Overview
Much of our understanding of the biological processes that underlie cellular functions in humans, such as cell-cell communication, intracellular signaling, and transcriptional and post-transcriptional control of gene expression, has been acquired from studying cells in a two-dimensional (2D) tissue culture environment. However, it has become increasingly evident that the 2D environment does not support certain cell functions. The need for more physiologically relevant models prompted the development of three-dimensional (3D) cultures of epithelial, endothelial and neuronal tissues (Shamir and Ewald 2014). These models afford investigators with powerful tools to study the contribution of spatial organization, often in the context of relevant extracellular matrix and stromal components, to cellular and tissue homeostasis in normal and disease states.
Keywords: Desmoglein, Organotypic, Keratinocyte, Epidermis
1. Introduction
In this chapter we will review methodology associated with one of the longest standing and best-characterized examples of 3D culture models – that of the living skin equivalent. Epidermal organotypic “raft” cultures, grown on an air-medium interface, were originally developed in the 1980s as an ex vivo approach for understanding human skin function (Asselineau and Prunieras 1984; Prunieras, Regnier and Woodley 1983) and continue to provide an important alternative to animal models (Getsios, Simpson, Kojima, Harmon, Sheu, Dusek et al. 2009). The cultures are typically generated using primary keratinocytes isolated from readily available human foreskins; however, adult keratinocytes and keratinocytes derived from iPS cells (Bilousova and Roop 2013; Kogut, Roop and Bilousova 2014) are emerging as additional cell sources. These isolated keratinocytes can be passaged multiple times in culture and genetically manipulated using viral vectors or RNAi. Outcomes of genetic or pharmacologic manipulation can be assessed in weeks rather than months, and cultures are easily harvested and analyzed. Here we discuss how this 3D tissue model can be used to interrogate the role of the intermediate filament-desmosome scaffold in cytoarchitectural and signaling pathways driving epidermal differentiation.
1.1 The Skin
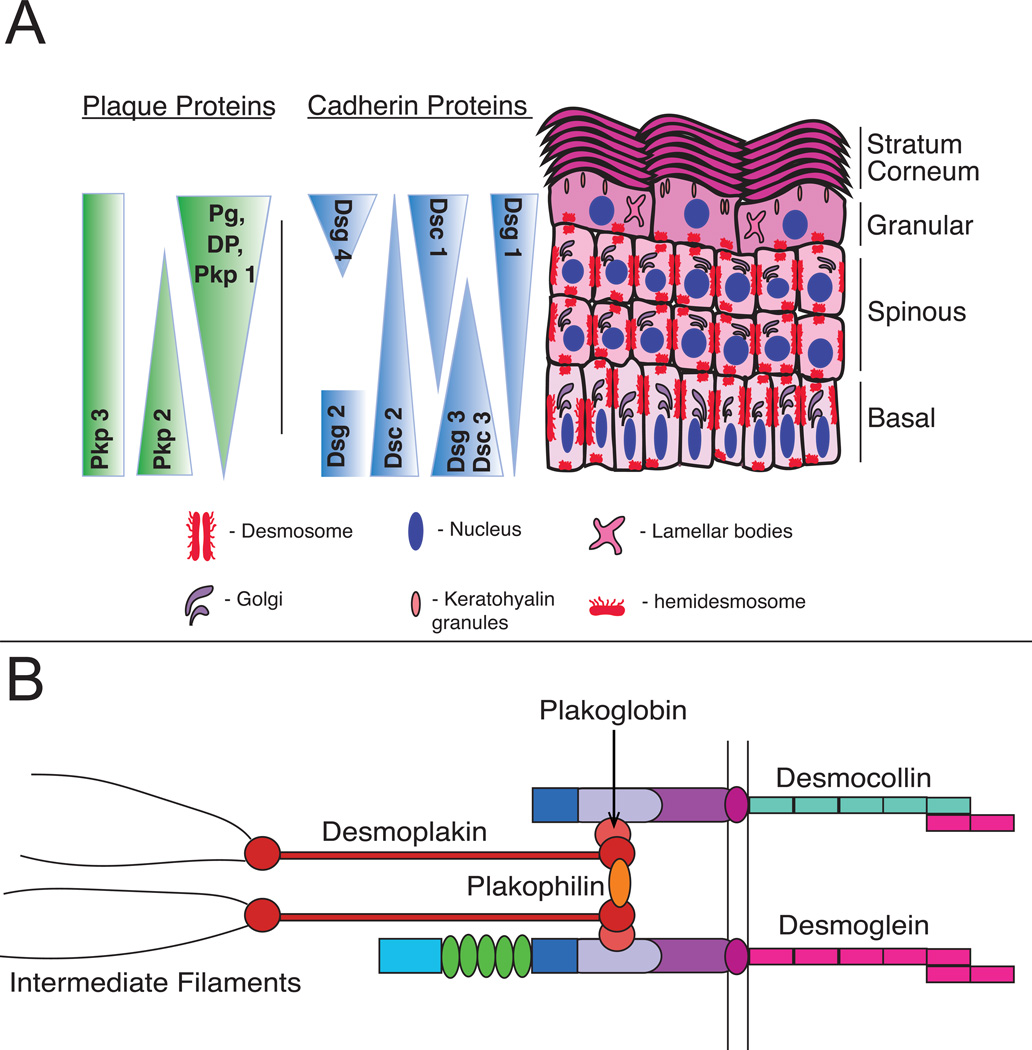
3D organotypic cultures of human epidermis mimic the outermost layer of the skin. The epidermis is a constantly renewing, multilayered epithelium composed primarily of keratinocytes, which derive their name from the word keratin, the major cellular component that provides building blocks for intermediate filaments (IFs)(Coulombe and Fuchs 1990). Keratinocytes begin their life in the basal proliferative layer of the epidermis. Upon committing to undergo differentiation, they stop dividing and leave the basal layer, transiting through the spinous, granular and, ultimately, cornified layers (Figure 1). This process results in the creation of a barrier that protects against insults from the environment and prevents water loss. Throughout the differentiating tissue, the keratinocyte-IF scaffold associates with calcium-dependent, cell-cell adhesive structures known as desmosomes (See Figure 1B) (Harmon and Green 2013; Osmani and Labouesse 2015). Interaction of desmosomal components and IFs is dictated by a tightly regulated transcriptional program that results in differentiation-dependent patterning of both desmosomal components and IFs within the epidermis (Desai, Harmon and Green 2009; Kowalczyk and Green 2013). As cells commit to terminal differentiation, they switch from expression of basal keratins, keratin 5/keratin 14 (Fuchs and Green 1980; Nelson and Sun 1983), to more suprabasal keratins, keratin 1/keratin 10 (Fuchs and Green 1980; Kim, Marchuk and Fuchs 1984). Likewise, while desmogleins 2 and 3 concentrate within the proliferating basal layer, desmoglein 1 becomes concentrated in the suprabasal layers of the epidermis (See Figure 1A) (Green and Simpson 2007).
Figure 1. Organization and expression of desmosomal components within the skin.
