Abstract
The yeast Saccharomyces cerevisiae is ideal for systematic studies relying on collections of modified strains (libraries). Despite the significance of yeast libraries and the immense variety of available tags and regulatory elements, only a few such libraries exist as their construction is extremely expensive and laborious. To overcome these limitations we developed a SWAp-Tag method (SWAT), in which one parental library can be modified easily and efficiently to give rise to an endless variety of libraries of choice. We showcase the versatility of the SWAT approach by constructing and investigating a library of ~1,800 strains carrying a SWAT-GFP module at the amino termini of endomembrane proteins and then using it to create two new libraries (mCherry or seamless GFP). Our work demonstrates how the SWAT method enables fast and effortless creation of yeast libraries, opening the door for endless new ways to systematically study cell biology.
Introduction
One of the most important tools that the yeast model organism Saccharomyces cerevisiae offers are systematic collections of strains (termed libraries), where in each strain a different gene is modified in a similar manner to enable genome wide studies 1. Several libraries are currently available, such as a deletion library 2, a protein complementation assay library 3 and a Green Fluorescent Protein (GFP) library 4. Each established library has given rise to many biological insights 5–12. This argues for continuous construction of new libraries so that new genome or proteome wide studies can be conducted. Despite this, to date very few libraries have been made. This is because the construction of each yeast library is an extremely laborious and expensive procedure. Researchers are deterred by the planning and ordering of thousands of primer sets, transformations, clone picking and validation steps. This high energetic barrier for production of new libraries hinders many labs from the pursuit of a variety of open biological questions.
We devised and implemented a methodology to remove the major hurdles of library construction for future library assembly projects, making creativity the only limiting factor. Our methodology, termed SWAT (SWAp-Tag), is based on an initial acceptor library which serves as a template that can be “swapped” into other libraries of choice 13. This requires the one-time construction of an acceptor library of strains (still relying on traditional methods 14–16) each with an acceptor module integrated at a specific genomic location. The acceptor library can then be converted easily, rapidly and efficiently into any new library by replacement of the acceptor module with a new tag or genomic sequence of choice, introduced by crossing with a donor strain.
To establish this strategy we constructed a library containing ~1,800 strains where all proteins with known or predicted localization to the yeast endomembrane system are tagged with an amino terminal (N′) SWAT acceptor module. The used module also contains a constitutive promoter and a GFP tag. With this N′-tag library, which we call SWAT-GFP, we were able to uncover the sub-cellular localization of hundreds of proteins never visualized before, and to characterize several new peroxisomal and secreted proteins.
To demonstrate the ease of new library creation by the SWAT method we created two additional libraries: A seamless GFP library – where the natural regulatory sequences have been restored - and an mCherry library. These three N′ tag libraries (the SWAT-GFP acceptor, seamless GFP, and mCherry) as well as a variety of donor and acceptor modules are now freely available to the yeast community. We believe that the SWAT methodology will promote the construction of many new yeast libraries, opening the door to a flood of possibilities for systematic exploration of open questions in cell biology.
Results
SWAp-Tag (SWAT) strategy enables rapid library creation
We designed a method to rapidly and efficiently create yeast libraries. Our method, which we call SWAT, relies on one “parental” library that specifies the insertion point (e.g. the 3` or 5` end of each gene). This library then provides an easy gateway for the generation of any “daughter” library of choice. SWAT essentially allows anyone to create a customized, high-throughput tool to investigate a biological question.
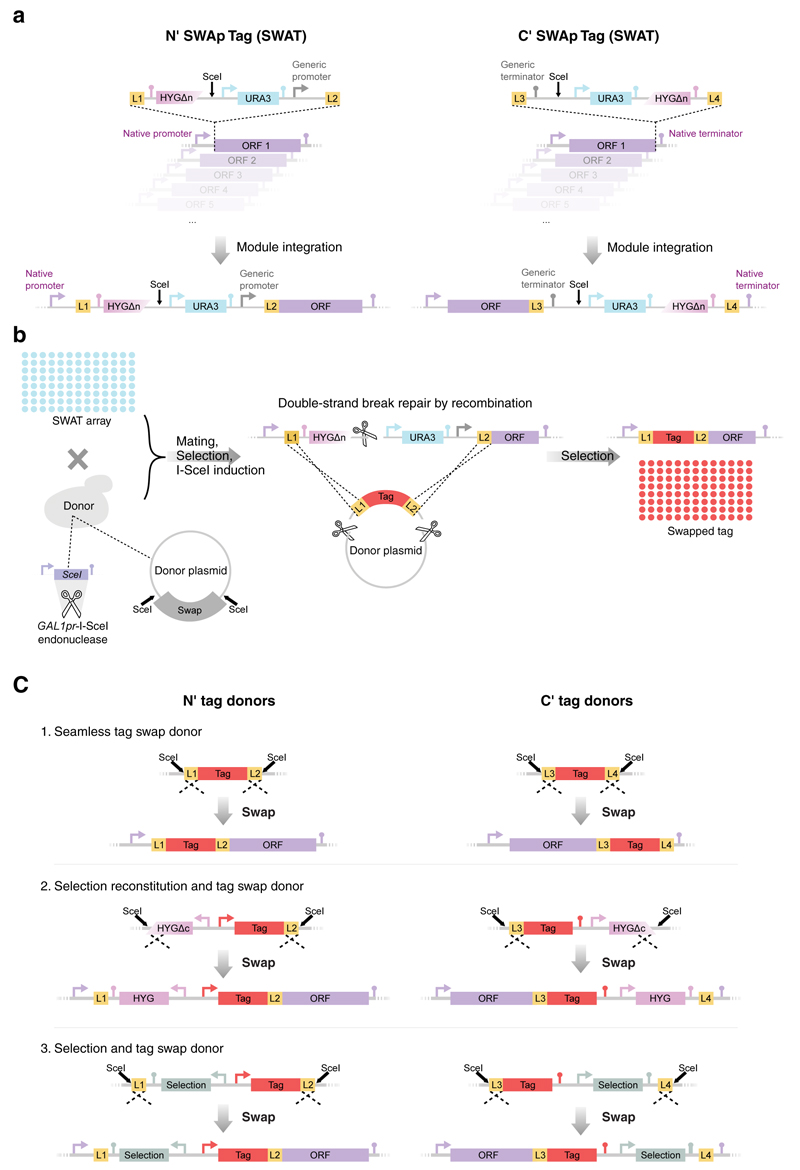
The SWAT method relies on the genomic integration of an acceptor tagging module (using classical PCR mediated transformation 14,15). We designed and made acceptor modules for tagging proteins at the N`- or C`-terminus, namely N′-SWAT and C′-SWAT. The SWAT modules include a restriction site for the endonuclease I-SceI (that is not found elsewhere within the yeast genome) 17, and are flanked by two short (45 base-pair (bp)) sequences that serve as homology arms for recombination. These homology arms also act as flexible protein linkers in the case where protein fusions are desired (L1, L2 for N′ tagging or L3, L4 for C′ tagging). Upon I-SceI induction, a genomic double strand break is created within the SWAT module, facilitating its replacement by homologous recombination of any donor sequence that contains corresponding flanking regions. To select for such desired recombination events we included two features in the SWAT tags. First, the URA3 selection marker that allows both positive selection (on medium lacking Uracil) for initial integration, and negative selection (on medium containing 5-Fluoroorotic Acid, 5-FOA) for module swap; Second, a truncated version of the HygromycinB resistance cassette (HYGΔn) that can be restored to the full resistance gene upon precise recombination (Fig. 1a and Supplementary Table 1).
Figure 1. The Swap-Tag (SWAT) strategy enables fast and easy creation of systematic yeast libraries.
(a) Swap-tag (SWAT) acceptor modules are used to tag proteins at either their N- terminus (left) or C- terminus (right). The modules contain the restriction site for the I-SceI endonuclease (SceI), a URA3 selection marker (URA3), a truncated Hygromycin B selection marker (HYGΔn), and a generic promoter/terminator (depending on the terminus tagged), and are flanked by two generic sequences for homologous recombination that also serve as linkers for protein fusion (L1, L2 or L3, L4). (b) An acceptor library of strains with SWAT tagged genes (SWAT array) can be crossed with a donor strain to create a new library of choice. The donor strain encodes a galactose induced I-SceI and harbors a donor plasmid with the desired tag flanked by two generic sequences (L1, L2 or L3, L4) and two SceI restriction sites for systematic insertion (left). Automated mating procedure is used to create haploids carrying all genetic traits. At this point, I-SceI expression is induced by a growth step on galactose, leading to double strand breaks in the tagged gene locus and the donor plasmid. The genomic break is mended by homologous recombination of the donor tag (middle). Correct swapping is selected for and a library featuring the new tag is created (right). (c) Donor plasmid features (top), which result in several types of tagged-gene libraries (bottom). This can be similarly performed on N′ (left) or C′ (right) tagged acceptor libraries (Online Methods).
