Abstract
Post-translational modification of proteins by ubiquitin (Ub) and Ub-like modifiers regulates various aspects of DNA replication. We previously showed that the chromatin around replisomes is rich in SUMO and depleted in Ub, whereas an opposite pattern is observed in mature chromatin. How this SUMO-rich/Ub-low environment is maintained at sites of DNA replication is not known. Here we identify USP7 as a replisome-enriched SUMO deubiquitinase that is essential for DNA replication. By acting on SUMO and SUMOylated proteins, USP7 counteracts their ubiquitination. Chemical inhibition or genetic deletion of USP7 leads to the accumulation of Ub on SUMOylated proteins, which are displaced to chromatin away from replisomes. Our findings provide a model to explain the differential accumulation of SUMO and Ub at replication forks, and identify an essential role of USP7 in DNA replication that should be taken into account for the use of USP7 inhibitors as anticancer agents.
Introduction
Post-translational modifications (PTM) play essential roles in the regulation of biological reactions. Among them, Ub and Ub-like modifiers such as SUMO have an increasingly prominent role in DNA replication and repair, where they work in a coordinated manner. This is best exemplified by the clamp loader PCNA, which is ubiquitinated and/or SUMOylated in response to DNA damage to mediate the loading of alternative DNA Polymerases or to promote template switching at replication forks1,2. Moreover, PCNA assembles an E3 ubiquitin ligase complex that regulates origin licensing and limits re-replication3. Finally, PCNA ubiquitination is counteracted by the action of the Ubiquitin Specific Protease (USP) USP1, which deubiquitinates PCNA to prevent excessive loading of translesion synthesis (TLS) polymerases4 and also acts on FANCD2-FANCI complexes to limit DNA damage responses5,6. Nevertheless, PCNA is just one example that illustrates the dynamic nature of Ub and SUMO modifications at sites of DNA replication.
SUMO targeted Ub ligases (STUbLs) are important mediators of the interplay between SUMO and Ub7. In mammals, RNF4 is the main chromatin-related STUbL with roles in double strand break (DSB) repair and DNA replication8–12. Of note, chromatin imposes a barrier for the access of PTM modifiers and readers13 and recent studies have revealed a role for the chaperone p97 in extracting ubiquitinated substrates from chromatin. Together with its cofactor DVC1/Spartan, p97 extracts TLS polymerases bound to ubiquitinated PCNA from chromatin during DNA replication14,15 and removes active FANCI-FANCD2 complexes after genotoxic stress9. In what regards SUMO deubiquitinases (SDUBs), and while writing this manuscript, USP11 has been identified as a SUMO2 deubiquitinase that acts on SUMOylated PML16. In contrast to STUbLs, no SDUBs with an active role on DNA replication or repair have been identified.
Besides the specific roles of individual PTMs, it has been proposed that SUMO regulates signaling networks through the collective modification of factors in a common pathway17–19. Supporting this view, proteomic analyses have revealed global changes in SUMOylation in response to stress conditions, including replication stress20–24. Through the method of isolation of proteins on nascent DNA (iPOND)25 coupled to Mass Spectrometry (MS), we recently showed that chromatin surrounding replisomes shows an overall enrichment of SUMO peptides, which occurs concomitantly with lower levels of ubiquitination26. A comprehensive iPOND-MS analysis has recently confirmed this observation27. How such a SUMO-rich/Ub-low chromatin is built around replisomes remains unknown, but we reasoned that at least the low levels of Ub could be related to the presence of deubiquitinating activities at replication forks.
Here we investigated the role of USPs, and USP7 in particular, in DNA replication. Early work showed that depletion of USP7 leads to a loss of MDM2 and a concomitant upregulation of p5328–30. Based on these findings, chemical inhibitors of USP7 are being developed as anticancer agents due to their capacity to stabilize p5331–33. However, USP7 is essential for mouse development in a manner that cannot be rescued by p53 deletion34, and there is a growing number of targets identified for USP7 besides MDM2 and p5335–46. In addition, recent evidence has also shown a role for USP7 during DNA replication termination through the unloading of the MCM complex47. Our data show that USP7 is enriched at sites of DNA replication, and necessary for replication fork progression and the firing of new origins. Proteomic identification of USP7 targets revealed several peptides on the surface of SUMO2, and our in vitro and in vivo data confirmed the role of USP7 as a replication-associated SDUB. Importantly, USP7 inhibition or genetic deletion abrogates the local concentration of SUMO at replication forks, identifying USP7 as the first factor that regulates the overall concentrations of Ub and SUMO PTMs at sites of DNA replication.
Results
Replisome-associated USP7 activity is essential for DNA replication
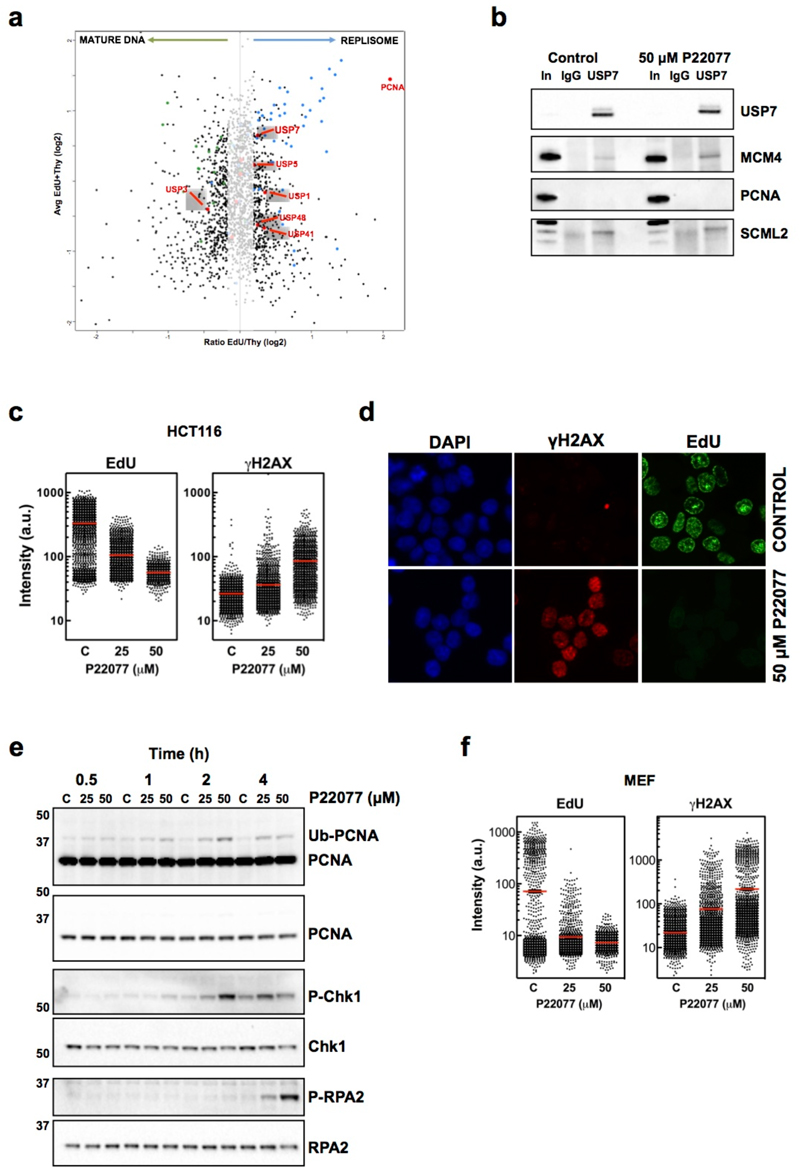
To determine the relative abundance of USPs around replisomes we performed iPOND experiments coupled to isobaric tag for absolute and relative quantitation (iTRAQ) and evaluated their presence in nascent and mature chromatin (Fig. 1a, red spots; Supplementary Table 1). Among the different USPs found enriched in the replisome fraction, USP7 called our attention for several reasons. As mentioned, deletion of USP7 is essential even at the embryonic stem (ES) cell stage, due to impaired proliferation34. Second, USP7 regulates the stability of known components of the replisome like UHRF1 and DNMT148–50, and proteomic analyses have revealed an interaction between USP7 and members of the MCM complex51. We confirmed that USP7 immunoprecipitation recovered MCM4, an interaction that was not affected by USP7 inhibition (Fig. 1b). The interaction was specific, since no PCNA was detected in USP7 pull-downs. As a control, USP7 interacted with SCML240. Together, these data show that USP7 is a replisome-associated USP.
FIG. 1. USP7 IS ENRICHED AROUND REPLISOMES AND ESSENTIAL FOR DNA REPLICATION.
(a) Nascent (EdU) and mature (Thy) chromatin fractions were isolated from HeLa cells by iPOND and analyzed by iTRAQ. The enrichment in the EdU vs the Thy fraction for each protein was plotted against the mean abundance of that protein in both samples. The shaded region includes proteins that are not significantly enriched in mature or nascent chromatin. Proteins previously identified in mature or nascent chromatin are represented as green and blue dots, respectively. USPs are highlighted as red dots, and the name of significantly enriched USPs in either fraction indicated.
(b) Nuclear extracts of control treated HCT116 cells or cells treated with 50 µM P22077 for 4h were immunoprecipitated with antibodies against USP7 or non-specific IgG. Pulled-down material was analyzed by WB with antibodies against USP7, MCM4, PCNA or SCML2. 5% of the input is shown as control (In).
(c) HTM of EdU and γH2AX levels per individual nucleus in response to USP7 inhibition. HCT116 cells were treated with increasing concentrations of P22077 for 4h or DMSO as a control (C). Nuclei were counterstained with DAPI. Representative images of the IF are shown in (d). The experiment was repeated 4 times and one representative experiment is shown.
(e) Whole cell extracts of HCT116 cells treated with increasing concentrations of P22077 or DMSO as a control (C) for the indicated time were analyzed by WB with antibodies against PCNA, total and phosphorylated Chk1 (S345), and total and phosphorylated RPA2 (S4/S8). The experiment was repeated three times and one representative example is shown.
(f) MEF were treated with increasing concentrations of P22077 for 4h or DMSO as a control (C) and the levels of EdU and γH2AX per nucleus were measured as in (c). The experiment was repeated three times and one representative example is shown.
