Abstract
Background
Preimplantation genetic testing (PGT) requires an invasive biopsy to obtain embryonic material for genetic analysis. The availability of a less invasive procedure would increase the overall efficacy of PGT. The aim of the study was to explore the potential of blastocoele fluid (BF) as an alternative source of embryonic DNA for PGT.
Methods
Collection of BF was performed by aspiration with a fine needle prior to vitrification. BF DNA was subjected to whole-genome amplification (WGA) and analyzed by high-resolution next-generation sequencing (NGS).
Results
A high-quality WGA product was obtained from 8 of 11 (72.7 %) samples. Comparison of matching BF and blastomere samples showed that the genomic representation of sequencing reads was consistently similar with respect to density and regional coverage across the 24 chromosomes. A genome-wide survey of the sample sequencing data also indicated that BF was highly representative of known single gene sequences, and this observation was validated by PCR analyses of ten randomly selected genes, with an overall efficiency of 84 %.
Conclusion
This study provides further evidence that BF is a promising alternative source of DNA for PGT.
Electronic supplementary material
The online version of this article (doi:10.1007/s10815-016-0667-7) contains supplementary material, which is available to authorized users.
Keywords: Blastocoele fluid, Blastomere, Next-generation sequencing, Preimplantation genetic testing, Bioinformatics
Introduction
Preimplantation genetic testing (PGT) is a commonly used technology for assessing the genetic health of embryos produced by in vitro fertilization (IVF). PGT encompasses both preimplantation genetic diagnosis (PGD) and preimplantation genetic screening (PGS) [1, 2]. PGD is the recommended procedure for couples at high risk for passing on a familial genetic disease [1–3], whereas PGS is mainly applicable to infertile patients for selecting euploid embryos for transfer [1–4]. Over the last 20 years, through the co-development of advanced assisted reproductive and diagnostic technologies, the practice of PGT has been an enormously successful program worldwide [5, 6].
In PGT, a biopsy is necessary for obtaining embryonic material for genetic analysis. Presently, three methods of biopsy are in current practice, namely, polar body biopsy of oocytes, blastomere biopsy of cleavage-stage embryos, and trophectoderm (TE) biopsy of blastocysts [3, 7]. Polar body biopsy, which involves removing the first and second polar bodies, is less invasive but can only be used to detect maternally derived aneuploidies or mutations [8–11]. Blastomere biopsy, which removes one to two cells from the six- to eight-cell embryos, is a more commonly methods to assess both the paternal and maternal contributions to the embryo. However, blastomere biopsy can adversely affect the embryos, particularly when two cells are removed [12–15]. More recently, TE cell biopsy has been more widely adopted for PGT because it can analyze multiple cells of the trophoblast lineage, avoiding the inner mass cells (ICM), which form the fetus proper [7, 12, 13].
While embryo biopsy is generally considered a safe procedure, the quest continues for the development and validation of less invasive methods as an alternative to the current invasive embryo biopsy methods. One promising source of embryonic DNA is blastocoele fluid which is normally removed prior to embryo vitrification to protect the blastocyst from membrane-damaging ice crystal formation [16]. Blastocoele fluid (BF) has been shown to contain small amounts of DNA which are amplifiable by PCR [17]. Three recent studies comparing BF with polar bodies or TE cells biopsied from the corresponding embryos suggest that BF might be predictive of the embryo ploidy [18–20]. If these initial findings can be verified, BF may provide a new source of embryonic DNA for noninvasive PGT. However, several questions remain, including the reliability of retrieving the sample, the nature, and source of BF DNA and whether the DNA is truly representative of the embryonic cells [21].
As an extension of these studies, we have applied next-generation sequencing (NGS) to better understand the nature of BF DNA and determine its genomic representation by comparison to a matching blastomere sample taken at an earlier stage of embryo development. Here, we show that BF DNA is an alternative source of embryonic DNA with potential for both PGD and PGS.
Results
The nature of BF DNA
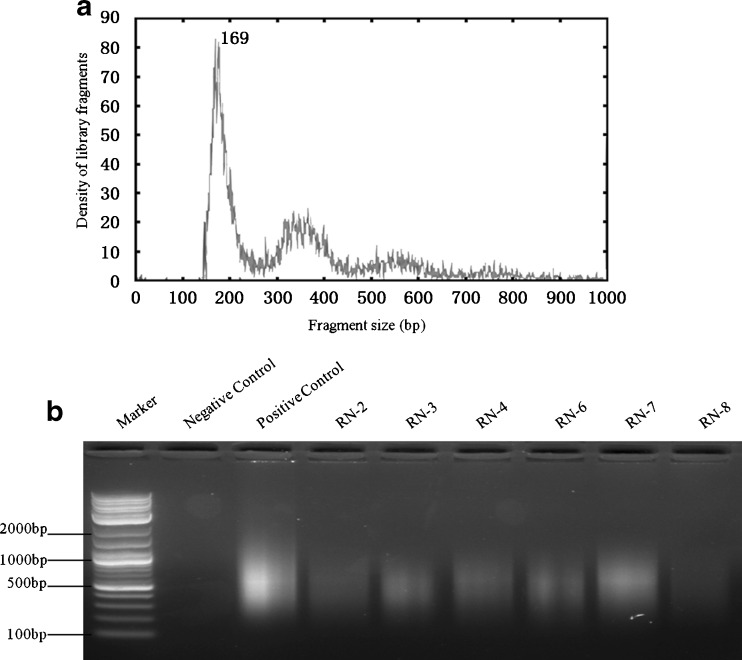
Pooled BF from three research blastocysts was directly analyzed without whole-genome amplification (WGA) by NGS to determine the native population size distribution of fragmented DNA. Fragments were end-modified, ligated with sequencing primer adaptors and libraries generated by PCR as described for analysis of fragmented plasma DNA [22]. Following massively parallel sequencing, the density of sequencing reads was plotted against fragment size after removing the contribution of adaptor sequences (Fig. 1a). Two populations of fragments sizes were observed. The first peak comprised fragments with a range of 160–220 bp and a dominant peak at 169 bp whereas the second peak was broader with fragment sizes ranging from 300 to 400 bp.
