Graphical abstract
Introduction
Basidiomycota and Ascomycota represent the major phyla of the fungal kingdom which is estimated to be comprised of more than 1.5 mio species [1]. Only a fraction (~100,000 species) of this diversity has been described and less than a 10th of the known species have been characterized in more detail and/or exploited by humans. Ascomycota (~70% of the described species) have so far received the greatest attention by scientists and for applications in biotechnology while Basidiomycota remain greatly understudied despite their importance for carbon recycling, ecosystem functioning and medicinal properties. The Basidiomycota are distinguished from the Ascomycota by the formation of cellular structures known as basidia on which haploid, sexual spores (basidiospores) are produced. Unlike Ascomycota, most Basidiomycota have long-lived dikaryotic states and their hyphae grow and divide as dikaryons with two genetically different nuclei. Mitotic division of growing hyphal cells involves passage of one nucleus back over a developing septum between parent and daughter cell via a readily observed clamp structure. Many Basidiomycota produce characteristic fruiting bodies with club-like spore-bearing basidia that line gills, pores or surfaces (e.g. gilled mushrooms, boletes, chanterelles, polypores, coral fungi) or are sometimes located inside of fruiting bodies (e.g. puffballs). Other Basidiomycota classes with different basidia and fruiting body structures include the jelly, smut and rust fungi.
Funding of bioenergy and biofuel research in the past decade, however, has greatly accelerated research aimed at characterizing the lignocellulolytic mechanisms and enzymes of Basidiomycota. Through efforts of the US Department of Energy’s (DOE) Fungal Genome Program, resources and information are being built as a foundation to tap into the unique biochemical abilities of these fungi for biotechnological applications. The DOE’s Joint Genome Institute’s (JGI) MycoCosm Portal (http://jgi.doe.gov/fungi) [2] now lists close to 200 genome sequences of Basidiomycota compared to fewer than 10 available genomes only a few years ago. The majority of genomes for basidiomycete fungi are of wood- [3–9] and leaf litter-decaying [10] mushroom-forming fungi (Agaricomycetes), including a few medicinally relevant fungi [6,11–13]. More recently, mushrooms with different nutritional modes such as mycorrhizal [14] and plant pathogenic fungi [15] have been added to the list of genera with sequenced genomes.
The steady influx of genome data has facilitated detailed comparative genomic studies aimed at understanding the evolution and function of fungal lignocellulose degradation [3,5,9,10]. These studies have uncovered a wealth of known and novel oxidoreductase and hydrolase families of which many have undergone lineage specific expansions and reductions to support specialized wood decay mechanisms and lifestyles. Although relatively few members of these families have been functionally studied (reviewed in: [16–19]), the characterized enzymes point to an intricately interwoven tapestry of lignocellulolytic activities that are useful for applications beyond lignocellulose breakdown, including future efforts at finding novel biotransformation activities. Little attention has so far been paid to enzymes involved in the biosynthesis of the diverse secondary metabolites made by Basidiomycota. Genome mining efforts [6,11–13,20] and the numerous natural products [21] isolated from cultures or fruiting bodies suggest a treasure trove of novel catalytic activities awaiting to be discovered.
This review will provide an overview of the biocatalytic portfolio of Basidiomycota characterized to date with the hope of motivation more research into the chemical toolkits of this diverse group of fungi. Emphasis will be placed on enzymatic activities useful for bioctalysis and biosynthesis. To appreciate the yet largely uncharacterized, biocatalytic abilities of Basidiomycota, it is necessary to understand their diverse lifestyles and ecological functions which have shaped their secondary metabolomes and enzyme repertoires.
Fungal life styles at a glance
Basidiomycota can roughly be divided into saprobic, symbiontic or parasitic/pathogenic groups based on their nutritional modes. Saprobic fungi decompose organic matter (wood, leaf litter or dung) while symbiotic fungi form a mutualistic relationship with another organism such for example plants where >80% of species engage in such symbiotic relationships. For example, ectomycorrhizal fungi exchange nutrients with trees and shrubs via mycorrhiza that envelop root tips to provide phosphorous, nitrogen and other nutrients to the plant in exchange for photosynthetic sugars. These fungi therefore play a critical role in the function of forest ecosystems. Pathogenic or parasitic fungi infect plants, animals and even other fungi.
Following the evolution of lignin biosynthesis in land plants, the ancestors of basidiomycete fungi evolved ~ 295 million years ago the ability to completely mineralize lignocellulose [5], thereby bringing an end to the carboniferous period by releasing CO2 from plant biomass back into the atmosphere. These white-rot (WR) fungi evolved high-redox potential class II peroxidases able to directly attack lignin to oxidize and break non-aromatic bonds of this complex and recalcitrant polymer. This family of class II peroxidases (PODs) was subsequently expanded, but also lost again in most brown-rot (BR) fungi and are absent or reduced in mycorrhizal fungi. Only WR fungi are able to simultaneously depolymerize and mineralize polysaccharide and lignin-polymers to a white mass, while BR fungi only modify lignin in order to access and aggressively degrade the polysaccharide polymers of lignocellulose, leaving a brownish discolored and brittle wood residue behind. On a genome levels, these two types of wood-decay fungi can typically be distinguished by the presence or absence of lignocellulose depolymerizing enzymes; only WR fungi have lignin degrading PODs as well as GH6 and GH7 family cellobiohydrolases [3,5]. Mycorrhizal life styles have evolved from different WR and BR fungi leading to a reduction of lineage specific lignocellulolytic enzyme repertoires as carbohydrates are derived from plant and the development of new symbiosis specific protein repertoires [14]. Non-woody litter degrading fungi have a reduced ability to degrade lignin, but have evolved instead a generalist approach to utilize diverse carbohydrates and deal with a variety of aromatic compounds present in litter and compost by expanding e.g. their heme-thiolate peroxygenase repertoires [10].
Fungal mycelial networks are continuously remodeled to access new nutrient sources. Many saprotrophic fungi scout for new nutrient sources by sending out long-range linear hyphal organs (cords) that can lead to the formation of extensive networks connecting multiple nutritient sources. Basidiomycota also form fruiting bodies for spore dispersal to establish new mycelial networks. Underground hyphae and fruiting bodies are vulnerable to a number of threats, including antagonistic and competing fungi and bacteria as well as grazing by invertebrates such as small arthropods, insects, nematodes, mollusks and larger animals [22]. In response, fungi have developed diverse defense mechanisms such as the production of secondary metabolites as defense and/or microbial and fauna control agents [23–25]. Hyphae invading wood or decomposing litter also encounter a range of potentially toxic lignin degradation products and plant defense compounds, including mono- and polyphenolic compounds and resins with high contents of terpenes. Consequently, Basidiomycota have evolved distinct secondary metabolite profiles and biochemical arsenals.
Masters of redox biocatalytic transformations
Wood- or litter decomposing fungi secrete suites of oxidoreductases to modify and breakdown lignocellulolytic polymers and/or deal with toxic compounds (Box 1 and Figure 1) [17,26]. Hundreds of cytochrome P450 monooxygenase (P450s) homologs are typically present in their genomes [27] along with large numbers flavin-dependent oxidoreductases. Many of these enzymes are found in secondary metabolic clusters [13,28], others, especially P450s, are assumed to be part of the fungal defense system against xenobiotics [29].
Box 1: Summary of major classes of oxidoreductases in Basidiomycota.
