Abstract
Ribosomally synthesized natural products are found in all forms of life. Their biosynthesis uses simple ribosomally synthesized peptides as starting materials that are transformed into complex structures via posttranslational modifications, enriched with elaborate chemical scaffolds that make them desirable as pharmacological tools. In addition, these natural products often exhibit combinatorial biosynthesis, making them attractive targets for engineering. An increasing knowledge of their biosynthetic machinery has provided key insights into their fascinating chemistry. Marine organisms have been a rich source of this class of natural products and here we review the lessons learned from marine life that enables exploitation of their potential for combinatorial engineering, opening up new routes for peptide-based drug discovery.
INTRODUCTION
The combinatorial nature of secondary metabolites, and their potential for combinatorial biosynthesis has long been recognized [1,2]. In nature, dedicated pathways carry the enzymatic machinery responsible for the biosynthesis of natural products. The question arises whether such biosynthetic pathways exhibit modularity, wherein each module can be used multiple times in different combinations with other modules, to enable combinatorial chemistry. If so, what are the degrees of freedom in their chemistry that can be exploited?
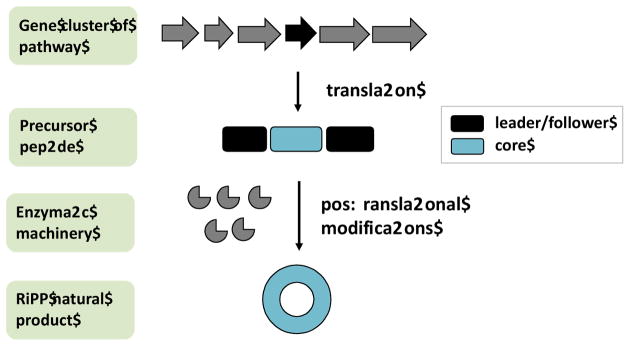
These questions have recently been addressed with the ribosomally synthesized and posttranslationally modified peptide (RiPP) class of natural products. RiPPs begin as precursor peptides that usually contain leader (or sometimes follower) and core peptide sequences. The leader peptide is cleaved off in the course of biosynthesis while the core directly encodes the natural products (Figure 1). Depending upon the RiPP, the leader and other non-core elements encode diverse functions, such as signal sequences dictating localization, enzyme recognition elements, and others [3]. Diverse posttranslational modifications (PTMs) decorate the core peptides to produce an astounding array of natural products [4], which are often bioactive and amenable to peptidomimetics in vivo and in vitro [5] [6–13].
Figure 1.
A simple schematic of RiPP biosynthesis, showing that the biosynthetic machinery is genetically encoded. The precursor peptide substrate gene is shown in black, which is flanked by multiple posttranslational modification enzymes and other proteins (shown in grey) responsible for the transformation of the precursor peptide substrate into the final product. The precursor peptide itself can be differentiated into the core sequence (that matures into the final product) and a leader/follower sequence that often carry enzyme recognition elements among others.
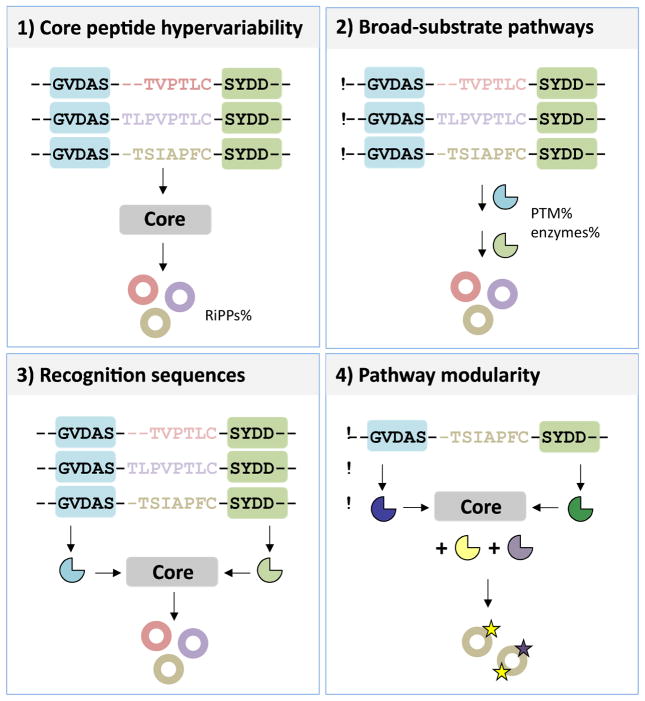
The marine environment has played an important role in RiPP engineering. Marine-derived RiPPs provided the first examples of 4 important concepts that make RiPP combinatorial chemistry a possibility: 1) core peptide hypervariability; 2) natural broad-substrate pathways; 3) enzyme recognition sequences; and 4) modularity of posttranslational elements (Figure 2). In addition, many novel PTMs were first observed in marine organisms. Below, we discuss how these features diversify RiPPs enabling combinatorial synthesis.
