Abstract
Bax plays a central role in the mitochondrial pathway of apoptosis. Upon activation, cytosolic Bax monomers oligomerize on the surface of mitochondria and change conformation concertedly to punch holes into the outer membrane. The subsequent release of cytochrome c initiates cell death. However, the structure of membrane-inserted Bax and its mechanism of action remain largely unknown. Here, we propose a 3D model of active Bax at the membrane based on double electron-electron resonance (DEER) spectroscopy in liposomes and isolated mitochondria. We show that active Bax is organized at the membrane as assemblies of dimers. In addition to a stable dimerization domain, each monomer contains a more flexible piercing domain involved in interdimer interactions and pore formation. The most important structural change during Bax activation is the opening of the hairpin formed by helices 5 and 6, which adopts a clamp-like conformation central to the mechanism of mitochondrial permeabilization.
Introduction
Bax and Bak are key regulators of apoptosis that mediate the permeabilization of the mitochondrial outer membrane (MOM) and the release of cytochrome c (Czabotar et al., 2014; García-Sáez, 2012). Double knockout mice lacking Bax and Bak die mostly during embryo development or shortly after birth (Lindsten et al., 2000), and the reduced programmed cell death induces severe abnormalities in the few mice that reach adulthood. Moreover, cells from Bax−/−Bak−/− mice are resistant to most forms of stress-induced apoptosis, including the artificial expression of BH3-only proteins (Wei et al., 2001). Therefore, these two proteins are essential for the induction of apoptosis downstream of the BH3-only proteins.
In healthy cells Bax is mainly a cytosolic, globular, and monomeric protein, which can also be loosely bound to the MOM (Schellenberg et al., 2013). The structure of monomeric Bax solved by nuclear magnetic resonance (NMR) shows an α-helical bundle with a central amphipathic hairpin motif surrounded by solvent-exposed helices (Suzuki et al., 2000). Upon induction of apoptosis, Bax permanently inserts into the MOM, where it oligomerizes and induces the release of cytochrome c and other apoptotic factors (Youle and Strasser, 2008). Activation is a multistep process induced by the interaction of Bax with an activator BH3 only protein (like tBid or Bim) (Edwards et al., 2013; Gavathiotis et al., 2008; Lovell et al., 2008), followed by the release of helix α9 from the BH groove (Bleicken and Zeth, 2009; Kim et al., 2009; Suzuki et al., 2000). Then, helices α2 to α4 build a symmetric dimer interface (Bleicken et al., 2010; Czabotar et al., 2013; Dewson et al., 2012). Once membrane-embedded, several amino acids in helices α5, α6, and α9 are inaccessible to water, suggesting that they become membrane-inserted (Annis et al., 2005; García-Sáez et al., 2004). Based on this and on the structural similarities with colicins, the “umbrella” model was introduced to represent active Bax in the membrane. This model proposes the insertion of helices α5 and α6 as a transmembrane hairpin into the lipid bilayer (Annis et al., 2005). However, the structure of full-length Bax in the membrane environment of the MOM remains elusive to date.
Here, we present a 3D model of a Bax dimer embedded in the membrane with a calculated accuracy of 8 Å. To build this model, we used a multilateration approach based on distance constraints gained from Q-band double electron-electron resonance (DEER) on spin-labeled Bax variants inserted into large unilamellar vesicles mimicking the MOM lipid composition (MOM-LUVs). The model proposed here retains the idea of a core and latch domain in active Bax and Bak (Brouwer et al., 2014; Czabotar et al., 2013, 2014), but describes the relative arrangement of the helices in the full-length oligomeric Bax at the membrane. We found that the Bax dimer assumes a clamp-like conformation at the membrane via a partial opening of helices α5 and α6 that is suggested to be central in the mechanism of membrane permeabilization. The DEER data show that in full-length active Bax the core domain (helices 2–5) builds a stable interaction interface with another analogous domain, in line with the crystallized truncated GFP-fused dimer found by Czabotar et al. (2013) (Protein Data Bank [PDB]: 4BDU). Based on their function in active Bax dimers, we named helices 2–5 the dimerization domain. Interestingly, we found that the helices beyond α5 adopt a more flexible conformation. Due to their structural features in active Bax dimers at the membrane, which we suggest to be essential for membrane destabilization, we named helices 6–9 the piercing domain. DEER performed on selected Bax mutants interacting with isolated mitochondria corroborated the distance information obtained in MOM-LUVs, which supports the physiological relevance of the structural model proposed.
Results
Spin-Labeled Bax Variants Reproduce the NMR Fold of Monomeric Bax
DEER is a powerful technique to extract dipolar interactions, and thus distance distributions, between spin-labeled probes in proteins (distance range from 1.5 to 6 nm in membrane-embedded proteins) (Jeschke, 2012). In order to apply DEER to Bax, we introduced cysteine mutations to engineer singly and doubly spin-labeled variants (Figure 1A). In total, we studied 42 single and double cysteine mutants of full-length Bax labeled with the nitroxide-based spin label (1-Oxyl-2,2,5,5-tetramethyl-Δ3-pyrroline-3-methyl) Methanethiosulfonate (MTSL). All spin-labeled Bax variants retained membrane-permeabilizing activity based on calcein release from LUVs (Figure 1B and Figure S1B available online; detailed information is given in Supplemental Information). Moreover, we checked that Bax cysteine variants were cytosolic in healthy cells and translocated into distinct foci at mitochondria after apoptosis induction (in line with Nechushtan et al., 2001), indicating that the mutants used for the EPR measurements are functionally active in cells (Figures 1C and S1C).
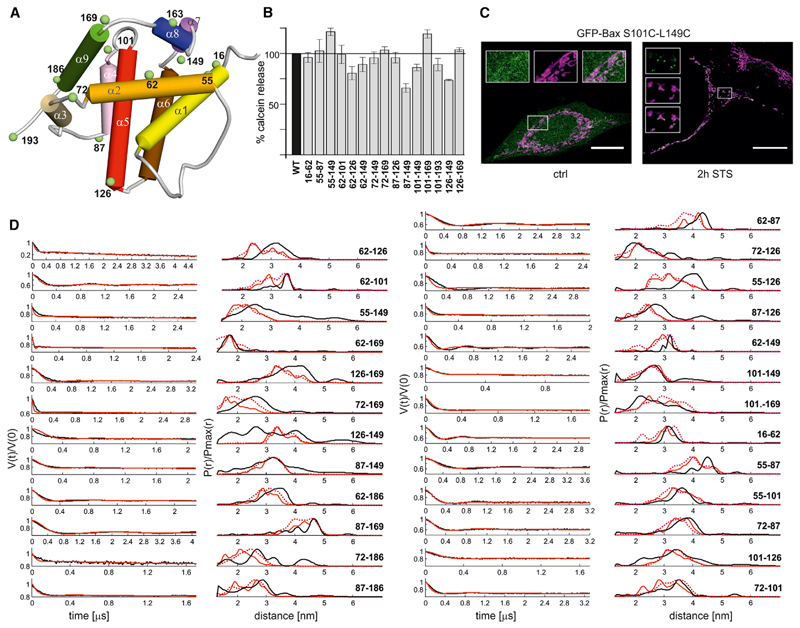
Figure 1. Activity and Folding of the Bax Mutants.
(A) Cartoon model of inactive Bax (NMR model 8, PDB 1F16) with the location of spin labels (green, Cα atoms). Color code of the helices: yellow (α1), orange (α2), gold (α3), pink (α4), red (α5), brown (α6), violet (α7), blue (α8), and green (α9).
