Abstract
Since the RNA world hypothesis was proposed, a large number of regulatory noncoding RNAs (ncRNAs) have been identified in many species, ranging from microorganisms to mammals. During the characterization of these newly discovered RNAs, RNAs having both coding and noncoding functions were discovered, and these were considered bifunctional RNAs. The recent use of computational and high-throughput experimental approaches has revealed increasing evidence of various sources of bifunctional RNAs, such as protein-coding mRNAs with a noncoding isoform and long ncRNAs bearing a small open reading frame. Therefore, the genomic diversity of Janus-faced RNA molecules that have dual characteristics of coding and noncoding indicates that the functional roles of RNAs have to be revisited in cells on a genome-wide scale. Such studies would allow us to further understand the complex gene-regulatory network in cells. In this review, we discuss three major genomic sources of bifunctional RNAs and present a handful of examples of bifunctional RNA along with their functional roles.
Keywords: lncRNA, sORF, bifunctional RNA
INTRODUCTION
Since the “one gene-one enzyme hypothesis” (Beadle and Tatum, 1941) and the “one gene-one polypeptide hypothesis” (Ingram, 1957) were proposed, the perception among geneticists and molecular biologists earlier on was that proteins were the only core molecules that controlled diverse cellular processes and gene expression regulation, and that RNAs were passive carriers of genetic information (Crick, 1970). However, this hypothesis was challenged by polypeptides arising from alternative splicing. In the 1980s, Thomas Cech’s group first demonstrated that the role of RNA molecules extended beyond that, to enzymatic regulation (Bussard, 2005; Kruger et al., 1982).
The idea of RNA as a primordial molecule was brewing in the 1960s. Subsequently, the “RNA world” hypothesis was proposed, according to which the earliest forms of genetic information and effector molecules were RNAs. This hypothesis has been supported by the observation of self-cleavage of viral RNAs with a structural model of the active sites (Forster and Symons, 1987) and additional enzymatic activities such as RNA-catalyzed RNA polymerization (Johnston et al., 2001) and peptide bond formation (Zhang and Cech, 1997). However, it was still believed that such transcriptional and translational roles of RNAs were later taken over by DNA and protein, respectively, and that RNA had no such regulatory roles in genomes.
Eventually, the protein- and DNA-centric view was challenged by the discovery of a small regulatory noncoding RNA (ncRNA) group, which included microRNAs (miRNAs) (Lee et al., 1993; Reinhart et al., 2000; Wightman et al., 1993) and the steroid receptor RNA activator (SRA; a ncRNA with both coding and noncoding function) (Chooniedass-Kothari et al., 2004; Lanz et al., 1999), which was considered a bifunctional RNA. miRNA downregulates hundreds of conserved mRNA targets by base-pairing to the 3′-untranslated region (UTR) of the target in a posttranscriptional and translational manner (Bartel, 2009). Thousands of miRNAs participate in modulating diverse biological processes by controlling the expression level of more than 50% of protein-coding genes (Bartel, 2009). The first mammalian bifunctional RNA discovered, SRA, mediates the transactivation of eukaryotic gene expression as an RNA (Lanz et al., 1999) and encodes a highly conserved SRA protein in breast cancer cells (Chooniedass-Kothari et al., 2004). The SRA protein Np95 mediates epigenetic upregulation of gene expression by recruiting Dnmt1 to methylated DNA (Sharif et al., 2007). In turn, VegT and Oskar have been classified as bifunctional RNA with coding and noncoding functions in animal development (Jenny et al., 2006; Kloc et al., 2005).
During the last decade, reproducible transcription across mammalian coding and noncoding genomes were extensively discovered using high-throughput sequencing technology (Consortium, 2012). In the human genome, approximately 62% of the genome has been reproducibly transcribed (Consortium, 2012), of which approximately 30% corresponds to exonic and intronic regions of protein-coding genes, and the remainder mostly comprise long ncRNAs (lncRNAs) transcribed in intergenic regions or antisense strands (Hangauer et al., 2013). Furthermore, protein-coding genes encode nine multiple isoforms on an average, very few of which lack coding potential and are regarded as noncoding isoforms (Mascarenhas et al., 2015). date, tens of thousands of lncRNAs have been discovered in genomes and found to be involved in diverse biological functions such as regulating gene expression at the level of transcription, RNA processing, or translation; protecting genomes from transposons or viral DNAs; and guiding DNA synthesis, genome rearrangement, or genome editing (Cech and Steitz, 2014). The versatile functions of lncRNAs suggest that RNAs actively participate in diverse cellular processes and play a regulatory role in the expression of genetic information.
Ever-increasing evidence of the existence of bifunctional RNAs with coding and noncoding potential is being provided by current high-throughput RNA sequencing (RNA-seq) and ribosome footprinting followed by sequencing (Ribo-seq) (Andrews and Rothnagel, 2014; Kumari and Sampath, 2015). The existence of lncRNAs with small open reading frames (sORFs) (Andrews and Rothnagel, 2014) and the intrinsic RNA function of protein-coding genes (Kumari and Sampath, 2015) in species ranging from bacteria to mammals indicates that the bifunctional mode of RNAs has been evolutionarily conserved. Although mono-functional RNAs might be major players in current genomes, bifunctional RNAs seem to have significantly contributed to the complexity and robustness of gene expression regulation over time.
In this mini-review, we discuss three major types of bifunctional RNAs with both coding and noncoding potential: lncRNA with sORFs, coding mRNAs with noncoding functions, and noncoding isoforms of protein-coding genes. We then go on to summarize the functional categories of RNAs and their corresponding proteins or peptides.
