Abstract
Skeletal dysplasias are highly heterogeneous disorders composed of 40 clinical subtypes that are part of 456 well-delineated syndromes in humans. Here, we enrolled consanguineous kindred from a remote area of Sindh province of Pakistan, with fourteen affected individuals suffering with short stature, kyphoscoliosis, joint dislocations, clubfoot, heart valve anomalies and progressive bilateral mixed hearing loss. To identify pathogenic variants in this family, whole exome sequencing (WES) was performed in one affected and one normal individual, which revealed a novel transversion mutation (c.802G>T; p.Glu268*) in CHST3 associated with the phenotype. CHST3 encodes a chondroitin 6-O-sulfotransferase-1 (C6ST-1) enzyme that is essential for the sulfation of proteoglycans found in cartilages. Previously, mutations in CHST3 have largely been reported in sporadic cases of skeletal dysplasia, primarily with severe spinal abnormalities, joint dislocations, joint contractures, and clubfoot. Clinical and radiological examination of the affected individuals in this family provides new insights into phenotypic spectrum of CHST3 alleles and disease progression with age.
Introduction
Genetic disorders that result in deficits of skeletal systems are collectively termed as skeletal dysplasias (SDs) or Osteochondrodysplasias. Their prevalence has been estimated to be 1:5000 (1). However, in children with congenital birth defects, SD prevalence is approximately 5% (2). SD has been classified into 40 sub-groups primarily on the basis of clinical, molecular, biochemical and radiological findings (3). In some cases, SD leads to lethality due to small chest, pulmonary hypoplasia, and respiratory problems (4).
Genetically, SD can manifest under a variety of inheritance models including autosomal dominant or recessive, and X-linked dominant or recessive (2, 3). Hundreds of genes have been found that subserve a variety of functions in bone development, mineralization and growth in humans. Deciphering the genetic causes of SDs often help in comprehending basic bone biology, disease prognosis and treatment options. For instance, in some cases of monogenic SDs, novel targeted therapies including Cathepsin K inhibitors and anti-sclerostin monoclonal antibodies are now in late phases of clinical trials (5–7).
Here, we describe a six-generation Pakistani family (LUAB01) with autosomal recessively inherited congenital skeletal dysplasia and hearing loss. Whole exome sequencing (WES) revealed a nonsense (p.Glu268*) mutation in the CHST3 (OMIM 603799) gene segregating with the phenotype. C6ST-1 enzyme, encoded by CHST3, belongs to the carbohydrate sulfotransferase family of fifteen enzymes that is involved in the transfer of sulfate groups to carbohydrates in glycoprotein and glycolipids. C6ST-1 catalyzes sulfation of chondroitin containing proteoglycan, an essential constituent of connective tissues (8). Previously, mutations in CHST3 have been associated with a rare phenotype of skeleton dysplasia, known as Spondyloepiphyseal dysplasia with congenital joint dislocations (OMIM 143095) (9, 10). The phenotypic presentation of the disease varies among ethnically and geographically divergent populations (9–12). Common features include large joint contractures, progressive kyphosis, clubfoot, and dislocation of hip joints (9–12). Hearing loss incidence in patients with CHST3 mutations has been anecdotal. Intriguingly, all the affected individuals of the family LUAB01 have bilateral symmetric mild-to-severe hearing impairment and thus provided statistically significant evidence of linkage of hearing loss phenotype with the CHST3 allele.
Subject and Methods
Enrollment and clinical evaluation
This study was approved by the Ethical Review Committees of the participating institutes. After obtaining written informed consent, blood samples were collected from the participating members of the family LUBA01. Anthropological measurements and earlier developmental milestone were recorded. Detailed clinical investigations including X-rays, echocardiograms, and audiometry were performed on available affected individuals.
Whole exome sequencing (WES)
WES was performed using genomic DNA samples from one affected and one normal individual and NimbleGen SeqCap EX Exome v2.0 kit (Roche Diagnostics, San Francisco, CA). Sequencing data analysis, alignment, variants calling, and filteration were performed as described previously (13).
