Abstract
Heterozygous deletion of RPS14 occurs in del(5q) MDS and has been linked to impaired erythropoiesis, characteristic of this disease subtype. We generated a murine model with conditional inactivation of Rps14 and demonstrated a p53-dependent erythroid differentiation defect with apoptosis at the transition from polychromatic to orthochromatic erythroblasts resulting in age-dependent progressive anemia, megakaryocyte dysplasia, and loss of hematopoietic stem cell (HSC) quiescence. Using quantitative proteomics, we identified significantly increased expression of proteins involved in innate immune signaling, particularly the heterodimeric S100a8/S100a9 proteins in purified erythroblasts. S100a8 expression was significantly increased in erythroblasts, monocytes and macrophages and recombinant S100a8 was sufficient to induce an erythroid differentiation defect in wild-type cells. We rescued the erythroid differentiation defect in Rps14 haploinsufficient HSCs by genetic inactivation of S100a8 expression. Our data link Rps14 haploinsufficiency to activation of the innate immune system via induction of S100A8/A9 and the p53-dependant erythroid differentiation defect in del(5q) MDS.
Introduction
Isolated, interstitial deletion of Chromosome 5q in patients with myelodysplastic syndrome (MDS) is associated with a clinical phenotype termed the 5q- syndrome that is characterized by a severe macrocytic anemia, a normal or elevated platelet count with hypolobated micromegakaryocytes and a low rate of progression to acute myelogenous leukemia1–3. The severe macrocytic anemia in del(5q) MDS patients has been linked to haploinsufficiency of the ribosomal protein small subunit 14 (RPS14)4. In a screen of the 5q33 common deleted region associated with the 5q- syndrome, only shRNAs targeting the RPS14 gene caused a severe block in erythroid differentiation, while forced overexpression of RPS14 in cells from MDS patients with the 5q deletion rescued erythropoeisis4.
Germline, heterozygous inactivating mutations or deletions of RPS19 and other ribosomal protein genes cause Diamond-Blackfan anemia (DBA), a disorder that, like del(5q) MDS, is characterized by macrocytic anemia5–9. Decreased expression of individual ribosomal proteins, including RPS19 and RPS14, increases p53 levels and p53 target gene expression in cell lines, primary human hematopoietic progenitor cells, and patient samples10–12. Pharmacologic or genetic inactivation of p53 rescues the differentiation defect of progenitor cells in multiple model systems7,8,10, 13.
Several in vivo models of ribosome dysfunction have been described14. A murine model with hematopoietic-specific heterozygous deletion of Rps6 recapitulated the erythroid phenotype of del(5q) MDS and DBA that is rescued by p53 inactivation, though RPS6 inactivation has not been described in either DBA or MDS7,8,15. To model del(5q) MDS, a mouse was generated wherein a series of DNA segments syntenic to the commonly deleted region on human chromosome 5, including Rps14, was conditionally deleted from the hematopoietic stem cell compartment13. Heterozygous loss of this region resulted in a macrocytic anemia and dysplastic megakaryocytes, thereby recapitulating aspects of the 5q–syndrome. The smallest deleted segment tested that maintained the erythroid defect included Rps14 and 7 other genes.
In order to investigate the hematologic phenotype and molecular consequences specific to Rps14 haploinsufficiency in vivo, and to explore the role of this gene in del(5q) MDS, we generated a murine model with conditional Rps14 inactivation.
Results
Rps14 haploinsufficiency induces a p53-dependent erythroid differentiation defect in late-stage erythroblasts
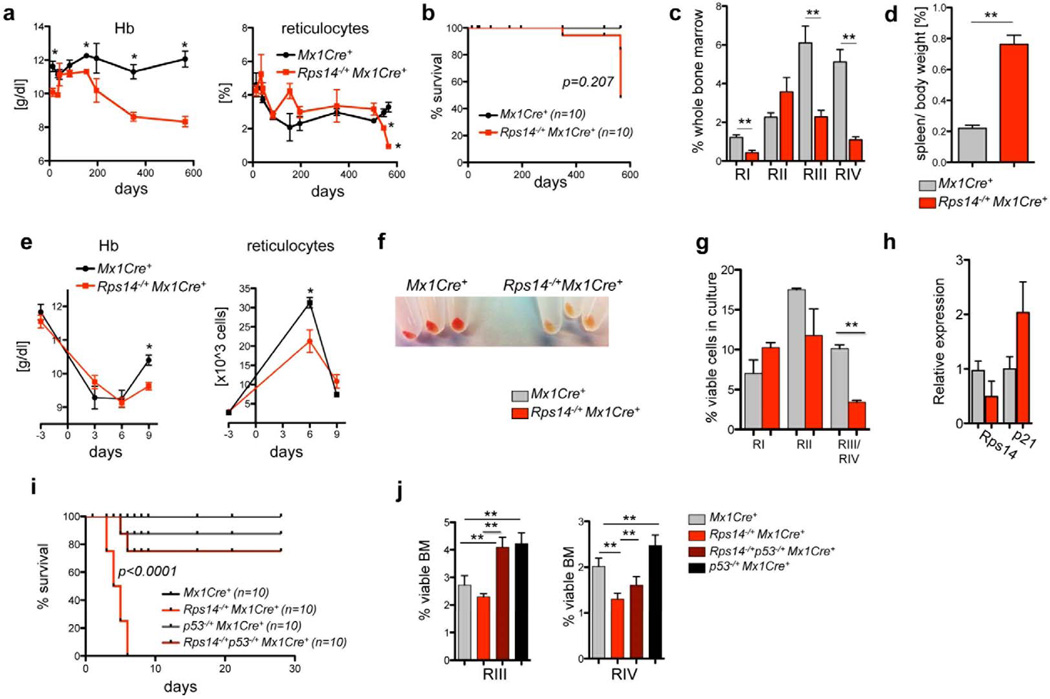
We generated a conditional Rps14 knockout model in which exons 2–4 are flanked by loxP sites (Suppl. Fig. 1a). Following crosses to Mx1Cre+ transgenic mice, we induced Rps14 excision in hematopoietic cells by poly(I:C) treatment and confirmed haploinsufficient expression of Rps14 (Suppl. Fig. 1b, c). Mice with Rps14 haploinsufficiency in hematopoietic cells developed a progressive anemia (Fig. 1a; Suppl. Fig. 1d, e). At approximately 550 days of age, the reticulocyte count of Rps14 haploinsufficient mice decreased precipitously and was associated with death in a subset of mice (Fig. 1a, b).
Figure 1. Rps14 haploinsufficiency results in a p53-mediated erythroid differentiation defect.
(a) Hemoglobin levels (Hb) and % of reticulocytes in the peripheral blood from Rps14−/+Mx1Cre+ mice in comparison to Mx1Cre+ wild-type controls. (mean±SD, n=10; *p<0.05). (b) Kaplan-Meier survival curve of Rps14−/+Mx1Cre+ (n=10) and Mx1Cre+ control mice (n=10). Time point 0 is the day of the first of three poly(I:C) inductions. (c) Frequency of RI-RIV erythroid progenitor populations (RI: CD71highTer119; RII: CD71highTer119+; RIII: CD71intermediateTer119+; RIV: CD71−Ter119+) among viable bone marrow cells in Mx1Cre+ and Rps14−/+Mx1Cre+ mice 18 months after poly(I:C); (mean±SD, n=5; **p<0.001). (d) Relative spleen to body weight [%] of Mx1Cre+ and Rps14−/+Mx1Cre+ mice 18 months after poly(I:C); (mean±SD, n=5; **p<0.001). (e) Hb level and reticulocyte counts in the peripheral time blood at serial time points before and after 25mg/kg Phenylhydrazine injection (mean±SD, n=8; *p<0.05). (f) Cell pellets of lineage-negative HSPCs subjected to erythroid differentiation in vitro for 5 days. (g) Quantification of different erythroid differentiation stages 5 days after induction of erythroid differentiation (mean±SD; n=3 biological replicates; **p<0.001). (h) Quantification of p21 and Rps14 transcript levels by quantitative real-time PCR in cells exposed for 5 days to erythroid differentiation. Data are normalized to expression in Mx1Cre+ control cells (n=5 biological replicates; mean±SD). (i) Kaplan-Meier survival curve after treatment with 35mg/kg Phenylhydrazine on two consecutive days (day 0 and day 1) of Rps14−/+Mx1Cre+ (n=10), p53−/+Mx1Cre+ (n=10), Rps14−/+p53−/+Mx1Cre+ (n=10) and Mx1Cre+ control mice (n=10). (j) Frequency of RIII and RIV erythroid progenitor populations among viable bone marrow cells in 10–12 week old Rps14−/+Mx1Cre+ (n=14), p53−/+Mx1Cre+ (n=5), Rps14−/+p53−/+Mx1Cre+ (n=5) and Mx1Cre+ control mice (n=8) characterized by differential CD71 and Ter119 expression 9 days after the first treatment with 25mg/kg Phenylhydrazine (mean±SD; **p<0.001). Unpaired two-sided t-test (a–i) or multiple group comparison (i, j) using analysis of variance with posthoc Tukey correction were applied for statistical analysis.
We next determined whether Rps14 haploinsufficiency causes a discrete, stage-specific defect in erythroid development. We characterized the stages of erythropoiesis by flow cytometry on the basis of Ter119 and CD71 expression (Supplementary Fig. 1d). Rps14 haploinsufficient mice had impaired erythropoiesis at the transition from CD71+Ter119+ basophilic and early chromatophilic erythroblasts (RII) to CD71intermediate/lowTer119+ poly/orthochromatophilic erythroblasts and enucleated erythrocytes (RIII/RIV), (Fig. 1c). Rps14 haploinsufficient mice had significant splenomegaly with repression of the white pulp due to an expansion of the early erythroid compartment (Fig. 1d; Suppl. Fig. 1i). Younger mice, 22 weeks after excision, also had impaired differentiation at the RIII/IV transition (p<0.001) with a decrease in quiescence of cells in the RI population (p>0.001); (Suppl. Fig. 1f, g), together suggesting that younger Rps14 haploinsufficient mice induce compensatory increase in erythropiesis causing a delay in development of severe anemia. To determine whether the anemia is driven by Rps14 haploinsufficiency in hematopoietic cells but not in the bone marrow stroma, we generated mixed bone marrow chimeras (Suppl. Fig. 1h). Wild-type mice transplanted with Rps14 haploinsufficient hematopoietic cells developed anemia with kinetics similar to untransplanted mice, confirming that the phenotype is caused by excision of Rps14 in hematopoietic cells.
We next examined the effect of Rps14 haploinsufficiency on stress erythropoiesis by inducing hemolysis with phenylhydrazine (PH) treatment in vivo. Following acute hemolytic stress, Rps14 haploinsufficient mice developed more severe anemia and had a delayed reticulocyte response compared to Mx1Cre control mice (Fig. 1e). The RI population (CD71+Ter119−) was significantly increased in Rps14−/+Mx1Cre+ mice, while the RIV population (CD71lowTer119high) was significantly decreased, highlighting a terminal erythroid differentiation defect consistent with induction of apoptosis in the RIII (CD71intermediateTer119+) population (Fig. 1h, Supp. Fig. 1j). Rps14−/+Mx1Cre+ mice were characterized by significant splenomegaly with effacement of the normal spleen architecture and expansion of the red pulp (p<0.05; Supp. Fig. 1k).
To analyze the effect of Rps14 haploinsufficiency on erythroid differentiation in the absence of in vivo compensatory mechanisms, we analyzed erythroid differentiation of lineage-negative hematopoietic stem and progenitor cells (HSPC) in vitro. After 5 days, Mx1Cre+ HSPCs differentiated into hemoglobinized CD71−Ter119+ cells while Rps14 haploinsufficient cells did not terminally differentiate (Fig. 1f–g). This terminal differentiation defect was accompanied by a significant reduction in cell proliferation, decrease in cell viability, and induction of the p53 downstream target p21 (Fig. 1h).
The p53 pathway is activated by decreased expression of ribosomal protein genes and has been linked to the erythroid defect in other models of ribosome dysfunction7,8,10,13. We found that compound haploinsufficiency for p53 and Rps14 prevented mortality from high dose PH (35mg/kg, Fig 1i). Following treatment with lower PH dose (25mg/kg), compound haploinsufficiency for p53 normalized the erythropoietic recovery and spleen size changes caused by Rps14 haploinsufficiency (Fig. 1j; Suppl. Fig. 1k, l). In aggregate, these experiments demonstrate that Rps14 haploinsufficiency causes a p53-dependent terminal erythroid differentiation defect.