Next, we decided to explore the function of USP7 in DNA replication using the specific inhibitor P22077, which has been shown to preferentially inhibit USP7 in vitro and in vivo, with some in vitro activity against USP4731. Treatment of HCT116 cells with P22077 led to a dose-dependent reduction in EdU incorporation together with an increase in γH2AX measured by high throughput microscopy (HTM) (Fig. 1c-d). Time-course experiments revealed a fast blockade of replication (Supplementary Fig. 1a) that precedes γH2AX accumulation (Supplementary Fig. 1b). These data suggest that USP7 inhibition generates replicative stress. Consistently, treatment with P22077 led to PCNA mono-ubiquitination followed by CHK1 phosphorylation (Fig. 1e). ATM-dependent phosphorylation of RPA was detected at later times, indicative of the formation of DSBs (Fig. 1e). Whereas, as expected, p53 ubiquitination decreased, no changes in the levels of p21, p53 or MCM4 were observed at these early times (Supplementary Fig. 1c). Moreover, USP7 inhibition led to a similar reduction of EdU and increase of γH2AX in wild-type (WT) and p53-deficient mouse embryonic fibroblasts (MEFs) (Fig. 1f and Supplementary Fig. 2), confirming that the effects of USP7 inhibition on DNA replication are independent of p53. Two independent USP7 inhibitors, P5091 (structurally related to P22077)32 and HBX19818 (structurally unrelated to P22077 but also selective towards USP7)33, reduced EdU incorporation and led to replication stress in HCT116 cells, confirming that the effect was not a particularity of P22077 (Supplementary Fig. 3). Importantly, USP7 inhibition generated replication stress in both WT and RAD18-deficient HCT116 cells that cannot ubiquitinate PCNA (Supplementary Fig. 4). Collectively, these data establish that USP7 is present in the vicinity of replisomes and that its activity is required to sustain active DNA replication and prevent replication stress, independently of PCNA ubiquitination or P53.
USP7 regulates fork progression and origin firing
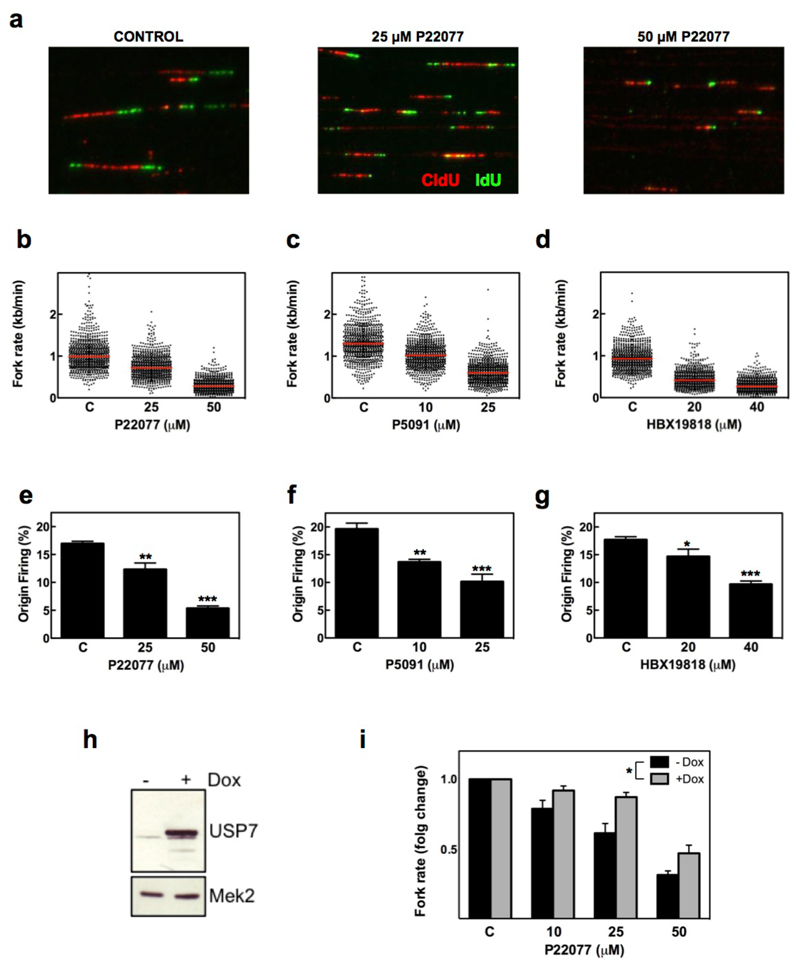
To understand how USP7 regulates DNA replication we analyzed fork progression and origin activity in stretched DNA fibers. HCT116 cells were cultured in the presence of USP7 inhibitors for 15 min, and then sequentially treated with CldU and IdU to label replicating DNA. Treatment with three independent USP7 inhibitors led to a strong reduction in the rate of ongoing replication forks (Fig. 2a-d). Challenges that reduce the speed of replication forks, such as oncogene overexpression or exposure to genotoxic drugs, lead to the activation of additional “dormant” origins as compensation52,53. In contrast, USP7 inhibition with any of the 3 drugs not only decreased fork speed but also reduced the firing of new origins (Fig. 2e-g). Moreover, using 293T-REx cells with doxycycline-inducible overexpression of USP7 (Fig. 2h), we observed that the percentage of cells in S phase and origin firing increased 24 h after USP7 induction (Supplementary Fig. 5). The use of these cells showed that USP7 expression significantly diminished the reduction in fork speed seen in response to P22077, further supporting that the effects of the inhibitors in DNA replication were due to USP7 (Fig. 2i). Hence, USP7 activity promotes origin firing and replication fork progression in human cells.
FIG. 2. USP7 INHIBITION REDUCES REPLICATION FORK SPEED AND ORIGIN FIRING.
(a-g) HCT116 cells were sequentially incubated with CldU and IdU in the presence of increasing concentrations of P22077, P5091, HBX19818 or DMSO as a control. Representative images of stretched DNA fibers visualized by IF for CldU (red) and IdU (green) are shown in (a). Fork rate (b-d) and percentage of new origin firing (e-g) were measured and quantified after the different treatments. All the experiments were repeated three times; the pool of the three experiments (fork rate) or the average (origin firing) is shown. * p<0.05; ** p<0.01; *** p<0.001, t- test.
(h) 293T-REx cells with inducible expression of OneStrep-FLAG-HA-USP7 were treated with 1 µg/ml Doxycycline (+) or EtOH as a control (-) for 1 day. Whole cell extracts were analyzed by WB with antibodies against USP7 or MEK2 as a loading control.
(i) DNA fibers were extracted from control (black bars) and doxycycline treated (grey bars) 293T-REx-USP7 cells treated with increasing amounts of P22077. Replication fork rates were measured in three independent experiments and the average normalized to the DMSO treated sample. *p<0.05, two-way ANOVA.
Proteomic analyses of ubiquitination identify SUMO2 as a USP7 target
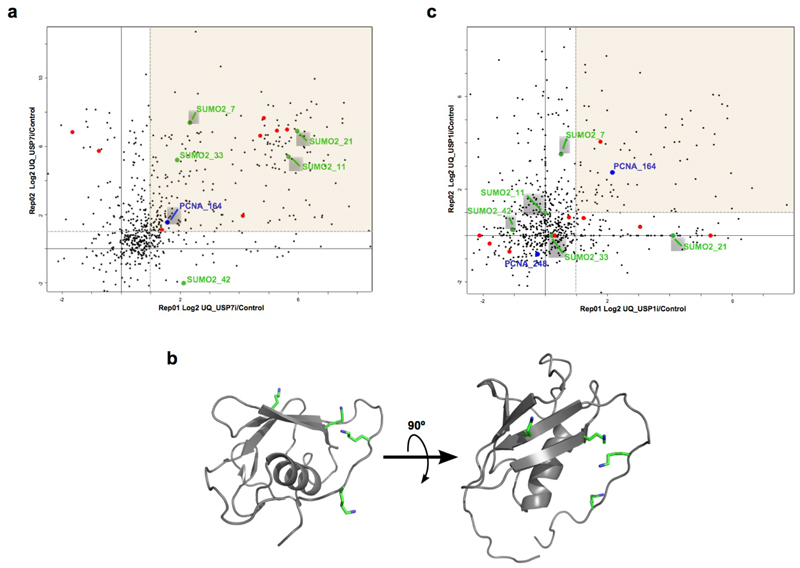
To investigate how USP7 regulates DNA replication we decided to look for new substrates of this DUB on chromatin. We employed a recently developed pipeline based on the immunoprecipitation of ubiquitinated peptides with an antibody that recognizes the di-Gly remnant left after tryptic digestion of the ubiquitin moiety (di-Gly method)54. We treated HCT116 cells with P22077 for 2 h, purified the chromatin fraction, and analyzed ubiquitinated peptides using di-Gly-proteomics (Supplementary Table 2). At this time point, while DNA replication is severely affected there is still no phosphorylation of RPA2, minimizing changes that are secondary to fork collapse and DSB formation. The shaded region in Fig. 3a shows the ubiquitinated peptides that are enriched upon USP7 inhibition in two different biological replicates. As expected, this region includes several known targets of USP7 (Fig. 3a, orange dots) and, consistent with our results, it also showed a mild increase of PCNA ubiquitination at K164 (Fig. 3a, blue dot). Interestingly, among the ubiquitination sites enriched upon USP7 inhibition we also detected 4 different lysines in SUMO2 (K7, K11, K21 and K33) (Fig. 3a, green dots), all of which mapped to the surface of the protein (Fig. 3b).
FIG. 3. IDENTIFICATION OF SUMO2 AS A USP7 TARGET.
(a) Scatter plot of changes in individual ubiquitinated peptide abundance from chromatin fractions of HCT116 cells treated with 50 µM P22077 for 2h compared to DMSO treated samples. Changes in two biological replicates are shown, with peptides showing a consistent trend being enclosed in the shaded region. Known USP7 substrate sites are highlighted in orange, PCNA is shown in blue and ubiquitination sites on SUMO2 are indicated in green.
(b) 3D structure of SUMO2 highlighting the lysines that are found ubiquitinated after USP7 inhibition.
(c) Same as in (b), but HCT116 cells were treated with 30 µM ML323 for 6h.
As a specificity control, we carried out similar experiments in chromatin extracts from HCT116 cells treated with the USP1 inhibitor ML32355. USP1, the DUb that removes PCNA mono-ubiquitination4, was also found enriched at replisomes in our iPOND experiments (Fig. 1a), consistent with previous findings56. Furthermore, USP1 inhibition strongly increased PCNA-K164 ubiquitination in two different biological replicates (Fig. 3c, blue dots), while no effect was observed on most known USP7 substrates (Fig. 3c, orange dots). Importantly, none of the lysines on SUMO2 with increased ubiquitination in response to P22077 were ubiquitinated after USP1 inhibition (Fig. 3c, green dots). These data indicate that the effect of USP7 inhibition on SUMO2 is selective and not a general action of USPs present in chromatin.