Once a library of strains is made in which the SWAT acceptor module is integrated into each gene, it is mated with a donor strain of the opposite mating type. The donor strain encodes for the I-SceI endonuclease under an inducible promoter (GAL1pr that is induced by galactose and repressed by glucose), and harbors a donor plasmid that contains the desired sequence to be “swapped-in”, also flanked by I-SceI sites. Following automated synthetic genetic array (SGA) 18 procedures for mating, sporulation and selection of haploid spores of choice, I-SceI expression is induced by galactose containing media and resulting recombination events are selected by one of various selection options (Online Methods). Starting with the acceptor library, the tag swapping procedure is completed in approximately three weeks, and a new library is formed and ready for experimentation (Fig. 1b).
A key advantage of the SWAT method is that any part of the acceptor module (promoter, tag and selection) can be replaced with any other of one’s needs, including replacement of the entire acceptor module to restore native gene regulation, allowing seamless tagging (Fig. 1c, Supplementary Fig. 1a,b,c, and Online Methods).
We measured the efficiency of swapping modules of the various options with the SWAT method and found that it is highly efficient and accurate, even for seamless swapping where no positive selection cassette is introduced (Supplementary Fig. 1d). To facilitate use by the community, we prepared an assortment of SWAT acceptor tagging modules and donor modules for either N′ or C′ tagging that would cater for a wide variety of needs and are freely available (Supplementary Table 1 and 2, Supplementary Data 1, and in our website: http://www.weizmann.ac.il/molgen/Maya/SWAT).
Creation of N` SWAT-GFP libraries
To demonstrate the SWAT approach we constructed an acceptor library using an acceptor module that included the constitutive SpNOP1 promoter 19 and a GFP tag (SWAT-GFP). We focused our efforts on endomembrane system proteins (residing on the surface or in the lumen of yeast secretory pathway organelles), nearly 30% of the yeast proteome 20.
We compiled a list of all endomembrane proteins and found that ~17% are predicted to contain an N′ cleavable signal peptide (SP) 21–24 that would be required for their correct targeting to the Endoplasmic Reticulum (ER) (Supplementary Fig. 2 and a complete list of SP predictions in Supplementary Table 3). To ensure that these proteins do not lose this essential targeting sequenc we created a unique SWAT acceptor module that harbors the generic and efficient SP of Kar2 before the GFP tag (termed SWAT-SP-GFP) (Fig. 2a). The SP module is integrated downstream of the predicted cleavage site of each SP containing protein (Online Methods).
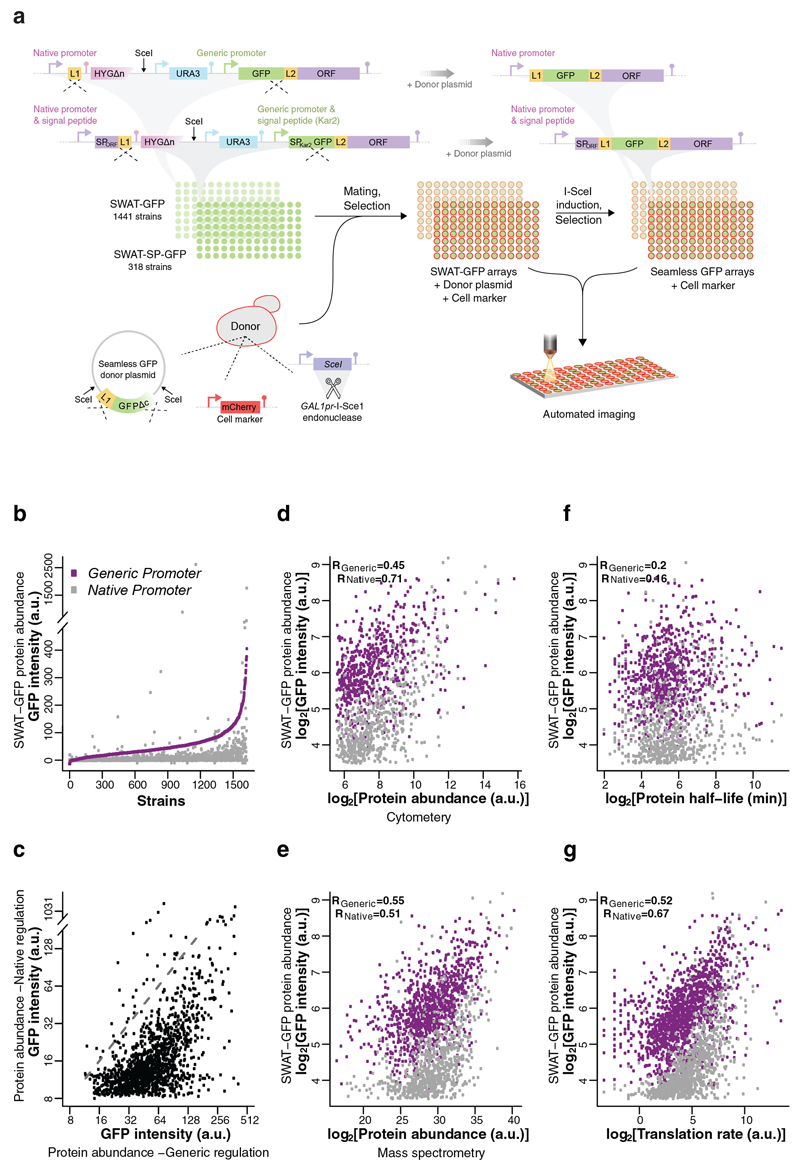
Figure 2. Utilizing SWAT strategy to rapidly create a seamless N′-tagged GFP library enables comparison of protein abundance under generic or native regulation.
(a)Two strain collections were assembled, in which proteins were tagged at their N′ with an acceptor SWAT module that contains the SpNOP1 constitutive promotor (generic) and a GFP. Proteins predicted to harbor a signal peptide (SP) were tagged with a similar acceptor SWAT module that contains a SP upstream to the GFP (SP of Kar2). The two libraries were crossed with a donor strain that harbors a plasmid with a seamless GFP swap donor plasmid, the inducible I-SceI enzyme and an mCherry cell marker. Imaging of strains was performed on both the parental SWAT-GFP library and the daughter seamless GFP library to compare protein abundance under generic or native regulation. (b) Expression levels of GFP-protein fusions under a single generic promoter spanned >2 orders of magnitude, and were almost exclusively higher than under native regulation. Purple: Generic regulation (SWAT-GFP); Grey: native regulation (seamless GFP). a.u - arbitrary units. (c) The correlation between protein abundance under generic and native regulation highlighted the effect of non-transcriptional regulation on protein abundance. Dashed line indicates the diagonal. (d-g) Comparison of N′ GFP tagged protein abundances, either under generic or native regulation with the: (d) abundance of these proteins as measured by flow cytometry analysis of C′ tagged GFP proteins27; (e) abundance of these proteins as measured by mass spectrometry 28; (f) protein half-lives 29; (g) protein translation rates as measured by ribosome profiling 30.
We used the SWAT-SP-GFP module to tag 336 proteins with predicted SPs and succeeded in making 318 of them (95%, SWAT-SP-GFP library). We used the SWAT-GFP module to tag the remaining 1,510 proteins that were not predicted to require a SP and succeeded in making 1441 strains (95%, SWAT-GFP library). Altogether our libraries contain 1,759 strains that were also validated by PCR (Fig. 2a and Supplementary Table 4. For further details on SP prediction, library construction and validation see Online Methods).
Once the SWAT-GFP libraries were assembled, we utilized them to performed high-throughput tag swapping with a “seamless GFP donor” plasmid. The donor plasmid contains the L1 linker and 45bp from the GFP start (GFPΔc), and is intended to excise out all acceptor features, leaving only the short protein linker and the GFP tag. In the case of SWAT-GFP strains this resulted in restoration of the native promoter, and in the case of the SWAT-SP-GFP strains this resulted in restoration of both the native promoter and native SP (Fig. 2a). The swapping procedure was highly efficient as >98% of the strains were recovered, 96% of those tested (70/73) were accurately swapped (as demonstrated by PCR, Supplementary Fig. 3), and a homogeneous population was observed in >98% of strains tested (Supplementary Fig. 4 and Online Methods). The end result of this procedure was an N` seamless GFP library.
We used the GFP libraries before and after seamless swapping to measure how protein abundance is affected by regulatory sequences (native vs generic). Specifically, we measured GFP intensity at single cell resolution using high content microscopy 25,26. Individual cells were identified by a cytosolic mCherry marker introduced by the donor strain (Fig. 2a and Online Methods). Surprisingly, we found that even when all proteins were under control of the SpNOP1 constitutive promoter, each strain had a unique expression level spanning two orders of magnitude (Fig. 2b, purple).