Enzymes are grouped into five classes (gray boxes) based on their reactions. EC numbers and auxillary activity (AA) family numbers if in the carbohydrate-enzymes database CaZy (www.cazy.org).
| Peroxidases EC.1.11.X, AA2 | Single electron oxidation without oxygen transfer: [S]H2 +H2O2 ⇔ [P] + 2H2O |
| Class II heme peroxidases (PODs) EC 1.11.1 | Secreted by lignin depolymerizing WR fungi; enzymes share a conserved helical fold with 2 domains forming a heme-binding cleft. |
| Lignin Peroxidase (LiP) EC 1.11.1.14 | High redox potential (RP) (1.4–1.5 V) of LiP facilitates oxidation of non-aromatic compounds without a mediator; LiP exhibit broad substrate specificity with (non)aromatic compounds and catalyze cleavage of the Cα-Cβ linkages of lignin polymers; Substrate oxidation occurs via long-range electron transfer to the heme from a hydroxylated Trp residue located on the protein surface. |
| Manganese Peroxidase (MnP) EC 1.11.1.13 | MnP (RP: 1.0–1.2 V) transfer one electron to Mn2+ to generate Mn3+ which upon chelation by carboxylic acids (e.g. oxalates) becomes a diffusible oxidant that abstracts electrons from aromatic compounds to generate radicals. |
| Versatile Peroxidase (VP) EC 1.11.1.16, | VP (RP: 1.4–1.5 V) exhibit broad substrate specificity and combine activities of both LiP and MnP such as the high-redox potential of LiP and the Mn2+ binding pocket of MnP. |
| Dye-decolorizing Peroxidases (DyP) EC 1.11.1.19 | DyP (RP: 1.2–1.5 V) are secreted peroxidases that can oxidize a broad range of aromatic compounds and compounds with conjugated double bonds; DyP are particularly known for their oxidation of various recalcitrant anthraquinone dyes; DyP natural substrates and functions are not known but they may play a role in lignin degradation despite having a lower redox potential than LiP. The structural fold of DyPs is distinct from other peroxidases as their heme binding pocket is located in a C-terminal domain. DyPs are evolutionary related to chlorite dismutases; their catalytic mechanism has yet to be fully elucidated. |
| Heme-thiolate Peroxygenase EC 1.11.2 | Incorporation of one oxygen from H2O2 into substrate: [S]H + H2O2 ⇔ [P]-OH + H2O |
| Unspecific Peroxygenase (UPO) EC 1.11.2.1 | UPOs (RP: 0.9–1.2 V) are heme-containing enzymes with a P450 spectrum. They catalyze similar reactions than P450 monooxygenases by using the P450 “peroxide shunt” pathway. UPOs oxidize many aromatic, aliphatic and heterocyclic substrates. |
| Chloroperoxygenase (CPO) EC 1.11.1.10 | CPOs are heme-containing enzymes with P450 characteristics that catalyzes one and two electron oxidations of halides, hydroxylation/epoxidation of benzylic carbons, double bonds of linear and cyclic alkanes, organic sulfides and halogenated substrates CPOs unlike UPOs do not oxidize aromatic ring carbons or alkanes. |
| Peroxide generating enzymes | Transfer of two electrons to O2: [S] + O2 ⇔ [S]ox+ H2O2 Generate H2O2 for i) non-enzymatic generation of diffusible oxidants via Fenton chemistry and ii) various peroxidases and peroxygenases. |
| Glucose-methanol-choline (GMC) oxidoreductase EC 1.1.X.X, AA3 | GMCs are flavin containing enzymes that catalyze the oxidation of a wide range of substrates in a first half reaction and reduce O2 to H2O2 in a second half reaction to regenerate the oxidized flavin co-factor: [S]OH +FAD + O2 ⇔ [S]O + FAD+ H2O2 GMC are a widely distributed family of extra- and intracellular enzymes that includes the aryl-alcohol -, pyranose – and methanol oxidases which generate H2O2 for lignin depolymeration. |
| Aryl-Alcohol oxidase(AAO) EC 1.1.3.7 | AAO oxidize aromatic alcohols. |
| Pyranose oxidase (Pox) EC1.1.3.10 | Pox xatalyzes the regioselective oxidation of aldopyranoses at the C2-position to form the corresponding keto-aldoses. Located in the periplasmic space of fungal hyphae. |
| Copper radical oxidase (CRO) EC 1.1.3, AA5 | Reaction: [S]O + 2O2 ⇔ [S]OOH + H2O2 CROs fall into several subfamilies that are widely distributed in Basidiomycota. Functions of most subfamilies uncharacterized |
| Glyoxyl oxidase (GLX) EC 1.1.3.1 | GLC oxidize simple aldehydes to hydroxyl-carbonyls to generate small organic acids as Mn3+ chelators. Their active site is similar to those of galactose oxidase |
| Multi-copper oxidase EC 1.10.3.X, AA1 | Catalyze 4 electron reduction of O2 to H2O: 4 [S]OH + O2 ⇔ 4[S]O + 2H2O |
| Laccase (LAC) EC 1.10.3.2 | LAC contain 4 copper ions of three different types; three copper ions form a trinuclear cluster LACs have moderate redox potentials (RP: 0.4–0.8 V) allowing them to abstract electrons from phenolic compounds to generate phenoxy radicals. LACs catalyze the oxidation of non-phenolic compounds in the presence of mediators |
| Benzoquinone reductase (BQR) EC 1.6.5.6, AA6 | FAD-containing intracellular oxidoreductase that reduces benzoquinones to the corresponding alcohols to fuel extracellular quinone redox cycling for H2O2 and radical generation: benzoquinone + NADPH + H+ ⇔ hydroquinone+ NADP++ H2O |
| P450 monoxygenase | P450s incorporate oxygen from molecular oxygen into a wide range of substrates: [S] + O2 + NADPH2 ⇔ [P]O + NADP + H2O P450s oxidize unreactive carbons and a play major role in the detoxification of xenobiotics and secondary metabolism. P450 families have been greatly expanded in Basidiomyctoa. |
| Dioxygenases | Dioxygenases transfer two oxygens from molecular oxygen into substrates: [S] + O2 + NADPH2 ⇔ [P]O2 Lipoxygenases (LOX) are a class of non-heme iron containing oxidoreductases that typically catalyze the regio- and stereoselective insertion of molecular oxygen into cis-polyunsaturated fatty acids to produce compounds involved in signaling and defense. LOX in Basidiomycota are responsible for the generation of oxygenated C8 volatiles that generate the typical mushroom odor. |
Figure 1. Oxidative biotransformations involved in lignin degradation.
