Abstract
Infertility is a complex disorder with multiple genetic and environmental causes. Although some specific mutations have been identified, other factors responsible for sperm defects remain largely unknown. Despite considerable efforts to identify the pathophysiology of the disease, we cannot explain the underlying mechanisms of approximately half of infertility cases. This study reviews current data on epigenetic regulation and idiopathic male infertility. Recent data have shown an association between epigenetic modifications and idiopathic infertility. In this regard, epigenetics has emerged as one of the promising research areas in understanding male infertility. Many studies have indicated that epigenetic modifications, including DNA methylation in imprinted and developmental genes, histone tail modifications and short non-coding RNAs in spermatozoa may have a role in idiopathic male infertility.
Keywords: Infertility, Epigenetics, DNA methylation, miRNA
Introduction
Infertility is described as the inability to conceive after at least a year of unprotected intercourse [1] and influences about 15 % of couples worldwide [2]. Male infertility affects approximately 7 % of men [3]. Infertility is a heterogeneous disorder that may be a result of genetic or environmental factors or both. Karyotypic abnormalities [4, 5], microdeletions on Y chromosome, and cystic fibrosis transmembrane conductance regulator (CFTR) gene mutations are well known genetic causes of infertility in azoospermic or severely oligozoospermic men. Recently, some copy number variations (CNV) have been described to be associated with severe oligozoospermia or Sertoli-cell-only (SCO) syndrome or both [6]. In addition, some autosomal deletions, rare X-linked CNV, DNA repair mechanism defects, Y-linked syndromes, and some single nucleotide polymorphisms (SNPs) have been found to be associated with male factor infertility [7–13]. Although some specific mutations have been identified, other factors responsible for the sperm defects remain unknown. Known genetic causes of male infertility make up approximately 30 % of infertility cases [4, 14, 15]. The causes of approximately 50 % of male factor infertility cases are still unexplained [4, 16]. These numbers highlight an urgent need for reliable diagnostic tools to identify the underlying mechanisms of male infertility.
Epigenetics refers to heritable and reversible forms of gene activity and expression without any modification of DNA sequences. These epigenetic modifications can be inherited through both mitotic and meiotic divisions. Recent studies demonstrate that aberrant DNA methylation of imprinted genes and reproduction-related genes in particular might be helpful to explain unknown infertility cases [17–22]. Therefore, epigenetics appears to be a promising research area for studying idiopathic male infertility.
Male infertility is a heterogeneous disorder that can result from aberrations in and interactions of multiple genes. Many studies have shown an association between idiopathic infertility and aberrant DNA methylation of whole genome or some genes in spermatozoa. Moreover, several recent studies have indicated a role for short non-coding RNAs and different histone tail modifications in infertility. In this review, we aim to summarize the current data on epigenetic mechanisms that are involved in idiopathic male infertility.
DNA methylation and male infertility
DNA methylation is the addition of a methyl group from S-adenosil-methionine to the fifth position of the cytosine ring (5meC) in the CpG islands (CGIs). CGIs are short interspersed C + G-rich DNA sequences and are localized in the promoters or regulatory regions of almost all housekeeping genes, developmental genes, and some tissue-specific genes [23], [24]. Methylation of these cytosines is correlated with inactivation or silencing of the associated promoter, whereas hypomethylation usually leads to activation of gene expression [24, 25]. Silencing of gene expression is either due to inhibition of transcription factor binding to methylated cytosines or repression mediated by methyl-CpG-binding proteins [24, 26].
DNA methylation is catalyzed by maintenance DNA methyltransferases (DNMTs) (DNMT1) and de novo DNMTs (DNMT3A, DNMT3B, DNMT3L) [27]. DNMT1 is responsible for maintenance of DNA methylation during DNA replication and termed as maintenance methyltransferase. DNMT3A, DNMT3B, and DNMT3L mediate de novo methylation of genomic DNA during early phase of embryonic development specifically in germ cells, and their activities are strictly essential for proper spermatogenesis. Indeed, conditional Dnmt3a knockout mice studies revealed impaired spermatogenesis and aberrant paternal imprinting in spermatogonia [28]. However, Dnmt3l null male mice germ cells showed hypomethylation at imprinted maternally expressed transcript (H19) DMR and several CG poor regions and delayed entry into meiosis [29]. Hypomethylation of paternally imprinted H19 DMR reported in this study may be explained with deficiency of DNMT3L. Recently, Cheng and colleagues reported a significant association between DNMT1 polymorphisms (rs16999593, rs2228612, and rs2228611) and oligozoospermia compared to fertile controls in a Chinese population. The role of these polymorphisms in the pathogenesis of oligozoospermia remains unknown but these polymorphisms may lead to a decrease in DNMT1 expression levels and impaired spermatogenesis [30].
Methylation of imprinted genes
Sperm cells have unique DNA methylation patterns that are formed during early stages of spermatogenesis and are essential for proper sperm production and spermatogenesis [31]. Some genes are imprinted differentially by DNA methylation depending on which parent they are inherited from, which causes alterations in gene expression depending on the allele transmitted from the father or the mother. All methylation marks of primordial germ cells (PGCs) are erased during the development of embryo between 8 and 13.5 days post coitum (dpc) [31–33]. DNA demethylation can be achieved by two mechanisms: passive and active demethylation. Passive demethylation is replication-dependent and may occur during cell division and mammalian development as a result of loss or erasure of 5-methylcytosines (5mC) caused by loss of maintenance by defective DNMTs [34]. However, active demethylation involves conversion of 5mC to 5-hydroxymethylcytosine (5hmC), 5-formylcytosine (5fC), and 5-carboxylcytosine (5caC) in three consecutive oxidation reactions catalyzed by a family of DNA hydroxylases called ten-eleven-translocation (TET) proteins. Following TET oxidation, multiple subsequent enzymatic and DNA repair reactions complete the process of active DNA demethylation, resulting in generation of unmethylated cytosines [35]. After this demethylation process, a specific remethylation program starts at around 15.5 dpc in spermatogonia and type I spermatocytes; hence, spermatozoa transmit the paternal imprint [36, 37]. Genomic regions that exhibit differential methylation depending on parental origin are called differentially methylated regions (DMRs). In humans, ejaculated and mature spermatozoal DNA should be methylated in the paternal DMRs, but unmethylated in the maternal DMRs [38]. Igf2/H19, Rasgrf1, Dlk1-Gtl2, and Zdbf2 loci of spermatozoal genome are methylated only in male germ cells and not expressed in male cells [39]. Many imprinted genes are involved in the regulation of growth and development [40]. On the other hand, several genes are methylated in female germ cells and expressed only in males. Mesodermal-specific transcript (MEST), also known as paternally expressed gene 1 (PEG1), encodes a member of the alpha/beta hydrolase family and maps to 7q32 and is imprinted during fetal development with monoallelic paternal expression. The MEST gene functions in alpha/beta hydroxylase folding and is important in the development of fetal mesoderm [41]. ZAC protein I is another paternally expressed gene and induces G1 cell-cycle arrest and apoptosis [42]. Paternally expressed 3 (PEG3) is a paternally expressed gene and may play role in the p53-mediated apoptotic pathway [43]. Paternally expressed small nuclear ribonucleoprotein polypeptide N (SNRPN) plays role in pre-messenger RNA (mRNA) processing likely through tissue-specific alternative splicing and is located on chromosome 15q11-13 [44]. Methylated region of long QT intronic transcript 1 (DMR-LIT1) is an imprinting control region, and its demethylation is associated with Beckwith-Wiedemann Syndrome [45]. Several studies have demonstrated a significant association between methylation statuses of both maternally and paternally imprinted genes and sperm abnormalities and are summarized in Table 1 [22, 38, 57, 58, 62–65] (Table 1).
Table 1.
Summary of studies investigating the relationship between spermatozoal DNA methylation and male infertility
| Gene | Study group (sample size) |
Summary of findings | Reference |
|---|---|---|---|
| Genome-wide DNA methylation | |||
| Global | Normospermic fertile men (17) and normospermic infertile patients (29) | • Aberrant methylation regions are locus-specific • Genome of spermatozoa is hypomethylated |
[17] |
| Global | Male (20) “High quality sperm” vs. “Poor quality sperm” |
• No global methylation • No differential methylation in CGIs 772 significant regional methylation alternations |
[46] |
|
H19 and DAZL
LINE1 |
Oligozoospermic men (20) Asthenozoospermic men (20) Normozospermic men (20) |
• No association between asthenozoospermia and normozospermia in H19-DMR
• Severe hypomethylation pattern at H19-DMR CTCF-binding site 6 DAZL promoter methylated only in infertile patents |
[47] |
| Global | Infertile males (38) | • Hypermethylation in spermatogenesis- related genes • Loss of methylation in inflammation and immune response-related genes |
[18] |
| Global | NOA (65) and oligoastenozoospermic (29) | • 78 hypomethylated sites • 143 hypermethylated sites in NOA compared oligoastenozoospermia |
[19] |
| Global | Cell-free seminal DNA of Normozospermic men (12) and post vasectomy (11) | • 367 testis and epididymis specific hypomethylated genes • 134 hypermethylated |
[11] |
| Global | OAT (69) | • Broad epigenetic defects associated with abnormal semen parameters | [20] |
| Global | Infertile males (7) | • 5/7 infertile men had non-programmatic histone • No difference in localization of H3 lysine 4 methylation (H3K4me) or H3 lysine 27 methylation (H3K27me) in the gametes of infertile men compared with fertile men • No single locus displays a complete change in chromatin packaging or DNA modification • Reduction in the amount of H3K4me or H3K27me retained at developmental transcription factors and certain imprinted genes |
[21] |
| Gene-specific DNA methylation | |||
| DDR1 | NOA (16) and fertile normospermic men (5) | • Aberrant DNA methylation and expression of DDR1 | [48] |
| MTHFR, SNRPN | OAT (27) and control (11) | • Low motility • Poor morphology |
[49] |
|
MTHFR
H19 |
Infertile males (20) | • Methylation defects at the H19 locus associated with hypermethylation at MTHFR promoter | [22] |
| GTF2A1L | NOA (86): Normozospermia (26) Hypospermatogenesis (17) SCO (26) |
• Aberrant TDMR methylation at the GTF2A1L promoter associated with hypospermatogenesis • Testicular sperm extraction (TESE) technique may be used to overcome male infertility due to aberrant TDMR methylation |
[50] |
| MEST | 212 consecutive infertile patients: Normozoospermic (31) volunteers (single samples) and Normozoospermic volunteers (10) | • 23 % of patient cohort displayed an aberrant MEST DNA methylation • MEST DNA methylation associated with oligozoospermia, decreased bi-testicular volume and increased FSH levels • DNA methylation in normozoospermic volunteers was stable over a time period of up to 951 days in contrast to classical semen parameters |