Sanger sequencing and LOD score calculation
Sequencing reaction was carried out by using a Big Dye Terminator v3.1 (Applied Biosystems) as described previously (14). To confirm the association with the phenotype, two point LOD score for the CHST3 variant was calculated with Superlink by EasyLinkage 5.02 v GUI (15). An autosomal recessive inheritance and a disease allele frequency of 0.001 were used for linkage analysis.
Molecular modeling
Molecular modeling for CHST3 was performed using Phyre2 (16) with the Sulfotransferase domain from the Curacin biosynthetic pathway as the template (Protein Data Bank (PDB), 4GBM).
Results
Clinical findings
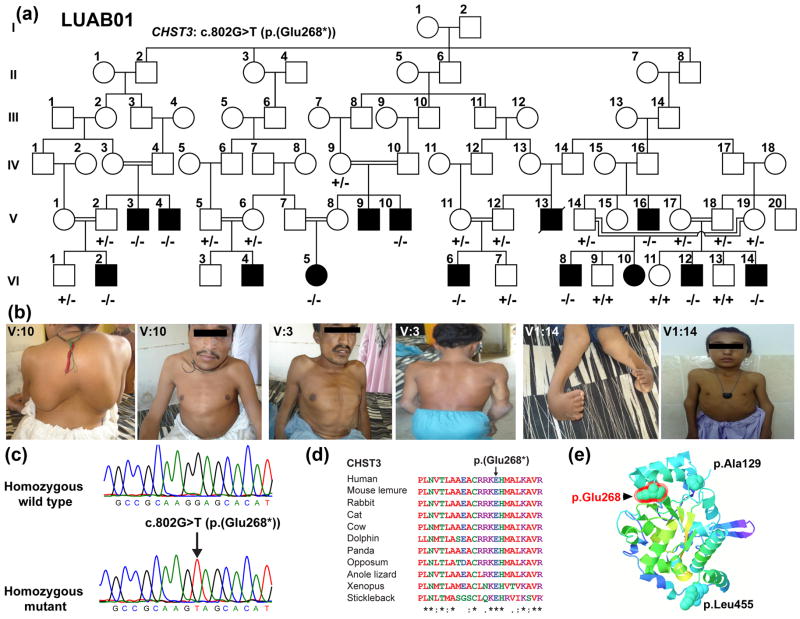
We enrolled a six-generation family (LUAB01) from Sindh province of Pakistan with fourteen individuals in ten sibships who were affected with recessively inherited congenital skeletal dysplasia (Fig. 1a). The neonatal clinical records of the affected individuals were not available at the time of enrollment. According to the family history, the affected individuals exhibited restricted movement at the elbow and knee joints at birth and progressively developed kyphoscoliosis and joint contractures in adulthood (Fig. 1b). The mean age of the patients at the time of enrollment was 13.4 years ranging from 5 to 25 years (Table 1). All affected family members were of short stature for age (Table 1). The mean height of the four adult males examined was 84±2.27 cm (SEM). Examination of the 5 pre-pubertal male children indicated that stature was significantly greater in childhood (mean height 101.2±1.16 cm; p-value 0.0016). This suggests that the short stature phenotype is progressive with age.
Fig. 1.
Congenital skeletal dysplasia and hearing loss phenotypes in family LUAB01 co-segregate CHST3 mutation. (a) Family LUBA01 pedigree. Filled and empty symbols represent affected and unaffected individuals, respectively. A double line uniting two individuals represents consanguineous marriage. Genotypes of all the participating individuals are shown. (b) Photographs of individuals homozygous for CHST3 allele. (c) Wild type and homozygous mutant nucleotide sequence chromatograms of exon 2 of CHST3 illustrating homozygosity for the c.802G>T (p.Glu268*) mutation. (d) ClustalW multiple sequence alignment of the 23 amino acids of CHST3 shows that p.Glu268 and downstream residues are conserved across species. Amino acids are numbered with reference to GenBank Accession number NP_004264.2. (e) A homology model of the sulfotransferase domain of CHST3 constructed using Phyre2 and c4gbmA as a template. The location of p.Glu268 residue (red) is shown along with the terminal amino-acids.
