Abstract
A long-sought goal in the hemoglobin field has been an improved understanding of the mechanisms that regulate the switch from fetal (HbF) to adult (HbA) hemoglobin during development. With such knowledge, the hope is that strategies for directed reactivation of HbF in adults could be devised as an approach to therapy for the β-hemoglobinopathies thalassemia and sickle cell disease. Recent genome-wide association studies led to identification of three loci (BCL11A, HBS1L-Myb, and the β-globin cluster itself) in which natural genetic variation is correlated with different HbF levels in populations. Here, the central role of BCL11A in control of HbF is reviewed from the perspective of how findings may be translated to gene therapy in the not-too-distant future. This summary traces the evolution of recent studies from the initial recognition of BCL11A through GWAS to identification of critical sequences in an enhancer required for its erythroid-specific expression, thereby highlighting an Achilles heel for genome editing.
Keywords: thalassemia, BCL11A, gene editing, enhancer, CRISPR
Introduction
The field of hemoglobin research has often revealed basic principles in biology and genetics, as stated in the prescient remarks of Arno Motulsky at a symposium in 1974: “Hemoglobin research plays a role in human biochemical genetics similar to that of Drosophila research in formal genetics. Many fundamental concepts have become clarified by investigations on human hemoglobins.”1 The molecular era of hemoglobin research was ushered in with the application of newly developed methods for gene cloning soon thereafter, work that set the stage for the elucidation of the diverse mutations in the β-thalassemias. Haplotype analysis in thalassemia, pioneered by Kazazian and colleagues,2 facilitated selection of β-thalassemia alleles for cloning and DNA sequencing,3 ultimately leading to the nearly complete molecular description of these disorders. With the mutations in hand, prenatal diagnosis by DNA analysis could be implemented in major centers located in high-incidence geographic areas. The reduction in births of newly affected individuals in locations such as Sardinia where prenatal diagnosis has been performed is an enormous public health success. Despite these genetic triumphs, the β-thalassemias remain global health challenges, along with sickle cell disease (SCD), particularly in the less developed world.4 Clinical management remains supportive care and iron chelation. Bone marrow transplantation (and possibly gene therapy with globin-expressing lentiviruses) achieves genetic cures but cannot be widely applied, especially in those regions where medical resources are limited. An attractive approach to management of the hemoglobin disorders is the reactivation of fetal hemoglobin (HbF), as we have known for more than 50 years that elevated HbF is beneficial in the β –hemoglobinopathies.5
For many years, a major obstacle to developing novel therapies based on HbF reactivation in adult erythroid cells was our relative ignorance regarding mechanisms by which the relative balance of HbF and HbA expression is regulated. In 1961, Ingram proposed the first sensible model for switching from fetal to adult globin in humans, namely the turning-down of fetal (or γ-globin) expression and the upregulation of adult (β- globin) expression.6 Perhaps this hypothesis seems obvious to us now, but at the time various other models were put forward, as the organization of globin genes was not well defined, nor were mechanisms of gene expression. Investigators in the globin field expended many years of research in the attempt to identify stage-selective globin regulators, albeit with little success. Our own efforts in the late 1980s led to the discovery of GATA1 binding at the site of one of the classical hereditary persistence of fetal hemoglobin mutations in a γ-gene promoter.7 Progress stalled for a long period of time.
