Abstract
The intestinal overgrowth of Clostridium difficile, often after disturbance of the gut microbiota by antibiotic treatment, leads to C. difficile infection (CDI) which manifestation ranges from mild diarrhea to life-threatening conditions. The increasing CDI incidence, not only in compromised subjects but also in traditionally considered low-risk populations, together with the frequent relapses of the disease, has attracted the interest for prevention/therapeutic options. Among these, probiotics, prebiotics, or synbiotics constitute a promising approach. In this study we determined the potential of selected Bifidobacterium strains for the inhibition of C. difficile growth and toxicity in different carbon sources. We conducted co-cultures of the toxigenic strain C. difficile LMG21717 with four Bifidobacterium strains (Bifidobacterium longum IPLA20022, Bifidobacterium breve IPLA20006, Bifidobacterium bifidum IPLA20015, and Bifidobacterium animalis subsp. lactis Bb12) in the presence of various prebiotic substrates (Inulin, Synergy, and Actilight) or glucose, and compared the results with those obtained for the corresponding mono-cultures. C. difficile and bifidobacteria levels were quantified by qPCR; the pH and the production of short chain fatty acids was also determined. Moreover, supernatants of the cultures were collected to evaluate their toxicity using a recently developed model. Results showed that co-culture with B. longum IPLA20022 and B. breve IPLA20006 in the presence of short-chain fructooligosaccharides, but not of Inulin, as carbon source significantly reduced the growth of the pathogen. With the sole exception of B. animalis Bb12, whose growth was enhanced, the presence of C. difficile did not show major effects upon the growth of the bifidobacteria. In accordance with the growth data, B. longum and B. breve were the strains showing higher reduction in the toxicity of the co-culture supernatants.
Keywords: probiotics, prebiotics, inhibition, Clostridium difficile, Bifidobacterium, toxin, HT29, RTCA
Introduction
Clostridium difficile is often present in the intestinal microbiota of both infants and adults, where it may be found in about 70 and 17% of the subjects, respectively (Ozaki et al., 2004; Jangi and Lamont, 2010). However, this microorganism is also the main causative agent of antibiotic associated diarrhea in nosocomial environments (Leffler and Lamont, 2015). The epidemiology of C. difficile infection (CDI) is changing, with an increasing occurrence in populations traditionally considered of low-risk (Carter et al., 2012), likely due to the appearance of hipervirulent strains (Rupnik et al., 2009; Yakob et al., 2015). CDI is treated with antibiotics but a high rate of recurrence is present. In this context, new therapeutic alternatives for treating or preventing CDI are being continuously explored, among them the inhibition of C. difficile growth by the use of probiotics or prebiotics has been tested (Ambalam et al., 2015; Auclair et al., 2015; Forssten et al., 2015).
In general, probiotics and prebiotics have been proposed as biotherapeutic agents to prevent the dysbiosis caused by antibiotics or infections, and to help the microbiota restoration after it (Reid et al., 2011). The development of food products targeting at the inhibition of C. difficile constitutes an interesting approach in the context of the marketing of products bearing health claims. Reducing the intestinal levels of specific pathogens, such as C. difficile, has been considered by the European Food Safety Authority (EFSA) as a beneficial physiological effect [(EFSA Panel on Dietetic Products, Nutrition and Allergies (NDA), 2011)]. Therefore, such an effect would constitute an opportunity for the development of food products bearing a health claim in the area of gastrointestinal health.
To date, different probiotic strains and prebiotic substrates have been reported to increase colonization resistance against C. difficile (Hopkins and Macfarlane, 2003; Kondepudi et al., 2014; Auclair et al., 2015; Forssten et al., 2015). In addition to their microbiota-modulatory properties, probiotics have been found to protect against infections by other mechanisms, such as production of antimicrobial compounds or competition by adhesion sites or nutrients (Servin, 2004). The ability of certain probiotics, mainly bifidobacteria and lactobacilli, to inhibit in vitro the adhesion of C. difficile to intestinal epithelial cells or intestinal mucus is well established (Collado et al., 2005; Banerjee et al., 2009). Similarly, the ability to produce antimicrobials inhibiting the growth of C. difficile in vitro has been repeatedly reported (Lee et al., 2013; Schoster et al., 2013). However, other potential targets of probiotics and prebiotics on CDI, such as their impact on toxin production by the pathogen, and/or toxin activity, have been explored to a lesser extent and have not attracted attention until recently (Kondepudi et al., 2014; Yun et al., 2014; Andersen et al., 2015). Ambalam et al. (2015) recently reported the ability of cell-free supernatants from some Lactobacillus strains, and a probiotic mix, to inhibit the growth of C. difficile strains in variable way depending on the carbon source used. Moreover, the authors observed a reduction of toxin titers in those C. difficile cultures with inhibitory cell-free supernatants added. Moreover, we have demonstrated that incubation of toxigenic C. difficile cell-free culture supernatants with specific bifidobacterial strains reduces the cytotoxic effect upon human epithelial intestinal cells (Valdés et al., 2016). However, the influence of prebiotic substrates upon C. difficile growth and toxicity when co-cultured with bifidobacteria remains largely unknown.
In this context the aim of this study was to evaluate in vitro the potential of four bifidobacterial strains for inhibiting the growth of C. difficile when co-cultured with different prebiotics as carbon source. Moreover, the effect of the strains and prebiotics on the toxicity of the co-culture supernatants upon human intestinal epithelial cells (HT29) was also determined.