A) The are four layers within the stratified epidermis: basal, spinous, granular, and cornified. Each layer contains a complement of differentiation-specific junctional proteins critical to support epidermal cytoarchitecture and drive tissue morphogenesis. B) Desmosomes are specialized, calcium-dependent adhesive structures that are composed of three family of proteins: desmosomal cadherins, armadillo proteins, and plakin proteins. These structures link to the intermediate filament cytoskeleton to maintain tissue integrity.
Tissue level integrity provided by intermediate filaments relies on connections to intercellular desmosomes (Saito, Tucker, Kohlhorst, Niessen and Kowalczyk 2012). This supracellular IF-desmosome scaffold allows the epidermis to maintain its structural stability in the face of constant remodeling, resulting in a highly dynamic but organized tissue that balances epidermal cell renewal with cell removal. Here we discuss how 3D organotypic cultures provide an important tool for investigating the functions of desmosomes as mediators not only of mechanical tissue integrity, but also as spatially distinct regulators of keratinocyte differentiation.
2. Historical Perspective
2.1 Technical History of Model Development
Up until 1975, attempts to serial culture keratinocytes were hindered by a lack of understanding of the necessary components to prolong cellular lifetimes in culture. James Rheinwald and Howard Green overcame these conditions, establishing a method by which isolated keratinocytes could be grown as sheets on a feeder layer of lethally irradiated 3T3 fibroblasts to support and maintain keratinocyte colony growth and stratification (Rheinwald and Green 1975). Both the ability to isolate and stratify keratinocytes in a 3D culture provided a new avenue for probing questions related to skin biogenesis.
In 1976, Aaron Freeman and colleagues successfully grew 3D organotypic cultures of keratinocytes on raised metal grids to support maintenance of keratinocytes at the air-liquid interface to induce differentiation and stratification. Unlike Rheinwald and Green, Freeman et al used porcine skin dermis as the extracellular matrix (ECM) component; however, an artifact of this in vitro method was the presence of membrane granules, which are not found in native tissue. Nonetheless, the composition of the cultures closely resembled normal skin, allowing for successful grafting back into human patients (Freeman, Igel, Herrman and Kleinfeld 1976).
Building upon the use of alternative ECM substrates, Prunerias, Regnier, and Woodley turned to a more physiological substrate by growing keratinocytes on de-epidermized dermis (DED) derived from human skin (Prunieras et al. 1983). While this method allowed for differentiation, it lacked proper keratin patterning. Then, in 1984, Asselineau and Prunieras developed a system using a collagen-fibroblast plug as the substrate to support keratinocyte growth (Asselineau and Prunieras 1984). Yet, while the cells on this collagen-fibroblast stratified, keratinocyte-specific differentiation markers were mis-expressed; however, Kopan and colleagues found that they could induce proper expression of differentiation-specific keratins by removing vitamin A from the medium of submerged cultures or growing 3D organotypic cultures in the absence of retinoic acid (Kopan, Traska and Fuchs 1987), permitting proper stratification and differentiation marker expression.
Continuous improvements on the original method have made this a common technique and also led to the development of alternative systems, such as the transwell system described below. While both of these methods (grid and transwell) are viable options for creating 3D raft cultures, the transwell system has the advantage of being contained and not requiring a physical lifting of the collagen plug. However both have improved our understanding of the organization and function of the cells of the skin and the skin as a whole have been greatly improved by the development of the 3D organotypic culture system.
3. Cellular Constituents of 3D raft cultures
3.1 Cell Lines Used
The protocol described below utilizes keratinocytes and fibroblasts to make 3D raft cultures. Normal Human Epidermal Keratinocytes (NHEKs) are typically isolated from human neonatal foreskin, but can also be isolated from biopsies taken from patient skin of non-pathological or pathological origin. The lethally irradiated J2 clone of 3T3 mouse fibroblasts, which serve as a feeder layer and secrete paracrine factors to support keratinocyte growth (Rheinwald and Green 1975), will be used. The methods to isolate and culture these two cell types are described below (Table 1).
Table 1.
Cell Lines Used
| Cell Line | Media | Media Supplements | Source |
|---|---|---|---|
| Normal Human Keratinocytes |
Medium 154 (M154) | Human Keratinocyte Growth Supplement (HKGS), 0.07mM Ca2+, and antibiotics (1X Gentamicin/Amphotericin B) |
Human neonatal foreskin |
| 3T3 J2 Fibroblasts | DMEM | 10% Fetal Bovine Serum and 1% Penicillin/Streptomycin |
Originally derived from Howard Green (Rheinwald and Green, 1975) |
3.2 Isolation of Keratinocytes from Neonatal Foreskin
Note: Under Institutional Review Board (IRB) approval, neonatal foreskins are collected in transport media (Hanks Balanced Salt Solution containing no calcium or magnesium). The foreskin can be stored at 4 °C and processed within 48 h after receipt.
Materials Needed:
Hanks Balanced Salt Solution (HBSS)
Dispase Solution (2.4 U/mL) (Roche, Catalog #04942078001)
0.05% Trypsin/1 mM EDTA (Corning, Catalog #30-002-CI)
Fetal Bovine Serum (FBS)
Medium 154 (M154; Life Technologies, Catalog #M-154CF-500)
Sterile Forceps and Scissors
40µm nylon cell strainers (Corning, Catalog #352340)
3.2.1 Isolation Protocol
Day 1
Carefully remove HBSS and replace it with PBS containing no Ca2+ or Mg2+.
Rinse foreskin with PBS by pipetting up and down to remove any excess blood 2X).
Using sterile forceps, transfer foreskin to a 10 cm dish containing 3 mL of PBS.
Cut foreskin on one side so that it lays flat with the epidermis side down.
Remove excess fat and vessels using sterile forceps and scissors.
Cut remaining tissue into small pieces (0.8 cm×0.8 cm).
Place tissue epidermal side up in 4 mL of dispase solution (2.4 U/mL; prepared according to manufacturer’s instructions in a 60 mm dish).
Incubate overnight at 4 °C.
Day 2
Using sterile forceps, separate the epidermis from the dermis from each piece of foreskin.
Transfer epidermal pieces to a 10 cm dish containing 3–4 mL of 0.25%Trypsin/1 mM EDTA.
Incubate 10–15 minutes at 37 °C to dissociate the cells.
Inactivate trypsin by adding 0.5 mL FBS to the dish.
With sterile forceps, scrape any remaining large epidermal pieces vigorously against the bottom of the dish for 10–30 seconds/piece. This further helps create a single cell suspension by breaking up any remaining larger sheets of cells as leaving the cells in trypsin too long can be detrimental to the cells causing cell lysis and a lower cell yield.
Using a pipet, transfer the cell suspension and debris through a 40-micron nylon cell strainer placed into a 50 mL conical tube. Rinse the scraped dish once with 5 mL of PBS and also filter through the same 40-micron cell strainer used for filtering the cell suspension. By running the cells and the rinse through a cell strainer, the larger debris are removed.
Transfer the cell suspension and the rinse (collected in same tube after straining) to a 15 mL conical tube.
Centrifuge at 200–250×g for 5 minutes at room temperature to pellet cells.
Discard supernatant. Re-suspend the cells in 7–8 mL of M154 and plate in a 10 cm dish. Cell yield will vary based on the size of the tissue.