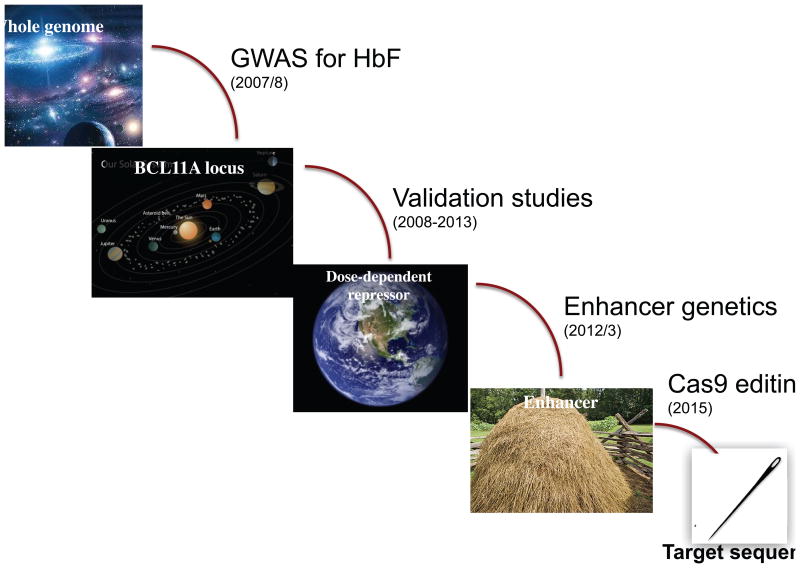
The first genome-wide association studies (GWAS) for hemoglobin F (HbF) cell number and HbF level8–10 provided the major breakthroughs that identified at least two principal regulators in humans, BCL11A and c-Myb. In sum, the three major hits of these studies were the BCL11A locus on chromosome 2p, the HBS1L-Myb region on chromosome 6q, and the β-globin cluster on chromosome 11p. Genetic variation in the globin locus itself that influenced HbF levels has been recognized for a long time (although the specific mechanisms remain obscure). Genetic and functional studies subsequent to the original GWAS have pinpointed c-Myb as the relevant gene in the HBS1L-Myb region. Over the past 7 years, my group has focused on the BCL11A locus, and now can confidently assert that it is a principal regulator of the fetal-to-adult switch, BCL11A, established by human genetics and surrogate genetics in cells and mice. Work from our laboratory has progressed from GWAS implicating BCL11A in HbF control to very recent studies in which an Achilles heel within an erythroid enhancer within BCL11A has been identified through saturating CRISPR/Cas9 mutagenesis11 (Fig. 1). This region is being evaluated for therapeutic gene editing as enhanced gene therapy for hemoglobin disorders. In this brief summary, some of the critical findings will be reviewed and discussed in the context of potential clinical application.
Figure 1.
Path from GWAS to target sequences in the BCL11A enhancer. After the GWAS reports identifying BCL11A as a potential regulator of HbF level, a series of studies validated the critical role of the factor, and more recently led to pinpointing target sequences in the BCL11A enhancer required for expression in erythroid cells. These target sequences have been proposed as a suitable region for therapeutic genome editing.
Functional validation of BCL11A as a critical regulator of the fetal-to-adult globin switch and HbF silencing
In initial experiments in primary human CD34+ stem/progenitor cells in culture, we showed that small hairpin RNA (shRNA)–mediated knockdown of BCL11A mRNA leads to increased γ-globin mRNA expression and increased HbF.12 As many manipulations of CD34+ cells in this system lead to apparent increases in HbF, often due to alteration of cell proliferation or differentiation, these data were reassuring but not definitive. Genetic studies demonstrated that knockout of BCL11A in the mouse leads to a striking failure to repress embryonic β-globins in the fetal liver.13 Moreover, BCL11A knockout embryos at this stage fail to turn off a human γ-globin gene in the context of a β-globin yeast artificial chromosome (YAC) transgene. Through study of erythroid-specific knockout of BCL11A, we observed that HbF expression in adult YAC mice was 500- to 1000-fold higher than in control mice (where human γ-globin is highly repressed).14 Finally, erythroid-specific knockout of BCL11A nearly completely rescues the phenotypes of genetically engineered SCD mouse models, due to high-level, pancellular HbF expression.14 It is important to recognize that the effects of BCL11A on HbF far exceed that reported for any other regulatory factors in mouse genetic experiments. Often, published results have been expressed as fold induction, rather than the actual amount of HbF production, thereby leading to overestimates of true contribution of some factors.