Materials and methods
Bacterial strains and culture conditions
The widely used probiotic strain Bifidobacterium animalis subsp. lactis Bb12 and three strains of bifidobacteria from IPLA culture collection, two of them isolated from infant's feces (Bifidobacterium longum IPLA20022 and Bifidobacterium bifidum IPLA20015) (Solís et al., 2010) and the other one from breast-milk (Bifidobacterium breve IPLA20006) (Arboleya et al., 2011), were used. These last three strains were selected based on the good ability to reduce toxicity of C. difficile supernatants (Valdés et al., 2016). With regard to C. difficile we used the strain LMG21717, known to produce TcdA toxin and also, although at lower quantities, TcdB. This strain belongs to ribotype 001, which is one of the most common ones found in Europe (Martin et al., 2016). The Bifidobacterium strains were routinely grown in MRS (Biokar Diagnostics, Beauvois, France) supplemented with 0.25% L-cysteine (Sigma-Chemical Co., St. Louis, MO, USA) in an anaerobic chamber MG500 (Don Whitley Scientific, Yorkshire, UK) and C. difficile was grown in Reinforced Clostridial Medium (RCM, Oxoid, Thermo Fisher Scientific Inc., Waltham, MA) in Hungate tubes as previously described (Valdés et al., 2016). Overnight cultures (18 h) of the bifidobacterial strains and 13 h-old cultures of C. difficile were used to inoculate the batch culture fermentations.
For the batch mono- and co-culture fermentations a defined medium with the following composition was used: proteose peptone (10 g/L) (BD-Difco, New Jersey, EE.UU.), beef extract (10 g/L) (BD-Difco), yeast extract (5 g/L) (BD-Difco), polysorbate 80 (1 mL/L) (Sigma), ammonium citrate (2 g/L) (Sigma), sodium acetate (5 g/mL) (Sigma), magnesium sulfate (0.2 g/L) (Probus, Barcelona, Spain), manganese sulfate (0.056 g/L) (Panreac, Barcelona, Spain), and dipotassium phosphate (2 g/L) (Merck, New Jersey, EE.UU). Pairwise combinations of the C. difficile strain with the different Bifidobacterium strains, as well as the corresponding monocultures, were performed in the medium described above with a 2% (w/v) of different commercial prebiotic substrates added [Synergy 1 (Beneo-Orafti, Barcelona, Spain), Inulin (Sigma) and Actilight (Beghin Meiji and Tereos Syral, Marckolsheim, France)], glucose or without adding any carbon source (used as control). Each media was distributed into Hungate tubes which were inoculated with different Bifidobacterium strains at a final level of about 105 CFU/ml in case of B. longum/B. breve and 104 CFU/ml in case of B. bifidum/B. animalis, with C. difficile strain at final level of 106 CFU/ml or with both of them, in the case of the co-culture. The bifidobacteria were inoculated at a different level depending on the strain with the aim of allowing a balanced growth of both microorganisms (bifidobacteria and clostridia). The appropriate inoculum size was determined in previous experiments (data not shown).
Co-cultures, and the corresponding mono-cultures, in different carbon sources were carried out in triplicate under anaerobic conditions at 37°C for 24 h. Samples were taken at 0 and 24 h for bacterial growth assessment by quantitative PCR (qPCR), quantification of SCFA by Gas Chromatography (GC), pH measurements (pH meter Basic 20+, Crison Instruments S.A., Barcelona, Spain), and toxigenicity determination. One milliliter of each mono-culture or co-culture was centrifuged (16,000 × g for 10 min), and pellets and supernatants were collected. For toxigenic activity upon HT29 cells, the pH of 0.7 ml cell-free supernatant from each batch culture was adjusted to 7.55 ± 0.05 with 1 and 0.1 N NaOH. All supernatants and pellets were immediately frozen at −80°C until use.
Quantification of bacterial growth by qPCR
DNA was extracted from pellets of batch cultures using the GenElute Bacterial Genomic DNA Kit (Sigma) and kept at −80°C until analyzed. The levels of C. difficile and bifidobacteria in the cultures were determined as DNA copies per ml by qPCR using previously described primers and conditions (Arboleya et al., 2012). Reactions were performed on MicroAmp optical plates sealed with MicroAmp optical caps (Applied Biosystems, Foster City, CA, USA) with a 7500 Fast Real-Time PCR System (Applied Biosystems) using SYBR Green PCR Master Mix (Applied Biosystems). One microlitre of template DNA was used in the 25 mL PCR mixture. Standard curves were made with pure cultures of B. longum NCIMB8809 and C. difficile LMG 21717. In all cultures the levels of the microorganisms were above the corresponding detection limit of the technique (1 × 103 and 3 × 103 for bifidobacteria and C. difficile, respectively). Samples were analyzed by duplicate in at least two independent PCR runs.
Determination of the production of short chain fatty acids by GC-MS
Cell-free supernatant (0.1 mL) from each batch culture was mixed with 1 ml methanol, 0.1 ml internal standard solution (2-ethylbutyric 1.05 mg/ml), and 0.1 ml 20% formic acid. This mixture was centrifuged and the supernatant obtained was used for quantification of SCFA by GC in a system composed of a 6890NGC injection module (Agilent Technologies Inc., Palo Alto, Ca, USA) connected to a flame injection detector (FID) and a mass spectrometry (MS) 5973N detector (Agilent) as described previously (Salazar et al., 2011).
Monitoring the cytotoxic effect of the culture supernatants upon intestinal epithelial cells
The intestinal cell line HT29 (ECACC 91072201) was purchased from the “European Collection of Cell Cultures” (Salisbury, UK) and stored under liquid N2. McCoy's Medium (MM) supplemented with 10% fetal serum bovine, 3 mM L-glutamine and a mixture of antibiotics (50 μg/ml streptomycin-penicillin, 50 μg/ml gentamicin, and 1.25 μg/ml amphotericin B) was used for HT29 cultivation. All media and reagents were purchased from Sigma-Aldrich. Maintenance of the cell line, between passages 145 and 149, was performed under standard conditions at 37°C 5% CO2 atmosphere, in a CO2-Series Shel-Lab incubator (Sheldon Manufacturing Inc., OR, USA). The experimental procedures were carried out with the cell passage 149.
We used an RTCA (real time cell analyser) xCelligence (ACEA Bioscience Inc., San Diego, CA) system, introduced in a Heracell-240 Incubator (Thermo Electron LDD GmbH, Langenselbold, Germany) set at 37°C with 5% CO2 atmosphere, to monitor HT29 cells behavior. A method previously described, allowing the assessment of the damage caused by C. difficile supernatants, was used (Valdés et al., 2015). This method is based in the real-time monitoring of the cell index (CI). This CI is an arbitrary unit that measures the impedance, in gold-microelectrodes coating the surface of E-plates, which changes as consequence of the HT29 cells attachment and growth.