BIFUNCTIONAL RNAs AND GENES
The first type of bifunctional RNA is lncRNA encoding small peptides. Recent studies have reported that a number of lncRNAs contain sORFs that extensively translate small peptides (Anderson et al., 2015; Aspden et al., 2014; Mackowiak et al., 2015; Olexiouk et al., 2016; Ruiz-Orera et al., 2014). Analysis of sequence conservation (Anderson et al., 2015; Mackowiak et al., 2015) and high-throughput sequencing of ribosome-protected fragments (RPFs) (Aspden et al., 2014; Olexiouk et al., 2016; Ruiz-Orera et al., 2014) have shown that small peptides from a considerable number of well-studied lncRNAs are actively synthesized (Andrews and Rothnagel, 2014). Although the majority of functional proteins are composed of more than 100 amino acids, a considerable number of small peptides, comprising fewer than 100 amino acids, attributed to lncRNA, are also detectable from cells by ribosome footprinting and proteomics approaches. In addition, some sORFs but not their host lncRNAs are evolutionarily conserved in other genomes (Calviello et al., 2016; Ji et al., 2015), indicating that they might play a functional role not related to the lncRNA functions in cells (Fig. 1A).
Fig. 1.
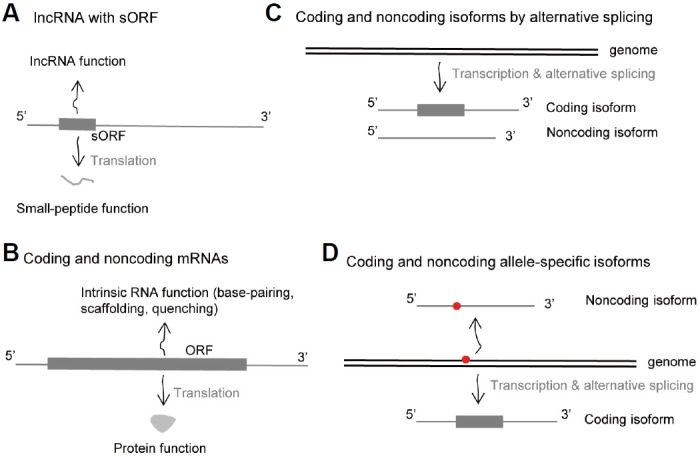
Four relevant scenarios of the generation of RNAs or genes with coding and noncoding functions. (A) lncRNAs with sORF. (B) Protein-coding mRNAs having non-coding functions. (C) Coding and noncoding isoforms of protein-coding genes by alternative splicing. (D) Allele-specific expression of coding and noncoding isoforms.
The second type of bifunctional RNA is coding mRNA having noncoding functions. So far, a dozen protein-coding mRNAs were revealed to have an intrinsic noncoding function, independent of the protein that they encode (Fig. 1B) (Karapetyan et al., 2013; Kumari and Sampath, 2015). The intrinsic noncoding function of mRNAs is involved in core molecular processes such as transcriptional, posttranscriptional, and translational regulation, as well as protein scaffolding in cis- and trans-acting manners (Kumari and Sampath, 2015).
The third type of bifunctional RNA is protein-coding RNA having either noncoding isoforms by alternative splicing or allele-specific transcripts with variants that destroy coding potential (Figs. 1C and 1D). The noncoding isoforms of protein-coding genes are often regarded as potential targets of nonsense-mediated mRNA decay (NMD); however, a considerable number appear to be sufficiently stable to have functional roles in cells (Mayba et al., 2014).
CODING POTENTIAL OF LncRNAs
LncRNA is defined as a long RNA molecule with no coding potential and no sequence similarity to protein-coding genes; however, several annotated lncRNAs appear to be highly associated with ribosomes (Table 1), indicating that lncRNAs contain a coding region that translates a peptide. Of the lncRNAs manually curated in VEGA (http://vega.sanger.ac.uk/), a few display strong ribosomal association, indicating a high translation efficiency, in HeLa and mouse neutrophil cells (Table 1). Intriguingly, the ribosome association is mostly restricted to sORFs, and this association rapidly decays at the location of ribosome release at the stop codon, estimated as the ribosome release score (RRS) (Guttman et al., 2013). For example, MALAT1 is highly expressed in the nucleus during cancer metastasis (Ji et al., 2003) and has a putative sORF that can encode about 71 amino acids. In fact, the sORFs are strongly associated with ribosomes (Fig. 2A). Although in vitro translation experiments have failed to detect peptides from the putative sORF (Ji et al., 2003), the strong ribosome association supports translation activity on the sORF. In addition, the movement of the 80S ribosome along the mRNA makes it possible to discern whether the sORF is actively translated. For example, RP11-21N3_1 lncRNA includes three putative sORFs strongly associated with ribosomes within a sORF with a subcodon positional bias of the 5′ termini of RPFs (Fig. 2B).
Table 1.
lncRNAs with ribosome association
| Gene_ID | Species | Transcript (nt) | Putative ORF (nt) | TE* | RPM** | RRS*** |
|---|---|---|---|---|---|---|
| RP5-875O13.1 | Human | 1548 | 78 | 3.26 | 122.76 | 0.15 |
| CGAT1 | Human | 2131 | 210 | 0.40 | 45.71 | 42.01 |
| SNHG8 | Human | 672 | 159 | 1.62 | 33.40 | 52.62 |
| MALAT1 | Human | 8708 | 213 | 0.01 | 26.99 | 1184.00 |
| CTD-2651B20.6 | Human | 308 | 72 | 3.99 | 21.92 | 0.12 |
| GAS5 | Human | 1698 | 502 | 0.10 | 21.82 | 170.13 |
| AC074183.4 | Human | 782 | 186 | 5.44 | 20.37 | 78.00 |
| DANCR | Human | 878 | 180 | 0.23 | 14.47 | 116.37 |
| RP11-21N3.1 | Human | 918 | 300 | 1.05 | 12.51 | 488.00 |
| RP11-220I1.1 | Human | 4375 | 192 | 0.18 | 11.58 | 371.20 |
| RP11-354P17.9 | Human | 745 | 246 | 0.93 | 11.38 | 69.54 |
| RPPH1 | Human | 638 | 108 | 0.22 | 10.34 | 8.00 |
| RP23-381B19.2 | Mouse | 1428 | 279 | 0.16 | 25.85 | 70.33 |
| Malat1 | Mouse | 6983 | 195 | 0.01 | 17.40 | 1998.00 |
| RP23-73N16.7 | Mouse | 475 | 162 | 0.81 | 12.82 | 0.42 |
| Gas5 | Mouse | 2556 | 177 | 0.05 | 8.44 | 80.66 |
Translation Efficiency (TE)
Read per million mapped reads (RPM)
Ribosome Release Score (RRS)
Fig. 2.