BCL11A as a quantitative regulator of HbF silencing
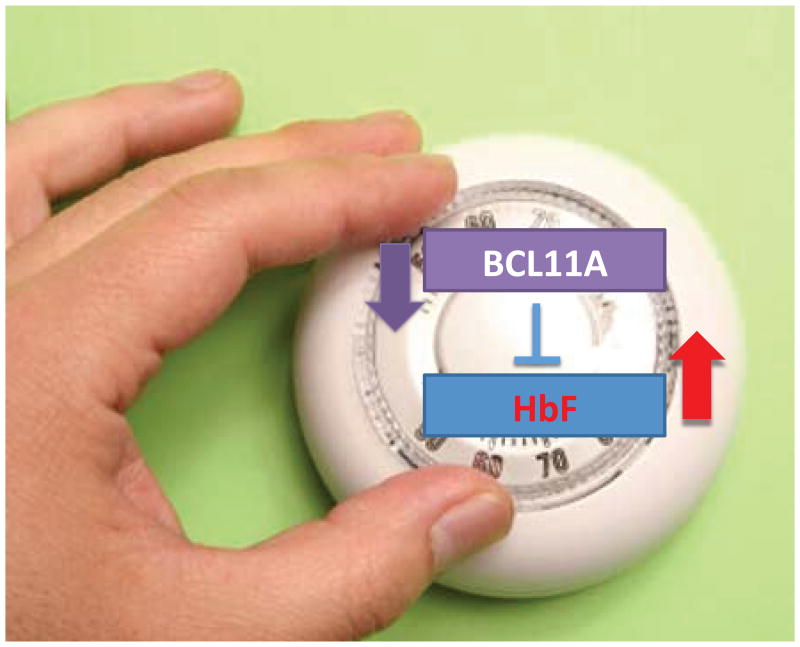
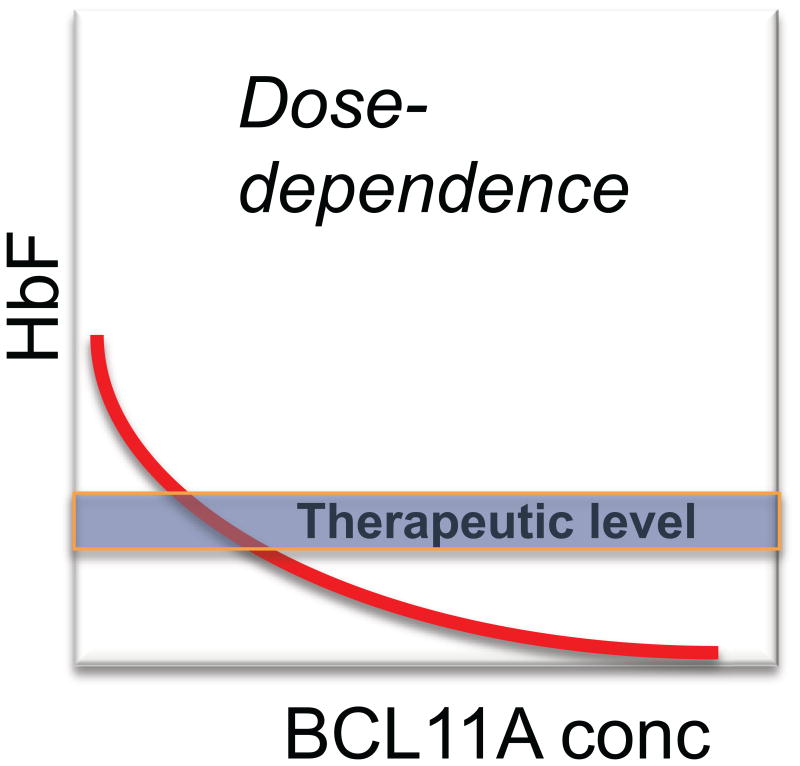
An important concept arising from the BCL11A knockout studies is that repression of HbF expression is sensitive to the level of BCL11A13 (Fig. 2). For example, in mouse fetal liver, the repression of endogenous embryonic globins of the mouse or HbF from a human transgene is less in homozygote as compared with heterozygote embryos, and less in heterozygote versus normal embryos. The data we have obtained in various experimental settings is consistent with the notion that BCL11A is a “rheostat” for HbF repression. The precise shape of the dosage curve of HbF versus BCL11A levels is unknown (Fig. 3). Nonetheless, recent insights are provided from rare, haploinsufficient patients that have chromosomal deletions involving one chromosome 2p region (including BCL11A).15 These individuals are ascertained by neurologists and geneticists and present with a severe autism-like syndrome. In three patients for whom we have hematological data, the level of HbF is quite remarkable: ∼15–30%. Thus, loss of one copy of BCL11A is sufficient to lead to considerable relief of HbF silencing. These clinical observations have important implications for considering the extent to which BCL11A levels might need to be reduced to cause a therapeutic effect in the hemoglobinopathies. An additional finding from the study of the haploinsufficient patients was normal numbers of B lymphocytes.15 Given that BCL11A is essential for B cell development,16 this observation suggests that B cells are relatively spared in the setting of a modest reduction in BCL11A levels.