Figure 2.
Features of RiPP pathways that enables combinatorial chemistry: 1) core peptide hypervariability (represented by variation in color of the core sequences), which allows to diversify the RiPP pathway products by simple mutations in the core sequence only; 2) broad-substrate pathways, which requires the respective PTM enzymes (shown in blue and green shapes corresponding to distinct PTMs) to be promiscuous enabling them to accept the diversity in the core substrate; 3) recognition sequences (shaded in blue and green boxes along with their respective PTM enzymes), which provide docking sites for these promiscuous enzymes, thus preserving chemistry, despite the susbtrate diversity; 4) pathway modularity, wherein certain units of the pathway act as modules that can be interchanged with other distinct pathways further diversifying PTM chemistry. For instance, recognition sequences from one pathway can be interchanged with PTM enzymes from another (represented by differently colored PTM enzymes deviant from the original) to generate new modifications (shown by stars) in the same core sequence.
Strategy one: Introduce variety (by core peptide hypervariability)
Hypervariability in protein sequences allows organisms to adapt. For example, pathogens often exhibit hypervariability in cell-surface molecules that likely help to evade host immune responses [14]. Likewise, hypervariability of RiPP core peptide sequences creates natural product diversity. The phenomenon of hypervariability was first described in marine cone snail toxins, the conopeptides [15]. Hypervariability exists only in the core sequence, which allows the cone snail to produce an army of mature toxins or “cabals” that act synergistically to paralyze prey by targeting different ion channels simultaneously [16].
While some core peptides were known to be hypervariable, the question remained as to whether the corresponding PTM enzymes were similarly hypervariable. This question was answered with marine peptides, the cyanobactins [17]. In a series of cyanobactin pathways encompassing diverse core peptides from across the tropical Pacific Ocean, essentially sequence identical enzymes were found, implying that libraries of compounds can be created using the same set of enzymes. Indeed, with the cyanobactins later engineering evidence revealed that certain pathways could produce potentially millions of derivatives [18]. Another example lies in the highly mutable marine lanthipeptide pathway to prochlorosins that was discovered by genome mining, and in vitro work reconstituted all the observed variants [19,20]. More recently, similar RiPP hypervariability is also starting to be discovered in terrestrial organisms [21], implying that this feature may be widespread.
However, there are limitations to this strategy since some pathways carry cores that are less mutable. For example, unlike the cyanobactin tru pathway that is mutable for PTMs at all positions excepting the C-terminal residue [22], the linear azoles/azoline peptide (LAP) family of RiPPs that include the microcins and plantazolicin are more restricted in their core sequence [23,24]. Nevertheless, in many RiPP pathways hypervariability has been exploited extensively to create unnatural natural product libraries. For example, not only have RiPPs been engineered with sequences not found in nature, but also highly engineered derivatives carrying non-proteinogenic amino acids can be encoded as has been successfully demonstrated in the lanthipeptides [25], cyanobactins [26] and more recently in the lasso peptides [27].
Strategy two: Pair variety with promiscuity (using broad-substrate pathways)
Enzymes are often remarkably selective, with strong affinities for their native substrates. But, this is not true for the hypervariable RiPPs, which necessitates the PTM enzymes to be promiscuous to be functional. Coupled with hypervariability, such broad substrate-tolerance of the PTM enzymes allows combinatorial synthesis in nature [15,17,19]. Since most RiPP pathways are multi-step, the noteworthy feature that stands out here is that due to the substrate variability ensuing each PTM event, it is not one PTM enzyme, but every PTM enzyme in the pathway that is promiscuous. Thus, the entire multi-step pathway as a whole unit displays remarkable promiscuity. This has led to successful creation of artificial compound libraries by heterologous expression of the entire RiPP pathways as in the microviridins [28], lasso peptides [11], thiopeptides [12] and cyanobactins [18] among others, in addition to libraries carrying non-canonical amino acids, already detailed above.