Fig. 1.
Analysis of BF DNA before and after WGA. a Size distribution of native BF DNA collected from three blastocysts. Major and minor populations of fragments were observed, ranging in size from 160 to 220 bp and 300 to 400 bp. b Agarose gel electrophoresis of WGA products of six of the eight successful amplification reactions. Products ranged in size between 200 and 1000 bp, and yields were variable between samples
Genomic representation of blastocoele fluid DNA by density maps
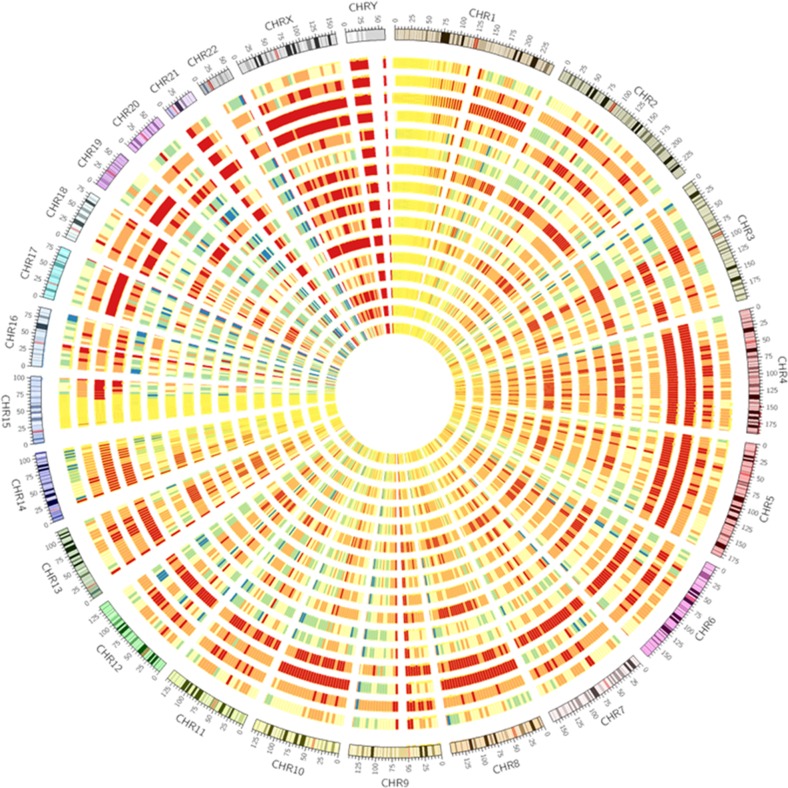
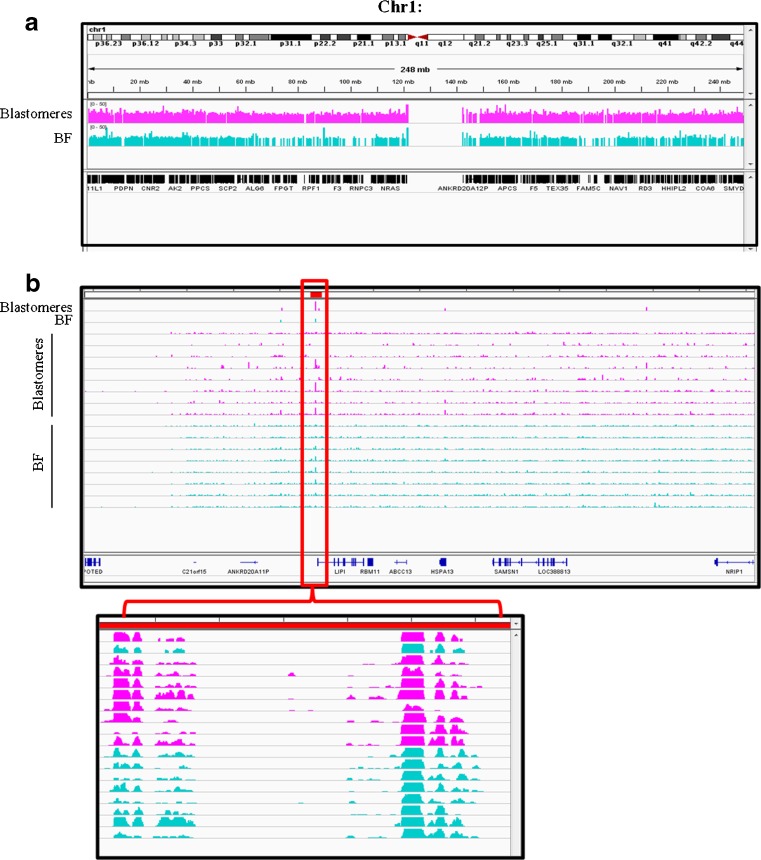
A total of 11 sample pairs with matching blastomere and BF biopsies were collected and subjected to WGA. All 11 blastomere samples produced a high-quality WGA product whereas only 8 of 11 (72.7 %) of BF samples amplified (Fig. 1b) with variable yields after purification (Table 1). There was no correlation between embryo quality and the amount of BF DNA recovered. The eight co-amplified matching samples were then subjected to NGS. Genomic coverage in BF was then benchmarked against genome coverage in blastomeres by dividing the sequencing reads into 5-Mb bins across the 24 chromosomes (Fig. 2). Overall, the density patterns were very similar within each group and between the two groups. There was a relatively consistent coverage of sequencing reads across most regions of the genome, although some regions contained a lower density of sequencing reads. To examine genome coverage in finer detail, chromosome 1 was selected as the representative model chromosome to examine sequencing read distribution at a much higher resolution (Fig. 3). The localized pattern of sequencing reads in BF and blastomere was seen as a series of “islands” with similar density and regional positions, indicating that the WGA coverage was consistently reproducible between the two different starting DNA templates.
