Abstract
Endoplasmic reticulum (ER) stress is a major contributor to inflammatory diseases, such as Crohn’s disease and type 2 diabetes1,2. ER stress induces the unfolded protein response (UPR), which involves activation of three transmembrane receptors, ATF6 (activating transcription factor 6), PERK (protein kinase RNA-like endoplasmic reticulum kinase) and IRE1α (inositol-requiring enzyme 1α)3 (Extended Data figure 1a). Once activated, IRE1α recruits TRAF2 (TNF receptor-associated factor 2) to the ER membrane to initiate inflammatory responses via the nuclear factor kappa B (NF-κB) pathway4. Inflammation is commonly triggered when pattern recognition receptors (PRRs), such as Toll-like receptors (TLRs) or nucleotide-binding oligomerization domain (NOD)-like receptors (NLRs), detect tissue damage or microbial infection. However, it is not clear which PRRs play a major role in inducing inflammation during ER stress. Here we show that NOD1 and NOD2, two members of the NLR family of PRRs, are important mediators of ER stress-induced inflammation. The ER stress inducers thapsigargin and dithiothreitol (DTT) triggered production of the pro-inflammatory cytokine interleukin (IL)-6 in a NOD1/2-dependent fashion. Inflammation and IL-6 production triggered by infection with Brucella abortus, which induces ER stress by injecting the type IV secretion system (T4SS) effector protein VceC into host cells5, was TRAF2, NOD1/2 and RIP2-dependent and could be blunted by treatment with the ER-stress inhibitor tauroursodeoxycholate (TUDCA) or an IRE1α kinase inhibitor. The association of NOD1 and NOD2 with pro-inflammatory responses induced by the IRE1α/TRAF2 signaling pathway provides a novel link between innate immunity and ER stress-induced inflammation.
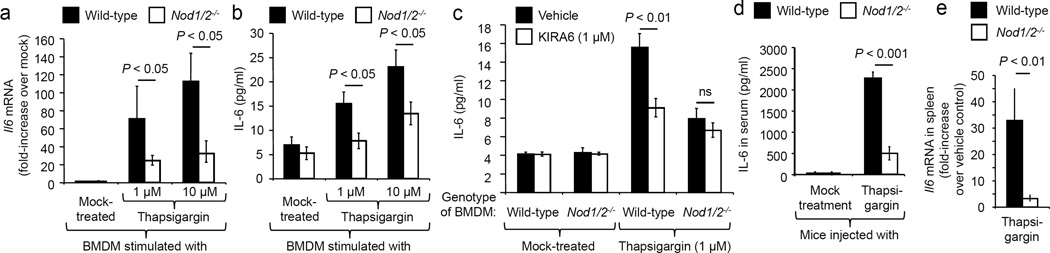
We investigated a potential role for NOD1 and NOD2 in the UPR, because this response triggers inflammation by activating IRE1α, a receptor that recruits TRAF2 to activate NF-κB (Extended Data figure 1a)4,6. NOD1 and NOD2 are two NLRs that contain major TRAF2 binding motifs7. Consistent with previous reports8,9, TRAF2 was required for NF-κB activation triggered by stimulation with canonical NOD1/2 ligands or mediated by auto-activation of NOD1, NOD2 or the NOD1/2 adaptor protein RIP2 (receptor-interacting serine/threonine-protein kinase 2, also known as RIPK2) (Extended Data figures 1b and 1c). To investigate whether NOD1 and NOD2 contribute to inflammatory responses during ER stress, we stimulated bone marrow-derived macrophages (BMDMs) of wild-type (C57BL/6) mice or mice deficient for NOD1 and NOD2 (Nod1/2−/− mice) with the ER stress inducer thapsigargin, a specific inhibitor of the SERCA (sarco/endoplasmic reticulum Ca2+-ATPase) channel10. Thapsigargin treatment induced the UPR, as indicated by a NOD1/2-independent elevation in transcript levels of Hsp5a and Chop (Extended Data figures 2a and 2b), two genes controlled by the ER stress sensors ATF6 and PERK (Extended Data figure 1a). Thapsigargin treatment also significantly (P < 0.05) induced Il6 transcription (Fig. 1a) and IL-6 synthesis (Fig. 1b), while synthesis of TRAF2 and the chaperones SGT1 and HSP90 remained unchanged (Extended Data figure 2c). Both Il6 transcription and IL-6 synthesis were blunted in BMDMs from Nod1/2−/− mice compared to wild-type mice (Fig. 1a–b), which was not due to differences in cell death (Extended Data figure 2d). Similarly, IL-6 synthesis induced by the ER stress inducer DTT was abrogated in BMDMs from Nod1/2−/− mice (Extended Data figure 2e) which was not due to differences in cell death (Extended Data figure 2d). Thapsigargin-induced IL-6 synthesis could be blunted by treatment with the ER stress inhibitor TUDCA (Extended Data figure 2f). In contrast, stimulation of BMDM with the canonical NOD2 ligand muramyldipeptide (MDP) did not result in increased transcript levels of Hsp5a and Chop (Extended Data figures 2a and 2b), but led to increased synthesis of IL-6 (Extended Data figure 2e), which could not be inhibited by TUDCA treatment (Extended Data figure 2f). In conclusion, the TUDCA-resistant pathway of IL-6 production induced by stimulation with a canonical NOD2 ligand was distinct from the TUDCA-sensitive pathway of IL-6 production induced by thapsigargin (Extended Data figure 1a).
Figure 1. Thapsigargin induced IL-6 production is dependent on NOD1 and NOD2.
(a–c) BMDM from Nod1/2−/− mice and wild type littermates (n = 8) were stimulated with thapsigargin (a and b) and/or KIRA6 (c) and Il6 mRNA expression (a) and IL-6 protein synthesis (b and c) were measured. (d and e) Nod1/2−/− mice and wild type littermates (n = 7) were injected with thapsigargin and IL-6 production in the serum (d) and Il6 mRNA expression in the spleen (e) were determined. Data are presented as mean ± s.e.m.