(B) Calcein release assay from LUVs with Bax wild-type and spin-labeled mutants (the positions of the spin labels are indicated below the bars; data normalized to WT Bax (black bar); shown are mean values and SEM (n = 6). All mutants are shown in Figures S1A and S1B.
(C) Fixed HeLa cells cotransfected with Mito-DsRed2 (magenta) and GFP-Bax S101C; L149C (green, wild-type Bax and additional mutants are shown in Figure S1C). Control conditions compared to 2 hr incubation with 1 μM staurosporine. Scale bar represents 20 μm. Insets show zooms of the different channels.
(D) Experimental Q-band DEER traces V(t)/V(0) (black, left panels) and distance distributions obtained via Tikhonov regularization with DeerAnalysis2013 (Jeschke et al., 2006) (black, right panels). The V(t)/V(0) traces are the raw DEER data and they show the modulation of the normalized echo intensity versus dipolar evolution time. Superimposed to the experimental time traces and distance distributions, those simulated with MMM2013.2 (Polyhach et al., 2011) on NMR model 8. The average distance distributions simulated using all NMR models are also presented in dotted red (the extracted mean distances are presented in Figure S1E).
See also Figure S1.
Finally, the correct fold of all doubly spin-labeled monomeric proteins was monitored by comparing the intramonomeric distances obtained for 25 doubly spin-labeled mutants in water with those simulated on the NMR structures (Suzuki et al., 2000) with the MTSL rotamer library approach implemented in the software MMM2013.2 (Polyhach et al., 2011). The NMR model 8 best fitted our experimental primary DEER traces (Figure S1D). Figure 1D shows DEER traces and distance distributions of all spin pairs compared to those simulated on model 8. The statistical analysis of the deviation between experimental mean distances and those simulated with MMM2013.2 on model 8 (Figure S1E) gave a root-mean-square deviation (rmsd) of 3.16 Å, close to the 3.5 Å value calculated using a training set of experimental and simulated distances on water soluble proteins and membrane proteins (Jeschke, 2013). This proves the very good agreement between the experimental distances extracted from spin-labeled Bax and the NMR structure (see also Supplemental Information).
The Core Helices 2–5 Build a Stable Dimerization Domain
To study the active, membrane-embedded Bax conformation, we incubated the spin-labeled variants with cBid in the presence of MOM-LUVs, which led to Bax insertion and oligomerization in the membrane (Bleicken et al., 2010, 2013b; Lovell et al., 2008). Figure 2 shows the distance distributions obtained in membrane-embedded Bax compared to those obtained in the water-soluble conformation (corresponding DEER traces in Figures S2A–S2D). Notably, the doubly labeled variants were spin diluted with unlabeled wild-type Bax (molar ratio 1:4) prior to incubation with the MOM-LUVs. This allows enrichment of the signal of the intramonomer distance, while suppressing the intermonomeric distances and maintaining a sufficient signal-to-noise ratio (see Figures S3A–S3C and Supplemental Information). By analyzing the intramonomeric distance changes from soluble to membrane-embedded Bax, we grouped the data in two sets showing minor (Figures 2B and 2C) or major (Figures 2D–2G) changes.
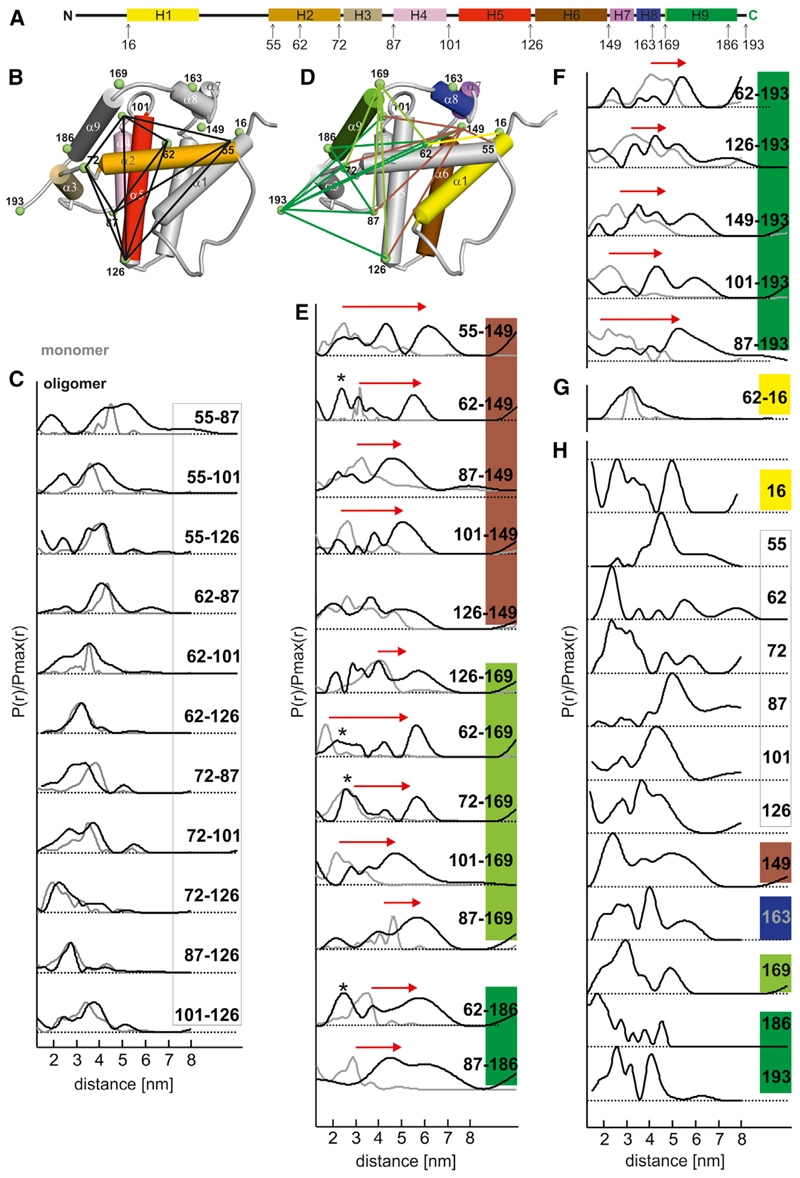
Figure 2. Conformational Changes Induced by Bax Activation.
(A) Location of the spin-labeled residues in a schematic view of the protein. Color code of the helices and corresponding spin-labeled sites: yellow (α1), orange (α2), gold (α3), pink (α4), red (α5), brown (α6), violet (α7), blue (α8), and green (α9). Site 169, in the loop connecting helices α8-α9, is associated with the light green color and the C-terminal site 193 with the green color (as for sites in helix α9).
(B) Model of inactive Bax (PDB: 1F16, model 8) with helices 2–5 colored according to (A) and the rest of the protein colored in gray. Black lines connect spin-labeled pairs within the core helices 2–5, which show minor distance changes upon Bax activation.
(C) Amplitude-normalized distance distributions in the soluble monomeric (gray) and membrane-embedded oligomeric (black) state in the dimerization domain.
(D) Model of inactive Bax (PDB: 1F16, model 8) with helices 1 and 6–9 colored according to (A) and the rest of the protein colored in gray. Lines connect spin labels from the dimerization domain to positions 149 (brown lines), 169 (light green), 193–186 (green), and 16 (yellow), which show major distance changes upon Bax activation.
(E–G) Corresponding amplitude-normalized distance distributions from doubly labeled mutants in the soluble monomeric (gray) and membrane-embedded oligomeric (black) state. Asterisks denote peaks assigned to intermonomer distances (see Figure S4). Red arrows highlight the more relevant distance changes.