In short, 16-well E-plates were seed with 2 × 105 HT29 cells (in 100 μl), hold in the RTCA equipment, incubated for 22 h to ensure the formation of a monolayer (confluent state) and the CI was monitored (recording signal every 15 min). After this incubation the medium was removed from the wells and the methodology followed was slightly different depending on the experiment. To determine the effect of the carbon source on the toxicity of C. difficile, 200 μL of MM containing different concentrations (from 0.63 to 40%, v/v) of cell-free neutralized-supernatants from C. difficile mono-cultures were added to the wells. EC50 values (concentration at which half of the maximum damage was detected) for the cultures, in the different carbon sources tested, were then calculated as previously described (Valdés et al., 2015). To determine the effect of bifidobacteria on the toxigenic capability of C. difficile in the different carbon sources, 200 μL of MM containing a 5% (v/v) of the neutralized supernatant from each mono- and co-culture were added to the wells. Additionally, wells filled with 200 μl of MM (non-cytotoxic control) were included in each experiment. Then, monitoring continued (recording signal every 10 min) up to 20 h under standard incubation conditions. The data analyses were carried out through RTCA software 1.2.1 (ACEA Bioscience). The CI values were normalized as previously described (Valdés et al., 2015) by dividing the CI at every point by the CI at time zero (the time of the supernatant addition, thus making the CI equal to 1 at this time) and then referred to the normalized CI of the control sample (MM) (the normalized-CI of the control sample is then the “0 line” shown in figures).
Toxin A concentration in the supernatant of C. difficile mono-cultures in different carbon sources was determined by ELISA test (tgcBIOMICS GmbH, Bingen, Germany).
Statistical analysis
To asses differences among carbon sources or between mono- and co-cultures, one-way ANOVAs followed by SNK (Student-Newman-Keuls, p < 0.05) mean comparison test were performed. The statistical package IBM SPSS Statistics for Window Version 22.0 (IBM Corp., Armonk NY) was used to carry out these analyses.
Results and discussion
Inhibition of C. difficile growth when co-cultured with Bifidobacterium strains in different carbon sources
There is a great scientific interest on the development of interventions for preventing or treating CDI, including vaccines (Senoh et al., 2015), antimicrobials (Gebhart et al., 2015; Vickers et al., 2015), anti-toxin antibodies (Yang et al., 2015), or genetically engineered bacteria producing them (Andersen et al., 2015), among others. Fecal transplants have demonstrated a high efficacy to treat recurrent CDI (Lee et al., 2016), underlining the importance of the gut microbiota in this disease. Probiotics and prebiotics constitute another interesting option although differences among strains and substrates seem to exist (Allen et al., 2013).
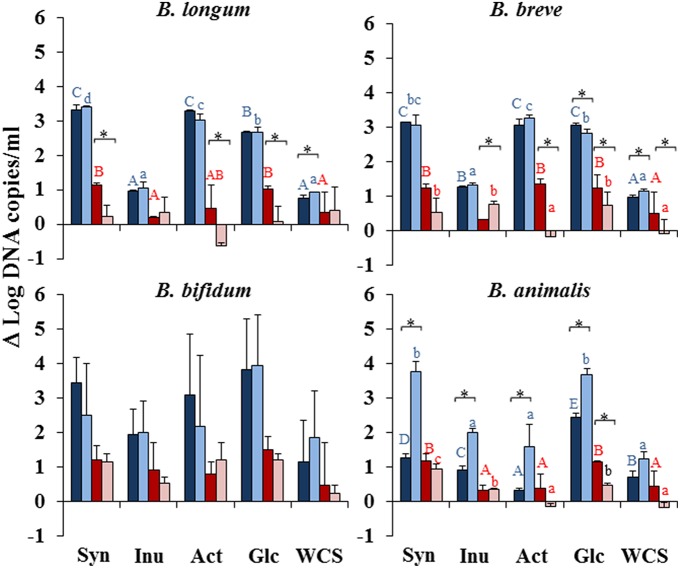
In our study the mono-culture of the Bifidobacterium strains (Figure 1) in different substrates (dark colored bars) showed that all the strains grew well in glucose. In agreement with previous reports (Rossi et al., 2005; Kondepudi et al., 2012), the strains showed the ability to grow in short-chain fructooligosaccharides (Synergy and Actilight) (scFOS) but they were not able to grow, or did it poorly, in Inulin (Figure 1). This observation was further supported by the production of bacterial metabolites (Figure 2, Supplementary File) and the pH (Supplementary Figure 1), which in the case of Inulin remained similar to those of the negative control without carbon source added (WCS). Interestingly, B. longum IPLA20022 showed a significantly higher growth (p < 0.05) in the prebiotics Synergy and Actilight than in glucose (Figure 1), whereas no statistically significant differences were observed for B. breve IPLA20006 or B. bifidum IPLA20015 between glucose and these two prebiotics. The mono-cultures of B. animalis Bb12 showed a significantly lower (p < 0.05) growth in all prebiotics than in glucose. This strain exhibited the lowest growth of all bifidobacteria in glucose, Synergy, and Actilight (Figure 1), which correlates with the limited drop in pH observed for this strain after 24 h of incubation (Supplementary Figure 1). With regard to the pathogen, C. difficile grew well in Synergy, not differing significantly from glucose, and to a lower extent in Actilight (Figure 1). Therefore, in spite of generally claimed high specific fermentation of prebiotic substrates, some intestinal pathogens may also be able to ferment and grow in some of them. This underlines the importance of a careful selection of the most appropriate strains, substrates, and combinations.
Figure 1.
Increments, with respect to time zero, on the levels (Log CFU/mL) of the strains when grown in mono-culture (Bifidobacterium dark-blue column and C. difficile dark-red) or co-culture (Bifidobacterium light-blue and C. difficile light-red column) in the prebiotics Synergy (Syn), Inulin (Inu), and Actilight (Act), in glucose (Glc) or without any carbon source added (WCS). Different capital letters above columns denote statistically significant differences (p < 0.05) among carbon sources in the mono-cultures of the corresponding bacterial strain, whereas different lowercase letters indicate differences in the co-cultures (either Bifidobacterium in blue letters or C. difficile in red letters). *Indicates statistically significant differences (p < 0.05) for the corresponding bacterial strain between mono- and co-culture within the same substrate.