USP7 deubiquitinates SUMO2 in vitro and in vivo
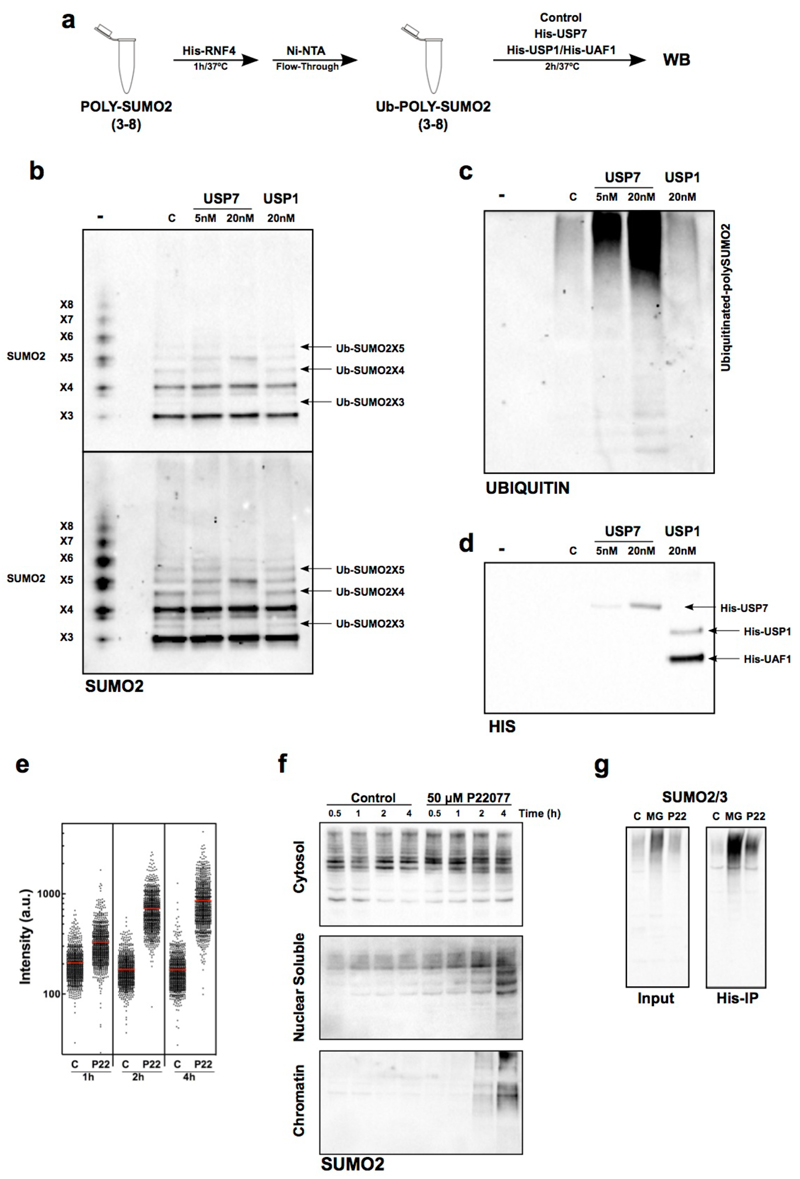
We next evaluated whether USP7 was able to deubiquitinate SUMO2 chains in vitro. Poly-SUMO2 (X3-8) chains were ubiquitinated using recombinant His-RNF4 (Fig. 4a). After purifying SUMO2 chains with Ni-NTA agarose chromatography, ubiquitinated poly-SUMO2 was incubated with recombinant USP7 or USP1/UAF1 and the products of the reactions analyzed by Western blot (WB) (Fig. 4a-d). His-RNF4 preferentially polyubiquitinated long poly-SUMO2 chains as previously described57 (Fig. 4b, compare lanes – to C), but also catalyzed mono-ubiquitination of chains with 3 to 5 SUMO2 molecules (Ub-SUMO2X3-5) (Fig. 4b, arrows). Increasing amounts of USP7 efficiently deubiquitinated Ub-SUMO2X3-5, while no effect was seen in the presence of USP1 (Fig. 4b). Importantly, the identity of ubiquitinated SUMO2 bands was confirmed by mass spectrometry, which identified SUMO2 peptides containing the diGly remnant of ubiquitin that arises upon tryptic digestion (Supplementary Fig. 6). Moreover, some of the lysines of SUMO2 that underwent USP7-dependent deubiquitination in vitro corresponded to the ones identified as increased in ubiquitination upon USP7 inhibition in vivo (K11 and K33).
FIG. 4. USP7 DEUBIQUITINATES SUMO2 IN VITRO AND IN VIVO.
(a) Scheme of the biochemical assay used to test SUMO2 deubiquitination in vitro.
(b-d) In vitro ubiquitinated poly-SUMO2 chains were incubated with His-USP7, His-USP1/His-UAF1 or no enzyme (C). Non-ubiquitinated poly-SUMO2 chains are shown as reference (-). Reaction products were analyzed by WB with antibodies against SUMO2/3 (b), ubiquitin (c) or His (d). One out of two representative experiments is shown. Note that the loss of ubiquitinated poly-SUMO2 bands occurs concomitantly to the increase in signal in the bands corresponding to unmodified SUMO2 polymers.
(e) The intensity of chromatin-bound SUMO2/3 per individual nucleus upon USP7 inhibition as quantified by HTM. U2OS cells treated with 50 µM P22077 or DMSO (C) for the indicated times were fixed and stained for SUMO2/3 after pre-extraction of the nuclear-soluble proteins. DAPI was used to stain nuclei.
(f) Cytosolic, nuclear-soluble and chromatin fractions were purified from HCT116 cells treated with 50 µM P22077 for the indicated times. The presence of SUMOylated proteins was measured by WB with antibodies against SUMO2/3. The experiment was repeated twice and one representative image is shown.
(g) 293T cells were transiently transfected with a plasmid encoding His-ubiquitin and treated with 5 µM MG132 (MG), 50 µM P22077 (P22) or DMSO (C) for 4 h. Nuclear extracts were obtained and ubiquitin containing proteins were purified using a Ni-NTA resin. The presence of SUMOylated proteins in the input or the His-IP was followed by WB with specific antibodies for SUMO2/3.
Highly ubiquitinated poly-SUMO2 chains accumulated as high molecular weight species that poorly entered the gel (Fig. 4c, lane C). The incubation with USP7, but not USP1, also reduced the size of these highly ubiquitinated species (Fig. 4c), indicating that USP7 can target both mono- and polyubiquitinated SUMO2 chains. Consistently, recombinant USP7 (and USP1) cleaved K48 and K63 poly-Ub chains (Supplementary Fig. 7a). In contrast, USP7 had no activity against non-ubiquitinated poly-SUMO2 chains (Supplementary Fig. 7b). Thus, our results confirm that USP7 is a SUMO2 deubiquitinase that removes mono and poly-ubiquitin from SUMO2 chains.
Finally, to determine whether USP7 also deubiquitinates SUMO2 in vivo, we first treated U2OS cells with P22077 and measured the levels of chromatin-bound SUMO2/3 by IF after removal of the nuclear-soluble proteins by extraction with a mild detergent. These analyses revealed an increase in chromatin-bound SUMO2 upon USP7 inhibition (Fig. 4e, Supplementary Fig. 7c). Subcellular fractionation of HCT116 extracts followed by WB confirmed that the action of USP7 is restricted to chromatin. While no change was detected in the cytosol, an accumulation of SUMO2-modified proteins was observed on the nuclear soluble and, particularly, chromatin fractions after USP7 inhibition (Fig. 4f). Importantly, this accumulation of chromatin-bound SUMO2 is not observed in response to hydroxyurea, indicating that it is a specific effect of USP7 inhibition and not a general response to replication stress (Supplementary Fig. 8). To determine whether SUMO2-modified proteins that accumulated upon USP7 inhibition were also ubiquitinated, we transfected 293T cells with a plasmid encoding His-Ubiquitin. After immunoprecipitation of ubiquitinated proteins using a Ni-NTA resin, we measured the levels of SUMOylated proteins in the input and the pull-down (Fig. 4g). As expected, the inhibition of the proteasome with MG132 led to a strong accumulation of SUMOylated proteins in the input, while inhibition of USP7 had a much milder effect (Fig. 4g, left). When the pull-down material was analyzed, we observed a strong increase in SUMOylated and ubiquitinated proteins both after the inhibition of the proteasome or USP7 (Fig. 4g, right). Together, these data show that USP7 is a SUMO DUB, and its inhibition results in the accumulation of chromatin-bound SUMOylated proteins that are also ubiquitinated.
The segregase p97 cooperates with USP7 on chromatin
Whereas ubiquitination usually targets proteins for degradation we observed that USP7 inhibition leads to the accumulation of ubiquitinated proteins on chromatin. Recent evidence has shown that in order for chromatin-bound ubiquitinated proteins to be degraded, they first need to be extracted by the p97 segregase58. We thus analyzed if p97 inhibition could further increase the accumulation of SUMOylated and ubiquitinated proteins on chromatin that occurs upon USP7 inhibition. Exposure of HCT116 cells to the recently described p97 inhibitor NMS87359 did not affect the levels of SUMOylated proteins by itself but, as expected, increased the levels of chromatin-bound ubiquitinated proteins (Supplementary Fig. 9a-b). In contrast, USP7 inhibition increased the levels of both SUMOylated and ubiquitinated proteins on chromatin, and the combined inhibition of USP7 and p97 enhanced this accumulation (Supplementary Fig. 9a-b). In addition, treatment with P22077 and NMS873 resulted in a further reduction in EdU incorporation compared to either treatment alone (Supplementary Fig. 9c). Together, these data suggest that USP7 targets SUMOylated proteins at replisomes to limit their ubiquitination. Ubiquitinated factors then become substrates for p97, which subsequently extracts them from chromatin. Consistent with this view, p97 inhibition results in a further accumulation of chromatin-bound SUMO2 upon exposure to USP7 inhibitors. This model suggests that USP7 facilitates the concentration of SUMOylated proteins around replisomes, by limiting their ubiquitination and subsequent extraction by the p97 segregase.