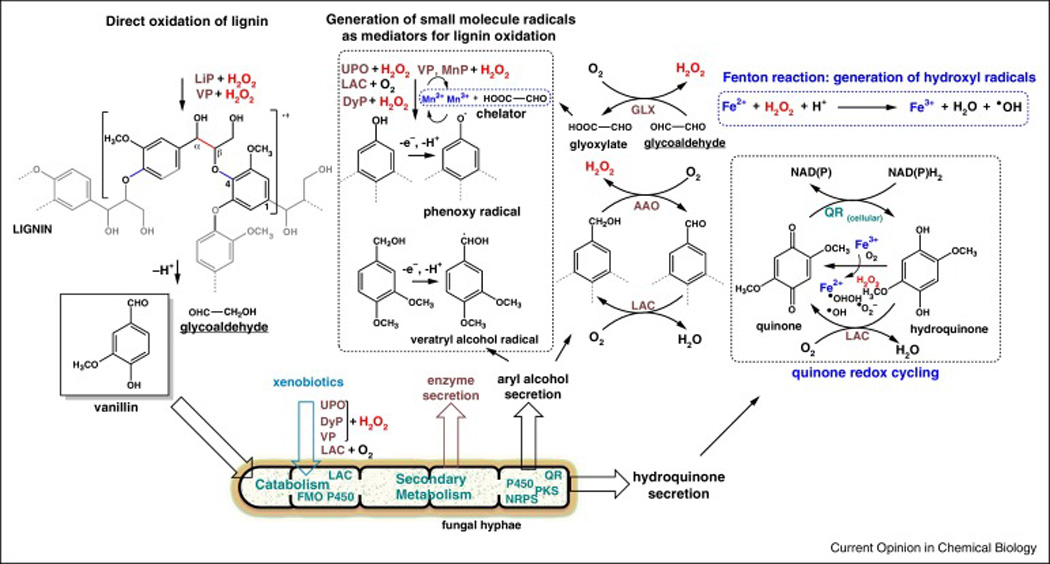
Secreted and hyphal enzymes are shown in maroon and green, respectively. See Table 1 for enzyme abbreviations and functions. Lignin peroxidases (LiP, VP and MnP) are characteristic for white-rot fungi, while brown-rot fungi rely on extracellular Fenton chemistry for lignin modification. LiP and VP are high-redox potential peroxidases capable of direct lignin oxidation that result for example in Cα-Cβ side chain cleavage products. Aromatic and non-aromatic redox mediators such as phenoxy and aryl alcohol radicals (e.g. veratryl alcohol radical), respectively are generated by fungal oxidases to expand their substrate scope and to potentially infiltrate cell wall polymers that are inaccessible to the enzymes themselves. MnP oxidizes Mn2+ to the short lived Mn3+ cation which upon complexation with glyoxylate becomes a diffusible oxidant for lignin degradation. Quinones and aryl alcohols are actively secreted by the fungi to generate hydrogen peroxide and to drive a hydroquinone-driven Fenton system for the generation of diffusible hydroxyl radicals from hydrogen peroxide as well as perhydroxyl and superoxide radicals during quinone redox cycling.
Biocatalysts of oxidative lignin degradation
WR fungi secret three types of lignin degrading class II heme containing peroxidases: lignin (LiP), manganese (MnP) and versatile (VP) peroxidase [17]. LiP and VP have high redox potentials compared to laccases and other peroxidases that allows them to oxidize wide range of aromatic compounds and cleave the non-phenolic Cα-Cβ linkages of lignin polymers in the absence of a small molecule mediator (Figure 1). MnP catalyzes the oxidation of Mn2+ to Mn3+ which functions as diffusible oxidant upon complexation with small organic acids such as e.g. oxalic acid. VP combines both catalytic activities of LiP and MnP. Basidiomycota secrete additional oxidases that modify lignin through the generation of reactive aromatic radicals like e.g. phenoxy radicals. Dye decolorizing heme binding PODs (DyP) [18] can oxidize a range of polycyclic aromatic compounds, phenols and some lignin model compounds [30–32] and may play a role both in lignin depolymerization and in the oxidation of xenobiotics. Secreted laccases also oxidize phenolic compounds and generate radicals, but also play a role in the reoxidation of hydroquinones and aryl-alcohols. The latter serves as substrate for alcohol aryl oxidase (AAO) which generates the H2O2 for peroxidases and radical generating Fenton chemistry (Figure 1). AAO belongs to the large group of catalytically versatile FAD binding GMC-type oxidoreductases [33,34] and reduces a range of aryl aclohols to the corresponding aldehydes. BR fungi rely exclusively on an extracellular Fenton chemistry that uses Fe2+ and H2O2 to produce hydroxyl radicals to attack lignin polymers. Quinone metabolites are reduced to hydroquinones by a quinone reductase prior to secretion to drive quinone redox cycling (Figure 1) for the generation of Fe2+ and hydroxyl-, perhydroxy-radicals and H2O2 [35].
Heme thiolate peroxygenases
Basidiomycota secrete two types of multi-talented oxygenases with cytochrome P450-like spectral properties: the classic and well-studied chloroperoxygenases (CPO) and the more recently discovered unspecific peroxygenases (UPO, formerly aromatic peroxygenase APO) [36]. Both enzymes catalyze the transfer of oxygen from H2O2 into a range of substrate mechanistically akin to the inefficient peroxide shunt pathway of P450s [37,38]. Yet unlike P450s, these enzymes are secreted, highly stable and do not require a reductase partner or NADPH as co-factor, which makes them very attractive for biocatalytic applications. CPO is a well characterized and already widely used biocatalyst that has a preference for unspecific halogenation and oxygenation of activated carbons like those in aromatic rings. UPO’s, however, are capable of catalyzing a spectrum oxygenation reactions, including the oxygenation of unactivated carbons, catalyzed by P450 monooxygenases [16]. Three UPO enzymes have been characterized that catalyze many useful reactions, including the epoxidation of alkenes, styrenes [39,40], selective hydroxylation of long-chain fatty acids, alkanes and alcohols, cyclic alkanes and flavonoids [41–44] (Figure 2A). UPO’s utility for biocatalysis has been demonstrated with several one-pot syntheses of pharmaceutically relevant compounds [45–48] and with a multi-enzyme process for 5-hydroxymethylfurfural conversion [49]. Recent insights into UPO structure and reaction mechanism [50–53] as well as optimization of heterologous secretion by yeast [54] have laid the foundation for the production of rational designed enzymes for biocatalysis. BLAST searches of fungal genomes identifies hundreds of additional UPO homologs that may be screened as potential biocatalysts.
Figure 2. Representative reactions catalyzed by versatile Basidiomycota oxygenases.
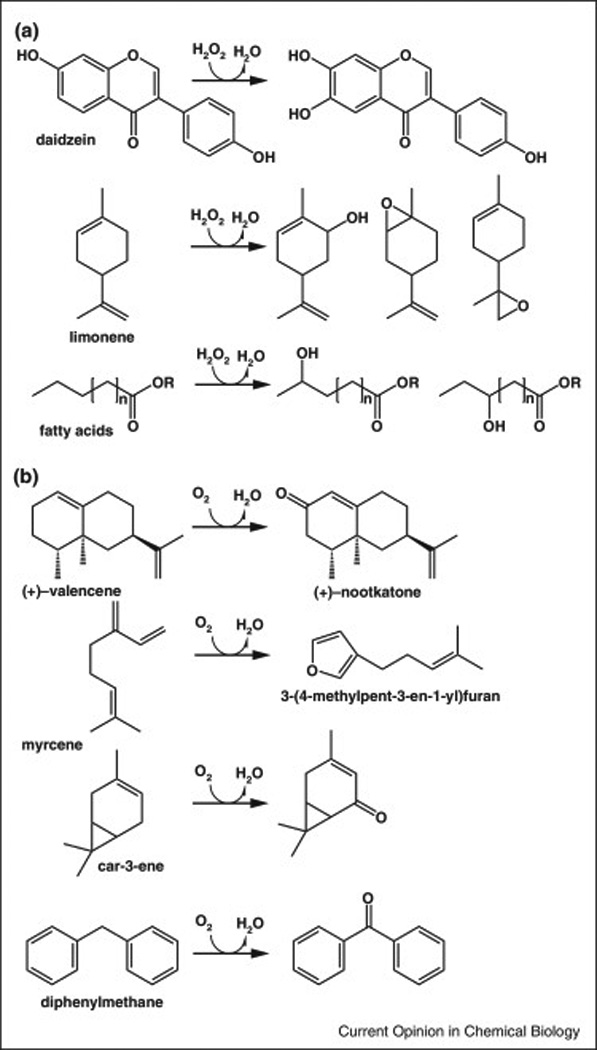
Biotransformations of (A) heme-thiolate peroxygenases UPO’s [41,43,44] and (B) non-heme iron lipoxygenase-type dioxygenases (LOX) [60,61,79,80].