Rps14 haploinsufficiency alters hematopoietic stem and progenitor cells
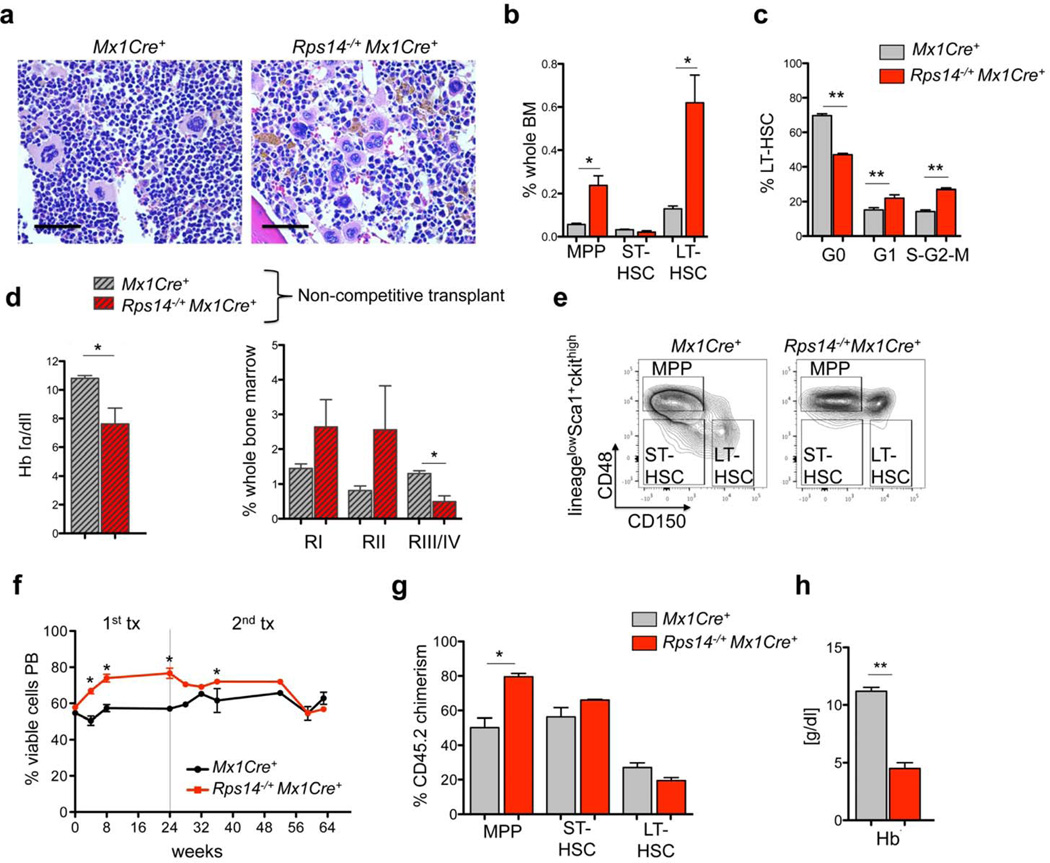
We next evaluated whether Rps14 haploinsufficiency alters hematopoiesis more broadly. In a histopathological analysis of the bone marrow, we found that 18-month old Rps14 haploinsufficient mice had a slightly decreased cellularity, diffuse hemosiderin deposition, and significantly increased numbers of hypolobulated micro-megakaryocytes, consistent with the pathognomonic morphology in del(5q) MDS patients (Fig. 2a; Suppl. Fig. 2a, b). Rps14 haploinsufficient mice had a mild thrombocytosis and platelet dysplasia in peripheral blood smears (Suppl. Fig. 2c). The white blood cell counts (WBC) were normal (Suppl. Fig. 2d). While the HSPC compartment was not altered in young Rps14 haploinsufficient mice (Suppl. Fig. 2e), long-term hematopoietic stem cells (LT-HSCs; lineagelowckit+Sca1+CD48−CD150+) and multipotent progenitor cells (MPPs; lineagelowckit+Sca1+CD48+CD150−) were significantly increased in the bone marrow at 18 months (Fig. 2c). To analyze whether the expansion of LT-HSCs might be due to exit from quiescence and enhanced LT-HSC cycling, we performed cell cycle analysis on HSPCs. In comparison with Mx1Cre+ controls, Rps14 haploinsufficient LT-HSCs had a significantly lower percentage of cells in the G0 phase of cell cycle, and a significantly higher percentage of cells in the cycling G1 as well as S-G2-M fraction, consistent with exit from quiescence (Fig. 2c). As the telomere length reflects the replicative history of a cell, we measured telomere length in bone marrow cells from Rps14 haploinsufficient and Mx1Cre+ control mice. Rps14 haploinsufficient bone marrows had significantly shorter telomeres than Mx1Cre+ control mice (Suppl. Fig. 2i; p<0.001).
Figure 2. Rps14 haploinsufficiency alters hematopoietic stem and progenitor cells.
(a) Bone marrow histopathology in HE-stained bone marrow sections (40x magnification; scale bar 50µm) 18 months after the first poly(I:C) injection in Rps14−/+Mx1Cre+ (n=5) and Mx1Cre+ mice (n=4), representative pictures are shown. (b) Frequency of MPPs (lineagelowckit+Sca1+CD48+CD150−), ST-HSCs (lineagelowckit+Sca1+CD48−CD150−) and LT-HSCs (lineagelowckit+Sca1+CD48−CD150+) 18 months after the first poly(I:C) injection in Rps14−/+Mx1Cre+ (n=5) and Mx1Cre+ mice (n=4); (mean±SD; *p<0.05). (c) Cell cycle was analyzed by combined proliferation (Ki67) and cell cycle (Hoechst33342) staining in permeabilized LT-HSCs from bone marrow (GO: Ki67−Hoechst; G1: Ki67+Hoechst; S-G2-M: Ki67+Hoechst+); (mean±SD; n=5; **p<0.001). (d) Whole bone marrow cells from primary 18 months old mice were transplanted in 6–8 week old SJL/CD45.1 recipient mice (n=5). Hemoglobin levels (Hb) in the peripheral blood from chimeric Rps14−/+Mx1Cre+ mice in comparison to Mx1Cre+ wild-type controls 6 weeks after transplantation and frequency of the RI-RIV erythroid progenitor populations (RI: CD71highTer119−; RII: CD71highTer119+; RIII/IV: CD71intermediate/lowTer119+) in CD45.1 mice transplanted with aged Mx1Cre+ and Rps14−/+Mx1Cre+ bone marrow cells 6 weeks after transplantation. (e) Representative flow plots of the HSC compartment of populations defined as MPPs, ST-HSCs and LT-HSCs in CD45.1 mice transplanted with old Mx1Cre+ (n=4) and Rps14−/+Mx1Cre+ (n=5) bone marrow cells 6 weeks after transplantation. ((f) Identical numbers of Rps14−/+Mx1Cre+ and Mx1Cre+ controls were mixed in equal ratios (approximately 50:50) and transplanted in lethally irradiated CD45.1 recipients. Time point 0 reflects the first bleeding 4 weeks after transplantation (engraftment) before inducing the excision of Rps14 by poly(I:C) injections. After 24 weeks, bone marrow was harvested and transplanted for secondary transplants. (mean±SD, n=5; *p<0.05). (g) Donor chimerism (CD45.2) of the HSC (MPP, ST-HSC, LT-HSC) compartment (mean±SD, n=5; *p<0.05). (h) Hemoglobin levels (Hb) in the peripheral blood from chimeric Rps14−/+Mx1Cre+ or Mx1Cre+ mice 64 weeks after engraftment (40 weeks after secondary transplantation). (mean±SD, n=5; *p<0.05). Unpaired two-sided t-test was applied for statistical analysis.
We next examined the capacity of HSPCs to reconstitute hematopoiesis following transplantation. We transplanted whole bone marrow cells from 18 month old Rps14−/+Mx1Cre+ and Mx1Cre+ controls into 6–8 week old CD45.1 recipient mice. Rps14−/+Mx1Cre+ mice died 6–7 weeks after transplantation with reduced hemoglobin (Hb) levels and an erythroid differentiation block from RII to RIII/IV, comparable to the differentiation block observed in the primary mice (Fig. 2d). The Rps14−/+Mx1Cre+ mice had a significant decrease in both LT- and ST-HSCs and an increase in MPPs (Fig. 2e and Suppl. Fig. 2f), but no decrease in the chimerism in the bone marrow (Suppl. Fig. 2g).
Since Rps14 haploinsufficiency leads to a significant increase in LT-HSCs in primary mice, we examined the functional capacity of Rps14 haploinsufficient cells in a competitive repopulation assay. We transplanted Rps14fl/+Mx1Cre+ or Mx1Cre+ bone marrow cells in competition with an equal number of age-matched CD45.1 competitor BM cells into lethally irradiated CD45.1 recipient mice (Fig. 2f). Four weeks after transplantation, mice were treated with poly(I:C) to induce hemizygous Rps14 inactivation. Rps14 haploinsufficient cells out-competed CD45.1 competitor cells in the primary transplant, while Mx1Cre+ control cells had no competitive advantage (Fig. 2f). To determine the long-term repopulation potential, whole bone marrow cells from primary recipients were transplanted into secondary recipients. In the first transplant, the contribution of Rps14 haploinsufficient bone marrow cells was stable and showed a significantly higher chimerism than Mx1Cre+ controls. By 32 weeks in the secondary transplant, we observed a progressive decrease in the Rps14 chimerism to levels comparable to Mx1Cre+ control cells in the bone marrow.
Given the progressive decrease in Rps14 chimerism, we evaluated HSC abundance. In multipotent progenitors cells, the frequency of Rps14 haploinsufficient cells was significantly higher than WT counterparts, whereas no differences were observed in ST- and LT-HSCs (Fig. 2g). In differentiated lineages, we observed significant myeloid skewing of Rps14 haploinsufficient cells, reflected by increased chimerism within the Gr1+CD11b+ population. In contrast, chimerism within lymphoid populations did not differ significantly from Mx1Cre+ control chimerism; (Suppl. Fig. 2h). The chimerism of Ter119+ erythroid cells was significantly decreased in mice transplanted with Rps14 haploinsufficient bone marrow (Suppl. Fig. 2h), highlighting the distinct erythroid phenotype. Mice transplanted with equivalent numbers of Rps14 haploinsufficient and wild type bone marrow cells succumbed to a severe anemia (Fig. 2h) in the setting of relatively preserved whole blood chimerism suggesting that Rps14 haploinsufficiency might have cell-extrinsic effects that suppress erythropoiesis.
In aggregate, Rps14 haploinsufficiency causes impaired terminal erythroid differentiation, an increase in hematopoietic stem cell frequency with myeloid skewing, and age-dependant loss of the HSC function. Overall, this hematopoietic phenotype resembles alterations during HSC aging and inflammation in the bone marrow16–18.
Ribosomal haploinsufficiency leads to global reduction in protein synthesis
Having established a mouse model that faithfully recapitulates the cardinal features of del(5q) MDS, we sought to understand how heterozygous deletion of Rps14, a component of the 40S ribosomal subunit, alters ribosome assembly and protein synthesis.
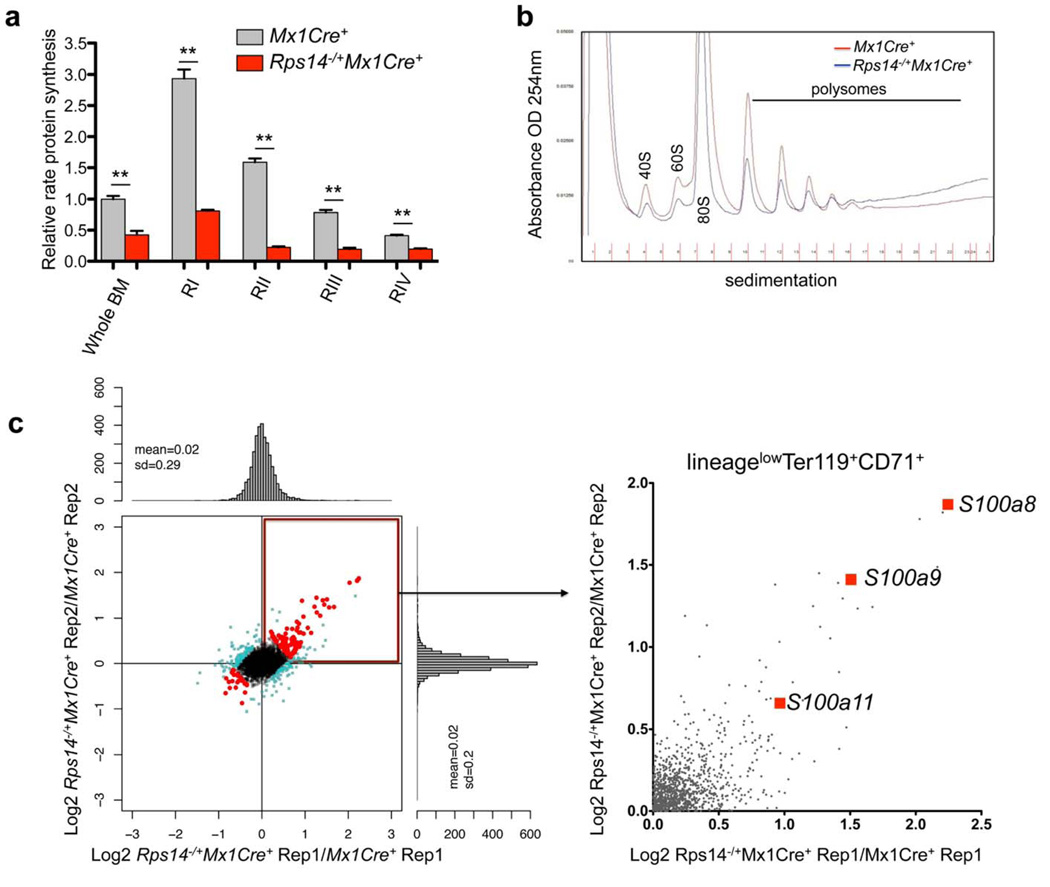
We first analyzed whether Rps14 haploinsufficiency induces quantitative changes in protein synthesis using a fluorogenic assay (O-propargyl-puromycin; OP-Puro) to visualize protein synthesis in vivo19. We administered a single intraperitoneal injection of OP-Puro (50 mg/kg) and measured OP-Puro incorporation as a reflection of total protein translation in a defined period of time. Overall, OP-Puro incorporation was significantly reduced in Rps14−/+Mx1Cre+ cells relative to wild-type cells. The reduction in protein synthesis was most striking in erythroid progenitor cells (Fig. 3a; Suppl. Fig. 3a–b), consistent with the erythroid differentiation block in these cells and the erythroid-specific requirement for high levels of ribosome biogenesis, ribosome activity, and protein translation20.