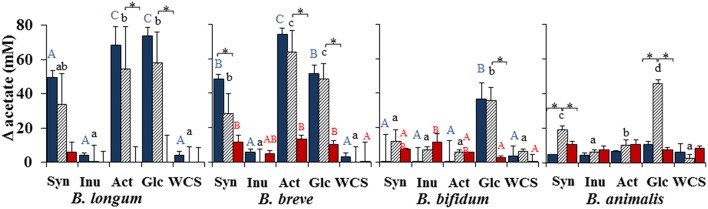
Figure 2.
Increments, with respect to time zero, in the concentration of acetate on the bacterial cultures when grown in mono-culture (Bifidobacterium blue-bars and C. difficile red-bars) or in co-culture (stripped bars) in the prebiotics Synergy (Syn), Inulin (Inu), and Actilight (Act), in glucose (Glc) or without any carbon source added (WCS). Different letters above columns denote statistically significant differences (p < 0.05) among carbon sources in the corresponding bacterial cultures, either mono-cultures (capital letter; red color for bifidobacteria and blue color for C. difficile) or co-cultures (lowercase letters). *Indicates statistically significant differences (p < 0.05) for the corresponding bacterial strain between mono- and co-culture.
When co-cultured with C. difficile in the different carbon sources, the behavior of the bifidobacteria was, in general, similar to that observed in the mono-cultures. We observed increases in bifidobacterial counts in glucose, Synergy, and Actilight and poor grow in Inulin. Regarding C. difficile, it grew better in glucose, followed by Synergy, which is in agreement with the mono-culture data, but the growth in Actilight was, in general, significantly (p < 0.05) worse in co-culture, the contrary being true for Inulin (Figure 1). This growth behavior of C. difficile in the different carbon sources was further confirmed by the metabolites production pattern (Supplementary File), showing in general a lower production of C. difficile metabolites, such as propionate or branched-SCFA, in co-culture with Actilight as carbon source than in the corresponding mono-culture, whilst the contrary was observed for Inulin.
When co- and mono-cultures were compared within the same carbon source, the growth of C. difficile was significantly reduced (p < 0.05) by B. longum IPLA20022, B. breve IPLA20006, or B. animalis Bb12 in glucose. The first two microorganisms also reduced C. difficile growth when co-cultured in Actilight and, in the case of B. longum also when Synergy was used as carbon source (Figure 1). On the contrary, no statistically significant differences between mono- and co-cultures were observed for B. bifidum in any carbon source. These results showed a good correlation with the pattern of production of C. difficile metabolites and the drop in pH (Supplementary File). This suggests the production of organic acids, with the concomitant reduction of the pH, as an important mechanism of inhibition (Tejero-Sariñena et al., 2012).
These results point out at B. longum IPLA20022 and B. breve IPLA20006, and the prebiotics Synergy and Actilight, as the most promising alternatives for inhibiting the growth of C. difficile. Moreover, they suggest that the pathogen inhibition is strain and substrate specific, which is in agreement with previous reports (Kondepudi et al., 2012; Tejero-Sariñena et al., 2013; Ambalam et al., 2015). Interestingly, the growth of C. difficile was significantly increased (p < 0.05) by B. breve in the presence of Inulin, indicating a potential risk of such combination and underlining the importance of a careful strain and substrate specific assessment.
Interestingly, effects of the co-culture with C. difficile on the growth of the bifidobacterial strains were also observed. Whilst in glucose the co-culture with the clostridia did not affect the growth of B. longum, it significantly (p < 0.05) reduced that of B. breve but increased that of B. animalis. Moreover, the growth of the latter microorganism was also increased by the presence of C. difficile in the three prebiotics tested, mainly Synergy (Figure 1) which was further confirmed by an enhanced production of acetate in the co-culture than in the corresponding monoculture (Figure 2) and a higher drop in pH (Supplementary Figure 1).
The carbon source determines the toxicity of C. difficile supernatants
In addition to bacterial growth, inhibiting the toxicity caused by C. difficile, for example by reducing toxin production or toxic activity, represents another target in CDI (Trejo et al., 2010, 2013). The toxicity of C. difficile culture supernatants has been found to be dependent on the culture media used (Valdés et al., 2015), suggesting a potential role of the carbon source available. Therefore, it is important to know whether the availability of different prebiotics as carbon source may have an impact on the toxicity of C. difficile. To clarify this point we determined the toxicity of neutralized cell-free supernatants, obtained from C. difficile monocultures after 24 h of incubation in the different carbon sources, upon the human epithelial cell line HT29 by using a real-time monitoring system (RTCA). To this end the EC50 values, defined as the concentration of supernatant causing 50% of the maximum cell damage, were calculated (Valdés et al., 2015). Supernatants obtained from the mono-cultures carried out without any carbon source or with Actilight added were significantly (p < 0.05) more toxic than the others (Figure 3). They showed EC50 values below 2%, which means that a concentration of monoculture supernatant lower than 2% already produced half of the maximum cell damage. On the contrary, the supernatant of the mono-culture in glucose resulted significantly (p < 0.05) less toxic than all the others (EC50 value over 6%), followed by that on Synergy and the one carried out with Inulin as carbon source (Figure 3). The method used (Valdés et al., 2015) allowed us to determine that the C. difficile supernatants' toxicity was higher when no carbon source was added or when the available carbon source supported only a limited growth of the pathogen, such as in the case of Actilight. On the contrary, the supernatant obtained when the pathogen was grown in glucose, in spite of the good growth of C. difficile, resulted less toxic. The availability of rapidly metabolizable sugars has been reported to inhibit toxin synthesis in C. difficile (Bouillaut et al., 2015). This inhibition is mediated through repression of treR (also known as tdcR), an alternative sigma factor responsible for the positive regulation of toxA and toxB genes (Mani et al., 2002). Our results seem to confirm the higher production of toxins by C. difficile under nutrient limitation or stress conditions in which readily fermentable sugars are not available. Moreover, in C. difficile a co-induction of metabolic pathways, such as that of butyrate production, and toxin production has been reported (Karlsson et al., 2000). In our study the C. difficile monoculture grown in glucose showed, in general, lowest butyrate production than those carried out with Synergy, Actilight or WCS added, which is in good agreement with the lower toxin production in glucose. However, the culture of the strain in Inulin, in spite of a lower production of butyrate than that in glucose, showed higher toxin concentrations, comparable to those found WCS or in the other prebiotics tested. These results indicate that, at least in some circumstances, toxin production by C. difficile is uncoupled from the production of metabolites such as butyrate.