The question of how promiscuity is evolutionarily favored in broad-substrate enzymes was elegantly investigated in the study of the promiscuous marine lanthionine synthetase ProcM from the prochlorosin pathway, which can install lanthionines on 29 different substrates [19]. A comparison of ProcM with its narrower substrate tolerant counterpart LanM showed that ProcM pays a “kinetic price” for its relaxed specificity [29]. Another example lays in cyanobactin biosynthesis that has been known to carry upto six or more PTMs. The initial enzymatic steps encounter comparatively similar substrates due to the presence of the leader, and exhibit comparatively faster reaction completion times in vitro, which is in contrast to the latter PTM steps that are progressively slower as the enzymes encounter greater structural diversity [22,30].
Strategy three: Provide roots to promiscuous partnership (through conserved recognition sequences)
The molecular basis of PTM promiscuity relies on the use of recognition sequences (RSs). In this strategy, short sequence motifs (RSs) within the precursor peptide substrate act as docking sites for the PTM enzymes. The idea that enzymes dock with substrates at specific sequence motifs is well known and has been documented for a large number of proteases, kinases, and methyltransferases, among others [31].
Within RiPPs, the presence of RSs is widespread [4]. Because of the unique evolutionary strategy employed in several marine cyanobactin biosynthetic pathways, conserved peptide sequence elements could be directly observed, leading to the concept of RSs in RiPP pathways [7]. Reports of crystal structures of many RiPP PTM enzymes have confirmed presence of specific contacts with between the enzyme and RSs [32–34]. Interestingly, homologs of certain RSs motifs are sometimes found in completely unrelated RiPP pathways, but carrying the same PTM, suggesting that the RS is likely serving the same purpose. For example, a homolog of the cyanobactin heterocyclization RS motif is also found in the bottromycin family of RiPPs, which also contain the same type of heterocycles [35].
The conservation of RSs allows PTM enzymes to be fairly well conserved in function between different pathways. Although, hypervariability of substrate core sequences implies that the substrates are rapidly evolving. This phenomenon of substrate evolution, wherein core peptide hypervariability is coupled with conservation of RSs, has allowed RiPP pathways to diversify the final natural products they encode, which in turn may endow evolutionary advantages [36]. Substrate evolution is yet another mechanism that has enabled creation of both natural and artificial combinatorial biosynthesis, examples of which have already been discussed.
This has further enabled in vitro engineering of individual PTM steps, wherein many approaches such as use of non-native cores, non-proteinogenic cores, chimeric substrates among others have been extensively exploited [12,22,36–39]. In addition, these short RSs are portable, wherein the RS only without the leader can be imported to non-native sequences, and still function to recruit the expected PTM [36]. More recently, engineered PTM enzymes fused to RSs could also successfully modify stand-alone core peptides. This was first shown in the lanthionine synthetase LctM [40] and later in the cyanobactin heterocyclase LynD [41], although cyanobactin heterocyclization is capable of proceeding without its RS altogether at the price of lower efficiencies [36,42,43]. Interestingly, such “leaderless” PTMs that do not require RSs outside the core sequence have also been reported in the cyanobactin prenyltransferases [44] and more recently in a lanthipeptide dehydrogenase [45].
Strategy four: Swap partners (by pathway modularity)
RiPP pathways are often modular, which was not appreciated prior to studies in marine organisms. In nature, a clear example of modularity is observed between the pat and tru cyanobactin pathways [46]. Both encode very different cyanobactins; the patellamides from the pat pathway are thiazole-oxazole carrying cyclic peptides, whereas the tru pathway produces thiazoline containing prenylated cyclic peptides called patellins. The only major differences between their gene clusters lie in regions that encode specific PTM enzymes, which are responsible for the structural differences between them. Otherwise, they share about 98% gene sequence identity over half of their gene clusters, implying that pat and tru elements maybe interchangeable.
This implied that by maintaining the same core sequences, different PTM enzymes between pathways could be swapped (as long as the RSs are preserved), leading to hybrid natural products. This was recently demonstrated in the multi-step cyanobactin pathways, wherein swapping PTM enzymes from unrelated pathways at each step led to complete in vitro reconstitution of both natural and unnatural products. Additionally, different combinations of PTM enzymes were used to create an unnaturally large macrocycle [30].
Strategy five: Achieve novelty (creating unique PTMs)
Structurally, RiPP diversity is limited due to the constraint of only twenty possibilities from the amino acid pool. Nonetheless, the structural complexity of RiPPs often rivals those of the nonribosomal peptides due to the presence of extensive posttranslational processing [5]. RiPPs are rich in such novel PTM chemistry [4], and detailed below are a few examples of PTMs discovered in marine organisms (Figure 3).