Table 1.
Quantification of DNA in BF after WGA
| Sample ID | Blastocyst classification [23] | Day of development to blastocyst | Concentration (ng/μL) |
|---|---|---|---|
| Negative controla | – | – | 0.19 |
| Positive control 1b | – | – | 11.6 |
| Positive control 2b | – | – | 14.5 |
| RN-1 | 4BC | D5 | 19.5 |
| RN-2 | 3AA | D6 | 12.9 |
| RN-3 | 4AA | D5 | 21.0 |
| RN-4 | 5AB | D7 | 20.0 |
| RN-5 | 4BB | D7 | 2.0 |
| RN-6 | 5BA | D5 | 25.3 |
| RN-7 | 4AA | D5 | 25.9 |
| RN-8 | 3AA | D5 | 16.3 |
| RN-9 | 5AA | D7 | 18.7 |
| RN-10 | 5AB | D6 | 2.4 |
| RN-11 | 5BA | D5 | 5.3 |
Samples RN-1 to RN-5 were aspirated from discarded embryos. Samples RN-6 to RN-11 were aspirated from embryos donated by couples undergoing PGD
aConcentration of DNA in negative control <1.0 ng/μL
bConcentration of DNA in positive control >10 ng/μL
Fig. 2.
Chromosome density plots of mapped sequencing reads from matching BF and blastomeres. The outer most group of eight concentric circles represents blastomere samples, and the inner most group of eight concentric circles represent matching BF samples. Sample order for each group from outer to inner was RN-4, RN-9, RN-7, RN-8, RN-2, RN-1, RN-3, and RN-6. Color coding signifies the density of sequencing reads, with red and yellow the highest and lowest densities, respectively
Fig. 3.
High-density plots of blastomere and BF sequencing reads mapped to chromosome 1. The position and density of sequencing read “islands” were similar between blastomere and BF samples
Gene coverage of amplified BF DNA
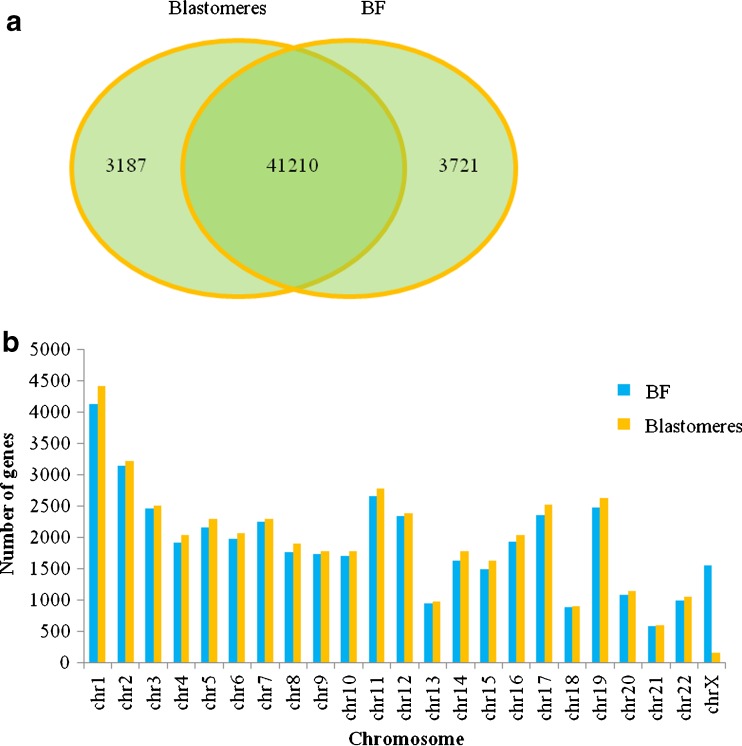
To assess the degree of sequence coverage across gene regions, all mapped BF and blastomere sequencing reads were annotated against the Ensembl database. Across the eight matching samples, sequences for 44,397 genes were detected in BF samples and 44,931 genes were detected in the blastomere samples. Among the detected genes, 41,210 genes were co-detected in both the BF and blastomere samples, suggesting similar gene coverage after WGA (Fig. 4a). Further, the genes distributed across each of the 24 chromosomes were also similar between BF and blastomere samples (Fig. 4b). Lastly, we explored the association of sequencing reads with known OMIM disease genes that are targeted for single gene PGD. Of the 6968 known OMIM genes, 4682 (67.2 %) were detected amongst the sequencing reads. Taken together, these findings suggested that the WGA product derived from the BF may be a suitable template for analyzing single genes.
Fig. 4.