Change the media 24 hours later. Media should be changed every other day.
3.3 Culturing NHEKs
NHEKs are cultured in M154 (Life Technologies, Catalog #M-154CF-500) supplemented with Human Keratinocyte Growth Supplement (HKGS; Life Technologies, Catalog# S-001-5), Gentamicin/Amphotericin solution (Life Technologies, Catalog# R-015-10), and a final Ca2+ concentration of 0.07 mM. Maintaining the culture in 0.07 mM Ca2+ will keep the cells in a relatively “undifferentiated” state with limited expression of differentiation markers such as desmoglein 1, keratin 10, or loricrin (Denning, Guy, Ellerbroek, Norvell, Kowalczyk and Green 1998; Hohl, Lichti, Breitkreutz, Steinert and Roop 1991). It must be noted that primary keratinocytes have a finite lifespan in culture that limits their proliferative capacity, however, methods to extend their lifetime in culture, such as the presence of a fibroblast feeder layer, have been explored (Green, Rheinwald and Sun 1977; Ramirez, Morales, Herbert, Rohde, Passons, Shay et al. 2001). Recently, it has also been found that addition of Rho kinase inhibitor, Y-27632, in combination with a fibroblast feeder layer, increases NHEK proliferation and results in efficient immortalization of NHEKs genetically indistinguishable from their freshly isolated counterparts (Chapman, Liu, Meyers, Schlegel and McBride 2010).
Note: After isolating NHEKs from foreskin, cultures should be checked daily for contamination. Like any cultured cells, they too are susceptible to bacterial and fungal contamination.
Comment: NHEK cultures will contain a population of dermal fibroblasts. To remove fibroblasts from the culture following attachment to the culture dish, add 0.02% EDTA and incubate at room temperature for 1–5 minutes then vigorously pipet against the dish surface to dislodge fibroblasts. Remove EDTA and add fresh medium to keratinocytes. Alternatively, removal of dermal fibroblasts from the culture typically occurs after 2–3 passages of NHEKs, as keratinocyte growth will outpace fibroblast growth in M154.
3.3.1 NHEK passage protocol
Wash cells 1X with PBS.
Add 1–2 mL of 0.05% Trypsin/1 mM EDTA and incubate at 37 °C for 5–10 minutes. Note: Different cell lines require different trypsinization times. To avoid over-trypsinization, which can severely damage the cells, it is essential to check them every few minutes.
To inactivate trypsin, add 2–3X the volume of DMEM supplemented with 10% FBS (the FBS will neutralize the Trypsin).
Centrifuge at 200–250×g for 5 minutes to pellet the cells.
Discard the supernatant.
Re-suspend the cells in M154 and re-plate 0.5 – 1.0×106 cells per 10 cm. Cell number will depend on experimental conditions.
3.4 3T3 Fibroblasts
The J2 clone of 3T3 mouse fibroblasts can be used as a feeder layer when culturing NHEKs and as a component of the collagen plug scaffold for organotypic raft cultures (Rheinwald and Green 1975). The use of a feeder layer provides a longer keratinocyte lifespan by secretion of paracrine factors necessary to support growth (Barreca, De Luca, Del Monte, Bondanza, Damonte, Cariola et al. 1992) and through reorganization of ECM components within the plug.
3.4.1 Generation of 3T3 Feeder Layers for Growth of Human Keratinocytes
Materials Needed
Dulbecco’s Modification of Eagle’s Medium (DMEM, 1X with 4.5 g/L glucose, L-glutamine and sodium pyruvate; Corning, Reference #10-013-CV)
Phosphate Buffered Saline (PBS; Corning Reference #21-040-CV)
Trypsin (Corning, Catalog #30-002-C1)
Mitomycin C (VWR, Catalog#80055-338)
FBS
Penicillin/Streptomycin (Corning, Reference #30-002-CI)
Grow 3T3 cells to 75–80% confluence using DMEM supplemented with 10% FBS and 1% Penicillin/Streptomycin. Never allow 3T3 cells to become confluent.
Treat with 4 µg/mL of mitomycin C for 2–4 hours at 37 °C. Note: mitomycin C is a cytotoxic agent and appropriate personal protective equipment (PPE) should be worn when handling it.
Aspirate off mitomycin C-containing medium and wash cells 3X with PBS to remove any trace of this cytotoxic agent.
Trypsinize the 3T3 cells for ~2 minutes and neutralize with DMEM containing 10% FBS. Centrifuge cells at 960×g for 5 minutes to pellet and re-suspend in FAD+ keratinocyte growth media.
Plate 5.0×104 cells/10 cm dish with re-suspended keratinocytes (see Section 3.3.1) (see Table 2).
Table 2.
Media Recipe
| E-Media Recipe | ||||
|---|---|---|---|---|
| Reagent | Vendor | Catalog | Storage | Volume |
| DMEM/F12 | Corning | 10-090-CV | 4°C | 500mL |
| DMEM | Corning | 10-013-CV | 4°C | 500 mL |
| Note: This give 3:1 DMEM: F12 Combine and remove 83ml |
||||
| Gentamycin/Amphotericin B |
Life Technologies | R01510 | −20°C | 0.500 mL |
| E-cocktail (see below) | −20°C | 10 mL | ||
| 0.4µg/ml Hydrocortisone | Sigma | H0888 | Room Temperature |
|
| 10ng/ml Cholera Toxin | Sigma | C8052 | 4°C | |
| E-Cocktail Recipe | ||||
|---|---|---|---|---|
| Reagent | Vendor | Catalog # | Storage Temperature |
How to prepare |
| 180µm Adenine | Sigma | A2786 | Room Temperature | 486mg in 20ml of deionized water + 500µl 6N HCl |
| 5µg/ml Human Recombinant Insulin |
Sigma | −20°C | 100mg in 20ml 0.1N HCl |
|
| 5µg/ml Human
Apo- Transferrin |
Sigma | T1147 | 4°C | 100mg in 20ml PBS |
| 5µg/ml Triiodothyronine (T3) |
Sigma | T6397 | −20°C |
|
| ||||
| FAD+ Media, 250mL | |
|---|---|
| Reagent | Amount |
| E-Media | 237.5 mL |
| Supplement with extra 5% FBS | 12.5 mL |
| 10ng/mL EGF | 2.5 µL of 1mg/mL stock |
Note: NHEKs can also be cultured using a feeder layer of 3T3 fibroblasts and maintained in FAD+ media (see Table 2) or alone in the presence of M154. See section 3.3 for NHEK culture information using M154.
4. 3D raft cultures
4.1 Utilization of 3D raft cultures
3D in vitro models offer an important complement to 2D models, providing a more physiologically relevant context, while lending itself to numerous applications for studying biological processes (Elliott and Yuan 2011).