Figure 3. Reduced protein synthesis in Rps14 haploinsufficient cells.
(a) OP-Puro incorporation in bone marrow (BM) cells in vivo 1 h after administration in 20 week old Mx1Cre+ control cells or Rps14−/+Mx1Cre+ cells [(16 weeks after the first injection of poly(I:C)]. Quantification of OP-Puro fluorescence reflecting protein synthesis rate in hematopoietic stem and progenitor cells relative to unfractionated bone marrow. Relative protein synthesis and quantification of OP-Puro fluorescence in erythroid RI-RIV progenitor populations relative to unfractionated bone marrow (mean±SD, n=5; **p<0.001; Unpaired two-sided t-test was applied for statistical analysis). (b) Polysome profiles from sort-purified lineagelowCD71highTer119+ erythroid progenitor cells in Mx1Cre+ control cells or Rps14−/+Mx1Cre+ cells. The x-axis shows the distance along the gradient. The arbitrary Y-axis shows the relative absorbance. Data are representative of 3 independent experiments (each n=3 biological replicates). (c) Proteomic analysis of induced changes in protein expression of Rps14 haploinsufficient sort-purified lineagelowCD71highTer119+ erythroid progenitor cells relative to Mx1Cre+ cells (300µg protein for each technical replicate=4 biological replicates). Log2 ratios and scatter plot for individual proteins for replicate 1 and 2, where each dot represents a unique protein. The upper right quadrant represents proteins that are significantly up-regulated by Rps14 haploinsufficiency in replicate 1 and 2 (Rep 1 and 2) relative to Mx1Cre+ control cells. Detailed statistical methods for the proteomic analysis are described in the methods section.
To determine whether the Rps14 haploinsufficiency specifically influences ribosomal subunit/polysome formation, we performed sucrose gradient analysis of intact polysomes from FACS-purified erythroid progenitor cells (lineagelowCD71high-intermediateTer119+, RII-III). We observed intact formation of polysome subunits (Fig. 3b), as expected given the rather mild, age-dependent phenotype. These findings indicate that while protein synthesis is impaired, the ribosomes that are assembled function normally. Ribosomal subunits that fail to assemble into an intact 80S ribosome may be rapidly degraded, as has been described previously 21–23.
Ribosomal haploinsufficiency induces increased levels of proteins involved in innate immune function
To determine whether specific proteins are differentially expressed in Rps14 haploinsufficient erythroid progenitor cells and to elucidate a mechanism for the severe erythroid differentiation defect, we performed quantitative proteomics. We purified erythroid progenitor cells (lineagelowCD71high-intermediateTer119+, RII-III) and used isobaric tags for relative and absolute quantification (iTRAQ)-based mass spectrometry (Fig. 3c). We detected 3,524 proteins with at least two unique peptides and ratios per replicate. Of these, 26 proteins were differentially expressed (adjusted p-value of <0.05; Supplementary table 1). Ribosome-associated proteins had decreased expression, consistent with global reduction in protein synthesis but intact polysome formation (Suppl. Fig. 3c). In Rps14 haploinsufficient cells, proteins involved in innate immune system activation were most enriched, including S100a8 and S100a9. In MDS patients, S100A9/CD33 pathway and innate immune activation have been implicated in disease pathogenesis 24,25.
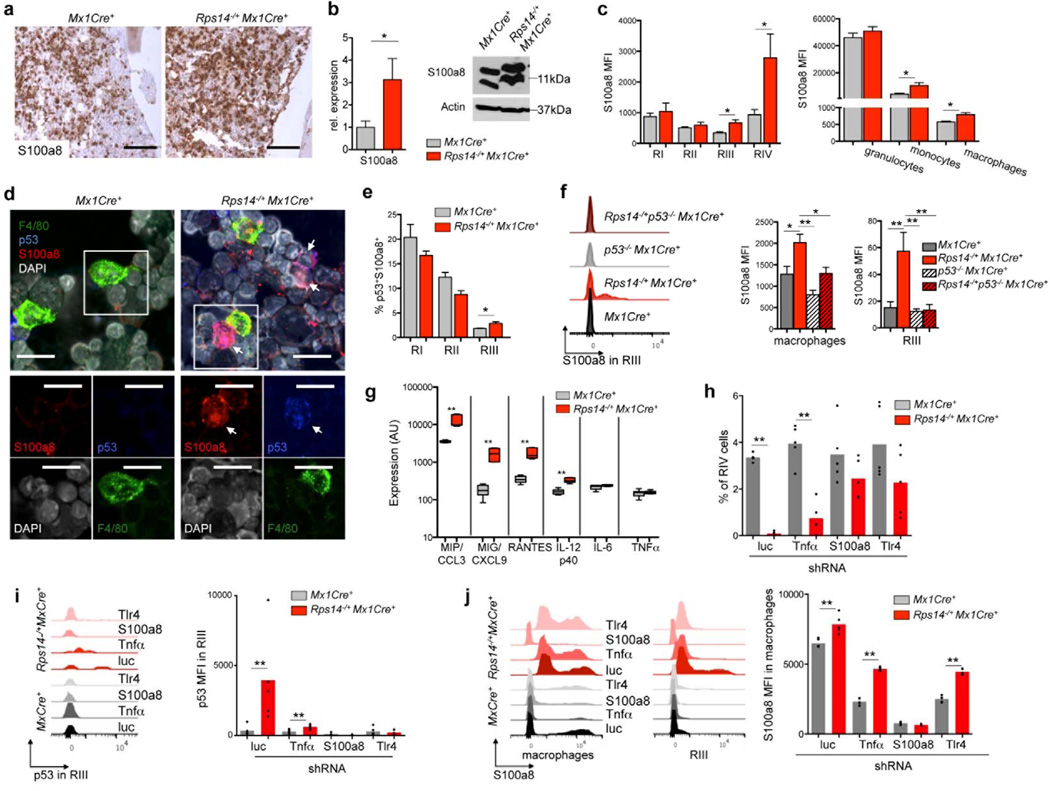
To validate that Rps14 haploinsufficiency induces expression of innate immune response proteins, we examined S100a8 expression by immunohistochemistry in bone marrows of Rps14 heterozygote knockout mice and Mx1Cre+ controls (Fig. 4a; Suppl. Fig. 4d). S100a8 expression was significantly increased in Rps14 haploinsufficient bone marrows and distinct clusters of S100A8 expressing cells were identified (Fig. 4a). We confirmed that S100a8 protein and mRNA expression were increased in lineage-negative bone marrow cells (Fig. 4b). We simultaneously analyzed surface marker staining and intracellular S100a8 staining to quantify S100a8 expression in defined cell populations. We found significantly increased S100a8 in the erythroid progenitor populations affected by the differentiation block (Gr1−CD11b−CD71highTer119high and Gr1−CD11b−CD71lowTer119high; RIII-RIV population), and in monocytes and macrophages of Rps14 haploinsufficient bone marrows (Fig. 4d–e; Suppl. Fig. 4a–b).
Figure 4. S100a8 is significantly up-regulated in Rps14 haploinsufficient bone marrows, is regulated by p53 induction, and is necessary for the erythroid differentiation defect.
(a) Immunohistochemical staining of S100a8 in bone marrows from Mx1Cre+ and Rps14−/+Mx1Cre+ mice 8 weeks after the induction of the Rps14 excision with poly(I:C); representative pictures are shown (n=5). Scale bar 100µm. (b) Quantification of S100a8 in lineage-negative bone marrow cells by quantitative real-time PCR (mean±SD, n=5; *p<0.05). Data are normalized to expression in Mx1Cre+ control cells 8 weeks post poly(I:C). Western blot on protein lysates from Mx1Cre+ and Rps14−/+Mx1Cre+ lineage-negative bone marrow cells. Data are representative of 3 independent experiments. (c) Mean fluorescence intensity (MFI) in CD11b−Gr1− erythroid progenitor populations characterized by CD71 and Ter119 expression (RI-RIV). (mean±SD, n=5; *p<0.05). Mean fluorescence intensity (MFI) of S100a8 expression in Gr1+CD11+ granulocytes, Gr1−CD11b+ monocytes and F4/80+ macrophages (mean±SD, n=5; *p<0.05). (d) Co-immunofluorescent staining of F4/80 (green), p53 (dark blue) and S100a8 (red) on cytospins of Rps14−/+Mx1Cre+ and Mx1Cre+ bone marrow cells 12 weeks after the induction of the Rps14 excision with poly(I:C). Inserts highlight areas of magnification shown in the lower panel. Scale bar: 20µm. Data are representative of 3 independent experiments. (e) Percentage of p53 and S100a8 co-expressing cells in RI-III erythroid progenitor cells from Rps14−/+Mx1Cre+ and Mx1Cre+ mice 12 weeks after the induction of the Rps14 excision with poly(I:C), (mean±SD, n=5; *p<0.05). (f) Representative histograms showing S100a8 expression in the RIII erythroid progenitor cell population and mean fluorescence intensity (MFI) of S100a8 expression in F4/80+ macrophages and RIII erythroblasts in Mx1Cre+, p53−/−, Rps14−/+Mx1Cre+ and Rps14−/+p53−/−Mx1Cre+ mice 6 days after induction of hemolysis with Phenylhydrazine (mean±SD, n=5; *p<0.05; *p*<0.001). (g) Fluorescence intensity normalized to background signals of inflammatory cytokines in bone marrows from Mx1Cre+ and Rps14−/+Mx1Cre+ mice 12 weeks after the induction of the Rps14 excision with poly(I:C). Log10 scale. (mean±SD, n=4; **p<0.001). (h) Ckit+ HSPCs were transduced with lentiviral shRNAs (n=5 each) targeting Tnfα, S100a8 and Tlr4 and a luc control. Transduced cells were selected with puromycin and induced to undergo erythroid differentiation for 5 days in vitro. The frequency of RIV erythroid progenitor populations in the culture is shown (mean±SD, **p<0.001). Circles represent the median of three replicates for each individual shRNA (n=5). The mean of all shRNAs targeting a given gene is shown with a grey bar or red bar. (i) Representative histogram presentation and mean fluorescence intensity (MFI) of p53 expression in the RIII population of HSPCs transduced with shRNA targeting Tnfα, S100a8 and Tlr4 and luc control after 5 days of erythroid differentiation in vitro. (mean±SD, **p<0.001, n=5). Circles represent the median of three replicates for each individual shRNA (n=5). The mean of all shRNAs targeting a given gene is shown with a grey bar or red bar. (j) Representative histograms in F4/80+ macrophages and in the RIII population and mean fluorescence intensity (MFI) of S100a8 expression in macrophages of HSPCs transduced with shRNA targeting Tnfα, S100a8 and Tlr4 and luc control after 5 days of erythroid differentiation in vitro. (mean±SD, **p<0.05, **p<0.001, n=5). Circles represent the median of three replicates for each individual shRNA (n=5). The mean of all shRNAs targeting a given gene is shown with a grey bar or red bar. Unpaired two-sided t-test (b, c, e; h-j) or multiple group comparison by analysis of variance with posthoc Tukey correction (f) were applied for statistical analysis.