[51] |
| RHOX homeobox genes | Infertile males (140) | • Aberrantly regulated in infertile patients | [52] |
|
PEG1/MEST
and H19 |
Normospermic (119) and azoospermic 175 | • PEG1/MEST methylation in 20 % and H19-DMR in 3 % of oligozoospermic men | [53] |
| MTHFR | NOA (50) and fertile control (50) peripheral blood: NOA (32) obstructive azoospermia (5) TESE |
• No difference in peripheral blood • Hypermethylation of MTHFR in testis biopsies (53 %), 0 % in obstructive azoospermia |
[54] |
| MTHFR | RSA couples (20) Non-RSA couples (147) Fertile men (20) |
• Methylated MTHFR epigenotype detected in 75 % of RSA men, 54 % of NRSA men and 15 % of fertile men | [55] |
| MTHFR | Idiopathic infertile (94): Idiopathic infertile men with normozoospermia (30) Idiopathic infertile men with oligozoospermia (64) Fertile controls (54) |
• Hypermethylation of MTHFR in 45 % of idiopathic infertile males, 15 % of fertile males • Higher methylation pattern was found in the group with oligozoospermia |
[11] |
| H19-ICR, KvDMR, SNRPN-ICR, IG-DMR and MEG3-DMR | Infertile men (107): Normozoospermic (15) Oligozoospermic (1) Astenozoospermic (8) Teratozoospermic (30) Oligoastenozoospermic (1) Oligoteratozoospermic (5) Astenoteratozoospermic (31) OAT (16) Fertile men (30) |
• Altered methylation patterns associated with H19-ICR, SNRPN-ICR and MEG3 • Methylation anomalies in at least 20 % of the CGIs • Significant inverse relation between the percent of altered CGIs and the number of affected individuals decreased |
[38] |
| CREM | Abnormal protamine 1/protamine 2 (P1/P2) ratio (60) Oligozoospermia (32) Fertile controls (40) |
• Significantly higher rate of methylation of CREM in patients with abnormal protamination and oligozoospermia • Sperm concentration, sperm motility, and normal head morphology negatively correlated with the amount of CGI methylation |
[56] |
| H19, IG-GTL2 and MEST | Oligoastenozoospermic (10), NOA (5), and unknown pathology (3) vasectomy reversal (17) |
• A significant decrease in DNA methylation at the H19 DMR in testicular sperm of azoospermic (NOA and oligoastenozoospermic) men and vasectomy reversal compared with fertile men, suggesting that aberrant DNA methylation may be associated with obstruction • No association between G-GTL2 and MEST DMRs and studied groups |
[57] |
|
H19, GTL2
LIT1, MEST, NESPAS, PEG3 and SNRPN ALU and LINE1 |
Couples with strictly male-factor infertility (106) | • Significant association between aberrant methylation imprints and abnormal semen parameters, but not with ART outcome • Significant repeat methylation difference between sperm samples from infertile and presumably fertile males |
[58] |
| OCT4, SOX2, NANOG, HOXC11, miR-17 and CREM | Patients with a high P1/P2 ratio (10) and patients with a low P1/P2 ratio (10) Normozoospermic controls with normal P1/P2 ratio (10) |
• No significant quantitative differences between groups of patients with either an abnormally high or low P1/P2 ratio compared to normal controls • No extreme methylation defects in severely infertile men • Two patients exhibited altered methylation of the CREM gene |
[59] |
| DAZL | OAT men (5) and Normozoospermic (5) |
• Increased methylation defects in the DAZL promoter CGI in OAT patients compared with NZ controls | [60] |
| DAZ | Idiopathic infertile patients (174) and fertile controls (58) | • No differences between DAZ gene methylation patterns among groups with different spermatogenic status and somatic cells, completely methylated except for the group with AZ | [61] |
| H19, MEST | Normozoospermic controls (27) Oligozoospermic patients (96) | • No aberration in MEST DNA methylation • Aberrant DNA methylation in H19 in oligozoospermic patients |
[62] |
| H19, MEST and LINE1 | Normozoospermic (5) and OAT (20) | • Hypomethylation of H19
• Hypermethylation of MEST • High level of global methylation |
[63] |
| H19 and MEST | Anejaculation (5) Secondary (5) Primary obstructive azoospermia (5) Secretory azoospermia (9) |
• Hypomethylation of MEST in all patients
• Significantly reduced DNA methylation of H19 in secretory azoospermia patients |
[64] |
| H19, GTL2, PEG1 (MEST), LIT1 (KCNQ1OT1), ZAC (PLAGL1), PEG3and SNRPN | Normozoospermic (79) and oligozoospermic patients (18) | • H19 unmethylation • MEST methylation |
[65] |
NOA non-obstructive azoospermia, OAT oligoastenoteratozoospermic, TESE testicular sperm extraction
Houshdaran and colleagues reported aberrant DNA methylation in semen samples with poor quality and suggested that both imprinted genes and other epigenetic defects were associated with sperm abnormalities [20]. Marques and colleagues compared DMRs in paternally methylated H19 and paternally unmethylated MEST in sperm samples from fertile and infertile males. They found a loss of methylation of H19 DMR in patients with oligo-astheno-teratozoospermia (OAT). In addition, abnormal DNA methylation patterns were observed in oligozoospermic patients [63]. Insulin-like growth factor 2 (IGF2)/H19 has function in growth regulation and mitogenic activities during gestation. Therefore, aberrant methylation of IGF2/H19 gene may cause abnormalities in mitotic cycles of spermatogenesis and result in OAT or oligozoospermia. These findings are in parallel with the results of Kobayashi and colleagues where abnormal methylation of the paternal DNA at H19 and GTL2, and abnormalities of maternal DMRs LIT1, PEG1/MEST, PEG3, SNRPN, and ZAC, which encodes a zinc finger protein regulating apoptosis and cell cycle arrest, were reported in oligozoospermic patients along with unchanged global DNA methylation [66]. Boissonnas and colleagues analyzed 47 CGIs located at the DMR0 and DMR2 of the IGF2 gene and in the third and sixth CTCF-binding sites of the H19 DMR in normal semen samples and patients with teratozoospermia (T) and/or OAT. Their results showed high global methylation level for all CGIs analyzed in all normal semen samples. Loss of methylation at variable CGI positions either in the IGF2 DMR2 or in both the IGF2 DMR2 and the sixth CTCF of the H19 DMR was observed in the teratozoospermic patients (11/19). In the OAT group (16/22), a severe loss of methylation of the sixth CTCF correlated with sperm concentration was indicated. On the other hand, they reported no correlation between methylation state of DMR0 and the third CTCF and status of semen samples [67]. In idiopathic infertile man, the paternally imprinted IGF2/H19 hypomethylation and maternally imprinted MEST hypermethylation were found to be significantly associated with decreased sperm counts and decreased sperm motility and abnormal morphology, respectively. Loss of methylation of paternally imprinted genes may be explained with the deficiency of DNA methyltransferases but hypermethylation of maternally imprinted genes might be result of erroneous de novo methylation or a failure to erase maternal imprint in the male germ cell genomes [68]. Consequently, the etiology of these aberrant imprinting patterns in paternally or maternally imprinted genes in idiopathic infertility remains elusive.
Global/genome-wide and gene-specific DNA methylation
Although most studies have focused on aberrant epigenetic marks of imprinted genes [47, 58, 63–67], several studies have shown associations between aberrant DNA methylation of non-imprinted genes and oligozoospermia, abnormal sperm morphology, and motility. Recently, Urdinguio and colleagues showed alterations in methylation pattern of 2752 CGIs of genomic sperm DNA in idiopathic infertile males compared with fertile men. In addition, they found statistically significant associations between DNA hypomethylation and corresponding regions in somatic cells enriched in the repressive histone mark H3K9me3 and between DNA hypermethylation and corresponding regions enriched in H3K4me1 and CTCF, suggesting a locus-dependent aberrant DNA methylation of sperm in infertile men. Furthermore, they also showed that DNA methylation of spermatozoa was lower in several repetitive sequences (LINE-1, Alu Yb8, NBL2, D4Z4) compared to that of somatic cells [17].
Methylenetetrahydrofolate reductase (MTHFR) is one of the main regulatory enzymes involved in folate metabolism, DNA synthesis, and methylation reactions. Inactivation of MTHFR gene results in hyperhomocysteinemia and infertility in male mice [69]. Khazamipour and colleagues were the pioneering group who investigated the methylation profiles of MTHFR gene in NOA infertile patients and reported a significant association between methylation status MTHFR gene and infertility in azoospermic males [54]. Recently, DNA methylation aberrations of MTHFR gene promoter in paternal spermatozoa were described in small cohorts of oligozoospermic men and in patients with poor sperm morphology and recurrent spontaneous aberrations [49, 55, 61] (Table 1). However, in our study, we did not observe a significant association between methylation profiles of MTHFR gene, neither in patients with NOA nor in patients with oligozoospermia, in a Turkish population (Kulaç et al., unpublished data). Ramasamy and colleagues reported a significant association between discoidin domain receptor 1 (DDR1) promoter DNA methylation and DDR1 expression levels in NOA patients when compared to fertile controls. As DDR1 is a receptor tyrosine kinase expressed in human postmeiotic germ cells and involved in proliferation, apoptosis, cell morphogenesis, and differentiation, abnormal expression of DDR1 in NOA patients may prevent primordial germ cell migration and development [48].
Table 1 gives a summary of the studies to date that analyze potential association of spermatozoal DNA methylation with male infertility. Further studies are needed to elucidate the mechanisms leading to the alterations of DNA methylation profiles and their significance and functional consequences for male infertility.
Role of histone modifications in male infertility
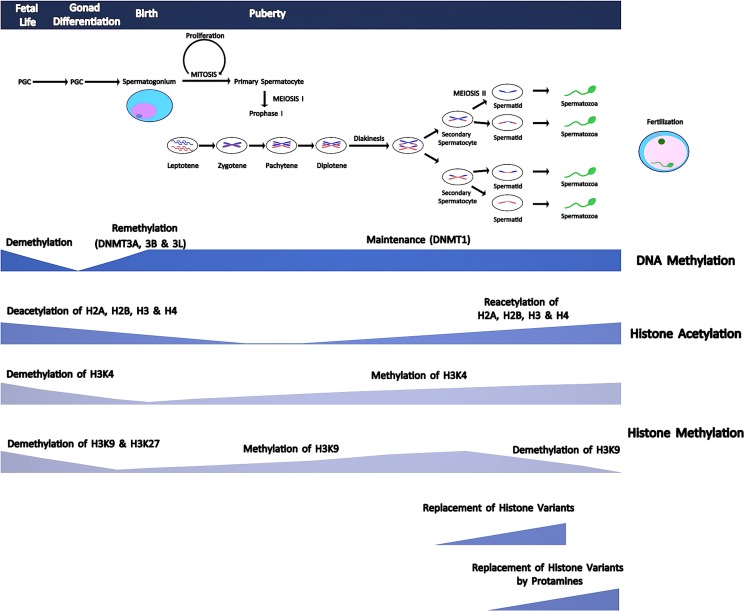
Histones are basic proteins rich in lysine and arginine located in nucleus and are subject to post-translational modifications on their N- and C-terminal tails via acetylation, methylation, phosphorylation, and ubiquitination [70]. These chemical modifications change binding capacity of regulatory factors to DNA and thus lead to alterations in gene activity and expression. Generally, acetylation of lysine (K) residues of histone 3 (H3) and histone 4 (H4) leads to active transcription through inducing open chromatin configuration and facilitating transcription factor binding in spermatogonial stem cells [71, 72]. On the contrary, deacetylation causes inactivation of transcription and generally correlates with methylation of histones [73]. Trichostatin A (TSA) is a histone deacetylase (HDAC) inhibitor and is able to induce cell cycle arrest in immortal somatic cells. Interestingly, TSA-treated mice showed no significant effects on either proliferation or apoptosis in mitotically active spermatogonia compared to controls. Furthermore, withdrawal of TSA led to complete regeneration of the seminiferous epithelium in fertility assays [74]. On the contrary, apoptosis of both spermatocytes and spermatids significantly increased with increasing TSA doses, suggesting for an inhibitory role of TSA mainly on meiosis but not mitosis [75, 76]. During spermatogenesis, methylation of H3K and H4K histone tails is regulated by histone methyltransferases (HTM) and histone demethylases (HDM) [77, 78]. Acetylation of H2A, H2B, H3, and H4 was shown to be high in mouse spermatogonia, and these histones were deacetylated throughout meiosis in round spermatids and reacetylated in elongating spermatids [72] (Fig. 1). Hyperacetylation of H4K has been shown to be responsible for histone to protamine change in elongating spermatids [72].