As expected, in the vast majority of cases the generic SpNOP1 promoter produced higher expression than the native one. While we could not detect a fluorescent signal above background auto-fluorescence for 2% of the proteins that were under regulation of the generic SpNOP1 promoter, in the seamless version of the library this number increased dramatically to 48% (Supplementary Table 4). While our analysis of swapping (above) suggests that ~4% of these strains simply did not undergo swapping and while additional cases may represent inaccurate swapping, the majority represent cases whereby the native promoter simply confers low expression levels. This emphasizes the importance of having two versions of an N′ library – one that enables studying proteins with low natural abundance, and another that represents native regulatory context.
The distinct correspondence between the abundance of proteins under the native vs the SpNOP1 promoter highlighted the importance of non-transcriptional events in regulating steady-state protein levels (Fig. 2c). When comparing protein abundance, both under generic or native regulation, to other, non-related, datasets that have measured protein abundance such as the C′ GFP library measured by flow cytometry 27 (Fig. 2d) and mass spectrometry 28 (Fig. 2e), a high correlation was seen. While systematic data on protein half-lives 29 (Fig. 2f) did not seem to be highly correlated, data on protein translation rates by ribosome profiling 30 was (Fig. 2g), suggesting that translational regulation is the major source of abundance regulation for these proteins.
New condition-specific secreted proteins discovered
Our SWAT-SP-GFP library contains proteins with a predicted SP and therefore would possibly include uncharacterized secreted proteins. To uncover such cases we utilized a simple secretion blotting assay on both the acceptor SWAT-SP-GFP as well as the SP-seamless-GFP libraries (Supplementary Fig. 5 and Supplementary Table 5 and Online Methods).
Clustering the strains in the SWAT-SP-GFP library by both secretion level and protein abundance highlighted several groups of proteins (Supplementary Fig. 6a). An interesting group of proteins seemed to be secreted only under the constitutive regulation of SpNOP1pr and the Kar2 SP. This could indicate that the overexpression of such proteins saturates retention and targeting mechanisms thus causing aberrant secretion (as in the case of the vacuolar protein Prc1, Supplementary Table 5). However, another possibility is that some of the secreted proteins are bona fide secreted proteins that are usually not expressed under normal yeast growth conditions and are therefore secreted in a condition specific fashion. Such condition-specific secreted proteins would have not been identified in the past since mapping of secreted proteins has not been done under all possible yeast growth conditions. We therefore focused on genes that were not expressed when under regulation of their native promoter, but once expression was driven by the generic promoter were very efficiently secreted (Supplementary Fig. 6b and Supplementary Table 5). In this group, we saw that some of the proteins are indeed known “condition specific” secreted proteins such as Pho5, which is secreted only during growth in low phosphate 31 and Bar1 which is secreted only from MATa cells 32. Systematic analysis of GO term enrichment in the proteins of this cluster indeed showed over representation of the “response to stimulus” category (p= 2E-4) 33. Three proteins found in this cluster were previously uncharacterized, did not have a vacuolar localization (so were not secreted simply because of saturation of retention machineries) and were not predicted to have transmembrane domains: Yil169c, Yfr020w, and Yol159c. We have therefore now named them Css1-3 (for Condition Specific Secretion 1-3) (Supplementary Fig. 5b). Importantly, these proteins may have never been identified by traditional assays for finding secreted yeast proteins since the specific conditions required for their secretion remain to be uncovered.
Uncovering localization of hundreds of proteins
Reasoning that constitutive expression in the SWAT-GFP libraries may help visualize many proteins, we next annotated protein localization in all strains in the SWAT-GFP and SWAT-SP-GFP libraries, for both the acceptor and seamless versions (in at least two independently created clones to ensure reproducibility) (Fig. 2a. For complete localization assignments see Supplementary Table 4). Indeed, using the constitutively expressed SWAT-GFP library we could now visualize 599 proteins for the first time (Fig. 3a). Nearly 200 of these proteins did not have any prior information on localization in the literature, neither computational predictions, manual annotations or high throughput ones (Fig. 3b). Although the newly visualized proteins were found in all possible sub cellular structures (Fig. 3c), we found that most new localization assignments were to the ER, punctate structures and vacuole (membrane and lumen) (Fig. 3d).
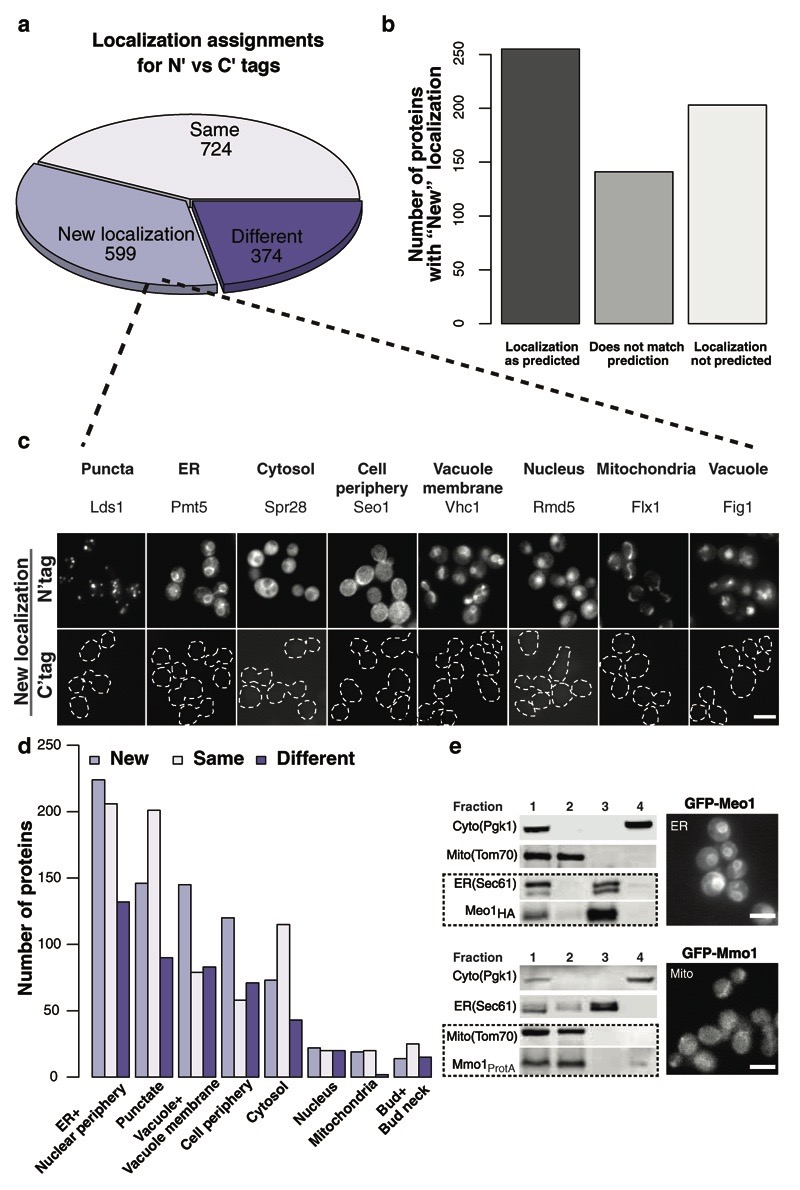
Figure 3. Creation of an N′ SWAT library sheds new light on hundreds of endomembrane proteins.
(a) Comparing protein N′ tagging localization assignments to assignments made from the C′ tagged library 4 showed that 724 proteins are visualized in the same localization (Same), 374 show different (Different) localization, and 599 proteins were only visualized using N′ tagging (New localization). For all localization assignments please see (Supplementary Table 4). (b) Comparing the 599 proteins that were only visualized using N′ tagging to GO ontology indicates that for nearly 30% no GO term was ever annotated. (c) A representative image for proteins that reside in each sub-cellular localization from the 599 proteins (New localization) that could be visualized by N′ tagging with the SWAT-GFP module (top) as opposed to C′ tagging with GFP (Bottom). Scale bars, 5 µm. (d) New assignments of protein localization by N′ tagging in the SWAT-GFP and SWAT-SP-GFP collections were mostly to the ER, punctate structures, the vacuole and cell periphery. Scale bars, 5 µm. (e) Two examples for the 78 small open reading frame proteins (smORFs) that were visualized for the first time using the SWAT-GFP libraries. Ybr126w-a was found to localize to the ER (top, newly named Meo1), and Ykl044w to mitochondria (bottom, named Mmo1). Their localization was validated by a cell fractionation assay in which Meo1 co-fractioned with the ER protein Sec61, and Mmo1 with the mitochondrial protein Tom70. Fraction numbers: 1-Post nuclear supernatant (PNS); 2-Pellet (P13); 3-Pellet (P100); 4-Supernatant (S100) (Online Methods).
