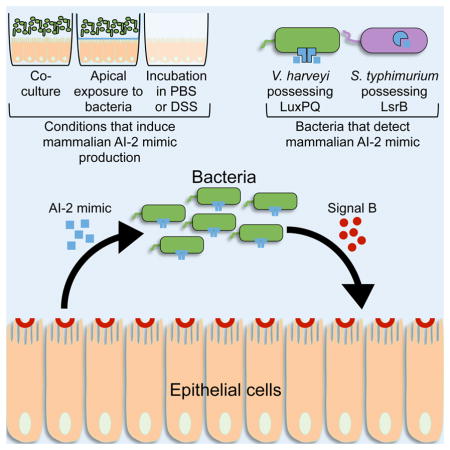
Summary
Host-microbial symbioses are vital to health; nonetheless, little is known about the role cross-kingdom signaling plays in these relationships. In a process called quorum sensing, bacteria communicate with one another using extracellular signal molecules called autoinducers. One autoinducer, AI-2, is proposed to promote inter-species bacterial communication, including in the mammalian gut. We show that mammalian epithelia produce an AI-2 mimic activity in response to bacteria or tight-junction disruption. This AI-2 mimic is detected by the bacterial AI-2 receptor, LuxP/LsrB, and can activate quorum-sensing-controlled gene expression, including in the enteric pathogen Salmonella typhimurium. AI-2 mimic activity is induced when epithelia are directly or indirectly exposed to bacteria, suggesting that a secreted bacterial component(s) stimulates its production. Mutagenesis revealed genes required for bacteria to both detect and stimulate production of the AI-2 mimic. These findings uncover a potential role for the mammalian AI-2 mimic in fostering cross-kingdom signaling and host-bacterial symbioses.
Graphical Abstract
Introduction
Mammals have coevolved with vast populations of commensal bacteria, the majority of which are located in the gut. It is estimated that one hundred trillion bacteria, consisting of ~800 species, are present in the gut and in intimate contact with the host (Backhed et al., 2005). Commensal bacteria can profoundly influence aspects of host physiology, including maturation of the immune system, digestion of food, and absorption of nutrients (Chinen and Rudensky, 2012, Brestoff and Artis, 2013). Furthermore, differences in the makeup of the microbial population in the gut have been linked to human diseases, including inflammatory bowel disease, obesity, diabetes, and colon cancer (Wen et al., 2008, Han and Lin, 2014, Tomasello et al., 2014). It is not clear how hosts maintain beneficial relationships with their symbionts, despite their importance to human health. One possibility is that commensal bacteria communicate with each other and with their hosts, and information from these interactions is used to influence commensal bacterial population densities, species composition, and host cell physiology. In a process called quorum sensing, bacteria communicate with one another using extracellular signal molecules called autoinducers. Quorum sensing relies on the production, release, and subsequent population-wide detection and response to autoinducers. Quorum sensing enables bacteria to synchronize behavior at the population level and, as a collective, successfully carry out tasks that would be unproductive if a single bacterium undertook them alone (Rutherford and Bassler, 2012).
Quorum-sensing autoinducers can confer species-specific communication, genera-wide communication, and species-non-specific communication, suggesting that autoinducers encode information about the number of bacteria in the vicinal community, as well as information about whether neighboring cells are closely or distantly related (Bassler et al., 1993, Schauder et al., 2001, Henke and Bassler, 2004, Ng and Bassler, 2009, Ke et al., 2015). Enteric bacteria engage in quorum-sensing activities including polysaccharide matrix production, biofilm formation, and exo-enzyme production, hallmark behaviors deployed by bacteria when colonizing complex environments such as the gut (Taga and Bassler, 2003, Xavier et al., 2007). Indeed, significant concentrations of bacterial autoinducers are present in vivo and manipulation of the levels of the inter-species quorum-sensing autoinducer called AI-2 in the mouse gut can alter the balance of bacterial species present (Thompson et al., 2015). It is also known that some host-microbial relationships in the gut depend on bacterial responses to host molecules, and conversely, host responses to bacterial molecules. For example, the mammalian stress hormones catecholamine and norepinephrine influence growth and virulence in Salmonella enterica, Escherichia coli, Pseudomonas aeruginosa, and Yersinia enterocolitica (Freestone et al., 1999, Lyte and Bailey, 1997, Lyte and Ernst, 1992, Pullinger et al., 2010). The host responds to bacterial-produced indoles by increasing epithelial tight junctions and strengthening barrier function (Bansal et al., 2010). The host also detects bacterial-produced molecules to promote maturation of immune cells and tissues (Chu and Mazmanian, 2013, Mazmanian et al., 2005, Schnupf et al., 2015). Beyond these initial findings, little else is known about quorum sensing at the host-microbial interface or about the possibility of host-bacterial communication. Cross-kingdom signaling is, however, widely appreciated as a mediator of plant-bacterial relationships, and quorum sensing is vital for these associations (Subramoni et al., 2011, Ferluga and Venturi, 2009, Venturi and Fuqua, 2013). We wonder about the possibility of bacterial- and/or mammalian-produced quorum-sensing signals being involved in maintaining homeostasis in mammals via intra- and inter-kingdom communication.
Most quorum-sensing autoinducers promote intra-species communication, but as mentioned, the autoinducer called AI-2, which is synthesized by LuxS, functions as a universal quorum-sensing autoinducer that enables inter-species communication (Surette et al., 1999). AI-2 is produced by over 50% of sequenced bacterial species and it regulates niche-specific behaviors such as biofilm formation, cell division, virulence, and motility in commensal and pathogenic bacteria (Federle, 2009, Hammer and Bassler, 2003). In Vibrio harveyi, our model bacterium, AI-2 regulates bioluminescence and hundreds of other genes (Henke and Bassler, 2004).
Here, we show that mammalian host tissues produce an activity during bacterial co-culture and following tight-junction disruption that acts analogously to AI-2. The AI-2 mimic is specific to the bacterial receptor responsible for AI-2 detection, because the mimic does not agonize a set of other previously characterized autoinducer receptors. This cross-kingdom communication occurs between eukaryotic cells and bacteria through production of two molecules: a bacteria-derived soluble molecule that stimulates the host to make the AI-2 mimic and the host-derived AI-2 mimic that stimulates bacterial quorum sensing. Mutagenesis reveals that the apt and VIBHAR_02470 genes are involved in bacterial stimulation of host production of the AI-2 mimic, and the luxP, tkt, and hldE genes are required for the bacteria to detect the host AI-2 mimic. Host production of an AI-2 mimic could have important ramifications for host-microbial cross-kingdom signaling in the maintenance of normal host-microbial relationships.
Results
Mammalian cells produce an autoinducer-2 mimic (AI-2 mimic)
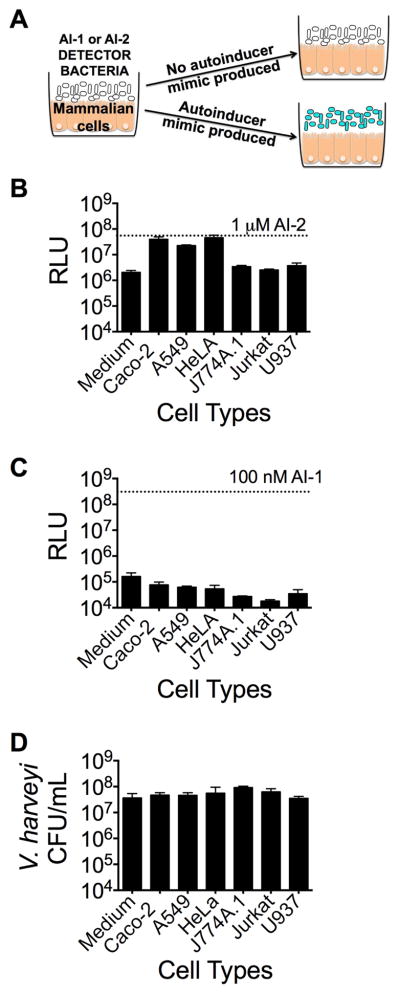
To explore whether mammals produce bacterial-like quorum-sensing molecules, we developed a host-bacterial co-culture system in which different mammalian cell-lines were grown in contact with bacteria, and subsequently tested for stimulation of V. harveyi quorum sensing. Our strategy relied on our ability to monitor the activities of the two dominant V. harveyi quorum-sensing autoinducers, designated AI-1 and AI-2 that are detected by LuxN and LuxPQ, respectively. AI-1 is 3OH-C4-homoserine lactone and AI-2 is (2S, 4S)-2-methyl-2,3,3,4-tetrahydroxytetrahydrofuran-borate (Cao and Meighen, 1989, Chen et al., 2002, Ng and Bassler, 2009). To monitor the two activities, two V. harveyi bioluminescent detector strains were used: V. harveyi TL25 that cannot synthesize AI-1 and cannot respond to AI-2, and V. harveyi TL26 that cannot synthesize AI-2 and cannot respond to AI-1 (Long et al., 2009). Thus, V. harveyi TL25 makes light only when AI-1 or an AI-1-mimic is provided, and V. harveyi TL26 makes light only when AI-2 or an AI-2 mimic is provided (Figure 1a).