Fig. 1.
A diagram showing comparative timing of meiotic phases, DNA methylation patterns, histone tail modifications, and protamination during spermatogenesis. Meiosis starts with puberty. Methylation marks of primordial germ cells (PGCs) are erased during the embryogenesis. After this demethylation process, a specific remethylation program starts in spermatogonia and type I spermatocytes. Acetylation of H2A, H2B, H3, and H4 is high in spermatogonia, and these histones are deacetylated throughout meiosis, and round spermatids are reacetylated in elongating spermatids. Hyperacetylation of histone tails causes loosening of chromatin structure and stimulates DNA strand breaks by topoisomerase enzyme, which in turn facilitates separation of histones and replacement by transition proteins that are later replaced by protamines
Recently, a few studies have investigated the role of histone tail modification in spermatogenesis. La Spina and colleagues evaluated the methylation of H3K4Me, H3K4Me3, H3K9Me2, H3K79Me2, H3K36Me3, and acetylation of H3K4Ac and H4K5Ac in normal and abnormal human sperm. They reported the presence of heterogeneous histone modifications, and the presence of H3K4Me1, H3K9Me2, H3K4Me3, H3K79Me2, and H3K36Me3 marks in poorly functional human sperm [79]. Yuen and colleagues made a knockout mouse model for the histone variant H3.3, H3f3b, which is involved in various biological processes including development, transcriptional memory, and transcriptional reprogramming. They showed that loss of H3f3b gene induced abnormalities in sperm and testes morphology leading to infertility. Additionally, their results indicated H3f3b-null testes exhibited abnormal chromatin organization in germ cells with reduced protamine incorporation and increased apoptosis [80]. Further studies are required to evaluate the impact of histone tail modifications in human infertility.
Role of protamination in male infertility
Sperm chromatin packaging is a critical process that serves to accommodate enormous amounts of DNA into a small sperm cell. Fertilization requires many physiological events including movement of sperm cells all along the female reproductive system, attachment to zona pellucida, and penetration into the oocyte. For accomplishment of all these phases, a regulatory mechanism controlling the replacement of 85–95 % of histones by protamines becomes effective [81]. Protamines are small proteins rich in arginine. They are located in sperm nucleus and synthesized during later stages of spermatogenesis. Protamination of sperm chromatin facilitates compaction of nucleus required for sperm motility and also protects sperm genome from oxidation and harmful molecules within the female reproductive system [81].
Replacement of histones by protamines involves translocation of histones by selected histone variants which are expressed during spermatogenesis. Hyperacetylation of histone tails causes loosening of chromatin structure and stimulates DNA strand breaks by topoisomerase enzyme that in turn facilitates separation of histones and replacement by transition proteins (TPs) [82, 83]. TP1 and 2 bind to DNA and are completely replaced by protamines. Transition proteins play a critical role in separation of histones and facilitate the condensation of sperm DNA by protamines at later stages [83] (Fig. 1).
The two protamines, P1 and P2, are equally expressed in human beings, and P1/P2 protamine ratio is equal to one in fertile men [56]. Erroneous processing of protamine transcripts leads to increase in the production of immature P2 precursors associated with subfertility [84, 85]. Despite controversial publications, deviations in protamine ratio might be associated with various phenotypic features including decreased sperm counts and function and poor embryonic quality [86, 87].
Post-transcriptional epigenetics: short non-coding RNAs and male infertility
Short non-coding RNAs are a class of functional RNA molecules that regulate gene expression at the post-transcriptional level via epigenetic mechanisms. As the name implies, these RNA molecules are shorter than 30 nucleotides, and they do not code for a particular protein. Short non-coding RNAs can be classified into three main groups called microRNAs (miRNAs), small-interfering RNAs (siRNAs), and piwi-interacting RNAs (piRNAs).
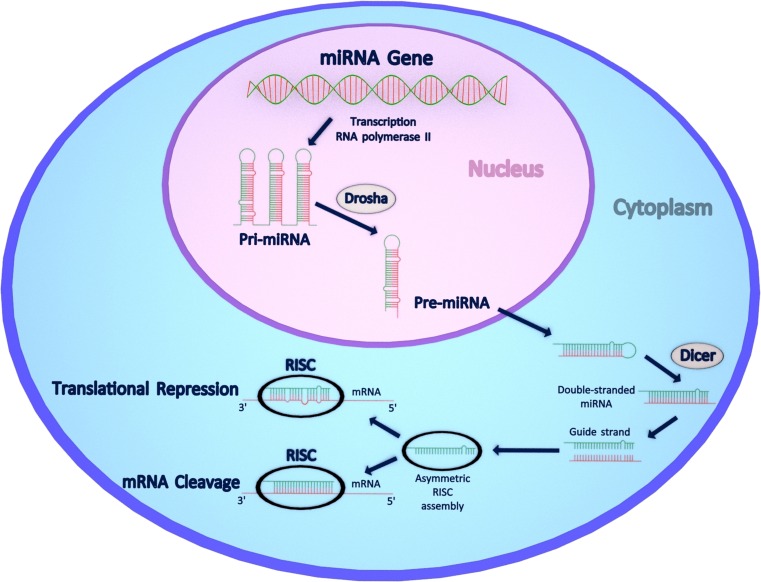
miRNAs are 21–25 nucleotide long, endogenous non-coding RNAs that downregulate gene expression by binding to their target mRNAs, causing either mRNA cleavage/degradation or translational repression (Fig. 2). In mammals, miRNAs are estimated to control about 50 % of all protein-coding genes and are involved in nearly all cellular, developmental, and pathological processes [88, 89]. miRNA biogenesis is a multistep process starting with RNA polymerase II-driven transcription of large precursor RNA molecules called pri-miRNAs (Fig. 2). The pri-miRNAs are then processed in the nucleus by Drosha, a type III RNase, to become pre-miRNAs. Pre-miRNAs are exported into the cytoplasm where they are further processed by another type III RNase called Dicer. Dicer cleavage results in formation of approximately 22 nucleotide-long double-stranded miRNA molecules. Of the two strands, only one is incorporated into a multiprotein complex termed as RNA-induced silencing complex (RISC). RISC uses the so-called “guide strand” to downregulate its target mRNA by complementary base pairing. The degree of miRNA-mRNA complementarity is considered to be a key factor in the choice of post-transcriptional mechanism employed by miRNA [90]. Perfect base-pairing results in mRNA cleavage and its subsequent degradation, whereas imperfect pairing mostly at the 3’ untranslated regions (3’UTRs) leads to translational repression. siRNAs function in a similar manner as miRNAs and use the same Dicer and RISC complexes to mediate their post-transcriptional gene-silencing effect that in contrast to miRNAs operates only by mRNA cleavage and degradation. piRNAs are a more specialized group of short non-coding RNAs, and they exert their silencing effect by interacting with P-element-induced wimpy testis (PIWI) family of proteins. In comparison to miRNAs and siRNAs, piRNAs are longer (approximately 26–31 nucleotides in length), and their biogenesis, although not well understood, is independent of Dicer [91]. piRNAs and PIWI proteins are known to be essential for germ cell development and silencing of repetitive elements such as transposons [92].
Fig. 2.
miRNA biogenesis pathway and its functional consequences for the mammalian cell. In the nucleus, miRNA genes are transcribed by RNA polymerase II to generate large precursor pri-miRNAs that are then processed by type III RNase Drosha to pre-miRNAs. Pre-miRNAs are then exported into the cytoplasm where they are further processed by the enzyme Dicer to form a mature, 21-25 nucleotide long, duplex miRNA. Of its two strands, the so-called guide strand is incorporated into the RISC complex where it base-pairs with its target mRNA sequence. Perfect base pairing results in mRNA cleavage and degradation, while imperfect pairing, mostly at 3’UTRs, causes translational repression
Short non-coding RNAs are required for normal spermatogenesis: animal studies and studies from fertile men
Investigating the role of short non-coding RNAs in male infertility has increasingly been an attractive research area. Recent studies have indicated that miRNAs, endogenous siRNAs (endo-siRNAs), and piRNAs are all expressed in the male germ cells and are required for spermatogenesis in animals [93, 94]. miRNAs and endo-siRNAs are abundantly expressed in male germ cells throughout spermatogenesis, whereas piRNAs are only present in spermatocytes at the pachytene stage and in round spermatids [92, 94]. The absolute requirement for miRNAs and endo-siRNAs for spermatogenesis has been shown by two initial studies where Dicer1 gene was knocked out in two different mouse models [95, 96]. Germ-cell specific deletion of Dicer1 in these models has led to complete male infertility due to alterations in meiotic progression, increased spermatocyte apoptosis, and failure of haploid male germ cell differentiation. Remarkably, Romero and colleagues also showed that Dicer1 is not required for spermatogonial stem cell renewal and mitotic proliferation, but is indispensible for meiotic and haploid stages of spermatogenesis [96]. To distinguish between the specific effects of miRNAs and endo-siRNAs, a following study has used Drosha and Dicer conditional knockout mouse models and reported that both knockout males were infertile due to impaired spermatogenesis characterized by depletion of spermatocytes and spermatids leading to oligoteratozoospermia or azoospermia [97]. Interestingly, when compared to ones from Dicer knockouts, the testes from Drosha knockouts were more severely disrupted in terms of spermatogenesis, which further highlights the significance of miRNAs for normal spermatogenesis and male fertility [97].
There have been numerous studies investigating the role of miRNAs in human male fertility, and results have so far supported the above-mentioned observations obtained from animal models. In a recent study where the expression levels of 736 miRNAs were tested in spermatozoa from 10 normozoospermic fertile men, 221 miRNAs were found to be consistently present in all individuals [98] (Table 2). Potential targets of these miRNAs were found to be enriched in processes involved in development, morphogenesis, spermatogenesis, and embryogenesis. In the same study, three most stably expressed miRNAs, namely, miR-532-5p (meaning from the 5’ arm), miR-374b-5p, and miR-564, have also been proposed by the authors to be used as fertility biomarkers [98]. Next generation sequencing analysis of short RNA transcriptome from testes of three normal men identified 775 miRNAs and 20121 piRNAs, indicating the abundance and complexity of short non-coding RNAs in the human testis [112]. The most abundant miRNAs detected in this study were let-7 family members, miR-34c-5p, miR-103a-3p (meaning a part from the 3’ arm), miR-202-5p, miR-508-3p, and miR-509-3-5p, which target gene transcripts involved in regulation of meiosis, spermatogenesis, germ cell apoptosis, testicular development, p53-related pathways, and homologous recombination pathways [112].
