Abstract
Low birth weight and rapid postnatal growth increases the risk of developing insulin resistance and type 2 diabetes in later life. However, underlying mechanisms and potential intervention strategies are poorly defined. Here we demonstrate that male Wistar rats exposed to a low-protein diet in utero that had a low birth weight but then underwent postnatal catch-up growth (recuperated offspring) had reductions in the insulin signaling proteins p110-β (13% ± 6% of controls [P <.001]) and insulin receptor substrate-1 (39% ± 10% of controls [P <.05]) in adipose tissue. These changes were not accompanied by any change in expression of the corresponding mRNAs, suggesting posttranscriptional regulation. Recuperated animals displayed evidence of a proinflammatory phenotype of their adipose tissue with increased IL-6 (139% ± 8% [P <.05]) and IL1-β (154% ± 16% [P <.05]) that may contribute to the insulin signaling protein dysregulation. Postweaning dietary supplementation of recuperated animals with coenzyme Q (CoQ10) (1 mg/kg of body weight per day) prevented the programmed reduction in insulin receptor substrate-1 and p110-β and the programmed increased in IL-6. These findings suggest that postweaning CoQ10 supplementation has antiinflammatory properties and can prevent programmed changes in insulin-signaling protein expression. We conclude that CoQ10 supplementation represents an attractive intervention strategy to prevent the development of insulin resistance that results from suboptimal in utero nutrition.
Many epidemiological studies have demonstrated that a suboptimal fetal environment, resulting in low birth weight (LBW) can result in increased risk of glucose intolerance (1, 2), insulin resistance (3), and type 2 diabetes (4, 5) in later life. Animal models of intrauterine growth restriction have very similar phenotypes to LBW humans. Using a well-established model of maternal protein restriction that generates LBW offspring, we have demonstrated that the growth-restricted pups develop an insulin-resistant and diabetic phenotype in later life (6). These offspring show alterations in key insulin signaling molecules in skeletal muscle and adipose tissue that are markedly similar to those observed in men with LBW (7–9). The risk for development of poor glucose tolerance (10), insulin resistance (11, 12), and type 2 diabetes (13, 14) is exacerbated when LBW is combined with rapid postnatal growth. We have recently demonstrated that the dysregulation of insulin signaling protein expression in adipose tissue occurs prior to the development of whole-body insulin resistance and is a very early consequence of LBW and catch-up growth (15).
Oxidative stress plays an important role in the etiology of insulin resistance (16), and in adipose tissue this is associated with increased inflammation (17). Oxidative stress accumulation is a common underlying consequence of many suboptimal in utero environments (18). Consequently, several studies have used antioxidant therapy during pregnancy to attempt to prevent the observed deleterious phenotypes resulting from a suboptimal in utero environment (19–21). These demonstrate proof of principle that antioxidants can prevent detrimental effects of developmental programming; however, the doses used in these studies far exceed those suitable for use in pregnant women. Furthermore, it is also important to address the potential beneficial effects of targeted postnatal antioxidant supplementation because often evidence is not present for suboptimal in utero exposure until at the time of, or just after, delivery and supplementation could be detrimental to those who do not need it.
Previously we demonstrated that postnatal supplementation of coenzyme Q (CoQ), also known as ubiquinone, at a clinically relevant dose can prevent a programmed accelerated aging phenotype in the aorta (22) and heart (23) of rats that were born small and underwent catch-up growth. CoQ exists in various isoforms, which differ in concentration between organisms. In humans, the most abundant form of CoQ is CoQ10 (because it contains 10 isoprenoid units attached to a benzoquinone ring); in rodents, however, CoQ9 (containing nine isoprenoid units) is the most abundant form. It is also known that rodents can convert dietary CoQ10 into CoQ9 (23). CoQ is the most abundant and potent antioxidant in the body (24) with additional antiinflammatory properties, including decreasing the production of proinflammatory cytokines (25). Clinical trials have confirmed the safety of CoQ up to doses of 1200 mg/kg · d (26, 27).
This study therefore aimed to establish whether a postnatal dietary supplementation of CoQ could alter any observed dysregulation of insulin signaling in rats exposed to a suboptimal environment in utero and postnatal catch-up growth and then to investigate the potential underlying mechanisms, including oxidative stress status and inflammatory phenotypes.
Materials and Methods
Animal experimentation
All procedures involving animals were conducted under the British Animals (Scientific Procedures) Act (1986). Pregnant Wistar rats were maintained on a 20% protein diet (control) or an isocaloric low-protein (8%) diet fed ad libitum, as previously described (28). Both diets were purchased from Arie Blok. The day of birth was recorded as day 1 of postnatal life. Pups born to low protein diet-fed dams were cross-fostered to control-fed mothers on postnatal day 3 to create a recuperated litter. Each recuperated litter was culled to four male pups at random to maximize their plane of nutrition. The control group constituted offspring of mothers fed the 20% protein diet and suckled by 20% protein-fed dams. Each control litter was culled to eight pups as a standard. To prevent any stress to the animals during cross-fostering, pups were transferred with their own bedding. At 21 days, two males per litter were weaned onto standard laboratory chow (Special Diet Services), and the other two were weaned onto the same diet supplemented with CoQ to give a dose of 1 mg/kg · d body weight. Animals were maintained on these diets until 3 months of age. Body weights were recorded at postnatal days 3, 7, 14, and 21 and 3 months of age. All animals were killed by CO2 asphyxiation. At postmortem, epididymal fat pads, liver, and vastus lateralis skeletal muscle were removed, weighed, and snap frozen in liquid nitrogen and then stored at −80°C until analysis. For all measurements, one pup per litter was used; thus, the n indicated throughout represents the number of litters. Only male animals were used in this study.
CoQ diet preparation
A dose of 1 mg/kg CoQ of body weight per day was used in this study. This was achieved by appropriate CoQ supplementation of laboratory chow as described previously (22, 23). Briefly, CoQ was impregnated into the diet pellets by dissolving CoQ10 in acetone and mixing this with the diet pellets. The mix was then left in a fume hood overnight to allow evaporation of acetone. The diet was prepared twice a week throughout the study.
CoQ, lipid profile, glucose, and insulin analysis
Serum was obtained from blood collected from the tail vein after overnight fasting. The blood clotted for 30 minutes before centrifugation for 3 minutes at 845 × g. Fasted blood glucose measurements were obtained using a blood glucose analyzer (Hemocue). The lipid profile and fasted serum insulin analysis was performed using an autoanalyzer (Clinical Chemistry Laboratory, Medical Research Council Centre for Obesity and Related Metabolic Diseases, Cambridge, United Kingdom). Blood from the tail vein was collected into heparin tubes and centrifuged to isolate plasma. Plasma CoQ10 levels were determined by reverse-phased HPLC with UV detection at 275 nm as previously described (22).
