Abstract
Alcohol dehydrogenases (ADH), encoded by multigene family in plants, play a critical role in plant growth, development, adaptation, fruit ripening and aroma production. Thirteen ADH genes were identified in melon genome, including 12 ADHs and one formaldehyde dehydrogenease (FDH), designated CmADH1-12 and CmFDH1, in which CmADH1 and CmADH2 have been isolated in Cantaloupe. ADH genes shared a lower identity with each other at the protein level and had different intron-exon structure at nucleotide level. No typical signal peptides were found in all CmADHs, and CmADH proteins might locate in the cytoplasm. The phylogenetic tree revealed that 13 ADH genes were divided into three groups respectively, namely long-, medium-, and short-chain ADH subfamily, and CmADH1,3-11, which belongs to the medium-chain ADH subfamily, fell into six medium-chain ADH subgroups. CmADH12 may belong to the long-chain ADH subfamily, while CmFDH1 may be a Class III ADH and serve as an ancestral ADH in melon. Expression profiling revealed that CmADH1, CmADH2, CmADH10 and CmFDH1 were moderately or strongly expressed in different vegetative tissues and fruit at medium and late developmental stages, while CmADH8 and CmADH12 were highly expressed in fruit after 20 days. CmADH3 showed preferential expression in young tissues. CmADH4 only had slight expression in root. Promoter analysis revealed several motifs of CmADH genes involved in the gene expression modulated by various hormones, and the response pattern of CmADH genes to ABA, IAA and ethylene were different. These CmADHs were divided into ethylene-sensitive and –insensitive groups, and the functions of CmADHs were discussed.
Keywords: melon, alcohol dehydrogenases, bioinformatic analysis, gene identification, expression pattern of gene
Introduction
Alcohol dehydrogenases (ADH, EC 1.1.1.1) belong to dehydrogenase enzymes superfamily, and are widely distributed in all types of organisms (Chase, 1999; Jönvall et al., 2010; Strommer, 2011; Alka et al., 2013). ADH enzyme has long been the subject of molecular studies and encoded by multigene family in eukaryotic and prokaryotic kingdoms, catalyzing the interconversion between alcohols and aldehydes (Höög et al., 2003; Thompson et al., 2007). In humans, animals, yeast and bacterias, ADHs have been widely investigated (Khan et al., 2010; Kumar et al., 2012; Alka et al., 2013; Çelik and Aktas, 2013; Jönvall et al., 2013; Plapp et al., 2013; Quaglia et al., 2013), and ADH genes were involved in an astonishingly wide range of metabolic processes (Höög et al., 2003; Thompson et al., 2007; Strommer, 2011). Hence, these ADH genes were classed into several main superfamilies respectively, namly medium- (∼350 amino acid residues), short- (∼250 residues) and long-chain ADH or Iron-ADH genes superfamilies (600–750 residues or approximately 385 amino acid residues up to almost 900 residues) (Chase, 1999; Deng et al., 2002; Alka et al., 2013; Jönvall et al., 2013), and the medium-chain ADHs clustered into eight classes in vertebrates, based on sequence similarity, catalytic features and gene expression patterns (Thompson et al., 2007). But functional roles of these ADHs are not fully understood.
Sets of ADH genes or ADH-like genes have been identified in the genomes of poaceae, rosaceae, brassicaceae, fabaceae, and pinaceae plants (Thompson et al., 2010; Zheng et al., 2011). So far, most members of the ADHs in plants characterized at the gene level belonged to the medium-chain ADH protein superfamily (including ADH1, EC 1.1.1.1 and FDH, Class III ADH, EC 1.2.1.1; Chase, 1999), which usually contains zinc ligands in their active site (Tesnière and Verriès, 2000; Garabagi et al., 2005; ManrÍquez et al., 2006; Koutsompogeras et al., 2010; Singh et al., 2010; Komatsu et al., 2011; Pathuri et al., 2011; Bukh et al., 2012; Iaria et al., 2012; Min et al., 2012; Cheng F.F. et al., 2013), and the short-chain ADH protein superfamily, which lacks zinc-liganding cysteine residues in their coenzyme binding regions, and the molecular functions of only a few short-chain ADH genes were known (ManrÍquez et al., 2006; Gonzalez-Agüero et al., 2009; Kim et al., 2009; Strommer, 2011; Moummou et al., 2012). Other ADHs have been not noted in plants, such as long-chain ADHs or Iron-ADH genes. On the basis of phylogenetic analyses, the medium-chain ADH genes in plant were distributed in different subgroups (Perry and Furnier, 1996; Gaut et al., 1999; Small and Wendel, 2000; Komatsu et al., 2011; Zheng et al., 2011). Previous studies highlighted that the medium-chain ADHs in plant were involved in response to abiotic and biotic stress which induced the specific expression of these ADHs in different tissues of soybean, wheat and barley, implying that they may participate in different tissues development in stresses (Komatsu et al., 2011; Pathuri et al., 2011; Yamauchi et al., 2014). Moreover, regardless of medium- chain ADHs or short-chain ADHs, ADH genes have been shown to play a major role in fruit ripening and aroma synthesis (Tesnière and Verriès, 2000; ManrÍquez et al., 2006; Singh et al., 2010; Zhang et al., 2011; Zheng et al., 2011; Bukh et al., 2012; Iaria et al., 2012; Moummou et al., 2012). However, the actual participation of medium-chain ADHs in aroma volatile production in vivo has been only clearly demonstrated in tomato fruit by over-expressing or down-regulating the LeADH2 gene (Speirs et al., 1998). Other detailed studies related to role of ADH genes (including medium- and short-chain ADHs) in relation to aroma synthesis have been proved in tomato, melon and mango by recombinant protein (ManrÍquez et al., 2006; Singh et al., 2010; Moummou et al., 2012). In addition, persimmon DkADH, Artemisia annua AaADH2, Sedum sarmentosum SsADH (belonging to medium-chain ADH family) and other short-chain ADHs have been implicated in other primary and secondary metabolisms (Polichuk et al., 2010; Tonfack et al., 2011; Sung et al., 2012). In panax ginseng, the expression pattern of PgADH under abiotic stimuli suggested that a short-chain ADH, PgADH, was involved in responses to hormone-related environmental stresses (Kim et al., 2009). Meanwhile, the tissue-specific patterns of these ADH gene expressions among different tissue under various environmental stresses and during fruit ripening and aroma synthesis also provided a few evidences of functional specialization. We conclude further investigation is required to make functional annotation for the majority of predicted ADH in plant genomes.
In melon, two ADH genes, CmADH1 and CmADH2, have been identified and characterized (ManrÍquez et al., 2006). The release of melon genome provides an easy way to identify new members of ADH gene family. This study is a continuation of CAD genes (belonging to dehydrogenase enzymes superfamily) research in melon (Jin et al., 2014). In this study, we identified 12 CmADHs, including CmADH1 and CmADH2, and one FDH gene from the melon genome, and compared ADH sequences from a wide variety of plants, making full use of the available plant genome sequences (Arabidopsis, mango, tomato, grape et al.). Alignment and phylogenetic analysis of the ADH gene family with related ADH proteins from other species indicated that these ADH genes were classified into three groups and the medium-chain melon ADHs were clustered into six subgroups separately. We analyzed the structure and the promoter of melon ADH genes, and also investigated these ADH genes transcripts in response to various fruit development stages and various plant hormones, and monitored tissue-specific expression. We reported here the results of these analyses, suggesting that the CmADHs may be involved in melon fruit development induced by various hormones and during development and ripening. Information gained from this investigation will significantly advance the understanding of the function of predicted ADH genes in Melon.
Materials and Methods
Plant Materials
All the experiments were conducted on a commercial oriental melons (Cucumis melo var. makuwa Makino) cultivar, ‘CaiHong7.’ All melon plants were grown in a greenhouse under standard cultural practices at Shenyang Agricultural University, Shenyang, China, from March to June in 2012, and the melon fruit were harvested at different development stages (1, 5, 10, 15, 20, 25, 30, 33, 36, 39, 42, 45, and 48 days after anthesis) for expression analysis as previously described (Jin et al., 2014). Physiological maturity of this melon is about 36 days after anthesis. To allow analysis of tissue-specific gene expression, mature leaf, developing leaf, pistillate and staminate flower, young stems and root tissues were collected from melon plants grown in a greenhouse, then frozen with liquid nitrogen and kept in -80°C refrigerator for further use.
Hormone Treatments
Mature unripe oriental melons were harvested at 30 days after anthesis (pre-climacteric stage) with the same node of the plant at a mature green stage, before the onset of ripening. No blemished or diseased Fruits were chosen for treatment. Exogenous ethylene (100 μL/L), 1-methylcyclopropene (1-MCP, an ethylene perception inhibitor) (100 μL/L) and control (water) treatments were performed according to our previous report (Jin et al., 2014). Then melon fruit of three treatments were allowed to ripen for 12 days at 23°C in air only. Three replications were carried out for treatments, and each replicate consisted of 15 fruits unless indicated otherwise. Fresh tissue was sampled every 48 h, frozen in liquid nitrogen and stored at -80°C until further use. Furthermore, for abscisic acid (ABA;100 μM) and auxin (IAA;100 μM) treatment, the fruit disks were dipped in a solution containing ABA or IAA in 0.2% teepol (detergent) and vacuum infiltrated for 2 h as described previously (Jin et al., 2014). Infiltrated melon fruits with 0.2% teepol were used as control. After treatment, fruit flesh were frozen in liquid nitrogen and stored at -80°C. Three replications were carried out for treatments or control groups.
Identification of Melon ADH Genes
The keyword ‘alcohol dehydrogenase’ was used to search for the melon ADH sequences from the melon (Cucumis melon L.) genome1 (Garcia-Mas et al., 2012). In addition, to confirm the accuracy of these genes, the predicted ADH-like gene sequences were compared to ADH proteins in other species by a BLASTp retrieve. Only those sequences with a high score (>200) were selected.
Sequence Analysis
The sequence analysis of CmADHs, including ADH protein prediction, functional domains and promoter analysis, signal peptides and disulfide bond prediction and CmADHs subcellular localization prediction, were performed according to the method described by Jin et al. (2014). Multiple alignments were performed with other known plant ADH proteins using Clustal Omega program and GENEDOC. Based on the neighbor-joining method (minimum evolution criterion, bootstrap values performed on 1000 replicates), phylogenetic analyses of putative melon ADH proteins were performed using MEGA52 program.
RNA Isolation and cDNA Synthesis
Total RNA from fruit samples, leaves, stem, root, and flower material, were extracted by the method described by Jin et al. (2014), quantified by the NanoDrop spectrophotometer ND-1000 and checked for integrity by electrophoresis (28S rRNA/18S rRNA ratios). RNA was treated by DNAse I (Promega, Madison, WI, USA) at 37°C for 50 min, re-precipitated and concentrated (40 μL) to remove any trace of genomic DNA. cDNA synthesis was initiated from DNase I-treated RNA using M-MLV RTase cDNA Synthesis Kit following the manufacturer’s instructions (Cat#D6130, TaKaRa, Tokyo, Japan).
Semi-quantitative PCR and Real-Time PCR
For semi-quantitative PCR and real-time PCR, gene-specific oligonucleotide primers for each ADH gene were designed by Primer33 and were listed in Table 1. The specificity of each pair of primers was determined by agarose gel electrophoresis and PCR products resequencing, then semi-quantitative PCR and Real time PCR were performed for gene expression studies according to the method described by Jin et al. (2014). 18SrRNA DNA fragment (148 bp) of melon as an internal control.
Table 1.