Monoxygenases and dioxygenases
The genomes of Basidiomycota contain large numbers of 100 to more than 250 P450 homologs. Several catalytically versatile cytochrome P450 (CYP) families such as CYP63 and CYP5144 are enriched in most Basidiomycota and are presumed to participate in the degradation of xenobiotic compounds [27,29,55]. CYP5144 members are also frequently found in secondary metabolic gene clusters [12,55], including sesquiterpenoid gene clusters described by us [13,28,56]. Other CYP families (e.g. CYP5348, CYP5359) have been expanded in a species specific manner, probably as an adaptation to specific ecological niches. Members of several of these CYP families have been shown to oxidize diverse compounds including various hydrocarbons, plant metabolites, pharmaceuticals and steroids [12,55]. A genome-wide functional screening in S. cerevisiae was carried out with a total of 300 P450s amplified from cDNA of the brown rotter Postia placenta and white rotter Phanerochaete chrysosporium [57,58]. A significant fraction of the cloned P450s was functional in S. cerevisia and showed a wide range of activities against various model substrates, demonstrating that the Basidiomycota CYPome is a rich source for the identification of novel P450 activities. Nevertheless, although a considerable number of P450s from these two Basidiomycota were functional in S. cerevisiae, in our experience functional expression of these ER membrane-bound enzymes is still largely a hit or miss undertaking often requiring extensive optimization of expression conditions, including testing different hosts (e.g. S. cerevisiae, P. pastoris, E. coli) and/or optimization or truncation of the N-terminal transmembrane domain and co-expression of a fungal reductase partner. Even after exhaustive optimization attempts, functional expression may not be achieved using current strategies and hosts.
In addition to P450s, two lipoxygenase-type dioxygenase (LOX) have been identified in Pleurotus mushrooms [59,60]. These enzymes are considered to be responsible for the formation of oxidized octane volatiles that create the typical mushroom aroma. One of the characterized LOX enzymes was found to accept bulky terpenoids as substrates. This enzyme catalyzes the selective hydroperoxidation of several sesqui- and monterpenes, including the biotransformation of (+)-valencene into the valuable aroma compound (+)-nootkatone [60,61] (Figure 2B).
Biocatalysts of secondary metabolism
Homology-based analysis of fungal genomes for known biosynthetic genes involved in secondary metabolism together with information on isolated natural products suggests that the secondary metabolome of Basidiomycota differs from those of other prolific microbial natural products producers [20,62]. Sesquiterpene synthases appear to be the most abundant class of natural products scaffold generating enzymes (see [20] for details) in these fungi. Compared to Ascomycota that may have 20-50 PKS or NRPS genes, Basidiomycota have comparatively few (on average fewer than 15) of these genes. So far, only a handful of these synthases have been studied [63–68]. Two NRPS lacking the peptide-bond forming condensation domain have been biochemically characterized and shown to condense two aromatic α-keto acids to produce compounds with a quinone core as pigments or antimicrobials [63,65]. These secondary metabolites are also secreted as iron reductants to drive quinone redox cycling for lignin degradation [69]. Nevertheless, some intriguing PKS derived bioactive metabolites have been isolated from Basidiomycota. These include the aromatic acetylenic compounds like e.g. the antrocamphins from the medicinal mushroom Antrodia camporata [11,70] and the choropropynyl bearing gymnopalynes from Gymnopus sp. [71]. Biosynthesis of these compounds likely involves either a fatty acid desaturase/acetenylase system with shorter chain length specificity like the recently characterized acetenylase from Cantharellus formosus [72], or an acyl carrier protein/acyl-CoA-ligase/desaturase system analogous to the cyanobacterial enzymes that install the terminal alkyne in jamaicamide [73].
Hundreds of terpenoids have been isolated from Basidiomycota, many with unique hydrocarbon skeletons not made by other organisms [62]. Sesquiterpene synthases (STS) catalyze the 1,11-cyclization of farnesyl diphosphate (FPP) into characteristic scaffolds that can undergo further rearrangement reactions; giving rise to additional structures (Figure 3). These scaffolds are often associated with specific fungal genera such as e.g. the lactaranes made by the milk-cap Lactarius mushrooms [74]. We have biochemically characterized twenty STS from several Basidiomycota, including the six STS genes in C. cinerea [56] and ten STS genes in the anti-cancer illudin producer Omphalotus olearius [13]. We identified two 1,11-cyclizing Δ6-protoilludene synthases located in two gene clusters that synthesize the illudin scaffold. Based on the product specificities of these enzymes, we developed a predictive framework for sesquiterpenoid pathway discovery in Basidiomycota. Phylogenetic analysis of STS homologs identified in Basidiomycota genomes resulted in five clades predicted to include STS catalyzing different initial cyclization reactions of FPP. About 30% of all STS homologs identified in Basidiomycota genomes are predicted to catalyze the 1,11-cyclization of FPP to generate the bioactive sesquiterpenoid scaffolds known from Basidiomycota. We applied this framework for the functional prediction of 18 STS in S. hirsutum which is known to produces diverse bioactive sesquiterpenoids [62]. We identified and characterized three additional protoilludene synthases located in large gene clusters [28,75]. With these functionally characterized STS sequences at hand, we noticed that automated natural products annotation programs [76] only find a fraction of the actual STS genes encoded in Basidiomycota genomes. For example, out of the 18 STS genes from S. hirsutum, only six STS are included in a recent genome analysis [3].
Figure 3. Basidiomycota produces a plethora of protoilludene derived sesquiterpenoid natural products.
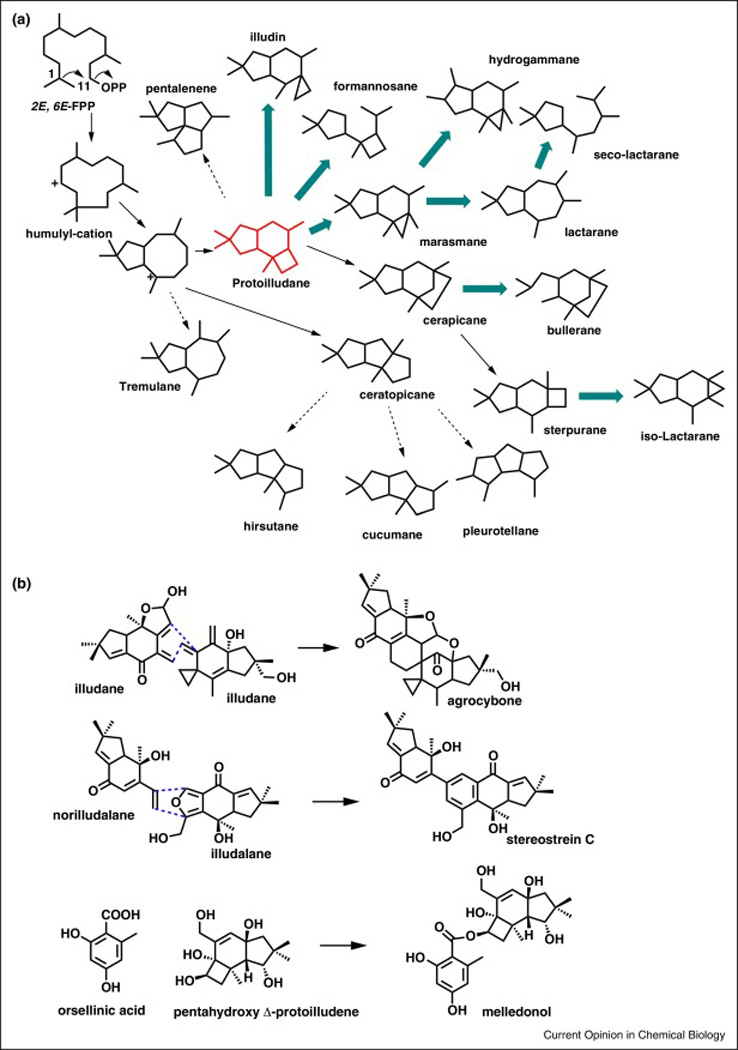
(A) 1,11 cyclization of FPP by sesquiterpene synthases gives rise to diverse sesquiterpenoid scaffolds, including protoilludene, which is further rearranged into additional scaffolds via uncharacterized secondary oxidation reactions (bold arrows)[62]. (B) Protoilludane-type sesquiterpenoids are coupled via a proposed Diels-Alder reaction in the biosynthesis of agrocybone [78] and stereostrein C [77]. Biosynthesis of melledonol involves cross-coupling of orsellenic acid with a hydroxylated protoilludene catalyzed by a polyketide synthase[66].