Protein analysis
Protein was extracted and assayed as described previously (29), and 20 µg protein was loaded onto 10%–15% polyacrylamide gels, dependent on the molecular weight of the target protein. The samples were then electrophoresed and transferred to polyvinylidene fluoride membranes (22), and antibodies to the following proteins were detected: IL-6 and IL-1β (Abcam); insulin receptor substrate (IRS)-1, p110-β, and p85α (Merck Millipore); Akt-1 (Cell Signaling Technology); and Akt-2 and pAkt(ser473) (New England Bio-labs) (please see Table 5 for further details). Antirabbit and antimouse IgG horseradish peroxidase-linked secondary antibodies were from Jackson ImmunoResearch Laboratories. Equal protein loading was confirmed by staining electrophoresed gels with Coomassie Blue (Bio-Rad Laboratories) to visualize the total protein. To ensure the chemiluminescent signal changed in a linear manner, the ratio between loading controls (50% and 100% pooled sample) was confirmed for each detected protein. Protein bands were detected using a West Pico chemiluminescence reagent (Pierce, Thermo Scientific) and analyzed using Alphaease Imaging Software (Alpha Innotech).
Table 5.
Antibody Table
| Peptide/Protein Target | Antigen Sequence (If Known) | Name of Antibody | Manufacturer, Catalog Number, or Name of Source | Species Raised (Monoclonal or Polyclonal) | Dilution Used |
|---|---|---|---|---|---|
| IRS-1 | Anti-IRS1 antibody | Merck Millipore; catalog number 06-248 | Rabbit polyclonal | 1:1000 | |
| PI3 kinase (p110-β) | PI3 kinase p110-β (C33D4) | Cell Signaling; catalog number 3011S | Rabbit monoclonal | 1:1000 | |
| PI3 kinase (p85-α) | Anti-PI3 kinase antibody, p85 | Upstate Biotechnology; catalog number 06-195 | Rabbit polyclonal | 1:5000 | |
| IR-β | Insulin Rβ antibody (C19) | Santa Cruz Biotechnology; catalog number sc-711 | Rabbit polyclonal | 1:200 | |
| PKC-ζ | PKC-ζ antibody (C20) | Santa Cruz Biotechnology; catalog number sc-216 | Rabbit polyclonal | 1:200 | |
| Akt-1 | Akt1 (2H10) | Cell Signaling; catalog number 2967 | Mouse monoclonal | 1:1000 | |
| Akt-2 | Akt2 (D6G4) | New England Biolabs; catalog number 3063 | Rabbit monoclonal | 1:1000 | |
| pAkt (ser473) | Phospho-Akt (Ser473) | New England Biolabs; catalog number 4058 | Rabbit monoclonal | 1:5000 | |
| GLUT4 | Antiglucose transporter (GLUT4) antibody | Abcam; catalog number Ab 654 | Rabbit polyclonal | 1:1000 | |
| IL-6 | Anti-IL6 antibody | Abcam; catalog number Ab 6672 | Rabbit polyclonal | 1:1000 |
Abbreviation: IR, insulin receptor; PI3 kinase, phosphatidylinositol 3-kinase.
Gene expression
RNA was extracted using a miRNeasy minikit (QIAGEN) following the manufacturer’s instructions (22), and a deoxyribonuclease digestion step was performed to ensure no genomic DNA contamination. RNA (1 µg) was used to synthesize the cDNA using oligodeoxythymidine primers and Moloney murine leukemia virus reverse transcriptase (Promega). Gene expression was determined using custom-designed primers (Sigma) and SYBR Green reagents (Applied Biosystems). (Primer sequences are described in Table 1.)
Table 1.
Primer Sequences
| Primer | Sequence (Forward) | Sequence (Reverse) |
|---|---|---|
| Irs-1 | TGGCAGTGAGGATGTGAAAC | CTTGGATGC TCCCCC TAGAT |
| p110-β | TGAGGTTGTGAGCACCTCTG | CTTTGTTGAAGGCTGCTGTG |
| Pkc-ζ | GGGTGGATGGGATCAAAATC | GGAGGACCTTGGCATAGCTT |
| Tnf-α | CCTCCTCTCTGCCATCAAGA | TGGAAGACTCCTCCCAGGTA |
| Tgfβ1 | TGCCCTCTACAACCAACACA | CTTGCGACCCACGTAGTAGA |
| Leptin | AAGCCTCGCTCTACTCCACA | CATTCAGGGCTAAGGTCCAA |
| Mcp1 | TGGACCAGAACCAAGTGAGA | TGCTGAAGTCCTTAGGGTTGA |
| Gp91phox | CGAAGCCTTGGCTAAAACTCT | TCCTTGTTGAAGATGAAGTGGA |
| P22phox | GTGAGCAGTGGACTCCCATT | GTAGGTGGCTGCTTGATGGT |
| P47phox | TGTGACACCCTCTCACAGACA | GTCGCATTTTCCCTCCTTTA |
Quantification of gene expression (expressed as average copy number) was performed using the Step One Plus quantitative PCR machine (Applied Biosystems). A melting curve analysis was performed to confirm the absence of primer-dimers. Equal efficiency of the reverse transcription of RNA from all groups was confirmed through quantification of expression of the housekeeping gene β-actin. Expression did not differ between groups (effect of maternal diet, P = .72; [control: 153 ± 32; recuperated: 134 ± 35 average copy number]).
microRNA (miRNA) analysis
Putative miRNAs targeting the 3′ untranslated regions of p110β and Irs-1 were identified using the miRanda/mirSVR 1.2 (release 2010) and miRmap 3 (release 1.1) prediction algorithms, respectively. Candidates were ranked according to the strength of the predicted interaction, high conservation across species, and target site position along the mRNA (higher scores for proximal and distal locations). The final selection of the most highly ranked candidates to assess was based on common target sites within both the p110β and Irs-1 3′ untranslated regions as well as information regarding their expression in white adipose tissue. RNA was extracted using a Direct-zol RNA miniprep kit (Zymo Research) to retain RNA species with a minimal length of 17 nucleotides. RNA purity and concentration were determined by a spectrophotometric analysis on a NanoDrop ND-1000. RNA integrity was confirmed by denaturing agarose gel electrophoresis.
