Abstract
SUMOylation is a post‐translational modification that regulates a multitude of cellular processes, including replication, cell‐cycle progression, protein transport and the DNA damage response. Similar to ubiquitin, SUMO (small ubiquitin‐like modifier) is covalently attached to target proteins in a reversible process via an enzymatic cascade. SUMOylation is essential for nearly all eukaryotic organisms, and deregulation of the SUMO system is associated with human diseases such as cancer and neurodegenerative diseases. Therefore, it is of great interest to understand the regulation and dynamics of this post‐translational modification. Within the last decade, mass spectrometry analyses of SUMO proteomes have overcome several obstacles, greatly expanding the number of known SUMO target proteins. In this review, we briefly outline the basic concepts of the SUMO system, and discuss the potential of proteomic approaches to decipher SUMOylation patterns in order to understand the role of SUMO in health and disease.
Keywords: cross‐talk, group modification, mass spectrometry, post‐translational modification, proteomics, site‐specific, small ubiquitin‐like modifier, SUMO, ubiquitin
Abbreviations
- RNF
RING finger protein
- SAE
SUMO activating enzyme
- SENP
SUMO‐specific protease
- SIM
SUMO interaction motif
- SUMO
small ubiquitin‐like modifier
Introduction
Nearly 20 years have elapsed since the discovery of the small ubiquitin‐like modifier (SUMO), which is attached to a multitude of target proteins and thereby governs essential cellular processes such as DNA replication, cell division 1, the DNA damage response 2, transcription, and nuclear trafficking 3. SUMOylation has variable effects on the half‐life, binding partners or localization of target proteins, and is a crucial mechanism to allow cells to adapt to stress stimuli. While SUMO1‐deficient mice or SUMO3‐deficient mice have no overt phenotype because of compensation (most likely by the dominant SUMO isoform SUMO2) 4, 5, mice deficient for SUMO2 die during embryonic development 5. Mice deficient for the SUMO conjugating enzyme UBC9 likewise die during embryonic development, with severe chromosomal defects 6. Additionally, deregulation of SUMOylation has been implicated in various human disorders and diseases, such as neurodegenerative diseases 7, 8, heart failure 9 and cancer 10. Deciphering the cellular SUMO network may therefore help us in further understanding the pathology of these diseases and in developing potent drugs. In this review, we briefly summarize the biochemistry of the SUMOylation machinery, and highlight the difficulties and great potential of proteomic approaches to uncover new SUMO targets, SUMOylation sites and cross‐talk between post‐translational modifications. The roles of SUMO during specific cellular processes have been discussed elsewhere [11, 12, 13, 14, 15, 16, 17].
The biochemistry of the SUMO system
SUMO proteins: similarities and differences
In contrast to yeast, which has a single SUMO protein called Suppressor of MIF2 mutations 3 (SMT3) 18, the human genome includes four SUMO genes, designated SUMO1, SUMO2, SUMO3 and SUMO4. While SUMO1 is distinct from the other family members (47% sequence identity between SUMO1 and SUMO2/3), mature SUMO2 and SUMO3 share a sequence identity of 97%, making them indistinguishable to currently available antibodies. Therefore, this sub‐group is mostly referred to as SUMO2/3. All members of the SUMO family are expressed as precursor proteins, and first need to be processed by specific SUMO proteases, resulting in a free C‐terminal di‐glycine motif that is attached via an isopeptide bond to the ε‐amino group of an internal lysine residue of the target protein. SUMO4, however, contains a specific proline residue (Pro90), preventing it from being processed by the known SUMO proteases 19. It is currently unknown if and by what means mature SUMO4 is produced. The mature forms of SUMO1–3 are ubiquitously expressed. SUMO1 and SUMO2/3 have unique and overlapping target proteins 20, 21. In contrast to SUMO2/3, a major fraction of SUMO1 is conjugated to Ran GTPase‐activating protein 1 under normal conditions, a mechanism that is essential for efficient nucleocytoplasmic transport 22, 23. Conversely, SUMO2/3 is mostly conjugated under cellular stress conditions such as heat shock 24. Without stress, the cell contains a pool of unconjugated SUMO2/3, indicative of a cellular mechanism facilitating quick adaption of cells to stress arising from the extracellular environment. To further boost the SUMO signal during stress responses, the flexible N‐terminal tail of SUMO2/3 contains a lysine residue (K11) situated within a SUMO consensus motif that is preferentially recognized by the SUMO conjugation machinery. This residue is the major acceptor site for SUMO2/3 chain formation 25. SUMO1 lacks this regular consensus site, and therefore cannot efficiently form chains, but is instead considered as a chain terminator when incorporated in SUMO2/3 chains 26, 27. However, proteomic analyses have identified additional non‐consensus acceptor sites within SUMO1 and SUMO2/3, suggesting a very complex chain formation pattern in cells 28, 29. To what extent these linkages are used in comparison with K11 linkages is currently not understood.
SUMO activating enzyme and SUMO conjugating enzyme
Similar to other ubiquitin‐like modifiers, SUMO proteins are attached to target proteins via an enzymatic cascade (Fig. 1). In the first step, the C‐terminus of the mature SUMO moiety is activated via ATP hydrolysis, resulting in a SUMO adenylate that is further attacked by the catalytic site of the SUMO activating enzyme dimer (SAE1/2, E1), forming a thioester with a cysteine residue in SAE2. In a second step, the SUMO conjugating enzyme UBC9 (E2) binds to SAE2, and SUMO is transferred to its catalytic cysteine residue. In contrast to the ubiquitylation machinery, which comprises 35 conjugating enzymes with distinct functions and substrates, the SUMOylation cascade only contains UBC9 as a single E2. This highlights the important role of UBC9 as a key player in the SUMO signalling network. It is therefore not surprising that changes in the expression and activity of this enzyme have an extensive effect on regulation of the entire SUMO system, and must be fine‐tuned via a multitude of additional signals. UBC9 has a preference for binding to proteins carrying a specific SUMO consensus motif (ΨKxE, where Ψ is a bulky hydrophobic amino acid) 30. Consequently, in contrast to ubiquitin, bioinformatics tools for prediction of SUMO attachment sites in target proteins are available 31, 32.
Figure 1.