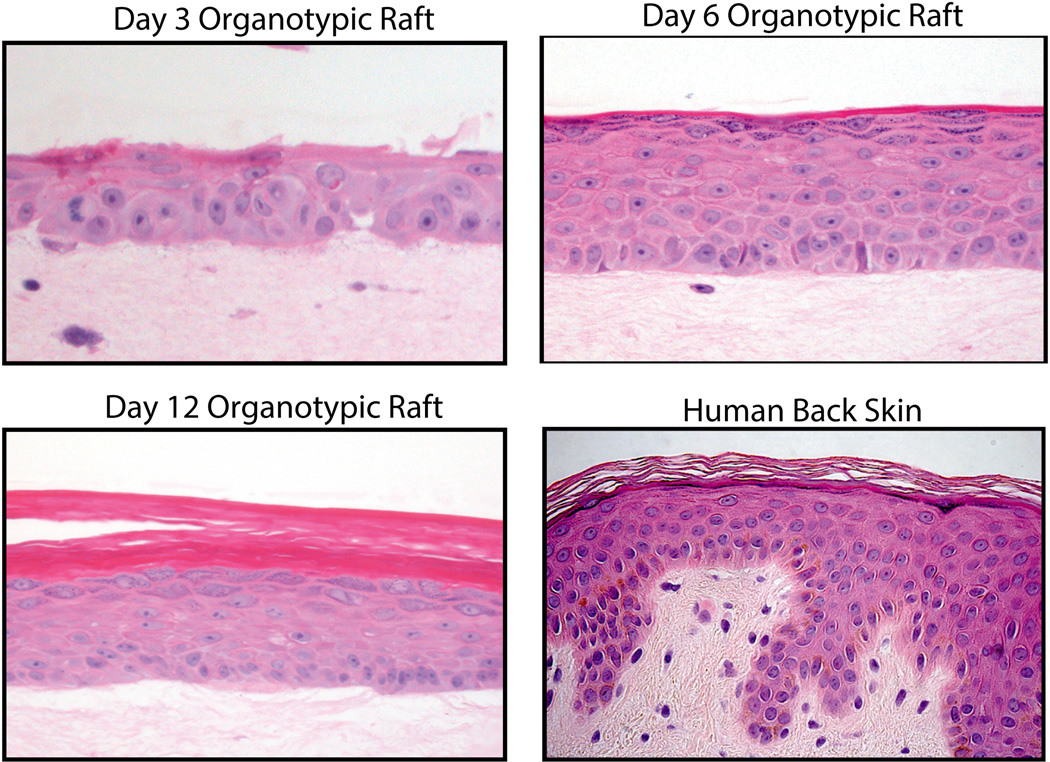
The epidermal organotypic cultures discussed in this chapter bear a striking resemblance to normal human epidermis (Figure 2), replicating the polarized protein distribution and architectural features characteristic of each cell layer seen in vivo (See Figure 2B). However, while the architecture and development of a stratified epidermis can be fully achieved using 3D organotypic raft cultures, the formation of a well-defined basement membrane within this system remains an issue (S. Getsios, personal communication, June 26, 2015). Nevertheless, 3D organotypic raft cultures still represent a powerful tool to study the epidermis and are amenable to genetic manipulation by CRISPR/Cas9-mediated gene ablation (Lopez-Pajares, Qu, Zhang, Webster, Barajas, Siprashvili et al. 2015), RNA interference(Simpson, Kojima and Getsios 2010), or viral delivery of silencing constructs or expression vectors, which allows one to analyze how loss of protein function contributes to a process or to mimic disease conditions(Brooks, Ostano, Jo, Dai, Getsios, Dziunycz et al. 2014). Furthermore, keratinocytes derived from patient biopsies can be expanded in culture and utilized in 3D models, affording the opportunity to ask directly how genetic defects impact skin structure and function (Brooke, Etheridge, Kaplan, Simpson, O'Toole, Ishida-Yamamoto et al. 2014).
Figure 2. Comparison to human epidermis and time course of 3D raft development.
A) Cellular architecture of human skin and 3D organotypic raft culture. B) Keratinocytes were seeded on collagen plugs and were grown submerged conditions for 2 days. The cultures were then lifted to the air-liquid interface. Organotypic rafts were harvested 3, 6, 9, or 12 days after being lifted to air.
Representative images of keratinocyte stratification in 3D raft cultures (Provided by Paul Hoover of the Northwestern University Skin Disease Research Core).
Here, we describe protocols for the isolation of human epidermal keratinocytes and a transwell system for generating 3D organotypic cultures (Figure 3). We outline genetic manipulation and analysis tools that allow for an understanding of the biology of the 3D culture. As an illustration of the utility of these methods, we present data showing how desmoglein 1 knockdown via retroviral transduction impacts differentiation of 3D organotypic cultures.
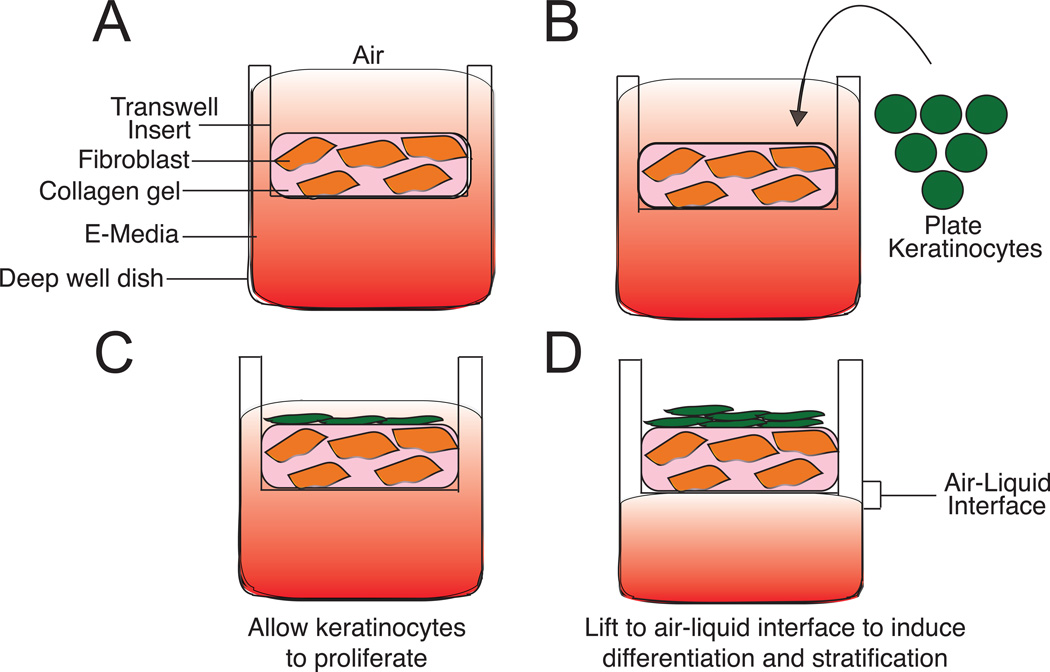
Figure 3. Generation of 3D organotypic cultures using the transwell method.
A) A collagen-fibroblast plug is made in the upper chamber (transwell). Media is supplied from the top and bottom. B and C) Keratinocytes are seeded onto the collagen-fibroblast plug and kept under submerged conditions: media is supplied from the top and bottom chambers. Keratinocytes are allowed to proliferate. D) Organotypic rafts are raised to air-liquid interface. Media is only supplied from the bottom chamber.