Semi-quantitative PCR and Real-time PCR primers.
| Gene | Primer | Sequence (5-3) |
|---|---|---|
| CmFDH1 | CmFDH1-F | GACATTTGAGTTAGGATGG |
| CmFDH1-R | GGTGATAGTTACAATCTTGG | |
| CmADH1 | CmADH1-F | CTAATGAAGTCCGATTGAAG |
| CmADH1-R | ATGATCTCCTGGTTGAAG | |
| CmADH2 | CmADH2-F | GCCATTGTTGATTCACTCA |
| CmADH2-R | CATTCGCAGTCACTTGTAA | |
| CmADH3 | CmADH3-F | GCTGGTGTCCATCGCTGTT |
| CmADH3-R | CCAACTCCGCTTGGTAATGG | |
| CmADH4 | CmADH4-F | ACCACCACAAGCCAATGAAG |
| CmADH4-R | TGACGACATTCTCCACATTCC | |
| CmADH5 | CmADH5-F | GGACTCAACTTCCTCATT |
| CmADH5-R | GTAACTCCGTAGAAACATAC | |
| CmADH6 | CmADH6-F | CAGAACTCTTGGCGATAA |
| CmADH6-R | TGGTCTACTCCTAACACTA | |
| CmADH7 | CmADH7-F | CACACTATTCACTGGAGA |
| CmADH7-R | CCACGGTATATTCACTGA | |
| CmADH8 | CmADH8-F | CCAATCATCCATCTGTATC |
| CmADH8-R | GGTTAATGCGAGTAAGTG | |
| CmADH9 | CmADH9-F | TCCGATCTTCCAATGCTTAT |
| CmADH9-R | ATGAGGCAACGAAGTGAT | |
| CmADH10 | CmADH10-F | TCAGATATTCCCACTCTTCT |
| CmADH10-R | ACAACACAACCGATGATG | |
| CmADH11 | CmADH11-F | CACTGGTAATTGAGGAAG |
| CmADH11-R | CTACACTCTCCACAACTA | |
| CmADH12 | CmADH12-F | TATGGAGGATTAGACATCTG |
| CmADH12-R | CCTTATCGCAAGTTGAGT | |
| 18srRNA | 18srRNA-F | AAACGGCTACCACATCCA |
| 18srRNA-R | CACCAGACTTGCCCTCCA |
Statistical Analysis
Data are expressed as mean values ± standard deviation of three independent experiments (n = 3). The data were analyzed by the analysis of variance (ANOVA) using the SPSS 13.0 statistics program, and statistical significance of differences was were calculated by a one-way ANOVA following Duncan’s multiple range tests for each experiment at a P < 0.05 level. Origin 8.0 (OriginLab, Northampton, MA, USA) was used to draw the figures.
Results
Identification of Melon ADH Genes
All the predicted ADH genes in the melon genome were collected and compared with ADH genes in other species. Hence, the presence of functional domains was checked via NCBI’s Conserved Domain Database (CDD)4, and 13 genes (including 12ADHs and 1 FDH), encoding full-or nearly full-length functional proteins, were identified in the melon genome.
By ExPASy tools, we found that the longest ADHs protein consisted of 635 amino acid residues, and the shortest consisted of 266 amino acid residues. The ORF length ranged from 801 to 1908 nucleotides. The predicted molecular weight and isoelectric points of all ADHs proteins ranged from 29.0 kDa/5.19–68.5 kDa/9.06, respectively (Table 2). Of these ADH genes, two of them have been reported previously and were labeled with their published names (ManrÍquez et al., 2006) (Table 2). For the other 11 unreported genes, we gave them names by adding a number to their family name in the order which they were searched, in which one ADH was named as CmFDH1 on account of the presence of same functional domains with reported FDH genes in other species (Dolferus et al., 1997).
Table 2.
The information of ADH genes in melon.
| Gene | Gene accession No. | Published name | ORF length | amino acid (aa) | Melon predicted protein No. | Molecular weight (kD) | Location | Isoelectric Point |
|---|---|---|---|---|---|---|---|---|
| CmFDH1 | MELO3C022399T1 | 1014 | 337 | MELO3C022399P1 | 36.18 | CM3.5_scaffold00052 | 6.74 | |
| CmADH1 | ABC02081 | CmADH1 | 1140 | 379 | ABC02081.1 | 41.01 | CM3.5_scaffold00061 | 6.83 |
| CmADH2 | ABC02082 | CmADH2 | 801 | 266 | ABC02082.1 | 29.09 | CM3.5_scaffold00023 | 8.72 |
| CmADH3 | MELO3C026552T1 | 1239 | 412 | MELO3C026552P1 | 45.02 | CM3.5_scaffold00096 | 7.38 | |
| CmADH4 | MELO3C027151T1 | 1146 | 381 | MELO3C027151P1 | 41.44 | CM3.5_scaffold00483 | 5.55 | |
| CmADH5 | MELO3C005792 T1 | 1179 | 392 | MELO3C005792 P1 | 42.24 | CM3.5_scaffold00005 | 6.17 | |
| CmADH6 | MELO3C026553 T1 | 1164 | 387 | MELO3C026553 P1 | 42.02 | CM3.5_scaffold00096 | 5.88 | |
| CmADH7 | MELO3C002189 T1 | 1143 | 380 | MELO3C002189 P1 | 40.67 | CM3.5_scaffold00001 | 6.55 | |
| CmADH8 | MELO3C003251 T1 | 1272 | 423 | MELO3C003251 P1 | 45.81 | CM3.5_scaffold00002 | 8.62 | |
| CmADH9 | MELO3C011043 T1 | 1104 | 367 | MELO3C011043 P1 | 40.29 | CM3.5_scaffold00014 | 5.19 | |
| CmADH10 | MELO3C026554 T1 | 864 | 287 | MELO3C026554 P1 | 42.04 | CM3.5_scaffold00096 | 6.61 | |
| CmADH11 | MELO3C023687 T1 | 984 | 327 | MELO3C023687 P1 | 36.59 | CM3.5_scaffold00061 | 6.76 | |
| CmADH12 | MELO3C019503 T1 | 1908 | 635 | MELO3C019503 P1 | 68.51 | CM3.5_scaffold00038 | 9.06 |
Intron-Exon Structure of Melon ADH Genes
Structural analysis of the identified melon ADH genes revealed different intron-exon patterns both in relation to position and number of introns which ranged from 2 to 17 per gene (Figure 1). Furthermore, the substantial differences in the size between the exons were observed. The standard number of introns in plant ADH genes in the common ancestor is 9, located at equivalent positions in ADH genes throughout the plant kingdom (Strommer, 2011). However, plant ethanol-active ADH genes have evolved from class III ADH genes by gene duplication and acquisition of new substrate specificities (Dolferus et al., 1997). Based on Figure 1, CmFDH1 (Class III ADH;GSH-FDH) contained nine exons and eight introns, while the intron-exon structures of other 12 CmADHs were relatively complex. CmADH1, CmADH8, CmADH10 and CmADH11 had seven introns, and CmADH3, CmADH5, CmADH6 and CmADH7 had nine introns, and CmADH4 and CmADH9 contain eight introns; CmADH2 had two introns, moreover CmADH12 contained 17 introns (Figure 1). For ADH-plant genes, introns patterns contributed more evidences for gene acquisition through duplications, insertion or loss of introns (Charlesworth et al., 1998; Small and Wendel, 2000; Strommer, 2011). Additionally, both 76-bp and 83-bp sequence were found in six sequences, and a 76-bp sequence was found in these sequences, except CmADH2,8,12 (Figure 1). Nevertheless, it was unclear whether these sequence encoded the putative functional region of ADH genes.
FIGURE 1.
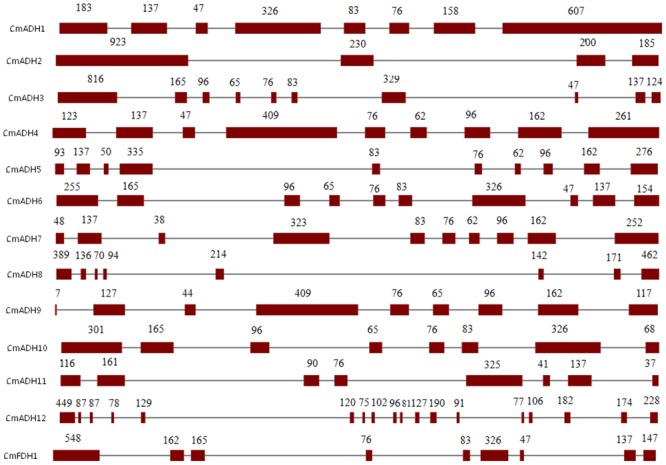
Intron-exon structures of ADH genes from melon. Exons and introns were indicated by open boxes and lines respectively. Numbers above boxes indicated the exon sizes. The intron sizes were not to scale. The names of ADH genes and intron-exon structure were indicated at the left and right sides respectively.
Sequence Characterization of CmADH Genes
To understand the possible functional divergence of the individual members of melon ADH genes superfamily, the sequences of a set of ADH family members from other plant species, including cucumber, Arabidopsis, grape, potato, tomato, mango, apricot, peach, soybean, cocoa, were analyzed using a Neighbor-joining (NJ) phylogenetic tree by MAGA5. As shown in Figure 2, the phylogenetic tree divided these ADHs into three clades, namely medium-, short-, and long-chain ADHs, and the medium-chain ADHs in melon were clustered into six medium-chain ADHs subgroups, called as midum-chain groups I–VI, based on their amino acids sequence (Supplementary Figure S1). In Cantaloupe, CmADH1 and CmADH2, two highly divergent genes, have been isolated, and it has been characterized that CmADH1 belonged to the medium-chain zinc-binding type of ADHs and CmADH2 was positioned in the short-chain ADHs (Singh et al., 2010). Moreover, CmADH3-11 belonged to medium-chain zinc-binding type of ADHs and have not been characterized, in which CmADH1, Pa-ADH1 (apricot) and Le-ADHs (tomato), having been characterized, were involved in fruit-ripening and aroma volatiles biosynthesis (ManrÍquez et al., 2006; Gonzalez-Agüero et al., 2009; Moummou et al., 2012). CmADH12 belonged to long-chain ADHs which have not been characterized in plant; CmFDH1 clustered into medium-chain zinc-binding type of FDHs, known as Class III ADH, and AtFDH1 (Arabidopsi) and OsFDH1 (rice) have been cloned (Dolferus et al., 1997); In addition, it is now recognized that FDHs are involved in plant development and stress response (Feechan et al., 2005; Rustérucci et al., 2007; Lee et al., 2008; Espunya et al., 2012).
FIGURE 2.
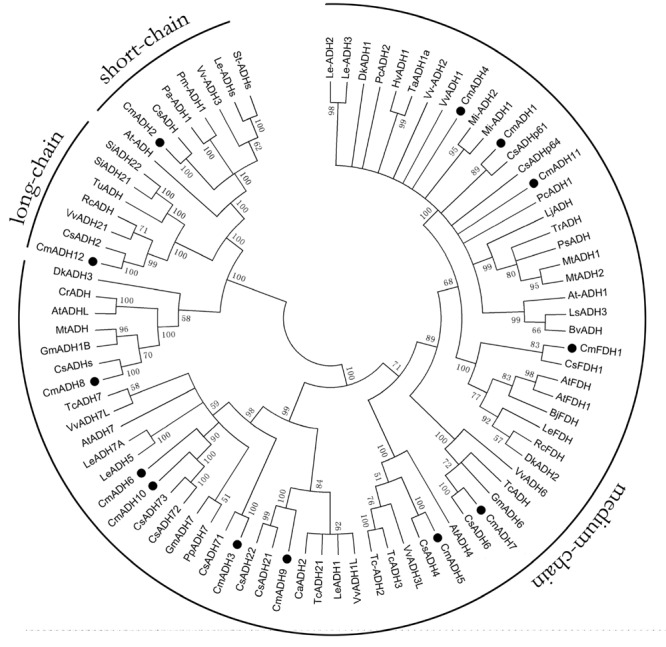
Phylogenetic relationship among CmADHs and ADHs from other plant species. The amino acid sequences were aligned by the Clustal Omega program, and the neighborjoining tree was drawn with MEGA5. The corresponding GenBank and the melon genome (https://melonomics.net/) were noted in the phylogenetic tree, and the accession number in the melon genome were CmADH1 (MELO3C023685P4), CmADH2 (MELO3C014897P1), CmADH3 (MELO3C026552P1), CmADH4 (MELO3 C027151P1), CmADH5 (MELO3C005792P1), CmADH6 (MELO3C026553P1), CmADH7 (MELO3C002189P1), CmADH8 (MELO3C003251P1), CmADH9 (MELO3C0110 43P1), CmADH10 (MELO3C026554P2), CmADH11 (MELO3C023687P1), CmADH12 (MELO3C019503P1), and CmFDH1 (MELO3C022399P1). The number for each interior branch was the percentage of bootstraps value (1000 replicates). Black circle denoted 13 CmADHs. ADHs from other plants in our paper were in supporting information (Supplementary Table S3).
Multiple alignments revealed that the CmADH genes showed lower identity in protein level with each other, as noted previously in Cantaloupe (ManrÍquez et al., 2006). Among the medium-chain zinc-binding type of CmADHs, there was lower identity between CmADH1 and other medium-chain zinc-binding CmADHs in the range of 30–88.51% (Figure 3), whereas CmADH1 showed the highest sequence identities to medium-chain ADH proteins from cucumber, mango, grape, tomato and Arabidopsis in range of 80–96.08% (Supplementary Figure S2), and other medium-chain CmADHs also showed higher sequence identities with other plants (Supplementary Figures S3–S9). All of the medium-chain zinc-binding CmADHs had the highly conserved Zn1 -binding signature [GHE(X)2G(X)5G(X)2V], the Zn2 structural motif [GD(X)9,10C(X)2C(X)2C(X)7C], and the NADPH-binding domain [GXG(X)2G] motif (so-called Rossmann fold) (Figure 3; Supplementary Figures S2–S9), which was the same with that of CmADH1 proved (Singh et al., 2010), suggesting that these proteins appear to be zinc-dependent ADHs and members of the plant ADH protein family and the medium-chain dehydrogenase/reductase (MDR) superfamily (Persson et al., 1991; McKie et al., 1993). Nevertheless, CmADH11, a medium-chain melon ADH, lacked the key determinant residues of NADPH-binding domain at protein level (Supplementary Figure S9).