The IRE1α kinase domain is important for TRAF2 binding4, while the IRE1α RNase domain functions in splicing a 26-nucleotide intronic sequence from the Xbp1 mRNA11. We thus investigated whether thapsigargin-induced IL-6 synthesis could be blunted by treatment with the IRE1α kinase inhibitor KIRA6 (1-[4-[8-Amino-3-tert-butylimidazo{1,5-a}pyrazin-1-yl]naphthalen-1-yl]-3-[3-{trifluoromethyl}phenyl]urea)12, the IRE1α RNase inhibitor STF-083010 (N-[{2-hydroxynaphthalen-1-yl}methylidene]thiophene-2-sulfonamide)13 or the PERK inhibitor GSK265615714. Thapsigargin-induced IL-6 synthesis in BMDM was blunted by treatment with the IRE1α kinase inhibitor KIRA6 (Fig. 1c) which was not due to differences in cell death (Extended Data figure 2d). The PERK inhibitor GSK2656157 and the IRE1α RNase inhibitor STF-083010 inhibited expression of Chop, and Hsp5a, respectively, but did not reduce thapsigargin-induced IL-6 synthesis (Extended Data figure 2g–j). Collectively, these data suggested that the IRE1α kinase branch of the ER stress response was most important for inducing IL-6 synthesis (Extended Data figure 1a).
To investigate the in vivo relevance of our observations, we injected mice with thapsigargin, which resulted in elevated levels of IL-6, KC (CXCL1) and MIP-1β (CCL4) in the serum (Fig. 1d, Extended Data figure 3a–c) and increased transcription of Il6 in the spleen, liver and kidney (P < 0.01) (Fig. 1e, Extended Data figure 3d–e) of wild type mice, while induction of these pro-inflammatory responses was significantly blunted in Nod1/2−/− mice. Furthermore, thapsigargin-induced cytokine synthesis could be blunted by treatment with the ER stress inhibitor TUDCA (Extended Data figure 3a–c). Collectively, these data suggested that ER stress induced NOD1 and/or NOD2 signaling independently of peptidoglycan and that this pathway was only required for orchestrating the pro-inflammatory branch of the UPR, which leads to IL-6 production (Extended Data figure 1a).
ER stress has recently emerged as a response induced during infection with some bacterial pathogens (summarized in3). B. abortus is an ideal model organism to investigate induction of the UPR in vivo, because this pathogen has developed mechanisms to evade recognition by TLRs (reviewed in15,16). As a result, inflammatory responses induced during brucellosis are almost entirely dependent on a functional T4SS17, a virulence factor that injects effector proteins, including VceC, into the host cell cytosol18. VceC translocates to the ER where it binds the ER chaperone BiP and induces an IRE1α-dependent induction of IL-6 production5. Profiling of host responses elicited by intraperitoneal infection of mice with B. abortus revealed elevated circulating levels of IL-6 (Extended Data figure 4a), IL-12p40 (Extended Data figure 4b), gamma interferon (IFNγ) (Extended Data figure 4c), KC (Extended Data figure 4d), MIP-1β (Extended Data figure 4e), G-CSF (Extended Data figure 4f) and RANTES (Extended Data figure 4g). A blunting of these cytokine levels after treatment with TUDCA confirmed that inflammatory responses were triggered by ER stress (Extended Data figure 4). Intraperitoneal infection of mice with B. abortus induced elevated transcript levels of Il6, but this response was significantly (P < 0.05) blunted in mice infected with a B. abortus vceC mutant. The VceC-induced pro-inflammatory response was caused by ER stress, because elevated Il6 transcript levels induced during infection with the B. abortus wild type, but not those triggered by the vceC mutant, were significantly (P < 0.05) blunted after treatment with the ER stress inhibitor TUDCA (Fig. 2a), which was not due to altered bacterial numbers in the spleen (Extended Data figure 5a). Furthermore, Il6 transcript levels induced in the spleen and IL-6 synthesis induced in the serum of mice infected with the B. abortus wild type was significantly (P < 0.05) blunted by treatment with the IRE1α kinase inhibitor KIRA6 (Extended Data figure 5b and figure 2b), which was not due to altered bacterial numbers in the spleen (Extended Data figure 5c).
Figure 2. The ER-stress associated pro-inflammatory response induced by B. abortus VceC is dependent on the NOD1/NOD2 signaling pathway.
(a) Il6 mRNA expression in the spleen of vehicle-treated or TUDCA-treated mice infected with the indicated B. abortus strains (n=5). (b, c, f, g and h) Expression of Il6 (g) or synthesis of IL-6 (b, c, f and h) induced after infection of BMDM (n=3) (c) or mice (n=5; b, f, g and h) with the indicated B. abortus strains. (d and e) NF-κB activation induced by ectopic expression of VceC in HEK293 cells (n=4) transfected with dominant negative forms of TRAF2 (d), NOD1, NOD2 or RIP2 (e). Data are presented as mean ± s.e.m.
We next studied whether VceC-induced ER stress triggered NF-κB activation and IL-6 production through a pathway requiring NOD1 and/or NOD2 activity. B. abortus infection elicited significantly (P < 0.05) higher production of IL-6 (Fig. 2c) and a significant elevation of Il6 transcript levels (Extended Data figure 5d) in BMDMs from wild type mice compared to those from Nod1/2−/− mice. In contrast, infection with a vceC mutant elicited similar responses in BMDMs from wild type and Nod1/2−/− mice. Differences in IL-6 synthesis were not caused by differences in bacterial numbers associated with BMDMs (Extended Data figure 5e). Similar results were obtained with BMDM from RIP2-deficient (Rip2−/−) mice (Extended Data figure 5f). Ectopic expression of VceC in HEK293 cells induced NF-κB activation, which could be significantly blunted by transfecting cells with dominant negative forms of TRAF2 (P < 0.005) (Fig. 2D), NOD1 (P < 0.001), NOD2 (P < 0.001) or RIP2 (P < 0.001) (Fig. 2e). In contrast, transfection with a control protein (a dominant negative form of cell division control protein 42 [CDC42]) did not reduce VceC-induced NF-κB activation (Fig. 2e). Collectively, these data suggested that VceC-triggered NF-κB activation required NOD1, NOD2 and RIP2 activity.