As we observed a rapidly developing anemia in chimeric mice transplanted with aged Rps14 haploinsufficient whole bone marrow cells (compare Fig. 2), we analyzed S100a8 expression in the bone marrow of these mice. Both monocytes and Gr1−CD11b−CD71lowTer119high (RIV) erythroblasts in Rps14 haploinsufficient mice had dramatically increased expression of S100a8, suggesting that additional stress in the bone marrow, like transplantation or aging, potentiates the expression of S100a8 with consequent effects on hematopoiesis (Suppl. Fig. 4c; p<0.01)..
The functional unit of mammalian erythropoiesis, the erythroblastic island, consists of a central macrophage that extends cytoplasmic protrusions to a ring of surrounding erythroblasts. As we specifically found increased S100a8 expression in both erythroblasts and macrophages, we examined expression of S100a8 and p53 in the erythroblastic island. We performed confocal imaging on Rps14 haploinsufficient bone marrow spins and Mx1Cre+ control cells (Fig. 4d). F4/80-positive macrophages co-expressed S100a8. In close proximity to macrophages, we detected a significant increase in S100a8-expressing cells in Rps14 haploinsufficient bone marrows, some of which had induction of p53.
To validate that p53 was induced in erythroid progenitor cells with S100a8 induction, we analyzed co-expression of p53 and S100a8 in erythroid progenitor cells by flow cytometry (Fig. 4e). In the RIII population, we identified a significant induction of p53 in S100a8-expressing cells. Since p53 is a known regulator of S100a826–29, we analyzed the expression of S100a8 in Rps14 haploinsufficiency in the presence and absence of p53, as well as during steady-state (Suppl. Fig. 4e) and stress erythropoiesis induced by PH treatment (Fig. 4f). We detected significantly decreased expression of S100a8 in Rps14−/+p53−/−Mx1Cre+ macrophages and RIII erythroid progenitor cells, compared to p53 WT controls, indicating that the induction of p53 by ribosomal haploinsufficiency regulates the expression of S100a8 (Fig. 4f, Suppl. Fig. 4e).
Having shown that Rps14 haploinsufficiency affects not only erythroblasts but also monocytes/macrophages, we measured S100a8 mRNA expression in purified lineagelowCD11b−CD71high/intermediateTer119+ (RII/RIII) erythroblasts and lineagelowF4/80+ macrophages. Both erythroblasts and macrophages from Rps14 haploinsufficient cells had decreased Rps14 and increased S100a8 gene expression (Supplementary Fig. 4f; p<0.05 F4/80+; p>0.001 RII/RIII). In addition, both erythroblasts and macrophages had elevated expression of the Tnfα gene, a down-stream target of the heterodimeric S100A8/A9 complex26,30 and a powerful repressor of erythropoiesis25,31. Consistent with the impaired erythroid differentiation, expression of the erythroid transcription factors Klf1 and Gata1 was decreased.
To investigate whether Rps14 haploinsufficiency induces an inflammatory environment in the bone marrow that represses erythropoiesis and hematopoiesis by cell-extrinsic mechanisms, we profiled levels of 40 cytokines in bone marrow serum (Fig. 4j; Suppl. Fig. 4g). We found 4 cytokines to be significantly increased in Rps14 heterozygous mice: macrophage-inflammatory protein (MIP/CCL3), CXCL9, RANTES, and IL12(p40). These cytokines are predominantly expressed in activated macrophages/monocytes, have negative effects on erythropoiesis and hematopoiesis and are significantly elevated in patients with low-risk MDS32–35. Recent studies have demonstrated that S100A8 and S100A9 are endogenous activators of Toll-like receptor-4 (TLR4)36. We found that Tlr4 expression was activated on both macrophages and monocytes in the bone marrow of Rps14 haploinsufficient mice but not on erythroblasts, suggesting that S100a8 activates macrophages and monocytes and drives an inflammatory environment (Suppl. Fig. 4h; p<0.05).
We examined the roles of S100a8, Trl4, and Tnfα in causing abnormal erythropoiesis in Rps14 haploinsufficient mice using validated small hairpin RNAs (shRNAs) (Suppl. Fig 4i). We transduced ckit+ HSPCs with test and control shRNAs, induced erythroid differentiation in vitro, and evaluated effects using flow cytometry and hemoglobinization (Fig. 4h; Suppl. Fig. 4j). Knockdown of S100a8 and Tlr4 improved erythroid differentiation and reduced p53 induction (Fig. 4i), indicating that S100a8/Tlr4 is central to the erythropoietic defect of Rps14 haploinsufficiency. Tnfα knockdown had a similar, although less robust effect on erythropoiesis and had no effect on S100a8 expression, suggesting a pro-inflammatory role downstream of S100a8 (Fig. 4i–j).
S100a8 is necessary and sufficient for the erythroid differentiation defect due to Rps14 haploinsufficiency
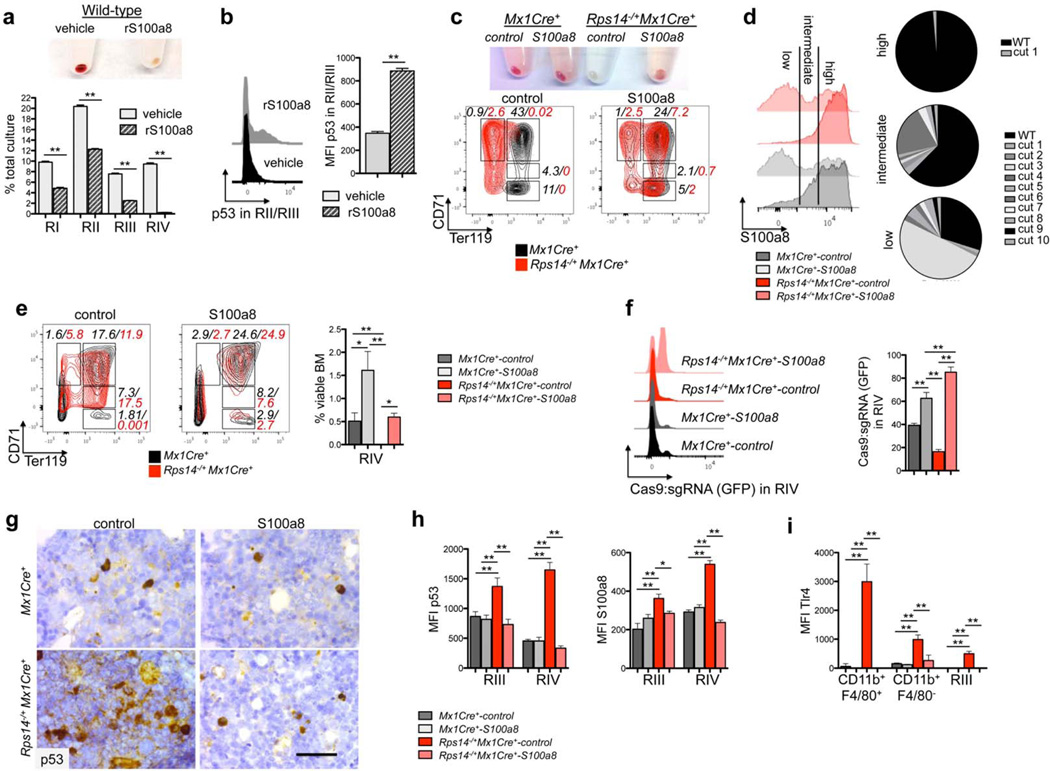
To explore a cell non-autonomous mechanism of the erythroid differentiation defect by S100a8, we induced erythroid differentiation of wild-type lineage-negative HSPCs in the presence of recombinant S100a8 protein (rS100a8). HSPCs cultured in the presence of rS100a8 had a differentiation block at the RIII/RIV transition (Fig. 5a, Suppl. Fig. 5a), consistent with the differentiation defect in Rps14 haploinsufficient cells, with significantly increased p53-induction in the RII/III population (Fig. 5b).
Figure 5. S100a8 is essential for the erythroid differentiation defect due to Rps14 haploinsufficiency.
(a) Lineage negative wild-type HSPCs were subjected to erythroid differentiation for 5 days in the presence of vehicle or S100a8 recombinant protein (rS100a8). Cell pellets 5 days after induction of differentiation (representative picture of 5 biological replicates is shown) and frequency of RI-RIV erythroid progenitor populations in the culture. (mean±SD, n=5 biological replicates; **p<0.001). (b) Intracellular flow cytometry for p53 expression within the RII/RIII populations in erythroid differentiation culture in vitro in presence or absence of recombinant S100a8. Representative histograms of p53 expression and mean fluorescence intensity of p53 within this population (mean±SD, n=5 biological replicates; **p<0.001). (c) Cell pellets of ckit+ HSPCs from Rps14−/+Mx1Cre+ or Mx1Cre+ mice expressing S100a8 or control sgRNAs following erythroid differentiation in vitro. Representative flow plots of the erythroid progenitor populations characterized by CD71 and Ter119 expression (RI-RIV); (data representative from n=3 biological replicates are shown). (d) Representative histogram presentation of S100a8 expression in Gr1lowCD11b+ monocytes in the erythroid differentiation culture in vitro after 5 days from Rps14−/+Mx1Cre+ or Mx1Cre+ cells which were transduced with either S100a8 or control sgRNAs. The pie chart represents the mutations introduced by the S100a8 sgRNA:Cas9 transduced cells after sequencing of S100a8 high, intermediate, and low fractions. (e) ckit+ HSPCs were transduced with a lentiviral vector expressing Cas9 and an sgRNA targeting S100a8 or control sgRNAs (non-targeting guide, NTG) and were transplanted 24 hours after infection into lethally irradiated wild-type recipients. 6 weeks after transplantation, recovered hematopoiesis in the transplanted mice was confirmed and PH was injected to induce hemolysis. 6 days after the first dose of PH, the bone marrow was harvested. Representative flow plots of the RI - RIV erythroid progenitor populations (mean±SD, n=5; **p<0.001; *p<0.05). (f) Representative histograms depicting GFP expression, representing cells transduced with control- or S100a8 sgRNA:Cas9 in the RIV population (mean±SD, n=5; **p<0.001). (g) p53 immunohistochemistry in bone marrows from Rps14−/+Mx1Cre+ or Mx1Cre+ mice transduced with either control control or S100a8 sgRNA:Cas9. Scale bar: 50µm. (h) Quantification of the mean fluorescence intensity (MFI) of intracellular staining with p53 and S100a8 in erythroid progenitor populations (RIII and IV) in bone marrows from Rps14−/+Mx1Cre+ or Mx1Cre+ mice transduced with either control or S100a8 sgRNA:Cas9. (mean±SD, n=5; *p<0.05; **p<0.001). (i) Quantification of the mean fluorescence intensity (MFI) of intracellular staining with Tlr4 in macrophages (CD11b+F4/80+), monocytes (CD11b+F4/80−) and in erythroid progenitor populations (RIII) in bone marrows from Rps14−/+Mx1Cre+ or Mx1Cre+ mice transduced with either control or S100a8 sgRNA:Cas9. (mean±SD, n=5; *p<0.05; **p<0.001). Unpaired two-sided t-test (a–b) or multiple group comparison (e–i) using analysis of variance with posthoc Tukey correction were applied for statistical analysis.
We next used CRISPR/Cas9-mediated genetic inactivation S100a8 to validate the requirement for S100a8 expression in the erythroid differentiation defect of Rps14 haploinsufficient cells. We transduced HSPCs with a lentiviral vector expressing the Cas9 nuclease and a small guide RNA (sgRNA) targeting S100a8, then analyzed in vitro erythroid differentiation as above. Compared to control non-targeting sgRNA (NTG-sgRNA:Cas9), CRISPR-mediated inactivation of S100a8 (S100a8-sgRNA:Cas9) in Rps14 haploinsufficient cells rescued terminal differentiation of hemoglobinized RIV erythroid cells (Fig. 5c). We confirmed efficient introduction of frameshift insertion/deletion mutations and reduced protein-level expression of S100a8 in myeloid and erythroid cells (Fig. 5d; Suppl. Fig. 5c).
We next determined whether S100a8 is necessary for the erythroid differentiation defect in Rps14 haploinsufficiency in vivo after PH-induced hemolysis (Fig. 5e–i). We transplanted HSPCs expressing either S100a8-sgRNA:Cas9 or control NTG-sgRNA:Cas9 into lethally irradiated wild-type recipients and treated with PH 6 weeks later. In contrast to controls, S100a8-sgRNA:Cas9 expression in Rps14+/−Mx1Cre+ hematopoietic cells rescued the ablation of the RIV population (Fig. 5e; Suppl. Fig. 5e–f), leading to a marked positive selection of Cas9:sgRNA-GFP+ RIV cells (Fig. 5f); reduced induction of p53 and S100a8 in erythroid cells (Fig. 5g, h; Suppl. Fig. 5g); decreased the frequency of Tlr4-expressing macrophages (Fig. 5i; Suppl. Fig. 5d, e, g); and reduced the induction of Tnfα (Suppl. Fig. 5h). These data demonstrate the role of S100a8 in the induction of a p53-dependent erythroid differentiation defect and the increase in Tlr4-expressing macrophages in Rps14 haploinsufficiency.