USP7 inhibition abrogates the concentration of SUMO at replisomes
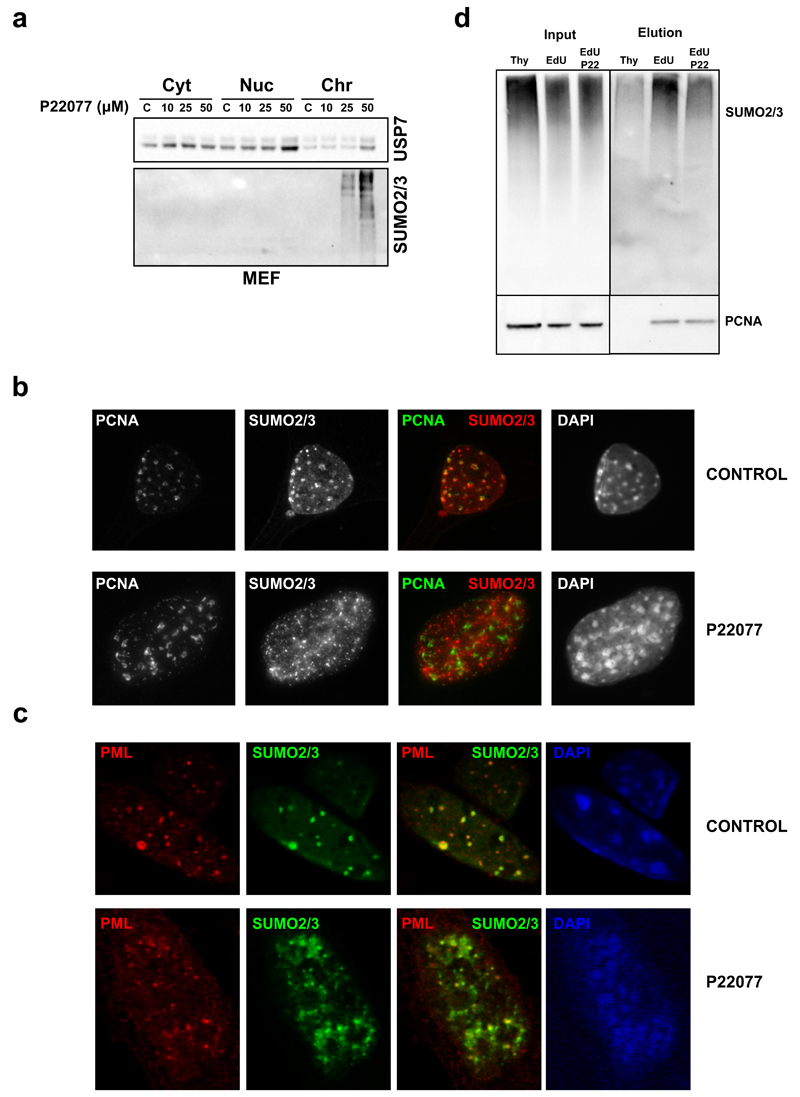
As mentioned above, and using iPOND, we recently described that the chromatin surrounding replisomes is enriched in SUMO26. Consistently, previous IF studies showed that SUMO2/3 co-localizes with PCNA during S phase60. We thus tested whether USP7 inhibition affected the enrichment of SUMO at sites of DNA replication. Similar to its effects on human cells, treatment of primary MEF with P22077 led to the accumulation of chromatin-bound SUMO2/3 conjugates (Fig. 5a). Strikingly, while IF confirmed that USP7 inhibition increases the amount of SUMOylated proteins on chromatin, it disrupted the localization of these factors to PCNA foci (Fig. 5b). In contrast, USP7 inhibition did not alter the co-localization that is also observed between SUMO2/3 and PML (Fig. 5c), supporting that the delocalization of SUMOylated proteins is specific of replication factories. In order to confirm this observation we carried out iPOND experiments in the presence of USP7 inhibitors, using conditions that did not completely stop DNA replication. Whereas, as expected, SUMO2/3 was enriched in nascent DNA (EdU fraction) when compared to mature chromatin (Thy fraction), USP7 inhibition reduced the enrichment of SUMO2/3 around replisomes (Fig. 5d, compare EdU and EdU-P22). Together with our previous results, these data show that USP7 inhibition leads to the ubiquitination of SUMOylated proteins, which drives these factors away from the surroundings of the replication fork.
FIG. 5. USP7 INHIBITION LEADS TO THE LOSS OF SUMOYLATED PROTEINS FROM REPLISOMES.
(a) MEF were treated with increasing concentrations of P22077 for 4h and cytosolic, nuclear-soluble and chromatin fractions were analyzed by WB with antibodies against USP7 and SUMO2/3.
(b) IF of PCNA (green) and SUMO2/3 (red) in MEF treated with DMSO (CONTROL) or 50 µM P22077 (P22077) for 4h. DAPI (blue) was used to stain nuclei.
(b) IF of PCNA (green) and SUMO2/3 (red) in MEF treated as in (b). DAPI (blue) was used to stain nuclei.
(d) Effect of USP7 inhibition on SUMO2/3 concentration at replisomes was evaluated by iPOND. Mature (Thy) and nascent (EdU) fractions from HeLa cells were obtained in control conditions and compared to the nascent fraction after treatment with 50 µM P22077 for 15 min prior to EdU incubation (EdU-P22). Samples were analyzed by WB with antibodies against SUMO2/3 and PCNA. One out of two independent experiments are shown.
Genetic deletion of USP7 recapitulates the effects of USP7 inhibitors
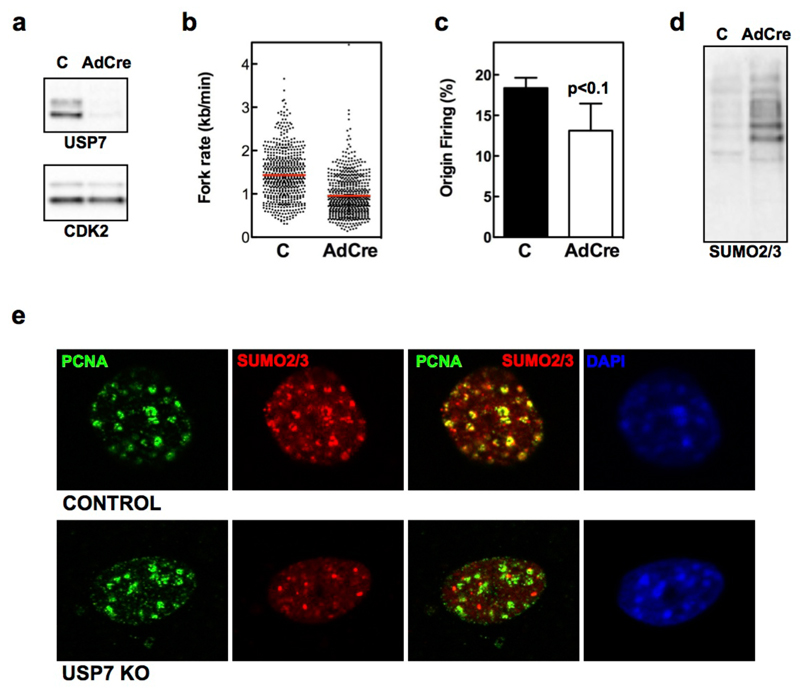
Finally, we sought to obtain genetic confirmation of our findings. To this end, we used previously described USP7 conditional knockout (USP7lox/lox) MEFs34. Infection of immortalized USP7lox/lox MEFs with adenoviruses expressing Cre recombinase (AdCre) led to the loss of USP7 protein by 4 days (Fig. 6a). DNA fiber assays revealed reduced fork rate and origin firing upon USP7 deletion (Fig. 6b-c). Importantly, and similar to the effects of USP7 inhibitors, infection of USP7lox/lox MEFs with AdCre led to the accumulation of SUMO2/3 modified chromatin-bound proteins measured by WB (Fig. 6d). Moreover, IF experiments in USP7 deleted MEF confirmed the increase of chromatin-bound SUMO2/3-conjugates, as well as the loss of co-localization of SUMO2/3 with PCNA (Fig. 6e). As a control, AdCre infection of WT MEF did not significantly affect USP7 levels, fork progression, origin firing or the levels of chromatin-bound SUMO2/3 (Supplementary Fig. 10). Together with the use of multiple USP7 inhibitors, these data provide genetic proof to support that USP7 is a SUMO DUB essential for mammalian DNA replication, and identify USP7 as the first factor required to maintain a SUMO-rich environment in the surroundings of replication forks.
FIG. 6. USP7 DELETION PHENOCOPIES THE EFFECTS OF USP7 INHIBITORS.
(a) WB analysis of USP7 and CDK2 levels in whole-cell extracts from USP7lox/lox MEF mock infected (C) or infected with AdCre for 4 days.
(b-c) DNA fibers were extracted 4 days after mock or AdCre infection of USP7lox/lox MEF and fork rate (b) and percentage of origin firing (c) were measured. The experiments were repeated three times; the pool of the three experiments (fork rate) or the average (origin firing) is shown.
(d) Chromatin fractions of mock or AdCre infected USP7lox/lox MEF assayed by WB with antibodies against SUMO2/3. The experiments were repeated three times and one representative example is shown.
(e) IF of PCNA (green) and SUMO2/3 (red) from mock infected (top) and AdCre infected (bottom) USP7lox/lox MEF 4 days after infection. DAPI was used to stain nuclei.
Discussion
Here we present the first proteomic analysis of the USP7 targetome and compared it to that of USP1, another DUB essential for DNA replication due to its role in PCNA deubiquitination4. Our results illustrate that USPs concentrated around replisomes have distinct specific substrates, rather than acting indiscriminately on ubiquitinated proteins. On one hand, USP1 acts on PCNA with no effect on USP7 targets. On the other hand, inhibition of USP7 increases the ubiquitination of its substrates with only a mild increase in PCNA ubiquitination that is secondary to the stalling of replication forks. Our diGlyproteomics also led to the identification of SUMO2 as a USP7 target, which was further confirmed both in vivo as well as in vitro using purified components. While the existence of STUbL has been known for some years7, there was no known SDUB until recently. While writing this manuscript, USP11 has been identified as a SUMO2 deubiquitinase that acts on SUMOylated PML16. However, USP11 is not found enriched at replisomes, and the reported role of USP11 seems to be restricted to the modulation of PML. In contrast, USP7 inhibition leads to a generalized accumulation of SUMOylated and ubiquitinated proteins on chromatin, which identifies USP7 as the main chromatin-bound SDUB.