Statistical analysis
Data were analyzed using a two-way ANOVA with maternal diet and CoQ supplementation as the independent variables. Data are represented as mean ± SEM. A value of P < .05 was considered statistically significant. All statistical analyses were performed using Statistica 7 software (Statsoft Inc) except for the miRNA analysis in which GraphPad Prism 6.0 (GraphPad) was used. In all cases, n refers to the number of litters.
Results
Physical parameters, plasma CoQ, lipid profile, and insulin data
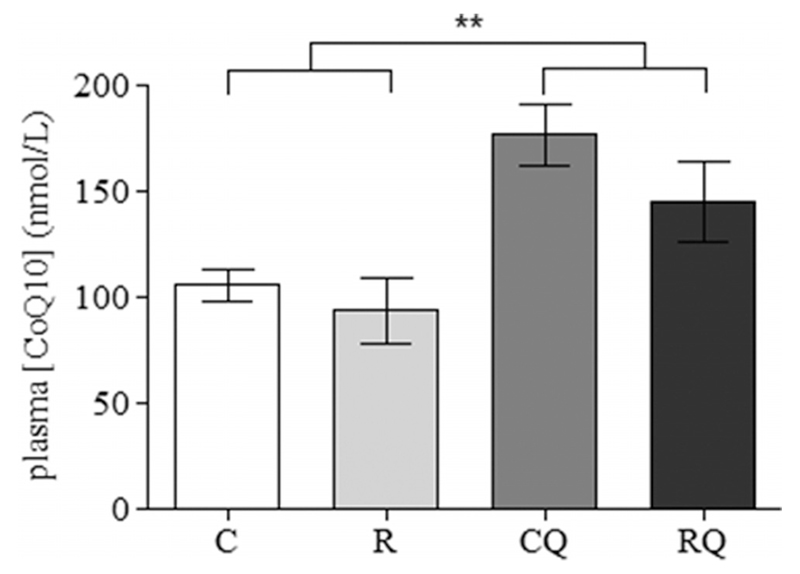
Recuperated offspring were born smaller ([6.3 ± 0.2 g vs 7.4 ± 0.2 g] [P < .001]) than control animals and remained smaller at postnatal day 7 ([13.4 ± 0.4 g vs 16.4 ± 0.4 g] [P < .001]). By postnatal days 14 (33.7 ± 0.5 g vs 33.3 ± 0.8 g) and 21 (52.2 ± 3.7 g vs 50.8 ± 1.2 g), the recuperated offspring had undergone accelerated catch-up growth and were therefore of similar weight to the control offspring. At 3 months of age, there was no significant effect of CoQ supplementation and maternal diet on offspring body or liver weights (Table 2). However, maternal diet increased (P < .05) epididymal fat pad weight (Table 2). There was no significant effect of CoQ supplementation on epididymal fat pad weight. Furthermore, no significant effect of maternal diet upon offspring plasma CoQ levels was observed. However, plasma CoQ levels were significantly (P < .01) increased by CoQ supplementation (Figure 1). Serum levels of fasting insulin, triglycerides, free fatty acids, total cholesterol, and fasting plasma glucose were unaltered by maternal diet or CoQ supplementation, and there was no interaction between these two variables on any of the serum levels (Table 3).
Table 2.
Effect of Poor Maternal Nutrition and Accelerated Postnatal Growth on Body and Organ Weights in 3-Month Male Rats
| Group | Body Weight, g | Epididymal Fat Pad Weight, g | Liver Weight, g |
|---|---|---|---|
| Control | 483 ± 12 | 8.0 ± 0.5 | 17.0 ± 0.6 |
| Recuperated | 478 ± 8.8 | 9.0 ± 0.6a | 17.2 ± 0.4 |
| Control CoQ | 476 ± 12.3 | 8.2 ± 0.5 | 16.7 ± 0.6 |
| Recuperated CoQ | 482 ± 6.0 | 9.6 ± 0.5a | 18.0 ± 0.6 |
P < .05 (overall effect of maternal diet).
Figure 1.
The effect of in utero protein restriction, accelerated postnatal growth, and CoQ supplementation upon plasma CoQ levels in 3-month-old male rats. Results are expressed as mean ± SEM.*, P < .01 (C and R vs CQ and RQ) (n = 10 per group). C, control; CQ, control CoQ; R, recuperated; RQ, recuperated CoQ.
Table 3.
Effect of Poor Maternal Nutrition and Accelerated Postnatal Growth on Blood Glucose, Insulin, and Lipid Profile in 3-Month Male Rats
| Group | FPG, mmol/L | Fasting Serum Insulin, mmol/L | Total Cholesterol, mmol/L | FFAs, mmol/L | Triglycerides, mmol/L |
|---|---|---|---|---|---|
| Control, g | 5.6 ± 0.2 | 95 ± 26 | 2.3 ± 0.1 | 1052 ± 99 | 1.7 ± 0.3 |
| Recuperated, g | 5.5 ± 0.2 | 49 ± 13 | 2.0 ± 0.1 | 990 ± 60 | 1.6 ± 0.1 |
| Control CoQ, g | 5.4 ± 0.1 | 47 ± 13 | 2.1 ± 0.1 | 1051 ± 94 | 1.8 ± 0.2 |
| Recuperated CoQ, g | 5.4 ± 0.1 | 47 ± 18 | 2.1 ± 0.2 | 976 ± 44 | 2.0 ± 0.4 |
Abbreviations: FFA, free fatty acid; FPG, fasting plasma glucose.
