic Ras44. In summary, we believe that much of the potential of this drug for cancer chemotherapy could be due to its potent effect on ATR, and this information should be used for a rational design of clinical studies using NVP-BEZ235.
The fact that some cancer cells carry high levels of RS might also expose them to therapies directed to further compromise genomic integrity, as already suggested in previous works5,45. In fact, targeting DNA Repair activities has emerged as a promising approach for cancer chemotherapy, a concept that was importantly fuelled by the discovery of a selective toxicity of PARP inhibitors for tumors carrying BRCA1 or BRCA2 mutations46,47. However, in what specifically regards to the DDR signaling, most of the efforts have been focused in understanding how the oncogene-induced DDR might protect us from cancer through the activation of p53-dependent apoptosis and senescence. In this context, targeting the DDR kinases could be understood as counterproductive, since it could fuel cancer development by eliminating this anti-cancer barrier. In agreement with this, mutations in p53 pathway genes are mutually exclusive with ATM mutations in human cancers48. In contrast to ATM, we here show that ATR inhibition is particularly toxic for cells presenting oncogene-induced RS, eliminating these cells through p53-independent apoptosis. We do not believe that all tumors, or all cancer-associated mutations, concur with the same levels of RS. However, measuring RS in tumor biopsies is affordable (checking the levels of pan-nuclear γH2AX staining), and we believe it might provide a predictive biomarker of the potential efficacy of ATR and Chk1 inhibitors in the clinic.
METHODS
Cells, reagents and treatments
MEF were isolated from 12.5d.p.c embryos. p53 deficient MEF49 and the 3T3TopBp1-ER clone10 were previously described. U2OS, HCT116 and 293T cells were obtained from the American Type Culture Collection. All treatments were done on cells that were seeded from a confluent plate 14 hrs before the treatment to maximize the number of S-phase cells at the time of the treatment. 4-Hydroxytamoxifen (Sigma) was used at 500 nM. Hydroxyurea (Sigma), Caffeine (Sigma), ATM inhibitor (KU5593, KuDOS Pharmaceuticals), DNA-PKcs inhibitor (NU7026, Sigma), Chk1 inhibitor (UCN-01, Sigma) and mTOR inhibitors (Rapamycin,Sigma) were used at the indicated concentrations. The PI3Kα specific, the dual PI3K-mTOR and the screening compounds were part of an in house library described in Link et al12.
Compound Screening
The screening was carried out on 96 well plates. A maximum of 80 wells were used for compound testing. The remaining 16 wells of each plate were used for internal controls: 6 negative controls (not 4-OHT treated), 6 positive controls (only 4-OHT treated) and 4 inhibition controls (5mM caffeine and 4-OHT treated). The day before a compound library was screened, 3T3TopBP1-ER cells (15000 per well) were seeded on Greiner (Greinerbioone) 96 well plates. Wells were pre-treated with 1% (w/v) gelatine. For the initial screening, compounds and control inhibitors were delivered directly to cell media (100 microliters per well) with a multi-well pipette at a final concentration of 10 μM. Media content was homogenized by carefully vortexing plates at 500rpm. Prior to 4-OHT addition, compounds were incubated at 37ºC for 15 minutes. Next, to induce ATR activity, 4-OHT was added to all wells and incubated for 60 minutes at 37ºC. Finally, cells were fixed with paraformaldehyde and processed for IF as described below. Every compound was analyzed at least in three independent experiments.
High-Content Analysis
For quantitative measurement of γH2AX levels, pictures were automatically acquired from each well by a BD-Pathway High-Throughput microscope (Beckton-Dickinson). A dry 10x magnification lens was used and pictures were taken at non-saturating conditions. At least 300 cells were analyzed for each well. Attovision software was then used to extract the average γH2AX intensity from each cell nucleus. Briefly, images were segmented using the DAPI staining to generate masks matching cell nuclei from which the average γH2AX was calculated. Within each plate, γH2AX levels were normalized using the average of negative and positive controls as minimum and maximum reference levels. The number obtained was taken as relative ATR activity for each condition or compound.