The 1,11-cyclizing protoilludene synthases are frequently associated with large biosynthetic gene clusters, while the large majority of the STS catalyzing other cyclization reactions typically are not [13,28]. These gene clusters usually contain several P450s and GMC oxidoreductases presumably responsible for the scaffold rearrangements and oxygenations that lead to the bioactive products (Figure 3). Interestingly, Stereum makes protoilludene-derived bis-sesquiterpenoid stereostreins via a postulated Diels-Alder condensation reaction [77]. Agrocybone is another bis-sesquiterpenoid hypothesized to involve a Diels-Alder condensation reaction [78]. One of the protoilludene synthase associated gene clusters in Stereum may therefore encode a much sought after Diels-Alderase. The biosynthesis of melleolide antibiotics by the honey mushroom Armillaria mellea involves another unique modification of the protoilludene scaffold; esterification with a PKS derived orsellinic acid moiety (Figure 3 catalyzed by a cross-coupling orsellinic acid PKS [66].
Conclusions and Perspective
The past few years have seen a dramatic increase in genome sequence data available for Basdiomycota. Comparative genomic studies have provided important insights into the evolution and function of the complex lifestyles of this group of fungi. These studies and data give us access to a mostly unexplored source for new enzymes and biosynthetic pathways. Unlocking this resource requires significant efforts to overcome several challenges that currently slow down functional characterization of individual enzymes and the clustered secondary metabolic pathways of Basidiomycota. Most of these fungi are genetically intractable, preventing functional genetic studies feasible in other microbial organisms, including many Ascomycota. This leaves heterologous expression of genes and biosynthetic gene clusters as the only practical alternative. While genes can now be inexpensively synthesized and entire gene clusters rapidly assembled for heterologous expression, this approach from gene prediction to recombinant product is more often than not quite challenging for Basidiomycota. In our own experience, automated gene prediction pipelines frequently mis-annotate or fail to annotate genes altogether. Further, prediction of introns and exons using existing models are often incorrect, leading to non-functional genes if synthetically obtained rather than amplified from cDNA. The latter of course requires conditions under which a gene or pathway of interest is expressed. The development of accurate gene prediction and annotation pipelines will be required to take advantage of current synthetic biology capabilities to go from in silico gene/pathway prediction to recombinant product. In the future, automated high-throughput pipelines may become available to synthesize, assemble, express and screen libraries of oxidoreductases and pathways identified in fungal genomes. The development of alternative fungal hosts that are not yeasts and are amenable to rapid genetic engineering may also be required for functional expression biosynthetic pathways and enzymes from Basidiomycota.
Highlights.
Unique lifestyles and ecologies of Basidiomycota are reflected in distinct portfolios of biocatalytic activities and secondary metabolic pathways.
Basidiomycota are masters of oxidative biocatalysis involving novel and expanded families of many different types of oxidoreductases.
Accelerated genome sequencing efforts provide resources to explore the biocatalytic capabilities of Basidiomycota.
Basidiomycota are prolific producers of bioactive sesquiterpenoid natural products and their genomes contain expanded families of sesquiterpene synthases.
Acknowledgments
Support by the National Institute of Health (grant GM080299) is acknowledged.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References (annotated reference list separately attached)
- 1.Blackwell M. The Fungi: 1, 2, 3 … 5.1 million species? American Journal of Botany. 2011;98:426–438. doi: 10.3732/ajb.1000298. [DOI] [PubMed] [Google Scholar]
- 2.Grigoriev IV, Nikitin R, Haridas S, Kuo A, Ohm R, Otillar R, Riley R, Salamov A, Zhao X, Korzeniewski F, et al. MycoCosm portal: gearing up for 1000 fungal genomes. Nucleic Acids Res. 2014;42:D699–D704. doi: 10.1093/nar/gkt1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Riley R, Salamov AA, Brown DW, Nagy LG, Floudas D, Held BW, Levasseur A, Lombard V, Morin E, Otillar R, et al. Extensive sampling of basidiomycete genomes demonstrates inadequacy of the white-rot/brown-rot paradigm for wood decay fungi. Proc Natl Acad Sci U S A. 2014;111:9923–9928. doi: 10.1073/pnas.1400592111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Levasseur A, Lomascolo A, Chabrol O, Ruiz-Duenas FJ, Boukhris-Uzan E, Piumi F, Kues U, Ram AF, Murat C, Haon M, et al. The genome of the white-rot fungus Pycnoporus cinnabarinus: a basidiomycete model with a versatile arsenal for lignocellulosic biomass breakdown. BMC Genomics. 2014;15:486. doi: 10.1186/1471-2164-15-486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Floudas D, Binder M, Riley R, Barry K, Blanchette RA, Henrissat B, Martinez AT, Otillar R, Spatafora JW, Yadav JS, et al. The Paleozoic origin of enzymatic lignin decomposition reconstructed from 31 fungal genomes. Science. 2012;336:1715–1719. doi: 10.1126/science.1221748. [DOI] [PubMed] [Google Scholar]
- 6.Chen S, Xu J, Liu C, Zhu Y, Nelson DR, Zhou S, Li C, Wang L, Guo X, Sun Y, et al. Genome sequence of the model medicinal mushroom Ganoderma lucidum. Nat Commun. 2012;3:913. doi: 10.1038/ncomms1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Martinez D, Challacombe J, Morgenstern I, Hibbett D, Schmoll M, Kubicek CP, Ferreira P, Ruiz-Duenas FJ, Martinez AT, Kersten P, et al. Genome, transcriptome, and secretome analysis of wood decay fungus Postia placenta supports unique mechanisms of lignocellulose conversion. Proc Natl Acad Sci U S A. 2009;106:1954–1959. doi: 10.1073/pnas.0809575106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fernandez-Fueyo E, Ruiz-Duenas FJ, Ferreira P, Floudas D, Hibbett DS, Canessa P, Larrondo LF, James TY, Seelenfreund D, Lobos S, et al. Comparative genomics of Ceriporiopsis subvermispora and Phanerochaete chrysosporium provide insight into selective ligninolysis. Proc Natl Acad Sci U S A. 2012;109:5458–5463. doi: 10.1073/pnas.1119912109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Eastwood DC, Floudas D, Binder M, Majcherczyk A, Schneider P, Aerts A, Asiegbu FO, Baker SE, Barry K, Bendiksby M, et al. The plant cell wall-decomposing machinery underlies the functional diversity of forest fungi. Science. 2011;333:762–765. doi: 10.1126/science.1205411. [DOI] [PubMed] [Google Scholar]