To investigate the biological significance of NOD1/2-mediated responses to VceC, we infected Nod1/2−/− mice and wild type littermate controls with either the B. abortus wild type or a vceC mutant. The B. abortus wild type elicited significantly higher levels of circulating IL-6 (Fig. 2f) and splenic Il6 gene transcription (Fig. 2g) in wild-type mice compared to Nod1/2−/− mice, although the latter animals did not exhibit reduced bacterial tissue load (Extended Data figure 5g). TUDCA treatment blunted IL-6 synthesis in wild-type mice but not in Nod1/2−/− mice (Fig. 2f). Responses triggered by the vceC mutant were not significantly different in wild type and Nod1/2−/− mice (Fig.2f and 2g). Similarly, the B. abortus wild type elicited significantly higher IL-6 synthesis (Fig. 2h) and splenic Il6 expression (Extended Data figure 5h) in wild-type mice compared to Rip2−/− mice, which was not due to differences in bacterial load (Extended Data figure 5i). These data supported the idea that the NOD1/NOD2/RIP2-signaling pathway was required for the VceC-dependent induction of pro-inflammatory responses during B. abortus infection in vivo.
B. abortus generally causes mild chronic inflammation during its persistence in tissue of experimentally infected mice or within its natural bovine reservoir. However, in pregnant cows, the pathogen causes severe acute inflammation in the placenta, which leads to abortion. We next wanted to investigate the contribution of ER stress to acute placentitis using the pregnant mouse model19. Bacterial numbers in the placenta of mice infected with B. abortus wild type at 5 days of pregnancy increased to 109 cfu/g tissue by day 13 after infection (corresponding to day 18 of gestation) (Extended Data figure 6a), which was accompanied by a significant (P < 0.05) increase in Il6 transcript levels in the placenta (Extended Data figure 6b) and acute placentitis (Extended Data figures 6c, 6d and 6e). This time point was chosen to investigate a potential role for VceC in causing placentitis. Although the B. abortus wild type and the vceC mutant were recovered in similar numbers from the placenta (Extended Data figure 7a), the severity of placentitis (Fig. 3a and 3b) and the Il6 transcript levels in the placenta (Fig. 3c) were significantly (P < 0.05) reduced in mice infected with the vceC mutant. Strikingly, while none of the pups survived an infection with the B. abortus wild type, in mice infected with the vceC mutant approximately 30% of the pups were viable at day 18 of gestation (Fig. 3d). These data suggested that VceC-induced inflammation contributed to both placentitis and reduction of pup viability in this model.
Figure 3. B. abortus VceC-induced ER stress activates the NOD1/NOD2-dependent pro-inflammatory response, thereby contributing to abortion during B. abortus infection.
(a and b) Histopathology in the placenta of mice infected with B. abortus. Whisker plots in (a) represent the second and third quartiles (boxes) and the first and fourth quartiles (lines). (b) Representative images of histopathological changes. Arrow, neutrophil infiltration; N, necrosis. (c and e) Il6 expression in the placenta of vehicle-treated or TUDCA-treated mice infected with the indicated B. abortus strains. (d) Viability of pups. Each dot represents one animal. (e) Il6 expression in the placenta of Nod1/2−/− mice and wild type littermates infected with B. abortus. The N for all experiments is indicated in panel d.
Injecting B. abortus infected pregnant mice with the ER stress inhibitor TUDCA decreased Il6 transcript levels in the placenta (P < 0.001) (Fig. 3c), reduced the severity of placentitis (P < 0.01) (Fig. 3a and 3b), and increased the viability of pups to approximately 40% (Fig. 3d), while recovery of bacteria from organs remained similar (Extended Data figure 7b). To examine whether the NOD1/2 signaling pathway contributed to placentitis, pregnant Nod1/2−/− deficient mice and wild type littermate control mice were infected with B. abortus. Compared to infection of pregnant wild-type mice, B. abortus infection of pregnant Nod1/2−/− deficient mice produced diminished transcript levels of Il6 in the placenta (P < 0.05) (Fig. 3e), reduced the severity of placentitis (P < 0.005) (Fig. 3a and 3b), and increased the viability of pups to approximately 30% (Fig. 3d), while recovery of bacteria from organs remained unchanged (Extended Data figure 7c). To conclude, our data suggest that VceC triggers ER stress, thereby inducing the NOD1/2-dependent pro-inflammatory branch of the UPR, which contributes to abortion during B. abortus infection.
To investigate whether the proposed pathway was induced during infections with other pathogens, we infected HeLa cells with Chlamydia muridarum, a known ER stress inducer20. Infection of HeLa cells with C. muridarum or treatment with thapsigargin induced Il6 mRNA expression, which could be blunted by treatment with the IRE1α kinase inhibitor KIRA6 (Extended Data figure 8). Consistent with our model (Extended Data figure 1a), KIRA6 treatment did not blunt Il6 expression induced by stimulation of HeLa cells with the NOD2-ligand MDP. Inhibition of NOD1 and NOD2 signaling by transfecting HeLa cells with a dominant negative form of RIP2 (RIP2DN) blunted Il6 expression induced by C. muridarum infection, thapsigargin treatment or stimulation with MDP (Extended Data figure 8). These results suggest that the NOD1/NOD2/RIP2 signaling pathway also detects ER stress generated during Chlamydia infection.
NOD1 and NOD2 are traditionally viewed as cytosolic sensors of bacterial peptidoglycan fragments. A clue that NOD2 may fulfill functions in addition to sensing canonical bacterial peptidoglycan ligands comes from the finding that this PRR activates innate immune responses during infection with influenza virus21, a pathogen that triggers ER stress in infected host cells22. Our results suggest that pro-inflammatory responses induced by ER stress are mediated through a NOD1/2-dependent, TUDCA/KIRA6-sensitive pathway, which differs from the TUDCA/KIRA6-resistant pathways induced by bacterial peptidoglycan fragments (Extended Data figure 1a). This finding provides a new link between NOD1, NOD2 and inflammatory diseases involving ER stress, such as Crohn’s disease, ulcerative colitis, obesity and type 2 diabetes1,2.