Table1.
Clinical findings of affected members of LUAB01
| ID | Age | Sex | Height (cm) | Height (SD)* | Weight (kg) | Weight (SD)* | Audiological findings | Radiological Findings | Echo cardiograms |
|---|---|---|---|---|---|---|---|---|---|
| V:10 | 25 | M | 80 | --- | 30 | --- | Mild to moderate | Severe scoliosis, kyphosis, fused vertebra, coxa vara and dislocated hips, club foot | Mitral valve and tricuspid valve regurgitation |
| V:3 | 23 | M | 81 | --- | 29 | --- | Mild to moderate | Scoliosis, fused vertebra, coxa vara and dislocated hips | Mitral valve and tricuspid valve regurgitation |
| V:16 | 21 | M | 85 | --- | 28 | --- | Mild to moderate | Scoliosis, kyphosis, fused vertebra, dislocated hips, Club foot | Mitral valve and tricuspid valve regurgitation, mild mitral stenosis and prolapse, mild tricuspid prolapse |
| V:4 | 20 | M | 90 | --- | 29 | --- | Not available | Not available | Not available |
| VI:6 | 11 | M | 105 | −5.98 | 27 | −1.49 | Mild to moderate | Kyphosis, fused vertebra, coxa vara and dislocated hips, club foot | Mitral valve and tricuspid valve regurgitation |
| VI:5 | 8 | F | 99 | −4.65 | 20 | −1.43 | Mild to moderate | Kyphosis, pes cavus, equinus hip and ankylosed elbow, dislocated hips, club foot | Mitral valve and tricuspid valve regurgitation, mild mitral stenosis and prolapse, mild tricuspid prolapse |
| VI:8 | 8 | M | 101 | −4.88 | 23.5 | −0.61 | Mild to moderate | Kyphosis, fused vertebra, coxa vara and dislocated hips, club foot | Mitral valve and tricuspid valve regurgitation, mild mitral stenosis and prolapse, mild tricuspid prolapse |
| VI:12 | 7 | M | 102 | −3.75 | 22 | −0.36 | Not available | Not available | Not available |
| VI:2 | 6 | M | 98 | −3.55 | 18.5 | −0.97 | Not available | Not available | Not available |
| VI:14 | 5 | M | 100 | −2.07 | 19 | 0.16 | Not available | Kyphosis, coxa vara and dislocated hips, Club foot | Mitral valve and tricuspid valve regurgitation, mild mitral stenosis and prolapse, mild tricuspid prolapse |
Based on Growth Calculator 2.01 (http://growthcalc.chip.org/GrowthCalc (ages 0–20)).
Radiological findings
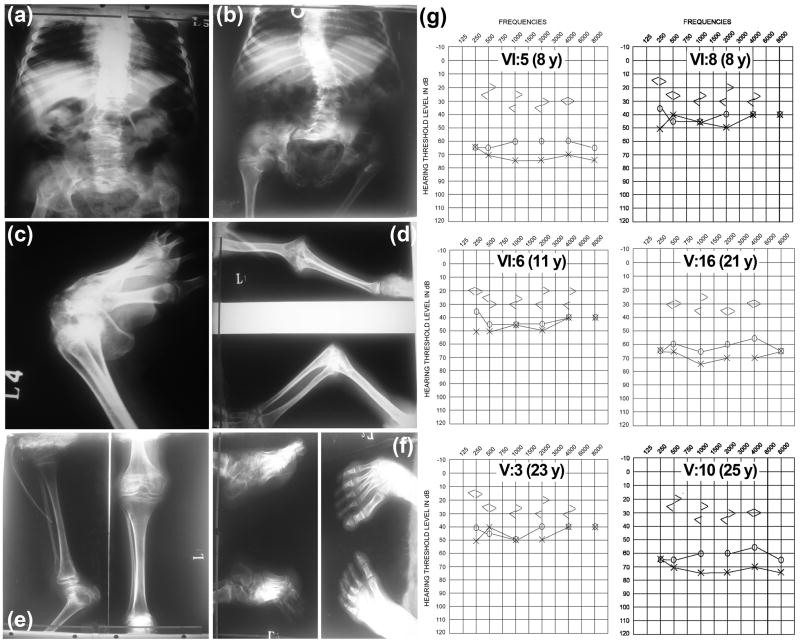
X-ray analysis of available affected individuals (Fig. 2a–f) revealed kyphosis and scoliosis along with bilateral fixed flexion deformities at knee, elbow, hip and shoulder joints. Vertebrae were fused, and distinct widening between T12 and L1 was observed (Fig. 2). Ribs were flattened and distorted (Fig. 2a–b). Femoral head was hypoplastic and femoral neck was short. Epiphysis (under-mineralization) was found in upper tibial and femoral bones. Hip joints were dislocated and dysplastic.