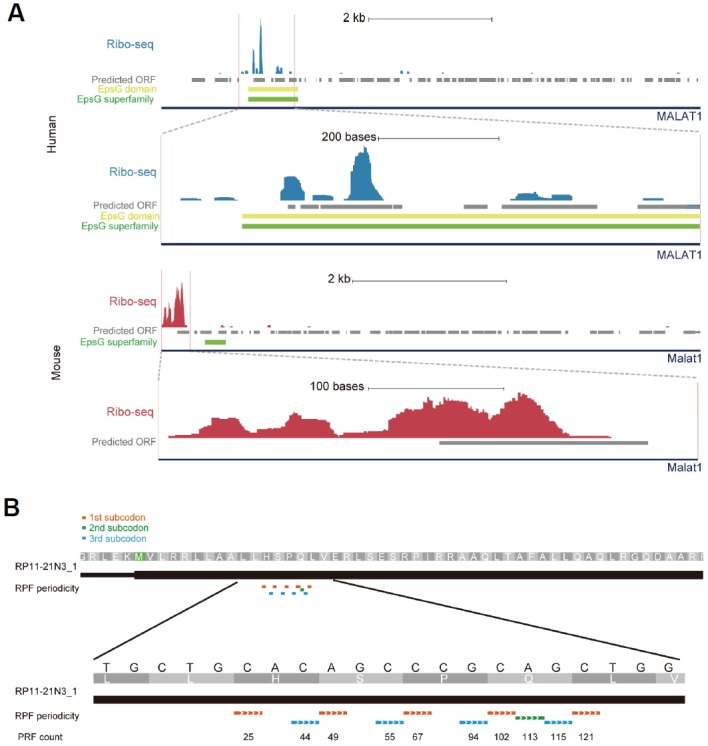
Ribosome-association signals on lncRNAs. (A) Strong RPF signals in a sORF of MALAT1 in HeLa cells and Malat1 in mouse neutrophils are shown. Yellow and green bars are EpsG protein and EpsG superfamily domains, respectively, detected from the NCBI Conserved Domain Database. (B) RPF signals in the lncRNA RP11-21N3_1 in HeLa cells.
Another type of lncRNA, the miRNA host transcript, sometimes includes ribosome footprints on the RNA. For example, pri-miR171b in Arabidopsis thaliana is processed into miR171b and downregulates several solute carrier family members. Its primary transcript, called miPEP171b, has two sORFs, which encode five and twenty amino acids, respectively. These peptides specifically enhance the expression of miR171b, affecting lateral root development (Lauressergues et al., 2015).
CODING mRNAs WITH NONCODING FUNCTIONS
Besides their protein-coding functions, mRNAs play functional roles as RNA, such as transcriptional and translational gene regulation and localization and scaffolding of proteins (Table 2). In general, the intrinsic RNA function is mediated by either cis- or trans-acting elements on the RNA. In fact, the UTR of protein-coding mRNAs often harbors cis-acting factors that control their own stability, localization, and translation efficiency, which could be regarded as intrinsic functions of the mRNAs. For example, the 3′-UTRs of DMPK, Oskar, and Sqt include motifs recognized by RNA-binding proteins (RBPs), induce the formation of scaffolds of the RBPs during embryo development, and also encode DNA sequences of proteins composed of hundreds of amino acids (Jansen et al., 1996; Jenny et al., 2006; Lim et al., 2012; Mahadevan et al., 1992).
Table 2.