To examine the cell-intrinsic effect of S100a8 in Rps14 haploinsufficiency, we purified RII erythroblasts and measured the effect of S100a8 inactivation on terminal differentiation in the absence of myeloid cells (Suppl. Fig. 5i). Purified Rps14 haploinsufficient RII progenitor cells expressing a control sgRNA did not terminally differentiate and underwent apoptosis, while those expressing S100a8-sgRNA:Cas9 differentiated into hemoglobinized, terminally differentiated erythroid cells, demonstrating that S100a8 directly affects erythroid progenitor cells by intrinsic mechanisms.
To examine whether S100a8 has cell-extrinsic effects, we analyzed S100a8 and p53 induction in wild-type competitor cells (CD45.1), as well as Mx1Cre+ or Rps14 (CD45.2) haploinsufficient cells, in competitive transplant studies (Suppl. Fig. 5j). Induction of S100a8 expression was restricted to CD45.2 Rps14 haploinsufficient cells (p<0.05). In contrast, we observed p53 induction in both the CD45.1 competitor cells and CD45.2 Rps14 haploinsufficient cells (p<0.05), indicating that increased S100a8 expression in Rps14 haploinsufficient exerts a pro-apoptotic effect on wild-type cells that may drive progressive anemia in a cell-extrinsic fashion, even in the setting of preserved whole bone marrow chimerism. In summary, these data demonstrate that S100a8 exerts both cell-intrinsic cell-extrinsic effects on erythropoiesis.
Frequency of S100A8-positive cells in del(5q) MDS human bone marrows positively correlates with disease severity
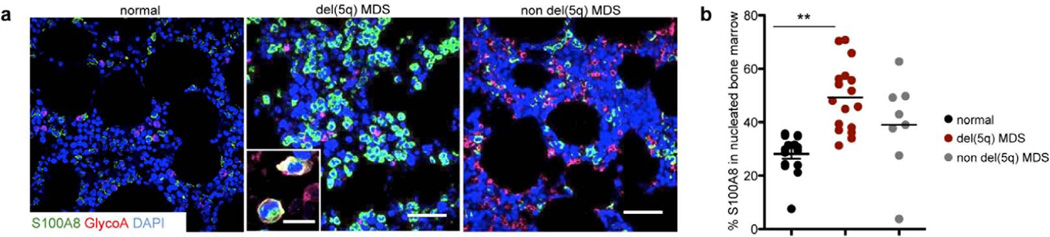
To examine whether ribosomal haploinsufficiency leads to activation of S100A8 in patients with del(5q) MDS, we performed immunofluorescence for S100A8 and the erythroid marker glycophorin A (GlyA) on bone marrow biopsies from non-MDS controls (normal) and MDS patients with and without del(5q) (Fig. 6; Suppl. Table 2; Suppl. Fig. 6a, b). In non-MDS controls, rare S100A8 expressing cells were interspersed in the hematopoietic marrow. In del(5q) MDS, S100A8-positive were positioned in the marrow in groups of greater than 5 cells. The frequency of S100A8-expressing nucleated cells was significantly increased in del(5q) MDS bone marrow biopsies compared to non-MDS bone marrows (Suppl. Fig. 6b). GlyA-positive early erythroid progenitor cells occasionally co-expressed S100A8 (Fig. 6a, insert), just as seen in the Rps14 haploinsufficient murine model. In aggregate, these data indicate that impaired erythropoiesis in the bone marrow of del(5q) MDS patients is associated with aberrant expression of S100A8.
Figure 6. S100A8-frequency is significantly increased in del(5q) MDS human bone marrows compared to non-MDS controls.
(a) Representative images of co-immunoflurescent staining of Glycophorin A (GlyA), S100A8 and DAPI as a nuclear staining in healthy (non-MDS; n=15), del(5q) MDS patients (n=21) and normal karyotype MDS (non del(5q), n=9). DAPI (blue), S100A8 (green), GlyA (magenta). Insert depicts S100A8 and GlyA expressing cells. Scale bars: 100µm. Immunofluorescence for all patients is shown in Supplementary Figure 6. (b) Quantification of S100A8+ cells as a percentage of DAPI-positive nucleated bone marrow cells (S100A8 frequency in the nucleated marrow). (mean±SD, p<0.001). Unpaired two-sided t-test was applied for statistical analysis.
Discussion
Anemia is the most common hematologic manifestation of MDS, particularly in patients with del(5q) MDS. In a novel conditional knockout mouse model of Rps14, we found that Rps14 haploinsufficiency is sufficient to cause a p53-dependent erythroid differentiation defect with apoptosis resulting in age-dependent progressive anemia, megakaryocyte dysplasia, and loss of hematopoietic stem cell (HSC) quiescence. In an unbiased proteomic analysis, we identified a link between Rps14 haploinsufficiency and induction of the danger associated molecular pattern (DAMP) heterodimer S100A8/S100A9 in monocytes, macrophages, and erythroblasts. Our studies link the p53-dependent erythroid differentiation defect to induction of S100A8 and an inflammatory environment produced by monocytes and macrophages, demonstrating that ribosomal haploinsufficiency exerts both cell intrinsic and extrinsic effects on erythroblasts and on hematopoiesis.
S100A8/S100A9 is induced during inflammatory processes including infection, autoimmunity, and cancer26,36,37. In addition to granuloyctes and macrophages, which produce these proteins under steady-state conditions, other cell types induce S100A8/S100A9 expression in response to stress30. We found significant induction of S100A8 in both CD11b+ monocytes and F4/80+ macrophages and concomitant increased expression of S100A8 in late-stage erythroblast, which was associated with p53 induction. Spatially, erythroid progenitor cells interact with a central macrophage in the erythroblastic island. Our data indicate that disruption of the regulatory mechanisms in the erythroblastic island contribute to the erythroid differentiation defects in Rps14 haploinsufficient bone marrows. S100A9 activation has been described in MDS patient samples24,38 and myeloid-derived suppressor cells (MDSC) driven by the S100A9/CD33 pathway perturb hematopoiesis24. Based on our data we posit that S100A8/S100A9 are induced as a stress response upon ribosomal haploinsufficiency in monocytes, macrophages, and erythroblasts and contribute to the MDS phenotype.
S100A8/A9 is an endogenous TLR4 ligand upstream of TNFα leading to NF-kB activation and secretion of pro-inflammatory cytokines37. Our data demonstrate that S100a8 acts upstream of Tlr4 and Tnfα in the Rps14 haploinsufficiency erythroid differentiation defect. Of note, RPS14 and miR-145 are universally co-deleted in the 5q- syndrome, and both converge on TLR4 signaling25,40–44, highlighting the cooperating effects of genes on 5q, as shown in Suppl. Figure 7.
S100 proteins participate in an autoregulatory feedback loop with p53, serving both as upstream drivers of p53 transcription and as direct downstream p53 transcriptional targets26–29. Consistent with these reports, our data demonstrate that p53 is required for induction of S100a8 expression in Rps14 haploinsufficient cells, and that recombinant S100a8 is sufficient to induce p53 activity in erythroid progenitor cells, leading to a block in terminal erythroid differentiation.
Specific cytokines are elevated in the serum of MDS patients33 and these inflammatory signals can alter proliferation and apoptosis of MDS HSPCs 45,46. Indeed, chronic immune stimulation, coupled with senescence-dependent changes48 in both HSPCs and the BM microenvironment49 may be central to disease pathogenesis25,41,50. Our analysis of Rps14 haploinsufficient HSCs suggests that inflammatory cues induced by Rps14 haploinsufficiency impact HSC aging and quiescence17,18,43,47. In patients with chronic inflammation, cytokines in bone marrow have been associated with inhibition of erythropoiesis51. In particular, TNFα expression has been implicated in erythroid defects observed in patients with DBA31 and is up-regulated in the bone marrow serum of MDS patients 52,53. TNF-receptor associated factor 6 (TRAF6) activation, as a target of miRNA145 and miRNA146a, also induces myelodysplasia in a mouse model.
Our data indicate an unexpected link between haploinsufficiency for a ribosomal gene, Rps14, activation of S100A8/S100A9 and other inflammatory molecules, and inhibition of erythropoiesis. Inhibition of this process, potentially through pharmacologic targeting of S100A8/S100A9, could improve red blood cell production in del(5q) MDS. These findings underscore a molecular link between the genetic abnormalities in MDS patients, activation of the innate immune system, and ineffective hematopoiesis that characterizes the disease.
Online Methods
Generation of Rps14 conditional knockout mouse and mouse experiments
The Rps14 target region is 2.12 Kb and includes exon 2–4. Briefly, a 10.64 Kb region used to construct the targeting vector was first subcloned from a positively identified C57BL/6 BAC clone (RP23: 205B18). The region was designed such that the 5’ homology arm extends about 5.76 Kb 5’ to the single LoxP. The 3’ homology arm ends 3’ to the loxP/FRT flanked Neo cassette and is 2.76 Kb long. The loxP/FRT flanked Neo cassette was inserted 255 bp downstream of exon 4. The single loxP site, containing engineered ApaLI and Bcl I sites for southern blot analysis, was inserted 379 bp upstream of exon 2. The targeting vector was confirmed by restriction analysis after each modification step. P6 and T7 primers anneal to the backbone vector sequence and read into the 5’ and 3’ ends of the BAC sub-clone. N1 and N2 primers anneal to the 5’ and 3’ ends of the LoxP/FRT Neo cassette and sequence the SA and LA, respectively. The BAC was subcloned into a ∼2.4kb backbone vector (pSP72, Promega) containing an ampicillin selection cassette for retransformation of the construct prior to electroporation. A pGK-gb2 loxP/FRT Neo cassette was inserted into the gene. The targeting construct was linearized using NotI prior to electroporation into ES cells. The total size of the targeting construct (including vector backbone and Neo cassette) is 14.74 Kb. Targeted iTL IC1 (C57BL/6N) embryonic stem cells were microinjected into Balb/c blastocysts. Resulting chimeras with a high percentage black coat color were mated to wild-type C57BL/6N mice to generate F1 heterozygous offspring. Tail DNA was analyzed from pups with black coat color. A PCR was performed to detect presence of the distal LoxP site using the ‘LEVI 3’ (5’- GTG ATC TCA ACG CAG GTG TGT AGC −3’) and ‘SDL2’ (5’-TAA CAG CAT GGA AGT CGG GTC TCA −3’) primers. This reaction amplifies a wild type product 474 bp in size. The presence of a second PCR product 73 bp greater than the wild type product indicates a positive LoxP PCR.
Chimeric mice were generated by standard methods. The Neo cassette was deleted by crossing with transgenic FLP1 recombinase mice purchased from Jackson Lab (Strain:B6.Cg-Tg(ACTFLPe)9205Dym/J). Rps14 littermate mice were genotyped by PCR with primers reverse (5’–GTG ATC TCA ACG CAG GTG TGT AGC-3’) and forward (5’-TAA CAG CAT GGA AGT CGG GTC TCA-3’) using the following parameters: 95°C for 3 min, followed by 35 cycles of 95°C for 30 sec, 60°C for 1 min, and 72°C for 1 min. Following confirmation of germline transmission, mice were crossed with the Mx1-cre mouse strain (Jackson: 002527). To excise Rps14 exon 2–4, 6–8 week old Rps14 conditional mice (and Mx1Cre+ mice as controls) were given three rounds of 200 µg of poly(I:C) (GE Healthcare Life Sciences) using intraperitoneal injections. Excision after Cre recombination was confirmed by PCR with primers to detect a floxed portion of the construct (Ndel 2: 5’-GTA TCT CCA ATG GTC AGC AAT CAC GG-3’ and LEVI: 5’- GTG ATC TCA ACG CAG GTG TGT AGC −3’). The knockout size is 511bp in size and a WT size of 623bp. For Qt-RT-PCR, I use the following Taqman probes were used: Rps14 mCG6028 Taqman expression Assay (Assay ID Mm00849906_g1; Life Technologies # 4331182). Rps14 haploinsufficient mice were further bred to p53 null mice (Jackson stock number 00813). Animals were monitored two to three times a week for the presence of disease by general inspection and palpation. Peripheral blood was collected from the retro-orbital cavity using an EDTA- treated glass capillary and automated total and differential blood cell counts were determined using Hemavet 950 (Drew Scienfitic). Following sacrifice, mice were examined for the presence of abnormalities, and organs were collected for further cell and histopathological analysis. Due to the experimental design, the genotypes of the mice could not be blinded or randomized. The group size was chosen based on our experience with conditional murine knockout models of genes on 5q in order to detect a disease typical fold change as significant54. All groups of mice (controls and studied genotypes) were age- and sex-matched.