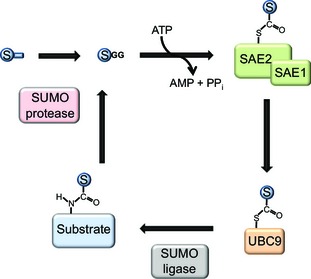
The SUMOylation cycle. First, the SUMO precursor protein is cleaved to its mature form by SUMO proteases, exposing a C‐terminal di‐glycine motif. In an ATP‐dependent reaction, the C‐terminus of SUMO is attached via a thioester bond to the SUMO activating enzyme, consisting of the two subunits SAE1 and SAE2. It is then further transferred to an internal cysteine of the SUMO conjugating enzyme (UBC9). In the third step, SUMO is attached to a lysine residue of a target protein, a process that is facilitated by the presence of a SUMO ligase. Finally, SUMO proteases cleave SUMO from its substrate, resulting in free SUMO that re‐enters the SUMOylation cycle.
SUMO ligases
In vitro studies have shown that SUMO‐loaded UBC9 alone is able to SUMOylate proteins bearing a SUMO consensus site 30. However, under physiological conditions, this process is facilitated by SUMO ligases (E3), which confer specificity to the substrate and are able to promote SUMOylation of substrates even without the presence of a SUMOylation consensus motif. While the human genome includes approximately 600 genes encoding ubiquitin ligases 33, only very few SUMO ligases have been described so far. The first group to be discovered was the Siz/PIAS RING family, containing a characteristic zinc finger domain structurally related to the RING domain of ubiquitin ligases. Siz/PIAS SUMO ligases are involved in a multitude of cellular processes, such as the DNA damage response, cell‐cycle control and transcriptional regulation 34, 35. However, their substrate specificity has been shown to be rather low 15. Uncovering the control mechanisms regulating the expression, localization and activity of these ligases may provide insights into the larger picture of the SUMO response after certain stimuli such as heat shock and DNA damage, and is therefore currently an important area of research.
In contrast to the Siz/PIAS family, the unrelated large Ran‐binding protein 2 specifically stabilizes the SUMOylated moiety of Ran GTPase‐activating protein 1, and forms a highly stable complex with UBC9, which is essential for nucleoplasmic transport 36. Ran‐binding protein 2 promotes SUMOylation of other SUMO target proteins, such as SP100 and topoisomerase IIα, and therefore has many additional functions, specifically during mitosis 37, 38.
Increasing numbers of proteins are thought to promote SUMO conjugation, such as the human polycomb protein Pc2/CBX4 39, histone deacetylase 4 40 and the tumor suppressor p14Arf 41. These findings suggest that additional SUMO ligases remain to be uncovered, consistent with the current understanding of the ubiquitin system and the fact that many hundreds of SUMO target proteins have been identified so far.
Very recently, the Fanconi anaemia protein SLX4 was suggested to be a SUMO ligase that is essential for the response to global replication stress, and it was found to bind to UBC9 and SUMOylate both itself and the DNA repair factor XPF in vitro 42, 43. Interestingly, the functions of SLX4 are dependent on so‐called SUMO interaction motifs (SIMs) present within the protein 43, 44. Such SIMs have been shown to promote SUMOylation of proteins even without the presence of a SUMO consensus site 45, and consist of large hydrophobic residues flanked by unstructured and exposed regions. Many proteins have already been shown to contain SIMs, and a recently developed bioinformatics tool facilitates the prediction of both SUMO consensus sites and SIMs in proteins of interest 32.
SUMO proteases
SUMO modification is a reversible process. Deconjugation of SUMO is catalysed by specific SUMO proteases. Only a small number of SUMO proteases have been identified so far, all of which are classified as cysteine isopeptidases. In mammals, a family of six SUMO‐specific proteases (SENP1, 2, 3, 5, 6 and 7) are involved in the maturation and deconjugation of SUMO moieties 46, 47. While SENP5 has a preference for the SUMO3 precursor protein 48, SENP1 and SENP2 have been reported to process all SUMO isoforms to their mature forms with varying efficiency 46, 49, 50. SENP1 has an additional unique feature, as it is mostly required for deconjugation of SUMO1 from substrates 51, whereas the other family members exhibit a strong preference for SUMO2/3 deconjugation, and SENP6 and SENP7 show particular specificity for disassembly of SUMO2/3 chains 52, 53. However, similar to SUMO ligases, the specificity of the SENPs for certain substrates is thought to be regulated in a spatial and temporal manner via regulation of protein amounts, localization and activity. Other SUMO proteases, such as Ubiquitin‐Specific Peptidase‐Like protein 1 54, DeSumoylating Isopeptidase 1 and DeSumoylating Isopeptidase 2 55, have been identified but do not appear to be involved in changing global SUMOylation patterns.
SUMO‐targeted ubiquitin ligases
As post‐translational modifications represent a fast and reversible mechanism to alter the characteristics of a protein, it is logical to assume that these processes and the machinery required for modification are themselves subject to regulation via post‐translational modifications. More than 500 post‐translational modifications are known to date 56, demonstrating the immense potential of these signals in fine‐tuning even minor biochemical processes within the cell. The picture becomes even more complicated when considering the plethora of post‐translational modifications that work in concert to orchestrate various essential cellular processes.
The identification of a novel class of ubiquitin E3 ligases that specifically enhance ubiquitylation of SUMOylated proteins revealed an essential cellular mechanism involving cross‐talk between two major post‐translational modifications 57. In humans, two such SUMO‐targeted ubiquitin ligases have been identified, each one containing characteristic SIMs to specifically recognize SUMOylated proteins. Both SUMO‐targeted ubiquitin ligases form SUMO–ubiquitin hybrid chains. RING finger protein 4 (RNF4), the smallest member of this enzyme class, functions as a homodimer and contains at least three SIMs, explaining its preference for target proteins modified by poly‐SUMO chains comprising at least three SUMO moieties 58. In addition to SUMO polymers, closely spaced mono‐SUMOylation events may also form binding sites for SUMO‐targeted ubiquitin ligases. Several substrates of RNF4 have been described to date, with promyelocytic leukemia protein (PML) and its oncogenic fusion variant PML–RARα being the most prominent 58, 59. PML is degraded in an arsenic trioxide‐induced manner, resulting in disassembly of PML nuclear bodies. These sub‐nuclear compartments contain many other SUMOylated proteins, such as Sp100 and Daxx, suggesting that these proteins are also subject to RNF4‐mediated proteolysis [60, 61, 62].
The second and more recently identified human SUMO‐targeted ubiquitin ligase is RING finger protein 111 (RNF111), or Arkadia, which contains at least three SIMs for recognition of poly‐SUMO signals 63. While RNF111 has also been implicated in PML degradation, it also catalyses very distinct non‐proteolytic processes during the DNA damage response, where it has been shown to form K63‐linked ubiquitin chains on SUMOylated Xeroderma Pigmentosum group C‐complementing protein 64.
