Abstract
Toxoflavin is a major virulence factor of the rice pathogen Burkholderia glumae. The tox operon of B. glumae contains five putative toxoflavin biosynthetic genes toxABCDE. ToxA is a predicted S-adenosylmethionine dependent methyltransferase and toxA knockouts of B. glumae are less virulent in plant infection models. In the present study, we show that ToxA performs two consecutive methylations to convert the putative azapteridine intermediate, 1,6-didemethyltoxoflavin, to toxoflavin. In addition, we report a series of crystal structures of ToxA complexes that reveals the molecular basis of the dual methyltransferase activity. The results suggest sequential methylations with initial methylation at N6 of 1,6-didemethyltoxoflavin followed by methylation at N1. The two azapteridine orientations that position N6 or N1 for methylation are coplanar with a 140° rotation between them. The structure of ToxA contains a Class I methyltransferase fold having an N-terminal extension that either closes over the active site or is largely disordered. The ordered conformation places Tyr7 at a position of a structurally conserved tyrosine site of unknown function in various methyltransferases. Crystal structures of ToxA-Y7F consistently show a closed active site whereas structures of ToxA-Y7A consistently show an open active site, suggesting that the hydroxyl group of Tyr7 plays a role in opening and closing the active site during the multistep reaction.
Toxoflavin was first isolated in the 1930s through investigations into fatal food poisonings in Indonesia.1, 2 The structure of toxoflavin was later characterized3, 4 and the toxicity of toxoflavin was attributed to its redox properties.5 Recently, toxoflavin has been shown to be a key virulence factor of some bacteria that cause bacterial panicle blight of rice and bacterial wilt of other field crops.6–12 Burkholderia glumae is the major causal agent in the United States and is currently considered a threat to global rice production.13 The role of toxoflavin as a major virulence factor has stimulated investigations into the function and utilization of the toxoflavin-degrading enzyme, toxoflavin lyase, from Paenibacillus polymyxa.14–17
The molecular mechanism by which B. glumae synthesizes toxoflavin is currently unknown. However, initial insights were drawn from studies of the tox operon and comparisons made with homologous proteins involved in riboflavin biosynthesis.18 The tox operon contains five putative biosynthetic genes, toxABCDE.18–20 ToxB and ToxE show high sequence similarity to guanosine triphosphate (GTP) cyclohydrolase II proteins21–23 (e.g., RibA from Escherichia coli) and deaminase/reductase proteins24–27 (e.g., RibD from E. coli) involved in riboflavin biosynthesis. Since homologs of ToxB and ToxE catalyze the first steps of riboflavin biosynthesis,28, 29 the initial steps of toxoflavin biosynthesis were predicted to be analogous with GTP being the initial substrate.18 This hypothesis is consistent with early studies utilizing labeled precursors.30 More recent work involving gene deletions in Pseudomonas protegens Pf-531 and in vivo work utilizing B. glumae32 have suggested that the biosynthesis of toxoflavin proceeds through 5-amino-6-d-ribitylaminouracil, a common intermediate in riboflavin and F0 biosynthesis. However, the subsequent steps, involvement and order of ToxA, ToxC, and ToxD action are less clear, although toxC and toxD are essential for toxoflavin biosynthesis in P. protegens Pf-5.31
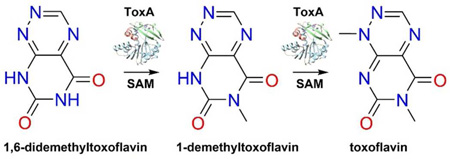
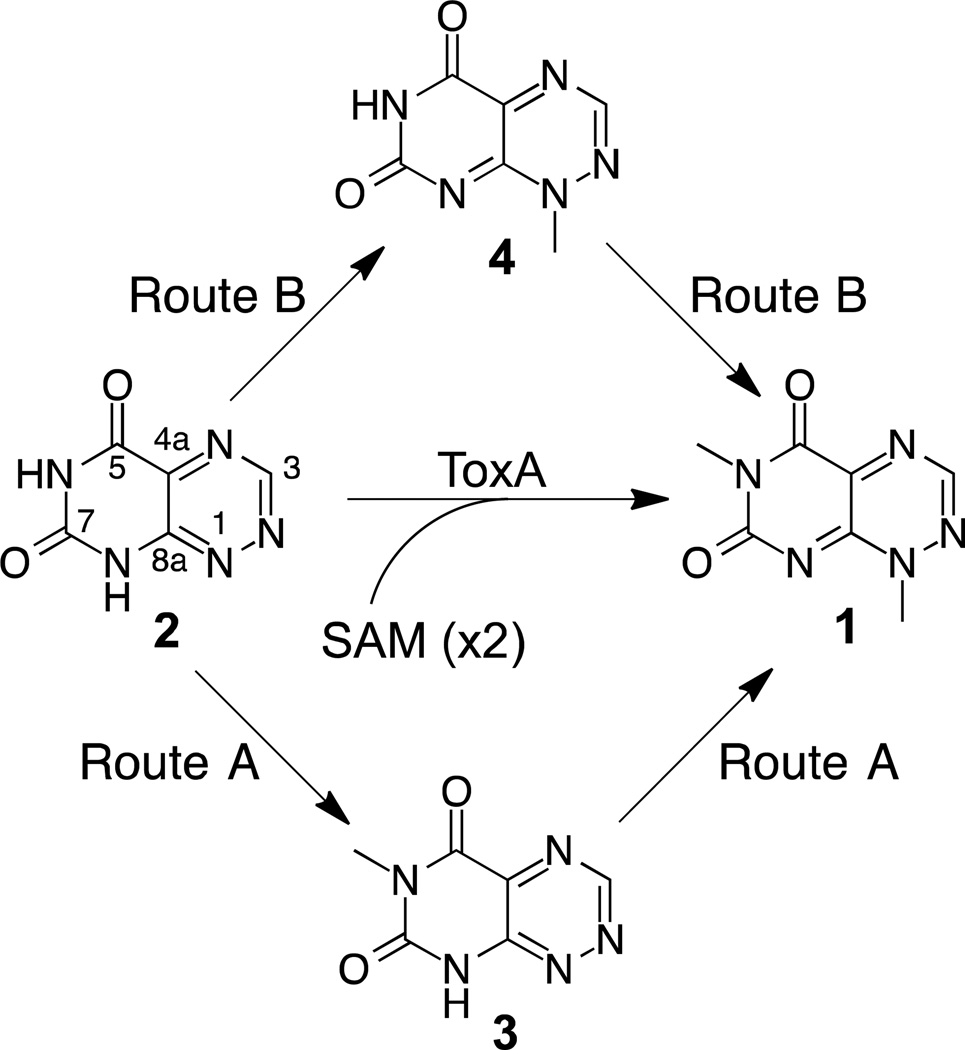
Mutant strains of B. glumae in which toxA is disrupted by insertional mutagenesis are unable to produce toxoflavin.20 In addition, toxA-deficient mutants are unable to induce chlorosis in rice leaves three days after inoculation and induce significantly less chlorosis in rice panicles seven days after inoculation.18 ToxA displays amino acid sequence similarity to Class I S-adenosylmethionine (SAM)-dependent methyltransferases and is the only protein product of the tox operon predicted to have methyltransferase activity.18 Furthermore, since toxoflavin contains two methyl groups derived from SAM,30 ToxA was predicted to catalyze both methylations during toxoflavin biosynthesis.18 The two methyl groups of toxoflavin are located at positions N1 and N6 of an azapteridine ring system and a plausible intermediate in toxoflavin biosynthesis is 1,6-didemethyltoxoflavin (1,6-DDMT).18 Conversion of 1,6-DDMT into toxoflavin by ToxA can be envisioned to proceed via one of three routes (Figure 1): (1) methylation of N6 to generate 1-demethyltoxoflavin (1-DMT, or reumycin) followed by methylation at N1, Route A; (2) methylation of N1 to produce 6-demethyltoxoflavin (6-DMT) followed by methylation at N6, Route B; (3) methylation of N1 or N6 without preference to give both intermediates 1-DMT and 6-DMT.
Figure 1.
Methylation routes for the conversion of 1,6-DDMT 2 to toxoflavin 1.
In the present study, we report biochemical results showing complete conversion of 1,6-DDMT to toxoflavin by ToxA and under some conditions accumulation of 1-DMT, but never 6-DMT, confirming that ToxA catalyzes the final two steps of toxoflavin biosynthesis. We also report crystal structures of complexes of ToxA with substrates and products. The results support sequential methylations in which methylation first takes place at N6 of 1,6-DDMT followed by methylation at N1 of the product 1-DMT. Our structural results also suggest that significant structural changes take place during this conversion, including a major repositioning of 1-DMT required for the second methylation and an order-disorder transition in an N-terminal segment of ToxA that opens and closes the active site. Biochemical and structural studies of ToxA mutations suggest that Tyr7 plays a role in the conformational change.
MATERIALS AND METHODS
ToxA Expression and Purification
ToxA of B. glumae18, 19, 33 and the variants ToxA-Y7A and ToxA-Y7F were expressed from a modified pET-28 vector that supplies an N-terminal fusion tag MGSDKIHHHHHHSSGENLYFQGH.
ToxA, ToxA-Y7A, and ToxA-Y7F were prepared as follows. E. coli BL21(DE3) cells were transformed with the recombinant vector and selected on Luria-Bertani (LB) agar plates supplemented with kanamycin (40 mg/L) after overnight growth at 37 °C. A few colonies containing the recombinant plasmid were inoculated into 50 mL LB medium supplemented with kanamycin (40 mg/L) and grown at 37 °C at 200 rpm for 14–18 h. A portion of the overnight culture (10 mL) was inoculated into shaker flasks containing 1 L of LB medium supplemented with kanamycin (40 mg/L). The cells were grown at 37 °C, 200 rpm until an optical density at 600 nm of 0.5–1.0 was reached (approximately 3 h). The cultures were then cooled to 22 °C for 30 min and then to 15 °C for 1 h. Isopropyl 1-β-d-galactopyranoside was then added to the cultures to a final concentration of 0.1 mM and the flasks were shaken at 15 °C at 200 rpm for 18 h. Following induction, the cells were pelleted and stored at −80 °C. Frozen cells were defrosted at room temperature and then resuspended in lysis buffer (50 mM tris(hydroxymethyl)aminomethane (Tris), 300 mM NaCl, 5 mM imidazole, pH 8.0), lysed via sonication on ice (8 × 30 second rounds consisting of 1.5 s on/off cycles), and subjected to centrifugation at 20,000 × g (4 °C, 20 min). The supernatant was gently mixed with 2 mL Ni-nitrilotriacetic acid resin (pre-equilibrated with lysis buffer) for 30 min on ice. The resin was loaded onto a polypropylene column and allowed to drain. The resin was then washed with 10 mL lysis buffer containing 5 mM β-mercaptoethanol, washed with 4 × 2 mL fractions of wash buffer (50 mM Tris, 300 mM NaCl, 25 mM imidazole, 5 mM β-mercaptoethanol, pH 8.0) and the protein was eluted in 0.5 mL fractions with elution buffer (50 mM Tris, 300 mM NaCl, 250 mM imidazole, 5 mM β-mercaptoethanol, pH 8.0). Fractions were tested for the presence of protein using the Bradford assay and sodium dodecyl sulfate polyacrylamide gel electrophoresis analysis. Fractions containing ToxA were then combined and the protein was desalted into 50 mM Tris, 50 mM NaCl, 10% (v/v) glycerol, 5 mM β-mercaptoethanol, pH 8.0, using an Econo-Pac 10DG column (Bio-Rad, Hercules, CA) according to the manufacturer’s supplied protocol. Fractions containing protein as judged by the Bradford assay were combined, the protein concentration was determined by absorbance at 280 nm and aliquots were flash frozen in liquid nitrogen and stored at −80 °C until used.