Figure 3.
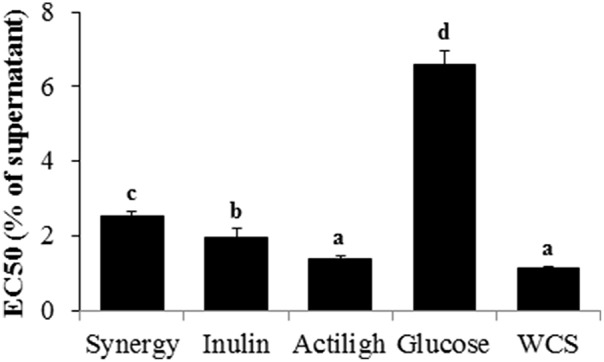
Concentration (% v/v) of supernatants of C. difficile mono-cultures, in the different carbon sources tested, showing 50% of the maximum cell damage (EC50). To calculate EC50s the cell indexes obtained after 12 h of incubation of the HT29 cells with supernatants were used. Different letters above the columns denote statistically significant differences (p < 0.05).
In accordance with the above mentioned toxicity data, the concentration of C. difficile toxin A showed the lowest value in the supernatant from the culture in glucose (Figure 4). The supernatants obtained from cultures grown in Synergy, Actilight showed the highest toxin concentrations, whilst those from the growth on C. difficile in Inulin or WCS showed intermediate levels.
Figure 4.
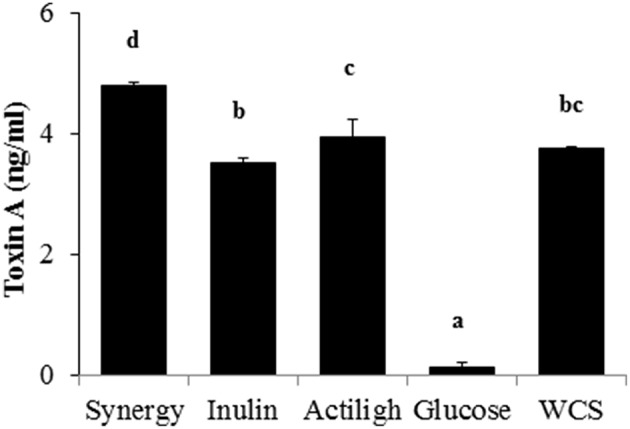
Toxin A concentration in the different C. difficile supernatants obtained when the microorganism was growth in the different carbon sources. Different letters above the columns denote statistically significant differences (p < 0.05).
Co-culture with bifidobacteria in different carbon sources reduces C. difficile toxicity
The ability of certain bifidobacterial strains, such as B. longum IPLA20022, to remove toxins from C. difficile cell-free supernatants, then diminishing their cytotoxicity, has been recently reported (Valdés et al., 2016). Now we compared the toxicity of the Clostridium-Bifidobacterium co-culture supernatants with that of the pathogen monoculture. In general we observed a significant reduction on the toxicity of the supernatants in co-culture. However, differences depending on the strain and the carbon source used were also observed, confirming the high specificity of these interactions (Trejo et al., 2010). The toxicities obtained for the co-cultures in the different carbon sources were compared by using the normalized cell index (CI) obtained after 12 h of incubation of HT29 cells with a 5% of the culture supernatants. As it was the case for the monocultures, supernatants from co-cultures carried out on the different carbon sources showed differences among them (p < 0.05) (Table 1). Similarly to the mono-cultures, supernatants obtained in glucose showed the lowest toxicity whilst those in Inulin, or without any carbon source added, resulted the most toxic. When the supernatants of the co-cultures with the different bifidobacteria were compared with the C. difficile monoculture no statistically significant differences (p > 0.05) were obtained in media WCS added. However, in all the carbon sources tested, either glucose or prebiotics, statistically significant differences (p < 0.05) were observed depending on the bifidobacterial strain used (Table 1). Co-culture in Synergy or Actilight of C. difficile with B. longum IPLA20022 or B. breve IPLA20006 significantly (p < 0.05) inhibited the toxicity of the supernatant (i.e., higher normalized CI) when compared with the mono-culture of C. difficile. However, B. bifidum IPLA20015 only was able to reduce (p < 0.05) the toxicity of the pathogen with Actiligh as carbon source whilst B. animalis Bb12 did not produce toxicity inhibition in any prebiotic. The four bifidobacteria tested were able to reduce (p > 0.05) the toxicity of the supernatant when co-cultured in glucose, in comparison to the C. difficile mono-culture, but none of them did it when the carbon source was Inulin. In the latter case, even, an increase in the toxicity was observed when the pathogen was co-incubated with B. bifidum (Table 1), suggesting a potential risk for such combination.
Table 1.