One group of proteins that was never visualized before systematically is of the Small Open Reading Frame (smORF) proteins (fewer than 110 amino acids in length. For a complete list of such smORFs see Supplementary Table 6). This is because initial annotation of the yeast genome 34 did not include such ORFs and they were annotated only later 35. Our library contained 98 smORFs, of which only 39 had been visualized before. We were now able to assign a localization to 78 of them (80%). To verify that these proteins have the capacity to be correctly localized with the GFP tag, we chose two examples of proteins that were never previously studied: One from the 36 smORFs that localized to the ER (Ybr126w-a) and the other from the eight smORFs that localized to mitochondria (Ykl044w). We tagged the selected proteins at their C′ with a small epitope tag (HA or ProtA) and used sub-cellular fractionation to verify their assigned localization. Indeed Ybr126w-a was verified as a new ER protein which we now name Meo1 (Mini ER ORF1), whereas Ykl044w was verified to be a new mitochondrial constituent that we now name Mmo1 (Mini Mitochondria ORF1) (Fig. 3e). In the past, smORFs have been underrepresented in functional systematic libraries and therefore are less studied than other yeast genes. We hope that the new information brought forward here for both smORFs and other previously uncharacterized proteins (that now have a localization assignment), as well as the presence of these genes in our new libraries will promote their study and the investigation of their function.
The N′ and C′ GFP libraries are complementary
Tagging proteins at either terminus can mask targeting sequences as well as regulatory sequences. Indeed, 374 proteins were localized differently when tagged at their N′ (in the SWAT-GFP libraries) vs. their C′ (as previously annotated 4,26) (Fig. 3a and Supplementary Fig 7a). Our endomembrane library encompasses many proteins that are known to be affected by masking of C′ localization signals. As expected, focusing on such proteins (Peroxisomal matrix proteins, Tail- or GPI- Anchored proteins, lipidated proteins and proteins with retrieval motifs) we found that many only localize correctly when N′ tagged (Supplementary Fig. 7b and see below in Fig. 4b). Comparing the 374 differing localization assignments with manually curated GO annotations suggests that in about one third of the cases the localization seen with the N′ tag matches the GO annotation, in another third it is the C′ tag that does, and for a third of the proteins it is inconclusive (Supplementary Fig. 8). This demonstrates again how important it is to have a variety of complementary libraries to explore yeast cell biology.
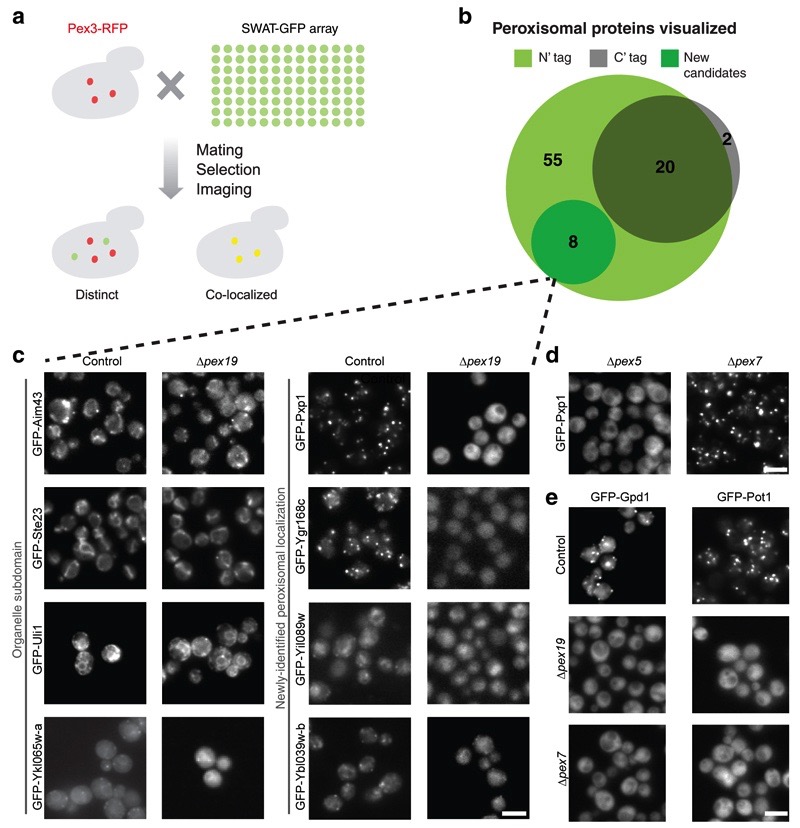
Figure 4. Correct targeting of peroxisomal proteins is maintained by the N′ GFP tagged strains enabling characterization of new peroxisomal proteins.
(a) The SWAT-GFP array was mated with a strain containing the peroxisomal marker Pex3-mCherry to query for peroxisome localized proteins. (b) Pex3 co-localization was observed in 55 proteins from the SWAT-GFP libraries, eight of which were never described before as peroxisomal (New candidates). Only 22 of these proteins were previously visualized by C′ GFP tagging, exemplifying the importance of an N′ GFP library. (c) Peroxisomal targeting of the eight new candidate proteins was validated by deletion of PEX19, which abolishes peroxisome biogenesis. Four proteins (left) remained in punctate formations, and are therefore most likely not targeted to peroxisomes, but rather to organelle sub-domains that reside near peroxisomes. Four other proteins (Right) dispersed to cytosolic localization upon PEX19 deletion validating them as bona fide peroxisomal proteins. Scale bars, 5 µm. (d) The prediction that Yel020c (now named Pxp1) is targeted to peroxisomes by a PTS1 was validated by deletion of the PTS1 receptor PEX5, in which Pxp1 no longer showed a peroxisomal localization (left), in contrast to deletion of the PTS2 receptor PEX7. Scale bars, 5 µm. (e) Protein targeting to peroxisomes via the N′ PTS2 signal was maintained despite the N′ SWAT-GFP tag. Gpd1 and Pot1 show proper localization to peroxisomes with the N′ SWAT-GFP tag (top). Their reliance on PTS2 for correct targeting is demonstrated by dispersion to the cytosol once the PTS2 receptor PEX7 is deleted (bottom), similar to the phenotype generated when peroxisomes are depleted completely by deletion of PEX19 (middle). Scale bars, 5 µm.
Discovering new peroxisomal proteins
One organelle that was under-represented in the C′ GFP library is the peroxisome. This is because many of the soluble proteins inside this organelle are targeted by virtue of the tri-peptide targeting sequence, PTS1, on the most C-terminus of the protein 36, which becomes masked upon addition of a C′ GFP tag. To uncover new peroxisomal proteins we mated a strain expressing the peroxisomal marker, Pex3-mCherry, with the haploid SWAT-GFP and SWAT-SP-GFP libraries, and assayed for co-localization in the emergent diploids in glucose (Fig. 4a). We found 55 proteins that co-localized with Pex3 (Supplementary Table 7). In contrast, only 22 proteins were co-localized with Pex3 in the C′ GFP library 4, and we could identify all but two of them in the N′ library as well (Fig. 4b).
Eight proteins co-localized with Pex3 that were never before assigned as peroxisomal proteins (Fig. 4b). We have recently shown that peroxisomes sit juxtaposed to the ER/Mitochondria contact site 37. Since the resolution of light microscopy is not enough to discriminate between bona-fide co-localization or juxta-positioning, we deleted Pex19, an essential protein for peroxisome biogenesis 38,39 from all eight GFP tagged candidate peroxisomal proteins. Four proteins, Ykl065w-a, Uli1, Ste23, and Aim43, remained in a punctate structure even in the absence of mature peroxiseoms (Fig. 4c, left) and therefore probably sit at peroxisomal contact sites on other organelles 40. The other four proteins were redistributed to the cytosol upon Pex19 depletion as expected from bona fide residents (Fig. 4c, right). These four new peroxisomal proteins have never been studied before and only have systematic names (Yel020c, Ygr168c, Yil089w, Ybl039w-b (that is also localized to another membrane)). One of the genes, Yel020c, has previously been suggested to be peroxisomal by computational predictions of PTS1 variants 41 and due to its presence in peroxisomal fractions during growth in oleic acid 42. Yel020c is homologous to the main yeast pyruvate decarboxylase (Pdc1). We deleted the receptor for PTS1 targeting (Pex5) and, as a control, the receptor for PTS2 targeting (Pex7) and could verify that Yel020c is targeted to Peroxisomes by Pex5 (Fig. 4d). We have therefore named this protein Pxp1 (PeroXisomal Protein 1).
An intriguing observation from our data was that the two proteins that are targeted to peroxisomes by the PTS2 system, Gpd1 and Pot1 36, localized properly to peroxisomes (Fig. 4e). This was surprising because PTS2 targeting sequences are N-terminal and were thought to be dependent on position 43. We deleted the PTS2 targeting receptor, Pex7 and found that both proteins completely lost peroxisomal localization, proving that both proteins still use Pex7 to correctly target to peroxisomes, and that the N′ tag doesn’t interfere with the PTS2 signal. The ability to now visualize the majority of peroxisomal proteins will enable future systematic efforts to study these organelles.