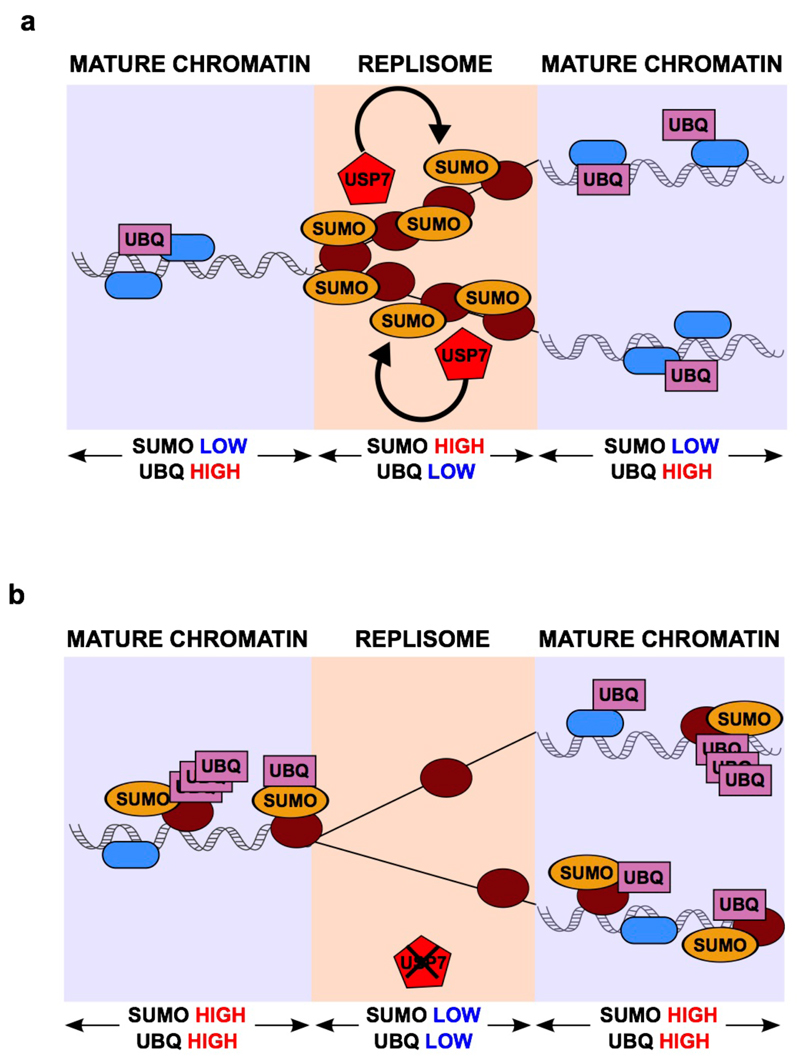
As mentioned, we recently described that chromatin nearby replisomes is overall rich in SUMO- and depleted for Ub-modified proteins26. This general changes in many modified factors is consistent with previous studies that revealed how SUMO-based regulation of biological pathways operates through the collective modification of multiple proteins at several sites17–19. How a localized PTM environment is maintained at replication forks was however not understood. Based on our data we propose the following model (Fig. 7). Consistent with recent proteomic reports20,22,24, several factors involved in DNA replication are SUMOylated during S- phase, which could facilitate their concentration around replication forks. Our data now show that the continuous action of USP7, enriched near replication forks, maintains a low level of ubiquitination of these factors (Fig. 7a). Following USP7 inhibition, SUMOylated factors are quickly ubiquitinated leading to their accumulation on chromatin away from replisomes (Fig. 7b). Identifying the individual targets of USP7 that are both SUMOylated and ubiquitinated will demand further work, but regardless of any specific target, the collective displacement of DNA replication factors from replication forks can explain the dramatic effect of USP7 inhibition on both fork rate as well as origin firing. Moreover, the discovery of USP7 as a replisome-associated SDUB provides an explanation for the SUMO-rich and Ub-low environment that is found around replisomes, and constitutes the first example of a factor involved in the regulation of this coordinated PTM balance.
FIG. 7. A ROLE FOR USP7 AT SITES OF DNA REPLICATION.
(a) Model depicting the action of USP7 on SUMOylated DNA replication factors, which mantains a SUMO-rich and Ub-low environment around replication forks.
(b) Upon inhibition of USP7, replisome-associated SUMOylated proteins rapidly increase their ubiquitination, which leads to their displacement to Ub-rich mature chromatin and away from nascent DNA molecules.
Besides the relevance of our findings for basic understanding of how DNA replication works, they may also be important in a biomedical context. Based on the finding that USP7 depletion leads to the stabilization of p5328,30, several specific USP7 inhibitors have been developed31–33 and their clinical potential is currently being assessed61. However, the embryonic lethality of USP7 deletion is not rescued by the absence of p53 and correlates with a loss of proliferation rather than with an increase in apoptosis34. Together with previous reports that showed a role for USP7 in S phase in processes such as the unloading of the MCM helicase47 or the regulation of DNMT148,49,62, we now show that USP7 plays an essential role in the basic progress of DNA replication by regulating fork speed and origin firing. In addition, we also show that USP7 inhibitors generate replication stress, which we believe can significantly contribute to the cytotoxic effects of these drugs. Importantly, the effects of USP7 inhibitors on DNA replication and replication stress are unaffected by the loss of p53. In fact, the toxic effects of reagents that generate replication stress such as ATR inhibitors are actually higher in p53-deficient cells, due to the enhanced S-phase entry of these cells63. Considering all of the above, we propose that the toxicity of USP7 inhibitors is, at least in part, due to their effects on DNA replication which, if anything, would only be aggravated in cells lacking p53.
In summary, we here reveal USP7 as a replisome-associated DUB, which counteracts ubiquitination at sites of DNA replication. The action of USP7 helps to maintain a high concentration of SUMOylated factors around replication forks. Upon USP7 inhibition, SUMOylated proteins are collectively displaced away from the replisome, which fully abrogates DNA replication both by limiting fork progression and the firing of new origins. These findings shed light onto the essential nature of USP7 in mammals, and should be taken into account when considering the use of USP7 inhibitors as anticancer therapies.
Online Methods
Cell lines, extract preparation, transfections and treatments
HCT116, HeLa and U2OS cells (ATCC); 293T-Rex (Invitrogen) cells were grown in DMEM with 10% FBS, penicillin (100 IU/ml), streptomycin (100 mg/ml) and glutamine (300 mg/ml). USP7 cKO MEF (kind gift of Dr. Wei Gu) were grown in 15% FBS, penicillin (100 IU/ml), streptomycin (100 mg/ml) and glutamine (300 mg/ml). P22077 (Tocris), P5091 (Tocris), HBX19818 (Chem Scene), ML323 (Axon), MG132 (Sigma), were dissolved in DMSO and cells were incubated for the indicated time in the presence of the inhibitor or an equivalent amount of DMSO. All cell lines were routinely tested for Mycoplasma. Whole cell extracts were prepared by lysing cells in 50 mM Tris, pH 7.5, 8 M Urea, and 1% Chaps. Cytosolic and nuclear extracts were prepared as described64 and the chromatin fraction was then extracted in 50 mM Tris, pH 7.5, 8 M Urea, and 1% Chaps. For immunoprecipitation assays, nuclei were resuspended in high-salt buffer containing 600 mM NaCl, and short sonication was applied to shear the chromatin.
Stable transfection of 293T-REx cells was carried out using Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions. One day after trasnfection the medium was replaced, and the following day the cells were trypsinized and seeded at a 1:10 dilution. The next day 5 µg/ml blasticidin, 2 µg/ml puromycin (InvivoGen), 100 µg/ml Zeocin (Invitrogen) were added to select resistant clones. The expression of the recombinant proteins was induced by adding 1 mg/ml doxycycline for 24 h.
Antibodies and plasmids
The antibodies against USP7 (Bethyl, A300-033A), MCM4 and MCM7 (provided by Juan Méndez), PCNA (Santa Cruz, sc-56), SCML265, Chk1 (Novocastra), Chk1-S345P (Cell Signaling 2348S), RPA2 (Abcam, ab2175), RPA2-S4/S8P (Bethyl, A300-245A), MEK2 (BD Biosciences Pharmingen 610236), SUMO2/3 (MBL, M114-3), Ubiquitin (Cell Signaling, #3933), His (Rockland, 600-401-382), H2A (Cell Signaling, #3636), γH2AX (Millipore, 05-636), CDK2 (Santa Cruz, M2, sc-163), p21 (Cell Signaling, #2947), PML (Santa Cruz, DO-1, sc-126) and p53 (Santa Cruz, DO-1, sc-126) were used for Western blot analysis, immunoprecipitation and immunofluorescence. The cDNA encoding for human USP7 was cloned in the pINTO plasmid66 using BamHI and Xhol restriction sites.
iPOND
We followed the iPOND protocol described in26. 1.5 × 108 HeLa cells (per condition) were plated the day before the experiment. On the following day, 10 μM EdU (Invitrogen) was added to the medium for 10 min. To assay USP7 inhibition we incubated the cells for 15 min with 50 µM P22077 before the addition of EdU. For the chase condition, the EdU treatment was followed by three thorough washes in PBS and a subsequent incubation for 1 h in medium containing 10 µM Thymidine (Sigma- Aldrich). Then, the cells were fixed with 1% formaldehyde for 15 min and the reaction was stopped by the addition of glycine. Cells were scraped from the plate, permeabilized (0.25% Triton X-100 in PBS, 30 min at room temperature), and subjected to the Click-iT reaction with the use of Biotin Azide (Invitrogen). Cell lysis (1% SDS in 50 mM Tris pH 8) and sonication followed. For each condition, 35 × 108 cells were resuspended in 7 ml of lysis buffer and sonicated with a Bioruptor (Diagenode) for 20 min in 30 s on/off cycles at high intensity. Finally, EdU-biotin-labeled DNA was pulled down with streptavidin-agarose beads (Novagen). We added 250 µl of beads to each sample and incubated them at 4°C for 20 hr. Then, the beads were washed twice with lysis buffer and NaCl 1M and eluted in 100 µl of 25× NuPAGE LDS Sample Buffer (Invitrogen) containing 10% β-mercaptoethanol (25 min at 95°C).
DNA fiber analysis
Exponentially growing cells were pulse-labeled with 50 µM CldU (15 min) followed by 250 µM IdU (15 min). Labeled cells were collected and DNA fibers were spread in buffer containing 0.5% SDS, 200 mM Tris pH 7.4 and 50 mM EDTA. For immunodetection of labeled tracks, fibers were incubated with primary antibodies (for CldU, rat anti-BrdU; for IdU, mouse anti-BrdU) and developed with the corresponding secondary antibodies conjugated to Alexa dyes. Mouse anti-ssDNA antibody was used to assess fiber integrity. Slides were examined with a Leica DM6000 B microscope, as described previously67. The conversion factor used was 1 μm = 2.59 kb68. In each assay, 200-300 tracks were measured to estimate fork rate and 300-500 tracks were analyzed to estimate the frequency of origin firing (first label origins - green-red-green - are shown as percentage of all red - CldU - labeled tracks)69.
In vitro ubiquitination of poly-SUMO2 chains
2.5 µg of poly-SUMO2 chains (3-8) (Boston Biochem) were incubated with 50 nM His-UBE1 (Boston Biochem), 1 µM His-HA-UbcH5a (Millipore), 1 µM His-RNF4 (Genwaybio) and 3 µM Ubiquitin (Boston Biochem) in a buffer containing 100 mM Tris pH 7.5, 400 mM NaCl, 10 mM MgCl2, 6 mM ATP, 0.2% Igepal CA630 and 0.2 mM DTT, and a total volume of 100 µl. After 30 min at 37°C, 10 µl of Ni-NTA agarose resin (Qiagen) were added to the mixture. The reactions were incubated for 1 h at 4°C, then centrifuged at 3000 g for 1 min. The supernatant was incubated again with 10 µl of Ni-NTA agarose resin for 30 min at 4°C. After centrifugation the supernatant containing the ubiquitinated poly-SUMO2 chains was employed in deubiquitinase assays or frozen at -80°C.
In vitro deubiquitinase assay
5 µg or poly-ubiquitin (3-7) K48 or K63 chains (Boston Biochem) or 15 µl of ubiquitinated poly-SUMO2 chains were incubated with His-USP7 (5-20 nM, Boston Biochem), His-USP1/His-UAF1 (20 nM, Boston Biochem) or no enzyme, in 100 mM Tris pH 7.5, 200 mM NaCl, 6 mM EDTA, 10 mM DTT and 0.1% Tween-20 at 37°C for 2 h. The assays were performed in a final volume of 40 µl. The reaction was stopped adding loading buffer and heating at 70°C for 10 min.