(H) Distance distributions in membrane-embedded Bax obtained with singly labeled mutants. All DEER traces are shown in Figure S2. The mean distances extracted from the presented distance distributions are shown in Table 1 and Figure S2E.
See also Figures S3 and S4.
In the membrane-embedded conformation, spin pairs within the core helices α2–α5 showed minor changes in the mean intramonomer distances with respect to the monomeric inactive conformation (Figure 2C), indicating that the protein’s tertiary structure was mostly retained upon Bax activation. Most distances were monomodal and relatively narrow, typical of stable, well-defined conformation. However, Bax’s quaternary structure was affected, in fact two core domains built a stable dimerization interface, as we detected defined distances between singly labeled mutants of this region (from C55 to C126; Figure 2H). Considering the functional role of this region of the protein with respect to the C-terminal part (see below), we named helices 2–5 the dimerization domain of active Bax.
The distances within α2–α5 in membrane-embedded Bax (Figure 2C) are compared in Figure 3 with those simulated on the X-ray structure of truncated dimeric Bax fused to GFP (Czabotar et al., 2013) (PDB: 4BDU). The structure was elongated in silico up to residue 126, using helix α5 of monomeric Bax (PDB: 1F16, model 8). The 4.01 Å rmsd between experimental and simulated mean distances (Figure S2E; Supplemental Information) is close to the reference value of 3.5 Å (Jeschke, 2013), which proves that the X-ray structure is a reliable representation of the dimerization domain of active full-length Bax, which had been questioned due to the protein truncation and crystallization in absence of membranes (detailed information on the methodology is given in the Supplemental Information).
Figure 3. The Crystal Structure of the Truncated α2-5 Dimer in Detergent Represents the Dimerization Domain of Full-Length Bax at the Membrane.
(A) Truncated dimeric structure of Bax (PDB: 4BDU) with helix 5 elongated up to residue C126 using as template the helix of monomeric Bax (PDB: 1F16, model 8). As an example the calculated MTSL rotamers at C62 in the BH3 domains (helices α2) and at 126 at the end of helices α5 are shown in stick representation. The colored spheres represent the population density of the nitroxide radical.
(B) Left, simulated (gray, red) versus experimental (black, as in Figure S2) DEER traces of the singly labeled variants after background correction (F(t)/F(0)). Right, simulated (gray, red) versus experimental (black, as in Figure 2) distance distributions. Color code: gray, pairs in the crystal structure (PDB: 4BDU); red: pairs involving C126 (in the elongated helix α5).
See also Figures S2 and S3.
Additionally, we found that the distance between position 16 in α1 and position 62 in the BH3 domain became broadly distributed upon Bax activation (Figure 2G). A broad distance distribution was found also for Bax singly labeled at position 16 (Figure 2H), indicating flexibility of α1 at the membrane (in agreement with Bleicken and Zeth, 2009; Gavathiotis et al., 2010; Hsu and Youle, 1998; Kim et al., 2009; Yethon et al., 2003).
The peak assignment of spin-diluted doubly labeled proteins having one label in the dimerization domain and the other in the piercing domain was complicated due to the broad distance distributions and unavoidable residual intermonomer peaks (Figures 2E and 2F). To suppress intermonomer distances we used the spin dilution approach mentioned before. This was combined with the assignment of residual intermonomer peaks using the experimentally obtained distances on singly labeled proteins. Additionally, to further corroborate the data, we measured spin-diluted doubly labeled mutants in detergent to increase the signal to noise and prolong the dipolar evolution time (Figures S3 and S4). Table 1 presents the final distance constraints obtained (see also the Supplemental Information).
Table 1. DEER Distance Constraints.
| Spin Pair | Mean Distance (nm) | σ (nm) |
|---|---|---|
| 149-62 | 5.43 | 0.48 |
| 149-101 | 4.42 | 1.01 |
| 149-55 | 4.60 | 0.86 |
| 149-72 (detergent) | 5.25 | 0.58 |
| 149-126 | 3.86 | 0.39 |
| 149-87 | 4.33 | 0.37 |
| 149-149a | 2.39 | 0.34 |
| 169-62 | 5.7 | 0.6 |
| 169-72 | 6.0 | 0.5 |
| 169-87 | 5.1 | 1.4 |
| 169-101 | 4.6 | 0.7 |
| 169-126 | 4.3 | 1.0 |
| 169-169b | 1.6 (3.0) | 0.2 (0.5) |
| 163-163b | 2.6 (4.5) | 0.5 (0.9) |
| 186-62 | 5.6 | 1.6 |
| 186-87 | 4.7 | 1.0 |
| 186-149 | 4.12 | 0.92 |
| 186-186c | <2.5 | NA |
| 193-62 | 5.6 | 0.45 |
| 193-101 | 4.8 | 0.7 |
| 193-149 | 4.5 | 0.9 |
| 193-126 | 4.7 | 0.9 |
| 193-87 | 5.4 | 0.7 |
| 193-193b | 2.6 (4.5) | 0.5 (0.9) |
NA, not assigned.
The 149-149 constraints at 2.4 nm could be assigned to the intradimer distance based on the symmetric model of helices 6 in the dimer obtained solely with the doubly labeled mutants. However, it was not used to create the isosurface shown in Figure 4A.
For 163-163, 169-169, and 193-193, the bimodal broad distribution was analyzed in terms of two mean distances and widths. The longer distances are given in parentheses.
For 186-186, distances <2.5 nm could be detected in agreement with the crosslinking experiments.
It is to be noted that due to the oligomeric nature of active Bax at the membrane, both intra- and interdimer distances are detected with singly spin-labeled Bax variants, therefore their assignment is difficult (Figure 2H). However, in the dimerization domain, the distance distributions were mainly monomodal and using the available information from the truncated and GFP-fused Bax-dimer (Czabotar et al., 2013), we could annotate them without problems. In contrast, for the broadly distributed distances in the C-terminal part of Bax the assignment was only possible in a few cases by performing crosslinking experiments or by analysis of the model obtained solely with doubly labeled proteins, as described later in the text.
Opening of the α5-α6 Hairpin Assembles a Flexible Piercing Domain
When one spin label was positioned in the dimerization domain and the second one in the C-terminal region of Bax, we detected changes of the mean distances up to 2–4 nm upon membrane insertion (Figures 2E and 2F), indicating large rearrangements in the C-terminal region of the protein. As helices α6 to α9 are involved in membrane-insertion and pore formation (Annis et al., 2005; García-Sáez et al., 2006; Nechushtan et al., 1999), we refer to this C-terminal half of active Bax as the piercing domain. It is to be noted that helix α5, which belongs to the dimerization domain is necessary for both Bax dimerization (George et al., 2007) and pore formation (Annis et al., 2005; García-Sáez et al., 2006).
In the “umbrella” model, the central hairpin of helices 5 and 6 acts as a membrane insertion domain around which the rest of the protein helices unfold on the membrane surface. Strikingly, we detected conformational changes incompatible with a persistent hairpin fold when we considered the spin pair connecting the beginning and the end of the hairpin (residues 101 and 149). In monomeric Bax, this pair shows a 2.5 nm distance fingerprint of the hairpin fold, but upon Bax activation this distance increased to 5 nm (Figure 2E). Moreover, other distances between spin pairs connecting the dimerization domain to position 149 became longer and broadly distributed upon Bax membrane insertion (Figure 2E). Therefore, the “umbrella” model does not adequately represent Bax conformation in the membrane.