Figure 2.
BCL11A as a quantitative regulator of HbF. Functional studies indicate that BCL11A functions as a continuous, quantitative regulator of HbF levels, such that downregulation of BCL11A expression (or protein) is accompanied by a concomitant increase in HbF expression. The relevance of this relationship is depicted in Figure 3.
Figure 3.
Concept of dose dependence of BCL11A in relation to a therapeutic level. Of greatest relevance to clinical translation of BCL11A as a target for therapeutic reactivation of HbF is the precise relationship between the level of BCL11A and expression of HbF in vivo. The shape of the curve is unknown. Based on cellular models11 and the level of HbF in haploinsufficient individuals,15 it is likely that the in vivo level of HbF in the absence of BCL11A would exceed the level needed to correct β-thalassemia and SCD. It will be important in any clinical studies, whether using shRNA knockdown21 of BCL11A or gene editing,11 to determine the extent of BCL11A reduction needed to achieve correction.
Genetic variation in the BCL11A locus within an erythroid enhancer element
Identification of the BCL11A locus as a potential regulator of HbF through GWAS suggests that genetic variation might influence protein structure or expression. The presence of the most highly correlated GWAS single-nucleotide polymorphisms (SNPs) in the second intron of BCL11A and the status of BCL11A as a “hyper-conserved” gene are consistent with the latter possibility, as is the dosage sensitivity of HbF repression noted above. The intragenic, sentinel SNPs within the BCL11A locus are intermingled with erythroid-specific DNase-hypersensitivity sites (HS), chromatin marks, and transcription factor binding indicative of regulatory potential. Through transient transgenesis, we demonstrated that the human SNP-dense region of ∼10 kb is a potential adult-stage, erythroid-specific enhancer that is active in mouse fetal liver and inactive in the B lymphoid lineage.17
The human enhancer region is comprised of three HS sites, +55, +58, and +62 kb from the transcription start site of the BCL11A gene. Further in-depth analysis of genetic variation, coupled with DNA sequencing, implicated a potential causal SNP in the +62 HS site. Allele-specific chromatin immunoprecipitation (ChIP) for GATA1 and TAL1 and allele-specific RNA analysis in CD34+-derived erythroid cells from individuals heterozygous for high and low BCL11A alleles provided strong support for this SNP as causal.17 A minor contribution of genetic variation in the +55 HS site was also observed. Remarkably, although the +58 region is the only HS that is autonomously active in the transgenic assay, no obvious SNPs were obtained through GWAS. Allele-specific RNA transcript analysis suggested that the SNP associated with the high-HbF BCL11A allele leads to ∼ 40% reduction in expression.
Requirement of the BCL11A enhancer for erythroid-specific gene expression
The functional data to this point demonstrated that the enhancer can drive erythroid-specific gene expression, but did not define a requirement in the context of the otherwise intact, endogenous chromosomal locus. As a preliminary test of necessity, we first turned to the corresponding mouse enhancer in cultured erythroleukemia (MEL) cells, and used a pair of engineered zinc fingers (TALENS) to delete the entire region. MEL cells lacking the enhancer expressed <1% of the level of BCL11A as wild-type cells, indicating that the enhancer is essential for BCL11A expression in this cellular context.17
The specificity of the enhancer is critical to considerations regarding the potential of BCL11A to serve as a target for HbF reactivation in patients. Genetic manipulations restricted to the enhancer would limit the effects on BCL11A expression to the erythroid lineage and thereby spare HSCs or other lineages any deleterious consequences.