Immunoprecipitation
500 µg of nuclear extract at 1 mg/ml in 50 mM Tris, pH 7.9, and 200 mM NaCl was centrifuged for 10 min at 20000 g at 4°C, and the supernatant was incubated with specific antibodies at 4°C overnight. The samples were then centrifuged for 10 min at 20000 g at 4°C, and Protein G-Dynabeads (Invitrogen), pre-blocked in the presence of 1mg/ml BSA, were added to the supernatant. After 1 h incubation at 4°C, the beads were washed 5 times with 50 mM Tris, pH 7.9, 200 mM NaCl and 0.05% Igepal CA630 (Sigma). Proteins were eluted in SDS loading buffer.
Fluorescence microscopy and high throughput microscopy
For imunofluorescence of PCNA and SUMO2/3 soluble proteins were preextracted by mild detergent permeabilization with CSKI buffer (10 mM Pipes, pH 6.8, 100 mM NaCl, 300 mM sucrose, 3 mM MgCl2, 1 mM EGTA, and 0.5% Triton X-100) before fixation.
For high throughput microscopy, cells were grown on µCLEAR bottom 96-well plates (Greiner Bio-One) and immunofluorescence was performed using standard procedures. Analysis of DNA Replication by EdU incorporation was done using Click-It (Invitrogen) following manufacturers' instructions. In all cases, images were automatically acquired from each well using an Opera High-Content Screening System (Perkin Elmer). A 20x magnification lens was used and images were taken at non-saturating conditions. Images were segmented using DAPI signals to generate masks matching cell nuclei from which the mean signals for the rest of the stainings were calculated. Data were represented with the use of the Prism software (GraphPad Software)
Quantitative proteomics for Factors Enriched at Nascent DNA Molecules using iTRAQ
Filter-aided sample preparation (FASP) and peptide labeling
Eluates were dissolved in 8 M urea in 0.1 M triethylammonium bicarbonate TEAB (UA) and samples were digested using the FASP method70 with some modifications. Briefly, of each sample was loaded onto centrifugal devices Microcon-30 (Merck Millipore). Filters were washed with UA and proteins were alkylated using 50 mM iodoacetamide for 20 min in the dark. The excess of alkylation reagents was washed out with UA. Proteins were digested overnight with the use of endoproteinase Lys-C from Acromobacter lyticus M497-1 (Wako). Finally, trypsin (Promega, Madison, WI) was added and samples were subjected to a second digestion for 5 h. Each tryptic digest was labeled according to the manufacturer's instructions (ABSciex) with one isobaric amine-reactive tag as follows: Tag114, Thymidine fraction; Tag116, EdU fraction; Tag117, EdU+USP7i fraction. After one-hour incubation, labeled samples were pooled and evaporated to dryness. The iTRAQ sample was cleaned up using a Sep-Pak C18 cartridge for solid phase extraction (Waters) and eluted peptides were vacuum-dried.
Peptide fractionation by strong anion exchange (SAX)
We separated iTRAQ-labeled peptides by SAX as described previously71, with some modifications. Briefly, the column was assembled by stacking three layers of 3M Empore Anion Exchange disk (Varian) into a 200 µl micropipette tip. Equilibration and elutions were done in Britton & Robinson buffer, composed of 20 mM acetic acid, 20 mM phosphoric acid and 20 mM boric acid titrated with NaOH to the following pH values: 11, 8, 6, 5, 4 and 2. Peptides were loaded onto the columns in 100 µl of pH 11 and subsequently eluted with the buffers of six different pH values. Eluted peptides were bound to StageTips as described previously72. Peptides were eluted from the StageTips before LC-MS/MS using 80% acetonitrile. After SAX separation, the 6 fractions were evaporated to dryness and resuspended in 10 µl of 0.1% formic acid (FA).
Mass Spectrometry Analysis
All samples were analyzed using a QqTOF MaXis Impact (Bruker Daltonics) coupled online to a nanoLC Ultra system (Eksigent), equipped with a CaptiveSpray nanoelectrospray ion source supplemented with a CaptiveSpray nanoBooster operated at 0.2 bar/minute with isopropanol as dopant. Two technical replicates (5 µl) from each SAX fraction were injected into the mass spectrometer. Samples were loaded onto a reversed-phase C18, 5 µm, 0.1 x 20 µm trapping column (NanoSeparations) and washed for 15 min at 2.5 µl/min with 0.1% FA. The peptides were eluted at a flow rate of 300 nl/min onto a home-made analytical column packed with ReproSil-Pur C18-AQ beads, 3 µm, 75 µm x 50 cm, heated to 45 °C. Solvent A was 4% ACN in 0.1% FA and Solvent B acetonitrile in 0.1% FA. The following gradient was used: 0–2 min 2% B, 2–133 min 6–30% B, 133–133.5 min 30-98% B, 133.5–143.5 min 98% B, 144–150 min 2% B.
The MS acquisition time used for each sample was 150 min. The QqTOF Impact was operated in a data dependent mode. The spray voltage was set to 1.35 kV (1868 nA) and the temperature of the source was set to 180°C. The MS survey scan was performed at a spectra rate of 2.5 Hz in the TOF analyzer scanning a window between 80 and 1600 m/z. The minimum MS signal for triggering MS/MS was set to a normalized threshold of 500 counts. The 30 most abundant isotope patterns with charge ≥2 and m/z > 350 from the survey scan were sequentially isolated with an isolation window of 2 m/z and fragmented in the collision cell by collision induced dissociation (CID) using a collision energy of 23 – 56 eV as function of the m/z value. The m/z values triggering MS/MS with a repeat count of 1 were put on an exclusion list for 30 s using the rethinking option: the precursor intensities were re-evaluated in the scan (n) regarding their values in the previous scan (n-1). Any m/z whit intensity exceeding 5 times the measured value in the preceding survey scan was reconsidered for MS/MS. Data acquired were transformed to MGF format using the Compass DataAnalysis program. For each MS/MS spectra, the 400 most abundant non-deconvoluted ions exceeding a threshold of 100 counts were exported and recorded.
MS data analysis
The MGF files were processed using the Proteome Discoverer (version 1.4.1.14) against a human database (SwissProt canonical, 20584 sequences, Dec_2014 release) including common laboratory contaminant proteins. Oxidation of methionines and Tyrosine labelling with iTRAQ-4plex reagent were set as variable modifications, whereas Lysine and peptide N-termini labelling with iTRAQ-4plex reagent as well as carbamidomethylation of cysteine were considered as fixed modifications in the SequestHT search engine. Precursor and fragment mass tolerances were set to 20 ppm and 0.025 Da, respectively. Minimal peptide length was set to six aminoacids and a maximum of two missed cleavages were allowed. Peptides were filtered at 1% false discovery rate (FDR) by using Percolator. Results were then exported into Excel for manual data interpretation and data visualisation was carried out using Perseus software.
K-ε-GG profiling and proteome analysis by liquid chromatography mass spectrometry (LC-MS)
Sample preparation
Preparation of proteins for mass spectrometry analysis was completed as previously described73 with some modifications. Briefly, pellets containing chromatin-bound proteins were lysed in an ice cold urea lysis buffer containing 8 M urea, 50 mM Tris HCl, pH 8 and 150 mM NaCl plus protease inhibitors (Roche Applied Science). The lysate was cleared by centrifugation at 20,000g for 10min at 4°C. A BSA protein assay (Pierce) was used to determine the concentration of each sample. Extracts containing 1 mg of total protein per condition were used. Proteins were reduced for 1 h with 1 mM DTT at 40°C and subsequently alkylated for 30 min with 5.5 mM 2- chloroacetamide (Sigma) at RT in the dark. Lysates were diluted 1:1 with 50 mM Tris HCl, pH 8, and proteins were digested for 2h with Lys-C from Acromobacter lyticus M497-1 (Wako) using an enzyme to substrate ratio of 1:100 at 25°C. Samples were further diluted 1:5 with 25 mM Tris HCl, pH 8, and proteins were digested O/N at 37°C with sequencing grade trypsin (Promega). Digests were quenched with TFA and the peptide solutions were cleared by centrifugation prior to desalting. Peptides were desalted using 50 mg C18 SepPak SPE cartridges (Waters). Samples were dried completely using vacuum centrifugation.
Immunoprecipitation of diGly Containing Peptides
For enrichment of K-ε-GG peptides, we used the PTMScan ubiquitin remnant motif (K-ε-GG) kit (Cell Signaling Technology, cat. no. 5562) containing anti-K-ε-GG antibody cross-linked to protein A beads. Digested proteins were resuspended in 750 μl of immunoaffinity purification (IAP) buffer (50 mM MOPS [pH 7.4], 10 mM Na2HPO4, 50 mM NaCl) and centrifuged at max speed for 5 min to remove any insoluble material. Enrichment was completed exactly as previously described73. Briefly, the supernatants were mixed with approximately 20 µl of anti-K-ε-GG antibody beads and incubated for 1 h at 4 °C with rotation. Beads were washed twice with 1 ml of ice-cold IAP buffer followed by three washes with ice-cold PBS. K-ε-GG peptides were eluted from the antibody with 2 × 50 μl of 0.15% TFA. For proteome analysis, 1% of the flowthrough from each IAP was taken.
All samples were desalted using StageTips. StageTips were conditioned by washing with 50 μl of 50% MeCN/0.1% formic acid (FA) followed by 2 × 50 μl of 0.1% FA. Samples for K-ε-GG and proteome analysis were then loaded on StageTips, washed 2 × with 50 μl of 0.1% FA and eluted with 50 μl of 50% MeCN/0.1% formic acid (FA). Eluted peptides were dried completely using vacuum centrifugation. Samples were reconstituted in 3% MeCN/0.1% FA.
Mass Spectrometry Analysis
All samples were analyzed using an LTQ-Orbitrap Velos (Thermo Scientific) coupled online to a nanoLC Ultra system (Eksigent), equipped with a nanoelectrospray ion source (Proxeon Biosystems). For K-ε-GG and proteome samples, 5/10 μl and 5/20 μl were injected into the mass spectrometer, respectively. Samples were loaded onto a reversed-phase C18, 5 μm, 0.1 × 20 mm trapping column (NanoSeparations) and washed for 15 min at 2.5 μl/min with 0.1% FA. The peptides were eluted at a flow rate of 300 nl/min onto a home-made analytical column packed with ReproSil-Pur C18-AQ beads, 3 μm, 75 μm × 50 cm, heated to 45 °C. Solvent A was 4% ACN in 0.1% FA and Solvent B acetonitrile in 0.1% FA. For K-ε-GG analysis, the following gradient was used: 0–2 min 2% B, 2–103 min 6–35% B, 103–103.5 min 35–98% B, 103.5–113.5 min 98% B, 114–120 min 2% B. For proteome analysis, the following gradient was used: 0–2 min 2% B, 2–133 min 6–30% B, 133–133.5 min 30–98% B, 133.5–143.5 min 98% B, 144–150 min 2% B.