The recently published structure of a domain-swapped dimer of Bax triggered by detergent (Czabotar et al., 2013) captured an off-pathway full opening of helices 5 and 6. We simulated intramonomer distances between the dimerization domain and position 149 or 169 in the swapped dimer and found that they resemble our experimental DEER distances. However, the intermonomer distances within the swapped dimer are incompatible with our data (Figure S3D), confirming that the domain-swapped dimer does not represent the conformation of active membrane-embedded Bax. Notably, by treating soluble Bax with low detergent concentration we obtained experimental hints pointing to a measurable fraction of domain swapped dimer in the sample (Figures S3E and S3F), suggesting that the presence of detergent molecules in low amounts is responsible for the formation of this nonphysiological structure.
Next, we analyzed the distances between residue 169 (beginning of C-terminal helix α9) and the dimerization domain in membrane-embedded Bax. The most drastic distance change upon Bax activation (4 nm) was found between position 62 in helix α2 and position 169 (Figure 2E), indicating that the opening of the hairpin α5–α6 pulled the C-terminal region away from the dimerization domain. A significant distance increase was also detected between 101 or 87 and 169 (Figure 2E). The end of helix α9 was mapped with spin labels at residues 193 (C terminus) and 186 (end of helix α9). We observed that the mean distances to the dimerization domain also increased up to 3 nm (e.g., 87–193 and 101–193, Figure 2E) upon membrane insertion, corroborating the idea that the globular fold in aqueous buffer is lost upon Bax activation. The large width of the distance distributions demonstrates that helix α9 in membrane-inserted Bax is structurally dynamic. This is illustrated by the distances between 62, 87, and 186. The 62–87 distance in the dimerization domain is narrowly distributed (Figure 2C), but spin pairs containing one of these two sites paired with 186 in helix α9 gave a broad distance distribution (62–186 and 87–186, Figure 2E), indicating that the flexibility comes from the label in α9.
Modeling Active Bax at the Membrane
From the collected DEER constraints, we built a structural model of full-length membrane-embedded Bax. As a starting point we used the X-ray structure of helices α2–α5 extended up to position 126, which agrees with our experimental constraints within this domain. We localized the spin labels at positions 149, 169, 186, and 193 with respect to the dimerization domain by applying the nanoscale multilateration process available in the software MMM2013.2 (Polyhach et al., 2011). This approach uses the basic principles of GPS localization, but adapted to the nitroxide midpoint of spin labels in proteins (see Supplemental Information). We validated this multilateration approach on monomeric Bax in water, for which we had six sites constrained by more than four DEER distances. The highest location probability obtained for each labeled site shows a 4.9 Å rmsd with respect to the mean location of the nitroxide labels simulated with MMM2013.2 on the available NMR structure (model 8) (see Figure S5C and Supplemental Information). We did the same with the coordinates of three sites (62, 87, and 126) localized via multilateration on the crystal structure of the dimerization domain (PDB: 4BDU) and found an rmsd of 7.8 Å (Figure S5D and Supplemental Information). For membrane-inserted Bax, we therefore consider 8 Å to be the accuracy of this approach in localizing the spin-labeled sites in active Bax at the membrane.
Using the DEER constraints obtained solely from the doubly labeled mutants, we found that the spin label 149 is localized ~4 nm below the BH3-BH3 dimerization helices (Figures 4A and S5B). To model the two helices 6 in the dimer, we assumed that all helices maintained their length and shape, based on the invariant circular dichroism spectra of inactive versus active Bax (Bleicken and Zeth, 2009; Yethon et al., 2003) and that the C2 symmetry axis in the dimerization domain applies to at least helix α6.
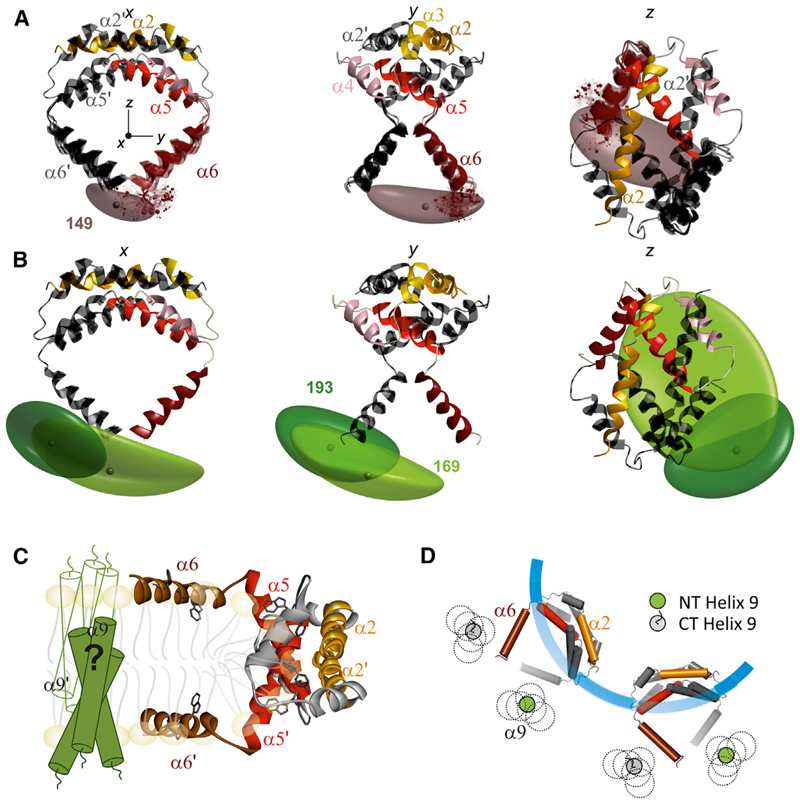
Figure 4. Model of Active Dimeric Bax at the Membrane.
(A) Three orthogonal views of Bax dimerization domain (PDB: 4BDU; with helix 5 elongated up to C126) and with helix 6 modeled using the DEER constraints and a multilateration approach (Table 1; see also Figure S5A and Supplemental Information). Chain A is color-coded as in Figure 1A and chain B is in dark gray. The z axis corresponds to the C2 symmetry axis, and the origin of the coordinate system is placed at the center of mass of the BH3-BH3 dimerization interface. The brown cloud is the isosurface presenting the localization of the spin label at position 149 of chain A determined only by the intramonomeric distance constraints. Four models of helices 6 in chain A (colored) and B (dark gray) obtained applying the C2-symmetry transformation and the additional 149-149 constraint (mean distance 2.4 nm) are shown. Solid ribbons, model with highest probability (64%); transparent ribbons, three additional models with >50% relative probability.
(B) The most probable model from (A) is shown together with the isosurfaces for the localization of the spin labels at position 169 (light green) and 193 (dark green) in chain A based only on the intramonomeric constraints (Table 1; the isosurface of the label at 186 is shown in Figure S5B).
(C) Hypothetical clamp model for the topology of active Bax dimers at the MOM: the dimerization domain is at the rim of a pore induced by Bax in the membrane, with helices 5 and 6 lying on the membrane surface. The hypothetical symmetric and transmembrane location of the dynamic helices 9 is sketched.
(D) Schematic top view of two adjacent Bax dimers in the pore based on the proposed structural model (C) and compatible with a transmembrane orientation of the dynamic helices 9. The colored helix α6 in the dimer is above the membrane, while the α6 depicted in gray is laying spatially below the one colored, so that the corresponding helix 6 is below the membrane plane. The possible locations of the dynamic N and C termini (NT, CT) of helices 9 are shown.
See also Figure S5.
