Abstract
Pore forming proteins (PFPs) share the ability of creating pores that allow the passage of ions, proteins or other constituents through a wide variety of target membranes, ranging from bacteria to humans. They often cause cell death, as pore formation disrupts the membrane permeability barrier required for maintaining cell homeostasis. The organization into supramolecular complexes or oligomers that pierce the membrane is a common feature of PFPs. However, the molecular pathway of self-assembly and pore opening remains unclear. Here, we review the most recent discoveries in the mechanism of membrane oligomerization and pore formation of a subset of PFPs, the α-PFPs, whose pore-forming domains are formed by helical segments. Only now we are starting to grasp the molecular details of their function, mainly thanks to the introduction of single molecule microscopy and nanoscopy techniques.
Keywords: Pore forming proteins (PFPs), Pore forming toxins (PFTs), Protein oligomerization, Pore structure, Membrane
1. Introduction
Membrane pore formation is an ancient but yet efficient mechanism used by a wide range of organisms, spanning from bacteria to humans, to allow the passage of ions or other constituents through the membrane cell barrier [1]. This function is carried out by the so-called pore forming proteins (PFPs) and can be triggered as part of attack/defense mechanisms such as in the case of pore forming toxins [2], during the defensive immune response of vertebrates to pathogens such as in the case of perforin [3], or as a regulatory step in a signaling pathway such as in the case of apoptosis activation by Bcl2 proteins [4,5]. Consequently, pore opening by PFPs is usually lethal for the cell by either directly altering the cell homeostasis or cooperating with a killing counterpart to allow its delivery into the cell, as in the case of anthrax or diphtheria toxins [1,6].
PFPs have the unique feature, compared to other membrane proteins, to “adapt” to the surrounding environment through conformational changes. This characteristic allows them to be produced in a water-soluble conformation and to acquire a more suitable structure for membrane insertion. The general mode of action of these proteins requires: 1) binding to the lipid membrane, which may be triggered by specific proteins or lipids acting as membrane receptors [7] and often occurs through a conformational change of the PFP soluble-form; 2) oligomerization and membrane insertion [1]; and finally 3) formation of a pore in the membrane [6].
PFPs are generally classified into two groups, α- and β-PFPs, according to the secondary structures of their pore-forming domains. β-PFPs usually have a small hydrophobic insertion segment and oligomerization is then required to insert and span the lipid membrane. The pore is formed by a transmembrane β-barrel where each subunit generally contributes with 1–2 β-strands spanning back and forth the lipid membrane to create a wall with a cylindrical structure. This specific configuration of the β-strands where all hydrogen bonds are satisfied by interchain interactions explains why β pores have a higher stability and their stoichiometry and structure have been better characterized compared to the α-PFPs ones [6]. β-PFPs include staphylococcal α-toxin [8], the protective antigen of anthrax toxin [9], the aerolysin family [10], and the family of cholesterol-dependent cytolysins [11,12].
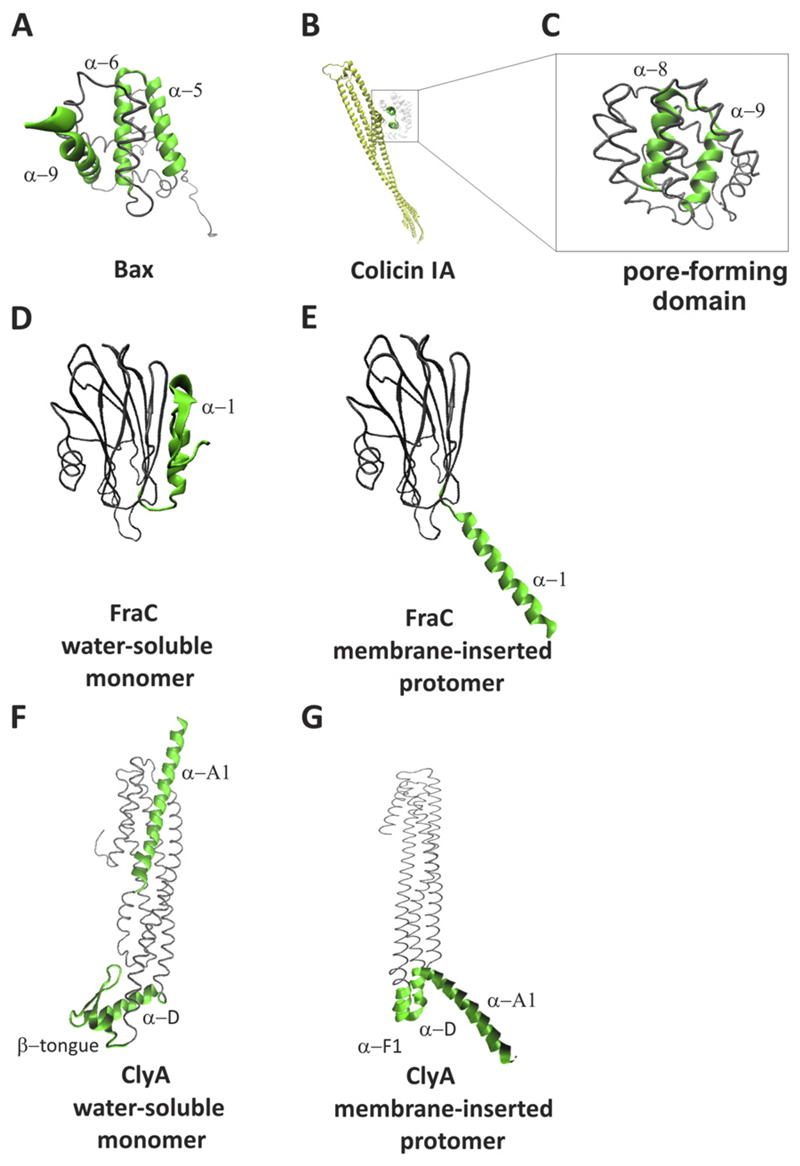
α-PFPs include a broad, inhomogeneous group of proteins with very different structures. Examples of α-PFPs are colicins and cytolysin A (ClyA), both from Escherichia coli [13], actinoporins from sea anemones [14], and the apoptotic Bcl-2 protein Bax [15] (Fig. 1). Other well-studied members of this family are: the exotoxin A produced by Pseudomonas aeruginosa [16], the diphtheria toxin from Corynebacterium diphtheria [17] and the δ-endotoxin Cry from Bacillus thuringiensis [18]. Their common feature is the presence of α-helices inserted in the membrane during pore formation. Commonly, the soluble form of the pore forming domain consists of a sandwich-like configuration where a hydrophobic helical hairpin is buried between two amphipathic layers of α-helices, as in the case of Bax and colicins (Fig. 1A, B and C). In other cases, the water-soluble structure of the protein is mainly composed by β-strands, as in the case of actinoporins (Fig. 1D), or contains a small β-tongue, like in ClyA (Fig. 1F). As a consequence of a structural change that exposes the hydrophobic or amphipathic segment to the aqueous environment, PFPs change from their soluble to the active, pore-forming conformation (Fig. 1E and G). This process can be triggered in several ways, such as cleavage of the protein at its C or N terminus, a change in the protein environment (e.g. pH change), or interaction with other proteins or lipids.
Fig. 1. 3D structure of some representative α-PFPs.
A) water-soluble structure of Bax (PDB: 1F16), B) water-soluble structure of full-length colicin IA (PDB: 1CII) and C) its pore-forming domain, D) water-soluble structure of FraC (PDB: 3ZWG) and E) its protomer conformation (PDB: 4TSY), F) water-soluble structure of ClyA (PDB: 1QOY) and E) its protomer conformation (PDB: 2WCD). The pore-forming domains are shown in green and highlighted as thicker structures.
In contrast to β-PFPs, the pore-forming domains of α-PFPs comprise a greater number of residues with a larger hydrophobic content compared to the β-strands. In the membrane, the polypeptides adopt a helical structure that is kept together by stable intra-chain hydrogen bounds. These prerequisites allow some PFPs to penetrate the membrane even in their monomeric form, which results in a molecular organization that may involve lipids and that is in general more dynamic and less defined than β-pores. Likely, because of this, the oligomerization mechanism of these PFPs has remained elusive for a long time. However, recent studies have disclosed significant steps of the assembly pathway of these proteins. This is mainly thanks to the introduction of advanced micro- and nano-single molecule techniques, which in contrast to the ensemble information provided by classical biophysical methods, have allowed resolving the heterogeneity of α-PFPs structures.
Here, we review the most recent findings about the characterization of pores formed by α-PFPs, including the Bcl-2 family and the structurally related colicins, as well as actinoporins and ClyA. In light of these new discoveries, we propose a general classification of the membrane insertion and assembly mechanisms of α-PFPs. In addition, we discuss the current models for α-pore structures and propose a more general description of protein–lipid pores. Finally, we provide an overview of the single molecule techniques currently used to elucidate the molecular mechanisms of PFPs from the assembly to the pore formation.
2. The lipid bilayer as an ideal environment to trigger PFP conformational changes and oligomerization
PFPs bind to their target membrane via interaction with a receptor exposed on the cell surface. These interactions ensure high selectivity of the PFPs towards their target, which is essential for their function. Specific proteins can act as receptors in some cases, as reported for colicins [19], but more often selectivity is achieved by interaction with individual lipids or even clusters of lipids. On one hand, binding to a protein receptor restricts targeting to a narrow set of host membranes [7]. On the other hand, interaction with lipids allows targeting a broad range of host cells with similar membrane composition. As examples of lipid receptors, sphyngomyelin (SM) is believed to promote membrane association of actinoporins [14] and cardiolipin (CL) has been suggested to be essential for the recruitment of the Bcl-2 family member Bid to the membrane during apoptosis [5]. Interestingly, in order to interact with Bax, the truncated form of Bid, tBid, requires the presence of a lipid environment [20]. Some PFPs possess more than a single target in the membrane, as in the case of colicins and diphtheria toxin that bind to a specific protein receptor, but accelerate their binding process by interaction with negatively charged lipids [19,21].