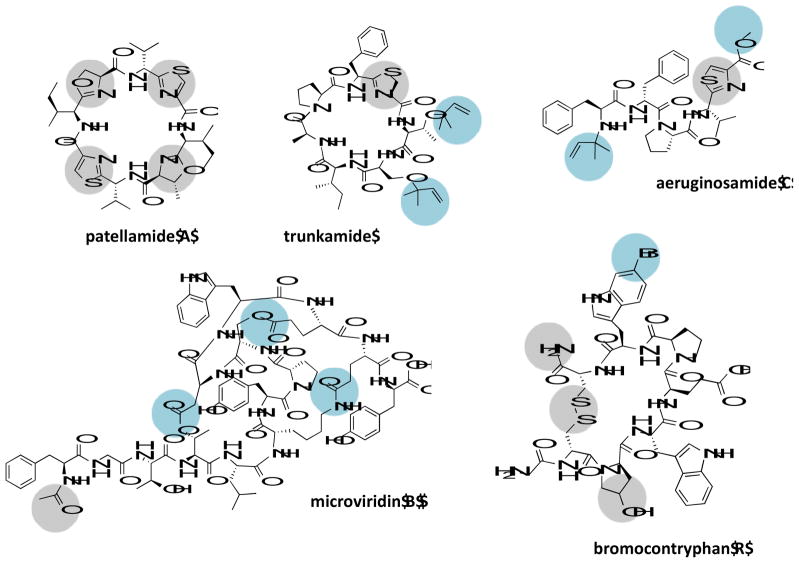
Figure 3.
Representatives of novel PTMs observed in marine derived RiPPs. Shown in grey circles are all PTMs present in the particular structure, whereas shown in blue circles are specific PTMs first observed in marine RiPPs.
The combination of N-C peptide macrocyclization with the presence of azol(in)es was observed first in the cyanobactin RiPPs from tunicates living in coral reefs [47], in addition to dimethylallylation or geranylation of a peptide side-chain [44,48] and prenylation of a peptide amino terminus [49]. The microviridins from limnal cyanobacterial blooms of Japan were the first to show presence of ester and amide linkages via functional groups of lysine or aspartate/glutamate, leading to rigid cage-like cyclic peptides with lactones and lactams [50]. Halogenation of tryptophan residues was first reported in the conopeptides [51]. Among recently discovered RiPPs, the most striking example of novel PTMs was reported in the new family of proteusins represented by the giant natural products polytheonamides derived from marine sponge associated symbiosis. They carry up to 48 PTMs, including an unprecedented N-terminal acyl unit derived from threonine [52,53]. Other marine peptides too exist that resemble RiPPs structurally, but their biosynthetic route is unknown, an interesting example of which is the lanthionine carrying vitilevuamide, isolated from marine ascidians [54].
Much ongoing research is aimed at understanding the PTM machinery of RiPPs. Such knowledge will further the engineering efforts for the creation of these enzymes as tools to generate PTM diversity in artificial combinatorial libraries. For example, ProcM was manipulated to act as a kinase instead of lanthionine synthetase, using principles of RiPP RSs [55]. Since many PTMs resemble those found in nonribosomal pathways in terms of complexity or structure, knowledge of RiPP enzymes will aid the increasing repertoire of chemical scaffolds for designer peptide motifs [5].
CONCLUSIONS AND PERSPECTIVES
Questions of specific biological problems in the oceans has led to the discovery of many of the principles of RiPP evolution and design described above. When taxonomic relationships are close, precise evolutionary relationships develop. For example, cone snails synthesize the conotoxin family of RiPPs to paralyze their prey. Research into these peptides was initiated to discover the active principles behind these ecological interactions, leading to the FDA approved drug Prialt. Because cone snails are closely related to each other and occupy different ecological niches, a compelling evolutionary story came to light that greatly impacted the design and discovery of conopeptides and analogs [56].
Similarly, compounds in a wide range of organisms contain close chemical relationships that were proposed to be of use in biosynthetic engineering [57]. Although most of these compounds are not RiPPs, examples of applying this evolution/function strategy have focused on the RiPPs. More recently, such features are beginning to emerge in cases like the cyclotides, found in plants [21]. As sequencing data continues to arise, these and other stories will continue to enable the designed enzymatic synthesis of modified peptides.
Highlights.