FIGURE 3.
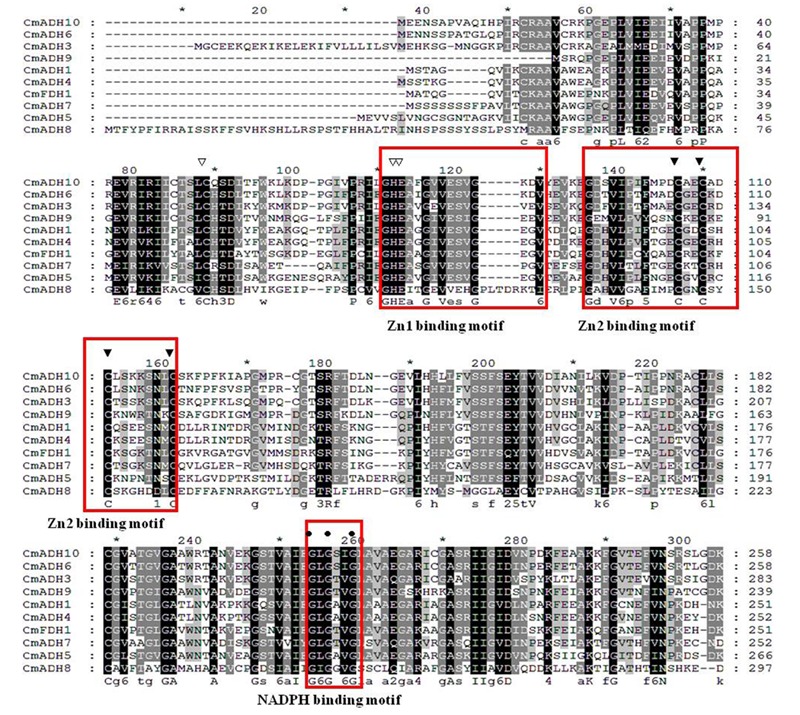
Alignment of amino acid sequences of CmADHs. Conserved important regions identified previously were marked as follows: white arrows denoted catalytic zinc ion coordinating residue, black arrows denoted structural Zn ion coordinating residue, the black circle denoted key residues for substrate specificity. The black square denoted key Phe299/Gly(300) residues for substrate specificity. The white square denoted key Trp199 and Asp123 residues for substrate binding. Locations of the Zn1, Zn2, and NADPH binding domains were shown in boxes. The alignment was performed with the Clustal Omega program.
Furthermore, CmADH2, a short-chain ADH protein, and CmADH12, a long-chain ADH protein, had highly homology with corresponding ADHs from other plants (Supplementary Figures S10 and S11). The sequence alignment showed that the CmADH12 protein sequence was found most similar to corresponding ADH-like proteins from other plants, with identities close to 95.43% (Supplementary Figure S11), and contained an N-terminal cofactor binding pattern of typically TGXXXGXG and an active-site pattern of YXXXK which are two conserved regions around amino acids of short-chain dehydrogenases in plants (Joërnvall et al., 1999). Additionally, we used the protein database in NCBI as a source of amino acids sequence bioinformatics for CmADH12, and found that CmADH12 possessed a coenzyme (NADPH) binding domain AAAGGTG, suggesting that CmADH12 may belong to long-chain ADH subfamily, but may have the same catalytic function as shot-chain dehydrogenases. Interestingly, the analysis of the amino acids sequence of CmADH12 showed that it did not contain structurally and functionally important iron-binding motif (G-X-X-H-X-X-A-H-X-X-GX-X-X-X-X-P-H-G), and NAD(H)-or NADP(H)-binding fingerprint patterns G-X-G-X-X-G or G-X-G-X-X-A, GGGSXXD and A/GXXATGG, and Trp docking motif (A-x-[DN]-x-x-T-G-[DEK]-x-x-W), possessed in long-chain or iron-dependent ADH (group III ADH) protein of prokaryotes, human and other mammals (Inoue et al., 1989; Deng et al., 2002; Lyon et al., 2009; Elleuche et al., 2013, 2014). Additionally, Filling et al. (2002) described several conserved residues of short-chain dehydrogenase, involved in coenzyme binding and catalysis, such as Thr12, Asp60, Asn86, Asn87, Ala88, Gly89, Gly131, Gly183, Asn111, Ser151 and Tyz157. In melon, CmADH2 only contains five residues of these conserved residues, namely Thr12, Asp60, Gly183, Ser151 and Tyz157 (Supplementary Figure S10). These findings suggested that the biological functions of plant short-chain ADH proteins might be related to the diversity in amino acid sequence.
No typical signal peptides were found in all CmADHs after analyzing their N-terminals using the SignalP software. CmADHs subcellular localization prediction showed that these ADH genes might exist in the cytoplasm (Supplementary Table S2). Moreover, the transmembrane topology predictions of 13 CmADHs using TMHMM 2.0 software and ABTMpro showed that there was no internal transmembrane segment in CmADHs. We also found that there were 4 and 3 disulfide bonds in 12 melon ADH proteins by Scratch Protein Predictor, respectively, but there were 6 disulfide bonds in CmADH12 protein.
Promoter Sequence Analysis
Analysis of promoter sequences for the melon ADH genes allowed us to identify several motifs that were known to be involved in the regulation of gene expression in various developmental and physiological processes. A promoter motif search showed that CmADHs promoter contained putative regulatory elements corresponding to known cis-elements of eukaryotic genes (Barakat et al., 2009; Kim et al., 2010; Cheng H. et al., 2013). In melon ADH genes promoter, there were mainly two kinds of motifs, namely, cis-acting element involved in defense and stress responsiveness (such as hypoxia stress, heat stress) and cis-acting regulatory element involved in the response to various hormones, such as auxin (IAA), ethylene (ETH), ABA, salicylic acid (SA), and methyl jasmonate (MeJA) (Supplementary Table S1) (Barakat et al., 2009). Nevertheless, the most abundant motifs detected were the cis-elements involved in the response to biotic and abiotic stresses, including TC-rich repeats, LTR, W-BOX, ARE, HSE and MBS (Jin et al., 2014). At the same time, we still found that there were putative cis-acting regulatory elements involved in the MeJA-responsiveness, namely CGTA-motif, TGACG-motif and CCAAT-motif (Matton et al., 1990; Prashant et al., 2012; Cheng H. et al., 2013). TCA-element clustered in the promoters of CmFDH1, CmADH1, CmADH3, CmADH5, CmADH7-9 and CmADH12, and SARE were also present in CmADH3 promoter, both of which were cis-acting regulatory element involved in the SA-responsiveness (Prashant et al., 2012; Cheng H. et al., 2013). ERE, ethylene-responsive element, was found in the promoter of CmFDH1, CmADH1, CmADH2, CmADH3, CmADH5, CmADH6, CmADH8 and CmADH12. TGA (Auxin responsiveness), ABRE (ABA responsiveness), p-box and GARE (gibberellin responsiveness) were also present in several CmADHs promoter. Additionally, promoter sequence analysis revealed several conserved motifs, such as MBS-I and MBS-II (MYB binding site involved in flavonoid biosynthesis and drought-inducibility), HSE (heat stress responsiveness) and LTR (Low temperature responsiveness) (Prashant et al., 2012). CmFDH1, CmADH3 and CmADH9 possessed elicitor responsive element and enhancer. These results indicated that transcriptional regulation of these CmADH genes might be involved in fruit development and ripening and senescence and in the response to various stresses.
CmADHs Expression in Vegetative Tissues
In order to investigate the transcript levels of these 13 CmADH genes in different organs and tissues in melon plant, we collected samples of root, developing leaf, mature leaf, young stem, pistillate and staminate flower petal. Expression analysis using semi-quantitative PCR and real-time PCR showed that 12 CmADH and one CmFDH were constitutively expressed in these parts, but greatly varied in different tissues, and there were differences in same tissues between CmADHs (Figure 4; Supplementary Figure S12). Of the 12 ADH genes identified in melons, CmADH2 and CmADH10 were expressed most abundantly in root, developing leaf, mature leaf, young stem, pistillate and staminate flower. Because of the higher expression of CmADH2 and CmADH10 seleted as “1,” real-time PCR analysis of CmADH2 and CmADH10 showed a lower relative expression level (Figure 4). The expression of CmFDH1 was higher in root, young stem and pistillate flower. CmADH9 and CmADH11 were either not expressed or expressed at very low levels in these tissues. Of all analyzed tissues, the CmADH4 gene was highly expressed in the root, and only rarely expressed in other vegetative organs. CmADH1 expression was the greatest in root and young stem, and CmADH3 was expressed in vegetative organs, with the highest expression in the young leaf and stem. Furthermore, CmADH5, CmADH6 and CmADH7 showed similar expression patterns between vegetative organs, with lower transcript levels, and without significant differences except root (P < 0.05). CmADH8 and CmADH12 were also expressed in all tissues except mature leaf, showing lower expression in young leaf.
FIGURE 4.
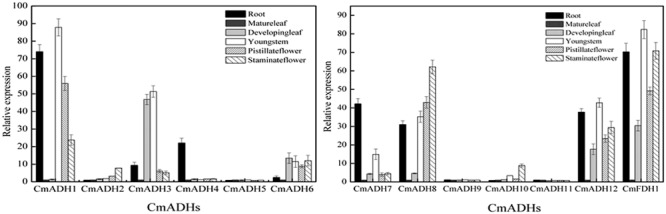
Transcript levels of CmADHs in different melon organs. Gene expressions of CmADHs in different organs in melon plants were determined by real-time PCR in root, developing leaves, mature leaves, young stems, pistillate flower petals and staminate flower petals in melon plants. 18s was used as internal control. The expression level of the genes in mature leaves was set as “1.0.” Data represented the means ± SD (n = 3) of three biological samples. The experiments were carried out in triplicate.
CmADHs Expressions during Melon Fruit Development
Transcript analysis indicated that these 12 CmADH genes and one CmFDH gene studied here were specifically expressed in fruit of different development stages, with remarkable differences in fruit (Figure 5; Supplementary Figure S12). The pattern of changes in transcript levels was similar for CmADH1, CmADH3, CmADH8, CmADH12 and CmFDH1 from 1 to 15 days after anthesis with gradual or sharp decrease at 15 days. While the expression of CmADH3, CmADH8 and CmADH12 showed an increase after 15 days, and the levels of these 3 ADH genes transcript were always maintained up to 48 days after anthesis. However, CmADH1 had higher expression in fruit during development, except from 30 to 32 days and 48 days after anthesis. In contrast, CmADH2 and CmADH10 were consistently expressed with an increase in transcript abundance, which subsequently remained at a relatively constant level through to harvest. Additionally, CmADH4, CmADH5, CmADH7, CmADH9 and CmADH11 were either not expressed or expressed at very low levels, and the transcript levels of CmADH6 were always weakly maintained during fruit development.
FIGURE 5.
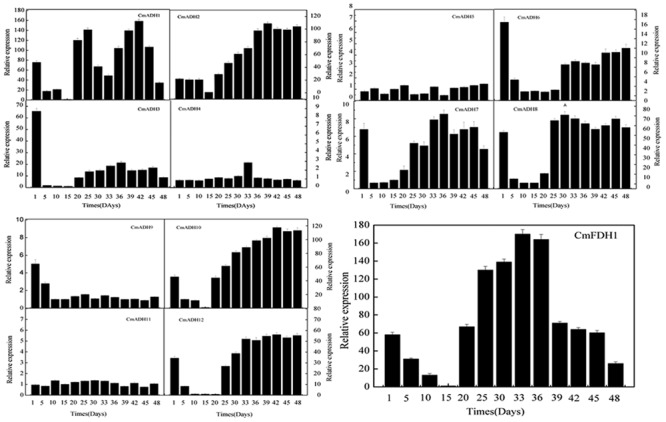
CmADHs relative expression in developing stages of melon fruit after anthesis were determined by real-time PCR. 18s was used as internal control. The expression level of CmADHs in melon fruit at 15 days after pollination was set as “1.0.” Data represented the means ± SD (n = 3) of three biological samples. The experiments were carried out in triplicate.
Effects of Hormones on CmADH Genes Expressions
The real-time PCR and semi-quantitative PCR results revealed that CmADHs were induced by IAA and ABA (though expression was lower in case of ABA), and showed different responses to IAA and ABA, except CmADH11 (Figure 6; Supplementary Figure S13). The CmADH1-4, CmADH10 and CmFDH1 were strongly induced by ABA, and the relative expression varied between these genes. Moreover, CmADH2 showed highest transcript levels. In contrast, IAA treatment strongly induced CmADH6, CmADH9 and CmADH12 expression, while other CmADHs were seemed to be insensitive to treatment with IAA and continued to be weakly expressed at a basal level.