Gene coverage of blastomere and BF samples a Venn diagram showing the overlap of genes identified in the two sample types. b Bar plot comparing the number of observed genes from the two sample types located to each chromosome. Across each chromosome, the gene coverage in BF samples was similar to that in blastomere samples
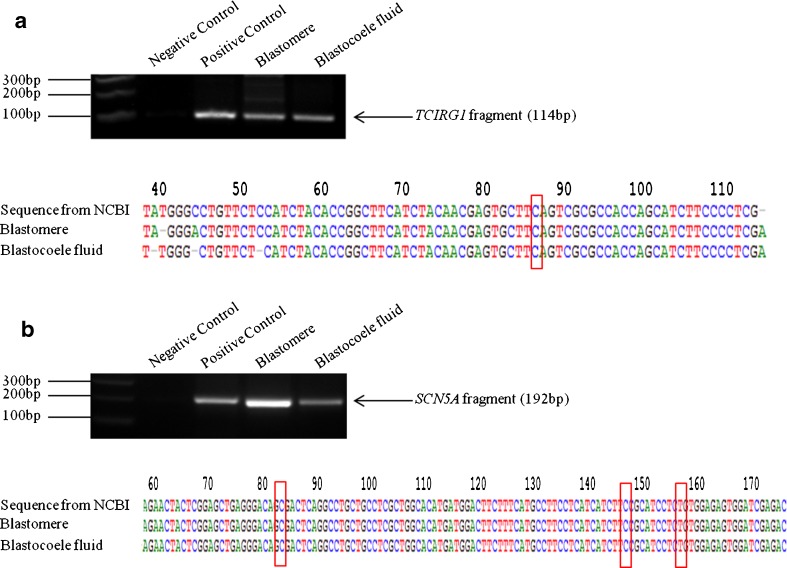
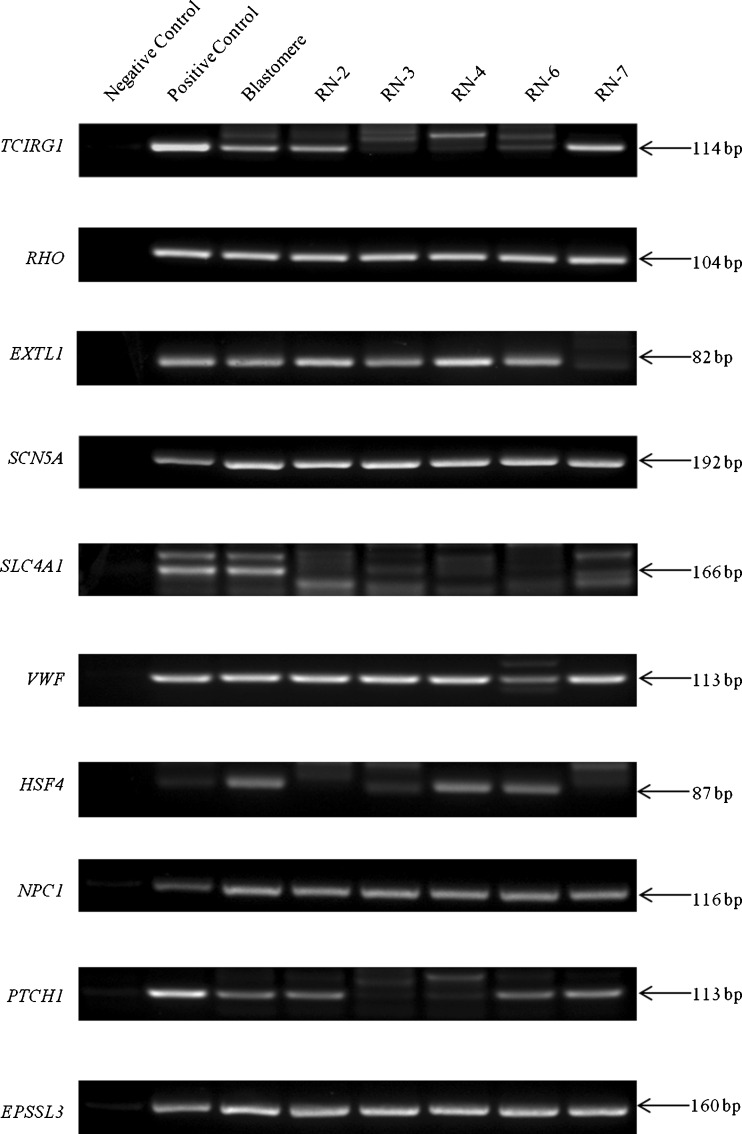
To validate the bioinformatics gene data, we initially performed gene-specific PCR for genes, TCIRG1 and SCN5A associated with autosomal recessive osteopetrosis and long QT syndrome type 3, respectively. Primers were designed to cover the common mutation sites (Table S1). In matching BF and blastomere samples, a band of the expected size for the TCIRG1 and SCN5A genes was amplified and confirmed by Sanger sequencing (Fig. 5). Using the remaining WGA template available from five of the eight BF samples, we extended the gene validation experiments to encompass a further eight randomly selected disease genes, namely, RHO, EXTL1, SLC4A1, VWF, HSF4, NPC1, PTCH1, and EPS8L3 (Fig. 6). The majority of PCR reactions (42 out of 50) produced positive bands for the ten test genes, giving an overall gene amplification efficiency of 84 %.
Fig. 5.
PCR validation for the presence of single gene DNA in a matching blastomere and BF sample. a TCIRG1 gene fragment (114 bp). b SCN5A fragment (192 bp). The red boxes mark the position of a common hotspot mutation in the two genes. Sanger sequencing confirmed that the two amplified fragments had the correct nucleotide sequence
Fig. 6.