Rapid creation of a new N′ mCherry library
The most exciting feature of the SWAT technology is that the parental library can be rapidly swapped into any library with a modification of choice to address specific biological questions of interest. As an example we decided to use the acceptor SWAT-GFP and SWAT-SP-GFP arrays to efficiently and easily switch the green fluorophore and create a new library that contains mCherry tagged proteins (Fig. 5a and Online Methods).
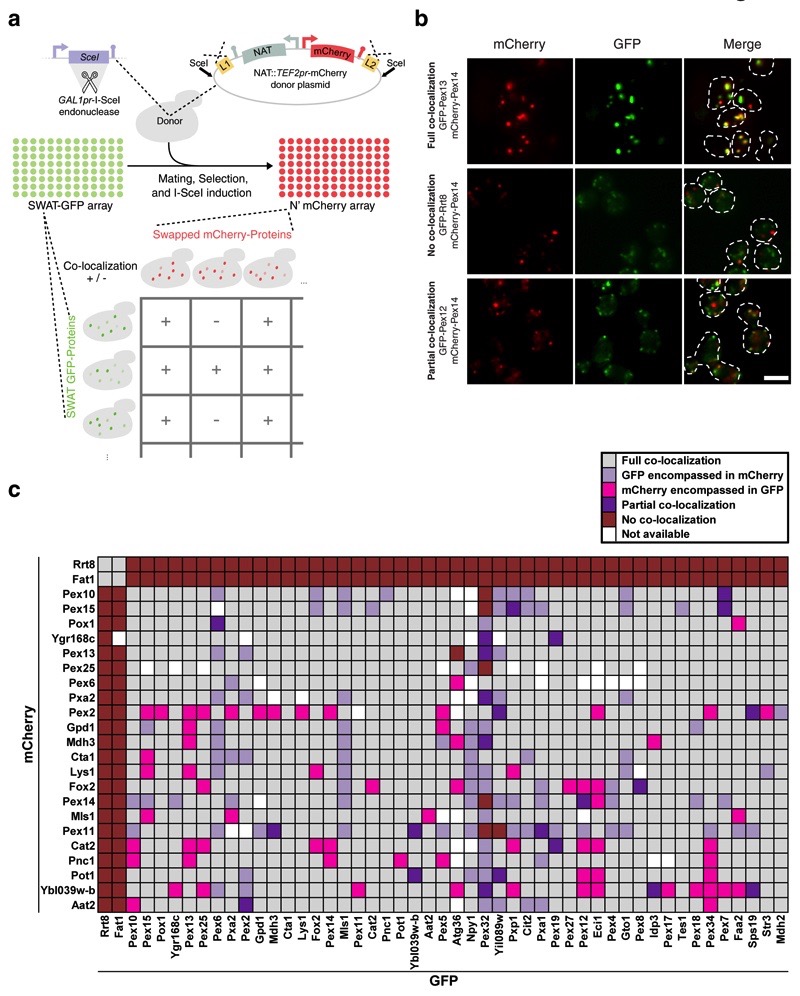
Figure 5. Rapid creation of a new N′ mCherry library exemplifies the swap-tag technology and opens up opportunities for systematic co-localization studies.
(a) An N′-mCherry tagged library was made by crossing the SWAT-GFP arrays with a donor strain containing a donor plasmid with the swap cassette NAT::TEF2pr-mCherry (Online Methods). Strains from the mCherry library, selected to be MAT alpha, can now be crossed with MATa SWAT-GFP strains to perform systematic co-localization experiments. (b) The co-localization experiment yielded several different patterns. Examples of three types of co-localization that were seen are given: a comprehensive overlap of the two proteins (top), distinct localization for each protein (middle), and a partial overlap (bottom). Scale bars, 5 µm. (c) Peroxisomal protein co-localizations were assayed in a systematic manner by mating of 24 mCherry tagged strains with 49 GFP tagged strains. While most peroxisomal proteins where visualized in the same peroxisomes (gray), in many instances proteins were visualized in separate groups of peroxisomes. The two lipid droplet proteins (Rrt8 & Fat1), which served as a negative control, indeed never co-localize with any of the peroxisomal proteins but did co-localize with each other.
Creation of the N′ mCherry library again demonstrated the efficiency of the SWAT approach as 98% of strains were retrieved following this procedure (Supplementary Table 4). Since 83% of strains retained the same localization as the SWAT-GFP/SWAT-SP-GFP parental libraries, it shows that the process was not only efficient but also accurate. We did, however, witness that the use of a stronger promoter (TEF2pr) in the donor module, and the enhanced stability of the mCherry in acidic environments creates higher vacuolar background fluorescence 44.
We chose to use the N′ mCherry library to elucidate an open question regarding peroxisome heterogeneity, asking whether peroxisome subpopulations exist (Fig. 5a). To do this we selected 24 ‘mCherry strains’ (22 peroxisomal proteins and two lipid droplet proteins serving as negative controls) and mated each of them with 49 ‘GFP strains’ (47 peroxisomal proteins, and the two lipid droplet proteins). We visualized the resulting 1,176 diploid strains and scored the extent of GFP-mCherry overlap (Fig. 5a, Supplementary Table 8). We found that indeed peroxisomal populations were heterogeneous (Fig. 5b,c). For example, many peroxisomal proteins (e.g. Pex10) appeared only in a subset of cellular peroxisomes (Fig. 5c). The mCherry library now enables similar studies to be performed on other organelles that may have heterogeneous populations such as mitochondria and lipid droplets, and is generally another freely available tool.
Discussion
The SWAT methodology opens a world of opportunities for studying cell biology by creation of yeast libraries. The SWAT acceptor library can be manipulated to give rise to libraries harboring new regulatory sequences, RNA tags, protein processing signals, affinity purification tags or protein fragment complementation tags. The ease and speed of this approach, coupled with high efficiency and accuracy, should enable any yeast lab to create their own “tailor made” strain collection in a matter of a few weeks. Although robotic manipulation of libraries facilitates such efforts, all of these experiments can easily be undertaken using hand-operated pinning tools 25 – thus removing technological barriers for SWAT library utilization. We hope that the SWAT method will advance the future of yeast research by boosting the number of systematic experimental possibilities.
Online Methods
Plasmid construction
Plasmids were constructed using restriction free cloning methods 45. For a complete list of plasmids see Supplementary Tables 1 and 2, and Supplementary Data 1. The I-SceI restriction site sequence is: agttacgctagggataacagggtaatatag. The protein linker sequences (that also serve as the generic recombination sites) are:
L1- 5′ cgtacgctgcaggtcgacggtggcggttctggcggtggcggatcc 3′
L2- 5′ ggcggttcctctggtggtggtggtgcgacagagaattcatcgatg 3′
L3- 5′ cgtacgctgcaggtcgacggtggcggttctggcggtggcggatcc 3′ (identical to L1)
L4- 5′ ggcggttcctctggtggtggtggtgcgagctcgaattcatcgat 3′
Underlined are the primer sequences used for amplification of the tagging module, corresponding to the pYM series sequences (S1, S4, S3, and S2 respectively) 46. The use of these sequences ensures compatibility with already existing oligo collections for these popular module sets.
The tagging modules include the constitutive SpNOP1pr 19 promoter to drive the fusion tag-protein expression. This promoter confers medium level expression compared to stronger promoters such as ScTEF1pr or ScGPDpr.
The Kar2 signal peptide (SP) sequence used in the SWAT-SP-GFP module is:
atgtttttcaacagactaagcgctggcaagctgctggtaccactctccgtggtcctgtacgcccttttcgtggtaatattacctttacagaattctttccactcctccaatgttttagttagaggtgccgat.
The codon altered (to avoid altered recombination) Kar2 signal peptide sequence used in the Donor NAT::TEF2pr- SPKar2-mCherry plasmid is :
atgttcttcaatagattgtcagctgggaagcttcttgtgccactgtctgtagttctttacgcactgttcgtagtgatactacccctgcaaaactcctttcactcttctaatgtcctggtcagaggcgcagac.
Types of possible swapping options
“Seamless tag swap donor”: Contains the tag of choice flanked by the homologous recombination sequences (L1, L2 for N′ tag or L3, L4 for C′ tag). Use of this cassette eliminates all the basic acceptor components giving rise to a seamless fusion of the new tag (Fig. 1c ; For an example see Supplementary Fig. 1a). This swap is selected for using 5-FOA, and leaves no positive selection marker.
“Selection reconstitution and tag swap donor”: Contains a new tag of choice flanked by one recombination sequence (L2 or L3 depending on whether N′ or C′ cassettes are used, respectively) and the N′ portion of the HygromycinB resistance cassette that serves both as homology for recombination as well as creates a new selection marker (Fig. 1c ; For an example see Supplementary Fig. 1b).