FIGURE 6.
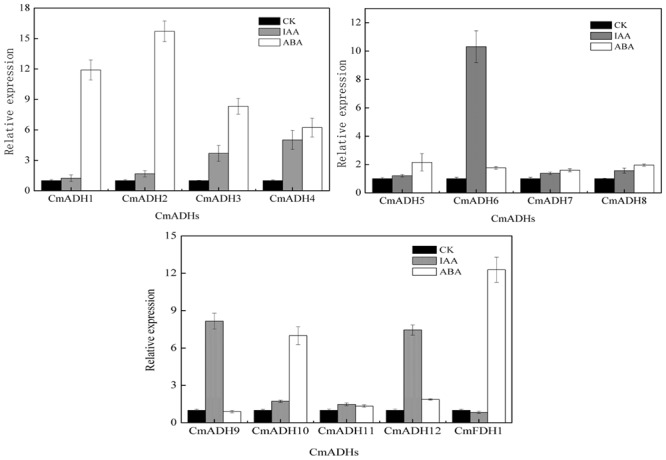
The expression of CmADHs in melon fruit after treatments with IAA and ABA. IAA and ABA (100 μM) treatments were given for 2 h as described in Section “Materials and Methods.” Expression analysis was carried out by real-time PCR. For each gene, the relative abundance of mRNA was normalized against the 18S in the corresponding samples. The expression level of the genes in untreated melon fruit by IAA and ABA was set as “1.0.” Data represented the means ± SD (n = 3) of three biological samples. The experiments were carried out in triplicate.
Since oriental melons is a climacteric fruit which requires ethylene for initiation of ripening, we checked whether these CmADHs were regulated by ethylene. Transcript accumulation of different CmADHs in melon were investigated during the course of ethylene induced ripening of harvested melon for 12 days at 23°C. A basal level of expression of CmADAHs transcript was detected in all of 12 ADH genes and one FDH gene in ethylene untreated fruit on day 1. The transcript levels of CmADH1-3, CmADH8, CmADH12 and CmFDH1 shot up on day 1 in ethylene treated fruit, apart from CmADH4-7, CmADH9 and CmADH11 (Figure 7; Supplementary Figure S14). However, levels of CmADH1, CmADH2 and CmADH3 transcript gradually decreased from day 1 levels in subsequent days, but levels of CmADH8, CmADH12 and CmFDH1 gradually increased from day 1 levels, and reduced from day 6 thereafter. Of the 12 ADH genes, CmADH4-7, CmADH9 and CmADH11 transcripts showed the lowest steady state levels in all of treatments during the course of melon ripening. Since these 6 CmADHs clustered into the medium-chain ADH subfamily, we checked the expression of CmADH4-7, CmADH9 and CmADH11 transcripts by semi-quantitative PCR at high annealing temperature using gene-specific primers to ensure that the transcript patterns were not due to cross hybridization. We found that these 6 CmADHs transcript levels were still lower and did not give clear results when 20 cycles of PCR were carried out as in case of other ADH genes. So for all semi-quantitative PCR experiments related to CmADH4-7, CmADH9 and CmADH11, 35 cycles of PCR were carried out. Our results of semi-quantitative PCRs matched with real time qPCR analysis, and confirmed that all 12 ADHs and one FDH were expressed at different levels during ripening. The expressions of CmADHs were also studied in 1-MCP + ethylene treated fruits. All 12 ADHs and one FDH showed transcript accumulation in 1-MCP + ethylene treated fruits, but the levels were far lower compared to the levels in ethylene treated fruits except CmADH4-7, CmADH9, CmADH10 and CmADH11, especially on day 1 post treatment (Figure 7; Supplementary Figure S14). Furthermore, CmADH10 always showed stable and strong expression in all of treatments during the course of melon ripening. Therefore, these finding implied that CmADH4-7 and CmADH9-11 appeared to be not induced by ethylene. Since the patterns were same in real-time qPCR and semi-quantitative PCR experiments, further studies will be required.
FIGURE 7.
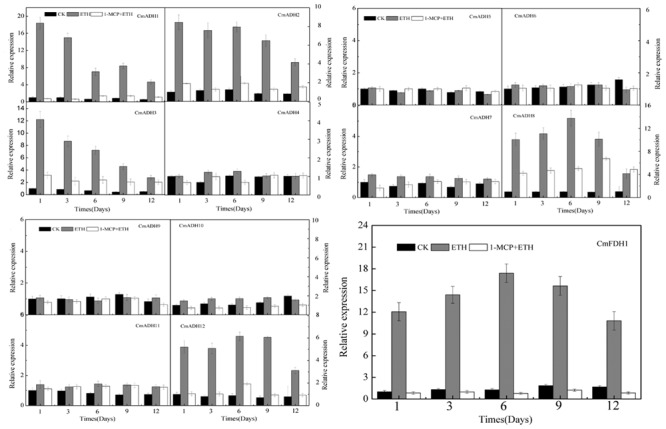
The expression levels of CmADHs in melon fruit after treatments with ethylene and 1-MCP. The transcript levels of CmADHs were measured by real-time PCR in melon fruit treated, and 18S was used as internal control. The expression of the genes in untreated melon fruit after 1 day of storage was set to 1.0. Data represented the means ± SD (n = 3) of three biological samples. The experiments were carried out in triplicate.
Discussion
Alcohol dehydrogenases, belonging to a group of dehydrogenase/reductases superfamily, are broadly distributed in all types of organisms and represent one of the most abundant classes of enzymes. More importantly, the ADH genes are involved in stress responses, elicitors and ABA (Matton et al., 1990; Kim et al., 2009; Strommer, 2011), and also closely associated with seedling, pollen development, root tissues, fruit ripening and aroma synthesis (Dolferus et al., 1994; Chung and Ferl, 1999; ManrÍquez et al., 2006; Singh et al., 2010; Kubienova et al., 2014). The ADH gene family has been investigated in a number of plant species, such as grape (Tesniere et al., 2004), Mango (Singh et al., 2010), tomato (Speirs et al., 1998), peaches (Zhang et al., 2010), apple (Defilippi et al., 2005a), soybean (Komatsu et al., 2011) and so on. The aim of the present study was to identify and analyze genes coding for the ADH enzyme in melon.
Bioinformatic Analysis
In the melon genome database, we identified 13 ADH/ADH-like genes (including 12 ADHs and 1 FDH) coding for putative ADH enzymes, in which CmADH1 and CmADH2 have been cloned and characterized (ManrÍquez et al., 2006). Our research results also demonstrated that these melon ADH genes had very low sequence identity at protein level (ranging from 4.8 to 52.48%) and different intron-exon patterns at nucleotide level (Figure 3). Up to the present, research on ADH in plants have been focused on the medium- and short- chain ADH superfamilies (ManrÍquez et al., 2006; Kim et al., 2009; Singh et al., 2010; Iaria et al., 2012; Yamauchi et al., 2014; Chen et al., 2015). Furthermore, plant FDH, with hydroxylmethyl-glutathione or formaldehyde as preferred substrates, belonged to the medium-chain ADHs (Strommer, 2011), while it is weakly active or inactive on n-octanol or ethanol (Chase, 1999); Hence, in regard to substrate specifity, FDH was different from ADH1 in plant. In our study, CmFDH1 clustered in medium-chain ADH superfamily, but the amino acid sequence of CmFDH1 shared 28.33% identity with CmADH1. Based on the convergent evolution and structure of ADH gene-families in plant and animal, the plant ADH family was deemed to originate from a glutathione-dependent formaldehyde dehydrogenase gene (GSH-FDH) (Strommer, 2011). In maize, rice, pea and Arabidopsis, FDH genes were cloned and purified, but their functions in plants were not yet clear (Chase, 1999). As of present, there is still insufficient information on the function of FDHs in higher plants (Espunya et al., 2006; Strommer, 2011; Nian et al., 2013), hence it would be widely concerned by researchers in future.
The phylogenetic tree showed that ADH genes in melon clustered into 3 groups (Figure 2), indicating that ADH genes in melon exist in multiple isoforms. However, the phylogenetic tree obtained differed from previous analyses which grouped the plant ADH genes into two main classes (Chase, 1999; Duester et al., 1999; Thompson et al., 2007; Borràs et al., 2014), without the long-chain or Iron-activated ADH reported in plants. At the same time, we found that CmADHs sequences clustered close to that of other various species, suggesting that there are species- or lineage-specific ADH gene duplications (Komatsu et al., 2011) or a number of separate duplication events within melon which may be not homologous (Thompson et al., 2007). Additionally, these medium-chain CmADHs, divided into six subgroups (Supplementary Figure S1 and S4–S8), also suggested that it seems possible to be different substrate specificities among melon medium-chain ADH genes and to be the same with that of vertebrates in which the medium-chain ADHs clustered in different subgroups (Duester et al., 1999). Hitherto, melon CmADH1 (ManrÍquez et al., 2006), tomato Le-ADH2 and LeADH3 (Speirs et al., 1998), mango MiADH1,2 (Singh et al., 2010), grape Vv-ADH2 (Tesniere et al., 2004) and olive OeADH (Iaria et al., 2012), clustering a same class, belonged to the medium-chain zinc-binding type of ADHs (Supplementary Figure S1), and in vitro, the functional verification results revealed that this type of plant ADHs were implicated in plant development, fruit ripening and aroma production, and response to stresses.
Interestingly, contrary to medium-chain ADHs, CmADH11 only retained a conserved functional protein domain, Zn2-binding region [Cys(102), Cys(105), Cys(108) and Cys(116)] (Supplementary Figure S9). Although CmADH1, CmADH4 and CmADH11 were grouped into a class in the phylogenetic tree (Figure 2), CmADH11 showed a lower degree of homology to CmADH1 and CmADH4, implying that CmADH11 may be a member of zinc-medium-chain ADHs subfamily, and that these 3 CmADHs probably have similar functions, but more researches are required to examine this speculation. In addition, short-chain ADHs were found to be present in all living kingdoms. In tomato, a short-chain ADH, SlscADH1 protein contained motifs characteristic of classical short-chain dehydrogenases (Persson et al., 1991; Moummou et al., 2012). Nevertheless, in melon and ginseng, the protein sequence of short-chain ADH only encompassed some of these conserved regions, which were demonstrated to be related to substrate binding and active sites (ManrÍquez et al., 2006; Kim et al., 2009; Moummou et al., 2012). Furthermore, we found that the deduced protein of CmADH12, encoded by 635 amino acids, appeared to belong to long-chain ADH subfamily. In microorganisms, the sequences of the so-called long-chain or iron-dependent ADH genes were encoded by 600–750 amino acids, or up to over 900 amino acids (Williamson and Paquin, 1987; Goodlove et al., 1989; Chase, 1999). Whereas, CmADH12 did not has the typical protein domains of long-chain or iron-dependent ADH proteins, possessed in corresponding ADHs of prokaryotes, human and other mammals (Inoue et al., 1989; Deng et al., 2002; Lyon et al., 2009; Elleuche et al., 2013, 2014). Interestingly, CmADH12 protein sequence exhibited two motifs typical of classical SDR-ADHs, namely an N-terminal coenzyme-binding pattern of typically GXXXGXG and an active-site pattern of YXXXK (Supplementary Figure S11); Simultaneously, this sequence possessed a NADPH-binding domain AXXGGTG that appears to be motifs typical of classical medium-chain ADHs by NCBI search, implying that CmADH12 may be consisted of a short-chain dehydrogenase protein and a medium-chain dehydrogenase. Up to now, for the long-chain ADH genes or iron-activated ADHs, no member of this type ADHs has been described in plant. Here, we reported a novel melon ADH, and the majority of predicted CmADH12 in melon genome still awaits functional annotation. With the report of melon, CmADH1 and CmADH2, belonging to the medium-chain ADHs and short-chain ADHs respectively (ManrÍquez et al., 2006), the identification of a long-chain ADH, CmADH12, will make a bridge to connect the gap in the distribution of medium- and short-chain ADHs, and will represent significant improvements on the understanding of the evolution of ADH in higher plants.