Functional mapping of the erythroid enhancer by CRISPR/Cas9 editing
Gene modification has become easier and more efficient through the development of the CRISPR/Cas9 platform.18 In evaluating parameters for gene deletion in hematopoietic cell lines, we found that creation of targeted deletions of moderate size (e.g., ∼10 kb) with pairs of guide RNAs (gRNAs) led to multiple outcomes, including inversions and “scarred” loci.19 Moreover, as the length of DNA to be deleted was increased, the average frequency of deletion decreased. With the hope of defining critical features of the BCL11A enhancer that might be leveraged for potential clinical gene editing, we explored the use of the CRISPR/Cas9 system for unbiased, high-resolution functional mapping of the enhancer.11 The concept was to perform a screen of the enhancer with a library of gRNAs and read out the results by scoring enrichment of gRNAs in cells fluorescence-assisted cell sorting (FACS) sorted for increased HbF. To establish the method, we employed a novel cell line, HUDEP-2, which was established through oncogenic immortalization of human CD34+ cells.20 The HUDEP-2 cells predominantly express HbA, and will express reactive HbF upon downregulation or knockout of BCL11A. The advantage of these cells, as compared with primary CD34+ cells, is that they can be expanded and cloned, and therefore handled as a typical cell line. The gRNA library was introduced into HUDEP-2 cells, HbF high-expressing cells were isolated by FACS, and the distribution of gRNAs was deconvoluted by DNA sequencing. The results were remarkably robust and identified a discrete region within +58, neighboring a GATA1-binding site, that can be targeted by individual gRNAs, leading to HbF reactivation.11 At the critical region, which corresponds to the peak DNase sensitivity of +58, single cleavages and repair via non-homologous end-joining (NHEJ) reactivates HbF expression nearly to the level of the BCL11A knockout itself.
A similar screen was performed in MEL cells in which a reporter was inserted into the embryonic εy gene to monitor loss of BCL11A function.11 Although the enhancer region of the mouse is superficially similar to that of the human, in detail it is quite distinct. For one, the +58 site is not hypersensitive in the mouse. The functional CRISPR/Cas9 screen identified a discrete region within the +62 site of the mouse that is required for maximal enhancer activity, though its contribution to overall enhancer function appeared less that than of the human +58 site. The differences between the functional maps of the mouse and human enhancers reflect considerable evolutionary changes that occurred between the species and may contribute to their different hemoglobin switching patterns.
Modifying the modifier as an approach to treatment of the hemoglobinopathies
As a major repressor of HbF expression, BCL11A is a bona fide target for genetic manipulation as a form of somatic gene therapy. Given the broader roles of BCL11A in the hematopoietic system, knockout of BCL11A in HSCs is disfavored. Remarkably, red blood cell development is quite unperturbed in the absence of BCL11A, which is unlike the majority of transcription factors that control erythroid gene expression. Two approaches relying on BCL11A as a target are currently being brought forward for clinical investigation. Williams and colleagues are exploring application of lentiviral-mediated transfer of erythroid-expressed shRNA directed to BCL11A into HSCs as gene therapy reactivation of HbF in SCD.21 This strategy follows the emerging use of globin gene transfer in LCR-based lentiviral vectors for therapy of β-thalassemias or SCD. Sangamo Biosciences is developing zinc-finger nucleases directed to the erythroid enhancer of BCL11A for treatment of hemoglobinopathies.22 Conditions have been established for high-efficiency ex vivo gene editing of the enhancer in CD34+ stem/progenitor cells. A potential advantage of editing is the absence of integration of foreign DNA, as occurs in current gene therapy approaches. It is hoped that phase I trials for these approaches will be initiated within the coming year. If carried out, such trials should establish whether manipulation of BCL11A alone elicits sufficient HbF reactivation to correct the hemoglobin disorders. In principle, knockdown of BCL11A with shRNA could be coupled with delivery of a globin chain, thereby maximizing the potential for full correction. Manipulation of BCL11A expression as a means of therapy is appealing in that reactivation of HbF is associated with concomitant downregulation of β-globin expression, thereby reducing expression of the defective gene and maintaining proper globin balance. Altering BCL11A expression for therapy differs conceptually from conventional gene therapy in that the strategy takes advantage of a genetic modifier of phenotype to ameliorate disease, essentially improving on what natural genetic variation achieves.