Insulin signaling: protein, mRNA, and miRNA analysis
Effect of maternal diet
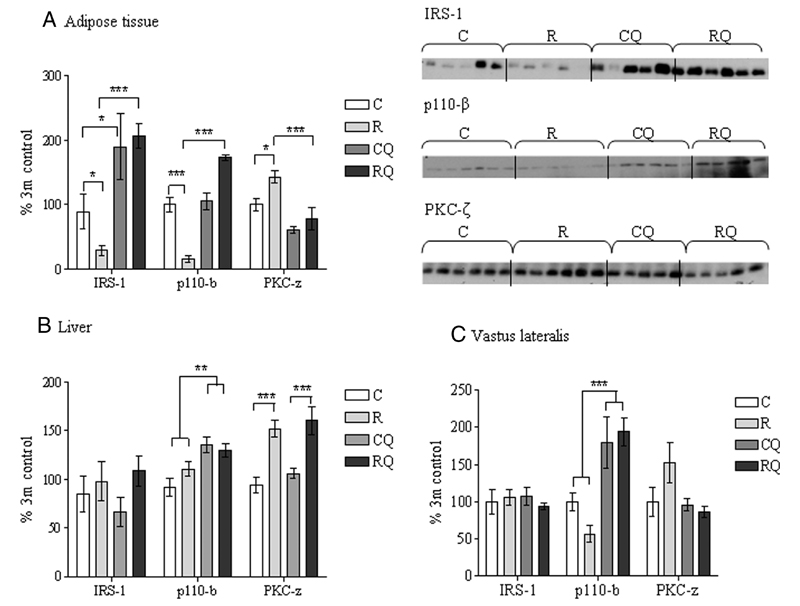
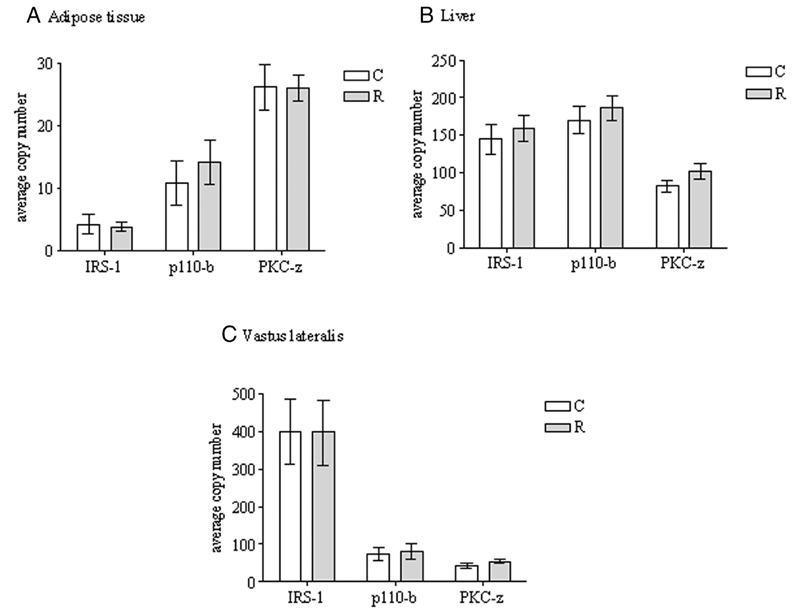
Protein expression of IRS-1 (P <.05) and p110-β (P <.001) were reduced in the epididymal fat pads of recuperated offspring compared with controls (Figure 2A). In contrast, protein kinase C (PKC)-ζ protein levels were significantly (P <.05) increased in the recuperated group (Figure 2A). Gene (mRNA) expression of these molecules were unaffected by maternal diet (Figure 3A). In liver and vastus lateralis skeletal muscle, IRS-1 and p110-β protein expression remained unaltered by maternal diet. However, liver PKC-ζ protein levels displayed an increase (P <.001) (Figure 2, B and C). The mRNA levels of these three genes were not different in either the liver or muscle of recuperated offspring when compared with controls (Figure 3, B and C). Protein expression of other insulin signaling molecules including IRβ, p85α, Akt-1, Akt-2, pAkt(Ser473), and glucose transporter (GLUT)-4 were unaffected by maternal diet in the epididymal fat, liver, and vastus lateralis muscle (Table 4).
Figure 2.
The effect of in utero protein restriction, accelerated postnatal growth, and CoQ supplementation upon protein expression of insulin signaling molecules in epididymal adipose tissue (A), liver (B), and vastus lateralis skeletal muscle (C) in 3-month-old male rats. Results are expressed as mean ± SEM. *, P < .01, ***, P < .001 (C vs R); *, P < .01, ***, P < .001 (R vs RQ) (n = 5–6 per group). C, control; CQ, control CoQ; R, recuperated; RQ, recuperated CoQ.
Figure 3.
The effect of in utero protein restriction and accelerated postnatal growth upon gene expression of insulin signaling molecules in epididymal adipose tissue (A), liver (B), and vastus lateralis skeletal muscle (C) in 3-month-old male rats. Results are expressed as mean ± SEM (n = 8 per group). C, control; R, recuperated.
Table 4.
Effect of Poor Maternal Nutrition and Accelerated Postnatal Growth on Insulin Signaling Molecule Protein Expression in 3-Month Male Rats
| Epididymal Fat |
Liver |
Vastus Lateralis |
||||
|---|---|---|---|---|---|---|
| Control | Recup | Control | Recup | Control | Recup | |
| IR-β | 100 ± 6 | 97 ± 9 | 100 ± 17 | 139 ± 17 | 100 ± 7 | 119 ± 10 |
| p85-α | 100 ± 7 | 101 ± 6 | 100 ± 11 | 111 ± 8 | 100 ± 7 | 112 ± 4 |
| pAkt (ser473) | 100 ± 20 | 130 ± 25 | 100 ± 10 | 110 ± 9 | 100 ± 17 | 141 ± 21 |
| GLUT-4 | 100 ± 25 | 146 ± 21 | Not detected | Not detected | 100 ± 17 | 107.7 ± 21 |
| Akt-1 | 100 ± 3 | 96 ± 3 | 100 ± 8 | 103 ± 6 | 100 ± 8 | 85 ± 11 |
| Akt-2 | 100 ± 5 | 96 ± 3 | 100 ± 4 | 115 ± 7 | 100 ± 6 | 107 ± 6 |
Abbreviation: Recup, recuperated.
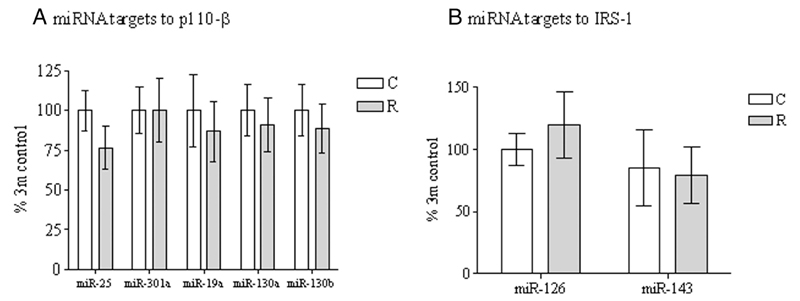
The programmed reduction in adipose tissue IRS-1 and p110-β in the absence of any differences in the corresponding mRNAs suggested that this effect was mediated by a posttranscriptional mechanism. However, we saw no effects of maternal diet on the five miRNAs predicted to regulate p110-β (miR-25, miR-301a, miR-19a, miR-130a, and miR-130b) or the two predicted to regulate IRS-1 (miR-126 and miR-143) (Figures 4, A and B).
Figure 4.