“Selection and tag swap donor”: Contains a new tag of choice and a new selection marker (not HygromycinB), flanked by the homologous recombination sequences (L1, L2 for N′ tag or L3, L4 for C′ tag). Use of this cassette enables replacement of all constituents of the original SWAT module (Selection cassette, regulatory sequences and tag) except for the linkers (Fig. 1c ; For an example see Supplementary Fig. 1c).
Signal peptide predictions of yeast proteins
Three algorithms were used to predict the presence of a signal peptide in each yeast protein, SignalP 4.1 22, Phobius 1.01 23, and Philius 24. We considered a protein as SP bearing for this study if all three programs predicted a SP in it, or if at least two programs predicted a SP with the exact same length (Supplementary Table 3 and Supplementary Fig. 2).
Primer Design
Primers for amplification of transformation cassettes and gene specific targeting were designed using the Primers-4-Yeast web tool 16 (http://wws.weizmann.ac.il/Primers-4-Yeast) using the pYM plasmid type 46. All tagging primers include a 40bp homology sequence followed by 20bp or 18bp of cassette amplification sequence. The homology sequences were upstream and downstream to the protein start codon for normal N′ tagging and to the protein stop codon for C′ tagging, as described in the Primers-4-Yeast web tool. For N′ tagging of signal peptide containing proteins, homology sequences were designed to insert the cassette five amino acids downstream to the predicted signal peptide cleavage point. Primers for validation of tagging and deletion transformations were also designed by the Primers-4-Yeast web tool, using the appropriate “Check primers” option. Primers were manufactured by Sigma-Aldrich in 96 well plates. For a full list of primers used in this study see Supplementary Table 9.
High throughput yeast transformations
The BY4741 laboratory strain 47, which is the basis for most systematic yeast libraries to date, was used as the master strain for the collection. The SWAT-GFP and SWAT-SP-GFP acceptor modules (Supplementary Table 1, pST-N2 and pST-N3) were PCR amplified (KAPA Hi-Fi or KOD Hot Start DNA Polymerase) in 96 well plates (Thermo Fisher Scientific), and were transformed into BY4741. Transformations were carried out in a modified PEG/LiAc protocol 48 in a high throughput manner, were each reaction was composed of 2.1 O.D600 of cells (3 ml of cells at 0.7-0.8 O.D600), 120 µl of PEG 3500 50% w/v, 18 µl of LiAc 1M, 25 µl of boiled SS-carrier DNA, 7 µl of ddW and 20 µl of PCR-amplified transformation cassette DNA. Heat shock was applied in a PCR machine, 15 min at 30 °C followed by 30 min at 42 °C. Transformed cultures were plated on SD-URA media in 48-well divided agar plates (Bioassay X6029), and were incubated for 2-3 days at 30 °C. All procedures were carried out utilizing an automated liquid handler (Janus by Perkin Elmer).
Yeast strain validation and collection assembly
Four clones at least were picked from each transformation. Validation PCR was performed using a common forward primer from the 3′ end of the SWAT modules (S4 reverse complement), and a gene specific reverse primer from the gene coding sequence. All four clones were imaged via a high-content screening platform in bright-field and GFP channels (see below). Images of all clones were reviewed manually, and up to three localizations were assigned to each clone. Assignments were: Ambiguous; Below threshold; Bud; Bud neck; Cell periphery; Cytosol; ER; Mitochondria; Nuclear periphery; Nucleolus; Nucleus; Punctate; Vacuole; Vacuole membrane. Only strains with a duplicate repeating localization assignment and validated by PCR were chosen to compose the SWAT-GFP and SWAP-SP-GFP collections.
Analysis of swapping procedure efficiency
We performed several tests in order to ascertain the consistency and efficiency of the SWAT strategy. First, we measured mCherry fluorescence levels by flow cytometry in SWAT tagged strains (both C′ and N′ SWAT modules) that underwent seamless mCherry swapping before and after I-SceI induction, and after 5-FOA counter selection. In all cases mCherry introduction was demonstrated in >99% of cells (Supplementary Fig. 1d). Second, we performed PCR analysis of the tagging state in several colonies of SWAT-GFP strains that underwent seamless GFP swapping before and after I-SceI induction, and after 5-FOA counter selection. The SWAT module was completely lost at the end of the process resulting in seamless tagging (performed in a high-throughput manner) (See an example in Supplementary Fig. 3). Last, swapping efficiency of SWAT-GFP strains was measured by single cell GFP expression analysis, as measured by microscopy. A homogenous distribution of GFP expression was seen for >98% of strains after seamless GFP swapping (bimodal distribution test performed using the R dip test function, see examples visualized in Supplementary Fig. 4). It is worth noting that seamless swapping is expected to be impaired when the gene tagged with the SWAT module is located between identical or highly similar stretches of genomic DNA19. In such cases the double strand break introduced by I-SceI within the SWAT module could be repaired by homologous recombination between the identical/similar genomic stretches, leading to excision of the entire genomic region instead of seamless swapping. Nevertheless, less than 250 of all yeast genes are expected to be affected by this, therefore SWAT can be used for high-throughput seamless tagging of most of the proteome.
Donor strain construction
Donor strains were constructed on the background of a synthetic genetic array (SGA) 18 compatible query strain, and contain a galactose induced I-SceI endonuclease and a donor plasmid. In order to spare a selection marker in the donor strain, a K. lactis URA3 selection marker was introduced into the can1Δ locus, upstream to the STE2pr-SpHIS5 fragment (used for selection of MATa). A Gal1pr-I-SceI fragment was then introduced to replace the URA3 selection resulting in the following: can1Δ::GAL1pr-SceI::STE2pr-SpHIS5 (strain yMS2085). In cases where automated cell recognition software was required, a NAT::TEF2pr-mCherry cassette was introduced to the hoΔ locus. Examples of donor strain utilization can be found in Fig. 2a and 5a.
Automated yeast library manipulations
Automated strain collection maintenance and manipulation was performed using a RoToR bench-top colony arrayer (Singer Instruments)25. SGA procedures 18 were performed for the mating of the parental SWAT-GFP and SWAT-SP-GFP collections with donor strains bearing the seamless GFP donor (Supplementary Table 2, pSD-N9), the mCherry donor (Supplementary Table 2, pSD-N15/6), and strain with the co-localization marker Pex3-RFP (Fig. 4a). Following double mutant selection, tag swapping was prompted by two cycles of 1-2 day growth on YEPGalactose (2%) media to induce I-SceI expression. Tag swapping was then selected by two cycles of growth on either SD+5-FOA (1g/L) media for seamless GFP swap, YEPD+NAT (Neurseothricin, 200µg/mL) for the N′ mCherry swap, or YEPD+HYG (HygromycinB, 200µg/mL) in the case of HYG selection reconstitution swap.
Creation of mCherry library
In order to create the mCherry library we built a donor plasmid of the “selection and tag swap” type. The plasmid included the mCherry coding region under control of the TEF2 promoter and a nourseothricin (NAT) selection marker, flanked by the homology linkers and the I-SceI cut sites. For SP proteins we used a plasmid also containing a SP (Fig. 5a). We introduced the donor plasmids into the entire parental SWAT library and promoted tag swapping by induction of I-SceI and selection for 5FOA and NAT resistant colonies. Using the SGA markers, the resulting library of mCherry tagged proteins was selected such that all strains were of the opposite mating type from the original SWAT library (MAT alpha).
High-throughput microscopy
High content screening of strain collections was performed using an automated microscopy set-up (ScanR system, Olympus) as previously described 26. Images were acquired using a 60× air lens using GFP (Excitation 490/20 nm, Emission 535/50 nm), mCherry (Excitation 572/35 nm, Emission 632/60 nm), and bright-field channels. When a cytosolic mCherry cell marker was used (Fig. 2), images were analyzed using the ScanR Analysis software (Olympus), and single cells were recognized based on the mCherry channel. Measures of cell size, shape and fluorescent signals were extracted. For localization assignments, images were manually reviewed using ImageJ. Since no co-localization markers were used we only assigned localizations that could be easily discriminated by eye: ER, nuclear periphery, cytosol, cell periphery, vacuole lumen, vacuole membrane, mitochondria, nucleus, bud/bud neck and punctate (that would include the Golgi apparatus, peroxisomes, endosomes, other vesicular structures and subdomain compartments) (Fig. 3c).
Computational quantification of single cell GFP intensity
The median GFP intensity was measured for each strain using a single cell recognition software (scanR Analysis software ,Olympus) as previously described 26. Strains with less than 30 recognized cells were excluded. Baseline auto fluorescence level was obtained from strains not expressing GFP, and any strain whose median GFP was below a cutoff of Meanauto-fluorescence+2.58*SD Meanauto-fluorescence was classified as “below threshold” (of detection).