- 10.Morin E, Kohler A, Baker AR, Foulongne-Oriol M, Lombard V, Nagy LG, Ohm RA, Patyshakuliyeva A, Brun A, Aerts AL, et al. Genome sequence of the button mushroom Agaricus bisporus reveals mechanisms governing adaptation to a humic-rich ecological niche. Proc Natl Acad Sci U S A. 2012;109:17501–17506. doi: 10.1073/pnas.1206847109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lu MY, Fan WL, Wang WF, Chen T, Tang YC, Chu FH, Chang TT, Wang SY, Li MY, Chen YH, et al. Genomic and transcriptomic analyses of the medicinal fungus Antrodia cinnamomea for its metabolite biosynthesis and sexual development. Proc Natl Acad Sci U S A. 2014;111:E4743–E4752. doi: 10.1073/pnas.1417570111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yap HY, Chooi YH, Firdaus-Raih M, Fung SY, Ng ST, Tan CS, Tan NH. The genome of the Tiger Milk mushroom, Lignosus rhinocerotis, provides insights into the genetic basis of its medicinal properties. BMC Genomics. 2014;15:635. doi: 10.1186/1471-2164-15-635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wawrzyn GT, Quin MB, Choudhary S, Lopez-Gallego F, Schmidt-Dannert C. Draft genome of Omphalotus olearius provides a predictive framework for sesquiterpenoid natural product biosynthesis in basidiomycota. Chem Biol. 2012;19:772–783. doi: 10.1016/j.chembiol.2012.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kohler A, Kuo A, Nagy LG, Morin E, Barry KW, Buscot F, Canback B, Choi C, Cichocki N, Clum A, et al. Convergent losses of decay mechanisms and rapid turnover of symbiosis genes in mycorrhizal mutualists. Nat Genet. 2015;47:410–415. doi: 10.1038/ng.3223. [DOI] [PubMed] [Google Scholar]
- 15.Kues U, Nelson DR, Liu C, Yu GJ, Zhang J, Li J, Wang XC, Sun H. Genome analysis of medicinal Ganoderma spp. with plant-pathogenic and saprotrophic life-styles. Phytochemistry. 2015;114:18–37. doi: 10.1016/j.phytochem.2014.11.019. [DOI] [PubMed] [Google Scholar]
- 16.Hofrichter M, Kellner H, Pecyna MJ, Ullrich R. Fungal unspecific peroxygenases: heme-thiolate proteins that combine peroxidase and cytochrome p450 properties. Adv Exp Med Biol. 2015;851:341–368. doi: 10.1007/978-3-319-16009-2_13. [DOI] [PubMed] [Google Scholar]
- 17.Pollegioni L, Tonin F, Rosini E. Lignin-degrading enzymes. FEBS J. 2015;282:1190–1213. doi: 10.1111/febs.13224. [DOI] [PubMed] [Google Scholar]
- 18.Linde D, Ruiz-Duenas FJ, Fernandez-Fueyo E, Guallar V, Hammel KE, Pogni R, Martinez AT. Basidiomycete DyPs: Genomic diversity, structural-functional aspects, reaction mechanism and environmental significance. Arch Biochem Biophys. 2015;574:66–74. doi: 10.1016/j.abb.2015.01.018. [DOI] [PubMed] [Google Scholar]
- 19.Kersten P, Cullen D. Copper radical oxidases and related extracellular oxidoreductases of wood-decay Agaricomycetes. Fungal Genet Biol. 2014;72:124–130. doi: 10.1016/j.fgb.2014.05.011. [DOI] [PubMed] [Google Scholar]
- 20.Schmidt-Dannert C. Biosynthesis of terpenoid natural products in fungi. Adv Biochem Eng Biotechnol. 2015;148:19–61. doi: 10.1007/10_2014_283. [DOI] [PubMed] [Google Scholar]
- 21.Zhong JJ, Xiao JH. Secondary metabolites from higher fungi: discovery, bioactivity, and bioproduction. Adv Biochem Eng Biotechnol. 2009;113:79–150. doi: 10.1007/10_2008_26. [DOI] [PubMed] [Google Scholar]
- 22.Crowther TW, Boddy L, Hefin Jones T. Functional and ecological consequences of saprotrophic fungus-grazer interactions. ISME J. 2012;6:1992–2001. doi: 10.1038/ismej.2012.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Spiteller P. Chemical ecology of fungi. Nat Prod Rep. 2015;32:971–993. doi: 10.1039/c4np00166d. [DOI] [PubMed] [Google Scholar]
- 24.Rohlfs M. Fungal secondary metabolite dynamics in fungus-grazer interactions: novel insights and unanswered questions. Front Microbiol. 2014;5:788. doi: 10.3389/fmicb.2014.00788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nielsen MT, Klejnstrup ML, Rohlfs M, Anyaogu DC, Nielsen JB, Gotfredsen CH, Andersen MR, Hansen BG, Mortensen UH, Larsen TO. Aspergillus nidulans synthesize insect juvenile hormones upon expression of a heterologous regulatory protein and in response to grazing by Drosophila melanogaster larvae. PLoS One. 2013;8:e73369. doi: 10.1371/journal.pone.0073369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kellner H, Luis P, Pecyna MJ, Barbi F, Kapturska D, Kruger D, Zak DR, Marmeisse R, Vandenbol M, Hofrichter M. Widespread occurrence of expressed fungal secretory peroxidases in forest soils. PLoS One. 2014;9:e95557. doi: 10.1371/journal.pone.0095557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Syed K, Shale K, Pagadala NS, Tuszynski J. Systematic identification and evolutionary analysis of catalytically versatile cytochrome p450 monooxygenase families enriched in model basidiomycete fungi. PLoS One. 2014;9:e86683. doi: 10.1371/journal.pone.0086683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Quin MB, Flynn CM, Wawrzyn GT, Choudhary S, Schmidt-Dannert C. Mushroom hunting by using bioinformatics: application of a predictive framework facilitates the selective identification of sesquiterpene synthases in basidiomycota. Chembiochem. 2013;14:2480–2491. doi: 10.1002/cbic.201300349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ichinose H. Cytochrome P450 of wood-rotting basidiomycetes and biotechnological applications. Biotechnol Appl Biochem. 2013;60:71–81. doi: 10.1002/bab.1061. [DOI] [PubMed] [Google Scholar]
- 30.Strittmatter E, Liers C, Ullrich R, Wachter S, Hofrichter M, Plattner DA, Piontek K. First crystal structure of a fungal high-redox potential dye-decolorizing peroxidase: substrate interaction sites and long-range electron transfer. J Biol Chem. 2013;288:4095–4102. doi: 10.1074/jbc.M112.400176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Salvachua D, Prieto A, Martinez AT, Martinez MJ. Characterization of a novel dye-decolorizing peroxidase (DyP)-type enzyme from Irpex lacteus and its application in enzymatic hydrolysis of wheat straw. Appl Environ Microbiol. 2013;79:4316–4324. doi: 10.1128/AEM.00699-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Strittmatter E, Serrer K, Liers C, Ullrich R, Hofrichter M, Piontek K, Schleicher E, Plattner DA. The toolbox of Auricularia auricula-judae dye-decolorizing peroxidase - Identification of three new potential substrate-interaction sites. Arch Biochem Biophys. 2015;574:75–85. doi: 10.1016/j.abb.2014.12.016. [DOI] [PubMed] [Google Scholar]