Table 2.
Summary of studies investigating the relationship between expression levels of testicular, spermatozoal, and seminal fluidal miRNAs and idiopathic male infertility in humans
| Study group (sample size) and Type of analyzed samples |
Brief results and miRNAs displaying the greatest fold changes | Reference |
|---|---|---|
| Patients with NOA (3) Testicular tissues |
154 downregulated miRNAs such as miR-17-92 and miR-371/2/3 clusters, miR-1, miR-181a, miR-221, miR-9*, miR-145, miR-383, let-7f, let-7f-2*, let-7i*, miR-19a, miR-20b, miR-29c, miR-30a*, miR-30d*, miR-34b*, miR-449a, miR-652, miR-92a
17 up-regulated miRNAs: miR-129-5p, miR-193a-3p, miR-193a-5p, miR-554, miR-423-3p, miR-491-3p, miR-557, miR-210, miR-23a, miR-302a, miR-371-5p, miR-374a, miR-654-5p, miR-663, miR-638, miR-572, miR-744 |
[99] |
| Patients with azoospermia and oligozoospermia (490) Genomic DNA |
A SNP in the 3’UTR of HIWI2 gene (rs508485T > C) exhibited a significantly increased oligozoospermia risk, whereas HIWI3 variant rs11703684C > T displayed a significantly reduced oligozoospermia risk. | [100] |
| Patients with azoospermia (266) or severe oligozoospermia (228) Genomic DNA |
A SNP (rs6631A > T) in CGA encoding glycoprotein hormone α-subunit resulted in decreased binding affinity of miR-1302 and overexpression of CGA in vitro, and is associated with increased risk for idiopathic male infertility. | [101] |
| Patients with NOA (118), asthenozoospermia (137) and oligoospermia (34) Seminal plasma |
7 miRNAs (miR-34c-5p, miR-122, miR- 146b-5p, miR-181a, miR-374b, miR-509–5p and miR-513a-5p) were found to be markedly decreased in azoospermia but increased in asthenozoospermia. | [102] |
| Patients with NOA (48) and oligozoospermia (48) Seminal plasma |
Expression levels of miR-19b and let-7a in both seminal plasmas and testicular tissues (n = 5) were significantly increased in idiopathic infertile males with NOA. No significant differences were found between the fertile controls and infertile males with oligozoospermia. | [103] |
| Infertile normospermic, oligozospermic and asthenospermic men (667) Genomic DNA |
SNPs in the 3’UTR (rs10719T > C and rs642321C > T) and promoter (rs12323635T > C) regions of Dicer1 were found to be significantly associated with oligozoospermia, and were proposed to result in global changes in miRNA processing through affecting Dicer1 expression levels. | [104] |
| Patients with NOA (100) Seminal plasma |
miR-141, miR-429 and miR-7-1-3p were significantly upregulated in NOA compared to fertile controls. | [129] |
| Patients with asthenozoospermia (9) and oligoasthenozoospermia (9) Spermatozoa |
50 miRNAs upregulated (such as miR-30a, miR-363, miR-26a, miR-200a, miR-141, miR-429, miR-193b, miR-29a, miR-1274a, miR-24, miR-4286, miR-99a) and 27 miRNAs downregulated (such as miR-34b, miR-122, miR-1973) in asthenozoospermic males 42 miRNAs upregulated (such as miR-141, miR-193b, miR-26a, miR-200c, miR-29a, miR-429, miR-200a, miR-99a, miR-363) and 44 miRNAs downregulated (such as miR-34b*, miR-34b, miR-34c-5p, miR-15b, miR-122, miR-449a, miR-1973, miR-16, miR-19a) in oligoasthenozoospermic males. |
[105] |
| Normozoospermic fertile men (10) Spermatozoa |
221 miRNAs were found to be consistently present in all individuals. 48 miRNA pairs displayed a stable expression. miR-532-5p, miR-374b-5p and miR-564 were the best normalizing miRNA candidates, and were proposed to be used as fertility biomarkers. | [98] |
| Oligospermic infertile patients (43) Spermatozoa |
mir-100 and let-7b levels were significantly higher, and ERα (estrogen receptor α) expression was significantly decreased in oligospermic groups. | [106] |
| Oligospermic infertile men (43) Spermatozoa |
mir-21 and mir-22 levels were significantly higher, and ERβ (estrogen receptor β) expression was significantly decreased in oligospermic males. | [13] |
| Azoospermic men (40): SCO (12), MA (12) and GCA (16) subgroups Testicular tissues |
197, 68, and 46 miRNAs were found to be differentially expressed in SCO, MA and GA groups, respectively. | [107] |
| SCO down | miR-34b*, miR-34c-5p, miR-449a, miR-574–5p, miR-15b, miR-125b, miR-125a-5p, miR-16, miR-204, miR-1260, miR-23a, miR-145, miR-1260b, miR-30b, miR-25, miR-1274a, miR-22, miR-34b, miR-19a, miR-574–3p, miR-92a | |
| SCO up | miR-3925, miR-135a*, miR-1471, miR-642b, miR-617, miR-3180–3p, miR-718, miR-3200–5p, miR-99b*, miR-3945, miR-3648, miR-575, miR-936, miR-3137, miR-548q, miR-4322, miR-1181, miR-125a-3p, miR-371–5p, miR-373*, miR-3197, miR-3656, miR-3194 | |
| MA down | miR-34c-5p, miR-34b*, miR-449a, miR-509–5p, miR-514, miR-34b, miR-517a, miR-506, miR-514b-5p, miR-129–3p | |
| MA up | miR-127–3p, miR-410, miR-199a-5p, miR-379 | |
| GCA down | miR-449a, miR-34b*, miR-34c-5p, miR-34b, miR-449b* | |
| GCA up | miR-135a*, miR-3137, miR-99b*, miR-3692* | |
| Patients with oligoospermia / oligoasthenozoospermia (80) and NOA (40) Spermatozoa and testicular tissues |
miR-429 was significantly increased, whereas miR-34b*, miR-34b, miR-34c-5p and miR-122 were decreased in both tested groups compared to normal control subjects. | [108] |
| Infertile men of various subgroups (30) Genomic DNA |
Two allele-specific methylation-sensitive SNPs in PIWIL1 and PIWIL2, rs10773767 and rs6982089 respectively, were found to be associated with idiopathic male infertility. | [109] |
| Azoospermic men with SCO (5) Testicular tissues |
miR-34c-5p, miR-126, miR-191, miR-10b, miR-202-5p, miR-103, miR-514, miR-204 expressions were markedly reduced in testicular tissues from SCO men compared to normal fertile men. | [110] |
| Infertile men with high sperm DNA damage (94) Seminal plasma |
miR-424 was found to be significantly downregulated in the study group compared to the control group. | [111] |
SCO Sertoli cell only, NOA non-obstructive azoospermia, MA mixed atrophy, GCA germ cell arrest
Testicular miRNAs and male infertility
One of the earliest studies linking miRNAs with any pathological condition leading to male infertility has been conducted with NOA patients by using microarray technology [99]. Analysis of the testicular tissues obtained from three NOA patients has revealed 154 differentially downregulated and 17 upregulated miRNAs compared to controls (Table 2). Of the downregulated group, miR-17-92 and miR-371/2/3 clusters are noteworthy to mention as they might act as potential oncogenes by inhibiting apoptosis through E2F1, oncogenic RAS, and p53 pathway in models of testicular cancer [113, 114]. Therefore, low expression of these miRNA clusters may explain increased apoptosis observed in the testes of NOA patients [99]. A more recent and comprehensive miRNA microarray analysis of testicular tissue samples from 40 azoospermic men of different histopathologic subgroups has revealed a total of 311 differentially expressed miRNAs when compared to samples with normal spermatogenesis [107]. The numbers of differentially expressed miRNAs were 197, 68, and 46 for SCO, mixed atrophy (MA), and germ cell arrest (GA) groups, respectively, in comparison with normal spermatogenesis (Table 2). Among all tested groups, the highest fold changes were observed with only seven miRNAs, all of them downregulated including miR-449 family members (miR-449a, miR-449b*), miR-34 family members (miR-34b*, miR-34b, miR-34c-5p), miR-517b, and miR-129-3p. Notably, potential targets of these miRNAs are involved in spermatogenesis process, apoptosis, cell proliferation, differentiation, and testicular development [107]. Of special interest are miR-34b and miR-34c, which were previously shown to target deleted in azoospermia-like (DAZL) transcript that is essential for gametogenesis in mice [115, 116]. Among the potential targets, insulin-like growth factor-binding protein 5 (IGFBP5) gene transcript, which was reported to be highly expressed in NOA patients, stands out as predicted to be targeted by both miR-449a and miR-34c-5p [117].
In a more recent study conducted with testicular tissue samples from five azoospermic men with SCO syndrome, miRNAs with the highest fold reductions in comparison to controls were found to be miR-34c-5p, miR-126, miR-191, miR-10b, and miR-202-5p [110]. Further experimentation with immunohistochemistry showed a specific localization of miR-202-5p to Sertoli cells of normal fertile men, but not in those of infertile men with SCO. Germ cell-dependent expression observed for this miRNA has been proposed to indicate a functional role in Sertoli cell maturation and/or regulation of spermatogenesis [110].
Spermatozoal miRNAs and male infertility
Apart from testicular tissue samples, spermatozoa from infertile men have also been analyzed in terms of their miRNA profiles. A comprehensive miRNA microarray analysis of spermatozoa from nine asthenozoospermic and nine oligoasthenozoospermic men has revealed 50 upregulated, 27 downregulated miRNAs, and 42 upregulated, 44 downregulated miRNAs, respectively, when compared to normozoospermic men [105] (Table 2). The miRNAs that exhibited the highest fold changes were the ones that are downregulated, with most of them more than 10 times decreased in their expression levels. To name a few, miR-34b, miR-122, and miR-1973 were found to be downregulated in asthenozoospermic men, whereas miR-34b, miR-34b*, miR-34c-5p, miR-15b, miR-122, miR-449a, miR-1973, and miR-16 were downregulated in oligoasthenozoospermic men [105] (Table 2). Although bioinformatics analyses have predicted putative target genes of these miRNAs, detailed in vitro and in vivo studies linking most of them to their specific target genes within the context of male infertility are largely missing. Two recent reports have demonstrated increased expression levels of several miRNAs (i.e., miR-100, let-7b, and miR-21, miR-22) along with a concurrent decrease in their respective predicted targets, namely, estrogen receptor-α and -β, in spermatozoa from 43 oligospermic infertile men [13, 106]. Given the crucial roles of estrogen receptor-α and -β in maintenance of male reproductive tract function, sperm metabolism and Sertoli cell proliferation, it would not be surprising if there exists a miRNA-based mechanism for downregulation of these two receptors in infertile men [118, 119]. Using qRT-PCR, a validation study has tested the potential of a set of five differentially expressed miRNAs selected from the two aforementioned miRNA microarray analyses [105, 107], to be used as biomarkers for the assessment of male infertility [108]. Spermatozoa from 80 subfertile (mostly oligospermic and oligoasthenospermic) men and testicular tissues from 40 men with non-obstructive azoospermia were analyzed along with their appropriate controls, and with the exception of miR-429, the expressions of all other four miRNAs (miR-34b*, miR-34b, miR-34c-5p, miR-122) were found to be decreased in both tested groups [108]. The miR-34 family members are known to be direct transcriptional targets of p53, and they appear to play a vital role as mediators of tumor suppression by p53 via induction of apoptosis, cell cycle arrest, or senescence [120]. Although how the inactivation of this family in a cancer setting reconciles with its downregulation in infertile males is yet not clear, miR-34c seems to relate through a novel pathway by promoting germinal lineage differentiation [121]. Interestingly, miR-34c has also been shown to promote murine male germ cell apoptosis by targeting the transcription factor ATF1 in a p53-independent manner [122]. In mouse models, miR-34c and miR-34b were found to be highly expressed in post-mitotic male germ cells from primary spermatocytes up to round spermatids [121, 123], and deletion of this locus along with miR-449 led to OAT and infertility [124]. Remarkably, miR-34c seems to extend its reproductive role into the zygote, as sperm-borne miR-34c has been shown to have a post-fertilization function in mice by promoting the first cleavage division via modulation of anti-apoptotic and anti-proliferative Bcl-2 expression [125]. As being the most abundant miRNA in human spermatozoa, miR-34c may also play a similar role in humans and may shape early embryonic development at the post-transcriptional and/or transcriptional level [126].