Where do we go from here?
Over the past several years, there has been a revolution in our understanding of the factors responsible for globin switching. In addition to BCL11A, another transcription factor, LRF, has emerged as an important regulator through similar transgenic mouse studies and gene deletion in human erythroid cells.23 HUDEP-2 cells in which both BCL11A and LRF and inactivated express nearly 100% HbF. Thus, in some sense the two factors encompass the entirety of HbF silencing. Of note, BCL11A and LRF each interacts physically with the NuRD chromatin complex, yet are not present in the same multiprotein complexes. These observations indicate that they share some common features, yet play distinct roles. How any of the known HbF repressors function in detail is unknown.
The cell-restricted role of the BCL11A erythroid enhancer and its discrete region of vulnerability, taken together with the limited consequences of BCL11A loss on red blood cell production, lie at the foundation of current efforts to target BCL11A for therapeutic reactivation of HbF. Unless unforeseen obstacles arise, the true in vivo effects of manipulating the BCL11A enhancer by gene editing or reducing BCL11A by shRNA expression are likely to be defined. The hope is that these efforts will lead to safe and effective gene therapy for patients with the β-hemoglobinopathies, and justify, as if such justification is needed, the investments that have been made over the years in basic and translational research.
We must remember that the health burden of the hemoglobin disorders is global in scope.4 Unless unimagined advances occur in how genetic therapy can be achieved in geographic areas with meager or modest medical resources, we will still need improved small molecule (drug) approaches for either reactivation of HbF or prevention of the sickling process. With improved understanding of the factors controlling globin switching and how they act, there are no theoretical barriers to the development of active small molecules. Achievement of this long-sought, elusive goal necessitates a concerted investment and effort, both by academic and pharmaceutical entities, to apply leading-edge chemistry and chemical biology to the problem. Only then will the full promise of molecular medicine be realized.
Acknowledgments
I am grateful to those who have contributed to the work reviewed here, including Vijay Sankaran, Guillaume Lettre, Jian Xu, Dan Bauer, Sophia Kamran, Crew Smith, Yuko Fujiwara, and Matt Canver. Work from my laboratory is supported by the National Heart, Lung, and Blood Institute (NHLBI) and the National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK) of the National Institutes of Health and the Howard Hughes Medical Institute.