Figure 1.
Mammalian epithelial cells produce an autoinducer-2 mimic (AI-2 mimic). (A) Schematic of the co-culture assay. Co-cultures were performed with the following mammalian cell lines for 5 h at 37°C, 5% CO2: Caco-2, A549, HeLa, J774A.1, Jurkat, or U937 cells. (B and C) Bioluminescence from the V. harveyi reporter strain (B) TL26 (ΔluxN, ΔluxS, ΔcqsS) or (C) TL25 (ΔluxM, ΔluxPQ, ΔcqsS) following co-culture with the indicated mammalian cell line. Saturating, 1 μM AI-2 or 100 nM AI-1 were used as positive controls for V. harveyi TL26 and TL25, respectively. We note that the V. harveyi TL26 AI-2 detector strain displays intrinsically higher background bioluminescence than does the V. harveyi TL25 AI-1 detector strain. Relative light units (RLU) are counts per minute per mL per OD600. (D) Total surviving V. harveyi after co-culture (CFU/mL). In all panels, error bars represent standard deviations (SD) for three replicates. See also Fig. S1 and S2.
Mammalian cells of epithelial origin produced an AI-2 activity, but not an AI-1 activity during co-culture (Figure 1b,c; Figure S1a,b). Production of the AI-2 mimic occurred within 5 h of co-culture of V. harveyi with mammalian epithelial cells. Epithelial cells from colon tissues (Caco-2), lung tissues (A549), and cervical tissues (HeLa) all produced 10–50 times more AI-2 mimic activity than did cells of hematopoietic origin including T cells (Jurkat), monocytes (U937), and macrophages (J774A.1). Indeed, the light levels produced by the reporter strains during co-culture with hematopoietic cells were equivalent to the background bioluminescence levels demonstrating that hematopoietic cells make neither AI-1 nor AI-2 mimic activity. The number of bacteria recovered following co-culture with hematopoeitic cell lines was equal to the number recovered following co-culture with epithelial cell lines, showing that hematopoietic cells did not kill the bacterial reporter strains, but rather, hematopoietic cells did not produce significant AI-2 mimic during co-culture (Figure 1d).
The V. harveyi TL26 and TL25 reporter strains are exquisitely specific for detection of only their cognate autoinducers (Long et al., 2009). We therefore assayed for additional host-produced activities using bacterial strains that report on other autoinducers. Co-cultured epithelial cells did not make an activity that stimulated strains that detect unmodified C4-homoserine lactone (C4-HSL) or 3O-C12-homoserine lactone (3O-C12-HSL), the two autoinducers from P. aeruginosa (Figure S1c,d). The Chromobacterium violaceum CviR quorum-sensing receptor is promiscuous and responds to several homoserine lactone autoinducers (McClean et al., 1997, Swem et al., 2009). However, in our model, epithelial cells did not make an activity that stimulated CviR signaling (Figure S1e). Finally, epithelial cells did not make an activity that induced a Vibrio cholerae reporter strain that detects (S)-3-hydroxytridecan-4-one, the vibrio genera autoinducer called CAI-1 (Figure S1f) (Miller et al., 2002, Higgins et al., 2007). Thus, we only find an AI-2 mimic. Our results do not preclude the possibility that homoserine lactone or other classes of autoinducer mimics are produced by eukaryotic cells. If so, such molecules were either not detected by our reporter strains or were not produced under the culturing conditions we tested.
A-2 is a universal inter-species autoinducer and organisms beyond vibrios respond to AI-2 to control gene expression. For example, gut-associated bacteria including E. coli and S. typhimurium activate transcription of the lsr operon in response to AI-2. Lsr stands for LuxS-regulated (Taga et al., 2003, Xavier et al., 2007). To explore the generality of our discovery of a host-produced AI-2 mimic, we assayed whether S. typhimurium could react to the mimic. We co-cultured Caco-2 cells with a ΔluxS S. typhimurium strain carrying an AI-2 inducible lsr-luxCDABE transcriptional reporter. 100-fold more light was produced by the reporter strain in co-culture with Caco-2 cells than in control wells (Figure S2a). We confirmed our results using PCR of lsr genes following co-culture or AI-2 addition (Figure S2b).
It was possible that mammalian epithelia constitutively produce the AI-2 mimic, irrespective of bacterial co-culture. To address this possibility, conditioned medium from Caco-2 cells cultured in the absence of bacteria was assayed for the AI-2 mimic activity. None was present, suggesting a requirement for the presence of bacteria to stimulate AI-2 mimic production by the epithelial cells (Figure S2c).
Two-way signaling between epithelial cells and bacterial cells occurs in co-culture
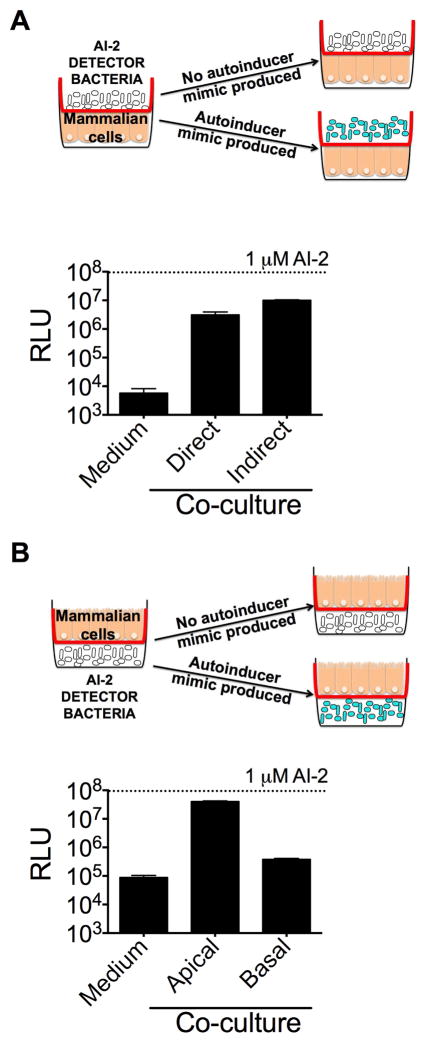
To investigate the requirements for AI-2 mimic production, we tested whether direct host-bacterial contact was required. To do this, we exposed the AI-2 detector V. harveyi TL26 strain grown in the upper chamber of a transwell to the Caco-2 line cultured as a monolayer beneath the transwell (Figure 2a). The transwell barrier physically separates bacteria from the epithelial cells while allowing soluble components to transit the barrier. Similar transwell strategies have been used to identify soluble factors involved in host responses to bacteria (Zargar et al., 2015). V. harveyi TL26 produced an equal amount of light in response to Caco-2 cells irrespective of whether the bacteria were in direct or indirect contact with the epithelial cells. Thus, Caco-2 cells do not require direct bacterial contact to produce the AI-2 mimic (Figure 2a).
Figure 2.
Two-way signaling between epithelial and bacterial cells occurs during co-culture. Bioluminescence responses of V. harveyi TL26 during (A) direct or indirect co-culture and (B) basal or apical incubation with Caco-2 cells. Culture conditions as in Fig. 1. 1 μM AI-2 was included as a positive control. In all panels, error bars represent SD for three replicates. See also Fig. S3 and S4.
AI-2 mimic production was not specific to incubation with V. harveyi on the far side of the barrier as identical experiments with ΔluxS (i.e., AI-2−) strains of E. coli and Salmonella typhimurium, two gut-associated species, also led to AI-2 mimic production by Caco-2 cells (Figure S3a,b). In those cases, we collected the conditioned medium from the upper chamber of the transwell and assayed for AI-2 mimic activity using the V. harveyi TL26 detector strain. Finally, live but not dead bacteria were required to induce production of the AI-2 mimic during co-culture with epithelial cells (Figure S3c).
A key feature of epithelia compared to other cell types, is that they form sheets that line tissues and are polarized with apical, lateral, and basal membrane domains (Roignot et al., 2013). Polarity is necessary for normal epithelial functions, including maintaining a barrier against bacteria colonizing apical surfaces of host tissues (Peterson and Artis, 2014, Roignot et al., 2013). In the above co-culture transwell experiments, our goal was to accurately reproduce the in vivo host-microbial association. Thus, the epithelial cells in the bottom chamber of the transwell were polarized with their apical face exposed to the V. harveyi TL26 detector strain grown in the upper chamber of the transwell. To assess whether epithelial orientation plays a role in production of the mammalian AI-2 mimic during co-culture, we next exposed V. harveyi TL26 grown in the lower chamber of transwells to epithelial cells cultured as a monolayer in the upper chamber of transwells (Figure 2b). Our rationale was that, in this arrangement, Caco-2 cells would detect bacterial signals from the basal face, thus, reversing the host-microbial polarization present in colonized tissues. In this setup, Caco-2 cells produced 100-fold less AI-2 mimic activity than in the reverse setup, showing that AI-2 mimic production occurs from the apical side (Figure 2b). Collectively, our data suggest that a secreted bacterial component stimulates the host to produce the AI-2 mimic from the apical surface. One possible candidate, lipopolysaccharide (LPS), is a component of bacterial cell walls that modulates epithelial cell behavior (Ruemmele et al., 2002, Cario et al., 2000, Panja et al., 1995). However, addition of LPS to epithelial cells failed to stimulate AI-2 mimic production (Figure S4a).