Materials Needed:
Transwell plates (Corning, Catalog #355467)
Transwell inserts (3 µm pore size) (Corning, Catalog #353091). One insert is placed into one well of the transwell plate.
Media: E-Media: see Table 2.
Collagen plug: Rat Tail Collagen Type I, high concentration (100 mg/bottle; Corning, Catalog#354249)
Reconstitution Buffer: (1.1 g NaHCO3, 2.3 g HEPES re-suspended in 50 mL of 0.05 N NaOH and sterile filtered)
10X DMEM (Sigma, Catalog #D2429)
0.5 N Sodium Hydroxide (NaOH)
E-media (see Table 2 for media recipe)
Trypsin (Corning, Catalog #30-002-CI)
DMEM (Corning, Reference #10-013-CV)
4.2 3-D Organotypic Raft Culture Protocol
4.2.1 Making the Collagen-Fibroblast Plugs
The transwell system requires 2.0 mL collagen mix/ raft and 7.5×105 J2 3T3 fibroblasts per 6 well plug. Each collagen plug contains 1×106 NHEKs (see section 4.2.3 for plating NHEKs onto the plug).
4.2.2 Generation of Collagen Plugs Protocol (for 10 plugs)
Trypsinize the 3T3 cells in 4 mL of 0.05% Trypsin/1 mM EDTA to dissociate the cells.
Inactivate trypsin by adding serum-containing media (e.g. DMEM).
Count the number of cells using a hemocytometer.
Plate the exact number of cells required for rafts (e.g. for 10 rafts, one would need 7.5×106 cells) by centrifugation at 960×g for 5 minutes.
Discard supernatant.
Re-suspend the 3T3 cells in 2 mL (1/10 of the final volume) of 10X Reconstitution Buffer.
Add 2 mL (1/10 of final volume) of 10X DMEM.
Add collagen to a final concentration of 4 mg/mL.
Add sterile water to bring the final volume to 20 mL for 10 plugs.
Neutralize pH with 0.5 N NaOH and mix thoroughly without generating air bubbles. Note: The ideal color of the collagen plug is faint red/light pink. If orange, it is too acidic. If bright pink, it is too basic.
Pipet 2.0 mL of the collagen-fibroblast mixture into the top chamber of the transwell.
Incubate dishes in a 37 °C, humidified CO2 incubator for 30 minutes to allow the collagen plug to polymerize.
Add 2 mL of DMEM supplemented with 10 % FBS and antibiotics to top chamber.
Add 13 mL of DMEM supplemented with 10 % FBS and antibiotics to the bottom chamber. Incubate for 24–48 hours at 37 °C prior to plating NHEKs on top. This process will allow the formation of a smooth, flat surface for when the keratinocytes are seeded.
4.2.3 Seeding NHEKs onto Collagen-Fibroblast Plugs and creating an “Air-Liquid” Interface
Note: NHEKs are cultured in a keratinocyte growth media (e.g. M154) prior to seeding onto the collagen plugs. Once on the collagen plugs, the cells are cultured in E-Media supplemented with EGF (5ng/mL) (Keratinocyte Growth Media; (Wu, Parker, Binder, Beckett, Sinard, Griffiths et al. 1982). Once introduced to the air-liquid interface, the raft cultures are fed with only E-Media that has not been supplemented with EGF.
Trypsinize NHEKs as described in section 3.3.1.
Count the cells using a hemocytometer. For an experiment with 10 plugs, 10×106 cells would be required for the experiment.
Remove the media from both the top and bottom chamber of the transwell plate.
Plate 1.0×106 NHEKS per plug in 2 mL of E-media supplemented with EGF (5 ng/mL) in the top chamber of the transwell. The addition of EGF to the media supports cell proliferation to better ensure a confluent keratinocyte sheet when cells are introduced to the air-liquid interface.
Fill the bottom chamber with 13 mL of E-Media supplemented with EGF (5 ng/mL).
Allow cells to grow to confluency as described above.
To create an air-liquid interface, remove the media from the top chamber of the transwell without disturbing the collagen plug or the NHEK sheet.
Remove the media from the bottom chamber and replace with E-Media (not supplemented with EGF). Approximately 9–10 ml are required to fill the lower chamber up to the bottom of the insert. The media level should not exceed past the insert.
Remove the media from the bottom chamber every 2 days and replace with fresh E-Media containing no EGF. Take care not to disturb the top chamber of the transwell when the media in the bottom chamber is replaced
Comment: Culture methods typically allow for maintenance of NHEKs at the air-liquid interface for up to three weeks. Stratification can be observed within 1–2 days of introduction to an air-liquid interface(Sugihara, Toda, Yonemitsu and Watanabe 2001) and morphological differentiation can be seen as early as 1 week(Popov, Kovalski, Grandi, Bagnoli and Amieva 2014). Maintenance of 3D raft cultures at the air-liquid interface for 2 weeks or longer is typically necessary for development of a more mature raft with well-defined layers (Hildebrand, Hakkinen, Wiebe and Larjava 2002).
5. Experimental Manipulation of the Organotypic Raft Cultures
5.1 Genetic Manipulation of Raft Cultures
3D organotypic cultures provide researchers with a more complex model of the skin that is amenable to genetic manipulation. For example, to identify the function of Dsg1 in normal skin processes and how those functions may be altered in disease states, such as pemphigus foliaceus, Dsg1 or its associated binding partner, Erbin, were genetically silenced in keratinocytes. 3D organotypic rafts made from these infected cells allowed Getsios et al and Harmon et al to understand the normal functions of Dsg1 within the epidermis, as silencing resulted in impaired morphogenesis and differentiation of the 3D cultures as compared to control (Getsios et al. 2009; Harmon, Simpson, Johnson, Koetsier, Dubash, Najor et al. 2013). Alternatively, the use CRISPR/Cas9-mediated gene ablation technique in primary keratinocytes is also an option. Using this gene-targeting approach, the Khavari group showed that loss of transcription factors, MAF and MAFB, severely impaired expression of differentiation markers and differentiation genes in 3D organotypic raft cultures (Lopez-Pajares et al. 2015). While this gene-editing technique represents a powerful tool, editing in primary cells is generally more challenging. Although the exact cause of the increased difficulty in primary cells is unknown, differences in factors such as transfection efficiency, promoter activity, or DNA repair mechanisms may contribute (Hendel, Bak, Clark, Kennedy, Ryan, Roy et al. 2015). Ultimately, the ability to readily silence proteins within this system provides a practical tool to understand how the 3D structure is connected to protein function.
5.1.1 Knockdown using RNA interference via electroporation
While several methods exist to silence gene function in 3D cultures, the use of electroporation has become a common method for this purpose. Electroporation can increase transfection efficiency for difficult cell types (Escobar-Chavez, Bonilla-Martinez, Villegas-Gonzalez and Revilla-Vazquez 2009). However, lack of optimization for specific cell types can result in the formation of pores that may become too large or fail to close after membrane discharge, resulting in cellular damage or rupture (Weaver 1995), thus, care must be taken to find the proper conditions. Once optimized, electroporation represents a quick and efficient option for gene silencing.