Independently of its biochemical nature, binding to a receptor provides advantages for PFPs not only by guaranteeing specificity but also by promoting conformational changes that are necessary for membrane insertion [2]. In this sense, the lipid bilayer is a biologically relevant component as it is the region of the cell that PFPs contact first. In fact, the lipid environment is a strong catalyzer of such conformational changes since the non-polar and interfacial features of the membrane trigger the exposition of amphipathic and hydrophobic portions of the proteins required for pore-formation. These regions are commonly buried in solution to avoid contact with the water environment. That is why the membrane form of α-PFPs corresponds to a rearrangement of the pore-forming domains into a fold energetically compatible with the membrane (Fig. 1E and G) [7]. Conformational states showing the characteristics of a molten globule have also been proposed to be important for the mechanism of different PFPs, such as colicins [22], diphtheria toxin [23], equinatoxin II (EqtII) [24] and ClyA [25]. This feature might seem intuitively as a denatured intermediate state that facilitates the transition between the two different structures found in solution and in membranes.
When PFPs associate with the membrane, they effectively increase their concentration by lowering their diffusion space from 3D in the extracellular medium to 2D on the cell surface [7]. Many PFP receptors are also intrinsically concentrated, pre-clustered, or associated with membrane domains [26], which have the additional benefit of promoting an even higher increase in the local protein concentration. In this sense, lipid-packing defects at the edges of lipid domains might also be adequate places for membrane oligomerization and insertion, as they confine the protein molecules to a more limited space [27]. Consequently, membrane features like fluidity, lipid domains and domain edges have been shown to strongly promote monomer–monomer interaction and the association into an oligomer [7,28,29].
3. The assembly pathway: how PFPs oligomerize to form a pore
After binding to the membrane, α-PFPs follow a series of steps that lead to pore formation. Because the α-helices of the pore forming domain have preexisting intra-chain hydrogen bonds, they are able to interact with the hydrophobic core of the lipid bilayer without the need to associate with other proteins. This explains why α-PFPs do not necessarily need to oligomerize, but can also form monomeric pores [30,31]. Although this characteristic may justify the formation of pores that involve lipids, the high variability in size of the pores formed by these proteins, imply, however, the presence of higher order structures. How these structures assemble together and form pores is still poorly understood. Indeed, for many α-PFPs it is not easy to separate oligomerization from pore formation. Recent efforts have shed light on the series of events that characterize the assembly mechanism of some α-PFPs on their way to execute pore formation [25,32]. In this section we discuss the assembly mechanisms currently proposed for some α-PFPs and point out the most recent findings in the field. In Section 4 we will provide a detailed description of all the current pore models proposed up to now for α-PFPs.
3.1. Mechanisms of membrane insertion: concerted vs non-concerted
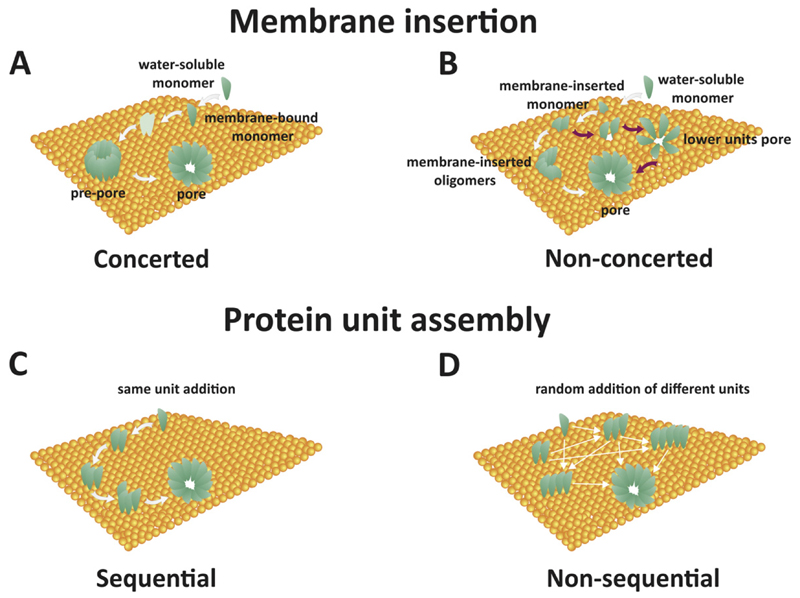
Upon reaching the target membrane, α-PFPs have two possible pathways leading to pore formation (Table 1 and Fig. 2A and B). In one case scenario, they first oligomerize at the membrane interface and consequently insert into the lipid bilayer (Fig. 2A). In this case, initially the protein units assemble together to form a ring on the membrane surface. This intermediate structure has been called “pre-pore” [33]. Then, membrane insertion occurs through a concerted conformational change that involves the membrane-interacting domain of each unit penetrating the bilayer. This “concerted” model (Fig. 2A), which in α-PFPs has only been seen for ClyA so far [34], shows strong similarities with the pre-pore structure that precedes pore formation in β-PFPs [8,35,36] and currently seems to apply only to pure protein pores [34]. In the second case scenario, they insert into the membrane, often through a deep conformational change, before undergoing oligomerization and pore formation, like in the case of Bax [37], and actinoporins [38]. This model of membrane insertion has been called “non-concerted” (Fig. 2B). According to the current state of the art, this model seems to be associated with the formation of lipid–protein pores and is characteristic of heterogeneous and flexible architectures (see Section 4).
Table 1.
Characteristics of the assembly for different PFPs.
| PFPs | Source | Nature of the pore | Molecularity of the oligomers | Molecularity of the pore | Mode of insertion/oligomerization |
|---|---|---|---|---|---|
| Bax | Humans | α-Toroidal [4] | Based on dimer units [60] | Unknown | Non-concerted/sequential by the addition of dimer units [60] |
| Colicins | E. coli | α-Toroidal[19] | Based on monomeric or dimer units [45] | Unknown | Non-concerted/sequential [45] |
| Actinoporins | Sea anemones | α-Protein–lipid hybrid [32] | Based on dimer units [72] | Octamer [32] | Non-concerted/sequential by the addition of dimer units [38,72] |
| ClyA | E. coli | α-Barrel [34] | Tetramers to octamers and dodecamer (pre-pore) [25,34] | Dodecamer [34] | Concerted/non-sequential [25,34] |
| Anthrax's protective antigen | C. anthracis | β-Barrel [116] | Heptamer (pre-pore) [74] | Heptamer [116] | Concerted/sequential [74] |
| α-Hemolysin | E. coli | β-Barrel [8] | Heptamer (pre-pore) [8] | Heptamer [8] | Concerted/sequential [75] |
Fig. 2. Mechanisms of PFPs membrane insertion and protein assembly.
A) Concerted mechanism of membrane insertion. Water-soluble monomers bind to the membrane. Oligomerization and pre-pore formation take place in a fast step. Low-stoichiometry oligomers (shown in light green) are commonly not detected. Membrane insertion occurs after pre-pore formation. B) Non-concerted mechanism of membrane insertion. Water-soluble monomers bind to the membrane in a first fast step. Membrane insertion may take place before or concomitantly with oligomerization. Intermediate pore stages with lower stoichiometry can be detected (magenta arrows pathway). C) Sequential model of protein assembly in the membrane. Addition of units of fixed molecularity (i.e. monomers or dimers) takes place. D) Non-sequential mechanism of protein assembly in the membrane. Random addition of units of different molecularities takes place.
3.2. Mechanisms of assembly: sequential vs non-sequential
Regardless of the membrane insertion mechanism, PFPs can follow two alternative pathways of assembly (Table 1). One possibility involves the addition of units with the same stoichiometry, which is known as “sequential” model (Fig. 2C). This is the mechanism that has been proposed for most α-PFPs so far. Whether the building blocks are monomers, dimers or even higher order protomers depend on the specific PFP (see Section 3.3). However, this has not been clarified for most α-PFPs yet. Alternatively, PFP assembly can proceed via the addition of units with random stoichiometry, which is referred as the “non-sequential” model (Fig. 2D). This model has been recently proposed for the pore assembly mechanism of ClyA [25]. In this case, the assembly pathway follows a kinetic mechanism initiated with the formation of n-mers that continue to associate in a combinatorial fashion. Because this approach brings the clear advantage of greatly enhancing assembly efficiency, it is tempting to speculate that it could apply to other PFPs.
3.3. The membrane insertion and assembly mechanism of different α-PFPs
3.3.1. Colicins
Colicins are pore forming toxins produced by different species of bacteria (including E. coli) in order to kill other bacteria [30]. These toxins are secreted to the extracellular medium, where they bind to the host cell via specific receptors present on the outer membrane. Then, they are translocated to the periplasm. Colicins cause cell death either by forming pores in the cytoplasmic membrane, by inhibiting protein synthesis in the cytoplasm, or by behaving as a DNAse [19,30]. A common feature of all the colicins for which the atomic structure has been solved, such as Colicin Ia, N A and E1, is the presence of three domains corresponding to three different steps of the killing mechanism: the receptor-binding, the translocation and the pore forming domains [19] (Fig. 1B and C). Activation seems to be triggered by low pH [30] and induces a conformational change that destabilizes the globular domain composed of 10 helices in which the hydrophobic hairpin (helices 8 and 9) is hidden (Fig. 1C). The final pore, however, involves the additional insertion of adjacent helices in the membrane [39] and may require oligomerization.