Mouse experiments were performed according to an IACUC approved protocol at Children’s Hospital Boston.
Flow cytometry and cell isolation
Bone marrow (BM) cells were isolated by flushing and crushing pelvis, hind leg bones and vertebrae with mortar and pestle in PBS (GIBCO) supplemented with 2% heat inactivated fetal bovine serum (FBS) and Penicillin/Streptomycin (GIBCO). Whole bone marrow was lysed on ice with red blood cell (RBC) lysis solution (Invitrogen/Life Technologies), and washed in PBS (GIBCO) with 2% FBS. Single-cell suspensions of spleen were prepared by pressing tissue through a 70µm cell strainer followed by red blood cell lysis. Cells were labeled with monoclonal antibodies in 2% FBS/PBS for 30 min on ice. For flow cytometric analysis and isolation of specific hematopoietic progenitors, cells were incubated with combinations of antibodies zo the following cell surface markers, conjugated to FITC, Pe, APC, PercP-Cy5.5, APC-Cy7 (APC-efluor780), Pe-Cy7, Alexa Fluor 700, Pacific blue (efluor450) or biotin: CD3 (17A2), CD5 (53–7.3), CD11b (M1/70), Gr1 (RB6–8C5), B220 (RA3–6B2), Ter119 (TER119), CD71 (C2), ckit (2B8), Sca1 (D7), CD34 (RSM34), CD16/32 (93), CD150 (TC15–12F12.2), CD48 (HM48-1), CD45.1 (A20), CD45.2 (104). For sorting of lineage-negative cells, lineage markers included CD3, CD5, CD11b, Gr1 and Ter119. For sorting of erythroid progenitor cells, the lineage cocktail did not include Ter119. All reagents were acquired from BD Biosciences, eBiosciences, or BioLegend. To increase the sorting efficiency, whole bone marrow samples were either lineage-depleted or ckit enriched using paramagnetic microbeads and an autoMACS magnetic separator (Miltenyi Biotec). Cell Sorting was performed on a FACSAria flow cytometer (BD Biosciences), data acquisition was performed on an LSR II or Canto II (BD Biosciences). Data were analyzed by FlowJo (Tree Star) software.
Long-term Competitive repopulation assays
In competitive bone marrow transplantation studies, 2×106 freshly isolated bone marrow cells were transplanted in competition with 2×106 freshly isolated CD45.1+ bone marrow cells via tail vein injection into female lethally irradiated 6–8 week old (10.5 Gy) CD45.1+ recipient mice. The donor cell chimerism was determined in the peripheral blood four weeks after transplantation before the excision of Rps14 was induced by poly(I:C) injection (week 0) as well as every four to eight weeks. Red blood cells were lysed (Invitrogen/Life Technologies) and the remaining cells were stained with antibodies against CD45.2, CD45.1, Gr1, CD11b, CD3, CD19 (eBio1D3) to assess the donor cell engraftment. For secondary transplants, 5×106 bone marrow cells collected from primary recipients were transplanted into lethally irradiated CD45.1 recipient mice.
In vivo measurement of protein synthesis
One hundred microliter of a 20mM solution of O-Propargyl-Puromycin (OP-Puro; life technologies) were injected intraperitoneally and mice injected with PBS were used as controls. Bone marrow and spleen were harvested after one hour and then kept on ice. 3×106 cells were stained with antibodies against cell surface markers, fixed in 1% paraformaldehyde, and permeabilized in PBS with 3% fetal bovine serum and 0.1% saponin. The azide-alkyne cyclo-addition was performed using the Click-iT Cell Reaction Buffer Kit (Life Technologies) and azide conjugated to Alexa Fluor 488 (Life Technologies) at 5µM final concentration for 30 minutes. Cells were washed twice again and then analyzed by flow cytometry. ‘Relative rates of protein synthesis’ were calculated by normalizing OP-Puro signals to whole bone marrow after subtracting autofluorescence background as described by Singer et al19.
Methylcellulose Assays
48 hours after viral transduction, 15,000 ckit+ cells were sorted and plated in semi-solid methylcellulose culture medium (M3434, StemCell Technologies) and incubated at 37°C in a humidified atmosphere. Colony formation was assessed 7 days after plating.
shRNA and sgRNA:CRISPR Cas9 vector construction, virus production and transduction
Lentivirally expressed shRNAs in the pLKO.1 backbone vector (puromycin resitance) were obtained from the RNAi Consortium at the Broad Institute. S100A8 guide RNA (gRNA; forward 5’-CAC CGA ATT GTG GTA GAC ATC AAT G-3’; reverse 5’-AAA CCA TTG ATG TCT ACC ACA ATT C-3’) were cloned into pL-CRISPR.EFS.GFP (http://www.addgene.org/57827) using BsmBI restriction digestion. Lentiviral particles were produced by transient transfection of 293T cells with lentivirus plasmid together with pSPAX and VSVG packaging plasmids using TransIT-LT (Mirus). Lentiviral particles were concentrated using ultracentrifugation. Ckit+ or lineage-negative cells were cultured in StemSpan SFEM (StemCell Technologies) supplemented with 50 ng/ml murine Thpo and 50 ng murine Scf (both Peprotech) for 24 h and then transduced with concentrated lentiviral supernatant in presence of 2 µg/ml Polybrene using spin-infection for 90 minutes at 2,200rpm at 37C.
Sequencing analysis of CRISPR variants
To assess the proportion and diversity of CRISPR variants, we performed PCR amplicon deep sequencing of bulk genomic DNA at the S100a8 sgRNA target site (chr3:90669574–90669596; mm9 genome build) using the primers S100a8-g1-F: GGACACTCAGTAGTGACCATTT and S100a8-g1-R: GAGTAACTGCAGCTCCCATC. After addition of sample indexes and sequencing adaptors, the product was subjected to 150 nucleotide paired-end sequencing on an Illumina MiSeq, producing approximately 100,000 reads per sample. Reads were then aligned to the genomic reference and grouped according to specific insertion and deletion sequences.
In vitro erythroid differentiation
Total bone marrow cells were labeled with biotin-conjugated α-lineage antibodies, consisting of α-CD3e, α-CD11b, α-CD45R/B220, α-Ly6G/Ly6C and α-TER-119 (BD Pharmingen, San Diego, CA) and purified using anti-biotin beads and negative selection on the AutoMACS (Miltenyi). Purified cells were then seeded in fibronectin-coated (2µg/cm2) tissue-culture treated polystyrene wells (BD Discovery Labware, Bedford, MA) at a cell density of 105 per ml. Erythroid differentiation was carried out according to modified, published protocols55. The erythropoietic medium consisted of IMDM supplemented with Epo at 10 units/ml, 10ng/ml SCF (Peprotech), 10µM dexamethasone (Sigma), 15% FBS, 1% detoxified BSA, 200µg/ml holotransferrin (Sigma, St. Louis, MO), 10µg/ml recombinant human insulin (Sigma), 2mM L-glutamine, 10−4 β-mercaptoethanol and penicillin/streptomycin. After 48 hours the medium was replaced by maintenance medium consisting of IMDM with 20% FBS, 2mM L-glutamine and 10−4 β-mercaptoethanol. Recombinant S100a8 (mouse, Abnova P4345) protein was used where indicated in concentrations of 1µg/ml.
Phenylhydrazine treatment
Phenylhydrazine (PH) was purchased from Sigma and injected subcutaneously at two consecutive days (day 0 and 1) at the dose of 35 or 25 mg/kg as previously described56. Peripheral blood was collected 4 days before the start of treatment and at day 3, 6 and 9. PH treatment experiments were carried out in 8–12 week old mice.
Polysome profiling
For polysome profiling of primary erythroid progenitor cells, whole bone marrow was lineage-depleted (CD3, CD5, B220, Gr1, CD11b) using microbeads and the autoMACS as described above and stained for Ter119 and CD71. Erythroid progenitor cells (lineagenegativeCD71highTer119intermediate/high) were sort-purified in pure FCS, cultured for 1 hours in IMDM with 20% FCS and 2mM L-glutamine at 37°C at humidified conditions. Cells (ca. 3×106) were then harvested, spun down and incubated with 100µg/ml of cycloheximide for 10 minutes at 37°C, washed with ice-cold PBS containing 100µg/ml of cycloheximide and lysed in 300ul 5mM Tris (pH7.4), 2.5mM MgCl2, 1.5mM KCl. Gradients were poured using a Biocomp Gradient Station. Polysomes were separated on a 10–50% linear sucrose gradient containing 20mM HEPES-KOH (pH 7.4). 5mM MgCl2, 100mM KCL, 2mM DTT and 100µh/ml cycloheximide and centrifuged at 36,000 rpm for 2 hours in a Beckman Coulter L8-M centrifuge with SW40Ti rotor. Gradients were fractionated using a Gilson FC-203B fractionator. Absorbance at 254nm was used to visualize the gradients using a BioRad EM-1 Econo UV monitor.
iTRAQ labeling of peptides and basic reversed phase (brp) fractionation
20 million RII/RIII erythroblasts (4 biological replicates per 1 technical/process replicate) were sort-purified and resuspended in 500µl lysis buffer (8M Urea, 50 mM Tris HCl, 1 mM EDTA, 75 mM NaCl, aprotinin, leupeptin, PMSF, NaF, PIC2, and PIC3; all Sigma), vortexed on ice and spun at 20,000xg for 10 min at 4 oC. 300ug protein of each sample were reduced (2 uL 500 mM DTT, 30 minutes, RT), alkylated (4 uL 500 mM IAA, 45 minutes, dark) and digested using 2ug of sequencing grade trypsin overnight with shaking at room temperature (Suppl. Table 3). The samples were then quenched with 20 uL 10% FA and desalted on 10 mg SepPak columns. Desalted peptides were labeled with iTRAQ reagents according to the manufacturer’s instructions (AB Sciex, Foster City, CA). Peptides were dissolved in 30 µl of 0.5 M TEAB pH 8.5 solution and labelling reagent was added in 70 ul of ethanol. After 1 h incubation the reaction was stopped with 50 mM Tris/HCl pH 7.5. Differentially labelled peptides were mixed and subsequently desalted on a 30 mg SepPak column. Basic Reversed Phase fractionation and subsequent concatenation of the differentially labelled and combined peptides was performed as described by Mertins et al.57 with the following amendments: the Zorbax 300 Å Extended-C18 column used was 2.1×150mm with a 3.5µm bead size (Agilent).
Mass Spectrometry Analysis
Reconstituted peptides were separated on an online nanoflow EASY-nLC 1000 UHPLC system (Thermo Fisher Scientific) and analyzed on a benchtop Orbitrap Q Exactive mass spectrometer (Thermo Fisher Scientific). The peptide samples were injected onto a capillary column (Picofrit with 10 µm tip opening / 75 µm diameter, New Objective, PF360-75-10-N-5) packed in-house with 20 cm C18 silica material (1.9 µm ReproSil-Pur C18-AQ medium, Dr. Maisch GmbH, r119.aq). The UHPLC setup was connected with a custom-fit microadapting tee (360 µm, IDEX Health & Science, UH-753), and capillary columns were heated to 50 °C in column heater sleeves (Phoenix-ST) to reduce backpressure during UHPLC separation. Injected peptides were separated at a flow rate of 200 nL/min with a linear 80 min gradient from 100% solvent A (3% acetonitrile, 0.1% formic acid) to 30% solvent B (90% acetonitrile, 0.1% formic acid), followed by a linear 6 min gradient from 30% solvent B to 90% solvent B. Each sample was run for 120 min, including sample loading and column equilibration times. The Q Exactive instrument was operated in the data-dependent mode acquiring HCD MS/MS scans (R=17,500) after each MS1 scan (R=70,000) on the 12 top most abundant ions using an MS1 ion target of 3× 106 ions and an MS2 target of 5×104 ions. The maximum ion time utilized for the MS/MS scans was 120 ms; the HCD-normalized collision energy was set to 27; the dynamic exclusion time was set to 20s, and the peptide match and isotope exclusion functions were enabled.