Normalized cell index (mean ± sd) obtained after 12 h of incubation of HT29 cells with the supernatants (5%) of the C. difficile mono-culture or C. difficile-Bifidobacterium co-cultures grown in different prebiotics, glucose or without any carbon source added (WCS).
| Culture | Normalized cell index | ||||
|---|---|---|---|---|---|
| Carbon source | |||||
| Synergy | Inulin | Actilight | Glucose | WCS | |
| C. difficile | −0.39±0.03a,1 | −0.30±0.03a,2 | −0.35±0.04a,1 | −0.23±0.01b,1 | −0.34±0.04a |
| C. difficile–B. longum | −0.06±0.04b,2 | −0.32±0.07a,2 | −0.13±0.05b,3 | 0.01±0.02b,3 | −0.43±0.13a |
| C. difficile–B. breve | −0.02±0.02d,2 | −0.32±0.03b,2 | −0.07±0.01c,4 | 0.00±0.01d,3 | −0.37±0.01a |
| C. difficile–B. bifidum | −0.40±0.08b,1 | −0.56±0.02a,1 | −0.24±0.01c,2 | 0.00±0.01d,3 | −0.34±0.01b |
| C. difficile–B. animalis | −0.35±0.02a,1 | −0.31±0.03a,2 | −0.32±0.02a,1 | −0.03±0.00b,2 | −0.34±0.02a |
*Different superscripts letters within the same row indicate statistically significant differences (p < 0.05) among carbon sources, whereas different superscript numbers within the same column denote differences (p < 0.05) among cultures.
Our results show that B. longum IPLA20022 and B. breve IPLA20006 reduced the toxicity of the co-cultures with sc-FOS as carbon source. Interestingly these two strains have previously shown the ability to remove C. difficile toxins from solution (Valdés et al., 2016). Although the putative mechanism behind toxin inactivation remains to be elucidated, it has been demonstrated that certain microorganisms produce compounds able to degrade C. difficile toxins or to reduce their toxicity (Castagliuolo et al., 1996; Banerjee et al., 2009; Carasi et al., 2012; Valdés et al., 2016). These mechanisms may be involved in the effect observed by us. However, given that in our case both microorganisms are co-incubated, the direct inhibition of the growth of the pathogen and/or an modulation of the expression of the toxin genes in C. difficile by the presence of bifidobacteria, similarly to that previously reported for Lactobacillus acidophilus (Yun et al., 2014), may also be involved. Previous studies pointed out a role of organic acids, such as lactic acid, in the inhibition of both growth and toxin production by C. difficile (Kolling et al., 2012; Yun et al., 2014). Therefore, the ability of bifidobacteria to produce acids, mainly acetic and lactic acids, and the pH drop caused by them may partially explain our observations. However, the role of other interactions cannot be overruled, especially since behaviors not explained by the acids, such as the increased toxicity of the co-culture C. difficile-B. bifidum in Inulin, were also observed.
Conclusion
Co-culture with B. longum IPLA20022 or B. breve IPLA20006 in the presence of scFOS, but not of Inulin, reduces significantly the growth of C. difficile. Moreover, co-culture with these two strains in Synergy or Actilight reduced the toxicity of the C. difficile supernatants. Therefore, B. longum IPLA20022 and B. breve IPLA20006, in combination with Synergy or Actilight, are the most promising strains and compounds for the development of probiotic, prebiotic, or synbiotic products targeting at the reduction of CDI. However, future in vitro studies aiming at other clinically relevant C. difficile strains, as well as in vivo evaluation of the efficacy of the products, would be needed before drawing firm conclusions.
Author contributions
MG and PR contributed with the conception, experimental design, and results interpretation of this study. LV carried out all experiments, AH performed chromatographic analyses. MG was in charge of writing the drafted manuscript. All authors performed a critical revision of the manuscript and approved the final version.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was fund by FEDER European Union funds through the projects AGL2012-33278 and AGL2013-43770R from the Spanish Ministry of Economy and Competitiveness (MINECO), and through the grants EQUIP11 and GRUPIN14-043 from the Program of Science, Technology and Innovation from the “Principado de Asturias.” LV acknowledges her JAE-Pre fellowship to CSIC.
Supplementary material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2016.00738
References
- Allen S. J., Wareham K., Wang D., Bradley C., Hutchings H., Harris W., et al. (2013). Lactobacilli and bifidobacteria in the prevention of antibiotic-associated diarrhoea and Clostridium difficile diarrhoea in older inpatients (PLACIDE): a randomised, double-blind, placebo-controlled, multicentre trial. Lancet 382, 1249–1257. 10.1016/S0140-6736(13)61218-0 [DOI] [PubMed] [Google Scholar]
- Ambalam P., Kondepudi K. K., Balusupati P., Nilsson I., Wadström T., Ljungh A. (2015). Prebiotic preferences of human lactobacilli strains in co-culture with bifidobacteria and antimicrobial activity against Clostridium difficile. J. Appl. Microbiol. 119, 1672–1682. 10.1111/jam.12953 [DOI] [PubMed] [Google Scholar]
- Andersen K. K., Strokappe N. M., Hultberg A., Truusalu K., Smidt I., Mikelsaar R. H., et al. (2015). Neutralization of Clostridium difficile Toxin B mediated by engineered Lactobacilli that produce single-domain antibodies. Infect. Immun. 84, 395–406. 10.1128/IAI.00870-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arboleya S., Binetti A., Salazar N., Fernández N., Solís G., Hernández-Barranco A., et al. (2012). Establishment and development of intestinal microbiota in preterm neonates. FEMS Microbiol. Ecol. 79, 763–772. 10.1111/j.1574-6941.2011.01261.x [DOI] [PubMed] [Google Scholar]
- Arboleya S., Ruas-Madiedo P., Margolles A., Solis G., Salminen S., de los Reyes-Gavilan C. G., et al. (2011). Characterization and in vitro properties of potentially probiotic Bifidobacterium strains isolated from breast-milk. Int. J. Food Microbiol. 149, 28–36. 10.1016/j.ijfoodmicro.2010.10.036 [DOI] [PubMed] [Google Scholar]