PCR validation for detection of randomly selected genes in WGA products from five BF samples. The majority of samples showed a PCR fragment of the expected size for the ten test genes
Discussion
The aim of PGT is to obtain biopsy material that is representative of the genetic constitution of the whole embryo under test, without compromising developmental potential. Currently, invasive methods such as blastomere and TE biopsy are used for this purpose and, with training, can be safely performed with high precision in a clinical embryology laboratory [24, 25]. Nonetheless, a less invasive method such as blastocentesis would be preferable as an alternative source of embryonic DNA because it is routinely harvested prior to vitrification [16, 26]. In our study, we report the application of NGS to evaluate the nature and genomic representation of the DNA found in the BF of embryos donated to research. As a benchmark, we directly compared the sequencing profiles of BF with sister blastomeres removed prior to preimplantation development to the blastocyst stage. Overall, at the chromosome and gene level, the sequencing profiles generated from the WGA products were remarkably similar, suggesting that BF has potential as an alternative template for both PGS and PGD.
It has been proposed that the BF DNA originates from cell death by apoptosis of trophoblastic or ICM of the developing blastocyst [27, 28]. Therefore, understanding the nature of BF DNA is critical for the development of reliable and accurate PGT methods. By sequencing, we identified two fragmentation patterns of the native DNA, comprising a dominant population of 160–220 bp (major peak at 169 bp) and a minor population of 300–400 bp. The dominant population of DNA fragments is very similar in size range to that seen in the circulating plasma of human blood [22]. This DNA size is believed to mimic the nucleosome spacing on the DNA strand which then undergoes cleavage by blood DNA nucleases to release the exposed DNA strand and create a population of fragmented DNA [29]. However, although the BF DNA was found to be highly fragmented with significant size heterogeneity, it was possible to generate a typical WGA product similar to that derived from a single blastomere, where the starting material is intact genomic DNA. From the analysis of 11 BF samples, we achieved a WGA efficiency of 72.5 %, although the quality and yield were variable based on gel analysis. In four recent studies of BF, the reliability of WGA using PCR-based methods was 60 of 96 (63 %) [20], 39 of 51 (76.5 %) [18], 4 of 5 (80 %) [17], and 95 of 116 (82 %) [19], respectively. We speculate that WGA failure observed in these initial studies was either due to technical problems in retrieving a very small volume (in the order of 1 nL) or that some blastocysts were actually devoid of BF DNA. Accordingly, these collective studies suggest that it may not always be possible to obtain reliable genetic results on all embryos using BF.
Through precise mapping of sequencing reads derived from BF DNA, we identified chromosome density read profiles that were very similar to those produced from corresponding sister blastomeres. In addition, coverage of sequencing reads was relatively uniform across the 24 chromosomes and patterns were reproducible from one BF sample to another. On the basis of our bioinformatic analyses, the BF WGA products should in theory be suitable as a template for chromosomal analysis by techniques such as array comparative genomic hybridization (CGH) and NGS. In support of this notion, recent array CGH studies provide the first clinical evidence that interpretable 24 chromosomal profiles can be generated from amplified BF DNA [18–20]. Based on comparison to polar body [18] or TE biopsy [19, 20] results, there was a high degree of diagnostic concordance for euploid and aneuploid embryos using the corresponding BF. However, there were significant numbers of discordant embryo profiles. The reasons for the discordance are debatable but, at a biological level, may involve self-correction of existing errors or the generation of new chromosomal errors during preimplantation development [30]. At a technical level, difficulties in sample retrieval or sub-optimal WGA BF reactions for array analysis may have contributed in part to the discordant results. Further comparative studies of BF and TE biopsies taken from the same PGS embryos, preferably using a much higher-resolution diagnostic technique such as NGS, are now required to identify the biological and physical limitations of BF as a DNA template for PGS.
Based on gene annotations, we found that the BF WGA product contained sequences of the majority of known genes and, specifically, disease genes, which are commonly the subject of different single gene PGD cases. In a limited survey, we showed that hotspot mutation regions of ten randomly selected genes could be amplified with a relatively high efficiency of 84 %. However, this efficiency is significantly lower than 90–95 % which can be consistently achieved following WGA of a single or few cells [31, 32]. Further, when amplification efficiency is low for a given set of primers, concomitantly, allelic dropout (ADO) rates are also generally higher [33]. Although ADO rates were not investigated in this study, it is therefore possible that ADO rates from a BF WGA product could be consistently higher than that from a blastomere WGA product. On this basis, the application of PCR-based methods involving mutation detection and linked STRs may be a sub-optimal approach for single gene PGD using BF. Nonetheless, a more genome-wide technology such as karyomapping [34–37] that can analyze a large number of heterogeneous intragenic and intergenic linked SNPs might serve as a more robust approach to single gene PGD using a BF template.
In conclusion, BF represents a promising noninvasive approach to PGT. Future studies are warranted before BF can be considered for clinical PGD, including determining the source of BF DNA, improving the methods of BF collection and amplification by WGA, and determining the diagnostic concordance between BF and the embryo at the chromosomal and single gene level. These studies are best suited in a clinical PGD setting where standard methods of embryo biopsy, molecular diagnosis, and vitrification are routinely performed. Based on the key findings of the study, we propose that NGS and karyomapping would seem to be the ideal technologies with the highest potential to extract the vital chromosome and single gene diagnostic information from a BF WGA product.
Materials and methods
Blastomere biopsy
Blastomere biopsy was performed on eight-cell cleavage-stage embryos as previously described [38]. In brief, the embryo was removed from the culture and transferred to a droplet of the biopsy medium (COOK Medical) overlayed with paraffin oil. The embryo was immobilized by a holding pipette connected to an oil-filled syringe and mounted on a micromanipulator (Research Instruments), and a slit was created in the zona pellucida using a Saturn Laser System (Research Instruments). A single blastomere was gently aspirated using a blunt flame-polished biopsy pipette, washed three times in phosphate-buffered saline (PBS), transferred to a PCR tube with 2.5 μL of PBS, and stored at −80 °C until further processing.