Based on the C2 symmetry axis assumption, we found that in the dimer the experimental 2.4 nm 149-149 distance constraint was consistent with this probability density. However, we cannot exclude that also the closest interdimer distance is in the 2 nm range overlapping with the intradimer distance, supporting the notion that helix 6 is involved in interdimer contacts (Dewson et al., 2009, 2012), while the long experimental distances arise from other pairwise interactions within the oligomers.
By modeling the orientation of helix 6 (residues 130–149), allowing residues 126–129 to freely adopt their respective Ramachandran dihedral angles, we found that residues 127–129 between helices 5 and 6 introduce a kink (the four most probable models are shown in Figure 4A), likely favored by a conserved proline at the beginning of helix 6. Bak contains a glycine in an equivalent position, which may induce the same effect.
Based on the model up to 149, we calculated the isosurfaces describing the location probability of the spin labels at positions 169, 186, and 193 (Figures 4B and S4B). Both 169 and 193 are found ~4 nm below the BH3-BH3 dimerization domain. The broad distance distribution of the constraints obtained (Table 1) resulted in extended isosurfaces. Importantly, this indicates that helix 9 has large structural flexibility and that the C-terminal region of Bax in its membrane-embedded structure should not be described by a unique conformation, but rather by a conformational ensemble.
Validation of the Structural Model for Active Bax
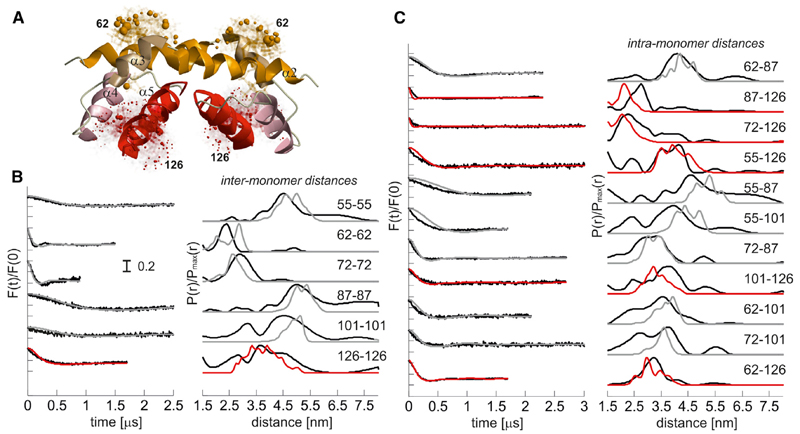
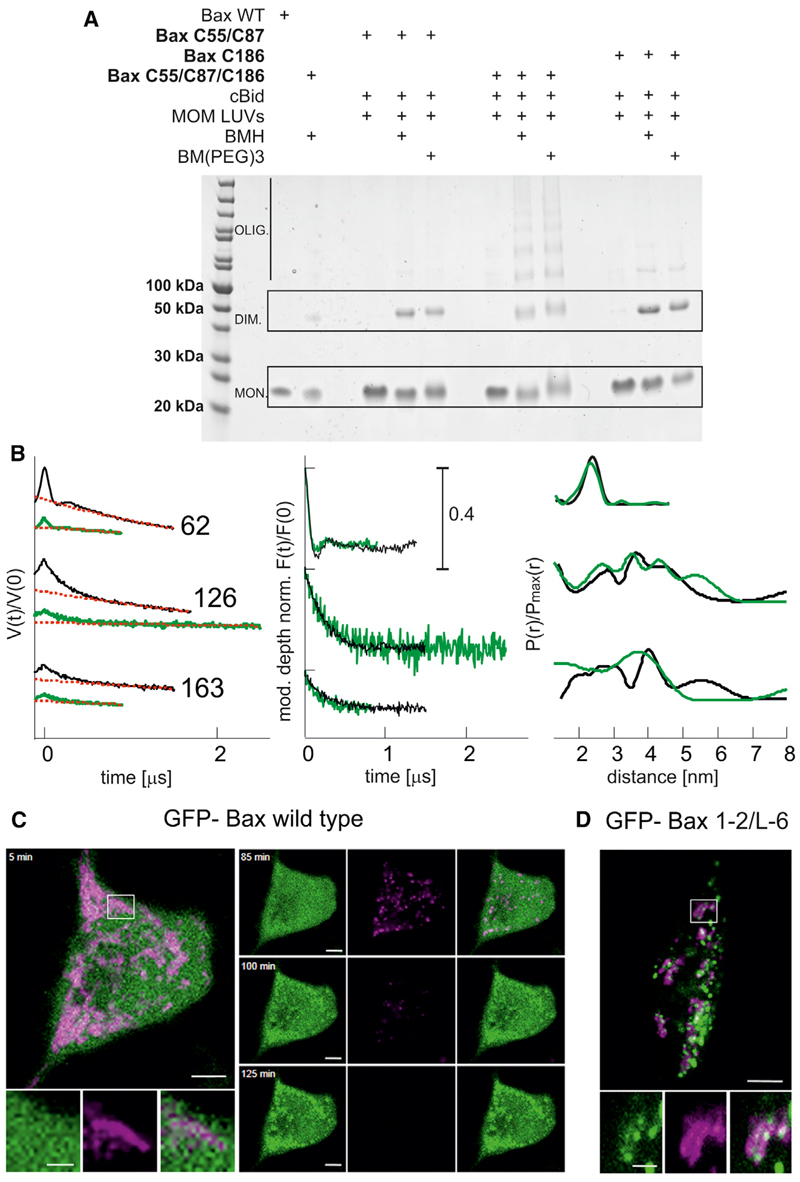
We found short distances for all spin labels placed in the piercing domain (149, 163, 169, 186, and 193), suggesting their involvement in dimer-dimer interactions. The broad localization probability (169, 186, and 193) supports a flexible interdimer interface, which, in the context of Bax forming pores of tunable size (Bleicken et al., 2013a), allows the oligomers to grow during pore formation. To validate the concept that the structure of active Bax in membranes contains a flexible piercing domain involved in interdimer interactions, we performed crosslinking experiments (Figure 5A). We crosslinked the two monomers in a dimer via positions 55 and 87. This produced dimers but not higher order oligomers (Figure 5A), in line with literature data (Dewson et al., 2012). When using the triple mutant C55/C87/C186, bigger complexes were crosslinked (Figure 5A), demonstrating that the multimerization occurs via an interface involving the piercing domain up to helix α9. The involvement of α9 in Bax homo- and hetero-oligomerization was proposed previously in crosslink experiments (Ding et al., 2010, 2014; Zhang et al., 2010).
Figure 5. Validation of the Proposed Model.
(A) Helix 9 is involved in interdimer interactions, as proven by crosslinking. Three different Bax mutants are used to selectively connect the proposed intradimer interface (Bax C55, C87) and the proposed interdimer interface (Bax C186) or both interfaces (Bax C55, C87, C186) within membrane-embedded Bax oligomers. The bismaleimide crosslinkers BMH (1.3 nm) and BM (PEG)3 (1.8 nm) were used.
(B) Bax adopts the same conformation in liposomes and mitochondria. Left: experimental Q-band DEER traces V(t)/V(0) with background function (dotted red) performed with DeerAnalysis2013 (Jeschke et al., 2006). Color code: black, data in liposomes; green, data in isolated mitochondria. Middle: modulation depth normalized background corrected traces (F(t)/F(0)). Right: comparison of the obtained distance distributions.
(C) Bax-GFP is soluble in healthy HeLa cells and translocates to mitochondria concomitant with potential loss upon apoptosis induction (control experiment).