- Auclair J., Frappier M., Millette M. (2015). Lactobacillus acidophilus CL1285, Lactobacillus casei LBC80R, and Lactobacillus rhamnosus CLR2 (Bio-K+): characterization, manufacture, mechanisms of action, and quality control of a specific probiotic combination for primary prevention of Clostridium difficile infection. Clin. Infect. Dis. 60, S135–S143. 10.1093/cid/civ179 [DOI] [PubMed] [Google Scholar]
- Banerjee P., Merkel G. J., Bhunia A. K. (2009). Lactobacillus delbrueckii ssp. bulgaricus B-30892 can inhibit cytotoxic effects and adhesion of pathogenic Clostridium difficile to Caco-2 cells. Gut Pathog. 1:8. 10.1186/1757-4749-1-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouillaut L., Dubois T., Sononshein A. L., Dupuy B. (2015). Integration of metabolism and virulence in Clostridium difficile. Res. Microbiol. 166, 375–383. 10.1016/j.resmic.2014.10.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carasi P., Trejo F. M., Pérez P. F., De Antoni G. L., Serradell M. A. (2012). Surface proteins from Lactobacillus kefir antagonize in vitro cytotoxic effect of Clostridium difficile toxins. Anaerobe 18, 135–142. 10.1016/j.anaerobe.2011.11.002 [DOI] [PubMed] [Google Scholar]
- Carter G. P., Rood J. I., Lyras D. (2012). The role of toxin A and toxin B in the virulence of Clostridium difficile. Trends Microbiol. 20, 21–29. 10.1016/j.tim.2011.11.003 [DOI] [PubMed] [Google Scholar]
- Castagliuolo I., Lamont J. T., Nikulasson S. T., Pothoulakis C. (1996). Saccharomyces boulardii protease inhibits Clostridium difficile Toxin A effects in the rat ileum. Infect. Immun. 64, 5225–5232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collado M. C., Gueimonde M., Hernández M., Sanz Y., Salminen S. (2005). Adhesion of selected Bifidobacterium strains to human intestinal mucus and its role in enteropathogen exclusion. J. Food Protect. 68, 2672–2678. [DOI] [PubMed] [Google Scholar]
- EFSA Panel on Dietetic Products Nutrition Allergies (NDA). (2011). Guidance on the scientific requirements for health claims related to gut and immune function. EFSA J. 9:1984 10.2903/j.efsa.2011.1984 [DOI] [Google Scholar]
- Forssten S. D., Röytió H., Hibberd A. A., Ouwehand A. C. (2015). The effect of polydextrose and probiotic lactobacilli in a Clostridium difficile-infected human colonic model. Microb. Ecol. Health Dis. 26, 27988. 10.3402/mehd.v26.27988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gebhart D., Lok S., Clare S., Tomas M., Stares M., Scholl D., et al. (2015). A modified R-type bacteriocin specifically targeting Clostridium difficile prevents colonization of mice without affecting gut microbiota diversity. MBio. 6, e02368–e02314. 10.1128/mBio.02368-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopkins M. J., Macfarlane G. T. (2003). Nondigestible oligosaccharides enhance bacterial colonization resistance against Clostridium difficile in vitro. Appl. Environ. Microbiol. 69, 1920–1927. 10.1128/AEM.69.4.1920-1927.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jangi S., Lamont J. T. (2010). Asyntomatic colonization by Clostridium difficile in infants: implications for disease in later life. J. Pediatr. Gastroenterol. Nutr. 51, 2–7. 10.1097/MPG.0b013e3181d29767 [DOI] [PubMed] [Google Scholar]
- Karlsson S. A., Lindberg A., Norin E., Burman L. G., Akerlund T. (2000). Toxins, butyric acid, and other short-chain fatty acids are coordinatedly expressed and down-regulated by cysteine in Clostridium difficile. Infect. Immun. 68, 5881–5888. 10.1128/IAI.68.10.5881-5888.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolling G. L., Wu M., Warren C. A., Durmaz E., Klaenhammer T. R., Guerrant R. L. (2012). Lactic acid production by Streptococcus thermophilus alters Clostridium difficile infection and in vitro Toxin A production. Gut Microbes. 3, 523–529. 10.4161/gmic.21757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondepudi K. K., Ambalam P., Karagin P. H., Nilsson I., Wadström T., Ljungh A. (2014). A novel multi-strain probiotic and synbiotic supplement for prevention of Clostridium difficile infection in a murine model. Microbiol. Immunol. 58, 552–558. 10.1111/1348-0421.12184 [DOI] [PubMed] [Google Scholar]
- Kondepudi K. K., Ambalam P., Nilsson I., Wadstrom T., Ljungh A. (2012). Prebiotic-non-digestible oligosaccharides preference of probiotics bifidobacteria and antimicrobial activity against Clostridium difficile. Anaerobe 18, 489–497. 10.1016/j.anaerobe.2012.08.005 [DOI] [PubMed] [Google Scholar]
- Lee C. H., Steiner T., Petrof E. O., Smieja M., Roscoe D., Nematallah A., et al. (2016). Frozen vs fresh fecal microbiota transplantation and clinical resolution of diarrhea in patients with recurrent Clostridium difficile infection: a randomized clinical trial. J. Am. Med. Assoc. 315, 142–149. 10.1001/jama.2015.18098 [DOI] [PubMed] [Google Scholar]
- Lee J.-S., Chung M.-J., Seo J.-G. (2013). In vitro evaluation of antimicrobial activity of lactic acid bacteria against Clostridium difficile. Toxicol. Res. 29, 99–106. 10.5487/TR.2013.29.2.099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leffler D. A., Lamont J. T. (2015). Clostridium difficile infection. N. Engl. J. Med. 372, 1539–1548. 10.1056/NEJMra1403772 [DOI] [PubMed] [Google Scholar]
- Mani N., Lyras D., Barroso L., Howarth P., Wilkins T., Rood J. I., et al. (2002). Environmental response and autoregulation of Clostridium difficile TxeR, a sigma factor for toxin gene expression. J. Bacteriol. 184, 5971–5978. 10.1128/JB.184.21.5971-5978.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin J. S., Monaghan T. M., Wilcox M. H. (2016). Clostridium difficile infection: epidemiology, diagnosis and understanding transmission. Nat. Rev. Gastroenterol. Hepatol. 13, 206–216. 10.1038/nrgastro.2016.25 [DOI] [PubMed] [Google Scholar]