BF collection
Following blastomere biopsy, embryos were cultured to the blastocyst stage. The method for aspirating BF was similar to that described in other studies [17, 39], with minor modifications. The expanded blastocyst was fixed by a holding pipette, and BF was completely aspirated from the blastocyst with a microneedle, which was inserted into the point of contact between two TE cells (Fig. S1). During this process, special attention was given to avoid aspirating any cellular material or debris. The aspirated fluid was transferred into a 2.5-μL droplet of PBS that was prepared prior to the operation. The droplet containing BF was finally transferred to a PCR tube and then stored at −80 °C until further processing.
WGA and next-generation sequencing
PicoPLEX WGA Kit (New England Biolabs) was used to amplify embryonic DNA from blastomere and BF samples, according to the manufacturer’s instructions. The WGA products were purified from unincorporated primers using the Agencourt AMpure XP Kit (BECKMAN COULTER). The amplified DNA was analyzed on 1.5 % (w/v) agarose gel electrophoresis to determine the quality and size distribution of the WGA products. In addition, quantification of the amplified DNA was performed using a NanoDrop 2000 spectrophotometer (Thermo Scientific, Germany). Successfully amplified DNA was used to construct sequencing libraries and high-throughput sequencing performed on the HiSeq 2500 Illumina platform [38] to generate 45-bp reads with 36 bp of genomic DNA sequence.
Bioinformatics analysis of sequencing data
The adapter sequence from the total raw reads of each sample was removed and sequences with a base quality score greater than 20 selected using Cutadapt software (version 1.2.1) [40]. Processed reads with a length of less than 18 nucleotides or with ambiguous nucleotides were discarded using Trimmomatic software (version 0.30) [41]. The remaining high-quality reads were aligned to the hg19 human genome by BWA software, with up to two mismatches allowed [42]. Mapped locations were only reported for those with the minimum number of observed mismatches for each read. The base depth of the sequencing reads was calculated by BEDTools [43].
PCR for disease-associated genes
Primers sequences for amplification of specific regions of the genes TCIRG1, SCN5A, RHO, EXTL1, SLC4A1, VWF, HSF4, NPC1, PTCH1, and EPS8L3 are shown in Table S1. PCR was performed in a final volume of 30 μL with DNA template (10–40 ng) and primers added to a 2× Taq PCR Master Mix (Aidlab). The DNA template comprised WGA products of BF or blastomeres. Human cDNA was used as the positive amplification control. Nuclease-free water was used as a negative control. The amplification conditions for TCIRG1, SCN5A, RHO, EXTL1, SLC4A1, VWF, HSF4, NPC1, PTCH1, and EPS8L3 were one cycle of 95 °C for 5 min and 40 cycles of 95 °C for 30 s, 53–55 °C for 30 s, and 72 °C for 15–20 s and one cycle of 72 °C for 5 min. PCR products were analyzed employing 2 % (w/v) agarose gel electrophoresis to determine the presence of the predicted amplicons. Sanger sequencing was used to validate the PCR products.
Electronic supplementary material
Below is the link to the electronic supplementary material.
(GIF 66 kb)
(DOCX 27 kb)
Acknowledgments
The study was supported by the grant awarded to Yuanqing Yao by the Key Program of the “Twelfth Five-Year Plan” of the People’s Liberation Army (No. BWS11J058) and the National High Technology Research and Development Program (2015AA020402).
Compliance with ethical standards
Ethical approval
This study was approved by the Institutional Review Board of Chinese PLA General Hospital (S2013-092-02). All embryos donated to research were obtained after obtaining informed written consent by couples undertaking PGS.
Contributor Information
Baofa Sun, Phone: +8610 84097656, Email: sunbf@big.ac.cn.
Yuanqing Yao, Phone: +861066939258, Email: yuanqingy@sina.cn.