- 33.Ferreira P, Carro J, Serrano A, Martinez AT. A survey of genes encoding H2O2-producing GMC oxidoreductases in 10 Polyporales genomes. Mycologia. 2015 doi: 10.3852/15-027. [DOI] [PubMed] [Google Scholar]
- 34.Hernandez-Ortega A, Ferreira P, Martinez AT. Fungal aryl-alcohol oxidase: a peroxide-producing flavoenzyme involved in lignin degradation. Appl Microbiol Biotechnol. 2012;93:1395–1410. doi: 10.1007/s00253-011-3836-8. [DOI] [PubMed] [Google Scholar]
- 35.Gomez-Toribio V, Garcia-Martin AB, Martinez MJ, Martinez AT, Guillen F. Induction of extracellular hydroxyl radical production by white-rot fungi through quinone redox cycling. Appl Environ Microbiol. 2009;75:3944–3953. doi: 10.1128/AEM.02137-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ullrich R, Nuske J, Scheibner K, Spantzel J, Hofrichter M. Novel haloperoxidase from the agaric basidiomycete Agrocybe aegerita oxidizes aryl alcohols and aldehydes. Appl Environ Microbiol. 2004;70:4575–4581. doi: 10.1128/AEM.70.8.4575-4581.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Poulos TL. Heme enzyme structure and function. Chem Rev. 2014;114:3919–3962. doi: 10.1021/cr400415k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shoji O, Watanabe Y. Peroxygenase reactions catalyzed by cytochromes P450. J Biol Inorg Chem. 2014;19:529–539. doi: 10.1007/s00775-014-1106-9. [DOI] [PubMed] [Google Scholar]
- 39.Peter S, Kinne M, Ullrich R, Kayser G, Hofrichter M. Epoxidation of linear, branched and cyclic alkenes catalyzed by unspecific peroxygenase. Enzyme Microb Technol. 2013;52:370–376. doi: 10.1016/j.enzmictec.2013.02.013. [DOI] [PubMed] [Google Scholar]
- 40.Kluge M, Ullrich R, Scheibner K, Hofrichter M. Stereoselective benzylic hydroxylation of alkylbenzenes and epoxidation of styrene derivatives catalyzed by the peroxygenase of Agrocybe aegerita. Green Chemistry. 2012;14:440–446. [Google Scholar]
- 41.Babot ED, del Rio JC, Kalum L, Martinez AT, Gutierrez A. Oxyfunctionalization of aliphatic compounds by a recombinant peroxygenase from Coprinopsis cinerea. Biotechnol Bioeng. 2013;110:2323–2332. doi: 10.1002/bit.24904. [DOI] [PubMed] [Google Scholar]
- 42.Gutierrez A, Babot ED, Ullrich R, Hofrichter M, Martinez AT, del Rio JC. Regioselective oxygenation of fatty acids, fatty alcohols and other aliphatic compounds by a basidiomycete heme-thiolate peroxidase. Arch Biochem Biophys. 2011;514:33–43. doi: 10.1016/j.abb.2011.08.001. [DOI] [PubMed] [Google Scholar]
- 43.Peter S, Kinne M, Wang X, Ullrich R, Kayser G, Groves JT, Hofrichter M. Selective hydroxylation of alkanes by an extracellular fungal peroxygenase. FEBS J. 2011;278:3667–3675. doi: 10.1111/j.1742-4658.2011.08285.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Barkova K, Kinne M, Ullrich R, Hennig L, Fuchs A, Hofrichter M. Regioselective hydroxylation of diverse flavonoids by an aromatic peroxygenase. Tetrahedron. 2011;67:4874–4878. [Google Scholar]
- 45.Kiebist J, Holla W, Heidrich J, Poraj-Kobielska M, Sandvoss M, Simonis R, Grobe G, Atzrodt J, Hofrichter M, Scheibner K. One-pot synthesis of human metabolites of SAR548304 by fungal peroxygenases. Bioorg Med Chem. 2015;23:4324–4332. doi: 10.1016/j.bmc.2015.06.035. [DOI] [PubMed] [Google Scholar]
- 46.Peter S, Karich A, Ullrich R, Grobe G, Scheibner K, Hofrichter M. Enzymatic one-pot conversion of cyclohexane into cyclohexanone: Comparison of four fungal peroxygenases. Journal of Molecular Catalysis B-Enzymatic. 2014;103:47–51. [Google Scholar]
- 47.Poraj-Kobielska M, Atzrodt J, Holla W, Sandvoss M, Grobe G, Scheibner K, Hofrichter M. Preparation of labeled human drug metabolites and drug-drug interaction-probes with fungal peroxygenases. Journal of Labelled Compounds & Radiopharmaceuticals. 2013;56:513–519. doi: 10.1002/jlcr.3103. [DOI] [PubMed] [Google Scholar]
- 48.Poraj-Kobielska M, Peter S, Leonhardt S, Ullrich R, Scheibner K, Hofrichter M. Immobilization of unspecific peroxygenases (EC 1.11.2.1) in PVA/PEG gel and hollow fiber modules. Biochemical Engineering Journal. 2015;98:144–150. [Google Scholar]
- 49.Carro J, Ferreira P, Rodriguez L, Prieto A, Serrano A, Balcells B, Arda A, Jimenez-Barbero J, Gutierrez A, Ullrich R, et al. 5-hydroxymethylfurfural conversion by fungal aryl-alcohol oxidase and unspecific peroxygenase. FEBS J. 2015;282:3218–3229. doi: 10.1111/febs.13177. [DOI] [PubMed] [Google Scholar]
- 50.Piontek K, Strittmatter E, Ullrich R, Grobe G, Pecyna MJ, Kluge M, Scheibner K, Hofrichter M, Plattner DA. Structural basis of substrate conversion in a new aromatic peroxygenase: cytochrome P450 functionality with benefits. J Biol Chem. 2013;288:34767–34776. doi: 10.1074/jbc.M113.514521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang X, Peter S, Kinne M, Hofrichter M, Groves JT. Detection and kinetic characterization of a highly reactive heme-thiolate peroxygenase compound I. J Am Chem Soc. 2012;134:12897–12900. doi: 10.1021/ja3049223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang X, Peter S, Ullrich R, Hofrichter M, Groves JT. Driving force for oxygen-atom transfer by heme-thiolate enzymes. Angew Chem Int Ed Engl. 2013;52:9238–9241. doi: 10.1002/anie.201302137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang X, Ullrich R, Hofrichter M, Groves JT. Heme-thiolate ferryl of aromatic peroxygenase is basic and reactive. Proc Natl Acad Sci U S A. 2015;112:3686–3691. doi: 10.1073/pnas.1503340112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Molina-Espeja P, Garcia-Ruiz E, Gonzalez-Perez D, Ullrich R, Hofrichter M, Alcalde M. Directed evolution of unspecific peroxygenase from Agrocybe aegerita. Appl Environ Microbiol. 2014;80:3496–3507. doi: 10.1128/AEM.00490-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Syed K, Nelson DR, Riley R, Yadav JS. Genomewide annotation and comparative genomics of cytochrome P450 monooxygenases (P450s) in the polypore species Bjerkandera adusta, Ganoderma sp and Phlebia brevispora. Mycologia. 2013;105:1445–1455. doi: 10.3852/13-002. [DOI] [PubMed] [Google Scholar]