- Ozaki E., Kato H., Kita H., Karasawa T., Maegawa T., Koino Y., et al. (2004). Clostridium difficile colonization in healthy adults: transient colonization and correlation with enterococcal colonization. J. Med. Microbiol. 53, 167–172. 10.1099/jmm.0.05376-0 [DOI] [PubMed] [Google Scholar]
- Reid G., Younes J. A., Van der Mei H. C., Gloor G. B., Knight R., Busscher H. J. (2011). Microbiota restoration: natural and supplemented recovery of human microbial communities. Nat. Rev. Microbiol. 9, 27–38. 10.1038/nrmicro2473 [DOI] [PubMed] [Google Scholar]
- Rossi M., Corradini C., Amaretti A., Nicolini M., Pompei A., Zanoni S., et al. (2005). Fermentation of fructooligosaccharides and inulin by bifidobacteria: a comparative study of pure and fecal cultures. Appl. Environ. Microbiol. 71, 6150–6158. 10.1128/AEM.71.10.6150-6158.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rupnik M., Wilcox M. H., Gerding D. N. (2009). Clostridium difficile infection: new developments in epidemiology and pathogenesis. Nat. Rev. Microbiol. 7, 526–536. 10.1038/nrmicro2164 [DOI] [PubMed] [Google Scholar]
- Salazar N., Binetti A., Gueimonde M., Alonso A., Garrido P., Gonzalez del Rey C., et al. (2011). Safety and intestinal microbiota modulation by the exopolysaccharide-producing strains Bifidobacterium animalis IPLA R1 and Bifidobacterium longum IPLA E44 orally administered to Wistar rats. Int. J. Food Microbiol. 144, 342–351. 10.1016/j.ijfoodmicro.2010.10.016 [DOI] [PubMed] [Google Scholar]
- Schoster A., Kokotovic B., Permin A., Pedersen P. D., Dal Bello F., Guardabassi L. (2013). In vitro inhibition of Clostridium difficile and Clostridium perfringens by commercial probiotic strains. Anaerobe. 20, 36–41. 10.1016/j.anaerobe.2013.02.006 [DOI] [PubMed] [Google Scholar]
- Senoh M., Iwaki M., Yamamoto A., Kato H., Fukuda T., Shibayama K. (2015). Inhibition of adhesion of Clostridium difficile to human intestinal cells after treatment with serum and intestinal fluid isolated from mice immunized with nontoxigenic C. difficile membrane fraction. Microb. Pathog. 81, 1–5. 10.1016/j.micpath.2015.03.001 [DOI] [PubMed] [Google Scholar]
- Servin A. L. (2004). Antagonistic activities of lactobacilli and bifidobacteria against microbial pathogens. FEMS Microbiol. Rev. 28, 405–440. 10.1016/j.femsre.2004.01.003 [DOI] [PubMed] [Google Scholar]
- Solís G., de los Reyes-Gavilán C. G., Fernández N., Margolles A., Gueimonde M. (2010). Establishment and development of lactic acid bacteria and bifidobacteria microbiota in breast-milk and the infant gut. Anaerobe 16, 307–310. 10.1016/j.anaerobe.2010.02.004 [DOI] [PubMed] [Google Scholar]
- Tejero-Sariñena S., Barlow J., Costabile A., Gibson G. R., Rowland I. (2012). In vitro evaluation of the antimicrobial activity of a range of probiotics against pathogens: evidence for the effects of organic acids. Anaerobe 18, 530–530. 10.1016/j.anaerobe.2012.08.004 [DOI] [PubMed] [Google Scholar]
- Tejero-Sariñena S., Barlow J., Costabile A., Gibson G. R., Rowland I. (2013). Antipathogenic activity of probiotics against Salmonella Typhimurium and Clostridium difficile in anaerobic batch culture systems: is it due to synergies in probiotic mixtures or the specificity of single strains? Anaerobe 24, 60–65. 10.1016/j.anaerobe.2013.09.011 [DOI] [PubMed] [Google Scholar]
- Trejo F. M., De Antoni G. L., Pérez P. F. (2013). Protective effect of bifidobacteria in an experimental model of Clostridium difficile associated colitis. J. Dairy Res. 80, 263–269. 10.1017/S0022029913000216 [DOI] [PubMed] [Google Scholar]
- Trejo F. M., Pérez P. F., De Antoni G. L. (2010). Co-culture with potentially probiotic microorganisms antagonises virulence factors of Clostridium difficile in vitro. Antonie Van Leeuwenhoek. 98, 19–29. 10.1007/s10482-010-9424-6 [DOI] [PubMed] [Google Scholar]
- Valdés L., Alonso-Guervos M., García-Suárez O., Gueimonde M., Ruas-Madiedo P. (2016). Selection of bifidobacteria and lactobacilli able to antagonise the cytotoxic effect of Clostridium difficile upon intestinal epithelial HT29 monolayer. Front. Microbiol. 7:577. 10.3389/fmicb.2016.00577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valdés L., Gueimonde M., Ruas-Madiedo P. (2015). Monitoring in real time the cytotoxic effect of Clostridium difficile upon the intestinal epithelial cell line HT29. J. Microbiol. Methods 119, 66–73. 10.1016/j.mimet.2015.09.022 [DOI] [PubMed] [Google Scholar]
- Vickers R., Robinson N., Best E., Echols R., Tillotson G., Wilcox M. (2015). A randomised phase 1 study to investigate safety, pharmacokinetics and impact on gut microbiota following single and multiple oral doses in healthy male subjects of SMT19969, a novel agent for Clostridium difficile infections. BMC Infect. Dis. 15:91. 10.1186/s12879-015-0759-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yakob L., Riley T. V., Paterson D. L., Marquess J., Soares-Magalhaes R. J., Furuya-Kanamori L., et al. (2015). Mechanisms of hypervirulent Clostridium difficile ribotype 027 displacement of endemic strains: an epidemiological model. Sci. Rep. 5:12666. 10.1038/srep12666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z., Ramsey J., Hamza T., Zhang Y., Li S., Yfantis H. G., et al. (2015). Mechanisms of protection against Clostridium difficile infection by the monoclonal antitoxin antibodies actoxumab and bezlotoxumab. Infect. Immun. 83, 822–831. 10.1128/IAI.02897-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yun B., Oh S., Griffiths M. W. (2014). Lactobacillus acidophilus modulates the virulence of Clostridium difficile. J. Dairy Sci. 97, 4745–4758 10.3168/jds.2014-7921 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.