References
- 1.Brezina PR, Brezina DS, Kearns WG. Preimplantation genetic testing. BMJ. 2012;345 doi: 10.1136/bmj.e5908. [DOI] [PubMed] [Google Scholar]
- 2.Brezina PR, Kutteh WH. Clinical applications of preimplantation genetic testing. BMJ. 2015;350:g7611. doi: 10.1136/bmj.g7611. [DOI] [PubMed] [Google Scholar]
- 3.Yan L, Wei Y, Huang J, Zhu X, Shi X, Xia X, et al. Advances in preimplantation genetic diagnosis/screening. Sci Chin Life Sci. 2014;57(7):665–71. doi: 10.1007/s11427-014-4683-5. [DOI] [PubMed] [Google Scholar]
- 4.Brezina PR, Ke RW, Kutteh WH. Preimplantation genetic screening: a practical guide. Clin Med Insights Reprod Health. 2013;7:37–42. doi: 10.4137/CMRH.S10852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Simpson JL. Preimplantation genetic diagnosis at 20 years. Prenat Diagn. 2010;30(7):682–95. doi: 10.1002/pd.2552. [DOI] [PubMed] [Google Scholar]
- 6.SenGupta SB, Delhanty JD. Preimplantation genetic diagnosis: recent triumphs and remaining challenges. Expert Rev Mol Diagn. 2012;12(6):585–92. doi: 10.1586/erm.12.61. [DOI] [PubMed] [Google Scholar]
- 7.Milachich T. New advances of preimplantation and prenatal genetic screening and noninvasive testing as a potential predictor of health status of babies. BioMed Res Int. 2014;2014:306505. doi: 10.1155/2014/306505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fiorentino F, Biricik A, Nuccitelli A, De Palma R, Kahraman S, Sertyel S, et al. Rapid protocol for pre-conception genetic diagnosis of single gene mutations by first polar body analysis: a possible solution for the Italian patients. Prenat Diagn. 2008;28(1):62–4. doi: 10.1002/pd.1905. [DOI] [PubMed] [Google Scholar]
- 9.Capalbo A, Bono S, Spizzichino L, Biricik A, Baldi M, Colamaria S, et al. Reply: questions about the accuracy of polar body analysis for preimplantation genetic screening. Hum Reprod. 2013;28(6):1733–6. doi: 10.1093/humrep/det070. [DOI] [PubMed] [Google Scholar]
- 10.Hou Y, Fan W, Yan L, Li R, Lian Y, Huang J, et al. Genome analyses of single human oocytes. Cell. 2013;155(7):1492–506. doi: 10.1016/j.cell.2013.11.040. [DOI] [PubMed] [Google Scholar]
- 11.Salvaggio CN, Forman EJ, Garnsey HM, Treff NR, Scott RT., Jr Polar body based aneuploidy screening is poorly predictive of embryo ploidy and reproductive potential. J Assist Reprod Genet. 2014;31(9):1221–6. doi: 10.1007/s10815-014-0293-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dahdouh EM, Balayla J, Audibert F, Wilson RD, Audibert F, Brock JA, et al. Technical update: preimplantation genetic diagnosis and screening. J Obstet Gynaecol Can: JOGC = J D’obstet Gynecol Du Can: JOGC. 2015;37(5):451–63. doi: 10.1016/s1701-2163(15)30261-9. [DOI] [PubMed] [Google Scholar]
- 13.Scott KL, Hong KH, Scott RT., Jr Selecting the optimal time to perform biopsy for preimplantation genetic testing. Fertil Steril. 2013;100(3):608–14. doi: 10.1016/j.fertnstert.2013.07.004. [DOI] [PubMed] [Google Scholar]
- 14.Scott RT, Jr, Upham KM, Forman EJ, Zhao T, Treff NR. Cleavage-stage biopsy significantly impairs human embryonic implantation potential while blastocyst biopsy does not: a randomized and paired clinical trial. Fertil Steril. 2013;100(3):624–30. doi: 10.1016/j.fertnstert.2013.04.039. [DOI] [PubMed] [Google Scholar]
- 15.Goossens V, De Rycke M, De Vos A, Staessen C, Michiels A, Verpoest W, et al. Diagnostic efficiency, embryonic development and clinical outcome after the biopsy of one or two blastomeres for preimplantation genetic diagnosis. Hum Reprod. 2008;23(3):481–92. doi: 10.1093/humrep/dem327. [DOI] [PubMed] [Google Scholar]
- 16.Mukaida T, Oka C, Goto T, Takahashi K. Artificial shrinkage of blastocoeles using either a micro-needle or a laser pulse prior to the cooling steps of vitrification improves survival rate and pregnancy outcome of vitrified human blastocysts. Hum Reprod. 2006;21(12):3246–52. doi: 10.1093/humrep/del285. [DOI] [PubMed] [Google Scholar]
- 17.Palini S, Galluzzi L, De Stefani S, Bianchi M, Wells D, Magnani M, et al. Genomic DNA in human blastocoele fluid. Reprod Biomed Online. 2013;26(6):603–10. doi: 10.1016/j.rbmo.2013.02.012. [DOI] [PubMed] [Google Scholar]
- 18.Gianaroli L, Magli MC, Pomante A, Crivello AM, Cafueri G, Valerio M, et al. Blastocentesis: a source of DNA for preimplantation genetic testing. Results from a pilot study. Fertil Steril. 2014;102(6):1692–9. doi: 10.1016/j.fertnstert.2014.08.021. [DOI] [PubMed] [Google Scholar]
- 19.Magli MC, Pomante A, Cafueri G, Valerio M, Crippa A, Ferraretti AP, et al. Preimplantation genetic testing: polar bodies, blastomeres, trophectoderm cells, or blastocoelic fluid? Fertil Steril. 2015 doi: 10.1016/j.fertnstert.2015.11.018. [DOI] [PubMed] [Google Scholar]
- 20.Tobler KJ, Zhao Y, Ross R, Benner AT, Xu X, Du L, et al. Blastocoel fluid from differentiated blastocysts harbors embryonic genomic material capable of a whole-genome deoxyribonucleic acid amplification and comprehensive chromosome microarray analysis. Fertil Steril. 2015;104(2):418–25. doi: 10.1016/j.fertnstert.2015.04.028. [DOI] [PubMed] [Google Scholar]
- 21.Cohen J, Grudzinskas G, Johnson MH. Embryonic DNA sampling without biopsy: the beginnings of non-invasive PGD? Reprod Biomed Online. 2013;26(6):520–1. doi: 10.1016/j.rbmo.2013.03.001. [DOI] [PubMed] [Google Scholar]