Quantification and identification of peptides and proteins
All mass spectra were processed using the Spectrum Mill software package v4.2 pre-release (Agilent Technologies) which includes modules developed by us for iTRAQ -based quantification. Precursor ion quantification was done using extracted ion chromatograms (XIC’s) for each precursor ion. The peak area for the XIC of each precursor ion subjected to MS/MS was calculated automatically by the Spectrum Mill software in the intervening high-resolution MS1 scans of the LC-MS/MS runs using narrow windows around each individual member of the isotope cluster. Peak widths in both the time and m/z domains were dynamically determined based on MS scan resolution, precursor charge and m/z, subject to quality metrics on the relative distribution of the peaks in the isotope cluster vs. theoretical. Similar MS/MS spectra acquired on the same precursor m/z in the same dissociation mode within +/− 60 sec were merged. MS/MS spectra with precursor charge >7 and poor quality MS/MS spectra, which failed the quality filter by not having a sequence tag length > 1 (i.e., minimum of 3 masses separated by the in-chain mass of an amino acid) were excluded from searching. For peptide identification MS/MS spectra were searched against mouse Uniprot database to which a set of common laboratory contaminant proteins was appended. Search parameters included: ESI-QEXACTIVE-HCD scoring parameters, trypsin enzyme specificity with a maximum of two missed cleavages, 40% minimum matched peak intensity, +/− 20 ppm precursor mass tolerance, +/− 20 ppm product mass tolerance, and carbamidomethylation of cysteines and iTRAQ labeling of lysines and peptide n-termini as fixed modifications. Allowed variable modifications were oxidation of methionine, N-terminal acetylation, Pyroglutamic acid (N-termQ), deamidated (N), Pyro Carbamidomethyl Cys (N-termC), with a precursor MH+ shift range of −18 to 64 Da. Identities interpreted for individual spectra were automatically designated as valid by optimizing score and delta rank1-rank2 score thresholds separately for each precursor charge state in each LC-MS/MS while allowing a maximum target-decoy-based false-discovery rate (FDR) of 1.0% at the spectrum level. In calculating scores at the protein level and reporting the identified proteins, redundancy is addressed in the following manner: the protein score is the sum of the scores of distinct peptides. A distinct peptide is the single highest scoring instance of a peptide detected through an MS/MS spectrum. MS/MS spectra for a particular peptide may have been recorded multiple times, (i.e. as different precursor charge states, isolated from adjacent brp fractions, modified by oxidation of Met) but are still counted as a single distinct peptide. When a peptide sequence >8 residues long is contained in multiple protein entries in the sequence database, the proteins are grouped together and the highest scoring one and its accession number are reported. In some cases when the protein sequences are grouped in this manner there are distinct peptides which uniquely represent a lower scoring member of the group (isoforms or family members). Each of these instances spawns a subgroup and multiple subgroups are reported and counted towards the total number of proteins. iTRAQ ratios were obtained from the protein-comparisons export table in Spectrum Mill. To obtain iTRAQ protein ratios the median was calculated over all distinct peptides assigned to a protein subgroup in each replicate58. To assign differential expressed proteins we used the Limma package in the R environment to calculate moderated t-test p, as described previously. We also added Blandt-Altman testing to filter out proteins for which the CI for reproducibility was below 95%. Normalized iTRAQ ratios for the 2 biological replicates were filtered to retain only those deemed reproducible. Reproducible replicates were then subjected to a moderated t-test to assess statistical significance. This statistic is similar to the ordinary t-statistic, with the exception that the standard errors are calculated using an empirical Bayes method utilizing information across all proteins, thereby making inference about each individual protein more robust. The nominal p-values arising from the moderated t-statistic are corrected for multiple testing by controlling the false discovery rate (FDR), as proposed by Benjamini and Hochberg. Proteins with an FDR adjusted p-value of less than 0.01 were deemed to be reproducibly regulated. Statistical significance was assessed using only reproducible data points.
Cytokine Array
To detect inflammatory cytokines in the bone marrow serum, we applied the mouse Inflammation Array G1 (CODE: AAM-INF-G1, 8 sample size) according to manusfacturer’s instructions. For detection of TNFa in the bone marrow serum, we used the TNF-alpha quantikine ELISA assay (R&D systems). The long bones of mice were kept on ice and flushed with a total of 200µl pure PBS. Cell-serum suspensions were spun down twice at 4°C at 2,200rpm and the supernatant was harvested and then spun at 13,000rpm for 10min. The protein content was determined using the Pierce protein content BCA assay kit and 100µg protein in 100µl were used per sample.
Western blots
Western blots were performed according to standard protocols. In brief, cell lysis was performed in RIPA buffer with protease/phosphatase inhibitors. After protein quantification, lysates were resuspended in Laemmli Sample Buffer, and loaded to gradient gels (Criterion Tris-HCl Gel, 8–16%). Proteins were transferred onto Immobilon polyvinyl difluoride (PVDF) membranes. As primary antibodies S100A8 (Abcam ab92331), Rps14 (Abcam ab199273), Actin (EPR8484) were used. Blots were incubated with HRP-conjugated secondary antibody and developed using SuperSignal West Pico Chemiluminescent Substrate (Pierce).
Histopathology, immunohistochemistry and immunofluorescence
For histological and immunohistochemical analyses, organs and bones were fixed in 3.7% formaldehyde overnight, dehydrated and prepared for paraffin embedding. Hematoxylin-Eosin (H&E) staining was done according to routine protocols. For immunohistochemical stainings, the Avidin-Biotin Complex (ABC)/HRP (Dako Cytomation, K5001) was applied for color development using an Autostainer platform (Dako Cytomation, Glostrup, Denmark). Peripheral blood smears were stained with May-Grünwald-Giemsa (Sigma-Aldrich). For immunofluorescence studies, sternum sections were fixed in 4% paraformaldehyde on ice for 1 hour, then incubated in 30% sucrose in PBS at 4°C overnight. Cytospins were air-dried first and then fixed in 4% paraformaldehyde for 10 minutes at room temperature. OCT-embedded (Sakura Finetek) tissues were cryosectioned into 7 µm sections and mounted on Superfrost slides (Fisher Scientific). For immunofluorescence stainings, sections were washed in 1X PBS, blocked in 10% normal goat serum (Vector Labs) and incubated with primary antibodies specific for S100A8 (Abcam ab92331) or p53 (Cell Signaling, 1C12) or CD68 (DAKO, clone PGM1). Secondary antibodies were FITC-, Cy3, or Cy5- conjugated (Jackson ImmunoResearch). Nuclei were then stained with DAPI (4′,6′- diamidino-2-phenylindole) and mounted in Prolong Gold (Life Technologies). Images were obtained on a Nikon Eclipse E400 microscope (Nikon, Tokyo, Japan) equipped with a SPOT RT color digital camera model 2.1.1 (Diagnostic Instruments) or obtained on a confocal microscope (Nikon C1 eclipse, Nikon, Melville, NY).
Confocal Q-FISH
Deparaffinization and antigen retrieval paraffin-embedded bone marrow specimens was carried out using standard protocols. Telomere Q-FISH staining was performed as described previously59,60. Briefly, Telomeres were stained with Cy3-(C3TA2) PNA (Panagene, South Corea) followed by six further washing steps and DNA staining was done using DAPI solution (Sigma, US). All images of BM sections were captured within 48h after sample processing and stored at 4°C. Telomere length analysis was performed using the LSM710 (Zeiss, Jena, Germany) confocal microscope. Images were captured using 63x optical magnification with additional 1.2x digital zoom and multi-tracking mode on 0.5 µm steps was used to acquire images of DAPI and Cy3 staining. Maximum projection of 5 single consecutive steps was done and acquired images were used for further digital image analysis. 5 representative images of randomly chosen areas were captured of each bone marrow. Telomere length detection was carried out using Definiens software (Definiens, Germany). Nuclei and telomeres were detected based on the respective DAPI and Cy3 intensity. Mean nuclear background of the Cy3 staining nuclei was calculated and subtracted of each detected telomere within the respective nucleus. Median value of all detected telomeres was used for analysis.
Primary human samples and confocal microscopy
Patient samples originated from different study centers in Germany and this study was approved by according institutional review boards (University of Technology Dresden, Heinrich-Heine-University Düsseldorf, University Hospital RWTH Aachen, all Germany). Samples were deidentified at the time of inclusion. All patients provided informed consent and the data collection was performed in accordance with the Declaration of Helsinki. Criteria for inclusion of del(5q) MDS patients were: cytogenetic isolated del(5q), blast counts <5% in the bone marrow and International Prognostic Scoring System (IPSS) of low risk or intermediate-1. Patient data are summarized in supplementary table 1. Bone marrow biopsies were fixed for 24 hours using the Hannover Solution (12 % buffered formaldehyde plus 64 % methanol), decalcified (EDTA), dehydrated and embedded in paraffin. For immunofluorescence, samples were deparaffinized, hydrated using decreasing ethanol series and subject to heat-induced antigen retrieval using citrate buffer pH 6.0 before proceeding to the immunofluorescence co-stainings. Sections were first blocked with 1x Roti®-block (Carl Roth, Karlsruhe, Germany) for 1h at RT, washed with PBS-Tween and then incubated at 4°C overnight with a primary rabbit monoclonal anti-S100A8 antibody (Abcam, ab92331, 1:250). After washing steps, section were incubated for 30 minutes at RT with a goat anti-rabbit secondary antibody Alexa Fluor®488 conjugate (Life Technologies, Darmstadt, A-11034). Sections were then incubated at 4°C overnight with a primary mouse monoclonal anti-Glycophorin A antibody (Dako, M0819, 1:50) or anti-MPO (DAKO, A0398, 1:300) followed by incubation for 30 minutes at RT with a donkey anti-mouse secondary antibody Alexa Fluor®633 or Alexa Fluor®555 conjugate (Life Technologies, Darmstadt). Nuclei were stained with DAPI and mounted with Vectashield® mounting media (Vector Labs, CA, USA). All antibodies were diluted in 0.1x Roti®-block. Fluorescence was acquired with a confocal laser-scanning microscope (LSM 710, Zeiss, Germany) running Zen 2012 software (Zeiss). A 405 nm diode laser (DAPI) and a 488 nm/ 633 nm argon laser (Alexa Fluor®488/ Alexa Fluor®633) were used for fluorescence excitation. Maximal intensity projections of 3 z-stacks of 1µm each were acquired. Quantification of S100A8 in nucleated cells was done using Image J open source software.
Statistical analysis
Data are presented as mean±SEM. Comparison of two groups was performed using unpaired t-test. For multiple group comparison, analysis of variance with posthoc Tukey correction was applied. Statistical analyses were performed using GraphPad Prism 5.0c (GraphPad Software Inc., San Diego, CA). A p value of less than 0.05 was considered significant.
Supplementary Material
Acknowledgments
This work was supported by the NIH (R01HL082945), a Gabrielle’s Angel Award, and Leukemia and Lymphoma Society Scholar and SCOR Awards to B.L.E. R.K.S was supported by the German Research Foundation (DFG1188/3-1), a Max Eder fellowship to RKS provided by the German Cancer Aid (Deutsche Krebshilfe) and the Edward P. Evans Foundation. G.B. was supported by the German Cluster of Excellence REBIRTH. We thank D. Haase (Georg-August-University Goettingen) for the cytogenetic (karyotype) analysis in del(5q) MDS patients. This work was supported by the confocal microscope facility, a core facility of the Interdisciplinary Center for Clinical Research (IZKF) Aachen within the Faculty of Medicine at RWTH Aachen University.
Footnotes
Author Contributions: R.K.S, R.C.L, S.A.C. and B.L.E. designed experiments. R.K.S, M.S., R.K., M.V.F., C.E.J., C.H., F.B. M.C.C., E.C., C.S.W., C.D.N. and L.P.C. performed experiments and analyzed data. F.B., T.H.B., U.G., U.P. R.K. and G.B. collected patient samples and clinical information, reviewed bone marrow biopsies and analyzed data. R.K.S and B.L.E. wrote the manuscript. All authors provided critical review of the manuscript.