List of bifunctional RNAs
| Bifunctional RNA | Species | RNA length | Peptide length | Coding function | Intrinsic function of RNA | RNA function category | References |
|---|---|---|---|---|---|---|---|
| RNAIII | Staphylococcus aureus | 514 nt | 26 aa | Delta-hemolysin that targets host cell membranes, causing lysis | Regulation of stability and translation of virulence factors by base pairing | Translation | Verdon et al., 2009 |
| SgrS | Escherichia coli | 227 nt | 43 aa | SgrT, relieving glucose stress through a base pairing-independent mechanism | Negative regulation of the stability and translation of the major gulcose transporters ptsG and manXYZ via base pairing | Translation | Wadler and Vanderpool, 2007 |
| SR1 | Bacillus subtilis | 117 nt | 39 aa | SR1P, stabilizing gapA operon RNA by binding to gapA protein but not RNA | Repression of transcription activator ahrC that regulates the arginine catabolic operons | Transcription | Gimpel et al., 2010 |
| ENOD40 | Medicago truncatula, Oryza sativa, Arabidopsis thaliana | ∼700 nt | I: 13 aa, II: 27 aa | Two ORFs, cell divisions at high frequency. Two peptides binds to sucrose synthase | MtRBP1 interacting with a secondary structure of interORF region, which is involved in translation regulation | Translation | Hanada et al., 2007; Okamoto et al., 2014; Sousa et al., 2001 |
| VegT | Xenopus laevis | 2703 nt | 455 aa | VegT codes for a T-box TF that patterns the mesendoderm | VegT mRNA and ncRNA, Xlsirts play structural roles in the organization of the cytoskeleton at the vegetal cortex | Scaffolding | Kloc et al., 2005 |
| Oskar | Drosophila melanogaster | 2869 nt | 606 aa | Involved in late oogenesis function | Oskar 3′UTR, scaffolding RBPs, regulating early oogenesis function | Scaffolding | Jenny et al., 2006 |
| Sqt | Danio rerio | 1480 nt | 392 aa | Nodal signaling in mesendoderm induction and patterning, and specification of the neural tube | Sqt 3′UTR, scaffolding RBPs, initiation of dorsal axis specification | Scaffolding | Lim et al., 2012 |
| c-Myc (potential), Brca1, Klf4, and Dnmt1 | Homo sapiens, Mus musculus | - | - | Oncogenic and tumor suppressor function | Cis-trascriptional regulation by binding to PRC2 and EZH2, polycomb repressive complex | Transcription | Karapetyan et al., 2013; Zhao et al., 2010 |
| DMPK | Homo sapiens | 3243 nt | 639 aa | Delayed onset of mild myotonic dystrophy symptoms | DMPK 3′UTR, CUG repeat forms hairpin structure that sequesters developmentally regulated splicing factors like MBNL, leading to severe myotonic dystrophy symptoms | Scaffolding | Jansen et al., 1996; Mahadevan et al., 1992 |
| TP53 (potential) | Homo sapiens | 2591 nt | 393 aa | Transcription factor in response to DNA-damage, hypoxia, nutrient deprivation, and telomere damage, which can lead to cancer. Initiating a program of cell cycle arrest and apoptosis | Enhance p53 translation and stabilization by interacting with Mdm2 | Translation | Candeias et al., 2008 |
| H2B | Homo sapiens | 431 nt | 126 aa | Replication-dependent histone gene | Nucleation of nuclear bodies (histone locus bodies, nuclear speckles, paraspeckles, and nuclear stress bodies) | Scaffolding | Shevtsov and Dundr, 2011 |
| HIC | Homo sapiens | 5312 nt | 152 aa | Involved in transcriptional regulation of viral genome expression | HIC 3′UTR hairpin structure, binds to and activates P-TEFb by displacing 7SK RNA from inhibitory complex | Translation | Young et al., 2007 |
| BCMA-AS | Homo sapiens | >345 nt | 115 aa | p12, ? | BCMA translation repression | Translation | Hatzoglou et al., 2002 |
| P23/TCTP | Homo sapiens | 4814 nt | 197 aa | Control of homeostasis | Inhibits translation by interacting with the RNA binding domain of PKR | Translation | Bommer et al., 2002 |
| SMD targets | Homo sapiens | varying | varying | varying | Stability control of interacting SMD targets and lncRNA via Alu repeats | Post-transcription | Gong and Maquat, 2011 |
| SRA | Homo sapiens | 2015 nt | 236 or 208 aa | RNA binding and related to breast cancer | Nuclear receptor coactivator | Coactivator | Chooniedass-Kothari et al., 2004; Hube et al., 2011; Lanz et al., 1999 |
In one sense, all protein-coding RNAs that have spatially separable functions of coding in coding sequences and noncoding in UTRs can be regarded as bifunctional RNAs. Intriguingly, the base compositions, structures, and functions of 3′-UTRs are similar to those of lncRNA genes (Niazi and Valadkhan, 2012; Wan et al., 2012), which indicates that UTRs might be traces (or fossils) of ancient regulatory RNAs in the view of the RNA world hypothesis. Of course, UTRs also mediate trans-gene regulation by competing with other endogenous RNAs. For example, the hairpin structure of the HIC 3′-UTR binds to and translationally activates P-TEFb by displacing 7SK RNA from the inhibitory complex (Young et al., 2007) (Table 2). In addition, the UTRs interact with lncRNAs via imperfect base pairing based on Alu repeats, facilitating the formation of STAU1-binding sites (Gong and Maquat, 2011). This recruitment of STAU1 directs staufen-mediated mRNA decay (SMD), which controls the stability of the SMD target mRNAs (Table 2). Notably, mRNAs with functional UTRs tend to have either sORFs or long UTRs, resembling lncRNAs with sORFs. For example, SgrS and ENOD40 mRNAs, which encode 43 and 27 amino acids, respectively, and yet have UTRs ∼100 nt and ∼600 nt in length, have noncoding functions of translational regulation and RNA stability (Hanada et al., 2007; Okamoto et al., 2014; Sousa et al., 2001; Wadler and Vanderpool, 2007) (Table 2).
In some cases, the region responsible for the noncoding function, however, is barely separable from that required for its coding function. For example, RNAIII in Staphylococcus aureus encodes a delta-hemolysin comprising 26 amino acids, which induces the lysis of the host cell membranes. The RNA sequence includes the CDS and has an intrinsic function of regulating the stability and translation of virulence factors by base pairing (Verdon et al., 2009). Hence, the regions for coding and noncoding functions might not necessarily be distinct. VegT mRNA, which encodes a T-box transcription factor that patterns the mesendoderm of Xenopus laevis, plays a structural role in the organization of the cytoskeleton at the vegetal cortex (Kloc et al., 2005) (Table 2). In addition, c-Myc, H2B, TP53, BCMAAS, and P23 have regulatory roles at the RNA level, but the regions for these functions are not clearly separable from the CDS regions (Table 2). For spatially inseparable functional elements on RNA, it is difficult to pinpoint the functional roles of the elements by simply removing them. Rather, the functional role of these regions can be determined by introducing mutations in the element but with a minimal effect on the RNA sequence. Thus, at present, we classify these groups as potential bifunctional RNAs.