Fig. 2.
X-Ray and audiometric findings of affected individuals of LUBA01. (a) Patient V: 10 and (b) patient V: 3 imaging revealed severe scoliosis, fused vertebra, Coxa vara and dislocated hips. (c–d) Patient V: 3 imaging revealed clubfoot, Pes cavus, equinuship and ankylosed elbow. (e–f) Patient VI: 4 had sub-luxated knee and clubfoot. (g) Representative air and bone conduction audiograms of patients with CHST3 mutation.
Audiometry
Six individuals (VI:5, VI:8, VI:6, V:16, V:3 and V:10), underwent pure tone air and bone conduction audiometry (Fig. 2g). All of them demonstrated bilateral symmetric mild-to-severe hearing impairment (Fig. 2g). Additionally, three (VI:5, V:16 and V:10) of the six individuals tested had mixed hearing impairment (Fig. 2g). Median threshold values from all audiograms indicated moderate sensorineural hearing impairment at 2–4 kHz and moderate mixed hearing loss at the low frequencies (Fig. 2g). These results indicated that inner ear function at all frequencies was affected, with greater loss of function at low frequencies (Fig. 2g; Table 1). We did not observe any obvious correlation between age and hearing thresholds (Fig. 2g).
Echocardiograms
Echocardiography in seven affected individuals revealed multiple heart valve anomalies (Table 1). Mitral valve regurgitation and tricuspid valve regurgitation was observed in all patients. In addition, mild mitral stenosis, mild mitral prolapse and mild tricuspid prolapse were noticed in four patients (Table 1). Mild to moderate aortic regurgitation was found in two patients and one patient had bicuspid aortic valve (Table 1).
Molecular Analysis
WES followed by segregation analysis through Sanger sequencing (Supplementary Table 1) revealed a novel homozygous single base substitution (c.802G>T;p.Glu268*) in CHST3, segregating with the skeletal dysplasia phenotype in family LUAB01 (Fig 1a, 1c). This variant is present in an evolutionary conserved region (Fig. 1d) and absent in the 1000 Genomes (see web resource), the NHLBI 6500 exome variants, and the ExAC databases as well as in 140 ethnically matched chromosomes. Parametric linkage analysis was performed using the CHST3 variant genotype, which resulted in a two-point LOD score of 10.5 at θ=0. In the ExAC database, 10 loss-of-function (truncating) alleles are present, of which none are homozygous. The carrier frequency is approximately 1 in 3800, meaning that the likelihood of bi-allelic loss-of-function mutations in CHST3 is less than 1 in 14 million individuals based on Hardy-Weinberg calculations. The CHST3 gene consists of two coding exons and encodes a 479 amino acid long chondroitin 6-O-sulfotransferase-1 (C6ST-1) enzyme. The stop-gained mutation is predicted to remove 211 residues from the encoded protein, which would result in truncation of sulfotransferase domain (Fig. 1e).
Discussion
Mutations in CHST3 gene have been associated with skeleton dysplasia, primarily among patients with short stature, joint dislocation and kyphoscoliosis (12). Other associated phenotypes include microdontia and cardiac valve anomalies (10, 12). As of August 2015, 30 disease-associated mutations in CHST3 have been found in 45 patients, including familial cases from Oman and Tanzania, with congenital dislocations and vertebral changes (9, 12, 17).