(D) Crosslinked GFP-Bax 1-2/L6 is trapped in a globular protein fold and is constitutively bound to the mitochondrial membrane without inducing permeabilization. Color code: GFP-Bax, green; membrane potential sensitive dye TMRE, magenta. Scale bar represents 5 μm and 2 μm for zoomed images. At t = 0, 1 μM staurosporine was added to the cells.
To validate the physiological relevance of the data obtained in LUVs, we measured distances in spin-labeled Bax inserted into isolated mitochondria (Figure 5B). The reducing agents released from mitochondria during Bax incorporation induce spin label dissociation and reduction of the nitroxide, decreasing signal intensity and DEER modulation depth. Nevertheless, due to the increased signal-to-noise ratio of the high-power Q-band instrument (Polyhach et al., 2012), we could obtain DEER traces of acceptable quality for three mutants in the dimerization and piercing domains in mitochondria-embedded Bax. The distances showed good agreement with those obtained in liposomes (Figure 5B), demonstrating their physiological relevance.
Here, we demonstrate the unfolding of the hairpin of helices 5 and 6 in active Bax into an unexpected clamp-like structure contrary to the “umbrella” model. Accordingly, a Bax mutant with internal crosslinking hindering the unfolding of the piercing domain (Edlich et al., 2011) was constitutively bound to mitochondria, but did not induce loss of mitochondrial potential, indicating that it is unable to permeabilize the MOM (Figure 5C). Moreover, crosslinking helices 5–6 in Bax blocked cytochrome c release from isolated mitochondria (Czabotar et al., 2013). These data indicate that the clamp-like structure formed by helices 5 and 6 is critical for Bax-induced MOM permeabilization.
Discussion
Here we propose a structural model of active Bax inserted in the membrane. It is based on 42 distance constraints between 12 different spin-labeled positions of full-length Bax in lipid bilayers mimicking the MOM. We found that the core helices α2–α5 maintain a similar conformation in the inactive water-soluble and active membrane-embedded states. In the membrane, this region is directly involved in the formation of a well-defined dimerization domain similar to that adopted by truncated Bax-GFP in detergent (Czabotar et al., 2013). The relatively narrow distributions of intradimer distances within the dimerization domain indicate that this part of the protein assembles a stable basic dimeric unit of Bax at the membrane, in agreement with previous studies (Bleicken et al., 2010; Czabotar et al., 2013; Dewson et al., 2012).
We found that the C-terminal half of Bax (beyond helix 5), undergoes dramatic reorganization and adopts a dynamic configuration at the membrane. Czabotar et al. (2013) found that Bax activation is characterized by the displacement of the latch (helices 6–9 and eventually 1) from the core of the protein (helices 2–5) via opening of the hairpin. Whether this opening is transient and occurs in an early stage of the activation or it persists in the oligomeric state after the pore is formed, was left as an open question (Czabotar et al., 2013). Additionally, the conformation of helices 5 and 6 in the active, membrane-embedded Bax was so far unknown. Here, we prove that the α5-α6 hairpin is not stably present in the active Bax oligomers at the membrane, a notion recently confirmed with EPR on activated truncated Bak as well (Aluvila et al., 2014). Importantly these two helices assemble in the dimer into a clamp-like structure with a kink around proline 130. This disproves previous models for Bax activation, like the umbrella or the penknife models, both of which assumed the permanence of the hairpin upon Bax activation (Annis et al., 2005). Accordingly, a crosslinked mutant that cannot disengage helix 6 from the core of Bax was found not to induce MOM permeabilization even when it was constitutively bound to mitochondria (Figure 5C).
DEER measurements involving positions 169, 186, and 193 yielded complex distance patterns with mainly broad distributions. This shows that the C-terminal part of Bax exhibits conformational dynamics and is involved in interdimer interactions. In the context of Bax pores of different size (Bleicken et al., 2013a), these C-terminal helices provide a flexible interface where the distribution of conformations is the natural consequence of the different species in the membrane. Importantly, these findings challenge the paradigm of a single fully defined atomistic structure as a prerequisite for understanding the function of Bax.
The proposed structural model of active Bax dimers at the membrane suggests a speculative mechanism for membrane permeabilization through destabilization of the lipid bilayer, which is presented in Figures 4C and 4D. Bax activation is triggered by the interaction with a BH3-only protein close to the membrane that induces a conformational change within Bax. This leads to the formation of a stable Bax dimer via the dimerization domain and to the opening of the α5-α6 hairpin that assembles the flexible piercing domain on the membrane. In Bax dimers four helices of the dimerization domain (4,-5,4′-5′) build a surface containing 12 aromatic and 16 hydrophobic amino acids, to which each helix 6 adds one more tryptophan and six hydrophobic residues. The resulting surface is therefore a good candidate for membrane contacts. In the model proposed the two arms of the “clamp” (helices 6) are separated from each other by ~3 nm, which fits well with the membrane thickness. The piercing domain thus assembled is proposed to pinch the membrane, possibly stabilizing a toroidal pore in the MOM as proposed in Figures 4C and 4D. This model is in line with the accessibility data on which the umbrella/hairpin model was based (Annis et al., 2005; García-Sáez et al., 2006), showing helices 5 and 6 shielded from the bulk water. Finally, the piercing domain, and in particular helix 9, adopt a flexible configuration and have a role in Bax multimerization. Nevertheless, and despite the role of helix 9 in the early steps of membrane targeting and anchoring (George et al., 2010; Nechushtan et al., 1999), Bax lacking this helix retains ability to oligomerize and permeabilize the MOM (Antonsson et al., 2000; Czabotar et al., 2013), confirming the central role of helices 5–6 to create the pore in the MOM, as suggested here.
To summarize, we propose a model for the 3D structure of full-length, active Bax in the membrane calculated from DEER distance constraints. In the lipid bilayer, the N-terminal half of Bax forms a well-defined dimerization surface with a structure similar to the crystallized soluble truncated protein. In contrast, the C-terminal region of Bax shows conformational dynamics in agreement with its role in interdimer interactions. The most significant reorganization of Bax involves the partial opening of the hairpin of helices 5 and 6 that assembles into a clamp-like structure, which is suggested to play an essential role in the stabilization of the membrane pore.
Experimental Procedures
Protein Production, Spin Labeling, and Sample Preparation
cBid, Bax wild-type, and all the Bax mutants were purified and labeled as described in Bleicken et al. (2010, 2012). The Bax mutants were cloned by site-specific mutagenesis. MOM-LUVs and isolated mitochondria were prepared as in Bleicken et al. (2012) and the calcein assays were performed as in (Bleicken et al. (2013b). Spin labeling was performed with the MTSL spin label (1-Oxyl-2,2,5,5-tetramethyl-Δ3-pyrroline-3-methyl Methanethiosulfonate; Toronto Research Chemicals) as described in Bleicken et al. (2010).
For the membrane-embedded samples, doubly labeled Bax (3–4 μM mixed with 10–15 μM unlabeled WT Bax; final concentrations) or singly labeled Bax (5–10 μM) was incubated for 2 min on ice with cBid (1–2 μM). Freshly prepared LUVs were added to the solution (final concentrations: ~10 mM lipids in a total volume of 250–400 μl) and the suspension was incubated for 1 hr at 37°C. The mixture was spun down for 30 min at 120,000 × g to pellet the LUVs. The pellet was separated from the supernatant and resuspended with 20 μl buffer (150 mM NaCl, 20 mM TRIS, pH 7.5). In this way the spin-labeled Bax variants were concentrated 5- to 10-fold and residual soluble proteins were removed. Deuterated glycerol (10% v/v) was added to the sample as cryoprotectant. The sample was inserted into a quartz tube and snap frozen in liquid nitrogen.