Immunofluorescence
Cells were washed once with PBS (phosphate buffered saline) and fixed for 10 min at room temperature in 2% (w/v) paraformaldehyde. After 2 more PBS washes, cells were permeabilized with 0.1% (w/v) sodium citrate, 0.1% (v/v) Triton X-100 for 8 minutes at RT. After two more washes in PBS, cells were blocked in PBS containing 3% (w/v) BSA and 0.1% (v/v) Tween20 for 30 minutes at RT. Next, primary antibodies were incubated in the same blocking solution. After incubation, cells were washed 3 times for 5 minutes at RT with PBS containing 0.25% (w/v) BSA and 0.1% (v/v) Tween20. Secondary antibodies (see below) were subsequently incubated in blocking solution for 45 minutes at RT. After 3 washes in PBS, 96 well plates were stored in PBS containing 200 ng ml-1 DAPI at 4ºC. When cells were grown on coverslips the preparations were mounted in mowiol, air-dried and stored at 4ºC. Images were taken with a Zeiss Imager Z1 fluorescence microscope using 40x or 63x magnification lenses, and with oil as immersion media.
Protein extracts and western blotting
For total protein extracts, cells were washed once with PBS, collected by adding directly 2x NuPAGE LDS Sample buffer and incubated for 5 min at 95 °C. For soluble protein extracts, cells were lysed in RIPA buffer (Tris-HCl 50 mM pH 7.4, 1% (v/v) NP-40, 0.25% (w/v) sodium deoxycholate, NaCl 150 mM, EDTA 1 mM) containing protease and phosphatase inhibitors (Sigma). Samples were resolved by SDS-PAGE and analyzed by standard western blotting techniques. The following primary antibodies were used: γH2AX (Millipore), Chk1-S345P and Chk2-T68P (Cell Signalling), Chk1 (Leica-Microsystems), DNA-PKcs-S2506P (Abcam), ATR (Serotec), ATM-1981P (Rockland) and ATM (Calbiochem). Alexa 680- and Alexa 800-conjugated secondary antibodies (Molecular Probes) were used for detection with a LI-COR Odyssey infrared imaging system from LI-COR Biosciences.
Cell cycle arrest and proliferation analysis
For G2/M checkpoint analysis cells were incubated with the different compounds 15 minutes prior to ionizing radiation. Concentrations were as indicated. One hour later cells were harvested and fixed in suspension in ice-cold 70% (v/v) Ethanol in PBS. After a wash in PBS, cells were incubated with an H3-S10P recognizing antibody (Millipore) in PBS containing 1% (w/v) BSA and 0.1% (v/v) Tween20 for 2h at RT. After one wash with the same solution and incubation with Alexa-488 secondary antibody, cells were resuspended in a PBS solution containing 1% (w/v) BSA, propidium iodide (10 μg ml−1) and RNase A (0.5 mg ml−1), and analyzed by flow citometry in a BD FACSCalibur machine. To monitor the HU-induced reversible replication block, cells were incubated with EdU for 1h at 37ºC. After fixation with 4% (w/v) paraformaldehyde for 30 minutes, cells were stained with 488-Azide as recommended by kit manufacturers (Applied Biosciences). Finally, cells were resuspended cells were resuspended in a PBS solution containing 1% (w/v) BSA, propidium iodide (10 μg ml−1) and RNase A (0.5 mg ml−1), and analyzed by flow citometry in a BD FACSCalibur machine.
Retroviral infections
Viruses were generated by contransfection of the retroviral plasmids and the packaging plasmid (pCL-Eco) in 293T cells. 48h after transfection, MEF were exposed to viral supernatant complemented with 8 μg ml−1 polibrene for 8 to 16 hours. 36 to 48 hours after infection, cells were selected with 2 μg ml−1 puromycin for another 48 hours. Experiments were carried out immediately after selection with cells in exponential growth. pBabe-cyclin E and pSM2-hTP5350 were kind gifts from M. Barbacid and J.M. Silva, respectively (see Acknowledgments).
Additional Methods
A detailed description of all other methods is given in Supplementary Methods.
Supplementary Material
ACKNOWLEDGEMENTS
We thank M. Barbacid for providing reagents, M. Serrano for critical comments on the manuscript and M.I. Albarran and P. Alfonso for help in the kinase assays (Spanish National Cancer Research Centre, Madrid, Spain). We also thank J.M. Silva (Irving Cancer Research Centre, Columbia University, New York, USA) for providing reagents. M. M. is supported by a grant from Fondo de Investigaciones Sanitarias (PI05945). Work in O. F.´s laboratory is supported by grants from the Spanish Ministry of Science (CSD2007-00017 and SAF2008-01596), Pfizer Foundation Award, EMBO Young Investigator Programme and the European Research Council (ERC-210520).