Unlike miR34b/c, miR-122a is predominantly expressed in late-stage, post-meiotic germ cells and has been reported to downregulate translation of murine TNP2, a testis-specific protein acting in histone-to-protamine transition during spermiogenesis [127]. It is thus tempting to speculate that the above-mentioned decrease in this miRNA reported for subfertile and infertile men [108] may suggest a role for failure of miR-122-driven spatiotemporal control of chromatin remodeling in male infertility. Interestingly, miR-122 has been shown to promote differentiation of human-induced pluripotent stem cells into spermatozoa-like cells in vitro by suppressing TNP2 expression [128].
Seminal fluidal miRNAs and male infertility
Besides spermatozoa, seminal plasma has also been proposed to have a potential to provide researchers with another noninvasive source for the assessment of male infertility. A genome-wide low density miRNA array has analyzed seminal plasmas from 20 patients with NOA and found three miRNAs, miR-141, miR-429, and miR-7-1-3p, to be significantly upregulated compared to fertile controls [129]. Notably, increased expression patterns for these miRNAs were also observed in testicular tissues of patients with NOA, implicating the potential use of seminal miRNAs as noninvasive biomarkers for the diagnosis of male infertility. However, care must be taken to utilize these findings to make a differential diagnosis, as a previous study has also found similar fold inductions in miR-141 and miR-429 expression in spermatozoa from asthenozoospermic and oligoasthenozoospermic patients [105]. Moreover, miR-429 appears to be highly upregulated in spermatozoa from oligospermic males as well and to a lesser extent in testicular tissues of patients with NOA [108], consistent with the above finding [129]. In silico analysis of these miRNAs and subsequent immortalized spermatocyte cell culture experiments have identified candidate target genes such as PIK3R3, RB1, CBL, and TGFβ2, which have been suggested to have critical roles in the cell cycling and apoptotic process of germ cells [129].
Using qRT-PCR, another study performed with seminal plasmas from 96 patients with NOA and oligozoospermia has detected two miRNAs, namely, miR-19b and let-7a, to be upregulated only in males with NOA compared to fertile controls [103]. Testicular tissues of these patients also showed higher levels of the two miRNAs, whereas no significant changes were observed in men with oligozoospermia, introducing these miRNAs as potentially good biomarkers for NOA in particular [103]. Remarkably, these two miRNAs are predicted to target a fibronectin-like adhesion protein FNDC3A that functions in mediating spermatid and Sertoli cell adhesion during spermatogenesis [130]. Recently, altered expression levels of some miRNAs have also been correlated with the extent of sperm DNA damage observed in male infertility. In an infertile knock-out mouse model, increase in unrepaired DNA breaks has been linked with reduced expression of miR-16 and miR-19b in testes [131]. Moreover, analysis of seminal plasmas from 94 infertile men with high sperm DNA fragmentation index has revealed significant downregulation of miR-424 in this group, implicating a role for miR-424 in repair of double-strand breaks during spermatogenesis [111].
Another miRNA profiling study conducted with seminal plasma samples has identified seven miRNAs to be oppositely regulated in patients with NOA and asthenozoospermia, presenting a noninvasive approach for the differential diagnosis of these two pathological conditions [102]. Seven miRNAs that were significantly decreased in NOA patients but markedly increased in asthenozoospermia patients were found to be miR-34c-5p, miR-122, miR-146b-5p, miR-181a, miR-374b, miR-509-5p, and miR-513a-5p (Table 2). Although testicular tissues of the patients were not analyzed in this study, miR-34c-5p, miR-122, miR-181a, and miR-509-5p have also been shown to be decreased in testicular tissues of NOA patients in several different studies, confirming the utility of these miRNAs as noninvasive diagnostic tools for NOA [99, 107, 108, 110]. Interestingly, in patients with asthenozoospermia, miR-122 has later been reported to be downregulated in spermatozoa samples [105], by contrast with its abovementioned upregulation observed in seminal plasmas [102]. Although such a discrepancy may result from a passive leakage or an active secretion of some miRNAs from apoptotic spermatozoa into the seminal plasma, comparative studies are needed to simultaneously analyze spermatozoa, seminal plasma, and testicular tissues from diverse subgroups of subfertile and infertile men. Such studies will enable researchers to assess whether miRNA expression profiles from different sources, particularly testicular tissues versus spermatozoa and seminal plasmas, should reflect each other and elucidate the usefulness of select miRNAs as noninvasive biomarkers for the diagnosis of male infertility.
SNPs in genes involved in miRNA and piRNA pathways and male infertility
Apart from expression studies, there have been several other miRNA studies focusing on the role of SNPs in male infertility. SNPs in DICER and DROSHA, the key enzymes of miRNA biogenesis, have been reported to be associated with semen quality in infertile men of Han-Chinese descent [104]. Out of seven potentially functional SNPs analyzed by real-time PCR, rs10719T > C, rs12323635T > C, and rs642321C > T were found to be significantly associated with oligozoospermia (Table 2). As these SNPs are located in the 3’ UTR (rs10719 and rs642321) and promoter (rs12323635) regions of DICER1, it has been suggested that these genetics variants may alter the binding sites of regulatory miRNAs and essential transcription factors, resulting in global changes in miRNA processing through affecting Dicer1 expression levels [104]. A more comprehensive study has analyzed all SNPs in the 3’UTR of 140 spermatogenesis-related genes from a total of 494 infertile men and found a single nucleotide polymorphism (rs6631) in the miRNA-binding site of glycoprotein hormone α subunit-encoding gene CGA is associated with an increased risk of idiopathic male infertility [101]. Further in vitro cell culture experiments have revealed that A substituted by T in rs6631 causes reduced binding of miR-1302 to its target site in CGA mRNA, leading to CGA overexpression. Therefore, the variant allele of rs6631 has been suggested to increase the risk of idiopathic male infertility through upregulation of CGA expression and subsequent deregulation in the assembly of essential glycoprotein hormones like TSH, FSH and LH [101]. A possible association between SNPs in piRNA pathway genes and male infertility has also been investigated. A study conducted with 490 patients with idiopathic azoospermia or oligozoospermia has reported that an SNP in the 3’ UTR of human PIWI gene HIWI2/PIWIL4, rs508485T > C, exhibited a significantly increased oligozoospermia risk in Han-Chinese population [100]. Given the position of this variant and the importance of piRNAs in germline development, it is tempting to speculate there might be a miRNA-driven control over the expression of this particular PIWI protein, and loss of such a potential crosstalk between miRNAs and piRNAs might lead to impairment of spermatogenesis. A recent array-based study performed with peripheral blood samples from 30 infertile men of various subgroups has identified two allele-specific methylation-sensitive SNPs in PIWIL1 and PIWIL2, rs10773767 and rs6982089 respectively, indicating DNA methylation differences in these key genes of piRNA pathway are associated with impaired spermatogenesis [109]. These studies indicate that non-coding RNAs may play a crucial role in the etiology of male infertility.
Conclusion
The conventional andrological diagnostic process involves clinical and endocrinological examination of patient and semen analysis and provides little information about fertilizing capacity [132]. Severe spermatogenic impairment is most likely a genetic abnormality but usually the genetic cause cannot be identified in many infertile males. In about half of the infertility cases, the underlying cause remains unknown and the risk of transmitting genetic disorders to the offspring increases when artificial reproductive technology (ART) is used to treat the infertile couple. Moreover, infertility is a complex disorder with multiple genetic and other factors, including aging and exposure to environmental factors such as chemicals, diet, and personal lifestyle. As described in this review, recent studies have revealed epigenetics as one of the promising research areas in understanding male infertility.
Spermatozoa from infertile men have shown to be associated with aberrant methylation, including imprinted genes and developmental genes, histone tail modifications, and non-coding RNAs. Epimutations in spermatozoa can be associated with oligozoospermia, abnormal sperm morphology, and decreased progressive motility. The epigenetic status of spermatozoa may also affect the health of offspring since epigenetic aberrations are heritable. However, the recent discovery of endogeneous TET-mediated conversion of 5-methylcytosine (5mC) back to the unmethylated state through 5hmC, 5fC, and 5caC intermediates has raised some concerns over the interpretation of the past methylation data produced by bisulfite modification technique, since this technique is unable to distinguish between 5mC and 5hmC, leaving them both as cytosines [133]. Although some new techniques such as oxidative bisulfite sequencing have been developed to make this distinction possible, the number of studies using such methods yet remains limited [134, 135]. Taken together, although the exact cause and effect relationship between epigenetics and male infertility has not been elucidated, further investigation of this area holds a significant potential and great promise for understanding the molecular mechanisms of infertility.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Funding
This research did not receive any specific grant from any funding agency in the public, commercial, or not-for-profit sector.
Footnotes
Capsule Aberrant DNA methylation, including imprinted genes and developmental genes, histone tail modifications, and short non-coding RNAs are discussed with respect to their association with idiopathic male infertility.
Sezgin Gunes and Mehmet Alper Arslan contributed equally to this work.
Contributor Information
Sezgin Gunes, Phone: +90 362 312 19 19/3164, Email: sgunes@omu.edu.tr.
Mehmet Alper Arslan, Phone: +90 362 312 19 19/2278, Email: alpera55@gmail.com.
Gulgez Neslihan Taskurt Hekim, Phone: +90 362 312 19 19/3245, Email: gntkurt@gmail.com.
Ramazan Asci, Phone: +90 362 312 19 19/2255, Email: rasci@omu.edu.tr.