Expression Analysis in Different Tissue
In plants, multiple homologous ADH genes, thought to have distinct roles, have been implicated in the response to biotic and abiotic stresses, particularly, in the tissues development under stresses (Abe et al., 2003; Delessert et al., 2004; Kumar et al., 2010; Frazier et al., 2011; Komatsu et al., 2011; Qi et al., 2012). At the same time, many previous studies indicated that plant ADHs have been involved in seedling, pollen development and fruit development (Kim et al., 2010; Thompson et al., 2010), suggesting that ADH genes play a important role in progress of the vegetative and reproductive organs development. In melon, CmADHs also showed different expression patterns in tissues and fruits during different development stages (Figures 4 and 5; Supplementary Figure S12). CmADH1 and CmADH2 were weakly expressed or not expressed in leaf, stem, seed, root, and flower of Cantaloupe (ManrÍquez et al., 2006), without the same as our results (Figure 4; Supplementary Figure S12). Hence, we speculated that it seems to be related to sample type or other, which are worthy of further study. In mango, all of three MiADHs were weakly expressed in flower, leaf, and stem, with different transcript levels between MiADHs (Singh et al., 2010). In petunia hybrid, three functional ADHs had different expressions in immature pollen grains, anther, stigma, petal, ovary and hypoxic root (Garabagi and Strommer, 2004); Moreover, particularly in the nectary, ADH3 was remarkably expressed, suggesting the association of ADH genes with production of aromatic compounds in the nectary of petunia hybrid (Garabagi et al., 2005). In the present study, CmADH1, CmADH2, CmADH10 and CmFDH1 were strongly expressed in petal, with a higher corresponding enzymatic activity in pistillate and staminate flower petal respectively (Supplementary Figure S15). Therefore, we infered that these four CmADHs participated in the flowers development and the production of volatile substances in melon flowers, and based on these findings, the previously assumed functional redundancy of these genes in the ADH network needs to be questioned (Strommer, 2011).
In addition, plant ADH genes were also implicated in fruit tissue development and fruit ripening, and were closely related to aroma volatile production. For example, mango flesh undergone softening during ripening, and MiADH1 and MiADH2 transcripts particularly accumulated at the onset of ripening in mango fruit, and gradually reduced thereafter; whereas, MiADH3 accumulated during early development of fruit (Singh et al., 2010). In tamato, SlscADH1, a fruit-ripening-associated short-chain ADH, has been related to the formation of aroma volatiles (Gaut et al., 1999). In the present study, the ADH enzyme activity increased at first and then decreased, and had a peak at 36 days after the anthesis during fruit development (Supplementary Figure S16). Nevertheless, only CmADH2 and CmADH10 were strongly expressed during the progress of fruit development except 15 days after the anthesis, suggesting that CmADH2 and CmADH10 may participate in fruit development of the whole development period of melon. In olea europaea, OeADH transcript guadually rose along with olea fruit development and peaked at fruit ripening, and the change of OeADH expression was coherent with the olea fruit growth curve (Iaria et al., 2012), and the similar pattern of ADH gene expression was found in apricot fruits (Gonzalez-Agüero et al., 2009). Additionally, in Cantaloupe, the transcript levels of CmADH1 and CmADH2 gradually increased at first and then decreased from 32 to 42 days with a peak at 39 days (ManrÍquez et al., 2006). However, our data showed CmADH1 was strongly expressed at the late stage of development; CmADH2 transcripts always maintained higher levels during fruit development (Supplementary Figure S12). In addition, VvADH1 and VvADH3 in grape were temporarily expressed in young and developing berry, whereas VvAdh2 transcript was strongly accumulated at later developmental stages, from the onset of ripening (Tesnière and Verriès, 2000). In melon, CmADH1, CmADH3, CmADH8 and CmFDH1 were highly expressed in fruit at 1 day after anthesis, suggesting that these 4 ADH genes likely participate in the ovary development of melon fruit. Of three functional petunia ADHs, PhADH2 and PhADH3 were expressed in ovary (Garabagi et al., 2005). As for melons, little information was available on the role of ADH in relation to the ovary development of fruit. Furthermore, expression analysis also indicated that CmADH8 and CmFDH1 were prominently expressed later in fruit development. However, little information was available on the role of another medium-chain ADH in relation to melon fruit development and ripening. These findings implied that CmADH8 and CmFDH1 might also be involved in fruit development or in other unknown functions at different developmental stages, and also suggested that medium-chain CmADHs might have evolved individual unique functions in fruit development, but it appears that a complex network, consisted of the members of ADH superfamily, plays a important role to modulate the fruit development and ripening. There is much to learn about medium-chain ADHs in melon.
There were significant expression differences between CmADH11 and other medium-chain CmADHs in different tissues and during development and ripening. These observed differences could partly be explained by the amino acids differences at the Zn1-binding sequences and the coenzyme binding position (Supplementary Figure S9). Based on our analysis of medium-chan CmADHs, CmADH11 had little key residues, and had Phe, Ala, and Tyr instead of three Gly in the coenzyme binding domain GXG(X)2G of all functional medium-chain ADHs. Furthermore, some substituted residues in soybean ADH2 protein might retain the function of amino acids, and it is not clear in other substitutions (Komatsu et al., 2011). Accordingly, these findings implied difference in enzymatic properties between CmADH11 and CmADH1 and other plant ADHs. On the basis of the above results, it appears that CmADH11 is a pseudogene. But we used the expressed sequence tag (EST) database in NCBI as a source of mRNA sequence bioinformatics for CmADH11, and we found that CmADH11 showed the highest homology with a Cucumis melo cDNA clone (GenBank no.JG552191.1) (89% identity), obtained from callus, from EST database of melon (CM-DEa library). Hence, it appears that CmADH11 is specifically expressed in callus. Further research is required to confirm this observation.
Induced Expression Analysis
We also studied the effect of ABA and IAA on CmADHs expression. ABA is known to be extensively involved in plant developmental processes and the plant’s response to abiotic stresses, and can regulate the expression of relevant genes to increase plant adaptability and to regulate tissues development (Skriver and Mundy, 1990; Matsukura et al., 2010; Su et al., 2013). ABA has been shown to induce ADH gene expression in Arabidopsis (de Bruxelles et al., 1996; Su et al., 2013), barley (Macnicol and Jacobsen, 2001) and mango (Singh et al., 2010); In particular, maize (Hwang and Vantoai, 1991) and lettuce (Kato-Noguchi, 2000) ADH genes were strongly induced under abiotic stresses. Purportedly, ABA-mediated plant responses to drought stress might be related to the regulation of relevant genes by key regulator in ABA signaling, including MYB, MYC, NAC, PP2Cs, DREB family and so on (Abe et al., 2003; Fujita et al., 2011; Su et al., 2013). However, to the best of our information, there were little reports on effect of ABA or IAA on ripening specific ADHs in melon. Promoter analysis of the 12 melon ADH genes and one FDH gene suggested the presence of ABA responsive ABRE motif in the promoter of CmFDH1, CmADH2, CmADH3 and CmADH10 (Su et al., 2013) (Supplementary Table S1). Additionally, the promoter of mango ADHs has the ABA responsive ABRE motif (Gomez-Porras et al., 2007; Singh et al., 2010), while MiADH1 was induced by ABA, and MiADH2 was repressed. In the present study, ABA treatment boosted the expressions of CmADH1, CmADH2, CmADH3, CmADH4, CmADH10 and CmFDH1, but ABA did not obviously inhibit other ADHs transcripts (Figure 6; Supplementary Figure S13). Therefore, the ABA-induced these CmADHs expression observed in present study may be related to the upstream key transcription factors. However, this speculation has to be demonstrated by further cloning and function analyses of the promoter of CmADHs. Furthermore, co-treatment with ABA and ethylene enhanced MiADH1 expression which was higher than those by either of the hormone alone, and ethylene could relieve the inhibition of ABA on MiADH2 transcript, suggesting that there were likely mutual promotion between ABA and ethylene, or that ethylene was in some manner able to repress ABA action (Singh et al., 2010). All of these derived results also implied that it would be important to investigate whether ABA triggers different hormonal signal pahways, particularly the possible crosstalk between ABA and ethylene, because ABA and ethylene are key hormones in fruit ripening and senescence, as hormone crosstalk has been shown to be important for biotic and abiotic stresses (Bari and Jones, 2009; Su et al., 2013). More further work is needed to investigate the crosstalk in response to fruit ripening. Auxin (IAA) also plays a role in fruit development and ripening (Singh et al., 2010), and there was an active crosstalk between IAA and ethylene during fruit ripening (Trainotti et al., 2007). However, in the case of melon, there were not reports about the inducing of ripening in mature fruit by IAA as done by ethylene. In tamato, the mutual effect between IAA and ethylene boosted tomato fruit development at early developmental stages (Balbi and Lomax, 2003). Three mango ADH genes expressions were not obviously induced by IAA treatment, and IAA did not induced the mango fruit ripening, moreover there are not IAA responsive TGA motif in promoter in mango ADHs (Singh et al., 2010). CmADH6, CmADH9 and CmADH12 promoters contained IAA responsive TGA motif, and IAA treatment induced these three ADHs transcripts (Figure 6; Supplementary Figure S13). Other ADH genes were expressed at basal level or not expressed, but did not appear to be significantly regulated by IAA, which were consistent with observations in mango (Singh et al., 2010).
Our results also indicated that 12 CmADHs and 1 CmFDH showed different patterns of transcripts during ethylene induced ripening (Figure 7; Supplementary Figure S14), which could be explained by the presence of ethylene responsive ERE motifs in CmADH1, CmADH2, CmADH3, CmADH8, CmADH12 and CmFDH1 (Tesniere et al., 2004; Jin et al., 2014) (Supplementary Table S1). According to previous reports, there are ethylene responsive ERE motifs in Arabidopsis AtADH1 (Peng et al., 2001), mango MiADH1 (Singh et al., 2010) and tomato LeADH2 (Speirs et al., 1998) promoters, and these ADH genes were responsive to ethylene. So, in view of melon ADH genes responses to ethylene, CmADH1, CmADH2, CmADH3, CmADH8, CmADH12 and CmFDH1 were classed into ethylene-sensitive group, and other CmADHs clustered ethylene-insensitive group. Furthermore, Manråquez et al.(2006) also found that ethylene increased the levels of CmADH1 and CmADH2 transcripts in Cantaloupe during fruit ripening, while 1-MCP treatment inhibited the inducing effect of ethylene to CmADH1 and CmADH2. This was similar to the ripening related expression observed in mango ADHs (Singh et al., 2010), tomato LeADH2 (climacteric fruit) (Defilippi et al., 2005b; Barakat et al., 2009) and VvADH2 in grape (non-climacteric fruit) (Tesniere et al., 2004). Simultaneously, ethylene treatment enhanced the activity of ADH enzyme in mango and ethylene production and accelerated mango fruit ripening and senescence, but the increase effect of ethylene was weakened by 1-MCP (Singh et al., 2010). Whereas, in tomato, ADH enzymatic activity increased during fruit ripening, and LeADH2 was involved in fruit ripening and aroma compounds synthesis, but was not regulated by ethylene (Speirs et al., 1998, 2002); Ethylene induced the increases of ADH enzymatic activity and VvADH2 transcript in grape (a climacteric fruit) during fruit ripening (Tesniere et al., 2004), but did not clearly enhanced the activity of ADH enzyme in ACC oxidase antisense transgenic apple, without influence on the esters production in this kind of apple (Defilippi et al., 2005a). These findings showed that the ADH genes superfamily included ethylene non-dependent ADH gene members in climacteric kinds of fruit and ethylene dependent ADH gene members in non-climacteric kinds of fruit. In this paper, the changes of ADH enzyme activity in fruit treated by ethylene and 1-MCP (Supplementary Figure S17) and CmADH10 expression indicated that CmADH10 may be a non-dependent ADH gene member (Figure 7; Supplementary Figure S14). Whereas, more experimental works are needed to further elucidate the function of CmADH10 in aroma compounds synthesis and during fruit ripening and senescence.
Additionally, we also identified several hormone-responsive cis-regulatory elements in the CmADHs promoter region, such as GARE, TATC-box, P-box (gibberellin), TCA-element (SA), CCAAT-box, TGACG-motif and CGTCA-motif (MeJA) (Barakat et al., 2009; Kim et al., 2010; Jin et al., 2014) (Supplementary Table S3). Several previous studies showed a complex interplay of hormones was known to affect fruit development and ripening, with auxin and GA being important during fruit expansion and ABA and ethylene being important for ripening (Srivastava and Handa, 2005; Singh et al., 2010). In addition, oriental melon (Cucumis melo var. makuwa Makino) has distinct ripening patterns within the fruit (Jin et al., 2014). Therefore, based on our derived results in this study, we speculated that the different regulation of melon fruit ripening by ABA, IAA and ethylene might selectively affect the expression of one- or multi- ADHs, which prompted CmADHs to regulate fruit ripening and senescence. Maybe, it will be possible to retain a crosstalk between ABA/IAA/JA/GA and ethylene to regulate the ADH genes network involved in fruit development and ripening, as hormone crosstalk has been shown to be important for biotic and abiotic stresses (Bari and Jones, 2009; Su et al., 2013). Further research is required to confirm this hypothesis.