Footnotes
AUTHOR CONTRIBUTIONS
O.F. designed the study and experiments and wrote the paper. L.I.T. performed most of the experiments presented. M.M. helped in the work with oncogenes and CDC25A. R.Z. and R.S. provided technical help. J.O., A.R., S.M., J.P. and J.R.B provided the chemicals, and helped in the development of the small molecule screening.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
References
- 1.Lempiainen H, Halazonetis TD. Emerging common themes in regulation of PIKKs and PI3Ks. EMBO J. 2009;28:3067–73. doi: 10.1038/emboj.2009.281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lopez-Contreras AJ, Fernandez-Capetillo O. The ATR barrier to replication-born DNA damage. DNA Repair (Amst) 2010;9:1249–55. doi: 10.1016/j.dnarep.2010.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bartkova J, et al. DNA damage response as a candidate anti-cancer barrier in early human tumorigenesis. Nature. 2005;434:864–70. doi: 10.1038/nature03482. [DOI] [PubMed] [Google Scholar]
- 4.Gorgoulis VG, et al. Activation of the DNA damage checkpoint and genomic instability in human precancerous lesions. Nature. 2005;434:907–13. doi: 10.1038/nature03485. [DOI] [PubMed] [Google Scholar]
- 5.Halazonetis TD, Gorgoulis VG, Bartek J. An oncogene-induced DNA damage model for cancer development. Science. 2008;319:1352–5. doi: 10.1126/science.1140735. [DOI] [PubMed] [Google Scholar]
- 6.Bartkova J, et al. Oncogene-induced senescence is part of the tumorigenesis barrier imposed by DNA damage checkpoints. Nature. 2006;444:633–7. doi: 10.1038/nature05268. [DOI] [PubMed] [Google Scholar]
- 7.Di Micco R, et al. Oncogene-induced senescence is a DNA damage response triggered by DNA hyper-replication. Nature. 2006;444:638–42. doi: 10.1038/nature05327. [DOI] [PubMed] [Google Scholar]
- 8.Mallette FA, Gaumont-Leclerc MF, Ferbeyre G. The DNA damage signaling pathway is a critical mediator of oncogene-induced senescence. Genes Dev. 2007;21:43–8. doi: 10.1101/gad.1487307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cimprich KA, Cortez D. ATR: an essential regulator of genome integrity. Nat Rev Mol Cell Biol. 2008;9:616–27. doi: 10.1038/nrm2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Toledo LI, Murga M, Gutierrez-Martinez P, Soria R, Fernandez-Capetillo O. ATR signaling can drive cells into senescence in the absence of DNA breaks. Genes Dev. 2008;22:297–302. doi: 10.1101/gad.452308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kumagai A, Lee J, Yoo HY, Dunphy WG. TopBP1 activates the ATR–ATRIP complex. Cell. 2006;124:943–55. doi: 10.1016/j.cell.2005.12.041. [DOI] [PubMed] [Google Scholar]
- 12.Link W, et al. Chemical interrogation of FOXO3a nuclear translocation identifies potent and selective inhibitors of phosphoinositide 3-kinases. J Biol Chem. 2009;284:28392–400. doi: 10.1074/jbc.M109.038984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cliby WA, et al. Overexpression of a kinase-inactive ATR protein causes sensitivity to DNA-damaging agents and defects in cell cycle checkpoints. EMBO J. 1998;17:159–69. doi: 10.1093/emboj/17.1.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bakkenist CJ, Kastan MB. DNA damage activates ATM through intermolecular autophosphorylation and dimer dissociation. Nature. 2003;421:499–506. doi: 10.1038/nature01368. [DOI] [PubMed] [Google Scholar]
- 15.Cuadrado M, et al. ATM regulates ATR chromatin loading in response to DNA double-strand breaks. J Exp Med. 2006;203:297–303. doi: 10.1084/jem.20051923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jazayeri A, et al. ATM- and cell cycle-dependent regulation of ATR in response to DNA double-strand breaks. Nat Cell Biol. 2006;8:37–45. doi: 10.1038/ncb1337. [DOI] [PubMed] [Google Scholar]