Although a high resolution structure of the membrane-inserted form of the colicin pore forming domain is still missing, much evidence [40, 41] points out to a mechanism in which the central hairpin (Fig. 1C) inserts perpendicularly into the membrane, while the other helices spread on the membrane surface in an “umbrella”-like configuration. In this model, pore opening occurs only after further insertion of other helices [42]. Additional evidence showing a hairpin conformation lying parallel to the membrane has led to the proposal of the “penknife” model [43]. It is also unclear how many colicin units are required in the pore configuration. More precisely, despite evidence pointing to the existence of monomeric channels [44], the requirement for pores big enough to allow the passage of large ions is the basis for an oligomerization hypothesis for colicin pore formation [19]. Recent cryo-electron microscopy data in liposomes and E. coli cells have confirmed this assumption, suggesting the assembly of colicin A in dimeric units [45,46].
3.3.2. Bcl-2 family
The proteins of the Bcl-2 family play a key role in the mitochondrial pathway of apoptosis [4,5]. Their mechanism of action requires the formation of pores in the mitochondrial outer membrane (MOM), which allow the release of apoptotic factors into the cytosol, like cytochrome c, to induce caspase activation and cell death [47]. The proteins of the Bcl-2 family are classified according to their function and the number of Bcl-2 homology (BH) domains they contain. The pro-apoptotic members of the family promote MOM permeabilization. They include the executioner proteins Bax and Bak, which contain domains BH1-3 and are believed to participate directly in MOM permeabilization, and the BH3-only proteins, such as Bid, PUMA or Noxa, which contain only the BH domain 3 and induce Bax and Bak activation [4]. The antiapoptotic or pro-survival Bcl-2 proteins, like Bcl-2 and Bcl-xL, contain all four BH domains and inhibit the action of the pro-apoptotic proteins [48,49]. All the members of the family for which the soluble structure has been solved, as for example Bcl-xL [50], Bcl-2 [51], Bax [52] and Bid [53], show a strong structural homology to colicins (Fig. 1A and C). Bax and Bak are formed by 9 α-helices that include a central hydrophobic hairpin surrounded by amphipathic helices exposed to the aqueous environment [52]. In both proteins, activation involves a conformational transition from a globular fold into an extended conformation that is embedded in the membrane [20,54–56]. Activated Bax or Bak then recruit other Bax/Bak molecules, oligomerize and induce pore formation [55,57]. However, the assembly mechanism and the final pore structure of Bax and Bak are still poorly understood.
Upon binding to the membrane, Bax exposes helix α9, which acts as a C-terminal anchor to the membrane, while the rest of the protein retains the globular fold. Bak, which is constitutively inserted into the membrane, is believed to be in the same conformation. Activation proceeds through the unfolding of the N-terminal helix and the rearrangement of the α2/BH3 domain [58,59] in a nucleation phase that is considered the rate-limiting step for Bax/Bak oligomerization [55]. Recent crystallographic data of truncated Bax in detergent solution show that the α2/BH3 domain of a Bax molecule binds to the canonical groove (α2–α5) of another Bax and viceversa to form a stable dimerization domain, while the C-terminal part of the protein detaches from the core [58]. This conformational change proceeds via the partial unfolding of the hairpin of helices 5 and 6, which disproves the previously accepted “umbrella” model for Bax in favor of the “clamp” model [60]. This hypothesis is supported by low-resolution structural studies [57] and by studies with Bak in native conditions [61]. However, these structural approaches were so far unable to provide further insight into the nature of Bax oligomers. Similarly, cross-linking and gel filtration studies have detected oligomeric forms of Bax, from dimers to higher oligomers [59], but none of these experimental approaches have allowed a precise estimation of Bax/Bak stoichiometry.
A recent single particle imaging study from our group has provided unprecedented information on the assembly mechanism and stoichiometry of individual Bax oligomers in the membrane [37]. Initially, Bax binds to the membrane as a monomer, but it quickly converts into dimers and higher order oligomers. This process, which appears to happen in seconds, induces the formation of a mixture of oligomeric species based on dimer units. Interestingly, the anti-apoptotic protein Bcl-xL is able to dissociate previously activated Bax oligomers and therefore inhibit its function. Even though the formation of pores by monomeric Bax has been shown in lipid nanodiscs [31], the idea of an active dimerization unit is quite attractive. It implies that the inter-dimer interactions are less strong than the intra-dimer ones. This is in agreement with a reversible, dynamic vision of the assembly mechanism of Bax that would explain the difficulties in the structural characterization and that is consistent with our findings that Bax forms pores tunable in size [62,63]. Our data also highlight the importance of Bax autoactivation, in which active Bax molecules recruit additional proteins, according to a sequential mechanism, for effective oligomerization [37] (Fig. 2C). This mechanism would explain the linear dependency of MOM permeabilization with increasing concentrations of Bax in MOM vesicles [64].
3.3.3. Actinoporins
Actinoporins are cytolytic proteins produced by sea anemones. They form cation-selective pores of around 2 nm in diameter on their target membranes, which cause colloid-osmotic shock and cell death [65]. To date, the most studied actinoporins have been EqtII from Actinia equina, sticholysins I and II from Stichodactyla helianthus, and fragaceatoxin C (FraC) from Actinia fragacea. They are produced as single cysteine-less polypeptide chains of around 175 amino acids with molecular weight around 20 kDa. The 3D structures of the soluble forms of the four actinoporins mentioned above show a high level of conservation and a common folding [66–69]. These toxins contain a hydrophobic β-sandwich core flanked on opposite sides by two α-helices (Fig. 1D). Actinoporins' pore-forming domain consists of an amphipathic α-helix located at the N-terminus. SM has been proposed as the actinoporin lipid receptor in the membrane [14], although some studies have found that this phospholipid is not essential for permeabilizing activity in liposomes [29,70]. Even though the exact sequence of events for actinoporins pore-formation is still under debate, the most recent model explaining their assembly in membranes assumes a pore structure without well-defined stoichiometry [71,72].
The mechanism of action of actinoporins was in part revealed thanks to the determination of the crystal structure of different conformations of FraC upon the lytic process: the water-soluble state, the monomeric lipid-bound form, an assembly intermediate, and the fully assembled transmembrane pore [32]. This study was exceptionally enlightening since it provided for the first time a high-resolution structure of a protein–lipid pore at different stages (further details in Section 4.3). Once bound to the membrane, actinoporins transfer their N-terminal helical region towards the membrane hydrophobic core. Theoretically, this passage must take place in a non-concerted way (Fig. 2B) before oligomerization, as there is not enough space to coordinately transfer the helices through the lumen of a pore of 2 nm in diameter. Moreover, this process seems to occur [38] through the addition of dimeric units (Fig. 2C) [32,72,73]. Finally, the N-terminal α-helix spans the entire thickness of the membrane and lines the wall of a pore formed by molecules of proteins and lipids [32,68].
Yet, the exact sequence of events taking place during the assembly of actinoporins in the membrane remains under debate. It is very likely that during pore structuration each pore passes through different states via the incorporation of growing number of α-helices and lipids [71]. Recent data highlight the relevance of dimers as an assembly intermediate [72]. In the monomeric form of FraC, the N-terminus is attached to the main body of the protein with the side chain of Phe16 inserted in a hydrophobic cavity of the β-core. During dimerization, the Val60 of a protein unit displaces the Phe16 of the other unit from its original position. This interaction leads to the partial unfolding and further detachment of the N-terminus towards the membrane. This is in agreement with our recent model for actinoporins assembly in membranes based on the sequential addition of dimeric units [72] (Fig. 2C). We determined the spatiotemporal organization of EqtII in living cells by single-molecule imaging and found that the protein exists as a mixture of oligomeric species mostly including monomers, dimers, tetramers and hexamers. Importantly, an EqtII version containing a mutation in the N-terminal helix of the protein that likely hinders membrane insertion [38] oligomerized more slowly via consecutive addition of monomers [72]. This suggests that actinoporin dimer formation is relevant for the proper conformational change required for N-terminal insertion in membranes and more efficient than simple sequential monomer addition. A mechanism of sequential dimer addition has been also proposed for β-PFPs like anthrax toxin [74] and γ-hemolysin [75]. However, the presence of lower order stable intermediates with functional relevance in the assembly model of actinoporins contrasts with the presence of pre-pores in the concerted mechanism proposed for β-PFPs (Fig. 2A). In fact, although a non-lytic 9-mer oligomer assembly initially assumed as a pre-pore has been obtained for FraC in detergent [69], it seems that it does not evolve into functional pores due to unfavorable packing [32]. The strong evidence that actinoporins` N-terminal helix can exist at the lipid–water interface in a protein–lipid pore (for details see Section 4) may explain the non-concerted mode of insertion–oligomerization in their assembly pathway (Fig. 2B).