Data processing
The subcellular localization annotations of the SWAT-GFP Swap libraries (with the generic promoter and signal peptide) were compared with those of the C′ tag library (comprising data from two previous datasets 4,26). The pairwise comparisons between N′-tagging and C′-tagging annotations were classified in the following manner: ‘Same’ was assigned when at least one N′ annotation corresponded to a C′ one. ‘New localization’ was assigned if the C′ localization was classified as below threshold, ambiguous, or no assignment existed. All other cases were classified as ‘Different’ (Fig. 3a). Localization annotations of both N′ (SWAT-GFP) and C′ tagged proteins were compared with GO cellular component, manually curated annotations, after applying the localization terminology to each GO term (Fig. 3b and Supplementary Fig. 8). All of the calculations were performed with either MATLAB or Rstudio. GO terms were retrieved from the SGD website (http://www.yeastgenome.org/cgi-bin/GO/goTermFinder.pl).
Cluster analysis
We implemented agglomerative hierarchical clustering using MATLAB for the analysis of secretion levels (Supplementary Fig. 7) with a Euclidean or Cityblock distance metric for calculating the similarity between pairs of proteins.
Manual Microscopy
Imaging of specific strains in follow-up experiments were obtained by an Olympus IX71 microscope, connected to a PhoetometricsCoolsnap HQ camera, controlled by the Delta Vision SoftWoRx 3.5.1 software with 60× and 100× oil lenses using GFP (Excitation 490/20 nm ,Emission 528/38 nm), mCherry (Excitation 555/28 nm , Emission 617/73 nm), and bright-field channels.
Protein secretion assay
Analysis of protein secretion was performed as previously described 49. Specifically, yeast collections in either 384 or 1536 colony array format were pinned by the RoToR colony arrayer onto YEPD plates and a nitrocellulose membrane was applied onto the plate immediately thereafter. Overnight incubation at 30 °C allowed cell growth and ensured that secreted proteins were transferred to the nitrocellulose membrane. The membrane was removed from the plate and washed (0.5 M NaCl 10 mM Tris-HCl pH 7.5) to release remaining cells. Secreted GFP fused proteins were probed with a primary rabbit antibody against the GFP (Made by Ineke Braakman). Next, a secondary goat-anti-rabbit antibody conjugated to IRDye800 (LI-COR Biosciences) was used, and membranes were scanned for infrared signal (Including colony auto-fluorescence at 700nm as a background control) using the Odyssey Imaging System (LI-COR Biosciences).
Subcellular fractionation
Yeast cells were grown to an O.D600 of 1, and a total of 70 O.D600 were harvested and washed in dH2O. The pellet was suspended in 2 ml of DTT buffer (100 mM Tris-H2SO4 pH 9.4, 10mM DTT) and incubated for 20 min at 30 °C. Spheroplasts were generated by incubation in 1 ml of Zymolyase buffer (1.2 M sorbitol, 20 mM potassium phosphate, pH 7.4, 0.5% w/v Zymolyase) for 45 min at 30 °C and harvested by centrifugation for 5 min at 3000 × g. After washing with 1 ml of 1.2 M sorbitol at 4 °C, spheroplasts were suspended in 2 ml of homogenization buffer (0.6 M sorbitol, 10 mM Tris-HCl, 1 mM EDTA, 1 mM PMSF, pH 7.4) and dounced 20 times in a cooled glass-teflon potter on ice. The samples were subjected to a clearing spin for 5 min at 300 × g (4 °C) and half of the supernatant was used as post nuclear supernatant (PNS-fraction 1), while the other half was further centrifuged for 15 min at 13,000 × g (4 °C) to generate a pellet (P13-fraction 2). The supernatant (S13) was centrifuged for 60 min at 100,000 × g, generating the P100-fraction 3 pellet and the S100-fraction 4 supernatant. Proteins in the PNS and S100 fractions were precipitated using StrataClean Resin (Agilent Technologies).
SDS-PAGE, Western Blotting and Immunodecoration
After Tricine–SDS polyacrylamide gel electrophoresis, proteins were transferred to Polyvinylidendifluorid (PVDF) membranes using the semi dry transfer method 50. Proteins blotted onto PVDF membranes were detected by immunodecoration with specific primary antibodies (Tom70- GR657-5, Pgk1- GR753-7, Sec61- GR760-1) and horseradish peroxidase coupled secondary antibodies (HA- Roche-12013819001, Peroxidase- Sigma-P1291, Rabbit IgG - Jackson-111-035-003) after incubation with chemiluminescence solution and recorded using the LAS-4000 CCD camera system and MultiGauge (Fujifilm).
Obtaining the libraries, plasmids and protocols
All strains, plasmids and libraries presented in this manuscript are freely available upon request (email: maya.schuldiner@weizmann.ac.il). All protocols for using the SWAT strategy can be downloaded from our lab website: http://www.weizmann.ac.il/molgen/Maya/SWAT
Supplementary Material
Acknowledgements
We wish to thank N. Steinberg for his enormous contribution to the graphical design of this manuscript. E. Levy for exciting scientific discussions. This work was supported by an ERC CoG grant (Peroxisystem 646604 to MS), the Deutsche Forschungsgemeinschaft, Collaborative Research Centre (SFB 1140 to NW), the Excellence Initiative of the German Federal & State Governments (EXC 294 BIOSS to NW), an ERC CoG grant (MITOsmORFs 648235 to NW), and the Sonderforschungsbereich 1036 from the Deutsche Forschungsgemeinschaft (DFG) (SFB1036, TP10, to MK). We also acknowledge the generous support of the Mitzutani Foundation for Glycosciences (MS), Adelis foundation (MS), the Kahn center for systems biology (MS).
Footnotes
Author contributions
AK, MM and MK conceived the SWAT strategy. MM, AK, IY and UW developed and implemented the method. SB performed computational prediction of signal peptides. UW, IY and SC constructed the SWAT libraries (with help from OG) and performed all of the experiments and analysis on the systematic libraries. SC performed all automated robotic procedures. EZ, CS and UW performed the low throughput follow up experiments. AK, NW, MK and MS supervised the work. IY, UW, SC and MS wrote the manuscript with input from all other authors.
Competing Financial Interests
The authors declare no competing financial interests.
References
- 1.Botstein D, Fink GR. Yeast: an experimental organism for 21st Century biology. Genetics. 2011;189:695–704. doi: 10.1534/genetics.111.130765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Giaever G, et al. Functional profiling of the Saccharomyces cerevisiae genome. Nature. 2002;418:387–91. doi: 10.1038/nature00935. [DOI] [PubMed] [Google Scholar]
- 3.Tarassov K, et al. An in vivo map of the yeast protein interactome. Science. 2008;320:1465–70. doi: 10.1126/science.1153878. [DOI] [PubMed] [Google Scholar]
- 4.Huh W-K, et al. Global analysis of protein localization in budding yeast. Nature. 2003;425:686–91. doi: 10.1038/nature02026. [DOI] [PubMed] [Google Scholar]
- 5.Ben-Aroya S, et al. Toward a comprehensive temperature-sensitive mutant repository of the essential genes of Saccharomyces cerevisiae. Mol Cell. 2008;30:248–58. doi: 10.1016/j.molcel.2008.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li Z, et al. Systematic exploration of essential yeast gene function with temperature-sensitive mutants. Nat Biotechnol. 2011;29:361–7. doi: 10.1038/nbt.1832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mnaimneh S, et al. Exploration of essential gene functions via titratable promoter alleles. Cell. 2004;118:31–44. doi: 10.1016/j.cell.2004.06.013. [DOI] [PubMed] [Google Scholar]
- 8.Schuldiner M, et al. Exploration of the function and organization of the yeast early secretory pathway through an epistatic miniarray profile. Cell. 2005;123:507–19. doi: 10.1016/j.cell.2005.08.031. [DOI] [PubMed] [Google Scholar]
- 9.Breslow DK, et al. A comprehensive strategy enabling high-resolution functional analysis of the yeast genome. Nat Methods. 2008;5:711–8. doi: 10.1038/nmeth.1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kanemaki M, Sanchez-Diaz A, Gambus A, Labib K. Functional proteomic identification of DNA replication proteins by induced proteolysis in vivo. Nature. 2003;423:720–4. doi: 10.1038/nature01692. [DOI] [PubMed] [Google Scholar]
- 11.Sopko R, et al. Mapping pathways and phenotypes by systematic gene overexpression. Mol Cell. 2006;21:319–30. doi: 10.1016/j.molcel.2005.12.011. [DOI] [PubMed] [Google Scholar]