- 17.Matsuoka S, Huang M, Elledge SJ. Linkage of ATM to cell cycle regulation by the Chk2 protein kinase. Science. 1998;282:1893–7. doi: 10.1126/science.282.5395.1893. [DOI] [PubMed] [Google Scholar]
- 18.Chen BP, et al. Cell cycle dependence of DNA-dependent protein kinase phosphorylation in response to DNA double strand breaks. J Biol Chem. 2005;280:14709–15. doi: 10.1074/jbc.M408827200. [DOI] [PubMed] [Google Scholar]
- 19.Stiff T, et al. ATM and DNA-PK function redundantly to phosphorylate H2AX after exposure to ionizing radiation. Cancer Res. 2004;64:2390–6. doi: 10.1158/0008-5472.can-03-3207. [DOI] [PubMed] [Google Scholar]
- 20.Maira SM, et al. Identification and characterization of NVP-BEZ235, a new orally available dual phosphatidylinositol 3-kinase/mammalian target of rapamycin inhibitor with potent in vivo antitumor activity. Mol Cancer Ther. 2008;7:1851–63. doi: 10.1158/1535-7163.MCT-08-0017. [DOI] [PubMed] [Google Scholar]
- 21.Garcia-Echeverria C, et al. In vivo antitumor activity of NVP-AEW541-A novel, potent, and selective inhibitor of the IGF-IR kinase. Cancer Cell. 2004;5:231–9. doi: 10.1016/s1535-6108(04)00051-0. [DOI] [PubMed] [Google Scholar]
- 22.Petermann E, Orta ML, Issaeva N, Schultz N, Helleday T. Hydroxyurea-stalled replication forks become progressively inactivated and require two different RAD51-mediated pathways for restart and repair. Mol Cell. 2010;37:492–502. doi: 10.1016/j.molcel.2010.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Murga M, et al. A mouse model of ATR-Seckel shows embryonic DNA replicative stress and accelerated ageing. Nat Genet. 2009 doi: 10.1038/ng.420. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Barlow C, et al. Atm-deficient mice: a paradigm of ataxia telangiectasia. Cell. 1996;86:159–71. doi: 10.1016/s0092-8674(00)80086-0. [DOI] [PubMed] [Google Scholar]
- 25.Elson A, et al. Pleiotropic defects in ataxia-telangiectasia protein-deficient mice. Proc Natl Acad Sci U S A. 1996;93:13084–9. doi: 10.1073/pnas.93.23.13084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xu Y, Baltimore D. Dual roles of ATM in the cellular response to radiation and in cell growth control. Genes Dev. 1996;10:2401–10. doi: 10.1101/gad.10.19.2401. [DOI] [PubMed] [Google Scholar]
- 27.Taccioli GE, et al. Targeted disruption of the catalytic subunit of the DNA-PK gene in mice confers severe combined immunodeficiency and radiosensitivity. Immunity. 1998;9:355–66. doi: 10.1016/s1074-7613(00)80618-4. [DOI] [PubMed] [Google Scholar]
- 28.Brown EJ, Baltimore D. ATR disruption leads to chromosomal fragmentation and early embryonic lethality. Genes Dev. 2000;14:397–402. [PMC free article] [PubMed] [Google Scholar]
- 29.de Klein A, et al. Targeted disruption of the cell-cycle checkpoint gene ATR leads to early embryonic lethality in mice. Curr Biol. 2000;10:479–82. doi: 10.1016/s0960-9822(00)00447-4. [DOI] [PubMed] [Google Scholar]
- 30.Liu Q, et al. Chk1 is an essential kinase that is regulated by Atr and required for the G(2)/M DNA damage checkpoint. Genes Dev. 2000;14:1448–59. [PMC free article] [PubMed] [Google Scholar]
- 31.Takai H, et al. Aberrant cell cycle checkpoint function and early embryonic death in Chk1(-/-) mice. Genes Dev. 2000;14:1439–47. [PMC free article] [PubMed] [Google Scholar]
- 32.Syljuasen RG, et al. Inhibition of human Chk1 causes increased initiation of DNA replication, phosphorylation of ATR targets, and DNA breakage. Mol Cell Biol. 2005;25:3553–62. doi: 10.1128/MCB.25.9.3553-3562.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Beck H, et al. Regulators of cyclin-dependent kinases are crucial for maintaining genome integrity in S phase. J Cell Biol. 188:629–38. doi: 10.1083/jcb.200905059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ruzankina Y, et al. Tissue regenerative delays and synthetic lethality in adult mice after combined deletion of Atr and Trp53. Nat Genet. 2009;41:1144–9. doi: 10.1038/ng.441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Koniaras K, Cuddihy AR, Christopoulos H, Hogg A, O'Connell MJ. Inhibition of Chk1-dependent G2 DNA damage checkpoint radiosensitizes p53 mutant human cells. Oncogene. 2001;20:7453–63. doi: 10.1038/sj.onc.1204942. [DOI] [PubMed] [Google Scholar]