References
- 1.Boivin J, et al. International estimates of infertility prevalence and treatment-seeking: potential need and demand for infertility medical care. Hum Reprod. 2007;22(6):1506–12. doi: 10.1093/humrep/dem046. [DOI] [PubMed] [Google Scholar]
- 2.Rives N. Y chromosome microdeletions and alterations of spermatogenesis, patient approach and genetic counseling. Ann Endocrinol. 2014;75(2):112–4. doi: 10.1016/j.ando.2014.04.001. [DOI] [PubMed] [Google Scholar]
- 3.Roy A, Lin YN, Matzuk M. Genetics of idiopathic male infertility. In: Carrell DT, editor. The genetics of male ınfertility. Totowa: Humana Press Inc; 2007. pp. 99–111. [Google Scholar]
- 4.Hotaling JM. Genetics of male infertility. Urol Clin North Am. 2014;41(1):1–17. doi: 10.1016/j.ucl.2013.08.009. [DOI] [PubMed] [Google Scholar]
- 5.Gunes S, et al. Two males with SRY-positive 46, XX testicular disorder of sex development. Syst Biol Reprod Med. 2013;59(1):42–7. doi: 10.3109/19396368.2012.731624. [DOI] [PubMed] [Google Scholar]
- 6.Tuttelmann F, et al. Copy number variants in patients with severe oligozoospermia and Sertoli-cell-only syndrome. PLoS One. 2011;6(4):e19426. doi: 10.1371/journal.pone.0019426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lopes AM, et al. Human spermatogenic failure purges deleterious mutation load from the autosomes and both sex chromosomes, including the gene DMRT1. PLoS Genet. 2013;9(3):e1003349. doi: 10.1371/journal.pgen.1003349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.He XJ, et al. PRM1 variant rs35576928 (Arg > Ser) is associated with defective spermatogenesis in the Chinese Han population. Reprod Biomed Online. 2012;25(6):627–34. doi: 10.1016/j.rbmo.2012.09.005. [DOI] [PubMed] [Google Scholar]
- 9.Gupta N, et al. Strong association of 677 C > T substitution in the MTHFR gene with male infertility—a study on an indian population and a meta-analysis. PLoS One. 2011;6(7):e22277. doi: 10.1371/journal.pone.0022277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Teng YN, et al. A single-nucleotide polymorphism of the DAZL gene promoter confers susceptibility to spermatogenic failure in the Taiwanese Han. Hum Reprod. 2012;27(9):2857–65. doi: 10.1093/humrep/des227. [DOI] [PubMed] [Google Scholar]
- 11.Wu W, et al. GSTM1 and GSTT1 null polymorphisms and male infertility risk: an updated meta-analysis encompassing 6934 subjects. Sci Rep. 2013;3:2258. doi: 10.1038/srep02258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gunes S, Al-Sadaan M, Agarwal A. Spermatogenesis, DNA damage and DNA repair mechanisms in male infertility. Reprod Biomed Online. 2015;31(3):309–19. doi: 10.1016/j.rbmo.2015.06.010. [DOI] [PubMed] [Google Scholar]
- 13.Abhari A, et al. Altered of microRNA expression level in oligospermic patients. Iran J Reprod Med. 2014;12(10):681–6. [PMC free article] [PubMed] [Google Scholar]
- 14.Jungwirth A, et al. European Association of Urology guidelines on male infertility: the 2012 update. Eur Urol. 2012;62(2):324–32. doi: 10.1016/j.eururo.2012.04.048. [DOI] [PubMed] [Google Scholar]
- 15.Harton GL, Tempest HG. Chromosomal disorders and male infertility. Asian J Androl. 2012;14(1):32–9. doi: 10.1038/aja.2011.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Krausz C. Polymorphisms and male infertility. In: Carrell DT, editor. The genetics of male ınfertility. Totowa: Humana Press Inc; 2007. pp. 275–89. [Google Scholar]
- 17.Urdinguio RG, et al. Aberrant DNA methylation patterns of spermatozoa in men with unexplained infertility. Hum Reprod. 2015;30(5):1014–28. doi: 10.1093/humrep/dev053. [DOI] [PubMed] [Google Scholar]
- 18.Schutte B, et al. Broad DNA methylation changes of spermatogenesis, inflammation and immune response-related genes in a subgroup of sperm samples for assisted reproduction. Andrology. 2013;1(6):822–9. doi: 10.1111/j.2047-2927.2013.00122.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ferfouri F, et al. A genome-wide DNA methylation study in azoospermia. Andrology. 2013;1(6):815–21. doi: 10.1111/j.2047-2927.2013.00117.x. [DOI] [PubMed] [Google Scholar]
- 20.Houshdaran S, et al. Widespread epigenetic abnormalities suggest a broad DNA methylation erasure defect in abnormal human sperm. PLoS One. 2007;2(12):e1289. doi: 10.1371/journal.pone.0001289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hammoud SS, et al. Genome-wide analysis identifies changes in histone retention and epigenetic modifications at developmental and imprinted gene loci in the sperm of infertile men. Hum Reprod. 2011;26(9):2558–69. doi: 10.1093/humrep/der192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rotondo JC, et al. Methylation loss at H19 imprinted gene correlates with methylenetetrahydrofolate reductase gene promoter hypermethylation in semen samples from infertile males. Epigenetics. 2013;8(9):990–7. doi: 10.4161/epi.25798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhu J, et al. On the nature of human housekeeping genes. Trends Genet. 2008;24(10):481–4. doi: 10.1016/j.tig.2008.08.004. [DOI] [PubMed] [Google Scholar]
- 24.Deaton AM, Bird A. CpG islands and the regulation of transcription. Genes Dev. 2011;25(10):1010–22. doi: 10.1101/gad.2037511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jones PA. Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nat Rev Genet. 2012;13(7):484–92. doi: 10.1038/nrg3230. [DOI] [PubMed] [Google Scholar]
- 26.Klose RJ, Bird AP. Genomic DNA methylation: the mark and its mediators. Trends Biochem Sci. 2006;31(2):89–97. doi: 10.1016/j.tibs.2005.12.008. [DOI] [PubMed] [Google Scholar]
- 27.Eden S, Cedar H. Role of DNA methylation in the regulation of transcription. Curr Opin Genet Dev. 1994;4(2):255–9. doi: 10.1016/S0959-437X(05)80052-8. [DOI] [PubMed] [Google Scholar]
- 28.Kaneda M, et al. Essential role for de novo DNA methyltransferase Dnmt3a in paternal and maternal imprinting. Nature. 2004;429(6994):900–3. doi: 10.1038/nature02633. [DOI] [PubMed] [Google Scholar]
- 29.La Salle S, et al. Loss of spermatogonia and wide-spread DNA methylation defects in newborn male mice deficient in DNMT3L. BMC Dev Biol. 2007;7:104. doi: 10.1186/1471-213X-7-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cheng P, et al. Polymorphism in DNMT1 may modify the susceptibility to oligospermia. Reprod Biomed Online. 2014;28(5):644–9. doi: 10.1016/j.rbmo.2014.01.003. [DOI] [PubMed] [Google Scholar]
- 31.Santos F, Dean W. Epigenetic reprogramming during early development in mammals. Reproduction. 2004;127(6):643–51. doi: 10.1530/rep.1.00221. [DOI] [PubMed] [Google Scholar]
- 32.Hajkova P, et al. Epigenetic reprogramming in mouse primordial germ cells. Mech Dev. 2002;117(1-2):15–23. doi: 10.1016/S0925-4773(02)00181-8. [DOI] [PubMed] [Google Scholar]
- 33.Li E. Chromatin modification and epigenetic reprogramming in mammalian development. Nat Rev Genet. 2002;3(9):662–73. doi: 10.1038/nrg887. [DOI] [PubMed] [Google Scholar]
- 34.Ficz G. New insights into mechanisms that regulate DNA methylation patterning. J Exp Biol. 2015;218(Pt 1):14–20. doi: 10.1242/jeb.107961. [DOI] [PubMed] [Google Scholar]
- 35.Wu H, Zhang Y. Mechanisms and functions of Tet protein-mediated 5-methylcytosine oxidation. Genes Dev. 2011;25(23):2436–52. doi: 10.1101/gad.179184.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Davis TL, et al. The H19 methylation imprint is erased and re-established differentially on the parental alleles during male germ cell development. Hum Mol Genet. 2000;9(19):2885–94. doi: 10.1093/hmg/9.19.2885. [DOI] [PubMed] [Google Scholar]
- 37.Boissonnas CC, Jouannet P, Jammes H. Epigenetic disorders and male subfertility. Fertil Steril. 2013;99(3):624–31. doi: 10.1016/j.fertnstert.2013.01.124. [DOI] [PubMed] [Google Scholar]
- 38.Camprubi C, et al. Semen samples showing an increased rate of spermatozoa with imprinting errors have a negligible effect in the outcome of assisted reproduction techniques. Epigenetics. 2012;7(10):1115–24. doi: 10.4161/epi.21743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Arnaud P. Genomic imprinting in germ cells: imprints are under control. Reproduction. 2010;140(3):411–23. doi: 10.1530/REP-10-0173. [DOI] [PubMed] [Google Scholar]
- 40.Horsthemke B. In brief: genomic imprinting and imprinting diseases. J Pathol. 2014;232(5):485–7. doi: 10.1002/path.4326. [DOI] [PubMed] [Google Scholar]
- 41.Zheng HY, et al. Assisted reproductive technologies do not increase risk of abnormal methylation of PEG1/MEST in human early pregnancy loss. Fertil Steril. 2011;96(1):84–9. doi: 10.1016/j.fertnstert.2011.04.021. [DOI] [PubMed] [Google Scholar]
- 42.Kamiya M, et al. The cell cycle control gene ZAC/PLAGL1 is imprinted—a strong candidate gene for transient neonatal diabetes. Hum Mol Genet. 2000;9(3):453–60. doi: 10.1093/hmg/9.3.453. [DOI] [PubMed] [Google Scholar]
- 43.Broad KD, Curley JP, Keverne EB. Increased apoptosis during neonatal brain development underlies the adult behavioral deficits seen in mice lacking a functional paternally expressed gene 3 (Peg3) Dev Neurobiol. 2009;69(5):314–25. doi: 10.1002/dneu.20702. [DOI] [PubMed] [Google Scholar]
- 44.Jing J, et al. Effect of small nuclear ribonucleoprotein-associated polypeptide N on the proliferation of medulloblastoma cells. Mol Med Rep. 2015;11(5):3337–43. doi: 10.3892/mmr.2015.3148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Higashimoto K, et al. Loss of CpG methylation is strongly correlated with loss of histone H3 lysine 9 methylation at DMR-LIT1 in patients with Beckwith-Wiedemann syndrome. Am J Hum Genet. 2003;73(4):948–56. doi: 10.1086/378595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jenkins TG, Aston KI, Trost C, Farley J, Hotaling JM, Carrell DT. Intrasample heterogeneity of sperm DNA methylation. Mol Hum Reprod. 2015;21:313–9. [DOI] [PubMed]