ToxA, ToxA-Y7A, and ToxA-Y7F were prepared for crystallization using similar procedures with the following additional steps. After immobilized-nickel affinity chromatography, the sample was subjected to size exclusion chromatography using a HiLoad Superdex 200 26/60 column (GE Healthcare) equilibrated with 15 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES), 125 mM NaCl, 1 mM dithiothreitol (DTT), pH 7.5. ToxA was isolated and then incubated for two days at 4 °C with tobacco etch virus protease to cleave the hexahistidine tag. The reaction mixture was then subjected to subtractive immobilized-nickel affinity chromatography and a second round of size exclusion chromatography, and the purified ToxA was concentrated to approximately 25 mg/mL in either 10 mM sodium citrate and 40 mM NaCl, pH 6.5, 10 mM Tris and 30 mM NaCl, pH 7.5, or 5 mM HEPES and 20 mM NaCl, pH 7.0. Selenomethionine substituted ToxA was prepared similarly except was overexpressed in E. coli B834 cells in minimal medium containing 11.3 g/L M9 salts, 4 g/L dextrose, 50 mg/L L-selenomethionine, 40 mg/L of the remaining 19 L-amino acids, 2 mM MgSO4, 0.1 mM CaCl2, 25 mg/L FeSO4·7H2O, 1 × minimal essential medium vitamin solution, and 30 mg/L kanamycin.
Protein Crystallization
ToxA crystals were grown using the hanging drop vapor diffusion method with drops made from 1:1 mixtures of protein and well solutions. Plate-like crystals of apo ToxA were grown at 18 °C using well solutions containing 19% (w/v) polyethylene glycol (PEG) 3000 and 100 mM sodium citrate, pH 5.5. The cryoprotectant contained an additional 16% (w/v) PEG 3000. Thicker plate-like crystals of ToxA/SAH, ToxA/SAH/1,6-DDMT, ToxA/SAH/toxoflavin, ToxA/SAH/1-DMT, ToxA-Y7A/SAH and ToxA-Y7A/SAH/toxoflavin, thin rod crystals of ToxA/SAH/toxoflavin, ToxA-Y7F/SAH and ToxA-Y7F/SAH/toxoflavin, and hexagonal rod-shaped crystals of ToxA/SAH/1-DMT were grown at 22 °C using well solutions containing 18–23% (w/v) PEG monomethyl ether 2000 and 100 mM Tris, pH 6.1–6.8. The cryoprotectant contained an additional 12–17% (w/v) PEG monomethyl ether 2000. 1,6-DDMT, 1-DMT, toxoflavin, and S-adenosylhomocysteine (SAH) (in dimethylsulfoxide) were added to different ToxA samples for cocrystallization at the final concentrations of approximately 10 mM, 2 mM, 1 mM, and 6 mM, respectively.
Data Collection and Processing
X-ray diffraction experiments were conducted at beamline NE-CAT 24-ID-C of the Advanced Photon Source (APS) and beam line A1 of the Cornell High Energy Synchrotron Source (CHESS). These beamlines were equipped with ADSC Q315 and ADSC Q210 CCD detectors, respectively. Crystals were irradiated at 100 K using X-rays having wavelength λ = 0.98 Å. X-ray diffraction images were collected using 1° oscillations per image and were processed using HKL2000.34 Crystal to detector distances at 24-ID-C ranged from 210 to 300 mm with exposure times of 1.0 to 1.5 s. Crystal to detector distances at A1 ranged from 140 to 180 mm with exposure times of 3 to 9 s. Table 1 summarizes the data collection statistics.
Table 1.
X-ray Data Collection Statistics.
| ToxA SeMet |
ToxA | ToxA SAH |
ToxA SAH DDMT |
ToxA SAH Tox |
ToxA Y7F SAH |
ToxA Y7F SAH Tox |
ToxA Y7A SAH |
ToxA Y7A SAH Tox |
ToxA SAH 1-DMT |
|
|---|---|---|---|---|---|---|---|---|---|---|
| Beamline | 24-ID-C | A1 | 24-ID-C | A1 | 24-ID-C | 24-ID-C | A1 | 24-ID-C | 24-ID-C | A1 |
| λ (Å) | 0.97918 | 0.97700 | 0.97926 | 0.97590 | 0.97918 | 0.97918 | 0.97670 | 0.97918 | 0.97918 | 0.97670 |
| Space group | P1 | C2 | P212121 | P212121 | P21 | P21 | P21 | P212121 | P212121 | P32 |
| a(Å) | 56.5 | 153.0 | 46.8 | 46.9 | 68.7 | 44.9 | 45.2 | 47.1 | 46.5 | 45.5 |
| b (Å) | 60.8 | 56.4 | 65.6 | 78.6 | 45.6 | 71.0 | 71.2 | 67.3 | 66.6 | |
| c (Å) | 81.4 | 60.8 | 144.7 | 144.3 | 74.9 | 77.8 | 78.2 | 144.8 | 145.1 | 96.2 |
| α (°) | 68.3 | - | - | - | - | - | - | - | - | - |
| β(°) | 70.6 | 91.3 | - | - | 99.0 | 100.5 | 100.8 | - | - | - |
| γ (°) | 78.6 | - | - | - | - | - | - | - | - | - |
| Resolution of highest shell (Å) | 1.82–1.76 | 1.63–1.57 | 1.66–1.60 | 1.61–1.55 | 2.07–2.00 | 1.57–1.52 | 1.83–1.77 | 1.87–1.81 | 2.00–1.93 | 1.62–1.56 |
| Reflections | 387,131 | 233,166 | 220,469 | 316,139 | 81,745 | 225,528 | 128,027 | 198,852 | 179,049 | 73,303 |
| Unique reflections | 91,078, 89,746a |
69,075 | 56,573 | 76,116 | 30,077 | 73,380 | 46,227 | 43,444 | 32,710 | 31,081 |
| Redundancy | 4.3 (4.1) b | 3.4 (2.9) | 3.9 (3.5) | 4.2 (4.0) | 2.7 (2.4) | 3.1 (2.8) | 2.8 (2.1) | 4.6 (4.3) | 5.5 (5.5) | 2.4 (1.9) |
| % Complete | 97.1 (91.4) | 95.6 (89.3) | 94.6 (84.8) | 98.0 (100.0) | 96.0 (91.7) | 99.1 (93.9) | 97.2 (92.3) | 99.9 (100.0) | 94.5 (97.5) | 97.9 (89.2) |
| <I>/<σI>c | 18.8 (4.2) | 34.7 (4.1) | 21.4 (4.5) | 17.1 (3.0) | 10.9 (2.2) | 30.0 (9.5) | 16.0 (4.1) | 27.2 (3.4) | 18.0 (3.7) | 34.9 (5.4) |
| Rmerge (%) | 8.8 (30.0) | 2.7 (21.5) | 5.2 (29.6) | 8.1 (45.6) | 9.5 (42.2) | 3.6 (10.9) | 6.3 (16.4) | 6.0 (46.5) | 9.9 (49.5) | 3.6 (16.9) |
Number of Bijvoet pairs
Values in parentheses are associated with the highest resolution shell
Average intensity divided by average error in intensity.
Crystals of apo ToxA belong to space group C2 with 48% solvent content and two monomers of ToxA in the asymmetric unit. Crystals of selenomethionine substituted apo ToxA belong to space group P1 with 44% solvent content, and four monomers of ToxA in the asymmetric unit. Crystals of ToxA/SAH, ToxA/SAH/1,6-DDMT, ToxA-Y7A/SAH, and ToxA-Y7A/SAH/toxoflavin belong to space group P212121 with solvent contents ranging from 39 to 49% and two monomers of ToxA in the asymmetric unit. Crystals of ToxA/SAH/toxoflavin, ToxA-Y7F/SAH, and ToxA-Y7F/SAH/toxoflavin belong to space group P21 with solvent contents ranging from 41 to 45% and two monomers of ToxA in the asymmetric unit. Finally, crystals of ToxA/SAH/1-DMT belong to space group P32 with 41% solvent content and one monomer of ToxA in the asymmetric unit.
Structure Determination and Refinement
The structure of selenomethionine substituted ToxA was determined using single wavelength anomalous diffraction phasing. The selenium substructure was determined using SHELXD35, 36 and more than 90% of protein residues were built using the Autosol module of PHENIX.37 ToxA structures with other space groups were determined by molecular replacement using MOLREP38 or PHENIX. Automated structural refinement was performed using PHENIX and model building was performed using COOT.39Later rounds of refinement took into account translation, libration, and screw vibrational motion of partitioned chains40 of ToxA using the TLS module in PHENIX. Difference electron density (Fo-Fc) for each ligand is shown in Supplementary Figure 1.
Synthesis of 1,6-DDMT [Pyrimido[5,4-e]-as-Triazine-5,7(6H,8H)-dione]
The synthesis of 1,6-DDMT was performed as previously described41 with the exception that triphosgene (0.3 eq) was substituted for phosgene gas. The product was then purified using an Agilent 1200 HPLC instrument, which consisted of a quaternary pump, vacuum degasser, autosampler, column thermostat (set to 30 °C), diode array detector, fluorescence detector, and fraction collector. Separation of reaction components was achieved using an SPLC-18-DB column (10 × 250 mm, 5 µm, Supelco, Bellefonte, PA) with a flow rate of 2 mL/min where line A was water, line C was methanol, and line D was 10 mM ammonium acetate with the following program. The column was pre-equilibrated in 90% A/10% D and upon injection this composition was held for 2 min. The composition of mobile phase was then changed to 55% A/35% C/10% D over 8 min utilizing a linear gradient followed by changing to 40% A/50% C/10% D over the next 5 min. The composition of mobile phase was then changed to 10% A/80% C/10% D over 2 min and this composition was held for 5 min followed by changing to 90% A/10% D over 2 min. The column was equilibrated in 90% A/10% D for 12 min prior to the next injection.
Synthesis of 1-DMT
1-DMT was synthesized as previously described.42
ToxA and ToxA Variant In Vitro Activity Assays
For the full conversion of 1,6-DDMT to toxoflavin, reaction mixtures consisted of 100 mM Tris, pH 7.5, 1 mM SAM, 1 mM DTT, and 100 µM 1,6-DDMT in a total volume of 90 µL. Reactions were initiated by the addition of 10 µL ToxA (15 µM final concentration) with gentle mixing, followed by incubation at room temperature for 1 h. The proteins were then removed using a PALL protein concentrator (10,000 MWCO, 14,000 × g, 60 min, 4 °C). A portion of the reaction mixture (25 µL) was then analyzed by HPLC as described below.