METHODS
Bacterial strains, tissue culture cells and culture conditions
E. coli and B. abortus strains were routinely cultured aerobically at 37°C in Luria-Bertani (LB) broth and on LB agar plates and in tryptic soy broth and on tryptic soy agar (TSA) plates, respectively. Chlamydia muridarum strain Nigg II was purchased from ATCC (Manassas, VA). Bacteria were cultured in HeLa 229 cells in DMEM supplemented with 10% FBS. Elementary bodies (EBs) were purified by discontinuous density gradient centrifugations as described previously1 and stored at −80 °C. The HEK293 cell line was maintained in Dulbecco’s modified Eagle’s medium (DMEM) containing 10% FBS at 37°C in a 5% CO2 atmosphere.
Transfections
HEK293 cells (ATCC CRL-1573) were obtained from ATCC and were grown in a 48-well tissue culture plates in DMEM containing 10% FBS until ~40% of confluency was reached. HEK293 cells were transfected with a total of 250 ng of plasmid DNA per well, consisting of 25 ng of the reporter construct pNF-κB-luc, 25 ng of the normalization vector pTK-LacZ, and 200 ng of the different combinations of mammalian expression vectors carrying the indicated gene (empty control vector, pCMV-HA-VceC2, pCMV-HA-TRAF2DN (this study), hNOD1-3xFlag, hNOD2-3xFlag, pCMV-HA-hRip2, hNOD1DN-3xFlag, hNOD2DN-3xFlag or pCMV-HA-Rip2DN3 and pCMV-myc-CDC42DN4. The dominant-negative form of TRAF2, lacking an amino-terminal RING finger domain5, was PCR amplified from cDNA prepared from HEK293 cells and cloned into the mammalian expression vector pCMV-HA (BD Biosciences Clontech). Forty-eight hours after transfection, cells were lysed either without any treatment, or stimulated with C12-iE-DAP (1000 ng/mL, InvivoGen) and MDP (10 µg/mL, InvivoGen). After five hours of treatment the cells were lysed and analysed for β-galactosidase and luciferase activity (Promega). FuGene HD (Roche) was used as a transfection reagent according to the manufacturer’s instructions. Cell lines were monitored for Mycoplasma contamination.
Bone marrow derived macrophage infection
Bone marrow-derived macrophages (BMDM) were differentiated from bone marrow precursors from femur and tibiae of C57BL/6 mice obtained from The Jackson Laboratory (Bar Harbor), Nod1+/−Nod2+/− (wild-type littermates) and Nod1−/−Nod2−/− (NOD1/NOD2-deficient) mice (generated at UC Davis) as described previously6. For BMDM experiments, 24-well microtiter plates were seeded with macrophages at a concentration of 5 × 105 cells/well in 0.5 mL of RPMI media (Invitrogen, Grand Island, NY) supplemented with 10% FBS and 10 mM L-glutamine (RPMI supl) and incubated for 48 h at 37°C in 5% CO2. BMDM were stimulated with C12-iE-DAP (1000 ng/mL, InvivoGen), MDP (10 µg/mL, InvivoGen), thapsigargin (1 µM and 10 µM, Sigma-Aldrich), dithiothreitol (DTT) (1 mM, Sigma-Aldrich), and LPS (10 ng/mL, InvivoGen) with or without pretreatment (30 min) of the cells with IRE1α kinase inhibitor KIRA6 (1 µM, Calbiochem), IRE1α endonuclease inhibitor STF-083010 (50 µM, Sigma-Aldrich), PERK inhibitor GSK2656157 (500 nM, Calbiochem) and tauroursodeoxycholate TUDCA (200 µM, Sigma-Aldrich) in the presence of 1 ng/mL of recombinant mouse IFN-γ (BD Bioscience, San Jose, CA). After 24 hours of stimulation, samples for ELISA and gene expression analysis were collected as described below. Preparation of the B. abortus wild type strain 2308 and the ΔvceC mutant inoculum and BMDM infection was performed as previously described6. Approximately 5 × 107 bacteria in 0.5 mL of RPMI supl were added to each well containing 5 × 105 BMDM. Microtiter plates were centrifuged at 210 × g for 5 min at room temperature in order to synchronize infection. Cells were incubated for 20 min at 37°C in 5% CO2, and free bacteria were removed by three washes with phosphate-buffered saline (PBS), and the zero-time-point sample was taken as described below. After the PBS wash, RPMI supl plus 50 mg/mL gentamicin and 1 ng/mL of recombinant mouse IFN-γ (BD Bioscience, San Jose, CA) was added to the cells, and incubated at 37°C in 5% CO2. For cytokine production assays, supernatant for each well was sampled at 24h after infection. In order to determine bacterial survival, the medium was aspirated at the time-point described above, and the BMDM were lysed with 0.5 mL of 0.5% Tween 20, followed by rinsing each well with 0.5 mL of PBS. Viable bacteria were quantified by serial dilution in sterile PBS and plating on TSA. For gene expression assays, BMDM were suspended in 0.5 mL of TRI-reagent (Molecular Research Center, Cincinnati) at the time-points described above and kept at −80°C until further use. At least three independent assays were performed with triplicate samples, and the standard error of the mean for each time point was calculated.
Animal experiments
All mouse experiments were approved by the Institutional Animal Care and Use Committees at the University of California, Davis, and were conducted in accordance with institutional guidelines. Sample sizes were determined based on experience with infection models and were calculated to use the minimum number of animals possible to generate reproducible results. C57BL/6 wild-type mice and Rip2−/− mice (The Jackson Laboratory), Nod1+/−Nod2+/− (wild type littermates) and Nod1−/−Nod2−/− (NOD1/NOD2-deficient) mice (generated at UC Davis) were injected intraperitoneally (i.p.) with 100 µl of 2.5 mg/kg body weight of thapsigargin (Sigma-Aldrich) at 0 and 24 hours, and 4 hours after the second injection the mice were sacrificed and serum and tissues collected for gene expression analysis and detection of cytokines. Where indicated, mice were treated i.p. at 12 hours before the first thapsigargin dose and 12 hours before the second thapsigargin dose with the ER stress inhibitor TUDCA (250 mg/kg body weight).