- 47.Li B, Li JB, Xiao XF, Ma YF, Wang J, Liang XX, Zhao HX, Jiang F, Yao YQ, Wang XH. Altered DNA methylation patterns of the H19 differentially methylated region and the DAZL gene promoter are associated with defective human sperm. PLoS One. 2013;8:e71215. [DOI] [PMC free article] [PubMed]
- 48.Ramasamy R, et al. Integrative DNA methylation and gene expression analysis identifies discoidin domain receptor 1 association with idiopathic nonobstructive azoospermia. Fertil Steril. 2014;102(4):968–73. doi: 10.1016/j.fertnstert.2014.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Botezatu A, et al. Methylation pattern of methylene tetrahydrofolate reductase and small nuclear ribonucleoprotein polypeptide N promoters in oligoasthenospermia: a case-control study. Reprod Biomed Online. 2014;28(2):225–31. doi: 10.1016/j.rbmo.2013.10.010. [DOI] [PubMed] [Google Scholar]
- 50.Sugimoto K, Koh E, Iijima M, Taya M, Maeda Y, Namiki M. Aberrant methylation of the TDMR of the GTF2A1L promoter does not affect fertilisation rates via TESE in patients with hypospermatogenesis. Asian J Androl. 2013;15:634–9. [DOI] [PMC free article] [PubMed]
- 51.Klaver R, Tuttelmann F, Bleiziffer A, Haaf T, Kliesch S, Gromoll J. DNA methylation in spermatozoa as a prospective marker in andrology. Andrology. 2013;1:731–40. [DOI] [PubMed]
- 52.Richardson ME, Bleiziffer A, Tuttelmann F, Gromoll J, Wilkinson, MF. Epigenetic regulation of the RHOX homeobox gene cluster and its association with human male infertility. Hum Mol Genet. 2014;23:12–23. [DOI] [PMC free article] [PubMed]
- 53.Montjean D, Ravel C, Benkhalifa M, Cohen-Bacrie P, Berthaut I, Bashamboo A, Mcelreavey K. Methylation changes in mature sperm deoxyribonucleic acid from oligozoospermic men: assessment of genetic variants and assisted reproductive technology outcome. Fertil Steril. 2013;100:1241–7. [DOI] [PubMed]
- 54.Khazamipour N, et al. MTHFR promoter hypermethylation in testicular biopsies of patients with non-obstructive azoospermia: the role of epigenetics in male infertility. Hum Reprod. 2009;24(9):2361–4. doi: 10.1093/humrep/dep194. [DOI] [PubMed] [Google Scholar]
- 55.Rotondo JC, et al. Methylenetetrahydrofolate reductase gene promoter hypermethylation in semen samples of infertile couples correlates with recurrent spontaneous abortion. Hum Reprod. 2012;27(12):3632–8. doi: 10.1093/humrep/des319. [DOI] [PubMed] [Google Scholar]
- 56.Nanassy L, Carrell DT. Abnormal methylation of the promoter of CREM is broadly associated with male factor infertility and poor sperm quality but is improved in sperm selected by density gradient centrifugation. Fertil Steril. 2011;95(7):2310–4. doi: 10.1016/j.fertnstert.2011.03.096. [DOI] [PubMed] [Google Scholar]
- 57.Minor A, Chow V, Ma S. Aberrant DNA methylation at imprinted genes in testicular sperm retrieved from men with obstructive azoospermia and undergoing vasectomy reversal. Reproduction. 2011;141(6):749–57. doi: 10.1530/REP-11-0008. [DOI] [PubMed] [Google Scholar]
- 58.El Hajj N, et al. Methylation status of imprinted genes and repetitive elements in sperm DNA from infertile males. Sex Dev. 2011;5(2):60–9. doi: 10.1159/000323806. [DOI] [PubMed] [Google Scholar]
- 59.Nanassy L, Carrell DT. Analysis of the methylation pattern of six gene promoters in sperm of men with abnormal protamination. Asian J Androl. 2011;13:342–6. [DOI] [PMC free article] [PubMed]
- 60.Navarro-Costa P, Nogueira P, Carvalho M, Leal F, Cordeiro I, Calhaz-Jorge C, Goncalves J, Plancha, CE. Incorrect DNA methylation of the DAZL promoter CpG island associates with defective human sperm. Hum Reprod. 2010;25:2647–54. [DOI] [PMC free article] [PubMed]
- 61.Wu W, et al. Idiopathic male infertility is strongly associated with aberrant promoter methylation of methylenetetrahydrofolate reductase (MTHFR) PLoS One. 2010;5(11):e13884. doi: 10.1371/journal.pone.0013884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Marques CJ, et al. Genomic imprinting in disruptive spermatogenesis. Lancet. 2004;363(9422):1700–2. doi: 10.1016/S0140-6736(04)16256-9. [DOI] [PubMed] [Google Scholar]
- 63.Marques CJ, et al. Abnormal methylation of imprinted genes in human sperm is associated with oligozoospermia. Mol Hum Reprod. 2008;14(2):67–74. doi: 10.1093/molehr/gam093. [DOI] [PubMed] [Google Scholar]
- 64.Marques CJ, et al. Methylation defects of imprinted genes in human testicular spermatozoa. Fertil Steril. 2010;94(2):585–94. doi: 10.1016/j.fertnstert.2009.02.051. [DOI] [PubMed] [Google Scholar]
- 65.Kobayashi H, et al. Aberrant DNA methylation of imprinted loci in sperm from oligospermic patients. Hum Mol Genet. 2007;16(21):2542–51. doi: 10.1093/hmg/ddm187. [DOI] [PubMed] [Google Scholar]
- 66.Kobayashi H, et al. DNA methylation errors at imprinted loci after assisted conception originate in the parental sperm. Eur J Hum Genet. 2009;17(12):1582–91. doi: 10.1038/ejhg.2009.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Boissonnas CC, et al. Specific epigenetic alterations of IGF2-H19 locus in spermatozoa from infertile men. Eur J Hum Genet. 2010;18(1):73–80. doi: 10.1038/ejhg.2009.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Poplinski A, et al. Idiopathic male infertility is strongly associated with aberrant methylation of MEST and IGF2/H19 ICR1. Int J Androl. 2010;33(4):642–9. doi: 10.1111/j.1365-2605.2009.01000.x. [DOI] [PubMed] [Google Scholar]
- 69.Kelly TL, et al. Infertility in 5,10-methylenetetrahydrofolate reductase (MTHFR)-deficient male mice is partially alleviated by lifetime dietary betaine supplementation. Biol Reprod. 2005;72(3):667–77. doi: 10.1095/biolreprod.104.035238. [DOI] [PubMed] [Google Scholar]
- 70.Campos EI, Reinberg D. Histones: annotating chromatin. Annu Rev Genet. 2009;43:559–99. doi: 10.1146/annurev.genet.032608.103928. [DOI] [PubMed] [Google Scholar]
- 71.Cairns BR. The logic of chromatin architecture and remodelling at promoters. Nature. 2009;461(7261):193–8. doi: 10.1038/nature08450. [DOI] [PubMed] [Google Scholar]
- 72.Hazzouri M, et al. Regulated hyperacetylation of core histones during mouse spermatogenesis: involvement of histone deacetylases. Eur J Cell Biol. 2000;79(12):950–60. doi: 10.1078/0171-9335-00123. [DOI] [PubMed] [Google Scholar]
- 73.Peng L, Seto E. Deacetylation of nonhistone proteins by HDACs and the implications in cancer. Handb Exp Pharmacol. 2011;206:39–56. doi: 10.1007/978-3-642-21631-2_3. [DOI] [PubMed] [Google Scholar]
- 74.de Rooij DG. Proliferation and differentiation of spermatogonial stem cells. Reproduction. 2001;121(3):347–54. doi: 10.1530/rep.0.1210347. [DOI] [PubMed] [Google Scholar]
- 75.Fenic I, et al. In vivo effects of histone-deacetylase inhibitor trichostatin-A on murine spermatogenesis. J Androl. 2004;25(5):811–8. doi: 10.1002/j.1939-4640.2004.tb02859.x. [DOI] [PubMed] [Google Scholar]
- 76.Fenic I, et al. In vivo application of histone deacetylase inhibitor trichostatin-a impairs murine male meiosis. J Androl. 2008;29(2):172–85. doi: 10.2164/jandrol.107.003848. [DOI] [PubMed] [Google Scholar]
- 77.Lachner M, Jenuwein T. The many faces of histone lysine methylation. Curr Opin Cell Biol. 2002;14(3):286–98. doi: 10.1016/S0955-0674(02)00335-6. [DOI] [PubMed] [Google Scholar]
- 78.Carrell DT, Emery BR, Hammoud S. The aetiology of sperm protamine abnormalities and their potential impact on the sperm epigenome. Int J Androl. 2008;31(6):537–45. doi: 10.1111/j.1365-2605.2008.00872.x. [DOI] [PubMed] [Google Scholar]
- 79.La Spina FA, et al. Heterogeneous distribution of histone methylation in mature human sperm. J Assist Reprod Genet. 2014;31(1):45–9. doi: 10.1007/s10815-013-0137-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Yuen BT, et al. Histone H3.3 regulates dynamic chromatin states during spermatogenesis. Development. 2014;141(18):3483–94. doi: 10.1242/dev.106450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Torregrosa N, et al. Protamine 2 precursors, protamine 1/protamine 2 ratio, DNA integrity and other sperm parameters in infertile patients. Hum Reprod. 2006;21(8):2084–9. doi: 10.1093/humrep/del114. [DOI] [PubMed] [Google Scholar]
- 82.Rousseaux S, et al. Molecular models for post-meiotic male genome reprogramming. Syst Biol Reprod Med. 2011;57(1-2):50–3. doi: 10.3109/19396368.2010.498076. [DOI] [PubMed] [Google Scholar]
- 83.Meistrich ML, et al. Roles of transition nuclear proteins in spermiogenesis. Chromosoma. 2003;111(8):483–8. doi: 10.1007/s00412-002-0227-z. [DOI] [PubMed] [Google Scholar]
- 84.Belokopytova IA, et al. Human male infertility may be due to a decrease of the protamine P2 content in sperm chromatin. Mol Reprod Dev. 1993;34(1):53–7. doi: 10.1002/mrd.1080340109. [DOI] [PubMed] [Google Scholar]
- 85.Aoki VW, Liu L, Carrell DT. Identification and evaluation of a novel sperm protamine abnormality in a population of infertile males. Hum Reprod. 2005;20(5):1298–306. doi: 10.1093/humrep/deh798. [DOI] [PubMed] [Google Scholar]
- 86.Nasr-Esfahani MH, et al. Effect of protamine-2 deficiency on ICSI outcome. Reprod Biomed Online. 2004;9(6):652–8. doi: 10.1016/S1472-6483(10)61776-2. [DOI] [PubMed] [Google Scholar]
- 87.de Mateo S, et al. Protamine 2 precursors (Pre-P2), protamine 1 to protamine 2 ratio (P1/P2), and assisted reproduction outcome. Fertil Steril. 2009;91(3):715–22. doi: 10.1016/j.fertnstert.2007.12.047. [DOI] [PubMed] [Google Scholar]
- 88.Ha M, Kim VN. Regulation of microRNA biogenesis. Nat Rev Mol Cell Biol. 2014;15(8):509–24. doi: 10.1038/nrm3838. [DOI] [PubMed] [Google Scholar]
- 89.Su Z, et al. MicroRNAs in apoptosis, autophagy and necroptosis. Oncotarget. 2015;6(11):8474–90. doi: 10.18632/oncotarget.3523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Krol J, Loedige I, Filipowicz W. The widespread regulation of microRNA biogenesis, function and decay. Nat Rev Genet. 2010;11(9):597–610. doi: 10.1038/nrg2843. [DOI] [PubMed] [Google Scholar]
- 91.Meister G. Argonaute proteins: functional insights and emerging roles. Nat Rev Genet. 2013;14(7):447–59. doi: 10.1038/nrg3462. [DOI] [PubMed] [Google Scholar]