Observations of marine peptide evolution have enabled combinatorial biosynthesis
Marine RiPP peptides contain hypervariable cores and conserved leaders
Mutability of core sequences enabled combinatorial library production
Marine RiPPs led to the discovery of conserved enzyme recognition sequences
Portability of conserved recognition sequences enables hybridization of RiPP pathways
Several posttranslational modifications were discovered in marine RiPPs
Acknowledgments
Our work on RiPPs is funded by NIH GM102602.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
Papers of particular interest, published within the period of review, have been highlighted as:
* of special interest
** of outstanding interest
- 1.Olivera MB, Hillyard DR, Marsh M, Yoshikami D. Combinatorial peptide libraries in drug design: lessons from venomous cone snails. Trends Biotechnol. 1995;13:422–426. doi: 10.1016/S0167-7799(00)88996-9. [DOI] [PubMed] [Google Scholar]
- 2.Cane DE, Walsh CT, Khosla C. Harnessing the biosynthetic code: combinations, permutations, and mutations. Science. 1998;282:63–68. doi: 10.1126/science.282.5386.63. [DOI] [PubMed] [Google Scholar]
- 3.Oman TJ, van der Donk WA. Follow the leader: the use of leader peptides to guide natural product biosynthesis. Nat Chem Biol. 2010;6:9–18. doi: 10.1038/nchembio.286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Arnison PG, Bibb MJ, Bierbaum G, Bowers AA, Bugni TS, Bulaj G, Camarero JA, Campopiano DJ, Challis GL, Clardy J, et al. Ribosomally synthesized and post-translationally modified peptide natural products: overview and recommendations for a universal nomenclature. Nat Prod Rep. 2013;30:108–160. doi: 10.1039/c2np20085f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.McIntosh JA, Donia MS, Schmidt EW. Ribosomal peptide natural products: bridging the ribosomal and nonribosomal worlds. Nat Prod Rep. 2009;26:537. doi: 10.1039/b714132g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Widdick DA, Dodd HM, Barraille P, White J, Stein TH, Chater KF, Gasson MJ, Bibb MJ. Cloning and engineering of the cinnamycin biosynthetic gene cluster from Streptomyces cinnamoneus cinnamoneus DSM 40005. Proc Natl Acad Sci U S A. 2003;100:4316–4321. doi: 10.1073/pnas.0230516100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schmidt EW, Nelson JT, Rasko DA, Sudek S, Eisen JA, Haygood MG, Ravel J. Patellamide A and C biosynthesis by a microcin-like pathway in Prochloron didemni, the cyanobacterial symbiont of Lissoclinum patella. Proc Natl Acad Sci U S A. 2005;102:7315–7320. doi: 10.1073/pnas.0501424102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ziemert N, Ishida K, Liaimer A, Hertweck C, Dittmann E. Ribosomal synthesis of tricyclic depsipeptides in bloom-forming cyanobacteria. Angew Chem Inter Ed. 2008;47:7756–7759. doi: 10.1002/anie.200802730. [DOI] [PubMed] [Google Scholar]
- 9.Knappe TA, Linne U, Zirah S, Rebuffat S, Xie X, Marahiel MA. Isolation and structural characterization of capistruin, a lasso peptide predicted from the genome sequence of Burkholderia thailandensis E264. J Am Chem Soc. 2008;130:11446–11454. doi: 10.1021/ja802966g. [DOI] [PubMed] [Google Scholar]
- 10.Claesen J, Bibb M. Genome mining and genetic analysis of cypemycin biosynthesis reveal an unusual class of posttranslationally modified peptides. Proc Natl Acad Sci U S A. 2010;107:16297–16302. doi: 10.1073/pnas.1008608107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pan SJ, Link AJ. Sequence diversity in the lasso peptide framework: discovery of functional microcin J25 variants with multiple amino acid substitutions. J Am Chem Soc. 2011;133:5016–5023. doi: 10.1021/ja1109634. [DOI] [PubMed] [Google Scholar]
- 12.Li C, Zhang F, Kelly WL. Heterologous production of thiostrepton A and biosynthetic engineering of thiostrepton analogs. Mol Biosyst. 2011;7:82–90. doi: 10.1039/c0mb00129e. [DOI] [PubMed] [Google Scholar]