3.3.4. ClyA
ClyA is a cytolytic toxin responsible for the hemolytic phenotype of several E. coli and Salmonella enterica strains [76]. ClyA lyses erythrocytes from different species, shows cytotoxicity towards cultured mammalian cells and induces apoptosis of macrophages [77] via the formation of cation-selective pores of 2–7 nm of diameter [34]. The X-ray structure of the water-soluble ClyA shows that the 34 kDa protein consists of a bundle of four long α-helices (Fig. 1F). In the tail domain, which contains the N- and C-terminus of the protein, a fifth, shorter helix is found packed against the four long helices. A short hydrophobic ß-hairpin, termed the ß-tongue, is buried at the head of the protein [78]. Pore formation of ClyA does not require any receptor proteins and monomers become assembly-competent only after binding to the membrane [34]. Sequence comparison suggests that ClyA forms a small isolated family of virulence factors restricted to closely related organisms. In fact, structural work has revealed a 3D fold resemblance between ClyA and a family of pore-forming toxins from the Gram positive bacteria Bacillus cereus. These bacteria possess three enterotoxins: hemolysin BL (Hbl), the non-hemolytic enterotoxin (Nhe) and cytotoxin K. Hbl and Nhe are tripartite toxins showing sequence homology among their components. The functional and 3D structural similarity of these components with ClyA have allowed to group them into a superfamily of PFPs [79].
The pathway proposed for the transition from a monomer to a pore complex of ClyA is based on the comparison of the soluble monomeric [78] and the dodecameric membrane-inserted crystal structures [34]. Upon membrane binding, ClyA undergoes dramatic conformational changes that involve about 50% of its full sequence and converts from a four- to a three-helix bundle. Such conformational changes mainly encompass the α-helical N-terminus (αA) and the β-tongue. This last segment is suggested to be the first region to get in contact with the membrane bilayer and to convert to α-helical on binding. The β-tongue then acts as a spring segment that triggers subsequent conformational changes required to extend the adjacent two helices, allowing them to enter the bilayer. Thereafter, the top of the elongated helix located in the N-terminus remains extended lying parallel to the membrane. At this stage, the subunits of ClyA come together to form a dodecameric pre-pore ring and finally the N-terminal helices of the 12 units span the bilayer to form a functional pore (Fig. 2A). The suggested route of assembly for ClyA resembles the mechanism elucidated for β-PFPs [35], in which the non-penetrating ring has been termed the prepore because it is followed by concerted, irreversible penetration into the bilayer (Fig. 2A). It is tempting to speculate that the delay introduced on the pre-pore formation promotes an effective concentration of amphipathic subunits on the membrane surface to favor its penetration [35]. The mechanism of pore formation and the architecture of the membrane pore described for ClyA may also apply to the related PFPs produced by B. cereus (Hbl and Nhe) [80,81], although the formation of heteroligomers composed by different components of the tripartite toxins is still unclear [82].
Recent kinetic data by single-molecule spectroscopy have revealed interesting aspects of the mechanism of assembly of this protein into a pore [25]. It is very likely that once the protomer is formed, pre-pore assembly can be explained via a mechanism in which all formed oligomers contribute to the complex. In this non-sequential mechanism (Fig. 2D), units with different molecularity can be added to preexisting oligomers. An important step for the formation of the pre-pore by ClyA is the dimer formation, which takes place without detectable conformational adjustments. This strategy is more efficient compared with the previous model based on simple monomer addition, commonly referred as sequential model (Fig. 2C). Despite the similarity in the mechanism of pore formation through the existence of a pre-pore, the assembly mechanism proposed for ClyA differs from the one proposed for the β-PFP anthrax toxin [74] and γ-hemolysin [75] that are based on the addition of units with fixed molecularity (i.e. dimers or monomers) (Fig. 2C). However, owing its higher effectiveness, it seems intuitive that other PFPs would follow a similar complex mechanism. Future work will assess if other α-PFPs are able to associate through such competent pathway.
4. Pore architecture: protein vs protein/lipid pores
4.1. Protein-lined pores: ClyA α-barrel pores
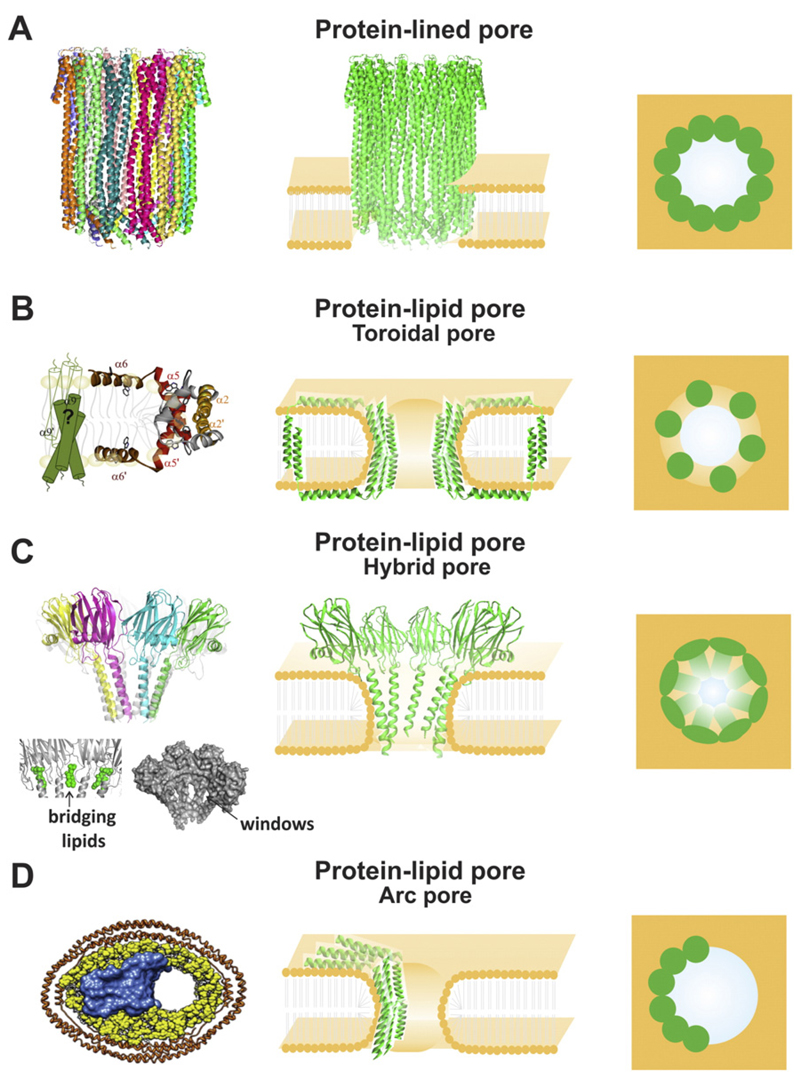
Attempts to obtain the 3D structure of α-helical pores either by X-ray diffraction or by electron microscopy have often failed, probably because α-PFPs form weakly associated oligomers without a fixed stoichiometry [35]. In this sense, the description of the structure of the oligomeric pore of ClyA in detergent was considered exceptionally enlightening [34]. ClyA is, together with FraC, the only α-PFP for which the structure of both the soluble monomer and the membrane-bound protomer are currently available (Fig. 1F and G) [34,78]. The structure of the monomer units in the pore (Fig. 1G) is substantially different from the water soluble monomer (Fig. 1F). The ClyA pore is formed by 12 subunits, each containing three long helices. The tips of the helices span the bilayer partially or completely (Fig. 3A). In the assembled pore almost all helices contribute to protomer–protomer contacts, which generates a relatively large (2400 Å) protein–protein interface [34], similar to those observed in β-barrel pores (i.e. pores formed by α and γ hemolysins) and larger than in protein–lipid pores (i.e. FraC pores) [32]. A key feature of the ClyA pore structure is its assembly in a compact α-helical bundle without lipids, which makes this toxin an atypical α-PFP representative. Therefore, the visualization of the pore formed by ClyA demonstrated a novel concept: a pore can be constructed exclusively with protein molecules arranged in an α-helical bundle [34], changing the paradigm according to which such types of pores were exclusive of β-PFPs. However, since this pore structure has been obtained in presence of detergents, questions arise about their “native” nature. In fact, different stoichiometries ranging from 8- to 13-mers have been obtained by different groups depending on the lipid membrane system employed [83–85].
Fig. 3. Different models of pore-formation by α-PFPs.
A) Protein-lined pores. Left panel: 3D structure of the pore formed by ClyA, determined in detergents (PDB: 2WCD). Central panel: side view graphical representation. Right panel: top view graphical representation. B) Protein–lipid pores: toroidal pores. Left panel: clamp model proposed for Bax pore (adapted from [60]). Central panel: side view graphical representation. Right panel: top view graphical representation. C) Protein–lipid pores: hybrid pores. Left panel: 3D structure of the pore formed by FraC and liposomes (PDB: 4TSY). The bridging lipids connecting two adjacent molecules of FraC are shown in the bottom left side (green). Windows in the pore are also shown in the bottom right side. Central panel: side view graphical representation. Right panel: top view graphical representation. B) Protein–lipid pores: arc pores. Left panel: pore formed by Bax in nanodisc (adapted from [117]). Central panel: side view graphical representation. Right panel: top view graphical representation. In all the graphical representations protein molecules are shown in green and lipids are shown in orange.