Female and male C57BL/6, Nod1+/−Nod2+/−, Nod1−/−Nod2−/− mice, and Rip2−/− mice aged 6–8 weeks, were held in microisolator cages with sterile bedding and irradiated feed in a biosafety level 3 laboratory. Groups of 5 mice were inoculated i.p. with 0.2 mL of phosphate-buffered saline (PBS) containing 5 × 105 CFU of B. abortus 2308 or its isogenic mutant ΔvceC, as previously described7. At 3 days post infection, mice were euthanized by CO2 asphyxiation and their serum and spleens were collected aseptically at necropsy. The spleens were homogenized in 2 mL of PBS, and serial dilutions of the homogenate were plated on TSA for enumeration of CFU. Spleen samples were also collected for gene expression analysis as described below. When necessary, mice were treated i.p. at day one and two post infection with a daily dose of 250 mg/kg body weight of the ER stress inhibitor TUDCA (Sigma-Aldrich), or 10mg/kg body weight of the IRE1α kinase inhibitor KIRA6 (Calbiochem) or vehicle control.
For the placentitis mouse model, C57BL/6, Nod1+/−Nod2+/− and Nod1−/−Nod2−/− mice, aged 8–10 weeks, were held in microisolator cages with sterile bedding and irradiated feed in a biosafety level 3 laboratory. Female Nod1+/−Nod2+/− mice were mated with male C57BL/6 mice (control mice) and female Nod1−/−Nod2−/− mice were mated with male Nod1−/−Nod2−/− mice (Nod1/Nod2-deficient), and pregnancy was confirmed by presence of a vaginal plug. At 5 days of gestation, groups of pregnant mice were mock infected or infected i.p. with 105 CFU of Brucella abortus 2308 or its isogenic mutant ΔvceC (Day 0). At 3, 7, and 13 days after infection mice were euthanized by CO2 asphyxiation and the spleen and placenta were collected aseptically at necropsy. At day 13 after infection (corresponding to day 18 of gestation), viability of pups was evaluated based on the presence of fetal movement and heartbeat, and fetal size and skin color. Fetuses were scored as viable if they exhibited movement and a heartbeat, visible blood vessels, bright pink skin, and were of normal size for their gestational period. Fetuses were scored as non-viable if fetal movement, heartbeat, and visible blood vessels were absent, skin was pale or opaque, and their size for gestational period or compared to littermates was small, or they showed evidence of fetal reabsorption. Percentage of viability was calculated as [(number viable pups per litter/total number pups per litter) × 100]. At each time point, the placenta samples were collected for bacteriology, gene expression analysis and histopathological analysis (Extended Data figure 6d). When indicated, mice were treated i.p. at days 5, 7 and 9 post infection with a daily dose of 250 mg/kg body weight of the ER stress inhibitor tauroursodeoxycholate TUDCA (Sigma-Aldrich) or vehicle control.
Real-Time PCR
RNA was isolated from BMDMs and mouse tissues using Tri-reagent (Molecular Research Center) according to the instructions of the manufacturer. Reverse transcription was performed on 1 µg of DNase-treated RNA with Taqman reverse transcription reagent (Applied Biosystems). For each real-time reaction, 4 µl of cDNA was used combined with primer pairs for mouse β-actin, Il6, Hsp5a and Chop. Real time transcription-PCR was performed using Sybr green and an ABI 7900 RT-PCR machine (Applied Biosystems). The fold change in mRNA levels was determined using the comparative threshold cycle (Ct) method. Target gene transcription was normalized to the levels of β-actin mRNA.
Cytokine detection
Cytokine levels in mouse serum and supernatants of infected BMDMs were measured using either a multiplex cytokine/chemokine assay (Bio-Plex 23-plex Mouse cytokine assay; Bio-Rad), or via an enzyme-linked immunosorbent assay (IL-6 ELISA; eBioscience), according to the manufacturer’s instructions.
LDH release assay
Cytotoxicity was determined by using a LDH release assay in supernatant of BMDM treated as described above. LDH release assay was performed using a CytoTox 96® Non-Radioactive Cytotoxicity Assay (Promega), following manufacture’s protocol. The percentage of LDH release was calculated as follows: % LDH release = 100× (absorbance reading treated well – absorbance reading untreated control) / (absorbance reading maximum LDH release control – absorbance reading untreated control). The kit provided lysis buffer was used to achieve complete cell lysis and the supernatant from lysis buffer treated cells was used to determine maximum LDH release control.
Chlamydia infection (line 545)
Hela 229 cells (ATCC CCL-2.1) were cultured in 96-well tissue culture plates at a concentration of 4 × 104 cells/well in Dulbecco’s Modified Eagle Medium (DMEM) (Life Technologies, Grand Island, NY) supplemented with 10% FBS. HeLa 229 cells were transfected with a total of 125 ng of pCMV-HA-Rip2DN or empty control vector per well. 24 hours post transfection Hela 229 cells were treated with Dextran to enhance infection efficacy before they were infected with 1.7 × 105 Chlamydia bacteria/well. The plates were centrifuged at 2000rpm for 60 min at 37°C, then incubated for 30 min at 37°C in 5% CO2. Supernatant was discarded and replaced with DMEM containing 1ug/mL cyclohexine (Sigma Aldrich) and where indicated, 1µM KIRA6, 10µM thapsigargin or 10µg/ml MDP, was added to cultures before incubation at 37°C in 5% CO2 for 40 hours. For gene expression assays, HeLa 229 cells were suspended in TRI-reagent (Molecular Research Center, Cincinnati) and RNA was isolated. Infection efficiency was confirmed in separate plates by staining Chlamydia-infected Hela 229 cells with anti-Chlamydia MOMP antibody and counting bacteria under fluorescent microscope. Four independent assays were performed and the standard error of the mean calculated.