References for the main text
- 1.Ebert BL. Deletion 5q in myelodysplastic syndrome: a paradigm for the study of hemizygous deletions in cancer. Leukemia. 2009;23:1252–1256. doi: 10.1038/leu.2009.53. [DOI] [PubMed] [Google Scholar]
- 2.Ebert BL. Molecular dissection of the 5q deletion in myelodysplastic syndrome. Seminars in oncology. 2011;38:621–626. doi: 10.1053/j.seminoncol.2011.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Komrokji RS, Padron E, Ebert BL, List AF. Deletion 5q MDS: molecular and therapeutic implications Best practice & research. Clinical haematology. 2013;26:365–375. doi: 10.1016/j.beha.2013.10.013. [DOI] [PubMed] [Google Scholar]
- 4.Ebert BL, et al. Identification of RPS14 as a 5q- syndrome gene by RNA interference screen. Nature. 2008;451:335–339. doi: 10.1038/nature06494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Choesmel V, et al. Impaired ribosome biogenesis in Diamond-Blackfan anemia. Blood. 2007;109:1275–1283. doi: 10.1182/blood-2006-07-038372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ruggero D, Shimamura A. Marrow failure: a window into ribosome biology. Blood. 2014;124:2784–2792. doi: 10.1182/blood-2014-04-526301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McGowan KA, et al. Ribosomal mutations cause p53-mediated dark skin and pleiotropic effects. Nature genetics. 2008;40:963–970. doi: 10.1038/ng.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.McGowan KA, et al. Reduced ribosomal protein gene dosage and p53 activation in low-risk myelodysplastic syndrome. Blood. 2011;118:3622–3633. doi: 10.1182/blood-2010-11-318584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Matsson H, et al. Erythropoiesis in the Rps19 disrupted mouse: Analysis of erythropoietin response and biochemical markers for Diamond-Blackfan anemia. Blood cells, molecules & diseases. 2006;36:259–264. doi: 10.1016/j.bcmd.2005.12.002. [DOI] [PubMed] [Google Scholar]
- 10.Dutt S, et al. Haploinsufficiency for ribosomal protein genes causes selective activation of p53 in human erythroid progenitor cells. Blood. 2011;117:2567–2576. doi: 10.1182/blood-2010-07-295238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pellagatti A, et al. Induction of p53 and up-regulation of the p53 pathway in the human 5q–syndrome. Blood. 2010;115:2721–2723. doi: 10.1182/blood-2009-12-259705. [DOI] [PubMed] [Google Scholar]
- 12.Zhou X, Hao Q, Liao J, Zhang Q, Lu H. Ribosomal protein S14 unties the MDM2-p53 loop upon ribosomal stress. Oncogene. 2013;32:388–396. doi: 10.1038/onc.2012.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Barlow JL, et al. A p53-dependent mechanism underlies macrocytic anemia in a mouse model of human 5q- syndrome. Nature medicine. 2010;16:59–66. doi: 10.1038/nm.2063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Raiser DM, Narla A, Ebert BL. The emerging importance of ribosomal dysfunction in the pathogenesis of hematologic disorders. Leukemia & lymphoma. 2014;55:491–500. doi: 10.3109/10428194.2013.812786. [DOI] [PubMed] [Google Scholar]
- 15.Volarevic S, et al. Proliferation, but not growth, blocked by conditional deletion of 40S ribosomal protein S6. Science. 2000;288:2045–2047. doi: 10.1126/science.288.5473.2045. [DOI] [PubMed] [Google Scholar]
- 16.Morrison SJ, Wandycz AM, Akashi K, Globerson A, Weissman IL. The aging of hematopoietic stem cells. Nature medicine. 1996;2:1011–1016. doi: 10.1038/nm0996-1011. [DOI] [PubMed] [Google Scholar]
- 17.Pang WW, et al. Human bone marrow hematopoietic stem cells are increased in frequency and myeloid-biased with age. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:20012–20017. doi: 10.1073/pnas.1116110108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Du W, et al. Inflammation-mediated notch signaling skews fanconi anemia hematopoietic stem cell differentiation. J Immunol. 2013;191:2806–2817. doi: 10.4049/jimmunol.1203474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Signer RA, Magee JA, Salic A, Morrison SJ. Haematopoietic stem cells require a highly regulated protein synthesis rate. Nature. 2014;509:49–54. doi: 10.1038/nature13035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lajtha LG, Oliver R. A kinetic model of the erythron. Proceedings of the Royal Society of Medicine. 1961;54:369–371. [PubMed] [Google Scholar]
- 21.Karbstein K. Inside the 40S ribosome assembly machinery. Current opinion in chemical biology. 2011;15:657–663. doi: 10.1016/j.cbpa.2011.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Strunk BS, Karbstein K. Powering through ribosome assembly. RNA. 2009;15:2083–2104. doi: 10.1261/rna.1792109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Strunk BS, et al. Ribosome assembly factors prevent premature translation initiation by 40S assembly intermediates. Science. 2011;333:1449–1453. doi: 10.1126/science.1208245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen X, et al. Induction of myelodysplasia by myeloid-derived suppressor cells. The Journal of clinical investigation. 2013;123:4595–4611. doi: 10.1172/JCI67580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Starczynowski DT, et al. Identification of miR-145 and miR-146a as mediators of the 5q–syndrome phenotype. Nature medicine. 2010;16:49–58. doi: 10.1038/nm.2054. [DOI] [PubMed] [Google Scholar]
- 26.Bresnick AR, Weber DJ, Zimmer DB. S100 proteins in cancer. Nature reviews. Cancer. 2015;15:96–109. doi: 10.1038/nrc3893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li C, et al. A novel p53 target gene, S100A9, induces p53-dependent cellular apoptosis and mediates the p53 apoptosis pathway. The Biochemical journal. 2009;422:363–372. doi: 10.1042/BJ20090465. [DOI] [PubMed] [Google Scholar]
- 28.Tan M, Heizmann CW, Guan K, Schafer BW, Sun Y. Transcriptional activation of the human S100A2 promoter by wild-type p53. FEBS letters. 1999;445:265–268. doi: 10.1016/s0014-5793(99)00135-0. [DOI] [PubMed] [Google Scholar]
- 29.Mueller A, et al. The calcium-binding protein S100A2 interacts with p53 and modulates its transcriptional activity. The Journal of biological chemistry. 2005;280:29186–29193. doi: 10.1074/jbc.M505000200. [DOI] [PubMed] [Google Scholar]
- 30.Hiratsuka S, Watanabe A, Aburatani H, Maru Y. Tumour-mediated upregulation of chemoattractants and recruitment of myeloid cells predetermines lung metastasis. Nature cell biology. 2006;8:1369–1375. doi: 10.1038/ncb1507. [DOI] [PubMed] [Google Scholar]
- 31.Bibikova E, et al. TNF-mediated inflammation represses GATA1 and activates p38 MAP kinase in RPS19-deficient hematopoietic progenitors. Blood. 2014;124:3791–3798. doi: 10.1182/blood-2014-06-584656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schepers K, et al. Myeloproliferative Neoplasia Remodels the Endosteal Bone Marrow Niche into a Self-Reinforcing Leukemic Niche. Cell stem cell. 2013 doi: 10.1016/j.stem.2013.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kordasti SY, et al. IL-17-producing CD4(+) T cells, pro-inflammatory cytokines and apoptosis are increased in low risk myelodysplastic syndrome. British journal of haematology. 2009;145:64–72. doi: 10.1111/j.1365-2141.2009.07593.x. [DOI] [PubMed] [Google Scholar]
- 34.Su S, et al. Inhibition of immature erythroid progenitor cell proliferation by macrophage inflammatory protein-1alpha by interacting mainly with a C-C chemokine receptor, CCR1. Blood. 1997;90:605–611. [PubMed] [Google Scholar]
- 35.Frisch BJ, et al. Functional inhibition of osteoblastic cells in an in vivo mouse model of myeloid leukemia. Blood. 2012;119:540–550. doi: 10.1182/blood-2011-04-348151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vogl T, et al. Mrp8 and Mrp14 are endogenous activators of Toll-like receptor 4, promoting lethal, endotoxin-induced shock. Nature medicine. 2007;13:1042–1049. doi: 10.1038/nm1638. [DOI] [PubMed] [Google Scholar]
- 37.Ehrchen JM, Sunderkotter C, Foell D, Vogl T, Roth J. The endogenous Toll-like receptor 4 agonist S100A8/S100A9 (calprotectin) as innate amplifier of infection, autoimmunity, and cancer. Journal of leukocyte biology. 2009;86:557–566. doi: 10.1189/jlb.1008647. [DOI] [PubMed] [Google Scholar]
- 38.Wei Y, et al. Global H3K4me3 genome mapping reveals alterations of innate immunity signaling and overexpression of JMJD3 in human myelodysplastic syndrome CD34+ cells. Leukemia. 2013;27:2177–2186. doi: 10.1038/leu.2013.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sahin E, et al. Telomere dysfunction induces metabolic and mitochondrial compromise. Nature. 2011;470:359–365. doi: 10.1038/nature09787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chang KH, et al. p62 is required for stem cell/progenitor retention through inhibition of IKK/NF-kappaB/Ccl4 signaling at the bone marrow macrophage-osteoblast niche. Cell reports. 2014;9:2084–2097. doi: 10.1016/j.celrep.2014.11.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Starczynowski DT, Karsan A. Deregulation of innate immune signaling in myelodysplastic syndromes is associated with deletion of chromosome arm 5q. Cell Cycle. 2010;9:855–856. doi: 10.4161/cc.9.5.11156. [DOI] [PubMed] [Google Scholar]
- 42.Starczynowski DT, et al. TRAF6 is an amplified oncogene bridging the RAS and NF-kappaB pathways in human lung cancer. The Journal of clinical investigation. 2011;121:4095–4105. doi: 10.1172/JCI58818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Reynaud D, et al. IL-6 controls leukemic multipotent progenitor cell fate and contributes to chronic myelogenous leukemia development. Cancer Cell. 2011;20:661–673. doi: 10.1016/j.ccr.2011.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rhyasen GW, et al. Targeting IRAK1 as a therapeutic approach for myelodysplastic syndrome. Cancer Cell. 2013;24:90–104. doi: 10.1016/j.ccr.2013.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kristinsson SY, et al. Chronic immune stimulation might act as a trigger for the development of acute myeloid leukemia or myelodysplastic syndromes. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2011;29:2897–2903. doi: 10.1200/JCO.2011.34.8540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Takizawa H, Boettcher S, Manz MG. Demand-adapted regulation of early hematopoiesis in infection and inflammation. Blood. 2012;119:2991–3002. doi: 10.1182/blood-2011-12-380113. [DOI] [PubMed] [Google Scholar]
- 47.Rossi DJ, et al. Deficiencies in DNA damage repair limit the function of haematopoietic stem cells with age. Nature. 2007;447:725–729. doi: 10.1038/nature05862. [DOI] [PubMed] [Google Scholar]
- 48.Verschoor CP, et al. Blood CD33(+)HLA-DR(−) myeloid-derived suppressor cells are increased with age and a history of cancer. Journal of leukocyte biology. 2013;93:633–637. doi: 10.1189/jlb.0912461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Raaijmakers MH, et al. Bone progenitor dysfunction induces myelodysplasia and secondary leukaemia. Nature. 2010;464:852–857. doi: 10.1038/nature08851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fang J, et al. Cytotoxic effects of bortezomib in myelodysplastic syndrome/acute myeloid leukemia depend on autophagy-mediated lysosomal degradation of TRAF6 and repression of PSMA1. Blood. 2012;120:858–867. doi: 10.1182/blood-2012-02-407999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Means RT., Jr Pathogenesis of the anemia of chronic disease: a cytokine-mediated anemia. Stem Cells. 1995;13:32–37. doi: 10.1002/stem.5530130105. [DOI] [PubMed] [Google Scholar]
- 52.Sawanobori M, et al. Expression of TNF receptors and related signaling molecules in the bone marrow from patients with myelodysplastic syndromes. Leukemia research. 2003;27:583–591. doi: 10.1016/s0145-2126(02)00095-4. [DOI] [PubMed] [Google Scholar]
- 53.Jacobs-Helber SM, et al. Tumor necrosis factor-alpha expressed constitutively in erythroid cells or induced by erythropoietin has negative and stimulatory roles in normal erythropoiesis and erythroleukemia. Blood. 2003;101:524–531. doi: 10.1182/blood-2001-11-0084. [DOI] [PubMed] [Google Scholar]
Methods only references
- 54.Schneider RK, et al. Role of casein kinase 1A1 in the biology and targeted therapy of del(5q) MDS. Cancer Cell. 2014;26:509–520. doi: 10.1016/j.ccr.2014.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shuga J, Zhang J, Samson LD, Lodish HF, Griffith LG. In vitro erythropoiesis from bone marrow-derived progenitors provides a physiological assay for toxic and mutagenic compounds. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:8737–8742. doi: 10.1073/pnas.0701829104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gritsman K, et al. Hematopoiesis and RAS-driven myeloid leukemia differentially require PI3K isoform p110alpha. The Journal of clinical investigation. 2014;124:1794–1809. doi: 10.1172/JCI69927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mertins P, et al. Integrated proteomic analysis of post-translational modifications by serial enrichment. Nature methods. 2013;10:634–637. doi: 10.1038/nmeth.2518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rappsilber J, Mann M. Analysis of the topology of protein complexes using cross-linking and mass spectrometry. CSH protocols. 2007;2007 doi: 10.1101/pdb.prot4594. pdb prot4594. [DOI] [PubMed] [Google Scholar]
- 59.Beier F, et al. Telomere length analysis in monocytes and lymphocytes from patients with systemic lupus erythematosus using multi-color flow-FISH. Lupus. 2007;16:955–962. doi: 10.1177/0961203307084299. [DOI] [PubMed] [Google Scholar]
- 60.Beier F, et al. Accelerated telomere shortening in glycosylphosphatidylinositol (GPI)-negative compared with GPI-positive granulocytes from patients with paroxysmal nocturnal hemoglobinuria (PNH) detected by proaerolysin flow-FISH. Blood. 2005;106:531–533. doi: 10.1182/blood-2004-10-3996. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.