Materials Needed
Lonza Amaxa Nucleofector™ 2b Device (Lonza, Catalog #AAB-1001)
Cell line Nucleofector™ Kit V (Lonza, Catalog #VCA-1003; Kit includes: Buffer V, electroporation cuvettes and cell droppers)
Medium 154 (M154; Life Technologies, Catalog #M-154CF-500)
0.05% Trypsin (Corning, Catalog #30-002-CI)/1 mM EDTA
Dulbecco’s Modification of Eagle’s Medium (DMEM, 1X with 4.5 g/L glucose, L-glutamine and sodium pyruvate; Corning, Reference #10-013-CV)
siRNA (commonly used vendors: Dharmacon, Sigma, IDT)
Note: Prior to beginning, allow Buffer V and M154 to warm to room temperature.
Grow NHEKS to ~90% confluence in a 10 cm culture dish.
Aspirate media and wash cells 3X in PBS.
Add 1–2 mL of 0.05% Trypsin/1 mM EDTA and incubate cells at 37 °C for 5–10 minutes to dissociate cells.
To inactivate Trypsin/EDTA, add 2–3X the volume of DMEM supplemented with 10% FBS (the FBS will neutralize the Trypsin).
Pipette dissociated cell solution up and down to break apart any cellular clumps that may have formed during trypsinization.
Count the number of cells in solution using a hemocytometer. Typically, a 10 cm dish will yield ~3 million cells at confluence, however, this can vary between 2–4 million depending on the clone.
Centrifuge the correct number of cells needed for the experiment at 200–250×g for 5 minutes to pellet the cells. Note: To seed cells at confluency, plate 1.0×106 cells per collagen plug. Scale up or down as determined by total number of cells required for experiment.
Aspirate off supernatant and resuspend cells in Buffer V solution by adding 100 µL per 1 million cells
Add appropriate amount of siRNA (concentration range varies between 10 nM – 50 nM, but concentration for each siRNA should be empirically determined) per 1 million cells to the cell/Buffer V solution and mix well.
Add 100 µL of the cell/siRNA/Buffer V solution to each electroporation cuvette.
Select the correct electroporation program on the Lonza Amaxa nucleoporator and place the cuvette into the machine. Note: The correct program for specific cell types will need to be determined for each application, however, recommendations for programs can be found on the Lonza website: http://bio.lonza.com/resources/product-instructions/protocols/#ops_k. For Buffer V, the program is X-001.
Run electroporation program.
Remove electroporated cells from the cuvette using the cell dropper and add to fresh media. Note: Cuvettes can be pooled if necessary. If 4 million cells are needed, use 2 cuvettes with 2 million cells/cuvette and plate in the same dish.
Allow cells to attach for 24 hours, and then replace media with fresh M154 the following day.
5.1.2 Knockdown and Overexpression using Retroviral Systems
Utilization of viral systems to silence or overexpress a particular gene represent a feasible alternative to traditional transfection or siRNA methods in 3D organotypic rafts (Getsios et al. 2009), including electroporation (Figure 4). Concentrated virus can be added directly to NHEK media to infect them prior to 3D organotypic raft formation. For a more detailed description of designing plasmids and viral production, see Simpson et al. 2010.
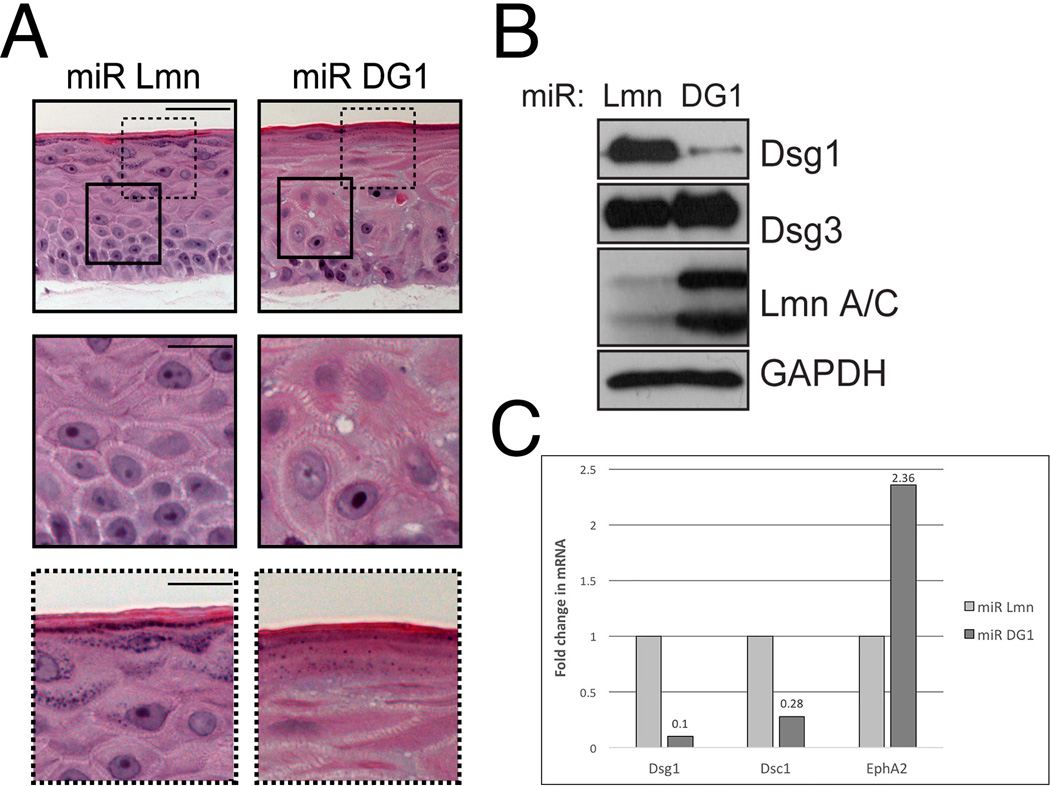
Figure 4. Manipulation and analysis of Dsg1 in 3D organotypic raft cultures.
A) H&E-stained sections of 6-day-old rafts expressing either miR Lmn or miR DG1. Silencing of DG1 results in altered suprabasal morphology (insets with continuous lines) and impaired differentiation (insets with dashed lines) compared to control. B) Western blot analysis of desmosomal proteins, desmoglein 1 and 3, lamin A/C (Lmn A/C), and GAPDH (glyceraldehyde-3-phosphate dehydrogenase) in 6-day-old 3D raft cultures. Either Dsg1 or Lamin expression was silenced in raft cultures using retroviruses engineered to express microRNA (miRNA)-like sequences specific for each protein. KD reveals ~90% reduction of protein levels under each condition. C) Real-time PCR analysis show decreased levels of desmocollin 1 mRNA levels following Dsg1 knockdown in 3D raft cultures, while a gene target of EGFR, EphA2, mRNA levels are increased.
Note: Figure originally published in Getsios et al (© 2009 Getsios et al. Journal of Cell Biology. 185:1243-1258. doi:10.1083/jcb.200809044); Used with permission.