For the conversion of 1,6-DDMT to 1-DMT and toxoflavin (incomplete reaction), reaction mixtures consisted of 100 mM Tris, pH 7.5, 1 mM SAM, 1 mM DTT, and 100 µM 1,6-DDMT in a total volume of 90 µL. Reactions were initiated by the addition of 10 µL ToxA (1.5 µM final concentration) with gentle mixing, followed by incubation at room temperature for 1 h, followed by the addition of 30 µL 1 M potassium citrate, pH 5.0, or 1 M Tris, pH 7.5. The reactions were mixed by pipetting gently and allowed to stand at room temperature for an additional 30 min. The proteins were then removed using a PALL protein concentrator (10,000 MWCO, 14,000 × g, 60 min, 4 °C) followed by incubation at room temperature for 2–4 h. A portion of the reaction mixture (60 µL) was then analyzed by HPLC as described below. To investigate the conversion of 1-DMT to toxoflavin, an identical reaction substituting 1-DMT for 1,6-DDMT was used. An identical procedure was also used for the ToxA variant activity assays.
HPLC Analysis of In Vitro Activity Assays
HPLC analyses were performed using an Agilent 1200 HPLC instrument, which consisted of a quaternary pump, vacuum degasser, autosampler, column thermostat (set to 30 °C), diode array detector and fluorescence detector. Separation of reaction components was achieved using an Eclipse XDB-C18 column (4.6 × 150 mm, 5 µm, Agilent, Santa Clara, CA) with a flow rate of 1 mL/min where line A was water, line C was methanol and line D was 10 mM ammonium acetate with the following program. The column was pre-equilibrated in 90% A/10% D and upon injection this composition was held for 2 min. The composition of mobile phase was then changed to 66% A/24% C/10% D over 10 min utilizing a linear gradient followed by changing to 25% A/65% C/10% D over the next 1.5 min. This composition was held for 5 min followed by changing to 90% A/10% D over 1 min. The column was equilibrated in 90% A/10% D for 6 min prior to the next injection. Data were processed with ChemStation ver. B.04.01 SP1 (Agilent Technologies).
Figure Preparation
Illustrations were made using Chimera43 and ChemDraw (Perkin Elmer).
RESULTS
Conversion of 1,6-DDMT and 1-DMT into Toxoflavin by ToxA
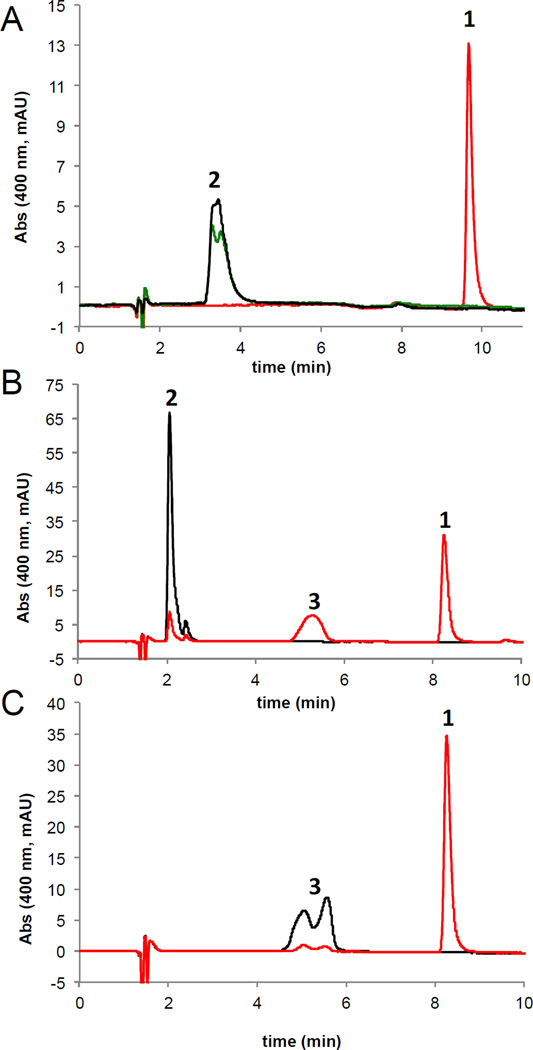
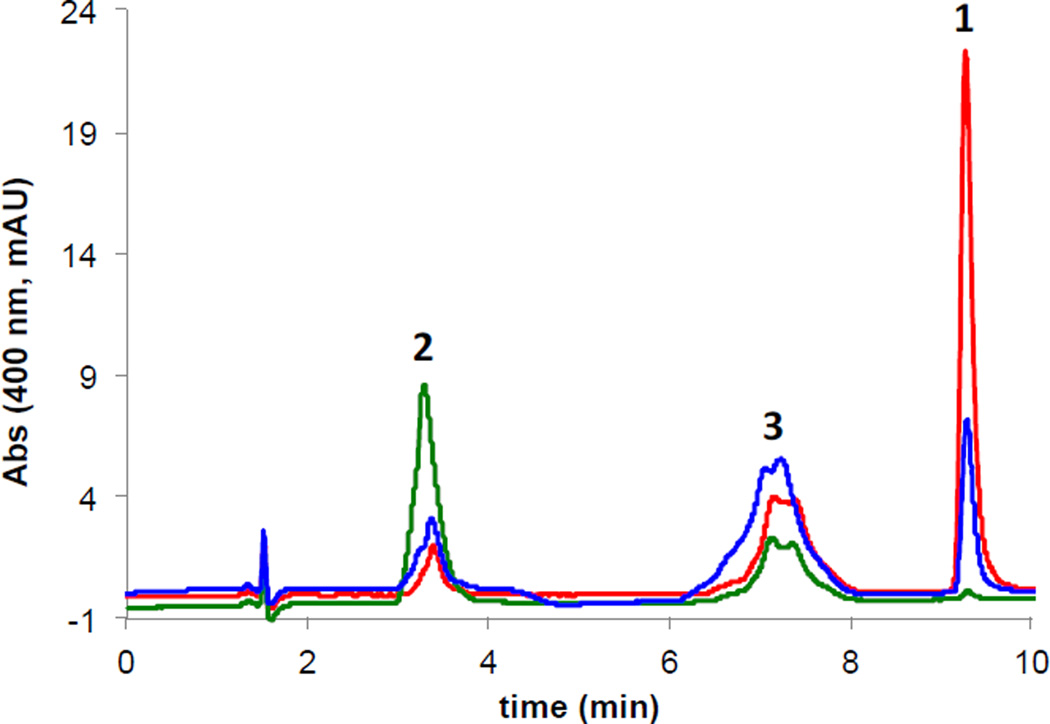
Incubation of 1,6-DDMT with ToxA and SAM resulted in clean conversion to toxoflavin (Figure 2A). Examination of incomplete reaction mixtures allowed us to observe an intense peak at 5.5 min when the reaction was performed at pH 7.5 (Figure 2B). This compound could be shifted to 7 min when the reaction was quenched with potassium citrate, pH 5.0, suggesting a structure capable of conversion between tautomer forms. We confirmed the identity of this compound as 1-DMT by co-injection with a synthetic standard. Incubation of 1-DMT with ToxA led to efficient conversion to toxoflavin (Figure 2C). Under no conditions were we able to observe a second peak in the reaction mixture that would correspond to 6-DMT.
Figure 2.
HPLC analysis of in vitro ToxA reactions monitored at 400 nm. (A) Full conversion of 1,6-DDMT 2 to toxoflavin. Red trace: reaction containing 1,6-DDMT 2, SAM and ToxA (15 µM). Green trace: reaction containing 1,6-DDMT 2 and ToxA (15 µM). Black trace: reaction containing 1,6-DDMT 2 and SAM. (B) Chromatogram of a reaction quenched before completion showing the accumulation of 1-DMT 3. Red trace: reaction containing 1,6-DDMT 2, SAM and ToxA (1.5 µM). Black trace: reaction containing 1,6-DDMT 2 and SAM. (C) Chromatogram of a reaction showing the conversion of 1-DMT 3 to toxoflavin 1. Red trace: reaction containing 1-DMT 3, SAM and ToxA (1.5 µM). Black trace: reaction containing 1-DMT 3 and SAM. The peak for 1-DMT 3 is broadened/doubled due to a mixture of tautomers.
Overall Structure of ToxA
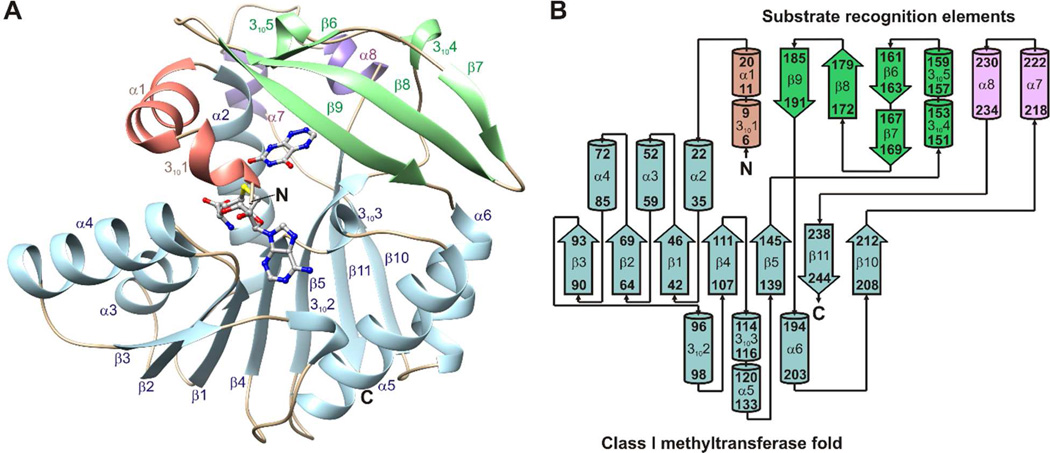
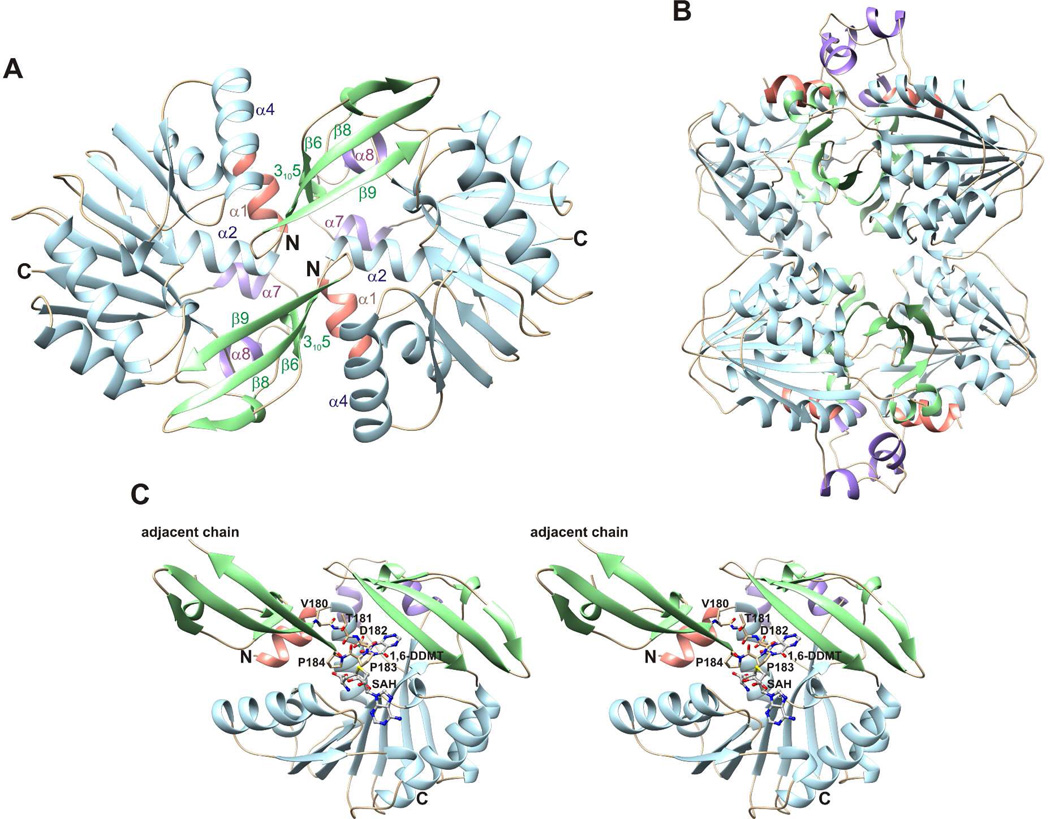
Crystal structures were determined for ToxA and ToxA variants with different combinations of bound substrates and products (Table 1). The structures of ToxA display a canonical Class I methyltransferase fold with three extensions: an N-terminal segment that contains a 310 helix followed by a 10-residue α-helix, a flap domain between β5 and α6 that contains a three-stranded antiparallel β-sheet and a 15 residue segment having two 310 helices, and a 25-residue segment between β10 and β11 that contains two 5-residue α-helices (Figure 3).