The MS acquisition time used for each K-ε-GG and proteome sample was 120min and 150 min, respectively. The LTQ Orbitrap Velos was operated in a data dependent mode. The spray voltage was set to 1.8 kV and the temperature of the heated capillary was set to 325°C. The MS survey scan was performed in the FT analyzer scanning a window between 350 and 1500 m/z. The resolution was set to 60 000 FWHM at m/z 400. The m/z values triggering MS/MS with a repeat count of 1 were put on an exclusion list for 60 s. The minimum MS signal for triggering MS/MS was set to 800 counts. In all cases, one microscan was recorded. The 15 most abundant isotope patterns with charge ≥2 from the survey scan were sequentially isolated with an isolation window of 1.5 m/z and fragmented in the linear ion trap (LTQ) by collision induced dissociation (CID) using a normalized collision energy of 35%. The Q value to 0.25 and an activation time to 10 ms. The maximum ion injection times for the survey scan and the MS/MS scans were 500 ms and 100 ms respectively and the ion target values were set to 1e6 and 5000, respectively for each scan mode.
MS data analysis
Both K-ε-GG enriched data and proteome data were processed using MaxQuant (version 1.5.1.2) software package. Peak lists were searched against a human database (SwissProt canonical, 20187 sequences, Febr14_2014 release), and Andromeda was used as a search engine. The enzyme specificity was set to trypsin, the maximum number of missed cleavages was set to 2, the precursor mass tolerance was set to 20 ppm for the first search, and the tolerance was set to 6 ppm for the main search. Carbamidomethylation of cysteines was searched as a fixed modification and addition of glycine-glycine to lysine, oxidation of methionines and N terminal acetylation of proteins were searched as variable modifications. For identification, the minimum peptide length was set to 6, and false discovery rate for peptide, protein, and side identification was set to 1%. Only peptide identifications with K-ε-GG localization probabilities ≥0.75 and score differences ≥5 were considered. Also, di-glycine sites localized to a C-terminal lysine residue of a peptide were considered false positives and manually removed from the data set. Log2 Ratios between intensities of quantified UQ sites were estimated for USP7i/DMSO and USP1i/DMSO for two biological replicates. Global protein expression levels were used to normalize changes in the abundance of ubiquitin sites (Log2 protein-normalized K-ε-GG (USP7i/DMSO) and Log2 protein-normalized K-ε-GG (USP1i/DMSO)).
Supplementary Material
Acknowledgements
Research was funded by Fundación Botín, Banco Santander, through its Santander Universities Global Division and by grants from the Spanish Ministry of Economy and Competitiveness (MINECO) (SAF2011-23753; SAF2014-57791-REDC), Worldwide Cancer Research (12-0229), Fundació La Marato de TV3, Howard Hughes Medical Institute and the European Research Council (ERC-617840) to OF; by a Marie-Curie International Outgoing Fellowshp (IOF) from the FP7 Marie Curie Actions and a grant from MINECO (BFU2014-55168-JIN) that was co-funded by European Regional Development Funds (FEDER) to EL; by grants from the Danish Council for Independent Research (DFF) and the Danish National Research Foundation to AL and by a PhD fellowship from MINECO to JS (BES-2012-05 2030). We would like to thank Dr. Wei Gu for providing USP7lox/lox MEF, Dr. Junjie Chen for providing Rad18-deficient HCT116 cells and Dr. Jorge Alegre-Cebollada for his help in the use of the PyMOL software.
Footnotes
Data Dissemination
Mass spectrometry proteomics data related to this manuscript have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository (http:// http://www.ebi.ac.uk/pride/archive/) with the dataset identifier PXD002408.
Author contributions
EL participated in most of the experiments of this study and wrote the MS. SR and JM performed DNA fiber analyses. IR and JM helped with proteomics. JS and MM helped with IF and HTM analyses. AJL helped with iPOND experiments. OF coordinated the study and wrote the MS.
References
- 1.Lehmann AR. Ubiquitin-family modifications in the replication of DNA damage. FEBS Lett. 2011;585:2772–9. doi: 10.1016/j.febslet.2011.06.005. [DOI] [PubMed] [Google Scholar]
- 2.Zhang W, Qin Z, Zhang X, Xiao W. Roles of sequential ubiquitination of PCNA in DNA-damage tolerance. FEBS Lett. 2011;585:2786–94. doi: 10.1016/j.febslet.2011.04.044. [DOI] [PubMed] [Google Scholar]
- 3.Havens CG, Walter JC. Mechanism of CRL4(Cdt2), a PCNA-dependent E3 ubiquitin ligase. Genes Dev. 2011;25:1568–82. doi: 10.1101/gad.2068611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Huang TT, et al. Regulation of monoubiquitinated PCNA by DUB autocleavage. Nat Cell Biol. 2006;8:339–47. doi: 10.1038/ncb1378. [DOI] [PubMed] [Google Scholar]
- 5.Cohn MA, et al. A UAF1-containing multisubunit protein complex regulates the Fanconi anemia pathway. Mol Cell. 2007;28:786–97. doi: 10.1016/j.molcel.2007.09.031. [DOI] [PubMed] [Google Scholar]
- 6.Nijman SM, et al. The deubiquitinating enzyme USP1 regulates the Fanconi anemia pathway. Mol Cell. 2005;17:331–9. doi: 10.1016/j.molcel.2005.01.008. [DOI] [PubMed] [Google Scholar]
- 7.Sriramachandran AM, Dohmen RJ. SUMO-targeted ubiquitin ligases. Biochim Biophys Acta. 2014;1843:75–85. doi: 10.1016/j.bbamcr.2013.08.022. [DOI] [PubMed] [Google Scholar]
- 8.Galanty Y, Belotserkovskaya R, Coates J, Jackson SP. RNF4, a SUMO-targeted ubiquitin E3 ligase, promotes DNA double-strand break repair. Genes Dev. 2012;26:1179–95. doi: 10.1101/gad.188284.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gibbs-Seymour I, et al. Ubiquitin-SUMO circuitry controls activated fanconi anemia ID complex dosage in response to DNA damage. Mol Cell. 2015;57:150–64. doi: 10.1016/j.molcel.2014.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hendriks IA, Treffers LW, Verlaan-de Vries M, Olsen JV, Vertegaal AC. SUMO-2 Orchestrates Chromatin Modifiers in Response to DNA Damage. Cell Rep. 2015 doi: 10.1016/j.celrep.2015.02.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ragland RL, et al. RNF4 and PLK1 are required for replication fork collapse in ATR-deficient cells. Genes Dev. 2013;27:2259–73. doi: 10.1101/gad.223180.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yin Y, et al. SUMO-targeted ubiquitin E3 ligase RNF4 is required for the response of human cells to DNA damage. Genes Dev. 2012;26:1196–208. doi: 10.1101/gad.189274.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ulrich HD. Two-way communications between ubiquitin-like modifiers and DNA. Nat Struct Mol Biol. 2014;21:317–24. doi: 10.1038/nsmb.2805. [DOI] [PubMed] [Google Scholar]
- 14.Davis EJ, et al. DVC1 (C1orf124) recruits the p97 protein segregase to sites of DNA damage. Nat Struct Mol Biol. 2012;19:1093–100. doi: 10.1038/nsmb.2394. [DOI] [PubMed] [Google Scholar]
- 15.Mosbech A, et al. DVC1 (C1orf124) is a DNA damage-targeting p97 adaptor that promotes ubiquitin-dependent responses to replication blocks. Nat Struct Mol Biol. 2012;19:1084–92. doi: 10.1038/nsmb.2395. [DOI] [PubMed] [Google Scholar]
- 16.Hendriks IA, Schimmel J, Eifler K, Olsen JV, Vertegaal AC. Ubiquitin-specific Protease 11 (USP11) Deubiquitinates Hybrid Small Ubiquitin-like Modifier (SUMO)-Ubiquitin Chains to Counteract RING Finger Protein 4 (RNF4) J Biol Chem. 2015;290:15526–37. doi: 10.1074/jbc.M114.618132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Golebiowski F, et al. System-wide changes to SUMO modifications in response to heat shock. Sci Signal. 2009;2:ra24. doi: 10.1126/scisignal.2000282. [DOI] [PubMed] [Google Scholar]
- 18.Psakhye I, Jentsch S. Protein group modification and synergy in the SUMO pathway as exemplified in DNA repair. Cell. 2012;151:807–20. doi: 10.1016/j.cell.2012.10.021. [DOI] [PubMed] [Google Scholar]
- 19.Tatham MH, Matic I, Mann M, Hay RT. Comparative proteomic analysis identifies a role for SUMO in protein quality control. Sci Signal. 2011;4:rs4. doi: 10.1126/scisignal.2001484. [DOI] [PubMed] [Google Scholar]
- 20.Bursomanno S, et al. Proteome-wide analysis of SUMO2 targets in response to pathological DNA replication stress in human cells. DNA Repair (Amst) 2015;25:84–96. doi: 10.1016/j.dnarep.2014.10.011. [DOI] [PubMed] [Google Scholar]
- 21.Bursomanno S, McGouran JF, Kessler BM, Hickson ID, Liu Y. Regulation of SUMO2 target proteins by the proteasome in human cells exposed to replication stress. J Proteome Res. 2015;14:1687–99. doi: 10.1021/pr500997p. [DOI] [PubMed] [Google Scholar]
- 22.Schimmel J, et al. Uncovering SUMOylation dynamics during cell-cycle progression reveals FoxM1 as a key mitotic SUMO target protein. Mol Cell. 2014;53:1053–66. doi: 10.1016/j.molcel.2014.02.001. [DOI] [PubMed] [Google Scholar]
- 23.Tammsalu T, et al. Proteome-wide identification of SUMO2 modification sites. Sci Signal. 2014;7:rs2. doi: 10.1126/scisignal.2005146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xiao Z, et al. System-wide Analysis of SUMOylation Dynamics in Response to Replication Stress Reveals Novel Small Ubiquitin-like Modified Target Proteins and Acceptor Lysines Relevant for Genome Stability. Mol Cell Proteomics. 2015;14:1419–34. doi: 10.1074/mcp.O114.044792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sirbu BM, et al. Analysis of protein dynamics at active, stalled, and collapsed replication forks. Genes Dev. 2011;25:1320–7. doi: 10.1101/gad.2053211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lopez-Contreras AJ, et al. A proteomic characterization of factors enriched at nascent DNA molecules. Cell Rep. 2013;3:1105–16. doi: 10.1016/j.celrep.2013.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dungrawala H, et al. The Replication Checkpoint Prevents Two Types of Fork Collapse without Regulating Replisome Stability. Mol Cell. 2015;59:998–1010. doi: 10.1016/j.molcel.2015.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cummins JM, et al. Tumour suppression: disruption of HAUSP gene stabilizes p53. Nature. 2004;428 doi: 10.1038/nature02501. 1 p following 486. [DOI] [PubMed] [Google Scholar]