The mammalian AI-2 mimic is produced following PBS-treatment
We wondered whether the presence of bacteria was absolutely essential for AI-2 mimic production by epithelial cells or whether other conditions could also induce the Caco-2 cells to produce the mimic. To test this, we cultured Caco-2 cells in different media for 48 h, collected the conditioned medium, and tested for AI-2 mimic activity. Caco-2 cells were grown in rich medium (DMEM), serum-free medium (FBS), medium lacking glucose and/or glucosamine, and phosphate buffered saline (PBS). Only conditioned medium from Caco-2 cells incubated in PBS contained significant AI-2 mimic activity (Figure 3a). This result suggests that, in addition to co-culture with bacteria, stressing the Caco-2 cells promotes AI-2-mimic production.
Figure 3.
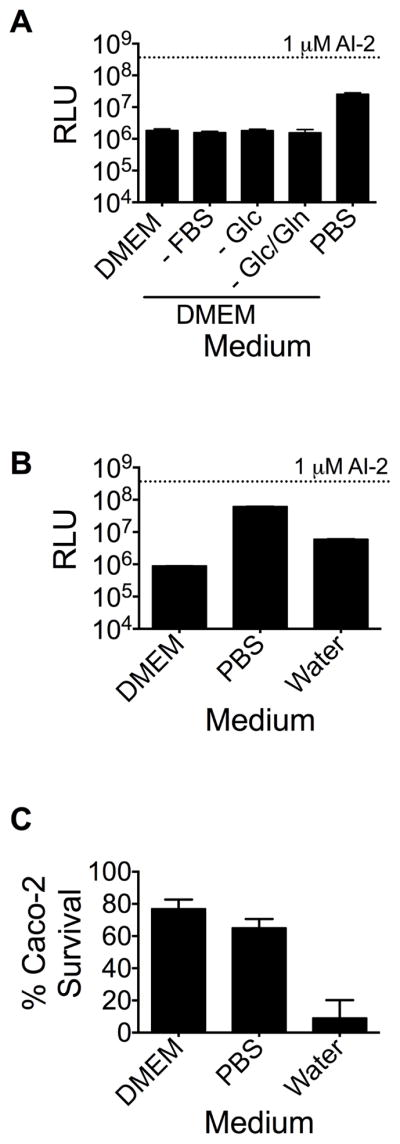
Caco-2 cells produce the AI-2 mimic when subjected to PBS-treatment. (A) Caco-2 cells were cultured for 48 h at 37°C, 5% CO2 in the specified media. AI-2 mimic activity was measured using the V. harveyi TL26 bioluminescence assay. FBS is fetal bovine serum, Glc is glucose, Gln is glucosamine. (B) Caco-2 cells were cultured in DMEM, PBS, and water. AI-2 mimic activity was analyzed as in (A). (C) Caco-2 survival was assessed through Trypan blue staining. In panels (A) and (B), 1 μM AI-2 was included as the positive control. In all panels, error bars represent SD for three replicates. See also Fig. S4 and S5.
One concern with respect to PBS-cultured Caco-2 cells was the possibility of autolysis, which could result in non-specific release of cellular components, including, possibly, the AI-2 mimic. To address this issue, Caco-2 cells were incubated in water for 48 h, which resulted in >90% Caco-2 cell death. PBS-treated Caco-2 cells, by contrast, suffered minimal cell death (<25%). Conditioned medium collected from the water-treated cells contained only 10% of the AI-2 mimic activity present in conditioned medium from PBS-treated Caco-2 cells, suggesting that production of the AI-2 mimic occurs under specific conditions and by metabolically active epithelial cells (Figure 3b,c). This in vitro method of producing AI-2 mimic, in the absence of bacteria, provided us a means to simplify our procedure to access the AI-2 mimic activity for our studies. Dose response curves for AI-2 and for PBS-produced mimic are provided in Figure S5.
The mammalian AI-2 mimic is not produced from an intermediate in the bacterial AI-2 biosynthesis pathway
AI-2 is produced from S-adenosylmethionine (SAM) as follows: SAM-dependent methylation of substrates converts SAM into S-adenosylhomocysteine (SAH) which is subsequently converted into adenine and S-ribosylhomocysteine (SRH) by the enzyme Pfs. LuxS acts on SRH to make the AI-2 precursor called DPD (4,5-dihydroxy-2,3-pentanedione) and homocysteine (Schauder et al., 2001). DPD spontaneously rearranges into the family of active AI-2 signaling molecules.
We considered the possibility that Caco-2 cells make the mimic by releasing an enzymatic activity that acts on an intermediate in the bacterial AI-2 biosynthetic pathway that accumulates in the ΔluxS strains used in this work. SAH is unlikely to accumulate because Pfs is present in our bacterial strains. Thus, SRH is the most plausible candidate. To test whether SRH could be made into the mimic by Caco-2 cells, we added SRH at concentrations up to 1 mM to mammalian-bacterial cocultures and to AI-2 mimic preparations acquired from PBS-treated Caco-2 cells. Our rationale was that, if a mammalian-produced activity were present that could convert SRH into the AI-2 mimic, increased AI-2 mimic would be produced. Increased mimic would, in turn, induce increased light output from the V. harveyi TL26 reporter strain. No increase in bioluminescence emission occurred upon SRH supplementation suggesting that the mimic is not made by Caco-2 cells from a bacterial-produced intermediate in the AI-2 biosynthesis pathway (Figure S4b,c).
The mammalian AI-2 mimic functions through the bacterial AI-2 receptor LuxP
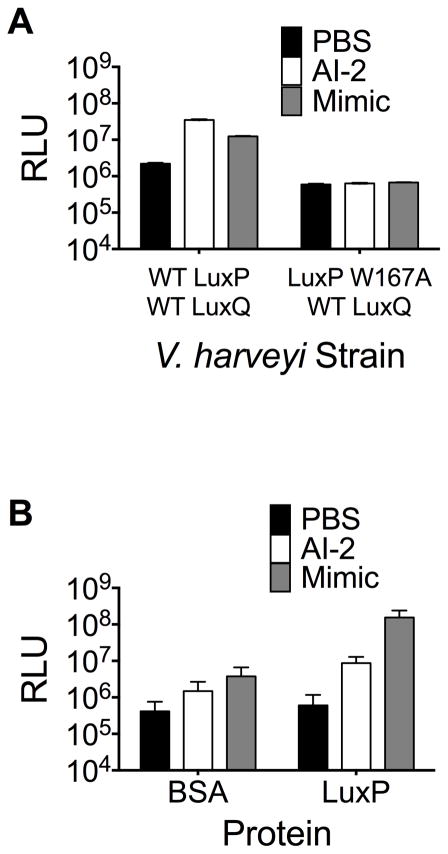
We wondered whether AI-2 mimic detection required the known bacterial AI-2 detection apparatus. In V. harveyi, AI-2 binds to the periplasmic protein LuxP which functions in conjunction with the transmembrane two-component receptor LuxQ, to transduce the AI-2 signal internally and elicit the quorum-sensing response (Bassler et al., 1994, Neiditch et al., 2005, Neiditch et al., 2006). V. harveyi strain FED119 (ΔluxN, ΔluxPQ, ΔluxS) carrying a plasmid harboring wild-type LuxQ and a LuxP variant with an alteration in a residue essential for AI-2 binding (LuxP W167A) did not respond to the AI-2 mimic whereas the strain carrying a plasmid with both wild-type LuxP and LuxQ produced a robust response (Figure 4a). The presence of boric acid in the medium is crucial for V. harveyi detection of AI-2 because DPD, the AI-2 precursor, binds borate to make the active AI-2 autoinducer, (2S, 4S)-2-methyl-2,3,3,4-tetrahydroxytetrahydrofuran-borate (Chen et al., 2002, Miller et al., 2002). We found that addition of 0.1 mM boric acid to AI-2 mimic preparations was also required for full activity (Figure S6). Thus, the AI-2 mimic indeed functions to control V. harveyi gene expression through the canonical AI-2 quorum-sensing pathway.
Figure 4.
Detection of the AI-2 mimic requires the LuxP receptor in V. harveyi. (A) Preparations from PBS-cultured Caco-2 cells (denoted Mimic) were incubated with V. harveyi FED119 (ΔluxN, ΔluxPQ, ΔluxS) harboring wild-type LuxPQ (expressed from pFED368) or LuxP W167A and wild-type LuxQ (expressed from pFED408), and bioluminescence was measured. (B) Assessment of AI-2 mimic bound by recombinant LuxP. AI-2 mimic activity was assayed with V. harveyi TL26 as in Fig. 3. In both panels, additions to the protein (BSA or LuxP) are as follows: PBS, black; 1 μM AI-2, white; 10% v/v preparations from PBS-cultured Caco-2 cells (Mimic), gray. In all panels, error bars represent SD for three replicates. See also Fig. S6.