Conclusion
We identified 13 CmADHs (including 12 ADHs and 1 FDH) in melon genome, exhibiting lower identity in protein level with each other, and they were expressed differentially in different vegetative tissues and during fruit development and ripening. Phylogenetic analysis indicated that they belonged to three different ADH subfamilies and the medium-chain ADHs were clustered into six subgroups. On the transcript level, differential CmADHs expression suggested tight adaptation of the fruit to the developmental events. Promoter sequence analysis, intron-exon structure and the different responses of these ADH genes to ABA, IAA and ethylene implied that ADH genes had different functions or distinct metabolic roles in melon fruit development and ripening, and the different ADH genes expression at transcript induced by ethylene indicated that CmADH10 was an ethylene non-dependent ADH gene member in CmADH genes. Further and detailed research efforts should be made to find the detailed function of these ADH proteins in addition to CmADH1 and CmADH2 during melon fruit development and ripening. Therefore, selective silencing of the CmADH genes and more studies related to biochemical characterization of these CmADHs will be needed.
Author Contributions
Conceived and designed the experiments: YJ, HQ, CZ; Performed the experiments: YJ, CZ, WL, HC, SC; Analyzed the data: YJ, CZ, HQ, HC; Contributed reagents/materials/analysis tools: YJ, CZ, YT; Wrote the paper: YJ, CZ, HQ.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Qiang Chen, Deqing Lv, and Tao Xu for excellent technical assistance.
Funding. This work was supported by grants from the National Natural Science Foundation of China (31272154) and the Excellent Talent Project of Liaoning Provincial Department of Education (LR2014020).
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2016.00670
Phylogenetic tree of melon medium-chain ADH proteins. The amino acid sequences were aligned by the Clustal Omega program, and the neighborjoining tree was drawn with TreeView. The corresponding GenBank and the melon genome (https://melonomics.net/) were noted in the phylogenetic tree and the accession number in the melon genome were CmADH1 (MELO3C023685P4), CmADH3 (MELO3C026552P1), CmADH4 (MELO3C027151P1), CmADH5 (MELO3C00579 2P1), CmADH6 (MELO3C026553P1), CmADH7 (MELO3C002189P1), CmADH8 (ME LO3C003251P1), CmADH9 (MELO3C011043P1), CmADH10 (MELO3C026554P2), CmADH11 (MELO3C023687P1), and CmFDH1 (MELO3C022399P1). The number for each interior branch was the percentage of bootstraps value (1000 replicates). Black circle denoted 13 CmADHs. ADHs from other plants in our paper are in Supplementary Table S3.
Amino acid sequence alignment of melon CmADH1 (MELO3C023685P4) and CmADH4 (MELO3C027151 P1) with closely related sequences of other plants. GenBank accession numbers are in Supplementary Table S3.
Amino acid sequence alignment of melon CmFDH1 (MELO3C022399P1) with closely related sequences of other plants. GenBank accession numbers are in Supplementary Table S3.
Amino acid sequence alignment of melon CmADH3 (MELO3C026552P1), CmADH6 (MELO3C026553P1) and CmADH10 (MELO3C026554P2) with closely related sequences of other plants. GenBank accession numbers are in Supplementary Table S3.
Amino acid sequence alignment of melon CmADH5 (MELO3C005 792P1) with closely related sequences of other plants. GenBank accession numbers are in Supplementary Table S3.
Amino acid sequence alignment of melon CmADH7 (MELO3C002 189P1) with closely related sequences of other plants. GenBank accession numbers are in Supplementary Table S3.
Amino acid sequence alignment of melon CmADH8 (MELO3C003 251P1) with closely related sequences of other plants. GenBank accession numbers are in Supplementary Table S3.
Amino acid sequence alignment of melon CmADH9 (MELO3C011043 P1) with closely related sequences of other plants. GenBank accession numbers are in Supplementary Table S3.
Amino acid sequence alignment of melon CmADH11 (MELO3C023 687P1) with closely related sequences of other plants. GenBank accession numbers are in Supplementary Table S3.
Amino acid sequence alignment of melon CmADH2 (MELO3C014 897P1) with closely related sequences of other plants. GenBank accession numbers are in Supplementary Table S3.
Amino acid sequence alignment of melon CmADH12 (MELO3C019 503P1) with closely related sequences of other plants. GenBank accession numbers are in Supplementary Table S3.
Transcript abundance of CmADHs in vegetative tissues and developing stages of melon fruit (1–48). R, root; ML, mature leaf; DL, developing leaf; YS, young stem; PF, pistillate flower petal; SF, staminate flower petal. RNA isolated from the samples. Expression analysis was carried out by semi-quantitative PCR using 18S as an internal control.
mRNA abundance of CmADHs in mature green fruit after treatment with ABA and IAA. IAA (Auxin) and ABA (100μM) treatments were given for 2 h as described in Section “Materials and Methods.” Expression analysis was carried out by semi-quantitative PCR as described in section of Hormone treatments using actin as 18S internal control.
CmADHs accumulation during different stages of ethylene and 1-MCP treatment induced ripening of melon by semi-quantitative PCR. 1–12 indicate days after ethylene and 1-MCP treatment. The accumulation level of the genes in untreated melon fruit was control. 18S was used as an internal control.
Alcohol dehydrogenases activity in different melon organs. R, root; ML, mature leaf; DL, developing leaf; YS, young stem; PF, pistillate flower petal; SF, staminate flower petal.
Alcohol dehydrogenases activity in fruits during melon development. 1, 5, 10, 15, 20, 25, 30, 33, 36, 39, 42, 45, and 48 indicate days after anthesis.
Alcohol dehydrogenases activity in fruits during different stages of ethylene and 1-MCP treatment induced ripening of melon. 1, 3, 6, 9, and 12 indicate days after ethylene and 1-MCP treatment. All the treatments have been described in section of Hormone treatments.
References
- Abe H., Urao T., Ito T., Seki M., Shinozaki K., Yamaguchi-Shinozaki K. (2003). Arabidopsis AtMYC2 (bHLH) and AtMYB2 (MYB) function as transcriptional activators in abscisic acid signaling. Plant Cell 15 63–78. 10.1105/tpc.006130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alka K., Windle H. J., Cornally D., Ryan B. J., Henehan G. T. M. (2013). A short chain NAD(H)-dependent alcohol dehydrogenase (HpSCADH) from Helicobacter pylori: a role in growth under neutral and acidic conditions. Int. J. Biochem. Cell Biol. 45 1347–1355. 10.1016/j.biocel.2013.04.006 [DOI] [PubMed] [Google Scholar]
- Balbi V., Lomax T. L. (2003). Regulation of early tomato fruit development by the diageotropica gene. Plant Physiol. 131 186–197. 10.1104/pp.010132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barakat A., Bagniewska-Zadworna A., Choi A., Plakkat U., DiLoreto D., Yellanki P., et al. (2009). The cinnamyl alcohol dehydrogenase gene family in Populus: phylogeny, organization, and expression. BMC Plant Biol. 9:26 10.1186/1471-2229-9-26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bari R., Jones J. D. (2009). Role of plant hormones in plant defence responses. Plant Mol. Biol. 69 473–488. 10.1007/s11103-008-9435-0 [DOI] [PubMed] [Google Scholar]
- Borràs E., Albalat R., Duester G., Parés X., Farrés J. (2014). The Xenopus alcohol dehydrogenase gene family: characterization and comparative analysis incorporating amphibian and reptilian genomes. BMC Genomics 15:216 10.1186/1471-2164-15-216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bukh C., Nord-Larsen P. H., Rasmussen S. K. (2012). Phylogeny and structure of the cinnamyl alcohol dehydrogenase gene family in Brachypodium distachyon. J. Exp. Bot. 63 6223–6236. 10.1093/jxb/ers275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Çelik A., Aktas F. A. (2013). A new NADH-dependent, zinc containing alcohol dehydrogenase from Bacillus thuringiensis serovar israelensis involved in oxidations of short to medium chain primary alcohols. J. Mol. Catal. B Enzym. 89 114–121. 10.1016/j.molcatb.2013.01.005 [DOI] [Google Scholar]
- Charlesworth D., Liu F. L., Zhang L. (1998). The evolution of the alcohol dehydrogenase gene family by loss of introns in plants of the genus Leavenworthia (Brassicaceae). Mol. Biol. Evol. 15 552–559. 10.1093/oxfordjournals.molbev.a025955 [DOI] [PubMed] [Google Scholar]
- Chase T. (1999). Alcohol dehydrogenases: identification and names for gene families. Plant Mol. Biol. Rep. 17 333–350. 10.1023/A:1007620627083 [DOI] [Google Scholar]
- Chen F. F., Wang P., An Y. A., Huang J. Q., Xu Y. W. (2015). Structural insight into the conformational change of alcohol dehydrogenase from Arabidopsis thaliana L. during coenzyme binding. Biochimie 108 33–39. 10.1016/j.biochi.2014.10.023 [DOI] [PubMed] [Google Scholar]
- Cheng F. F., Hu T., An Y., Huang J. Q., Xu Y. W. (2013). Purification and enzymatic characterization of alcohol dehydrogenase from Arabidopsis thaliana. Protein Expr. Purif. 90 74–77. 10.1016/j.pep.2013.05.004 [DOI] [PubMed] [Google Scholar]
- Cheng H., Li L. L., Xu F., Cheng S. Y., Cao F. L., Wang Y., et al. (2013). Expression patterns of a cinnamyl alcohol dehydrogenase gene involved in lignin biosynthesis and environmental stress in Ginkgo biloba. Mol. Biol. Rep. 40 707–721. 10.1007/s11033-012-2111-0 [DOI] [PubMed] [Google Scholar]
- Chung H. J., Ferl R. J. (1999). Arabidopsis alcohol dehydrogenase expression in both shoots and roots is conditioned by root growth environment. Plant Physiol. 121 429–436. 10.1104/pp.121.2.429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Bruxelles G. L., Peacock W. J., Dennis E. S., Dolferus R. (1996). Abscisic acid induces the alcohol dehydrogenase gene in Arabidopsis. Plant Physiol. 111 381–391. 10.1104/pp.111.2.381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Defilippi B. G., Dandekar A. M., Kader A. A. (2005a). Relationship of ethylene biosynthesis tovolatile production, related enzymesand precursor availability in apple peel and flesh tissues. J. Agric. Food Chem. 53 3133–3141. 10.1021/jf047892x [DOI] [PubMed] [Google Scholar]
- Defilippi B. G., Kader A. A., Dandekar A. M. (2005b). Apple aroma: alcohol acyltransferase, a rate limiting step for ester biosynthesis, is regulated by ethylene. Plant Sci. 168 199–210. 10.1016/j.plantsci.2004.12.018 [DOI] [Google Scholar]
- Delessert C., Wilson I. W., Van Der Straeten D., Dennis E. S., Dolferus R. (2004). Spatial and temporal analysis of the local response to wounding in Arabidopsis leaves. Plant Mol. Biol. 55 165–181. 10.1007/s11103-004-0112-7 [DOI] [PubMed] [Google Scholar]
- Deng Y., Wang Z., Gu S., Ji C., Ying K., Xie Y., et al. (2002). Cloning and characterization of a novel human alcohol dehydrogenase gene (ADHFe1). DNA Seq. 13 301–306. 10.1080/1042517021000011636 [DOI] [PubMed] [Google Scholar]
- Dolferus R., Jacobs M., Peacock W. J., Dennis E. S. (1994). Differential interactions of promoter elements in stress responses of the Arabidopsis Adh gene. Plant Physiol. 105 1075–1087. 10.1104/pp.105.4.1075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolferus R., Osterman J., Peacock W. J., Dennis E. S. (1997). Cloning of the Arabidopsis and rice class III Adh genes: implications for the origin of plant ADH enzymes. BMC Genet. 146:1131–1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duester G., Farrés J., Felder M. R., Holmes R. S., Höög J. O., Parés X., et al. (1999). Recommended nomenclature for the vertebrate alcohol dehydrogenase gene family. Biochem. Pharmacol. 58 389–395. 10.1016/S0006-2952(99)00065-9 [DOI] [PubMed] [Google Scholar]