Proteomic approaches to decipher the SUMO code
Difficulties and pitfalls in identifying SUMO target proteins
Due to constant improvements in proteomic approaches, the number of known SUMO target proteins is greatly increasing, but still lags behind the number of target proteins found for some other post‐translational modifications, such as phosphorylation and ubiquitylation. Several major difficulties impede efficient identification of SUMOylated proteins on a global scale. First, SUMO expression levels are much lower compared to ubiquitin, and the amount of SUMOylated target protein usually only represents a minor fraction of the entire pool of the protein. Therefore, SUMO targets must first be enriched via immunoprecipitation or pulldown experiments. Second, SUMO proteases are highly potent and must be inactivated in denaturing buffers containing SDS, guanidine hydrochloride or urea. However, the use of antibodies during enrichment requires partial refolding of proteins after lysis, potentially reactivating SUMO proteases and allowing them to remove some of the SUMO moieties. Third, identification of SUMOylation sites via mass spectrometry is highly challenging, due to the cumbersome C‐terminal tryptic remnants of mammalian SUMO proteins that are as large as 32 amino acids for human and mouse SUMO2 and SUMO3, and 19 amino acids for human and mouse SUMO1, which are too large for efficient identification by mass spectrometry (Fig. 2E,F). Several strategies have been developed to circumvent these obstacles (Fig. 2), and each method has its clear advantages and disadvantages as detailed below.
Figure 2.
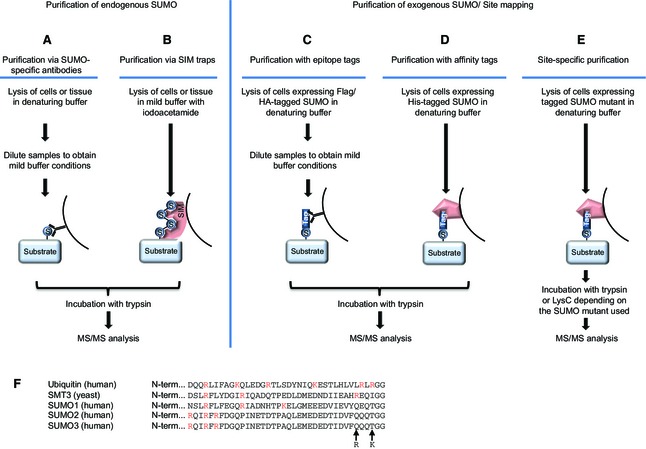
Proteomic approaches to identify SUMO targets. (A) Purification via SUMO‐specific antibodies. Cells are lysed under denaturing conditions to inactivate SUMO proteases. Afterwards, samples are diluted to obtain mild buffer conditions and SUMOylated proteins are purified using SUMO‐specific antibodies. Proteins are subsequently trypsinized and subjected to mass spectrometry. (B) Purification via SIM traps. Cells are lysed in a mild buffer supplemented with iodoacetamide, and SUMOylated proteins are purified using the SIM‐containing protein RNF4 as bait. Proteins are subsequently trypsinized and subjected to mass spectrometry. (C) Purification with epitope tags. Cells expressing a tagged SUMO fusion protein are lysed in denaturing buffer. For subsequent immunoprecipitation of the SUMO target proteins using antibodies targeting the protein tag, samples are diluted to obtain mild buffer conditions. Finally, they are trypsinized and analysed via mass spectrometry. (D) Purification with affinity tags. Cells expressing SUMO tagged with affinity tags are lysed in denaturing buffer, and SUMO targets are purified using affinity matrices that specifically bind to the tag. Subsequently, proteins are trypsinized and analysed via mass spectrometry. (E) After trypsin digestion, the C‐terminal fragments of mammalian SUMO family members are too large to efficiently map the SUMO‐conjugated lysines in target proteins. To enable site‐specific purification, protease cleavage sites are introduced in the C‐termini of mammalian SUMO family members. SUMO target proteins modified with these mutant versions of SUMOs are fused to specific protein tags and purified as previously described for epitope or affinity tags. Subsequent digestion with either trypsin or the endoproteinase LysC, depending on the SUMO mutant employed, results in shorter SUMO peptides to facilitate identification of SUMO sites via mass spectrometry. (F) Alignment of the C‐termini of mature SMT3 from yeast and mature human ubiquitin, SUMO1, SUMO2 and SUMO3, demonstrating the various lengths of the tryptic remnants remaining after cleavage. Arginine and lysine residues are highlighted in red. The mutations used to facilitate identification of SUMO2 sites are indicated by arrows.
Identification of endogenous SUMO target proteins
Almost 600 potential SUMO target proteins were identified under completely endogenous conditions, making use of monoclonal antibodies that specifically purify endogenous SUMO moieties 21. The biggest advantage of this method is the possibility of applying it to a broad range of samples, such as primary cell lines, entire organs or rare patient material. Similarly, enrichment of SUMO target proteins using SIM traps may be used to identify endogenous SUMO targets, but is mostly limited to poly‐SUMOylated and multi‐SUMOylated proteins 28, 65. However, both methods are relatively costly, require large amounts of sample material, and the number of SUMO targets identified under these conditions is considerably less compared to exogenous purification systems. Additionally, the endogenous methods are not suitable to efficiently identify SUMOylation sites, making it challenging to distinguish between real SUMO targets and fortuitously co‐purifying proteins, although attempts to chemically shorten the tryptic remnant of endogenous SUMO2 have been described 66.
Use of tagged SUMO variants and the identification of SUMO sites
For more efficient SUMO target enrichment, the N‐terminus of SUMO is most commonly fused to an epitope tag or tandem tags, such as His6, FLAG, Myc, His6‐FLAG, His6‐HA, FLAG‐TEV and ProtA‐TEV‐CBP [24, 67, 68, 69, 70, 71, 72]. Use of a His10 tag instead of the commonly used His6 tag increases the yield and purity of the samples, and has allowed us to identify more than 1600 SUMOylated proteins in human cells 29. SUMO fusion proteins are either exogenously expressed from transgenes 69, 73 or endogenously expressed using elegant knock‐in approaches 8, 72. Therefore, these methods are restricted to specific cell types and organisms. Additionally, exogenous expression of SUMO fusion proteins may lead to higher SUMOylation levels of target proteins compared to endogenous conditions. Identification of SUMO target proteins via this method should ideally be confirmed using an endogenous approach. Several model organisms expressing tagged SUMO proteins have been developed, including yeast 69, Arabidopsis 73 and mouse 8. Simultaneous application of tagged SUMO variants, together with quantification methods such as label‐free quantification and stable isotope labeling with amino acids in cell culture, has uncovered major dynamics in the SUMOylation pattern 24, 60, 68, 74.