We next exploited the interaction between LuxP and the AI-2 mimic in an attempt to trap the AI-2 mimic in the LuxP protein and purify it. This strategy is analogous to the one we originally used to capture and identify AI-2 (Chen et al., 2002, Miller et al., 2004). We incubated recombinant His-tagged LuxP protein with conditioned medium prepared from Caco-2 cells grown under PBS-treatment conditions. We released bound AI-2 mimic from LuxP by heating the complex, followed by centrifugation to remove denatured LuxP protein. Released mammalian AI-2 mimic activity was quantified using the V. harveyi TL26 bioluminescence reporter assay. This procedure yielded a 50-fold enrichment in AI-2 mimic activity compared to background controls in which LuxP was incubated with PBS, or when a non-specific protein (BSA) was incubated with conditioned medium from PBS-treated Caco-2 cells (Figure 4b). We are currently attempting to purify the mammalian AI-2 mimic using this strategy.
Screen to identify bacterial genes required for stimulation and detection of the mammalian AI-2 mimic
Our results suggest that two molecules are involved in the Caco-2-bacterial interaction we are studying: one, the AI-2 mimic made by the Caco-2 cells, and another, a soluble factor made by the bacteria that stimulates the Caco-2 cells to produce the AI-2 mimic. With respect to the bacteria, we suspect that two types of genes are involved: one type required for producing the soluble factor(s) that stimulates mammalian AI-2 mimic production during co-culture, and another type that is required for the bacteria to detect the AI-2 mimic. We know that quorum-sensing signal relay components including LuxPQ are among the second class. We do not know if additional factors are required for AI-2 mimic detection. We performed a Tn5 mutagenesis of V. harveyi TL26 to identify the two putative classes of genes. We screened 30,000 mutants for those producing less light than the V. harveyi TL26 parent strain during co-culture with Caco-2 cells. We reasoned that V. harveyi TL26 mutants disabled in the release of the factor that stimulates Caco-2 cells to produce the AI-2 mimic would cause reduced release of the AI-2 mimic from the Caco-2 cells, which, in turn, would cause the detector bacteria themselves to exhibit a reduced bioluminescence emission response during co-culture. V. harveyi TL26 insertion mutants disabled in detection of the AI-2 mimic would also display reduced bioluminescence in co–culture with AI-2 mimic producing Caco-2 cells. We reasoned that we could distinguish between these two types of defects with subsequent secondary assays.
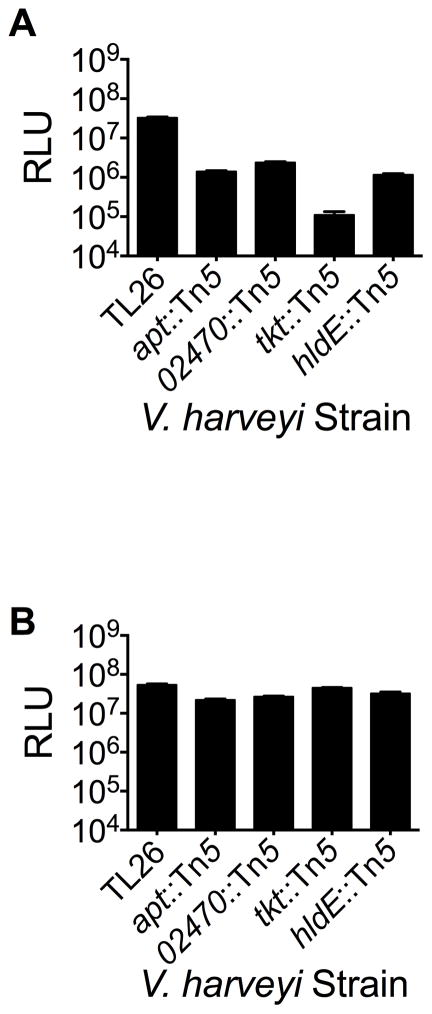
We isolated ~100 V. harveyi TL26 Tn5 insertion mutants exhibiting reduced bioluminescence. Beyond the two classes of genes we hoped to identify, reduced bioluminescence could also be a consequence of insertions in quorum-sensing genes we know are required to detect and relay the AI-2 and AI-2 mimic signals or in genes required to produce light. We therefore performed a secondary screen in which we supplied exogenous AI-2 to eliminate mutants defective in AI-2 detection (i.e, luxPQ mutants) or that were otherwise generally deficient in bioluminescence. We went forward with mutants that exhibited wild-type bioluminescence when AI-2 was added. This strategy yielded four Tn5 insertion mutants displaying at least 10-fold reductions in bioluminescence during co-culture with Caco-2 cells but which retained the ability to detect exogenously added AI-2 (Figure 5a, b).
Figure 5.
V. harveyi mutants defective in stimulation or detection of the AI-2 mimic. (A) Bioluminescence from V. harveyi TL26 Tn5 insertions mutants during co-culture with Caco-2 cells. (B) Bioluminescence of the same strains in response to 100 nM AI-2 was used to verify that mutants could quantitatively detect AI-2 at non-saturating levels. In all panels, error bars represent SD for three replicates. See also Fig. S7
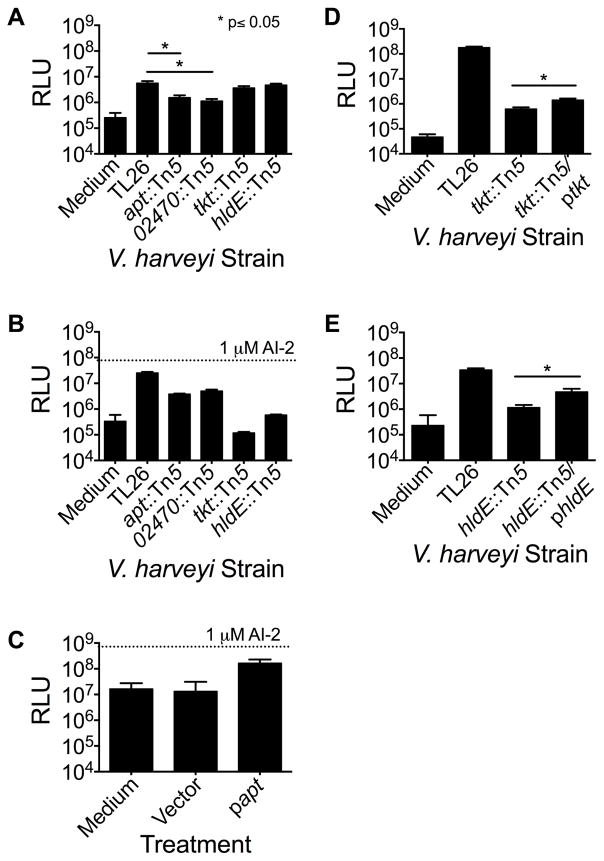
The genes identified in our screen are: VIBHAR_02472, VIBHAR_02470, VIBHAR_03567, and VIBHAR_00868. VIBHAR_02472 encodes aerolysin (apt), a cytolytic pore-forming toxin exported by aeromonads and vibrios (Parker et al., 1994) that punctures the mammalian membrane causing osmotic lysis. VIBHAR_02470 is a hypothetical protein with a putative DNA-binding domain that is located immediately upstream of apt, suggesting a role for VIBHAR_02470 in apt expression. Indeed, quantitative PCR revealed that VIBHAR_02470 mutants displayed a 100-fold decrease in apt expression, whereas mutation of apt did not affect expression of VIBHAR_02470 (Figure S7a,b). Thus, VIBHAR_02470 likely modulates AI-2 mimic production/detection through regulation of apt. VIBHAR_03567 encodes a transketolase (tkt) that is conserved among many Gram-negative bacteria, and catalyzes the formation of ribose-5-phosphate from fructose 6-phosphate (Schenk et al., 1998). Finally, VIBHAR_00868 encodes a bifunctional heptose 1-phosphate adenyltransferase (hldE), that catalyzes the phosphorylation of D-glycero-D-manno-heptose 7-phosphate to form D,D-heptose-1,7- bisphosphate (Kneidinger et al., 2002, McArthur et al., 2005).
To distinguish V. harveyi mutants defective in mammalian AI-2 mimic detection from those defective in production of the factor that stimulates AI-2 mimic production in Caco-2 cells, we measured the level of AI-2 mimic produced by Caco-2 cells following co-culture with each of the above four V. harveyi mutants. Our expectation was that co-incubation of Caco-2 cells with V. harveyi mutants defective in making the factor that stimulates AI-2-mimic production would result in Caco-2 cells producing less AI-2 mimic. By contrast, incubation with V. harveyi mutants defective in detection of the AI-2 mimic would not affect AI-2 mimic production by Caco-2 cells. The levels of AI-2 mimic produced in each case could be assessed using the V. harveyi TL26 bioluminescence assay. The V. harveyi apt and VIBHAR_02470 mutants caused 5-fold decreases in the amount of mimic produced by the Caco-2 cells (Figure 6a). These mutants also showed a slight deficiency in their ability to detect the AI-2 mimic (Figure 6b).