Western Blots
BMDM stimulated where indicated with 10µM thapsigargin for 24 hours were lysed in lysis buffer (4% SDS, 100mM Tris, 20% glycerol) and 10 µg of protein was analyzed by Western blot using antibodies raised against rabbit TRAF2 (C192, #4724, Cell signaling), rabbit HSP90 (E289, # 4875, Cell signaling), mouse SGT1 (ab60728, Abcam) and rabbit α/β-Tubulin (#2148, Cell signaling).
Statistical analysis
For tissue culture experiments, statistical differences were calculated using a paired Student’s t-test. To determine statistical significance in animal experiments, an unpaired Student’s t-test was used. To determine statistical significance of differences in total histopathology scores, a Mann-Whitney U-test was used. A two-tailed P value of <0.05 was considered to be significant.
Extended Data
Extended Data figure 1. Schematic of ER stress and NOD1/2 signaling.

(a) Model of how ER stress induces a NOD1/2-dependent pro-inflammatory response through a TUDCA/KIRA6-sensitive pathway, which differs from the TUDCA/KIRA6-resistant pathways induced by bacterial peptidoglycan fragments (MDP or C12-iE-DAP). (b) NF-κB activation induced by ectopic expression of VceC in HEK293 cells transfected with a dominant negative form of TRAF2 or a vector control. (c) NF-κB activation mediated by expression-induced auto-activation of NOD1, NOD2 or RIP2 in HEK293 cells that were transfected with a dominant negative form of TRAF2 or a vector control. Data are expressed as mean luciferase activity ± s.e.m. from five independent experiments.
Extended Data figure 2. Only the pro-inflammatory arm of the UPR requires NOD1 and NOD2.

(a and b) BMDM from Nod1/2−/− mice and wild type littermates were stimulated with thapsigargin or MDP, and mRNA abundance for Hsp5a (a) and Chop (b) was quantified (n = 4). (c) Expression of SGT1, HSP90 and TRAF2 was detected by Western blot in lysates of thapsigargin-stimulated BMDM from wild-type mice and Nod1/2−/− mice (n = 3). Detection of tubulin served as a loading control. A representative image for BMDM from one wild-type and one Nod1/2−/− animal is shown. (d) LDH release induced by treatment of BMDM from wild-type mice and Nod1/2−/− mice (n = 5) with thapsigargin, DTT or KIRA6. (e) Stimulation with MDP or DTT induced IL-6 production in BMDM from C57BL/6 mice (wild type) but not in BMDM from Nod1/2−/− mice (n = 8). (f) IL-6 secretion induced by thapsigargin, but not by the canonical NOD2 ligand MDP, was significantly inhibited by ER stress inhibitor TUDCA in BMDM (n = 8). BMDM did not respond to stimulation with a canonical NOD1 ligand (C12-iE-DAP). (g–k) BMDM from wild-type mice and Nod1/2−/− mice (n = 4) were treated with the PERK inhibitor GSK2656157 (GSK) (g and i) or the IRE1α RNase inhibitor STF-083010 (STF) (h and k) and IL-6 synthesis measured by ELISA (g and h) or mRNA analyzed by real-time PCR (i and j). Data are presented as mean ± s.e.m.
Extended Data figure 3. Proinflammatory responses induced by thapsigargin are NOD1/2-dependent.

(a–c) Groups (n = 5) of wild-type mice and Nod1/2−/− mice were treated with thapsigargin and received either vehicle control of TUDCA. Synthesis of IL-6 (a), KC (b) and MIP-1β (c) in the serum was determined using a Bio-plex cytokine assay. (d and e) Wild-type (C57BL/6) mice and Nod1/2−/− mice (n = 4) were treated with thapsigargin and transcript levels of Il6 determined by quantitative real-time PCR. Data are expressed as fold-increases over vehicle control-treated animals. Data are presented as mean ± s.e.m.
Extended Data figure 4. B. abortus-induced inflammatory responses in mice are blunted by TUDCA treatment.

Mice (n ≥ 4) were mock infected or infected with the B. abortus wild type and were treated with TUDCA or vehicle control. Three days after infection, circulating levels of IL-6 (a), IL-12p40 (b), IFNγ (c), KC (d), MIP-1β (e), G-CSF (f) and RANTES (g) were profiled in serum using a Bio-Plex cytokine assay. Data are presented as mean ± s.e.m.
Extended Data figure 5. Bacterial burden and host responses during infection with B. abortus.

(a, c, e, g and i) Bacterial burden in the spleen and in BMDM of wild type and Nod1/2−/− mice (a, c, e, and g) or Rip2−/− mice (i). No statistically significant differences in colony forming units (CFU) recovered from the spleen (a, c, g and i) or from BMDM (e) of wild type and Nod1/2−/− or Rip2−/− mice (n ≥ 4) infected with B. abortus wild type or the vceC mutant were observed. (b, d, f and h) Host responses elicited during B. abortus infection. (b) Groups of mice (n=5) were infected with the indicated B. abortus strains and treated with KIRA6. (d and f) BMDM from wild-type mice and Nod1/2−/− mice (d) or wild-type mice and Rip2−/− mice (n ≥ 4) (f) were infected with the indicated B. abortus strains. (h) Groups (n = 5) of wild-type mice and Rip2−/− mice were infected with the indicated B. abortus strains. (b, d and h) Il6 mRNA levels were determined by quantitative real-time PCR. (f) IL-6 synthesis was determined by ELISA. Data are presented as mean ± s.e.m.
Extended Data figure 6. The B. abortus placentitis model.