Point mutations of these SUMO fusions, such as insertion of an additional cleavage site situated near the C‐terminus (Q87R, T90R and T90K), result in shorter proteolytic fragments, which facilitate efficient mass spectrometric analysis. These SUMO mutants have been successfully used to identify SUMO attachment sites [68, 72, 75, 76, 77, 78]. Introduction of additional mutations to specifically enrich for SUMOylated peptides in a two‐step purification protocol allowed identification of over 4300 SUMOylation sites that dynamically change in response to various cellular treatments 29. It has been confirmed that these mutations do not alter the overall conjugation efficiency of the SUMO proteins. However, it cannot be excluded that use of these SUMO mutant variants may alter SUMO conjugation to specific target proteins, particularly as one of the SUMO mutants used is deficient in forming SUMO chains. Therefore, it is recommended that SUMO sites are individually verified for each target protein before proceeding with further experiments. To circumvent the use of SUMO mutant variants, our laboratory has recently established a new site‐specific SUMO proteomics methodology using His10‐tagged wild‐type SUMO and SENP2, leading to identification of more than 750 wild‐type SUMOylation sites in HeLa cells 79. Despite its great advantage in identifying wild‐type SUMO2 sites, this method still utilizes an exogenously expressed SUMO fusion protein. The levels of exogenously expressed SUMO fusion proteins must be tightly controlled to prevent over‐expression artefacts. To circumvent these pitfalls completely, development of similar methods using endogenous SUMO proteins to identify SUMO sites would be of considerable value, but this remains a major challenge.
The various site‐specific analyses described above have allowed us to refine the canonical SUMO consensus motif and identify additional residues promoting SUMOylation. Several sites match the so‐called inverted SUMO consensus motif or lie within a hydrophobic cluster SUMOylation motif bearing hydrophobic amino acids in close proximity to the modified lysine 29, 75. Interestingly, the percentage of sites situated within a SUMO consensus motif decreases after cellular stress, indicating that SUMO ligases that are active under these conditions may facilitate SUMOylation of non‐consensus sites 29. Alternatively, inactivation of SUMO proteases after cellular stress may lead to stabilization of SUMOylation of non‐consensus sites.
Proteomic analyses to unravel protein group modification
The proteomic analyses of SUMO target proteins performed to date clearly support the concept of protein group modification (Fig. 3). This model suggests that, after specific stimuli, an entire group of functionally related proteins is SUMOylated, allowing a quick and efficient cellular response 14, 80. Accordingly, treatment with DNA damaging agents such as methyl methanesulfonate and hydroxyurea induces SUMOylation of entire clusters of functionally related DNA repair proteins, chromatin modifiers and replication factors 60, 81, 82, while arresting cells in mitosis increases SUMOylation of a protein sub‐group required for accurate chromosomal alignment and segregation 68. This collective modification is mostly achieved via the presence of SUMO ligases and proteases that are differentially regulated in a spatial and temporal manner. Proteomic analyses of SUMO target proteins after depletion of these enzymes may provide additional insights into their substrate specificity and function in distinct cellular pathways. In further agreement with the concept of protein group modification, mass spectrometric analyses identified several protein complexes that contain two or more SUMOylated subunits, including chromatin remodeling complexes, histone deacetylases and spliceosomes 29. It is believed that formation of many of these complexes is triggered and stabilized by the presence of SUMOylated subunits and additional SIMs, representing a key mechanism to maintain genome stability and nuclear integrity 14, 17, 80.
Figure 3.
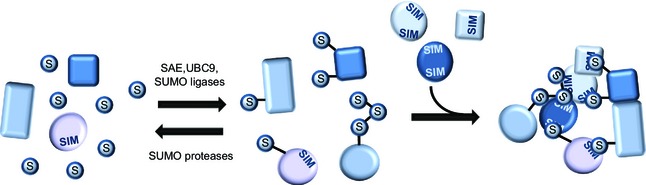
Protein group modification via SUMO. In response to specific cellular or external stimuli, the activity and localization of the SUMO conjugation machinery is altered, leading to SUMOylation of target proteins with similar functions during the cellular response. This protein group modification triggers the formation of larger protein complexes via specific SUMO–SIM interactions. Increased activity of SUMO proteases reverses this process, leading to disassembly of these protein complexes.
Post‐translational modification cross‐talk identified by proteomic analyses
In addition to providing a comprehensive view of the SUMOylated proteome, proteomic approaches are a valuable tool in deciphering the vast network of post‐translational modifications that work in concert to regulate specific cellular pathways (Fig. 4). Many components of the enzymatic machinery required to modify proteins are subject to SUMOylation, such as kinases, phosphatases, ubiquitin ligases and proteases, demethylases, and acetyltransferases 83. Accordingly, the SUMO machinery itself has been found to be post‐translationally regulated. SUMOylation and phosphorylation of UBC9 have both been shown to promote its enzymatic activity 45, 84. In contrast, acetylation of UBC9 specifically inhibits modification of substrates containing a so‐called negatively charged SUMO consensus motif (ΨKxExxEEEE), providing a clear example of how post‐translational modifications may confer substrate specificity to a relatively promiscuous enzyme 85. Interestingly, several lysine residues of ubiquitin have been shown to be SUMOylated, indicative of the formation of hybrid chains consisting of the two post‐translational modifications 29.
Figure 4.