4.2. Protein–lipid pores: toroidal pores
A key feature of toroidal pores is that the membrane curves to form a torus-like channel thanks to the assistance of PFPs. In this model, the walls of the channel are formed by both polypeptide chains and lipid head groups (Fig. 3B). To avoid the high energetic cost of exposing their hydrophobic acyl chains to the aqueous environment, lipids bend and form a highly curved non-bilayer structure at the pore edge that connects the two monolayers of the membrane with a continuous surface (Fig. 3B, central panel). Thus, the toroidal name mainly refers to the torus shape adopted by the lipids in the pore structure. The main role of the protein components is to help reduce the stress caused by membrane distortion and curvature formation. Because of this, pore-forming domains are not required to span completely the bilayer [33,86]. Probably, the most direct observation of a toroidal pore was obtained by using X-ray diffraction to monitor the electron density distribution of Br atoms in a pore induced by a Bax-derived peptide [87]. This study visualized the lining of the pore as an extension of the water–lipid interface and validated for the first time the toroidal pore concept, although the distribution of the peptides with respect to the pore was not detected [87]. A toroidal pore complex has been suggested to explain the relatively big pore formed by monomeric colicins [21] and it has been assumed over the years as the putative structure for Bax-induced pores based on in vitro results with little structural information [60,88,89]. Recently, we have proposed a 3D structural model for active, membrane-inserted Bax (the “clamp” model), based on double electron–electron resonance spectroscopy data, which provides a physical–chemical basis for the stabilization of the pore edge by Bax oligomers at the membrane (Fig. 3B) [60].
However, up to date, the toroidal pore still remains a model and definitive structural evidence is still missing. Indeed, very recent studies [32,90] have provided details of the structure of protein–lipid pores. As depicted in the toirodal model, these structures suggest the involvement of membrane bending, reinforcing the notion that this is probably a common characteristic of protein–lipid pores. Due to their specific features we will discuss these examples separately in Sections 4.3 and 4.4.
4.3. Protein–lipid pores: actinoporin hybrid pores
During decades of efforts, pores formed by both lipids and proteins were difficult to visualize due to the high dynamism and low stability of these structures. Very recently, the crystal structure of the transmembrane pore of FraC was determined at 3.1 Å resolution in lipid mesophases (Fig. 3C) [32]. This is the only α-PFP for which a high-resolution structure of a pore based on both protein and lipids has been solved so far. These data revealed a critical role of lipids in the activation and in the architecture of α-PFPs. FraC forms an octameric pore where each protein molecule is associated with 3 molecules of lipids. It has been proposed that SM not only acts as a lipid receptor of actinoporins on the membrane surface, but also as a structural element of the pore, where it plays an assembly co-factor role [32]. Notably, one of these lipid molecules is located between two adjacent molecules of proteins (Fig. 3C, bottom pictures on the left panel). A critical difference with respect to the tetrameric toroidal model initially proposed for the actinoporins (where non-interacting proteins were glued together only through the lipids) [68], is the presence of protein-protein interactions in the N-terminal helices constructing the pore that, together with the lipids, help stabilizing the channel (Fig. 3C, central and right panel). To emphasize this aspect, Tanaka et al. [32] called the structure formed by FraC “hybrid protein–lipid” pore, since it resembles properties of the hypothetical toroidal pore (Fig. 3B) and the protein-lined pore (Fig. 3A). However, protein–protein interactions in the oligomeric interface are small compared with oligomers exclusively formed by proteins (i.e. ClyA pores). The main limitation of this model is that it is based on a high resolution structure of the protein chains in the pore but it does not provide any information about the bending of the membrane. However, the visualized channel exhibits windows, partially occupied by the acyl-chains of the lipid molecules connecting the pore (Fig. 3C, bottom pictures on the left panel). Hypothetically, such fenestrations favor the local disruption of the membrane lamellar structure by catalyzing the transbilayer movement of the lipids [32].
4.4. Protein–lipid pores: arc pores
Another kind of protein–lipid pore structure is the so called arc-shaped pore [91] (Fig. 3D). In this architecture protein oligomers form an arc on one side of the pore and lipids are located on the opposite side with a toroidal shape completing the structure [33]. Thus, this pore configuration can be understood as another hybrid structure between the protein-lined and toroidal models. The arc model has been extensively proposed for different β-PFPs during the recent years [33]. Pores formed, for instance, by perforin and pneumolysin are representative of arc pores [90,92]. Arc structures seem to be relatively stable since they have been observed using atomic force microscopy (AFM) [93,94] and electron microscopy [95,96]. The assumption that arcs are thermodynamically stable structures is also supported by theoretical calculations [97]. Recently, the structure and assembly pathway for suilysin, a bacterial β-PFP was mapped combining real-time AFM and electron microscopy with atomic structure fitting [36]. Suilysin oligomers show a broad distribution of both arc- and ring- shapes. Interestingly, kinetically trapped arc-shaped assemblies are able to perforate the membrane [36]. Thus, based on this model it could be hypothesized that the toroidal conformation of the lipids in the arc pore architecture may be an intermediate stage during the assembly of full ring protein-lined pores. Even though most of the data supporting arc pore formation have been obtained with β-PFPs so far, a study based on cryo-electron microscopy showed that monomeric Bax is able to induce pores in lipid nanodiscs (Fig. 3D left panel). This supports the idea that α-PFPs can also act according to the arc model. Future work should be addressed to visualize protein–lipid arc pores formed by other PFPs and to generalize the concept that they might be intermediate pore structures with high stability.
5. On the way to see a pore: methods for studying pore formation in membranes
The difficulty in defining the assembly pathway of PFPs is partially due to the fact that this is a highly dynamic process. Therefore, intermediates are difficult to trap by static techniques, such as X-ray crystallography or classical AFM. Fluorescence microscopy methods have greatly helped to overcome this problem. Techniques like FRET (Förster resonance energy transfer) or FCS (fluorescence correlation spectroscopy) are useful tools to measure real-time interactions of proteins complexes and their assembly in higher structures [25,98]. These techniques provide relevant kinetics information, along with a quantitative estimate of the stoichiometry [99]. Therefore, they have been largely employed as a more elegant alternative to classical biochemical methods, like cross-linking, gel filtration and blue native PAGE [59,61,100] to study the stoichiometry of protein complexes. However, these methods do not allow us to resolve the heterogeneity of the ensemble and are unable to provide a precise estimation of the molecularity. To account for this, stoichiometry needs to be determined at the single molecule level [101]. In the last decades, the application of single particle techniques to life sciences has experienced a rapid evolution [102]. The ability of these techniques to visualize individual events enlarges their applicability to all those studies where information is lost by either spatial or/and temporal averaged measurements.
High resolution TIRF (total internal reflection) or super-resolution Palm/STORM microscopies have turned out to be very versatile tools to either follow the dynamics or the structure of PFPs assemblies [72, 103–105]. In particular, photobleaching counting of single subunits [106] and brightness analysis [107,108] are two alternative single molecule imaging-based tools to determine the precise stoichiometry of protein complexes. These methods have allowed, for example, revealing the stoichiometry of γ-hemolysin [104] and EqtII [72] in cells. However, it is worth noting that these techniques might present several experimental challenges that make data difficult to interpret. For example, the determination of the labeling efficiency might be problematic: according to the fluorophore of choice, the molecules of interest might only be partially labeled with the fluorophore, inducing an underestimation of the stoichiometry, or contrarily, monovalent labeling may be difficult to achieve [109–111]. Additionally, low photostability of the fluorophore can induce quick photobleaching of the molecule, thus leading to a wrong estimate of the stoichiometry [110]. Finally, data analysis might be laborious and can get complicated by the need to find proper models for data processing [111].
While fluorescent-single-molecule-based methods have provided unanticipated information about the molecular mechanisms of pore formation, with these techniques it is not possible to get any conclusion regarding the existence of pore structures associated to the oligomers. Structural methods like AFM, cryo-electron microscopy and X-ray diffraction have proved to be suitable for revealing the presence of pores and characterizing the position of the proteins with respect to the rim [32, 34,87,112]. To enhance spatial and temporal resolution simultaneously, these structural and dynamic techniques are often combined on the same device. FLIM/FRET [113], TIRF/AFM [114] and single-molecule imaging/single-channel recording [115] are only few examples of the advances made in this direction. Big efforts have also been done in order to improve existing techniques, like in the case of high speed AFM [36].
6. Concluding remarks
The complex picture that we have presented to describe how α-PFPs behave on their way to form a pore has still many obscure aspects. A correct mechanistic analysis requires to work at four different levels. In the first place, which pathway do proteins follow to assemble together? The proposed sequential/non-sequential modes have not been unequivocally demonstrated for any α-PFP so far. Second, which is the stoichiometry required to open a pore in the membrane? While for a few PFPs this question has already got an answer, for many others it still remains a crucial point. Most likely, some PFPs can have more than one functionally relevant oligomeric species. This aspect has greatly contributed to the delay in solving the oligomerization pathway of these proteins. Third, in the case of protein–lipid pores, how do proteins locate with respect to the pore rim? Answering this question is crucial to understand how proteins contribute to the stability of the pores. For protein-lined pores the role of the proteins is clear – they form a wall that buries the hydrophobic membrane core from the aqueous environment. In contrast, for lipid–protein pores the protein contribution might consist in reducing the membrane curvature stress at the pore edge. Finally, and most fundamental, it is important to assess the functionality of the pore structures. Many efforts have been done in this direction, but the way to solve the puzzle it is still long.
Acknowledgments
The authors thank Joseph Unsay and Raquel Salvador-Gallego for carefully reading the manuscript. The work in our laboratory is supported by the Max Planck Society, the Bundesministerium für Bildung und Forschung (grant N.0312040) and the European Research Council (ERC-2012-StG-309966) and by the Deutsche Forschungsgemeinschaft (DFG FOR2036). Work of K.C. is partially supported by the Institutional Strategy of the University of Tübingen (Deutsche Forschungsgemeinschaft, ZUK 63). U.R. acknowledges funding support from the Alexander von Humboldt Foundation.