Figure 6.
Bacterial genes required for stimulation and detection of the mammalian AI-2 mimic. (A) AI-2 mimic activity in conditioned medium following co-culture of mutant V. harveyi strains with Caco-2 cells. (B) Bioluminescence from mutant V. harveyi strains in response to preparations from PBS-treated Caco-2 cells. (C) Cell-free culture fluids from LB-grown ΔluxS E. coli harboring the cloned apt gene or the empty vector were incubated with Caco-2 cells. We note that LB medium causes high endogenous background bioluminescence. In panels A, B, and C, AI-2 mimic activity was assessed using the V. harveyi TL26 bioluminescence assay, as in Fig. 3. (D and E) Bioluminescence of the specified V. harveyi strains following co-culture with Caco-2 cells: (D) V. harveyi TL26 tkt::Tn5, +/− ptkt and (E) V. harveyi TL26 hldE::Tn5 +/− phldE. We were unable to complement the V. harveyi apt and VIBHAR_02470 mutants because introduction of apt or VIBHAR_02470 on plasmids caused severe growth defects. In panels (B) and (C), 1 μM AI-2 was included as a positive control. In all panels, error bars represent SD of three replicates. See also Fig. S7.
To test the role of aerolysin in activation of mammalian AI-2 mimic production, we introduced a plasmid carrying the cloned apt gene into ΔluxS E. coli. Conditioned medium was collected from this recombinant E. coli and added to Caco-2 cells for 5 h. 10-fold more AI-2 mimic activity was present in conditioned medium prepared from Caco-2 cells that had been incubated with the preparations made from recombinant E. coli expressing apt than from Caco-2 cells that had been incubated with the preparations made from E. coli containing the vector alone (Figure 6c). Quantitative PCR confirmed high-level expression of the cloned apt gene (Figure S7c). These results imply that secreted aerolysin is sufficient to activate mammalian AI-2 mimic production. The number of Caco-2 cells recovered after incubation with aerolysin-containing culture fluids was equal to that recovered following incubation with fluid from the control preparation showing that aerolysin does not kill the Caco-2 cells. While inactivation of apt and VIBHAR_02470 decreased the ability of Caco-2 cells to produce the AI-2 mimic, they did not completely eliminate AI-2 mimic production. This result suggests that V. harveyi possesses more than one mechanism to stimulate AI-2 mimic production by Caco-2 cells. We are currently mutagenizing a Δapt V. harveyi strain and screening for mutants that are completely defective in stimulation of AI-2 mimic production by Caco-2 cells.
The V. harveyi tkt and hldE mutants stimulated maximal production of the AI-2 mimic by Caco-2 cells during co-culture (Figure 6a). We therefore tested the ability of these mutants to detect exogenously added AI-2 mimic collected from PBS-treated Caco-2 cells. The tkt and hldE mutants produced 100-fold and 50-fold less light, respectively, than did the V. harveyi TL26 parent strain in response to this preparation suggesting that these genes are required for detection of the AI-2 mimic (Figure 6b). Complementation of the tkt and hldE mutants with the corresponding genes on plasmids partially restored the mutants’ ability to respond to the AI-2 mimic produced by Caco-2 cells during co-culture, 3- and 5-fold, respectively (Figure 6d,e).
The mammalian AI-2 mimic is produced following tight-junction disruption
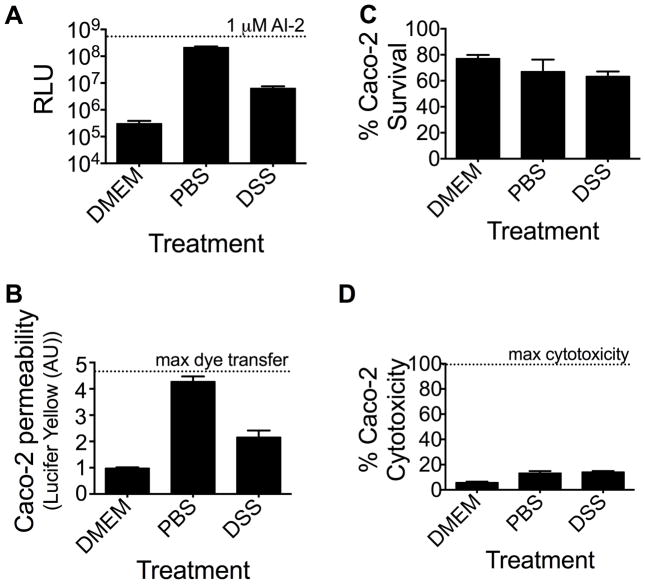
Our PBS-treated Caco-2 cells do not form intact monolayers in tissue culture. Additionally, aerolysin is a known disrupter of epithelial tight-junctions (Abrami et al., 2003, Bucker et al., 2011). Thus, we hypothesized that disruption of the integrity of Caco-2 monolayers by either PBS or aerolysin treatment could promote AI-2 mimic production. Dextran sulfate sodium (DSS) induces colitis in mice in vivo and disrupts Caco-2 cell monolayers in vitro (Bjorck et al., 1997, Samak et al., 2015). We treated Caco-2 cells with a 2.5% v/v solution of DSS for 48 h, collected the conditioned medium, and tested for AI-2 mimic activity. Indeed, similar to exposure to bacteria and PBS-treatment, conditioned medium from Caco-2 cells treated with DSS contained significant AI-2 mimic activity (Figure 7a). We next assessed whether PBS- and DSS-treatment of Caco-2 cells caused increased epithelial permeability. To do this, we supplied the transepithelial marker, Lucifer Yellow (Molecular Probes), to Caco-2 cells grown in the upper chamber of a transwell and measured dye transfer to the bottom chamber. Intact Caco-2 monolayers (grown in DMEM) prevented dye transfer to the bottom chamber, whereas Caco-2 cells grown in PBS or that had been treated with DSS allowed 4- and 2-fold higher dye transfer respectively (Figure 7b). We observed no significant differences in the survival of Caco-2 cells following the different treatments for 48 h (Figure 7c). We also measured lactate dehydrogenase (LDH) release following PBS or DSS treatment of Caco-2 monolayers as a biomarker of cytotoxicity. There is minimal cytotoxicity (<15%) under our conditions, suggesting that disruption of epithelial tight junctions, not Caco-2 cell death, leads to AI-2 mimic production (Figure 7d).
Figure 7.
Caco-2 tight junction disruption promotes production of AI-2 mimic. (A) AI-2 mimic activity in conditioned medium following incubation in DMEM, PBS, and 2.5% DSS. 1 μM AI-2 was included as the positive control. (B) Lucifer Yellow transfer and (C) Caco-2 survival was assessed as in Fig. 3. (D) Lactate dehydrogenase (LDH) release from Caco-2 cells cultured as in (A). In all panels, error bars represent SD for three replicates.
Discussion
Epithelial cells are faced with the unique challenge of shielding host tissues from the environment while properly interacting with microbes including both pathogens and symbionts (Peterson and Artis, 2014). Intestinal epithelial cells detect their beneficial bacterial counterparts and, in response, produce antibacterial peptides, mucins, and immunoglobulins, which maintain proper host-microbial symbioses by limiting the ability of bacteria to penetrate host tissues (Peterson and Artis, 2014). We discuss here one additional strategy that may contribute to maintaining host-microbial symbioses through cross-kingdom quorum-sensing-mediated communication. Previous studies suggest that quorum sensing is involved in bacterial-host interactions (Visick et al., 2000, Smith et al., 2002, Bearson and Bearson, 2008, Hughes and Sperandio, 2008, Thompson et al., 2015). Likewise, plants produce autoinducer mimics that influence quorum sensing among their bacterial colonizers (Teplitski et al., 2004, Teplitski et al., 2000). Similarly, mammals could also synthesize autoinducer mimics.
Our results suggest that cross-kingdom communication occurs between eukaryotic cells and bacteria via the AI-2 bacterial quorum-sensing system. Eukaryotic cells lack the luxS gene encoding the AI-2 synthase, and they also apparently lack the ability to convert intermediates in the bacterial AI-2 biosynthesis pathway into the mimic. Thus, an independent process must make the AI-2 mimic. We do not know the identity of the AI-2 mimic. Our initial purification studies suggest that it is resistant to heat-denaturation, as incubation of conditioned medium prepared from PBS-treated Caco-2 cells at temperatures above 95°C did not result in marked loss of activity. The AI-2 mimic activity passes through 10,000 MWCO filters, suggesting that the AI-2 mimic is a small molecule. We can also detect AI-2 mimic activity following organic extraction in acetonitrile and separation using Hydrophilic Interaction Chromatography suggesting it is polar. We are currently attempting to purify the AI-2 mimic from preparations of PBS-treated Caco-2 cells followed by trapping of the mimic in LuxP.