DEER Measurements
DEER measurements were performed on a home-built or commercial (Bruker ELEXSYS-II E580) Q-band spectrometer (34–35 GHz) equipped with a TWT amplifier (150–200 W) and a home-made rectangular resonator enabling the insertion of 30–40 μl of sample in quartz tubes with 3 mm outer diameter. Dipolar time evolution data were acquired using the four-pulse DEER experiment at 50 K. All pulses were set to 12 ns with the pump frequency set at the maximum of the echo-detected field swept spectrum, 100 MHz higher than the observer frequency, as in Polyhach et al. (2012). Data analysis was performed with DeerAnalysis2013 (Jeschke et al., 2006). Spin-label positions in the unknown piercing domain of Bax were localized by multilateration from reference points in the dimerization domain and determination of a probability density distribution. Its application to spinlabeled proteins is available in the freely available open-source software used here (MMM2013.2).
Crosslink Experiments
Bax C55-C87, Bax C186, or Bax C55-C87-C186 were linked with the maleimide-based crosslinkers BMH or BM(PEG)3 (Thermo Scientific). For each Bax variant a 100 μl reaction mix was prepared containing LUVs (2 mM lipid), cBid (0.5 μM), and Bax (6.5 μM) and incubated for 45 min at 37°C. The sample was separated into two equal fractions and each fraction was mixed with one of the two crosslinkers (in 5× excess over Bax concentration) and incubated 45 min at RT. Afterward, each mixture was incubated for 5 min at 95°C with SDS loading buffer and the samples were loaded on a Gradient SDS-PAGE (Nu-PAGE, Invitrogen).
In-Cell Experiments
HeLa cells were maintained in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% heat-inactivated fetal calf serum (FCS) (Invitrogen) and 1% penicillin/streptomycin in 5% CO2 at 37°C. Cells were seeded and transfected with Lipofectamine 2000 (Invitrogen) typically with 100 ng of GFP-Bax or GFP-Bax 1-2/L-6 constructs or 100 ng of GFP-Bax mutants and 100 nM pDS-Red-Mito (Clontech) according to the manufacturer’s instructions. Cells were maintained at 37°C and 5% CO2 on DMEM without phenol red supplemented with FCS and containing 1 μM staurosporine for apoptosis induction. Experiments were performed on a Zeiss LSM 710 microscope and images were captured over 2 hr at 2 min intervals and analyzed using Fiji.
Please see the Supplemental Information for complete methods.
Supplemental Information
Supplemental Information includes Supplemental Experimental Procedures and five figures and can be found with this article online at http://dx.doi.org/10.1016/j.molcel.2014.09.022.
Acknowledgments
We thank J. Suckale for careful reading of the manuscript and E. Conti and F. Megli for providing the isolated mitochondria. This work was supported by ETH Zurich, the Max Planck Society, the German Cancer Research Center, the German Ministry for Education and Research (BMBF, grant 0312040), DFG grant FOR2036, SNF grant 200020_144441, and ERC-2012-StG_20111109.
References
- Aluvila S, Mandal T, Hustedt E, Fajer P, Choe JY, Oh KJ. Organization of the mitochondrial apoptotic BAK pore: oligomerization of the BAK homodimers. J Biol Chem. 2014;289:2537–2551. doi: 10.1074/jbc.M113.526806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Annis MG, Soucie EL, Dlugosz PJ, Cruz-Aguado JA, Penn LZ, Leber B, Andrews DW. Bax forms multispanning monomers that oligomerize to permeabilize membranes during apoptosis. EMBO J. 2005;24:2096–2103. doi: 10.1038/sj.emboj.7600675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antonsson B, Montessuit S, Lauper S, Eskes R, Martinou JC. Bax oligomerization is required for channel-forming activity in liposomes and to trigger cytochrome c release from mitochondria. Biochem J. 2000;345:271–278. [PMC free article] [PubMed] [Google Scholar]
- Bleicken S, Zeth K. Conformational changes and protein stability of the pro-apoptotic protein Bax. J Bioenerg Biomembr. 2009;41:29–40. doi: 10.1007/s10863-009-9202-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bleicken S, Classen M, Padmavathi PV, Ishikawa T, Zeth K, Steinhoff HJ, Bordignon E. Molecular details of Bax activation, oligomerization, and membrane insertion. J Biol Chem. 2010;285:6636–6647. doi: 10.1074/jbc.M109.081539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bleicken S, García-Sáez AJ, Conte E, Bordignon E. Dynamic interaction of cBid with detergents, liposomes and mitochondria. PLoS ONE. 2012;7:e35910. doi: 10.1371/journal.pone.0035910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bleicken S, Landeta O, Landajuela A, Basañez G, García-Sáez AJ. Proapoptotic Bax and Bak proteins form stable protein-permeable pores of tunable size. J Biol Chem. 2013a;288:33241–33252. doi: 10.1074/jbc.M113.512087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bleicken S, Wagner C, García-Sáez AJ. Mechanistic differences in the membrane activity of Bax and Bcl-xL correlate with their opposing roles in apoptosis. Biophys J. 2013b;104:421–431. doi: 10.1016/j.bpj.2012.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brouwer JM, Westphal D, Dewson G, Robin AY, Uren RT, Bartolo R, Thompson GV, Colman PM, Kluck RM, Czabotar PE. Bak Core and Latch domains separate during activation, and freed Core domains form symmetric homodimers. Mol Cell. 2014;55:938–946. doi: 10.1016/j.molcel.2014.07.016. [DOI] [PubMed] [Google Scholar]
- Czabotar PE, Westphal D, Dewson G, Ma S, Hockings C, Fairlie WD, Lee EF, Yao S, Robin AY, Smith BJ, et al. Bax crystal structures reveal how BH3 domains activate Bax and nucleate its oligomerization to induce apoptosis. Cell. 2013;152:519–531. doi: 10.1016/j.cell.2012.12.031. [DOI] [PubMed] [Google Scholar]
- Czabotar PE, Lessene G, Strasser A, Adams JM. Control of apoptosis by the BCL-2 protein family: implications for physiology and therapy. Nat Rev Mol Cell Biol. 2014;15:49–63. doi: 10.1038/nrm3722. [DOI] [PubMed] [Google Scholar]
- Dewson G, Kratina T, Czabotar P, Day CL, Adams JM, Kluck RM. Bak activation for apoptosis involves oligomerization of dimers via their alpha6 helices. Mol Cell. 2009;36:696–703. doi: 10.1016/j.molcel.2009.11.008. [DOI] [PubMed] [Google Scholar]