The effect of in utero protein restriction and accelerated postnatal growth upon miRNA targets to p110β (A) and IRS-1 (B). Results are expressed as mean ± SEM (n = 10 per group). C, control; R, recuperated.
Effect of CoQ supplementation
CoQ supplementation significantly increased both IRS-1 and p110-β protein levels in recuperated offspring epididymal fat (Table 5). In control offspring, CoQ supplementation had no effect on p110-β levels; however, supplementation increased IRS-1 protein expression (Figure 2A). CoQ supplementation increased (P < .001) p110-β protein expression in both liver and vastus lateralis skeletal muscle (Figure 2, B and C). CoQ supplementation significantly (P < .001) decreased the PKC-ζ protein levels in the recuperated adipose tissue compared with the effect on the control offspring (Figure 2A); however, PKC-ζ expression remained similar in the liver and vastus lateralis muscle (Figure 2, B and C).
Cytokine analysis
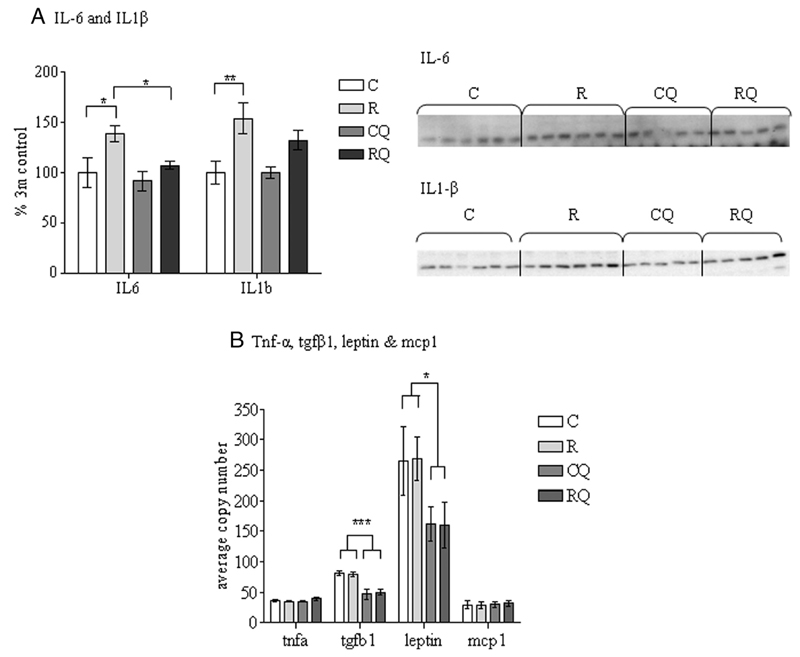
Adipose tissue protein expression of IL-6 (P <.05) and IL-β (P < .01) were increased in recuperated offspring (Figure 5A). CoQ supplementation significantly (P <.05) reduced IL-6 levels back to that of the controls; however, the IL-1β levels were unchanged by CoQ supplementation (Figure 5A). The mRNA levels of Tgfb1 and Lep were unaltered by maternal diet; however, CoQ supplementation reduced Tgf-β1 (P < .001) and Lep (P < .05) levels (Figure 5B). Tnf-α and Mcp-1 gene expression were similar between all groups (Figure 5B). There was no significant effect of maternal diet or CoQ10 supplementation on adiponectin (adipoq) mRNA in epididymal fat (control: 100% ± 26.6%; recuperated: 107% ± 18%; control CoQ: 91% ± 32%; recuperated CoQ: 73% ± 9%).
Figure 5.
The effect of in utero protein restriction, accelerated postnatal growth and CoQ supplementation upon cytokine protein and gene expression in epididymal adipose tissue in 3-month-male rats. Results are expressed as mean ± SEM (n = 5–6 per group). *, P < .05 (C vs R); *, P < .05 (R vs RQ). C, control; CQ, control CoQ; R, recuperated; RQ, recuperated CoQ. A, IL-6 and IL1β protein expression. B, Tnfα, Tgfβ1, Leptin, and Mcp1 gene expression.
Reactive oxygen species (ROS) and antioxidant defense capacity
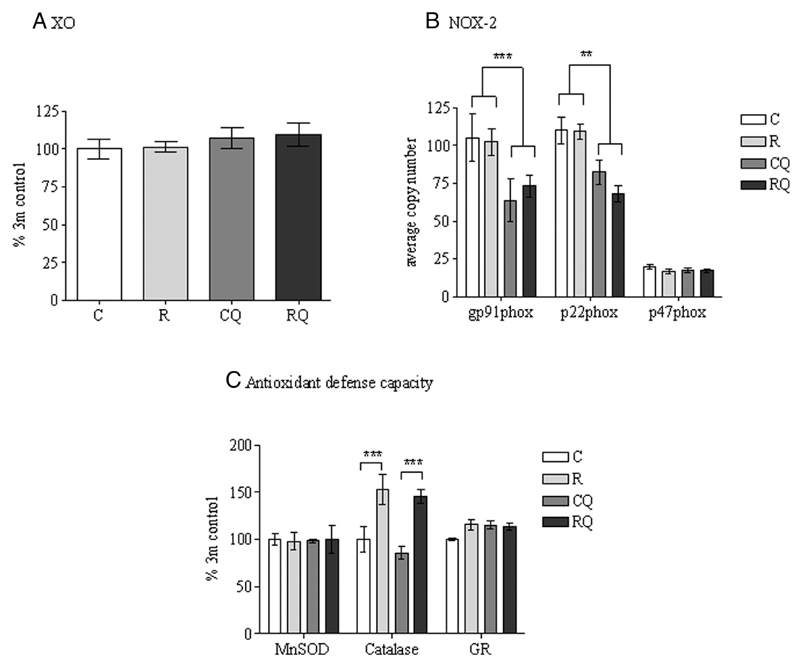
4-Hydroxynoneal (a marker of lipid peroxidation) and 3-nitrotyrosine (a measure of protein tyrosine nitration) were both undetectable in epididymal adipose tissue (data not shown). No significant effect of maternal diet or CoQ supplementation was observed upon xanthine oxidase protein expression (Figure 6A). The mRNA levels of components of nicotinamide adenine dinucleotide phosphate oxidase-2 (Gp91phox and P22phox) were unchanged by the maternal diet; however, CoQ supplementation reduced Gp91phox (P <.001) and P22phox (P <.01) levels, (Figure 6B). No significant effect of maternal diet or CoQ supplementation was observed upon manganese superoxide dismutase or glutathione reductase protein expression (Figure 6C). Catalase protein expression was significantly (P < .01) increased in recuperated offspring compared with controls (Figure 6C), and this effect remained unaffected by CoQ supplementation.