Figure 3.
Structure and topology of ToxA. (A) The crystal structure of ToxA displays a canonical Class I methyltransferase fold (light blue) and three substrate recognition elements: an N-terminal segment (salmon), a flap domain (green), and a 25 residue segment connecting β10 and β11 (purple). SAH and 1,6-DDMT are shown as balls and sticks. (B) ToxA topology diagram. β-strands are represented with thick arrows and α- and 310 helices are represented with cylinders.
Unliganded ToxA forms a homodimer with extensive contacts in two different crystal forms (triclinic and monoclinic) (Figure 4A). Two dimers further join to form a homotetramer with 222 symmetry (Figure 4B), and this tetramer is predicted to be stable in solution by the PISA website.44 After complex formation, the tetramer is disrupted by large conformational changes. In the dimer, residues 180–184 (VTDPP) insert intothe substrate-binding site and prevent the N-terminal segment from folding over the active site (Figure 4C). While some complexes contain two monomers in the asymmetric unit, no assembly is predicted to be stable in solution by PISA. Furthermore, each monomer contains a complete active site. Therefore we conclude that the complexes are most likely to be monomeric.
Figure 4.
Crystal structure of apo ToxA. (A) Apo ToxA forms a homodimer having noncrystallographic twofold symmetry. (B) Homotetramic arrangement of apo ToxA observed within the crystal packing. Two dimers having the structure shown in panel A form a homotetramer with 222 symmetry. (C) A flexible loop (Val180-Thr181-Asp182-Pro183-Pro184) from the adjacent chain inserts into the azapteridine binding site and prevents the N-terminal segment from folding over the active site. 1,6-DDMT and SAH are superimposed onto the apo structure to indicate potential steric clashes.
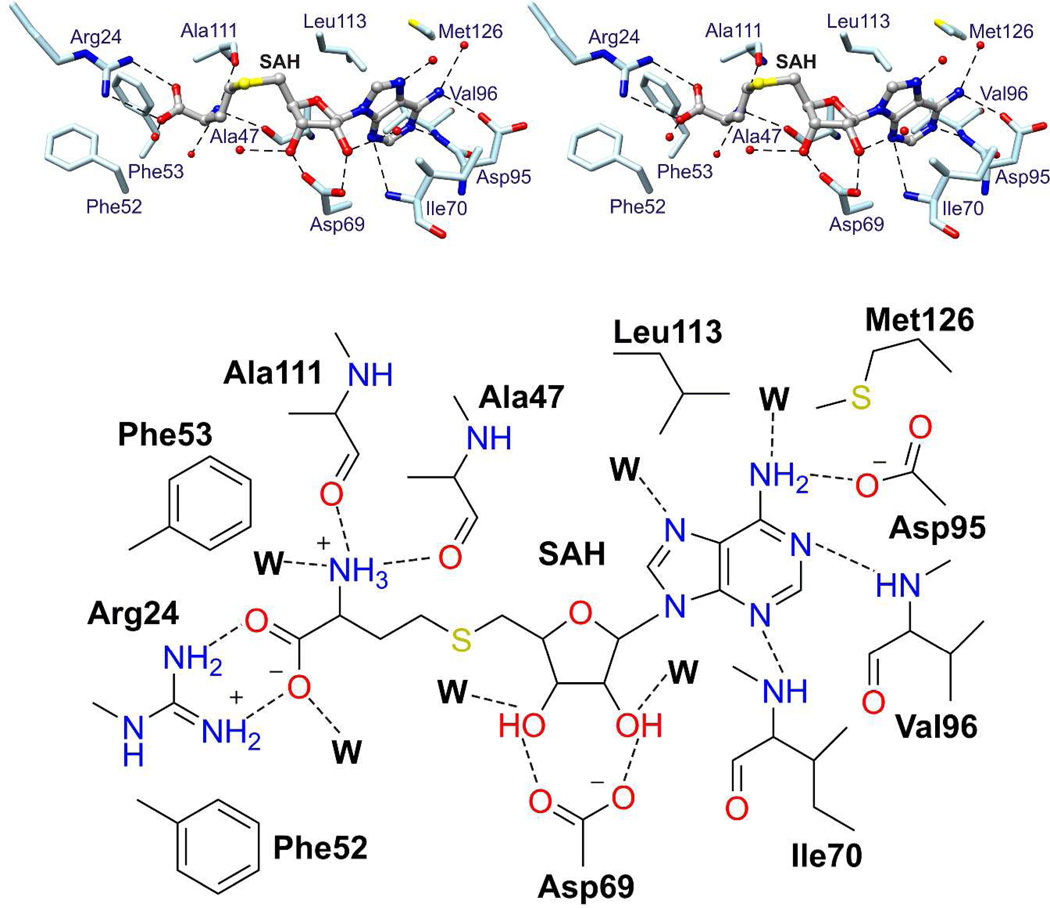
SAH/SAM Binding Site
SAH binds in a groove near the C-terminal ends of β1 - β4 (Figure 5). The carboxylate group of SAH forms a salt bridge with Arg24. The amino group of homocysteine hydrogen bonds to the backbone carbonyls of Ala47 and Ala111. The ribose hydroxyl groups hydrogen bond to the carboxylate of Asp69. The adenine moiety is sandwiched between Leu113 and Ile70 and its amino group hydrogen bonds to the carboxylate of Asp95. Finally, the adenine N1 atom hydrogen bonds to the amide NH of Val96 and the adenine N3 atom hydrogen bonds to the amide NH of Ile70.
Figure 5.
SAH binding site. Stereo diagram (top) and schematic drawing (bottom) of the SAH binding site based on the crystal structure of ToxA/SAH. SAH interacts with residues of the Class I methyltransferase fold (light blue sticks). Waters are shown as red spheres (in stereo diagram) or W (in schematic drawing), and broken lines indicate potential hydrogen bonds.
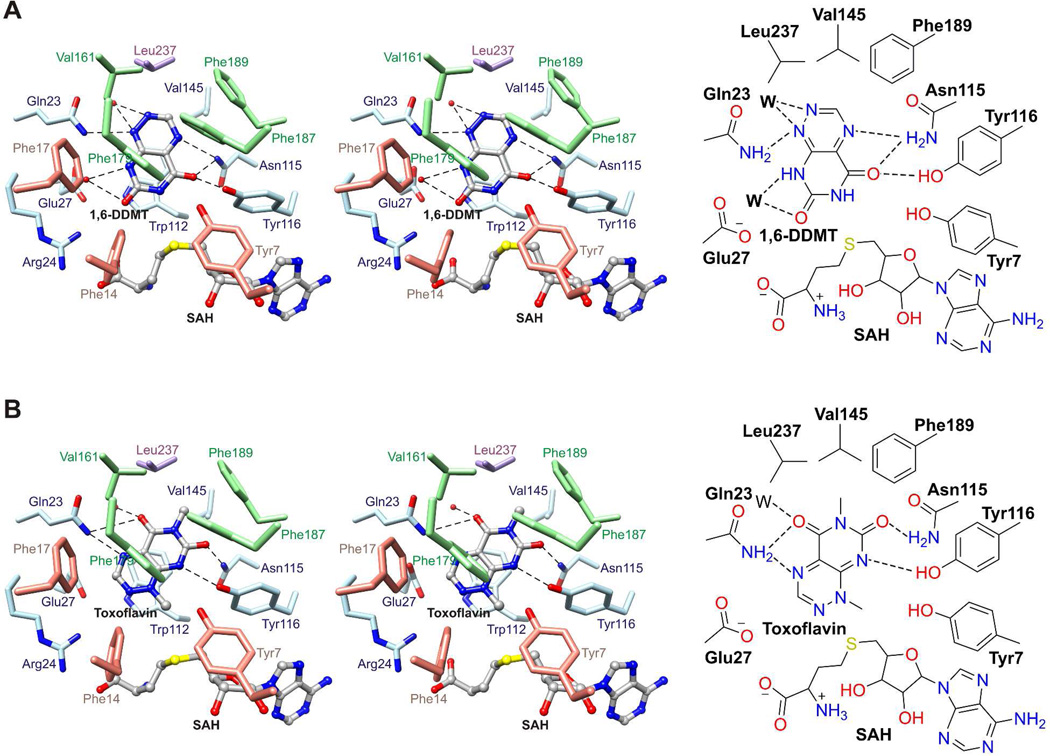
1,6-DDMT and Toxoflavin Binding Sites
1,6-DDMT and toxoflavin bind in a deep slot between the Class I methyltransferase domain and the flap domain. The two azapteridines are coplanar but rotated by about 140° with respect to each other. The azapteridine ring system is stacked face-to-face with the side chain of Trp112 and face-to-edge with the side chains of Phe179 and Phe187. 1,6-DDMT binds in an orientation that positions N6 3.6 Å away from Sδ of SAH in an alignment that would be favorable for methyl transfer from SAM (Figure 6A). N1 hydrogen bonds to the side chain amide of Gln23 and O5 hydrogen bonds to the side chain amide of Asn115 and the hydroxyl of Tyr116. In contrast, toxoflavin binds in an orientation that positions N1 4.5 Å away from Sδ of SAH in an alignment that would also be favorable for methyl transfer from SAM (Figure 6B). N8 hydrogen bonds to the hydroxyl group of Tyr116 and the N6 methyl group packs in a hydrophobic pocket formed by Val145, Phe189, and Leu237. DFT calculations (for the ligand in H2O) and the ToxA structure indicate that the aromatized tautomer of 1-DMT 3 is the stable tautomer (Figure 1).