Abbreviations
- PFPs
pore forming proteins
- ClyA
cytolysin A
- SM
sphingomyelin
- CL
cardiolipin
- EqtII
equinatoxin II
- MOM
mitochondrial outer membrane
- BH
Bcl-2 homology
- FraC
fragaceatoxin C
- Hbl
hemolysin BL
- Nhe
non-hemolytic enterotoxin
- AFM
atomic force microscopy
- FRET
Förster resonance energy transfer
- FCS
fluorescence correlation spectroscopy
- TIRF
total internal reflection fluorescence
Footnotes
The authors declare no competing financial interest.
References
- 1.Iacovache I, Bischofberger M, van der Goot FG. Structure and assembly of pore-forming proteins. Curr Opin Struct Biol. 2010;20:241–246. doi: 10.1016/j.sbi.2010.01.013. [DOI] [PubMed] [Google Scholar]
- 2.Bischofberger M, Iacovache I, Gisou van der Goot F. Pathogenic pore-forming proteins: function and host response. Cell Host Microbe. 2012;12:266–275. doi: 10.1016/j.chom.2012.08.005. [DOI] [PubMed] [Google Scholar]
- 3.Pipkin ME, Lieberman J. Delivering the kiss of death: progress on understanding how perforin works. Curr Opin Immunol. 2007;19(3):301–308. doi: 10.1016/j.coi.2007.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.García-Sáez AJ. The secrets of the Bcl-2 family. Cell Death Differ. 2012;19:1733–1740. doi: 10.1038/cdd.2012.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cosentino K, García-Sáez AJ. Mitochondrial alterations in apoptosis. Chem Phys Lipids. 2014;181:62–75. doi: 10.1016/j.chemphyslip.2014.04.001. [DOI] [PubMed] [Google Scholar]
- 6.Fradin C, Satsoura D, Andrews DW. Punching Holes in Membranes: How Oligomeric Pore-Forming Proteins and Lipids Cooperate to Form Aqueous Channels in Membranes. In: Faller R, et al., editors. Biomembrane Frontiers. Humana press; totowa NJ: 2009. pp. 223–262. [Google Scholar]
- 7.Ros U, Garcia-Saez AJ. More than a pore: the interplay of pore-forming proteins and lipid membranes. J Membr Biol. 2015 doi: 10.1007/s00232-015-9820-y. [DOI] [PubMed] [Google Scholar]
- 8.Song L, et al. Structure of staphylococcal α-hemolysin, a heptameric transmembrane pore. Science. 1996;274(5294):1859–1865. doi: 10.1126/science.274.5294.1859. [DOI] [PubMed] [Google Scholar]
- 9.Young JA, Collier RJ. Anthrax toxin: receptor binding, internalization, pore formation, and translocation. Annu Rev Biochem. 2007;76:243–265. doi: 10.1146/annurev.biochem.75.103004.142728. [DOI] [PubMed] [Google Scholar]
- 10.Gurcel L, et al. Caspase-1 activation of lipid metabolic pathways in response to bacterial pore-forming toxins promotes cell survival. Cell. 2006;126(6):1135–1145. doi: 10.1016/j.cell.2006.07.033. [DOI] [PubMed] [Google Scholar]
- 11.Tweten RK. Cholesterol-dependent cytolysins, a family of versatile pore-forming toxins. Infect Immun. 2005;73(10):6199–6209. doi: 10.1128/IAI.73.10.6199-6209.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hotze EM, Tweten RK. Membrane assembly of the cholesterol-dependent cytolysin pore complex. Biochim Biophys Acta. 2012;1818(4):1028–1038. doi: 10.1016/j.bbamem.2011.07.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lakey JH, Slatin SL. Pore-Forming Colicins and Their Relatives. In: van der Goot FG, editor. Pore-Forming Toxins. Springer; Berlin Heidelberg: 2001. pp. 131–161. [DOI] [PubMed] [Google Scholar]
- 14.Álvarez C, et al. Sticholysins, two pore-forming toxins produced by the Caribbean Sea anemone Stichodactyla helianthus: their interaction with membranes. Toxicon. 2009:1135–1147. doi: 10.1016/j.toxicon.2009.02.022. [DOI] [PubMed] [Google Scholar]
- 15.García-Sáez A, Fuertes G, Suckale J, Salgado J. Permeabilization of the Outer Mitochondrial Membrane by Bcl-2 Proteins. In: Anderluh G, Lakey J, editors. Proteins Membrane Binding and Pore Formation. Vol. 677. Springer; New York: 2010. pp. 91–105. [DOI] [PubMed] [Google Scholar]
- 16.Menestrina G, et al. Lipid interaction of Pseudomonas aeruginosa exotoxin A. Acid-triggered permeabilization and aggregation of lipid vesicles. Biophys J. 1991;60(6):1388. doi: 10.1016/S0006-3495(91)82176-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Collier RJ. Diphtheria toxin: mode of action and structure. Bacteriol Rev. 1975;39(1):54. doi: 10.1128/br.39.1.54-85.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wünn J, et al. Transgenic indica rice breeding line IR58 expressing a synthetic crylA (b) gene from Bacillus thuringiensis provides effective insect pest control. Nat Biotechnol. 1996;14(2):171–176. doi: 10.1038/nbt0296-171. [DOI] [PubMed] [Google Scholar]
- 19.Cascales E, et al. Colicin biology. Microbiol Mol Biol Rev. 2007;71(1):158–229. doi: 10.1128/MMBR.00036-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shamas-Din A, et al. tBid undergoes multiple conformational changes at the membrane required for Bax activation. J Biol Chem. 2013;288(30):22111–22127. doi: 10.1074/jbc.M113.482109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sobko AA, et al. Effect of lipids with different spontaneous curvature on the channel activity of colicin E1: evidence in favor of a toroidal pore. FEBS Lett. 2004;576(1):205–210. doi: 10.1016/j.febslet.2004.09.016. [DOI] [PubMed] [Google Scholar]
- 22.van der Goot FG, et al. A ‘molten-globule’ membrane-insertion intermediate of the pore-forming domain of colicin A. Nature. 1991;354(6352):408–410. doi: 10.1038/354408a0. [DOI] [PubMed] [Google Scholar]
- 23.Man P, et al. Accessibility changes within diphtheria toxin T domain when in the functional molten globule state, as determined using hydrogen/deuterium exchange measurements. FEBS J. 2010;277(3):653–662. doi: 10.1111/j.1742-4658.2009.07511.x. [DOI] [PubMed] [Google Scholar]
- 24.Poklar N, et al. pH and temperature-induced molten globule-like denatured states of equinatoxin II: a study by UV-melting, DSC far- and near-UV CD spectroscopy, and ANS fluorescence. Biochemistry. 1997;36(47):14345–14352. doi: 10.1021/bi971719v. [DOI] [PubMed] [Google Scholar]
- 25.Benke S, et al. The assembly dynamics of the cytolytic pore toxin ClyA. Nat Commun. 2015;6:6198. doi: 10.1038/ncomms7198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Garcia-Saez AJ, et al. Oligomerization and pore formation by equinatoxin II inhibit endocytosis and lead to plasma membrane reorganization. J Biol Chem. 2011;286:37768–37777. doi: 10.1074/jbc.M111.281592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Barlic A, et al. Lipid phase coexistence favors membrane insertion of equinatoxin-II, a pore-forming toxin from Actinia equina . J Biol Chem. 2004;279:34209–34216. doi: 10.1074/jbc.M313817200. [DOI] [PubMed] [Google Scholar]
- 28.Pedrera L, et al. The presence of sterols favors sticholysin I-membrane association and pore formation regardless of their ability to form laterally segregated domains. Langmuir. 2015 doi: 10.1021/acs.langmuir.5b01687. [DOI] [PubMed] [Google Scholar]
- 29.Barlič A, et al. Lipid phase coexistence favors membrane insertion of equinatoxin-II, a pore-forming toxin from Actinia equina . J Biol Chem. 2004;279(33):34209–34216. doi: 10.1074/jbc.M313817200. [DOI] [PubMed] [Google Scholar]
- 30.Parker MW, Feil SC. Pore-forming protein toxins: from structure to function. Prog Biophys Mol Biol. 2005;88:91–142. doi: 10.1016/j.pbiomolbio.2004.01.009. [DOI] [PubMed] [Google Scholar]
- 31.Xu X, et al. Three-dimensional structure of Bax-mediated pores in membrane bilayers. Cell Death Dis. 2013;4(6):e683. doi: 10.1038/cddis.2013.210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tanaka K, et al. Structural basis for self-assembly of a cytolytic pore lined by protein and lipid. Nat Commun. 2015;6:6337. doi: 10.1038/ncomms7337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gilbert RJC, et al. Membrane pore formation at protein–lipid interfaces. Trends Biochem Sci. 2014;39:510–516. doi: 10.1016/j.tibs.2014.09.002. [DOI] [PubMed] [Google Scholar]
- 34.Mueller M, et al. The structure of a cytolytic alpha-helical toxin pore reveals its assembly mechanism. Nature. 2009;459(7247):726–730. doi: 10.1038/nature08026. [DOI] [PubMed] [Google Scholar]
- 35.Bayley H. Membrane-protein structure: Piercing insights. Nature. 2009;459(7247):651–652. doi: 10.1038/459651a. [DOI] [PubMed] [Google Scholar]
- 36.Leung C, et al. Stepwise visualization of membrane pore formation by suilysin, a bacterial cholesterol-dependent cytolysin. Elife. 2014;3:e04247. doi: 10.7554/eLife.04247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Subburaj Y, et al. Bax monomers form dimer units in the membrane that further self-assemble into multiple oligomeric species. Nat Commun. 2015;6 doi: 10.1038/ncomms9042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rojko N, et al. Membrane damage by an alpha-helical pore-forming protein, Equinatoxin II proceeds through a succession of ordered steps. J Biol Chem. 2013;288(33):23704–23715. doi: 10.1074/jbc.M113.481572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Slatin S, et al. Gating movements of colicin A and colicin Ia are different. J Membr Biol. 2004;202(2):73–83. doi: 10.1007/s00232-004-0720-9. [DOI] [PubMed] [Google Scholar]