CODING AND NONCODING ISOFORMS OF PROTEIN-CODING GENES
A major isoform analysis of human transcriptomes has revealed that 15.26–20.64% of protein-coding genes in primary tissues express the major isoform that lacks an annotated CDS in GENCODE (Gonzalez-Porta et al., 2013). The major noncoding isoforms are recurrently expressed across tissues and cell lines, and their subcellular localization showed that they are enriched in the nucleus compared to other major coding isoforms. Although some of the noncoding isoforms enriched in the nucleus are likely to be retained introns, about half are processed transcripts, and their abundant levels indicate their functional roles in the primary tissues.
Processed noncoding transcripts are often generated by alternative splicing, which either introduces a premature stop codon (PTC) or disrupts the ORF (Gonzalez-Porta et al., 2013). Because transcripts with a PTC are easily damaged by the NMD pathway, processed noncoding transcripts with high expression levels are more likely to be non-NMD targets. Major noncoding isoforms mainly arise by alternative splicing or skipping the first or last exon, including the start or stop codons. For example, the SRA gene expresses coding isoforms produced by alternative start codons in the first exon and noncoding isoforms with a truncated or missed first exon (Chooniedass-Kothari et al., 2004; Hube et al., 2011; Lanz et al., 1999) (Fig. 3). The ratio of coding and noncoding isoforms is dynamically altered across different cell states or types. In fact, the fraction of noncoding isoforms increases during myogenic differentiation of primary human satellite cells, while the fraction decreases in myotonic dystrophy muscles (Hube et al., 2011). The production of coding and noncoding isoforms of SRA is also observed in the mouse transcriptome, indicating that the dual function of coding and noncoding isoforms of SRA has been evolutionary conserved (Fig. 3).
Fig. 3.
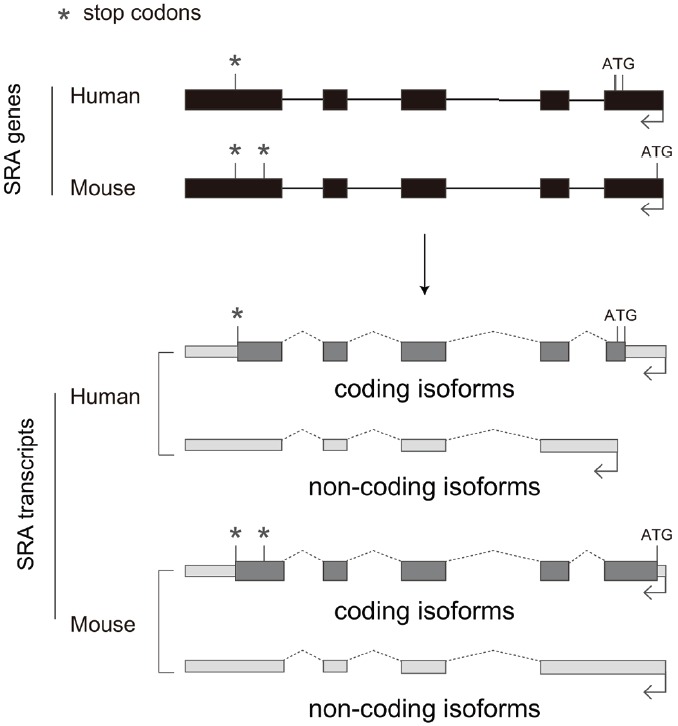
An example of coding and noncoding isoforms from the human SRA gene with 5 exons, which is conserved in the mouse. The human SRA gene has 2 alternative start codons, whereas mouse SRA has 1 start codon and 2 stop codons. The gene produces both coding and noncoding isoforms, with the first intron retained in both human and mouse.
An alternative source of noncoding transcripts of protein-coding genes could be allele-specific isoforms generated by heterozygous mutations that cause a loss of coding potential. In many cases, mutations in splice sites cause the truncation or retention of introns of transcripts, some of which are damaged by the NMD pathway (Rivas et al., 2015). In particular, these allele-specific isoforms are frequently discovered in cancer cells (Mayba et al., 2014), suggesting that the allelic imbalance of coding and noncoding transcripts of a protein-coding gene might be involved in the origin of cancer.
CONCLUSION
The discovery of bifunctional genes raises several questions. How prevalent are genes with dual functions in genomes? How can we distinguish genuine coding RNA, ncRNA, and bifunctional RNA at the genome level? How do we examine the coding and noncoding functions of RNA? Finally, why do cells maintain the evolutionarily conserved dual functions of RNAs across species? To address these questions, genome-wide experimental and computational approaches to identify noncoding functions of protein-coding genes, detect putative sORFs in noncoding genes, and reconstruct noncoding isoforms in coding loci could be useful.
The coding potential of lncRNAs has been predicted computationally by comparing phylogenetic codon models across species, such as PhyloCSF (Lin et al., 2011), or by experimentally detecting RNA regions associated with ribosomes (Guttman et al., 2013; Ingolia et al., 2014). Despite the successful detection of putative sORFs in lncRNAs by the above methods, we need to further examine whether the peptides are productively generated, whether the peptides are stable in cells, and whether peptides from sORFs are functionally relevant.
A more prevalent source of bifunctional RNAs might be the noncoding isoforms of protein-coding genes. However, detecting the noncoding isoforms of protein-coding genes at the genome level is a challenging task. For this, the isoform-level analysis of ribosome footprinting might be helpful. In addition, to verify the function of the noncoding isoform, we can examine whether the function is maintained after removing the coding potential by introducing a frameshift mutation or nonsense mutation in the coding isoform.
Recently, many truncated proteins encoded from NMD targets and noncoding isoforms of protein-coding genes have been shown to have a stable form in NMD-deficient conditions, and to be involved in cancer development (Liu et al., 2014; Wang et al., 2011). This highlights the need to understand how distinct RNAs and proteins of bifunctional RNAs are pathologically relevant, which will in turn provide interesting biological insights with potential medical implications.