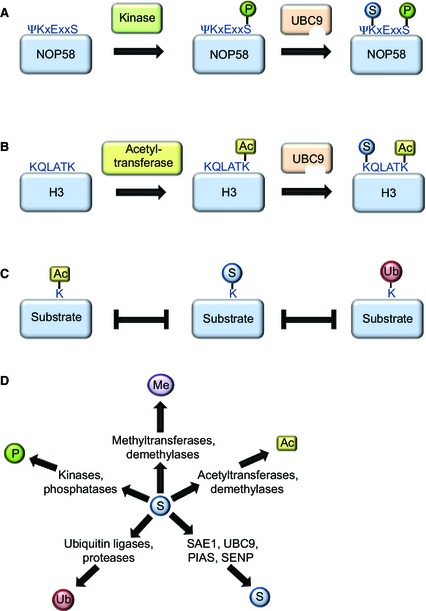
Cross‐talk between post‐translational modifications as identified via mass spectrometry. (A) Several SUMO target proteins contain a so‐called phosphorylation‐dependent SUMO motif, in which the modified lysine residue is followed by a phosphorylated serine, usually five amino acids further downstream. A serine residue situated in a phosphorylation‐dependent SUMO motif of the nuclear protein NOP58 is phosphorylated via casein kinase 2 (CK2), promoting UBC9 binding and subsequent SUMOylation of the indicated lysine residue. (B) Similar to phosphorylation, acetylation via specific acetyl transferases may induce SUMOylation of a protein, as described for histone H3. (C) Many lysine residues that were found as SUMO acceptor sites have also been shown to be ubiquitylated or acetylated, suggesting extensive competition between these modifiers. (D) Dozens of enzymes regulating post‐translational protein modifications have been identified as SUMO target proteins in proteomic screens, including SUMO pathway components (S), or enzymes regulating other post‐translational modifications, such as phosphorylation (P), ubiquitylation (Ub), methylation (Me) or acetylation (Ac).
The possibility of identifying SUMO sites has enabled us to identify the strong dependency of SUMOylation events on other post‐translational modifications. An obvious example of cross‐talk between phosphorylation and SUMOylation is the existence of a phosphorylation‐dependent SUMO consensus motif, in which a phosphorylated serine residue is generally situated five residues downstream of a lysine within a SUMO consensus motif 86. Dozens of target proteins bearing such phosphorylation‐dependent SUMO consensus motifs have been identified via mass spectrometry, and, for some of these proteins, both modifications were simultaneously present on the same peptide, indicative of a direct dependency. Indeed, mutagenesis experiments have shown that phosphorylation of these residues strongly increases SUMOylation of the relevant lysines 29, 75. Accordingly, an acetylation‐dependent SUMOylation motif has recently been shown to be present in histones H3 and H4 29. Furthermore, many lysine residues identified as SUMOylated have also been shown to be acetylated or ubiquitylated, indicating competitive usage of a subset of lysines by these post‐translational modifications.
Conclusions and outlook
After 20 years of SUMO research, we are only beginning to grasp the enormous potential of this post‐translational modification for regulating a vast number of cellular processes. Proteomic approaches have allowed us to identify hundreds of SUMO target proteins, but the number is still lagging behind findings for other essential post‐translational modifications such as phosphorylation and ubiquitylation. However, the current proteomic analyses greatly help in understanding the network of SUMO targets within the cell, and further underline the concept of protein group modification. In addition, identification of specific SUMO sites and other post‐translational modifications regulating SUMOylation events is facilitated by these mass spectrometric approaches. New purification methods that consecutively enrich SUMO and other post‐translational modifications would further expand our knowledge about cross‐talk between post‐translational modifications, but are highly challenging and have not been successfully applied. Finally, it is of great importance to further improve purification methods to enrich endogenous SUMO proteins, to study relevant samples such as human patient material. Being able to identify SUMO target proteins, SUMO sites and components of the SUMO machinery that are deregulated in specific human diseases, such as cancer and neurodegenerative disorders, is an important step in developing new therapies and potent drugs.
Author contributions
KE and ACOV wrote the manuscript.
Acknowledgements
This work was supported by the Nederlandse Organisatie voor Wetenschappelijk Onderzoek (Netherlands Organisation for Scientific Research) and the European Research Council. We would like to thank Ivo A. Hendriks for helpful discussions and critically reading the manuscript.
References
- 1. Dasso M (2008) Emerging roles of the SUMO pathway in mitosis. Cell Div 3, 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Jackson SP & Durocher D (2013) Regulation of DNA damage responses by ubiquitin and SUMO. Mol Cell 49, 795–807. [DOI] [PubMed] [Google Scholar]
- 3. Melchior F, Schergaut M & Pichler A (2003) SUMO: ligases, isopeptidases and nuclear pores. Trends Biochem Sci 28, 612–618. [DOI] [PubMed] [Google Scholar]
- 4. Evdokimov E, Sharma P, Lockett SJ, Lualdi M & Kuehn MR (2008) Loss of SUMO1 in mice affects RanGAP1 localization and formation of PML nuclear bodies, but is not lethal as it can be compensated by SUMO2 or SUMO3. J Cell Sci 121, 4106–4113. [DOI] [PubMed] [Google Scholar]
- 5. Wang L, Wansleeben C, Zhao S, Miao P, Paschen W & Yang W (2014) SUMO2 is essential while SUMO3 is dispensable for mouse embryonic development. EMBO Rep 15, 878–885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Nacerddine K, Lehembre F, Bhaumik M, Artus J, Cohen‐Tannoudji M, Babinet C, Pandolfi PP & Dejean A (2005) The SUMO pathway is essential for nuclear integrity and chromosome segregation in mice. Dev Cell 9, 769–779. [DOI] [PubMed] [Google Scholar]
- 7. Droescher M, Chaugule VK & Pichler A (2013) SUMO rules: regulatory concepts and their implication in neurologic functions. Neuromolecular Med 15, 639–660. [DOI] [PubMed] [Google Scholar]