Here, we described a large Pakistani consanguineous family, LUAB01, segregating CHST3-associated skeleton dysplasia and hearing loss. The prominent phenotypic features, consistent with spondyloepiphyseal dysplasia with congenital joint dislocations syndrome, were progressive short stature likely due to progressive kyphoscoliosis and congenital joint contractures including clubfoot and restricted movement at elbow and knee joints (Table 1). The severe adult short stature, likely related to progressive spinal curvature, is worse in this family (average 84 cm) than typically reported (110–130 cm). Notably, children in this family were on average significantly taller than adults. Within the group of male children (age range 5–11), heights were essentially the same, and as such, older children exhibited further deviation from normative stature than their younger relatives. Thus, there may be a halting of growth that occurs sometime in childhood. However, we do not have access to individual heights across time, thus we will be following these children prospectively to understand whether the growth deficits during childhood are attributable to long bone length, spinal curvature, or endocrinologic etiologies. Importantly, we will determine whether their post-pubertal height is less than their pre-pubertal height, as suggested by comparison to the adult average male height.
The affected individuals also had bilateral mixed hearing loss. Hearing loss incidence in patients with CHST3 mutations has been anecdotal. One of two Turkish siblings, described previously, had unilateral mixed hearing loss (18). Similarly one of two Somalian half siblings with mutation in CHST3 had conductive hearing loss (8). However, hearing loss observed in these two individuals was attributed to environmental cause, especially for Somalian proband who had frequent ear infections requiring surgical intervention (8). To date, limited data are available about the association of sensorineural hearing loss with skeleton dysplasia. So far, only one patient with Larsen syndrome (OMIM 150250) has been documented in detail with presentation of mixed hearing loss with dominant sensorineural portion (8, 11, 18, 19). In this study, six affected individuals evaluated through audiometry demonstrated mixed hearing loss. We did not observe any obvious co-relation of age with hearing loss severity segregating in family LUAB01 (Fig. 2g). The conductive portion of the hearing loss may be due to ossicular malformation; however, the sensorineural hearing loss requires further exploration. In summary, our results support a role of CHST3 in the middle and inner ear functioning. The identification of p.(Glu268*) nonsense mutation in CHST3 expands the repertoire of the known genetic causes of hearing loss and the skeletal dysplasia phenotypic spectrum of CHST3 alleles.
Supplementary Material
Acknowledgments
We thank the family members for their participation and cooperation. This work was supported by the NIDCD/NIH (grant numbers R01 DC011803 and R01 DC012564 to S.R. and Z.M.A., respectively) and LUMHS intramural funds to A.M.W.
Footnotes
Conflict of Interest:
No conflict to declare.
Web Resources
1000 Genomes: http://www.1000genomes.org NHLBI_EPS: http://evs.gs.washington.edu/EVS/ ExAC: http://exac.broadinstitute.org
References
- 1.Krakow D, Rimoin DL. The skeletal dysplasias. Genet Med. 2010;12:327–341. doi: 10.1097/GIM.0b013e3181daae9b. [DOI] [PubMed] [Google Scholar]
- 2.Krakow D. Skeletal dysplasias. Clin Perinatol. 2015;42:301–319. viii. doi: 10.1016/j.clp.2015.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Warman ML, Cormier-Daire V, Hall C, et al. Nosology and classification of genetic skeletal disorders: 2010 revision. Am J Med Genet A. 2011;155A:943–968. doi: 10.1002/ajmg.a.33909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.al Gazali LI, Devadas K, Hall CM. A new lethal neonatal short limb dwarfism. Clin Dysmorphol. 1996;5:159–164. [PubMed] [Google Scholar]