- 22.Liang D, Lv W, Wang H, Xu L, Liu J, Li H, et al. Non-invasive prenatal testing of fetal whole chromosome aneuploidy by massively parallel sequencing. Prenat Diagn. 2013;33(5):409–15. doi: 10.1002/pd.4033. [DOI] [PubMed] [Google Scholar]
- 23.Gardner DK, Lane M, Stevens J, Schlenker T, Schoolcraft WB. Blastocyst score affects implantation and pregnancy outcome: towards a single blastocyst transfer. Fertil Steril. 2000;73(6):1155–8. doi: 10.1016/S0015-0282(00)00518-5. [DOI] [PubMed] [Google Scholar]
- 24.Lukaszuk K, Pukszta S, Wells D, Cybulska C, Liss J, Plociennik L, et al. Routine use of next-generation sequencing for preimplantation genetic diagnosis of blastomeres obtained from embryos on day 3 in fresh in vitro fertilization cycles. Fertil Steril. 2015;103(4):1031–6. doi: 10.1016/j.fertnstert.2014.12.123. [DOI] [PubMed] [Google Scholar]
- 25.Chang LJ, Huang CC, Tsai YY, Hung CC, Fang MY, Lin YC, et al. Blastocyst biopsy and vitrification are effective for preimplantation genetic diagnosis of monogenic diseases. Hum Reprod. 2013;28(5):1435–44. doi: 10.1093/humrep/det048. [DOI] [PubMed] [Google Scholar]
- 26.Vanderzwalmen P, Bertin G, Debauche C, Standaert V, van Roosendaal E, Vandervorst M, et al. Births after vitrification at morula and blastocyst stages: effect of artificial reduction of the blastocoelic cavity before vitrification. Hum Reprod. 2002;17(3):744–51. doi: 10.1093/humrep/17.3.744. [DOI] [PubMed] [Google Scholar]
- 27.Hardy K, Handyside AH, Winston RM. The human blastocyst: cell number, death and allocation during late preimplantation development in vitro. Development. 1989;107(3):597–604. doi: 10.1242/dev.107.3.597. [DOI] [PubMed] [Google Scholar]
- 28.Hardy K, Stark J, Winston RM. Maintenance of the inner cell mass in human blastocysts from fragmented embryos. Biol Reprod. 2003;68(4):1165–9. doi: 10.1095/biolreprod.102.010090. [DOI] [PubMed] [Google Scholar]
- 29.Mouliere F, Rosenfeld N. Circulating tumor-derived DNA is shorter than somatic DNA in plasma. Proc Natl Acad Sci U S A. 2015;112(11):3178–9. doi: 10.1073/pnas.1501321112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Taylor TH, Gitlin SA, Patrick JL, Crain JL, Wilson JM, Griffin DK. The origin, mechanisms, incidence and clinical consequences of chromosomal mosaicism in humans. Hum Reprod Update. 2014;20(4):571–81. doi: 10.1093/humupd/dmu016. [DOI] [PubMed] [Google Scholar]
- 31.Zong C, Lu S, Chapman AR, Xie XS. Genome-wide detection of single-nucleotide and copy-number variations of a single human cell. Sci (New York, NY) 2012;338(6114):1622–6. doi: 10.1126/science.1229164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu D, Liu C, DeVries S, Waldman F, Cote RJ, Datar RH. LM-PCR permits highly representative whole genome amplification of DNA isolated from small number of cells and paraffin-embedded tumor tissue sections. Diagn Mol Pathol: Am J Surg Pathol, Part B. 2004;13(2):105–15. doi: 10.1097/00019606-200406000-00007. [DOI] [PubMed] [Google Scholar]
- 33.Lam CW, Mak CM. Allele dropout caused by a non-primer-site SNV affecting PCR amplification—a call for next-generation primer design algorithm. Clinica Chimica Acta. Int J Clin Chem. 2013;421:208–12. doi: 10.1016/j.cca.2013.03.014. [DOI] [PubMed] [Google Scholar]
- 34.Natesan SA, Bladon AJ, Coskun S, Qubbaj W, Prates R, Munne S, et al. Genome-wide karyomapping accurately identifies the inheritance of single-gene defects in human preimplantation embryos in vitro. Genet Med: Off J Am Coll Med Genet. 2014;16(11):838–45. doi: 10.1038/gim.2014.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gimenez C, Sarasa J, Arjona C, Vilamajo E, Martinez-Pasarell O, Wheeler K, et al. Karyomapping allows preimplantation genetic diagnosis of a de-novo deletion undetectable using conventional PGD technology. Reprod Biomed Online. 2015;31(6):770–5. doi: 10.1016/j.rbmo.2015.08.017. [DOI] [PubMed] [Google Scholar]
- 36.Konstantinidis M, Prates R, Goodall NN, Fischer J, Tecson V, Lemma T, et al. Live births following Karyomapping of human blastocysts: experience from clinical application of the method. Reprod Biomed Online. 2015;31(3):394–403. doi: 10.1016/j.rbmo.2015.05.018. [DOI] [PubMed] [Google Scholar]
- 37.Berger VK, Baker VL. Preimplantation diagnosis for single gene disorders. Semin Reprod Med. 2014;32(2):107–13. doi: 10.1055/s-0033-1363552. [DOI] [PubMed] [Google Scholar]
- 38.Wang L, Cram DS, Shen J, Wang X, Zhang J, Song Z, et al. Validation of copy number variation sequencing for detecting chromosome imbalances in human preimplantation embryos. Biol Reprod. 2014;91(2):37. doi: 10.1095/biolreprod.114.120576. [DOI] [PubMed] [Google Scholar]
- 39.D’Alessandro A, Federica G, Palini S, Bulletti C, Zolla L. A mass spectrometry-based targeted metabolomics strategy of human blastocoele fluid: a promising tool in fertility research. Mol BioSyst. 2012;8(4):953–8. doi: 10.1039/C1MB05358B. [DOI] [PubMed] [Google Scholar]
- 40.Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 2011;17:10–2. doi: 10.14806/ej.17.1.200. [DOI] [Google Scholar]
- 41.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–20. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li H, Durbin R. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics. 2009;25:1754–60. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26(6):841–2. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(GIF 66 kb)
(DOCX 27 kb)