- 13.Jagadish K, Gould A, Borra R, Majumder S, Mushtaq Z, Shekhtman A, Camarero JA. Recombinant expression and phenotypic screening of a bioactive cyclotide against alpha-synuclein-induced cytotoxicity in baker's yeast. Angew Chem Int Ed Engl. 2015;54:8390–8394. doi: 10.1002/anie.201501186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rando OJ, Verstrepen KJ. Timescales of genetic and epigenetic inheritance. Cell. 2007;128:655–668. doi: 10.1016/j.cell.2007.01.023. [DOI] [PubMed] [Google Scholar]
- 15.Woodward SR, Cruz LJ, Olivera BM, Hilyard DR. Constant and hypervariable regions in conotoxin propeptides. EMBO J. 1990;9:1015–1020. doi: 10.1002/j.1460-2075.1990.tb08204.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Twede VD, Miljanich G, Olivera BM, Bulaj G. Neuroprotective and cardioprotective conopeptides: An emerging class of drug leads. Curr Opin Drug Discov Devel. 2009;12:231–239. [PMC free article] [PubMed] [Google Scholar]
- 17.Donia MS, Hathaway BJ, Sudek S, Haygood MG, Rosovitz MJ, Ravel J, Schmidt EW. Natural combinatorial peptide libraries in cyanobacterial symbionts of marine ascidians. Nat Chem Biol. 2006;2:729–735. doi: 10.1038/nchembio829. [DOI] [PubMed] [Google Scholar]
- 18.Ruffner DE, Schmidt EW, Heemstra JR. Assessing the combinatorial potential of the RiPP cyanobactin tru pathway. ACS Synth Biol. 2015;4:482–492. doi: 10.1021/sb500267d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li B, Sherb D, Kelly L, Shic Y, Huang K, Knerrc PJ, Joewonod I, Rusche D, Chisholmb SW, van der Donk WA. Catalytic promiscuity in the biosynthesis of cyclic peptide secondary metabolites in planktonic marine cyanobacteria. Proc Natl Acad Sci U S A. 2010;107:10430–10435. doi: 10.1073/pnas.0913677107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang Q, Yang X, Wang H, van der Donk WA. High divergence of the precursor peptides in combinatorial lanthipeptide biosynthesis. ACS Chem Biol. 2014;9:2686–2694. doi: 10.1021/cb500622c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21*.Mollica A, Costante R, Stefanucci A, Novellino E. Cyclotides: a natural combinatorial peptide library or a bioactive sequence player? J Enzyme Inhib Med Chem. 2014;30:575–580. doi: 10.3109/14756366.2014.954108. this discusses how plant derived RiPPs share similar combinatorial chemistry features as in the marine environment derived RiPPs. [DOI] [PubMed] [Google Scholar]
- 22.McIntosh JA, Robertson CR, Agarwal V, Nair SK, Bulaj GW, Schmidt EW. Circular logic: Nonribosomal peptide-like macrocyclization with a ribosomal peptide catalyst. J Am Chem Soc. 2010;132:15499–15501. doi: 10.1021/ja1067806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Melby JO, Dunbar KL, Trinh NQ, Mitchell DA. Selectivity, directionality, and promiscuity in peptide processing from a Bacillus sp. Al Hakam cyclodehydratase. J Am Chem Soc. 2012;134:5309–5316. doi: 10.1021/ja211675n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Deane CD, Melby JO, Molohon KJ, Susarrey AR, Mitchell DA. Engineering unnatural variants of plantazolicin through codon reprogramming. ACS Chem Biol. 2013;8:1998–2008. doi: 10.1021/cb4003392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25*.Shi Y, Yang X, Garg N, van der Donk WA. Production of lantipeptides in Escherichia coli. J Am Chem Soc. 2011;133:2338–2341. doi: 10.1021/ja109044r. this along with reference 26 is the first report of heterologous epression of RiPP pathways carrying non-canonical amino acids. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26*.Tianero MD, Donia MS, Young TS, Schultz PG, Schmidt EW. Ribosomal route to small-molecule diversity. J Am Chem Soc. 2012;134:418–425. doi: 10.1021/ja208278k. see reference 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Piscotta FJ, Tharp JM, Liu WR, Link AJ. Expanding the chemical diversity of lasso peptide MccJ25 with genetically encoded noncanonical amino acids. Chem Commun. 2015;51:409–412. doi: 10.1039/c4cc07778d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ziemert N, Ishida K, Weiz A, Hertweck C, Dittmann E. Exploiting the natural diversity of microviridin gene clusters for discovery of novel tricyclic depsipeptides. Appl Environ Microbiol. 