- Dewson G, Ma S, Frederick P, Hockings C, Tan I, Kratina T, Kluck RM. Bax dimerizes via a symmetric BH3:groove interface during apoptosis. Cell Death Differ. 2012;19:661–670. doi: 10.1038/cdd.2011.138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding J, Zhang Z, Roberts GJ, Falcone M, Miao Y, Shao Y, Zhang XC, Andrews DW, Lin J. Bcl-2 and Bax interact via the BH1-3 groove-BH3 motif interface and a novel interface involving the BH4 motif. J Biol Chem. 2010;285:28749–28763. doi: 10.1074/jbc.M110.148361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding J, Mooers BH, Zhang Z, Kale J, Falcone D, McNichol J, Huang B, Zhang XC, Xing C, Andrews DW, Lin J. After embedding in membranes antiapoptotic Bcl-XL protein binds both Bcl-2 homology region 3 and helix 1 of proapoptotic Bax protein to inhibit apoptotic mitochondrial permeabilization. J Biol Chem. 2014;289:11873–11896. doi: 10.1074/jbc.M114.552562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edlich F, Banerjee S, Suzuki M, Cleland MM, Arnoult D, Wang C, Neutzner A, Tjandra N, Youle RJ. Bcl-x(L) retrotranslocates Bax from the mitochondria into the cytosol. Cell. 2011;145:104–116. doi: 10.1016/j.cell.2011.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards AL, Gavathiotis E, LaBelle JL, Braun CR, Opoku-Nsiah KA, Bird GH, Walensky LD. Multimodal interaction with BCL-2 family proteins underlies the proapoptotic activity of PUMA BH3. Chem Biol. 2013;20:888–902. doi: 10.1016/j.chembiol.2013.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-Sáez AJ. The secrets of the Bcl-2 family. Cell Death Differ. 2012;19:1733–1740. doi: 10.1038/cdd.2012.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-Sáez AJ, Mingarro I, Pérez-Payá E, Salgado J. Membrane-insertion fragments of Bcl-xL, Bax, and Bid. Biochemistry. 2004;43:10930–10943. doi: 10.1021/bi036044c. [DOI] [PubMed] [Google Scholar]
- García-Sáez AJ, Coraiola M, Serra MD, Mingarro I, Müller P, Salgado J. Peptides corresponding to helices 5 and 6 of Bax can independently form large lipid pores. FEBS J. 2006;273:971–981. doi: 10.1111/j.1742-4658.2006.05123.x. [DOI] [PubMed] [Google Scholar]
- Gavathiotis E, Suzuki M, Davis ML, Pitter K, Bird GH, Katz SG, Tu HC, Kim H, Cheng EH, Tjandra N, Walensky LD. BAX activation is initiated at a novel interaction site. Nature. 2008;455:1076–1081. doi: 10.1038/nature07396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavathiotis E, Reyna DE, Davis ML, Bird GH, Walensky LD. BH3-triggered structural reorganization drives the activation of proapoptotic BAX. Mol Cell. 2010;40:481–492. doi: 10.1016/j.molcel.2010.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- George NM, Evans JJD, Luo X. A three-helix homooligomerization domain containing BH3 and BH1 is responsible for the apoptotic activity of Bax. Genes Dev. 2007;21:1937–1948. doi: 10.1101/gad.1553607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- George NM, Targy N, Evans JJD, Zhang L, Luo X. Bax contains two functional mitochondrial targeting sequences and translocates to mitochondria in a conformational change and homooligomerization-driven process. J Biol Chem. 2010;285:1384–1392. doi: 10.1074/jbc.M109.049924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu Y-T, Youle RJ. Bax in murine thymus is a soluble monomeric protein that displays differential detergent-induced conformations. J Biol Chem. 1998;273:10777–10783. doi: 10.1074/jbc.273.17.10777. [DOI] [PubMed] [Google Scholar]
- Jeschke G. DEER distance measurements on proteins. Annu Rev Phys Chem. 2012;63:419–446. doi: 10.1146/annurev-physchem-032511-143716. [DOI] [PubMed] [Google Scholar]
- Jeschke G. Conformational dynamics and distribution of nitroxide spin labels. Prog Nucl Magn Reson Spectrosc. 2013;72:42–60. doi: 10.1016/j.pnmrs.2013.03.001. [DOI] [PubMed] [Google Scholar]
- Jeschke G, Chechik V, Ionita P, Godt A, Zimmermann H, Banham J, Timmel CR, Hilger D, Jung H. DeerAnalysis2006—a comprehensive software package for analyzing pulsed ELDOR data. Appl Magn Reson. 2006;30:473–498. [Google Scholar]
- Kim H, Tu HC, Ren D, Takeuchi O, Jeffers JR, Zambetti GP, Hsieh JJ, Cheng EH. Stepwise activation of BAX and BAK by tBID, BIM, and PUMA initiates mitochondrial apoptosis. Mol Cell. 2009;36:487–499. doi: 10.1016/j.molcel.2009.09.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindsten T, Ross AJ, King A, Zong WX, Rathmell JC, Shiels HA, Ulrich E, Waymire KG, Mahar P, Frauwirth K, et al. The combined functions of proapoptotic Bcl-2 family members bak and bax are essential for normal development of multiple tissues. Mol Cell. 2000;6:1389–1399. doi: 10.1016/s1097-2765(00)00136-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovell JF, Billen LP, Bindner S, Shamas-Din A, Fradin C, Leber B, Andrews DW. Membrane binding by tBid initiates an ordered series of events culminating in membrane permeabilization by Bax. Cell. 2008;135:1074–1084. doi: 10.1016/j.cell.2008.11.010. [DOI] [PubMed] [Google Scholar]
- Nechushtan A, Smith CL, Hsu Y-T, Youle RJ. Conformation of the Bax C-terminus regulates subcellular location and cell death. EMBO J. 1999;18:2330–2341. doi: 10.1093/emboj/18.9.2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nechushtan A, Smith CL, Lamensdorf I, Yoon SH, Youle RJ. Bax and Bak coalesce into novel mitochondria-associated clusters during apoptosis. J Cell Biol. 2001;153:1265–1276. doi: 10.1083/jcb.153.6.1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polyhach Y, Bordignon E, Jeschke G. Rotamer libraries of spin labelled cysteines for protein studies. Phys Chem Chem Phys. 2011;13:2356–2366. doi: 10.1039/c0cp01865a. [DOI] [PubMed] [Google Scholar]
- Polyhach Y, Bordignon E, Tschaggelar R, Gandra S, Godt A, Jeschke G. High sensitivity and versatility of the DEER experiment on nitroxide radical pairs at Q-band frequencies. Phys Chem Chem Phys. 2012;14:10762–10773. doi: 10.1039/c2cp41520h. [DOI] [PubMed] [Google Scholar]
- Schellenberg B, Wang P, Keeble JA, Rodriguez-Enriquez R, Walker S, Owens TW, Foster F, Tanianis-Hughes J, Brennan K, Streuli CH, Gilmore AP. Bax exists in a dynamic equilibrium between the cytosol and mitochondria to control apoptotic priming. Mol Cell. 2013;49:959–971. doi: 10.1016/j.molcel.2012.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki M, Youle RJ, Tjandra N. Structure of Bax: coregulation of dimer formation and intracellular localization. Cell. 2000;103:645–654. doi: 10.1016/s0092-8674(00)00167-7. [DOI] [PubMed] [Google Scholar]
- Wei MC, Zong WX, Cheng EH, Lindsten T, Panoutsakopoulou V, Ross AJ, Roth KA, MacGregor GR, Thompson CB, Korsmeyer SJ. Proapoptotic BAX and BAK: a requisite gateway to mitochondrial dysfunction and death. Science. 2001;292:727–730. doi: 10.1126/science.1059108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yethon JA, Epand RF, Leber B, Epand RM, Andrews DW. Interaction with a membrane surface triggers a reversible conformational change in Bax normally associated with induction of apoptosis. J Biol Chem. 2003;278:48935–48941. doi: 10.1074/jbc.M306289200. [DOI] [PubMed] [Google Scholar]
- Youle RJ, Strasser A. The BCL-2 protein family: opposing activities that mediate cell death. Nat Rev Mol Cell Biol. 2008;9:47–59. doi: 10.1038/nrm2308. [DOI] [PubMed] [Google Scholar]
- Zhang Z, Zhu W, Lapolla SM, Miao Y, Shao Y, Falcone M, Boreham D, McFarlane N, Ding J, Johnson AE, et al. Bax forms an oligomer via separate, yet interdependent, surfaces. J Biol Chem. 2010;285:17614–17627. doi: 10.1074/jbc.M110.113456. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.