(a) Bacterial numbers of wild-type B. abortus (strain 2308) recovered from in the spleen and placenta (n = 5). (b–c), Il6 mRNA expression (b) and total histopathology scores (c) in the placenta of mice at days 3, 7 and 13 after infection with B. abortus. (d) Scoring criteria for blinded evaluation of H&E-stained sections from the placenta. (e) Representative images of the histopathology observed in the placenta of B. abortus infected mice at days 3, 7 and 13 after infection. Arrow, neutrophil infiltration; N, necrosis.
Extended Data figure 7. Bacterial burden in the spleen and placenta of wild type and Nod1/2−/− mice.

(a–c) no statistically significant differences in colony forming units (CFU) in the spleen and placenta of wild type and Nod1/2−/− mice infected with B. abortus wild type or the vceC mutant at 13 days post infection were observed. Data are presented as mean ± s.e.m., (n = 5).
Extended Data figure 8. Il6 expression induced by Chlamydia muridarum.

HeLa cells were stimulated with MDP, thapsigargin or infected with Chlamydia muridarum and treated with KIRA6 or transfected with RIP2DN. Expression of Il6 was determined by quantitative real-time PCR. Data are presented as mean ± s.e.m., (n = 4).
Acknowledgments
Work in R.M.T’s laboratory is supported by US Public Health Service (USPHS) Grants AI112258 and AI109799. Work in A.J.B.’s laboratory was supported by USPHS Grants AI044170, AI076246, and AI096528. Work in S.J.M’s laboratory is supported by USPHS Grants AI076278 and AI117303. S.A.C. was supported by USPHS Grant GM056765. A.M.K-G. is supported by the American Heart Association Grant 12SDG12220022. N.S. was supported by a CAPES Science without Borders fellowship.
Footnotes
Supplementary Information accompanies the paper on www.nature.com/nature.
Contributions
A.M.K-G. and M.X.B. performed and analysed the experiments. R.R., P.A.L., O.H.P., A.Y.T., S.A.C., C.R.T., N.B.S, B.M.Y., A.C-A., M.F.dJ and M.G.W. performed experiments. A.M.K-G., M.X.B., S.J.M., A.J.B. and R.M.T. were responsible for the overall study design and for writing the manuscript.
Competing financial Interests
The authors declare no competing financial interests
Contributor Information
A. Marijke Keestra-Gounder, Department of Medical Microbiology and Immunology, School of Medicine, University of California at Davis, One Shields Ave; Davis CA 95616, USA.
Mariana X. Byndloss, Department of Medical Microbiology and Immunology, School of Medicine, University of California at Davis, One Shields Ave; Davis CA 95616, USA
Núbia Seyffert, Department of Medical Microbiology and Immunology, School of Medicine, University of California at Davis, One Shields Ave; Davis CA 95616, USA.
Briana M. Young, Department of Medical Microbiology and Immunology, School of Medicine, University of California at Davis, One Shields Ave; Davis CA 95616, USA
Alfredo Chávez-Arroyo, Department of Medical Microbiology and Immunology, School of Medicine, University of California at Davis, One Shields Ave; Davis CA 95616, USA.
April Y. Tsai, Department of Medical Microbiology and Immunology, School of Medicine, University of California at Davis, One Shields Ave; Davis CA 95616, USA
Stephanie A. Cevallos, Department of Medical Microbiology and Immunology, School of Medicine, University of California at Davis, One Shields Ave; Davis CA 95616, USA
Maria G. Winter, Department of Medical Microbiology and Immunology, School of Medicine, University of California at Davis, One Shields Ave; Davis CA 95616, USA
Oanh H. Pham, Center for Comparative Medicine, School of Veterinary Medicine, University of California at Davis, One Shields Ave; Davis CA 95616, USA
Connor R. Tiffany, Department of Medical Microbiology and Immunology, School of Medicine, University of California at Davis, One Shields Ave; Davis CA 95616, USA
Maarten F. de Jong, Department of Medical Microbiology and Immunology, School of Medicine, University of California at Davis, One Shields Ave; Davis CA 95616, USA
Tobias Kerrinnes, Department of Medical Microbiology and Immunology, School of Medicine, University of California at Davis, One Shields Ave; Davis CA 95616, USA.
Resmi Ravindran, Center for Comparative Medicine, School of Veterinary Medicine, University of California at Davis, One Shields Ave; Davis CA 95616, USA.
Paul A. Luciw, Center for Comparative Medicine, School of Veterinary Medicine, University of California at Davis, One Shields Ave; Davis CA 95616, USA
Stephen J. McSorley, Center for Comparative Medicine, School of Veterinary Medicine, University of California at Davis, One Shields Ave; Davis CA 95616, USA
Andreas J. Bäumler, Department of Medical Microbiology and Immunology, School of Medicine, University of California at Davis, One Shields Ave; Davis CA 95616, USA
Renée M. Tsolis, Department of Medical Microbiology and Immunology, School of Medicine, University of California at Davis, One Shields Ave; Davis CA 95616, USA
LITERATURE
- 1.Kaser A, et al. XBP1 links ER stress to intestinal inflammation and confers genetic risk for human inflammatory bowel disease. Cell. 2008;134:743–756. doi: 10.1016/j.cell.2008.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Montane J, Cadavez L, Novials A. Stress and the inflammatory process: a major cause of pancreatic cell death in type 2 diabetes. Diabetes, metabolic syndrome and obesity : targets and therapy. 2014;7:25–34. doi: 10.2147/DMSO.S37649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Celli J, Tsolis RM. Bacteria, the endoplasmic reticulum and the unfolded protein response: friends or foes? Nature reviews. Microbiology. 2014 doi: 10.1038/nrmicro3393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Urano F, et al. Coupling of stress in the ER to activation of JNK protein kinases by transmembrane protein kinase IRE1. Science. 2000;287:664–666. doi: 10.1126/science.287.5453.664. [DOI] [PubMed] [Google Scholar]