- 8. Tirard M, Hsiao HH, Nikolov M, Urlaub H, Melchior F & Brose N (2012) In vivo localization and identification of SUMOylated proteins in the brain of His6‐HA‐SUMO1 knock‐in mice. Proc Natl Acad Sci USA 109, 21122–21127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kho C, Lee A, Jeong D, Oh JG, Chaanine AH, Kizana E, Park WJ & Hajjar RJ (2011) SUMO1‐dependent modulation of SERCA2a in heart failure. Nature 477, 601–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bawa‐Khalfe T & Yeh ET (2010) SUMO Losing balance: SUMO proteases disrupt SUMO homeostasis to facilitate cancer development and progression. Genes Cancer 1, 748–752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Sriramachandran AM & Dohmen RJ (2014) SUMO‐targeted ubiquitin ligases. Biochim Biophys Acta 1843, 75–85. [DOI] [PubMed] [Google Scholar]
- 12. Wan J, Subramonian D & Zhang XD (2012) SUMOylation in control of accurate chromosome segregation during mitosis. Curr Protein Pept Sci 13, 467–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Cubenas‐Potts C & Matunis MJ (2013) SUMO: a multifaceted modifier of chromatin structure and function. Dev Cell 24, 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Jentsch S & Psakhye I (2013) Control of nuclear activities by substrate‐selective and protein‐group SUMOylation. Annu Rev Genet 47, 167–186. [DOI] [PubMed] [Google Scholar]
- 15. Flotho A & Melchior F (2013) Sumoylation: a regulatory protein modification in health and disease. Annu Rev Biochem 82, 357–385. [DOI] [PubMed] [Google Scholar]
- 16. Bekker‐Jensen S & Mailand N (2011) The ubiquitin‐ and SUMO‐dependent signaling response to DNA double‐strand breaks. FEBS Lett 585, 2914–2919. [DOI] [PubMed] [Google Scholar]
- 17. Sarangi P & Zhao X (2015) SUMO‐mediated regulation of DNA damage repair and responses. Trends Biochem Sci 40, 233–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Chen A, Mannen H & Li SS (1998) Characterization of mouse ubiquitin‐like SMT3A and SMT3B cDNAs and gene/pseudogenes. Biochem Mol Biol Int 46, 1161–1174. [DOI] [PubMed] [Google Scholar]
- 19. Owerbach D, McKay EM, Yeh ET, Gabbay KH & Bohren KM (2005) A proline‐90 residue unique to SUMO‐4 prevents maturation and sumoylation. Biochem Biophys Res Commun 337, 517–520. [DOI] [PubMed] [Google Scholar]
- 20. Vertegaal AC, Andersen JS, Ogg SC, Hay RT, Mann M & Lamond AI (2006) Distinct and overlapping sets of SUMO‐1 and SUMO‐2 target proteins revealed by quantitative proteomics. Mol Cell Proteomics 5, 2298–2310. [DOI] [PubMed] [Google Scholar]
- 21. Becker J, Barysch SV, Karaca S, Dittner C, Hsiao HH, Berriel DM, Herzig S, Urlaub H & Melchior F (2013) Detecting endogenous SUMO targets in mammalian cells and tissues. Nat Struct Mol Biol 20, 525–531. [DOI] [PubMed] [Google Scholar]
- 22. Mahajan R, Delphin C, Guan T, Gerace L & Melchior F (1997) A small ubiquitin‐related polypeptide involved in targeting RanGAP1 to nuclear pore complex protein RanBP2. Cell 88, 97–107. [DOI] [PubMed] [Google Scholar]
- 23. Matunis MJ, Coutavas E & Blobel G (1996) A novel ubiquitin‐like modification modulates the partitioning of the Ran‐GTPase‐activating protein RanGAP1 between the cytosol and the nuclear pore complex. J Cell Biol 135, 1457–1470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Golebiowski F, Matic I, Tatham MH, Cole C, Yin Y, Nakamura A, Cox J, Barton GJ, Mann M & Hay RT (2009) System‐wide changes to SUMO modifications in response to heat shock. Sci Signal 2, ra24. [DOI] [PubMed] [Google Scholar]
- 25. Tatham MH, Jaffray E, Vaughan OA, Desterro JM, Botting CH, Naismith JH & Hay RT (2001) Polymeric chains of SUMO‐2 and SUMO‐3 are conjugated to protein substrates by SAE1/SAE2 and Ubc9. J Biol Chem 276, 35368–35374. [DOI] [PubMed] [Google Scholar]
- 26. Matic I, van Hagen M, Schimmel J, Macek B, Ogg SC, Tatham MH, Hay RT, Lamond AI, Mann M & Vertegaal AC (2008) In vivo identification of human small ubiquitin‐like modifier polymerization sites by high accuracy mass spectrometry and an in vitro to in vivo strategy. Mol Cell Proteomics 7, 132–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Vertegaal AC (2010) SUMO chains: polymeric signals. Biochem Soc Trans 38, 46–49. [DOI] [PubMed] [Google Scholar]
- 28. Bruderer R, Tatham MH, Plechanovova A, Matic I, Garg AK & Hay RT (2011) Purification and identification of endogenous polySUMO conjugates. EMBO Rep 12, 142–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Hendriks IA, D'Souza RC, Yang B, Verlaan‐de Vries M, Mann M & Vertegaal AC (2014) Uncovering global SUMOylation signaling networks in a site‐specific manner. Nat Struct Mol Biol 21, 927–936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Bernier‐Villamor V, Sampson DA, Matunis MJ & Lima CD (2002) Structural basis for E2‐mediated SUMO conjugation revealed by a complex between ubiquitin‐conjugating enzyme Ubc9 and RanGAP1. Cell 108, 345–356. [DOI] [PubMed] [Google Scholar]
- 31. Gnad F, Gunawardena J & Mann M (2011) PHOSIDA 2011: the posttranslational modification database. Nucleic Acids Res 39, D253–D260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Zhao Q, Xie Y, Zheng Y, Jiang S, Liu W, Mu W, Liu Z, Zhao Y, Xue Y & Ren J (2014) GPS‐SUMO: a tool for the prediction of sumoylation sites and SUMO‐interaction motifs. Nucleic Acids Res 42, W325–W330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Deshaies RJ & Joazeiro CA (2009) RING domain E3 ubiquitin ligases. Annu Rev Biochem 78, 399–434. [DOI] [PubMed] [Google Scholar]
- 34. Schmidt D & Muller S (2002) Members of the PIAS family act as SUMO ligases for c‐Jun and p53 and repress p53 activity. Proc Natl Acad Sci USA 99, 2872–2877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Palvimo JJ (2007) PIAS proteins as regulators of small ubiquitin‐related modifier (SUMO) modifications and transcription. Biochem Soc Trans 35, 1405–1408. [DOI] [PubMed] [Google Scholar]
- 36. Werner A, Flotho A & Melchior F (2012) The RanBP2/RanGAP1*SUMO1/Ubc9 complex is a multisubunit SUMO E3 ligase. Mol Cell 46, 287–298. [DOI] [PubMed] [Google Scholar]
- 37. Pichler A, Gast A, Seeler JS, Dejean A & Melchior F (2002) The nucleoporin RanBP2 has SUMO1 E3 ligase activity. Cell 108, 109–120. [DOI] [PubMed] [Google Scholar]