Both AI-2 and the mammalian AI-2 mimic are recognized by the bacterial LuxP receptor. It is noteworthy that in addition to a requirement for LuxP, we also identified a transketolase and a bifunctional heptose 1-phosphate adenyltransferase that are necessary for maximal AI-2 mimic detection by V. harveyi. Both of these enzymes are involved in sugar metabolism and are frequently present in genomes of gut-associated bacteria including E. coli and S. typhimurium. This is significant because DPD, the precursor to AI-2, is derived from the ribose moiety of SRH (Schauder et al., 2001). We have previously shown that borate reacts with a cyclized AI-2 precursor to produce the active furanosyl borate diester AI-2 signal recognized by V. harveyi (Chen et al., 2002, Miller et al., 2004). Similarly, the AI-2 mimic requires boric acid for detection by V. harveyi, implying that the AI-2 mimic shares key features with DPD and, possibly, the precursor to the AI-2 mimic reacts with borate to enable detection by LuxP (Chen et al., 2002). This line of reasoning suggests that the mammalian AI-2 mimic is a sugar derivative, possibly possessing cis-diols, which readily form adducts with borate (Loomis and Durst, 1992). We suspect the molecule requires additional processing by Tkt and/or HldE for detection by V. harveyi via LuxP.
Hosts respond to microbe-associated molecular patterns (MAMPs) during symbioses by producing secreted factors such as cytokines and chemokines that activate immune responses (Peterson and Artis, 2014). Our data suggest that epithelial cells respond to secreted bacterial products by producing the AI-2 mimic and we have identified a role for aerolysin. Epithelial cells of the intestine are the primary targets of bacterial aerolysin, which causes Caco-2 cells to disassemble tight junctions (Abrami et al., 2003). Our results show that an additional consequence of apical exposure to aerolysin is AI-2 mimic release. Thus, aerolysin, like the PBS- and DSS-treatments, may function as a stress signal that disrupts epithelial tight junctions and activates production of the AI-2 mimic. Gut microbes protect the host from epithelial damage, and following injury they accelerate host cell healing (Rakoff-Nahoum et al., 2004, Ismail et al., 2009). At least one bacterially-produced molecule, the polysaccharide called PSA, is known to have an ameliorative effect (Shen et al., 2012). Our observation that AI-2 mimic production increases following epithelial tight-junction damage, suggests a possible role for the mammalian AI-2 mimic during epithelial repair. Our findings furthermore suggest that the AI-2 mimic could be involved in host association with commensal bacteria rather than with pathogens since commensal bacteria are typically apically associated with epithelia, while bacterial pathogens are usually detected from the basolateral surface. Finally, our discovery that the mammalian AI-2 mimic is made in cells of epithelial lineage, but not in hematopoietic cell lines, suggests the mimic could have a role in host-microbial symbioses, since epithelial cells directly interact with colonizing bacteria.
It is curious that the mammalian-produced activity mimics AI-2, rather than any other autoinducer activity we tested. AI-2 is a universal species non-specific quorum-sensing signal. Exploiting this molecule, as opposed to a highly species-specific autoinducer, could be a strategy that enables the host to maximally manipulate bacterial behavior in mixed populations such as those that exist in the gut.
Experimental Procedures
Bacterial strains and media and molecular biology procedures
Strains, plasmids, and oligonucleotides used in this study are listed in Tables S1, S2, and S3, respectively. Detailed protocols in Supplemental Experimental Procedures.
Mammalian Cell culture
Caco-2 cells were purchased from ATCC (ATCC® HTB-37™). HeLa cells (ATCC® CCL-2™), A549 cells (ATCC® CCL-185™), Jurkat E6-1 cells (ATCC® TIB-152™), U937 cells (ATCC® CRL-1593.2™), and J774A.1 (ATCC® TIB-67™) cells were kindly donated.
Mammalian cell co-culture incubations with bacteria
Mammalian cells lines described above were co-cultured with V. harveyi TL26, V. harveyi TL25, or S. typhimurium MET687 for 5 h at 30°C, with 5% CO2. SRH was added to some co-cultures as specified in the text. Following incubation, co-cultured bacteria were assessed for bioluminescence using an EnVision® Multilabel Reader (Perkin Elmer).
AI-2 mimic production from PBS- and DSS-treated Caco-2 cells
Caco-2 cells were detached from tissue culture plates and incubated in either Dulbecco’s PBS (DPBS; Invitrogen) or 2.5% dextran sulfate sodium (DSS; Sigma Aldrich) for 48 h at 37°C, in the presence of 5% CO2. In some cases, SRH was added as specified in the text. 10% v/v conditioned medium was tested for AI-2 mimic activity using the V. harveyi TL26 bioluminescence assay described below. Caco-2 cell permeability and cytotoxicity were determined by Lucifer Yellow transport and lactate dehydrogenase release, respectively.
Bioluminescence assays
V. harveyi strains (Table S1) were grown overnight in LM medium and diluted 1:1000 into AB broth supplemented with 0.1 mM boric acid and dispensed into 96-well plates containing 10% v/v mammalian AI-2 mimic (produced from conditioned medium from PBS- or DSS-treated Caco-2 cells or from Caco-2/V. harveyi TL26 co-culture) or with synthetic DPD (i.e., AI-2) diluted to a final concentration of 1 μM. (Semmelhack et al., 2005). The cultures were allowed to grow for 48 h at 30°C with aeration, after which bioluminescence and optical density were measured with an Envision Multilabel plate reader. Relative light units (RLUs) are defined as counts per minute per mL per OD600. V. harveyi strains showed no differences in growth in the presence or absence of the AI-2 mimic preparations.
LuxP-AI-2 mimic binding assay
Expression and purification of LuxP were performed as described previously (Chen et al., 2002). The mammalian AI-2 mimic was trapped in recombinant LuxP during an overnight incubation in phosphate buffer. Resulting samples were concentrated, washed to remove unbound AI-2 mimic, and AI-2 mimic released by gentle heating of the LuxP/AI-2 mimic complex. Released AI-2 mimic activity was assessed using the V. harveyi TL26 bioluminescence assay.
Screen for V. harveyi TL26 mutants defective in stimulating AI-2 mimic production or in AI-2 mimic detection
Tn5 transposon mutagenesis was performed in V. harveyi TL26 followed by screening for decreased bioluminescence during co-culture with Caco-2 cells. These co-cultures were grown for 5 h at 30°C in the presence of 5% CO2. To screen for mutants in stimulation or detection of the mammalian AI-2 mimic, we assayed for V. harveyi TL26 mutants exhibiting low bioluminescence during co-culture, but high bioluminescence in the presence of 100 nM synthetic AI-2. The locations of the Tn5 insertions in mutants were mapped by cloning followed by sequencing.
Supplementary Material
Highlights.
Mammalian epithelial cells produce an autoinducer-2 (AI-2) mimic in response to bacteria
Direct and indirect bacterial contact induces AI-2 mimic production
Bacterial AI-2 receptor LuxP/LsrB detects the AI-2 mimic and activates quorum sensing
Mutagenesis reveals genes required for mimic production and detection
Acknowledgments
We thank members of the B.L.B laboratory for helpful discussion, Robert Scheffler for help with bacterial mutagenesis, and Dr. K. Xavier for suggesting the tight-junction possibility. This work was supported by the Howard Hughes Medical Institute, NIH Grant 5R01GM065859, and National Science Foundation Grant MCB-0948112 (to B.L.B.). A.S.I. was supported by NIH Fellowship 5F32GM100711-02 and a L’Oréal-AAAS USA Fellowship For Women in Science.
Footnotes
Supplemental Information includes seven figures, three tables, and extended experimental procedures.
Author Contributions
A.S.I., J.S.V., and B.L.B. designed research; A.S.I. and J.S.V. performed research; A.S.I. and J.S.V. contributed new reagents/analytic tools; A.S.I., J.S.V., and B.L.B. analyzed data; and A.S.I., J.S.V and B.L.B. wrote the paper.
The authors declare no conflict of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Host-bacterial symbioses are vital for host health, yet little is known about cross-kingdom signaling mechanisms that maintain their balance. Ismail et al. demonstrate that mammalian epithelial cells produce a mimic of the bacterial autoinducer, AI-2, in response to secreted bacterial factors and tight-junction disruption that activates quorum sensing in bacteria.