References
- 1.Motulsky AG. Genetics of human hemoglobins: an overview. Ann NY Acad Sci. 1974;241(0):7–11. doi: 10.1111/j.1749-6632.1974.tb21862.x. [DOI] [PubMed] [Google Scholar]
- 2.Kazazian HH, Jr, et al. Use of haplotype analysis in the beta-globin gene cluster to discover beta-thalassemia mutations. Prog Clin Biol Res. 1983;134:91–8. [PubMed] [Google Scholar]
- 3.Orkin SH, et al. Linkage of beta-thalassaemia mutations and beta-globin gene polymorphisms with DNA polymorphisms in human beta-globin gene cluster. Nature. 1982;296(5858):627–31. doi: 10.1038/296627a0. [DOI] [PubMed] [Google Scholar]
- 4.Weatherall DJ. The inherited diseases of hemoglobin are an emerging global health burden. Blood. 2010;115(22):4331–6. doi: 10.1182/blood-2010-01-251348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bauer DE, Kamran SC, Orkin SH. Reawakening fetal hemoglobin: prospects for new therapies for the beta-globin disorders. Blood. 2012;120(15):2945–53. doi: 10.1182/blood-2012-06-292078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Baglioni C, Ingram VM, Sullivan E. Genetic control of foetal and adult human haemoglobin. Nature. 1961;189:467–9. doi: 10.1038/189467a0. [DOI] [PubMed] [Google Scholar]
- 7.Tsai SF, et al. Cloning of cDNA for the major DNA-binding protein of the erythroid lineage through expression in mammalian cells. Nature. 1989;339(6224):446–51. doi: 10.1038/339446a0. [DOI] [PubMed] [Google Scholar]
- 8.Menzel S, et al. A QTL influencing F cell production maps to a gene encoding a zinc-finger protein on chromosome 2p15. Nat Genet. 2007;39(10):1197–9. doi: 10.1038/ng2108. [DOI] [PubMed] [Google Scholar]
- 9.Uda M, et al. Genome-wide association study shows BCL11A associated with persistent fetal hemoglobin and amelioration of the phenotype of beta-thalassemia. Proc Natl Acad Sci U S A. 2008;105(5):1620–5. doi: 10.1073/pnas.0711566105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lettre G, et al. DNA polymorphisms at the BCL11A, HBS1L-MYB, and beta-globin loci associate with fetal hemoglobin levels and pain crises in sickle cell disease. Proc Natl Acad Sci U S A. 2008;105(33):11869–74. doi: 10.1073/pnas.0804799105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Canver MC, et al. BCL11A enhancer dissection by Cas9-mediated in situ saturating mutagenesis. Nature. 2015;527(7577):192–7. doi: 10.1038/nature15521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sankaran VG, et al. Human fetal hemoglobin expression is regulated by the developmental stage-specific repressor BCL11A. Science. 2008;322(5909):1839–42. doi: 10.1126/science.1165409. [DOI] [PubMed] [Google Scholar]
- 13.Sankaran VG, et al. Developmental and species-divergent globin switching are driven by BCL11A. Nature. 2009;460(7259):1093–7. doi: 10.1038/nature08243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xu J, et al. Correction of sickle cell disease in adult mice by interference with fetal hemoglobin silencing. Science. 2011;334(6058):993–6. doi: 10.1126/science.1211053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Basak A, et al. BCL11A deletions result in fetal hemoglobin persistence and neurodevelopmental alterations. J Clin Invest. 2015;125(6):2363–8. doi: 10.1172/JCI81163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu P, et al. Bcl11a is essential for normal lymphoid development. Nat Immunol. 2003;4(6):525–32. doi: 10.1038/ni925. [DOI] [PubMed] [Google Scholar]
- 17.Bauer DE, et al. An erythroid enhancer of BCL11A subject to genetic variation determines fetal hemoglobin level. Science. 2013;342(6155):253–7. doi: 10.1126/science.1242088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hsu PD, Lander ES, Zhang F. Development and applications of CRISPR-Cas9 for genome engineering. Cell. 2014;157(6):1262–78. doi: 10.1016/j.cell.2014.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Canver MC, et al. Characterization of genomic deletion efficiency mediated by clustered regularly interspaced palindromic repeats (CRISPR)/Cas9 nuclease system in mammalian cells. J Biol Chem. 2014;289(31):21312–24. doi: 10.1074/jbc.M114.564625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kurita R, et al. Establishment of immortalized human erythroid progenitor cell lines able to produce enucleated red blood cells. PLoS One. 2013;8(3):e59890. doi: 10.1371/journal.pone.0059890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guda S, et al. miRNA-embedded shRNAs for Lineage-specific BCL11A Knockdown and Hemoglobin F Induction. Mol Ther. 2015;23(9):1465–74. doi: 10.1038/mt.2015.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vierstra J, et al. Functional footprinting of regulatory DNA. Nat Methods. 2015;12(10):927–30. doi: 10.1038/nmeth.3554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Masuda T, et al. The LRF transcription factor is a BCL11A-independent repressor of fetal hemoglobin. Science. 2015 in press. [Google Scholar]