Figure 6.
1,6-DDMT and toxoflavin binding sites. (A) Stereo structure (top) and schematic diagram (bottom) of the 1,6-DDMT binding site in the structure of ToxA/SAH/1,6-DDMT. (B) Stereo structure (top) and schematic diagram (bottom) of the toxoflavin binding site in the structure of ToxA/SAH/toxoflavin. 1,6-DDMT, toxoflavin, and SAH are represented with balls and sticks. Water molecules are represented with red spheres (in stereo diagrams) or W (in schematic drawings), and broken lines indicate potential hydrogen bonds.
1-DMT Binding Site
1-DMT binds in the same pocket as 1,6-DDMT and toxoflavin and is coplanar with both. However, the structure of ToxA/SAH/1-DMT shows 1-DMT in an orientation that is incompatible with a methyl transfer to a ring nitrogen atom (Supplementary Figure 2). Instead, C3 is nearest to Sδ of SAH with a separation distance of 3.5 Å. The C3 hydrogen atom would clash with the methyl group of SAM if SAM were present at the SAH site suggesting that in the presence of SAM the orientation of 1-DMT would be different (Supplementary Figure 3).
N-terminal Segment
We observe two types of monomeric wild-type ToxA structures: one in which the N-terminus is disordered and one in which residues 7–23, which make up a portion of the active site, are ordered. The ordered structure is observed for ToxA/SAH/1,6-DDMT and ToxA/SAH/toxoflavin (Figure 3A, Figure 6, and Table 3). In contrast, the structure of ToxA/SAH shows a poorly ordered N-terminal segment; one chain in the asymmetric unit shows complete disorder in this region and the other chain shows weak ordering (Table 3). In the ordered conformation, the side chain of Tyr7 packs near C3′ and C5′ of SAH within a cluster of hydrophobic residues and its hydroxyl group hydrogen bonds to the hydroxyl group of Tyr116; the hydroxyl group of Tyr7 is located 3.5–3.8 Å from Sδ of SAH. Phe14 interacts with the homocysteine portion of SAH and Phe17 interacts via π stacking with Phe179, which in turn interacts via π-stacking with Phe187.
Table 3.
Ordered State of the N-terminal Segment in ToxA Crystal Structures.
| Crystal | Chain | N-terminal segment | B-factorsa Residues 7–21 |
B-factorsa Residues 24–245 |
|---|---|---|---|---|
| ToxA/SAH | A B |
Poorly ordered Disordered |
66.7 - |
22.0 22.9 |
| ToxA/SAH/1,6DDMT | A B |
Ordered Ordered |
40.4 18.8 |
24.2 15.8 |
| ToxA/SAH/Toxoflavin | A B |
Ordered Ordered |
15.6 28.5 |
15.5 16.1 |
| ToxA-Y7F/SAH | A B |
Ordered Ordered |
6.1 7.3 |
6.0 5.5 |
| ToxA-Y7F/SAH/Toxoflavin | A B |
Ordered Ordered |
6.6 9.2 |
6.0 5.8 |
| ToxA-Y7A/SAH | A B |
Disordered Disordered |
- - |
27.4 27.5 |
| ToxA-Y7A/SAH/Toxoflavin | A B |
Disordered Disordered |
- - |
24.7 22.9 |
Average B-factor of main chain atoms.
ToxA-Y7F/SAH and ToxA-Y7F/SAH/toxoflavin show a highly ordered N-terminal segment similar to the ordered wild-type ToxA structures. For the toxoflavin complex, the electron density was strong for both SAH and toxoflavin, with toxoflavin oriented with N1 nearest to Sδ of SAH. ToxA-Y7A/SAH and ToxA-Y7A/SAH/toxoflavin show a disordered N-terminal segment. For the toxoflavin complex, the electron density was clear for SAH, but less distinct for toxoflavin (Supplementary Figure 1).
Biochemical Analysis of ToxA-Y7A and ToxA-Y7F
Activity assays using 1,6-DDMT as the substrate are shown in Figure 7. After 1 h total reaction time with 1.5 µM enzyme and 100 µM 1,6-DDMT, ToxA-Y7F produced approximately 55% more 1-DMT and 67% less toxoflavin compared with wild-type ToxA; ToxA-Y7A produced approximately 65% of 1-DMT and 2% of the toxoflavin produced by the wild-type enzyme.
Figure 7.
HPLC analysis of ToxA variant reactions. ToxA-red, ToxA-Y7A–green, ToxA-Y7F–blue. Reactions consisted of SAM, 1,6-DDMT 2, DTT and enzyme (1.5 µM) in pH 7.5 buffer. The peak for DMT 3 is broadened/doubled due to a mixture of tautomers.
DISCUSSION
Comparison of the Structure of ToxA with Other SAM-dependent Methyltransferases
ToxA contains a canonical Class I methyltransferase domain that has a conserved SAH/SAM binding site45, 46 and SAH/SAM conformation.47 A search for structural homologues using DALI48 showed that ToxA has unique substrate recognition elements, a hallmark of Class I methyltransferases, but which resemble ones in the methyltransferase CcbJ (PDB ID 4HH4),49the N,N-dimethyltransferase TylM1 (PDB ID 3PFG),50 mRNA cap (guanine N7) methyltransferases (PDB IDs 1RI3 and 2VDW),51, 52 and glycine N-methyltransferase (PDB code 1NBH).53 Like ToxA, these structures contain a flap domain that packs against the Class I methyltransferase domain to form the active site. Apo ToxA crystallized in a distinct homodimeric structure in which each flap domain inserts into the active site of a two-fold related monomer.
Dual Substrate Specificity of ToxA
The dual methylation of 1,6-DDMT 2 to form toxoflavin 1 by ToxA can be envisioned to proceed via one of three possible pathways (Figure 1). Our biochemical studies show that ToxA converts 1,6-DDMT 2 to toxoflavin 1 and that under some conditions 1-DMT 3 is produced but never 6-DMT 4. Furthermore, ToxA converts 1-DMT to toxoflavin. These results show that ToxA from B. glumae has dual specificity and catalyzes the last two steps in the biosynthesis of toxoflavin.
The structure of ToxA/SAH/1,6-DDMT is consistent with initial methylation at N6 (Route A). While we cannot exclude the possibility that the initial methylation occurs at N1 (Route B), the crystal structure with bound 1,6-DDMT suggests that the added methyl group would clash with the side chain of Gln23. Furthermore, the structure of ToxA/SAH/toxoflavin places the N1 methyl group nearest to Sδ of SAH consistent with the predicted order; however, the structure of ToxA/SAH/1-DMT places C3 nearest to Sδ of SAH. A model of ToxA/SAM/1-DMT based on the structure of ToxA/SAH/toxoflavin shows a plausible substrate arrangement for methyl transfer, with a 3.0 Å separation between Cε of SAM and N1 of 1-DMT and an Sδ-Cε…N1 angle of 155°. Furthermore, a model of ToxA/SAM/1-DMT based on the structure of ToxA/SAH/1-DMT shows a steric clash between the C3 hydrogen of 1-DMT and the SAM methyl group (Supplementary Figure 3). This result suggests that 1-DMT could partially reorder after methylation, but before product release.
Reorientation of 1-DMT
Toxoflavin biosynthesis by ToxA requires a 140° rotation of 1-DMT after 1,6-DDMT is methylated at N6. Dissociation of newly generated 1-DMT followed by binding to ToxA/SAM would prohibit 1-DMT binding in the 1,6-DDMT orientation due to clashes between the SAM methyl group and the newly added methyl group at N6. Although ToxA with bound SAH binds 1-DMT and toxoflavin differently, the methyl group at N6 packs in a hydrophobic pocket formed by Val145, Phe189, and Leu237 for both azapteridines. These results suggest that the additional hydrophobic interactions that become available after N6 methylation contribute to the reorientation of 1-DMT.
Role of N-terminal Segment
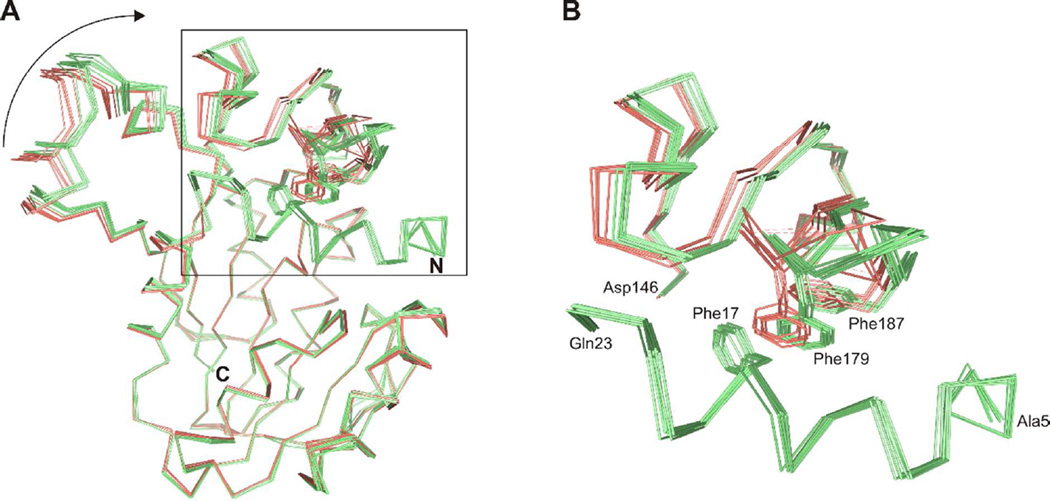
The ordered and disordered states of the N-terminal segment of ToxA are associated with a 1–2 Å difference in the positioning of the substrate recognition domain relative to the Class I methyltransferase domain (Figure 8A). In the disordered state, residues 181–183 in the flap domain are also poorly ordered. In the ordered state, primary and secondary interactions form between the N-terminal segment and the flap domain or azapteridine. First, Ile10, Phe14, and Phe17 interact with Phe179 within a large cluster of hydrophobic residues. The side chain of Phe17 interacts face-to-face with the side chain of Phe179 (Figure 8B), which, along with the side chain of Phe187, interacts edge-to-face with the bound azapteridine. Second, the side chain of Gln23 hydrogen bonds with the bound azapteridine (Figure 6A, B). Gln23 is located in helix α2 and the ordering of the N-terminal part of this helix is correlated with the ordering of the preceding residues. Finally, the ordered N-terminal segment places Tyr7 in the active site near the site of methyl transfer (Figure 6A, B). The hydroxyl group of Tyr7 packs against the ribose moiety of SAH and its hydroxyl group hydrogen bonds to the hydroxyl group of Tyr116, which interacts with the bound azapteridine.
Figure 8.