- Elleuche S., Fodor K., Klippel B., Heyde A., Wilmanns M., Antranikian G. (2013). Structural and biochemical characterisation of a NAD+-dependent alcohol dehydrogenase from Oenococcus oeni as a new model molecule for industrial biotechnology applications. Appl. Microbiol. Biotechnol. 97 8963–8975. 10.1007/s00253-013-4725-0 [DOI] [PubMed] [Google Scholar]
- Elleuche S., Fodor K., von der Heyde A., Klippel B., Wilmanns M., Antranikian G. (2014). Group III alcohol dehydrogenase from Pectobacterium atrosepticum: insights into enzymatic activity and organization of the metal ion-containing region. Appl. Microbiol. Biotechnol. 98 4041–4051. 10.1007/s00253-013-5374-z [DOI] [PubMed] [Google Scholar]
- Espunya M. C., De Michele R., Gomez-Cadenas A., Martinez M. C. (2012). S-nitrosoglutathione is a component of wound- and salicylic acid-induced systemic responses in Arabidopsis thaliana. J. Exp. Bot. 63 3219–3227. 10.1093/jxb/ers043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espunya M. C., Díaz M., Moreno-Romero J., Martínez M. C. (2006). Modification of intracellular levels of glutathione-dependent formaldehyde dehydrogenase alters glutathione homeostasis and root development. Plant Cell Environ. 29 1002–1011. 10.1111/j.1365-3040.2006.01497.x [DOI] [PubMed] [Google Scholar]
- Feechan A., Kwon E., Yun B. W., Wang Y., Pallas J. A., Loake G. J. (2005). A central role for S-nitrosothiols in plant disease resistance. Proc. Natl. Acad. Sci. U.S.A. 102 8054–8059. 10.1073/pnas.0501456102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filling C., Berndt K. D., Benach J., Knapp S., Prozorovski T., Nordling E., et al. (2002). Critical residues for structure and catalysis in short-chain dehydrogenases/reductases. J. Biol. Chem. 277 25677–25684. 10.1074/jbc.M202160200 [DOI] [PubMed] [Google Scholar]
- Frazier T. P., Sun G., Burklew C. E., Zhang B. (2011). Salt and drought stresses induce the aberrant expression of microRNA genes in tobacco. Mol. Biotechnol. 49 159–165. 10.1007/s12033-011-9387-5 [DOI] [PubMed] [Google Scholar]
- Fujita Y., Fujita M., Shinozaki K., Yamaguchi-Shinozaki K. (2011). ABA-mediated transcriptional regulation in response to osmotic stress in plants. J. Plant Res. 124 1–17. 10.1007/s10265-011-0412-3 [DOI] [PubMed] [Google Scholar]
- Garabagi F., Duns G., Strommer J. (2005). Selective recruitment of Adh genes for distinct enzymatic functions in Petunia hybrid. Plant Mol. Biol. 58 283–294. 10.1007/s11103-005-3545-8 [DOI] [PubMed] [Google Scholar]
- Garabagi F., Strommer J. (2004). Distinct genes produce the alcohol dehydrogenases of pollen and maternal tissues in Petunia hybrida. Biochem. Genet. 42 199–208. 10.1023/B:BIGI.0000026634.69911.2e [DOI] [PubMed] [Google Scholar]
- Garcia-Mas J., Benjak A., Sanseverino W., Bourgeois M., Mir G., González V. M., et al. (2012). The genome of melon (Cucumis melo L.). Proc. Natl. Acad. Sci. U.S.A. 109 11872–11877. 10.1073/pnas.1205415109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaut B. S., Peek A. S., Morton B. R., Clegg M. T. (1999). Patterns of genetic diversification within the Adh gene family in the grasses (Poaceae). Mol. Biol. Evol. 16 1086–1097. 10.1093/oxfordjournals.molbev.a026198 [DOI] [PubMed] [Google Scholar]
- Gomez-Porras J. L., Riano-Pachon D. M., Dreyer I., Mayer J. E. (2007). Mueller-Roeber B. Genome-wide analysis of ABA-responsive elements ABRE and CE3. BMC Genomics 8:260 10.1186/1471-2164-8-260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Agüero M., Troncoso S., Gudenschwager O., Campos-Vargas R., Moya-Leon M. A., Defilippi B. G. (2009). Differential expression levels of aroma-related genes during ripening of apricot (Prunusarmeniaca L.). Plant Physiol. Biochem. 47 435–440. 10.1016/j.plaphy.2009.01.002 [DOI] [PubMed] [Google Scholar]
- Goodlove P. E., Cunningham P. R., Parker J., Clark D. P. (1989). Cloning and sequence analysis of the fermentative alcohol-dehydrogenase-encoding gene of Escherichia coli. Gene 85 209–214. 10.1016/0378-1119(89)90483-6 [DOI] [PubMed] [Google Scholar]
- Höög J. O., Strömberg P., Hedberg J. J., Griffiths W. J. (2003). The mammalian alcohol dehydrogenases interact in several metabolic pathways. Chem. Biol. Interact. 144 175–181. [DOI] [PubMed] [Google Scholar]
- Hwang S. Y., Vantoai T. T. (1991). Abscisic acid induces anaerobiosis tolerance in corn. Plant Physiol. 97 593–597. 10.1104/pp.97.2.593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iaria D. L., Bruno L., Macchione B., Tagarelli A., Sindona G., Giannino D., et al. (2012). The aroma biogenesis-related olea europaea alcohol dehydrogenase gene is developmentally regulated in the fruits of two O. europaea L. cultivars. Food Res. Int. 49 720–727. 10.1016/j.foodres.2012.09.004 [DOI] [Google Scholar]
- Inoue T., Sunagawa M., Mori A., Imai M., Fukuda M., Takagi M., et al. (1989). Cloning and sequencing of the gene encoding the 72-kilodalton dehydrogenase subunit of the alcohol dehydrogenase from Acetobacter aceti. J. Bacteriol. 171 3115–3122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Y. Z., Zhang C., Liu W., Qi H. Y., Chen H., Cao S. X. (2014). The cinnamyl alcohol dehydrogenase gene family in melon (Cucumis melo L.): bioinformatic analysis and expression patterns. PLoS ONE 9:e101730 10.1371/journal.pone.0101730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joërnvall H., Hoëoëg J. O., Persson B. (1999). SDR and MDR: completed genome sequences show these protein families to be large, of old origin, and of complex nature. FEBS Lett. 445 261–264. 10.1016/S0014-5793(99)00130-1 [DOI] [PubMed] [Google Scholar]
- Jönvall H., Hedlund J., Bergman T., Kallberg Y., Cederlund E., Persson B. (2013). Origin and evolution of medium chain alcohol dehydrogenases. Chem. Biol. Interact. 202 91–96. 10.1016/j.cbi.2012.11.008 [DOI] [PubMed] [Google Scholar]
- Jönvall H., Hedlund J., Bergman T., Oppermann U., Persson B. (2010). Superfamilies SDR and MDR: from early ancestry to present forms. Emergence of three lines, a Zn-metalloenzyme, and distinct variabilities. Biochem. Biophys. Res. Commun. 396 125–130. 10.1016/j.bbrc.2010.03.094 [DOI] [PubMed] [Google Scholar]
- Kato-Noguchi H. (2000). Abscisic acid and hypoxic induction of anoxia tolerance in roots of lettuce seedlings. J. Exp. Bot. 51 1939–1944. 10.1093/jexbot/51.352.1939 [DOI] [PubMed] [Google Scholar]
- Khan A. J., Husain Q., Choudhuri G., Parmar D. (2010). Association of polymorphism in alcohol dehydrogenase and interaction with other genetic risk factors with alcoholic liver cirrhosis. Drug Alcohol Depend. 109 190–197. 10.1016/j.drugalcdep.2010.01.010 [DOI] [PubMed] [Google Scholar]
- Kim Y. H., Bae J. M., Huh G. H. (2010). Transcriptional regulation of the cinnamyl alcohol dehydrogenase gene from sweetpotato in response to plant developmental stage and environmental stress. Plant Cell Rep. 29 779–791. 10.1007/s00299-010-0864-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y. J., Shim J. S., Lee J. H., Jung S. Y., Sun H., In J. G., et al. (2009). Isolation and characterization of a novel short-chain alcohol dehydrogenase gene from Panax ginseng. BMB Rep. 42 673–678. 10.5483/BMBRep.2009.42.10.673 [DOI] [PubMed] [Google Scholar]
- Komatsu S., Thibaut D., Hiraga S., Kato M., Chiba M., Hashiguchi A., et al. (2011). Characterization of a novel flooding stress-responsive alcohol dehydrogenase expressed in soybean roots. Plant Mol. Biol. 7 309–322. 10.1007/s11103-011-9812-y [DOI] [PubMed] [Google Scholar]
- Koutsompogeras P., Kyriacou A., Zabetakis I. (2010). Characterization of NAD-dependent alcohol dehydrogenase enzymes of strawberry’s achenes (Fragaria x ananassa cv. Elsanta) and comparison with respective enzymes from Methylobacterium extorquens. Food Sci. Technol. 43 828–835. [Google Scholar]
- Kubienova L., Ticha T., Jahnova J., Luhova L., Mieslerova B., Petrivalsky M. (2014). Effect of abiotic stress stimuli on S-nitrosoglutathione reductase in plants. Planta 239 139–146. 10.1007/s00425-013-1970-5 [DOI] [PubMed] [Google Scholar]
- Kumar M. S., Hema R., Suryachandra T. R., Ramegowda H. V., Gopalakrishna R., Rama N., et al. (2010). Functional characterization of three water deficit stress-induced genes in tobacco and Arabidopsis: an approach based on gene down regulation. Plant Physiol. Biochem. 48 35–44. 10.1016/j.plaphy.2009.09.005 [DOI] [PubMed] [Google Scholar]
- Kumar S., Sandell L. L., Trainor P. A., Koentgend F., Duestera G. (2012). Alcohol and aldehyde dehydrogenases: retinoid metabolic effects in mouse knockout models. Biochim. Biophys. Acta 1821 198–205. 10.1016/j.bbalip.2011.04.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee U., Wie C., Fernandez B. O., Feelisch M., Vierling E. (2008). Modulation of nitrosative stress by S-nitrosoglutathione reductase is critical for thermotolerance and plant growth in Arabidopsis. Plant Cell 20 786–802. 10.1105/tpc.107.052647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyon R. C., Johnston S. M., Panopoulos A., Alzeer S., McGarvie G., Ellis E. M. (2009). Enzymes involved in the metabolism of γ-hydroxybutyrate in SH-SY5Y cells: identification of an iron-dependent alcohol dehydrogenase ADHFe1. Chem. Biol. Interact. 178 283–287. 10.1016/j.cbi.2008.10.025 [DOI] [PubMed] [Google Scholar]
- Macnicol P. K., Jacobsen J. V. (2001). Regulation of alcohol dehydrogenase gene expression in barley aleurone by gibberellin and abscisic acid. Plant Physiol. 111 533–539. 10.1034/j.1399-3054.2001.1110414.x [DOI] [PubMed] [Google Scholar]
- ManrÍquez D., El-Sharkawy I., Flores F. B., El-Yahyaoui F., Regad F., Bouzayen M., et al. (2006). Two highly divergent alcohol dehydrogenases of melon exhibit fruit ripening-specific expression and distinct biochemical characteristics. Plant Mol. Biol. 61 675–685. 10.1007/s11103-006-0040-9 [DOI] [PubMed] [Google Scholar]
- Matsukura S., Mizoi J., Yoshida T., Todaka D., Ito Y., Maruyama K., et al. (2010). Comprehensive analysis of rice DREB2-type genes that encode transcription factors involved in the expression of abiotic stress-responsive genes. Mol. Genet. Genomics 283 185–196. 10.1007/s00438-009-0506-y [DOI] [PubMed] [Google Scholar]
- Matton D. P., Constable P., Brisson N. (1990). Alcohol dehydrogenase gene expression in potato following elicitor and stress treatment. Plant Mol. Biol. 14 775–783. 10.1007/BF00016510 [DOI] [PubMed] [Google Scholar]
- McKie J. H., Jaouhari R., Douglas K. T., Goffner D., Feuillet C., Grima-Pettenati J., et al. (1993). A molecular model for cinnamyl alcohol dehydrogenase, a plant aromatic alcohol dehydrogenase involved in lignification. Biochim. Biophys. Acta 1202 61–69. 10.1016/0167-4838(93)90063-W [DOI] [PubMed] [Google Scholar]
- Min T., Yin X. R., Shi Y. N., Luo Z. R., Yao Y. C., Grierson D., et al. (2012). Ethylene-responsive transcription factors interact with promoters of ADH and PDC involved in persimmon (Diospyros kaki) fruit de-astringency. J. Exp. Bot. 63 6393–6405. 10.1093/jxb/ers296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moummou H., Tonfack L. B., Chervin C., Benichou M., Youmbi E., Giniese C., et al. (2012). Functional characterization of SlscADH1, a fruit-ripening-associated short-chain alcohol dehydrogenase of tomato. J. Plant Physiol. 169 1435–1444. 10.1016/j.jplph.2012.06.007 [DOI] [PubMed] [Google Scholar]
- Nian H. J., Meng Q. C., Zhang W., Chen L. M. (2013). Overexpression of the formaldehyde dehydrogenase gene from Brevibacillus brevis to enhance formaldehyde tolerance and detoxification of tobacco. Appl. Biochem. Biotechnol. 169 170–180. 10.1007/s12010-012-9957-4 [DOI] [PubMed] [Google Scholar]