Acknowledgments
This work was supported by the Basic Science Research Program through the NRF, funded by the Ministry of Science, ICT & Future Planning (NRF-2013R1A1010185, NRF-2014M3C9A 3063541, and NRF-2012M3A9D1054516).
REFERENCES
- Anderson D.M., Anderson K.M., Chang C.L., Makarewich C.A., Nelson B.R., McAnally J.R., Kasaragod P., Shelton J.M., Liou J., Bassel-Duby R., et al. A micropeptide encoded by a putative long noncoding RNA regulates muscle performance. Cell. 2015;160:595–606. doi: 10.1016/j.cell.2015.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews S.J., Rothnagel J.A. Emerging evidence for functional peptides encoded by short open reading frames. Nat. Rev. Genet. 2014;15:193–204. doi: 10.1038/nrg3520. [DOI] [PubMed] [Google Scholar]
- Aspden J.L., Eyre-Walker Y.C., Phillips R.J., Amin U., Mumtaz M.A., Brocard M., Couso J.P. Extensive translation of small open reading frames revealed by Poly-Ribo-Seq. Elife. 2014;3:e03528. doi: 10.7554/eLife.03528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel D.P. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beadle G.W., Tatum E.L. Genetic Control of Biochemical Reactions in Neurospora. Proc. Natl. Acad. Sci. USA. 1941;27:499–506. doi: 10.1073/pnas.27.11.499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bommer U.A., Borovjagin A.V., Greagg M.A., Jeffrey I.W., Russell P., Laing K.G., Lee M., Clemens M.J. The mRNA of the translationally controlled tumor protein P23/TCTP is a highly structured RNA, which activates the dsRNA-dependent protein kinase PKR. RNA. 2002;8:478–496. doi: 10.1017/s1355838202022586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bussard A.E. A scientific revolution? The prion anomaly may challenge the central dogma of molecular biology. EMBO Rep. 2005;6:691–694. doi: 10.1038/sj.embor.7400497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calviello L., Mukherjee N., Wyler E., Zauber H., Hirsekorn A., Selbach M., Landthaler M., Obermayer B., Ohler U. Detecting actively translated open reading frames in ribosome profiling data. Nat. Methods. 2016;13:165–170. doi: 10.1038/nmeth.3688. [DOI] [PubMed] [Google Scholar]
- Candeias M.M., Malbert-Colas L., Powell D.J., Daskalogianni C., Maslon M.M., Naski N., Bourougaa K., Calvo F., Fahraeus R. P53 mRNA controls p53 activity by managing Mdm2 functions. Nat. Cell Biol. 2008;10:1098–1105. doi: 10.1038/ncb1770. [DOI] [PubMed] [Google Scholar]
- Cech T.R., Steitz J.A. The noncoding RNA revolution-trashing old rules to forge new ones. Cell. 2014;157:77–94. doi: 10.1016/j.cell.2014.03.008. [DOI] [PubMed] [Google Scholar]
- Chooniedass-Kothari S., Emberley E., Hamedani M.K., Troup S., Wang X., Czosnek A., Hube F., Mutawe M., Watson P.H., Leygue E. The steroid receptor RNA activator is the first functional RNA encoding a protein. FEBS Lett. 2004;566:43–47. doi: 10.1016/j.febslet.2004.03.104. [DOI] [PubMed] [Google Scholar]
- Consortium E.P. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crick F. Central dogma of molecular biology. Nature. 1970;227:561–563. doi: 10.1038/227561a0. [DOI] [PubMed] [Google Scholar]
- Forster A.C., Symons R.H. Self-cleavage of plus and minus RNAs of a virusoid and a structural model for the active sites. Cell. 1987;49:211–220. doi: 10.1016/0092-8674(87)90562-9. [DOI] [PubMed] [Google Scholar]
- Gimpel M., Heidrich N., Mader U., Krugel H., Brantl S. A dual-function sRNA from B. subtilis: SR1 acts as a peptide encoding mRNA on the gapA operon. Mol. Microbiol. 2010;76:990–1009. doi: 10.1111/j.1365-2958.2010.07158.x. [DOI] [PubMed] [Google Scholar]
- Gong C., Maquat L.E. lncRNAs transactivate STAU1-mediated mRNA decay by duplexing with 3′ UTRs via Alu elements. Nature. 2011;470:284–288. doi: 10.1038/nature09701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Porta M., Frankish A., Rung J., Harrow J., Brazma A. Transcriptome analysis of human tissues and cell lines reveals one dominant transcript per gene. Genome Biol. 2013;14:R70. doi: 10.1186/gb-2013-14-7-r70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttman M., Russell P., Ingolia N.T., Weissman J.S., Lander E.S. Ribosome profiling provides evidence that large noncoding RNAs do not encode proteins. Cell. 2013;154:240–251. doi: 10.1016/j.cell.2013.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanada K., Zhang X., Borevitz J.O., Li W.H., Shiu S.H. A large number of novel coding small open reading frames in the intergenic regions of the Arabidopsis thaliana genome are transcribed and/or under purifying selection. Genome Res. 2007;17:632–640. doi: 10.1101/gr.5836207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hangauer M.J., Vaughn I.W., McManus M.T. Pervasive transcription of the human genome produces thousands of previously unidentified long intergenic noncoding RNAs. PLoS Genet. 