- 92.Bak CW, Yoon TK, Choi Y. Functions of PIWI proteins in spermatogenesis. Clin Exp Reprod Med. 2011;38(2):61–7. doi: 10.5653/cerm.2011.38.2.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Gou LT, Dai P, Liu MF. Small noncoding RNAs and male infertility. Wiley Interdiscip Rev RNA. 2014;5(6):733–45. doi: 10.1002/wrna.1252. [DOI] [PubMed] [Google Scholar]
- 94.Song R, et al. Male germ cells express abundant endogenous siRNAs. Proc Natl Acad Sci U S A. 2011;108(32):13159–64. doi: 10.1073/pnas.1108567108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Korhonen HM, et al. Dicer is required for haploid male germ cell differentiation in mice. PLoS One. 2011;6(9):e24821. doi: 10.1371/journal.pone.0024821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Romero Y, et al. Dicer1 depletion in male germ cells leads to infertility due to cumulative meiotic and spermiogenic defects. PLoS One. 2011;6(10):e25241. doi: 10.1371/journal.pone.0025241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Wu Q, et al. The RNase III enzyme DROSHA is essential for microRNA production and spermatogenesis. J Biol Chem. 2012;287(30):25173–90. doi: 10.1074/jbc.M112.362053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Salas-Huetos A, et al. New insights into the expression profile and function of micro-ribonucleic acid in human spermatozoa. Fertil Steril. 2014;102(1):213–22. doi: 10.1016/j.fertnstert.2014.03.040. [DOI] [PubMed] [Google Scholar]
- 99.Lian J, et al. Altered microRNA expression in patients with non-obstructive azoospermia. Reprod Biol Endocrinol. 2009;7:13. doi: 10.1186/1477-7827-7-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Gu A, et al. Genetic variants in Piwi-interacting RNA pathway genes confer susceptibility to spermatogenic failure in a Chinese population. Hum Reprod. 2010;25(12):2955–61. doi: 10.1093/humrep/deq274. [DOI] [PubMed] [Google Scholar]
- 101.Zhang H, et al. A single nucleotide polymorphism in a miR-1302 binding site in CGA increases the risk of idiopathic male infertility. Fertil Steril. 2011;96(1):34–9. doi: 10.1016/j.fertnstert.2011.04.053. [DOI] [PubMed] [Google Scholar]
- 102.Wang C, et al. Altered profile of seminal plasma microRNAs in the molecular diagnosis of male infertility. Clin Chem. 2011;57(12):1722–31. doi: 10.1373/clinchem.2011.169714. [DOI] [PubMed] [Google Scholar]
- 103.Wu W, et al. Seminal plasma microRNAs: potential biomarkers for spermatogenesis status. Mol Hum Reprod. 2012;18(10):489–97. doi: 10.1093/molehr/gas022. [DOI] [PubMed] [Google Scholar]
- 104.Qin Y, et al. Genetic variants in microRNA biogenesis pathway genes are associated with semen quality in a Han-Chinese population. Reprod Biomed Online. 2012;24(4):454–61. doi: 10.1016/j.rbmo.2012.01.006. [DOI] [PubMed] [Google Scholar]
- 105.Abu-Halima M, et al. Altered microRNA expression profiles of human spermatozoa in patients with different spermatogenic impairments. Fertil Steril. 2013;99(5):1249–55. doi: 10.1016/j.fertnstert.2012.11.054. [DOI] [PubMed] [Google Scholar]
- 106.Abhari A, et al. Significance of microRNA targeted estrogen receptor in male fertility. Iran J Basic Med Sci. 2014;17(2):81–6. [PMC free article] [PubMed] [Google Scholar]
- 107.Abu-Halima M, et al. MicroRNA expression profiles in human testicular tissues of infertile men with different histopathologic patterns. Fertil Steril. 2014;101(1):78–86. doi: 10.1016/j.fertnstert.2013.09.009. [DOI] [PubMed] [Google Scholar]
- 108.Abu-Halima M, et al. Panel of five microRNAs as potential biomarkers for the diagnosis and assessment of male infertility. Fertil Steril. 2014;102(4):989–97. doi: 10.1016/j.fertnstert.2014.07.001. [DOI] [PubMed] [Google Scholar]
- 109.Friemel C, et al. Array-based DNA methylation profiling in male infertility reveals allele-specific DNA methylation in PIWIL1 and PIWIL2. Fertil Steril. 2014;101(4):1097–103. doi: 10.1016/j.fertnstert.2013.12.054. [DOI] [PubMed] [Google Scholar]
- 110.Dabaja AA, et al. Possible germ cell-Sertoli cell interactions are critical for establishing appropriate expression levels for the Sertoli cell-specific MicroRNA, miR-202-5p, in human testis. Basic Clin Androl. 2015;25:2. doi: 10.1186/s12610-015-0018-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Zhao, K., et al., miR-424/322 is downregulated in the semen of patients with severe DNA damage and may regulate sperm DNA damage. Reprod Fertil Dev, 2015. [DOI] [PubMed]
- 112.Yang Q, et al. MicroRNA and piRNA profiles in normal human testis detected by next generation sequencing. PLoS One. 2013;8(6):e66809. doi: 10.1371/journal.pone.0066809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Novotny GW, et al. Translational repression of E2F1 mRNA in carcinoma in situ and normal testis correlates with expression of the miR-17-92 cluster. Cell Death Differ. 2007;14(4):879–82. doi: 10.1038/sj.cdd.4402090. [DOI] [PubMed] [Google Scholar]
- 114.Beauchamp JF, et al. Mechanism of the pressor response to intraperitoneal injection of bradykinin in guinea pigs. Peptides. 1991;12(3):513–21. doi: 10.1016/0196-9781(91)90094-6. [DOI] [PubMed] [Google Scholar]
- 115.Luo L, et al. Microarray-based approach identifies differentially expressed microRNAs in porcine sexually immature and mature testes. PLoS One. 2010;5(8):e11744. doi: 10.1371/journal.pone.0011744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Ruggiu M, et al. The mouse Dazla gene encodes a cytoplasmic protein essential for gametogenesis. Nature. 1997;389(6646):73–7. doi: 10.1038/37987. [DOI] [PubMed] [Google Scholar]
- 117.Okada H, et al. Genome-wide expression of azoospermia testes demonstrates a specific profile and implicates ART3 in genetic susceptibility. PLoS Genet. 2008;4(2):e26. doi: 10.1371/journal.pgen.0040026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Arao Y, et al. Transactivating function (AF) 2-mediated AF-1 activity of estrogen receptor alpha is crucial to maintain male reproductive tract function. Proc Natl Acad Sci U S A. 2012;109(51):21140–5. doi: 10.1073/pnas.1216189110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Hess RA, Bunick D, Bahr J. Oestrogen, its receptors and function in the male reproductive tract - a review. Mol Cell Endocrinol. 2001;178(1-2):29–38. doi: 10.1016/S0303-7207(01)00412-9. [DOI] [PubMed] [Google Scholar]
- 120.Hermeking H. The miR-34 family in cancer and apoptosis. Cell Death Differ. 2010;17(2):193–9. doi: 10.1038/cdd.2009.56. [DOI] [PubMed] [Google Scholar]
- 121.Bouhallier F, et al. Role of miR-34c microRNA in the late steps of spermatogenesis. RNA. 2010;16(4):720–31. doi: 10.1261/rna.1963810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Liang X, et al. MicroRNA-34c enhances murine male germ cell apoptosis through targeting ATF1. PLoS One. 2012;7(3):e33861. doi: 10.1371/journal.pone.0033861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Smorag L, et al. MicroRNA signature in various cell types of mouse spermatogenesis: evidence for stage-specifically expressed miRNA-221, -203 and -34b-5p mediated spermatogenesis regulation. Biol Cell. 2012;104(11):677–92. doi: 10.1111/boc.201200014. [DOI] [PubMed] [Google Scholar]
- 124.Comazzetto S, et al. Oligoasthenoteratozoospermia and infertility in mice deficient for miR-34b/c and miR-449 loci. PLoS Genet. 2014;10(10):e1004597. doi: 10.1371/journal.pgen.1004597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Liu WM, et al. Sperm-borne microRNA-34c is required for the first cleavage division in mouse. Proc Natl Acad Sci U S A. 2012;109(2):490–4. doi: 10.1073/pnas.1110368109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Krawetz SA, et al. A survey of small RNAs in human sperm. Hum Reprod. 2011;26(12):3401–12. doi: 10.1093/humrep/der329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Yu Z, Raabe T, Hecht NB. MicroRNA Mirn122a reduces expression of the posttranscriptionally regulated germ cell transition protein 2 (Tnp2) messenger RNA (mRNA) by mRNA cleavage. Biol Reprod. 2005;73(3):427–33. doi: 10.1095/biolreprod.105.040998. [DOI] [PubMed] [Google Scholar]
- 128.Liu T, et al. MicroRNA-122 influences the development of sperm abnormalities from human induced pluripotent stem cells by regulating TNP2 expression. Stem Cells Dev. 2013;22(12):1839–50. doi: 10.1089/scd.2012.0653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Wu W, Qin Y, Li Z, Dong J, Dai J, Lu C, Guo X, Zhao Y, Zhu Y, Zhang W, Hang B, Sha J, Shen H, Xia Y, Hu Z, Wang X. Genome-wide microRNA expression profiling in idiopathic non-obstructive azoospermia: significant up-regulation of miR-141, miR-429 and miR-7-1-3p. Hum Reprod. 2013;28(7):1827–36. [DOI] [PubMed]
- 130.Obholz KL, et al. FNDC3A is required for adhesion between spermatids and Sertoli cells. Dev Biol. 2006;298(2):498–513. doi: 10.1016/j.ydbio.2006.06.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Meng X, et al. Genetic deficiency of mtdh gene in mice causes male infertility via impaired spermatogenesis and alterations in the expression of small non-coding RNAs. J Biol Chem. 2015;290(19):11853–64. doi: 10.1074/jbc.M114.627653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Klaver R, Gromoll J. Bringing epigenetics into the diagnostics of the andrology laboratory: challenges and perspectives. Asian J Androl. 2014;16(5):669–74. doi: 10.4103/1008-682X.125412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Huang Y, et al. The behaviour of 5-hydroxymethylcytosine in bisulfite sequencing. PLoS One. 2010;5(1):e8888. doi: 10.1371/journal.pone.0008888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Yu M, et al. Base-resolution analysis of 5-hydroxymethylcytosine in the mammalian genome. Cell. 2012;149(6):1368–80. doi: 10.1016/j.cell.2012.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Booth MJ, et al. Oxidative bisulfite sequencing of 5-methylcytosine and 5-hydroxymethylcytosine. Nat Protoc. 2013;8(10):1841–51. doi: 10.1038/nprot.2013.115. [DOI] [PMC free article] [PubMed] [Google Scholar]