Structural change in ToxA after ordering of the N-terminal segment. (A) Cα traces of ToxA structures with an ordered (light green) or disordered (salmon) N-terminal segment after superimposition of the Class I methyltransferase domain. Phe17 in the N-terminal segment and Phe179 and Phe187 in the flap domain are also shown. The arrow highlights the displacement in the substrate recognition domain. (B) Close-up view of hydrophobic interactions between the N-terminal segment and flap domain. The position of Phe179 in the ToxA structures having a disordered N-terminal segment would lead to a steric clash with Phe17.
Tyr7 anchors the tail of the N-terminal segment to the active site and plays a role in orienting the azapteridine. Structures of ToxA-Y7A show a disordered N-terminal segment in both the presence and absence of bound toxoflavin and the electron density of the bound toxoflavin is indistinct and does not show a productive orientation. In contrast, structures of ToxA-Y7F show a highly ordered N-terminal segment in the presence and absence of bound toxoflavin, and toxoflavin is well-ordered and oriented in agreement with the expected methyl transfer (Supplementary Figure 1).
The functional importance of the N-terminus, in particular Tyr7, was probed by in vitro characterization of ToxA-Y7F and ToxA-Y7A. ToxA-Y7F showed modest reductions in activity when incubated with either 1,6-DDMT or 1-DMT when compared to wild-type ToxA. In contrast, ToxA-Y7A showed large reductions catalyzing the conversion of 1,6-DDMT to 1-DMT and 1-DMT to toxoflavin supporting the structural results suggesting that Tyr7 is important for azapteridine binding.
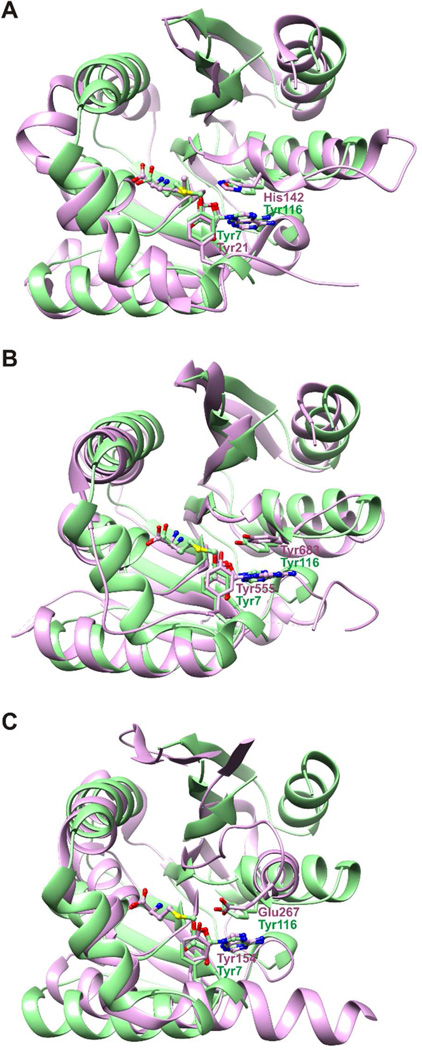
Intriguingly, similar arrangements involving a tyrosine at an equivalent active site as Tyr7 in ToxA are observed in the structures of other methyltransferases. Structural superimpositions show that Tyr7 and Tyr116 of ToxA occupy similar active site locations as Tyr21 and His142 in glycine N-methyltransferase (PDB code 1NBH),53 Tyr555 and Tyr683 in vaccinia virus mRNA cap (guanine N7) methyltransferase (PDB code 2VDW),51, 54 and Tyr154 and Glu267 in Type I protein arginine methyltransferases (PDB codes 3B3F and 2V74)55, 56(Figure 9). In each case, the tyrosine equivalent to Tyr7 in ToxA resides on a mobile structural element that displays order or disorder in different crystal structures. Mutation of these tyrosines leads to modest to complete loss of activity and in particular mutation to phenylalanine or alanine in vaccinia virus mRNA cap methyltransferase shows trends similar to ToxA51, 53, 54, 57.
Figure 9.
Tyr7 of ToxA occupies a structurally conserved tyrosine site in various methyltransferases. Superimposition of ToxA (green) onto (A) glycine N-methyltransferase (purple), (B) vaccinia virus mRNA cap methyltransferase (purple), and (C) a Type I protein arginine methyltransferase (purple). For clarity, the substrate recognition domains are not shown. Superimpositions were based on SAH/SAM coordinates. SAH and SAM are represented with balls and sticks.
Mechanistic Considerations
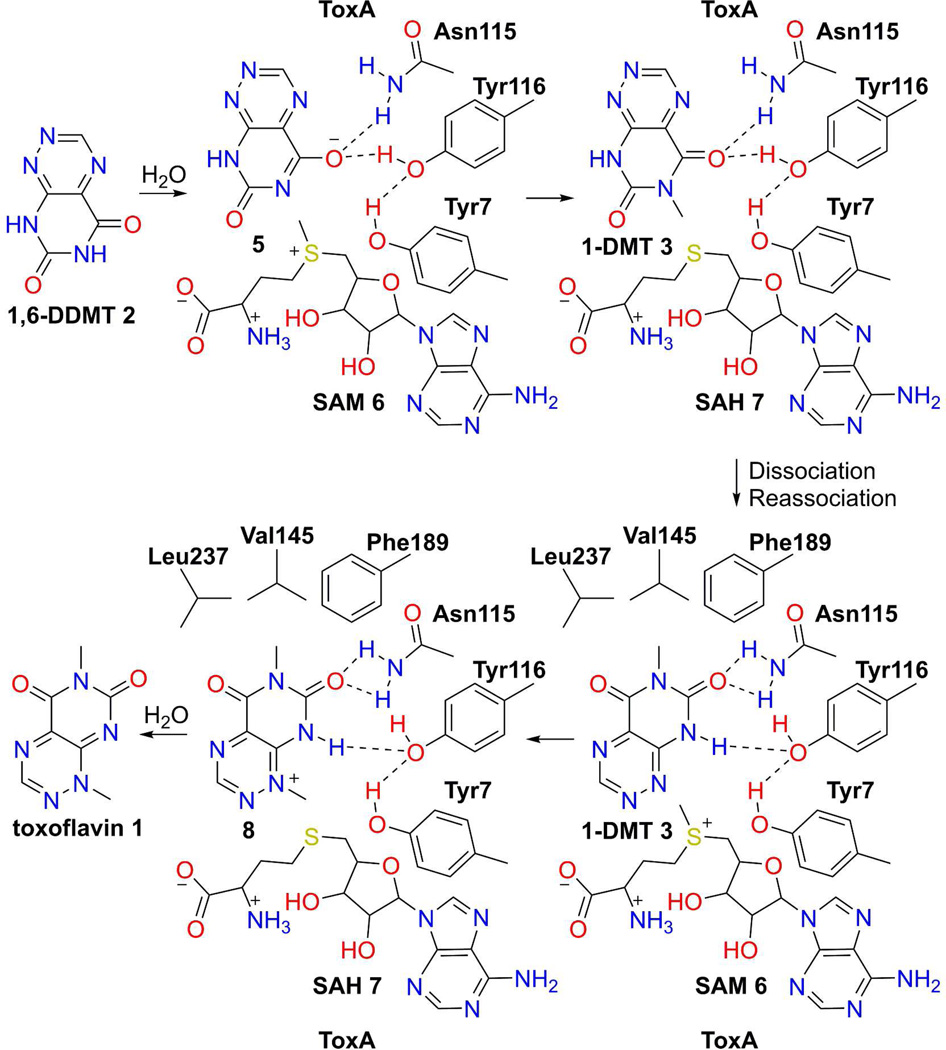
The structures suggest a mechanism for conversion of 1,6-DDMT 2 to toxoflavin 1 by ToxA (Figure 10). There is no obvious base close to N6 in the ternary complex so it is possible that either 1,6-DDMT 2 undergoes deprotonation prior to binding or that protonation and deprotonation occur via a dynamic water channel (Figure 11A). The resulting substrate anion is stabilized by hydrogen bonding to Asn115 and Tyr116. SAM binding, methyl transfer and product dissociation complete the first phase of toxoflavin formation. A base is not required for the second methyl transfer because N1 is nucleophilic with its lone pair oriented directly at the methyl group of SAM (Figure 11B). Overall, the active site of ToxA is relatively simple. It binds N6 and N1 of the substrate iteratively in close proximity to the methyl group of SAM but does not provide a base to assist the methyl transfer reactions. The order of the methylation reactions may be due to the greater nucleophilicity of deprotonated N6 over N1. This relative reactivity is also mirrored in the ease of demethylation of toxoflavin at the N1 position.
Figure 10.
Mechanistic proposal of the ToxA-catalyzed methylation of 1,6-DDMT 2 to toxoflavin 1 via 1-DMT 3. Either deprotonated 1,6-DDMT 5 binds to ToxA/SAM 6 or 1,6-DDMT 2 is deprotonated via a dynamic water channel. Oxyanion stabilization is provided by Tyr116 and Asn115. N6 methylation by SAM 6 makes 1-DMT 3 and SAH 7. For N1 methylation, 1-DMT 3 dissociates from ToxA and then reassociates with ToxA in a new orientation with SAM bound. N1 is positioned favorably for methylation and the N6 methyl is buried in a hydrophobic pocket. N1 of 1-DMT 3 is not protonated so no base is required for the second methylation that leads to toxoflavin formation.
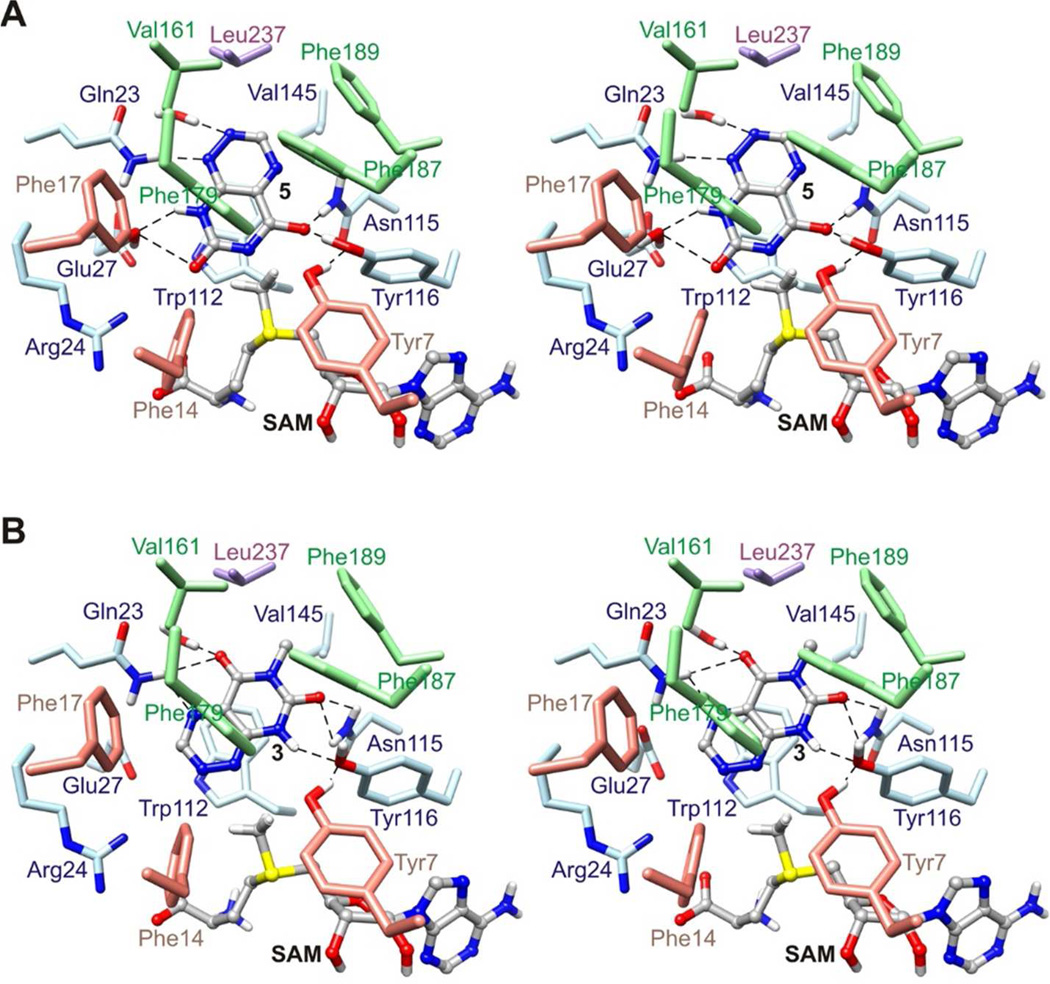
Figure 11.