Materials Needed
Medium 154 (Life Technologies, Catalog# M-154CF-500)
Viral supernatant(Simpson et al. 2010)
PBS
10% bleach solution
Grow NHEKs to ~20 – 30% confluence in a 10 cm dish prior to infection. Note: This is a BSL-2 level procedure. Proper handling of cultures must be followed according to institutional rDNA protocols. PPE should be worn at all times.
Aspirate off M154 media and wash cells 2X with PBS.
Add 4 mL of fresh M154 to cells.
Carefully add viral supernatant to the dish.
Incubate cells at 32 °C, which is a permissible temperature for retroviral infection, for 90 minutes.
Pipette off viral supernatant-containing media into a 10% bleach solution.
Wash cells 2X with PBS, discarding each wash in a 10% bleach solution.
Add 7–8 mL of M154 and incubate at 37 °C.
When cells have reached desired confluency, prepare cells for 3D organotypic raft formation as described in section 4.2.3.
Comment: Use of replication-deficient viruses for infection creates a more stable, long-term knockdown or overexpression in NHEKs.
6. Analysis of Organotypic Raft Cultures and other methods
Standard analysis tools commonly used for 2D cultures can also be applied to 3D raft cultures. 3D raft cultures can be harvested from the collagen plug wholly or in part for protein and/or RNA analysis (See Figure 4 B and C). Protein-protein interactions in situ can also be assessed by Bio-ID screens and proximity ligation assays (PLA) or via immunoprecipitation. Additionally, 3D organotypic cultures also allow for the addition of pharmacological agents directly to the media to interfere with protein function or modulate signaling pathways. The effect of modulating these signaling cascades, such as alterations in proliferation, can be measured through 5-Bromo-2’-deoxyuridine (BrdU) incorporation (Getsios et al. 2009). Standard histological methods and analysis can also be utilized to stain proteins of interest. The flexibility and ability to investigate several questions at once utilizing the 3D organotypic culture system makes them an attractive tool for investigating protein function.
7. Conclusions
The evolution of 3D organotypic raft culture technology has impacted a range of research areas from cancer biology to pharmacology to basic cell biology. One potential reason for this is that such a model lends itself to a number of applications. While our focus has been on the use of 3D models to study structural components of the skin, these 3D cultures are being used to model disease (Shamir and Ewald 2014), study drug delivery (Nam, Smith, Lone, Kwon and Kim 2015), and investigate mechanisms of human development (Ader and Tanaka 2014). Continued improvements of the system are likely to emerge in the future with the engineering of novel biocompatible scaffolding materials, as well as incorporation of increasingly complex cellular components to more accurately mimic the skin.
Acknowledgments
This work was supported in part by grants to K.J.G from the National Institute of Arthritis and Musculoskeletal and Skin Diseases (R37 AR043380 and RO1 AR041836), the National Cancer Institute (R01 CA122151), the Joseph L. Mayberry Endowment, and in part by a grant to S.G. from the National Institute of Arthritis and Musculoskeletal and Skin Diseases (RO1 AR062110) and the Northwestern University Skin Disease Research Core (P30 AR057216).
References
- Ader M, Tanaka EM. Modeling human development in 3D culture. Curr Opin Cell Biol. 2014;31:23–28. doi: 10.1016/j.ceb.2014.06.013. [DOI] [PubMed] [Google Scholar]
- Asselineau D, Prunieras M. Reconstruction of 'simplified' skin: control of fabrication. Br J Dermatol. 1984;111(Suppl 27):219–222. doi: 10.1111/j.1365-2133.1984.tb15608.x. [DOI] [PubMed] [Google Scholar]
- Barreca A, De Luca M, Del Monte P, Bondanza S, Damonte G, Cariola G, Di Marco E, Giordano G, Cancedda R, Minuto F. In vitro paracrine regulation of human keratinocyte growth by fibroblast-derived insulin-like growth factors. J Cell Physiol. 1992;151(2):262–268. doi: 10.1002/jcp.1041510207. [DOI] [PubMed] [Google Scholar]
- Bilousova G, Roop DR. Generation of functional multipotent keratinocytes from mouse induced pluripotent stem cells. Methods Mol Biol. 2013;961:337–350. doi: 10.1007/978-1-62703-227-8_22. [DOI] [PubMed] [Google Scholar]
- Brooke MA, Etheridge SL, Kaplan N, Simpson C, O'Toole EA, Ishida-Yamamoto A, Marches O, Getsios S, Kelsell DP. iRHOM2-dependent regulation of ADAM17 in cutaneous disease and epidermal barrier function. Hum Mol Genet. 2014;23(15):4064–4076. doi: 10.1093/hmg/ddu120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks YS, Ostano P, Jo SH, Dai J, Getsios S, Dziunycz P, Hofbauer GF, Cerveny K, Chiorino G, Lefort K, Dotto GP. Multifactorial ERbeta and NOTCH1 control of squamous differentiation and cancer. J Clin Invest. 2014;124(5):2260–2276. doi: 10.1172/JCI72718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman S, Liu X, Meyers C, Schlegel R, McBride AA. Human keratinocytes are efficiently immortalized by a Rho kinase inhibitor. J Clin Invest. 2010;120(7):2619–2626. doi: 10.1172/JCI42297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coulombe PA, Fuchs E. Elucidating the early stages of keratin filament assembly. J Cell Biol. 1990;111(1):153–169. doi: 10.1083/jcb.111.1.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denning MF, Guy SG, Ellerbroek SM, Norvell SM, Kowalczyk AP, Green KJ. The expression of desmoglein isoforms in cultured human keratinocytes is regulated by calcium, serum, and protein kinase C. Exp Cell Res. 1998;239(1):50–59. doi: 10.1006/excr.1997.3890. [DOI] [PubMed] [Google Scholar]
- Desai BV, Harmon RM, Green KJ. Desmosomes at a glance. J Cell Sci. 2009;122(Pt 24):4401–4407. doi: 10.1242/jcs.037457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elliott NT, Yuan F. A review of three-dimensional in vitro tissue models for drug discovery and transport studies. J Pharm Sci. 2011;100(1):59–74. doi: 10.1002/jps.22257. [DOI] [PubMed] [Google Scholar]
- Escobar-Chavez JJ, Bonilla-Martinez D, Villegas-Gonzalez MA, Revilla-Vazquez AL. Electroporation as an efficient physical enhancer for skin drug delivery. J Clin Pharmacol. 2009;49(11):1262–1283. doi: 10.1177/0091270009344984. [DOI] [PubMed] [Google Scholar]
- Freeman AE, Igel HJ, Herrman BJ, Kleinfeld KL. Growth and characterization of human skin epithelial cell cultures. In Vitro. 1976;12(5):352–362. doi: 10.1007/BF02796313. [DOI] [PubMed] [Google Scholar]
- Fuchs E, Green H. Changes in keratin gene expression during terminal differentiation of the keratinocyte. Cell. 1980;19(4):1033–1042. doi: 10.1016/0092-8674(80)90094-x. [DOI] [PubMed] [Google Scholar]