Figure 6.
The effect of in utero protein restriction, accelerated postnatal growth, and CoQ supplementation upon sources of ROS (A and B) and antioxidant defense capacity (C) in 3-month-old male rat epididymal adipose tissue. Results are expressed as mean ± SEM (n = 6 per group). *, P < .05 (C and R vs CQ and RQ). C, control; CQ, control CoQ; GR, glutathione reductase; MnSOD, manganese superoxide dismutase; NOX-2, nicotinamide adenine dinucleotide phosphate oxidase-2; R, recuperated; RQ, recuperated CoQ; XO, xanthine oxidase.
Discussion
Previous studies by us and others have demonstrated that adipose tissue is very vulnerable to the effects of developmental programming and is the site in which some of the earliest and most striking programming effects are observed (15, 30, 31). Consistent with these observations, in the current study, we have shown a reduction in IRS-1 and p110-β insulin signaling protein expression in response to suboptimal nutrition in utero and catch-up growth. In contrast, the maternal diet increased protein expression of PKC-ζ in adipose tissue and liver, which can negatively regulate insulin signaling via serine phosphorylation of IRS-1 (32). All of these differences are therefore consistent with an insulin-resistant phenotype. These alterations are present in adipose tissue prior to any such defects in liver or skeletal muscle. The differences in adipose tissue insulin signaling proteins are present at a time when animals display no difference in fasting glycemia or insulinemia. These defects are therefore not a consequence of metabolic dysfunction and more likely a contributor to the increased risk of insulin resistance in the recuperated offspring (33).
The changes in p110β and IRS-1 protein expression were not accompanied by altered mRNA expression, which suggests the involvement of posttranscriptional regulatory mechanisms. However, none of the seven candidate miRNAs predicted to regulate translation of IRS-1 and/or p110β were differentially expressed in offspring epididymal adipose tissue in response to the maternal diet. These could mean that a simple concentration change of complementary miRNAs is not the cause of altered protein expression of IRS-1 and p110-β or that other undefined miRNAs or other posttranscriptional mechanisms are involved.
A clinically relevant dietary supplementation of CoQ normalized the defects in adipose tissue insulin signaling protein expression in recuperated offspring. In addition, CoQ supplementation increased p110β protein expression in liver and skeletal muscle. This is consistent with previous studies showing that administration of a much higher dose of CoQ (20 mg/kg) affects insulin sensitivity and has antidiabetic properties via increasing the activity of phosphatidylinositol kinase in the liver and skeletal muscle of rats fed a high-fat, high-fructose diet (34). It is unclear how CoQ supplementation has its effects on insulin signaling protein expression. However, uptake of CoQ into most tissues is thought to be low (35) and a systemic effect is likely to be involved.
Inflammation is an important factor in the development of insulin resistance via inhibition of insulin signaling through activation of the inhibitory-κB kinase-β and c-Jun N-terminal kinase pathways (36). IL-6, IL-1β, TNF-α, monochemoattractant protein-1 (MCP-1), TGF-β1, and leptin are major cytokines and chemokines that can be secreted by dysfunctional adipocytes or infiltrated adipose tissue macrophages. These factors play a pivotal role in adipose tissue-induced, low-grade systemic inflammation and/or obesity. In the current study, we demonstrated increased protein expression of IL-6 and IL-1β in the epididymal fat pads of recuperated offspring. IL-6 is known to inhibit protein and mRNA expression of IRS-1 (37), suggesting that the increased IL-6 levels could, at least in part, explain the observed reduction in IRS-1 protein levels in recuperated animals. Furthermore, IL-1β is a key factor in mediating macrophage-induced insulin resistance in human adipocytes (38). However, Mcp-1 expression (a key chemokine that regulates migration and infiltration of monocytes and macrophages) was unaltered between groups, suggesting that any macrophage infiltration in recuperated adipose tissue could be via an Mcp-1-independent mechanism. Interestingly, this proinflammatory phenotype occurred in the absence of obesity, suggesting that maternal suboptimal nutrition is the driving factor for this effect in a manner that is independent of offspring adiposity. CoQ supplementation significantly reduced the IL-6 protein and Tgf-β1 and Lep mRNA levels, which is in agreement with findings in cardiac tissue (25), and CoQ is known to have antiinflammatory properties in mouse liver (39) and human plasma (40); however, to our knowledge this is the first time that CoQ has been demonstrated to have antiinflammatory properties in adipose tissue. This highlights the function of CoQ as an important antiinflammatory molecule and may partially explain some of our reported insulin-sensitizing effects of CoQ.
Overproduction of ROS is well known to contribute to the development of insulin resistance. In the current study, however, no evidence of an oxidative stress phenotype was observed in the adipose tissue of recuperated animals, given that Gp91phox, P22phox, and xanthine oxidase (all important sources of ROS) were unaltered by the maternal diet, and indicators of oxidative stress such as 4-hydroxynoneal and 3-nitrotyrosine were undetectable. This suggests that the manifestation of insulin resistance in recuperated offspring may be driven by inflammation and not oxidative stress at this age. Previously we have described an oxidative stress phenotype in other tissues from recuperated offspring adipose tissue at 3 months of age, including pancreatic islets (41) and the heart (23), and its absence in the adipose tissue highlights the tissue specificity of developmental programming on the manifestation of oxidative stress. mRNA levels of Gp91phox and P22phox were significantly decreased by CoQ supplementation, demonstrating that CoQ has a role as a potent antioxidant in adipose tissue.
In conclusion, a suboptimal maternal environment and rapid postnatal catch-up growth initiates the dysregulation of insulin signaling protein expression in epididymal adipose tissue that may contribute to later development of whole-body insulin resistance. This was associated with increased expression of inflammatory markers, independent of increased ROS generation and independent of obesity. CoQ supplementation ameliorated both insulin signaling dysfunction and inflammation in the adipose tissue of recuperated offspring, suggesting that CoQ’s antiinflammatory actions may play some role in modulating insulin resistance. Although the mechanism through which CoQ mediates these changes in adipose tissue is currently unknown, it is plausible that CoQ could act via antioxidant mechanisms to impact the activity of genes involved in the regulation of transcription or translation.
Acknowledgments
We thank Professor Ken Siddle for his helpful discussion.