References
- Abrami L, Fivaz M, Glauser PE, Sugimoto N, Zurzolo C, Van Der Goot FG. Sensitivity of polarized epithelial cells to the pore-forming toxin aerolysin. Infect Immun. 2003;71:739–46. doi: 10.1128/IAI.71.2.739-746.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Backhed F, Ley RE, Sonnenburg JL, Peterson DA, Gordon JI. Host-bacterial mutualism in the human intestine. Science. 2005;307:1915–20. doi: 10.1126/science.1104816. [DOI] [PubMed] [Google Scholar]
- Bansal T, Alaniz RC, Wood TK, Jayaraman A. The bacterial signal indole increases epithelial-cell tight-junction resistance and attenuates indicators of inflammation. Proc Natl Acad Sci U S A. 2010;107:228–33. doi: 10.1073/pnas.0906112107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassler BL, Wright M, Showalter RE, Silverman MR. Intercellular signalling in Vibrio harveyi: sequence and function of genes regulating expression of luminescence. Mol Microbiol. 1993;9:773–86. doi: 10.1111/j.1365-2958.1993.tb01737.x. [DOI] [PubMed] [Google Scholar]
- Bassler BL, Wright M, Silverman MR. Multiple signalling systems controlling expression of luminescence in Vibrio harveyi: sequence and function of genes encoding a second sensory pathway. Mol Microbiol. 1994;13:273–86. doi: 10.1111/j.1365-2958.1994.tb00422.x. [DOI] [PubMed] [Google Scholar]
- Bearson BL, Bearson SM. The role of the QseC quorum-sensing sensor kinase in colonization and norepinephrine-enhanced motility of Salmonella enterica serovar Typhimurium. Microb Pathog. 2008;44:271–8. doi: 10.1016/j.micpath.2007.10.001. [DOI] [PubMed] [Google Scholar]
- Bjorck S, Jennische E, Dahlstrom A, Ahlman H. Influence of topical rectal application of drugs on dextran sulfate-induced colitis in rats. Dig Dis Sci. 1997;42:824–32. doi: 10.1023/a:1018880501437. [DOI] [PubMed] [Google Scholar]
- Brestoff JR, Artis D. Commensal bacteria at the interface of host metabolism and the immune system. Nat Immunol. 2013;14:676–84. doi: 10.1038/ni.2640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bucker R, Krug SM, Rosenthal R, Gunzel D, Fromm A, Zeitz M, Chakraborty T, Fromm M, Epple HJ, Schulzke JD. Aerolysin from Aeromonas hydrophila perturbs tight junction integrity and cell lesion repair in intestinal epithelial HT-29/B6 cells. J Infect Dis. 2011;204:1283–92. doi: 10.1093/infdis/jir504. [DOI] [PubMed] [Google Scholar]
- Cao JG, Meighen EA. Purification and structural identification of an autoinducer for the luminescence system of Vibrio harveyi. J Biol Chem. 1989;264:21670–6. [PubMed] [Google Scholar]
- Cario E, Rosenberg IM, Brandwein SL, Beck PL, Reinecker HC, Podolsky DK. Lipopolysaccharide activates distinct signaling pathways in intestinal epithelial cell lines expressing Toll-like receptors. J Immunol. 2000;164:966–72. doi: 10.4049/jimmunol.164.2.966. [DOI] [PubMed] [Google Scholar]
- Chen X, Schauder S, Potier N, Van Dorsselaer A, Pelczer I, Bassler BL, Hughson FM. Structural identification of a bacterial quorum-sensing signal containing boron. Nature. 2002;415:545–9. doi: 10.1038/415545a. [DOI] [PubMed] [Google Scholar]
- Chinen T, Rudensky AY. The effects of commensal microbiota on immune cell subsets and inflammatory responses. Immunol Rev. 2012;245:45–55. doi: 10.1111/j.1600-065X.2011.01083.x. [DOI] [PubMed] [Google Scholar]
- Chu H, Mazmanian SK. Innate immune recognition of the microbiota promotes host-microbial symbiosis. Nat Immunol. 2013;14:668–75. doi: 10.1038/ni.2635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Federle MJ. Autoinducer-2-based chemical communication in bacteria: complexities of interspecies signaling. Contrib Microbiol. 2009;16:18–32. doi: 10.1159/000219371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferluga S, Venturi V. OryR is a LuxR-family protein involved in interkingdom signaling between pathogenic Xanthomonas oryzae pv. oryzae and rice. J Bacteriol. 2009;191:890–7. doi: 10.1128/JB.01507-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freestone PP, Haigh RD, Williams PH, Lyte M. Stimulation of bacterial growth by heat-stable, norepinephrine-induced autoinducers. FEMS Microbiol Lett. 1999;172:53–60. doi: 10.1111/j.1574-6968.1999.tb13449.x. [DOI] [PubMed] [Google Scholar]
- Hammer BK, Bassler BL. Quorum sensing controls biofilm formation in Vibrio cholerae. Mol Microbiol. 2003;50:101–4. doi: 10.1046/j.1365-2958.2003.03688.x. [DOI] [PubMed] [Google Scholar]
- Han JL, Lin HL. Intestinal microbiota and type 2 diabetes: from mechanism insights to therapeutic perspective. World J Gastroenterol. 2014;20:17737–45. doi: 10.3748/wjg.v20.i47.17737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henke JM, Bassler BL. Three parallel quorum-sensing systems regulate gene expression in Vibrio harveyi. J Bacteriol. 2004;186:6902–14. doi: 10.1128/JB.186.20.6902-6914.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins DA, Pomianek ME, Kraml CM, Taylor RK, Semmelhack MF, Bassler BL. The major Vibrio cholerae autoinducer and its role in virulence factor production. Nature. 2007;450:883–6. doi: 10.1038/nature06284. [DOI] [PubMed] [Google Scholar]
- Hughes DT, Sperandio V. Inter-kingdom signalling: communication between bacteria and their hosts. Nat Rev Microbiol. 2008;6:111–20. doi: 10.1038/nrmicro1836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ismail AS, Behrendt CL, Hooper LV. Reciprocal interactions between commensal bacteria and gamma delta intraepithelial lymphocytes during mucosal injury. J Immunol. 2009;182:3047–54. doi: 10.4049/jimmunol.0802705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ke X, Miller LC, Bassler BL. Determinants governing ligand specificity of the Vibrio harveyi LuxN quorum-sensing receptor. Mol Microbiol. 2015;95:127–42. doi: 10.1111/mmi.12852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kneidinger B, Marolda C, Graninger M, Zamyatina A, Mcarthur F, Kosma P, Valvano MA, Messner P. Biosynthesis pathway of ADP-L-glycero-beta-D-manno-heptose in Escherichia coli. J Bacteriol. 2002;184:363–9. doi: 10.1128/JB.184.2.363-369.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long T, Tu KC, Wang Y, Mehta P, Ong NP, Bassler BL, Wingreen NS. Quantifying the integration of quorum-sensing signals with single-cell resolution. PLoS Biol. 2009;7:e68. doi: 10.1371/journal.pbio.1000068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loomis WD, Durst RW. Chemistry and biology of boron. Biofactors. 1992;3:229–39. [PubMed] [Google Scholar]
- Lyte M, Bailey MT. Neuroendocrine-bacterial interactions in a neurotoxin-induced model of trauma. J Surg Res. 1997;70:195–201. doi: 10.1006/jsre.1997.5130. [DOI] [PubMed] [Google Scholar]
- Lyte M, Ernst S. Catecholamine induced growth of gram negative bacteria. Life Sci. 1992;50:203–12. doi: 10.1016/0024-3205(92)90273-r. [DOI] [PubMed] [Google Scholar]
- Mazmanian SK, Liu CH, Tzianabos AO, Kasper DL. An immunomodulatory molecule of symbiotic bacteria directs maturation of the host immune system. Cell. 2005;122:107–18. doi: 10.1016/j.cell.2005.05.007. [DOI] [PubMed] [Google Scholar]
- Mcarthur F, Andersson CE, Loutet S, Mowbray SL, Valvano MA. Functional analysis of the glycero-manno-heptose 7-phosphate kinase domain from the bifunctional HldE protein, which is involved in ADP-L-glycero-D-manno-heptose biosynthesis. J Bacteriol. 2005;187:5292–300. doi: 10.1128/JB.187.15.5292-5300.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mcclean KH, Winson MK, Fish L, Taylor A, Chhabra SR, Camara M, Daykin M, Lamb JH, Swift S, Bycroft BW, Stewart GS, Williams P. Quorum sensing and Chromobacterium violaceum: exploitation of violacein production and inhibition for the detection of N-acylhomoserine lactones. Microbiology. 1997;143( Pt 12):3703–11. doi: 10.1099/00221287-143-12-3703. [DOI] [PubMed] [Google Scholar]
- Miller MB, Skorupski K, Lenz DH, Taylor RK, Bassler BL. Parallel quorum sensing systems converge to regulate virulence in Vibrio cholerae. Cell. 2002;110:303–14. doi: 10.1016/s0092-8674(02)00829-2. [DOI] [PubMed] [Google Scholar]
- Miller ST, Xavier KB, Campagna SR, Taga ME, Semmelhack MF, Bassler BL, Hughson FM. Salmonella typhimurium recognizes a chemically distinct form of the bacterial quorum-sensing signal AI-2. Mol Cell. 2004;15:677–87. doi: 10.1016/j.molcel.2004.07.020. [DOI] [PubMed] [Google Scholar]
- Neiditch MB, Federle MJ, Miller ST, Bassler BL, Hughson FM. Regulation of LuxPQ receptor activity by the quorum-sensing signal autoinducer-2. Mol Cell. 2005;18:507–18. doi: 10.1016/j.molcel.2005.04.020. [DOI] [PubMed] [Google Scholar]