2013;9:e1003569. doi: 10.1371/journal.pgen.1003569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatzoglou A., Deshayes F., Madry C., Lapree G., Castanas E., Tsapis A. Natural antisense RNA inhibits the expression of BCMA, a tumour necrosis factor receptor homologue. BMC Mol. Biol. 2002;3:4. doi: 10.1186/1471-2199-3-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hube F., Velasco G., Rollin J., Furling D., Francastel C. Steroid receptor RNA activator protein binds to and counteracts SRA RNA-mediated activation of MyoD and muscle differentiation. Nucleic Acids Res. 2011;39:513–525. doi: 10.1093/nar/gkq833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingolia N.T., Brar G.A., Stern-Ginossar N., Harris M.S., Talhouarne G.J., Jackson S.E., Wills M.R., Weissman J.S. Ribosome profiling reveals pervasive translation outside of annotated protein-coding genes. Cell Rep. 2014;8:1365–1379. doi: 10.1016/j.celrep.2014.07.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingram V.M. Gene mutations in human haemoglobin: the chemical difference between normal and sickle cell haemoglobin. Nature. 1957;180:326–328. doi: 10.1038/180326a0. [DOI] [PubMed] [Google Scholar]
- Jansen G., Groenen P.J., Bachner D., Jap P.H., Coerwinkel M., Oerlemans F., van den Broek W., Gohlsch B., Pette D., Plomp J.J., et al. Abnormal myotonic dystrophy protein kinase levels produce only mild myopathy in mice. Nat. Genet. 1996;13:316–324. doi: 10.1038/ng0796-316. [DOI] [PubMed] [Google Scholar]
- Jenny A., Hachet O., Zavorszky P., Cyrklaff A., Weston M.D., Johnston D.S., Erdelyi M., Ephrussi A. A translation-independent role of oskar RNA in early Drosophila oogenesis. Development. 2006;133:2827–2833. doi: 10.1242/dev.02456. [DOI] [PubMed] [Google Scholar]
- Ji P., Diederichs S., Wang W., Boing S., Metzger R., Schneider P.M., Tidow N., Brandt B., Buerger H., Bulk E., et al. MALAT-1, a novel noncoding RNA, and thymosin beta4 predict metastasis and survival in early-stage non-small cell lung cancer. Oncogene. 2003;22:8031–8041. doi: 10.1038/sj.onc.1206928. [DOI] [PubMed] [Google Scholar]
- Ji Z., Song R., Regev A., Struhl K. Many lncRNAs, 5′UTRs, and pseudogenes are translated and some are likely to express functional proteins. Elife. 2015;4:e08890. doi: 10.7554/eLife.08890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston W.K., Unrau P.J., Lawrence M.S., Glasner M.E., Bartel D.P. RNA-catalyzed RNA polymerization: accurate and general RNA-templated primer extension. Science. 2001;292:1319–1325. doi: 10.1126/science.1060786. [DOI] [PubMed] [Google Scholar]
- Karapetyan A.R., Buiting C., Kuiper R.A., Coolen M.W. Regulatory roles for long ncRNA and mRNA. Cancers (Basel) 2013;5:462–490. doi: 10.3390/cancers5020462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kloc M., Wilk K., Vargas D., Shirato Y., Bilinski S., Etkin L.D. Potential structural role of non-coding and coding RNAs in the organization of the cytoskeleton at the vegetal cortex of Xenopus oocytes. Development. 2005;132:3445–3457. doi: 10.1242/dev.01919. [DOI] [PubMed] [Google Scholar]
- Kruger K., Grabowski P.J., Zaug A.J., Sands J., Gottschling D.E., Cech T.R. Self-splicing RNA: autoexcision and autocyclization of the ribosomal RNA intervening sequence of tetrahymena. Cell. 1982;31:147–157. doi: 10.1016/0092-8674(82)90414-7. [DOI] [PubMed] [Google Scholar]
- Kumari P., Sampath K. cncRNAs: Bi-functional RNAs with protein coding and non-coding functions. Semin. Cell Dev. Biol. 2015;47–48:40–51. doi: 10.1016/j.semcdb.2015.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanz R.B., McKenna N.J., Onate S.A., Albrecht U., Wong J., Tsai S.Y., Tsai M.J., O’Malley B.W. A steroid receptor coactivator, SRA, functions as an RNA and is present in an SRC-1 complex. Cell. 1999;97:17–27. doi: 10.1016/s0092-8674(00)80711-4. [DOI] [PubMed] [Google Scholar]
- Lauressergues D., Couzigou J.M., Clemente H.S., Martinez Y., Dunand C., Becard G., Combier J.P. Primary transcripts of microRNAs encode regulatory peptides. Nature. 2015;520:90–93. doi: 10.1038/nature14346. [DOI] [PubMed] [Google Scholar]
- Lee R.C., Feinbaum R.L., Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- Lim S., Kumari P., Gilligan P., Quach H.N., Mathavan S., Sampath K. Dorsal activity of maternal squint is mediated by a non-coding function of the RNA. Development. 2012;139:2903–2915. doi: 10.1242/dev.077081. [DOI] [PubMed] [Google Scholar]
- Lin M.F., Jungreis I., Kellis M. PhyloCSF: a comparative genomics method to distinguish protein coding and non-coding regions. Bioinformatics. 2011;27:i275–282. doi: 10.1093/bioinformatics/btr209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C., Karam R., Zhou Y., Su F., Ji Y., Li G., Xu G., Lu L., Wang C., Song M., et al. The UPF1 RNA surveillance gene is commonly mutated in pancreatic adenosquamous carcinoma. Nat. Med. 2014;20:596–598. doi: 10.1038/nm.3548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackowiak S.D., Zauber H., Bielow C., Thiel D., Kutz K., Calviello L., Mastrobuoni G., Rajewsky N., Kempa S., Selbach M., et al. Extensive identification and analysis of conserved small ORFs in animals. Genome Biol. 2015;16:179. doi: 10.1186/s13059-015-0742-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahadevan M., Tsilfidis C., Sabourin L., Shutler G., Amemiya C., Jansen G., Neville C., Narang M., Barcelo J., O’Hoy K., et al. Myotonic dystrophy mutation: an unstable CTG repeat in the 3′ untranslated region of the gene. Science. 1992;255:1253–1255. doi: 10.1126/science.1546325. [DOI] [PubMed] [Google Scholar]