Abbreviations
- CoQ
coenzyme Q
- GLUT
glucose transporter
- IRS
insulin receptor substrate
- LBW
low birth weight
- MCP-1
monochemoattractant protein-1
- miRNA
microRNA
- PKC
protein kinase C
- ROS
reactive oxygen species
Footnotes
This work was supported by The British Heart Foundation Grants PG/09/037/27387 and FS/09/029/27902; Medical Research Council Grant MC_UU_12012/4; and Diabetes United Kingdom Grant 12/0004508. S.E.O. is a member of the Medical Research Council Metabolic Diseases Unit. I.P.H. is supported by the Department of Health’s National Institute for Health Research Biomedical Research Centres funding scheme at University College London Hospitals/University College London.
Disclosure Summary: The authors have nothing to disclose.
References
- 1.Hales CN, Barker DJ, Clark PM, et al. Fetal and infant growth and impaired glucose tolerance at age 64. BMJ. 1991;303:1019–1022. doi: 10.1136/bmj.303.6809.1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ravelli ACJ, van der Meulen JHP, Michels RPJ, et al. Glucose tolerance in adults after prenatal exposure to famine. Lancet. 1998;351:173–177. doi: 10.1016/s0140-6736(97)07244-9. [DOI] [PubMed] [Google Scholar]
- 3.Ong KK, Petry CJ, Emmett PM, et al. Insulin sensitivity and secretion in normal children related to size at birth, postnatal growth and plasma insulin-like growth factor-I levels. Diabetologia. 2004;47:1064–1070. doi: 10.1007/s00125-004-1405-8. [DOI] [PubMed] [Google Scholar]
- 4.Barker DJ, Hales CN, Fall CH, Osmond C, Phipps K, Clark PM. Type 2 (non-insulin-dependent) diabetes mellitus, hypertension and hyperlipidaemia (syndrome X): relation to reduced fetal growth. Diabetologia. 1993;36:62–67. doi: 10.1007/BF00399095. [DOI] [PubMed] [Google Scholar]
- 5.Lithell HO, McKeigue PM, Berglund L, Mohsen R, Lithell UB, Leon DA. Relation of size at birth to non-insulin dependent diabetes and insulin concentrations in men aged 50–60 years. BMJ. 1996;312:406–410. doi: 10.1136/bmj.312.7028.406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Petry CJ, Dorling MW, Pawlak DB, Ozanne SE, Hales CN. Diabetes in old male offspring of rat dams fed a reduced protein diet. Int J Exp Diabetes Res. 2001;2:139–143. doi: 10.1155/EDR.2001.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ozanne SE, Jensen CB, Tingey KT, Storgaard H, Madsbad S, Vaag AA. Low birth weight is associated with specific changes in muscle insulin signalling protein expression. Diabetologia. 2005;48:547–552. doi: 10.1007/s00125-005-1669-7. [DOI] [PubMed] [Google Scholar]
- 8.Ozanne SE, Olsen GE, Hansen LL, et al. Early growth restriction leads to down regulation of protein kinase C ζ and insulin resistance in skeletal muscle. J Endocrinol. 2003;177:235–241. doi: 10.1677/joe.0.1770235. [DOI] [PubMed] [Google Scholar]
- 9.Ozanne SE, Jensen CB, Tingey KJ, et al. Decreased protein levels of key insulin signalling molecules in adipose tissue from young men with a low birth weight: potential link to increased risk of diabetes? Diabetologia. 2006;49:2993–2999. doi: 10.1007/s00125-006-0466-2. [DOI] [PubMed] [Google Scholar]
- 10.Crowther NJ, Cameron N, Trusler J, Gray IP. Association between poor glucose tolerance and rapid postnatal weight gain in seven year old children. Diabetologia. 1998;41:1163–1167. doi: 10.1007/s001250051046. [DOI] [PubMed] [Google Scholar]
- 11.Mericq V, Ong KK, Bazaes R, et al. Longitudinal changes in insulin sensitivity and secretion from birth to age three years in small- and appropriate-for-gestational-age children. Diabetologia. 2005;48:2609–2614. doi: 10.1007/s00125-005-0036-z. [DOI] [PubMed] [Google Scholar]
- 12.Dellschaft NS, Alexandre-Gouabau MC, Gardner DS, et al. Effect of pre- and postnatal growth and post-weaning activity on glucose metabolism in the offspring. J Endocrinol. 2015;224:171–182. doi: 10.1530/JOE-14-0600. [DOI] [PubMed] [Google Scholar]
- 13.Forsen T, Eriksson J, Tuomilehto J, Reunanen A, Osmond C, Barker D. The fetal and childhood growth of persons who develop type 2 diabetes. Ann Intern Med. 2000;133:176–182. doi: 10.7326/0003-4819-133-3-200008010-00008. [DOI] [PubMed] [Google Scholar]
- 14.Yajnik CS. Early life origins of insulin resistance and type 2 diabetes in India and other Asian countries. J Nutr. 2004;134:205–210. doi: 10.1093/jn/134.1.205. [DOI] [PubMed] [Google Scholar]
- 15.Berends LM, Fernandez-Twinn DS, Martin-Gronert MS, Ozanne SE. Catch-up growth following intrauterine growth-restriction programmes an insulin-resistant phenotype in adipose tissue. Int J Obes. 2013;8:1051–1057. doi: 10.1038/ijo.2012.196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Henrikse EJ, Diamond-Stanic MK, Marchionne EM. Oxidative stress and the etiology of insulin resistance and type 2 diabetes. Free Radic Biol Med. 2011;51:991–999. doi: 10.1016/j.freeradbiomed.2010.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Torres-Leal FL, Fonseca-Alaniz MH, Cunha de Oliveira A, Alonso-Vale MIC. Insulin resistance. In: Sarika A, editor. Adipose Tissue Inflammation and Insulin Resistance, Insulin Resistance. Rijeka, Croatia: InTech; 2012. [Google Scholar]
- 18.Tarry-Adkins JL, Ozanne SE. The impact of early nutrition on the ageing trajectory. Proc Nutr Soc. 2014;2:289–301. doi: 10.1017/S002966511300387X. [DOI] [PubMed] [Google Scholar]
- 19.Sen S, Simmons RA. Maternal antioxidant supplementation prevents adiposity in Western diet fed rats. Diabetes. 2010;59:3058–3065. doi: 10.2337/db10-0301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Giussani DA, Camm EJ, Nui Y, et al. Developmental programming of cardiovascular dysfunction by prenatal hypoxia and oxidative stress. PLoS One. 2012;2:e31017. doi: 10.1371/journal.pone.0031017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cambonie G, Comte B, Yzdorczyk C, et al. Antenatal oxidant prevents adult hypertension, vascular dysfunction, and microvascular rarefaction associated with in utero exposure to a low-protein diet. Am J Regul Interg Comp Physiol. 2007;292:R1236–R1245. doi: 10.1152/ajpregu.00227.2006. [DOI] [PubMed] [Google Scholar]