- 5.Clarke BL. Anti-sclerostin antibodies: utility in treatment of osteoporosis. Maturitas. 2014;78:199–204. doi: 10.1016/j.maturitas.2014.04.016. [DOI] [PubMed] [Google Scholar]
- 6.Langdahl B, Binkley N, Bone H, et al. Odanacatib in the Treatment of Postmenopausal Women With Low Bone Mineral Density: Five Years of Continued Therapy in a Phase 2 Study. Journal of Bone and Mineral Research. 2012;27:2251–2258. doi: 10.1002/jbmr.1695. [DOI] [PubMed] [Google Scholar]
- 7.Padhi D, Jang G, Stouch B, et al. Single-dose, placebo-controlled, randomized study of AMG 785, a sclerostin monoclonal antibody. J Bone Miner Res. 2011;26:19–26. doi: 10.1002/jbmr.173. [DOI] [PubMed] [Google Scholar]
- 8.van Roij MH, Mizumoto S, Yamada S, et al. Spondyloepiphyseal dysplasia, Omani type: further definition of the phenotype. Am J Med Genet A. 2008;146A:2376–2384. doi: 10.1002/ajmg.a.32482. [DOI] [PubMed] [Google Scholar]
- 9.Thiele H, Sakano M, Kitagawa H, et al. Loss of chondroitin 6-O-sulfotransferase-1 function results in severe human chondrodysplasia with progressive spinal involvement. Proc Natl Acad Sci U S A. 2004;101:10155–10160. doi: 10.1073/pnas.0400334101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tuysuz B, Mizumoto S, Sugahara K, et al. Omani-type spondyloepiphyseal dysplasia with cardiac involvement caused by a missense mutation in CHST3. Clin Genet. 2009;75:375–383. doi: 10.1111/j.1399-0004.2009.01167.x. [DOI] [PubMed] [Google Scholar]
- 11.Hermanns P, Unger S, Rossi A, et al. Congenital joint dislocations caused by carbohydrate sulfotransferase 3 deficiency in recessive Larsen syndrome and humero-spinal dysostosis. Am J Hum Genet. 2008;82:1368–1374. doi: 10.1016/j.ajhg.2008.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Unger S, Lausch E, Rossi A, et al. Phenotypic features of carbohydrate sulfotransferase 3 (CHST3) deficiency in 24 patients: congenital dislocations and vertebral changes as principal diagnostic features. Am J Med Genet A. 2010;152A:2543–2549. doi: 10.1002/ajmg.a.33641. [DOI] [PubMed] [Google Scholar]
- 13.Simon M, Richard EM, Wang X, et al. Mutations of human NARS2, encoding the mitochondrial asparaginyl-tRNA synthetase, cause nonsyndromic deafness and Leigh syndrome. PLoS Genet. 2015;11:e1005097. doi: 10.1371/journal.pgen.1005097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Waryah AM, Narsani AK, Sheikh SA, et al. The novel heterozygous Thr377Arg MYOC mutation causes severe Juvenile Open Angle Glaucoma in a large Pakistani family. Gene. 2013;528:356–359. doi: 10.1016/j.gene.2013.07.016. [DOI] [PubMed] [Google Scholar]
- 15.Lindner TH, Hoffmann K. easyLINKAGE: a PERL script for easy and automated two-/multi-point linkage analyses. Bioinformatics. 2005;21:405–407. doi: 10.1093/bioinformatics/bti009. [DOI] [PubMed] [Google Scholar]
- 16.Kelley LA, Sternberg MJ. Protein structure prediction on the Web: a case study using the Phyre server. Nat Protoc. 2009;4:363–371. doi: 10.1038/nprot.2009.2. [DOI] [PubMed] [Google Scholar]
- 17.Rajab A, Kunze J, Mundlos S. Spondyloepiphyseal dysplasia Omani type: a new recessive type of SED with progressive spinal involvement. Am J Med Genet A. 2004;126A:413–419. doi: 10.1002/ajmg.a.20606. [DOI] [PubMed] [Google Scholar]
- 18.Tanteles GA, Dixit A, Dhar S, et al. Two Somali half-siblings with CHST3-related chondrodysplasia illustrating the phenotypic spectrum and intrafamilial variability. Am J Med Genet A. 2013;161A:2588–2593. doi: 10.1002/ajmg.a.36094. [DOI] [PubMed] [Google Scholar]
- 19.Ott IO, Issing PR. Bilateral, mixed hearing loss with a predominant sensorineural component in Larsen’s syndrome. J Laryngol Otol. 2008;122:e6. doi: 10.1017/S0022215107001466. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.