- 12.Sung M-K, et al. Genome-wide bimolecular fluorescence complementation analysis of SUMO interactome in yeast. Genome Res. 2013;23:736–46. doi: 10.1101/gr.148346.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Storici F, Resnick MA. The delitto perfetto approach to in vivo site-directed mutagenesis and chromosome rearrangements with synthetic oligonucleotides in yeast. Methods Enzymol. 2006;409:329–45. doi: 10.1016/S0076-6879(05)09019-1. [DOI] [PubMed] [Google Scholar]
- 14.Baudin A, Ozier-Kalogeropoulos O, Denouel A, Lacroute F, Cullin C. A simple and efficient method for direct gene deletion in Saccharomyces cerevisiae. Nucleic Acids Res. 1993;21:3329–30. doi: 10.1093/nar/21.14.3329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wach A, Brachat A, Alberti-Segui C, Rebischung C, Philippsen P. Heterologous HIS3 marker and GFP reporter modules for PCR-targeting in Saccharomyces cerevisiae. Yeast. 1997;13:1065–75. doi: 10.1002/(SICI)1097-0061(19970915)13:11<1065::AID-YEA159>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 16.Yofe I, Schuldiner M. Primers-4-Yeast: a comprehensive web tool for planning primers for Saccharomyces cerevisiae. Yeast. 2014;31:77–80. doi: 10.1002/yea.2998. [DOI] [PubMed] [Google Scholar]
- 17.Colleaux L, et al. Universal code equivalent of a yeast mitochondrial intron reading frame is expressed into E. coli as a specific double strand endonuclease. Cell. 1986;44:521–33. doi: 10.1016/0092-8674(86)90262-x. [DOI] [PubMed] [Google Scholar]
- 18.Tong AHY, Boone C. Yeast Gene Analysis - Second Edition Methods Microbiol. Vol. 36. Elsevier: 2007. [Google Scholar]
- 19.Khmelinskii A, Meurer M, Duishoev N, Delhomme N, Knop M. Seamless gene tagging by endonuclease-driven homologous recombination. PLoS One. 2011;6:e23794. doi: 10.1371/journal.pone.0023794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fujita M, Kinoshita T. GPI-anchor remodeling: potential functions of GPI-anchors in intracellular trafficking and membrane dynamics. Biochim Biophys Acta. 2012;1821:1050–8. doi: 10.1016/j.bbalip.2012.01.004. [DOI] [PubMed] [Google Scholar]
- 21.Hegde RS, Bernstein HD. The surprising complexity of signal sequences. Trends Biochem Sci. 2006;31:563–71. doi: 10.1016/j.tibs.2006.08.004. [DOI] [PubMed] [Google Scholar]
- 22.Petersen TN, Brunak S, von Heijne G, Nielsen H. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat Methods. 2011;8:785–6. doi: 10.1038/nmeth.1701. [DOI] [PubMed] [Google Scholar]
- 23.Käll L, Krogh A, Sonnhammer ELL. A combined transmembrane topology and signal peptide prediction method. J Mol Biol. 2004;338:1027–36. doi: 10.1016/j.jmb.2004.03.016. [DOI] [PubMed] [Google Scholar]
- 24.Reynolds SM, Käll L, Riffle ME, Bilmes JA, Noble WS. Transmembrane topology and signal peptide prediction using dynamic bayesian networks. PLoS Comput Biol. 2008;4:e1000213. doi: 10.1371/journal.pcbi.1000213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cohen Y, Schuldiner M. Advanced methods for high-throughput microscopy screening of genetically modified yeast libraries. Methods Mol Biol. 2011;781:127–59. doi: 10.1007/978-1-61779-276-2_8. [DOI] [PubMed] [Google Scholar]
- 26.Breker M, Gymrek M, Schuldiner M. A novel single-cell screening platform reveals proteome plasticity during yeast stress responses. J Cell Biol. 2013;200:839–50. doi: 10.1083/jcb.201301120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Newman JRS, et al. Single-cell proteomic analysis of S. cerevisiae reveals the architecture of biological noise. Nature. 2006;441:840–6. doi: 10.1038/nature04785. [DOI] [PubMed] [Google Scholar]
- 28.Picotti P, et al. A complete mass-spectrometric map of the yeast proteome applied to quantitative trait analysis. Nature. 2013;494:266–70. doi: 10.1038/nature11835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Belle A, Tanay A, Bitincka L, Shamir R, O’Shea EK. Quantification of protein half-lives in the budding yeast proteome. Proc Natl Acad Sci U S A. 2006;103:13004–9. doi: 10.1073/pnas.0605420103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ingolia NT, Ghaemmaghami S, Newman JRS, Weissman JS. Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science. 2009;324:218–23. doi: 10.1126/science.1168978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Barbarić S, Münsterkötter M, Svaren J, Hörz W. The homeodomain protein Pho2 and the basic-helix-loop-helix protein Pho4 bind DNA cooperatively at the yeast PHO5 promoter. Nucleic Acids Res. 1996;24:4479–86. doi: 10.1093/nar/24.22.4479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Manney TR. Expression of the BAR1 gene in Saccharomyces cerevisiae: induction by the alpha mating pheromone of an activity associated with a secreted protein. J. Bacteriol. 1983;155:291–301. doi: 10.1128/jb.155.1.291-301.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Eden E, Navon R, Steinfeld I, Lipson D, Yakhini Z. GOrilla: a tool for discovery and visualization of enriched GO terms in ranked gene lists. BMC Bioinformatics. 2009;10:48. doi: 10.1186/1471-2105-10-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Basrai MA, Hieter P, Boeke JD. Small open reading frames: beautiful needles in the haystack. Genome Res. 1997;7:768–71. doi: 10.1101/gr.7.8.768. [DOI] [PubMed] [Google Scholar]
- 35.Kastenmayer JP, et al. Functional genomics of genes with small open reading frames (sORFs) in S. cerevisiae. Genome Res. 2006;16:365–73. doi: 10.1101/gr.4355406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hasan S, Platta HW, Erdmann R. Import of proteins into the peroxisomal matrix. Front Physiol. 2013;4:261. doi: 10.3389/fphys.2013.00261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cohen Y, et al. Peroxisomes are juxtaposed to strategic sites on mitochondria. Mol Biosyst. 2014;10:1742–8. doi: 10.1039/c4mb00001c. [DOI] [PubMed] [Google Scholar]
- 38.Götte K, et al. Pex19p, a farnesylated protein essential for peroxisome biogenesis. Mol Cell Biol. 1998;18:616–28. doi: 10.1128/mcb.18.1.616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hettema EH, Girzalsky W, van Den Berg M, Erdmann R, Distel B. Saccharomyces cerevisiae pex3p and pex19p are required for proper localization and stability of peroxisomal membrane proteins. EMBO J. 2000;19:223–33. doi: 10.1093/emboj/19.2.223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schuldiner M, Zalckvar E. Peroxisystem - Harnessing systems cell biology to study peroxisomes. Biol Cell. 2015 doi: 10.1111/boc.201400091. [DOI] [PubMed] [Google Scholar]
- 41.Schlüter A, Real-Chicharro A, Gabaldón T, Sánchez-Jiménez F, Pujol A. PeroxisomeDB 2.0: an integrative view of the global peroxisomal metabolome. Nucleic Acids Res. 2010;38:D800–5. doi: 10.1093/nar/gkp935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yi EC, et al. Approaching complete peroxisome characterization by gas-phase fractionation. Electrophoresis. 2002;23:3205–16. doi: 10.1002/1522-2683(200209)23:18<3205::AID-ELPS3205>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 43.Jung S, Marelli M, Rachubinski RA, Goodlett DR, Aitchison JD. Dynamic changes in the subcellular distribution of Gpd1p in response to cell stress. J Biol Chem. 2010;285:6739–49. doi: 10.1074/jbc.M109.058552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Khmelinskii A, Knop M. Analysis of protein dynamics with tandem fluorescent protein timers. Methods Mol Biol. 2014;1174:195–210. doi: 10.1007/978-1-4939-0944-5_13. [DOI] [PubMed] [Google Scholar]
- 45.Unger T, Jacobovitch Y, Dantes A, Bernheim R, Peleg Y. Applications of the Restriction Free (RF) cloning procedure for molecular manipulations and protein expression. J Struct Biol. 2010;172:34–44. doi: 10.1016/j.jsb.2010.06.016. [DOI] [PubMed] [Google Scholar]
- 46.Janke C, et al. A versatile toolbox for PCR-based tagging of yeast genes: new fluorescent proteins, more markers and promoter substitution cassettes. Yeast. 2004;21:947–62. doi: 10.1002/yea.1142. [DOI] [PubMed] [Google Scholar]
- 47.Brachmann CB, et al. Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast. 1998;14:115–32. doi: 10.1002/(SICI)1097-0061(19980130)14:2<115::AID-YEA204>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 48.Gietz RD, Woods RA. Transformation of yeast by lithium acetate/single-stranded carrier DNA/polyethylene glycol method. Methods Enzymol. 2002;350:87–96. doi: 10.1016/s0076-6879(02)50957-5. [DOI] [PubMed] [Google Scholar]
- 49.Copic A, et al. Genomewide analysis reveals novel pathways affecting endoplasmic reticulum homeostasis, protein modification and quality control. Genetics. 2009;182:757–69. doi: 10.1534/genetics.109.101105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schägger H. Tricine-SDS-PAGE. Nat Protoc. 2006;1:16–22. doi: 10.1038/nprot.2006.4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.