- Mascarenhas R., Pietrzak M., Smith R.M., Webb A., Wang D., Papp A.C., Pinsonneault J.K., Seweryn M., Rempala G., Sadee W. Allele-selective transcriptome recruitment to polysomes primed for translation: protein-coding and noncoding RNAs, and RNA isoforms. PLoS One. 2015;10:e0136798. doi: 10.1371/journal.pone.0136798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayba O., Gilbert H.N., Liu J., Haverty P.M., Jhunjhunwala S., Jiang Z., Watanabe C., Zhang Z. MBASED: allele-specific expression detection in cancer tissues and cell lines. Genome Biol. 2014;15:405. doi: 10.1186/s13059-014-0405-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niazi F., Valadkhan S. Computational analysis of functional long noncoding RNAs reveals lack of peptide-coding capacity and parallels with 3′ UTRs. RNA. 2012;18:825–843. doi: 10.1261/rna.029520.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto M., Higuchi-Takeuchi M., Shimizu M., Shinozaki K., Hanada K. Substantial expression of novel small open reading frames in Oryza sativa. Plant Signal. Behav. 2014;9:e27848. doi: 10.4161/psb.27848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olexiouk V., Crappe J., Verbruggen S., Verhegen K., Martens L., Menschaert G. sORFs.org: a repository of small ORFs identified by ribosome profiling. Nucleic Acids Res. 2016;44:D324–329. doi: 10.1093/nar/gkv1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhart B.J., Slack F.J., Basson M., Pasquinelli A.E., Bettinger J.C., Rougvie A.E., Horvitz H.R., Ruvkun G. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature. 2000;403:901–906. doi: 10.1038/35002607. [DOI] [PubMed] [Google Scholar]
- Rivas M.A., Pirinen M., Conrad D.F., Lek M., Tsang E.K., Karczewski K.J., Maller J.B., Kukurba K.R., DeLuca D.S., Fromer M., et al. Human genomics. Effect of predicted protein-truncating genetic variants on the human transcriptome. Science. 2015;348:666–669. doi: 10.1126/science.1261877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz-Orera J., Messeguer X., Subirana J.A., Alba M.M. Long non-coding RNAs as a source of new peptides. Elife. 2014;3:e03523. doi: 10.7554/eLife.03523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharif J., Muto M., Takebayashi S., Suetake I., Iwamatsu A., Endo T.A., Shinga J., Mizutani-Koseki Y., Toyoda T., Okamura K., et al. The SRA protein Np95 mediates epigenetic inheritance by recruiting Dnmt1 to methylated DNA. Nature. 2007;450:908–912. doi: 10.1038/nature06397. [DOI] [PubMed] [Google Scholar]
- Shevtsov S.P., Dundr M. Nucleation of nuclear bodies by RNA. Nat. Cell Biol. 2011;13:167–173. doi: 10.1038/ncb2157. [DOI] [PubMed] [Google Scholar]
- Sousa C., Johansson C., Charon C., Manyani H., Sautter C., Kondorosi A., Crespi M. Translational and structural requirements of the early nodulin gene enod40, a short-open reading frame-containing RNA, for elicitation of a cell-specific growth response in the alfalfa root cortex. Mol. Cell Biol. 2001;21:354–366. doi: 10.1128/MCB.21.1.354-366.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verdon J., Girardin N., Lacombe C., Berjeaud J.-M., Héchard Y. δ-hemolysin, an update on a membrane-interacting peptide. Peptides. 2009;30:817–823. doi: 10.1016/j.peptides.2008.12.017. [DOI] [PubMed] [Google Scholar]
- Wadler C.S., Vanderpool C.K. A dual function for a bacterial small RNA: SgrS performs base pairing-dependent regulation and encodes a functional polypeptide. Proc. Natl. Acad. Sci. USA. 2007;104:20454–20459. doi: 10.1073/pnas.0708102104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan Y., Qu K., Ouyang Z., Kertesz M., Li J., Tibshirani R., Makino D.L., Nutter R.C., Segal E., Chang H.Y. Genome-wide measurement of RNA folding energies. Mol. Cell. 2012;48:169–181. doi: 10.1016/j.molcel.2012.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D., Zavadil J., Martin L., Parisi F., Friedman E., Levy D., Harding H., Ron D., Gardner L.B. Inhibition of nonsense-mediated RNA decay by the tumor microenvironment promotes tumorigenesis. Mol. Cell Biol. 2011;31:3670–3680. doi: 10.1128/MCB.05704-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wightman B., Ha I., Ruvkun G. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell. 1993;75:855–862. doi: 10.1016/0092-8674(93)90530-4. [DOI] [PubMed] [Google Scholar]
- Young T.M., Tsai M., Tian B., Mathews M.B., Pe’ery T. Cellular mRNA activates transcription elongation by displacing 7SK RNA. PLoS One. 2007;2:e1010. doi: 10.1371/journal.pone.0001010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B., Cech T.R. Peptide bond formation by in vitro selected ribozymes. Nature. 1997;390:96–100. doi: 10.1038/36375. [DOI] [PubMed] [Google Scholar]
- Zhao J., Ohsumi T.K., Kung J.T., Ogawa Y., Grau D.J., Sarma K., Song J.J., Kingston R.E., Borowsky M., Lee J.T. Genome-wide identification of polycomb-associated RNAs by RIP-seq. Mol. Cell. 2010;40:939–953. doi: 10.1016/j.molcel.2010.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]