- 40.Yao X, Hong M. Effects of anionic lipid and ion concentrations on the topology and segmental mobility of colicin Ia channel domain from solid-state NMR. Biochemistry. 2006;45(1):289–295. doi: 10.1021/bi051540h. [DOI] [PubMed] [Google Scholar]
- 41.Wei Z, et al. Tilted, extended, and lying in wait: the membrane-bound topology of residues Lys-381-Ser-405 of the colicin E1 channel domain. Biochemistry. 2007;46(20):6074–6085. doi: 10.1021/bi700317k. [DOI] [PubMed] [Google Scholar]
- 42.Parker MW, et al. Insights into membrane insertion based on studies of colicins. Trends Biochem Sci. 1990;15(4):126–129. doi: 10.1016/0968-0004(90)90205-p. [DOI] [PubMed] [Google Scholar]
- 43.Parker MW, et al. Refined structure of the pore-forming domain of colicin A at 2.4 Å resolution. J Mol Biol. 1992;224(3):639–657. doi: 10.1016/0022-2836(92)90550-4. [DOI] [PubMed] [Google Scholar]
- 44.Anderluh G, Lakey JH. Proteins: membrane binding and pore formation. Springer Science & Business Media; 2011. [Google Scholar]
- 45.Dunkel S, et al. In vivo EPR on spin labeled colicin A reveals an oligomeric assembly of the pore-forming domain in E. coli membranes. Phys Chem Chem Phys. 2015;17(7):4875–4878. doi: 10.1039/c4cp05638h. [DOI] [PubMed] [Google Scholar]
- 46.Greig SL, Radjainia M, Mitra AK. Oligomeric structure of colicin ia channel in lipid bilayer membranes. J Biol Chem. 2009;284(24):16126–16134. doi: 10.1074/jbc.M900292200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wei MC. Proapoptotic BAX and BAK: a requisite gateway to mitochondrial dysfunction and death. Science. 2001;292(5517):727–730. doi: 10.1126/science.1059108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Youle RJ, Strasser A. The BCL-2 protein family: opposing activities that mediate cell death. Nat Rev Mol Cell Biol. 2008;9(1):47–59. doi: 10.1038/nrm2308. [DOI] [PubMed] [Google Scholar]
- 49.Shamas-Din A, et al. Mechanisms of action of Bcl-2 family poteins. Cold Spring Harb Perspect Biol. 2013;5(4) doi: 10.1101/cshperspect.a008714. a008714-a008714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Muchmore SW, et al. X-ray and NMR structure of human Bcl-xL, an inhibitor of programmed cell death. Nature. 1996;381(6580):335–341. doi: 10.1038/381335a0. [DOI] [PubMed] [Google Scholar]
- 51.Petros AM, et al. Solution structure of the antiapoptotic protein bcl-2. Proc Natl Acad Sci. 2001;98(6):3012–3017. doi: 10.1073/pnas.041619798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Suzuki M, Youle RJ, Tjandra N. Structure of Bax: coregulation of dimer formation and intracellular localization. Cell. 2000;103(4):645–654. doi: 10.1016/s0092-8674(00)00167-7. [DOI] [PubMed] [Google Scholar]
- 53.McDonnell JM, et al. Solution structure of the proapoptotic molecule BID: a structural basis for apoptotic agonists and antagonists. Cell. 1999;96(5):625–634. doi: 10.1016/s0092-8674(00)80573-5. [DOI] [PubMed] [Google Scholar]
- 54.García-Sáez AJ, et al. Membrane-insertion fragments of Bcl-xL, Bax, and Bid. Biochemistry. 2004;43(34):10930–10943. doi: 10.1021/bi036044c. [DOI] [PubMed] [Google Scholar]
- 55.Lovell JF, et al. Membrane binding by tBid initiates an ordered series of events culminating in membrane permeabilization by Bax. Cell. 2008;135(6):1074–1084. doi: 10.1016/j.cell.2008.11.010. [DOI] [PubMed] [Google Scholar]
- 56.Brouwer JM, et al. Bak core and latch domains separate during activation, and freed core domains form symmetric homodimers. Mol Cell. 2014;55(6):938–946. doi: 10.1016/j.molcel.2014.07.016. [DOI] [PubMed] [Google Scholar]
- 57.Bleicken S, et al. Molecular details of Bax activation, oligomerization, and membrane insertion. J Biol Chem. 2010;285(9):6636–6647. doi: 10.1074/jbc.M109.081539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Czabotar PE, et al. Bax crystal structures reveal how BH3 domains activate Bax and nucleate its oligomerization to induce apoptosis. Cell. 2013;152(3):519–531. doi: 10.1016/j.cell.2012.12.031. [DOI] [PubMed] [Google Scholar]
- 59.Annis MG, et al. Bax forms multispanning monomers that oligomerize to permeabilize membranes during apoptosis. EMBO J. 2005;24(12):2096–2103. doi: 10.1038/sj.emboj.7600675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bleicken S, et al. Structural model of active Bax at the membrane. Mol Cell. 2014;56:496–505. doi: 10.1016/j.molcel.2014.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ma S, et al. Assembly of the Bak apoptotic pore: a critical role for the bak protein α6 helix in the multimerization of homodimers during apoptosis. J Biol Chem. 2013;288(36):26027–26038. doi: 10.1074/jbc.M113.490094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gillies LA, et al. Visual and functional demonstration of growing Bax-induced pores in mitochondrial outer membranes. Mol Biol Cell. 2015;26(2):339–349. doi: 10.1091/mbc.E13-11-0638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bleicken S, et al. Proapoptotic Bax and Bak proteins form stable protein-permeable pores of tunable size. J Biol Chem. 2013;288(46):33241–33252. doi: 10.1074/jbc.M113.512087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kushnareva Y, et al. Bax activation initiates the assembly of a multimeric catalyst that facilitates Bax pore formation in mitochondrial outer membranes. PLoS Biol. 2012;10(9):e1001394. doi: 10.1371/journal.pbio.1001394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.García-Ortega L, et al. The behavior of sea anemone actinoporins at the water–membrane interface. Biochim Biophys Acta Biomembr. 2011;1808(9):2275–2288. doi: 10.1016/j.bbamem.2011.05.012. [DOI] [PubMed] [Google Scholar]
- 66.Athanasiadis A, et al. Crystal structure of the soluble form of equinatoxin II, a pore-forming toxin from the sea anemone Actinia equina . Structure. 2001;9(4):341–346. doi: 10.1016/s0969-2126(01)00592-5. [DOI] [PubMed] [Google Scholar]
- 67.Hinds MG, et al. Solution structure of the eukaryotic pore-forming cytolysin equinatoxin II: implications for pore formation. J Mol Biol. 2002;315(5):1219–1229. doi: 10.1006/jmbi.2001.5321. [DOI] [PubMed] [Google Scholar]
- 68.Mancheño JM, et al. Crystal and electron microscopy structures of sticholysin II actinoporin reveal insights into the mechanism of membrane pore formation. Structure. 2003;11:1319–1328. doi: 10.1016/j.str.2003.09.019. [DOI] [PubMed] [Google Scholar]
- 69.Mechaly AE, et al. Structural insights into the oligomerization and architecture of eukaryotic membrane pore-forming toxins. Structure. 2011;19(2):181–191. doi: 10.1016/j.str.2010.11.013. [DOI] [PubMed] [Google Scholar]
- 70.de Los Rios V, et al. Mechanism of the leakage induced on lipid model membranes by the hemolytic protein sticholysin II from the sea anemone Stichodactyla helianthus . Eur J Biochem. 1998;252(2):284–289. doi: 10.1046/j.1432-1327.1998.2520284.x. [DOI] [PubMed] [Google Scholar]
- 71.Antonini V, et al. Functional characterization of sticholysin I and W111C mutant reveals the sequence of the actinoporin's pore assembly. PLoS One. 2014;9:e110824. doi: 10.1371/journal.pone.0110824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Subburaj Y, et al. Toxicity of an alpha-pore-forming toxin depends on the assembly mechanism on the target membrane as revealed by single molecule imaging. J Biol Chem. 2015;290(8):4856–4865. doi: 10.1074/jbc.M114.600676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Valle A, et al. Cys mutants in functional regions of sticholysin I clarify the participation of these residues in pore formation. Toxicon. 2011;58(1):8–17. doi: 10.1016/j.toxicon.2011.04.005. [DOI] [PubMed] [Google Scholar]
- 74.Christensen KA, et al. Interaction of the 20 kDa and 63 kDa fragments of anthrax protective antigen: kinetics and thermodynamics. Biochemistry. 2005;44(3):1047–1053. doi: 10.1021/bi047791s. [DOI] [PubMed] [Google Scholar]