- 29.Li M, Brooks CL, Kon N, Gu W. A dynamic role of HAUSP in the p53-Mdm2 pathway. Mol Cell. 2004;13:879–86. doi: 10.1016/s1097-2765(04)00157-1. [DOI] [PubMed] [Google Scholar]
- 30.Li M, et al. Deubiquitination of p53 by HAUSP is an important pathway for p53 stabilization. Nature. 2002;416:648–53. doi: 10.1038/nature737. [DOI] [PubMed] [Google Scholar]
- 31.Altun M, et al. Activity-based chemical proteomics accelerates inhibitor development for deubiquitylating enzymes. Chem Biol. 2011;18:1401–12. doi: 10.1016/j.chembiol.2011.08.018. [DOI] [PubMed] [Google Scholar]
- 32.Chauhan D, et al. A small molecule inhibitor of ubiquitin-specific protease-7 induces apoptosis in multiple myeloma cells and overcomes bortezomib resistance. Cancer Cell. 2012;22:345–58. doi: 10.1016/j.ccr.2012.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Reverdy C, et al. Discovery of specific inhibitors of human USP7/HAUSP deubiquitinating enzyme. Chem Biol. 2012;19:467–77. doi: 10.1016/j.chembiol.2012.02.007. [DOI] [PubMed] [Google Scholar]
- 34.Kon N, et al. Inactivation of HAUSP in vivo modulates p53 function. Oncogene. 2010;29:1270–9. doi: 10.1038/onc.2009.427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Alonso-de Vega I, Martin Y, Smits VA. USP7 controls Chk1 protein stability by direct deubiquitination. Cell Cycle. 2014;13:3921–6. doi: 10.4161/15384101.2014.973324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Colleran A, et al. Deubiquitination of NF-kappaB by Ubiquitin-Specific Protease-7 promotes transcription. Proc Natl Acad Sci U S A. 2013;110:618–23. doi: 10.1073/pnas.1208446110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.de Bie P, Zaaroor-Regev D, Ciechanover A. Regulation of the Polycomb protein RING1B ubiquitination by USP7. Biochem Biophys Res Commun. 2010;400:389–95. doi: 10.1016/j.bbrc.2010.08.082. [DOI] [PubMed] [Google Scholar]
- 38.Faustrup H, Bekker-Jensen S, Bartek J, Lukas J, Mailand N. USP7 counteracts SCFbetaTrCP- but not APCCdh1-mediated proteolysis of Claspin. J Cell Biol. 2009;184:13–9. doi: 10.1083/jcb.200807137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Khoronenkova SV, Dianov GL. USP7S-dependent inactivation of Mule regulates DNA damage signalling and repair. Nucleic Acids Res. 2013;41:1750–6. doi: 10.1093/nar/gks1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lecona E, Narendra V, Reinberg D. USP7 cooperates with SCML2 to regulate the activity of PRC1. Mol Cell Biol. 2015;35:1157–68. doi: 10.1128/MCB.01197-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Maertens GN, El Messaoudi-Aubert S, Elderkin S, Hiom K, Peters G. Ubiquitin-specific proteases 7 and 11 modulate Polycomb regulation of the INK4a tumour suppressor. EMBO J. 2010;29:2553–65. doi: 10.1038/emboj.2010.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Qian J, et al. USP7 modulates UV-induced PCNA monoubiquitination by regulating DNA polymerase eta stability. Oncogene. 2014 doi: 10.1038/onc.2014.394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schwertman P, et al. UV-sensitive syndrome protein UVSSA recruits USP7 to regulate transcription-coupled repair. Nat Genet. 2012;44:598–602. doi: 10.1038/ng.2230. [DOI] [PubMed] [Google Scholar]
- 44.Song MS, et al. The deubiquitinylation and localization of PTEN are regulated by a HAUSP-PML network. Nature. 2008;455:813–7. doi: 10.1038/nature07290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.van der Horst A, et al. FOXO4 transcriptional activity is regulated by monoubiquitination and USP7/HAUSP. Nat Cell Biol. 2006;8:1064–73. doi: 10.1038/ncb1469. [DOI] [PubMed] [Google Scholar]
- 46.Zhang P, et al. ATM-mediated stabilization of ZEB1 promotes DNA damage response and radioresistance through CHK1. Nat Cell Biol. 2014;16:864–75. doi: 10.1038/ncb3013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jagannathan M, et al. A role for USP7 in DNA replication. Mol Cell Biol. 2014;34:132–45. doi: 10.1128/MCB.00639-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Du Z, et al. DNMT1 stability is regulated by proteins coordinating deubiquitination and acetylation-driven ubiquitination. Sci Signal. 2010;3:ra80. doi: 10.1126/scisignal.2001462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Felle M, et al. The USP7/Dnmt1 complex stimulates the DNA methylation activity of Dnmt1 and regulates the stability of UHRF1. Nucleic Acids Res. 2011;39:8355–65. doi: 10.1093/nar/gkr528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Qin W, Leonhardt H, Spada F. Usp7 and Uhrf1 control ubiquitination and stability of the maintenance DNA methyltransferase Dnmt1. J Cell Biochem. 2011;112:439–44. doi: 10.1002/jcb.22998. [DOI] [PubMed] [Google Scholar]
- 51.Sowa ME, Bennett EJ, Gygi SP, Harper JW. Defining the human deubiquitinating enzyme interaction landscape. Cell. 2009;138:389–403. doi: 10.1016/j.cell.2009.04.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ge XQ, Jackson DA, Blow JJ. Dormant origins licensed by excess Mcm2-7 are required for human cells to survive replicative stress. Genes Dev. 2007;21:3331–41. doi: 10.1101/gad.457807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ibarra A, Schwob E, Mendez J. Excess MCM proteins protect human cells from replicative stress by licensing backup origins of replication. Proc Natl Acad Sci U S A. 2008;105:8956–61. doi: 10.1073/pnas.0803978105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kim W, et al. Systematic and quantitative assessment of the ubiquitin-modified proteome. Mol Cell. 2011;44:325–40. doi: 10.1016/j.molcel.2011.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Liang Q, et al. A selective USP1-UAF1 inhibitor links deubiquitination to DNA damage responses. Nat Chem Biol. 2014;10:298–304. doi: 10.1038/nchembio.1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Alabert C, et al. Nascent chromatin capture proteomics determines chromatin dynamics during DNA replication and identifies unknown fork components. Nat Cell Biol. 2014;16:281–93. doi: 10.1038/ncb2918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tatham MH, et al. RNF4 is a poly-SUMO-specific E3 ubiquitin ligase required for arsenic-induced PML degradation. Nat Cell Biol. 2008;10:538–46. doi: 10.1038/ncb1716. [DOI] [PubMed] [Google Scholar]
- 58.Dantuma NP, Hoppe T. Growing sphere of influence: Cdc48/p97 orchestrates ubiquitin-dependent extraction from chromatin. Trends Cell Biol. 2012;22:483–91. doi: 10.1016/j.tcb.2012.06.003. [DOI] [PubMed] [Google Scholar]
- 59.Magnaghi P, et al. Covalent and allosteric inhibitors of the ATPase VCP/p97 induce cancer cell death. Nat Chem Biol. 2013;9:548–56. doi: 10.1038/nchembio.1313. [DOI] [PubMed] [Google Scholar]
- 60.Uwada J, et al. The p150 subunit of CAF-1 causes association of SUMO2/3 with the DNA replication foci. Biochem Biophys Res Commun. 2010;391:407–13. doi: 10.1016/j.bbrc.2009.11.071. [DOI] [PubMed] [Google Scholar]
- 61.Weinstock J, et al. Selective Dual Inhibitors of the Cancer-Related Deubiquitylating Proteases USP7 and USP47. ACS Med Chem Lett. 2012;3:789–92. doi: 10.1021/ml200276j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ma H, et al. M phase phosphorylation of the epigenetic regulator UHRF1 regulates its physical association with the deubiquitylase USP7 and stability. Proc Natl Acad Sci U S A. 2012;109:4828–33. doi: 10.1073/pnas.1116349109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Toledo LI, et al. A cell-based screen identifies ATR inhibitors with synthetic lethal properties for cancer-associated mutations. Nat Struct Mol Biol. 2011;18:721–7. doi: 10.1038/nsmb.2076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lecona E, et al. Upregulation of annexin A1 expression by butyrate in human colon adenocarcinoma cells: role of p53, NF-Y, and p38 mitogen-activated protein kinase. Mol Cell Biol. 2008;28:4665–74. doi: 10.1128/MCB.00650-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lecona E, et al. Polycomb protein SCML2 regulates the cell cycle by binding and modulating CDK/CYCLIN/p21 complexes. PLoS Biol. 2013;11:e1001737. doi: 10.1371/journal.pbio.1001737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gao Z, et al. PCGF homologs, CBX proteins, and RYBP define functionally distinct PRC1 family complexes. Mol Cell. 2012;45:344–56. doi: 10.1016/j.molcel.2012.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Mouron S, et al. Repriming of DNA synthesis at stalled replication forks by human PrimPol. Nat Struct Mol Biol. 2013;20:1383–9. doi: 10.1038/nsmb.2719. [DOI] [PubMed] [Google Scholar]
- 68.Jackson DA, Pombo A. Replicon clusters are stable units of chromosome structure: evidence that nuclear organization contributes to the efficient activation and propagation of S phase in human cells. J Cell Biol. 1998;140:1285–95. doi: 10.1083/jcb.140.6.1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Petermann E, Woodcock M, Helleday T. Chk1 promotes replication fork progression by controlling replication initiation. Proc Natl Acad Sci U S A. 2010;107:16090–5. doi: 10.1073/pnas.1005031107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wisniewski JR, Zougman A, Mann M. Combination of FASP and StageTipbased fractionation allows in-depth analysis of the hippocampal membrane proteome. J Proteome Res. 2009;8:5674–8. doi: 10.1021/pr900748n. [DOI] [PubMed] [Google Scholar]
- 71.Wisniewski JR, Zougman A, Nagaraj N, Mann M. Universal sample preparation method for proteome analysis. Nat Methods. 2009;6:359–62. doi: 10.1038/nmeth.1322. [DOI] [PubMed] [Google Scholar]
- 72.Rappsilber J, Ishihama Y, Mann M. Stop and go extraction tips for matrixassisted laser desorption/ionization, nanoelectrospray, and LC/MS sample pretreatment in proteomics. Anal Chem. 2003;75:663–70. doi: 10.1021/ac026117i. [DOI] [PubMed] [Google Scholar]
- 73.Udeshi ND, Mertins P, Svinkina T, Carr SA. Large-scale identification of ubiquitination sites by mass spectrometry. Nat Protoc. 2013;8:1950–60. doi: 10.1038/nprot.2013.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.