- Pathuri I. P., Reitberger I. E., Ckelhoven R., Proels R. K. (2011). Alcohol dehydrogenase 1 of barley modulates susceptibility to the parasitic fungus Blumeria graminis f.sp. hordei. J. Exp. Bot. 62 3449–3457. 10.1093/jxb/err017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng H. P., Chan C. S., Shih M. C., Yang S. F. (2001). Signalling events in the hypoxic induction of alcohol dehydrogenase gene in Arabidopsis. Plant Physiol. 126 742–749. 10.1104/pp.126.2.742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry D. J., Furnier G. R. (1996). Pinus banksiana has at least seven expressed alcohol dehydrogenase genes in two linked groups. Proc. Natl. Acad. Sci. U.S.A. 93 13020–13023. 10.1073/pnas.93.23.13020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Persson B., Krook M., Jörnvall H. (1991). Characteristics of short-chain alcohol dehydrogenases and related enzymes. Eur. J. Biochem. 200 537–543. 10.1111/j.1432-1033.1991.tb16215.x [DOI] [PubMed] [Google Scholar]
- Plapp B. V., Lee A. T., Khanna A., Pryor J. M. (2013). Bradykinetic alcohol dehydrogenases make yeast fitter for growth in the presence of allyl alcohol. Chem. Biol. Interact. 202 104–110. 10.1016/j.cbi.2012.11.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polichuk D. R., Zhang Y. S., Reed D. W., Schmidt J. F., Covello P. S. (2010). A glandular trichome-specific monoterpene alcohol dehydrogenase from Artemisia annua. Phytochemistry 711 1264–1269. 10.1016/j.phytochem.2010.04.026 [DOI] [PubMed] [Google Scholar]
- Prashant S., Sunita M. S. L., Sirisha V. L., Bhaskar V. V., Rao A. M., Narasu M. L., et al. (2012). Isolation of cinnamoyl CoA reductase and cinnamyl alcohol dehydrogenase gene promoters from Leucaena leucocephala, a leguminous tree species, and characterization of tissue-specific activity in transgenic tobacco. Plant Cell Tissue Organ Cult. 108 421–436. 10.1007/s11240-011-0053-1 [DOI] [Google Scholar]
- Qi X. H., Xu X. W., Lin X. J., Zhang W. J., Chen X. H. (2012). Identification of differentially expressed genes in cucumber (Cucumis sativus L.) root under waterlogging stress by digital gene expression profile. Genomics 99 160–168. 10.1016/j.ygeno.2011.12.008 [DOI] [PubMed] [Google Scholar]
- Quaglia D., Pori M., Galletti P., Emer E., Paradisi F., Giacomini D. (2013). His-tagged Horse Liver Alcohol Dehydrogenase: immobilization and application in the bio-based enantioselective synthesis of (S)-arylpropanols. Process Biochem. 48 810–818. 10.1016/j.procbio.2013.03.016 [DOI] [Google Scholar]
- Rustérucci C., Espunya M. C., Díaz M., Chabannes M., Martínez M. C. (2007). S-nitrosoglutathione reductase affords protection against pathogens in Arabidopsis, both locally and systemically. Plant Physiol. 143 1282–1292. 10.1104/pp.106.091686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh R. K., Sane V. A., Misra A., Ali S. A., Nath P. (2010). Differential expression of the mango alcohol dehydrogenase gene family during ripening. Phytochemistry 71 1485–1494. 10.1016/j.phytochem.2010.05.024 [DOI] [PubMed] [Google Scholar]
- Skriver K., Mundy J. (1990). Gene expression in response to abscisic acid and osmotic stress. Plant Cell 2 503–512. 10.2307/3869112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Small R. L., Wendel J. F. (2000). Copy number lability and evolutionary dynamics of the Adh gene family in diploid and tetraploid cotton (Gossypium). BMC Genet. 155:1913–1926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speirs J., Correll R., Cain P. (2002). Relationship between ADH activity, ripeness and softness in six tomato cultivars. Sci. Hortic. 93 137–142. 10.1016/S0304-4238(01)00316-8 [DOI] [Google Scholar]
- Speirs J., Lee E., Holt K., Kim Y. D., Scott N. S., Loveys B., et al. (1998). Genetic manipulation of alcohol deshydrogenase levels in ripening tomato fruit affects the balance of some flavour aldehydes and alcohols. Plant Physiol. 117 1047–1058. 10.1104/pp.117.3.1047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava A., Handa A. K. (2005). Hormonal regulation of tomato fruit development: a molecular perspective. J. Plant Growth Regul. 24 67–82. 10.1007/s00344-005-0015-0 [DOI] [Google Scholar]
- Strommer J. (2011). The plant ADH gene family. Plant J. 66 128–142. 10.1111/j.1365-313X.2010.04458.x [DOI] [PubMed] [Google Scholar]
- Su Z., Ma X., Guo H. H., Sukiran N. L., Guo B., Assmann S. M., et al. (2013). Flower development under drought stress: morphological and transcriptomic analyses reveal acute responses and long-term acclimation in Arabidopsis. Plant Cell 25 3785–3807. 10.1105/tpc.113.115428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung C. K., Kim S. M., Oh C. J., Yang S. A., Han B. H., Mo E. K. (2012). Taraxerone enhances alcohol oxidation via increases of alcohol dehyderogenase (ADH) and acetaldehyde dehydrogenase (ALDH) activities and gene expressions. Food Chem. Toxicol. 50 2508–2514. 10.1016/j.fct.2012.04.031 [DOI] [PubMed] [Google Scholar]
- Tesniere C., Pradal M., El-Kereamy A., Torregrosa L., Chatelet P., Roustan J. P., et al. (2004). Involvement of ethylene signalling in a non-climacteric fruit: new elements regarding the regulation of ADH expression in grapevine. J. Exp. Bot. 55 2235–2240. 10.1093/jxb/erh244 [DOI] [PubMed] [Google Scholar]
- Tesnière C., Verriès C. (2000). Molecular cloning and expression of cDNAs encoding alcohol dehydrogenases from Vitis vinifera L. during berry development. Plant Sci. 157 77–88. 10.1016/S0168-9452(00)00274-0 [DOI] [PubMed] [Google Scholar]
- Thompson C. E., Fernandes C. L., de Souza O. N., de Freitas L. B., Salzano F. M. (2010). Evaluation of the impact of functional diversification on Poaceae, Brassicaceae, Fabaceae, and Pinaceae alcohol dehydrogenase enzymes. J. Mol. Model. 16 919–928. 10.1007/s00894-009-0576-0 [DOI] [PubMed] [Google Scholar]
- Thompson C. E., Salzano F. M., de Souza O. N., Freitas L. B. (2007). Sequence and structural aspects of the functional diversification of plant alcohol dehydrogenases. Gene 396 108–115. 10.1016/j.gene.2007.02.016 [DOI] [PubMed] [Google Scholar]
- Tonfack L. B., Moummou H., Latché A., Youmbi E., Benichou M., Pech J. C., et al. (2011). The plant SDR superfamily: involvement in primary and secondary metabolism. Curr. Top. Plant Biol. 12 41–53. [Google Scholar]
- Trainotti L., Tadiello A., Casadoro G. (2007). The involvement of auxin in the ripening of climacteric fruits comes of age: the hormone plays a role of its own and has an intense interplay with ethylene in ripening peaches. J. Exp. Bot. 58 3299–3308. 10.1093/jxb/erm178 [DOI] [PubMed] [Google Scholar]
- Williamson V. M., Paquin C. E. (1987). Homology of Saccharomyces cerevisiæ ADH4 to an iron-activated alcohol dehydrogenase from Zymomonas mobilis. Mol. Gen. Genet. 209 374–381. 10.1007/BF00329668 [DOI] [PubMed] [Google Scholar]
- Yamauchi T., Watanabe K., Fukazawa A., Mori H., Abe F., Kawaguchi K., et al. (2014). Ethylene and reactive oxygen species are involved in root aerenchyma formation and adaptation of wheat seedlings to oxygen-deficient conditions. J. Exp. Bot. 65 261–273. 10.1093/jxb/ert371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B., Xi W. P., Wei W. W., Shen J. Y., Ferguson I., Chen K. S. (2011). Changes in aroma-related volatiles and gene expression during low temperature storage and subsequent shelf-life of peach fruit. Postharvest Biol. Technol. 60 7–16. 10.1016/j.postharvbio.2010.09.012 [DOI] [Google Scholar]
- Zhang D., Lopez-Reyes J. G., Spadaro D., Garibaldi A., Gullino M. L. (2010). Efficacy of yeast antagonists used individually or in combination with hot water dipping for control of postharvest brown rot of peaches. J. Plant Dis. Prot. 117 226–232. 10.1007/BF03356365 [DOI] [Google Scholar]
- Zheng X. Y., Hu C. Y., Spooner D., Liu J., Cao J. S., Teng Y. W. (2011). Molecular evolution of Adh and LEAFY and the phylogenetic utility of their introns in Pyrus (Rosaceae). BMC Evol. Biol. 11:255 10.1186/1471-2148-11-255 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Phylogenetic tree of melon medium-chain ADH proteins. The amino acid sequences were aligned by the Clustal Omega program, and the neighborjoining tree was drawn with TreeView. The corresponding GenBank and the melon genome (https://melonomics.net/) were noted in the phylogenetic tree and the accession number in the melon genome were CmADH1 (MELO3C023685P4), CmADH3 (MELO3C026552P1), CmADH4 (MELO3C027151P1), CmADH5 (MELO3C00579 2P1), CmADH6 (MELO3C026553P1), CmADH7 (MELO3C002189P1), CmADH8 (ME LO3C003251P1), CmADH9 (MELO3C011043P1), CmADH10 (MELO3C026554P2), CmADH11 (MELO3C023687P1), and CmFDH1 (MELO3C022399P1). The number for each interior branch was the percentage of bootstraps value (1000 replicates). Black circle denoted 13 CmADHs. ADHs from other plants in our paper are in Supplementary Table S3.
Amino acid sequence alignment of melon CmADH1 (MELO3C023685P4) and CmADH4 (MELO3C027151 P1) with closely related sequences of other plants. GenBank accession numbers are in Supplementary Table S3.
Amino acid sequence alignment of melon CmFDH1 (MELO3C022399P1) with closely related sequences of other plants. GenBank accession numbers are in Supplementary Table S3.
Amino acid sequence alignment of melon CmADH3 (MELO3C026552P1), CmADH6 (MELO3C026553P1) and CmADH10 (MELO3C026554P2) with closely related sequences of other plants. GenBank accession numbers are in Supplementary Table S3.
Amino acid sequence alignment of melon CmADH5 (MELO3C005 792P1) with closely related sequences of other plants. GenBank accession numbers are in Supplementary Table S3.
Amino acid sequence alignment of melon CmADH7 (MELO3C002 189P1) with closely related sequences of other plants. GenBank accession numbers are in Supplementary Table S3.
Amino acid sequence alignment of melon CmADH8 (MELO3C003 251P1) with closely related sequences of other plants. GenBank accession numbers are in Supplementary Table S3.
Amino acid sequence alignment of melon CmADH9 (MELO3C011043 P1) with closely related sequences of other plants. GenBank accession numbers are in Supplementary Table S3.
Amino acid sequence alignment of melon CmADH11 (MELO3C023 687P1) with closely related sequences of other plants. GenBank accession numbers are in Supplementary Table S3.
Amino acid sequence alignment of melon CmADH2 (MELO3C014 897P1) with closely related sequences of other plants. GenBank accession numbers are in Supplementary Table S3.
Amino acid sequence alignment of melon CmADH12 (MELO3C019 503P1) with closely related sequences of other plants. GenBank accession numbers are in Supplementary Table S3.
Transcript abundance of CmADHs in vegetative tissues and developing stages of melon fruit (1–48). R, root; ML, mature leaf; DL, developing leaf; YS, young stem; PF, pistillate flower petal; SF, staminate flower petal. RNA isolated from the samples. Expression analysis was carried out by semi-quantitative PCR using 18S as an internal control.
mRNA abundance of CmADHs in mature green fruit after treatment with ABA and IAA. IAA (Auxin) and ABA (100μM) treatments were given for 2 h as described in Section “Materials and Methods.” Expression analysis was carried out by semi-quantitative PCR as described in section of Hormone treatments using actin as 18S internal control.
CmADHs accumulation during different stages of ethylene and 1-MCP treatment induced ripening of melon by semi-quantitative PCR. 1–12 indicate days after ethylene and 1-MCP treatment. The accumulation level of the genes in untreated melon fruit was control. 18S was used as an internal control.
Alcohol dehydrogenases activity in different melon organs. R, root; ML, mature leaf; DL, developing leaf; YS, young stem; PF, pistillate flower petal; SF, staminate flower petal.
Alcohol dehydrogenases activity in fruits during melon development. 1, 5, 10, 15, 20, 25, 30, 33, 36, 39, 42, 45, and 48 indicate days after anthesis.
Alcohol dehydrogenases activity in fruits during different stages of ethylene and 1-MCP treatment induced ripening of melon. 1, 3, 6, 9, and 12 indicate days after ethylene and 1-MCP treatment. All the treatments have been described in section of Hormone treatments.