Structural models of SAM-bound ToxA complexes in the reaction pathway proposed in Figure 10. (A) Stereo structure of ToxA with bound SAM and deprotonated 1,6-DDMT 5. (B) Stereo structure of ToxA with bound SAM and 1-DMT 3, with 1-DMT 3 in an orientation favorable for N1 methylation. SAM was modeled by adding a methyl group to SAH. Deprotonated 1,6-DDMT 5, 1-DMT 3, and SAM are represented with balls and sticks. Broken lines indicate potential hydrogen bonds.
Formation of 1-DMT as a Discrete Intermediate on the Way to Toxoflavin
Our in vitro analysis identified 1-DMT (reumycin) as a discrete intermediate but not 6-DMT. This could be viewed as an adaptation by B. glumae to scavenge 1-DMT formed during chlorosis as a possible defense mechanism by the infected plant. The N1 methyl group is known to be labile42 and it is plausible that an infected plant could detoxify toxoflavin by simply removing the N1 methyl group through use of a nucleophile (e.g. glutathione). If the biosynthesis proceeded through 6-DMT, then removal of the methyl group at N1 would result in the accumulation of a less toxic shunt metabolite. By utilizing 1-DMT as an intermediate, any toxoflavin that is converted to 1-DMT either through the action of plant enzymes or non-enzymatic hydrolysis could be recovered back to toxoflavin, saving B. glumae significant metabolic resources.
Supplementary Material
Table 2.
Structure Refinement Statistics.
| ToxA SeMet |
ToxA | ToxA SAH |
ToxA SAH DDMT |
ToxA SAH Tox |
ToxA Y7F SAH |
ToxA Y7F SAH Tox |
ToxA Y7A SAH |
ToxA Y7A SAH Tox |
ToxA SAH 1-DMT |
|
|---|---|---|---|---|---|---|---|---|---|---|
| No. of reflections | 182,110 | 69,067 | 56,500 | 75,116 | 30,058 | 73,355 | 46,170 | 43,336 | 32,644 | 31,048 |
| No. of reflections in working set |
172,760 | 65,670 | 53,735 | 71,447 | 28,561 | 69,745 | 43,859 | 41,214 | 31,057 | 29,533 |
| Resolution (Å) | 45.0–1.76 | 19.4–1.57 | 39.3–1.60 | 17.7–1.55 | 38.8–2.0 | 29.1–1.52 | 19.8–1.77 | 34.0–1.79 | 38.1–1.93 | 18.6–1.56 |
| No. of protein atoms | 7,548 | 3,801 | 3,661 | 3,854 | 3,877 | 3,979 | 3,913 | 3,539 | 3,512 | 2,016 |
| No. of waters | 1,059 | 622 | 436 | 573 | 431 | 848 | 770 | 397 | 212 | 270 |
| No. of ligand atoms | - | - | 52 | 76 | 80 | 72 | 88 | 52 | 80 | 39 |
| RMSD bonds (Å) | 0.007 | 0.006 | 0.006 | 0.006 | 0.007 | 0.007 | 0.007 | 0.007 | 0.006 | 0.008 |
| RMSD angles (deg) | 1.022 | 1.066 | 1.041 | 1.051 | 1.071 | 1.139 | 1.089 | 1.029 | 1.010 | 1.164 |
| Rwork (%) | 16.4 | 16.1 | 18.0 | 21.6 | 15.1 | 13.4 | 15.4 | 17.6 | 17.4 | 15.3 |
| Rfree (%) | 20.1 | 18.5 | 21.7 | 25.2 | 20.5 | 15.9 | 19.9 | 21.3 | 23.1 | 17.6 |
| Ramachandran analysis | ||||||||||
| Most favored (%) | 93.8 | 92.4 | 93.2 | 94.5 | 93.2 | 94.0 | 93.3 | 94.8 | 93.7 | 94.8 |
| Additional allowed (%) | 6.2 | 7.6 | 6.8 | 5.5 | 6.8 | 6.0 | 6.7 | 5.2 | 6.3 | 5.2 |
Acknowledgments
Funding
This work was supported by NIH grant GM73220 (to SEE) and by the Robert A. Welch Foundation (A-0034 to TPB). This work is based upon research conducted at the Advanced Photon Source on the Northeastern Collaborative Access Team beamlines, which are supported by award GM103403 from the NIH. Use of the Advanced Photon Source is supported by the U.S. Department of Energy, Office of Basic Energy Sciences, under Contract No. DE-AC02-06CH11357. MacCHESS is supported by NIH grant GM103485 at the Cornell High Energy Synchrotron Source.
We thank the staff of NE-CAT at the Advanced Photon Source and the staff of the Cornell High Energy Synchrotron Source for assistance with data collection, Dr. Cynthia Kinsland for cloning ToxA, ToxA-Y7A, and ToxA-Y7F, and Leslie Kinsland for help in preparing the manuscript.
ABBREVIATIONS
- SAM
S-adenosylmethionine
- SAH
S-adenosylhomocysteine
- GTP
guanosine triphosphate
- SeMet
selenomethionine
- Tris
tris(hydroxymethyl)aminomethane
- DTT
dithiothreitol
- RMSD
root mean square deviation
- PEG
polyethylene glycol
- 1,6-DDMT
1,6-didemethyltoxoflavin
- 1-DMT
1-demethyltoxoflavin (reumycin)
- 6-DMT
6-demethyltoxoflavin
- HPLC
high pressure liquid chromatography
- HEPES
4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid
- LB
Luria-Bertani
Footnotes
ASSOCIATED CONTENT
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.bio-chem.6b00030.
Supplementary Figure 1 is the difference electron density for the ligand complexes.
Supplementary Figure 2 shows binding of 1-DMT to ToxA in a nonproductive orientation.
Supplementary Figure 3 shows 1-DMT in a nonproductive orientation plus modeled SAM.
Accession Codes
The coordinates of ToxA, ToxA/SAH, ToxA/SAH/1,6-DDMT, ToxA/SAH/toxoflavin, ToxA-Y7F/SAH, ToxA-Y7F/SAH/toxoflavin, ToxA-Y7A/SAH, ToxA-Y7A/SAH/toxoflavin, and ToxA/SAH/1-DMT have been deposited in the Protein Data Bankas entries 5JE6, 5JDZ, 5JE0, 5JE1, 5JE2, 5JDY, 5JE3, 5JE4, and 5JE5, respectively.
REFERENCES
- 1.van Veen AG, Mertens WK. On the isolation of a toxic bacterial pigment. Proc. Acad. Sci. Amsterdam. 1933;36:666–670. [Google Scholar]
- 2.van Veen AG, Mertens WK. Die giftstoff der sogenannten bongkrek vergiftungen auf Java. Rec. Trav. Chim. Pays-Bas. 1934;53:257–266. [Google Scholar]
- 3.Hellendoorn AS, Ten Cate-Dhont RM, Peerdeman AF. On the structure of toxoflavin. Rec. Trav. Chim. Pays-Bas. 1961;80:307–308. [Google Scholar]
- 4.van Damme PA, Miss AG, Johannes H, Cox C, Berends W. On toxoflavin, the yellow poison of Pseudomonas cocovenenans . Rec. Trav. Chim. Pays-Bas. 1960;79:255–267. [Google Scholar]
- 5.Latuasan HE, Berends W. On the origin of the toxicity of toxoflavin. Biochim. Biophys. Acta. 1961;52:502–508. doi: 10.1016/0006-3002(61)90408-5. [DOI] [PubMed] [Google Scholar]
- 6.Goto K, Ohata K. New bacterial diseases of rice (brown stripe and grain rot) Ann. Phytopathol. Soc. Jpn. 1956;21:46–47. [Google Scholar]
- 7.Iiyama K, Furuya N, Takanami Y, Matsuyama N. A role of phytotoxin in virulence of Pseudomonas glumae Kurita et Tabei. Ann. Phytopathol. Soc. Jpn. 1995;61:470–476. [Google Scholar]
- 8.Jeong Y, Kim J, Kim S, Kang Y, Nagamatsu T, Hwang I. Toxoflavin produced by Burkholderia glumae causing rice grain rot is responsible for inducing bacterial wilt in many field crops. Plant Disease. 2003;87:890–895. doi: 10.1094/PDIS.2003.87.8.890. [DOI] [PubMed] [Google Scholar]
- 9.Nandakumar R, Shahjahan AKM, Yuan XL, Dickstein ER, Groth DE, Clark CA, Cartwright RD, Rush MC. Burkholderia glumae and B. gladioli cause bacterial panicle blight in the southern United States. Plant Disease. 2009;93:896–905. doi: 10.1094/PDIS-93-9-0896. [DOI] [PubMed] [Google Scholar]
- 10.Sato Z, Koiso Y, Iwasaki S, Matsuda I, Shirata A. Toxins produced by Pseudomonas glumae . Ann. Phytopathol. Soc. Jpn. 1989;55:353–356. [Google Scholar]
- 11.Uematsu T, Yoshimura D, Nishiyama K, Ibaraki T, Fujii H. Pathogenic bacterium causing seedling rot of rice. Ann. Phytopathol. Soc. Jpn. 1976;42:464–471. [Google Scholar]
- 12.Yoneyama K, Kono Y, Yamaguchi I, Horikoshi M, Hirooka T. Toxoflavin is an essential factor for virulence of Burkholderia glumae causing rice seedling rot disease. Ann. Phytopathol. Soc. Jpn. 1998;64:91–96. [Google Scholar]
- 13.Ham JH, Melanson RA, Rush MC. Burkholderia glumae: next major pathogen of rice? Mol. Plant Pathol. 2011;12:329–339. doi: 10.1111/j.1364-3703.2010.00676.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fenwick MK, Philmus B, Begley TP, Ealick SE. Toxoflavin lyase requires a novel 1-His-2-carboxylate facial triad. Biochemistry. 2011;50:1091–1100. doi: 10.1021/bi101741v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jung WS, Lee J, Kim MI, Ma J, Nagamatsu T, Goo E, Kim H, Hwang I, Han J, Rhee S. Structural and functional analysis of phytotoxin toxoflavin-degrading enzyme. PloS One. 2011;6:e22443. doi: 10.1371/journal.pone.0022443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Philmus B, Abdelwahed S, Williams HJ, Fenwick MK, Ealick SE, Begley TP. Identification of the product of toxoflavin lyase: degradation via a Baeyer-Villiger oxidation. J. Am. Chem. Soc. 2012;134:5326–5330. doi: 10.1021/ja211759n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hwang IG, Moon JS, Jwa NS. In: TflA gene which can degrade toxoflavin and its chemical derivatives and transgenic organisms expressing TflA gene. Organization WIP, editor. 2009. [Google Scholar]
- 18.Suzuki F, Sawanda H, Azegami K, Tsuchiya K. Molecular characterization of the tox operon involved in toxoflavin biosynthesis of Burkholderia glumae . J. Gen. Plant Pathol. 2004;70:97–107. [Google Scholar]
- 19.Kim J, Kim JG, Kang Y, Jang JY, Jog GJ, Lim JY, Kim S, Suga H, Nagamatsu T, Hwang I. Quorum sensing and the LysR-type transcriptional activator ToxR regulate toxoflavin biosynthesis and transport in Burkholderia glumae . Mol. Microbiol. 2004;54:921–934. doi: 10.1111/j.1365-2958.2004.04338.x. [DOI] [PubMed] [Google Scholar]