- 38. Dawlaty MM, Malureanu L, Jeganathan KB, Kao E, Sustmann C, Tahk S, Shuai K, Grosschedl R & van Deursen JM (2008) Resolution of sister centromeres requires RanBP2‐mediated SUMOylation of topoisomerase IIα. Cell 133, 103–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Kagey MH, Melhuish TA & Wotton D (2003) The polycomb protein Pc2 is a SUMO E3. Cell 113, 127–137. [DOI] [PubMed] [Google Scholar]
- 40. Zhao X, Sternsdorf T, Bolger TA, Evans RM & Yao TP (2005) Regulation of MEF2 by histone deacetylase 4‐ and SIRT1 deacetylase‐mediated lysine modifications. Mol Cell Biol 25, 8456–8464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Woods YL, Xirodimas DP, Prescott AR, Sparks A, Lane DP & Saville MK (2004) p14 Arf promotes small ubiquitin‐like modifier conjugation of Werners helicase. J Biol Chem 279, 50157–50166. [DOI] [PubMed] [Google Scholar]
- 42. Guervilly JH, Takedachi A, Naim V, Scaglione S, Chawhan C, Lovera Y, Despras E, Kuraoka I, Kannouche P, Rosselli F et al (2015) The SLX4 complex is a SUMO E3 ligase that impacts on replication stress outcome and genome stability. Mol Cell 57, 123–137. [DOI] [PubMed] [Google Scholar]
- 43. Ouyang J, Garner E, Hallet A, Nguyen HD, Rickman KA, Gill G, Smogorzewska A & Zou L (2015) Noncovalent interactions with SUMO and ubiquitin orchestrate distinct functions of the SLX4 complex in genome maintenance. Mol Cell 57, 108–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Gonzalez‐Prieto R, Cuijpers SA, Luijsterburg MS, van Attikkum H & Vertegaal AC (2015) SUMOylation and PARylation cooperate to recruit and stabilize SLX4 at DNA damage sites. EMBO Rep 16, 512–519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Knipscheer P, Flotho A, Klug H, Olsen JV, van Dijk WJ , Fish A, Johnson ES, Mann M, Sixma TK & Pichler A (2008) Ubc9 sumoylation regulates SUMO target discrimination. Mol Cell 31, 371–382. [DOI] [PubMed] [Google Scholar]
- 46. Nayak A & Muller S (2014) SUMO‐specific proteases/isopeptidases: SENPs and beyond. Genome Biol 15, 422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Hickey CM, Wilson NR & Hochstrasser M (2012) Function and regulation of SUMO proteases. Nat Rev Mol Cell Biol 13, 755–766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Di BA, Ouyang J, Lee HY, Catic A, Ploegh H & Gill G (2006) The SUMO‐specific protease SENP5 is required for cell division. Mol Cell Biol 26, 4489–4498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Reverter D & Lima CD (2006) Structural basis for SENP2 protease interactions with SUMO precursors and conjugated substrates. Nat Struct Mol Biol 13, 1060–1068. [DOI] [PubMed] [Google Scholar]
- 50. Shen LN, Dong C, Liu H, Naismith JH & Hay RT (2006) The structure of SENP1–SUMO‐2 complex suggests a structural basis for discrimination between SUMO paralogues during processing. Biochem J 397, 279–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Sharma P, Yamada S, Lualdi M, Dasso M & Kuehn MR (2013) Senp1 is essential for desumoylating Sumo1‐modified proteins but dispensable for Sumo2 and Sumo3 deconjugation in the mouse embryo. Cell Rep 3, 1640–1650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Drag M, Mikolajczyk J, Krishnakumar IM, Huang Z & Salvesen GS (2008) Activity profiling of human deSUMOylating enzymes (SENPs) with synthetic substrates suggests an unexpected specificity of two newly characterized members of the family. Biochem J 409, 461–469. [DOI] [PubMed] [Google Scholar]
- 53. Lima CD & Reverter D (2008) Structure of the human SENP7 catalytic domain and poly‐SUMO deconjugation activities for SENP6 and SENP7. J Biol Chem 283, 32045–32055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Schulz S, Chachami G, Kozaczkiewicz L, Winter U, Stankovic‐Valentin N, Haas P, Hofmann K, Urlaub H, Ovaa H, Wittbrodt J et al (2012) Ubiquitin‐specific protease‐like 1 (USPL1) is a SUMO isopeptidase with essential, non‐catalytic functions. EMBO Rep 13, 930–938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Suh HY, Kim JH, Woo JS, Ku B, Shin EJ, Yun Y & Oh BH (2012) Crystal structure of DeSI‐1, a novel deSUMOylase belonging to a putative isopeptidase superfamily. Proteins 80, 2099–2104. [DOI] [PubMed] [Google Scholar]
- 56. Garavelli JS (2004) The RESID Database of Protein Modifications as a resource and annotation tool. Proteomics 4, 1527–1533. [DOI] [PubMed] [Google Scholar]
- 57. Uzunova K, Gottsche K, Miteva M, Weisshaar SR, Glanemann C, Schnellhardt M, Niessen M, Scheel H, Hofmann K, Johnson ES et al (2007) Ubiquitin‐dependent proteolytic control of SUMO conjugates. J Biol Chem 282, 34167–34175. [DOI] [PubMed] [Google Scholar]
- 58. Tatham MH, Geoffroy MC, Shen L, Plechanovova A, Hattersley N, Jaffray EG, Palvimo JJ & Hay RT (2008) RNF4 is a poly‐SUMO‐specific E3 ubiquitin ligase required for arsenic‐induced PML degradation. Nat Cell Biol 10, 538–546. [DOI] [PubMed] [Google Scholar]
- 59. Lallemand‐Breitenbach V, Jeanne M, Benhenda S, Nasr R, Lei M, Peres L, Zhou J, Zhu J, Raught B & de Thé H (2008) Arsenic degrades PML or PML‐RARα through a SUMO‐triggered RNF4/ubiquitin‐mediated pathway. Nat Cell Biol 10, 547–555. [DOI] [PubMed] [Google Scholar]