2010;76:3568–3574. doi: 10.1128/AEM.02858-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29**.Thibodeaux CJ, Ha T, van der Donk WA. A price to pay for relaxed substrate specificity: a comparative kinetic analysis of the class II lanthipeptide synthetases ProcM and HalM2. J Am Chem Soc. 2014;136:17513–17529. doi: 10.1021/ja5089452. this work provides kinetic evidence suporting the functioning of broad-substrate enzymes in contrast to narrow substrate counterparts. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30**.Sardar D, Lin Z, Schmidt EW. Modularity of RiPP Enzymes Enables Designed Synthesis of Decorated Peptides. Chem Biol. 2015;22:907–916. doi: 10.1016/j.chembiol.2015.06.014. this work demonstrates the modularity of entire RiPP pathways enabling formation of hybrid natural products from multiple PTMs. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fuchs SM. Chemically modified tandem repeats in proteins: natural combinatorial peptide libraries. ACS Chem Biol. 2013;8:275–282. doi: 10.1021/cb3005066. [DOI] [PubMed] [Google Scholar]
- 32.Koehnke J, Bent A, Houssen WE, Zollman D, Morawitz F, Shirran S, Vendome J, Nneoyiegbe AF, Trembleau L, Botting CH, et al. The mechanism of patellamide macrocyclization revealed by the characterization of the PatG macrocyclase domain. Nat Struct Mol Biol. 2012;19:767–772. doi: 10.1038/nsmb.2340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Agarwal V, Pierce E, McIntosh J, Schmidt EW, Nair SK. Structures of cyanobactin maturation enzymes define a family of transamidating proteases. Chem Biol. 2012;19:1411–1422. doi: 10.1016/j.chembiol.2012.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.van der Donk WA, Nair SK. Structure and mechanism of lanthipeptide biosynthetic enzymes. Curr Opin Struct Biol. 2014;29:58–66. doi: 10.1016/j.sbi.2014.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hou Y, Tianero MDB, Kwan JC, Wyche TP, Michel CR, Ellis GA, Vazquez-Rivera E, Braun DR, Rose WE, Schmidt EW, et al. Structure and Biosynthesis of the Antibiotic Bottromycin D. Org Lett. 2012;14:5050–5053. doi: 10.1021/ol3022758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36*.Sardar D, Pierce E, McIntosh JA, Schmidt EW. Recognition sequences and substrate evolution in cyanobactin biosynthesis. ACS Synth Biol. 2015;4:167–176. doi: 10.1021/sb500019b. this work shows that firstly RS elements are portable and secondly (along with references 42 and 43) that the leader peptide is dispensable. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kluskens LD, Kuipers A, Rink R, Boef E, Fekken S, Driessen AJM, Kuipers OP, Moll GN. Post-translational modification of therapeutic peptides by NisB, the dehydratase of the lantibiotic nisin. Biochemistry. 2005;44:12827–12834. doi: 10.1021/bi050805p. [DOI] [PubMed] [Google Scholar]
- 38.Levengood MR, Knerr PJ, Oman TJ, van der Donk WA. In vitro mutasynthesis of lantibiotic analogues containing nonproteinogenic amino acids. J Am Chem Soc. 2009;131:12024–12025. doi: 10.1021/ja903239s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang Q, Yu Y, Velasquez JE, van der Donk WA. Evolution of lanthipeptide synthetases. Proc Natl Acad Sci U S A. 2012;109:18361–18366. doi: 10.1073/pnas.1210393109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40**.Oman TJ, Knerr PJ, Bindman NA, Velasquez JE, van der Donk WA. An engineered lantibiotic synthetase that does not require a leader peptide on its substrate. J Am Chem Soc. 2012;134:6952–6955. doi: 10.1021/ja3017297. this work was the first to shows that RSs can be fused to their respective PTM enzymes for succesful chemistry on stand-alone cores. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Koehnke J, Mann G, Bent AF, Ludewig H, Shirran S, Botting C, Lebl T, Houssen WE, Jaspars M, Naismith JH. Structural analysis of leader peptide binding enables leader-free cyanobactin processing. Nat Chem Biol. 2015;11:558–563. doi: 10.1038/nchembio.1841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42*.Koehnke J, Bent AF, Zollman D, Smith K, Houssen WE, Zhu X, Mann G, Lebl T, Scharff R, Shirran S, et al. The cyanobactin heterocyclase enzyme: a processive adenylase that operates with a defined order of reaction. Angew Chem Int Ed Engl. 2013;52:13991–13996. doi: 10.1002/anie.201306302. see references 36 and 43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43**.Goto Y, Ito Y, Kato Y, Tsunoda S, Suga H. One-pot synthesis of azoline-containing peptides in a cell-free translation system integrated with a posttranslational cyclodehydratase. Chem Biol. 2014;21:766–774. doi: 10.1016/j.chembiol.2014.04.008. this work used an unique platform to recapitulate a RiPP PTM demonstrating leader peptide dispensability and extreme promiscuity of the PTM enzyme. [DOI] [PubMed] [Google Scholar]