- 5.de Jong MF, et al. Sensing of bacterial type IV secretion via the unfolded protein response. mBio. 2013;4:e00418–e00412. doi: 10.1128/mBio.00418-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kaneko M, Niinuma Y, Nomura Y. Activation signal of nuclear factor-κB in response to endoplasmic reticulum stress is transduced via IRE1 and tumor necrosis factor receptor-associated factor 2. Biological & pharmaceutical bulletin. 2003;26:931–935. doi: 10.1248/bpb.26.931. [DOI] [PubMed] [Google Scholar]
- 7.Schneider M, et al. The innate immune sensor NLRC3 attenuates Toll-like receptor signaling via modification of the signaling adaptor TRAF6 and transcription factor NF-κB. Nature immunology. 2012;13:823–831. doi: 10.1038/ni.2378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.McCarthy JV, Ni J, Dixit VM. RIP2 is a novel NF-κB-activating and cell death-inducing kinase. The Journal of biological chemistry. 1998;273:16968–16975. doi: 10.1074/jbc.273.27.16968. [DOI] [PubMed] [Google Scholar]
- 9.Li L, et al. TRIP6 is a RIP2-associated common signaling component of multiple NF-κB activation pathways. Journal of cell science. 2005;118:555–563. doi: 10.1242/jcs.01641. [DOI] [PubMed] [Google Scholar]
- 10.Lytton J, Westlin M, Hanley MR. Thapsigargin inhibits the sarcoplasmic or endoplasmic reticulum Ca-ATPase family of calcium pumps. The Journal of biological chemistry. 1991;266:17067–17071. [PubMed] [Google Scholar]
- 11.Calfon M, et al. IRE1 couples endoplasmic reticulum load to secretory capacity by processing the XBP-1 mRNA. Nature. 2002;415:92–96. doi: 10.1038/415092a. [DOI] [PubMed] [Google Scholar]
- 12.Ghosh R, et al. Allosteric inhibition of the IRE1α RNase preserves cell viability and function during endoplasmic reticulum stress. Cell. 2014;158:534–548. doi: 10.1016/j.cell.2014.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Papandreou I, et al. Identification of an Ire1α endonuclease specific inhibitor with cytotoxic activity against human multiple myeloma. Blood. 2011;117:1311–1314. doi: 10.1182/blood-2010-08-303099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Atkins C, et al. Characterization of a novel PERK kinase inhibitor with antitumor and antiangiogenic activity. Cancer Res. 2013;73:1993–2002. doi: 10.1158/0008-5472.CAN-12-3109. [DOI] [PubMed] [Google Scholar]
- 15.Tsolis RM, Young GM, Solnick JV, Baumler AJ. From bench to bedside: stealth of enteroinvasive pathogens. Nature reviews. Microbiology. 2008;6:883–892. doi: 10.1038/nrmicro2012. [DOI] [PubMed] [Google Scholar]
- 16.Martirosyan A, Moreno E, Gorvel JP. An evolutionary strategy for a stealthy intracellular Brucella pathogen. Immunol Rev. 2011;240:211–234. doi: 10.1111/j.1600-065X.2010.00982.x. [DOI] [PubMed] [Google Scholar]
- 17.Roux CM, et al. Brucella requires a functional Type IV secretion system to elicit innate immune responses in mice. Cell Microbiol. 2007;9:1851–1869. doi: 10.1111/j.1462-5822.2007.00922.x. [DOI] [PubMed] [Google Scholar]
- 18.de Jong MF, Sun YH, den Hartigh AB, van Dijl JM, Tsolis RM. Identification of VceA and VceC, two members of the VjbR regulon that are translocated into macrophages by the Brucella type IV secretion system. Mol Microbiol. 2008;70:1378–1396. doi: 10.1111/j.1365-2958.2008.06487.x. doi:MMI6487 [pii] 10.1111/j.1365-2958.2008.06487.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim S, et al. Interferon-γ promotes abortion due to Brucella infection in pregnant mice. BMC microbiology. 2005;5:22. doi: 10.1186/1471-2180-5-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Derre I. Chlamydiae interaction with the endoplasmic reticulum: contact, function and consequences. Cell Microbiol. 2015;17:959–966. doi: 10.1111/cmi.12455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sabbah A, et al. Activation of innate immune antiviral responses by Nod2. Nature immunology. 2009;10:1073–1080. doi: 10.1038/ni.1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Roberson EC, et al. Influenza induces endoplasmic reticulum stress, caspase-12-dependent apoptosis, and c-Jun N-terminal kinase-mediated transforming growth factor-β release in lung epithelial cells. American journal of respiratory cell and molecular biology. 2012;46:573–581. doi: 10.1165/rcmb.2010-0460OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
References for Materials and Methods
- 1.Scidmore MA. Cultivation and Laboratory Maintenance of Chlamydia trachomatis . Curr Protoc Microbiol. 2005;Chapter 11(Unit 11A):11. doi: 10.1002/9780471729259.mc11a01s00. [DOI] [PubMed] [Google Scholar]
- 2.de Jong MF, et al. Sensing of bacterial type IV secretion via the unfolded protein response. mBio. 2013;4:e00418–e00412. doi: 10.1128/mBio.00418-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Keestra AM, et al. A Salmonella virulence factor activates the NOD1/NOD2 signaling pathway. mBio. 2011;2 doi: 10.1128/mBio.00266-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Keestra AM, et al. Manipulation of small Rho GTPases is a pathogen-induced process detected by NOD1. Nature. 2013;496:233–237. doi: 10.1038/nature12025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rothe M, Sarma V, Dixit VM, Goeddel DV. TRAF2-mediated activation of NF-kappa B by TNF receptor 2 and CD40. Science. 1995;269:1424–1427. doi: 10.1126/science.7544915. [DOI] [PubMed] [Google Scholar]
- 6.Rolan HG, Tsolis RM. Mice lacking components of adaptive immunity show increased Brucella abortus virB mutant colonization. Infection and immunity. 2007;75:2965–2973. doi: 10.1128/IAI.01896-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rolan HG, Tsolis RM. Inactivation of the type IV secretion system reduces the Th1 polarization of the immune response to Brucella abortus infection. Infection and immunity. 2008;76:3207–3213. doi: 10.1128/IAI.00203-08. [DOI] [PMC free article] [PubMed] [Google Scholar]