- 60. Hendriks IA, Treffers LW, Verlaan‐de Vries M, Olsen JV & Vertegaal AC (2015) SUMO‐2 orchestrates chromatin modifiers in response to DNA damage. Cell Rep 10, 1778–1791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Sahin U, Ferhi O, Jeanne M, Benhenda S, Berthier C, Jollivet F, Niwa‐Kawakita M, Faklaris O, Setterblad N, de Thé H et al (2014) Oxidative stress‐induced assembly of PML nuclear bodies controls sumoylation of partner proteins. J Cell Biol 204, 931–945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Sahin U, de Thé H & Lallemand‐Breitenbach V (2014) PML nuclear bodies: assembly and oxidative stress‐sensitive sumoylation. Nucleus 5, 499–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Erker Y, Neyret‐Kahn H, Seeler JS, Dejean A, Atfi A & Levy L (2013) Arkadia, a novel SUMO‐targeted ubiquitin ligase involved in PML degradation. Mol Cell Biol 33, 2163–2177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Poulsen SL, Hansen RK, Wagner SA, van Cuijk L, van Belle GJ, Streicher W, Wikstrom M, Choudhary C, Houtsmuller AB, Marteijn JA et al (2013) RNF111/Arkadia is a SUMO‐targeted ubiquitin ligase that facilitates the DNA damage response. J Cell Biol 201, 797–807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Lang V, Aillet F, Da Silva‐Ferrada E, Xolalpa W, Zabaleta L, Rivas C & Rodriguez MS (2014) Analysis of PTEN ubiquitylation and SUMOylation using molecular traps. Methods 77, 112–118. [DOI] [PubMed] [Google Scholar]
- 66. Osula O, Swatkoski S & Cotter RJ (2012) Identification of protein SUMOylation sites by mass spectrometry using combined microwave‐assisted aspartic acid cleavage and tryptic digestion. J Mass Spectrom 47, 644–654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Vertegaal AC, Ogg SC, Jaffray E, Rodriguez MS, Hay RT, Andersen JS, Mann M & Lamond AI (2004) A proteomic study of SUMO‐2 target proteins. J Biol Chem 279, 33791–33798. [DOI] [PubMed] [Google Scholar]
- 68. Schimmel J, Eifler K, Sigurethsson JO, Cuijpers SA, Hendriks IA, Verlaan‐de Vries M, Kelstrup CD, Francavilla C, Medema RH, Olsen JV et al (2014) Uncovering SUMOylation dynamics during cell‐cycle progression reveals FoxM1 as a key mitotic SUMO target protein. Mol Cell 53, 1053–1066. [DOI] [PubMed] [Google Scholar]
- 69. Denison C, Rudner AD, Gerber SA, Bakalarski CE, Moazed D & Gygi SP (2005) A proteomic strategy for gaining insights into protein sumoylation in yeast. Mol Cell Proteomics 4, 246–254. [DOI] [PubMed] [Google Scholar]
- 70. Ganesan AK, Kho Y, Kim SC, Chen Y, Zhao Y & White MA (2007) Broad spectrum identification of SUMO substrates in melanoma cells. Proteomics 7, 2216–2221. [DOI] [PubMed] [Google Scholar]
- 71. Hannich JT, Lewis A, Kroetz MB, Li SJ, Heide H, Emili A & Hochstrasser M (2005) Defining the SUMO‐modified proteome by multiple approaches in Saccharomyces cerevisiae . J Biol Chem 280, 4102–4110. [DOI] [PubMed] [Google Scholar]
- 72. Wohlschlegel JA, Johnson ES, Reed SI & Yates JR III (2004) Global analysis of protein sumoylation in Saccharomyces cerevisiae . J Biol Chem 279, 45662–45668. [DOI] [PubMed] [Google Scholar]
- 73. Miller MJ, Barrett‐Wilt GA, Hua Z & Vierstra RD (2010) Proteomic analyses identify a diverse array of nuclear processes affected by small ubiquitin‐like modifier conjugation in Arabidopsis. Proc Natl Acad Sci USA 107, 16512–16517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Yang W, Thompson JW, Wang Z, Wang L, Sheng H, Foster MW, Moseley MA & Paschen W (2012) Analysis of oxygen/glucose‐deprivation‐induced changes in SUMO3 conjugation using SILAC‐based quantitative proteomics. J Proteome Res 11, 1108–1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Matic I, Schimmel J, Hendriks IA, van Santen MA, van de Rijke F, van Dam H, Gnad F, Mann M & Vertegaal AC (2010) Site‐specific identification of SUMO‐2 targets in cells reveals an inverted SUMOylation motif and a hydrophobic cluster SUMOylation motif. Mol Cell 39, 641–652. [DOI] [PubMed] [Google Scholar]
- 76. Tammsalu T, Matic I, Jaffray EG, Ibrahim AF, Tatham MH & Hay RT (2014) Proteome‐wide identification of SUMO2 modification sites. Sci Signal 7, rs2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Lamoliatte F, Bonneil E, Durette C, Caron‐Lizotte O, Wildemann D, Zerweck J, Wenshuk H & Thibault P (2013) Targeted identification of SUMOylation sites in human proteins using affinity enrichment and paralog‐specific reporter ions. Mol Cell Proteomics 12, 2536–2550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Wohlschlegel JA, Johnson ES, Reed SI & Yates JR III (2006) Improved identification of SUMO attachment sites using C‐terminal SUMO mutants and tailored protease digestion strategies. J Proteome Res 5, 761–770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Hendriks IA, D'Souza RC, Chang JG, Mann M & Vertegaal AC (2015) System‐wide identification of wild‐type SUMO‐2 conjugation sites. Nat Commun 6, 7289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Johnson ES & Blobel G (1999) Cell cycle‐regulated attachment of the ubiquitin‐related protein SUMO to the yeast septins. J Cell Biol 147, 981–994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Psakhye I & Jentsch S (2012) Protein group modification and synergy in the SUMO pathway as exemplified in DNA repair. Cell 151, 807–820. [DOI] [PubMed] [Google Scholar]
- 82. Xiao Z, Chang JG, Hendriks IA, Sigurdsson JO, Olsen JV & Vertegaal AC (2015) System‐wide analysis of SUMOylation dynamics in response to replication stress reveals novel SUMO target proteins and acceptor lysines relevant for genome stability. Mol Cell Proteomics 14, 1419–1434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Vertegaal AC (2011) Uncovering ubiquitin and ubiquitin‐like signaling networks. Chem Rev 111, 7923–7940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Su YF, Yang T, Huang H, Liu LF & Hwang J (2012) Phosphorylation of Ubc9 by Cdk1 enhances SUMOylation activity. PLoS One 7, e34250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Hsieh YL, Kuo HY, Chang CC, Naik MT, Liao PH, Ho CC, Huang TC, Jeng JC, Hsu PH, Tsai MD et al (2013) Ubc9 acetylation modulates distinct SUMO target modification and hypoxia response. EMBO J 32, 791–804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Hietakangas V, Anckar J, Blomster HA, Fujimoto M, Palvimo JJ, Nakai A & Sistonen L (2006) PDSM, a motif for phosphorylation‐dependent SUMO modification. Proc Natl Acad Sci USA 103, 45–50. [DOI] [PMC free article] [PubMed] [Google Scholar]