- 20.Suzuki F, Sawanda H, Matsuda I. Molecular characterization of toxoflavin biosynthesis-related gene in Pseudomonas (Burkholderia) glumae . Ann. Phytopathol. Soc. Jpn. 1998;64:276–281. [Google Scholar]
- 21.Foor F, Brown GM. Purification and properties of guanosine triphosphate cyclohydrolase II from Escherichia coli. J. Biol. Chem. 1975;250:3545–3551. [PubMed] [Google Scholar]
- 22.Ren J, Kotaka M, Lockyer M, Lamb HK, Hawkins AR, Stammers DK. GTP cyclohydrolase II structure and mechanism. J. Biol. Chem. 2005;280:36912–36919. doi: 10.1074/jbc.M507725200. [DOI] [PubMed] [Google Scholar]
- 23.Richter G, Ritz H, Katzenmeier G, Volk R, Kohnle A, Lottspeich F, Allendorf D, Bacher A. Biosynthesis of riboflavin: cloning, sequencing, mapping, and expression of the gene coding for GTP cyclohydrolase II in Escherichia coli . J. Bacteriol. 1993;175:4045–4051. doi: 10.1128/jb.175.13.4045-4051.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Burrows RB, Brown GM. Presence of Escherichia coli of a deaminase and a reductase involved in biosynthesis of riboflavin. J. Bacteriol. 1978;136:657–667. doi: 10.1128/jb.136.2.657-667.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen SC, Chang YC, Lin CH, Lin CH, Liaw SH. Crystal structure of a bifunctional deaminase and reductase from Bacillus subtilis involved in riboflavin biosynthesis. J. Biol. Chem. 2006;281:7605–7613. doi: 10.1074/jbc.M510254200. [DOI] [PubMed] [Google Scholar]
- 26.Richter G, Fischer M, Krieger C, Eberhardt S, Luttgen H, Gerstenschlager I, Bacher A. Biosynthesis of riboflavin: characterization of the bifunctional deaminase-reductase of Escherichia coli and Bacillus subtilis . J. Bacteriol. 1997;179:2022–2028. doi: 10.1128/jb.179.6.2022-2028.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stenmark P, Moche M, Gurmu D, Nordlund P. The crystal structure of the bifunctional deaminase/reductase RibD of the riboflavin biosynthetic pathway in Escherichia coli: implications for the reductive mechanism. J. Mol. Biol. 2007;373:48–64. doi: 10.1016/j.jmb.2006.12.009. [DOI] [PubMed] [Google Scholar]
- 28.Bacher A, Eberhardt S, Eisenreich W, Fischer M, Herz S, Illarionov B, Kis K, Richter G. Biosynthesis of riboflavin. Vitam. Horm. 2001;61:1–49. doi: 10.1016/s0083-6729(01)61001-x. [DOI] [PubMed] [Google Scholar]
- 29.Bacher A, Eberhardt S, Fischer M, Kis K, Richter G. Biosynthesis of vitamin B2 (riboflavin) Annu. Rev. Nutr. 2000;20:153–167. doi: 10.1146/annurev.nutr.20.1.153. [DOI] [PubMed] [Google Scholar]
- 30.Levenberg B, Linton SN. On the biosynthesis of toxoflavin, an azapteridine antibiotic produced by Pseudomonas cocovenenans . J. Biol. Chem. 1966;241:846–852. [PubMed] [Google Scholar]
- 31.Philmus B, Shaffer BT, Kidarsa TA, Yan Q, Raaijmakers JM, Begley TP, Loper JE. Investigations into the biosynthesis, regulation, and self-resistance of toxoflavin in Pseudomonas protegens Pf-5. Chembiochem. 2015;16:1782–1790. doi: 10.1002/cbic.201500247. [DOI] [PubMed] [Google Scholar]
- 32.Joo M, Yoo H-G, Kim H-J, Kwon H-J. ToxB encodes a canonical GTP cyclohydrolase II in toxoflavin biosynthesis and ribA expression restored toxoflavin production in a ΔtoxB mutant. J. Korean Soc. Appl. Biol. Chem. 2015;58:877–885. [Google Scholar]
- 33.Lim J, Lee TH, Nahm BH, Choi YD, Kim M, Hwang I. Complete genome sequence of Burkholderia glumae BGR1. J. Bacteriol. 2009;191:3758–3759. doi: 10.1128/JB.00349-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Otwinoski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Method. Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 35.Schneider TR, Sheldrick GM. Substructure solution with SHELXD. Acta. Crystallogr. 2002;D58:1772–1779. doi: 10.1107/s0907444902011678. [DOI] [PubMed] [Google Scholar]
- 36.Sheldrick GM. A short history of SHELX. Acta Crystallogr. 2008;A64:112–122. doi: 10.1107/S0108767307043930. [DOI] [PubMed] [Google Scholar]
- 37.Adams PD, Afonine PV, Bunkoczi G, Chen VB, Echols N, Headd JJ, Hung LW, Jain S, Kapral GJ, Grosse Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner RD, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH. The Phenix software for automated determination of macromolecular structures. Methods. 2011;55:94–106. doi: 10.1016/j.ymeth.2011.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vagin A, Teplyakov A. Molecular replacement with MOLREP. Acta Crystallogr. 2010;D66:22–25. doi: 10.1107/S0907444909042589. [DOI] [PubMed] [Google Scholar]
- 39.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr. 2010;D66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Painter J, Merritt EA. Optimal description of a protein structure in terms of multiple groups undergoing TLS motion. Acta Crystallogr. 2006;D62:439–450. doi: 10.1107/S0907444906005270. [DOI] [PubMed] [Google Scholar]
- 41.Temple C, Kussner CL, Montgome JA. Preparation of pyrimido[5,4-e]-as-Triazine-5,7(6H,8H)-dione. J. Heterocyclic Chem. 1968;5:581–582. [Google Scholar]
- 42.Black TH. An improved, large-scale synthesis of xanthothricin and reumycin. J. Heterocyclic Chem. 1987;24:1373–1375. [Google Scholar]
- 43.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera--a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 44.Krissinel E, Henrick K. Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 2007;372:774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- 45.Martin JL, McMillan FM. SAM (dependent) I AM: the S-adenosylmethionine-dependent methyltransferase fold. Curr. Opin. Struct. Biol. 2002;12:783–793. doi: 10.1016/s0959-440x(02)00391-3. [DOI] [PubMed] [Google Scholar]
- 46.Schubert HL, Blumenthal RM, Cheng X. Many paths to methyltransfer: a chronicle of convergence. Trends. Biochem. Sci. 2003;28:329–335. doi: 10.1016/S0968-0004(03)00090-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schubert HL, Blumenthal RM, Cheng X. Protein methyltransferases: their distribution among the five structural classes of AdoMet-dependent methyltransferases. In: Clarke SG, Tamanoi F, editors. The Enzymes. Burlington, MA: Academic Press; 2006. [DOI] [PubMed] [Google Scholar]
- 48.Holm L, Rosenstrom P. Dali server: conservation mapping in 3D. Nucleic Acids Res. 2010;38:W545–W549. doi: 10.1093/nar/gkq366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bauer J, Ondrovicova G, Najmanova L, Pevala V, Kamenik Z, Kostan J, Janata J, Kutejova E. Structure and possible mechanism of the CcbJ methyltransferase from Streptomyces caelestis . Acta Crystallogr. 2014;D70:943–957. doi: 10.1107/S139900471303397X. [DOI] [PubMed] [Google Scholar]
- 50.Carney AE, Holden HM. Molecular architecture of TylM1 from Streptomyces fradiae: an N,N-dimethyltransferase involved in the production of dTDP-D-mycaminose. Biochemistry. 2011;50:780–787. doi: 10.1021/bi101733y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.De la Pena M, Kyrieleis OJ, Cusack S. Structural insights into the mechanism and evolution of the vaccinia virus mRNA cap N7 methyl-transferase. EMBO J. 2007;26:4913–4925. doi: 10.1038/sj.emboj.7601912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fabrega C, Hausmann S, Shen V, Shuman S, Lima CD. Structure and mechanism of mRNA cap (guanine-N7) methyltransferase. Mol. Cell. 2004;13:77–89. doi: 10.1016/s1097-2765(03)00522-7. [DOI] [PubMed] [Google Scholar]
- 53.Takata Y, Huang Y, Komoto J, Yamada T, Konishi K, Ogawa H, Gomi T, Fujioka M, Takusagawa F. Catalytic mechanism of glycine N-methyltransferase. Biochemistry. 2003;42:8394–8402. doi: 10.1021/bi034245a. [DOI] [PubMed] [Google Scholar]
- 54.Zheng S, Shuman S. Mutational analysis of vaccinia virus mRNA cap (guanine-N7) methyltransferase reveals essential contributions of the N-terminal peptide that closes over the active site. RNA. 2008;14:2297–2304. doi: 10.1261/rna.1201308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Troffer-Charlier N, Cura V, Hassenboehler P, Moras D, Cavarelli J. Functional insights from structures of coactivator-associated arginine methyltransferase 1 domains. EMBO J. 2007;26:4391–4401. doi: 10.1038/sj.emboj.7601855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yue WW, Hassler M, Roe SM, Thompson-Vale V, Pearl LH. Insights into histone code syntax from structural and biochemical studies of CARM1 methyltransferase. EMBO J. 2007;26:4402–4412. doi: 10.1038/sj.emboj.7601856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Feng Q, He B, Jung SY, Song Y, Qin J, Tsai SY, Tsai MJ, O’Malley BW. Biochemical control of CARM1 enzymatic activity by phosphorylation. J. Biol. Chem. 2009;284:36167–36174. doi: 10.1074/jbc.M109.065524. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.