- 22.Tarry-Adkins JL, Fernandez-Twinn DS, Chen JH, et al. Nutritional programming of coenzyme Q: potential for prevention and intervention? FASEB J. 2014;28:5398–5405. doi: 10.1096/fj.14-259473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tarry-Adkins JL, Blackmore HL, Martin-Gronert MS, et al. Coenzyme Q prevents accelerated cardiac aging in a rat model of poor maternal nutrition and accelerated postnatal growth. Mol Metab. 2013;2:480–490. doi: 10.1016/j.molmet.2013.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Littarru GP, Tiano L. Bioenergetic and antioxidant properties of coenzyme Q10: recent developments. Mol Biotechnol. 2007;37:31–37. doi: 10.1007/s12033-007-0052-y. [DOI] [PubMed] [Google Scholar]
- 25.Lee BJ, Tseng YF, Yen CH, Lin PT. Effects of coenzyme Q10 supplementation (300 mg/day) on antioxidation and anti-inflammation in coronary artery disease patients during statins therapy: a randomized, placebo-controlled trial. Nutr J. 2013;12(1):142. doi: 10.1186/1475-2891-12-142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hidaka T, Fuji K, Funahashi I, Fukutomi N, Hoseo K. Safety assessment of coenzyme Q10 (CoQ10) Biofactors. 2008;32:199–208. doi: 10.1002/biof.5520320124. [DOI] [PubMed] [Google Scholar]
- 27.Ikematsu H, Nakamura K, Harashima S, Fujii K, Fukutomi N. Safety assessment of coenzyme Q10 (Kaneka Q10) in healthy subjects: a double-blind, randomized placebo controlled trial. Regul Toxicol Pharmacol. 2006;44:212–218. doi: 10.1016/j.yrtph.2005.12.002. [DOI] [PubMed] [Google Scholar]
- 28.Snoeck A, Remacle C, Reusens B, Hoett JJ. Effect of low protein diet during pregnancy on the fetal rat endocrine pancreas. Biol Neonate. 1990;57:107–118. doi: 10.1159/000243170. [DOI] [PubMed] [Google Scholar]
- 29.Tarry-Adkins JL, Joles JA, Chen JH, et al. Protein restriction in lactation confers neproprotective effects in the male rat and is associated with increased antioxidant protein expression. Am J Physiol Regul Integr Comp Physiol. 2007;293:R1259–R1266. doi: 10.1152/ajpregu.00231.2007. [DOI] [PubMed] [Google Scholar]
- 30.Mackay H, Khazall R, Patterson ZR, Wellman M, Abizaid A. Rats perinatally exposed to food restriction and high-fat diet show differences in adipose tissue gene expression under chronic caloric restriction. Adipocyte. 2013;4:237–245. doi: 10.4161/adip.24752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schenk S, Saberi M, Olefsky JM. Insulin sensitivity: modulation by nutrients and inflammation. J Clin Invest. 2008;118:2992–3002. doi: 10.1172/JCI34260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Guilherme A, Virbasius JV, Puri V, Czech MP. Adipocyte dysfunctions linking obesity to insulin resistance and type 2 diabetes. Nat Rev Mol Cell Biol. 2008;5:367–377. doi: 10.1038/nrm2391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ravichandran LV, Esposito DL, Chen J, Quon MJ. Protein kinase C-ζ phosphorylates insulin receptor substrate-1 and impairs its ability to activate phosphatidylinositol 3-kinase in response to insulin. J Biol Chem. 2001;276:3543–3549. doi: 10.1074/jbc.M007231200. [DOI] [PubMed] [Google Scholar]
- 34.Amin MM, Asaad GF, Abdel-Salem RM, El-Abhar HS, Arbid MS. Novel CoQ10 antidiabetic mechanisms underlie its positive effect: modulation of insulin and adiponectin receptors, tyrosine kinase, PI3K, glucose transporters, sRAGE, and visfatin in insulin resistant/diabetic rats. PLoS One. 2014;9:e89169. doi: 10.1371/journal.pone.0089169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lonnrot K, Holm P, Lagerstedt A, Huhtala H, Alho H. The effects of lifelong ubiquinone supplementation on the CoQ9 and CoQ10 tissue concentrations and lifespan of male rats and mice. Biochem Mol Biol Int. 1998;44:727–737. doi: 10.1080/15216549800201772. [DOI] [PubMed] [Google Scholar]
- 36.McNelis JC, Olefsky JM. Macrophages, immunity and metabolic disease. Immunity. 2014;41:36–48. doi: 10.1016/j.immuni.2014.05.010. [DOI] [PubMed] [Google Scholar]
- 37.Rotter V, Nagaev I, Smith U. Interleukin-6 (IL-6) induces insulin resistance in 3T3-L1 adipocytes and is, like IL-8 and tumor necrosis factor-α, overexpressed in human fat cells from insulin-resistant subjects. J Biol Chem. 2003;46:45777–45784. doi: 10.1074/jbc.M301977200. [DOI] [PubMed] [Google Scholar]
- 38.Gao D, Madi M, Ding C, et al. Interleukin-1-β mediates macrophage-induced impairment of insulin signaling in human primary adipocytes. Am J Physiol Endocrinol Metab. 2014;307:E289–E304. doi: 10.1152/ajpendo.00430.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sohet FM, Neyrinck AM, Pachikian BD, et al. Coenzyme Q10 supplementation lowers hepatic oxidative stress and inflammation associated with diet-induced obesity in mice. Biochem Pharmacol. 2009;78:1391–1400. doi: 10.1016/j.bcp.2009.07.008. [DOI] [PubMed] [Google Scholar]
- 40.Sanoobar M, Eghtesadi S, Azimi A, et al. Coenzyme Q10 supplementation ameliorates inflammatory markers in patients with multiple sclerosis: a double blind, placebo, controlled randomised clinical trial. Nutr Neurosci. 2015;18:169–176. doi: 10.1179/1476830513Y.0000000106. [DOI] [PubMed] [Google Scholar]
- 41.Tarry-Adkins JL, Chen JH, Smith NS, Jones RH, Cherif H, Ozanne SE. Poor maternal nutrition followed by accelerated postnatal growth leads to telomere shortening and increased markers of cell senescence in rat islets. FASEB J. 2009;23:1521–1528. doi: 10.1096/fj.08-122796. [DOI] [PubMed] [Google Scholar]