- 44.McIntosh JA, Donia MS, Nair SK, Schmidt EW. Enzymatic basis of ribosomal peptide prenylation in cyanobacteria. J Am Chem Soc. 2011;133:13698–13705. doi: 10.1021/ja205458h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yang X, van der Donk WA. Post-translational introduction of d-alanine into ribosomally synthesized peptides by the dehydroalanine reductase NpnJ. J Am Chem Soc. 2015 doi: 10.1021/jacs.5b05207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Donia MS, Ravel J, Schmidt EW. A global assembly line for cyanobactins. Nat Chem Biol. 2008;4:341–343. doi: 10.1038/nchembio.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ireland C, Scheuer J. Ulicyclamide and ulithiacyclamide, two new small peptides from a marine tunicate. J Am Chem Soc. 1980;102:5688–5691. [Google Scholar]
- 48.Leikoski N, Fewer DP, Jokela J, Alakoski P, Wahlsten M, Sivonen K. Analysis of an inactive cyanobactin biosynthetic gene cluster leads to discovery of new natural products from strains of the genus Microcystis. PLOS ONE. 2012;7 doi: 10.1371/journal.pone.0043002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Leikoski N, Liu L, Jokela J, Wahlsten M, Gugger M, Calteau A, Permi P, Kerfeld CA, Sivonen K, Fewer DP. Genome mining expands the chemical diversity of the cyanobactin family to include highly modified linear peptides. Chem Biol. 2013;20:1033–1043. doi: 10.1016/j.chembiol.2013.06.015. [DOI] [PubMed] [Google Scholar]
- 50.Ishitsuka M, Kusumi T, Kakisawa H. Microviridin: A novel tricyclic depsipeptide from the toxic cyanobacterium Microcystis viridis. J Am Chem Soc. 1990;112:8180–8182. [Google Scholar]
- 51.Olivera BM, Gray WR, Zeikus R, McIntosh M, Varga J, de Santos V, Cruz LJ. Peptide neurotoxins from fish-hunting cone snails. Science. 1985;230:1338–1343. doi: 10.1126/science.4071055. [DOI] [PubMed] [Google Scholar]
- 52**.Freeman MF, Gurgui C, Helf MJ, Morinaka BI, Uria AR, Oldham NJ, Sahl HG, Matsunaga S, Piel J. Metagenome mining reveals polytheonamides as posttranslationally modified ribosomal peptides. Science. 2012;338:387–390. doi: 10.1126/science.1226121. this work identified the polytheonamides to be ribosomal in origin, which was previously thought to be non-ribosomal due to their rich chemistry. [DOI] [PubMed] [Google Scholar]
- 53.Hamada T, Sugawara T, Matsunaga S, Fusetani N. Polytheonamides, unprecedented highly cytotoxic polypeptides from the marine sponge Theonella swinhoei. Tetrahedron Lett. 1994;35:609–612. [Google Scholar]
- 54.Edler MC, Fernandezb AM, Lassotac P, Ireland CM, Barrows LR. Inhibition of tubulin polymerization by vitilevuamide, a bicyclic marine peptide, at a site distinct from colchicine, the vinca alkaloids, and dolastatin 10. Biochem Pharmacol. 2002;63:707–715. doi: 10.1016/s0006-2952(01)00898-x. [DOI] [PubMed] [Google Scholar]
- 55*.Thibodeaux GN, van der Donk WA. An engineered lantipeptide synthetase serves as a general leader peptide-dependent kinase. Chem Commun (Camb) 2012;48:10615–10617. doi: 10.1039/c2cc34138g. this work shows how principles of RiPP RSs can be exploited to engineer a PTM enzyme to redirect its chemistry. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.McIntosh JM, Jones RM. Cone venom from accidental stings to deliberate injection. Toxicon. 2001;39:1447–1451. doi: 10.1016/s0041-0101(01)00145-3. [DOI] [PubMed] [Google Scholar]
- 57.Schmidt EW. Trading molecules and tracking targets in symbiotic interactions. Nat Chem Biol. 2008;4:466–473. doi: 10.1038/nchembio.101. [DOI] [PMC free article] [PubMed] [Google Scholar]