- 56.Agger S, Lopez-Gallego F, Schmidt-Dannert C. Diversity of sesquiterpene synthases in the basidiomycete Coprinus cinereus. Mol Microbiol. 2009;72:1181–1195. doi: 10.1111/j.1365-2958.2009.06717.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hirosue S, Tazaki M, Hiratsuka N, Yanai S, Kabumoto H, Shinkyo R, Arisawa A, Sakaki T, Tsunekawa H, Johdo O, et al. Insight into functional diversity of cytochrome P450 in the white-rot basidiomycete Phanerochaete chrysosporium: involvement of versatile monooxygenase. Biochem Biophys Res Commun. 2011;407:118–123. doi: 10.1016/j.bbrc.2011.02.121. [DOI] [PubMed] [Google Scholar]
- 58.Ide M, Ichinose H, Wariishi H. Molecular identification and functional characterization of cytochrome P450 monooxygenases from the brown-rot basidiomycete Postia placenta. Arch Microbiol. 2012;194:243–253. doi: 10.1007/s00203-011-0753-2. [DOI] [PubMed] [Google Scholar]
- 59.Tasaki Y, Toyama S, Kuribayashi T, Joh T. Molecular characterization of a lipoxygenase from the basidiomycete mushroom Pleurotus ostreatus. Biosci Biotechnol Biochem. 2013;77:38–45. doi: 10.1271/bbb.120484. [DOI] [PubMed] [Google Scholar]
- 60.Fraatz MA, Riemer SJL, Stober R, Kaspera R, Nimtz M, Berger RG, Zorn H. A novel oxygenase from Pleurotus sapidus transforms valencene to nootkatone. Journal of Molecular Catalysis B - Enzymatic. 2009;61:202–207. [Google Scholar]
- 61.Krugener S, Krings U, Zorn H, Berger RG. A dioxygenase of Pleurotus sapidus transforms (+)-valencene regio-specifically to (+)-nootkatone via a stereo-specific allylic hydroperoxidation. Bioresource Technology. 2010;101:457–462. doi: 10.1016/j.biortech.2009.08.087. [DOI] [PubMed] [Google Scholar]
- 62.Quin MB, Flynn CM, Schmidt-Dannert C. Traversing the fungal terpenome. Nat Prod Rep. 2014;31:1449–1473. doi: 10.1039/c4np00075g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Schneider P, Bouhired S, Hoffmeister D. Characterization of the atromentin biosynthesis genes and enzymes in the homobasidiomycete Tapinella panuoides. Fungal Genet Biol. 2008;45:1487–1496. doi: 10.1016/j.fgb.2008.08.009. [DOI] [PubMed] [Google Scholar]
- 64.Misiek M, Braesel J, Hoffmeister D. Characterisation of the ArmA adenylation domain implies a more diverse secondary metabolism in the genus Armillaria. Fungal Biol. 2011;115:775–781. doi: 10.1016/j.funbio.2011.06.002. [DOI] [PubMed] [Google Scholar]
- 65.Wackler B, Lackner G, Chooi YH, Hoffmeister D. Characterization of the Suillus grevillei quinone synthetase GreA supports a nonribosomal code for aromatic alpha-keto acids. Chembiochem. 2012;13:1798–1804. doi: 10.1002/cbic.201200187. [DOI] [PubMed] [Google Scholar]
- 66.Lackner G, Bohnert M, Wick J, Hoffmeister D. Assembly of melleolide antibiotics involves a polyketide synthase with cross-coupling activity. Chem Biol. 2013;20:1101–1106. doi: 10.1016/j.chembiol.2013.07.009. [DOI] [PubMed] [Google Scholar]
- 67.Kalb D, Lackner G, Hoffmeister D. Functional and phylogenetic divergence of fungal adenylate-forming reductases. Appl Environ Microbiol. 2014;80:6175–6183. doi: 10.1128/AEM.01767-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Winterberg B, Uhlmann S, Linne U, Lessing F, Marahiel MA, Eichhorn H, Kahmann R, Schirawski J. Elucidation of the complete ferrichrome A biosynthetic pathway in Ustilago maydis. Mol Microbiol. 2010;75:1260–1271. doi: 10.1111/j.1365-2958.2010.07048.x. [DOI] [PubMed] [Google Scholar]
- 69.Shah F, Schwenk D, Nicolas C, Persson P, Hoffmeister D, Tunlid A. Involutin is a Fe3+ reductant Secreted by the Ectomycorrhizal Fungus Paxillus involutus during Fenton-based Decomposition of Organic Matter. Appl Environ Microbiol. 2015 doi: 10.1128/AEM.02312-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Chen PY, Wu JD, Tang KY, Yu CC, Kuo YH, Zhong WB, Lee CK. Isolation and synthesis of a bioactive benzenoid derivative from the fruiting bodies of Antrodia camphorata. Molecules. 2013;18:7600–7608. doi: 10.3390/molecules18077600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Thongbai B, Surup F, Mohr K, Kuhnert E, Hyde KD, Stadler M. Gymnopalynes A and B, chloropropynyl-isocoumarin antibiotics from cultures of the basidiomycete Gymnopus sp. J Nat Prod. 2013;76:2141–2144. doi: 10.1021/np400609f. [DOI] [PubMed] [Google Scholar]
- 72.Blacklock BJ, Scheffler BE, Shepard MR, Jayasuriya N, Minto RE. Functional diversity in fungal fatty acid synthesis: the first acetylenase from the Pacific golden chanterelle, Cantharellus formosus. J Biol Chem. 2010;285:28442–28449. doi: 10.1074/jbc.M110.151498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zhu X, Liu J, Zhang W. De novo biosynthesis of terminal alkyne-labeled natural products. Nat Chem Biol. 2015;11:115–120. doi: 10.1038/nchembio.1718. [DOI] [PubMed] [Google Scholar]
- 74.Malagon O, Porta A, Clericuzio M, Gilardoni G, Gozzini D, Vidari G. Structures and biological significance of lactarane sesquiterpenes from the European mushroom Russula nobilis. Phytochemistry. 2014;107:126–134. doi: 10.1016/j.phytochem.2014.08.018. [DOI] [PubMed] [Google Scholar]
- 75.Quin MB, Michel SN, Schmidt-Dannert C. Moonlighting Metals: Insights into Regulation of Cyclization Pathways in Fungal Delta -Protoilludene Sesquiterpene Synthases. Chembiochem. 2015 doi: 10.1002/cbic.201500308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Blin K, Medema MH, Kazempour D, Fischbach MA, Breitling R, Takano E, Weber T. antiSMASH 2.0--a versatile platform for genome mining of secondary metabolite producers. Nucleic Acids Res. 2013;41:W204–W212. doi: 10.1093/nar/gkt449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Isaka M, Srisanoh U, Choowong W, Boonpratuang T. Sterostreins A-E, new terpenoids from cultures of the basidiomycete Stereum ostrea BCC 22955. Org Lett. 2011;13:4886–4889. doi: 10.1021/ol2019778. [DOI] [PubMed] [Google Scholar]
- 78.Zhu YC, Wang G, Yang XL, Luo DQ, Zhu QC, Peng T, Liu JK. Agrocybone, a novel bis-sesquiterpene with a spirodienone structure from basidiomycete Agrocybe salicacola. Tetrahedron Letters. 2010;51:3443–3445. [Google Scholar]
- 79.Lehnert N, Krings U, Sydes D, Wittig M, Berger RG. Bioconversion of car-3-ene by a dioxygenase of Pleurotus sapidus. J Biotechnol. 2012;159:329–335. doi: 10.1016/j.jbiotec.2011.06.007. [DOI] [PubMed] [Google Scholar]
- 80.Leonhardt RH, Plagemann I, Linke D, Zelena K, Berger RG. Orthologous lipoxygenases of Pleurotus spp. - A comparison of substrate specificity and sequence homology. Journal of Molecular Catalysis B-Enzymatic. 2013;97:189–195. [Google Scholar]