- 75.Thompson JR, et al. Rapid assembly of a multimeric membrane protein pore. Biophys J. 2011;101(11):2679–2683. doi: 10.1016/j.bpj.2011.09.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ludwig A, et al. Molecular analysis of cytolysin A (ClyA) in pathogenic Escherichia coli strains. J. Bacteriol. 2004;186(16):5311–5320. doi: 10.1128/JB.186.16.5311-5320.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lai XH, et al. Cytocidal and apoptotic effects of the ClyA protein from Escherichia coli on primary and cultured monocytes and macrophages. Infect Immun. 2000;68(7):4363–4367. doi: 10.1128/iai.68.7.4363-4367.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wallace AJ, et al. E. coli hemolysin E (HlyE, ClyA, SheA): X-ray crystal structure of the toxin and observation of membrane pores by electron microscopy. Cell. 2000;100(2):265–276. doi: 10.1016/s0092-8674(00)81564-0. [DOI] [PubMed] [Google Scholar]
- 79.Hunt S, Green J, Artymiuk PJ, Hemolysin E. (HlyE, ClyA, SheA) and related toxins. Adv Exp Med Biol. 2010;677:116–126. doi: 10.1007/978-1-4419-6327-7_10. [DOI] [PubMed] [Google Scholar]
- 80.Madegowda M, et al. X-ray crystal structure of the B component of hemolysin BL from Bacillus cereus. Proteins. 2008;71(2):534–540. doi: 10.1002/prot.21888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ganash M, et al. Structure of the NheA component of the Nhe toxin from Bacillus cereus: implications for function. PLoS One. 2013;8(9):e74748. doi: 10.1371/journal.pone.0074748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Lindback T, et al. Cytotoxicity of the Bacillus cereus Nhe enterotoxin requires specific binding order of its three exoprotein components. Infect Immun. 2010;78(9):3813–3821. doi: 10.1128/IAI.00247-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Hunt S, et al. The formation and structure of Escherichia coli K-12 haemolysin E pores. Microbiology. 2008;154(2):633–642. doi: 10.1099/mic.0.2007/011700-0. [DOI] [PubMed] [Google Scholar]
- 84.Tzokov SB, et al. Structure of the hemolysin E (HlyE, ClyA, and SheA) channel in its membrane-bound form. J Biol Chem. 2006;281(32):23042–23049. doi: 10.1074/jbc.M602421200. [DOI] [PubMed] [Google Scholar]
- 85.Eifler N, et al. Cytotoxin ClyA from Escherichia coli assembles to a 13-meric pore independent of its redox-state. EMBO J. 2006;25(11):2652–2661. doi: 10.1038/sj.emboj.7601130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Iacovache I, van der Goot FG, Pernot L. Pore formation: An ancient yet complex form of attack. Biochimica et Biophysica Acta (BBA) - Biomembranes. 2008;1778:1611–1623. doi: 10.1016/j.bbamem.2008.01.026. [DOI] [PubMed] [Google Scholar]
- 87.Qian S, et al. Structure of transmembrane pore induced by Bax-derived peptide: Evidence for lipidic pores. Proc Natl Acad Sci. 2008;105(45):17379–17383. doi: 10.1073/pnas.0807764105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Basanez G. Bax-type apoptotic proteins porate pure lipid bilayers through a mechanism sensitive to intrinsic monolayer curvature. J Biol Chem. 2002;277:49360–49365. doi: 10.1074/jbc.M206069200. [DOI] [PubMed] [Google Scholar]
- 89.García-Sáez AJ, et al. Peptides derived from apoptotic Bax and bid reproduce the poration activity of the parent full-length proteins. Biophys J. 2005;88:3976–3990. doi: 10.1529/biophysj.104.058008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Metkar SS, et al. Perforin oligomers form arcs in cellular membranes: a locus for intracellular delivery of granzymes. Cell Death Differ. 2015;22:74–85. doi: 10.1038/cdd.2014.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Dalla Serra M, Tejuca Martínez M. Pore-forming Toxins. eLS, John Wiley & Sons Ltd; 2001. [Google Scholar]
- 92.Sonnen AF-P, Plitzko JM, Gilbert RJ. Incomplete pneumolysin oligomers form membrane pores. Open Biol. 2014;4(4):140044. doi: 10.1098/rsob.140044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Czajkowsky DM, et al. Vertical collapse of a cytolysin prepore moves its transmembrane beta-hairpins to the membrane. EMBO J. 2004;23(16):3206–3215. doi: 10.1038/sj.emboj.7600350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Praper T, et al. Human perforin employs different avenues to damage membranes. J Biol Chem. 2011;286(4):2946–2955. doi: 10.1074/jbc.M110.169417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Palmer M. Staphyloccal alpha toxin. Symp Ser Soc Appl Microbiol. 1998;27:125S–126S. doi: 10.1046/j.1365-2672.1998.0840s1125s.x. [DOI] [PubMed] [Google Scholar]
- 96.Morgan PJ, et al. Modeling the bacterial protein toxin, pneumolysin, in its monomeric and oligomeric form. J Biol Chem. 1994;269(41):25315–25320. [PubMed] [Google Scholar]
- 97.Prieto L, He Y, Lazaridis T. Protein arcs may form stable pores in lipid membranes. Biophys J. 2014;106(1):154–161. doi: 10.1016/j.bpj.2013.11.4490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.García-Sáez AJ, et al. Membrane promotes tBID interaction with BCLXL. Nat Struct Mol Biol. 2009;16(11):1178–1185. doi: 10.1038/nsmb.1671. [DOI] [PubMed] [Google Scholar]
- 99.Nguyen TA, et al. Fluorescence Polarization and Fluctuation Analysis Monitors Subunit Proximity. Stoichiometry, and Protein Complex Hydrodynamics. 2012 doi: 10.1371/journal.pone.0038209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Schagger H, Cramer W, Vonjagow G. Analysis of molecular masses and oligomeric states of protein complexes by blue native electrophoresis and isolation of membrane protein complexes by two-dimensional native electrophoresis. Anal Biochem. 1994;217(2):220–230. doi: 10.1006/abio.1994.1112. [DOI] [PubMed] [Google Scholar]
- 101.Hallworth R, Nichols MG. Prestin in HEK cells is an obligate tetramer. J Neurophysiol. 2012;107(1):5–11. doi: 10.1152/jn.00728.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Moerner W. New directions in single-molecule imaging and analysis. Proc Natl Acad Sci. 2007;104(31):12596–12602. doi: 10.1073/pnas.0610081104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Baker MA, et al. Photobleaching reveals heterogeneous stoichiometry for equinatoxin II oligomers. Chembiochem. 2014;15(14):2139–2145. doi: 10.1002/cbic.201300799. [DOI] [PubMed] [Google Scholar]
- 104.Nguyen VT, Kamio Y, Higuchi H. Single-molecule imaging of cooperative assembly of γ-hemolysin on erythrocyte membranes. EMBO J. 2003;22(19):4968–4979. doi: 10.1093/emboj/cdg498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Heron AJ, Thompson JR, Cronin B, Bayley H, Wallace MI. Simultaneous Measurement of Ionic Current and Fluorescence from Single Protein Pores. Journal of the American Chemical Society. 2009;131:1652–1653. doi: 10.1021/ja808128s. [DOI] [PubMed] [Google Scholar]
- 106.Ulbrich MH, Isacoff EY. Subunit counting in membrane-bound proteins. Nat Methods. 2007;4(4):319–321. doi: 10.1038/NMETH1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Schmidt T, et al. Imaging of single molecule diffusion. Proc Natl Acad Sci. 1996;93(7):2926–2929. doi: 10.1073/pnas.93.7.2926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Anderluh A, et al. Single molecule analysis reveals coexistence of stable serotonin transporter monomers and oligomers in the live cell plasma membrane. J Biol Chem. 2014;289(7):4387–4394. doi: 10.1074/jbc.M113.531632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Jaiswal JK, Simon SM. Imaging single events at the cell membrane. Nat Chem Biol. 2007;3(2):92–98. doi: 10.1038/nchembio855. [DOI] [PubMed] [Google Scholar]
- 110.Xia T, Li N, Fang X. Single-molecule fluorescence imaging in living cells. Annu Rev Phys Chem. 2013;64:459–480. doi: 10.1146/annurev-physchem-040412-110127. [DOI] [PubMed] [Google Scholar]
- 111.Hallworth R, Nichols MG. The single molecule imaging approach to membrane protein stoichiometry. Microsc Microanal. 2012;18(4):771–780. doi: 10.1017/S1431927612001195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Epand RF, et al. Direct evidence for membrane pore formation by the apoptotic protein Bax. Biochem Biophys Res Commun. 2002;298(5):744–749. doi: 10.1016/s0006-291x(02)02544-5. [DOI] [PubMed] [Google Scholar]
- 113.Padilla-Parra S, Tramier M. FRET microscopy in the living cell: different approaches, strengths and weaknesses. BioEssays. 2012;34(5):369–376. doi: 10.1002/bies.201100086. [DOI] [PubMed] [Google Scholar]
- 114.Ramachandran S, et al. Structure and permeability of ion-channels by integrated AFM and waveguide TIRF microscopy. Sci Report. 2014;4 doi: 10.1038/srep04424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Weatherill EE, Wallace MI. Combining single-molecule imaging and single-channel electrophysiology. J Mol Biol. 2015;427(1):146–157. doi: 10.1016/j.jmb.2014.07.007. [DOI] [PubMed] [Google Scholar]
- 116.Collier RJ. Membrane translocation by anthrax toxin. Mol Asp Med. 2009;30(6):413–422. doi: 10.1016/j.mam.2009.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Volkmann N, et al. The rheostat in the membrane: BCL-2 family proteins and apoptosis. Cell Death Differ. 2014;21(2):206–215. doi: 10.1038/cdd.2013.153. [DOI] [PMC free article] [PubMed] [Google Scholar]