- Neiditch MB, Federle MJ, Pompeani AJ, Kelly RC, Swem DL, Jeffrey PD, Bassler BL, Hughson FM. Ligand-induced asymmetry in histidine sensor kinase complex regulates quorum sensing. Cell. 2006;126:1095–108. doi: 10.1016/j.cell.2006.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng WL, Bassler BL. Bacterial quorum-sensing network architectures. Annu Rev Genet. 2009;43:197–222. doi: 10.1146/annurev-genet-102108-134304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panja A, Siden E, Mayer L. Synthesis and regulation of accessory/proinflammatory cytokines by intestinal epithelial cells. Clin Exp Immunol. 1995;100:298–305. doi: 10.1111/j.1365-2249.1995.tb03668.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker MW, Buckley JT, Postma JP, Tucker AD, Leonard K, Pattus F, Tsernoglou D. Structure of the Aeromonas toxin proaerolysin in its water-soluble and membrane-channel states. Nature. 1994;367:292–5. doi: 10.1038/367292a0. [DOI] [PubMed] [Google Scholar]
- Peterson LW, Artis D. Intestinal epithelial cells: regulators of barrier function and immune homeostasis. Nat Rev Immunol. 2014;14:141–53. doi: 10.1038/nri3608. [DOI] [PubMed] [Google Scholar]
- Pullinger GD, Carnell SC, Sharaff FF, Van Diemen PM, Dziva F, Morgan E, Lyte M, Freestone PP, Stevens MP. Norepinephrine augments Salmonella enterica-induced enteritis in a manner associated with increased net replication but independent of the putative adrenergic sensor kinases QseC and QseE. Infect Immun. 2010;78:372–80. doi: 10.1128/IAI.01203-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rakoff-Nahoum S, Paglino J, Eslami-Varzaneh F, Edberg S, Medzhitov R. Recognition of commensal microflora by toll-like receptors is required for intestinal homeostasis. Cell. 2004;118:229–41. doi: 10.1016/j.cell.2004.07.002. [DOI] [PubMed] [Google Scholar]
- Roignot J, Peng X, Mostov K. Polarity in mammalian epithelial morphogenesis. Cold Spring Harb Perspect Biol. 2013;5 doi: 10.1101/cshperspect.a013789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruemmele FM, Beaulieu JF, Dionne S, Levy E, Seidman EG, Cerf-Bensussan N, Lentze MJ. Lipopolysaccharide modulation of normal enterocyte turnover by toll-like receptors is mediated by endogenously produced tumour necrosis factor alpha. Gut. 2002;51:842–8. doi: 10.1136/gut.51.6.842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutherford ST, Bassler BL. Bacterial quorum sensing: its role in virulence and possibilities for its control. Cold Spring Harb Perspect Med. 2012;2 doi: 10.1101/cshperspect.a012427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samak G, Chaudhry KK, Gangwar R, Narayanan D, Jaggar JH, Rao R. Calcium/Ask1/MKK7/JNK2/c-Src signalling cascade mediates disruption of intestinal epithelial tight junctions by dextran sulfate sodium. Biochem J. 2015;465:503–15. doi: 10.1042/BJ20140450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schauder S, Shokat K, Surette MG, Bassler BL. The LuxS family of bacterial autoinducers: biosynthesis of a novel quorum-sensing signal molecule. Mol Microbiol. 2001;41:463–76. doi: 10.1046/j.1365-2958.2001.02532.x. [DOI] [PubMed] [Google Scholar]
- Schenk G, Duggleby RG, Nixon PF. Properties and functions of the thiamin diphosphate dependent enzyme transketolase. Int J Biochem Cell Biol. 1998;30:1297–318. doi: 10.1016/s1357-2725(98)00095-8. [DOI] [PubMed] [Google Scholar]
- Schnupf P, Gaboriau-Routhiau V, Gros M, Friedman R, Moya-Nilges M, Nigro G, Cerf-Bensussan N, Sansonetti PJ. Growth and host interaction of mouse segmented filamentous bacteria in vitro. Nature. 2015;520:99–103. doi: 10.1038/nature14027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semmelhack MF, Campagna SR, Federle MJ, Bassler BL. An expeditious synthesis of DPD and boron binding studies. Org Lett. 2005;7:569–72. doi: 10.1021/ol047695j. [DOI] [PubMed] [Google Scholar]
- Shen Y, Giardino Torchia ML, Lawson GW, Karp CL, Ashwell JD, Mazmanian SK. Outer membrane vesicles of a human commensal mediate immune regulation and disease protection. Cell Host Microbe. 2012;12:509–20. doi: 10.1016/j.chom.2012.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith RS, Harris SG, Phipps R, Iglewski B. The Pseudomonas aeruginosa quorum-sensing molecule N-(3-oxododecanoyl)homoserine lactone contributes to virulence and induces inflammation in vivo. J Bacteriol. 2002;184:1132–9. doi: 10.1128/jb.184.4.1132-1139.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramoni S, Gonzalez JF, Johnson A, Pechy-Tarr M, Rochat L, Paulsen I, Loper JE, Keel C, Venturi V. Bacterial subfamily of LuxR regulators that respond to plant compounds. Appl Environ Microbiol. 2011;77:4579–88. doi: 10.1128/AEM.00183-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surette MG, Miller MB, Bassler BL. Quorum sensing in Escherichia coli, Salmonella typhimurium, and Vibrio harveyi: a new family of genes responsible for autoinducer production. Proc Natl Acad Sci U S A. 1999;96:1639–44. doi: 10.1073/pnas.96.4.1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swem LR, Swem DL, O’loughlin CT, Gatmaitan R, Zhao B, Ulrich SM, Bassler BL. A quorum-sensing antagonist targets both membrane-bound and cytoplasmic receptors and controls bacterial pathogenicity. Mol Cell. 2009;35:143–53. doi: 10.1016/j.molcel.2009.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taga ME, Bassler BL. Chemical communication among bacteria. Proc Natl Acad Sci U S A. 2003;100(Suppl 2):14549–54. doi: 10.1073/pnas.1934514100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taga ME, Miller ST, Bassler BL. Lsr-mediated transport and processing of AI-2 in Salmonella typhimurium. Mol Microbiol. 2003;50:1411–27. doi: 10.1046/j.1365-2958.2003.03781.x. [DOI] [PubMed] [Google Scholar]
- Teplitski M, Chen H, Rajamani S, Gao M, Merighi M, Sayre RT, Robinson JB, Rolfe BG, Bauer WD. Chlamydomonas reinhardtii secretes compounds that mimic bacterial signals and interfere with quorum sensing regulation in bacteria. Plant Physiol. 2004;134:137–46. doi: 10.1104/pp.103.029918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teplitski M, Robinson JB, Bauer WD. Plants secrete substances that mimic bacterial N-acyl homoserine lactone signal activities and affect population density-dependent behaviors in associated bacteria. Mol Plant Microbe Interact. 2000;13:637–48. doi: 10.1094/MPMI.2000.13.6.637. [DOI] [PubMed] [Google Scholar]
- Thompson JA, Oliveira RA, Djukovic A, Ubeda C, Xavier KB. Manipulation of the quorum sensing signal AI-2 affects the antibiotic-treated gut microbiota. Cell Rep. 2015;10:1861–71. doi: 10.1016/j.celrep.2015.02.049. [DOI] [PubMed] [Google Scholar]
- Tomasello G, Tralongo P, Damiani P, Sinagra E, Di Trapani B, Zeenny MN, Hussein IH, Jurjus A, Leone A. Dismicrobism in inflammatory bowel disease and colorectal cancer: changes in response of colocytes. World J Gastroenterol. 2014;20:18121–30. doi: 10.3748/wjg.v20.i48.18121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venturi V, Fuqua C. Chemical signaling between plants and plant-pathogenic bacteria. Annu Rev Phytopathol. 2013;51:17–37. doi: 10.1146/annurev-phyto-082712-102239. [DOI] [PubMed] [Google Scholar]
- Visick KL, Foster J, Doino J, Mcfall-Ngai M, Ruby EG. Vibrio fischeri lux genes play an important role in colonization and development of the host light organ. J Bacteriol. 2000;182:4578–86. doi: 10.1128/jb.182.16.4578-4586.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen L, Ley RE, Volchkov PY, Stranges PB, Avanesyan L, Stonebraker AC, Hu C, Wong FS, Szot GL, Bluestone JA, Gordon JI, Chervonsky AV. Innate immunity and intestinal microbiota in the development of Type 1 diabetes. Nature. 2008;455:1109–13. doi: 10.1038/nature07336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xavier KB, Miller ST, Lu W, Kim JH, Rabinowitz J, Pelczer I, Semmelhack MF, Bassler BL. Phosphorylation and processing of the quorum-sensing molecule autoinducer-2 in enteric bacteria. ACS Chem Biol. 2007;2:128–36. doi: 10.1021/cb600444h. [DOI] [PubMed] [Google Scholar]
- Zargar A, Quan DN, Carter KK, Guo M, Sintim HO, Payne GF, Bentley WE. Bacterial secretions of nonpathogenic Escherichia coli elicit inflammatory pathways: a closer investigation of interkingdom signaling. MBio. 2015;6:e00025. doi: 10.1128/mBio.00025-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.