- 36.Ekholm-Reed S, et al. Deregulation of cyclin E in human cells interferes with prereplication complex assembly. J Cell Biol. 2004;165:789–800. doi: 10.1083/jcb.200404092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Spruck CH, Won KA, Reed SI. Deregulated cyclin E induces chromosome instability. Nature. 1999;401:297–300. doi: 10.1038/45836. [DOI] [PubMed] [Google Scholar]
- 38.Myers K, Gagou ME, Zuazua-Villar P, Rodriguez R, Meuth M. ATR and Chk1 suppress a caspase-3-dependent apoptotic response following DNA replication stress. PLoS Genet. 2009;5:e1000324. doi: 10.1371/journal.pgen.1000324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sidi S, et al. Chk1 suppresses a caspase-2 apoptotic response to DNA damage that bypasses p53, Bcl-2, and caspase-3. Cell. 2008;133:864–77. doi: 10.1016/j.cell.2008.03.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hickson I, et al. Identification and characterization of a novel and specific inhibitor of the ataxia-telangiectasia mutated kinase ATM. Cancer Res. 2004;64:9152–9. doi: 10.1158/0008-5472.CAN-04-2727. [DOI] [PubMed] [Google Scholar]
- 41.Willmore E, et al. A novel DNA-dependent protein kinase inhibitor, NU7026, potentiates the cytotoxicity of topoisomerase II poisons used in the treatment of leukemia. Blood. 2004;103:4659–65. doi: 10.1182/blood-2003-07-2527. [DOI] [PubMed] [Google Scholar]
- 42.Nishida H, et al. Inhibition of ATR protein kinase activity by schisandrin B in DNA damage response. Nucleic Acids Res. 2009;37:5678–89. doi: 10.1093/nar/gkp593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Konstantinidou G, et al. Dual phosphoinositide 3-kinase/mammalian target of rapamycin blockade is an effective radiosensitizing strategy for the treatment of non-small cell lung cancer harboring K-RAS mutations. Cancer Res. 2009;69:7644–52. doi: 10.1158/0008-5472.CAN-09-0823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gilad O, et al. Combining ATR suppression with oncogenic Ras synergistically increases genomic instability, causing synthetic lethality or tumorigenesis in a dosage-dependent manner. Cancer Res. 70:9693–702. doi: 10.1158/0008-5472.CAN-10-2286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bartek J, Bartkova J, Lukas J. DNA damage signalling guards against activated oncogenes and tumour progression. Oncogene. 2007;26:7773–9. doi: 10.1038/sj.onc.1210881. [DOI] [PubMed] [Google Scholar]
- 46.Bryant HE, et al. Specific killing of BRCA2-deficient tumours with inhibitors of poly(ADP-ribose) polymerase. Nature. 2005;434:913–7. doi: 10.1038/nature03443. [DOI] [PubMed] [Google Scholar]
- 47.Farmer H, et al. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature. 2005;434:917–21. doi: 10.1038/nature03445. [DOI] [PubMed] [Google Scholar]
- 48.Negrini S, Gorgoulis VG, Halazonetis TD. Genomic instability--an evolving hallmark of cancer. Nat Rev Mol Cell Biol. 2010;11:220–8. doi: 10.1038/nrm2858. [DOI] [PubMed] [Google Scholar]
- 49.Donehower LA, et al. Mice deficient for p53 are developmentally normal but susceptible to spontaneous tumours. Nature. 1992;356:215–21. doi: 10.1038/356215a0. [DOI] [PubMed] [Google Scholar]
- 50.Silva JM, et al. Second-generation shRNA libraries covering the mouse and human genomes. Nat Genet. 2005;37:1281–8. doi: 10.1038/ng1650. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.