- Getsios S, Simpson CL, Kojima S, Harmon R, Sheu LJ, Dusek RL, Cornwell M, Green KJ. Desmoglein 1-dependent suppression of EGFR signaling promotes epidermal differentiation and morphogenesis. J Cell Biol. 2009;185(7):1243–1258. doi: 10.1083/jcb.200809044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green H, Rheinwald JG, Sun TT. Properties of an epithelial cell type in culture: the epidermal keratinocyte and its dependence on products of the fibroblast. Prog Clin Biol Res. 1977;17:493–500. [PubMed] [Google Scholar]
- Green KJ, Simpson CL. Desmosomes: new perspectives on a classic. J Invest Dermatol. 2007;127(11):2499–2515. doi: 10.1038/sj.jid.5701015. [DOI] [PubMed] [Google Scholar]
- Harmon RM, Green KJ. Structural and functional diversity of desmosomes. Cell Commun Adhes. 2013;20(6):171–187. doi: 10.3109/15419061.2013.855204. [DOI] [PubMed] [Google Scholar]
- Harmon RM, Simpson CL, Johnson JL, Koetsier JL, Dubash AD, Najor NA, Sarig O, Sprecher E, Green KJ. Desmoglein-1/Erbin interaction suppresses ERK activation to support epidermal differentiation. J Clin Invest. 2013;123(4):1556–1570. doi: 10.1172/JCI65220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendel A, Bak RO, Clark JT, Kennedy AB, Ryan DE, Roy S, Steinfeld I, Lunstad BD, Kaiser RJ, Wilkens AB, Bacchetta R, Tsalenko A, Dellinger D, Bruhn L, Porteus MH. Chemically modified guide RNAs enhance CRISPR-Cas genome editing in human primary cells. Nat Biotechnol. 2015 doi: 10.1038/nbt.3290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hildebrand HC, Hakkinen L, Wiebe CB, Larjava HS. Characterization of organotypic keratinocyte cultures on de-epithelialized bovine tongue mucosa. Histol Histopathol. 2002;17(1):151–163. doi: 10.14670/HH-17.151. [DOI] [PubMed] [Google Scholar]
- Hohl D, Lichti U, Breitkreutz D, Steinert PM, Roop DR. Transcription of the human loricrin gene in vitro is induced by calcium and cell density and suppressed by retinoic acid. J Invest Dermatol. 1991;96(4):414–418. doi: 10.1111/1523-1747.ep12469779. [DOI] [PubMed] [Google Scholar]
- Kim KH, Marchuk D, Fuchs E. Expression of unusually large keratins during terminal differentiation: balance of type I and type II keratins is not disrupted. J Cell Biol. 1984;99(5):1872–1877. doi: 10.1083/jcb.99.5.1872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kogut I, Roop DR, Bilousova G. Differentiation of human induced pluripotent stem cells into a keratinocyte lineage. Methods Mol Biol. 2014;1195:1–12. doi: 10.1007/7651_2013_64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopan R, Traska G, Fuchs E. Retinoids as important regulators of terminal differentiation: examining keratin expression in individual epidermal cells at various stages of keratinization. J Cell Biol. 1987;105(1):427–440. doi: 10.1083/jcb.105.1.427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowalczyk AP, Green KJ. Structure, function, and regulation of desmosomes. Prog Mol Biol Transl Sci. 2013;116:95–118. doi: 10.1016/B978-0-12-394311-8.00005-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Pajares V, Qu K, Zhang J, Webster DE, Barajas BC, Siprashvili Z, Zarnegar BJ, Boxer LD, Rios EJ, Tao S, Kretz M, Khavari PA. A LncRNA-MAF:MAFB transcription factor network regulates epidermal differentiation. Dev Cell. 2015;32(6):693–706. doi: 10.1016/j.devcel.2015.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nam KH, Smith AS, Lone S, Kwon S, Kim DH. Biomimetic 3D Tissue Models for Advanced High-Throughput Drug Screening. J Lab Autom. 2015;20(3):201–215. doi: 10.1177/2211068214557813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson WG, Sun TT. The 50- and 58-kdalton keratin classes as molecular markers for stratified squamous epithelia: cell culture studies. J Cell Biol. 1983;97(1):244–251. doi: 10.1083/jcb.97.1.244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osmani N, Labouesse M. Remodeling of keratin-coupled cell adhesion complexes. Curr Opin Cell Biol. 2015;32:30–38. doi: 10.1016/j.ceb.2014.10.004. [DOI] [PubMed] [Google Scholar]
- Popov L, Kovalski J, Grandi G, Bagnoli F, Amieva MR. Three-Dimensional Human Skin Models to Understand Staphylococcus aureus Skin Colonization and Infection. Front Immunol. 2014;5:41. doi: 10.3389/fimmu.2014.00041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prunieras M, Regnier M, Woodley D. Methods for cultivation of keratinocytes with an air-liquid interface. J Invest Dermatol. 1983;81(1 Suppl):28s–33s. doi: 10.1111/1523-1747.ep12540324. [DOI] [PubMed] [Google Scholar]
- Ramirez RD, Morales CP, Herbert BS, Rohde JM, Passons C, Shay JW, Wright WE. Putative telomere-independent mechanisms of replicative aging reflect inadequate growth conditions. Genes Dev. 2001;15(4):398–403. doi: 10.1101/gad.859201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rheinwald JG, Green H. Serial cultivation of strains of human epidermal keratinocytes: the formation of keratinizing colonies from single cells. Cell. 1975;6(3):331–343. doi: 10.1016/s0092-8674(75)80001-8. [DOI] [PubMed] [Google Scholar]
- Saito M, Tucker DK, Kohlhorst D, Niessen CM, Kowalczyk AP. Classical and desmosomal cadherins at a glance. J Cell Sci. 2012;125(Pt 11):2547–2552. doi: 10.1242/jcs.066654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shamir ER, Ewald AJ. Three-dimensional organotypic culture: experimental models of mammalian biology and disease. Nat Rev Mol Cell Biol. 2014;15(10):647–664. doi: 10.1038/nrm3873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson CL, Kojima S, Getsios S. RNA interference in keratinocytes and an organotypic model of human epidermis. Methods Mol Biol. 2010;585:127–146. doi: 10.1007/978-1-60761-380-0_10. [DOI] [PubMed] [Google Scholar]
- Sugihara H, Toda S, Yonemitsu N, Watanabe K. Effects of fat cells on keratinocytes and fibroblasts in a reconstructed rat skin model using collagen gel matrix culture. Br J Dermatol. 2001;144(2):244–253. doi: 10.1046/j.1365-2133.2001.04008.x. [DOI] [PubMed] [Google Scholar]
- Weaver JC. Electroporation theory. Concepts and mechanisms. Methods Mol Biol. 1995;55:3–28. doi: 10.1385/0-89603-328-7:3. [DOI] [PubMed] [Google Scholar]
- Wu YJ, Parker LM, Binder NE, Beckett MA, Sinard JH, Griffiths CT, Rheinwald JG. The mesothelial keratins: a new family of cytoskeletal proteins identified in cultured mesothelial cells and nonkeratinizing epithelia. Cell. 1982;31(3 Pt 2):693–703. doi: 10.1016/0092-8674(82)90324-5. [DOI] [PubMed] [Google Scholar]